Abstract
Telomere shortening is a hallmark of aging. Telomere length (TL) in blood cells has been studied extensively as a biomarker of human aging and disease; however, little is known regarding variability in TL in nonblood, disease-relevant tissue types. Here, we characterize variability in TLs from 6391 tissue samples, representing >20 tissue types and 952 individuals from the Genotype-Tissue Expression (GTEx) project. We describe differences across tissue types, positive correlation among tissue types, and associations with age and ancestry. We show that genetic variation affects TL in multiple tissue types and that TL may mediate the effect of age on gene expression. Our results provide the foundational knowledge regarding TL in healthy tissues that is needed to interpret epidemiological studies of TL and human health.
Graphical Abstract
TL in human tissues. Using a Luminex-based assay, TL was measured in DNA samples from >25 different human tissue types from 952 deceased donors in the GTEx project. TL within tissue types is determined by numerous factors, including zygotic TL, age, and exposures. TL differs across tissues and correlates among tissue types. TL in most tissues declines with age.
INTRODUCTION:
Telomeres are DNA-protein complexes located at the end of chromosomes that protect chromosome ends from degradation and fusion. The DNA component of telomeres shortens with each cell division, eventually triggering cellular senescence. Telomere length (TL) in blood cells has been studied extensively as a biomarker of human aging and risk factor for age-related diseases. The extent to which TL in whole blood reflects TL in disease-relevant tissue types is unknown, and the variability in TL across human tissues has not been well characterized. The postmortem tissue samples collected by the Genotype-Tissue Expression (GTEx) project provide an opportunity to study TL in many human tissue types, and accompanying data on inherited genetic variation, gene expression, and donor characteristics enable us to examine demographic, genetic, and biologic determinants and correlates of TL within and across tissue types.
RATIONALE:
To better understand variation in and determinants of TL, we measured relative TL (RTL, telomere repeat abundance in a DNA sample relative to a standard sample) in more than 25 tissue types from 952 GTEx donors (deceased, aged 20 to 70 years old). RTL was measured for 6391 unique tissue samples using a Luminex assay, generating the largest publicly available multitissue TL dataset. We integrated our RTL measurements with data on GTEx donor characteristics, inherited genetic variation, and tissue-specific expression and analyzed relationships between RTL and covariates using linear mixed models (across all tissues and within tissues). Through this analysis, we sought to accomplish four goals: (i) characterize sources of variation in TL, (ii) evaluate whole-blood TL as a proxy for TL in other tissue types, (iii) examine the relationship between age and TL across tissue types, and (iv) describe biological determinants and correlates of TL.
RESULTS:
Variation in RTL was attributable to tissue type, donor, and age and, to a lesser extent, race or ethnicity, smoking, and inherited variants known to affect leukocyte TL. RTLs were generally positively correlated among tissues, and whole-blood RTL was a proxy for RTL in most tissues. RTL varied across tissue types and was shortest in whole blood and longest in testis. RTL was inversely associated with age in most tissues, and this association was strongest for tissues with shorter average RTL. African ancestry was associated with longer RTL across all tissues and within specific tissue types, suggesting that ancestry-based differences in TL exist in germ cells and are transmitted to the zygote. A polygenic score consisting of inherited variants known to affect leukocyte TL was associated with RTL across all tissues, and several of these TL-associated variants affected expression of nearby genes in multiple tissue types. Carriers of rare, loss-of-function variants in TL-maintenance genes had shorter RTL (based on analysis of multiple tissue types), suggesting that these variants may contribute to shorter TL in individuals from the general population. Components of telomerase, a TL maintenance enzyme, were more highly expressed in testis than in any other tissue. We found evidence that RTL may mediate the effect of age on gene expression in human tissues.
CONCLUSION:
We have characterized the variability in TL across many human tissue types and the contributions of aging, ancestry, genetic variation, and other biologic processes to this variability. The correlation observed among TL measures from different tissues highlights the existence of host factors with effects on TL that are shared across tissue types (e.g., TL in the zygote). These results have important implications for the interpretation of epidemiologic studies of leukocyte TL and disease.
Telomeres are DNA-protein complexes located at the end of chromosomes that protect chromosome ends from degradation and fusion (1). The length of the DNA component of telomeres, a six-nucleotide repeat sequence, shortens as cells divide (2), with short telomeres eventually triggering cellular senescence (3, 4). In most human tissues, telomere length (TL) gradually shortens over time, and TL shortening is considered a hallmark (and a potential underlying cause) of human aging (5). In human studies, short TL measured in leukocytes is associated with increased risk of aging-related diseases, including cardiovascular disease (6) and type 2 diabetes (7), as well as overall mortality and human life span (8). However, long TL may increase the risks for some types of cancer (9–11). Leukocyte TL is influenced by inherited genetic variation [single-nucleotide polymorphisms (SNPs)], some of which reside near genes with known roles in telomere maintenance (12–15). Leukocyte TL is also associated with lifestyle factors (e.g., physical activity), health factors (e.g., obesity, cholesterol), and environmental exposures (e.g., cigarette smoking) (16, 17).
Epidemiologic studies of TL predominantly use blood (occasionally saliva) as a DNA source. Thus, our understanding of variation in TL, its determinants (e.g., demographic, lifestyle, and genetic factors), and its associations with disease phenotypes almost entirely rely on TL measured in leukocytes from whole blood (WB). Few studies have compared TL in leukocytes with TL in other human tissue types; those that have are relatively small (<100 participants; <5 tissue types) but provide evidence that TL differs across tissue types and that TL measurements from different tissue types are correlated (18, 19). Thus, larger studies of many additional tissue types are needed to gain a comprehensive understanding of variation in TL and its determinants within and across a wide range of human tissues and cell types.
To address these gaps in our understanding of TL and its role in disease risk and its relationship with age, we measured TL in >6000 unique tissue samples, representing >20 distinct tissue types and >950 individual donors from the Genotype-Tissue Expression (GTEx) project version 8 (v8) (20). In this work, we (i) characterize sources of variation in TL, (ii) evaluate leukocyte TL as a proxy for TL in other tissues, (iii) examine the relationship between age and TL across tissue types, and (iv) describe biological determinants and correlates of TL. This work presents results from tissue-specific and pan-tissue TL analyses that are crucial for improving our understanding of the etiologic role of TL in aging and chronic disease.
We attempted measurement of relative TL (RTL, the telomere repeat abundance relative to a standard reference DNA sample) for 7234 tissue samples from 962 GTEx donors using a Luminex-based assay (21). After removing 836 samples with failed RTL measurements and seven RTL measurements that were within-tissue outliers, our analytic dataset included 6391 tissue-specific RTL measurements from 952 donors, with 24 different tissue types having ≥25 RTL measurements (table S1). Each donor provided only one RTL measurement per tissue type, and on average, each donor had RTL measured in seven different tissue types (range: 1 to 26 tissue types) (fig. S1). The median donor age was 55 (range: 20 to 70) years. The majority of donors were male (67%) and of European descent (85%), and there were more postmortem donors (54%) than organ donors (table S1). Extensive validation and characterization of the Luminex-based RTL assay are described in (21).
TL varies across (and correlates among) human tissue types
We estimated the contribution of tissue type to the variation in RTL using linear mixed models (LMMs) adjusted for fixed effect covariates [age, sex, body mass index (BMI), race and ethnicity category, donor ischemic time, and technical factors, represented by plate (e.g., batch effects, DNA quality and concentration)] and with random effects representing tissue type and donor (table S2) (21). On average, RTL was the shortest in WB and longest in testis, with testis being an outlier tissue type [analysis of variance (ANOVA), p < 2 × 10−16 compared with all other tissues] (Fig. 1A). Tissue type explained 24.3% of the variation in RTL across all tissues but only 11.5% when testis was excluded, indicating that tissue type accounts for substantial variability in human TL.
Fig. 1. TLs differ across human tissue types but are correlated among tissues types.
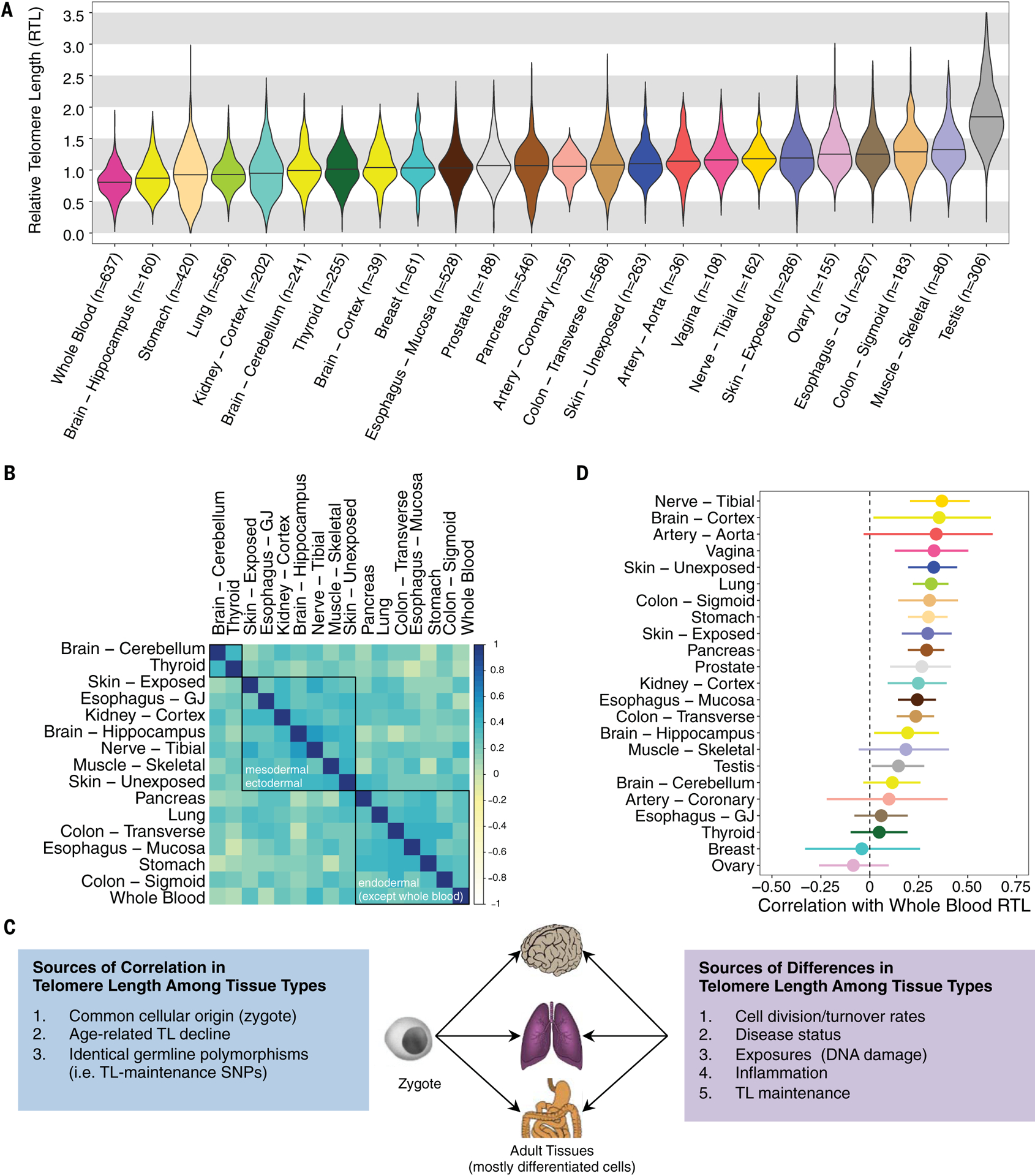
(A) Distribution of RTL across 24 GTEx tissue types (ordered by median RTL) (see table S1). Nine-hundred fifty-two donors contributed one or more tissue samples to the analysis, and the sample size for each tissue type corresponds to unique donors (i.e., no donors are represented twice for a given tissue type). (B) Pearson (r) correlations between RTL measures from different tissue types. Tissues included have ≥75 samples and were not sex specific. Red, yellow, and blue correspond to r = 1, 0, and −1, respectively. Black boxes are results from hierarchical clustering (three clusters). (Exact correlations are in table S3.) (C) Theoretical framework describing determinants of TL across human tissue types. (D) Pearson correlations between WB RTL and tissue-specific RTL measurements (with 95% confidence intervals).
