Summary
The continual emergence of novel coronaviruses (CoV), such as severe acute respiratory syndrome-(SARS)-CoV-2, highlights the critical need for broadly reactive therapeutics and vaccines against this family of viruses. From a recovered SARS-CoV donor sample, we identify and characterize a panel of six monoclonal antibodies that cross-react with CoV spike (S) proteins from the highly pathogenic SARS-CoV and SARS-CoV-2, and demonstrate a spectrum of reactivity against other CoVs. Epitope mapping reveals that these antibodies recognize multiple epitopes on SARS-CoV-2 S, including the receptor-binding domain, the N-terminal domain, and the S2 subunit. Functional characterization demonstrates that the antibodies mediate phagocytosis—and in some cases trogocytosis—but not neutralization in vitro. When tested in vivo in murine models, two of the antibodies demonstrate a reduction in hemorrhagic pathology in the lungs. The identification of cross-reactive epitopes recognized by functional antibodies expands the repertoire of targets for pan-coronavirus vaccine design strategies.
Keywords: SARS-CoV-2, COVID-19, antibody discovery, cross-reactivity, Fc effector function, LIBRA-seq, single-cell sequencing
Graphical abstract
Highlights
Applied LIBRA-seq to PBMCs from a recovered SARS-CoV donor
Identified six cross-reactive CoV mAbs that target distinct domains on SARS-CoV-2 spike
Characterized mAbs with effector functions in SARS-CoV-2 murine infection model
Shiakolas et al. demonstrate that cross-reactive coronavirus antibodies induced by natural infection display a spectrum of epitope specificities across the spike protein and exhibit in vitro and in vivo antiviral functions.
Introduction
The emergence of a novel coronavirus severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has resulted in a worldwide pandemic, threatening the lives of billions and imposing an immense burden on healthcare systems and the global economy. SARS-CoV-2, the seventh coronavirus known to infect humans, is a member of the Betacoronavirus genus, which includes the highly pathogenic SARS-CoV and Middle Eastern respiratory syndrome (MERS)-CoV, as well as endemic variants HCoV-OC43 and HCoV-HKU1.1 Recent coronavirus outbreaks and the threat of future emerging zoonotic strains highlight the need for broadly applicable coronavirus therapeutic interventions and vaccine design approaches.2
Coronaviruses use the homotrimeric spike (S) protein to engage with cell-surface receptors and enter host cells. S consists of two functional subunits: S1 and S2. S1 facilitates the attachment to target cells and is composed of the N-terminal domain (NTD) and the receptor-binding domain (RBD), whereas S2, which encodes the fusion peptide and heptad repeats, promotes viral fusion.3,4 To facilitate cell entry, human coronaviruses use different host factors; however, SARS-CoV and SARS-CoV-2 both use the cell-surface receptor angiotensin-converting enzyme 2 (ACE2).5 In addition, SARS-CoV-2 S shares 76% amino acid identity with SARS-CoV S.1 Furthermore, S serves as a dominant antibody target and is a focus of countermeasure development for the treatment and prevention of COVID-19 infection.6,7 S proteins from the Betacoronavirus genus share multiple regions of structural homology and thus could serve as targets for a cross-reactive antibody response.8 Identifying cross-reactive antibody epitopes can inform rational design strategies for vaccines and therapies that target multiple highly pathogenic coronaviruses.
Numerous potent neutralizing antibodies against SARS-CoV-2 have been discovered, including multiple candidates currently in clinical trials or approved for emergency use for the prophylactic and acute treatment of COVID-19.9, 10, 11, 12, 13, 14, 15, 16 Investigation of SARS-CoV-2/SARS-CoV cross-reactive antibodies has focused primarily on the RBD epitope, which has resulted in the identification of a number of SARS-CoV-2/SARS-CoV cross-reactive antibody candidates.12,17,18 However, the diversity of epitopes and functions beyond virus neutralization have not been extensively explored for cross-reactive antibodies.19, 20, 21 Evidence of Fc effector function contributing to protection in vivo against SARS-CoV22 and SARS-CoV-223 suggests that the role of antibodies beyond neutralization may be a crucial component of protection and an important consideration in vaccine design strategies for coronaviruses.20,24, 25, 26
In this study, we investigated antibody cross-reactivity across the Betacoronavirus genus at monoclonal resolution. To do this, we applied LIBRA-seq (linking B cell receptor to antigen specificity through sequencing)27 to a recovered SARS-CoV donor sample from >10 years after infection. We identified and characterized SARS-CoV-2/SARS-CoV cross-reactive human antibodies that target multiple, distinct structural domains of S, mediate phagocytosis and trogocytosis, and mitigate pathological burden in vivo. A better understanding of the genetic features, epitope specificities, and functional characteristics of cross-reactive coronavirus antibodies may translate into strategies for current vaccine design efforts and additional measures to counteract potential future pandemic strains.
Results
LIBRA-seq characterization of a SARS-CoV recovered donor
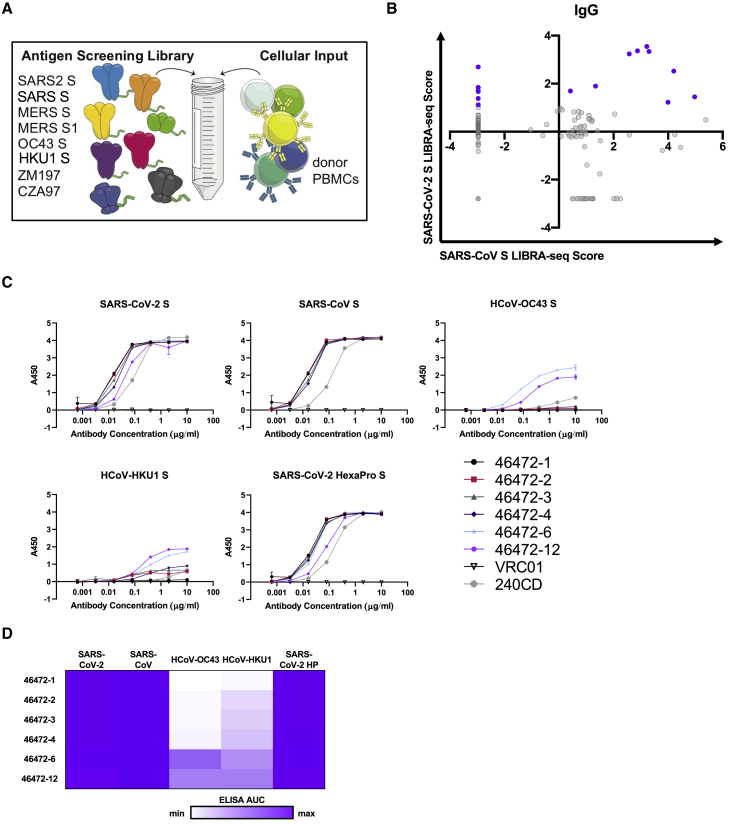
To identify cross-reactive antibodies to multiple coronavirus antigens, LIBRA-seq was applied to a peripheral blood mononuclear cell (PBMC) sample from a donor infected with SARS-CoV >10 years before sample collection. The antigen screening library consisted of eight oligo-tagged recombinant soluble antigens: six coronavirus trimer antigens (SARS-CoV-2 S, SARS-CoV S, MERS-CoV S, MERS-CoV S1 [with foldon domain], HCoV-OC43 S, and HCoV-HKU1 S) and two HIV trimer antigens from strains ZM197 and CZA97 as negative controls (Figure 1A). After the antigen screening library was mixed with donor PBMCs, antigen-positive B cells were enriched by fluorescence-activated cell sorting (FACS) and processed for single-cell sequencing (Figure S1A). After bioinformatic processing, we recovered 2,625 cells with paired heavy-/light-chain sequences and antigen reactivity information (Figure S1B), and from these cells, there were 2,368 unique VDJ (variability, diversity, and joining) sequences. Overall, LIBRA-seq enabled rapid screening of PBMCs from a patient sample, with recovery of paired heavy-/light-chain sequences and antigen reactivity for thousands of B cells at the single-cell level.
Figure 1.
Identification of coronavirus cross-reactive antibodies from SARS-CoV recovered PBMC sample using LIBRA-seq
(A) Schematic of DNA-barcoded antigens used to probe a SARS-CoV donor PBMC sample.
(B) LIBRA-seq scores for SARS-CoV (x axis) and SARS-CoV-2 (y axis) for all IgG+ B cells recovered from sequencing are shown as circles; 15 lead antibody candidates are highlighted in purple.
(C) Antibodies were tested for binding to CoV antigens by ELISA. HIV-specific antibody VRC01 was used as a negative control. Anti-SARS-CoV mouse antibody 240CD was also used. ELISAs were performed in technical duplicates with at least 2 biological duplicates. Data are represented as means ± SEMs.
(D) ELISA binding data are displayed as a heatmap of the AUC values calculated from data in (C), with an area under the curve (AUC) of 0 as white, and maximum AUC as purple.
See also Figure S1.
