Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causing COVID-19 is associated with excessive inflammation, as a main reason for severe condition and death. Increased inflammatory cytokines and humoral response to SARS-CoV-2 correlate with COVID-19 immunity and pathogenesis. Importantly, the levels of pro-inflammatory cytokines that increase profoundly in systemic circulation appear as part of the clinical pictures of two overlapping conditions, sepsis and the hemophagocytic syndromes. Both conditions can develop lethal inflammatory responses that lead to tissue damage, however, in many patients hemophagocytic lymphohistiocytosis (HLH) can be differentiated from sepsis. This is a key issue because the life-saving aggressive immunosuppressive treatment, required in the HLH therapy, is absent in sepsis guidelines. This paper aims to describe the pathophysiology and clinical relevance of these distinct entities in the course of COVID-19 that resemble sepsis and further highlights two effector arms of the humoral immune response (inflammatory cytokine and immunoglobulin production) during COVID-19 infection.
Abbreviations: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, 2019 novel coronavirus disease; CoV, coronavirus; WHO, World Health Organization; ACE2, Angiotensin-converting enzyme 2; IFNγ, Interferon gamma; IL-1β, interleukin-1β; IP-10, interferon-inducible protein 10; MCP1, monocyte chemoattractant protein 1; ICU, intensive care unit; TNF-α, tumor necrosis factor alpha; G-CSF, Granulocyte colony-stimulating factor; MIP1a, inflammatory protein 1a; CRS, Cytokine release syndrome; CAR T cells, Chimeric antigen receptor T cells; scRNA-seq, Single-cell RNA sequencing; NFIL3, ETS2, nuclear factor regulated by IL-3; PHLDA2, Pleckstrin Homology Like Domain Family A Member 2; HLH, hemophagocytic lymphohistiocytosis; ARDS, acute respiratory distress syndrome; NK cell, Natural killer cell; AST, aspartate aminotransferase; PB, plasmablast; CCL2, chemokine C–C motif ligand 2; SOFA, Sequential/Sepsis-related Organ Failure Assessment; SHLHOS, sepsis-HLH overlap syndrome; IgM, Immunoglobulin M; ELISA, enzyme-linked immunoassay; MERS-CoV, Middle Eastern Respiratory Syndrome Coronavirus
Keywords: Antibody response, Hemophagocytic lymphohistiocytosis (HLH), 2019 novel coronavirus disease (COVID-19), Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Sepsis
1. Introduction
The novel human coronavirus (CoV) designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first distinguished in infected patients with pneumonia in Wuhan, China, in December 2019. The respiratory illness derived from SARS-CoV-2 was termed by the World Health Organization (WHO) as the corona virus disease 2019 (COVID-19) that has become the most severe public health issue worldwide. Since the first reports of COVID-19 in Wuhan, there has been an exponential growth in the number of individuals diagnosed with COVID-19 all over the world. On March 11, 2020, the outbreak was declared a pandemic by WHO [1].
Several studies have now established that the COVID-19 is associated with excessive inflammation, as a main reason for severe condition and death in infected patients [2], [3], [4]. A key question for hospitalized patients with COVID-19, then, is how immune responses alter over time in the course of COVID-19. Complete and comprehensive clinical assessment focusing on the immunological characteristics is fundamental to the appropriate selection of treatment for the patient groups and for reliable analysis of experimental results [5]. A proper comprehension of viral immunopathogenesis may help with earlier management of the severe complications and clarify the best approach in managing this disease and better monitoring of the treatment response as well as its clinical course. These include both cellular immune responses, such as the induction of a high level of Th1 responses and cytotoxicity and humoral immune responses, mediated by increased antibody and cytokine levels [6]. Humoral responses have been associated with clinical outcome in patients with SARS-CoV-2 virus infection. Although the humoral immune responses induced by SARS-CoV-2 is rapid and is elicited by most infected individuals, its magnitude and time course kinetics correlates with COVID-19 disease severity [7]. This is the key issue in the management of the pandemic since the main target of current vaccine approaches is B cells that produce antibodies to target virus and infected cells. Biomedical data has also evidenced the association between COVID-19 clinical outcome and inflammatory cytokines. The level of pro-inflammatory cytokines that increase profoundly in systemic circulation appear as part of the clinical pictures of two distinct but overlapping conditions, where a robust inflammation is elicited affecting multiple organ damage, sepsis that usually presents with respiratory distress and multiple organ dysfunction and the hemophagocytic syndromes which are mostly caused by the activation of macrophages as a result of an infection [8]. The review may shed some lights on the understanding of the pathophysiology of the ambiguous and complex manifestations of COVID-19 and will additionally inform about the clinico-pathogenesis of acute lung inflammation caused by COVID-19, in a natural host-pathogen interaction. Given the key role of antibodies in protective immunity and immune pathogenesis of viral diseases, we focus this review to specific issues relating to a serological correlate of protection from SARS-CoV-2 for COVID-19 vaccine evaluation and discuss how they are related to viral load in acute infection and SARS-CoV-2-induced clinical illness severity. We further highlight the importance of screening in seroprevalence studies of the infection, seroconversion rate and the complications with rapid therapeutic intervention with immunoglobulin treatment.
2. COVID-19 pathology frameworks
2.1. COVID-19 pathology is substantially associated with perturbations in immune system compartments
It seems that COVID-19 illness drives two distinct but related pathologies triggered by the virus itself and by the host response, albeit in different levels of severity [9]. The early reports suggest that in the establishment phase of the infection the symptom expression is similar in immunocompetent and immunoquiescent states as in the elderly, or transplant recipients [10]. Owing to the concomitant use of anti-inflammatory therapy in heart transplantation, the COVID-19 disease tends to be milder in the second phase which is determined by the inflammatory host response [11], [10]. Accumulating evidence indicates a plausible link between the host immune response and disease progression during 2019-nCov infection and this plays an important role in shaping SARS-CoV-2 pathology. In this respect, at non-severe stages, development of COVID-19-induced immunity is thought to produce a classical two-phase immune profile that provides a protective response followed by pro-inflammatory damaging reactions at the severe stage [12]. Lung damage is a major source of morbidity and mortality limiting recovery in those severe patients and there are occasional serious complications associated with kidney failure or heart problems. It has been reported that severe COVID-19 disease is more likely in the elderly, who have weaker immune function. These population groups are considered to have an adverse outcome with regard to their general health status. This report establishes a role for good health in mounting a protective endogenous immune response that elicits specific antiviral immunity. When host-protective immune response is impaired, virus will propagate to a high extent causing massive destruction of the tissues with high expression of Angiotensin-converting enzyme 2 gene (ACE2) such as kidney and intestine. Impaired cells, as a result, contribute to innate immune activation leading to inflammation in the lungs [12]. Pulmonary inflammation is the most common feature of life-threatening respiratory disorders that afflict patients in the older age group/severe stages [13]. Although early adaptive immune response is needed to eradicate the virus in the early stages and likely contribute to the susceptibility of the host to infection but it may be even a causative factor in pulmonary pathologies. The first immune response emerges from innate immune cells including macrophages, neutrophils, and the NK cell activities initiated to try to eliminate the virus, which further activates the adaptive immune system [14]. The anti‐viral adaptive immune response resides on cytotoxicity by CD8 + CTL, Th1 subset of CD4 + T cells, and antibody-secreting plasma cells [14]. Many of the evidenced severe COVID-19 cases demonstrate a large number of proinflammatory cytokines in serum [9], [15]. Moreover, the existence of autoantibodies directed against a variety of proteins including cytokines, chemokines, and cell surface antigens in the serum of COVID-19 patients may contribute to the tissue damage by immune complex formation and activating complement [16]. Since an exacerbated immune response to the virus can aggravate a preexisting injury condition, being in a good overall health state may not be beneficial for those who have progressed to the severe stage of the disease. In the late phases, once severe lung damage occurs, treatment of virally driven hyperinflammation tailored to this demanding condition may be able to keep it from getting worse or stop it in order to reduce fatality rates. Yet identifying the immune mechanisms which determine the infection duration induced by the virus and discriminate between people with severe and non-severe (mild, moderate) COVID-19 infection has been the subject of debate.
