Abstract
Tuft cells—rare solitary chemosensory cells in mucosal epithelia—are undergoing intense scientific scrutiny fueled by recent discovery of unsuspected connections to type 2 immunity. These cells constitute a conduit by which ligands from the external space are sensed via taste-like signaling pathways to generate outputs unique among epithelial cells: the cytokine IL-25, eicosanoids associated with allergic immunity, and the neurotransmitter, acetylcholine. The classic taste cell transcription factor POU2F3 is lineage-defining, suggesting a conceptualization of these cells as widely distributed environmental sensors with effector functions interfacing type 2 immunity and neural circuits. Increasingly refined single-cell analytics have revealed diversity among tuft cells that extends from nasal epithelia and Type II taste cells, to ex-Aire-expressing medullary thymic cells and small intestinal cells that mediate tissue remodeling in response to colonizing helminths and protists.
Keywords: Tuft cells, POU2F3, type 2 immunity, IL-25, TRPM5, chemosensation
Introduction
Tuft cells are rare, primarily endoderm-derived, epithelial cells present predominantly at mucosal surfaces of vertebrates. Named for the iconic apical cluster of rigid microvilli that project into the lumen of hollow viscera, tuft cells were defined by their morphologic characteristics over 50 years ago. However, the function of tuft cells—whose aliases include microvillous (nasopharynx), multivesicular, fibrillovesicular, caveolated (stomach and gastrointestinal tract), and brush (airways) cells—remained unclear for decades. Aided by characterization of tuft cell transcriptional programs, recent work has focused on the development and effector functions of these chemosensory cells, whose signaling outputs include the capacity to produce interleukin-25 (IL-25), eicosanoids—including certain prostaglandins and leukotrienes—and acetylcholine (ACh). Elucidation of these programs has enabled improved tuft cell identification, facilitating the discovery of unexpected roles for tuft cells in biologic processes as disparate as allergy and type 2 immunity, airway tone in responses to bacterial metabolites, small intestinal remodeling, and dictating the cytokine mileau of the thymus. With excellent comprehensive reviews covering the early (1–3) and more recent (4, 5) history and functions of tuft cells, we will summarize these older findings and focus on recent advances that have expanded the repertoire of these enigmatic cells across diverse tissues. Finally, we suggest a more contextual biology for chemosensory-like cells, including tuft cells, in vertebrate physiology with speculations regarding opportunities for future discovery integrating tuft cells into immune and neural circuits critical for tissue health and homeostasis.
Tuft cells express a unique gene signature across diverse tissues
Tuft cells share a core gene signature with elements of chemosensory, immune and neural functions. First characterized using microarrays (6), the core signature was validated in tuft cells from diverse epithelia (6–10). Despite their rarity, tuft cells can be identified based on characteristic markers (Table 1). We here define tuft cells, minimally, as epithelial cells dependent on the transcription factor POU2F3 for development and expressing: IL-25; eicosanoid biosynthetic pathways characterized by COX1/2, and ALOX5; and transient receptor potential cation channel subfamily M member 5 (TRPM5). Additional common markers include choline acetyltransferase (ChAT) and doublecortin-like kinase 1 (DCLK1), although these can be expressed more broadly by other cells, including neurons. Although clearly a distinct lineage, the stem cell – tuft cell relationship remains incompletely defined (see below).
Table 1.
Characteristic tuft cell markers enable identification across distinct epithelia, and associated genetic tools.
| Markera | Expression and exceptions | Putative function | Useful mouse toolsb |
|---|---|---|---|
| Structural | |||
| DCLK1 | Most, not in Type II taste bud cells and olfactory epithelium | Microtubule polymerization |
Dclk1-CreERT2-IRES-EGFP (107), Dclk1-CreERT (93) Dclk1-CreGFP (93) |
| Acetylated α-tubulin | All, apically enriched | Protection of long-lived microtubules from mechanical aging | NA |
| Keratin 18 | All | Structural integrity | NA |
| Villin | Apically enriched in tuft cells, expressed by other cells | Actin-binding, microfilament bundling | Vil1-Cre, Vil1-CreERT (91) |
| Chemosensation | |||
| α-gustducin | Most, incomplete overlap with TRPM5 in intestine (143) | α subunit of heterotrimeric G protein |
Gnat3-GFP (144) Gnat3−/− (12) |
| PLCβ2 | Most, incomplete overlap with TRPM5 in intestine (143) | Catalyzing formation of IP3 and DAG from PIP2 |
Plcb2-GFP (145, 146) Plcb2−/− (147) |
| TRPM5 | All | Depolarizing Ca2+-activated cation channel |
Trpm5-GFP (6, 143) Trpm5−/− (12) |
| ChAT | Most, not in Type II taste bud cells (16) | Choline acetyltransferase catalyzing synthesis of acetylcholine |
Chat-eGFP (16) Chat-Cre (148) Other literature: Chat-CreERT2 (149) Chatfl/fl (150, 151) |
| T1Rs, T2Rs | Most, possibly in combinatorial pattern | Taste reception |
Tas1R1-BL-IRES-mCherry (152) Tas1R2-LacZ (153) Tas1R2-Cre (154) Tas1R3-GFP (155) Tas2r131-BL-IRES-hrGFP (152) Tas2r131-BL-IRES-Cre (156) Tas2r143-CreERT2 (40) Multiple KO alleles for Tas1Rs and Tas2Rs |
| Other | |||
| POU2F3 | All | Lineage-defining transcription factor | Pou2f3−/− (99) |
| GFI1B | All | Zinc finger transcriptional repressor |
Gfi1b-GFP (92) Gfi1bfl/fl (157, 158) |
| COX1, COX2 | All | Prostanoid synthesis | Multiple (conditional) KO alleles |
| PGDS | All? | Prostaglandin-D synthesis | Hpgds−/− |
| IL-25 | All | Type 2-associated cytokine, IL-17 family member | Il25fl/fl-IRES-RFP (11) |
Tuft cells engage with immune and neural networks via a limited repertoire of secreted molecules (Figure 1 a and b). Tuft cell secretion of IL-25 plays an important physiologic role in type 2 immune responses in small intestine (11–13) and modulates the thymic cytokine milieu (8, 10). Tuft cells express the molecular machinery for synthesis of ACh (ChAT), prostaglandins (COX1 and COX2) and leukotrienes (ALOX5) (6). In some tissues, however, tuft cells lack elements of the typical secretory cascade for these effector molecules, and further study is needed to define the physiologic role for these effector molecules. Due to their chemosensory properties, tuft cell production of ACh has been proposed as a major functional output in several organs, including in “classical” sensory nasal and olfactory epithelium. ACh from gastric, urethral, and tracheal tuft cells has been proposed to signal to neurons (14–16), although validation of ACh production specifically from tuft cells has proven difficult and genetic approaches for the conditional deletion of Chat are needed to support a relevant cholinergic role.
Figure 1:
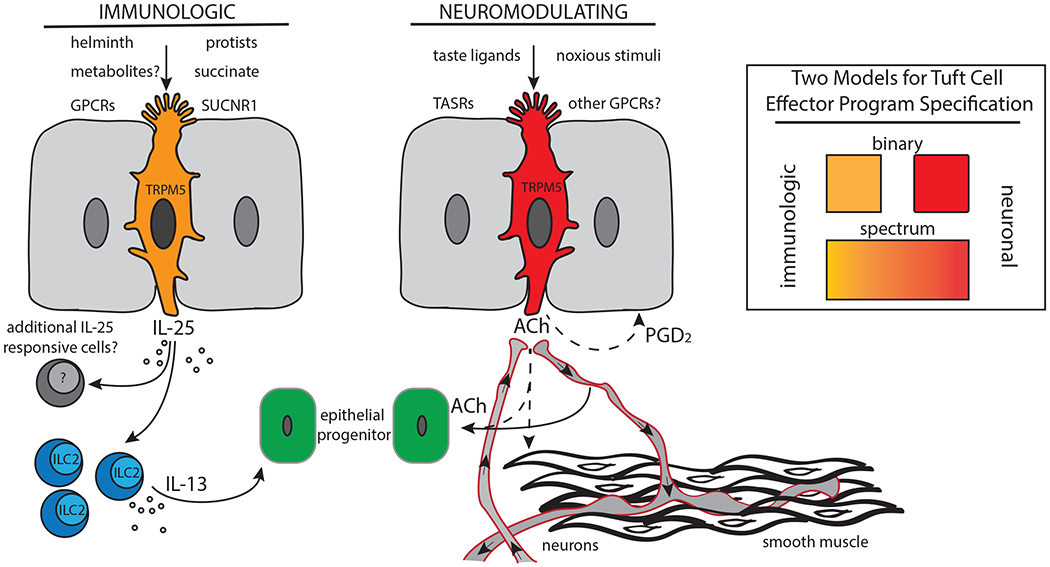
Tuft cells have been found in diverse epithelia where it is thought they play chemosensory and/or immunomodulatory roles enabled by expression of multiple GPCRs to sense luminal stimuli. Tuft cells in the small intestine, for example, are known to respond to luminal helminths and protists like Tritrichomonas. Presence of the latter is sensed by detection of a major metabolic endproduct, succinate, via a tuft cell GPCR, SUCNR1. Subsequently, tuft cells activate tissue resident ILC2s in a TRPM5- and IL-25-dependent fashion to produce effector cytokines like IL-13. Other tuft cell-derived immune mediators, including ALOX5-dependent eicosanoids, may be involved. IL-13 acts on epithelial progenitor cells in the intestinal crypt, promoting dynamic increases in tuft and goblet cell output. In other tissues such as the olfactory epithelium, tuft cells appear to respond to bitter taste ligands, although the physiological ligands are unknown. Here, tuft cells may enact a neuronal effector program, perhaps through secretion of ACh which could act on efferent neurons, smooth muscle, or epithelial progenitor cells. Additional tuft cell effectors, including HPGDS-dependent prostanoids like PGD2, may contribute. Inset: We propose that tuft cells exhibit immunologic and/or neuronal effector states. These effector phenotypes could be mutually exclusive, discreet cell populations, or could represent tuning of fully differentiated cells along a spectrum of function dependent on mutable luminal or niche signals.
The presence of differentially expressed taste receptors and other GPCRs, as well as the canonical taste signal transduction cascade proteins, α-gustducin (GNAT3), phospholipase C β2 (PLCβ2), and TRPM5, has led to the hypothesis that tuft cells “taste” luminal contents in various tissues and thus integrate responses to mucosal stimuli (SIDEBAR TRPM5) (4, 5, 17).
SIDEBARS.
Transient Receptor Potential Cation Channel Subfamily M Member 5 (TRPM5) and Type II taste cell signaling
TRPM5 is highly expressed by tuft cells, other solitary chemosensory cells, and Type II taste cells (17, 85, 127, 128). TRPM5 is a calcium-gated, inwardly rectifying nonspecific cation channel which displays both voltage and temperature modulation (129–134). TRPM5 in Type II taste cells of taste buds is activated downstream of the taste receptor (TASR) family of GPCRs in the response to bitter (T2Rs), sweet and umami (T1Rs) tastants (51). GPCR binding by distinct taste ligands results in G protein-mediated activation of PLCβ2, cleavage of PIP2 into IP3 and DAG, and IP3-mediated release of intracellular calcium. Intracellular calcium activates TRPM5, resulting in transient depolarization of the cell body and activation of additional cation gated channels involved in taste sensation (51, 128). Activation of TASRs also results in release of the Gα subunit; while primarily thought to regulate Gβγ activation of PLCβ2, Gα subunits can promote production of cyclic AMP (51). T1Rs and T2Rs are expressed in tuft cells and show tissue specificity (9), but physiological ligands remain to be defined.
POU domain, class 2, transcription factor 3 (POU2F3)
The POU (Pit-Oct-Unc) protein family of transcription factors control gene expression regulating cell fate determination and embryonic development. POU2F3 (SKN-1a, OCT11) is a subclass II Oct protein, characterized by their strong affinity for an 8 bp consensus sequence [ATGC(A/T)AAT] termed the ‘octamer motif’ and variants thereof (98, 135). The octamer sequence is bound by two linker-joined domains characteristic of the POU family, with the ~80 amino acid POU-specific (POUS) domain contacting the 5’ part of the motif, and the ~60 amino acid POU homeodomain (POUHD) binding to the 3’ region. Pou2f3 was initially found to encode two isoforms expressed in suprabasal cells of the neonatal epidermis, Skn-1a and the less abundant Skn-1i, which regulate gene expression involved in terminal differentiation of epidermal keratinocytes, including Krt10 (136–138). Subsequently, high Pou2f3 expression was discovered to be restricted to TRPM5+ cells in taste buds and TRPM5+ epithelial tuft cells in various tissue, which are all absent in Pou2f3−/− mice (13, 46, 99). POU2F3 is a crucial factor that controls lineage specification in basal progenitor cells towards highly differentiated cells with a chemosensory gene expression progam.