We examined Pearson pairwise correlations in RTL among tissue types with tissue pairs from same donor, restricting to 20 tissue types with TL data for ≥75 samples (Fig. 1B). Forty-one tissue-pair correlations passed a Bonferroni p value threshold (t tests, p < 3 × 10−4), and all 41 correlations were positive (table S3). Tissue pairs from the same organ were among the strongest correlations observed: sun-exposed and nonexposed skin [Pearson correlation coefficient (r) = 0.24, t test, p = 9 × 10−3, n = 112], transverse and sigmoid colon (Pearson r = 0.40, t test, p = 8 × 10−7, n = 139), and esophagus mucosa (EM) and gastric junction (EGJ) (Pearson r = 0.22, t test, p = 3 × 10−3, n = 188). After applying hierarchical clustering to these pairwise correlations with average linkage, tissue RTLs separated into three clusters (Fig. 1B and fig. S2). Two clusters were characterized by common developmental origin: (i) mesodermal and ectodermal (e.g., muscle and skin) and (ii) endodermal origin tissues (e.g., stomach and lung). Thyroid and brain cerebellum formed the third cluster. Similar clustering patterns among tissue types were observed for females (fig. S3) and males (fig. S4), where testis was also an outlying tissue type and clustered with thyroid. The positive correlations observed among most tissue types are likely due to the fact that the initial TL in the zygote affects TL in all adult tissues through mitotic inheritance. Differences in tissue-type TL and the extent of correlation among tissue-type TLs are likely attributable to variability in both intrinsic (e.g., cell division rate and history, telomere maintenance) and extrinsic (e.g., response to environmental exposures) factors across tissues (Fig. 1C). To assess the possibility that extrinsic factors could modify the correlation between TL in different tissues, we assessed the overall difference in the correlation matrix by smoking history and obesity (as an indicator of disease status and health). In this exploratory analysis, the observed pairwise correlations among tissue types did not substantially differ between obese and normal or overweight donors. However, among individuals with a history of smoking, the correlation among tissue types was somewhat stronger compared with never-smokers (Jennrich’s chi-square test, p = 0.003), but the underlying reason for this observation is unknown.
WB TL is a proxy for TL in other tissues
WB RTL was positively correlated (Pearson correlation, t test, p < 0.05) with tissue-specific RTL measurements from 15 out of 23 tissue types (n ≥ 25 for each test), with Pearson correlations ranging from 0.15 to 0.37 (Fig. 1D). These results demonstrate that WB TL is a proxy for TL in many tissue types. WB RTL captured between 2% (testis) and 14% (tibial nerve) of the variation in RTL measured in other tissue types. Adjustment for age, sex, BMI, and donor ischemic time did not have a major impact on the associations observed between WB RTL and tissue-type RTL in the 23 tissue types (fig. S5). Notably, tibial nerve RTL had the strongest correlation with WB RTL. The GTEx tibial nerve samples largely contain connective tissue, Schwann cells, and the axons of neuron cells (which do not contain the DNA from neuron cells), and the strong correlation between tibial nerve RTL and WB RTL is likely due to the fact that the tibial nerve tissue and WB have connective tissue origins. Breast and ovary RTL had negative point estimates for their correlations with WB RTL, but the 95% confidence intervals over-lapped zero. The relationships between the RTL from these tissue types and WB RTL require further investigation.
RTL measurements have inherent measurement error (22), including our Luminex assay (23), and this error can attenuate the strength of the correlation observed between RTL measurements taken from two different tissue types. To better understand this error, we conducted extensive validation and characterization of our Luminex-based assay, including comparisons to TL measured by Southern blot of terminal restriction fragments (TRFs) reported previously by Pierce et al. (23) and conducted within GTEx (21). Based on this validation work (23), we conclude that that the percentage of variation in our Luminex RTL measures that is due to (nondifferential) measurement error is <50%. The true percentage cannot be estimated because the extent of measurement error in our gold standard TL measure, Southern blot analysis of TRFs, is unknown. Therefore, we used simulated data to estimate the impact of measurement error (ranging from 0 to 50% of the variation in RTL) on the correlations between RTL measurements from different tissues (21). Our results show that the correlations observed in this study will be attenuated, and this attenuation will increase with increasing error in the RTL measurements (fig. S6).
In addition to validating our Luminex RTL measurements against TL measured using Southern blot, we have also validated these measurements against RTL measured using quantitative polymerase chain reaction (qPCR) (24), both in previous work (25) and using GTEx samples (21). Within GTEx, RTL measurements from qPCR (24) and TL measured from Southern blot (26) showed strong correlation with our Luminex RTL measurements and similar differences among tissue types as observed for the Luminex RTL measurements (Fig. 1A) (21).
TL varies among individuals and by participant characteristics
TL varied across individuals (donors) (Fig. 2A, top), with 8.7% of the variation in RTL attributable to variability among individuals (estimates obtained from an adjusted LMM) (table S2). This percentage increased to 11.2% when testis was excluded. After adjusting for tissue type and donor (as random effects), age explained 3.3% (among all tissues) and 4.4% (excluding testis) of variation in RTL, whereas BMI, TL-associated SNPs, smoking status, and race and ethnicity category each explained <1% of the variation across all tissues [marginal coefficient of determination (R2), likelihood-ratio test (LRT), p < 0.05] (Fig. 2B, top), demonstrating that these factors contribute to pan-tissue TL dynamics. We observed no clear association between sex and RTL across all tissues (table S2), and sex showed weak evidence of association with RTL in tissue-specific analyses (table S4). Multiple prior studies have reported an association between longer leukocyte TL and female sex (27). However, we may be underpowered to detect this association for WB RTL, considering some larger studies have failed to detect it (28) and the association may be less evident at younger ages (29). The lack of association across all tissue types points to the possibility that this sex difference for leukocyte TL may not be consistent across all tissue types. RTL was shorter among (ever) smokers compared with never-smokers in lung and in WB (LRT, p < 0.05) (fig. S7), consistent with prior studies of leukocyte TL (30).
Fig. 2. TL varies among individuals and by ancestry.
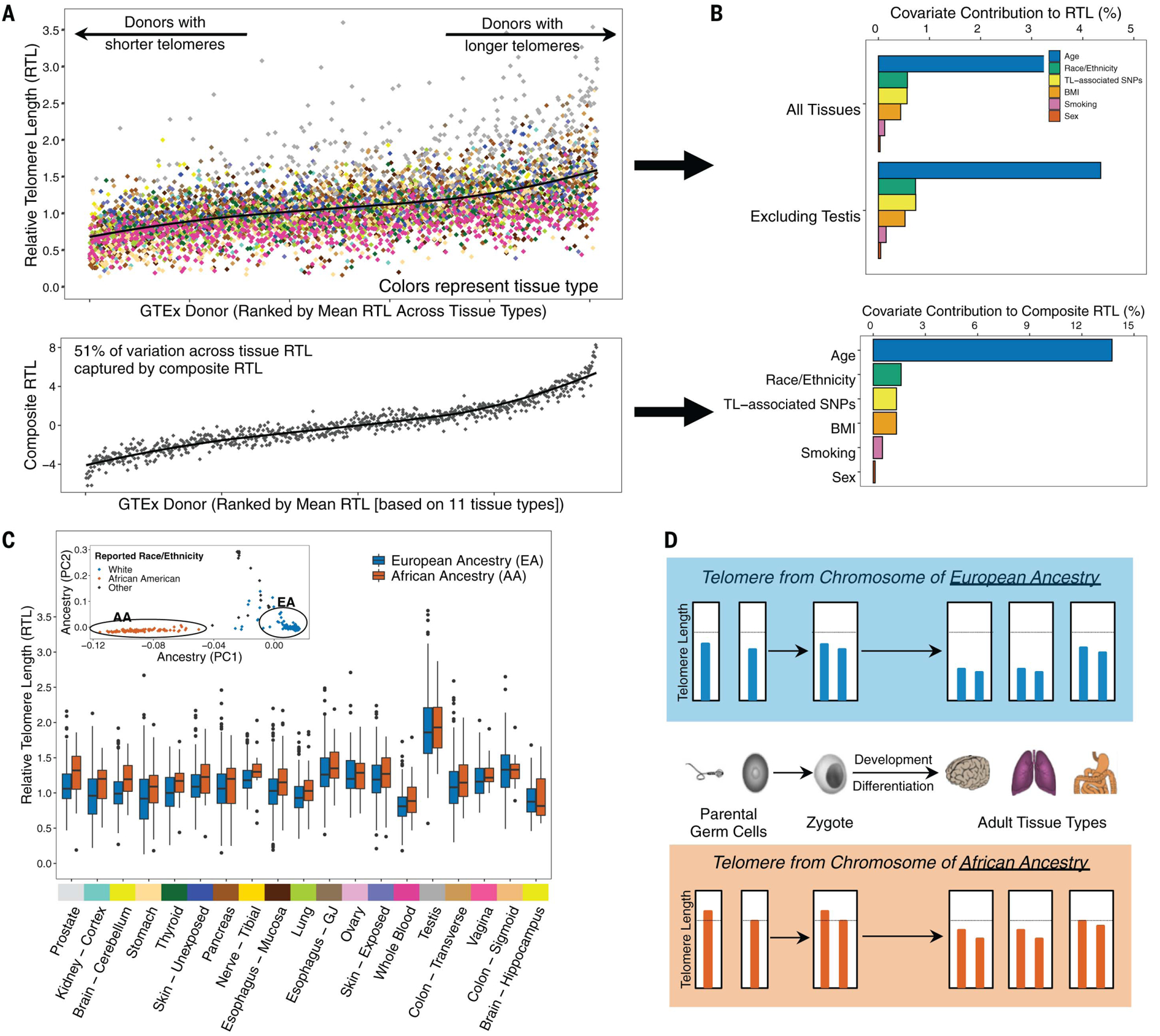
(A) Distribution of RTL across GTEx donors ranked by donors’ mean RTL across all measured tissue types (top) and distribution of a “composite RTL” measure (bottom), estimated as the first PC from a PC analysis (PCA) of 11 tissue types (21). Colors correspond to GTEx tissue type. (B) Contribution of selected covariates to variability in RTL across all tissues (top) and composite RTL (bottom). For the analysis across all tissues, estimates were extracted as marginal R2 values from LMMs adjusted for tissue type and donor as random effects. (C) Distribution of RTL measures for individuals of European ancestry (EA) and African ancestry (AA). Tissue types are ranked by the largest difference between median RTL of the two ancestry groups. The inset shows genotyping PCs, demonstrating consistent clustering of individuals by genetically predicted ancestry. Sample-size information and associations between African ancestry and RTL are reported in table S5. (D) Schematic describing the direct inheritance of TL from parental germ cells and expected relationship to TL across adult tissue types for individuals of African and European ancestry. Genetic (and reported race and ethnicity category) ancestry was color coded for African (red) and European (blue) in (C) and (D).
We conducted a principal component (PC) analysis of RTL from 11 nonreproductive tissue types (each with n ≥ 200 samples) from 750 participants (21) and generated a composite measure of TL for each donor on the basis of the first PC that explains 51% of the variation in TL among these tissue types (Fig. 2A, bottom). We observed that age, BMI, and smoking status were associated with shorter composite RTL and explained 13.7, 1.3, and 0.6%, respectively, of the variation in this composite TL measure (Fig. 2B, bottom). Race and ethnicity category was associated with longer composite TL in African Americans compared with European Americans and explained 1.6% of the variation in composite TL. This composite TL likely reflects variation in TL present in the zygote (and in tissues during early development) that is mitotically inherited by cells in adult tissues.