2.2. The molecular dynamics of cytokine production during SARS-CoV-2 infection
It would also be relevant to illuminate the molecular dynamics of cytokine response during the course of disease. Early studies have shown that 2019-nCoV infection induce increased concentrations of proinflammatory cytokines, including IFN-γ, IL-1β, IP-10, and MCP1 which is similar to features of infections caused by SARS-CoV [17] and MERS-CoV [18]. The role of these cytokines in pathophysiology of the disease is briefly explained as a diagram in Fig. 1 . At the same time, SARS-CoV-2 may antagonize the antiviral interferon response of the host and thus evade innate immunity [16]. Furthermore, similarities and differences of clinical features between severe and non-severe COVID-19 patients have been noted in regard to the kinetics of the immune response which is of major importance in the pathogenesis or progression of COVID-19 infection. In this regard, comparison between 2019-nCoV-infected patients admitted to the intensive care unit (ICU) and non-ICU patients has shown higher levels of specific cytokines (TNF-α, IL-2, IL-7, IL-10, MCP-1, G-CSF, MIP1A, and IP-10) in patients requiring ICU admission than did subgroups not requiring ICU admission [9] proposing that the cytokine storm strongly correlates with disease severity [19]. In the first critical COVID‐19 case in Zhejiang Province, Zhang et al. [20] showed that elevated circulating levels of IL‐6, IL‐10 and IFN‐γ decreased quickly while the levels of IL‐4 and TNF‐α increased when RT-PCR test for viral RNA returned negative. Since dynamics of the cytokine levels during SARS‐CoV‐2 infection appear to be related to disease severity, they may therefore serve as a potential biomarker for prognostic evaluation [21].
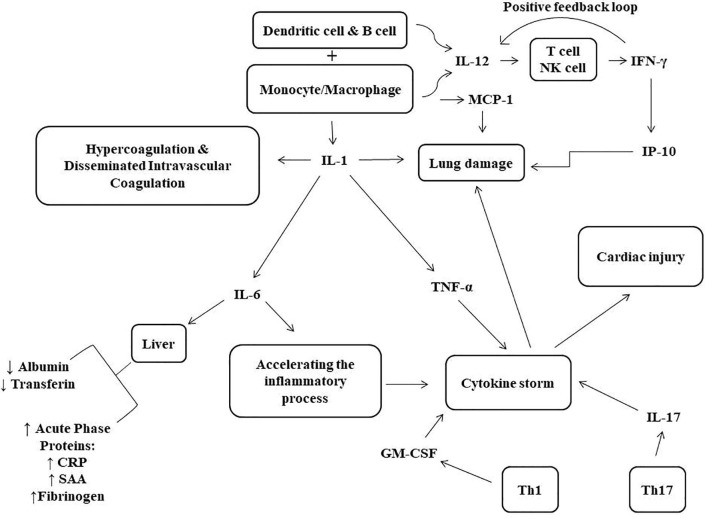
Fig. 1.
Schematic diagram of the pathological effects of immune system cytokines in COVID-19. During the disease caused by SARS-CoV-2, activated monocytes and macrophages produce various cytokines such as IL-1, IL-6, and TNFα which can cause cytokine storm and multiorgan damage. IL-6 also can induce liver cells to synthesize acute phase proteins and is associated with low albumin and transferrin concentrations. In addition, IL-12, produced by monocyte/macrophage, dendritic cell and B cells, may induce NK and T cells to secrete IFN-γ which in turn stimulates IL-12 production in a positive feedback loop. SAA; serum amyloid A, CRP; C reactive protein, Th; helper T cell, IL-; interleukin-, TNF-α; tumor necrosis factor alpha, GM-CSF; granulocyte–macrophage colony-stimulating factor, MCP-1; monocyte chemoattractant protein1, IP-10; Interferon-Inducible Protein 10, IFNγ; interferon γ protein.
2.3. The primary source of the cytokine storm in response to SARS-CoV-2 infection
Cytokine release syndrome (CRS) or cytokine storm is a complex hyperimmune response syndrome usually seen with T-cell activating therapeutics as in patients receiving CAR-T cell therapy which results in symptoms including fever, nausea, headache, and hypotension [22]. CRS can occur after a wide variety of infectious and non-infectious stimuli. Cytokine stimulation by infectious factors, or condition would exacerbate severity of the disease and an exaggerated cytokine response has been described as a driver of pathology in COVID-19 patients with advanced disease [23]. Since severe 2019-nCoV infection have been characterized with lymphocytopenia (indicating a state of immunosuppression), it is inferred that COVID-19-induced CRS may be the result of an overactive innate immune response mounted by other leukocytes [12]. Previous studies on SARS virus have shown the stronger host innate immune responses to viral infection in older animals inoculated with SARS-CoV compared to younger adults with a marked elevation in expression levels of inflammation related genes [24]. Regarding SARS-CoV-2, the innate immune response to the virus has been proposed to contribute to the development of acute respiratory distress syndrome (ARDS) due to the rapid onset of widespread inflammation in the lungs [13]. The pathological investigation of the lungs in fatal cases of COVID-19 reveals massive infiltration of alveolar macrophages with slight lymphocytic infiltration [25]. The composition of immune cells localized to the lung differs across patients ranging from mild to severe. In the severely injured lung, the predominant macrophage lineage is greatly inflammatory Fibronectin-like sequences within NC1+ (FCN1 + ) macrophages, a phenotype that associates with monocyte-derived macrophages [26], whereas, in both healthy subjects and mildly infected cases, the alveolar macrophages consisted the principle tissue-resident macrophages in the lungs. Indeed, in lung (bronchoalveolar lavage fluid) immune cell composition, FABP4+ (fatty acid-binding protein 4) alveolar macrophages with lipid metabolic functions replace the inflammatory monocyte-derived FCN1 + macrophages indicating a disturbed balance of lung macrophage subpopulations during the progression of severe COVID-19 [26]. Importantly, the depletion of alveolar macrophages, as effector cells for pulmonary cell-mediated immunity [27], in severely infected lungs is likely a leading cause of failed lung function. These data show that there is relationship between disease severity during the COVID-19 infection and the loss of resident alveolar macrophages accompanied by the accumulation of monocyte-derived inflammatory macrophages. Inflammatory macrophages having interferon signaling and monocyte-recruiting chemokine programs can lead to a macrophage excess and this may drive severe lethal pneumonia in SARS-CoV-2 infected individuals [28]. Profiling the peripheral immune cells in COVID-19 demonstrated a unique monocyte subset called as “severe stage-specific monocyte” which existed only in severe patients. Single-cell RNA sequencing (scRNA-seq) analysis of peripheral blood mononuclear cells suggest that distinct properties of these cells are dictated by a gene regulatory network consisted of ETS2, NFIL3 and PHLDA2 transcription factors that involved in regulating the monocyte inflammatory storm [29]. Taken together, the data convincingly propose that an excessive secretion of proinflammatory cytokines of macrophage origin is responsible for immunologically mediated adverse effects in SARS-CoV-2 infected patients. The critical issue is how to recognize and intervene early in those patients at increased risk of developing this complication. Fig. 2 depicts the local immune mechanisms and mediators of pulmonary hyperinflammation and impaired gas exchange in the lungs in patients with mild and severe COVID-19 illness.
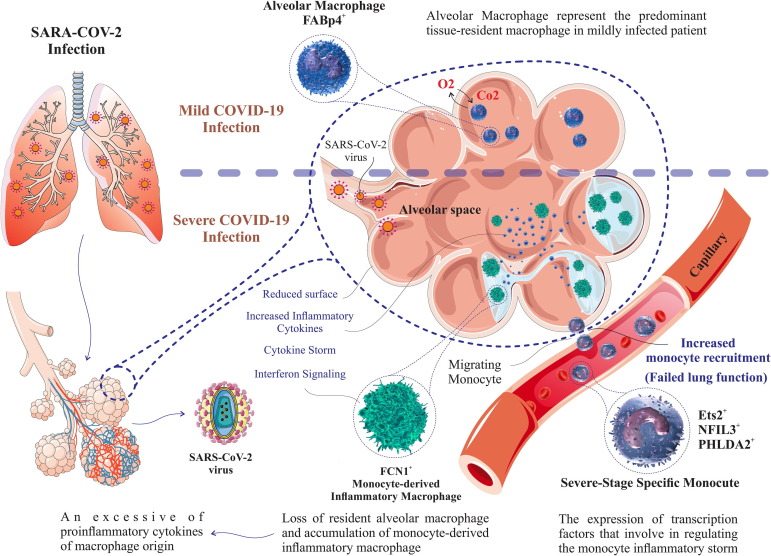
Fig. 2.