Doublecortin-like kinase-1 (DCLK1)
DCLK1 is a microtubule-associated protein with two N-terminal doublecortin (DCX) domains, which regulate microtubule polymerization, and a C-terminal serine-threonine kinase domain; functional splice variants lack one microtubule-binding domain (139). Biochemical and structural studies suggest the kinase domain and C-terminal tail autoinhibit the tubulin polymerization domains (115). The doublecortin microtubule-binding domain is homologous to doublecortin (DCX). Mutations in Dcx in humans that interfere with microtubule binding are associated with defects in neuronal migration, patterning defects with double cortical laminations and severe neurologic deficits (lissencephaly) (139, 140). In mice, deletion of both Dcx and Dclk1 recapitulate aspects of the human phenotype (141, 142). DCLK1 is expressed in mouse neurons during embryogenesis, in mature neurons, and in epithelial tuft cells during adult life (115). Expression of DCLK1 is observed in stem and stem-like cells in various tissues, and is considered a hallmark of stem-like cells found in cancer or after injury (114). Kinase domain inactivating human mutations are driver mutations in ~10% of gastric cancers and in lesser numbers of other gastrointestinal tumors (115), perhaps leading to loss of regulation in tubulin polymerization. Despite the prominent cytoskeletal architecture of tuft cells, the precise role of DCLK1 in tuft cells is unknown.
Despite similarities of tuft cells in distinct tissues, both in gene expression and morphology, tissue-specific differences are also apparent (9) (Figure 2 a and b). The expression of specific genes from anatomically distinct tuft cell populations suggests tissue-specific functions. Of particular interest may be the unique expression of surface GPCRs which converge upon TRPM5 channel activation, facilitating tissue-specific inputs to activate tuft cell function through a common signaling machinery, with stereotyped outputs. The ability of tuft cells to respond with immunologic or neuromodulating outputs may represent microenviroment-dependent, deterministic cell fate decisions, or a spectrum of function dependent on tissue-specific cell-surface receptor expression and ligand bioavailability (Figure 1).
Figure 2:
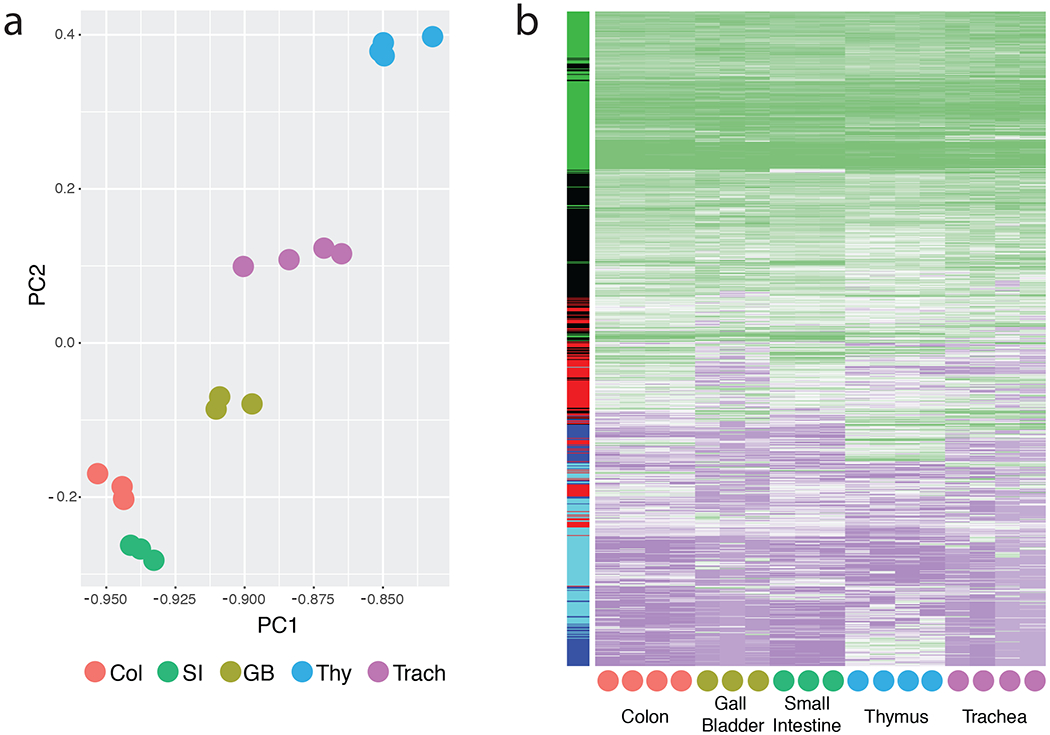
Recently published RNA-seq data (9) was reanalyzed to isolate heterogeneity of tuft cells among distinct tissues. Samples are sorted tuft cells (CD45low EPCAM+ IL-25+) from various tissues of IL-25 reporter mice, with mRNA prepared and analyzed as described in Nadjsombati et al. The following processing steps were applied in this order: exclusion of very large outliers, minimal cumulative read count per transcript cut off at 150, log 10 transformed, missing values set to minimum value of the sample, scaled to a mean of 0 with standard deviation of 1. (a) Bayesian principle component analysis with two components was performed on processed and filtered data, with each dot representing one biological replicate (see Nadjsombati et al.). (b) Hierarchical clustering was performed on the processed data using average linkage and Euclidean distance. The dendogram is not shown for clarity. The colored side bar represents gene membership in clusters determined by K-means (k = 5).
Tissue distribution and characterization
Tuft cells in distinct tissues share a pronounced, microtubule-rich apical tuft consisting of a thick cluster of microvilli, a morphologic feature that aided in their identification in early electron microscopy (EM) studies, as recently summarized (5). More recent advanced volumetric EM imaging validated existence of lateral projections (cytospinules) into neighboring cells, suggesting unappreciated ways for epithelial communication (18). However, studies to-date have focused largely on the secretory effector functions of tuft cells. Below, we review the localization, development and potential functions of tuft cells in the diverse tissues in which they reside.
Nasopharyngeal epithelia
The nasal passages represent a critical conduit to the outside world, lined by a complex epithelial layer extending from squamous and transitional zones at the anterior nares through the prominent anterior/inferior nasal, or respiratory, epithelium (NE), and posterior/superior olfactory epithelium (OE). OE constitutes a greater proportion of the epithelial surface in rodents (~40%) than in humans (~5%). The nasopharyngeal epithelia engage in numerous processes, including air humidification, particle filtration, odorant and toxin detection, and elaboration of mucus and antimicrobial peptides (19). The epithelia of both NE and OE includes TRPM5+, ChAT+, bitter taste receptor-expressing pear-shaped cells with an apical, luminal tuft, that are often designated solitary chemosensory cells in NE and microvillous cells in OE. In mice, these cells require POU2F3 for development (20), leading us to consider these bona fide tuft cells.
Application of T2R bitter ligands, including acyl-homoserine lactones associated with quorum sensing in Gram-negative bacteria, initiated α-gustducin- and TRPM5-dependent Ca2+ depolarization and ACh release from NE tuft cells, resulting in local neurogenic mast cell-mediated inflammation and activation of trigeminal sensory fibers, thereby initiating a protective apnea response (21, 22). Using human NE cell cultures, Lee et al. demonstrated that bitter taste receptor ligands initiated a propagating Ca2+ depolarization from tuft cells that stimulated robust antimicrobial peptide secretion from adjacent cells, suggesting a rapidly deployed method by which relatively few cells could control bacterial homeostasis across the epithelial surface (23). Conversely, activation of the sweet taste receptor T1R2/3 by bacterial D-amino acids from Staphylococcus aureus inhibited the pathway, leading to enhanced bacterial growth (24). These findings are consistent with a chemosensory role for nasal tuft cells in epithelial – microbial homeostasis, and possible activation of aversive behaviors, like coughing and sneezing.
The association of type 2 immunity with Group 2 innate lymphoid cells (ILC2s) prompted examination of patients with chronic rhinosinusitis with nasal polyps, which presents in an allergic form associated with asthma and eosinophilia, and more frequently among patients with worse disease (25). Elevated ILC2s, type 2 cytokines, and eosinophils were present in polyps (26), and blockade of IL-4Rα led to diminution in the polyp burden, implicating type 2 cytokines in the epithelial hyperplasia (27). Consistent with studies in the mouse, solitary chemosensory cells (tuft cells) were the major source of IL-25 in the NE from these patients (28), but whether IL-25 or additional epithelial cytokines, such as IL-33 (29), or both, drive the disease remains unknown. A recent single-cell RNA sequencing (scRNA-seq) analysis of human allergic nasal polyp tissues in NE revealed profound effects on epithelial diversity, with reductions in ciliated cells and basal cell hyperplasia (30).
Microvillous cells, here defined as tuft cells, are present within the complex OE, and are likely to arise from horizontal basal cells, which generate transit-amplifying-like globose basal cells competent to differentiate into olfactory sensory neurons, supporting sustentacular cells, and tuft cells (31). Computational analysis of scRNA-seq suggests the epithelial sustentacular and tuft cell populations diverge prior to cells committing to neural identity, although these trajectories were altered after epithelial injury (32). Tuft cells in OE express ChAT and the vesicular ACh transporter, VAChT, and modulate olfactory responses to xenobiotic challenges that activate Ca2+ depolarization and ACh release (33) and may play a role in maintaining olfactory function, in part via release of neuropeptides (34, 35). VAChT is notably missing from the tuft cell transcriptome in other tissues.
Eyes and ears
Tuft cells with typical morphology and expression of canonical markers of the taste receptor signaling cascade were present in the auditory tubes and nasal aspects of the conjunctival epithelia in mice (36, 37). In both tissues, tuft cells were in close proximity to cholinoreceptive sensory nerve fibers with the capacity for neuropeptide release, including CGRP, consistent with a tuft cell-mediated inflammatory circuit involved in restricting ingression of toxic substances into deeper tissues.
Thymus
Speculations regarding a role for cholinergic signaling in thymic function led Panneck et al. to investigate the sources of ACh using Chat-eGFP BAC transgenic mice (38). A subset of eGFP+ medullary thymic epithelial cells (mTECs) expressed α-gustducin, TRPM5 and PLCβ2, and was present as isolated cells and in loose aggregations associated with cornified, terminally-differentiated epithelia in Hassall’s corpuscle-like clusters. A taste receptor reporter mouse (Tas2r131) confirmed expression of several bitter taste-associated receptors (Tas2r105, Tas2r108, and Tas2r131) in these cells, although heterogeneity was evident (39). Thymic tuft cells were also labeled using BAC transgenic mice expressing CreERT2 from the Tas2r143 promoter, which is controlled by shared cis-regulatory elements in the bitter taste receptor cluster, which also includes Tas2r135 and Tas2r126 (40).
More recently, Miller et al, using inducible lineage-tracking of Aire expression in murine mTECs, identified a population of post-Aire cells that expressed the transcriptional signature of tuft cells: components of the taste receptor signal transduction cascade, ChAT, IL-25, DCLK1, and multiple bitter taste receptors. The latter were expressed heterogeneously among different cells (8). Post-Aire mTECs were absent in Pou2f3- and in Trpm5-deficient mice, corroborating their lineage specification as tuft cells and suggesting dependence on Ca2+-mediated depolarization. Mice deficient in post-Aire mTECs (tuft cells) had profound reductions in NKT2 cells that express IL-4 and Eomes-expressing CD8+ innate-like (“virtual memory”) thymocytes that require IL-4 for differentiation. Unlike peripheral tuft cells, thymic tuft cells expressed low levels of MHCII and CD74; transfer of Pou2f3-deficient thymi into athymic nude mice rendered animals non-tolerant to subsequent immunization with IL-25, suggesting that these cells enforce tolerance to tuft cell-derived antigens. A second scRNA-seq analysis of mouse thymic epithelia also found IL-25-expressing tuft cells and used ATAC-seq and ChIP-seq to define the landscape of POU2F3 binding sites in these cells (10).
Lower respiratory tract
The lower respiratory tract comprises the trachea and branching bronchi and bronchioles, which carry air to the distal alveoli for gas exchange. In humans, the trachea, bronchi, and larger bronchioles are lined by pseudostratified epithelium of multiciliated, secretory, and basal cells, followed by a simple cuboidal epithelium in the terminal bronchioles and squamous type 1 and cuboidal type 2 alveolar epithelial cells. In mice, only the trachea and main-stem bronchi are lined with pseudostratified epithelium; simple columnar epithelium lacking basal cells lines the smaller bronchi and bronchioles (reviewed in (41, 42)).
Despite early descriptions of tuft cells in respiratory epithelium, understanding remains incomplete beyond histological characterization. Tuft cells appear to comprise at least a subset of cells commonly termed “brush cells” due to their dense apical tuft morphology (43). Solitary α-gustducin, TRPM5, ChAT, villin, and bitter taste receptor-expressing pear-shaped cells with an apical, luminal tuft, were present in rodent airways with decreasing abundance from trachea to larger bronchioles, but not in smaller bronchioles and alveoli (44); a similar distribution was present in cattle (45). Some studies reported brush cells in the rat distal airways, including in alveolar epithelia, with increased frequencies under certain pathological conditions (reviewed in (43)).
