Significance
The importin α family is well known as a cargo transporter from the cytoplasm to the nucleus, with diverse functions in eukaryotes. However, some members of the importin α family also localize to the cell membrane. Here, we report that a cell membrane–associated importin α takes part in viral transmission by vector insects. Importin α2 of the small brown planthopper binds the rice stripe virus on the proteoglycans of the cell membrane and efficiently controls viral entry into insect salivary glands and subsequent transmission to rice. These findings not only demonstrate the specific function of importin α in the cell membrane utilized by viruses, but also provide a promising target gene for preventing outbreaks of rice stripe disease.
Keywords: importin α, plasma membrane, small brown planthopper, salivary gland, rice stripe virus
Abstract
The importin α family belongs to the conserved nuclear transport pathway in eukaryotes. However, the biological functions of importin α in the plasma membrane are still elusive. Here, we report that importin α, as a plasma membrane–associated protein, is exploited by the rice stripe virus (RSV) to enter vector insect cells, especially salivary gland cells. When the expression of three importin α genes was simultaneously knocked down, few virions entered the salivary glands of the small brown planthopper, Laodelphax striatellus. Through hemocoel inoculation of virions, only importin α2 was found to efficiently regulate viral entry into insect salivary-gland cells. Importin α2 bound the nucleocapsid protein of RSV with a relatively high affinity through its importin β–binding (IBB) domain, with a dissociation constant KD of 9.1 μM. Furthermore, importin α2 and its IBB domain showed a distinct distribution in the plasma membrane through binding to heparin in heparan sulfate proteoglycan. When the expression of importin α2 was knocked down in viruliferous planthoppers or in nonviruliferous planthoppers before they acquired virions, the viral transmission efficiency of the vector insects in terms of the viral amount and disease incidence in rice was dramatically decreased. These findings not only reveal the specific function of the importin α family in the plasma membrane utilized by viruses, but also provide a promising target gene in vector insects for manipulation to efficiently control outbreaks of rice stripe disease.
The importin α family belongs to the conserved importin α/β nuclear transport pathway in eukaryotes (1–3). It is well known that the importin α family plays an indispensable role in transporting proteins from the cytoplasm to the nucleus, with diverse functions in gene regulation, cell physiology, and differentiation (1, 4, 5). In addition to nucleocytoplasmic transport, some members of the importin α family localize to the plasma membrane with palmitoylation modification or through binding to heparin in heparan sulfate proteoglycan (HSPG) (6–8). Increased importin α levels in the plasma membrane are concomitant with decreased importin α levels in the cytoplasm, which affect the nuclear import of cargos and regulates intracellular scaling (7). However, the function of the importin α family in the plasma membrane is still elusive.
Many plant viruses are transmitted by vector insects in a persistent, circulative mode, which is characterized by systemic invasion of diverse tissues prior to entering salivary glands and release in saliva (9–13). The salivary glands are the last barriers for viral transmission to overcome (14–18). Unfortunately, the molecular mechanisms underlying viral entry into salivary-gland cells are not well known. The rice stripe virus (RSV) is a typical persistent, circulative plant virus and is capable of proliferating in the midgut epithelial cells and of being efficiently transmitted by the vector insect small brown planthopper (Laodelphax striatellus) (19). This virus causes one of the most destructive rice stripe diseases, showing an incidence of up to 80% and causing yield losses of 30 to 40% in the rice fields of Asian countries (20). RSV is a nonenveloped negative-strand RNA virus of the Tenuivirus genus (21, 22). The genome of RSV contains four single-stranded RNA segments, each of which is encapsidated by a viral nucleocapsid protein (NP). In addition to the NP, RSV encodes an RNA-dependent RNA polymerase and five nonstructural proteins (NS2, NSvc2, NS3, SP, and NSvc4) (23–25).
In our recent work, we found that three importin α proteins, importin α1 (GenBank registration number MT036051), α2 (MT036050), and α3 (MT036052), of the small brown planthopper participate in the nuclear entry of RSV through direct interactions with RSV NPs, triggering an antiviral caspase-dependent apoptotic reaction (26). Knockdown of the expression of all the three importin α genes retarded the nuclear entry of RSV and led to an increase in viral load in planthoppers (26). However, we did not determine the influence on viral transmission. In the present study, we surprisingly found that one of the importin α proteins, importin α2, is associated with the plasma membrane and efficiently facilitates viral entry into insect salivary glands and subsequent viral transmission.
Results
Effect of the Importin α Family on RSV Transmission by Planthoppers.
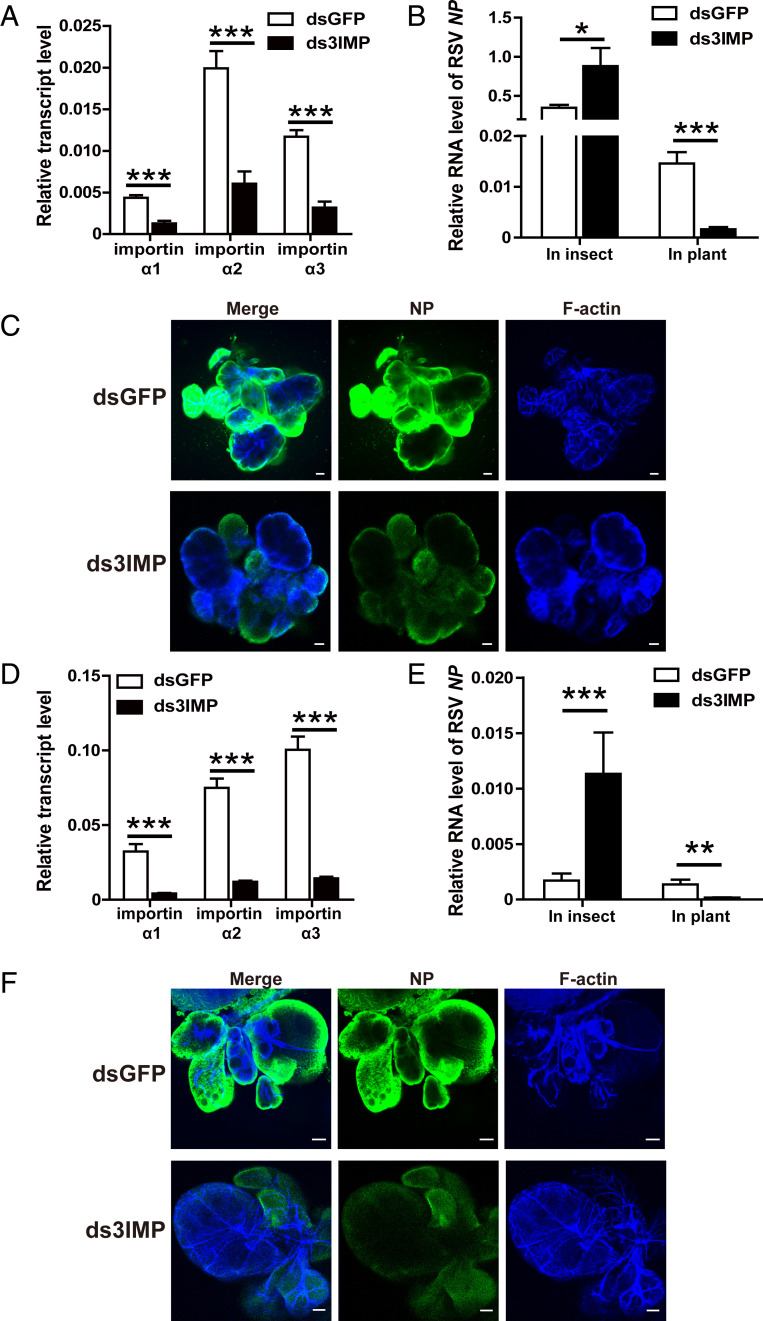
To test the effects of the three importin α proteins on RSV transmission, we knocked down all three importin α genes in viruliferous third-instar planthoppers by injecting a mixture of double-stranded RNAs (dsRNAs) for the three importin α genes (ds3IMP-RNA) (Fig. 1A and SI Appendix, Fig. S1A). As expected from previous work (26), the RSV amount in terms of the NP RNA level and viral genomic levels significantly increased in the whole bodies of viruliferous planthoppers at 8 d after ds3IMP-RNA delivery, measured with RT-qPCR (Fig. 1B and SI Appendix, Fig. S1B). However, the NP RNA level in rice plants fed to the three importin α–knockdown insects for 1 d was reduced by 89% compared to the plants fed to the dsGFP-RNA–injected insects (Fig. 1B). When the salivary glands were dissected, few virions entered the salivary glands of the three importin α–knockdown insects as shown by immunofluorescence with an NP monoclonal antibody (Fig. 1C).