Specific macrophage-monocyte lineage cells surrounding alveoli that cause local pulmonary inflammation after SARS-CoV-2 infection or COVID-19 disease. The composition of immune cells localized to the lung differs across patients ranging from mild to severe. The pathological investigation in mild cases of COVID-19 reveals massive infiltration of alveolar macrophages, while in the severely injured lung the predominant macrophage lineage is inflammatory FCN1 + macrophages, that associates with monocyte-derived macrophages. A unique monocyte subset called as “severe stage-specific monocyte” exists only in severe stage patients with COVID-19.
2.4. Immunopathogenesis of SARS-CoV-2-induced disease: A potential infection-associated hemophagocytic lymphohistiocytosis or viral sepsis?
Current data supports that hyper immune reaction, leading to cytokine storm in COVID-19, which is clinically specified by lymphopenia, pathological damage, respiratory failure, shock, and organ failure, is at least partially accounted for these poor outcomes. Two prominent immune dysregulation syndromes implicated as common causes of hyper inflammation associated with tissue injury include HLH and toxic shock syndrome/sepsis and thus it is crucial to think about it when facing a patient with fever, cytopenia, hepatosplenomegaly, and other systemic manifestations. Identification and characterization of the sepsis and HLH overlapping syndromes might be an obstacle to deal with in this setting, because both disorders cause a similar presentation. To explore better clinical care for critically COVID-19 ill patients with pneumonia, this section aimed to describe the clinical and laboratory manifestations of these patients to accurately define the immunopathogenesis derived from the systemic cytokine storm. To date, the underlying cause of hyperinflammation in patients with COVID-19 has remained elusive. Hemophagocytic lymphohistiocytosis (HLH; hemophagocytic syndrome), is known as a potentially fatal hyperinflammatory status that describes the phenomenon of activated macrophages which phagocytose hematopoietic cells such as leukocytes, platelets, erythrocytes, and their precursor cells in the bone marrow, lymph nodes, or liver, leading to the clinical symptoms. It is of two types - primary HLH (familial HLH) and secondary HLH (acquired HLH). The later occurs following strong immunologic activation such that occurs with systemic infection (virus, bacteria and protozoa), neoplasms, and autoimmune disease [30]. The clinical feature of the syndrome is mainly determined by prolonged fever, splenomegaly, and hemophagocytosis in the bone marrow and the major laboratory hallmarks include hyperferritinemia, hypertriglyceridemia, cytopenias, hypofibrinogenemia, decreased or absent activity of NK cells, and elevated sCD25 [31]. For SARS-CoV-1, HLH have been shown related to adverse clinical outcomes in a subset of fatal infections [32], [33], [34], [35]. Analysis of laboratory results in a large cohort of inpatients with COVID-19 showed that such abnormalities as anemia, thrombocytopenia, elevated ferritin and ALT are significantly more frequent in non-survivors compared to survivors [36]. There is a small case-series report in literature describing hypertriglyceridemia, high fever, and hyperferritinemia, which are helpful in combination with distincting HLH from non-HLH COVID-19 patients with ARDS [37]. Progression to ARDS , that is, the upregulation of pro-inflammatory cytokines and chemokines, in several severe COVID-19 patients is very similar to the pattern found in macrophage activation syndrome or secondary hemophagocytic lymphohistiocytosis (sHLH), a clinical condition presenting as a cytokine storm syndrome associated with multi-organ system dysfunction [38]. Laboratory and clinical features of a severe COVID-19 patient often resemble that of HLH including fever, cytopenias, and pulmonary involvement [39], [40]. A HLH-like cytokine profile involving enhanced the cytokine production, including IL-1β, IL-2, IL-6, IL-7, IL-8, TNF-α and chemokines such as CXCL10 and CCL2 predominate in the majority of severe COVID-19 infections [41], [42]. Cytokine storm with features akin to HLH, however, is associated with profound immunosuppression which is evident with pronounced lymphopenia, and decreased natural killer cell function [43], [42]. HLH is often diagnosed using clinical, laboratory, and histologic features [44]. Pathologic detection of hemophagocytosis plays an essential role in the diagnosis of HLH. Post-mortem findings in a series of 4 cases with laboratory-confirmed COVID-19 have documented histologic evidence of hemophagocytosis [37].
Sepsis as a distinct medical entity represents a state of uncontrolled inflammatory response [45]. Although bacterial infection has been the predominant cause of sepsis syndrome, viral infections can also elicit sepsis. This association has previously been described where it was shown that nearly 40% of community-acquired pneumonia adults had sepsis on account of viral infection [46]. Similarly, sepsis might be directly resulted from SARS-CoV-2 infection. A univariate and multivariate analysis for the risk factors of in-hospital death using retrospective data on 191 patients with COVID-19 detected the developed sepsis and no bacterial pathogens in more than half of patients [36]. According to the International Consensus definitions for sepsis and septic shock (Sepsis-3), the assessment of Sequential/Sepsis-related Organ Failure Assessment (SOFA) score is suggested as a measure of sepsis-associated organ dysfunction [47]. In addition to severe lung injury, many late phase COVID-19 patients satisfy several of the criteria required for sepsis diagnosis including cold extremities and weak peripheral pulses, severe metabolic acidosis, impaired liver [48] and kidney function [49] indicating possible recognition of sepsis in these patients [50].
Documentation of the mechanism of hyperimmune host reactions triggered by the virus that results in hypercytokinemia is found to be complicated because human COVID-19 disease has been associated with severe clinical manifestations in the form of sepsis and the overlapping disorder, HLH-like illness, as well. It seems possible that the inflammatory response elicited by SARS-CoV-2 virus may trigger a hyper-inflammatory disease course identified by HLH syndrome in at least a subset of patients. Experimental evidence in support of this concept has been given in a cohort of 16 fatal H1N1 adult patients where 81% exhibited HLH histologically and 36% were identified to carry heterozygous mutations in genes associated with familial HLH [51]. Intriguingly, others have reported that 14% of the patients who develop HLH in adulthood harbor hypomorphic mutations in familial HLH–causing genes and these mutations might have an assisting role in developing the late-onset HLH when challenged by viral infection or other stresses [52]. These data may explain that both genetic and immunologic diagnostic testing may be beneficial in forecasting which individuals are at highest risk of cytokine storm and that HLH-directed treatment can reduce mortality associated with HLH in a subset of COVID-19 patients. There are also characteristics of sepsis with cytokine storm that might argue against HLH as the major cause for increased mortality in this pandemic setting. In spite of significant similarity of HLH to in terms of clinical manifestations and pathophysiologic characteristics, it can be discriminated from sepsis in many patients. Since the aggressive immunosuppressive regimen required to treat HLH is absent in sepsis guidelines, differential diagnosis is critically essential between these two conditions [53]. However, the majority of physicians consider sepsis as a leading cause of critical illness for understanding of severe COVID-19 pathogenesis [54] mostly due to the fact that severe COVID-19 presents with hyper-cytokinemia [55], [9]. It is now evident that severe COVID-19 can cause sHLH [56], [37]. Finally, however final conclusions cannot be made, we propose that sepsis-HLH overlap syndrome (SHLHOS) which represents a severe form of sepsis or a subgroup of septic patients who are suffering from dysregulated immune hyperactivity where infection triggers macrophage activation, might explain a significant fraction of critically ill COVID-19 patients with no clear dividing line between sepsis and HLH [57]. Identifying these patients might allow us to select those who would benefit most from immunomodulation.