In the trachea of mice, tuft cells express IL-25 (11), and share a core gene expression program with tuft cells in other organs (9). Tracheal tuft cells are absent in Pou2f3−/− mice, whereas a cell population with similar morphology – marked by a villin-bright apical tuft, but of otherwise unknown identity and negative for ChAT and TRPM5 – remained intact (46), revealing heterogeneity among airway brush cells as previously noted (16). The tuft cell developmental trajectory and relationship with other lineages in the lower airways remain unknown. Lineage tracing in mice revealed regionally distinct epithelial progenitor pools (41, 42), including Trp63+ NGFR+ basal cells, Club cells, and type 2 alveolar epithelial cells. Using BrdU pulse-chase experiments, Saunders et al. determined that tracheal tuft cells, like those in other tissues, are post-mitotic. During post-natal tracheal growth, tuft cells differentiated with slow kinetics from BrdU-labelled proliferating precursors that expressed basal cell-associated cytokeratin 14 (47). However, precise lineage tracing during development, in adulthood, and in the context of tissue damage is lacking.
In addition to taste signaling components, tracheal tuft cells express various taste receptors, consistent with chemosensory functions (9, 16, 40, 44, 46). Tracheal tuft cells were in contact with peptidergic cholinoceptive vagal sensory neurons, and stimulation with the bitter ligands cycloheximide or 3-OxoC12-homoserine lactone, a P. aeruginosa-associated quorum sensing molecule, resulted in decreased respiratory rate that was inhibited by a nicotinic acetylcholine receptor antagonist (16, 48). Bitter taste receptors were also present on cultured human airway epithelia, which responded to agonist stimulation with increased ciliary beat frequency (49), and on human airway smooth muscle cells, which responded to bitter tastants with relaxation and airway dilation (50). Single cell analysis of airway tuft cells will clarify whether taste receptors are expressed individually or combinatorially (e.g.: T1Rs with T2Rs), distinct from the classical concept of taste segregation among taste bud cells (51), but consistent with the observed co-expression in nasal tuft cells (52) with antagonistic signaling characteristics (23, 24). Elucidating as-yet unexplored immunologic roles for airway tuft cells will be important to identify roles in chronic diseases like asthma and eosinophilic lung vasculitic syndromes (e.g.: eosinophilic granulomatosis with polyangiitis (EGPA), formerly Churg-Strauss syndrome).
The Gastrointestinal Tract
During embryogenesis, the gastrointestinal tract develops from the endodermal primitive gut tube, subsequently forming the esophagus, stomach, intestines, and their associated organs, the liver, gall bladder and pancreas (53). The specialized mucosal epithelia in these tissues differ in microanatomical organization, cellular composition and turnover rates, and can be broadly separated into stratified squamous non-keratinizing epithelia in the esophagus and anal canal, and simple columnar epithelia in the remaining gastrointestinal tract, organized into gastric pits and glands in the stomach, crypts in the colon and cecum, and crypt-villus units in the small intestine (54). Tuft cells are absent from the stratified squamous epithelium of the anal canal and esophagus but may increase following replacement with a metaplastic, intestinal-like columnar epithelium (55, 56). In contrast, tuft cells are present throughout the simple columnar epithelia of the pancreato-biliary system, stomach, small and large intestine, and cecum.
Pancreato-biliary system
The pancreato-biliary system includes the gallbladder, extrahepatic bile ducts (EHBDs), pancreas and pancreatic ducts (57). The liver, pancreas and biliary tree develop together from the foregut endoderm, with the pancreas and extrahepatic biliary tree sharing a precursor distinct from the intrahepatic ducts and the liver (58). The biliary tissue consists of simple columnar epithelium, a thin lamina propria, and smooth muscle, and is innervated at several levels (59). The morphologically similar epithelial cells of the intrahepatic and extrahepatic ducts are commonly termed cholangiocytes (59). In the pancreas, the larger interlobular ducts and the major pancreatic duct are lined by simple columnar epithelium; small intralobular pancreatic duct epithelium is cuboidal (60). During feeding, cholecystokinin stimulates gallbladder emptying and the release of pancreatic secretions into the duodenum (57, 61). Bile is a key route for excretion of xenobiotics, and both gallbladder and EHBDs can be sites of long-term bacterial colonization and persistent bacterial shedding (62). Thus, similar to the small intestine and the colon, the epithelium of the pancreato-biliary system, and particularly the biliary tree, is exposed to nutrients and hormones, as well as micro-organisms and soluble xenobiotics.
The presence of tuft cells in biliary epithelium was noted decades ago by EM of the mouse gallbladder and the rat bile duct, where the striking microvillus structure was noted among the homogeneous principal cells (63–65). Tuft cells in gallbladder and EHBDs are far more abundant than in small intestine (63). Similar cells are present in human tissues (66). More recent work using reporter mice for ChAT, TRPM5, and IL-25, and staining for DCLK1 has confirmed the abundance of tuft cells in the gallbladder and EHBDs (11, 55). IL-25+ tuft cells are not present in intrahepatic bile ducts (IHBDs) (our unpublished observations), consistent with their development from distinct epithelial precursors (58). Despite their abundance, tuft cell function in the gall bladder and EHBDs remains unknown. Given the involvement of type 2 cytokines in fibrotic diseases (67), it will be important to evaluate potential roles for tuft cells in this constellation of hepatobiliary diseases. Biliary epithelial cells are found in close proximity to neurons, which directly innervate biliary epithelium, and smooth muscle, both potential cell signaling partners (68, 69).
Tuft cells in the pancreatic duct were also noted in early studies (70, 71), but little information has since accrued. The high abundance of tuft cells – 22% in specific ductal locations – reported in rat pancreatic duct (72) has not been quantified in other studies, although tuft cells in the major pancreatic duct are POU2F3-dependent in mouse (46). Tuft-like cells have also been documented in some pancreatic cancers (73, 74), and may have a biliary phenotype (73). Although additional work is needed, this suggests that pancreatic ductal tissue may acquire a biliary phenotype during oncogenesis, marked by appearance of tuft-like cells in a Sox17-dependent fashion (73). However, the alternative hypothesis, that this represents expansion of an existing tuft cell population in the pancreatic ducts, as suggested by older literature, has not been tested. Staining of normal pancreas using DCLK1 revealed ductal tuft cells, but also peripheral islet cells suggested to be stem-like cells, perhaps complicating tuft cell studies in this tissue (75).
Stomach
Jawed vertebrates have a muscular stomach, derived from the foregut endoderm, used for food storage, mechanical disruption, and preliminary digestion by acid and proteases (76). The stomach is adapted to food intake, and thus varies substantially from species to species: rodents have an anatomically distinct forestomach and glandular stomach (the corpus and the antrum) while in humans the corpus is expanded (76). The simple columnar epithelium of the gastric mucosa in humans begins abruptly after the esophagus and continues throughout the stomach, while in rodents the forestomach epithelium, like the esophagus, is squamous.
Tuft cells are found in the gastric mucosa of the corpus and antrum in rodents and humans as solitary cells (6, 77). In mice, however, tuft cells are enriched along the limiting ridge at the transition from squamous to simple columnar epithelium separating the forestomach and the corpus (78, 79). Tuft cells appear in the stomach of mice at birth, but are rare and lack any enrichment at the limiting ridge (80). At this time, murine corpus and antrum tuft cells appear morphologically distinct. As the glandular stomach develops, tuft cells increase to achieve the densely packed organization of the limiting ridge observed after weaning (80). Although this stereotyped appearance of gastric tuft cells suggests that tuft cells are linked to digestive function of the stomach during postnatal feeding – perhaps delimiting a barrier between the acidic gastric juices and the storage function of the forestomach – the role for tuft cells remains unclear.
It has been proposed that bile duct and gastric tuft cells may play a role in regulating levels of bicarbonate and/or sensing acidic conditions; indeed, the high proportion of tuft cells that are observed in acidic locations suggests a potential specialized role in dealing with electrolyte-rich, acidic luminal content, similar to the role for chloride cells in fish (81). Ultrastructural analysis of rat bile duct and gastric groove following in vivo exposure to electrolytes induced visual morphological changes in tuft cells (81). Although previously suggested that tuft cells in rat gastric mucosa and bile duct express components of the bicarbonate secretion machinery (82, 83), this was not apparent in RNAseq data from gallbladder tuft cells (6). Eberle et al. proposed that secretion of prostaglandins from gastric tuft cells downstream of acid-sensitive GPCRs might act on neighboring bicarbonate-secreting cells (84). Although correlative, the relationship between tuft cells and acid sensing is consistent with the impact of acid on TRPM5 signaling (85), and further study is warranted.
Small and large intestine
Early studies, recently reviewed (5), described tuft cells in the small and large intestines as rare isolated cells similar to other secretory cell lineages, including goblet and enteroendocrine cells, interspersed between the more abundant enterocytes. Epithelial progenitors and signals promoting the differentiation of epithelial lineages are best characterized in small intestine, where dividing LGR5+ stem cells at the crypt base give rise to rapidly proliferating transit-amplifying cells and continuously renew the differentiated epithelium on the villus with a complete turnover rate of approximately 4-5 days (86). Despite increasing clarity regarding stem cell-epithelial cell differentiation in the small intestine, tuft cell differentiation remains controversial.
The fate of intestinal progenitor cells, i.e., specification into absorptive (enterocyte) vs. secretory (tuft, goblet, enteroendocrine, Paneth) lineages, is controlled in stem and progenitor cells by Notch signaling, which is activated by delta-like 1 and 4 expressed in intestinal secretory cells, including Paneth cells and crypt base goblet cells (87). Proteolytically-released Notch intracellular domain translocates into the nucleus resulting in transcription of target genes, including the transcriptional repressor Hes1. HES1 promotes enterocyte fate, in part through repression of Atoh1 (88), the transcriptional activator essential for intestinal secretory cell commitment (89, 90). However, the requirement for ATOH1 in tuft cell differentiation is unclear. Studies report both absence of tuft cells in the small intestine of mice with conditionally deleted Atoh1 alleles (91), and increased abundance of tuft cells, possibly because of the deficiency in other secretory lineages (92). The latter study reported only minor frequencies of fate-mapped tuft cells using Atoh1-Cre and Atoh1-CreER lineage tracing in adult mice. However, Bjerkenes et al did note absence of tuft cells from the small intestine, but not from the gastric epithelium, of global Atoh1 knockout mice on embryonic day E18.5. While the authors suggested this was related to developmental abnormalities in Atoh1 deficiency, an alternative interpretation is that fetal small intestinal tuft cell development is ATOH1-dependent (92). Another study verified intact small intestinal tuft cells in adult mice with conditional deletion of Atoh1 using Lgr5-CreERT2 (93). A third study used Vil1-CreERT2 and Lrig-CreERT2 to delete Atoh1 from the intestinal epithelium and observed increased tuft cells in the proximal and distal small intestine consistent with the results of Bjerknes et al.; in contrast, colonic tuft cells were strongly reduced after Atoh1 deletion, suggesting differential requirements for ATOH1 in small and large intestinal tuft cells (94). Using p-Creode, a novel computational trajectory-mapping algorithm for high-dimensional single-cell data, these authors also provided evidence for a tuft cell developmental trajectory that was distinct from ATOH1-dependent goblet and Paneth cells.
One explanation for the conflicting results from distinct laboratories and different developmental timepoints could be the existence of separate tuft cell differentiation pathways, one ATOH1-dependent and one ATOH1-independent, perhaps equivalent to the IL-4Rα-independent and IL-4Rα-/STAT6-dependent populations of tuft cells found in small intestine (11–13, 95). Indeed, using scRNA-seq analysis of small intestinal epithelial cells, Haber et al. suggested the existence of two tuft cell populations; tuft cell heterogeneity was also described based on protein expression analysis using multiplex immunofluorescence (96). This heterogeneity could be driven by the variable presence of type 2 immune stimuli in different mouse colonies caused by Tritrichomonas (described below). Notably, IL-13 can cause upregulation of SOX4 in crypt progenitors, and a recent study showed increased SOX4 promotes tuft and enteroendocrine fate in an ATOH1-independent manner (97). Vil1-Cre x Sox4fl/fl mice displayed a minor baseline decrease in tuft cells, but a more pronounced defect in tuft cell expansion after infection with the intestinal helminth N. brasiliensis, which potently activates the IL-13 – IL-4Rα axis. In vitro deletion of Atoh1 in organoid cultures, unexposed to exogenous IL-4/IL-13 and perhaps more analogous to the fetal state, suppressed tuft cell differentiation. This defect could be overcome by overexpression of Sox4. Goblet cell fate remained ATOH1-dependent.