TL is longer in genomes of African ancestry
To further explore differences in TL by race and ethnicity category, we first confirmed that PCs derived from genome-wide SNP data (n = 838 donors), representing genetic ancestry, showed clear clustering by reported race and ethnicity category among donors (Fig. 2C, inset). Genetic ancestry (European versus African) explained 0.6% of the variation in RTL across all tissues (marginal R2, LRT, p = 1 × 10−5) after adjusting for tissue type and donor as random effects and 2.3% of the variation in composite RTL (F test, p = 7 × 10−5). After including adjustments for age, sex, donor ischemic time, technical factors, and random effects of tissue type and donor, RTL was longer among individuals of African ancestry compared with individuals of European ancestry across all tissue types (LRT, p = 0.007), demonstrating that the effect of ancestry on TL, reported previously for leukocyte TL (31–34), extends to TL in other tissue types. The adjusted association between African ancestry and RTL was positive for 16 out of 19 tissues tested, with LRT p values <0.05 for brain cerebellum (p = 0.03), thyroid (p = 0.02), prostate (p = 0.03), lung (p = 0.02), and WB (p = 0.005) (Fig. 2C and table S5). The observation that individuals of African ancestry have longer TL in many tissue types is consistent with the hypothesis that ancestry-based differences in TL are present early in development (35) and potentially in germ cells (preconception). In other words, our results suggest that offspring (zygotes) inherit telomeres from germ cells that vary in TL because of ancestry, and these ancestry-based differences in TL are mitotically transmitted to daughter cells, and eventually to cells in many adult tissue types. This “direct transmission” of TL from parent to offspring (36) would result in the observed ancestry-based differences across many tissue types (summarized in Fig. 2D). One likely cause of this ancestry-based difference is natural selection on SNPs know to affect TL (37), although selection on TL itself could also contribute.
TL is correlated with age in most tissues
Of 24 tissues with ≥25 samples, RTL was negatively correlated (Pearson r < 0) with age in 21 tissue types (p < 0.05 in 14 tissue types from t test) (Fig. 3A and fig. S8), providing new evidence to support the hypothesis that age-related TL shortening occurs in most tissue types. The strongest correlations with age were observed for WB (Pearson r = −0.35, t test, p = 2 × 10−19, n = 637) and stomach (r = 0.37, t test, p = 7 × 10−15, n = 420) (table S6). Age explained more of the variation in RTL for tissues with shorter mean RTL [coefficient of determination (r2) = 0.23, F test, p = 0.02] (Fig. 3B). The association between age and RTL differed by sex for hippocampus (t test, pinteraction = 0.04), transverse colon (t test, pinteraction = 0.01), and lung (t test, pinteraction = 0.04), suggesting that TL shortening with age is greater in men compared with women in some tissues. Among tissue types for which RTLs did not have a clear correlation with age (t test, p > 0.05), we examined whether RTL differed among 5-year age groups, but we observed no age-related differences in RTL for testis, ovary, cerebellum, vagina, skeletal muscle, thyroid, and EGJ (ANOVA, p > 0.05). Although prior studies have observed longer TL in sperm from older men (38), we did not observe a clear increasing (or decreasing) trend for testis RTL with increasing age (fig. S9).
Fig. 3. Age is negatively correlated with TL in most tissues, and correlation is strongest in tissues with shorter telomeres.
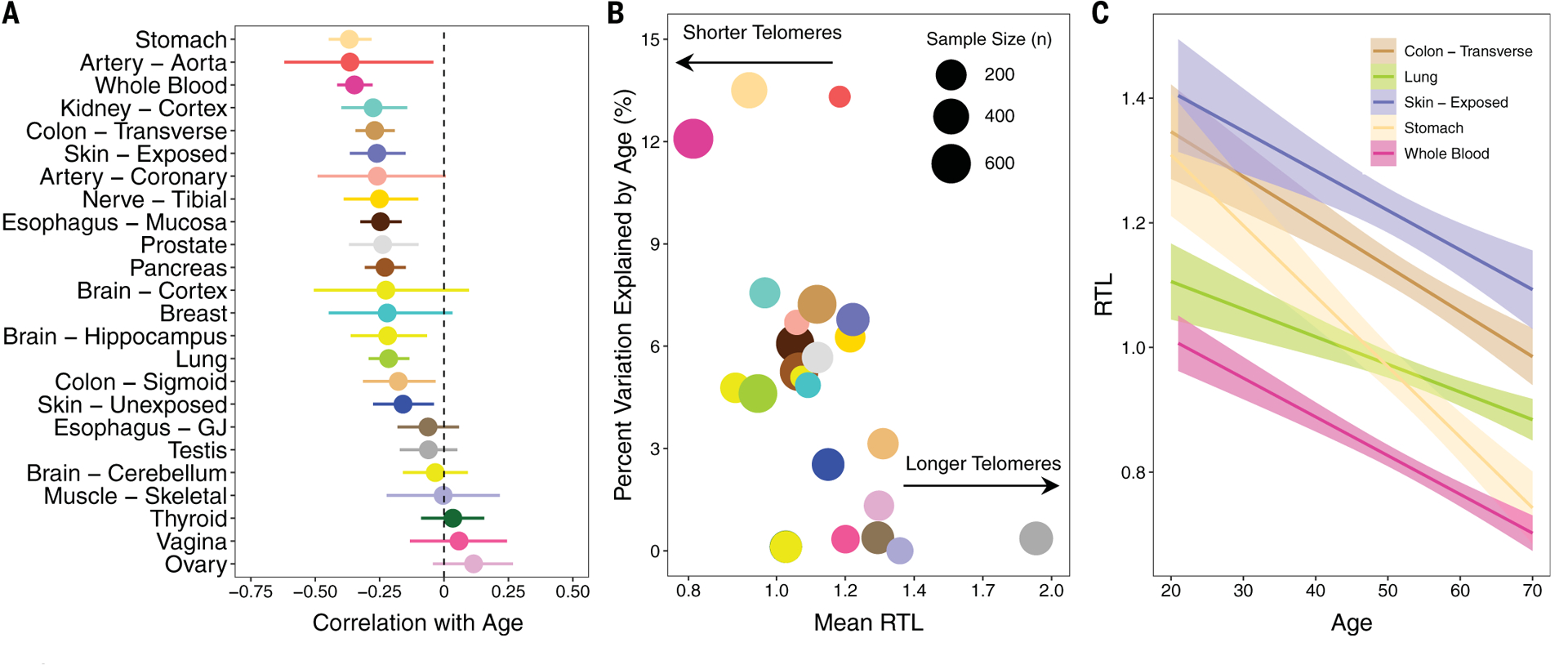
(A) Pearson correlations between age and tissue-specific RTL measures. (B) Scatterplot of mean RTL for each tissue versus the percent variation explained by age (r2) for each tissue. The size of each point is proportional to sample size for that tissue type. (C) Relationship between RTL and age for five selected tissue types [WB, lung, stomach, transverse colon, and skin (exposed)]. For all plots, colors correspond to tissue type.
Among tissue types for which RTL was correlated with age (t test, p < 0.05), the strength of association varied across tissue types (Fig. 3C and table S6). To further explore the hypothesis that TL shortens at different rates in different tissue types, we calculated the difference in RTL (ΔRTL) between all pairs of tissue types available for each donor. We constructed 155 ΔRTL variables, restricting to tissue pairs with complete data for ≥50 donors. The Pearson correlation between ΔRTL and age was estimated for each tissue-type pair to determine if the ΔRTL varies with age (fig. S10). Forty-two of the 155 ΔRTL variables were correlated with age (Pearson correlation, t test, p < 0.05), and the absolute values of these correlations ranged from 0.12 to 0.38 (table S7). Four of the ΔRTLs surpassed a Bonferroni p value of 3 × 10−4: EGJ and stomach (r = 0.32, t test, p = 1 × 10−5, n = 176), WB and thyroid (r = 0.30, t test, p = 3 × 10−5, n = 182), EM and stomach (r = 0.25, t test, p = 3 × 10−5, and n = 276), and WB and ovary (r = 0.33, t test, p = 2 × 10−4, n = 120). Our results indicate that age explains up to 14% of the variation in the difference in RTL between pairs of tissue types. A prior study of 87 adults reported that the rate of age-related TL shortening was similar for muscle, leukocytes, fat, and skin (i.e., no association between age and ΔRTLs), concluding that age-related TL loss within stem cells is consistent across adult tissue types (18). When we examined these tissue types among our ΔRTL pairs (n ≥ 50), age was correlated with ΔRTL for skeletal muscle and blood (r = 0.36, t test, p = 2 × 10−3, n = 68) but less for skin (unexposed) and blood (r = 0.09, t test, p = 0.20, n = 197) and skin (exposed) and blood (r = 0.08, t test, p = 0.24, n = 200).
Leukocyte TL–associated genetic variants and TL in other tissues
Prior genome-wide association studies (GWASs) have identified SNPs associated with leukocyte TL (12–15). We constructed a weighted polygenic SNP score for each donor using nine leukocyte TL–associated SNPs (21), with higher score reflecting longer TL (table S8) (39).We examined the association between this polygenic SNP score and RTL for tissue types with ≥100 samples. After adjustment for age, sex, genotyping PCs, donor ischemic time, and technical factors as a random effect, an association with the SNP score (LRT, p < 0.05) was observed for WB RTL (p = 0.007) (fig. S11), cerebellum RTL (p = 0.03), pancreas RTL (p = 0.04), and transverse colon RTL (p = 0.02) (Fig. 4A, fig. S12, and table S9). Among these 18 tissue types, 16 had positive association estimates [binomial test (p0 = 0.5), p = 0.001]. In analyses of all tissue types, RTL was positively associated with the SNP score (LRT, p = 0.01) after adjustments. These results indicate that at least some of the genetic variants (or regions) that affect leukocyte TL also affect TL in other tissue types.
Fig. 4. Inherited genetic variation affects telomere length in multiple tissue types and expression of nearby genes.
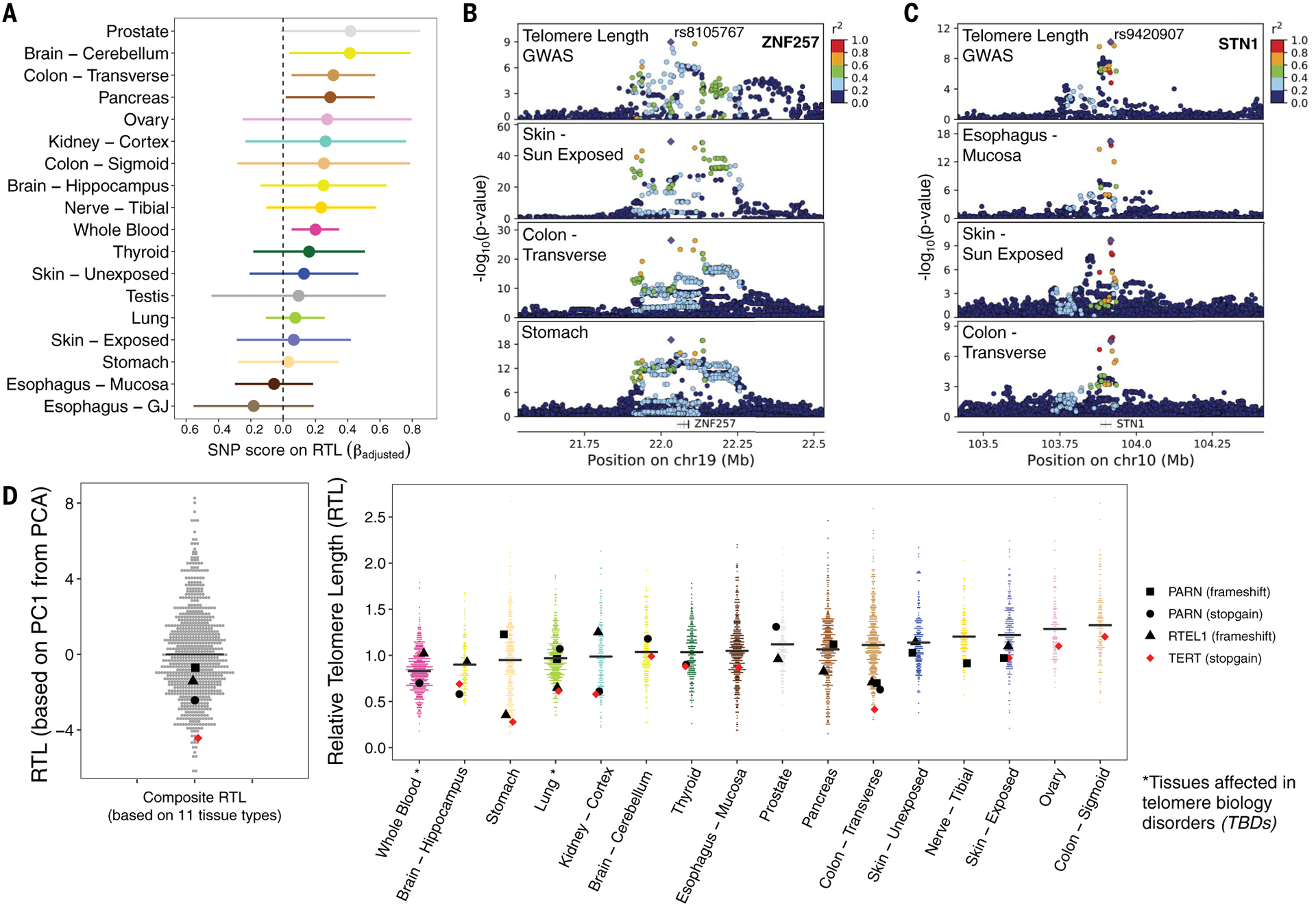
(A) Associations between a polygenic SNP score for leukocyte TL and tissue-specific RTL measures. Colors correspond to tissue type. (B) Leukocyte TL association signal from GWASs colocalizes with a cis-eQTL for ZNF257 (~40 kb upstream of ZNF208). The top plot shows results from the ENGAGE Consortium GWAS of leukocyte TL, and the bottom three plots correspond to cis-eQTL results from GTEx tissues: skin–sun exposed, colon–transverse, and stomach. chr19, chromosome 19. (C) Leukocyte TL association signal colocalizes with a cis-eQTL for STN1 (also known as OBFC1 in human genome reference hg19). The top plot corresponds to results from the ENGAGE Consortium GWAS of leukocyte TL, and the bottom three plots correspond to cis-eQTL results from GTEx tissues: skin–sun exposed, EM, and colon–transverse. (D) Distribution of composite RTL (based on PC1 from PCA of 11 tissue types) (left) and tissue type RTL (right), with highlighted dots representing GTEx donors carrying a rare LOF variant in a telomere maintenance gene previously implicated in TBDs. LOF variants are noted in the legend. The black horizontal line corresponds to median composite RTL and tissue type RTL. The tissue types presented contain one or more LOF carriers, and colors correspond to tissue type.