3. Humoral outlines in SARS-CoV-2 infection
3.1. B cell responses
Although the development of lymphopenia is mainly related to the decrease in absolute T cell counts, contribution of B lymphocytes in this setting in COVID-19 pneumonia remains controversial. There are a significant number of studies that indicate the absolute numbers of B cells were within normal range in most patients during the course of COVID-19 disease [58], [43]. Other reports suggest decrease in B cells in COVID-19 patients and that severe cases have a diminished level than mild cases [59], [43]. In contrast, in a comparison between severe, recovery and healthy stages, a distinct difference has been observed between the groups; while the absolute number of total lymphocytes was decreased in COVID-19 patients, the proportion of B lymphocytes was found to be higher in most patients, more profoundly in severe cases [60]. In addition, plasma B cells, the antibody-secreting cells, were found enriched at severe and recovery stages versus healthy controls indicating that humoral immunity is crucial to fight off viral infection [29]. However, it remains a matter of debate whether antibody-dependent enhancement play roles in disease exacerbation [61], [62]. Such a scenario has been considered especially based on the findings that COVID-19 ICU patients who had evidence of SARS-CoV2-specific antibodies were not protected yet, and may even be at increased risk for adverse outcome [63]. Meanwhile, in another study, it is suggested that B‐cell response might be nonessential based on the observation that the two patients with X‐linked agammaglobulinemia who were exposed to SARS‐CoV‐2 and developed pneumonia could recover from the COVID-19 disease [64] implying that the production of antibody is probably involving in disease progression. It also may reflect that normal T cell response may be sufficient in the immune response against SARS‐CoV‐2 infection.
The failure to document a definitive pattern of B cell kinetics in SARS-CoV-2 infection may be attributed to the analysis of the whole B cell population but not considering subpopulations. A deep profiling study of B cell populations has revealed several alterations in the distribution of B cell subsets in patients with COVID-19 [65]. Within the CD19 + B cells, plasmablast (PB) frequencies (CD19 + CD27 + CD38 + ) were often robustly increased, representing > 30% of circulating B cells in some cases, whereas IgD + CD27- naïve B cell counts were not. However, robust plasmablast populations were only observed in two third of cases, with the remaining patients presenting PB at similar frequencies to recovered cases and healthy subjects [65]. Conversely, class-switched (IgD − CD27 + ) and not-class-switched (IgD + CD27 + ) memory B cell subsets were decreased in COVID-19 patients compared to recovered patients and healthy controls. In following up patients longitudinally for temporal pattern of change in lymphocyte subpopulations, the prior study also found that COVID-19 patients maintain a stable frequency of PB cells at day 0 and day 7 of hospitalization, however there were significant changes in memory B cell subsets [65]. Findings from another study, using single-cell sequencing, found that naïve B cells expressing CD19, CD20 (MS4A1), TCL1A, IL4R, IGHD, and IGHM decreased significantly in the course of COVID-19 recovery stage [66], which contrasts with the previously mentioned report, using high dimensional cytometry, in patients who present with COVID-19 infection [65]. Overall, there is considerable inter-patient heterogeneity for circulating B cell responses, although it appears that both the proportion and number of B cells are not frequently decreased in both severe and non-severe patients [60]. Considering the dynamic acute immune response to SARS CoV-2 [67], a possible reason for the observed heterogeneity may rely in different sampling time points and different sample sizes in the discussed studies.
3.2. Antibody response dynamics in association with clinical manifestations
After SARS-CoV-2 virus exposure, adults are usually capable to mount strong, weak or no antibody response to SARS-CoV-2 nucleocapsid protein. Also, the magnitude of antibody responses produced in COVID-19-infected adults negatively correlates with clinical immunity [68]. It has been observed that the earlier response, and higher antibody titer is associated with disease severity, indicating that strong responders for IgM and IgG among patients with COVID-19 may be actually those with severe disease [69], [70]. A high antibody titers, therefore, is suggested to be an independent risk factor for a worse clinical prognosis in COVID-19 [70]. The potential contribution of antibody response to viral clearance must also be considered, as patients with COVID-19 who were poor IgG responders followed with higher viral clearance rate than that of strong responders [68] which resemble SARS-CoV [62] and MERS-CoV [71] infections. Alternatively, a short duration of viral shedding has been reported to occur in patients with positive anti-SARS-CoV-2 IgM results compared to those with the absence of anti-SARS-CoV-2 IgM antibodies [72]. It has been documented that both anti-SARS-CoV-2 IgM and IgG are produced during COVID-19 infection but their contribution to viral clearance remains to be elucidated. Antibodies specific for the viral spike protein, which facilitate the infection of human immune cells independent of ACE2 receptor, comprise an important fraction of antibodies elicited by SARS-CoV-2 infection [73]. The basic idea and theoretical concern of antibody-dependent enhancement (ADE) with SARS-CoV-2 coronavirus is based primarily on experimental findings and limited clinical evidence [74]. These data indicate a novel cell entry mechanism into immune cells known as antibody-mediated infection. Further evidence support the predominant role of Fcγ receptor (FcγR) in ADE of SARS-CoV-2. The immune cells expressing FcγR for IgG may be infected by IgG-FcγR interactions mediated by anti-SARS-CoV-2 spike protein IgG antibodies which are found in high levels in severe COVID-19 patients [75]. These findings may explain the reason of functional dichotomy between IgG and IgM in the pathogenesis of SARS-CoV-2. Prolonged virus shedding even after seroconversion has been demonstrated in an individual case report [76]. Fig. 3 illustrates the possible association between the specific antibody response in early infection which is mainly of IgM type and after seroconversion to IgG anti-SARS-CoV-2 antibodies with the viral load and shedding in COVID-19 patients.
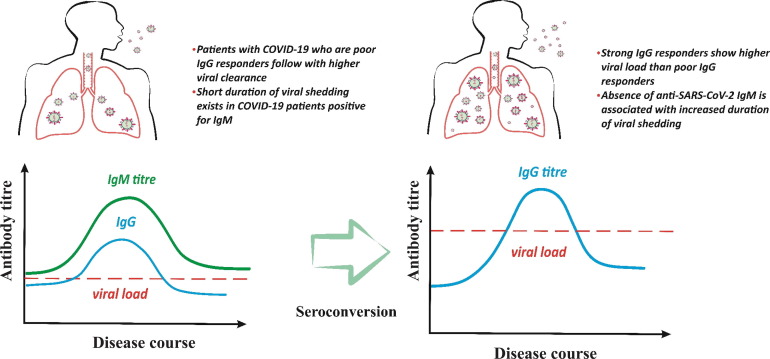
Fig. 3.
Antibody response associates with viral load and shedding of patients with COVID-19 infection. In the early infection the specific antibody responses against SARS-CoV-2 is mainly the IgM antibody response that is correlated to higher viral clearance whereas following seroconversion or in the individuals who produce IgG earlier than IgM, the higher viral load and longer duration of viral shedding has been detected.
The causal link between humoral response and critical illness is still poorly understood. Reasonable hypotheses can be made based on knowledge from MERS-CoV and SARS-CoV which indicate the possibility of antibody-dependent disease enhancement effects [77], [62]. Assessment of IgM and IgG antibody responses in patients who underwent seroconversion, show that IgG and IgM titers were raised in the severe group compared to non-severe group. The serological courses of COVID-19 infection in 285 patients suggest that all had detectable antiviral IgM or IgG within 19 days after symptom onset and the median day of seroconversion for both IgG and IgM was day 13 [78]. Moreover, sequential analysis revealed three models of seroconversion including IgM seroconversion earlier than that of IgG, IgM seroconverted later than or synchronously with IgG [78]. These results are in great contradiction with the assumed principles of sequential serum antibody response to the pathogens switching from an early IgM response to a later IgG response [79], and suggest that the total antibody is more sensitive and rises faster than IgM and IgG for detecting SARS-CoV-2 infection [70]. As a mucosal pathogen, SARS-CoV-2 virus infects individuals mainly through the mucosal routes and it would thus be expected to induce secretory IgA (sIgA). One major effector molecule of mucosal anti-viral immunity is sIgA [80]. IgA-mediated protection prevents pathogens from binding and invading the host cells. A role for antibody-dependent cellular cytotoxicity (ADCC) has also been proposed as a mechanism of effector immune responses mediated by sIgA [81]. In particular, sIgA is able to drive activating signals, leading to cytokine release [82]. A recent study evaluating the pattern of humoral immune response to SARS-CoV-2 showed that remarkably higher level of IgA and IgG were found in severe patients compared to non-severe patients [83]. The positive association between the level of SARS-CoV-2-specific IgA and the disease severity has been established in COVID-19 patients [83]. However, we cannot draw final conclusions, there is overall agreement in that the great majority of confirmed COVID19 patients seroconvert and antibody response vary with different clinical manifestations and disease severity [72].