Notably, SOX family proteins are well-characterized partners for heterodimerization with transcription factors of the POU homeodomain family (SIDEBAR POU2F3) (98), of which POU2F3 has emerged as the only lineage-defining transcription factor identified that is required for tuft cell specification. POU2F3 is expressed in TRPM5+ chemosensory cells, including taste receptor cells and tuft cells, which are all absent in Pou2f3-deficient mice (13, 20, 99, 100), with the only exception so far being certain TRPM5− Villinbright brush cells outside the intestinal tract, whose identity remains unknown (46). How Pou2f3 expression is regulated and its target genes in tuft cell fate determination are unknown. The role of the transcription factor GFI1B, which in the intestinal epithelium is expressed specifically by tuft cells, remains to be determined (92).
Based on existing literature, we propose the following model for tuft cell lineage specification in the small intestine (Figure 3a and b): under germfree/fetal conditions, epithelial progenitors acquire tuft cell fate in an ATOH1-dependent manner, like other secretory lineages; alternatively, upon activation of IL-4Rα signaling and SOX4, an ATOH1-independent pathway is engaged, possibly involving different progenitor cells, enabling a rapid and dynamic increase in small intestinal tuft cell differentiation in the context of colonization with helminths or certain protists.
Figure 3:
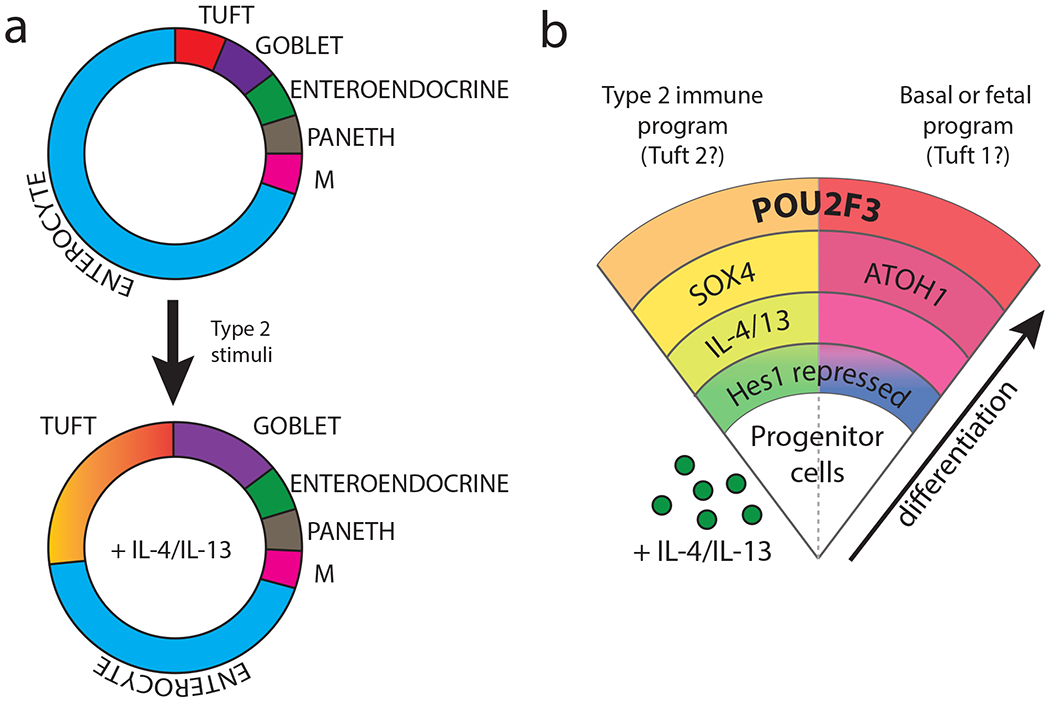
(a) Small intestinal epithelia comprise absorptive enterocytes , M cells, and secretory lineages, including tuft, goblet, enteroendocrine and Paneth cells, which derive from progenitor cells in the intestinal crypt. In the presence of type 2 immune stimulation, IL-4/13-dependent tuft and goblet cell hyperplasia is induced (not to scale). (b) In addition to the increased proportion of tuft cells, extrapolation from existing literature suggests that the tuft cells arising in this IL4Rα-dependent fashion could engage an alternative path of differentiation, schematized on the right, developing in a SOX4-dependent, ATOH1-independent fashion from an unknown epithelial progenitor, and in contrast to a basal and fetal trajectories involving an ATOH1-dependent development similar to other secretory lineages. Thus, observed tuft cell heterogeneity could be programmed early in lineage specification by impacts on epithelial progenitor cells.
While the roles of small intestinal tuft cells in type 2 immune responses have recently been elucidated (see below), both small and large intestinal tuft cells share a “neuronal” gene signature suggestive of roles in both immune and neuronal circuits. The emerging concept that tuft cells in the intestine are part of a diffuse cholinergic chemosensory system is supported by the expression of taste transduction signaling components and ChAT by intestinal tuft cells. However, intestinal tuft cells lack the high-affinity choline transporter (CHT) and the vesicular acetylcholine transporter (VAChT), responsible for loading acetylcholine into secretory organelles, with the exception of a VAChT+ subset of tuft cells in the ascending colon (55). The bitter ligand denatonium stimulates intracellular calcium flux in colon tuft cells (55). A Tas2r143/Tas2r135/Tas2r126 cluster fate-mapping mouse model marked tuft cell in the gastroinstinal tract and revealed Tas2r126 expression by gastric tuft cells (40). Further, tuft cell – neuronal cross talk has been suggested based on the co-localization of tuft cells with nerves (reviewed in (101)). Nonetheless, many proposed roles of intestinal tuft cells, including a receptive function—until recently (discussed below)—lacked supporting experimental evidence.
Urogenital tissues
Chat-eGFP reporter mice were used to identify urogenital tuft cells in the urethral duct epithelia and adjoining excretory gland ducts (15, 102). Urethral tuft cells are POU2F3-dependent (46) and express typical taste receptor sensing pathway constituents (15, 40); further, these cells fluxed Ca2+ in response to monosodium glutamate and denatonium, as well as heat-killed Escherichia coli, a common urinary tract pathogen, and ACh concentration in the media increased following denatonium-challenge of urethral cells in vitro (15). In vivo, denatonium induced activation of the bladder detrusor muscle, suggesting a role for tuft cells in restricting reflux of microbes into urogenital organs (15). Similar cells were present in humans and other placental mammals, consistent with an ancient evolutionary origin (103).
Intestinal tuft cell chemosensation and integration in immune circuits
In 2016, three groups converged on tuft cells as critical players in induction of intestinal type 2 immune responses to luminal helminths and protists (Tritrichomonas muris) (11–13). Using an IL-25-reporter-floxed allele, von Moltke et al. demonstrated that tuft cells are a constitutive and major, if not only, source of IL-25. After infection with intestinal helminths, IL-25 stimulated lamina propria ILC2s to secrete IL-13 (11). IL-13 further drives tuft and goblet cell differentiation, amplifying the circuit and expanding the numbers of IL-25-producing tuft cells. Using Pou2f3−/− mice, which lack tuft cells, Gerbe et al. also demonstrated the requirement for tuft cells in this cytokine relay (13). The resulting ‘weep-and sweep’ response that accompanies infection was dependent on IL-13, IL-4Rα, IL-25, and POU2F3, with deficiency resulting in delayed expulsion of the helminth N. brasiliensis. Howitt et al. made the important discovery that an unappreciated protozoan pathosymbiont of the genus Tritrichomonas also elicited expansion of the tuft cell compartment via this circuit, leading to variable frequencies of tuft cells among mice maintained in different facilities (12). Tritrichomonas-mediated tuft cell expansion was TRPM5- and α-gustducin-dependent, providing the first evidence that chemosensation-based detection of eukaryotic pathosymbionts by tuft cells is transmitted to ILC2s in the lamina propria.
The nature of the upstream tuft cell agonist remained unknown until 2018. Following the observation that small intestinal tuft cells differentially express the succinate receptor GPR91 (SUCNR1), evident in prior work (6, 7), three groups independently identified the metabolite succinate as a potent circuit agonist (9, 95, 104). In a TRPM5-, α-gustducin-, IL-25-, and POU2F3-dependent fashion, succinate in the drinking water was sufficient to promote small intestinal tuft cell expansion, ILC2 proliferation, and IL-13 expression. Germ-free mice monocolonized with Tritrichomonas accumulated luminal succinate (95), and Tritrichomonas activated the circuit in a Sucnr1-dependent manner (9). Altering the bacterial microbiota using streptomycin or polyethylene glycol 3350, which promotes a succinate-producing flora, also triggered SUCNR1 (104). Surprisingly, although succinate is excreted by N. brasiliensis, tuft cell expansion and worm clearance were normal in Sucnr1−/− mice, in contrast to Trpm5−/− mice, suggesting alternative or redundant pathways for helminth-driven circuit activation (9, 104). Circuit activation did not impact Tritrichomonas load, suggesting that this pathway for luminal detection may have evolved to facilitate mutualistic responses to luminal pathosymbionts, including adaptive tissue remodeling that facilitates energy homeostasis (95). Using a genetic model for chronic circuit activation, Schneider et al. demonstrated that the associated small intestinal remodeling impairs helminth parasitism, indicative of concomitant immunity (95). Tuft cells themselves are targets for murine norovirus strain CR6 in the mouse intestine due to their high expression of the entry receptor CD300lf (105). Notably, small intestinal circuit activity is also influenced by diet (95, 106), suggesting additional nuances awaiting further discovery.
Tuft cells in epithelial injury and cancer
In several tissues, including the small intestine, colon, pancreas and stomach, BrdU labeling and/or lineage tracing of DCLK1+ cells revealed that tuft cells are post-mitotic and do not serve as stem or lineage-committed progenitors (75, 93, 107, 108). Nevertheless, tuft cells have been implicated in small and large intestinal tissue regeneration in models of irradiation-induced injury and in DSS colitis, in which mice with Dclk1 deletion (109, 110) or inducible Dclk1-driven tuft cell depletion (93) exhibit poorer epithelial recovery and survival. Interpretation of these results is complicated by the expression of DCLK1 in cell types other than tuft cells, including neurons (111), requiring validation using more specific models. However, in support of a regenerative role for tuft cells, Westphalen et al. detected a small percentage of predominantly crypt-localized DCLK1+ tuft cells throughout the gastrointestinal tract, which, following fate-mapping via a high-sensitivity Dclk1-CreERT2 BAC transgenic, retained label for up to 18 months (93). These data are in contrast to a previous report using a knock-in/knock-out Dclk1-CreERT2 allele (107), but may be consistent with earlier descriptions of DCLK1low crypt resident, Ki67+, GF1b+ cells (92). The potential for de-differentiation of committed cells to replenish the stem cell compartment during regeneration has repeatedly been demonstrated for both uncommitted and mature intestinal epithelial cells, including quiescent label-retaining +4 cells—partially committed secretory progenitors—and enterocytes and enteroendocrine cells, which undergo dynamic chromatin remodeling and resemble bona fide LGR5+ stem cells after injury (112). Thus, it is reasonable to hypothesize that DCLK1+ cells share this potential, perhaps serving in a pool of uncommitted crypt-residing secretory precursors (113). This has led to speculation that, following inflammation, stem cell depletion, or activation of oncogenic mutations (APC, KRAS), post-mitotic cell fate may be altered, possibly explaining the association of DCKL1 expression with tumorigenesis and tissue injury (114).
A growing literature suggests a role for DCLK1 mutations—specifically inactivating mutations in autoinhibitory domains (115)—in tumor incidence and progression in human cancers (SIDEBAR DCLK1). Upregulation of DCLK1 occurs in some human tumors, most notably in pancreatic tumors and metastases where this negatively correlates with survival (73, 116, 117). In mouse models of both gastric (14, 118, 119) and pancreatic (74, 116) cancer, increased tuft cells were observed during initiation of tumorigenesis under inflammatory conditions, with a decrease noted upon progression to later tumor stages. These data, together with the possibility that a DCLK1+ tuft cell can participate in tissue regeneration, suggest a relationship between tuft cells and dysregulated pro-tumorigenic DCLK1+ cancer cells. In the adenomatosis polyposis coli (APC) mutant model, polyp formation were decreased when DCLK1+ cells were depleted (107). Tumorigenic potential of DCLK1+ cells was also observed in Dclk1-CreERT2 mice when the tumor suppressor gene Apc was deleted specifically in DCLK1+ cells in combination with DSS-induced inflammation (93). DCLK1 expression is not unique to tuft cells, however, and cleaner genetic tools are required. A recent study utilized the lineage-defining nature of POU2F3 to demonstrate that a small cell lung cancer variant—previously believed to be neuroendocrine—has tuft cell-like hallmarks: variant tumor cells expressed POU2F3, TRPM5, GFI1B, and ChAT, with a POU2F3-activated enhancer landscape (120). Immunofluorescence of mouse lung using POU2F3 staining revealed tuft cells in trachea, but also in primary and secondary bronchi, consistent with the central airway presentation of human small cell lung cancers.