TL-associated variants influence local gene expression
Among the nine regions known to harbor SNPs associated with leukocyte TL, we examined whether these SNPs also affect local gene expression in GTEx tissue types and cell lines (21). Colocalization analysis can be used to determine if a common causal variant affects a trait (e.g., TL) and expression of a nearby gene (40). If there is a common causal variant underlying both association signals, then we may infer that SNPs may influence TL via effects on gene expression. We used colocalization analysis to estimate the probability that a common causal variant underlies association signals for leukocyte TL (from GWASs) (12–15) and cis-eQTL (expression quantitative trait loci) association signals from GTEx (v8) analyses (20). Colocalization results indicated that at least six of the nine TL-associated regions shared a common causal variant with a cis-eQTL in at least one tissue type, on the basis of a posterior probability of colocalization of ≥80% across all three sets of priors tested (Fig. 4, B and C; fig. S13; and table S10).
The association signal for TL on chromosome 19 (represented by rs8105767) showed strong evidence of colocalization with an eQTL affecting expression of gene ZNF257 in eight tissue types, including skin (sun exposed), transverse colon, and stomach (Fig. 4B). ZNF257 encodes a zinc-finger protein that may be involved in transcriptional regulation. The association signal for TL on chromosome 10 (represented by rs9420907) colocalized with an eQTL affecting expression of STN1 in seven tissue types, including skin (sun exposed), transverse colon, and EM (Fig. 4C). Additional TL-associated loci showed colocalization with GTEx eQTLs for NAF1, MYNN, RP11-109N23.6, and TSPYL6 (fig. S13 and table S10). Although these colocalizations were observed for eQTLs in tissue types with largely differentiated cells, eQTLs observed in induced pluripotent stem cells have been shown to be largely shared with eQTLs in GTEx tissue types (41). This finding suggests that the observed evidence of colocalization may be pertinent to TL maintenance within stem and progenitor cells, which have active telomerase activity. Notably, NAF1 encodes a protein involved in telomere assembly, and loss-of-function (LOF) mutations in this gene are associated with shorter telomere length in pulmonary fibrosis (PF) patients (42). These results suggest that TL-associated loci influence TL within human tissues through regulation of the expression of genes known to be involved in telomere maintenance (e.g., STN1, NAF1) (12), as well as genes whose role in telomere maintenance is unclear (e.g., ZNF257).
Notably, we observed little evidence of colocalization of the TERT or TERC TL-associated regions with any cis-eQTLs. TERT and TERC are important components of telomerase. The telomerase enzyme can extend the telomere repeat sequence, typically in stem and/or progenitor cells, to compensate for TL shortening; however, TERT and TERC have low or undetectable expression in a majority of adult GTEx tissue samples. This suggests that eQTL studies of cells from stem and/or developmental tissues may be needed to understand the mechanisms underlying genetic regulation of TERT and TERC expression.
Carriers of rare LOF variants may have shorter TL
Telomere biology disorders (TBDs, e.g., PF, dyskeratosis congenita, aplastic anemia) are characterized by short TL in affected individuals owing to inherited LOF mutations in telomere maintenance genes (1, 43–45). Individuals with TBDs often present with early-onset aging-related phenotypes—such as immune dysfunction, bone failure, liver disease, and lung function decline—and these effects can inform our understanding of how TL contributes to aging in the general population. Using whole-genome sequencing data from GTEx donors, we searched for LOF rare variants in seven genes that have evidence of autosomal dominant (or partial dominant) inheritance in relation to TBDs (e.g., TERC, TERT, TINF2, RTEL1, PARN, ACD, and NAF1). We identified four donors carrying a rare exonic variant (minor allele frequency <1%) resulting in a predicted LOF frameshift insertion or deletion or a stop-gain mutation (Fig. 4D). These LOF carriers had shorter TL across all tissues (LRT, p = 0.04) and shorter composite TL (t test, p = 0.03). One donor carried a stop-gain variant in TERT, and their composite TL was among the lowest observed (~first percentile), consistent with prior studies of TERT mutations among individuals with PF (46, 47).
Our results suggest that rare variants in TL-maintenance genes may contribute to shorter TL in multiple tissues in the general population (i.e., primarily individuals without TBDs). However, the PARN and RTEL1 mutation carriers among the GTEx donors did not have RTL values in the (lower) extreme of the composite TL and tissue-specific RTL distribution(s). Although mutations in TL maintenance genes and very short TL are often found in individuals with TBDs (43–45), prior studies of individuals with TBDs have shown that TL can vary substantially among carriers (of mutations in PARN, RTEL1, and TERT), and some carriers have TL values similar to noncarriers (46, 48, 49). Prior studies of PF patients suggest that LOF TERT mutations may have a larger impact on TL than LOF mutations in PARN or RTEL1 (46, 47, 49).
TL is associated with telomerase subunit expression across tissues
The protein products of TERT, TERC, and DKC1 comprise the telomerase catalytic subunit. We examined the association between RTL and expression of these genes using 3885 GTEx tissue samples with both RTL and RNA sequencing (RNA-seq) gene expression data (v8). TERT and TERC expression was detectable [i.e., transcripts per million (TPM) >0.1] in 28% (n = 1089) and 20% (n = 783) of these samples, respectively, but DKC1 was ubiquitously expressed (n = 3885) in all samples (table S11). Whereas DKC1 showed correlation with both TERT (Pearson r = 0.30, t test, p < 2 × 10−16, n = 1089) and TERC (r = 0.23, t test, p = 3 × 10−11, n = 783) across all samples, the correlation between TERT and TERC expression across samples was stronger (r = 0.49, t test, p < 2 × 10−16, n = 364) (fig. S14). Testis had substantially higher mean expression of TERT and TERC compared with all other tissues (ANOVA, p < 2 × 10−16) (table S11), but there was no association between testis RTL and TERT or TERC expression. Across all tissues, RTL was positively correlated with TERT (r = 0.58, t test, p < 2 × 10−16, n = 1089), TERC (r = 0.33, t test, p < 2 × 10−16, n = 783), and DKC1 (r = 0.29, t test, p < 2 × 10−16, n = 3885) (Fig. 5A). When testis was removed, the correlation decreased substantially for both TERT (r = 0.14, p = 4 × 10−5, n = 890) and DKC1 (r = 0.23, p < 2 × 10−16, n = 3686) and disappeared for TERC (r = 0.02, p = 0.63, n = 617). After adjustment for covariates and random effect of tissue type, RTL showed a positive association with increasing quartiles of TERT expression (LRT, p = 0.005 including testis and p = 0.002 excluding testis) and of DKC1 expression (LRT, p = 0.001 including testis and p = 3 × 10−4 excluding testis) across all tissues. Overall these results support the following: (i) high telomerase activity in testis (i.e., spermatocytes) likely contributes to longer TL observed in that tissue, and (ii) GTEx tissue samples consist primarily of differentiated cells, which typically have little to no telomerase activity, resulting in minimal detectable association between telomerase activity in those cells and the observed TL (50, 51).
Fig. 5. TL is associated with telomerase subunit gene expression and may mediate the effect of age on gene expression.
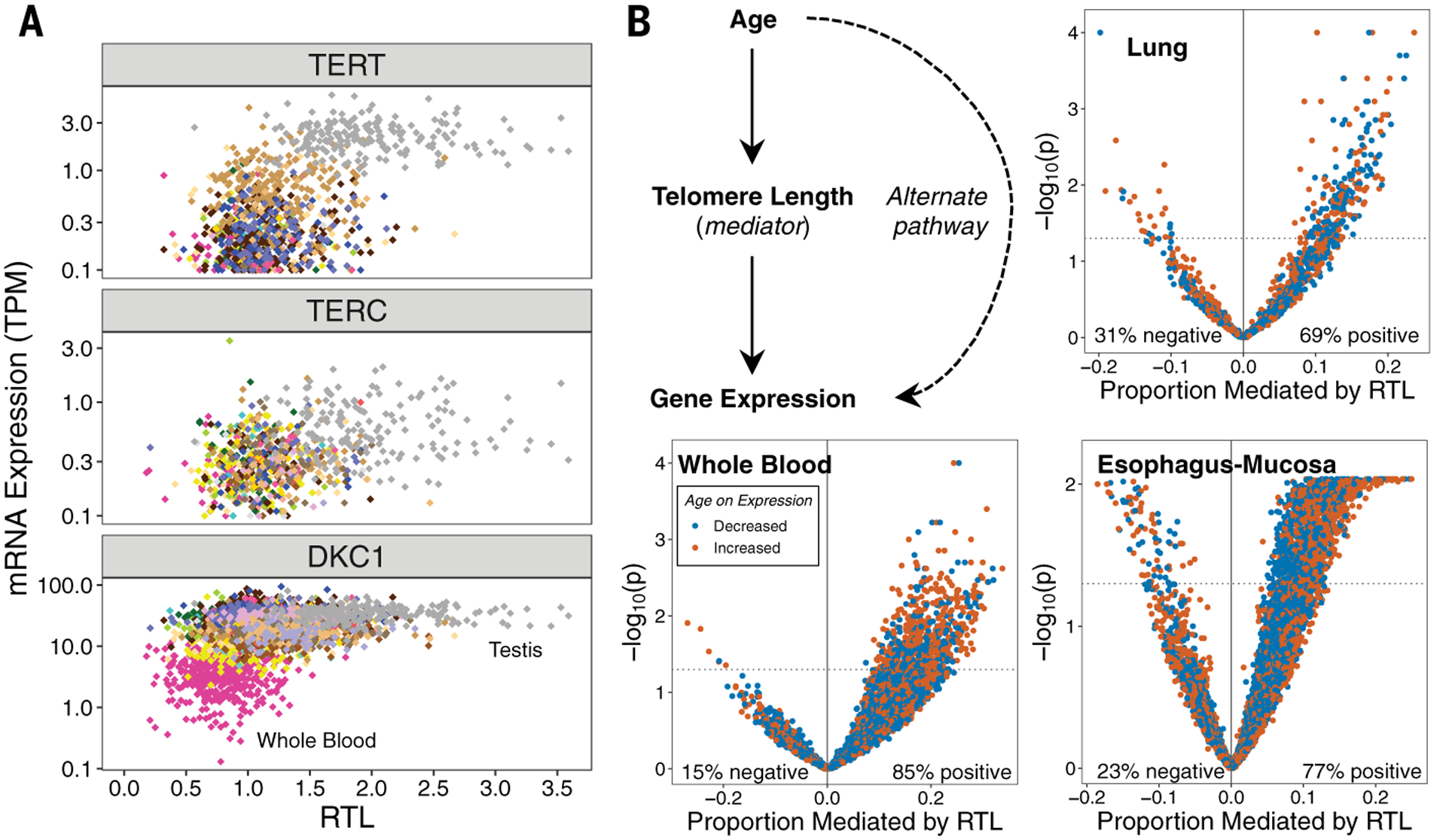
(A) RTL plotted against TERC, TERT, or DKC1 expression across tissue types. Colors correspond to GTEx tissue types. (B) Analyses addressing the hypothesis that TL mediates the effect of age on expression of specific genes. Scatterplots show estimates of the proportion of the effect of age on gene expression mediated by RTL (for each gene) and the −log10(p value) corresponding to the average causal mediation effect of RTL (for each gene). Results are presented for all age-associated genes in each of the three selected tissue types (WB, lung, and EM). The mediation p value was obtained using a nonparametric bootstrapping approach (n = 10,000 bootstraps).
