Boosterism could save lives
Postinfection immune protection against severe acute respiratory syndrome coronavirus 2 reinfection is not fully understood. It will be devastating if waves of new variants emerge that undermine natural immune protection. Stamatatos et al. investigated immune responsiveness 4 to 8 months after previously infected individuals were given a messenger RNA–based vaccine developed for the original Wuhan variant (see the Perspective by Crotty). Before vaccination, postinfection serum antibody neutralization responses to virus variants were variable and weak. Vaccination elevated postinfection serum-neutralizing capacity approximately 1000-fold against Wuhan-Hu-1 and other strains, and serum neutralization against the variant B.1.351 was enhanced. Although responses were relatively muted against the variant, they still showed characteristic memory responses. Vaccination with the Wuhan-Hu-1 variant may thus offer a valuable boost to protective responses against subsequent infection with variant viruses.
Science, abg9175, this issue p. 1413; see also abj2258, p. 1392
Previous infection results in enhanced vaccine-induced immune protection against variant B.1.351 and other variants.
Abstract
Emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants have raised concerns about resistance to neutralizing antibodies elicited by previous infection or vaccination. We examined whether sera from recovered and naïve donors, collected before and after immunizations with existing messenger RNA (mRNA) vaccines, could neutralize the Wuhan-Hu-1 and B.1.351 variants. Prevaccination sera from recovered donors neutralized Wuhan-Hu-1 and sporadically neutralized B.1.351, but a single immunization boosted neutralizing titers against all variants and SARS-CoV-1 by up to 1000-fold. Neutralization was a result of antibodies targeting the receptor binding domain and was not boosted by a second immunization. Immunization of naïve donors also elicited cross-neutralizing responses but at lower titers. Our study highlights the importance of vaccinating both uninfected and previously infected persons to elicit cross-variant neutralizing antibodies.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) betacoronavirus first emerged in the Hubei Province of China in late 2019 and has since infected more than 115 million people and caused more than 2.5 million deaths in 192 countries (1–3). Infection is mediated by the viral spike protein (S), which is composed of an S1 domain that contains an N-terminal domain (NTD), a C-terminal domain (CTD), and a receptor binding domain (RBD) that mediates attachment to the entry receptor angiotensin-converting enzyme 2 (ACE2) as well as an S2 domain that contains the fusion machinery (4–8).
Preexisting immunity to SARS-CoV-2 is associated with protection against reinfection in humans (9–11) and in nonhuman primates (12, 13). Although the correlates of protection in humans against repeat infection or after vaccination have not been firmly established, neutralizing antibodies (nAbs) are thought to be an important component of a protective immune response against SARS-CoV-2 (14, 15). In support of this, passive transfer of nAbs limits respiratory tract infection and protects against infection in animal models (16–20), and nAbs may contribute to protection against infection in humans (9). SARS-CoV-2 infection rapidly elicits nAbs (16, 21–24) that decline, but remain detectable, over several months (25–29).
Most serum nAb responses elicited during natural infection are directed at the RBD (21, 23, 30, 31). Numerous neutralizing anti-RBD monoclonal antibodies (mAbs) have been characterized, the most potent of which block the RBD-ACE2 interaction (16, 17, 22–24, 32–37). Neutralizing mAbs that bind regions of the viral spike have also been identified (24, 33, 38–42).
Two mRNA-based vaccines (Pfizer-BioNTech BNT162b2 and Moderna mRNA-1273) have received emergency use authorization in several countries. Both vaccines encode a stabilized ectodomain version of the S protein derived from the Wuhan-Hu-1 variant isolated in December 2019 (43), show >94% efficacy at preventing COVID-19 illness (44–47), and elicit nAbs (48, 49).
Because of the high global burden of SARS-CoV-2 transmission, viral evolution is occurring. Recently, viral variants of concern have emerged in the UK (B.1.1.7), South Africa (B.1.351), and Brazil (P.1) that harbor specific mutations in their S proteins that may be associated with increased transmissibility (50–55).
Of particular concern are mutations found in the B.1.351 lineage, which is defined by the D80A (amino acid substitution from aspartic acid to alanine at position 80) and D215G mutations in the NTD; the K417N, E484K, and N501Y mutations in the RBD; and the D614G mutation in S1 (52, 56). An A701V mutation in S2 is also observed at high frequencies, whereas deletions in residues 242 to 244 as well as R246I and L18F mutations in the NTD are present at lower frequencies (52). (Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.)
The B.1.1.7, B.1.351, and P.1 lineages all harbor a N501Y mutation in the RBD, which increases the affinity for the ACE2 receptor (57, 58), and a D614G mutation, which increases virion spike density, infectivity, and transmissibility (59, 60). The B.1.351 and P.1 lineages also share the E484K mutation in the RBD, and both variants are mutated at position 417 (K417T in P.1).
Mutations found in emergent S variants decrease sensitivity to neutralization by mAbs, convalescent plasma, and sera from vaccinated individuals (27, 37, 58, 61–70). As a result, there is concern that these and other emerging variants can evade nAb responses generated during infection with variants that were circulating earlier in the pandemic and also nAb responses elicited by vaccines based on the S protein of the Wuhan-Hu-1 variant. There is concern that these mutations are responsible for the reduced efficacy observed in ongoing trials of SARS-CoV-2 vaccines in South Africa (71, 72).
Here, we evaluated the neutralization susceptibility of spike variants harboring lineage-defining and prevalent B.1.351 mutations to sera from two groups. Sera were collected from 15 donors with previously confirmed SARS-CoV-2 infection [referred to as previously infected donors (PIDs)] before and after one or two immunizations with either mRNA vaccine and from 13 uninfected donors who received two doses of the above vaccines [referred to as naïve donors (NDs); tables S1 and S2].
Antibody neutralization experiments were performed with pseudoviruses expressing either the full-length Wuhan-Hu-1 S or either of two versions of the B.1.351 lineage S—one herein referred to as B.1.351, containing the lineage-defining S mutations D80A, D215G, K417N, E484K, N501Y, and D614G and the A701V mutation that is highly prevalent in this lineage, and a second variant that also includes a Δ242-243 deletion (B.1.351–Δ242-243). The viral stocks were appropriately diluted to achieve comparable entry levels during the neutralization experiments (fig. S1).
We first evaluated the neutralizing potency of several mAbs isolated from nonvaccinated patients infected early in the pandemic. These mAbs target different epitopes: three against the RBD (CV30, CV3-1, and CV2-75) and one against the NTD (CV1) (fig. S2). CV30 is a member of the VH3-53 class of antibodies that bind to the receptor binding motif (RBM) (22, 32, 73–78). It makes direct contact with the K417 and N501 residues in the RBM that are mutated in the B.1.351 and P.1 lineages; however, unlike other known VH3-53 mAbs, it does not contact E484 (78). The neutralization potency of this mAb was ~10-fold weaker toward both B.1.351 variants (Fig. 1A). Similarly, the non–VH3-53 mAb CV3-1 was three- to fourfold less potent against the B.1.351 variants (Fig. 1B), whereas CV2-75 was modestly less effective (Fig. 1C). By contrast, the anti-NTD CV1 mAb was unable to neutralize either B.1.351 variant (Fig. 1D). As expected, the control anti–Epstein-Barr virus mAb AMMO1 was nonneutralizing (79) (Fig. 1E). Collectively, these data indicate that the B.1.351 variants tested here are more resistant to neutralization by mAbs isolated from subjects infected by viral variants from early in the pandemic. We therefore examined whether the B.1.351 variants are resistant to nAb responses elicited by the Pfizer-BioNTech or Moderna mRNA vaccines in both PIDs and NDs.
Fig. 1. B.1.351 variants show decreased susceptibility to neutralizing mAbs.