Tuft cells comprise a systemically dispersed chemosensory cell system linking environmental signals with neural and immune circuits
Tuft cells are widely dispersed solitary chemosensory cells that utilize components of the Type II taste cell signaling cascade responsible for sensing sweet, bitter, and umami. Tuft cells express numerous additional GPCRs, including orphan receptors (7), but the connections between alternative G protein-coupled upstream signals and activation of the canonical taste transduction cascade, as defined for SUCNR1 in small intestinal tuft cells, remain undefined. The underlying logic of tuft cell “sensing” across different tissues is not clear, but distinct expression patterns of GPCRs among tissues is consistent with a sensory system modified to interact with ligands present in disparate microenvironments. Where tested, signals in all tuft cells propagate through the Ca2+-activated TRPM5 monovalent cation channel, universally expressed in these cells. Once activated, the output of these cells appears stereotyped, although not explicitly tested in all tissues, and experimental evidence and/or gene expression supports tuft cell production of IL-25, ACh and eicosanoids (certain prostaglandins and cysteinyl leukotrienes). Despite the diverse potential signals in distinct microenvironments, the functional output of tuft cells in all tissues examined is consistent with primary targets in the immune and nervous systems, relating specifically to type 2 immune responses and cholinergic signaling. Cellular partners in such tuft cell-nucleated circuits remain to be defined in the majority of tissues where tuft cells are found.
The use of components of the taste transduction cascade suggests evolutionary relationships with taste bud cells, particularly Type II cells, which share expression of sweet (T1R2/T1R3 components), umami (T1R1/T1R3 components) and bitter (a large family of T2Rs) taste receptors, their associated signaling components, and (perhaps) ChAT, although further study is needed (51, 121). Type II taste cells are also dependent on POU2F3 for their development and signal in a TRPM5-dependent fashion (99). Surprisingly, Type II taste cells express IL-25 as revealed using reporter mice (Fig. 4). The similarities between tuft cells and Type II taste cells suggests a shared cellular lineage precursor or evolutionary convergence of distinct linages. We propose the term “tuft cells” to describe a systemically dispersed chemosensory system (inclusive of Type II taste cells) comprised of epithelial cells sharing the following characteristics: POU2F3-dependent development; tuft and microvillous morphology; and expression of TRPM5, IL-25 and pathways of eicosanoid synthesis.
Figure 4:
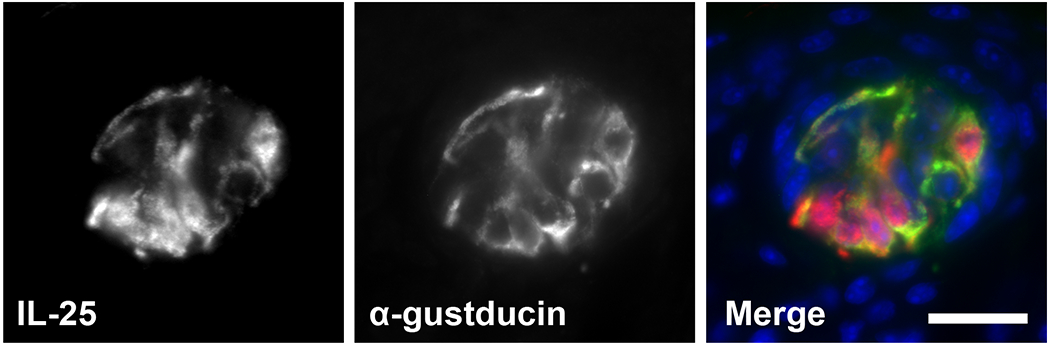
Expression of IL-25 (in red) in lingual taste buds of IL-25-RFP reporter mice; DAPI in blue. Taste bud structure is outlined. Scale bar, 20 μm.
Recent work suggests tuft cell heterogeneity in the small intestine, as noted earlier. Of particular interest in conceptualizing tuft cell functions is that these two types of small intestinal tuft cells associate with patterns of neuronal-associated genes (“type 1”) or innate immune genes (“type 2”), although both populations express IL-25, the IL-25 receptor IL-17RB, and the IL-13 receptor constituents, IL4Rα and IL-13Rα1 (7). This suggests that the small intestine—where immunological function of tuft cells is best characterized—may represent a tissue in which luminal signals robustly direct polarization or differentiation of “type 2” effector tuft cells, while in other tissues—in which tuft cells are better characterized by neuronal or sensory outputs—the “type 1” tuft cell may predominate. Whether a single tuft cell, particularly in tissues where these cells are long-lived, can switch between these outputs depending on signaling inputs, or whether these are true differentiation states remains unknown. Better fate-mapping tools will be necessary to define the relationships among these cells, particularly in comparing epithelia with distinct stem cell pools, stromal niches, and turnover rates.
The diversity of GPCRs expressed across tuft cells from distinct tissues suggest these cells may couple to distinct ligands in different compartments. In the nose, tuft cells respond to bitter ligand bacterial quorum-sensing molecules to promote aversive behaviors and antimicrobial defense; similar activities appear to occur in the urethra (15, 22). In small intestine, tuft cells use the succinate receptor in response to colonizing protists, driving extensive tissue remodeling associated with concomitant immunity (9, 95, 122). Analogously, Type II taste cells drive both positive (sweet/umami) and aversive (bitter) behaviors, depending on the taste receptor repertoire. Although the data set is small, tuft cells seem to rheostat both feed-forward positive and feedback aversive reactions, perhaps consistent with odorant and taste perceptions involved in nutrition and toxin detection, predator/prey relationships, and even access of volatile pheromones involved in mating behavior (123). In this way, the tuft cell chemosensory system resembles the solitary chemosensory system of fish and amphibians, that has seemingly evolved from externally arrayed cells that detect highly-diluted water-soluble molecules in fish to more internally arrayed mucosal epithelial cells for detection of both soluble and volatile molecules engaged at air-liquid interfaces in land animals; such sensory systems may have evolved to distinguish environmental cues involved in food finding, habitat recognition and avoidance behavior (124). Deep evolutionary roots are also suggested by selective effects on POU2F3 in human evolution (125).
We speculate that all tuft cells function through detection of endogenous or exogenous metabolites, including odorants and taste ligands, that coordinate positive (nutrient-rich, prey, etc.) or negative (toxins, predators, etc.) tissue and behavioral responses. As demonstrated by the capacity of tuft cell stimulation in mammals and solitary chemosensory cells in fish to cause intestinal tissue remodeling (95, 126), we further speculate that tuft cells are integrated upstream of signals important for postnatal tissue growth and differentiation, and that such inputs may be relevant in driving tissue receptive states in response to nutrients, but also pathologic states like allergy, chronic nasal sinusitis and polyposis, asthma, food reactivity, and potentially others diseases provoked by alterations in the environment in modernized civilizations.
SUMMARY POINTS.
Evolutionarily conserved, with similarity to chemosensory cells in fish, tuft cells share a unique gene program and are found as isolated cells in columnar epithelia throughout mammalian organ systems.
Tuft cells have properties of both immune- and neural-modulating cells, including a limited repertoire of secreted effector molecules that could interact with several distinct cellular partners.
Physiologic roles for tuft cells in the majority of tissues remain to be defined.
The relationship between tuft cells and neurons is best characterized in “sensory” epithelia like the oropharnyx.
The tuft cell/immune cell relationship has been conclusively demonstrated in small intestine, where IL-25 from tuft cells initiates ILC2 responses to intestinal metabolites and parasites.
Type II taste bud cells share a common gene signature and morphology with tuft cells, suggesting that these cells are a single lineage with distinct responsivities based on tissue location.
We propose that tuft cells be defined as epithelial cells sharing POU2F3-dependence and characterized by expression of IL-25, acetylcholine and eicosanoid biosynthetic pathways, and the canonical taste receptor signaling cascade.
FUTURE ISSUES.
The differentiation pathway of tuft cells remains unclear, even in small intestine where this has been most pursued.
In many tissues containing tuft cells, ILC2s have not been described or immune responses are thought to be limited, and investigations of tuft cell – immune (ILC2 or otherwise) cell interactions need further study.
Potential for tuft cell secretion of prostaglandins and leukotrienes remains unexplored.
The function of tuft cell acetylcholine - and whether ChAT expression is sufficient to infer functional acetylcholine synthesis - —remains unclear, particularly in the context of other acetylcholine-producing cells in close proximity to tuft cells.
The repertoire of cell surface receptors and possible tuft cell ligands, as well as tissue specificity and heterogeneity within tissues, will offer important clues to tuft cell function, and may explain how tuft cells acquire neuronal- or immune-biased effector functions.
Recognition of Type II taste bud cells as homologous in phenotype and function to tuft cells in other tissues will facilitate new understanding of tuft cell and taste cell function, including the impact of IL-25 in sensory function.
ACKNOWLEDGEMENTS
The authors appreciate comments from Locksley lab members and the photo of IL-25-positive taste bud cells by Benjamin Terrier while he was a Fulbright visiting scholar. This work was supported by grants from the NIH, HHMI and the SABRE Center at UCSF. CEO is supported by the Gastroenterology T32 Training Grant at UCSF. CS is supported by Fellowships from the Swiss National Science Foundation.