Identification of SARS-CoV-2 and SARS-CoV cross-reactive antibodies
With a goal of identifying antibodies that were cross-reactive to multiple coronavirus S proteins, we prioritized lead candidates based on their sequence features and LIBRA-seq scores (Figure S1C). We selected 15 antibody candidates that exhibited diverse sequence features and used a number of different variable genes for expression and characterization (Figures 1B and S1D). These antibodies displayed a broad range of percent identity to germline (83%–98%) and a variety of CDRH3 and CDRL3 lengths (6–24 and 5–13 amino acids, respectively) (Figure S1D). By ELISA, SARS-CoV-2 S and SARS-CoV S binding was confirmed for 6/15 of the tested antibodies (46472-1, 46472-2, 46472-3, 46472-4, 46472-6, and 46472-12), indicating that LIBRA-seq could successfully identify SARS-CoV-2 reactive B cells, but also suggesting potential differences in antigen-binding detection for primary B cells with a sequencing readout versus recombinant immunoglobulin G (IgG) by ELISA (Figures 1C, 1D, and S1E). Furthermore, antibodies 46472-6 and 46472-12 bound to S proteins from endemic HCoV-OC43 and HCoV-HKU1, albeit generally at lower levels (Figures 1C, 1D, and S1E). Although the six monoclonal antibodies (mAbs) showed reactivity by ELISA to the MERS-CoV antigen probe used in the LIBRA-seq screening library, antibody binding to other independent preparations of this protein was inconsistent, so MERS-CoV S reactivity could not be confirmed definitively (Figures S1F and S1G). Overall, the application of the LIBRA-seq technology enabled the identification of a panel of cross-reactive antibodies that recognize the S antigen from multiple coronaviruses.
Cross-reactive CoV antibodies target diverse epitopes on S
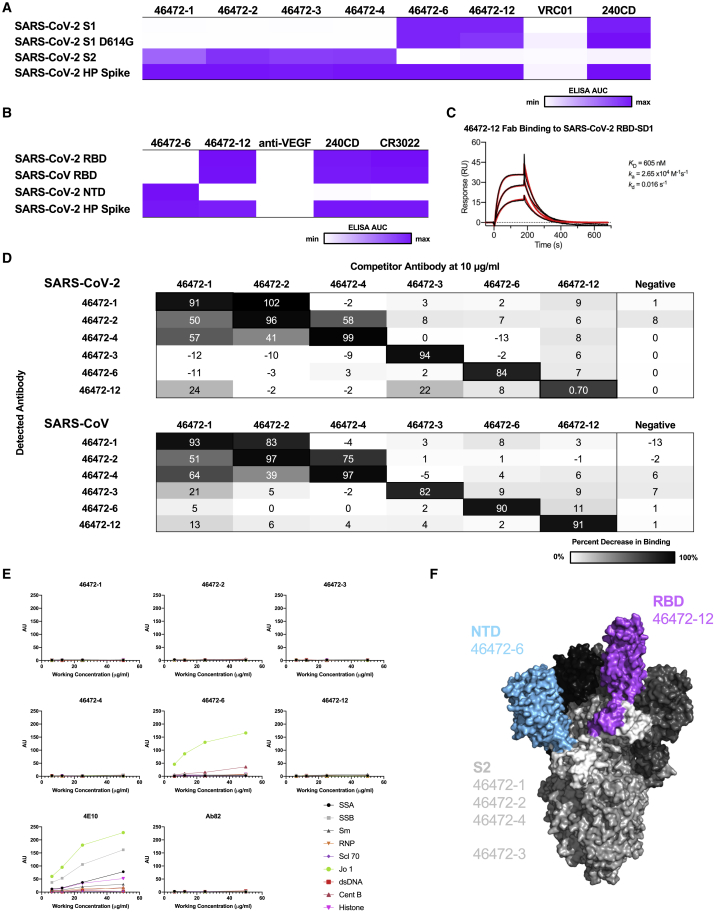
To elucidate the epitopes targeted by the cross-reactive antibodies, we performed binding assays to various structural domains of S as well as binding-competition experiments. We assessed antibody binding to the S1 and S2 subdomains of SARS-CoV-2. Antibodies 46472-1, 46472-2, 46472-3, and 46472-4 bound to the S2 domain, whereas 46472-6 and 46472-12 recognized the S1 domain but targeted different epitopes, the NTD and RBD, respectively (Figures 2A–2C, S2A, and S2B). Although 46472-12 bound to the RBD, it did not compete with ACE2 for binding to SARS-CoV-2 S and showed partial competition with RBD-directed antibody CR3022 (Figures S2C and S2D). To determine whether the antibodies targeted overlapping or distinct epitopes, we performed competition ELISA experiments and found that the S2-directed antibodies 46472-1, 46472-2, and 46472-4 competed for binding to S (Figure 2D). This pattern was observed for both SARS-CoV-2 and SARS-CoV S. Of note, this competition group did not include S2-directed antibody 46472-3, revealing the identification of multiple cross-reactive epitope targets on S2 (Figure 2D). Furthermore, antibody binding was not affected by two glycan knockout mutants (N165A or N709A) or mannose competition (Figures S2E and S2F). Lastly, we measured antibody autoreactivity and found that with the exception of 46472-6 binding to Jo-1, none of the antibodies showed autoreactivity against the tested antigens (Figure 2E). These data suggest that the identified cross-reactive antibodies are coronavirus specific and target multiple, diverse epitopes on the S protein (Figure 2F).
Figure 2.
Epitope mapping of cross-reactive antibodies
(A) For cross-reactive coronavirus antibodies, ELISA data against the antigens are displayed as a heatmap of the AUC values calculated from the data in Figure S2A.
(B) For SARS-CoV-2 S1-reactive antibodies, ELISA data against the RBD and NTD are displayed as a heatmap of the AUC values calculated from the data in Figure S2B. For (A) and (B), an AUC of 0 is displayed as white and maximum AUC as purple. ELISA data are representative of at least 2 independent experiments. Anti-HIV antibody VRC01 and anti-vascular endothelial growth factor (VEGF) antibody are shown as a negative control, and anti-SARS-CoV antibody 240CD is shown as a positive control.
(C) Surface plasmon resonance binding of 46472-12 Fab to SARS-CoV-2 RBD. Calculated binding constants are shown to the right of the graph.
(D) Cross-reactive antibodies were used in a competition ELISA to determine whether the binding of one antibody affected the binding of another. Competitor antibodies were added at 10 μg/mL, and then detected antibodies were added at 0.1 μg/mL. The percent reduction in binding compared to binding without a competitor is shown. An anti-HIV antibody was used as a negative control. ELISAs were performed in technical duplicates with at least 2 biological duplicates.
(E) Antibodies were tested for autoreactivity against a variety of antigens in the Luminex AtheNA assay. AU stands for Athena units. Anti-HIV antibody 4E10 was used as a positive control and Ab82 was used as a negative control.
(F) Cross-reactive coronavirus antibodies target a variety of epitopes on the SARS-CoV-2 S protein, including the RBD, NTD, and S2 domains, highlighted on the structure (PDB: 6VSB).
See also Figure S2.
Functional characterization of cross-reactive CoV antibodies
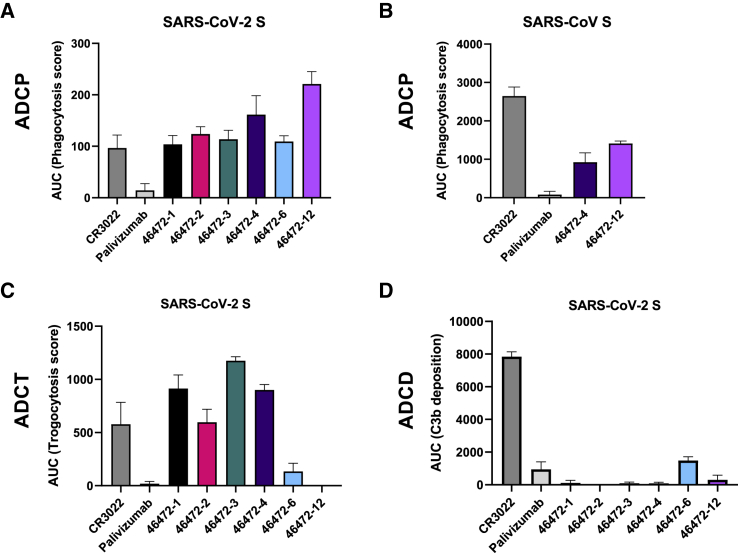
Next, we characterized our cross-reactive antibody panel for functional activity. Although none of the antibodies neutralized SARS-CoV or SARS-CoV-2 (Figures S3A and S3B), all of the antibodies showed antibody-dependent cellular phagocytosis (ADCP) in vitro for SARS-CoV-2 S (Figures 3A and S3C). In particular, the RBD-reactive antibody 46472-12 showed greater ADCP activity compared to the other cross-reactive antibodies and the SARS-CoV/SARS-CoV-2 cross-reactive RBD antibody control, CR302228 (Figures 3A and S3C). Furthermore, we tested and confirmed ADCP activity against SARS-CoV for two antibodies that mediated the highest phagocytotic activity against SARS-CoV-2, 46472-4, and 46472-12, illustrating that these antibodies have cross-coronavirus phagocytic ability (Figures 3B and S3D). We next tested the antibodies in a trogocytosis assay29 and found that four antibodies in our panel (46472-1, 46472-2, 46472-3, and 46472-4) mediated trogocytosis (Figures 3C and S3E). This warrants further investigation as this is the first description of trogocytosis performed by SARS-CoV-2-specific antibodies. Lastly, there was no antibody-dependent complement deposition (ADCD) (Figures 3D and S3F). These results revealed different profiles of Fc effector functionality within the panel of cross-reactive antibodies.