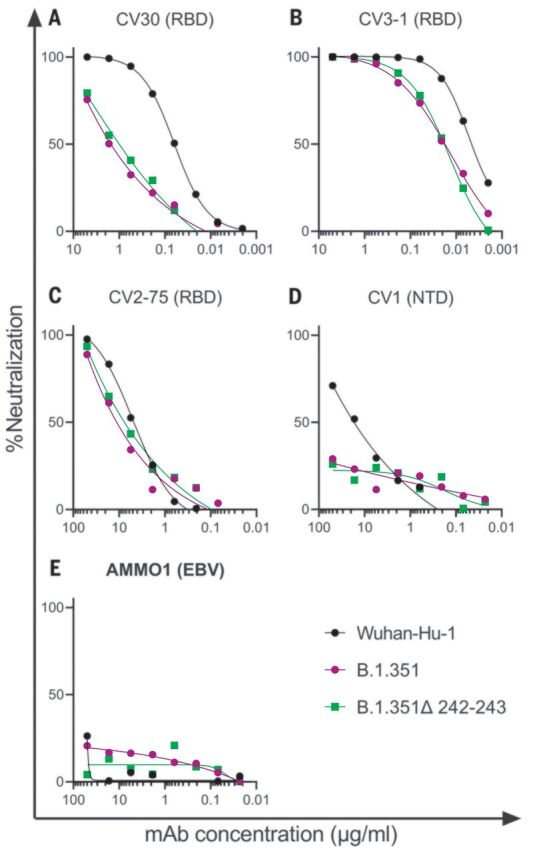
(A to E) The ability of the indicated mAbs to neutralize Wuhan-Hu-1, B.1.351, and B.1.351–Δ242-243 pseudovirus infectivity in 293T-hACE2 cells was measured as indicated. The epitope specificity of each mAb is shown in parentheses. EBV, Epstein-Barr virus. Data points represent the mean of two technical replicates. Data are representative of two independent experiments.
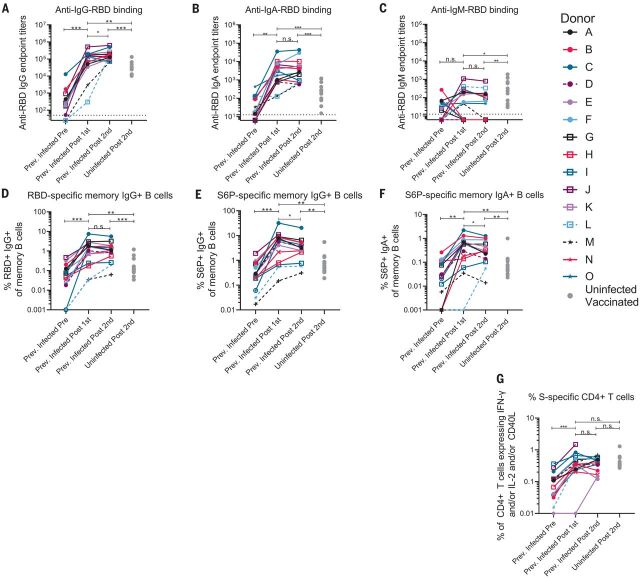
The RBD-specific immunoglobulin G (IgG), IgM, and IgA binding responses to the RBD from the Wuhan-Hu-1 variant were measured before (on average, 202 days after symptom onset; table S1) and either 5 to 29 days (table S1) after the first and second immunizations in the PIDs or 6 to 28 days after the second immunization in the NDs. Three PIDs experienced asymptomatic SARS-CoV-2 infection (donors D, L, and M; table S1), two of whom, L and M, did not have detectable anti-RBD IgG antibodies before immunization, whereas the third, D, had low but detectable serum anti-RBD IgG antibody titers (Fig. 2A). In the 13 PIDs with RBD-specific IgG antibodies before vaccination, a single dose of either vaccine boosted these titers ~500-fold (Fig. 2A). Across all PIDs, there was a 200-fold increase in median RBD-specific IgA titers after vaccination (Fig. 2B). Overall, in PIDs, a single vaccine dose elicited 4.5-fold higher IgG and 7.7-fold higher IgA titers compared with two vaccinations in NDs. RBD-specific IgM titers were generally lower and were not significantly boosted in response to vaccination in PIDs (Fig. 2C). In PIDs, a concomitant increase in RBD- (Fig. 2D) and S-specific IgG+ (Fig. 2E) memory B cell frequencies took place after vaccination. The two PIDs that lacked RBD-specific IgG titers before immunization (donors L and M) also lacked RBD-specific IgG+ memory B cells (Fig. 2D) and had lower frequencies of S-specific IgG+ memory B cells after vaccination. Consistent with the serology data, an increase in the frequency of IgA+ (Fig. 2F) but not IgM+ spike-specific memory B cells was observed (fig. S3). Vaccination also induced S-specific CD4+ T cell responses (Fig. 2G).
Fig. 2. A single dose of a spike-derived mRNA vaccine elicits a strong recall response.
(A to C) IgG (A), IgA (B), and IgM (C) end-point antibody titers specific to the RBD of the Wuhan-Hu-1 variant were measured in serum collected from PIDs before and after one or two immunizations with the Pfizer-BioNTech or Moderna mRNA vaccines by ELISA, as indicated. End-point titers measured in sera from NDs after two vaccine doses are shown for comparison (gray dots). (D) Frequency of Wuhan-Hu-1 RBD-specific IgG+ memory B cells (live, IgD−, CD19+, CD20+, CD3−, CD14, CD56−, singlet, and lymphocytes) in peripheral blood mononuclear cells (PBMCs) from PIDs was measured before and after one or two immunizations. (E and F) The frequency of S6P-specific IgG+ (E) and IgA+ (F) memory B cells in PBMCs from PIDs was measured before and after one or two immunizations. The frequencies of memory B cells from NDs after two vaccine doses are shown for comparison in (D) to (F) (gray dots). (G) The frequency of S-specific CD4+ T cells expressing interferon-γ (IFN-γ) and/or interleukin-2 (IL-2) and/or CD40L in PBMCs from PIDs was measured before and after one or two immunizations. The frequencies of S-specific CD4+ T cells in PBMCs from uninfected donors after two vaccine doses are shown for comparison (gray dots). Experiments were performed once. Significant differences in infected donors before or after vaccination [(A) to (G)] were determined using a Wilcoxon signed rank test (n.s., not significant; *P < 0.05; **P < 0.01; and ***P < 0.001). Significant differences between previously infected and uninfected donors [(A) to (G)] were determined using a Wilcoxon rank sum test (*P < 0.05; **P < 0.01; and ***P < 0.001).
Sera from 12 of 15 PIDs sampled before vaccination neutralized the Wuhan-Hu-1 SARS-CoV-2 variant (Fig. 3A and fig. S4). The nonneutralizing sera were from the three asymptomatic PIDs who had low or undetectable anti-RBD IgG titers (Fig. 3A, dashed lines, and fig. S4). Prevaccine sera from the NDs were also nonneutralizing (fig. S5). Consistent with the observed increase in binding antibodies after a single immunization in PIDs with preexisting RBD-specific IgG titers, the median half-maximal neutralizing titers [half-maximal inhibitory dilution (ID50)] were boosted ~1000-fold after the first dose, whereas the second dose had no effect (Fig. 3A). In the two PIDs lacking RBD-specific IgG titers before vaccination, the first vaccine dose elicited lower neutralizing titers (ID50 = ~30 in donor L and ~200 in donor M; Fig. 3A). In the NDs, two doses of the vaccine elicited ID50 titers that were ~10- and 5-fold lower than those elicited by one or two doses in the PIDs, respectively (Fig. 3A and fig. S6). Collectively, these data indicate that in PIDs who generate adequate immunological memory to the RBD, a single vaccine dose elicits an anamnestic response resulting in RBD-binding and nAb responses that are superior to a two-dose regimen in uninfected donors. A similar boost in binding and/or vaccine-matched neutralizing titers has been observed in PIDs who received a single mRNA vaccine dose in two recent studies (80, 81).
Fig. 3. Preexisting SARS-CoV-2 nAb responses are boosted by a single dose of a spike-derived mRNA vaccine.