TERMS AND DEFINITIONS
- Eicosanoids
polyunsaturated fatty acid oxidation products of arachidonic acid with signaling properties
- GPCR
G-protein coupled receptor
- Acetylcholine (ACh)
neurotransmitter/neuromodulator, acts on cells expressing muscarinic (GPCR) or nicotinic (ion channel) receptors
- Cyclooxygenase 1/2 (COX1/2)
constitutively (COX1) or inducibly (COX2) expressed prostaglandin-endoperoxide synthase; converts arachidonic acid to prostanoids (prostaglandins and thromboxins)
- Arachidonate 5-Lipoxygenase (ALOX5)
enzyme required in conversion of arachidonate to precursor of leukotrienes B4 and C4
- Type 2 immunity
immune response associated with activation of ILC2s, Th2 cells and TFH cells mediating B cell production of IgE antibodies
- IL-25
tuft cell-derived IL-17 family cytokine; previously designated IL-17E
- IL-17RB
forms the heterodimeric IL-25 receptor with IL-17RA
- PLCβ2
phosphodiesterase that catalyzes the formation of inositol 1,4,5-trisphosphate and diacylglycerol from phosphatidylinositol 4,5-bisphosphate
- Group 2 innate lymphocyte (ILC2)
innate immune cells producing IL-5 and IL-13 with early roles in Type 2 immune responses
- Autoimmune regulator (AIRE)
largely thymic-restricted transcription factor promoting ectopic expression of cell type specific antigens by medullary thymic epithelial cells
- Quorum sensing
bacterial sensing of population size through signaling molecules
- Leucine Rich Repeat Containing G Protein-Coupled Receptor 5 (LGR5)
binds R-spondins in canonical Wnt signaling; tissue-specific epithelial stem cell marker
- N. brasiliensis
rat-adapted parasitic roundworm, similar to human hookworms, that infects mice transiently and induces robust type 2 response
- Tritrichomonas muris
intestinal protozoan; likely many subtypes abundant in feral and laboratory mice; characteristic hydrogenosome-dependent anaerobic metabolism
- CD300lf
bile acid-binding co-receptor for mouse norovirus
- Cholinergic
involving the neurotransmitter acetylcholine
- Pathosymbiont
immune-stimulating organism with long-term host residency
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Sbarbati A, Osculati F. 2005. A new fate for old cells: brush cells and related elements. J Anat 206: 349–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato A 2007. Tuft cells. Anat Sci Int 82: 187–99 [DOI] [PubMed] [Google Scholar]
- 3.Gerbe F, Legraverend C, Jay P. 2012. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci 69: 2907–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee A, McKinley ET, von Moltke J, Coffey RJ, Lau KS. 2018. Interpreting heterogeneity in intestinal tuft cell structure and function. J Clin Invest 128: 1711–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Moltke J 2018. Chapter 31 - Intestinal Tuft Cells A2 - Said, Hamid M In Physiology of the Gastrointestinal Tract (Sixth Edition), pp. 721–33: Academic Press [Google Scholar]
- 6.Bezencon C, Furholz A, Raymond F, Mansourian R, Metairon S, Le Coutre J, Damak S. 2008. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. J Comp Neurol 509: 514–25 [DOI] [PubMed] [Google Scholar]; First expression profiling of tuft cells using Trpm5-GFP reporter revealing chemosensory-immune-neuronal programs
- 7.Haber AL, Biton M, Rogel N, Herbst RH, Shekhar K, Smillie C, Burgin G, Delorey TM, Howitt MR, Katz Y, Tirosh I, Beyaz S, Dionne D, Zhang M, Raychowdhury R, Garrett WS, Rozenblatt-Rosen O, Shi HN, Yilmaz O, Xavier RJ, Regev A. 2017. A single-cell survey of the small intestinal epithelium. Nature 551: 333–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent AV, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS. 2018. Thymic tuft cells promote an IL4-enriched medullary microenvironment and shape thymocyte development. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Erle DJ, Anderson MS, Locksley RM, Raftery D, Moltke J. 2018. Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity, under revision . [DOI] [PMC free article] [PubMed] [Google Scholar]; RNA-seq profiling of tuft cells and identification of Tritrichomonas-induced succinate-SUCNR1-mediated activation
- 10.Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Toth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann V, Abramson J, Amit I. 2018. Single cell mapping of the thymic stroma identifies IL25-producing tuft epithelial cells. Nature, in press [DOI] [PubMed] [Google Scholar]
- 11.von Moltke J, Ji M, Liang HE, Locksley RM. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529: 221–5 [DOI] [PMC free article] [PubMed] [Google Scholar]; Tuft cells are the major source of IL-25; first description of tuft cell–ILC2 circuit
- 12.Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, Gallini CA, Redding K, Margolskee RF, Osborne LC, Artis D, Garrett WS. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351: 1329–33 [DOI] [PMC free article] [PubMed] [Google Scholar]; Protist Tritrichomonas potently stimulates TRPM5-dependent ILC2-tuft cell circuit
- 13.Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, Bruschi M, Harcus Y, Zimmermann VS, Taylor N, Maizels RM, Jay P. 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529: 226–30 [DOI] [PMC free article] [PubMed] [Google Scholar]; Tuft cell deficiency in Pou2f3−/− mice abrogates helminth-induced small intestinal type 2 responses
- 14.Hayakawa Y, Sakitani K, Konishi M, Asfaha S, Niikura R, Tomita H, Renz BW, Tailor Y, Macchini M, Middelhoff M, Jiang Z, Tanaka T, Dubeykovskaya ZA, Kim W, Chen X, Urbanska AM, Nagar K, Westphalen CB, Quante M, Lin CS, Gershon MD, Hara A, Zhao CM, Chen D, Worthley DL, Koike K, Wang TC. 2017. Nerve Growth Factor Promotes Gastric Tumorigenesis through Aberrant Cholinergic Signaling. Cancer Cell 31: 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deckmann K, Filipski K, Krasteva-Christ G, Fronius M, Althaus M, Rafiq A, Papadakis T, Renno L, Jurastow I, Wessels L, Wolff M, Schutz B, Weihe E, Chubanov V, Gudermann T, Klein J, Bschleipfer T, Kummer W. 2014. Bitter triggers acetylcholine release from polymodal urethral chemosensory cells and bladder reflexes. Proc Natl Acad Sci U S A 111: 8287–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasteva G, Canning BJ, Hartmann P, Veres TZ, Papadakis T, Muhlfeld C, Schliecker K, Tallini YN, Braun A, Hackstein H, Baal N, Weihe E, Schutz B, Kotlikoff M, Ibanez-Tallon I, Kummer W. 2011. Cholinergic chemosensory cells in the trachea regulate breathing. Proc Natl Acad Sci U S A 108: 9478–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinnamon SC. 2012. Taste receptor signalling - from tongues to lungs. Acta Physiol (Oxf) 204: 158–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoover B, Baena V, Kaelberer MM, Getaneh F, Chinchilla S, Bohorquez DV. 2017. The intestinal tuft cell nanostructure in 3D. Sci Rep 7: 1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamanza R, Wright JA. 2015. A Review of the Comparative Anatomy, Histology, Physiology and Pathology of the Nasal Cavity of Rats, Mice, Dogs and Non-human Primates. Relevance to Inhalation Toxicology and Human Health Risk Assessment. J Comp Pathol 153: 287–314 [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi T, Yamashita J, Ohmoto M, Aoude I, Ogura T, Luo W, Bachmanov AA, Lin W, Matsumoto I, Hirota J. 2014. Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium. BMC Neurosci 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders CJ, Christensen M, Finger TE, Tizzano M. 2014. Cholinergic neurotransmission links solitary chemosensory cells to nasal inflammation. Proc Natl Acad Sci U S A 111: 6075–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tizzano M, Gulbransen BD, Vandenbeuch A, Clapp TR, Herman JP, Sibhatu HM, Churchill ME, Silver WL, Kinnamon SC, Finger TE. 2010. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc Natl Acad Sci U S A 107: 3210–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee RJ, Kofonow JM, Rosen PL, Siebert AP, Chen B, Doghramji L, Xiong G, Adappa ND, Palmer JN, Kennedy DW, Kreindler JL, Margolskee RF, Cohen NA. 2014. Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J Clin Invest 124: 1393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee RJ, Hariri BM, McMahon DB, Chen B, Doghramji L, Adappa ND, Palmer JN, Kennedy DW, Jiang P, Margolskee RF, Cohen NA. 2017. Bacterial d-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. Sci Signal 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunican EM, Elicker BM, Gierada DS, Nagle SK, Schiebler ML, Newell JD, Raymond WW, Lachowicz-Scroggins ME, Di Maio S, Hoffman EA, Castro M, Fain SB, Jarjour NN, Israel E, Levy BD, Erzurum SC, Wenzel SE, Meyers DA, Bleecker ER, Phillips BR, Mauger DT, Gordon ED, Woodruff PG, Peters MC, Fahy JV. 2018. Mucus plugs in patients with asthma linked to eosinophilia and airflow obstruction. J Clin Invest 128: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schleimer RP. 2017. Immunopathogenesis of Chronic Rhinosinusitis and Nasal Polyposis. Annu Rev Pathol 12: 331–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachert C, Mannent L, Naclerio RM, Mullol J, Ferguson BJ, Gevaert P, Hellings P, Jiao L, Wang L, Evans RR, Pirozzi G, Graham NM, Swanson B, Hamilton JD, Radin A, Gandhi NA, Stahl N, Yancopoulos GD, Sutherland ER. 2016. Effect of Subcutaneous Dupilumab on Nasal Polyp Burden in Patients With Chronic Sinusitis and Nasal Polyposis: A Randomized Clinical Trial. Jama 315: 469–79 [DOI] [PubMed] [Google Scholar]
- 28.Kohanski MA, Workman AD, Patel NN, Hung LY, Shtraks JP, Chen B, Blasetti M, Doghramji L, Kennedy DW, Adappa ND, Palmer JN, Herbert DR, Cohen NA. 2018. Solitary Chemosensory Cells are a Primary Epithelial Source of Interleukin-25 in Chronic Rhinosinusitis with Nasal Polyps. J Allergy Clin Immunol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanishi W, Yamaguchi S, Matsuda A, Suzukawa M, Shibui A, Nambu A, Kondo K, Suto H, Saito H, Matsumoto K, Yamasoba T, Nakae S. 2013. IL-33, but not IL-25, is crucial for the development of house dust mite antigen-induced allergic rhinitis. PLoS One 8: e78099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Deb C, Wadsworth MH, Hughes TK, Kazer SW, Yoshimoto E, Bhattacharyya N, Katz HR, Laidlaw TM, Boyce JA, Barrett NA, Shalek AK. 2017. Reduced cellular diversity and an altered basal progenitor cell state inform epithelial barrier dysfunction in human type 2 immunity. bioRxiv [Google Scholar]
- 31.Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J. 2017. Deconstructing Olfactory Stem Cell Trajectories at Single-Cell Resolution. Cell Stem Cell 20: 817–30.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Fletcher RB. 2017. Injury Activates Transient Olfactory Stem Cell States with Diverse Lineage Capacities. Cell Stem Cell 21: 775–90.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogura T, Szebenyi SA, Krosnowski K, Sathyanesan A, Jackson J, Lin W. 2011. Cholinergic microvillous cells in the mouse main olfactory epithelium and effect of acetylcholine on olfactory sensory neurons and supporting cells. J Neurophysiol 106: 1274–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemons K, Fu Z, Aoude I, Ogura T, Sun J, Chang J, Mbonu K, Matsumoto I, Arakawa H, Lin W. 2017. Lack of TRPM5-Expressing Microvillous Cells in Mouse Main Olfactory Epithelium Leads to Impaired Odor-Evoked Responses and Olfactory-Guided Behavior in a Challenging Chemical Environment. eNeuro 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle KL, Cunha C, Hort Y, Tasan R, Sperk G, Shine J, Herzog H. 2018. Role of neuropeptide Y (NPY) in the differentiation of Trpm-5-positive olfactory microvillar cells. Neuropeptides 68: 90–98 [DOI] [PubMed] [Google Scholar]
- 36.Krasteva G, Hartmann P, Papadakis T, Bodenbenner M, Wessels L, Weihe E, Schutz B, Langheinrich AC, Chubanov V, Gudermann T, Ibanez-Tallon I, Kummer W. 2012. Cholinergic chemosensory cells in the auditory tube. Histochem Cell Biol 137: 483–97 [DOI] [PubMed] [Google Scholar]
- 37.Wiederhold S, Papadakis T, Chubanov V, Gudermann T, Krasteva-Christ G, Kummer W. 2015. A novel cholinergic epithelial cell with chemosensory traits in the murine conjunctiva. Int Immunopharmacol 29: 45–50 [DOI] [PubMed] [Google Scholar]