3.3. Serological assays provide a means for sero-diagnosis, sero-epidemiology and evidence of naturally acquired or vaccine induced immunity
Although molecular diagnostic tests developed rapidly in the early phase of the pandemic, serologic assays are still somewhat limited. The role of adaptive immunity in the natural history of SARS-CoV-2 is particularly important. Adaptive immunity is expected to rise within one week from infection [84]. The use of serological assay as an indirect marker of infection is still debated in terms of its diagnostic values in SARS-CoV-2 infection [85]. Recently, interim guidance for laboratory testing are provided by the World Health Organisation (WHO) showing the strategic use of diagnostic testing in areas with different transmission/circulations of the COVID-19 outbreak [86]. That indicates where the COVID-19 virus is widely spread serological testing over time is recommended to support diagnosis. In areas with no established SARS-CoV-2 virus circulation, it is required to pay attention a case laboratory-confirmed by detecting the unique sequences of virus RNA by molecular testing such as real-time reverse-transcription polymerase chain reaction (rRT-PCR) for at least two different targets on the SARS-CoV-2 genome [86]. The reliability of RT-PCR depends on many factors, including the sample types (throat or nasopharyngeal swabs, sputum, blood, etc.), the quality of the sample (either during collection or shipment), and the quality and consistency of the PCR assays [87], [88]. Characterization of serological profiles also may provide support for the diagnosis of either reinfection or relapse in cases. Serological tests have low sensitivity in acute phase of the disease because of a 5- to 7-day delay in the IgM antibodies produced after exposure and thus give the correct diagnosis in certain phases of the disease. The serological testing represents the main examination in tracking the infection and identification of humoral immune response in vaccinated individuals. It also provides more accurate information regarding epidemiological aspects of the disease related to any previous exposure to SARS-CoV-2 in populations. As an example, the seroprevalence rate of COVID-19 in Wuhan was estimated 3.2%−3.9% in March 2020 and a ∼ 4.1% estimated seroprevalence rate has been recorded in California in April 2020 [16]. Another important aspect to consider is its use in those with mild symptoms or asymptomatic patients as an option for screening of populations including healthcare workers. Currently, there is a substantial group of people asymptomatically infected or very mild cases of COVID-19 infection who mask a population's true rate of infection [89]. This supports the view that screening is currently the strongest tool available in the fight against COVID-19 infection. A recent review of the literature describes the details of the various serological tests used for COVID-19 investigations including rapid antibody tests, and immunoenzymatic serological tests like indirect enzyme-linked immunosorbent assay (ELISA) [90]. Several ELISAs have been developed for the identification of individuals exposed to the virus and for the quantitation of IgG and IgM [90]. The diagnostic potential of the SARS-CoV-2 antibodies is an ongoing debate and further studies are needed to determine the best time to use them for disease assessment. Analysis of the humoral response of 140 cases diagnosed as confirmed (n = 82) and probable COVID-19 (n = 58) cases has shown that the early IgM and IgA antibodies increased both between days 8–14 but were not sustained between days 15–21 of infection or thereafter, whereas the IgG antibody titers increased on days 8–14 and tended to rise until days 15–21 peaking on day 21 [91]. That means that the lack of detection sensitivity at early time-points has limited this approach in early stage infection where the ELISA titer is virtually undetectable at days 0–7 [91]. Generally, however, such serological assays is not employed to make diagnosis of acute infections, they help support some relevant applications [92]. Up to date, serological testing for clinical diagnostic purposes is mostly requested in hospitalized patients when despite a strong clinical suspicion, RNA testing remains negative, or for patients whose samples are collected after the acute phase of the infection, as well as in patients who have low viral loads and await decision to end isolation in clinical practice [93]. One study testing ELISAs using the main immunogenic coronavirus proteins demonstrated that among the spike protein antigens tested, receptor binding domain (RBD), and the N protein antigen were more sensitive than S1 subunit of S protein, while S1 subunit specific IgG ELISA was more specific in detecting SARS-CoV-2 antibodies [94]. The specificity of serological testing for SARS-CoV-2 is of critical importance because cross reactions may occur due to the presence of antibodies against other circulating coronaviruses in the community.
Furthermore, serological tools should be considered to identify potential highly reactive human donors for generation of convalescent plasma/serum therapeutics [92]. Titration of neutralizing antibodies is effective prior to use convalescent plasma therapy. Neutralizing antibodies arise during the course of infection in some infected hosts to enable virus clearance and confer protection in an uninfected host that exposed to the virus [95]. For many viral infections, it is widely accepted that neutralizing antibodies are a main correlate of protection [96], [97], [98]. As an instance, testing for neutralizing antibodies has been an established gold standard for assessing individual protection from polioviruses [99]. In addition, the induction of neutralizing antibody is a crucial criterion of vaccine efficacy studies and can be used in the evaluation of population immunity [100]. Antibodies against SARS-CoV-2 S protein are likely most important to block binding of SARS-CoV-2 virus to the receptor [101]. Monoclonal antibodies against a series of immunodominant regions on the viral proteins—for example, the spike glycoprotein are serotype-specific, while other potential epitopes are not. Two immunodominant linear B-cell epitopes (S14P5 and S21P2) present on SARS-CoV-2 spike glycoprotein have been shown to be associated with a robust immune response; antibodies recognizing these two epitopes could result in a significant inhibition of virus infection, as demonstrated by using sera of convalescent COVID-19 patients and pseudotyped lentivirus assay [102]. While the optimal dose and time point for screening potential plasma donors needs further investigation, it is to be noted that a neutralizing response has been detected for SARS-CoV-2 in a case from day 9 onwards [103].
4. Passive immunization - antibody therapies in Covid-19
4.1. Antibody therapy: Possible benefits and limiting drawbacks
The great demand for the discovery of primary care-based therapeutic methods that combine the high specificity and accelerated development to control a serious viral outbreak often arise at times when vaccine and antivirals are not available. Therefore, it is urgent to consider rapid therapeutic interventions in order to enable emergency recovery from the severe condition of SARS-CoV-2 and its related consequences [104]. In view of the prior promising experience in treating other viral infections such as influenza, SARS, and MERS, great interest has been emphasized that passive immunotherapy and prophylaxis of SARS-CoV-2 infection would become possible by the potential utilization of antibodies [105]. Antibody therapy for infections includes plasma and monoclonal antibody therapies. To improve the emergency condition, passive immunotherapy in the form of convalescent sera represented a promising option where no other treatment was available. Immunotherapy by transferring the convalescent sera to infected patients may be capable of neutralizing the virus and prevent further infection. Early administration of convalescent plasma can be considered, although with some caution, for immunocompromised patients with suspected COVID-19 infection, a situation in which prolonged shedding of virus occurs frequently [106]. Treatment with passive antibody therapy can possibly reduce the viral load of infected patients and reduce the risk of subsequent mortality [107], [108], [109], [110], [111]. However, the challenges associated with availability of sufficient donors, viral kinetics, the influence of neutralizing antibodies on SARS-CoV-2 infection progression and underlying virus–host interactions are still under discussion. Also the challenges in developing these types of antibody-based treatments include the difficulties encountered with/in the viral safety of immunoglobulins preparations, the purity, and specificity. These factors have elicited the renew interest in applying antibody-based treatments to combat the COVID-19 virus. Chicken egg yolk antibodies (IgY), the main immunoglobulin present in avian blood (IgY), have proven useful for many biomedical applications [112]. IgY application as a non-invasive procedure has been successfully tested in human health. Specific anti-viral IgY monoclonal antibodies against SARS CoV-2 offers chances for rapid diagnosis and immunotherapy against COVID-19 [113]. It is more suitable than mammalian serum immunoglobulins because it does not react with components of the human immune system [114].