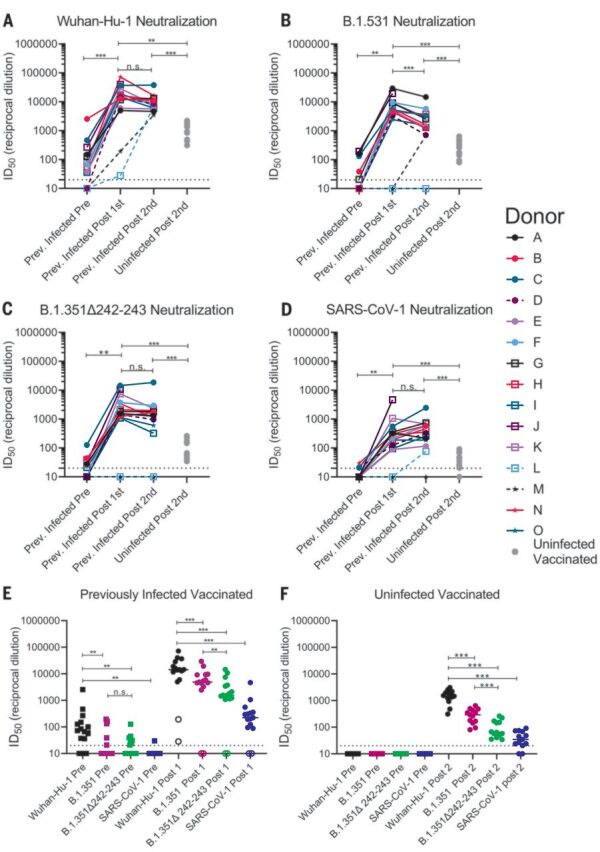
(A to D) The serum dilution resulting in 50% neutralization (ID50) of Wuhan-Hu-1 (A), B.1.351 (B), B.1.351–Δ242-243 (C), and SARS-CoV-1 (D) pseudoviruses was measured in PIDs before and after one or two immunizations with the Pfizer-BioNTech or Moderna vaccines and in NDs after two vaccine doses, as indicated. Data points between PIDs who were symptomatic and asymptomatic are connected by solid and dashed lines, respectively, in (A) to (D). (E) Serum dilution resulting in 50% neutralization (ID50) from PIDs before (squares) and after (circles) a single immunization with the Pfizer-BioNTech or Moderna vaccines against Wuhan-Hu-1, B.1.351, B.1.351–Δ242-243, and SARS-CoV-1 pseudoviruses, as indicated. PIDs who were asymptomatic and negative for anti-IgG RBD antibodies and RBD-specific IgG+ memory B cells before vaccination are shown as open circles. (F) Neutralizing potency (ID50) of serum from NDs after two immunizations with the Pfizer-BioNTech or Moderna vaccines against the indicated pseudoviruses. Each data point represents a different donor, and the horizonal bars represent the medians in (E) and (F). The dashed lines demarcate the lowest serum dilutions tested. Experiments were performed once. Significant differences in infected donors before or after vaccination, or from the same time point against different variants, were determined using a Wilcoxon signed rank test (*P < 0.05; **P < 0.01; and ***P < 0.001). Significant differences between previously infected and uninfected donors were determined using a Wilcoxon rank sum test (*P < 0.05; **P < 0.01; and ***P < 0.001).
We next evaluated the ability of sera collected before and after immunization in NDs and PIDs to neutralize the more resistant B.1.351 and B.1.351–Δ242-243 pseudoviruses. These variants are 0.5 and 0.7% divergent from the Wuhan-Hu-1 variant. We also included SARS-CoV-1 pseudoviruses in this analysis as a representative variant that is even more dissimilar to the vaccine. SARS-CoV-1 and SARS-CoV-2 are 24, 26, and 50% divergent in the overall S protein, RBD, and RBM, respectively (82). Consequently, several mAbs that potently neutralize SARS-CoV-2 fail to bind SARS-CoV-1 (16, 22–24).
Before vaccination, 5 of 15 sera from PIDs neutralized B.1.351, and only three had ID50 titers above 100 (Fig. 3, B and E, and fig. S4); 7 of 15 neutralized B.1.351–Δ242-243, and only one had titers above 100 (Fig. 3, C and E, and fig. S4). Only two prevaccine PID sera achieved 80% neutralization of B.1.351, and only one achieved 80% neutralization of B.1.351–Δ242-243 (fig. S7A). The median ID50 of the prevaccine sera against the Wuhan-Hu-1 variant was significantly higher than that against B.1.351 or B.1.351–Δ242-243 (Fig. 3E). Consistent with the high level of sequence disparity, sera from only one PID showed very weak neutralizing activity toward SARS-CoV-1 before vaccination (Fig. 3, D and E, and fig. S7).
A single immunization boosted the nAb titers against all three SARS-CoV-2 variants and SARS-CoV-1 in 13 of 15 PIDs (Fig. 3, A to D); however, the median ID50 titers were ~3-fold lower against B.1.351, ~10-fold lower against B.1.351–Δ242-243, and 100-fold lower against SARS-CoV-1 than against Wuhan-Hu-1 (Fig. 3E). A single immunization did not elicit nAbs against the B.1.351 variants or SARS-CoV-1 in the two asymptomatic donors who lacked RBD-specific IgG memory (donor L and M; Fig. 3, A to D, and Fig. 3E, open circles). The median ID80 values were also lower for the B.1.351 and B.1.351–Δ242-243 variants compared with the Wuhan-Hu-1 variant (fig. S7A).
The neutralizing titers elicited by a single immunization in PIDs were significantly higher than those elicited by two immunizations in NDs against all pseudoviruses tested—10-fold higher against Wuhan-Hu-1 (Fig. 3A), 20-fold higher against B.1.351 (Fig. 3B), 30-fold higher against B.1.351–Δ242-243 (Fig. 3C), and 7-fold higher against SARS-CoV-1 (Fig. 3D). Only 8 of 13 vaccinated NDs were able to achieve 80% neutralization of B.1.351–Δ242-243, and none could achieve 80% neutralization of SARS-CoV-1 (fig. S7B).
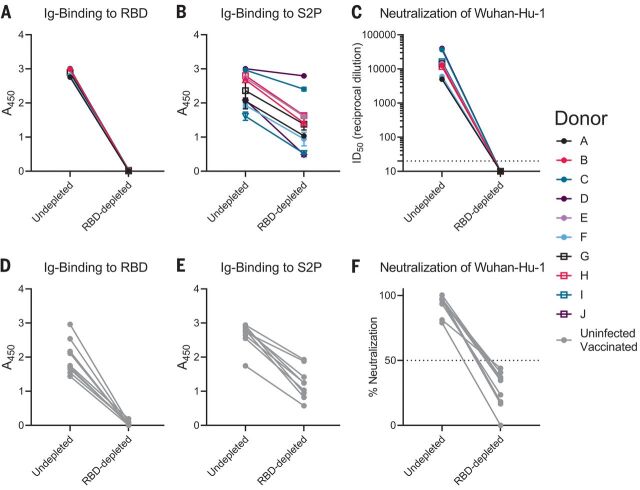
The B.1.351 and B.1.351–Δ242-243 variants contain three RBD mutations that affect the neutralization potency of anti-RBD mAbs (Fig. 1). Moreover, preexisting anti-RBD IgG memory appears to be important for a robust recall response to vaccination. To determine the relative contribution of anti-RBD antibodies to serum neutralization, we depleted RBD-specific antibodies from the sera of 10 PIDs after one vaccination and from nine NDs after two vaccinations. This approach efficiently removed RBD-specific (Fig. 4, A and C) but not anti-S2P–specific antibodies from sera, as measured by enzyme-linked immunosorbent assay (ELISA) (Fig. 4, B and D). This depletion abrogated serum neutralization of Wuhan-Hu-1 virus (Fig. 4, C and F), which suggests that most nAbs elicited or boosted by vaccination target this subdomain.
Fig. 4. Vaccine-elicited nAbs target the RBD.
RBD-binding antibodies were adsorbed from sera from PIDs after receiving a single vaccine dose or from NDs after receiving two vaccine doses using Wuhan-Hu-1 RBD immobilized to magnetic beads. (A and B) Antibody binding in undepleted or RBD-depleted sera from PIDs was measured to RBD at a 1:500 dilution (A) and S2P at a 1:4500 dilution (B) by ELISA, as indicated. A450, absorbance at 450 nM. (C) The serum dilution resulting in 50% neutralization (ID50) of the Wuhan-Hu-1 pseudovirus was measured in undepleted or RBD-depleted sera from the PIDs in (A) and (B). (D and E) Antibody binding in undepleted and RBD-depleted sera from NDs was measured to RBD at a 1:500 dilution (D) and S2P at a 1:500 dilution (E) by ELISA. (F) The percent neutralization of a 1:120 dilution of undepleted or RBD-depleted sera from the donors in (D) and (E) was measured against the Wuhan-Hu-1 pseudovirus. Experiments were performed once.
The above results indicate that in NDs, two doses of either the Pfizer-BioNTech or Moderna vaccines elicited nAb titers against the vaccine-matched Wuhan-Hu-1, lower titers against B.1.351, and even lower titers against B.1.351–Δ242-243. Reduced sensitivity to vaccine-elicited nAbs has been reported for other B.1.351 variants (66, 83, 84).