Fig. 1.
Effect of the importin α family on RSV transmission by L. striatellus. (A) Relative transcript levels of three importin α genes in viruliferous small brown planthoppers at 8 d after the injection of a mixture of dsRNAs for the three importin α genes (ds3IMP) as measured by RT-qPCR. The transcript levels of these genes were normalized to that of EF2. The control group was injected with dsRNA for GFP (dsGFP). (B) Relative RNA level of RSV NP in viruliferous planthoppers at 8 d after the injection of ds3IMP and in the rice fed to these insects for 1 d as measured by RT-qPCR. The transcript level of planthopper EF2 or rice tubulin was quantified to normalize the cDNA templates. (C) Immunohistochemistry showing the variation in RSV amounts in salivary glands of viruliferous planthoppers at 8 d after injecting ds3IMP compared to that of injecting dsGFP. RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.) (D) Relative transcript levels of three importin α genes in the nonviruliferous planthoppers after injection with ds3IMP and feeding on RSV-infected rice plants for 8 d measured by RT-qPCR. (E) Relative RNA level of RSV NP in nonviruliferous planthoppers after injection with ds3IMP and feeding on RSV-infected rice plants for 8 d and in the rice fed to these insects for 2 d measured by RT-qPCR. (F) Immunohistochemistry showing the variation in RSV amounts in salivary glands of nonviruliferous planthoppers after injection with ds3IMP and feeding on RSV-infected rice plants for 8 d using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). Insects injected with dsGFP were used as controls. F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.) *P < 0.05, **P < 0.01, ***P < 0.001.
In another group, nonviruliferous third-instar planthoppers were inoculated with RSV by feeding on RSV-infected rice plants after the expression of the three importin α genes was knocked down with injection of ds3IMP-RNA (Fig. 1D). The NP RNA level significantly increased in the whole bodies of planthoppers at 8 d postinoculation (dpi), while the amount of RSV secreted into rice plants was reduced by 87% after feeding for 2 d by the three importin α–knockdown insects (Fig. 1E), and fewer virions entered the salivary glands compared to the insects that were injected with dsGFP-RNA (Fig. 1F). These results demonstrated that the importin α family affects RSV entry into salivary glands of vector insects and subsequent viral transmission to rice.
Confirmation of the Effect of the Importin α Family on RSV Entry into Salivary Glands through Hemocoel Inoculation of RSV.
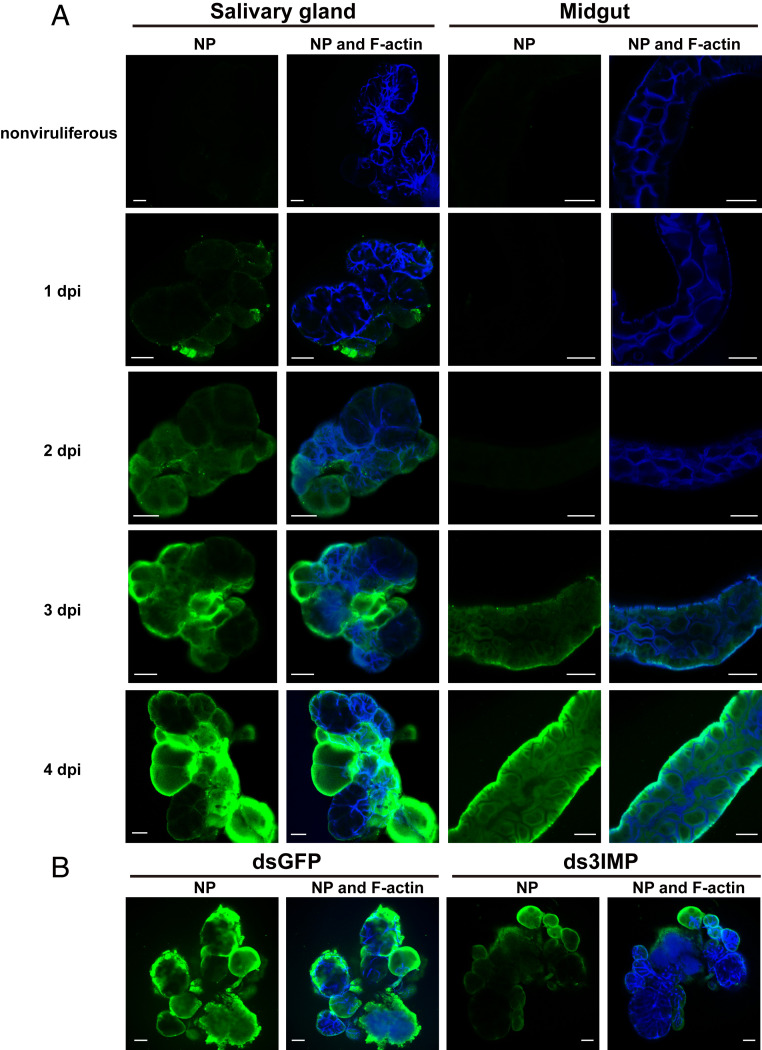
Under natural conditions, RSV infects planthoppers from midgut epithelial cells and then spreads from the midgut to the hemolymph and, finally, to the salivary glands (13). We utilized the method of hemocoel inoculation of RSV to verify the effects of the importin α family on RSV entry into salivary glands. First, the viral circulative route of hemocoel inoculation was explored. After the RSV crude extracts from viruliferous planthoppers were injected into the hemocoel of the nonviruliferous third-instar planthoppers, virions were detected in salivary glands at 1 dpi by immunofluorescence with an NP monoclonal antibody, and more virions entered salivary glands at 2, 3, and 4 dpi (Fig. 2A). In contrast, only 3 out of 20 midguts had RSV virions at 3 dpi, and virions entered the midgut universally at 4 dpi (Fig. 2A). Before RSV appeared in the midgut, RSV was detected in rice plants fed to the hemocoel-inoculated planthoppers at 1 dpi, and more virions were secreted in rice at 2 and 3 dpi (SI Appendix, Fig. S2). These results suggest that RSV enters salivary glands earlier than the midgut with hemocoel inoculation. Next, after ds3IMP-RNA and RSV crude extracts were injected together into the hemolymph of third-instar nymphs, few virions were detected by immunofluorescence at 4 dpi in the salivary glands compared to the injection of dsGFP-RNA and RSV crude extracts (Fig. 2B and SI Appendix, Fig. S3A). The hemocoel inoculation results confirmed that the importin α family plays a role in RSV entry into salivary glands.
Fig. 2.
Effect of the importin α family on RSV entry into salivary glands of L. striatellus through hemocoel inoculation with RSV. (A) Immunohistochemistry showing RSV virions in salivary glands and midguts of nonviruliferous small brown planthoppers from 1 to 4 dpi of RSV crude extracts from viruliferous planthoppers in the hemolymph. RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). The nonviruliferous insects were used as negative controls. (Scale bars, 40 μm.) (B) Immunohistochemistry showing the variation in RSV amounts in salivary glands of nonviruliferous planthoppers at 4 d after injection of dsRNAs for the three importin α genes (ds3IMP) and RSV crude extracts. The control group was injected with dsGFP and RSV crude extracts. (Scale bars, 40 μm.)
Importin α2 Regulates RSV Entry into Salivary Glands.
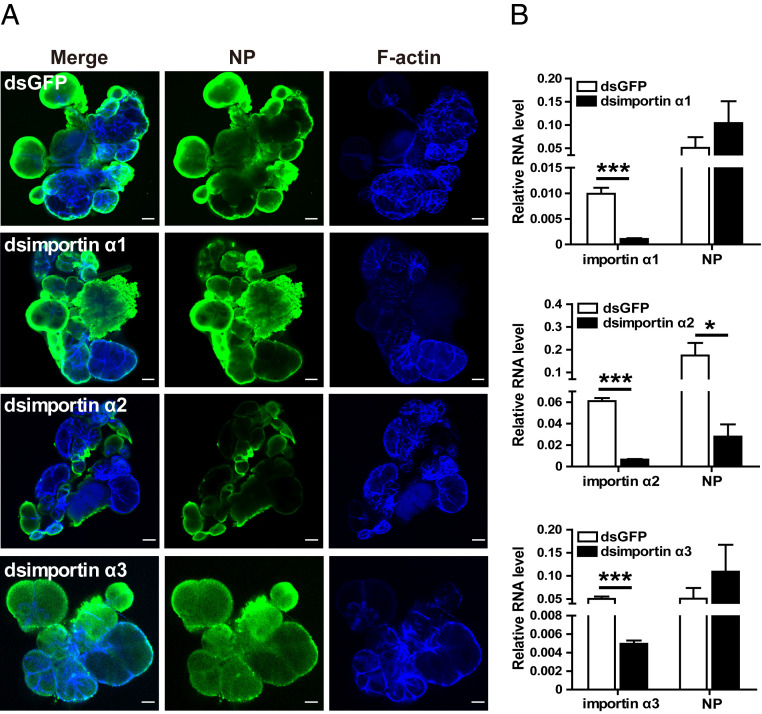
To determine which importin α regulates RSV entry into salivary glands, RSV crude extracts and dsRNA of each importin α were injected into the hemolymph. Immunofluorescence showed that interference with importin α2 resulted in a remarkable reduction in RSV in salivary glands at 4 dpi, while interference with importin α1 or α3 did not induce an obvious variation in RSV in salivary glands (Fig. 3A). Correspondingly, the viral amount in the whole bodies significantly decreased at 6 dpi only when the expression of importin α2 was knocked down, whereas no change was observed when importin α1 or α3 was knocked down (Fig. 3B). Therefore, importin α2 plays a key role in mediating RSV entry into salivary glands.