TL may mediate the effect of age on gene expression
Aging affects gene expression, so we examined whether TL mediates the association between age and expression of age-associated genes. We analyzed the association between age and RNA-seq–based gene expression levels among tissues with ≥150 samples and selected three tissue types with >1000 age-associated genes [false discovery rate (FDR) of 0.05] (21): WB (n = 5239), lung (n = 1366), and EM (n = 6024) (Fig. 5B). Using mediation analysis (52), we estimated the proportion of the effect of age on expression that was mediated by TL for each age-associated gene. For each tissue type, we observed substantially more positive than negative estimates of the “proportion mediated” (Fig. 5B), as expected under the hypothesis that TL is a mediator. (An equal number of positive and negative estimates are expected under the hypothesis of no mediation.) If TL is a mediator for a specific gene, then adjustment for TL will attenuate the association between age and gene expression. We observed evidence that RTL mediated the effect of age on expression for 607 genes (12%) in WB, 224 genes (16%) in lung, and 1177 genes (20%) in EM (pmediation < 0.05, and proportion mediated > 0) (tables S12 to S14). In these tissue types, RTL mediated between 4 and 34% of the effect of age on expression of individual genes; however, full mediation will be detected as partial mediation in the presence of measurement error (for either the mediator or the outcome) (53). We evaluated the enrichment of these RTL-mediated genes in gene ontology (GO) terms among the age-associated genes (Fisher’s exact test, FDR < 0.1). Enriched GO terms were identified for lung (5 terms), EM (30 terms), and WB (108 terms) (tables S15 to S17). No GO terms (FDR < 0.1) were common to WB, lung, and EM for any ontology. Among 108 enriched GO terms in WB, several terms related to apoptosis, cell death, and telomere DNA binding were identified. The results from this analysis provide evidence that TL is a potentially relevant biologic factor in the mediation of age on gene expression and may contribute to processes related to biologic aging.
Tissue-level stem cell features are associated with TL and TERT expression
After extracting tissue-specific estimates of the number of divisions per stem cell (per year) and the proportion of stem cells (among all cells) for specific tissue types from Tomasetti and Vogelstein et al. (54, 55), we examined their relationship with mean RTL and mean TERT expression among nonreproductive GTEx tissue types (n = 12; table S18). No associations were identified between mean TERC and DKC1 expression and these stem cell features. Mean RTL was positively correlated with estimated proportion of stem cells within a tissue type (r = 0.71, t test, p = 0.01) (Fig. 6A), and this association persisted after adjustment for number of divisions per stem cell (t test, p = 0.008) and mean TERT expression (t test, p = 0.02). We did not observe a clear association between mean TERT expression and the estimated proportion of stem cells within a tissue type. These results suggest that tissue types with a higher proportion of stem cells in their cellular composition may have longer TL measurements in bulk tissues as a consequence.
Fig. 6. TL and TERT expression are associated with estimated stem cell features.
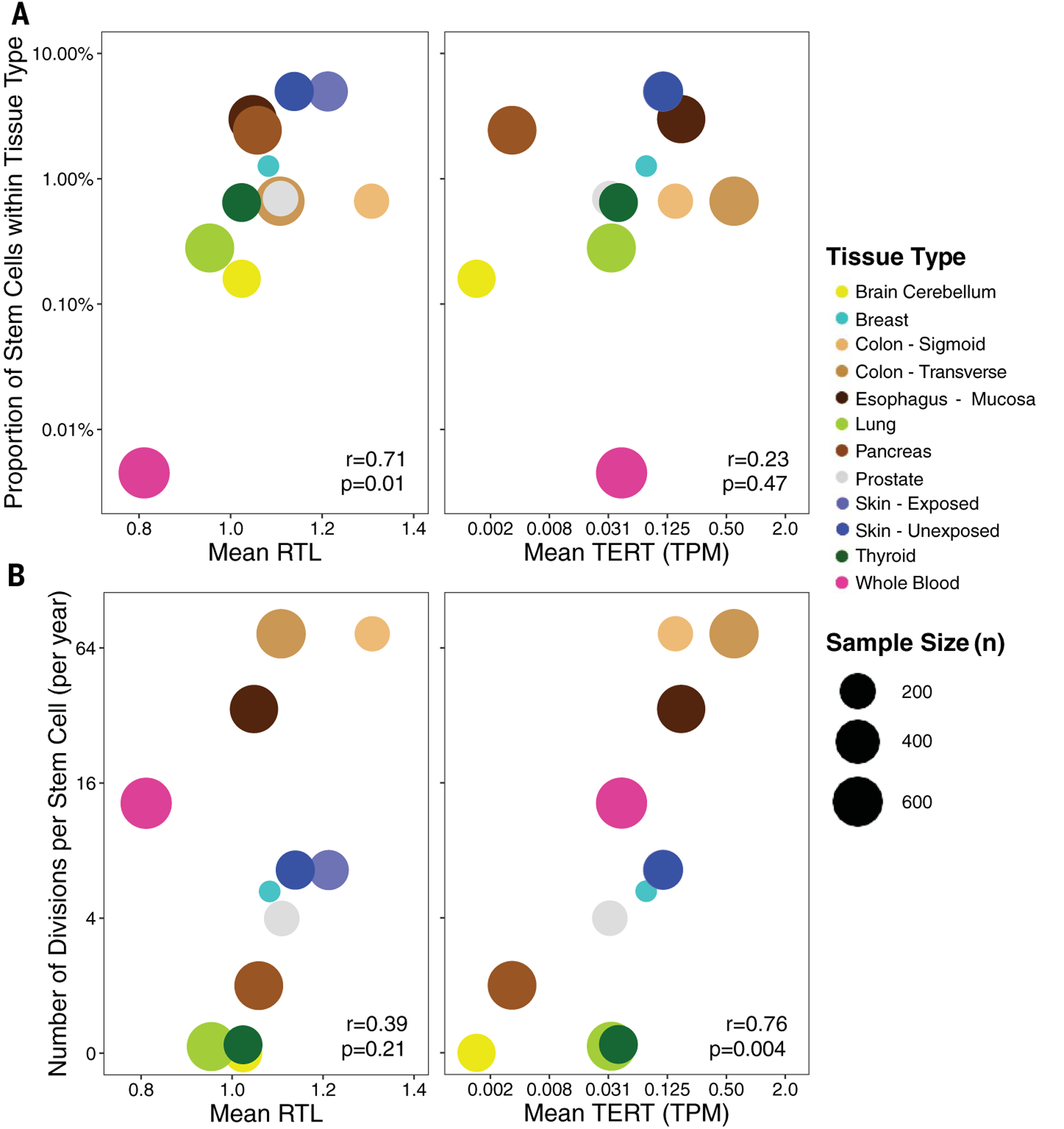
(A) Estimated proportion of stem cells within tissues and its relationship between mean RTL (left) and mean TERT expression (right). (B) Estimated number of divisions per stem cell (per year) within tissues and its relationship between mean RTL (left) and mean TERT expression (right). Colors correspond to GTEx tissue types, and the size of each point reflects the sample size of the tissue type. Pearson correlations and corresponding p values are reported. Analysis included nonreproductive tissues only.
We observed a positive correlation between mean TERT expression and the number of divisions per stem cell (r = 0.76, t test, p = 0.004) (Fig. 6B). This association persisted after adjustment for the proportion of stem cells within a tissue type (t test, p = 0.006) and mean RTL (t test, p = 0.01). Mean RTL showed suggestive evidence of correlation with the number of divisions per stem cell (r = 0.39, t test, p = 0.21), and when we restricted to nonblood tissue types, mean RTL was positively correlated with number of divisions per stem cell (r = 0.65, t test, p = 0.03). This finding suggests that tissue types that undergo more cellular turnover and replacement, such as colon, may have higher telomerase expression to maintain TL in the stem cell compartments.
Cell-type composition is associated with TL within tissues
To determine whether TL varies among the cell types within a given tissue sample, we examined the association between RTL and estimated cell-type enrichment scores (CTES) [generated using RNA-seq data and the xCell software (56)]. Seven CTES (for adipocytes, epithelial cells, hepatocytes, keratinocytes, myocytes, neurons, and neutrophils) were bench-marked by the GTEx Consortium (57), and we examined the association between these seven CTES and RTL in tissue types with ≥100 samples (n = 16 tissue types). After removing cell types not detected within a tissue type (n = 37 total CTES tested across 16 tissue types) and adjusting for age and sex, we identified eight associations (t test, p < 0.05) between CTES and RTL among 37 associations tested (fig. S15). In exploratory analyses, we examined all 64 CTES provided by xCell that had a detection p value <0.05 for >90% samples within a tissue type. Restricting to tissue types with ≥300 samples that had both CTES and RTL data (WB, lung, and EM), there were 27, 24, and 17 CTES detected in each tissue, respectively (fig. S16). EM and lung had 13 and 14 CTES that were associated with RTL, after adjustment for age and sex (t test, p < 0.05). RTL was positively associated with epithelial cell, smooth muscle cell, keratinocyte, and sebocyte CTES in both lung and EM (p < 0.05). Notably, five CTES were inversely associated with RTL (p < 0.05) in both lung and EM, including fibroblasts and endothelial cells. In WB, lymphoid and myeloid cell CTES accounted for 70% of the CTES detected, and eight CTES were associated with RTL (t test, p < 0.05). Neutrophil CTES were positively associated with RTL. Both CD8+ T cell CTES were inversely associated with RTL, consistent with prior work examining cell types and TL in blood (58). These results provide evidence that TL varies across cell types within a given tissue, and consequently, cell-type composition can affect TL measurement in human tissues.
TL across all tissues is associated with age-related chronic disease status
Using medical history data from GTEx donors, we examined the association between common age-related chronic diseases and RTL within and across tissues. A history of type 2 diabetes (22% of donors) was associated with shorter RTL across all tissues (LRT, p = 0.02) as well as shorter pancreas RTL (p = 0.07) and coronary artery RTL (p = 0.01) (fig. S17). Among all donors, 50% had no history of any chronic disease, and 30, 14, and 6% had a history of one, two, and three (or more) chronic diseases, respectively. Chronic disease burden (sum of chronic diseases from 0 to 5) was associated with shorter RTL across all tissues (LRT, p = 0.008) and in testis, coronary artery, kidney cortex, and cerebellum (LRT, p < 0.05 for each). When we excluded cancer from the chronic disease burden, these associations persisted across all tissues (LRT, p = 0.02) and in all tissues listed above except for kidney cortex (LRT, p = 0.09). These observations suggest that TL may capture some aspect of the biologic age-related health decline across tissues.