Figure 3.
Functional activity of cross-reactive coronavirus antibodies
(A) Cross-reactive coronavirus antibodies were tested for antibody-dependent cellular phagocytosis activity (ADCP) against SARS-CoV-2 S, compared to positive control CR3022 and negative control palivizumab, an anti-RSV antibody. AUC of the phagocytosis score is shown, calculated from data in Figure S3C. Data are represented as means ± SDs.
(B) 46472-4 and 46472-12 were tested for ADCP activity against SARS-CoV S, compared to CR3022 and anti-RSV palivizumab. AUC of the phagocytosis score is shown, calculated from data in Figure S3D. Data are represented as means ± SDs.
(C) Cross-reactive coronavirus antibodies were tested for antibody-dependent cellular trogocytosis (ADCT) activity against SARS-CoV-2 S displayed on transfected cells, compared to positive control CR3022 and anti-RSV palivizumab. AUC of the trogocytosis score is shown, calculated from data in Figure S3E. Data are represented as means ± SDs.
(D) Cross-reactive coronavirus antibodies were tested for antibody-dependent complement deposition (ADCD) activity against SARS-CoV-2 S, compared to positive control CR3022 and anti-RSV palivizumab. AUC of the C3b deposition score is shown, calculated from data in Figure S3F. Data are represented as means ± SDs.
See also Figure S3.
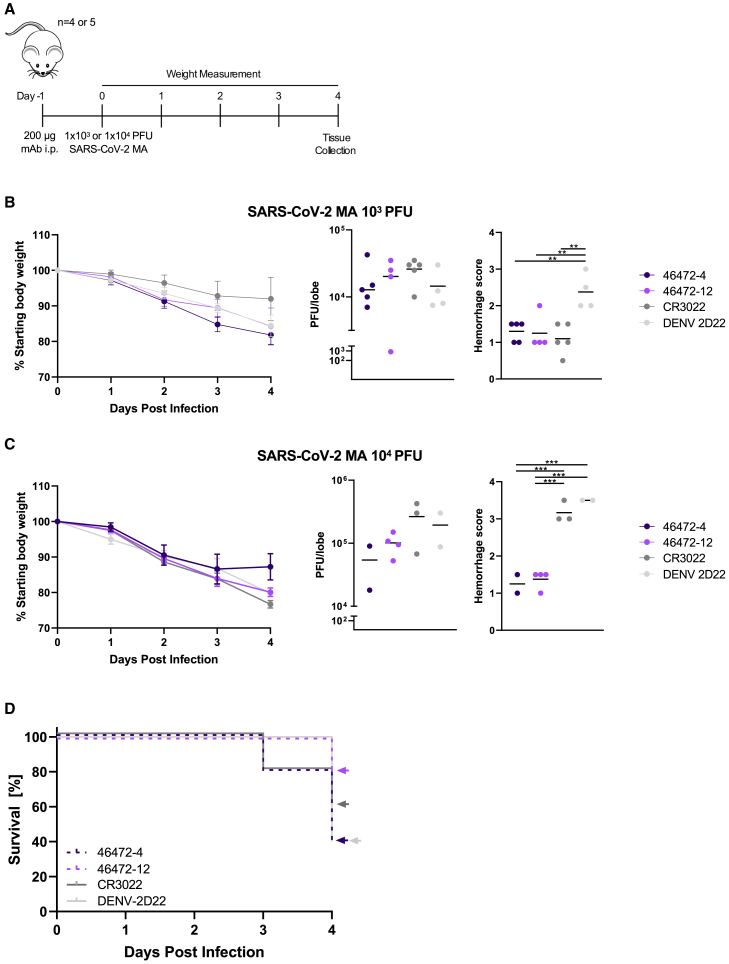
Since non-neutralizing SARS-CoV-2 antibodies with Fc effector function activity have not been extensively characterized in vivo, these results prompted us to test antibodies 46472-4 and 46472-12 for prophylaxis in a murine infection model using a mouse-adapted virus strain (SARS-CoV-2 MA)30,31 at a non-lethal dose of 1 × 103 plaque-forming units (PFU) (Figure 4A). Although there were no differences in survival and viral load between experimental and control groups, the lung hemorrhage scores (see Method details) for 46472-4 and 46472-12 were similar to antigen-specific control CR3022, and all three groups were significantly lower than the scores for isotype control DENV-2D22 (p < 0.01, ordinary one-way ANOVA with multiple comparisons) (Figures 4B and S4A). To evaluate the in vivo effect of these antibodies in a more stringent challenge model in 12-month-old female BALB/c mice, we increased the viral dose from 1 × 103 to 1 × 104 PFU. In this experiment, mice that received antibody 46472-12 exhibited the best survival rate (4/5 at day 4), compared to the other treatment groups that included CR3022 as an antigen-specific control and DENV-2D22 as a negative control, although statistical significance was not achieved (Figures 4C, 4D, and S4B). There were no significant differences in viral load between groups; however, the surviving animals from the 46472-4 and 46472-12 groups showed significantly lower hemorrhagic pathology scores in harvested mouse lungs compared to the negative control treatment group (p < 0.001, ordinary one-way ANOVA with multiple comparisons) (Figure 4C). Animals treated with the antigen-specific control, CR3022, had significantly higher hemorrhage scores than animals treated with 46472-4 and 46472-12 (p < 0.001, ordinary one-way ANOVA with multiple comparisons), although the statistical analysis may be limited by the small numbers of surviving animals for some of the groups (Figure 4C). While definitive evidence for protection is limited, the data from the in vivo experiments suggest that these cross-reactive antibodies could contribute to counteracting coronavirus infection in prophylaxis.
Figure 4.
In vivo effects of cross-reactive antibodies
(A) Timeline of the prophylactic antibody experiment in SARS-CoV-2 mouse adapted (MA) in vivo infection model.
(B and C) For each antibody treatment group for the experiment using (B) 1 × 103 PFU or (C) 1 × 104 PFU of SARS-CoV-2 MA, shown are daily body weight progression, and terminal qRT-PCR quantification of lung viral titer and lung hemorrhage scores of gross pathology. For viral titer values and the lung hemorrhage scores, an ordinary one-way ANOVA test with multiple comparisons was performed; ∗∗p < 0.01 and ∗∗∗p < 0.001.
(D) For the experiment with 1 × 104 PFU of SARS-CoV-2 MA, percent survival for each antibody group is shown.
See also Figure S4.
Discussion
Here, we described a set of cross-reactive Betacoronavirus antibodies isolated from a recovered SARS-CoV donor. The antibodies targeted diverse epitopes on S, including the S2 subdomain as well as the RBD and NTD on S1, and demonstrated Fc effector function in vitro. In addition, two of these antibodies were tested in vivo and displayed a reduction in lung hemorrhage score, while the effects on viral load were not definitive.
Given the similar effect of 46472-4 and 46472-12 on severe disease in the mouse model, their phagocytotic ability along with the inability to mediate neutralization suggests that the former may be a mechanism through which they function, and additional studies are under way to further assess this hypothesis. Phagocytosis has been shown to be associated with protection in a SARS-CoV-2 DNA vaccination in non-human primates32 as well as survival in natural infection,33 and as such could be an important mechanism for protection by antibodies.20 The role of trogocytosis in COVID-19 is unknown, as are the targets that may be important for this function. 46472-4 was able to mediate this membrane nibbling in contrast to 46472-12, suggesting that this function in addition to complement activity was not responsible for the in vivo effect on severe disease mediated by these antibodies. Although the precise in vivo effects of these antibodies have not been elucidated, the identification of multiple, cross-reactive antibodies highlights a potential role for Fc effector function activity, specifically phagocytosis, in coronavirus infection. Evidence of protection associated with Fc effector function in SARS-CoV,22 SARS-CoV-2,23,24,34 and other infectious diseases, including influenza, Ebola, and HIV, motivates further investigation into its contribution to the treatment of COVID-19.35, 36, 37, 38 Furthermore, the importance of Fc effector functionality of potently neutralizing candidate clinical SARS-CoV-2 antibodies in a therapeutic setting rather than prophylaxis highlights the potential benefit for investigation into non-neutralizing antibodies with phagocytic activity and their administration after infection onset.39 Elucidation of the functional roles of cross-reactive but non-neutralizing antibodies could have implications for understanding the factors involved in the protection or enhancement of disease.
Given the ongoing SARS-CoV-2 pandemic and the potential for future zoonotic coronavirus pathogens to emerge, coronavirus vaccine and therapeutic development is of paramount importance.40, 41, 42, 43 Antibodies that can cross-react with multiple coronavirus strains are primary targets as potential broadly reactive therapies. Such antibodies can further reveal cross-reactive epitopes that could serve as templates for the development of broadly protective vaccines. Understanding the spectrum of cross-reactive epitopes targeted by human antibodies, as well as the functional role that such antibodies have within coronavirus infection, are therefore a vital element of medical countermeasure development.