Fig. 3.
Importin α2 regulates RSV entry into salivary glands of L. striatellus. (A) Immunohistochemistry showing the variation in RSV amounts in salivary glands of nonviruliferous small brown planthoppers at 4 d after injection of RSV crude extracts and dsRNA of each importin α into the hemolymph. The control group was injected with dsGFP and RSV crude extracts. RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.) (B) Relative transcript levels of importin α genes and relative RNA level of RSV NP to that of EF2 in nonviruliferous planthoppers at 6 d after injection of RSV crude extracts and dsRNA of each importin α as measured by RT-qPCR. The control group was injected with dsGFP and RSV crude extracts. *P < 0.05, ***P < 0.001.
Importin α2 Binds Viral Nucleocapsid Protein with a Relatively High Affinity through the Importin β–Binding Domain.
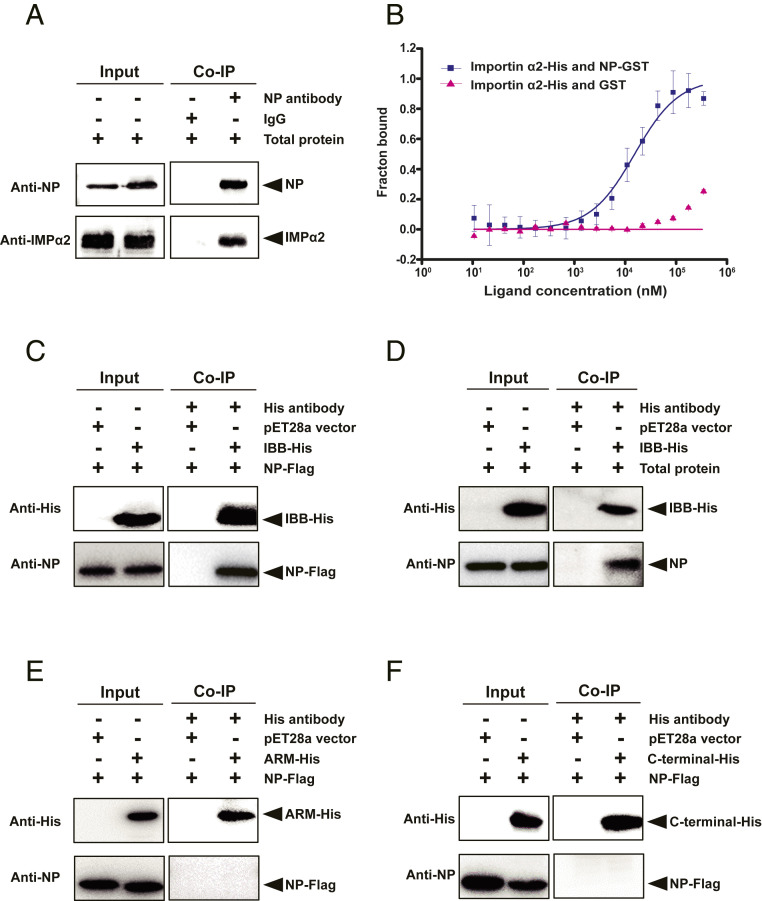
In our previous work, importin α2 was found to interact in vitro with RSV NP (26, 27). Here, we confirmed the contact of the two proteins in vivo through coimmunoprecipitation (Co-IP) assays using the monoclonal anti-NP antibody to pull down the complex of importin α2 and NP from the total protein of viruliferous planthoppers (Fig. 4A). Specific affinity activity between the two proteins was measured using the microscale thermophoresis (MST) assay. The binding of recombinant importin α2-His to NP-GST was successfully observed, and the dissociation constant (KD) was 9.1 ± 2.8 μM, indicating a relatively high affinity between the two proteins (Fig. 4B). Importin α proteins have three conserved structural features, including an N-terminal importin β–binding (IBB) domain, a series of armadillo (Arm) repeats domain, and a carboxyl-terminal short acidic amino acids cluster domain (1). The binding region of importin α2 with NP was further investigated through Co-IP assays using the recombinant IBB domain (from 1 to 69 aa), Arm domain (from 70 to 424 aa), and carboxyl terminus (from 425 to 498 aa) of importin α2 with His tags and Flag-tagged recombinant NP. The results showed that the IBB-His of importin α2 not only bound NP-Flag (Fig. 4C) but also pulled down NP from the total protein of viruliferous planthoppers (Fig. 4D), while Arm-His or carboxyl-terminal–His did not interact with NP-Flag (Fig. 4 E and F).
Fig. 4.
Importin α2 binds viral nucleocapsid protein with a relatively high affinity through the IBB domain. (A) NP and importin α2 (IMPα2) were coprecipitated from total protein of viruliferous small brown planthoppers in the Co-IP and Western blot assay using the anti-NP monoclonal antibody and anti–importin α2 polyclonal antibody. Mouse IgG was used as negative control. (B) MST assay to reveal specific binding between recombinant importin α2–His and NP-GST. The NP-GST (ligand) was gradient-diluted. The GST protein was used instead of NP-GST in the control group. The solid curve was fit to the standard KD-fit function. Bars represent SE. (C, E, and F) Interactions between recombinantly expressed Flag-tagged RSV NP and His-tagged IBB domain, Arm domain, and carboxyl-terminal domain of importin α2 in the Co-IP and Western blot assay using anti-His and anti-NP monoclonal antibodies. The expression products from the pET28a vector were used as a negative control. (D) The recombinantly expressed IBB-His of importin α2 pulled down the NP from total protein of viruliferous planthoppers in the Co-IP and Western blot assay using anti-His and anti-NP monoclonal antibodies. The expression products from the pET28a vector were used as a negative control.
Importin α2 Is Localized in the Plasma Membrane.
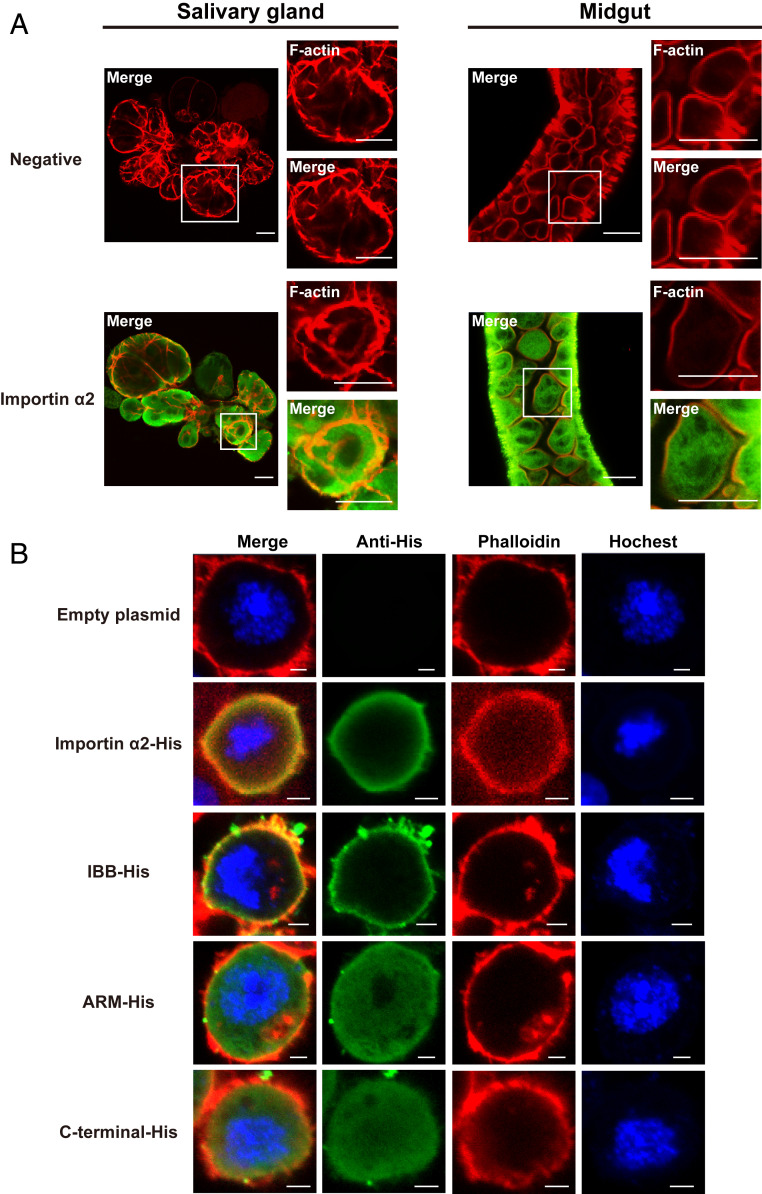
To explain the specific role of importin α2, we explored the localization of importin α2 in salivary-gland cells and midgut cells of viruliferous planthoppers with immunohistochemistry assays using polyclonal antibodies against importin α2 from our previous study (26). The orange was observed in the plasma membrane when the green importin α2 was merged with red F-actin, indicating that importin α2 was localized in the plasma membrane in addition to the cytoplasm in both tissues (Fig. 5A). When His-tagged recombinant importin α2 was expressed in Drosophila Schneider 2 cells, importin α2–His accumulated in the plasma membrane of S2 cells (Fig. 5B). The region of importin α2 binding to the plasma membrane was further explored through recombinant expression of IBB-His, Arm-His, and carboxyl-terminal–His of importin α2 in S2 cells. Immunohistochemistry assays showed that IBB-His was localized to the plasma membrane, while the other two fragments were diffusely localized to the cytoplasm and nucleus (Fig. 5B).