Similarly, sera from PIDs who experienced symptomatic SARS-CoV-2 infection and who had detectable anti-RBD IgG titers before vaccination displayed generally weak nAb titers against Wuhan-Hu-1 at 1 to 9 months after infection and lower or nonexistent titers against the B.1.351 variants, in agreement with another study (69). However, as long as RBD-specific IgG+ memory B cell and antibody responses were generated during infection, a single immunization with either mRNA vaccine elicited a robust recall response that boosted the autologous neutralizing titers by ~1000-fold, and these antibody responses cross-neutralized the B.1.351 variants, but at lower titers. In most of the previously infected vaccinees, the anti–B.1.351–Δ242-243 neutralizing titers were comparable to those against the vaccine-matched Wuhan-Hu-1 in uninfected vaccinees. This is notable, as these titers were associated with 95% protection from COVID-19 in phase 3 trials (44, 46, 48, 49). Moreover, vaccine-elicited antibody responses also neutralized SARS-CoV-1 but with much lower potencies. Collectively, our data suggest that the two mRNA vaccines that are based on the Wuhan-Hu-1 variant can elicit and/or boost nAb responses but that their potency is reduced against divergent variants.
Here, we show that the cross-nAb responses generated after immunization in previously infected subjects are a result of anti-RBD antibodies. Combined with the observation that the vaccines elicited nAb responses that are less potent against the B.1.351 variant with the Δ242-243 deletion in the NTD, this suggests that NTD mutations can modulate the sensitivity of emerging variants to anti-RBD nAbs. By contrast, the NTD region itself, which appears to tolerate antigenic variation in SARS-CoV-2 and other coronaviruses (50, 52, 55, 85), does not appear to be the target of cross-nAbs elicited by infection or vaccination. We note that there are other less-frequent mutations associated with this lineage, such as L18F, Δ244, L244H, and R246I, that were not examined here, which may further increase resistance to vaccine-elicited antibodies. In this study, a pseudovirus assay was used to measure nAbs. Several studies have now shown that authentic virus and pseudovirus neutralization correlate quite well (16, 86, 87). Although the absolute sensitivity of the authentic and pseudovirus assays may differ, we anticipate that the relative differences we report here will not vary between the two.
Although the correlates of protection for SARS-CoV-2 vaccines have not been established, studies in nonhuman primates indicate that even low titers of nAbs are sufficient to prevent experimental SARS-CoV-2 infection, particularly if CD8+ T cell responses are mounted (18). Our study suggests that most previously infected subjects will benefit from a single immunization with either the Pfizer-BioNTech or Moderna vaccines, as it will lead to significant increases in serum nAb responses against vaccine-matched and emerging variants. The observation that a second dose administered 3 to 4 weeks after the first did not further boost neutralizing titers in PIDs who have clear evidence of RBD-directed immunological memory before vaccination suggests that the second dose of an mRNA vaccine could be delayed in some persons who have previously been infected with SARS-CoV-2. Longitudinal monitoring of the nAb titers before and after the first dose should be used to determine the necessity or optimal timing of the second dose in the context of previous infection.
Acknowledgments
We thank the study participants for their dedication to this project, T. Bedford for assistance with the selection of spike mutations to include, L. Richert Spuhler for assistance with figure preparation, and T. Haight and the Seattle Vaccine Unit specimen processing laboratory and staff for their service. This work was conducted under Fred Hutchinson Cancer Research Center Institutional Review Boards IR10440 and IR5567. Funding: This work was supported by generous donations to the Fred Hutch COVID-19 Research Fund; funding to M.J.M. from the Paul G. Allen Family Foundation, the Joel D. Meyers Endowed Chair, and NIAID (UM1 AI068618-14S1, 2UM1 AI069481-15, and UM1A057266-S1); and funding from Sanofi Pasteur to Z.M. Author contributions: Conceptualization: M.J.M., A.T.M., and L.S.; Investigation: Y.-H.W., E.S., M.P.L., V.R., K.W.C., S.C.D.R., Z.M., M.N., L.J.H., A.J.M., M.F.J., J.F., G.M., H.G., and A.T.M.; Writing - Original Draft: A.T.M. and L.S.; Writing - Review & Editing: A.T.M., L.S., M.J.M., and E.S.; Funding Acquisition: L.S. and M.J.M.; Resources: M.J.M., J.C., and A.F.; Supervision: A.T.M. Competing interests: L.S. and A.T.M. have filed a provisional patent application on the CV1, CV30, and CV2-75 SARS-CoV-2–specific mAbs. L.S., A.T.M., and A.F. have filed a provisional patent application on the CV3-1 SARS-CoV-2–specific mAb. All other authors declare no competing interests. Data and materials availability: All data are available in the manuscript or the supplementary materials. The sequences of the CV3-1 and CV2-75 heavy- and light-chain variable regions have been deposited in GenBank under accession numbers: MW681558, MW681586, MW681758, and MW68175. The expression plasmids for the mAbs in this study are derived from the pTT3 vector, which requires a license from the National Research Council Canada (88). This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
science.sciencemag.org/content/372/6549/1413/suppl/DC1
Materials and Methods
Figs. S1 to S8
Tables S1 to S4
MDAR Reproducibility Checklist
References and Notes
- 1.Dong E., Du H., Gardner L., An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020). 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W., China Novel Coronavirus Investigating and Research Team , A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292.e6 (2020). 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280.e8 (2020). 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Letko M., Marzi A., Munster V., Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562–569 (2020). 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620 (2020). 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Addetia A., Crawford K. H. D., Dingens A., Zhu H., Roychoudhury P., Huang M.-L., Jerome K. R., Bloom J. D., Greninger A. L., Neutralizing Antibodies Correlate with Protection from SARS-CoV-2 in Humans during a Fishery Vessel Outbreak with a High Attack Rate. J. Clin. Microbiol. 58, e02107-20 (2020). 10.1128/JCM.02107-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pray I. W., Gibbons-Burgener S. N., Rosenberg A. Z., Cole D., Borenstein S., Bateman A., Pevzner E., Westergaard R. P., COVID-19 Outbreak at an Overnight Summer School Retreat - Wisconsin, July-August 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 1600–1604 (2020). 10.15585/mmwr.mm6943a4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lumley S. F., O’Donnell D., Stoesser N. E., Matthews P. C., Howarth A., Hatch S. B., Marsden B. D., Cox S., James T., Warren F., Peck L. J., Ritter T. G., de Toledo Z., Warren L., Axten D., Cornall R. J., Jones E. Y., Stuart D. I., Screaton G., Ebner D., Hoosdally S., Chand M., Crook D. W., O’Donnell A.-M., Conlon C. P., Pouwels K. B., Walker A. S., Peto T. E. A., Hopkins S., Walker T. M., Jeffery K., Eyre D. W., Oxford University Hospitals Staff Testing Group , Antibody Status and Incidence of SARS-CoV-2 Infection in Health Care Workers. N. Engl. J. Med. 384, 533–540 (2021). 10.1056/NEJMoa2034545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrashekar A., Liu J., Martinot A. J., McMahan K., Mercado N. B., Peter L., Tostanoski L. H., Yu J., Maliga Z., Nekorchuk M., Busman-Sahay K., Terry M., Wrijil L. M., Ducat S., Martinez D. R., Atyeo C., Fischinger S., Burke J. S., Slein M. D., Pessaint L., Van Ry A., Greenhouse J., Taylor T., Blade K., Cook A., Finneyfrock B., Brown R., Teow E., Velasco J., Zahn R., Wegmann F., Abbink P., Bondzie E. A., Dagotto G., Gebre M. S., He X., Jacob-Dolan C., Kordana N., Li Z., Lifton M. A., Mahrokhian S. H., Maxfield L. F., Nityanandam R., Nkolola J. P., Schmidt A. G., Miller A. D., Baric R. S., Alter G., Sorger P. K., Estes J. D., Andersen H., Lewis M. G., Barouch D. H., SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 369, 812–817 (2020). 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng W., Bao L., Liu J., Xiao C., Liu J., Xue J., Lv Q., Qi F., Gao H., Yu P., Xu Y., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Liu Y., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C., Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 369, 818–823 (2020). 10.1126/science.abc5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens D. S., McElrath M. J., COVID-19 and the Path to Immunity. JAMA 324, 1279–1281 (2020). 10.1001/jama.2020.16656 [DOI] [PubMed] [Google Scholar]
- 15.O’Callaghan K. P., Blatz A. M., Offit P. A., Developing a SARS-CoV-2 Vaccine at Warp Speed. JAMA 324, 437–438 (2020). 10.1001/jama.2020.12190 [DOI] [PubMed] [Google Scholar]