Limitations of the study
The present study focuses on the characterization of cross-reactive coronavirus antibodies, mostly in the context of SARS-CoV-2. Further characterization of this panel of antibodies against circulating endemic coronavirus strains would enhance the clinical relevance to less severe coronavirus-associated respiratory infections.
The present study used a dosing regimen in a prophylactic setting, and given the emerging evidence of survival benefit with effector function in antibodies provided after infection onset,39 antibody administration in a therapeutic setting may provide further insight into in vivo properties. Furthermore, additional effector function characterization such as ADCC and ADNP would strengthen the profile of this panel of non-neutralizing antibodies, considering their role in both human33 and mouse SARS-CoV-2 infection studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 mouse anti-human CD14 | BD | Cat#561709; RRID: RRID:AB_10893806 |
| FITC anti-human CD3 (OKT3) | Tonbo Biosciences | Cat#35-0037; RRD: RRID:AB_2621662 |
| PE-Cy5 mouse anti-human IgG | BD | Cat#551497; RRD: RRID: AB_394220 |
| BV711 mouse anti-human CD19 | BD | Cat#563036; RRID: AB_2737968 |
| FITC mouse anti-human IgG | BD | Cat#555786; RRID: AB_396121 |
| Anti-HIV-1 gp120 monoclonal (VRC01) | NIH AIDS Reagent Program | Cat#12033; RRID: AB_2491019 |
| CR3022 | Yuan et al.28 | N/A |
| Monoclonal anti-SARS-CoV 240CD | BEI Resources | NR-616 |
| Goat anti-human IgG (Fc specific) peroxidase | Sigma-Aldrich | Cat#A0170; RRID: AB_257868 |
| DENV-2D22 | James Crowe Jr. | N/A |
| Anti-VEGF | Baca et al.44 | Avastin (Bevacizumab) |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-1 | This paper | GenBank: MZ126644, MZ126659 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-2 | This paper | GenBank: MZ126645, MZ126660 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-3 | This paper | GenBank: MZ126646, MZ126661 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-4 | This paper | GenBank: MZ126647, MZ126662 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-6 | This paper | GenBank: MZ126649, MZ126664 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-12 | This paper | GenBank: MZ126655, MZ126670 |
| Monoclonal 46472-5 | This paper | GenBank: MZ126648, MZ126663 |
| Monoclonal 46472-7 | This paper | GenBank: MZ126650, MZ126665 |
| Monoclonal 46472-8 | This paper | GenBank: MZ126651, MZ126666 |
| Monoclonal 46472-9 | This paper | GenBank: MZ126652, MZ126667 |
| Monoclonal 46472-10 | This paper | GenBank: MZ126653, MZ126668 |
| Monoclonal 46472-11 | This paper | GenBank: MZ126654, MZ126669 |
| Monoclonal 46472-13 | This paper | GenBank: MZ126656, MZ126671 |
| Monoclonal 46472-14 | This paper | GenBank: MZ126657, MZ126672 |
| Monoclonal 46472-15 | This paper | GenBank: MZ126658, MZ126673 |
| Goat IgG anti-guinea pig complement C3, polyclonal antibody, FITC | MP Biomedicals | Cat# 11499934 |
| Palivizumab | MedImmmune | N/A |
| Goat anti-human IgG antibody (peroxidase) | Jackson ImmunoResearch | Cat#109-035-088; RRID:AB_2337584 |
| Avi Tag antibody, mAb, mouse | GenScript | Cat#A01738-100 |
| Monoclonal anti-influenza 3602-1707 | Setliff et al.27 | N/A |
| Monoclonal anti-MERS-CoV S1 | Sino Biological | Cat#40069-MM23; RRID: AB_2860454 |
| Monoclonal anti-MERS-CoV S2 | Sino Biological | Cat#40070-MM11; RRID: AB_2860457 |
| S230 | Walls et al.45 | N/A |
| 1F8 | Tang et al.46 | N/A |
| COV2-2196 | Zost et al.11 | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 | N. Thornburg | Centers for Disease Control and Prevention |
| SARS-CoV-2 (full length, Seattle Washington isolate) | Ralph Baric | N/A |
| SARS-CoV (full length, Urbani isolate) | Ralph Baric | N/A |
| SARS-CoV-2 MA10 | Ralph Baric | Dinnon et al.30 |
| Biological samples | ||
| PBMC from donor | Barney Graham | N/A |
| Low-tox guinea pig complement | Cedarlane | Cat #CL4051 |
| Chemicals, peptides, and recombinant proteins | ||
| CZA97.SOSIP.664 gp140 trimer | Ivelin Georgiev | N/A |
| ZM197.SOSIP.664 gp140 trimer | Ivelin Georgiev | N/A |
| SARS-CoV-2 S (S-2P) | PMID: 32075877 | N/A |
| SARS-CoV-2 S (Hexapro) | PMID: 32703906 | N/A |
| SARS-CoV S (S-2P) | PMID: 28807998 | N/A |
| MERS-CoV S (S-2P) | PMID: 28807998 | N/A |
| HCoV-OC43 S (S-2P) | McLellan Lab | N/A |
| HCoV-HKU1 S (S-2P) | PMID: 28807998 | N/A |
| SARS-CoV-2 S1 | Sino Biological | Cat#40591-V08H |
| SARS-CoV-2 S1 D614G | Sino Biological | Cat#40591-V08H3 |
| SARS-CoV-2 S2 | Sino Biological | Cat#40590-V08B |
| SARS-CoV-2 RBD | Sino Biological | Cat#40592-VNAH |
| SARS-CoV RBD | Sino Biological | Cat #40150-V08B2 |
| SARS-CoV-2 NTD | Sino Biological | Cat#40591-V49H |
| ACE2 | Sigma Aldrich | SAE0064-5UG |
| SARS-CoV-2 S HexaPro N165A | This paper | N/A |
| SARS-CoV-2 S HexaPro N709A | This paper | N/A |
| SARS-CoV-2 RBD-SD1 | PMID: 32075877 | N/A |
| MERS-CoV S1 Fd | PMID: 28807998 | N/A |
| Streptavidin HRP | Thermo Fisher Scientific | Cat#ENN100 |
| Streptavidin R-phycoerythrin (SA-PE) | Invitrogen | Cat#S866 |
| Carboxyfluorescein diacetate N-succinimidyl ester (CFSE) | Sigma-Aldrich | Cat#21888 |
| Protein A resin | GenScript | Cat# L00210 |
| Polyethyleniminine linear MW 25000 | Polysciences | Cat#23966-1 |
| Ghost dye red 780 | Tonbo biosciences | Cat#13-0865 |
| Live/Dead fixable aqua dead cell stain kit | Thermo Fisher Scientific | Cat#L34957 |
| Critical commercial assays | ||
| FluoSpheres neutrAvidin-labeled microspheres, 1.0 μm, yellow-green fluorescent | Thermo Fisher Scientific | Cat#F8776 |
| FluoSpheres NeutrAvidin-labeled microspheres, 1.0 μm, red fluorescent | Thermo Fisher Scientific | Cat#F8775 |
| Biacore X100 Sensorchip NTA | GE Healthcare | Cat# BR1004 |
| EZ link Sulfo-NHS-LC-biotin | Thermo Fisher Scientific | Cat#21327 |
| BirA-500: Bir A biotin-protein ligase standard reaction kit | Avidity | Cat# BirA500 |
| Solulink protein-oligonucleotide conjugation kit | TriLink Biotechnologies | Cat# S-9011 |
| B cell single cell V(D)J solution | 10X Genomics | N/A |
| AtheNA Multi-Lyte® ANA-II plus test kit | Zeus Scientific, Inc | Cat # A21101 |
| Deposited data | ||
| 46472 heavy chain sequences | This paper | GenBank: MZ126644- MZ126658 |
| 46472 light chain sequences | This paper | GenBank: MZ126659- MZ126673 |
| Raw next-generation sequencing data | This paper | SRA: PRJNA727275 |
| Experimental models: cell lines | ||
| Human THP-1 cells | AIDS Reagent Program | Cat#9942 |
| Human: Freestyle 293F cells | ThermoFisher Scientific | Cat#A14528 |
| Human: Expi293F cells | ThermoFisher Scientific | Cat#A14527 |
| ExpiCHO cells | Thermo Fisher Scientific | Cat#A29127 |
| Vero E6 USAMRID | N/A | N/A |
| HEK293T cells | Dr. George Shaw | N/A |
| Experimental models: organisms/strains | ||
| BALB/cAnHsd mice | Envigo | Stock number 047 |
| Oligonucleotides | ||
| Oligonucleotides for protein DNA-barcoding | Setliff et al.27 | N/A |
| Software and algorithms | ||
| Cell Ranger | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| HighV-Quest | Alamyar et al.47 | http://www.imgt.org/IMGTindex/IMGTHighV-QUEST.php |
| ChangeO | Gupta et al.48 | https://changeo.readthedocs.io/en/stable/ |
| Geneious 11.1.5 | https://www.geneious.com | N/A |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.3.5 Schrödinger, LLC | N/A |
| Flowjo v10 | TreeStar | https://www.flowjo.com/ |
| GraphPad Prism 8.0.0 | https://www.graphpad.com | N/A |
| Biacore X100 Evaluation Software | GE Healthcare | V2.0.1 |
| AtheNA Software | N/A | N/A |
| MUSCLE | Madeira et al.49 | N/A |
| BLOSUM-62 | Henikoff et al.50 | N/A |
| Other | ||
| Galanthus nivalis lectin | Vector Laboratories | Cat # AL-1243-5 |
| 1-Step Ultra TMB-ELISA substrate solution | ThermoFisher Scientific | Cat#34029 |
| Costar high binding microplates | Corning | Cat#9018 |
| D-(+)-Mannose | Sigma-Aldrich | Cat# M6020 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Ivelin Georgiev (Ivelin.Georgiev@Vanderbilt.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement. Please direct resource and reagent requests to the Lead Contact specified above, Ivelin Georgiev.