Fig. 5.
Importin α2 localizes to the plasma membrane of L. striatellus as assayed by immunohistochemistry. (A) Localization of importin α2 in salivary glands and midguts of viruliferous small brown planthoppers. The boxed region was enlarged and is shown in two panels on the right side. Importin α2 was observed using Alexa Fluor 594-conjugated anti–importin α2 polyclonal antibody (green). F-actin was stained with phalloidin (red). The negative control did not include the primary antibodies. (Scale bars, 40 μm.) (B) Immunofluorescence labeling of recombinantly expressed importin α2–His, IBB-His, Arm-His, and carboxyl-terminal–His of importin α2 in Drosophila S2 cells. The His-tagged proteins were labeled with an Alexa Fluor 488-conjugated anti-His monoclonal antibody (green). F-actin was labeled with phalloidin (red). Nuclei were stained with Hoechst (blue). Cells transfected with the empty pAc-5.1/V5-HisB plasmid were used as negative controls. (Scale bars, 2 μm.)
Importin α2 and RSV Nucleocapsid Proteins Bind Heparin in HSPG to Localize to the Plasma Membrane.
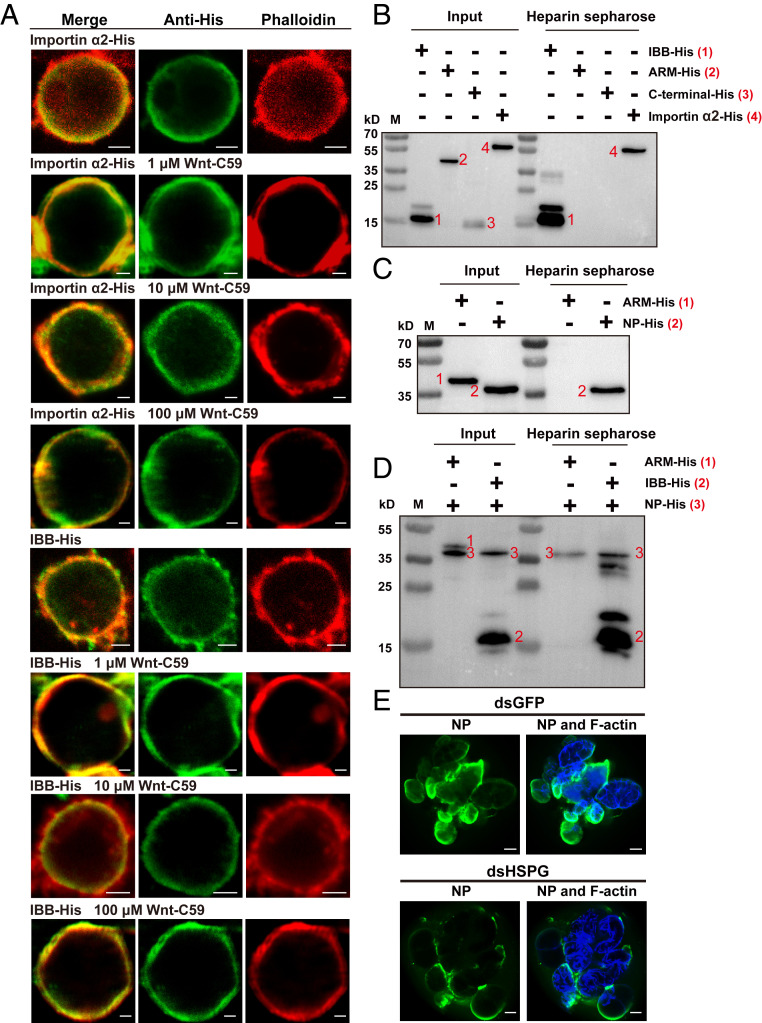
The amino-acid sequence of importin α2 was predicted not to contain an N-terminal signal peptide or transmembrane region in the PSORT server. Palmitoylation modification or binding heparin in HSPG could cause importin α proteins to localize to the plasma membrane (7, 8). To clarify the mechanism for the plasma membrane localization of importin α2, we tested the change in the localization of importin α2–His and the IBB-His domain in S2 cells treated with Wnt-C59, an inhibitor of palmitoylation. After treatment with different concentrations of Wnt-C59 for 48 h, importin α2-His and the IBB-His domain still localized to the plasma membrane of S2 cells (Fig. 6A), implying that palmitoylation is not responsible for the membrane association of importin α2.
Fig. 6.
Importin α2 and RSV nucleocapsid proteins bind heparin in HSPG to localize to the plasma membrane of L. striatellus. (A) Localization of recombinantly expressed importin α2–His and the IBB-His domain in Drosophila S2 cells treated with different concentrations of Wnt-C59 revealed by immunofluorescence. The His-tagged proteins were labeled with an Alexa Fluor 488-conjugated anti-His monoclonal antibody (green). F-actin was labeled with phalloidin (red). (Scale bars, 2 μm.) (B) Heparin-binding and Western blot assays for the recombinantly expressed importin α2–His, IBB-His, Arm-His, or carboxyl-terminal–His of importin α2. After elution from heparin beads, the supernatant was used for Western blotting using an anti-His monoclonal antibody. M, marker. (C) Heparin-binding and Western blot assays for recombinantly expressed RSV NP-His using an anti-His monoclonal antibody. The Arm-His was used as a negative control. (D) Heparin-binding and Western blot assays for IBB-His and NP-His using anti-His monoclonal antibody. The two proteins were mixed together before being loaded on heparin beads. A mixture of Arm-His and NP-His was used as a negative control. (E) Immunohistochemistry showing the variation in RSV amounts in salivary glands of nonviruliferous small brown planthoppers at 4 d after injection of RSV crude extracts and dsRNA of HSPG (dsHSPG) into the hemolymph. The control group was injected with dsRNA of GFP (dsGFP) and RSV crude extracts. RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.)
It was then determined whether importin α2 and its IBB domain can bind heparin in HSPG. We performed an in vitro heparin-binding assay in which recombinant importin α2 and its three His-tagged domains were mixed with heparin-conjugated beads. Western blot analysis showed that heparin directly bound importin α2–His and IBB-His but not Arm-His or carboxyl-terminal–His (Fig. 6B), consistent with the membrane association of importin α2 and the IBB domain. Furthermore, the recombinantly expressed RSV NP-His was also bound to heparin-conjugated beads (Fig. 6C). When IBB-His was first mixed with NP-His before loading the heparin-conjugated beads, both proteins were still able to bind heparin (Fig. 6D), showing that the interaction between importin α2 and RSV NP did not affect their membrane association.
Heparin in HSPG Mediates the Regulation of Importin α2 on RSV Entry into Salivary Glands.