Monoclonal antibodies are specific therapeutic molecules capable of serving as highly effective treatment candidates protective against particular disease [115], [116]. Accordingly, monoclonal antibodies against proteins present on the viral membrane or the receptor proteins located in the host cell surface can be used to restrain virus binding and can thus be useful in methods of treating or preventing viral infection. This can be achieved by using either an overall strategy of anti-SARS-CoV-2 neutralizing monoclonal antibodies, or anti-ACE2 monoclonal antibodies. The S protein in the viral membrane is the main mediator of virus entry into the target cells and plays a major role in determining host cell specificity of the virus. Two functional subunits consisting of the S1 subunit- the receptor interaction site- with RBD domain and S2 subunit responsible for fusion to host cell have been identified in the S protein. Multiple human neutralizing monoclonal antibodies against SARS-CoV-2 virus have been recognized that include 47D11 which has been shown to target the S1 RBD of SARS-CoV and SARS-CoV-2 spike proteins [117], and the B38, and H4 monoclonal antibodies that are capable of binding to SARS-CoV-2 RBD, but not to SARS-CoV RBD [118]. Of note, the identification of SARS-CoV-2 reactive antibodies has suggested both novel diagnostics and potentially better therapeutic tools for patients. Table 1 lists the clinical trials of immunotherapeutic approaches that provide passive humoral immunity against the COVID-19 disease registered in the ClinicalTrials.gov web site.
Table 1.
Recruited clinical trials of immune-based treatments in COVID-19a patients.
Abbreviations: aCoronavirus disease; bnot applicable; cimmunoglobulin; dProgrammed cell death protein-1; eJanus kinase.
5. Conclusion
Documentation of the mechanism of hyperimmune host reactions triggered by the virus that results in hypercytokinemia is found to be complicated because human COVID-19 disease has been associated with severe clinical manifestations in the form of sepsis and the overlapping disorder, HLH-like illness, as well. As defined on pathology data, it seems possible that the robust inflammatory response elicited by SARS-CoV-2 virus may trigger a hyper-inflammatory disease course identified by HLH syndrome, in at least a subset of patients. There are also characteristics of sepsis with cytokine storm that might argue against HLH as the major cause for increased mortality in this pandemic setting. Since the aggressive immunosuppressive regimen required to treat HLH is absent in sepsis guidelines, differential diagnosis is critically essential between these two conditions [53]. However, the majority of physicians consider sepsis as a leading cause of critical illness for understanding of severe COVID-19 pathogenesis [54] mostly due to the fact that severe COVID-19 presents with hyper-cytokinemia [55], [9], it is now evident that severe COVID-19 can cause sHLH [56], [37].
There is overall agreement in that the great majority of confirmed COVID19 patients seroconvert and antibody response vary with different clinical manifestations and disease severity [72]. Finally, there is considerable inter-patient heterogeneity for circulating B cell responses, although it appears that both the proportion and number of B cells are not frequently decreased in both severe and non-severe patients [60]. Considering the dynamic acute immune response to SARS CoV-2 [67], a possible reason for the observed heterogeneity may rely in different sampling time points and different sample sizes in the discussed studies.
6. Availability of data and materials
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank the Stem Cell Research Center (SCRC) at Tabriz University of Medical Sciences, Tabriz, Iran for the financial support of this project.
Funding
This work was supported by the Stem Cell Research Center (SCRC) at Tabriz University of Medical Sciences, Tabriz, Iran [No. 153645].
References:
- 1.Spinelli A., Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. The British journal of surgery. 2020 doi: 10.1002/bjs.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;1–8 doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Geng M., Peng Y., Meng L., Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020 doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., et al. Immunology of COVID-19: current state of the science. Immunity. 2020 doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghaebi M., Abdolmohammadi-Vahid S., Ahmadi M., Eghbal-Fard S., Dolati S., Nouri M., et al. T cell subsets in peripheral blood of women with recurrent implantation failure. J. Reprod. Immunol. 2019;131:21–29. doi: 10.1016/j.jri.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal transduction and targeted therapy. 2020;5(1):1–11. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo J., Izquierdo-Useros N., Ávila-Nieto C., Pradenas E., Clotet B., Blanco J. Humoral immune responses and neutralizing antibodies against SARS-CoV-2; implications in pathogenesis and protective immunity. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.10.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulert G.S., Grom A.A. Macrophage activation syndrome and cytokine-directed therapies. Best practice & research Clinical rheumatology. 2014;28(2):277–292. doi: 10.1016/j.berh.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F., Cai J., Dong N. First cases of COVID-19 in heart transplantation from China. The Journal of Heart and Lung Transplantation. 2020;39(5):496–497. doi: 10.1016/j.healun.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslam S., Mehra M.R. COVID-19: yet another coronavirus challenge in transplantation. The Journal of Heart and Lung Transplantation. 2020;39(5):408–409. doi: 10.1016/j.healun.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X et al. COVID-19 infection: the perspectives on immune responses. Nature Publishing Group; 2020. [DOI] [PMC free article] [PubMed]
- 13.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet respiratory medicine. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neagu M., Calina D., Docea A.O., Constantin C., Filippini T., Vinceti M., et al. Back to basics in COVID-19: Antigens and antibodies—Completing the puzzle. J. Cell Mol. Med. 2021 doi: 10.1111/jcmm.16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valizadeh H., Abdolmohammadi-Vahid S., Danshina S., Gencer M.Z., Ammari A., Sadeghi A., et al. Nano-curcumin therapy, a promising method in modulating inflammatory cytokines in COVID-19 patients. Int. Immunopharmacol. 2020;89 doi: 10.1016/j.intimp.2020.107088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.To K.-K.-W., Sridhar S., Chiu K.-H.-Y., Hung D.-L.-L., Li X., Hung I.-F.-N., et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerging Microbes Infect. 2021;10(1):507–535. doi: 10.1080/22221751.2021.1898291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong C., Lam C., Wu A., Ip W., Lee N., Chan I., et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahmasebi S., El-Esawi M.A., Mahmoud Z.H., Timoshin A., Valizadeh H., Roshangar L., et al. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe COVID-19 patients. J. Cell. Physiol. 2021;236(7):5325–5338. doi: 10.1002/jcp.30233. [DOI] [PubMed] [Google Scholar]
- 20.Zhang S., Gan J., Chen B.G., Zheng D., Zhang J.G., Lin R.H., et al. Dynamics of peripheral immune cells and their HLA-G and receptor expressions in a patient suffering from critical COVID-19 pneumonia to convalescence. Clin. Transl. Immunol. 2020;9(5) doi: 10.1002/cti2.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadeghi A., Tahmasebi S., Mahmood A., Kuznetsova M., Valizadeh H., Taghizadieh A., et al. Th17 and Treg cells function in SARS-CoV2 patients compared with healthy controls. J. Cell. Physiol. 2021;236(4):2829–2839. doi: 10.1002/jcp.30047. [DOI] [PubMed] [Google Scholar]
- 22.Stahl K., Schmidt B.M., Hoeper M.M., Skripuletz T., Möhn N., Beutel G., et al. Extracorporeal cytokine removal in severe CAR-T cell associated cytokine release syndrome. J. Crit. Care. 2020;57:124–129. doi: 10.1016/j.jcrc.2020.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Ye Q., Wang B., Mao J. Cytokine storm in COVID-19 and treatment. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smits SL, de Lang A, van den Brand JM, Leijten LM, van IJcken WF, Eijkemans MJ et al. Exacerbated innate host response to SARS-CoV in aged non-human primates. PLoS pathogens. 2010;6(2). [DOI] [PMC free article] [PubMed]