Data and code availability
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers MZ126644-MZ126658 (heavy chain) and MZ126659-MZ126673 (light chain). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number PRJNA727275. Custom scripts used to analyze data in this manuscript are available upon request to the corresponding author.
Experimental model and subject details
Human subjects
The donor had prior SARS-CoV infection during the 2004 outbreak in Hong Kong, and the PBMC sample was collected over 10 years post infection (20 million PBMCs). Additional information about the donor is not available.
Cell lines
A variety of cell lines were utilized for various assays in this study.
Expi293F mammalian cells (ThermoFisher) were maintained in FreeStyle F17 expression medium supplemented at final concentrations of 0.1% Pluronic Acid F-68 and 20% 4mM L-Glutamine. These cells were cultured at 37°C with 8% CO2 saturation and shaking.
FreeStyle293F cells were grown while shaking at 37 C in 8% CO2 and 80% humidity. Freestyle293F cells are derived from female human embryonic kidney epithelial cells.
THP-1 cells obtained from the AIDS Reagent Program (Division of AIDS, NIAID, NIH contributed by Dr. Li Wu and Vineet N. KewalRamani) were used for both the ADCP and ADCT assays. Cells were cultured at 37°C, 5% CO2 in RPMI containing 10% heat-inactivated fetal bovine serum (GIBCO, Gaithersburg, MD), 1% Penicillin Streptomycin (GIBCO, Gaithersburg, MD) and 2-mercaptoethanol to a final concentration of 0.05 mM. These cells were not allowed to exceed 4 × 105 cells/ml to prevent differentiation and are from a male donor.
HEK293T cells were obtained from Dr George Shaw and were used for the ADCT assay. These adherent cell lines were cultured at 37°C, 5% CO2, in DMEM containing 10% heat-inactivated fetal bovine serum (GIBCO BRL Life Technologies) and supplemented with 50 μg/ml gentamicin (Sigma). Cells were disrupted at confluence with 0.25% trypsin in 1 mM EDTA (Sigma) every 48–72 hours. HEK293F suspension cells were cultured in 293Freestyle media (GIBCO BRL Life Technologies) and grown in a shaking incubator at 37°C, 5% CO2, 70% humidity at 125rpm. Cells were diluted twice a week to between 0.2 and 0.5 million cells/ml. Both HEK293 derived cell lines are from female donors.
Murine model
12-month old female BALB/c mice (BALB/cAnHsd; Envigo, stock number 047) were used in a murine infection model for SARS-CoV-2 with a mouse adapted strain.
Eleven to twelve-month old female BALB/c mice (BALB/c AnNHsd, Envigo, stock# 047) were used for mouse-adapted SARS-CoV-2 (SARS-CoV-2 MA10) in vivo protection experiments as described previously.31 All mouse studies were performed at the University of North Carolina (Animal Welfare Assurance #A3410-01) using protocols (19-168) approved by the UNC Institutional Animal Care and Use Committee (IACUC) and were performed in a BSL3 facility at UNC.
Method details
Antigen purification
A variety of recombinant soluble protein antigens were used in the LIBRA-seq experiment and other experimental assays.
Plasmids encoding residues 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 986 and 987, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 S-2P); residues 1-1190 of the SARS-CoV spike with proline substitutions at positions 968 and 969, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV S-2P); residues 1-1291 of the MERS-CoV spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1060 and 1061, and a C-terminal T4-fibritin trimerization motif, an AviTag, an 8x HisTag, and a TwinStrepTag (MERS-CoV S-2P Avi); residues 1-751 of the MERS-CoV spike with a C-terminal T4-fibritin trimerization motif, 8x HisTag, and a TwinStrepTag (MERS-CoV S1); residues 1-1277 of the HCoV-HKU1 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1067 and 1068, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-HKU1 S-2P); residues 1-1278 of the HCoV-OC43 spike with proline substitutions at positions 1070 and 1071, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-OC43 S-2P); or residues 319–591 of SARS-CoV-2 S with a C-terminal monomeric human IgG Fc-tag and an 8x HisTag (SARS-CoV-2 RBD-SD1) were transiently transfected into FreeStyle293F cells (Thermo Fisher) using polyethylenimine. The coronavirus trimer spike antigens were in a prefusion-stabilized (S-2P) conformation that better represents neutralization-sensitive epitopes in comparison to their wild-type forms51. Two hours post-transfection, cells were treated with kifunensine to ensure uniform glycosylation. Transfected supernatants were harvested after 6 days of expression. SARS-CoV-2 RBD-SD1 was purified using Protein A resin (Pierce), SARS-CoV-2 S-2P, SARS-CoV S-2P, MERS-CoV S-2P Avi, MERS-CoV S1, HCoV-HKU1 S-2P and HCoV-OC43 S-2P were purified using StrepTactin resin (IBA). Affinity-purified SARS-CoV-2 RBD-SD1 was further purified over a Superdex75 column (GE Life Sciences). MERS-CoV S1 was purified over a Superdex200 Increase column (GE Life Sciences). SARS-CoV-2 S-2P, SARS-CoV S-2P, MERS-CoV S-2P Avi, HCoV-HKU1 S-2P and HCoV-OC43 S-2P were purified over a Superose6 Increase column (GE Life Sciences).
For the HIV-1 gp140 SOSIP variant from strain ZM197 (clade C) and CZA97 (clade C), recombinant, soluble antigens contained an AviTag and were expressed in Expi293F cells using polyethylenimine transfection reagent and cultured. FreeStyle F17 expression medium supplemented with pluronic acid and glutamine was used. The cells were cultured at 37°C with 8% CO2 saturation and shaking. After 5-7 days, cultures were centrifuged and supernatant was filtered and run over an affinity column of agarose bound Galanthus nivalis lectin. The column was washed with PBS and antigens were eluted with 30 mL of 1M methyl-a-D-mannopyranoside. Protein elutions were buffer exchanged into PBS, concentrated, and run on a Superdex 200 Increase 10/300 GL Sizing column on the AKTA FPLC system. Fractions corresponding to correctly folded protein were collected, analyzed by SDS-PAGE and antigenicity was characterized by ELISA using known monoclonal antibodies specific to each antigen. Avi-tagged antigens were biotinylated using BirA biotin ligase (Avidity LLC). Non-Avi-tagged antigens were biotinylated using the EZ-Link Sulfo-NHS-Biotin kits using a 50:1 biotin to protein molar ratio.
For binding studies, SARS-CoV-2 HexaPro S, SARS-CoV S, SARS-CoV-2 RBD, SARS-CoV RBD, and MERS-CoV RBD constructs were expressed in the transient expression system previously mentioned. S proteins were purified using StrepTrap HP columns and RBD constructs were purified over protein A resin, respectively. Each resulting protein was further purified to homogeneity by size-exclusion chromatography on a Superose 6 10/300 GL column.
SARS-CoV-2 S1, SARS-CoV-2 S1 D614G, SARS-CoV-2 S2, and SARS-CoV-2 NTD truncated proteins were purchased from the commercial vendor, Sino Biological.
DNA-barcoding of antigens
We used oligos that possess 15 bp antigen barcode, a sequence capable of annealing to the template switch oligo that is part of the 10X bead-delivered oligos, and contain truncated TruSeq small RNA read 1 sequences in the following structure: 5′-CCTTGGCACCCGAGAATTCCANNNNNNNNNNNNNCCCATATAAGA∗A∗A-3′, where Ns represent the antigen barcode. We used the following antigen barcodes: GCTCCTTTACACGTA (SARS-CoV-2 S), TGACCTTCCTCTCCT (SARS-CoV S), ACAATTTGTCTGCGA (MERS-CoV S), TCCTTTCCTGATAGG (MERS-CoV S1), CAGGTCCCTTATTTC (HCoV-HKU1 S), TAACTCAGGGCCTAT (HCoV-OC43 S), CAGCCCACTGCAATA (CZA97), and ATCGTCGAGAGCTAG (ZM197). Oligos were ordered from IDT with a 5′ amino modification and HPLC purified.
For each antigen, a unique DNA barcode was directly conjugated to the antigen itself. In particular, 5′amino-oligonucleotides were conjugated directly to each antigen using the Solulink Protein-Oligonucleotide Conjugation Kit (TriLink cat no. S-9011) according to manufacturer’s instructions. Briefly, the oligo and protein were desalted, and then the amino-oligo was modified with the 4FB crosslinker, and the biotinylated antigen protein was modified with S-HyNic. Then, the 4FB-oligo and the HyNic-antigen were mixed together. This causes a stable bond to form between the protein and the oligonucleotide. The concentration of the antigen-oligo conjugates was determined by a BCA assay, and the HyNic molar substitution ratio of the antigen-oligo conjugates was analyzed using the NanoDrop according to the Solulink protocol guidelines. AKTA FPLC was used to remove excess oligonucleotide from the protein-oligo conjugates, which were also verified using SDS-PAGE with a silver stain. Antigen-oligo conjugates were also used in flow cytometry titration experiments.