Because importin α2 and RSV NP bind heparin in HSPG to localize to the plasma membrane, it is possible that HSPG participates in the regulation of importin α2 on RSV entry into salivary glands. The open reading frame (ORF) of HSPG of the small brown planthoppers is 9,708 bp, putatively encoding a 356.1-kDa protein (MZ032031 in GenBank). HSPG had a higher transcript level in the gut than in the salivary glands (SI Appendix, Fig. S4A). When the expression of HSPG was knocked down (SI Appendix, Fig. S4B), fewer virions were detected by immunofluorescence in salivary glands at 4 d after injection of RSV crude extracts and dsHSPG-RNA into the hemolymph than those of the control group injected with dsGFP-RNA and RSV crude extracts (Fig. 6E), similar to the effect of importin α2 interference (Fig. 3A). However, when we recombinantly expressed HSPG protein in five fragments due to the huge molecular weight, Co-IP assays showed that NP-Flag did not bind any of the five His-tagged fragments of HSPG (SI Appendix, Fig. S4C), indicating that NP could not directly bind HSPG protein. Therefore, importin α2 mediates viral entry into salivary glands by cooperating with heparin in HSPG.
Importin β Has No Obvious Effects on RSV Entry into Salivary Glands.
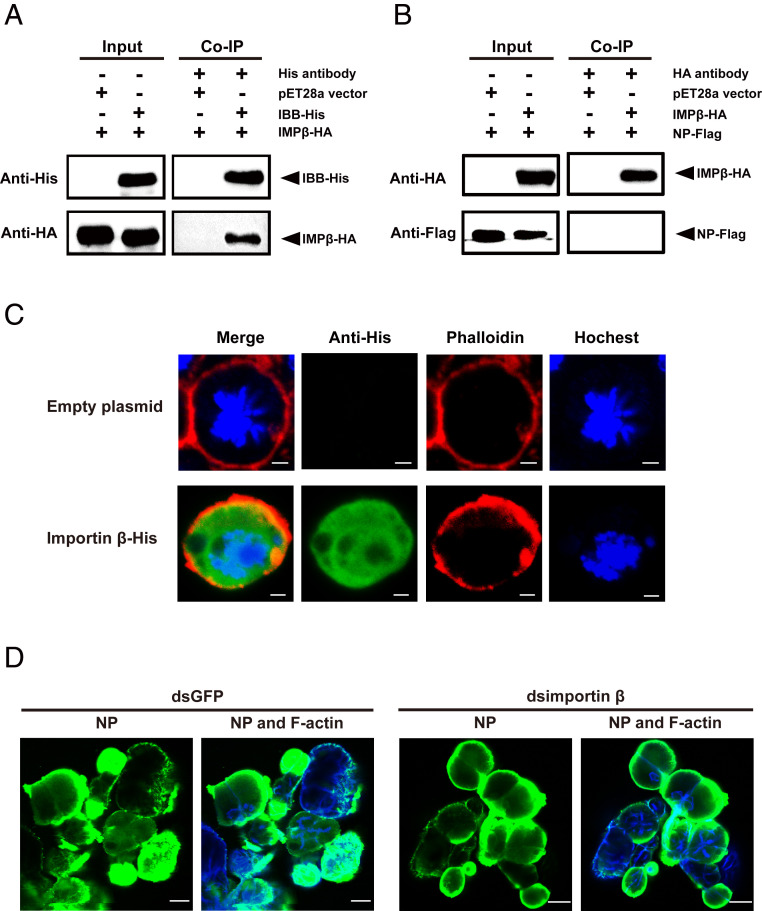
Because importin α usually binds importin β to form an import complex (1), whether importin β takes part in importin α2–mediated viral entry into salivary glands was further explored. There is only one importin β gene in the small brown planthopper. The ORF of importin β is 2,685 bp, putatively encoding a 98.9-kDa protein (MZ054426 in GenBank). The Co-IP assays with in vitro–expressed importin β–HA and IBB-His or NP-Flag showed that importin β bound the IBB domain of importin α2 but did not bind NP directly (Fig. 7 A and B). When importin β–His was expressed in S2 cells, it distributed in the whole cells, possibly including the plasma membrane (Fig. 7C). However, knockdown of importin β expression did not induce an obvious variation in RSV amount in salivary glands at 4 dpi, detected by immunofluorescence after injection of RSV crude extracts and dsimportin β–RNA into the hemolymph (Fig. 7D and SI Appendix, Fig. S3B). These results indicate that, although importin β could localize in the plasma membrane, it does not play a major role in mediating RSV entry into salivary-gland cells.
Fig. 7.
Importin β has no obvious effects on RSV entry into salivary glands of L. striatellus. (A and B) Interactions between recombinantly expressed importin β-HA (IMPβ-HA) and His-tagged IBB domain of importin α2 or Flag-tagged RSV NP in the Co-IP and Western blot assay using anti-His, anti-HA, and anti-Flag monoclonal antibodies. The expression products from the pET28a vector were used as a negative control. (C) Immunofluorescence labeling of recombinantly expressed importin β-His in Drosophila S2 cells. Importin β-His was labeled with an Alexa Fluor 488-conjugated anti-His monoclonal antibody (green). F-actin was labeled with phalloidin (red). Nuclei were stained with Hoechst (blue). Cells transfected with the empty pAc-5.1/V5-HisB plasmid were used as a negative control. (Scale bars, 2 μm.) (D) Immunohistochemistry showing the variation in RSV amounts in salivary glands of nonviruliferous small brown planthoppers at 4 d after injection of RSV crude extracts and dsRNA of importin β (dsimportin β) into the hemolymph. The control group was injected with dsRNA of GFP (dsGFP) and RSV crude extracts. RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.)
Effect of Importin α2 on the Viral Transmission Efficiency of Planthoppers.
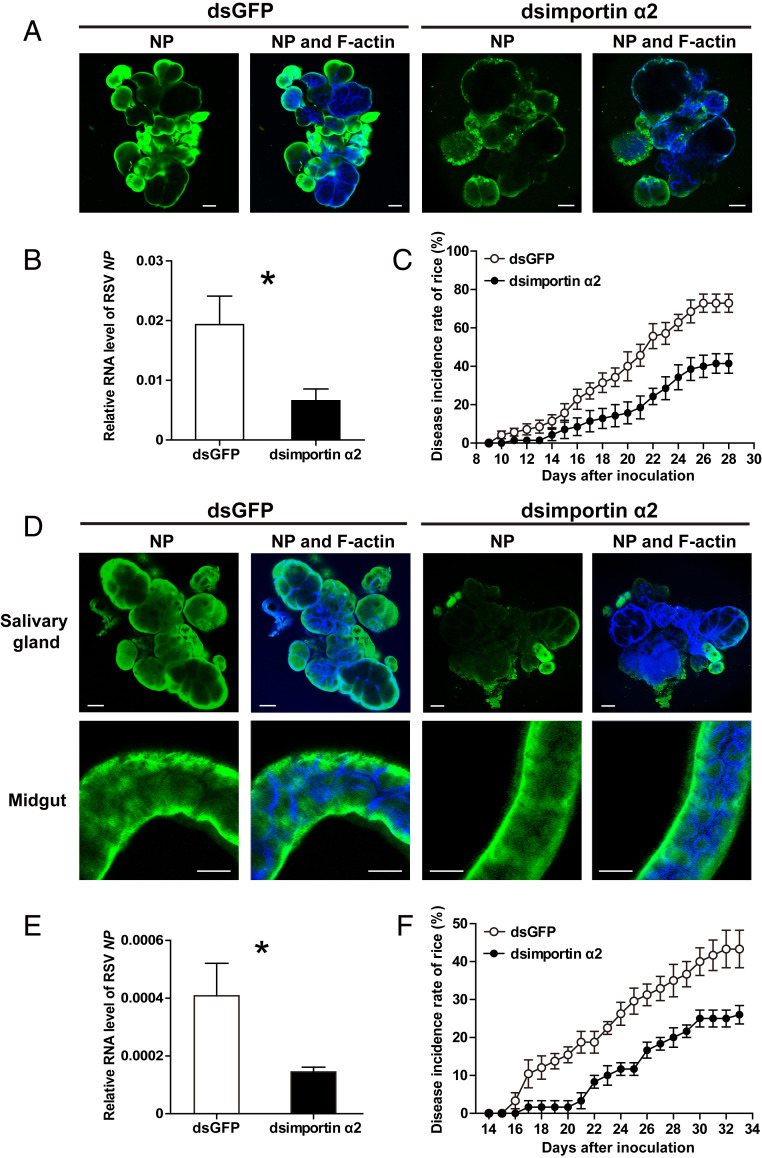
The effect of importin α2 on the viral transmission efficiency of small brown planthoppers was further evaluated. When the expression of importin α2 was knocked down in the viruliferous planthoppers, fewer RSV virions were observed in the salivary glands compared to the dsGFP-RNA injection group at 8 d (Fig. 8A). The viral amount in the rice fed to the importin α2–knockdown planthoppers for 2 d decreased by 66% (Fig. 8B), and the disease incidence of the rice dropped from 73% in the control group to 41% in the importin α2–knockdown group at 28 dpi (Fig. 8C). In another group, the expression of importin α2 was first knocked down in nonviruliferous planthoppers, and then the insects were allowed to acquire RSV from infected rice for 7 d before transfer to healthy rice for feeding for 3 d. Similarly, few virions were observed in the salivary glands, but no significant reduction was found in the midguts (Fig. 8D). With the viral load transmitted to rice decreasing by 65% (Fig. 8E), the disease incidence of the rice was remarkably reduced from 43 to 26% at 33 dpi (Fig. 8F). Therefore, importin α2 has a great impact on the viral transmission efficiency of small brown planthoppers.