- 16.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W. T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). 10.1126/science.abc7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zost S. J., Gilchuk P., Case J. B., Binshtein E., Chen R. E., Nkolola J. P., Schäfer A., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Martinez D. R., Williamson L. E., Chen E. C., Jones T., Day S., Myers L., Hassan A. O., Kafai N. M., Winkler E. S., Fox J. M., Shrihari S., Mueller B. K., Meiler J., Chandrashekar A., Mercado N. B., Steinhardt J. J., Ren K., Loo Y.-M., Kallewaard N. L., McCune B. T., Keeler S. P., Holtzman M. J., Barouch D. H., Gralinski L. E., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., ., Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). 10.1038/s41586-020-2548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahan K., Yu J., Mercado N. B., Loos C., Tostanoski L. H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., Bondzie E. A., Dagotto G., Gebre M. S., Jacob-Dolan C., Li Z., Nampanya F., Patel S., Pessaint L., Van Ry A., Blade K., Yalley-Ogunro J., Cabus M., Brown R., Cook A., Teow E., Andersen H., Lewis M. G., Lauffenburger D. A., Alter G., Barouch D. H., Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). 10.1038/s41586-020-03041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., Atwal G. S., Oyejide A., Goez-Gazi Y., Dutton J., Clemmons E., Staples H. M., Bartley C., Klaffke B., Alfson K., Gazi M., Gonzalez O., Dick E. Jr.., Carrion R. Jr.., Pessaint L., Porto M., Cook A., Brown R., Ali V., Greenhouse J., Taylor T., Andersen H., Lewis M. G., Stahl N., Murphy A. J., Yancopoulos G. D., Kyratsous C. A., REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science 370, 1110–1115 (2020). 10.1126/science.abe2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schäfer A., Muecksch F., Lorenzi J. C. C., Leist S. R., Cipolla M., Bournazos S., Schmidt F., Maison R. M., Gazumyan A., Martinez D. R., Baric R. S., Robbiani D. F., Hatziioannou T., Ravetch J. V., Bieniasz P. D., Bowen R. A., Nussenzweig M. C., Sheahan T. P., Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 218, e20201993 (2021). 10.1084/jem.20201993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suthar M. S., Zimmerman M. G., Kauffman R. C., Mantus G., Linderman S. L., Hudson W. H., Vanderheiden A., Nyhoff L., Davis C. W., Adekunle O., Affer M., Sherman M., Reynolds S., Verkerke H. P., Alter D. N., Guarner J., Bryksin J., Horwath M. C., Arthur C. M., Saakadze N., Smith G. H., Edupuganti S., Scherer E. M., Hellmeister K., Cheng A., Morales J. A., Neish A. S., Stowell S. R., Frank F., Ortlund E., Anderson E. J., Menachery V. D., Rouphael N., Mehta A. K., Stephens D. S., Ahmed R., Roback J. D., Wrammert J., Rapid Generation of Neutralizing Antibody Responses in COVID-19 Patients. Cell Rep. Med. 1, 100040 (2020). 10.1016/j.xcrm.2020.100040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seydoux E., Homad L. J., MacCamy A. J., Parks K. R., Hurlburt N. K., Jennewein M. F., Akins N. R., Stuart A. B., Wan Y.-H., Feng J., Whaley R. E., Singh S., Boeckh M., Cohen K. W., McElrath M. J., Englund J. A., Chu H. Y., Pancera M., McGuire A. T., Stamatatos L., Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity 53, 98–105.e5 (2020). 10.1016/j.immuni.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H.-H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr.., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). 10.1038/s41586-020-2456-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouwer P. J. M., Caniels T. G., van der Straten K., Snitselaar J. L., Aldon Y., Bangaru S., Torres J. L., Okba N. M. A., Claireaux M., Kerster G., Bentlage A. E. H., van Haaren M. M., Guerra D., Burger J. A., Schermer E. E., Verheul K. D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M. J., Bijl T. P. L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N. A., Wiersinga W. J., Vidarsson G., Haagmans B. L., Ward A. B., de Bree G. J., Sanders R. W., van Gils M. J., Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643–650 (2020). 10.1126/science.abc5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dan J. M., Mateus J., Kato Y., Hastie K. M., Yu E. D., Faliti C. E., Grifoni A., Ramirez S. I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S. A., Peters B., Krammer F., Simon V., Saphire E. O., Smith D. M., Weiskopf D., Sette A., Crotty S., Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodda L. B., Netland J., Shehata L., Pruner K. B., Morawski P. A., Thouvenel C. D., Takehara K. K., Eggenberger J., Hemann E. A., Waterman H. R., Fahning M. L., Chen Y., Hale M., Rathe J., Stokes C., Wrenn S., Fiala B., Carter L., Hamerman J. A., King N. P., Gale M. Jr.., Campbell D. J., Rawlings D. J., Pepper M., Functional SARS-CoV-2-Specific Immune Memory Persists after Mild COVID-19. Cell 184, 169–183.e17 (2021). 10.1016/j.cell.2020.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaebler C., Wang Z., Lorenzi J. C. C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T. Y., Cipolla M., Viant C., Barnes C. O., Bram Y., Breton G., Hägglöf T., Mendoza P., Hurley A., Turroja M., Gordon K., Millard K. G., Ramos V., Schmidt F., Weisblum Y., Jha D., Tankelevich M., Martinez-Delgado G., Yee J., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Robbiani D. F., Zhao Z., Gazumyan A., Schwartz R. E., Hatziioannou T., Bjorkman P. J., Mehandru S., Bieniasz P. D., Caskey M., Nussenzweig M. C., Evolution of antibody immunity to SARS-CoV-2. Nature 591, 639–644 (2021). 10.1038/s41586-021-03207-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K. J. A., Hemmings O., O’Byrne A., Kouphou N., Galao R. P., Betancor G., Wilson H. D., Signell A. W., Winstone H., Kerridge C., Huettner I., Jimenez-Guardeño J. M., Lista M. J., Temperton N., Snell L. B., Bisnauthsing K., Moore A., Green A., Martinez L., Stokes B., Honey J., Izquierdo-Barras A., Arbane G., Patel A., Tan M. K. I., O’Connell L., O’Hara G., MacMahon E., Douthwaite S., Nebbia G., Batra R., Martinez-Nunez R., Shankar-Hari M., Edgeworth J. D., Neil S. J. D., Malim M. H., Doores K. J., Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 5, 1598–1607 (2020). 10.1038/s41564-020-00813-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muecksch F., Wise H., Batchelor B., Squires M., Semple E., Richardson C., McGuire J., Clearly S., Furrie E., Greig N., Hay G., Templeton K., Lorenzi J. C. C., Hatziioannou T., Jenks S., Bieniasz P. D., Longitudinal Serological Analysis and Neutralizing Antibody Levels in Coronavirus Disease 2019 Convalescent Patients. J. Infect. Dis. 223, 389–398 (2021). 10.1093/infdis/jiaa659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccoli L., Park Y.-J., Tortorici M. A., Czudnochowski N., Walls A. C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L. E., Bowen J. E., Acton O. J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J. C., Mele F., Jovic S., Rodriguez B. F., Gupta S. V., Jin F., Piumatti G., Lo Presti G., Pellanda A. F., Biggiogero M., Tarkowski M., Pizzuto M. S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H. W., Lanzavecchia A., Corti D., Veesler D., Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e21 (2020). 10.1016/j.cell.2020.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. L. Steffen, E. T. Stone, M. Hassert, E. Geerling, B. T. Grimberg, A. M. Espino, P. Pantoja, C. Climent, D. F. Hoft, S. L. George, C. A. Sariol, A. K. Pinto, J. D. Brien, The receptor binding domain of SARS-CoV-2 spike is the key target of neutralizing antibody in human polyclonal sera. bioRxiv 2020.08.21.261727 [Preprint]. 22 August 2020. 10.1101/2020.08.21.261727. 10.1101/2020.08.21.261727 [DOI]
- 32.Barnes C. O., West A. P. Jr., Huey-Tubman K. E., Hoffmann M. A. G., Sharaf N. G., Hoffman P. R., Koranda N., Gristick H. B., Gaebler C., Muecksch F., Cetrulo Lorenzi J. C., Finkin S., Hägglöf T., Hurley A., Millard K. G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P. D., Caskey M., Robbiani D. F., Nussenzweig M. C., Bjorkman P. J., Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 182, 828–842.e16 (2020). 10.1016/j.cell.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., Wang P., Nair M. S., Yu J., Rapp M., Wang Q., Luo Y., Chan J. F.-W., Sahi V., Figueroa A., Guo X. V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.-Y., Kwong P. D., Sodroski J. G., Yin M. T., Sheng Z., Huang Y., Shapiro L., Ho D. D., Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). 10.1038/s41586-020-2571-7 [DOI] [PubMed] [Google Scholar]