We did not observe any associations between RTL and history of cancer; however, to test the hypothesis that normal tissues with relatively short (or long) TL are also short (or long) in tumors occurring in that tissue, we compared the mean tissue-to-WB TL ratio for each GTEx tissue with the mean tumor-to-WB TL ratio in corresponding cancer types from The Cancer Genome Atlas (TCGA) (21, 59). The mean cancer TL ratio from TCGA and normal TL ratio from GTEx were positively correlated (r = 0.44, t test, p = 0.04, n = 23) (fig. S18), providing support for this hypothesis.
After reviewing the medical and death report information for diseases and conditions related to TBDs (21), we identified six donors with a reported history of PF and/or interstitial lung disease (ILD). Five of these donors had TL measurements (n = 35 tissue-type samples). We observed that three of the donors with a history of PF or ILD had composite RTL below the fifth percentile (fig. S19). A history of PF or ILD was associated with shorter TL across all tissues (LRT, p = 0.02) and shorter composite RTL (t test, p = 0.01). Notably, we observed that within tissues, the median RTL was substantially shorter for WB (Mann-Whitney U test, p = 0.02), pancreas (p = 0.01), and EM (p = 0.05) among donors with a history of PF or ILD.
Discussion
This study provides a view of the substantial variation in human TL that exists across human tissue types and among individuals. We show that TL is generally positively correlated across human tissue types, and that WB TL is a proxy for tissue-specific TL for many tissues, a finding that may support the use of blood TL as a proxy for TL in some tissues in large epidemiological studies. TL was negatively associated with age in the majority of tissues studied, confirming the hypothesis of pervasive age-related telomere shortening in most human tissues. However, our results suggest that the rate of shortening can vary across tissues, and age explained more variation in TL in tissues with shorter mean TL. TERT and TERC expression were low or undetectable in most tissues and not associated with TL within any tissue, likely because progenitor cells, which express telomerase, are not present in large numbers in adult tissue samples, which consist primarily of differentiated cells. Notably, testicular TL was ~1.5- to 2.5-fold longer than TL in any other tissue type, and TERT was expressed in 100% of these samples and at higher levels than in any other tissue, consistent with the predominance of spermatogenic cells in testis (i.e., cells developing from germ cells into spermatozoa), which have high telomerase activity (51).
RTL measured in a tissue sample is an average of the TLs among all chromosomes within a heterogeneous population of cell types with different cell division rates and history, stem cell composition, and oxidative and inflammatory environments. To characterize variation in TL within specific cell types, cell type–specific and single-cell TL studies are needed, potentially using interphase quantitative fluorescence in situ hybridization approaches (60) and flow cell cytometry to isolate specific cell types, including stem cells.
A large proportion of the variation in RTL was unexplained across all tissue types, potentially attributed to sources such as cell-type composition (e.g., stem and progenitor cells), measurement error, and lifestyle and environmental factors with variable effects across tissues. From our simulation-based analysis of the impact of TL measurement error on our results, we show that random measurement error biases our estimate of the true correlation in TL between two tissues toward zero, suggesting that the correlations presented in this study are attenuated compared with their true associations.
We lack detailed exposure data (e.g., smoking and alcohol use) for GTEx donors; studies that can link human tissue samples to environmental and lifestyle histories are needed to better understand environmental determinants of TL across different tissues and cell types. As of now, all TL-associated SNPs have been identified in GWASs of leukocyte TL (12–15); our study suggests that some of these effects are also present in other tissue types, but larger studies of tissue-specific TL measurements are needed to characterize how these effects vary across tissues and cell types. Identifying variants that affect TL in all or most cell types (e.g., variants with effects on TL that may be present during development or in stem cells in multiple tissue types) may be ideal for evaluating the causal impact of TL on risk for a wide array of diseases (occurring in diverse tissues or cell types) using Mendelian randomization. TL shortening is an important hallmark of aging in human tissues, but TL should also be studied in conjunction with other hallmarks of aging. Characterizing the relationships among TL and other aging-related processes and biomarkers within and across tissues will improve our understanding of cellular aging and its impact on human health.
Methods summary
We measured RTL in 6391 samples from 952 GTEx donors using a Luminex-based method. These measurements were validated against other TL measurement methods, including TL measured using Southern blot of TRFs (fig. S20) (26), relative TL measured using qPCR (fig. S21) (24), and TL estimated from whole-genome sequencing data (fig. S22) (61). Publicly available GTEx donor covariate, genotyping, and RNA-seq gene expression data (all v8) were integrated into our analyses. We applied LMMs to examine the relationships of RTL with age, genetic ancestry, gene expression of telomerase components, estimates of cell types, and other covariates across and within tissue types. Using GTEx genotyping data, we constructed a weighted polygenic SNP score for each donor using nine leukocyte TL–associated SNPs identified from the ENGAGE GWAS of leukocyte TL (12) and examined colocalization of these GWAS association signals with local gene expression using summary statistics from the ENGAGE study and eQTL results from the GTEx Consortium. Mediation analyses were applied to examine the extent to which TL mediates the effect of age on gene expression. Estimates of stem cell division and proportion of stem cells were extracted from prior studies (54, 55) for corresponding GTEx tissues, and their relationship with average RTL and TERT expression was examined.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the GTEx donors and families for their generous participation in and contribution to the GTEx Consortium. GTEx Consortium members: We thank the donors and their families for their generous gifts of organ donation for transplantation and tissue donations for the GTEx research project; the Genomics Platform at the Broad Institute for data generation; and J. Struewing for his support and leadership of the GTEx project.
Funding:
This work was supported by the National Institute of Aging Specialized Demography and Economics of Aging Training Program (T32AG000243) (K.D. and C.Z.), NIH Research Supplement to Promote Diversity in Health-Related Research (associated with R35ES028379) (K.D. and D.D.), Marie-Skłodowska Curie Fellowship H2020 Grant 706636 (S.K.-H.), Susan G. Komen Fellowship (GTDR16376189) (M.C. and D.D.), Medical Scientist National Research Service Award (T32GM07281) (M.C.), Norwegian Research Council (NFR ES562296) (A.A.), active and past NIH grants (U01HG007601, R35ES028379, and R01ES020506 to B.L.P.; R01CA107431 and P30ES027792 to H.A.; R01GM108711 to L.S.C.; and R01HL134840 and U01AG066529 to A.A.), and the GTEx LDACC (HHSN268201000029C). GTEx Consortium members: This work was supported by the Common Fund of the Office of the Director, NIH, and by NCI, NHGRI, NHLBI, NIDA, NIMH, NIA, NIAID, and NINDS through NIH contracts HHSN261200800001E (Leidos Prime contract with NCI: A.M.S., D.E.T., N.V.R., J.A.M., L.S., M.E.B., L.Q., T.K., D.B., K.R., and A.U.), 10XS170 (NDRI: W.F.L., J.A.T., G.K., A.M., S.S., R.H., G.Wa., M.J., M.Wa., L.E.B., C.J., J.W., B.R., M.Hu., K.M., L.A.S., H.M.G., M.Mo., and L.K.B.), 10XS171 (Roswell Park Cancer Institute: B.A.F., M.T.M., E.K., B.M.G., K.D.R., and J.B.), 10X172 (Science Care Inc.), 12ST1039 (IDOX), 10ST1035 (Van Andel Institute: S.D.J., D.C.R., and D.R.V.), HHSN268201000029C (Broad Institute: F.A., G.G., K.G.A., A.V.S., X.Li., E.T., S.G., A.G., S.A., K.H.H., D.T.N., K.H., S.R.M., and J.L.N.), 5U41HG009494 (F.A., G.G., and K.G.A.), and through NIH grants R01 DA006227-17 (University of Miami Brain Bank: D.C.M. and D.A.D.), supplement to University of Miami grant DA006227 (D.C.M. and D.A.D.), R01 MH090941 (University of Geneva), R01 MH090951 and R01 MH090937 (University of Chicago), R01 MH090936 (University of North Carolina–Chapel Hill), R01MH101814 (M.M.-A., V.W., S.B.M., R.G., E.T.D., D.G.-M., and A.V.), U01HG007593 (S.B.M.), R01MH101822 (C.D.B.), U01HG007598 (M.O. and B.E.S.), U01MH104393 (A.P.F.), extension H002371 to 5U41HG002371 (W.J.K) as well as other funding sources: R01MH106842 (T.L., P.M., E.F., and P.J.H.), R01HL142028 (T.L., Si.Ka., and P.J.H.), R01GM122924 (T.L. and S.E.C.), R01MH107666 (H.K.I.), P30DK020595 (H.K.I.), UM1HG008901 (T.L.), R01GM124486 (T.L.), R01HG010067 (Y.Pa.), R01HG002585 (G.Wa. and M.St.), Gordon and Betty Moore Foundation GBMF 4559 (G.Wa. and M.St.), 1K99HG009916-01 (S.E.C.), R01HG006855 (Se.Ka. and R.E.H.), BIO2015-70777-P, Ministerio de Economia y Competitividad and FEDER funds (M.M.-A., V.W., R.G., and D.G.-M.), la Caixa Foundation ID 100010434 under agreement LCF/BQ/SO15/52260001 (D.G.-M.), NIH CTSA grant UL1TR002550-01 (P.M.), Marie-Skłodowska Curie fellowship H2020 Grant 706636 (S.K-H.), R35HG010718 (E.R.G.), FPU15/03635, Ministerio de Educación, Cultura y Deporte (M.M.-A.), R01MH109905, 1R01HG010480 (A.Ba.), Searle Scholar Program (A.Ba.), R01HG008150 (S.B.M.), 5T32HG000044-22, NHGRI Institutional Training Grant in Genome Science (N.R.G.), EU IMI program (UE7-DIRECT-115317-1) (E.T.D. and A.V.), FNS funded project RNA1 (31003A_149984) (E.T.D. and A.V.), DK110919 (F.H.), F32HG009987 (F.H.), and Massachusetts Lions Eye Research Fund Grant (A.R.H.).