Fig. 8.
Effect of importin α2 on the viral transmission efficiency of L. striatellus. (A) Immunohistochemistry showing the variation in RSV amounts in salivary glands of viruliferous small brown planthoppers at 8 d after injection of dsRNA of importin α2 (dsimportin α2) or (D) in salivary glands and midguts of nonviruliferous planthoppers after injection with dsimportin α2 and 7 d of feeding on RSV-infected rice. The control group was injected with dsRNA of GFP (dsGFP). RSV NP was observed using Alexa Fluor 488-conjugated anti-NP monoclonal antibody (green). F-actin was stained with phalloidin (blue). (Scale bars, 40 μm.) (B) Relative RNA level of RSV NP to that of tubulin in rice fed to dsimportin α2–injected viruliferous planthoppers for 2 d measured by RT-qPCR or (E) in rice fed to dsimportin α2–injected nonviruliferous planthoppers for 3 d after acquiring RSV from infected rice for 7 d. The control group was fed to dsGFP-injected planthoppers. (C) The disease incidence of rice fed to dsimportin α2–injected viruliferous planthoppers for 2 d or (F) rice fed to dsimportin α2–injected nonviruliferous planthoppers for 3 d after acquiring RSV from infected rice for 7 d. The control group was fed to dsGFP-injected planthoppers. A total of 10 rice seedlings per replicate and six replicates were applied. The values were reported as the mean ± SE. *P < 0.05.
Discussion
Our results reveal a specific function of importin α proteins in virus transmembrane transport. This function of importin α is apparently entirely distinct from its nucleocytoplasmic transport role (1, 6). Although the physiological function of importin α2 in the plasma membrane remains to be determined, this protein is clearly exploited by viruses. We provide evidence that importin α2 of small brown planthoppers associates in the plasma membrane and binds viral nucleocapsid proteins to regulate viral cell entry. The membrane association of importin α2 is fulfilled by the IBB domain, which usually participates in the unloading of cargo proteins in the nucleus (1, 28, 29). The IBB domain of importin α2 is not only localized to the plasma membrane but also binds RSV, while the Arm domain of importin α2 does not bind RSV, as it always functions as a cargo binding site (1, 30). These outstanding characteristics make importin α2 a specialized plasma-membrane receptor for RSV.
Our data indicate that importin α2 of small brown planthoppers associates with the plasma membrane through binding heparin in HSPG. Previous studies have shown that palmitoylation on two cysteine and two serine residues within the Arm domain is responsible for the membrane association of importin α in Xenopus eggs (7). Our data did not support the existence or influence of palmitoylation on the membrane localization of importin α2 in small brown planthoppers. Phosphorylation also affects the membrane association of importin α in Xenopus eggs as demonstrated by the fact that increased phosphorylation of importin α causes it to dissociate from membranes (6). In cancer cells, the cell-surface localization of importin α resulted from binding HSPG (8). HSPGs are conserved among vertebrates and invertebrates and are composed of a core protein that covalently links to unbranched, negatively charged heparan sulfate polysaccharides attached to a variety of cell surfaces (31, 32). They are broadly used by a range of pathogens, especially viruses, to attach to the cell surface by interacting electrostatically with the basic residues of viral surface glycoproteins or viral nucleocapsid proteins of nonenveloped viruses (33). Viruses exploit these weak interactions to increase their concentration at the cell surface and augment their chances of binding a more specific entry receptor (34). In rare cases, HSPGs serve directly as entry receptors through protein–protein interactions as described for the rice gall dwarf virus associated with the plasma membrane of sperm heads of green rice leafhoppers (35). But RSV NP does not bind HSPG through protein–protein interactions. Both importin α2 and RSV NP bind to heparin in HSPG, which provides a platform for the contact of virions and importin α2 to promote viral cell entry.
The roles of the three importin α proteins of small brown planthoppers differ in RSV transmission. Importin α1 and α3 did not affect RSV entry into salivary-gland cells. Our previous work showed that importin α3 was mainly localized in the nucleus and that the interaction of the three importin α proteins significantly affected RSV nuclear entry and replication, while single importin α did not have obvious effects in planthoppers (26). We speculate that importin α1 and α3 are the main players in regulating viral nuclear entry and that importin α2 is responsible for viral cell entry. Considering that viral nuclear entry inhibits RSV replication (26), the three importin α proteins exert contrary effects on RSV transmission.
It seems that importin α2 does not play a major role in viral entry into midgut cells. Although importin α2 is also localized to the plasma membrane of midgut cells and HSPG is highly expressed in the midgut, knockdown of importin α2 did not affect viral load in the midgut. RSV exploits other mechanisms to enter midgut cells. Sugar transporter 6 was reported to mediate the entrance of RSV into the midgut cells of planthoppers (36). The glycoprotein NSvc2 of RSV helps viruses overcome midgut barriers (37). It is possible that RSV utilizes different routes to enter midgut or salivary-gland cells.
In conclusion, an unexpected finding of our study was the discovery of a role for membrane-associated importin α in viral transmission through facilitating RSV virions to overcome the last barriers of the salivary glands in vector insects. This finding not only demonstrates that viruses can exploit an existing cellular pathway that evolved with a specific function but also provides a promising target gene for manipulation to efficiently control outbreaks of rice stripe disease in the future. Such objectives could possibly be achieved through genetically engineering rice plants that express small RNAs or exogenous topical application of small RNAs specifically targeting the planthopper importin α2.
Materials and Methods
Small Brown Planthoppers.
The viruliferous and nonviruliferous small brown planthopper strains were reared separately in the laboratory on seedlings of rice, Oryza sativa Huangjinqing, at 25 °C with 16 h of light per day. The viruliferous strain harbored the Jiangsu RSV isolate, and the frequency of RSV positivity was maintained at no less than 90% through purification selection performed every 3 mo via dot enzyme-linked immunosorbent assay with a monoclonal anti-NP antibody as described previously (38).
RNA Isolation and Complementary DNA (cDNA) Synthesis.
RNA was isolated from whole bodies, salivary glands, or midguts of planthoppers or rice leaves using TRIzol reagent (Invitrogen) following the manufacturer’s protocol. After being treated to remove genomic DNA contamination using a TURBO DNA-free kit (Ambion), 1 μg RNA was reverse-transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen) and random primers (Promega) in accordance with the manufacturer’s instructions.
dsRNA Synthesis and Injection.
dsRNAs for importin α1 (783 bp), α2 (472 bp) and α3 (880 bp), importin β (660 bp), HSPG (700 bp), and GFP (420 bp) were synthesized using the T7 RiboMAX Express RNAi System (Promega) following the manufacturer’s protocol. The corresponding PCR primers of the dsRNAs for these genes are shown in SI Appendix, Table S1. A 23-nL aliquot of a mixture of dsRNAs for the three importin α genes at 6 μg/μL for each gene was injected into viruliferous or nonviruliferous third-instar nymphs using a Nanoliter 2000 microinjector (World Precision Instruments). After injection, the nonviruliferous insects were fed RSV-infected rice for 8 d. The control group was injected with dsRNA for GFP or remained without injection. The RNA levels of RSV NP and four genomic segments and the transcript levels of the three importin α genes were measured in the whole bodies at 8 d after injection using RT-qPCR. For the viruliferous insects, five insects per replicate and eight biological replicates were prepared for RT-qPCR. For the nonviruliferous insects, 22 to 25 individual planthoppers were collected for RT-qPCR on each individual. Salivary glands were dissected for immunohistochemistry assays using an anti-NP monoclonal antibody with five to eight replicates.
A 23-nL aliquot of dsRNA for importin α2 at 6 μg/μL was injected into viruliferous or nonviruliferous third-instar nymphs. After injection, the nonviruliferous insects were fed RSV-infected rice for 8 d. The control group was injected with dsRNA for GFP. Salivary glands and midguts were dissected at 8 d after injection for immunohistochemistry assays using an anti-NP monoclonal antibody with 10 to 18 replicates for salivary glands and 6 to 10 replicates for midguts.