- 34.Hansen J., Baum A., Pascal K. E., Russo V., Giordano S., Wloga E., Fulton B. O., Yan Y., Koon K., Patel K., Chung K. M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W. Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D. C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020). 10.1126/science.abd0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L., Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- 36.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X. D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X. S., Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 73–84.e16 (2020). 10.1016/j.cell.2020.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greaney A. J., Starr T. N., Gilchuk P., Zost S. J., Binshtein E., Loes A. N., Hilton S. K., Huddleston J., Eguia R., Crawford K. H. D., Dingens A. S., Nargi R. S., Sutton R. E., Suryadevara N., Rothlauf P. W., Liu Z., Whelan S. P. J., Carnahan R. H., Crowe J. E. Jr.., Bloom J. D., Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe 29, 44–57.e9 (2021). 10.1016/j.chom.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W., A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020). 10.1126/science.abc6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. McCallum, A. De Marco, F. Lempp, M. A. Tortorici, D. Pinto, A. C. Walls, M. Beltramello, A. Chen, Z. Liu, F. Zatta, S. Zepeda, J. di Iulio, J. E. Bowen, M. Montiel-Ruiz, J. Zhou, L. E. Rosen, S. Bianchi, B. Guarino, C. S. Fregni, R. Abdelnabi, S.-Y. C. Foo, P. W. Rothlauf, L.-M. Bloyet, F. Benigni, E. Cameroni, J. Neyts, A. Riva, G. Snell, A. Telenti, S. P. J. Whelan, H. W. Virgin, D. Corti, M. S. Pizzuto, D. Veesler, N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv 2021.01.14.426475 [Preprint]. 14 January 2021. 10.1101/2021.01.14.426475. 10.1101/2021.01.14.426475 [DOI]
- 40.Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E. R., Yu J., Bahna F., Bimela J., Huang Y., Katsamba P. S., Liu L., Nair M. S., Rawi R., Olia A. S., Wang P., Zhang B., Chuang G.-Y., Ho D. D., Sheng Z., Kwong P. D., Shapiro L., Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. Cell Host Microbe 29, 819–833.E7 (2021). 10.1016/j.chom.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G. Song, W.-t. He, S. Callaghan, F. Anzanello, D. Huang, J. Ricketts, J. L. Torres, N. Beutler, L. Peng, S. Vargas, J. Cassell, M. Parren, L. Yang, C. Ignacio, D. M. Smith, J. E. Voss, D. Nemazee, A. B. Ward, T. Rogers, D. R. Burton, R. Andrabi, Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. bioRxiv 2020.09.22.308965 [Preprint]. 23 September 2020. 10.1101/2020.09.22.308965. 10.1101/2020.09.22.308965 [DOI] [PMC free article] [PubMed]
- 42.C. Wang, R. van Haperen, J. Gutiérrez-Álvarez, W. Li, N. M. A. Okba, I. Albulescu, I. Widjaja, B. van Dieren, R. Fernandez-Delgado, I. Sola, D. L. Hurdiss, O. Daramola, F. Grosveld, F. J. M. van Kuppeveld, B. L. Haagmans, L. Enjuanes, D. Drabek, B.-J. Bosch, Isolation of cross-reactive monoclonal antibodies against divergent human coronaviruses that delineate a conserved and vulnerable site on the spike protein. bioRxiv 2020.10.20.346916 [Preprint]. 20 October 2020. 10.1101/2020.10.20.346916. 10.1101/2020.10.20.346916 [DOI]
- 43.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study Group , Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 384, 403–416 (2021). 10.1056/NEJMoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbett K. S., Edwards D. K., Leist S. R., Abiona O. M., Boyoglu-Barnum S., Gillespie R. A., Himansu S., Schäfer A., Ziwawo C. T., DiPiazza A. T., Dinnon K. H., Elbashir S. M., Shaw C. A., Woods A., Fritch E. J., Martinez D. R., Bock K. W., Minai M., Nagata B. M., Hutchinson G. B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W.-P., Schmidt S. D., Wang L., Zhang Y., Phung E., Chang L. A., Loomis R. J., Altaras N. E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M. K., Shi W., Leung K., Yang E. S., West A., Gully K. L., Stevens L. J., Wang N., Wrapp D., Doria-Rose N. A., Stewart-Jones G., Bennett H., Alvarado G. S., Nason M. C., Ruckwardt T. J., McLellan J. S., Denison M. R., Chappell J. D., Moore I. N., Morabito K. M., Mascola J. R., Baric R. S., Carfi A., Graham B. S., SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). 10.1038/s41586-020-2622-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr.., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C., C4591001 Clinical Trial Group , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. B. Vogel, I. Kanevsky, Y. Che, K. A. Swanson, A. Muik, M. Vormehr, L. M. Kranz, K. C. Walzer, S. Hein, A. Güler, J. Loschko, M. S. Maddur, K. Tompkins, J. Cole, B. G. Lui, T. Ziegenhals, A. Plaschke, D. Eisel, S. C. Dany, S. Fesser, S. Erbar, F. Bates, D. Schneider, B. Jesionek, B. Sänger, A.-K. Wallisch, Y. Feuchter, H. Junginger, S. A. Krumm, A. P. Heinen, P. Adams-Quack, J. Schlereth, C. Kröner, S. Hall-Ursone, K. Brasky, M. C. Griffor, S. Han, J. A. Lees, E. H. Mashalidis, P. V. Sahasrabudhe, C. Y. Tan, D. Pavliakova, G. Singh, C. Fontes-Garfias, M. Pride, I. L. Scully, T. Ciolino, J. Obregon, M. Gazi, R. Carrion Jr., K. J. Alfson, W. V. Kalina, D. Kaushal, P.-Y. Shi, T. Klamp, C. Rosenbaum, A. N. Kuhn, Ö. Türeci, P. R. Dormitzer, K. U. Jansen, U. Sahin, A prefusion SARS-CoV-2 spike RNA vaccine is highly immunogenic and prevents lung infection in non-human primates. bioRxiv 2020.09.08.280818 [Preprint]. 8 September 2020. 10.1101/2020.09.08.280818. 10.1101/2020.09.08.280818 [DOI]
- 48.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A. 2nd, Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H., mRNA-1273 Study Group , An mRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 383, 1920–1931 (2020). 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh E. E., Frenck R. W. Jr.., Falsey A. R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M. J., Bailey R., Swanson K. A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K. R., Lyke K. E., Raabe V., Dormitzer P. R., Jansen K. U., Şahin U., Gruber W. C., Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 383, 2439–2450 (2020). 10.1056/NEJMoa2027906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. Rambaut, N. Loman, O. Pybus, W. Barclay, J. Barrett, A. Carabelli, T. Connor, T. Peacock, D. L. Robertson, E. Volz, COVID-19 Genomics Consortium UK (CoG-UK), “Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations” (Virological, 2021); https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 51.E. Volz, S. Mishra, M. Chand, J. C. Barrett, R. Johnson, L. Geidelberg, W. R. Hinsley, D. J. Laydon, G. Dabrera, Á. O’Toole, R. Amato, M. Ragonnet-Cronin, I. Harrison, B. Jackson, C. V. Ariani, O. Boyd, N. J. Loman, J. T. McCrone, S. Gonçalves, D. Jorgensen, R. Myers, V. Hill, D. K. Jackson, K. Gaythorpe, N. Groves, J. Sillitoe, D. P. Kwiatkowski, The COVID-19 Genomics UK (COG-UK) consortium, S. Flaxman, O. Ratmann, S. Bhatt, S. Hopkins, A. Gandy, A. Rambaut, N. M. Ferguson, Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034 [Preprint]. 4 January 2021. 10.1101/2020.12.30.20249034. 10.1101/2020.12.30.20249034 [DOI]
- 52.H. Tegally, E. Wilkinson, M. Giovanetti, A. Iranzadeh, V. Fonseca, J. Giandhari, D. Doolabh, S. Pillay, E. J. San, N. Msomi, K. Mlisana, A. von Gottberg, S. Walaza, M. Allam, A. Ismail, T. Mohale, A. J. Glass, S. Engelbrecht, G. Van Zyl, W. Preiser, F. Petruccione, A. Sigal, D. Hardie, G. Marais, M. Hsiao, S. Korsman, M.-A. Davies, L. Tyers, I. Mudau, D. York, C. Maslo, D. Goedhals, S. Abrahams, O. Laguda-Akingba, A. Alisoltani-Dehkordi, A. Godzik, C. K. Wibmer, B. T. Sewell, J. Lourenço, L. C. J. Alcantara, S. L. Kosakovsky Pond, S. Weaver, D. Martin, R. J. Lessells, J. N. Bhiman, C. Williamson, T. de Oliveira, Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640 [Preprint]. 22 December 2020. 10.1101/2020.12.21.20248640. 10.1101/2020.12.21.20248640 [DOI]
- 53.Davies N. G., Abbott S., Barnard R. C., Jarvis C. I., Kucharski A. J., Munday J. D., Pearson C. A. B., Russell T. W., Tully D. C., Washburne A. D., Wenseleers T., Gimma A., Waites W., Wong K. L. M., van Zandvoort K., Silverman J. D., CMMID COVID-19 Working Group, The COVID-19 Genomics UK (COG-UK) Consortium, Diaz-Ordaz K., Keogh R., Eggo R. M., Funk S., Jit M., Atkins K. E., Edmunds W. J., Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, eabg3055 (2021). 10.1126/science.abg3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabino E. C., Buss L. F., Carvalho M. P. S., Prete C. A. Jr.., Crispim M. A. E., Fraiji N. A., Pereira R. H. M., Parag K. V., da Silva Peixoto P., Kraemer M. U. G., Oikawa M. K., Salomon T., Cucunuba Z. M., Castro M. C., de Souza Santos A. A., Nascimento V. H., Pereira H. S., Ferguson N. M., Pybus O. G., Kucharski A., Busch M. P., Dye C., Faria N. R., Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 397, 452–455 (2021). 10.1016/S0140-6736(21)00183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.N. R. Faria, I. Morales Claro, D. Candido, L. A. Moyses Franco, P. S. Andrade, T. M. Coletti, C. A. M. Silva, F. C. Sales, E. R. Manuli, R. S. Aguiar, N. Gaburo, C. d. C. Camilo, N. A. Fraiji, M. A. Esashika Crispim, M. d. P. S. S. Carvalho, A. Rambaut, N. Loman, O. G. Pybus, E. C. Sabino, CADDE Genomic Network, “Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings” (Virological, 2021); https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586.