GTEx Consortium
Laboratory and Data Analysis Coordinating Center (LDACC): François Aguet1, Shankara Anand1, Kristin G. Ardlie1, Stacey Gabriel1, Gad A. Getz1,2,3, Aaron Graubert1, Kane Hadley1, Robert E. Handsaker4,5,6, Katherine H. Huang1, Seva Kashin4,5,6, Xiao Li1, Daniel G. MacArthur5,7, Samuel R. Meier1, Jared L. Nedzel1, Duyen T. Nguyen1, Ayellet V. Segrè1,8, Ellen Todres1
Analysis Working Group Funded by GTEx Project Grants: François Aguet1, Shankara Anand1, Kristin G. Ardlie1, Brunilda Balliu9, Alvaro N. Barbeira10, Alexis Battle11,12, Rodrigo Bonazzola10, Andrew Brown13,14, Christopher D. Brown15, Stephane E. Castel16,17, Donald F. Conrad18,19, Daniel J. Cotter20, Nancy Cox21, Sayantan Das22, Olivia M. de Goede20, Emmanouil T. Dermitzakis13,23,24, Jonah Einson16,25, Barbara E. Engelhardt26,27, Eleazar Eskin28, Tiffany Y. Eulalio29, Nicole M. Ferraro29, Elise D. Flynn16,17, Laure Fresard30, Eric R. Gamazon21,31,32,33, Diego Garrido-Martín34, Nicole R. Gay20, Gad A. Getz1,2,3, Michael J. Gloudemans29, Aaron Graubert1, Roderic Guigó34,35, Kane Hadley1, Andrew R. Hamel8,1, Robert E. Handsaker4,5,6, Yuan He11, Paul J. Hoffman16, Farhad Hormozdiari1,36, Lei Hou1,37, Katherine H. Huang1, Hae Kyung Im10, Brian Jo26,27, Silva Kasela16,17, Seva Kashin4,5,6, Manolis Kellis1,37, Sarah Kim-Hellmuth16,17,38, Alan Kwong22, Tuuli Lappalainen16,17, Xiao Li1, Xin Li30, Yanyu Liang10, Daniel G. MacArthur5,7, Serghei Mangul28,39, Samuel R. Meier1, Pejman Mohammadi16,17,40,41, Stephen B. Montgomery20,30, Manuel Muñoz-Aguirre34,42, Daniel C. Nachun30, Jared L. Nedzel1, Duyen T. Nguyen1, Andrew B. Nobel43, Meritxell Oliva10,44, YoSon Park15,45, Yongjin Park1,37, Princy Parsana12, Abhiram S. Rao46, Ferran Reverter47, John M. Rouhana1,8, Chiara Sabatti48, Ashis Saha12, Ayellet V. Segrè1,8, Andrew D. Skol10,49, Matthew Stephens50, Barbara E. Stranger10,51, Benjamin J. Strober11, Nicole A. Teran30, Ellen Todres1, Ana Viñuela13,23,24,52, Gao Wang50, Xiaoquan Wen22, Fred Wright53, Valentin Wucher34, Yuxin Zou54
Analysis Working Group Not Funded by GTEx Project Grants: Pedro G. Ferreira55,56,57,58, Gen Li59, Marta Melé60, Esti Yeger-Lotem61,62
Leidos Biomedical Project Management: Mary E. Barcus63, Debra Bradbury63, Tanya Krubit63, Jeffrey A. McLean63, Liqun Qi63, Karna Robinson63, Nancy V. Roche63, Anna M. Smith63, Leslie Sobin63, David E. Tabor63, Anita Undale63
Biospecimen Collection Source Sites: Jason Bridge64, Lori E. Brigham65, Barbara A. Foster66, Bryan M. Gillard66, Richard Hasz67, Marcus Hunter68, Christopher Johns69, Mark Johnson70, Ellen Karasik66, Gene Kopen71, William F. Leinweber71, Alisa McDonald71, Michael T. Moser66, Kevin Myer68, Kimberley D. Ramsey66, Brian Roe68, Saboor Shad71, Jeffrey A. Thomas71,70, Gary Walters70, Michael Washington70, Joseph Wheeler69
Biospecimen Core Resource: Scott D. Jewell72, Daniel C. Rohrer72, Dana R. Valley72
Brain Bank Repository: David A. Davis73, Deborah C. Mash73
Pathology: Mary E. Barcus63, Philip A. Branton74, Leslie Sobin63
ELSI Study: Laura K. Barker75, Heather M. Gardiner75, Maghboeba Mosavel76, Laura A. Siminoff75
Genome Browser Data Integration and Visualization: Paul Flicek77, Maximilian Haeussler78, Thomas Juettemann77, W. James Kent78, Christopher M. Lee78, Conner C. Powell78, Kate R. Rosenbloom78, Magali Ruffier77, Dan Sheppard77, Kieron Taylor77, Stephen J. Trevanion77, Daniel R. Zerbino77
eGTEx Group: Nathan S. Abell20, Joshua Akey79, Lin Chen44, Kathryn Demanelis44, Jennifer A. Doherty80, Andrew P. Feinberg81, Kasper D. Hansen82, Peter F. Hickey83, Lei Hou1,37, Farzana Jasmine44, Lihua Jiang20, Rajinder Kaul84,85, Manolis Kellis1,37, Muhammad G. Kibriya44, Jin Billy Li20, Qin Li20, Shin Lin86, Sandra E. Linder20, Stephen B. Montgomery20,30, Meritxell Oliva10,44, Yongjin Park1,37, Brandon L. Pierce44, Lindsay F. Rizzardi87, Andrew D. Skol10,49, Kevin S. Smith30, Michael Snyder20, John Stamatoyannopoulos84,88, Barbara E. Stranger10,51, Hua Tang20, Meng Wang20
NIH Program Management: Philip A. Branton74, Latarsha J. Carithers74,89, Ping Guan74, Susan E. Koester90, A. Roger Little91, Helen M. Moore74, Concepcion R. Nierras92, Abhi K. Rao74, Jimmie B. Vaught74, Simona Volpi93
1Broad Institute of MIT and Harvard, Cambridge, MA, USA. 2Cancer Center and Department of Pathology, Massachusetts General Hospital, Boston, MA, USA. 3Harvard Medical School, Boston, MA, USA. 4Department of Genetics, Harvard Medical School, Boston, MA, USA. 5Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA. 6Stanley Center for Psychiatric Research, Broad Institute of MIT and Harvard, Cambridge, MA, USA. 7Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, USA. 8Ocular Genomics Institute, Massachusetts Eye and Ear, Harvard Medical School, Boston, MA, USA. 9Department of Biomathematics, University of California, Los Angeles, CA, USA. 10Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, IL, USA. 11Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD, USA. 12Department of Computer Science, Johns Hopkins University, Baltimore, MD, USA. 13Department of Genetic Medicine and Development, University of Geneva Medical School, Geneva, Switzerland. 14Population Health and Genomics, University of Dundee, Dundee, Scotland, UK. 15Department of Genetics, University of Pennsylvania, Perelman School of Medicine, Philadelphia, PA, USA. 16New York Genome Center, New York, NY, USA. 17Department of Systems Biology, Columbia University, New York, NY, USA. 18Department of Genetics, Washington University School of Medicine, St. Louis, MO, USA. 19Division of Genetics, Oregon National Primate Research Center, Oregon Health & Science University, Portland, OR, USA. 20Department of Genetics, Stanford University, Stanford, CA, USA. 21Division of Genetic Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA. 22Department of Biostatistics, University of Michigan, Ann Arbor, MI, USA. 23Institute for Genetics and Genomics in Geneva (iGE3), University of Geneva, Geneva, Switzerland. 24Swiss Institute of Bioinformatics, Geneva, Switzerland. 25Department of Biomedical Informatics, Columbia University, New York, NY, USA. 26Department of Computer Science, Princeton University, Princeton, NJ, USA. 27Center for Statistics and Machine Learning, Princeton University, Princeton, NJ, USA. 28Department of Computer Science, University of California, Los Angeles, CA, USA. 29Program in Biomedical Informatics, Stanford University School of Medicine, Stanford, CA, USA. 30Department of Pathology, Stanford University, Stanford, CA, USA. 31Data Science Institute, Vanderbilt University, Nashville, TN, USA. 32Clare Hall, University of Cambridge, Cambridge, UK. 33MRC Epidemiology Unit, University of Cambridge, Cambridge, UK. 34Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, Barcelona, Catalonia, Spain. 35Universitat Pompeu Fabra (UPF), Barcelona, Catalonia, Spain. 36Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, USA. 37Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA, USA. 38Statistical Genetics, Max Planck Institute of Psychiatry, Munich, Germany 39Department of Clinical Pharmacy, School of Pharmacy, University of Southern California, Los Angeles, CA, USA. 40Scripps Research Translational Institute, La Jolla, CA, USA. 41Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA. 42Department of Statistics and Operations Research, Universitat Politècnica de Catalunya (UPC), Barcelona, Catalonia, Spain. 43Department of Statistics and Operations Research and Department of Biostatistics, University of North Carolina, Chapel Hill, NC, USA. 44Department of Public Health Sciences, The University of Chicago, Chicago, IL, USA. 45Department of Systems Pharmacology and Translational Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA. 46Department of Bioengineering, Stanford University, Stanford, CA, USA. 47Department of Genetics, Microbiology and Statistics, University of Barcelona, Barcelona, Spain. 48Departments of Biomedical Data Science and Statistics, Stanford University, Stanford, CA, USA. 49Department of Pathology and Laboratory Medicine, Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago, IL, USA. 50Department of Human Genetics, University of Chicago, Chicago, IL, USA. 51Center for Genetic Medicine, Department of Pharmacology, Northwestern University, Feinberg School of Medicine, Chicago, IL, USA. 52Department of Twin Research and Genetic Epidemiology, King’s College London, London, UK. 53Bioinformatics Research Center and Departments of Statistics and Biological Sciences, North Carolina State University, Raleigh, NC, USA. 54Department of Statistics, University of Chicago, Chicago, IL, USA. 55Department of Computer Sciences, Faculty of Sciences, University of Porto, Porto, Portugal. 56Instituto de Investigação e Inovação em Saúde, University of Porto, Porto, Portugal. 57Institute of Molecular Pathology and Immunology, University of Porto, Porto, Portugal. 58Laboratory of Artificial Intelligence and Decision Support, Institute for Systems and Computer Engineering, Technology and Science, Porto, Portugal. 59Mailman School of Public Health, Columbia University, New York, NY, USA. 60Life Sciences Department, Barcelona Supercomputing Center, Barcelona, Spain. 61Department of Clinical Biochemistry and Pharmacology, Ben-Gurion University of the Negev, Beer-Sheva, Israel. 62National Institute for Biotechnology in the Negev, Beer-Sheva, Israel. 63Leidos Biomedical, Rockville, MD, USA. 64Upstate New York Transplant Services, Buffalo, NY, USA. 65Washington Regional Transplant Community, Annandale, VA, USA. 66Therapeutics, Roswell Park Comprehensive Cancer Center, Buffalo, NY, USA. 67Gift of Life Donor Program, Philadelphia, PA, USA. 68LifeGift, Houston, TX, USA. 69Center for Organ Recovery and Education, Pittsburgh, PA, USA. 70LifeNet Health, Virginia Beach, VA. USA. 71National Disease Research Interchange, Philadelphia, PA, USA. 72Van Andel Research Institute, Grand Rapids, MI, USA. 73Department of Neurology, University of Miami Miller School of Medicine, Miami, FL, USA. 74Biorepositories and Biospecimen Research Branch, Division of Cancer Treatment and Diagnosis, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA. 75Temple University, Philadelphia, PA, USA. 76Virginia Commonwealth University, Richmond, VA, USA. 77European Molecular Biology Laboratory, European Bioinformatics Institute, Hinxton, UK. 78Genomics Institute, University of California, Santa Cruz, CA, USA. 79Carl Icahn Laboratory, Princeton University, Princeton, NJ, USA. 80Department of Population Health Sciences, The University of Utah, Salt Lake City, UT, USA. 81Departments of Medicine, Biomedical Engineering, and Mental Health, Johns Hopkins University, Baltimore, MD, USA. 82Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD, USA. 83Department of Medical Biology, The Walter and Eliza Hall Institute of Medical Research, Parkville, Victoria, Australia. 84Altius Institute for Biomedical Sciences, Seattle, WA, USA. 85Division of Genetics, University of Washington, Seattle, WA, USA. 86Department of Cardiology, University of Washington, Seattle, WA, USA. 87HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA. 88Genome Sciences, University of Washington, Seattle, WA, USA. 89National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA. 90Division of Neuroscience and Basic Behavioral Science, National Institute of Mental Health, National Institutes of Health, Bethesda, MD, USA. 91National Institute on Drug Abuse, Bethesda, MD, USA. 92Office of Strategic Coordination, Division of Program Coordination, Planning and Strategic Initiatives, Office of the Director, National Institutes of Health, Rockville, MD, USA. 93Division of Genomic Medicine, National Human Genome Research Institute, Bethesda, MD, USA.
Footnotes
Competing interests: J.A.D. is an affiliate investigator at the Fred Hutchinson Cancer Research Center, Seattle, WA, and an adjunct associate professor at The Geisel School of Medicine at Dartmouth, Hanover, NH. F.A. is an inventor on a patent application related to TensorQTL. GTEx Consortium members: S.E.C. is a co-founder, chief technology officer, and stock owner at Variant Bio; E.R.G. is on the Editorial Board of Circulation Research and does consulting for the City of Hope/Beckman Research Institute; E.T.D. is chairman and member of the board of Hybridstat, Ltd.; B.E.E. is on the scientific advisory boards of Celsius Therapeutics and Freenome; G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect, MSMutSig, POLYSOLVER, and TensorQTL. G.G. is a founder of and consultant to and holds privately held equity in Scorpion Therapeutics; S.B.M. is on the scientific advisory board of MyOme; D.G.M. is a co-founder with equity in Goldfinch Bio and has received research support from AbbVie, Astellas, Biogen, BioMarin, Eisai, Merck, Pfizer, and Sanofi-Genzyme; H.K.I. has received speaker honoraria from GSK and AbbVie; T.L. is a scientific advisory board member of Variant Bio with equity and Goldfinch Bio. P.F. is a member of the scientific advisory boards of Fabric Genomics, Inc., and Eagle Genomes, Ltd. P.G.F. is a partner of Bioinf2Bio.