Injection of RSV Crude Extracts.
A total of 50 viruliferous L. striatellus adults were homogenized with a disposable polypropylene pestle in 80 μL 10-mM Tris⋅HCl (pH 8.0) in a 1.5-mL tube. After centrifugation at 12,000 × g for 15 min at 4 °C, the supernatant was retained. Centrifugation was repeated five times in total, and the supernatant from the last centrifugation was used as the RSV crude extracts. A 23-nL aliquot of the RSV crude extracts was injected into the hemocoel of the nonviruliferous third-instar planthoppers using a Nanoliter 2000 microinjector (World Precision Instruments). Salivary glands and midguts were dissected at 1, 2, 3, and 4 dpi for immunohistochemistry assays using an anti-NP monoclonal antibody. At each time point, 20 to 28 salivary glands and 10 to 28 midguts were assayed.
Injection of dsRNAs and RSV Crude Extracts.
Equal aliquots of RSV crude extracts and the mixture of dsRNAs for the three importin α genes at 6 μg/μL for each gene or dsRNA of each importin α, importin β, or HSPG at 6 μg/μL were mixed, and 23 nL of the mixture was injected into nonviruliferous third-instar nymphs using a Nanoliter 2000 microinjector (World Precision Instruments). The control group was injected with RSV crude extracts and dsRNA for GFP. The RNA levels of RSV NP and the transcript levels of the three importin α genes, importin β, and HSPG in the whole bodies were measured at 6 dpi using RT-qPCR. Five insects per replicate and 8 or 10 biological replicates were prepared. Salivary glands were dissected at 4 dpi for immunohistochemistry assays using an anti-NP monoclonal antibody with 6 to 15 replicates.
Measurement of RSV Amount and Disease Incidence Rates in Rice Plants.
A total of 20 viruliferous third-instar nymphs were injected with a mixture of dsRNAs for the three importin α genes or dsRNA for GFP, and, after 8 d, they were fed two healthy rice seedlings for 1 d. A total of 20 nonviruliferous third-instar nymphs were injected with a mixture of dsRNAs for the three importin α genes or dsRNA for GFP and then raised on RSV-infected rice seedlings for 8 d before being transferred to two healthy rice seedlings and fed for 2 d. Thirty nonviruliferous third-instar nymphs were injected with RSV crude extracts and then raised on two healthy rice seedlings for 1, 2, and 3 d. The rice seedlings were collected, and the RNA levels of RSV NP were measured using RT-qPCR. A total of 6 to 13 biological replicates and two rice seedlings per replicate were prepared.
A total of 20 viruliferous third-instar nymphs were injected with dsRNA for importin α2, and, after 8 d, they were transferred to new healthy rice seedlings for feeding for 2 d. In another group, 20 nonviruliferous third-instar nymphs were injected with dsRNA for importin α2 and then raised on RSV-infected rice seedlings for 7 d before being transferred to two healthy rice seedlings and fed for 3 d. The control groups were injected with dsRNA for GFP. Partial rice seedlings were collected for measurement of RNA levels of RSV NP using RT-qPCR, and the remaining rice seedlings were cultured in a greenhouse at 30 °C with 16 h of light per day to observe disease symptoms. Eight to 10 biological replicates and two rice seedlings per replicate were prepared for RT-qPCR. Ten rice seedlings per replicate and six or seven replicates were used to calculate the disease incidence rates.
RT-qPCR.
RT-qPCR was used to measure the relative RNA levels of RSV NP, four genomic segments, and the relative transcript levels of importin α genes, importin β, and HSPG on a LightCycler 480 II instrument (Roche). RT-qPCR was performed in a volume of 20 μL comprising 3 μL template cDNA, 10 μL SYBR Green I Master Mix (Roche), and 0.25 μL of each primer (10 μM). The thermal cycling conditions were 95 °C for 10 s followed by 45 cycles of 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 20 s. The transcript level of planthopper translation elongation factor 2 (EF2) or rice tubulin (XP_015649724.2) was quantified to normalize the cDNA templates. The primers for each gene are listed in SI Appendix, Table S1. The relative transcript level of each gene or RNA levels of RSV NP and four genomic segments to that of EF2 or tubulin was reported as the mean ± SE. Differences were statistically evaluated using Student’s t test to compare the two means and one-way ANOVA followed by Tukey’s test for multiple comparisons in SPSS 17.0.
Recombinant Protein Expression in S2 Cells and Immunohistochemistry.
The full length, IBB domain, Arm domain, and carboxyl terminus of importin α2 and importin β were constructed in the plasmid pAc-5.1/V5-HisB (Invitrogen) with His tags. The primers used for cloning are listed in SI Appendix, Table S1. The recombinant plasmids (1 μg/well) were transfected into 500 μL Drosophila S2 cells in a 24-well plate using Lipofectamine 3000 (Invitrogen). In another group, recombinant plasmids containing importin α2 or the IBB domain were transfected together with 1, 10, or 100 μM Wnt-C59. After 48 h, the cells were fixed with 4% paraformaldehyde for 30 min at room temperature. After washing three times with 1× PBS buffer (0.01 M phosphate-buffered saline, pH 7.4), the cells were blocked with 1% bovine serum albumin for 30 min and then sequentially incubated with the anti-His monoclonal antibody (CWBiotech) at 4 °C overnight and the Alexa Fluor 488 AffiniPure goat anti-mouse immunoglobulin G (IgG) secondary antibody (Invitrogen) for 1 h at room temperature. After washing three times with 1× PBST buffer (0.01 M PBS containing 1% Tween-20, pH 7.4), F-actin was labeled with phalloidin (Abcam), and nuclei were labeled with Hoechst (Invitrogen). Cells transfected with the empty plasmid pAc-5.1/V5-HisB were used as negative controls. Fluorescence was viewed under a Zeiss LSM 710 confocal microscope (Carl Zeiss AG).
Immunohistochemistry on Salivary Glands and Midguts.
Salivary glands and midguts were dissected in cold 1×PBS buffer (pH 7.4) on a glass plate and fixed in 4% paraformaldehyde for 1 h at room temperature. The samples were permeabilized with Cell Penetrating Solution (Beyotime) for 1 h. After washing twice with 1× PBST buffer (pH 7.4), the samples were incubated with a homemade anti-NP monoclonal antibody or anti–importin α2 polyclonal antibody (26) overnight at 4 °C. After washing with 1× PBST buffer (pH 7.4), 5% goat serum was added for blocking for 1 h followed by the addition of Alexa Fluor 488 AffiniPure goat anti-mouse IgG or Alexa Fluor 594 AffiniPure goat anti-rabbit IgG secondary antibodies (Invitrogen). Phalloidin was used to label F-actin. The negative control did not include the primary antibodies. Fluorescence was viewed under a Zeiss LSM 710 confocal microscope (Carl Zeiss AG).
Recombinant Protein Expression in Escherichia coli Cells and Purification.
The ORF of RSV NP (DQ299151) was cloned into the pET28a vector between the NcoI and EcoRI or NcoI and XhoI restriction sites to generate Flag-tagged or His-tagged recombinant proteins. NP was also inserted into the pGEX-3X vector at the SmaI site to generate GST-tagged recombinant protein. The IBB domain, Arm domain, and carboxyl terminus of importin α2 and five fragments of HSPG (1 to 674 aa for HSPG-1, 675 to 1,396 aa for HSPG-2, 1,397 to 1,970 aa for HSPG-3, 1,971 to 2,500 aa for HSPG-4, and 2,501 to 3,235 aa for HSPG-5) were inserted into pET28a between the BamHI and XhoI restriction sites, and full-length importin α2 was inserted between NcoI and EcoRI restriction sites to generate His-tagged recombinant proteins. The full-length importin β was inserted between NcoI and XhoI to generate HA-tagged recombinant proteins. The corresponding primers are listed in SI Appendix, Table S1. The recombinant plasmids were transformed into E. coli BL21 (DE3) for expression. After 12 h of induction with 0.5 mM isopropyl b-d-thiogalactoside at 20 °C, the cells were pelleted by centrifugation and then sonicated for 30 min on ice. The supernatant was retained for Co-IP and heparin solution-binding assays. The importin α2–His, NP-GST, and GST proteins were purified from the supernatant using Ni Sepharose 6 Fast Flow (GE Healthcare) or glutathione Sepharose 4B beads (GE Healthcare) following the manufacturer’s instructions and dissolved in 10 mM Tris⋅HCl (pH 8.0) after filtration with a 10-kDa cutoff Amicon Ultra Centrifugal Filter (Millipore) for MST assay.