- 38.Panneck AR, Rafiq A, Schutz B, Soultanova A, Deckmann K, Chubanov V, Gudermann T, Weihe E, Krasteva-Christ G, Grau V, del Rey A, Kummer W. 2014. Cholinergic epithelial cell with chemosensory traits in murine thymic medulla. Cell Tissue Res 358: 737–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soultanova A, Voigt A, Chubanov V, Gudermann T, Meyerhof W, Boehm U, Kummer W. 2015. Cholinergic chemosensory cells of the thymic medulla express the bitter receptor Tas2r131. Int Immunopharmacol 29: 143–7 [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Lu S, Xu R, Atzberger A, Gunther S, Wettschureck N, Offermanns S. 2017. Members of Bitter Taste Receptor Cluster Tas2r143/Tas2r135/Tas2r126 Are Expressed in the Epithelium of Murine Airways and Other Non-gustatory Tissues. Front Physiol 8: 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swarr DT, Morrisey EE. 2015. Lung endoderm morphogenesis: gasping for form and function. Annu Rev Cell Dev Biol 31: 553–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, Rock J, Snitow M, Krummel M, Stripp BR, Vu T, White ES, Whitsett JA, Morrisey EE. 2014. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15: 123–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid L, Meyrick B, Antony VB, Chang LY, Crapo JD, Reynolds HY. 2005. The mysterious pulmonary brush cell: a cell in search of a function. Am J Respir Crit Care Med 172: 136–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tizzano M, Cristofoletti M, Sbarbati A, Finger TE. 2011. Expression of taste receptors in solitary chemosensory cells of rodent airways. BMC Pulm Med 11: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tizzano M, Merigo F, Sbarbati A. 2006. Evidence of solitary chemosensory cells in a large mammal: the diffuse chemosensory system in Bos taurus airways. J Anat 209: 333–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamashita J, Ohmoto M, Yamaguchi T, Matsumoto I, Hirota J. 2017. Skn-1a/Pou2f3 functions as a master regulator to generate Trpm5-expressing chemosensory cells in mice. PLoS One 12: e0189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saunders CJ, Reynolds SD, Finger TE. 2013. Chemosensory brush cells of the trachea. A stable population in a dynamic epithelium. Am J Respir Cell Mol Biol 49: 190–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krasteva G, Canning BJ, Papadakis T, Kummer W. 2012. Cholinergic brush cells in the trachea mediate respiratory responses to quorum sensing molecules. Life Sci 91: 992–6 [DOI] [PubMed] [Google Scholar]
- 49.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. 2009. Motile cilia of human airway epithelia are chemosensory. Science 325: 1131–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshpande DA, Wang WC, McIlmoyle EL, Robinett KS, Schillinger RM, An SS, Sham JS, Liggett SB. 2010. Bitter taste receptors on airway smooth muscle bronchodilate by localized calcium signaling and reverse obstruction. Nat Med 16: 1299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roper SD, Chaudhari N. 2017. Taste buds: cells, signals and synapses. Nat Rev Neurosci 18: 485–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohmoto M, Matsumoto I, Yasuoka A, Yoshihara Y, Abe K. 2008. Genetic tracing of the gustatory and trigeminal neural pathways originating from T1R3-expressing taste receptor cells and solitary chemoreceptor cells. Mol Cell Neurosci 38: 505–17 [DOI] [PubMed] [Google Scholar]
- 53.Chin AM, Hill DR, Aurora M, Spence JR. 2017. Morphogenesis and maturation of the embryonic and postnatal intestine. Semin Cell Dev Biol 66: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mowat AM, Agace WW. 2014. Regional specialization within the intestinal immune system. Nat Rev Immunol 14: 667–85 [DOI] [PubMed] [Google Scholar]
- 55.Schutz B, Jurastow I, Bader S, Ringer C, von Engelhardt J, Chubanov V, Gudermann T, Diener M, Kummer W, Krasteva-Christ G, Weihe E. 2015. Chemical coding and chemosensory properties of cholinergic brush cells in the mouse gastrointestinal and biliary tract. Front Physiol 6: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee MD, Lee Y, Friedman R, Asfaha S, Dubeykovskaya Z, Mahmood U, Figueiredo JL, Kitajewski J, Shawber C, Lightdale CJ, Rustgi AK, Wang TC. 2012. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell 21: 36–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Housset C, Chretien Y, Debray D, Chignard N. 2016. Functions of the Gallbladder. Compr Physiol 6: 1549–77 [DOI] [PubMed] [Google Scholar]
- 58.Spence JR, Lange AW, Lin SC, Kaestner KH, Lowy AM, Kim I, Whitsett JA, Wells JM. 2009. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell 17: 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrett KE. 2014. Chapter 12. Gallbladder Function. In Gastrointestinal Physiology, 2e. New York, NY: The McGraw-Hill Companies [Google Scholar]
- 60.Reichert M, Rustgi AK. 2011. Pancreatic ductal cells in development, regeneration, and neoplasia. J Clin Invest 121: 4572–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hundt M, John S. 2018. Physiology, Bile Secretion. In StatPearls. Treasure Island (FL): StatPearls Publishing StatPearls Publishing LLC. [PubMed] [Google Scholar]
- 62.Gunn JS, Marshall JM, Baker S, Dongol S, Charles RC, Ryan ET. 2014. Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol 22: 648–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luciano L, Castellucci M, Reale E. 1981. The brush cells of the common bile duct of the rat. This section, freeze-fracture and scanning electron microscopy. Cell Tissue Res 218: 403–20 [DOI] [PubMed] [Google Scholar]
- 64.Nevalainen TJ. 1977. Ultrastructural characteristics of tuft cells in mouse gallbladder epithelium. Acta Anat (Basel) 98: 210–20 [DOI] [PubMed] [Google Scholar]
- 65.Luciano L, Reale E. 1997. Presence of brush cells in the mouse gallbladder. Microsc Res Tech 38: 598–608 [DOI] [PubMed] [Google Scholar]
- 66.Dancygier H 1989. Endoscopic transpapillary biopsy (ETPB) of human extrahepatic bile ducts--light and electron microscopic findings, clinical significance. Endoscopy 21Suppl 1: 312–20 [DOI] [PubMed] [Google Scholar]
- 67.Gieseck RL 3rd, Wilson MS, Wynn TA. 2018. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol 18: 62–76 [DOI] [PubMed] [Google Scholar]
- 68.Luciano L, Groos S, Reale E. 2003. Brush cells of rodent gallbladder and stomach epithelia express neurofilaments. J Histochem Cytochem 51: 187–98 [DOI] [PubMed] [Google Scholar]
- 69.Gilloteaux J, Pomerants B, Kelly TR. 1989. Human gallbladder mucosa ultrastructure: evidence of intraepithelial nerve structures. Am J Anat 184: 321–33 [DOI] [PubMed] [Google Scholar]
- 70.Weyrauch KD. 1979. [Ultrastructure of the Tuft cell in some epithelia of the domestic ruminants (author’s transl)]. Anat Anz 146: 141–51 [PubMed] [Google Scholar]
- 71.Weyrauch KD, Schnorr B. 1976. [Ultrastructure of the epithelium of the major pancreatic duct in sheep]. Acta Anat (Basel) 96: 232–47 [PubMed] [Google Scholar]
- 72.Hofer D, Drenckhahn D. 1998. Identification of the taste cell G-protein, alpha-gustducin, in brush cells of the rat pancreatic duct system. Histochem Cell Biol 110: 303–9 [DOI] [PubMed] [Google Scholar]
- 73.Delgiorno KE, Hall JC, Takeuchi KK, Pan FC, Halbrook CJ, Washington MK, Olive KP, Spence JR, Sipos B, Wright CV, Wells JM, Crawford HC. 2014. Identification and manipulation of biliary metaplasia in pancreatic tumors. Gastroenterology 146: 233–44.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W, Maitra A, Leach SD. 2014. DCLK1 marks a morphologically distinct subpopulation of cells with stem cell properties in preinvasive pancreatic cancer. Gastroenterology 146: 245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.May R, Sureban SM, Lightfoot SA, Hoskins AB, Brackett DJ, Postier RG, Ramanujam R, Rao CV, Wyche JH, Anant S, Houchen CW. 2010. Identification of a novel putative pancreatic stem/progenitor cell marker DCAMKL-1 in normal mouse pancreas. Am J Physiol Gastrointest Liver Physiol 299: G303–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim TH, Shivdasani RA. 2016. Stomach development, stem cells and disease. Development 143: 554–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hofer D, Puschel B, Drenckhahn D. 1996. Taste receptor-like cells in the rat gut identified by expression of alpha-gustducin. Proc Natl Acad Sci U S A 93: 6631–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O’Neil A, Petersen CP, Choi E, Engevik AC, Goldenring JR. 2017. Unique Cellular Lineage Composition of the First Gland of the Mouse Gastric Corpus. J Histochem Cytochem 65: 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hass N, Schwarzenbacher K, Breer H. 2007. A cluster of gustducin-expressing cells in the mouse stomach associated with two distinct populations of enteroendocrine cells. Histochem Cell Biol 128: 457–71 [DOI] [PubMed] [Google Scholar]
- 80.Sothilingam V, Hass N, Breer H. 2011. Candidate chemosensory cells in the stomach mucosa of young postnatal mice during the phases of dietary changes. Cell Tissue Res 344: 239–49 [DOI] [PubMed] [Google Scholar]
- 81.Ogata T 2000. Mammalian Tuft (Brush) Cells and Chloride Cells of Other Vertebrates Share a Similar Structure and Cytochemical Reactivities. ACTA HISTOCHEMICA ET CYTOCHEMICA 33: 439–49 [Google Scholar]
- 82.Ogata T 2006. Bicarbonate secretion by rat bile duct brush cells indicated by immunohistochemical localization of CFTR, anion exchanger AE2, Na+/HCO3 -cotransporter, carbonic anhydrase II, Na+/H+ exchangers NHE1 and NHE3, H+/K+-ATPase, and Na+/K+-ATPase. Med Mol Morphol 39: 44–8 [DOI] [PubMed] [Google Scholar]
- 83.Ogata T 2005. Sodium bicarbonate secretion indicated by ultrastructural cytochemical localization of HCO3(−), Cl-, and Na+ ions on rat bile duct brush cells. Med Mol Morphol 38: 243–50 [DOI] [PubMed] [Google Scholar]
- 84.Eberle JA, Muller-Roth KL, Widmayer P, Chubanov V, Gudermann T, Breer H. 2013. Putative interaction of brush cells with bicarbonate secreting cells in the proximal corpus mucosa. Front Physiol 4: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liman ER. 2014. TRPM5. Handb Exp Pharmacol 222: 489–502 [DOI] [PubMed] [Google Scholar]
- 86.Beumer J, Clevers H. 2016. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143: 3639–49 [DOI] [PubMed] [Google Scholar]
- 87.Noah TK, Shroyer NF. 2013. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol 75: 263–88 [DOI] [PubMed] [Google Scholar]
- 88.Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. 2000. Control of endodermal endocrine development by Hes-1. Nat Genet 24: 36–44 [DOI] [PubMed] [Google Scholar]
- 89.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294: 2155–8 [DOI] [PubMed] [Google Scholar]
- 90.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. 2007. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology 132: 2478–88 [DOI] [PubMed] [Google Scholar]
- 91.Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer NF, Bourgaux JF, Pignodel C, Clevers H, Jay P. 2011. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol 192: 767–80 [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed characterization of small intestinal tuft cell markers and development
- 92.Bjerknes M, Khandanpour C, Moroy T, Fujiyama T, Hoshino M, Klisch TJ, Ding Q, Gan L, Wang J, Martin MG, Cheng H. 2012. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev Biol 362: 194–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, Chen X, May R, Houchen CW, Fox JG, Gershon MD, Quante M, Wang TC. 2014. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 124: 1283–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herring CA, Banerjee A, McKinley ET, Simmons AJ, Ping J, Roland JT, Franklin JL, Liu Q, Gerdes MJ, Coffey RJ, Lau KS. 2018. Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut. Cell Syst 6: 37–51.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schneider C, O’Leary CE, von Moltke J, Liang HE, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, Locksley RM. 2018. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell [DOI] [PMC free article] [PubMed] [Google Scholar]; Tritrichomonas-derived succinate triggers tuft cell – ILC2 circuit involved in adaptive remodeling and concomitant immunity
- 96.McKinley ET, Sui Y, Al-Kofahi Y, Millis BA, Tyska MJ, Roland JT, Santamaria-Pang A, Ohland CL, Jobin C, Franklin JL, Lau KS, Gerdes MJ, Coffey RJ. 2017. Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gracz A, Samsa LA, Fordham MJ, Trotier DC, Zwarycz B, Lo Y-H, Bao K, Starmer J, Shroyer NF, Reinhardt RL, Magness ST. 2018. Sox4 drives Atoh1-independent intestinal secretory differentiation toward tuft and enteroendocrine fates. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Malik V, Zimmer D, Jauch R. 2018. Diversity among POU transcription factors in chromatin recognition and cell fate reprogramming. Cell Mol Life Sci 75: 1587–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. 2011. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat Neurosci 14: 685–7 [DOI] [PMC free article] [PubMed] [Google Scholar]; First description of POU2F3-dependency in Type II taste bud cells
- 100.Ohmoto M, Yamaguchi T, Yamashita J, Bachmanov AA, Hirota J, Matsumoto I. 2013. Pou2f3/Skn-1a is necessary for the generation or differentiation of solitary chemosensory cells in the anterior nasal cavity. Biosci Biotechnol Biochem 77: 2154–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Middelhoff M, Westphalen CB, Hayakawa Y, Yan KS, Gershon MD, Wang TC, Quante M. 2017. Dclk1-expressing tuft cells: critical modulators of the intestinal niche? Am J Physiol Gastrointest Liver Physiol 313: G285–g99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deckmann K, Kummer W. 2016. Chemosensory epithelial cells in the urethra: sentinels of the urinary tract. Histochem Cell Biol 146: 673–83 [DOI] [PubMed] [Google Scholar]
- 103.Deckmann K, Krasteva-Christ G, Rafiq A, Herden C, Wichmann J, Knauf S, Nassenstein C, Grevelding CG, Dorresteijn A, Chubanov V, Gudermann T, Bschleipfer T, Kummer W. 2015. Cholinergic urethral brush cells are widespread throughout placental mammals. Int Immunopharmacol 29: 51–6 [DOI] [PubMed] [Google Scholar]