Data and materials availability: Luminex TL measurement, RNA-seq gene expression, and eQTL summary statistic data are available on the GTEx Portal (www.gtexportal.org) for future research use. All GTEx protected data are available through the database of Genotypes and Phenotypes (dbGaP) (accession no. phs000424.v8). Code has been deposited at Zenodo (62).
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/369/6509/eaaz6876/suppl/DC1
Materials and Methods
Figs. S1 to S23
Tables S1 to S18
References (63–71)
MDAR Reproducibility Checklist
REFERENCES AND NOTES
- 1.Blackburn EH, Epel ES, Lin J, Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198 (2015). doi: 10.1126/science.aab3389; [DOI] [PubMed] [Google Scholar]
- 2.Harley CB, Futcher AB, Greider CW, Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990). doi: 10.1038/345458a0; [DOI] [PubMed] [Google Scholar]
- 3.Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE, Does a sentinel or a subset of short telomeres determine replicative senescence? Mol. Biol. Cell 15, 3709–3718 (2004). doi: 10.1091/mbc.e04-03-0207; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shay JW, Role of telomeres and telomerase in aging and cancer. Cancer Discov. 6, 584–593 (2016). doi: 10.1158/2159-8290.CD-16-0062; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G, The hallmarks of aging. Cell 153, 1194–1217 (2013). doi: 10.1016/j.cell.2013.05.039; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haycock PC et al. , Leucocyte telomere length and risk of cardiovascular disease: Systematic review and meta-analysis. BMJ 349, g4227 (2014). doi: 10.1136/bmj.g4227; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willeit P et al. , Leucocyte telomere length and risk of type 2 diabetes mellitus: New prospective cohort study and literature-based meta-analysis. PLOS ONE 9, e112483 (2014). doi: 10.1371/journal.pone.0112483; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arbeev KG et al. , Association of leukocyte telomere length with mortality among adult participants in 3 longitudinal studies. JAMA Netw. Open 3, e200023 (2020). doi: 10.1001/jamanetworkopen.2020.0023; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rode L, Nordestgaard BG, Bojesen SE, Long telomeres and cancer risk among 95 568 individuals from the general population. Int. J. Epidemiol 45, 1634–1643 (2016). doi: 10.1093/ije/dyw179; [DOI] [PubMed] [Google Scholar]
- 10.Haycock PC et al. , Association between telomere length and risk of cancer and non-neoplastic diseases: A Mendelian randomization study. JAMA Oncol. 3, 636–651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang C et al. , Genetic determinants of telomere length and risk of common cancers: A Mendelian randomization study. Hum. Mol. Genet 24, 5356–5366 (2015). doi: 10.1093/hmg/ddv252; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Codd V et al. , Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet 45, 422–427, e1–e2 (2013). doi: 10.1038/ng.2528; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangino M et al. , DCAF4, a novel gene associated with leucocyte telomere length. J. Med. Genet 52, 157–162 (2015). doi: 10.1136/jmedgenet-2014-102681; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangino M et al. , Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet 21, 5385–5394 (2012). doi: 10.1093/hmg/dds382; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delgado DA et al. , Genome-wide association study of telomere length among South Asians identifies a second RTEL1 association signal. J. Med. Genet 55, 64–71 (2018). doi: 10.1136/jmedgenet-2017-104922; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel CJ, Manrai AK, Corona E, Kohane IS, Systematic correlation of environmental exposure and physiological and self-reported behaviour factors with leukocyte telomere length. Int. J. Epidemiol 46, 44–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehkopf DH et al. , Leukocyte telomere length in relation to 17 biomarkers of cardiovascular disease risk: A cross-sectional study of US adults. PLOS Med. 13, e1002188 (2016). doi: 10.1371/journal.pmed.1002188; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniali L et al. , Telomeres shorten at equivalent rates in somatic tissues of adults. Nat. Commun 4, 1597 (2013). doi: 10.1038/ncomms2602; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabharwal S et al. , Telomere length dynamics in early life: The blood-and-muscle model. FASEB J. 32, 529–534 (2018). doi: 10.1096/fj.201700630r; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium GTEx, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318 (2020). doi: 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Materials and methods are available as supplementary materials.
- 22.Barrett JH, Iles MM, Dunning AM, Pooley KA, Telomere length and common disease: Study design and analytical challenges. Hum. Genet 134, 679–689 (2015). doi: 10.1007/s00439-015-1563-4; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce BL et al. , Telomere length measurement by a novel Luminex-based assay: A blinded comparison to Southern blot. Int. J. Mol. Epidemiol. Genet 7, 18–23 (2016). [PMC free article] [PubMed] [Google Scholar]
- 24.Cawthon RM, Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). doi: 10.1093/nar/30.10.e47; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibriya MG, Jasmine F, Roy S, Ahsan H, Pierce BL, Novel luminex assay for telomere repeat mass does not show well position effects like qPCR. PLOS ONE 11, e0155548 (2016). doi: 10.1371/journal.pone.0155548; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura M et al. , Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc 5, 1596–1607 (2010). doi: 10.1038/nprot.2010.124; [DOI] [PubMed] [Google Scholar]
- 27.Gardner M et al. , Gender and telomere length: Systematic review and meta-analysis. Exp. Gerontol 51, 15–27 (2014). doi: 10.1016/j.exger.2013.12.004; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebreab SY et al. , Less than ideal cardiovascular health is associated with shorter leukocyte telomere length: The National Health and Nutrition Examination Surveys, 1999–2002. J. Am. Heart Assoc 6, e004105 (2017). doi: 10.1161/JAHA.116.004105; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapham K et al. , Automated assay of telomere length measurement and informatics for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics 200, 1061–1072 (2015). doi: 10.1534/genetics.115.178624; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Astuti Y, Wardhana A, Watkins J, Wulaningsih W; PILAR Research Network, Cigarette smoking and telomere length: A systematic review of 84 studies and meta-analysis. Environ. Res 158, 480–489 (2017). doi: 10.1016/j.envres.2017.06.038; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carty CL et al. , Leukocyte telomere length and risks of incident coronary heart disease and mortality in a racially diverse population of postmenopausal women. Arterioscler. Thromb. Vasc. Biol 35, 2225–2231 (2015). doi: 10.1161/ATVBAHA.115.305838; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbers CC et al. , Comparison between southern blots and qPCR analysis of leukocyte telomere length in the health ABC study. J. Gerontol. A Biol. Sci. Med. Sci 69, 527–531 (2014). doi: 10.1093/gerona/glt121; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunt SC et al. , Leukocyte telomeres are longer in African Americans than in whites: The National Heart, Lung, and Blood Institute Family Heart Study and the Bogalusa Heart Study. Aging Cell 7, 451–458 (2008). doi: 10.1111/j.1474-9726.2008.00397.x; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch SM et al. , Race, ethnicity, psychosocial factors, and telomere length in a multicenter setting. PLOS ONE 11, e0146723 (2016). doi: 10.1371/journal.pone.0146723; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rewak M et al. , Race-related health disparities and biological aging: Does rate of telomere shortening differ across blacks and whites? Biol. Psychol 99, 92–99 (2014). doi: 10.1016/j.biopsycho.2014.03.007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delgado DA et al. , The contribution of parent-to-offspring transmission of telomeres to the heritability of telomere length in humans. Hum. Genet 138, 49–60 (2019). doi: 10.1007/s00439-018-1964-2; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen ME et al. , Shorter telomere length in Europeans than in Africans due to polygenetic adaptation. Hum. Mol. Genet 25, 2324–2330 (2016). doi: 10.1093/hmg/ddw070; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aston KI et al. , Divergence of sperm and leukocyte age-dependent telomere dynamics: Implications for male-driven evolution of telomere length in humans. Mol. Hum. Reprod 18, 517–522 (2012). doi: 10.1093/molehr/gas028; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rode L, Nordestgaard BG, Bojesen SE, Peripheral blood leukocyte telomere length and mortality among 64,637 individuals from the general population. J. Natl. Cancer Inst 107, djv074 (2015). doi: 10.1093/jnci/djv074; [DOI] [PubMed] [Google Scholar]
- 40.Giambartolomei C et al. , Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLOS Genet. 10, e1004383 (2014). doi: 10.1371/journal.pgen.1004383; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBoever C et al. , Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells. Cell Stem Cell 20, 533–546.e7 (2017). doi: 10.1016/j.stem.2017.03.009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanley SE et al. , Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis-emphysema. Sci. Transl. Med 8, 351ra107 (2016). doi: 10.1126/scitranslmed.aaf7837; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savage SA, Beginning at the ends: Telomeres and human disease. F1000Res. 7, 524 (2018). doi: 10.12688/f1000research.14068.1; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shay JW, Wright WE, Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet 20, 299–309 (2019). doi: 10.1038/s41576-019-0099-1; [DOI] [PubMed] [Google Scholar]
- 45.Armanios M, Blackburn EH, The telomere syndromes. Nat. Rev. Genet 13, 693–704 (2012). doi: 10.1038/nrg3246; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart BD et al. , Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat. Genet 47, 512–517 (2015). doi: 10.1038/ng.3278; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dressen A et al. , Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: A candidate gene sequencing study. Lancet Respir. Med 6, 603–614 (2018). doi: 10.1016/S2213-2600(18)30135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alder JK et al. , Diagnostic utility of telomere length testing in a hospital-based setting. Proc. Natl. Acad. Sci. U.S.A 115, E2358–E2365 (2018). doi: 10.1073/pnas.1720427115; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton CA et al. , Telomere-related lung fibrosis is diagnostically heterogeneous but uniformly progressive. Eur. Respir. J 48, 1710–1720 (2016). doi: 10.1183/13993003.00308-2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Günes C, Rudolph KL, The role of telomeres in stem cells and cancer. Cell 152, 390–393 (2013). doi: 10.1016/j.cell.2013.01.010; [DOI] [PubMed] [Google Scholar]
- 51.Ozturk S, Telomerase activity and telomere length in male germ cells. Biol. Reprod 92, 53 (2015). doi: 10.1095/biolreprod.114.124008; [DOI] [PubMed] [Google Scholar]
- 52.Tingley D, Yamamoto T, Hirose K, Keele L, Imai K, mediation: R package for causal mediation analysis. J. Stat. Softw 59, 1–38 (2014). doi: 10.18637/jss.v059.i0526917999 [DOI] [Google Scholar]
- 53.Pierce BL et al. , Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: A genome-wide analysis among 1,800 South Asians. PLOS Genet. 10, e1004818 (2014). doi: 10.1371/journal.pgen.1004818; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tomasetti C, Vogelstein B, Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 347, 78–81 (2015). doi: 10.1126/science.1260825; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomasetti C, Li L, Vogelstein B, Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science 355, 1330–1334 (2017). doi: 10.1126/science.aaf9011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aran D, Hu Z, Butte AJ, xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017). doi: 10.1186/s13059-017-1349-1; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim-Hellmuth S et al. , Cell type–specific genetic regulation of gene expression across human tissues. Science 369, eaaz8528 (2020). doi: 10.1126/science.aaz8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin J et al. , Analyses and comparisons of telomerase activity and telomere length in human T and B cells: Insights for epidemiology of telomere maintenance. J. Immunol. Methods 352, 71–80 (2010). doi: 10.1016/j.jim.2009.09.012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barthel FP et al. , Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet 49, 349–357 (2017). doi: 10.1038/ng.3781; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharifi-Sanjani M, Meeker AK, Mourkioti F, Evaluation of telomere length in human cardiac tissues using cardiac quantitative FISH. Nat. Protoc 12, 1855–1870 (2017). doi: 10.1038/nprot.2017.082; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ding Z, Mangino M, Aviv A, Spector T, Durbin R; UK10K Consortium, Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 42, e75 (2014). doi: 10.1093/nar/gku181; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demanelis K, kdemanelis/gtex_telomerelength v1.1. Zenodo (2020); 10.5281/zenodo.3969532. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