Antigen specific B cell sorting
Cells were stained and mixed with DNA-barcoded antigens and other antibodies, and then sorted using fluorescence activated cell sorting (FACS). First, cells were counted and viability was assessed using Trypan Blue. Then, cells were washed three times with DPBS supplemented with 0.1% bovine serum albumin (BSA). Cells were resuspended in DPBS-BSA and stained with cell markers including viability dye (Ghost Red 780), CD14-APC-Cy7, CD3-FITC, CD19-BV711, and IgG-PE-Cy5. Additionally, antigen-oligo conjugates were added to the stain. After staining in the dark for 30 minutes at room temperature, cells were washed three times with DPBS-BSA at 300 g for five minutes. Cells were then incubated for 15 minutes at room temperature with Streptavidin-PE to label cells with bound antigen. Cells were washed three times with DPBS-BSA, resuspended in DPBS, and sorted by FACS. Antigen positive cells were bulk sorted and delivered to the Vanderbilt Technologies for Advanced Genomics (VANTAGE) sequencing core at an appropriate target concentration for 10X Genomics library preparation and subsequent sequencing. FACS data were analyzed using FlowJo.
Sample and library preparation, and sequencing
Single-cell suspensions were loaded onto the Chromium Controller microfluidics device (10X Genomics) and processed using the B cell Single Cell V(D)J solution according to manufacturer’s suggestions for a target capture of 10,000 B cells per 1/8 10X cassette, with minor modifications in order to intercept, amplify and purify the antigen barcode libraries as previously described.27
Sequence processing and bioinformatic analysis
We utilized and modified our previously described pipeline to use paired-end FASTQ files of oligo libraries as input, process and annotate reads for cell barcode, UMI, and antigen barcode, and generate a cell barcode - antigen barcode UMI count matrix.27 BCR contigs were processed using Cell Ranger (10X Genomics) using GRCh38 as reference. Antigen barcode libraries were also processed using Cell Ranger (10X Genomics). The overlapping cell barcodes between the two libraries were used as the basis of the subsequent analysis. We removed cell barcodes that had only non-functional heavy chain sequences as well as cells with multiple functional heavy chain sequences and/or multiple functional light chain sequences, reasoning that these may be multiplets. Additionally, we aligned the BCR contigs (filtered_contigs.fasta file output by Cell Ranger, 10X Genomics) to IMGT reference genes using HighV-Quest.47 The output of HighV-Quest was parsed using ChangeO48 and merged with an antigen barcode UMI count matrix. Finally, we determined the LIBRA-seq score for each antigen in the library for every cell as previously described.27
Antibody expression and purification
For each antibody, variable genes were inserted into custom plasmids encoding the constant region for the IgG1 heavy chain as well as respective lambda and kappa light chains (pTwist CMV BetaGlobin WPRE Neo vector, Twist Bioscience). Antibodies were expressed in Expi293F mammalian cells (ThermoFisher) by co-transfecting heavy chain and light chain expressing plasmids using polyethylenimine transfection reagent and cultured for 5-7 days. Cells were maintained in FreeStyle F17 expression medium supplemented at final concentrations of 0.1% Pluronic Acid F-68 and 20% 4mM L-Glutamine. These cells were cultured at 37°C with 8% CO2 saturation and shaking. After transfection and 5-7 days of culture, cell cultures were centrifuged and supernatant was 0.45 μm filtered with Nalgene Rapid Flow Disposable Filter Units with PES membrane. Filtered supernatant was run over a column containing Protein A agarose resin equilibrated with PBS. The column was washed with PBS, and then antibodies were eluted with 100 mM Glycine HCl at 2.7 pH directly into a 1:10 volume of 1M Tris-HCl pH 8.0. Eluted antibodies were buffer exchanged into PBS 3 times using Amicon Ultra centrifugal filter units and concentrated. Antibodies were analyzed by SDS-PAGE. Additionally, antibodies 46472-1, 46472-2, 46472-3, 46472-4, 46472-6 and 46472-12 were assessed by size exclusion chromatography on a Superdex 200 Increase 10/300 GL Sizing column with the AKTA FPLC system.
High-throughput antibody expression
For high-throughput production of recombinant antibodies, approaches were used that are designated as microscale. For antibody expression, microscale transfection were performed (∼1cml per antibody) of CHO cell cultures using the GIBCO ExpiCHO Expression System and a protocol for deep 96-well blocks (Thermo Fisher Scientific). In brief, synthesized antibody-encoding DNA (∼2cμg per transfection) was added to OptiPro serum free medium (OptiPro SFM), incubated with ExpiFectamine CHO Reagent and added to 800cμl of ExpiCHO cell cultures into 96-deep-well blocks using a ViaFlo 384 liquid handler (Integra Biosciences). The plates were incubated on an orbital shaker at 1,000crpm with an orbital diameter of 3cmm at 37c°C in 8% CO2. The next day after transfection, ExpiFectamine CHO Enhancer and ExpiCHO Feed reagents (Thermo Fisher Scientific) were added to the cells, followed by 4cd incubation for a total of 5cd at 37c°C in 8% CO2. Culture supernatants were collected after centrifuging the blocks at 450g for 5cmin and were stored at 4°C until use. For high-throughput microscale antibody purification, fritted deep-well plates were used containing 25cμl of settled protein G resin (GE Healthcare Life Sciences) per well. Clarified culture supernatants were incubated with protein G resin for mAb capturing, washed with PBS using a 96-well plate manifold base (QIAGEN) connected to the vacuum and eluted into 96-well PCR plates using 86cμl of 0.1cM glycine-HCL buffer pHc2.7. After neutralization with 14cμl of 1cM Tris-HCl pHc8.0, purified mAbs were buffer-exchanged into PBS using Zeba Spin Desalting Plates (Thermo Fisher Scientific) and stored at 4°C until use.
ELISA
To assess antibody binding, soluble protein was plated at 2 μg/ml overnight at 4°C. The next day, plates were washed three times with PBS supplemented with 0.05% Tween-20 (PBS-T) and coated with 5% milk powder in PBS-T. Plates were incubated for one hour at room temperature and then washed three times with PBS-T. Primary antibodies were diluted in 1% milk in PBS-T, starting at 10 μg/ml with a serial 1:5 dilution and then added to the plate. The plates were incubated at room temperature for one hour and then washed three times in PBS-T. The secondary antibody, goat anti-human IgG conjugated to peroxidase, was added at 1:10,000 dilution in 1% milk in PBS-T to the plates, which were incubated for one hour at room temperature. Goat anti-mouse secondary was used for SARS-CoV specific control antibody 240CD (BEI Resources). Plates were washed three times with PBS-T and then developed by adding TMB substrate to each well. The plates were incubated at room temperature for ten minutes, and then 1N sulfuric acid was added to stop the reaction. Plates were read at 450 nm.
Data are represented as mean ± SEM for one ELISA experiment. ELISAs were repeated 2 or more times. The area under the curve (AUC) was calculated using GraphPad Prism 8.0.0. For antibody 240CD, the following reagent was obtained through BEI Resources, NIAID, NIH: Monoclonal Anti-SARS-CoV S Protein (Similar to 240C), NR-616.
Competition ELISA
Competition ELISAs were performed as described above, with some modifications. After coating with antigen and blocking, 25 μL of non-biotinylated competitor antibody was added to each well at 10 μg/ml and incubated at 37°C for 10 minutes. Then, without washing, 75 μL biotinylated antibody (final concentration of 0.1 μg/ml) was added and incubated at 37°C for 1 hour. After washing three times with PBS-T, streptavidin-HRP was added at 1:10,000 dilution in 1% milk in PBS-T and incubated for 1 hour at room temperature. Plates were washed and substrate and sulfuric acid were added as described above. ELISAs were repeated at least 2 times. Data is shown as the % decrease in binding.
Autoreactivity
Monoclonal antibody reactivity to nine autoantigens (SSA/Ro, SS-B/La, Sm, ribonucleoprotein (RNP), Scl 70, Jo-1, dsDNA, centromere B, and histone) was measured using the AtheNA Multi-Lyte® ANA-II Plus test kit (Zeus scientific, Inc, #A21101). Antibodies were incubated with AtheNA beads for 30min at concentrations of 50, 25, 12.5 and 6.25 μg/mL. Beads were washed, incubated with secondary and read on the Luminex platform as specified in the kit protocol. Data were analyzed using AtheNA software. Positive (+) specimens received a score > 120, and negative (-) specimens received a score < 100. Samples between 100-120 were considered indeterminate.