- 25.Wang C, Cai J, Chen R, Shi Z, Bian X, Xie J et al. Aveolar Macrophage Activation and Cytokine Storm in the Pathogenesis of Severe COVID-19. 2020. [DOI] [PMC free article] [PubMed]
- 26.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., et al. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. 2020 [Google Scholar]
- 27.Pribul P.K., Harker J., Wang B., Wang H., Tregoning J.S., Schwarze J., et al. Alveolar macrophages are a major determinant of early responses to viral lung infection but do not influence subsequent disease development. J. Virol. 2008;82(9):4441–4448. doi: 10.1128/JVI.02541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salomé B, Magen A. Dysregulation of lung myeloid cells in COVID-19. Nature Publishing Group; 2020. [DOI] [PMC free article] [PubMed]
- 29.Guo C., Li B., Ma H., Wang X., Cai P., Yu Q., et al. Tocilizumab treatment in severe COVID-19 patients attenuates the inflammatory storm incited by monocyte centric immune interactions revealed by single-cell analysis. BioRxiv. 2020 [Google Scholar]
- 30.Mehta R.S., Smith R.E. Hemophagocytic lymphohistiocytosis (HLH): a review of literature. Med. Oncol. 2013;30(4):740. doi: 10.1007/s12032-013-0740-3. [DOI] [PubMed] [Google Scholar]
- 31.Henter J.I., Horne A., Aricó M., Egeler R.M., Filipovich A.H., Imashuku S., et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer. 2007;48(2):124–131. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 32.Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. The American journal of pathology. 2007;170(4):1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chong P.Y., Chui P., Ling A.E., Franks T.J., Tai D.Y., Leo Y.S., et al. Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis. Arch. Pathol. Lab. Med. 2004;128(2):195–204. doi: 10.5858/2004-128-195-AODDTS. [DOI] [PubMed] [Google Scholar]
- 34.Lang Z.W., Zhang L.J., Zhang S.J., Meng X., Li J.Q., Song C.Z., et al. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS) Pathology. 2003;35(6):526–531. doi: 10.1080/00313020310001619118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholls J.M., Poon L.L., Lee K.C., Ng W.F., Lai S.T., Leung C.Y., et al. Lung pathology of fatal severe acute respiratory syndrome. The Lancet. 2003;361(9371):1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prilutskiy A., Kritselis M., Shevtsov A., Yambayev I., Vadlamudi C., Zhao Q., et al. SARS-CoV-2 Infection Associated Hemophagocytic Lymphohistiocytosis: An autopsy series with clinical and laboratory correlation. medRxiv. 2020 doi: 10.1093/ajcp/aqaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGonagle D., O'Donnell J.S., Sharif K., Emery P., Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. The Lancet. Rheumatology. 2020 doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos-Casals M., Brito-Zerón P., López-Guillermo A., Khamashta M.A., Bosch X. Adult haemophagocytic syndrome. The Lancet. 2014;383(9927):1503–1516. doi: 10.1016/S0140-6736(13)61048-X. [DOI] [PubMed] [Google Scholar]
- 40.Seguin A., Galicier L., Boutboul D., Lemiale V., Azoulay E. Pulmonary involvement in patients with hemophagocytic lymphohistiocytosis. Chest. 2016;149(5):1294–1301. doi: 10.1016/j.chest.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L., et al. Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP) MedRxiv. 2020 [Google Scholar]
- 42.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan. Clinical Infectious Diseases: China. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergsten E., Horne A., Aricó M., Astigarraga I., Egeler R.M., Filipovich A.H., et al. Confirmed efficacy of etoposide and dexamethasone in HLH treatment: long-term results of the cooperative HLH-2004 study. Blood, The Journal of the American Society of Hematology. 2017;130(25):2728–2738. doi: 10.1182/blood-2017-06-788349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mello P., Gusmao-Flores D., Dellinger R.P. Sepsis. Surgical Intensive Care Medicine. Springer. 2016:373–387. [Google Scholar]
- 46.Zhou F., Wang Y., Liu Y., Liu X., Gu L., Zhang X., et al. Disease severity and clinical outcomes of community-acquired pneumonia caused by non-influenza respiratory viruses in adults: a multicentre prospective registry study from the CAP-China Network. Eur. Respir. J. 2019;54(2):1802406. doi: 10.1183/13993003.02406-2018. [DOI] [PubMed] [Google Scholar]
- 47.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Shi L., Wang F.-S. Liver injury in COVID-19: management and challenges. The lancet Gastroenterology & hepatology. 2020;5(5):428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020 doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Liu L., Zhang D., Xu J., Dai H., Tang N., et al. SARS-CoV-2 and viral sepsis: observations and hypotheses. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30920-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulert G.S., Zhang M., Fall N., Husami A., Kissell D., Hanosh A., et al. Whole-exome sequencing reveals mutations in genes linked to hemophagocytic lymphohistiocytosis and macrophage activation syndrome in fatal cases of H1N1 influenza. J. Infect. Dis. 2016;213(7):1180–1188. doi: 10.1093/infdis/jiv550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang K., Jordan M.B., Marsh R.A., Johnson J.A., Kissell D., Meller J., et al. Hypomorphic mutations in PRF1, MUNC13-4, and STXBP2 are associated with adult-onset familial HLH. Blood, The Journal of the American Society of Hematology. 2011;118(22):5794–5798. doi: 10.1182/blood-2011-07-370148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machowicz R., Janka G., Wiktor-Jedrzejczak W. Similar but not the same: differential diagnosis of HLH and sepsis. Critical reviews in oncology/hematology. 2017;114:1–12. doi: 10.1016/j.critrevonc.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 54.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan W-j, Ni Z-y, Hu Y, Liang W-h, Ou C-q, He J-x et al. Clinical characteristics of coronavirus disease 2019 in China. New England journal of medicine. 2020;382(18):1708-20. [DOI] [PMC free article] [PubMed]
- 56.Faguer S., Del Bello A., Abravanel F., Nicolau-Travers M.-L., Kamar N. Tocilizumab for hemophagocytic syndrome in a kidney transplant recipient with COVID-19. Ann. Intern. Med. 2020 doi: 10.7326/L20-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raschke R.A., Garcia-Orr R. Hemophagocytic lymphohistiocytosis: a potentially underrecognized association with systemic inflammatory response syndrome, severe sepsis, and septic shock in adults. Chest. 2011;140(4):933–938. doi: 10.1378/chest.11-0619. [DOI] [PubMed] [Google Scholar]
- 58.Liu J., Li S., Liu J., Liang B., Wang X., Wang H., et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;102763 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F., Nie J., Wang H., Zhao Q., Xiong Y., Deng L., et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J. Infect. Dis. 2020;221(11):1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020;130(5) doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94(5) doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., et al. Anti–spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI insight. 2019;4(4) doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. 2020;323(16):1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020 doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mathew D., Giles J.R., Baxter A.E., Greenplate A.R., Wu J.E., Alanio C., et al. Deep immune profiling of COVID-19 patients reveals patient heterogeneity and distinct immunotypes with implications for therapeutic interventions. bioRxiv. 2020 doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discovery. 2020;6(1):1–18. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ong E.Z., Chan Y.F.Z., Leong W.Y., Lee N.M.Y., Kalimuddin S., Mohideen S.M.H., et al. A dynamic immune response shapes COVID-19 progression. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv. 2020.