Co-IP Assay.
A total of 5 μg mouse anti-His, anti-HA, or anti-Flag monoclonal antibody (CWBiotech) was incubated with 50 μL Dynabeads Protein G (Novex, Thermo Fisher Scientific) for 15 min at room temperature. Then, 400 µL 1:1 mixture of recombinantly expressed importin α2 fragment (IBB-His, Arm-His or carboxyl-terminal–His) and NP-Flag, importin β–HA and NP-Flag, importin β–HA and IBB-His, or HSPG His-tagged five fragments and NP-Flag were added and incubated for 2 h at 4 °C. In another group, 400 µL IBB-His and 400 µL total protein from viruliferous planthoppers in 10 mM PBS buffer (pH 8.0) was added and incubated for 2 h at 4 °C. The total protein from E. coli–expressing empty pET28a was applied in the negative control groups. A total of 5 μg NP monoclonal antibody was first incubated with 50 μL Dynabeads Protein G (Novex) for 30 min at room temperature, after which 400 μL total protein from viruliferous planthoppers in 10 mM PBS buffer (pH 8.0) was added. Approximately 10% of the total protein was reserved as input. Mouse IgG (Merck Millipore) was used as a negative control. After washing three times with washing buffer (Novex), the antibody–protein complex was disassociated from the beads with elution buffer (Novex) for Western blot analysis with anti-His, anti-HA, anti-Flag, or anti-NP monoclonal antibodies or anti–importin α2 polyclonal antibodies.
Heparin-Binding Assay.
HiTrap heparin beads (GE Healthcare) were equilibrated with 1 mL equilibrium buffer (50 mM Tris⋅HCl, 10 mM sodium citrate, pH 7.4) for 5 min. Importin α2–His, IBB-His, Arm-His, carboxyl-terminal–His, or NP-His was added to heparin beads and incubated at 4 °C for 2 h, or IBB-His was first mixed with NP-His before loading on the heparin beads. After washing with equilibrium buffer five times, 50 μL elution buffer (50 mM Tris⋅HCl, 10 mM sodium citrate, 1 M NaCl, pH 7.4) was added, and the supernatant was used for Western blot analysis with anti-His monoclonal antibody (CWBiotech) after centrifugation for 2 min at 12,000 × g.
Microscale Thermophoresis Assay.
The affinity between importin α2–His and NP-GST was measured with an MST assay in a Monolith NT.115 instrument (NanoTemper Technologies). Importin α2–His was labeled with the blue fluorescent dye NHS-495. The concentration of importin α2–His was maintained at 10 μM, whereas the concentrations of NP-GST were gradient-diluted from 17.5 μM to 0.0417 nM. After a brief incubation, MST-standard glass capillaries (NanoTemper Technologies) were filled with the samples. The measurements were performed at 25 °C. The purified GST protein was used instead of NP-GST in the control group. The assays were repeated in three biological replicates. Data were analyzed to obtain KD using Monolith Affinity Analysis version 2.2.4 software.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 32090012), the State Key Research Development Program of China (No. 2019YFC1200504), the Collaborative Program between the Chinese Academy of Sciences (CAS) and Commonwealth Scientific and Industrial Research Organisation of Australia (No. 152111KYSB20190062), and the Youth Innovation Promotion Association, CAS (No. 2019086).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103393118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Miyamoto Y., Yamada K., Yoneda Y., Importin α: A key molecule in nuclear transport and non-transport functions. J. Biochem. 160, 69–75 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Miyamoto Y., Boag P. R., Hime G. R., Loveland K. L., Regulated nucleocytoplasmic transport during gametogenesis. Biochim. Biophys. Acta 1819, 616–630 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A., Importin α: A multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Yasuda Y., et al., Nuclear retention of importin α coordinates cell fate through changes in gene expression. EMBO J. 31, 83–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan C. K., Jans D. A., Synergy of importin alpha recognition and DNA binding by the yeast transcriptional activator GAL4. FEBS Lett. 462, 221–224 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Hachet V., Köcher T., Wilm M., Mattaj I. W., Importin alpha associates with membranes and participates in nuclear envelope assembly in vitro. EMBO J. 23, 1526–1535 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownlee C., Heald R., Importin α partitioning to the plasma membrane regulates intracellular scaling. Cell 176, 805–815.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamada K., et al., Cell surface localization of importin α1/KPNA2 affects cancer cell proliferation by regulating FGF1 signalling. Sci. Rep. 6, 21410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bragard C., et al., Status and prospects of plant virus control through interference with vector transmission. Annu. Rev. Phytopathol. 51, 177–201 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Ammar E.-D., Tsai C.-W., Whitfield A. E., Redinbaugh M. G., Hogenhout S. A.,Cellular and molecular aspects of rhabdovirus interactions with insect and plant hosts. Annu. Rev. Entomol. 54, 447–468 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Gray S. M., Banerjee N., Mechanisms of arthropod transmission of plant and animal viruses. Microbiol. Mol. Biol. Rev. 63, 128–148 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia D., et al., Vector mediated transmission of persistently transmitted plant viruses. Curr. Opin. Virol. 28, 127–132 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Dáder B., et al., Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Sci. 24, 929–946 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Hogenhout S. A., Ammar E.-D., Whitfield A. E., Redinbaugh M. G., Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46, 327–359 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Liu W., Hajano J. U., Wang X., New insights on the transmission mechanism of tenuiviruses by their vector insects. Curr. Opin. Virol. 33, 13–17 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Huang H. J., et al., Identification of salivary proteins in the whitefly Bemisia tabaci by transcriptomic and LC-MS/MS analyses. Insect Sci., 10.1111/1744-7917.12856 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Xu C., et al., Rice virus release from the planthopper salivary gland is independent of plant tissue recognition by the stylet. Pest Manag. Sci. 76, 3208–3216 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Dietzgen R. G., Mann K. S., Johnson K. N., Plant virus-insect vector interactions: Current and potential future research directions. Viruses 8, 303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitfield A. E., Falk B. W., Rotenberg D., Insect vector-mediated transmission of plant viruses. Virology 479-480, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Wei T. Y., et al., Genetic diversity and population structure of rice stripe virus in China. J. Gen. Virol. 90, 1025–1034 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Toriyama S., Rice stripe virus: Prototype of a new group of viruses that replicate in plants and insects. Microbiol. Sci. 3, 347–351 (1986). [PubMed] [Google Scholar]
- 22.He M., Guan S. Y., He C. Q., Evolution of rice stripe virus. Mol. Phylogenet. Evol. 109, 343–350 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Xiong R., Wu J., Zhou Y., Zhou X., Identification of a movement protein of the tenuivirus rice stripe virus. J. Virol. 82, 12304–12311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi M., Toriyama S., Hamamatsu C., Ishihama A., Nucleotide sequence and possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 74, 769–773 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Toriyama S., Takahashi M., Sano Y., Shimizu T., Ishihama A., Nucleotide sequence of RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. Gen. Virol. 75, 3569–3579 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Zhao W., et al., The nucleocapsid protein of rice stripe virus in cell nuclei of vector insect regulates viral replication. Protein Cell, 10.1007/s13238-021-00822-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J., et al., Characterization of protein-protein interactions between rice viruses and vector insects. Insect Sci., 10.1111/1744-7917.12840 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Lott K., Cingolani G., The importin β binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta 1813, 1578–1592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobe B., Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6, 388–397 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Pumroy R. A., Cingolani G., Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J. 466, 13–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J. P., Kusche-Gullberg M., Heparan sulfate: Biosynthesis, structure, and function. Int. Rev. Cell Mol. Biol. 325, 215–273 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Sarrazin S., Lamanna W. C., Esko J. D., Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cagno V., Tseligka E. D., Jones S. T., Tapparel C., Heparan sulfate proteoglycans and viral attachment: True receptors or adaptation bias? Viruses 11, 596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rusnati M., et al., Sulfated K5 Escherichia coli polysaccharide derivatives: A novel class of candidate antiviral microbicides. Pharmacol. Ther. 123, 310–322 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Mao Q., et al., Viral pathogens hitchhike with insect sperm for paternal transmission. Nat. Commun. 10, 955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin F., et al., Invasion of midgut epithelial cells by a persistently transmitted virus is mediated by sugar transporter 6 in its insect vector. PLoS Pathog. 14, e1007201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu G., et al., Tenuivirus utilizes its glycoprotein as a helper component to overcome insect midgut barriers for its circulative and propagative transmission. PLoS Pathog. 15, e1007655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao W., Yang P., Kang L., Cui F., Different pathogenicities of rice stripe virus from the insect vector and from viruliferous plants. New Phytol. 210, 196–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.