Mannose competition
Mannose competition ELISAs were performed as described above with minor modifications. After antigen coating and washing, nonspecific binding was blocked by incubation with 5% FBS diluted in PBS for 1 hour at RT. Primary antibodies were diluted in 5% FBS-PBST ± 1M D-(+)-Mannose starting at 10 μg/ml with a serial 1:5 dilution and then added to the plate for 1 hour at RT. After washing, antibody binding was detected with goat anti-human IgG conjugated to peroxidase and added at 1:10,000 dilution in 5% FBS in PBS-T to the plates. After 1 hour incubation, plates were washed and substrate and sulfuric acid were added as described above. Data shown is representative of three replicates.
Epitope mapping visualization
SARS-CoV-2 Spike (PDB-6VSB) was visualized using PyMOL software. Antibody epitopes were visualized on the SARS-CoV-2 spike using a structure of the pre-fusion stabilized SARS-CoV-2 S-2P construct5 modeled in the molecular graphics software PyMOL (The PyMOL Molecular Graphics System, Version 2.3.5 Schrödinger, LLC).
RTCA neutralization assay
To assess for neutralizing activity against SARS-CoV-2 strain 2019 n-CoV/USA_WA1/2020 (obtained from the Centers for Disease Control and Prevention, a gift from N. Thornburg), we used the high-throughput RTCA assay and xCelligence RTCA HT Analyzer (ACEA Biosciences) that has been described previously.11 After obtaining a background reading of a 384-well E-plate, 6,000 Vero-furin cells52 were seeded per well. Sensograms were visualized using RTCA HT software version 1.0.1 (ACEA Biosciences). One day later, equal volumes of virus were added to antibody samples and incubated for 1ch at 37°C in 5%cCO2. mAbs were tested in triplicate with a single (1:20) dilution. Virus–mAb mixtures were then added to Vero-furin cells in 384-well E-plates. Controls were included that had Vero-furin cells with virus only (no mAb) and media only (no virus or mAb). E-plates were read every 8–12 h for 72 h to monitor virus neutralization. At 32 h after virus-mAb mixtures were added to the E-plates, cell index values of antibody samples were compared to those of virus only and media only to determine presence of neutralization.
Nano-luciferase neutralization assay
A full-length SARS-CoV-2 virus based on the Seattle Washington isolate and a full-length SARS-CoV virus based on the Urbani isolate were designed to express luciferase and was recovered via reverse genetics and described previously.53,54 Viruses were titered in Vero E6 USAMRID cells to obtain a relative light units (RLU) signal of at least 10X the cell only control background. Vero E6 USAMRID cells were plated at 20,000 cells per well the day prior in clear bottom black walled 96-well plates (Corning 3904). Neutralizing antibody serum samples were tested at a starting dilution of 1:40 and were serially diluted 4-fold up to eight dilution spots. Antibody-virus complexes were incubated at 37C with 5% CO2 for 1 hour. Following incubation, growth media was removed and virus-antibody dilution complexes were added to the cells in duplicate. Virus-only controls and cell-only controls were included in each neutralization assay plate. Following infection, plates were incubated at 37C with 5% CO2 for 48 hours. After the 48 hour incubation, cells were lysed and luciferase activity was measured via Nano-Glo Luciferase Assay System (Promega) according to the manufacturer specifications. SARS-CoV and SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction in RLU was observed relative to the average of the virus control wells.
SPR
His-tagged SARS-CoV-2 RBD-SD1 was immobilized to a NiNTA sensorchip to a level of ∼150 RUs using a Biacore X100. Serial dilutions of purified Fab 46472-12 were evaluated for binding, ranging in concentration from 1 to 0.25 μM. The resulting data were fit to a 1:1 binding model using Biacore Evaluation Software.
Fc effector function assays
Antibody-dependent cellular phagocytosis (ADCP)
Antibody-dependent cellular phagocytosis (ADCP) was performed using biotinylated SARS-CoV-2 or SARS-CoV S coated fluorescent neutravidin beads as previously described.55 Briefly, beads were incubated for two hours with antibodies at a starting concentration of 50 μg/ml and titrated five fold. CR3022 was used as a positive control while Palivizumab was used as a negative control. Antibodies and beads were incubated with THP-1 cells overnight, fixed and interrogated on the FACSAria II. Phagocytosis score was calculated as the percentage of THP-1 cells that engulfed fluorescent beads multiplied by the geometric mean fluorescence intensity of the population in the FITC channel less the no antibody control.
Antibody-dependent cellular trogocytosis (ADCT)
ADCT was performed as described in and modified from a previously described study.29 HEK293T cells transfected with a SARS-CoV-2 spike pcDNA vector were surface biotinylated with EZ-Link Sulfo-NHS-LC-Biotin as recommended by the manufacturer. Fifty-thousand cells per well were incubated with antibody for 30 minutes starting at 25 μg/ml and titrated 5 fold. CR3022 was used as a positive control with Palivizumab as a negative. Following a RPMI media wash, these were then incubated with carboxyfluorescein succinimidyl ester (CFSE) stained THP-1 cells (5 X104 cells per well) for 1 hour and washed with 15mM EDTA/PBS followed by PBS. Cells were then stained for biotin using Streptavidin-PE and read on a FACSAria II. Trogocytosis score was determined as the proportion of CFSE positive THP-1 cells also positive for streptavidin-PE less the no antibody control.
Antibody-dependent complement deposition (ADCD)
Antibody-dependent complement deposition was performed as previously described.56 Briefly biotinylated SARS-Cov-2 S protein was coated 1:1 onto fluorescent neutravidin beads for 2 hours at 37 degrees. These beads were incubated with 100ug/ml of antibody for 1 hour and incubated with guinea pig complement diluted 1 in 50 with gelatin/veronal buffer for 15 minutes at 37 degrees. Beads were washed at 2000 g twice in PBS and stained with anti-guinea pig C3b-FITC, fixed and interrogated on a FACSAria II. Complement deposition score was calculated as the percentage of C3b-FITC positive beads multiplied by the geometric mean fluorescent intensity of FITC in this population less the no antibody or heat inactivated controls.
Antibody prophylaxis - murine model of infection
For evaluating the prophylactic efficacy of mAbs, 12-month old female BALB/c mice (BALB/cAnHsd; Envigo, stock number 047) were treated with 200 μg mAb intraperitoneally (i.p.) 12 hours prior to virus inoculation. The next day, mice were administered intranasally with 1x103 PFU or 1x104PFU of SARS-CoV-2 MA10, respectively. Mice were monitored daily for weight loss, morbidity, and mortality, and after four days, mice were sacrificed and lung tissue was harvested for viral titer as measured by plaque assays. One lung lobe was taken for pathological analysis and the other lobe was processed for qPCR and viral load determination as previously described.31 For viral plaque assays, the caudal lobe of the right lung was homogenized in PBS, and the tissue homogenate was then serial-diluted onto confluent monolayers of Vero E6 cells, followed by agarose overlay. Plaques were visualized with overlay of Neutral Red dye on day 2 post infection. Gross pulmonary hemorrhage was observed at time of tissue harvest and scored on a scale of 0 (no hemorrhage in any lobe, normal pink healthy lung) to 4 (complete hemorrhage in all lobes of the lung, completely dark red lung).
For viral titer and hemorrhage score comparisons, an ordinary one-way ANOVA test with multiple comparisons was performed using Prism software, GraphPad Prism version 8.0.
ACE2 binding inhibition assay
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S-2P ectoprotein at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS-T for 1 hr. Purified mAbs were diluted two-fold in blocking buffer starting from 10 μg/mL in triplicate, added to the wells (20 μL/well), and incubated at ambient temperature. Recombinant human ACE2 with a C-terminal FLAG tag protein was added to wells at 2 μg/mL in a 5 μL/well volume (final 0.4 μg/mL concentration of ACE2) without washing of antibody and then incubated for 40 min at ambient temperature. Plates were washed, and bound ACE2 was detected using HRP-conjugated anti-FLAG antibody and TMB substrate. ACE2 binding without antibody served as a control. Experiment was done in biological replicate and technical triplicates, shown is representative of one replicate with positive control mAb COV2-2196.11
Identification of residue-level mutants
Potential cross-reactive epitopes were identified based on sequence and structural homology. Reference sequences for each Coronavirus S were obtained either from NCBI for SARS-CoV-2 (YP_009724390.1) and MERS-CoV (YP_009047204.1) or from Uniprot for SARS-CoV (P59594) of the spikes was then obtained using MUSCLE49 and the amino acid similarity to SARS-CoV-2 at each residue position was calculated using the BLOSUM-62 scoring matrix.50 These scores were then used to color each residue position on the SARS-CoV-2 S structure (PDB ID: 6VSB) in PyMOL (Schrodinger, version 2.3.5) in order to visualize surface patches and linear epitopes with structural homology. These conserved regions were then visualized on the other human coronavirus spike structures by retrieving them from the Protein Databank (SARS-CoV: 5X5B, MERS-CoV: 5W9I) and aligning them to the SARS-CoV-2 S structure. Finally, the residue N165 was part of a conserved surface patches and was mutated to alanine and tested for binding with antibodies. The N709A mutant tested was previously described in Acharya et al., BioRxiv (2020).
Quantification and statistical analysis
ELISA error bars (standard error of the mean) were calculated using GraphPad Prism version 8.0.0. ANOVA analysis (ordinary one way ANOVA with multiple comparisons) was performed on viral load titers and hemorrhage scores from animal experiments using GraphPad Prism version 8.0.0. Details of the statistical analyses can be found in the main text and figure captions.