- 104.Lei W, Ren W, Ohmoto M, Urban JF Jr., Matsumoto I, Margolskee RF, Jiang P 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 115: 5552–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL Jr., McAllaster MR, Balce DR, Feehley T, Brestoff JR, Hickey CA, Yokoyama CC, Wang YT, MacDuff DA, Kreamalmayer D, Howitt MR, Neil JA, Cadwell K, Allen PM, Handley SA, van Lookeren Campagne M, Baldridge MT, Virgin HW. 2018. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360: 204–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aladegbami B, Barron L, Bao J, Colasanti J, Erwin CR, Warner BW, Guo J. 2017. Epithelial cell specific Raptor is required for initiation of type 2 mucosal immunity in small intestine. Sci Rep 7: 5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, Isomura A, Kawada K, Sakai Y, Yanagita M, Kageyama R, Kawaguchi Y, Taketo MM, Yonehara S, Chiba T. 2013. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet 45: 98–103 [DOI] [PubMed] [Google Scholar]
- 108.Westphalen CB, Takemoto Y, Tanaka T, Macchini M, Jiang Z, Renz BW, Chen X, Ormanns S, Nagar K, Tailor Y, May R, Cho Y, Asfaha S, Worthley DL, Hayakawa Y, Urbanska AM, Quante M, Reichert M, Broyde J, Subramaniam PS, Remotti H, Su GH, Rustgi AK, Friedman RA, Honig B, Califano A, Houchen CW, Olive KP, Wang TC. 2016. Dclk1 Defines Quiescent Pancreatic Progenitors that Promote Injury-Induced Regeneration and Tumorigenesis. Cell Stem Cell 18: 441–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Li L, Sureban SM, Houchen CW. 2014. Brief report: Dclk1 deletion in tuft cells results in impaired epithelial repair after radiation injury. Stem Cells 32: 822–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qu D, Weygant N, May R, Chandrakesan P, Madhoun M, Ali N, Sureban SM, An G, Schlosser MJ, Houchen CW. 2015. Ablation of Doublecortin-Like Kinase 1 in the Colonic Epithelium Exacerbates Dextran Sulfate Sodium-Induced Colitis. PLoS One 10: e0134212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ohmae S, Takemoto-Kimura S, Okamura M, Adachi-Morishima A, Nonaka M, Fuse T, Kida S, Tanji M, Furuyashiki T, Arakawa Y, Narumiya S, Okuno H, Bito H. 2006. Molecular identification and characterization of a family of kinases with homology to Ca2+/calmodulin-dependent protein kinases I/IV. J Biol Chem 281: 20427–39 [DOI] [PubMed] [Google Scholar]
- 112.Jadhav U, Saxena M, O’Neill NK, Saadatpour A, Yuan GC, Herbert Z, Murata K, Shivdasani RA. 2017. Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell 21: 65–77 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, Roelf K, Calderon RI, Cynn E, Hu X, Mandleywala K, Wilhelmy J, Grimes SM, Corney DC, Boutet SC, Terry JM, Belgrader P, Ziraldo SB, Mikkelsen TS, Wang F, von Furstenberg RJ, Smith NR, Chandrakesan P, May R, Chrissy MAS, Jain R, Cartwright CA, Niland JC, Hong YK, Carrington J, Breault DT, Epstein J, Houchen CW, Lynch JP, Martin MG, Plevritis SK, Curtis C, Ji HP, Li L, Henning SJ, Wong MH, Kuo CJ. 2017. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 21: 78–90.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Westphalen CB, Quante M, Wang TC. 2017. Functional implication of Dclk1 and Dclk1-expressing cells in cancer. Small GTPases 8: 164–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Patel O, Dai W, Mentzel M, Griffin MD, Serindoux J, Gay Y, Fischer S, Sterle S, Kropp A, Burns CJ, Ernst M, Buchert M, Lucet IS. 2016. Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1. Structure 24: 1550–61 [DOI] [PubMed] [Google Scholar]
- 116.Qu D, Johnson J, Chandrakesan P, Weygant N, May R, Aiello N, Rhim A, Zhao L, Zheng W, Lightfoot S, Pant S, Irvan J, Postier R, Hocker J, Hanas JS, Ali N, Sureban SM, An G, Schlosser MJ, Stanger B, Houchen CW. 2015. Doublecortin-like kinase 1 is elevated serologically in pancreatic ductal adenocarcinoma and widely expressed on circulating tumor cells. PLoS One 10: e0118933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nishio K, Kimura K, Amano R, Nakata B, Yamazoe S, Ohira G, Miura K, Kametani N, Tanaka H, Muguruma K, Hirakawa K, Ohira M. 2017. Doublecortin and CaM kinase-like-1 as an independent prognostic factor in patients with resected pancreatic carcinoma. World J Gastroenterol 23: 5764–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saqui-Salces M, Keeley TM, Grosse AS, Qiao XT, El-Zaatari M, Gumucio DL, Samuelson LC, Merchant JL. 2011. Gastric tuft cells express DCLK1 and are expanded in hyperplasia. Histochem Cell Biol 136: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mutoh H, Sashikawa M, Sakamoto H, Tateno T. 2014. Cyclooxygenase 2 in gastric carcinoma is expressed in doublecortin- and CaM kinase-like-1-positive tuft cells. Gut Liver 8: 508–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, Somerville TDD, Milazzo JP, Wilkinson JE, Demerdash OE, Spector DL, Egeblad M, Shi J, Vakoc CR. 2018. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sukumaran SK, Lewandowski BC, Qin Y, Kotha R, Bachmanov AA, Margolskee RF. 2017. Whole transcriptome profiling of taste bud cells. Sci Rep 7: 7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lei W, Ren W, Ohmoto M, Urban JF Jr., Matsumoto I, Margolskee RF, Jiang P 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of succinate-SUCNR1 mediated tuft cell activation by succinate-producing microbiota
- 123.Ogura T, Krosnowski K, Zhang L, Bekkerman M, Lin W. 2010. Chemoreception regulates chemical access to mouse vomeronasal organ: role of solitary chemosensory cells. PLoS One 5: e11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Finger TE. 1997. Evolution of taste and solitary chemoreceptor cell systems. Brain Behav Evol 50: 234–43 [DOI] [PubMed] [Google Scholar]
- 125.Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343: 1017–21 [DOI] [PubMed] [Google Scholar]
- 126.Mazzoni M, Bonaldo A, Gatta PP, Vallorani C, Latorre R, Canova M, Clavenzani P. 2015. alpha-Transducin and alpha-gustducin immunoreactive cells in the stomach of common sole (Solea solea) fed with mussel meal. Fish Physiol Biochem 41: 603–12 [DOI] [PubMed] [Google Scholar]
- 127.Kaske S, Krasteva G, Konig P, Kummer W, Hofmann T, Gudermann T, Chubanov V. 2007. TRPM5, a taste-signaling transient receptor potential ion-channel, is a ubiquitous signaling component in chemosensory cells. BMC Neurosci 8: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liman ER. 2007. TRPM5 and taste transduction. Handb Exp Pharmacol: 287–98 [DOI] [PubMed] [Google Scholar]
- 129.Perez CA, Huang L, Rong M, Kozak JA, Preuss AK, Zhang H, Max M, Margolskee RF. 2002. A transient receptor potential channel expressed in taste receptor cells. Nat Neurosci 5: 1169–76 [DOI] [PubMed] [Google Scholar]
- 130.Liu D, Liman ER. 2003. Intracellular Ca2+ and the phospholipid PIP2 regulate the taste transduction ion channel TRPM5. Proc Natl Acad Sci U S A 100: 15160–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prawitt D, Monteilh-Zoller MK, Brixel L, Spangenberg C, Zabel B, Fleig A, Penner R. 2003. TRPM5 is a transient Ca2+-activated cation channel responding to rapid changes in [Ca2+]i. Proc Natl Acad Sci U S A 100: 15166–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. 2003. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112: 293–301 [DOI] [PubMed] [Google Scholar]
- 133.Romanov RA, Rogachevskaja OA, Bystrova MF, Jiang P, Margolskee RF, Kolesnikov SS. 2007. Afferent neurotransmission mediated by hemichannels in mammalian taste cells. Embo j 26: 657–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Talavera K, Yasumatsu K, Voets T, Droogmans G, Shigemura N, Ninomiya Y, Margolskee RF, Nilius B. 2005. Heat activation of TRPM5 underlies thermal sensitivity of sweet taste. Nature 438: 1022–5 [DOI] [PubMed] [Google Scholar]
- 135.Tantin D 2013. Oct transcription factors in development and stem cells: insights and mechanisms. Development 140: 2857–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Andersen B, Schonemann MD, Flynn SE, Pearse RV 2nd, Singh H, Rosenfeld MG. 1993. Skn-1a and Skn-1i: two functionally distinct Oct-2-related factors expressed in epidermis. Science 260: 78–82 [DOI] [PubMed] [Google Scholar]
- 137.Yukawa K, Yasui T, Yamamoto A, Shiku H, Kishimoto T, Kikutani H. 1993. Epoc-1: a POU-domain gene expressed in murine epidermal basal cells and thymic stromal cells. Gene 133: 163–9 [DOI] [PubMed] [Google Scholar]
- 138.Andersen B, Weinberg WC, Rennekampff O, McEvilly RJ, Bermingham JR Jr., Hooshmand F, Vasilyev V, Hansbrough JF, Pittelkow MR, Yuspa SH, Rosenfeld MG 1997. Functions of the POU domain genes Skn-1a/i and Tst-1/Oct-6/SCIP in epidermal differentiation. Genes Dev 11: 1873–84 [DOI] [PubMed] [Google Scholar]
- 139.Fourniol F, Perderiset M, Houdusse A, Moores C. 2013. Structural studies of the doublecortin family of MAPs. Methods Cell Biol 115: 27–48 [DOI] [PubMed] [Google Scholar]
- 140.Sossey-Alaoui K, Hartung AJ, Guerrini R, Manchester DK, Posar A, Puche-Mira A, Andermann E, Dobyns WB, Srivastava AK. 1998. Human doublecortin (DCX) and the homologous gene in mouse encode a putative Ca2+-dependent signaling protein which is mutated in human X-linked neuronal migration defects. Hum Mol Genet 7: 1327–32 [DOI] [PubMed] [Google Scholar]
- 141.Friocourt G, Liu JS, Antypa M, Rakic S, Walsh CA, Parnavelas JG. 2007. Both doublecortin and doublecortin-like kinase play a role in cortical interneuron migration. J Neurosci 27: 3875–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sossey-Alaoui K, Srivastava AK. 1999. DCAMKL1, a brain-specific transmembrane protein on 13q12.3 that is similar to doublecortin (DCX). Genomics 56: 121–6 [DOI] [PubMed] [Google Scholar]
- 143.Bezencon C, le Coutre J, Damak S. 2007. Taste-signaling proteins are coexpressed in solitary intestinal epithelial cells. Chem Senses 32: 41–9 [DOI] [PubMed] [Google Scholar]
- 144.Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, Rosenzweig S, Spielman AI, Max M, Margolskee RF. 1999. Ggamma13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci 2: 1055–62 [DOI] [PubMed] [Google Scholar]
- 145.Kim JW, Roberts C, Maruyama Y, Berg S, Roper S, Chaudhari N. 2006. Faithful expression of GFP from the PLCbeta2 promoter in a functional class of taste receptor cells. Chem Senses 31: 213–9 [DOI] [PubMed] [Google Scholar]
- 146.Eberle JA, Richter P, Widmayer P, Chubanov V, Gudermann T, Breer H. 2013. Band-like arrangement of taste-like sensory cells at the gastric groove: evidence for paracrine communication. Front Physiol 4: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jiang H, Kuang Y, Wu Y, Xie W, Simon MI, Wu D. 1997. Roles of phospholipase C beta2 in chemoattractant-elicited responses. Proc Natl Acad Sci U S A 94: 7971–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gautron L, Rutkowski JM, Burton MD, Wei W, Wan Y, Elmquist JK. 2013. Neuronal and nonneuronal cholinergic structures in the mouse gastrointestinal tract and spleen. J Comp Neurol 521: 3741–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rotolo T, Smallwood PM, Williams J, Nathans J. 2008. Genetically-directed, cell type-specific sparse labeling for the analysis of neuronal morphology. PLoS One 3: e4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Misgeld T, Burgess RW, Lewis RM, Cunningham JM, Lichtman JW, Sanes JR. 2002. Roles of neurotransmitter in synapse formation: development of neuromuscular junctions lacking choline acetyltransferase. Neuron 36: 635–48 [DOI] [PubMed] [Google Scholar]
- 151.Lecomte MJ, Bertolus C, Santamaria J, Bauchet AL, Herbin M, Saurini F, Misawa H, Maisonobe T, Pradat PF, Nosten-Bertrand M, Mallet J, Berrard S. 2014. Selective disruption of acetylcholine synthesis in subsets of motor neurons: a new model of late-onset motor neuron disease. Neurobiol Dis 65: 102–11 [DOI] [PubMed] [Google Scholar]
- 152.Voigt A, Hubner S, Lossow K, Hermans-Borgmeyer I, Boehm U, Meyerhof W. 2012. Genetic labeling of Tas1r1 and Tas2r131 taste receptor cells in mice. Chem Senses 37: 897–911 [DOI] [PubMed] [Google Scholar]
- 153.Iwatsuki K, Nomura M, Shibata A, Ichikawa R, Enciso PL, Wang L, Takayanagi R, Torii K, Uneyama H. 2010. Generation and characterization of T1R2-LacZ knock-in mouse. Biochem Biophys Res Commun 402: 495–9 [DOI] [PubMed] [Google Scholar]
- 154.Huang AL, Chen X, Hoon MA, Chandrashekar J, Guo W, Trankner D, Ryba NJ, Zuker CS. 2006. The cells and logic for mammalian sour taste detection. Nature 442: 934–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Clapp TR, Medler KF, Damak S, Margolskee RF, Kinnamon SC. 2006. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Voigt A, Hubner S, Doring L, Perlach N, Hermans-Borgmeyer I, Boehm U, Meyerhof W. 2015. Cre-Mediated Recombination in Tas2r131 Cells-A Unique Way to Explore Bitter Taste Receptor Function Inside and Outside of the Taste System. Chem Senses 40: 627–39 [DOI] [PubMed] [Google Scholar]
- 157.Khandanpour C, Sharif-Askari E, Vassen L, Gaudreau MC, Zhu J, Paul WE, Okayama T, Kosan C, Moroy T. 2010. Evidence that growth factor independence 1b regulates dormancy and peripheral blood mobilization of hematopoietic stem cells. Blood 116: 5149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Foudi A, Kramer DJ, Qin J, Ye D, Behlich AS, Mordecai S, Preffer FI, Amzallag A, Ramaswamy S, Hochedlinger K, Orkin SH, Hock H. 2014. Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med 211: 909–27 [DOI] [PMC free article] [PubMed] [Google Scholar]