- 69.Okba N.M., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Abdely H.M., Midgley C.M., Alkhamis A.M., Abedi G.R., Lu X., Binder A.M., et al. Middle East respiratory syndrome coronavirus infection dynamics and antibody responses among clinically diverse patients, Saudi Arabia. Emerg. Infect. Dis. 2019;25(4):753. doi: 10.3201/eid2504.181595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee Y-L, Liao C-H, Liu P-Y, Cheng C-Y, Chung M-Y, Liu C-E et al. Dynamics of anti-SARS-Cov-2 IgM and IgG antibodies among COVID-19 patients. The Journal of Infection. 2020. [DOI] [PMC free article] [PubMed]
- 73.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yager E.J. Antibody-dependent enhancement and COVID-19: moving toward acquittal. Clinical Immunology (Orlando, Fla). 2020;217 doi: 10.1016/j.clim.2020.108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang B., Zhou X., Zhu C., Song Y., Feng F., Qiu Y., et al. Immune phenotyping based on the neutrophil-to-lymphocyte ratio and IgG level predicts disease severity and outcome for patients with COVID-19. Frontiers in molecular biosciences. 2020;7:157. doi: 10.3389/fmolb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu W.-D., Chang S.-Y., Wang J.-T., Tsai M.-J., Hung C.-C., Hsu C.-L., et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. The Journal of infection. 2020 doi: 10.1016/j.jinf.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peiris J.S.M., Chu C.-M., Cheng V.C.-C., Chan K., Hung I., Poon L.L., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. The Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;1–4 doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 79.Bermingham W.H., Wilding T., Beck S., Huissoon A. SARS-CoV-2 serology: Test, test, test, but interpret with caution! Clinical Medicine. 2020 doi: 10.7861/clinmed.2020-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Corthesy B. Role of secretory IgA in infection and maintenance of homeostasis. Autoimmun. Rev. 2013;12(6):661–665. doi: 10.1016/j.autrev.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 81.Shen L., Fanger M.W. Secretory IgA antibodies synergize with IgG in promoting ADCC by human polymorphonuclear cells, monocytes, and lymphocytes. Cell. Immunol. 1981;59(1):75–81. doi: 10.1016/0008-8749(81)90435-4. [DOI] [PubMed] [Google Scholar]
- 82.Arakawa S., Suzukawa M., Watanabe K., Kobayashi K., Matsui H., Nagai H., et al. Secretory immunoglobulin A induces human lung fibroblasts to produce inflammatory cytokines and undergo activation. Clin. Exp. Immunol. 2019;195(3):287–301. doi: 10.1111/cei.13253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu H-q, Sun B-q, Fang Z-f, Zhao J-c, Liu X-y, Li Y-m et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. European Respiratory Journal. 2020;56(2). [DOI] [PMC free article] [PubMed]
- 84.Infantino M., Damiani A., Gobbi F.L., Grossi V., Lari B., Macchia D., et al. Serological assays for SARS-CoV-2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J. 2020;22(4):203–210. [PubMed] [Google Scholar]
- 85.Tahmasebi S., Khosh E., Esmaeilzadeh A. The outlook for diagnostic purposes of the 2019-novel coronavirus disease. J. Cell. Physiol. 2020 doi: 10.1002/jcp.29804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Organization W.H. Vol. 19. World Health Organization; March 2020: 2020. (Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: interim guidance). [Google Scholar]
- 87.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? The Lancet. 2020 doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forouzesh M., Rahimi A., Valizadeh R., Dadashzadeh N., Mirzazadeh A. Clinical display, diagnostics and genetic implication of Novel Coronavirus (COVID-19) Epidemic. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4607–4615. doi: 10.26355/eurrev_202004_21047. [DOI] [PubMed] [Google Scholar]
- 90.Falzone L., Gattuso G., Tsatsakis A., Spandidos D.A., Libra M. Current and innovative methods for the diagnosis of COVID-19 infection. Int. J. Mol. Med. 2021;47(6):1–23. doi: 10.3892/ijmm.2021.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;1–4 doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.GeurtsvanKessel C.H., Okba N.M., Igloi Z., Embregts C.W., Laksono B.M., Leijten L., et al. Towards the next phase: evaluation of serological assays for diagnostics and exposure assessment. medRxiv. 2020 doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Y., Huang F., Xu J., Yang P., Qin Y., Cao M., et al. Anti-hypertensive Angiotensin II receptor blockers associated to mitigation of disease severity in elderly COVID-19 patients. medRxiv. 2020 [Google Scholar]
- 95.Prabakaran P., Zhu Z., Xiao X., Biragyn A., Dimitrov A.S., Broder C.C., et al. Potent human monoclonal antibodies against SARS CoV, Nipah and Hendra viruses. Expert Opin. Biol. Ther. 2009;9(3):355–368. doi: 10.1517/14712590902763755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stephenson I., Das R.G., Wood J.M., Katz J.M. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine. 2007;25(20):4056–4063. doi: 10.1016/j.vaccine.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 97.Spruth M., Kistner O., Savidis-Dacho H., Hitter E., Crowe B., Gerencer M., et al. A double-inactivated whole virus candidate SARS coronavirus vaccine stimulates neutralising and protective antibody responses. Vaccine. 2006;24(5):652–661. doi: 10.1016/j.vaccine.2005.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Handisurya A., Schellenbacher C., Haitel A., Senger T., Kirnbauer R. Human papillomavirus vaccination induces neutralising antibodies in oral mucosal fluids. Br. J. Cancer. 2016;114(4):409–416. doi: 10.1038/bjc.2015.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weldon W.C., Oberste M.S., Pallansch M.A. Standardized methods for detection of poliovirus antibodies. Poliovirus. Springer. 2016:145–176. doi: 10.1007/978-1-4939-3292-4_8. [DOI] [PubMed] [Google Scholar]
- 100.Ghaebi M., Osali A., Valizadeh H., Roshangar L., Ahmadi M. Vaccine development and therapeutic design for 2019-nCoV/SARS-CoV-2: Challenges and chances. J. Cell. Physiol. 2020 doi: 10.1002/jcp.29771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020 doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Poh C.M., Carissimo G., Wang B., Amrun S.N., Lee C.Y.-P., Chee R.S.-L., et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nature. Communications. 2020;11(1):1–7. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haveri A., Smura T., Kuivanen S., Österlund P., Hepojoki J., Ikonen N., et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Eurosurveillance. 2020;25(11):2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dauletova M., Hafsan H., Mahhengam N., Zekiy A.O., Ahmadi M., Siahmansouri H. Mesenchymal stem cell alongside exosomes as a novel cell-based therapy for COVID-19: A review study. Clinical Immunology. 2021;108712 doi: 10.1016/j.clim.2021.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 106.Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I., et al. preventive and control measures and recent developments to counter this pandemic virus. 2020. SARS-CoV-2: Jumping the species barrier, lessons from SARS and MERS, its zoonotic spillover, transmission to humans. [Google Scholar]
- 107.Mupapa K., Massamba M., Kibadi K., Kuvula K., Bwaka A., Kipasa M., et al. Treatment of Ebola hemorrhagic fever with blood transfusions from convalescent patients. J. Infect. Dis. 1999;179(Supplement_1):S18–S23. doi: 10.1086/514298. [DOI] [PubMed] [Google Scholar]
- 108.Yeh K.-M., Chiueh T.-S., Siu L., Lin J.-C., Chan P.K., Peng M.-Y., et al. Experience of using convalescent plasma for severe acute respiratory syndrome among healthcare workers in a Taiwan hospital. J. Antimicrob. Chemother. 2005;56(5):919–922. doi: 10.1093/jac/dki346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chan K-H, Chan JF-W, Tse H, Chen H, Lau CC-Y, Cai J-P et al. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. Journal of Infection. 2013;67(2):130-40. [DOI] [PMC free article] [PubMed]
- 110.Mair-Jenkins J., Saavedra-Campos M., Baillie J.K., Cleary P., Khaw F.-M., Lim W.S., et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J. Infect. Dis. 2015;211(1):80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arabi Y., Balkhy H., Hajeer A.H., Bouchama A., Hayden F.G., Al-Omari A., et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. Springerplus. 2015;4(1):1–8. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xiao Y., Gao X. Use of IgY antibodies and semiconductor nanocrystal detection in cancer biomarker quantitation. Biomarkers Med. 2010;4(2):227–239. doi: 10.2217/bmm.10.7. [DOI] [PubMed] [Google Scholar]
- 113.Constantin C., Neagu M., Supeanu T.D., Chiurciu V., Spandidos D.A. IgY-turning the page toward passive immunization in COVID-19 infection. Experimental and Therapeutic Medicine. 2020;20(1):151–158. doi: 10.3892/etm.2020.8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Somasundaram R., Choraria A., Antonysamy M. An approach towards development of monoclonal IgY antibodies against SARS CoV-2 spike protein (S) using phage display method: A review. Int. Immunopharmacol. 2020;106654 doi: 10.1016/j.intimp.2020.106654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sui J., Li W., Roberts A., Matthews L.J., Murakami A., Vogel L., et al. Evaluation of human monoclonal antibody 80R for immunoprophylaxis of severe acute respiratory syndrome by an animal study, epitope mapping, and analysis of spike variants. J. Virol. 2005;79(10):5900–5906. doi: 10.1128/JVI.79.10.5900-5906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Both L., Banyard A.C., van Dolleweerd C., Wright E., Ma J.K.-C., Fooks A.R. Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine. 2013;31(12):1553–1559. doi: 10.1016/j.vaccine.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang C., Li W., Drabek D., Okba N.M., van Haperen R., Osterhaus A.D., et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11(1):1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.