Acknowledgments
We thank Angela Jones, Latha Raju, and Jamie Roberson of Vanderbilt Technologies for Advanced Genomics for their expertise regarding next-generation sequencing (NGS) and library preparation; David Flaherty and Brittany Matlock of the Vanderbilt Flow Cytometry Shared Resource for help with flow panel optimization; and members of the Georgiev laboratory for comments on the manuscript. The Vanderbilt VANTAGE Core provided technical assistance for this work. VANTAGE is supported in part by CTSA grant 5UL1 RR024975-03, the Vanderbilt-Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956). This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University (Nashville, TN). Flow cytometry experiments were performed in the VUMC Flow Cytometry Shared Resource. The VUMC Flow Cytometry Shared Resource is supported by the Vanderbilt-Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). For work described in this manuscript, I.S.G., A.R.S., K.J.K., S.W., K.A.P., R.V., N.R., E.F.F., and C.M.H. were supported in part by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Disease (NIAID) award R01AI131722-S1, the Hays Foundation COVID-19 Research Fund, Fast Grants, and CTSA award no. UL1 TR002243 from the National Center for Advancing Translational Sciences. J.S.M and D.W. were supported in part by a NIH/NIAID grant awarded to J.S.M. (R01-AI127521). L.M. and S.I.R. acknowledge research funding from the South African Medical Research Council (MRC) Extramural Unit and SHIP-COVID19 programs and an H3 Africa grant (U01A136677). S.I.R. is supported by the South African Research Chairs Initiative of the Department of Science and Technology and the NRF (grant no. 98341) and is a L’Oreal/UNESCO South Africa Young Talents Award recipient. R.S.B., A.S., and D.R.M. were supported by NIH grants (U54CA260543 and R01AI157155). P.A. and K.J. were supported by NIH grant R01 AI14567. J.E.C., R.H.C., N.S., R.S.N., and R.E.S. were supported by the Defense Advanced Research Projects Agency (DARPA) grants HR0011-18-2-0001 and HR00 11-18-3-0001; NIH contracts 75N93019C00074 and 75N93019C00062; NIH grants U01 AI150739, R01 AI130591, and R35 HL145242; the Dolly Parton COVID-19 Research Fund at Vanderbilt; and NIH grant S10 RR028106 for the Next Generation Nucleic Acid Sequencer, housed in VANTAGE. M.S.D. and R.E.C. were supported by grants from NIH (R01 AI157155) and the Defense Advanced Research Projects Agency (HR001117S0019). B.F.H. and R.P. were supported by NC State funding for COVID research. B.S.G. was supported by intramural funding from the NIAID. C.M.H. was supported in part by NIH grant T32 GM008320-30. D.R.M. was supported by NIH F32 AI152296, a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award, and was previously supported by NIH/NIAID T32 AI007151.
Author contributions
Methodology, A.R.S., K.J.K., and I.S.G.; investigation, A.R.S., K.J.K., D.W., S.I.R., A.S., S.W., N.W., K.J., K.A.P., R.V., R.P., N.P.M., N.R., E.F.F., C.M.H., N.S., R.E.C., D.R.M., R.S.N., R.E.S., J.E.L., B.S.G., M.S.D., B.F.H., P.A., R.H.C., J.E.C., R.S.B., L.M., J.S.M., and I.S.G.; software, A.R.S., R.V., and N.R.; validation, A.R.S. and K.J.K.; writing – original draft, A.R.S. and K.J.K.; writing – review & editing, all authors; funding acquisition, I.S.G., B.S.G., M.S.D., B.F.H., P.A., R.H.C., J.E.C., R.S.B., L.M., J.S.M., A.R.S., and K.J.K.; resources, B.S.G., M.S.D., B.F.H., P.A., R.H.C., J.E.C., R.S.B., L.M., J.S.M., and I.S.G.; supervision, I.S.G.
Declaration of interests
A.R.S. and I.S.G. are co-founders of AbSeek Bio. A.R.S., K.J.K., I.S.G., D.W., N.W., and J.S.M. are listed as inventors on patents filed describing the antibodies mentioned here. D.W., J.S.M., B.S.G., and N.W. are also listed as inventors on US patent application no. 62/972,886 (2019-nCoV vaccine). M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has unrelated sponsored research agreements from Emergent BioSolutions, Moderna, and Vir Biotechnology. J.E.C. has served as a consultant for Eli Lilly, GlaxoSmithKline, and Luna Biologics; is a member of the Scientific Advisory Boards of CompuVax and Meissa Vaccines; and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from IDBiologics and AstraZeneca. R.S.B. has competing interests associated with Eli Lily, Takeda Pharmaceuticals, and Pfizer. The Georgiev laboratory at Vanderbilt University Medical Center has received unrelated funding from Takeda Pharmaceuticals.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100313.
Supplemental information
References
- 1.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang S., Hillyer C., Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 8.Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 13.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen M.S. Monoclonal Antibodies to Disrupt Progression of Early Covid-19 Infection. N. Engl. J. Med. 2021;384:289–291. doi: 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H. Cross-reactive Antibody Response between SARS-CoV-2 and SARS-CoV Infections. Cell Rep. 2020;31:107725. doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasui F., Kohara M., Kitabatake M., Nishiwaki T., Fujii H., Tateno C., Yoneda M., Morita K., Matsushima K., Koyasu S., Kai C. Phagocytic cells contribute to the antibody-mediated elimination of pulmonary-infected SARS coronavirus. Virology. 2014;454-455:157–168. doi: 10.1016/j.virol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2021;218:e20201993. doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J. Distinct Early Serological Signatures Track with SARS-CoV-2 Survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loos C., Atyeo C., Fischinger S., Burke J., Slein M.D., Streeck H., Lauffenburger D., Ryan E.T., Charles R.C., Alter G. Evolution of Early SARS-CoV-2 and Cross-Coronavirus Immunity. MSphere. 2020;5:e00622. doi: 10.1128/mSphere.00622-20. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell. 2019;179:1636–1646.e15. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson S.I., Crowther C., Mkhize N.N., Morris L. Measuring the ability of HIV-specific antibodies to mediate trogocytosis. J. Immunol. Methods. 2018;463:71–83. doi: 10.1016/j.jim.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M. Compromised Humoral Functional Evolution Tracks with SARS-CoV-2 Mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atyeo C., Slein M.D., Fischinger S., Burke J., Schäfer A., Leist S.R., Kuzmina N.A., Mire C., Honko A., Johnson R. Dissecting strategies to tune the therapeutic potential of SARS-CoV-2-specific monoclonal antibody CR3022. JCI Insight. 2021;6:e143129. doi: 10.1172/jci.insight.143129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bournazos S., DiLillo D.J., Goff A.J., Glass P.J., Ravetch J.V. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. USA. 2019;116:20054–20062. doi: 10.1073/pnas.1911842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards C.E., Yount B.L., Graham R.L., Leist S.R., Hou Y.J., Dinnon K.H., 3rd, Sims A.C., Swanstrom J., Gully K., Scobey T.D. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E., Graham R.L., Scobey T., Plante J.A., Royal S.R. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. USA. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menachery V.D., Yount B.L., Jr., Debbink K., Agnihothram S., Gralinski L.E., Plante J.A., Graham R.L., Scobey T., Ge X.-Y., Donaldson E.F. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., Zhu H., Zhao W., Han Y., Qin C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baca M., Presta L.G., O’Connor S.J., Wells J.A. Antibody humanization using monovalent phage display. J. Biol. Chem. 1997;272:10678–10684. doi: 10.1074/jbc.272.16.10678. [DOI] [PubMed] [Google Scholar]
- 45.Walls A.C., Xiong X., Park Y.-J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang X.-C., Agnihothram S.S., Jiao Y., Stanhope J., Graham R.L., Peterson E.C., Avnir Y., Tallarico A.S.C., Sheehan J., Zhu Q. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc. Natl. Acad. Sci. USA. 2014;111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alamyar E., Duroux P., Lefranc M.-P., Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 48.Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee S., Sirohi D., Dowd K.A., Chen Z., Diamond M.S., Kuhn R.J., Pierson T.C. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scobey T., Yount B.L., Sims A.C., Donaldson E.F., Agnihothram S.S., Menachery V.D., Graham R.L., Swanstrom J., Bove P.F., Kim J.D. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2013;110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yount B., Curtis K.M., Fritz E.A., Hensley L.E., Jahrling P.B., Prentice E., Denison M.R., Geisbert T.W., Baric R.S. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2003;100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ackerman M.E., Moldt B., Wyatt R.T., Dugast A.-S., McAndrew E., Tsoukas S., Jost S., Berger C.T., Sciaranghella G., Liu Q. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. J. Immunol. Methods. 2011;366:8–19. doi: 10.1016/j.jim.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischinger S., Fallon J.K., Michell A.R., Broge T., Suscovich T.J., Streeck H., Alter G. A high-throughput, bead-based, antigen-specific assay to assess the ability of antibodies to induce complement activation. J. Immunol. Methods. 2019;473:112630. doi: 10.1016/j.jim.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers MZ126644-MZ126658 (heavy chain) and MZ126659-MZ126673 (light chain). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number PRJNA727275. Custom scripts used to analyze data in this manuscript are available upon request to the corresponding author.