- 56.Á. O’Toole, V. Hill, O. G. Pybus, A. Watts, I. I. Bogoch, K. Khan, J. P. Messina, The COVID-19 Genomics UK (COG-UK) consortium, Network for Genomic Surveillance in South Africa (NGS-SA), Brazil-UK CADDE Genomic Network, H. Tegally, R. R. Lessells, J. Giandhari, S. Pillay, K. A. Tumedi, G. Nyepetsi, M. Kebabonye, M. Matsheka, M. Mine, S. Tokajian, H. Hassan, T. Salloum, G. Merhi, J. Koweyes, J. L. Geoghegan, J. de Ligt, X. Ren, M. Storey, N. E. Freed, C. Pattabiraman, P. Prasad, A. S. Desai, R. Vasanthapuram, T. F. Schulz, L. Steinbrück, T. Stadler, Swiss Viollier Sequencing Consortium, A. Parisi, A. Bianco, D. G. de Viedma, S. Buenestado-Serrano, V. Borges, J. Isidro, S. Duarte, J. P. Gomes, N. S. Zuckerman, M. Mandelboim, O. Mor, T. Seemann, A. Arnott, J. Draper, M. Gall, W. Rawlinson, I. Deveson, S. Schlebusch, J. McMahon, L. Leong, C. K. Lim, M. Chironna, D. Laconsole, A. Bal, L. Josset, E. Holmes, K. St George, E. Lasek-Nesselquist, R. S. Sikkema, B. B. Oude Munnink, M. Koopmans, M. Brytting, V. S. Rani, S. Pavani, T. Smura, A. Heim, S. Kurkela, M. Umair, M. Salman, B. Bartolini, M. Rueca, C. Drosten, T. Wolff, O. Silander, D. Eggink, C. Reusken, H. Vennema, A. Park, SEARCH Alliance San Diego, National Virus Reference Laboratory, SeqCOVID-Spain, Danish Covid-19 Genome Consortium (DCGC), Communicable Diseases Genomic Network (CDGN), Dutch National SARS-CoV-2 surveillance program, Division of Emerging Infectious Diseases KDCA, T. de Oliveira, N. R. Faria, A. Rambaut, M. U. G. Kraemer, “Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2” (Virological, 2021); https://virological.org/t/tracking-the-international-spread-of-sars-cov-2-lineages-b-1-1-7-and-b-1-351-501y-v2/592.
- 57.Chan K. K., Tan T. J. C., Narayanan K. K., Procko E., An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci. Adv. 7, eabf1738 (2021). 10.1126/sciadv.abf1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Starr T. N., Greaney A. J., Addetia A., Hannon W. W., Choudhary M. C., Dingens A. S., Li J. Z., Bloom J. D., Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371, 850–854 (2021). 10.1126/science.abf9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang L., Jackson C. B., Mou H., Ojha A., Peng H., Quinlan B. D., Rangarajan E. S., Pan A., Vanderheiden A., Suthar M. S., Li W., Izard T., Rader C., Farzan M., Choe H., SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 11, 6013 (2020). 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou B., Thi Nhu Thao T., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J. N., Portmann J., Halwe N. J., Ulrich L., Trüeb B. S., Fan X., Hoffmann B., Wang L., Thomann L., Lin X., Stalder H., Pozzi B., de Brot S., Jiang N., Cui D., Hossain J., Wilson M., Keller M., Stark T. J., Barnes J. R., Dijkman R., Jores J., Benarafa C., Wentworth D. E., Thiel V., Beer M., SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592, 122–127 (2021). 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 61.Z. Liu, L. A. VanBlargan, L.-M. Bloyet, P. W. Rothlauf, R. E. Chen, S. Stumpf, H. Zhao, J. M. Errico, E. S. Theel, M. J. Liebeskind, B. Alford, W. J. Buchser, A. H. Ellebedy, D. H. Fremont, M. S. Diamond, S. P. J. Whelan, Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv 2020.11.06.372037 [Preprint]. 11 January 2021. 10.1101/2020.11.06.372037. 10.1101/2020.11.06.372037 [DOI]
- 62.Baum A., Fulton B. O., Wloga E., Copin R., Pascal K. E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G. S., Murphy A. J., Stahl N., Yancopoulos G. D., Kyratsous C. A., Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020). 10.1126/science.abd0831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y., The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 182, 1284–1294.e9 (2020). 10.1016/j.cell.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J. C. C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C. D., Caskey M., Robbiani D. F., Rice C. M., Nussenzweig M. C., Hatziioannou T., Bieniasz P. D., Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife 9, e61312 (2020). 10.7554/eLife.61312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomson E. C., Rosen L. E., Shepherd J. G., Spreafico R., da Silva Filipe A., Wojcechowskyj J. A., Davis C., Piccoli L., Pascall D. J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O’Toole A., Pinto D., Colquhoun R. M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V. B., Nix J., Zhang I., Jarrett R. F., Glass W. G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T. I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A. M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M. G., Openshaw P. J. M., Fadda E., Baillie J. K., Chodera J. D., The ISARIC4C Investigators, COVID-19 Genomics UK (COG-UK) Consortium, Rihn S. J., Lycett S. J., Virgin H. W., Telenti A., Corti D., Robertson D. L., Snell G., Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 184, 1171–1187.e20 (2021). 10.1016/j.cell.2021.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.P. Wang, M. S. Nair, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P. D. Kwong, B. S. Graham, J. R. Mascola, J. Y. Chang, M. T. Yin, M. Sobieszczyk, C. A. Kyratsous, L. Shapiro, Z. Sheng, Y. Huang, D. D. Ho, Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. bioRxiv 2021.01.25.428137 [Preprint]. 12 February 2021. 10.1101/2021.01.25.428137. 10.1101/2021.01.25.428137 [DOI] [PubMed]
- 67.C. Graham, J. Seow, I. Huettner, H. Khan, N. Kouphou, S. Acors, H. Winstone, S. Pickering, R. P. Galao, M. J. Lista, J. M. Jimenez-Guardeno, A. G. Laing, Y. Wu, M. Joseph, L. Muir, W. M. Ng, H. M. E. Duyvesteyn, Y. Zhao, T. A. Bowden, M. Shankar-Hari, A. Rosa, P. Cherepanov, L. E. McCoy, A. C. Hayday, S. J. D. Neil, M. H. Malim, K. J. Doores, Impact of the B.1.1.7 variant on neutralizing monoclonal antibodies recognizing diverse epitopes on SARS-CoV-2 Spike. bioRxiv 2021.02.03.429355 [Preprint]. 3 February 2021. 10.1101/2021.02.03.429355. 10.1101/2021.02.03.429355 [DOI]
- 68.Shen X., Tang H., McDanal C., Wagh K., Fischer W., Theiler J., Yoon H., Li D., Haynes B. F., Sanders K. O., Gnanakaran S., Hengartner N., Pajon R., Smith G., Glenn G. M., Korber B., Montefiori D. C., SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 29, 529–539.E3 (2021). 10.1016/j.chom.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wibmer C. K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B. E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J. N., Moore P. L., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 27, 622–625 (2021). 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- 70.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C. O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J. A., Oliveira T. Y., Yang Z., Abernathy M. E., Huey-Tubman K. E., Hurley A., Turroja M., West K. A., Gordon K., Millard K. G., Ramos V., Da Silva J., Xu J., Colbert R. A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P. J., Casellas R., Hatziioannou T., Bieniasz P. D., Nussenzweig M. C., mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592, 616–622 (2021). 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson & Johnson, “Johnson & Johnson Announces Single-Shot Janssen COVID-19 Vaccine Candidate Met Primary Endpoints in Interim Analysis of its Phase 3 ENSEMBLE Trial” (2021); www.prnewswire.com/news-releases/johnson--johnson-announces-single-shot-janssen-covid-19-vaccine-candidate-met-primary-endpoints-in-interim-analysis-of-its-phase-3-ensemble-trial-301218035.html.
- 72.Novavax, Inc., “Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial” (2021); https://ir.novavax.com/news-releases/news-release-details/novavax-covid-19-vaccine-demonstrates-893-efficacy-uk-phase-3.
- 73.Yuan M., Liu H., Wu N. C., Lee C. D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T. F., Landais E., Sok D., Jardine J. G., Burton D. R., Wilson I. A., Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). 10.1126/science.abd2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu N. C., Yuan M., Liu H., Lee C. D., Zhu X., Bangaru S., Torres J. L., Caniels T. G., Brouwer P. J. M., van Gils M. J., Sanders R. W., Ward A. B., Wilson I. A., An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 33, 108274 (2020). 10.1016/j.celrep.2020.108274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G. F., Gao F., Liu L., A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368, 1274–1278 (2020). 10.1126/science.abc2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W. J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G. F., Yuan Z., Yan J., A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 77.Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., Liu J., Nie J., Zheng Y., Liang H., Liu R., Gong S., Xu H., Yisimayi A., Lv Q., Wang B., He R., Han Y., Zhao W., Bai Y., Qu Y., Gao X., Ji C., Wang Q., Gao N., Huang W., Wang Y., Xie X. S., Su X. D., Xiao J., Qin C., Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy. Cell 183, 1013–1023.e13 (2020). 10.1016/j.cell.2020.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hurlburt N. K., Seydoux E., Wan Y.-H., Edara V. V., Stuart A. B., Feng J., Suthar M. S., McGuire A. T., Stamatatos L., Pancera M., Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 11, 5413 (2020). 10.1038/s41467-020-19231-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Snijder J., Ortego M. S., Weidle C., Stuart A. B., Gray M. D., McElrath M. J., Pancera M., Veesler D., McGuire A. T., An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity 48, 799–811.e9 (2018). 10.1016/j.immuni.2018.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saadat S., Tehrani Z. R., Logue J., Newman M., Frieman M. B., Harris A. D., Sajadi M. M., Binding and Neutralization Antibody Titers After a Single Vaccine Dose in Health Care Workers Previously Infected With SARS-CoV-2. JAMA 325, 1467–1469 (2021). 10.1001/jama.2021.3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krammer F., Srivastava K., Alshammary H., Amoako A. A., Awawda M. H., Beach K. F., Bermúdez-González M. C., Bielak D. A., Carreño J. M., Chernet R. L., Eaker L. Q., Ferreri E. D., Floda D. L., Gleason C. R., Hamburger J. Z., Jiang K., Kleiner G., Jurczyszak D., Matthews J. C., Mendez W. A., Nabeel I., Mulder L. C. F., Raskin A. J., Russo K. T., Salimbangon A. T., Saksena M., Shin A. S., Singh G., Sominsky L. A., Stadlbauer D., Wajnberg A., Simon V., Antibody Responses in Seropositive Persons after a Single Dose of SARS-CoV-2 mRNA Vaccine. N. Engl. J. Med. 384, 1372–1374 (2021). 10.1056/NEJMc2101667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94, e00127-20 (2020). 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C. R., Swanson K. A., Cai H., Sarkar R., Chen W., Cutler M., Cooper D., Weaver S. C., Muik A., Sahin U., Jansen K. U., Xie X., Dormitzer P. R., Shi P.-Y., Neutralizing Activity of BNT162b2-Elicited Serum. N. Engl. J. Med. 384, 1466–1468 (2021). 10.1056/NEJMc2102017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu K., Werner A. P., Koch M., Choi A., Narayanan E., Stewart-Jones G. B. E., Colpitts T., Bennett H., Boyoglu-Barnum S., Shi W., Moliva J. I., Sullivan N. J., Graham B. S., Carfi A., Corbett K. S., Seder R. A., Edwards D. K., Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N. Engl. J. Med. 384, 1468–1470 (2021). 10.1056/NEJMc2102179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kistler K. E., Bedford T., Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. eLife 10, e64509 (2021). 10.7554/eLife.64509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pinto D., Park Y.-J., Beltramello M., Walls A. C., Tortorici M. A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J. B., Chen R. E., Havenar-Daughton C., Snell G., Telenti A., Virgin H. W., Lanzavecchia A., Diamond M. S., Fink K., Veesler D., Corti D., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). 10.1038/s41586-020-2349-y [DOI] [PubMed] [Google Scholar]
- 87.Yang R., Huang B., A R., Li W., Wang W., Deng Y., Tan W., Development and effectiveness of pseudotyped SARS-CoV-2 system as determined by neutralizing efficiency and entry inhibition test in vitro. Biosaf. Health 2, 226–231 (2020). 10.1016/j.bsheal.2020.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.National Research Council Canada, “HEK293SF-3F6 and HEK293-6E expression platforms (L-10894 / 11266 / 11565)” (2019); https://nrc.canada.ca/en/research-development/intellectual-property-licensing/hek293sf-3f6-hek293-6e-expression-platforms-l-10894-11266-11565.
- 89.Harris P. A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J. G., Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 42, 377–381 (2009). 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harris P. A., Taylor R., Minor B. L., Elliott V., Fernandez M., O’Neal L., McLeod L., Delacqua G., Delacqua F., Kirby J., Duda S. N., REDCap Consortium , The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inform. 95, 103208 (2019). 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crawford K. H. D., Eguia R., Dingens A. S., Loes A. N., Malone K. D., Wolf C. R., Chu H. Y., Tortorici M. A., Veesler D., Murphy M., Pettie D., King N. P., Balazs A. B., Bloom J. D., Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses 12, 513 (2020). 10.3390/v12050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsieh C. L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H.-C., Javanmardi K., Le K. C., Wrapp D., Lee A. G., Liu Y., Chou C.-W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). 10.1126/science.abd0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dintwe O., Rohith S., Schwedhelm K. V., McElrath M. J., Andersen-Nissen E., De Rosa S. C., OMIP-056: Evaluation of Human Conventional T Cells, Donor-Unrestricted T Cells, and NK Cells Including Memory Phenotype by Intracellular Cytokine Staining. Cytometry A 95, 722–725 (2019). 10.1002/cyto.a.23753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Horton H., Thomas E. P., Stucky J. A., Frank I., Moodie Z., Huang Y., Chiu Y.-L., McElrath M. J., De Rosa S. C., Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J. Immunol. Methods 323, 39–54 (2007). 10.1016/j.jim.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu X., Yang Z.-Y., Li Y., Hogerkorp C.-M., Schief W. R., Seaman M. S., Zhou T., Schmidt S. D., Wu L., Xu L., Longo N. S., McKee K., O’Dell S., Louder M. K., Wycuff D. L., Feng Y., Nason M., Doria-Rose N., Connors M., Kwong P. D., Roederer M., Wyatt R. T., Nabel G. J., Mascola J. R., Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861 (2010). 10.1126/science.1187659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
science.sciencemag.org/content/372/6549/1413/suppl/DC1
Materials and Methods
Figs. S1 to S8
Tables S1 to S4
MDAR Reproducibility Checklist