Abstract
The envelope glycoprotein, termed the spike protein, of severe acute respiratory syndrome coronavirus (SARS-CoV) is known to mediate viral entry. Similar to other class 1 viral fusion proteins, the heptad repeat regions of SARS-CoV spike are thought to undergo conformational changes from a prefusion form to a subsequent post-fusion form that enables fusion of the viral and host membranes. Recently, the structure of a post-fusion form of SARS-CoV spike, which consists of isolated domains of heptad repeats 1 and 2 (HR1 and HR2), has been determined by x-ray crystallography. To date there is no structural information for the prefusion conformations of SARS-CoV HR1 and HR2. In this work we present the NMR structure of the HR2 domain (residues 1141-1193) from SARS-CoV (termed S2-HR2) in the presence of the co-solvent trifluoroethanol. We find that in the absence of HR1, S2-HR2 forms a coiled coil symmetric trimer with a complex molecular mass of 18 kDa. The S2-HR2 structure, which is the first example of the prefusion form of coronavirus envelope, supports the current model of viral membrane fusion and gives insight into the design of structure-based antagonists of SARS.
In 2003, an atypical pneumonia, now known as severe acute respiratory syndrome (SARS)2 , caused a deadly worldwide outbreak originating in Guangdong province of China, eventually spreading to ∼30 countries and infecting ∼8000 people with a mortality rate of 10% (1). SARS is a respiratory illness caused by the newly discovered human coronavirus termed SARS-CoV, bearing low sequence homology to other members in the Coronaviridae family and therefore classified as a new group, group IV (2, 3, 4, 5).
Coronaviruses belong to a family of enveloped viruses, which use extracellular glycoproteins to mediate the fusion of the viral and cellular membranes. Other members of this group of viruses include: influenza virus hemagglutinin (HA), Ebola virus glycoprotein, human immunodeficiency virus (HIV) gp160, and parainfluenza virus HN and F (6, 7, 8). Such viral envelope glycoproteins consist of 2 extracellular domains: a receptor-binding domain and a domain that mediates fusion of the virus and target membranes (9). In SARS-CoV, the receptor-binding domain is termed S1, and the fusion domain is termed S2.
The viral fusion proteins, including the SARS-CoV S2, are believed to share a similar mechanism for viral entry and fusion (reviewed in Refs. 7, 10, and 11). Although the details of SARS-CoV entry are still unclear, it is believed that following binding to the host cell receptor, the angiotensin-converting enzyme 2 (12, 13), S2, the fusion domain, undergoes a series of structural changes from a prefusion conformation to a postfusion conformation. During these structural changes a fusion peptide within S2 is exposed and inserted into the membrane of the target cell. Within S2, there are two heptad repeat domains: HR1 and HR2. The post-fusion form of the fusion domain is relatively well characterized by structural biology. For example, high resolution structures have been determined for HIV gp41 (14, 15, 16), simian immunodeficiency virus gp41 (17, 18, 19), influenza virus HA (20), Ebola virus glycoprotein (21, 22), mouse hepatitis virus (23), SARS-CoV (24, 25), and parainfluenza virus F (26). Note that all of these structures represent the so called six-helix bundle form or fusion core, where the N-terminal helices, HR1, form a trimeric coiled coil core surrounded by the C-terminal helices, HR2, oriented in an antiparallel fashion. This conformation is thought to allow the target membrane to come into close contact with the viral membrane, ultimately promoting membrane fusion and viral entry. In the prefusion state, the HR2 region is thought to self-associate to form a trimeric coiled coil. Evidence for the prefusion state is based on studies of influenza HA (27, 28) and more recently parainfluenza F (29). Further evidence comes from studies of peptides from HIV gp41 and parainfluenza F corresponding to the HR1 and HR2 regions. These peptides are thought to inhibit viral fusion by binding to the prefusion conformation and thereby block the formation of the HR1/HR2 six-helix bundle that is necessary for membrane fusion (7, 30, 31, 32, 33). One of the peptides, corresponding to the HR2 region of gp41, termed T20 or fuzeon, is currently used as a drug against HIV (34, 35). Interestingly, analogous peptides from SARS-CoV have also been shown to exhibit antiviral activity, although at significantly higher concentrations than T20 (36, 37, 38, 39).
Recent studies have shown that SARS-CoV S2-HR2 in isolation, unlike the corresponding domain from HIV gp41, forms a helical structure in solution based on circular dichroism studies and, further, that the predominant species present is that of a trimer (40). However, a detailed three-dimensional structure of the SARS-CoV S2-HR2 has not yet been available. In this work, we present the solution structure of the timeric SARS-CoV-S2-HR2 in the presence of the co-solvent TFE, which was necessary to solubilize the construct. We find that S2-HR2 forms a symmetric coiled coil trimer in solution. Importantly, this structure provides the first structural evidence for the prefusion state of SARS-CoV envelope. Moreover, the S2-HR2 structure provides a framework for the design of novel peptides that inhibit viral entry.
EXPERIMENTAL PROCEDURES
Expression and Purification—The SARS-CoV S2-HR2 domain consisting of residues 1141-1193 was subcloned into the BamHI/HindIII restriction sites of a modified pQE30 expression vector (Qiagen). The resulting construct, termed His-PG-S2-HR2, consists of a N-terminal polyhistidine tag followed by protein G (the IgG-binding domain of streptococcus protein G) (41), a tobacco etch virus cleavage site (sequence, ENLYFQGS) (42) for removal of the expression tag, and S2-HR2. For simplicity, from here on S2-HR2 will be numbered 1-55 corresponding to residues 1141-1193 of the intact protein (the N-terminal glycine and serine residues are a cloning artifact). Protein expression of 15N-labeled and 13C/15N-labeled S2-HR2 was achieved by growing Escherichia coli strain SG13009 as previously described (43). Briefly, the cells were grown in 4 liters of LB medium at 37 °C until they reached an A 600 of 0.8. Subsequently, the cells were pelleted, washed one time with M9 minimal medium, resuspended in 1 liter of M9 minimal medium supplemented with 1 g/liter of 15NH4Cl (Martek Biosciences, Columbia, MD) and 4 g/liter of 13C-glucose (Isotec, Miamisburg, OH), set to recover for 1 h at 37°C, induced with 0.8 mm isopropyl β-d-thiogalactopyranoside, and grown for an additional 4-5 h at 37 °C. The His-PG-S2-HR2 fusion protein was purified from the soluble fraction using a Ni2+ fast flow Sepharose column (Qiagen). The protein was then cleaved using tobacco etch virus protease and run once more over the Ni2+ column to remove His-PG and tobacco etch virus protease, which also contains a polyhistidine tag. The flow through fraction containing S2-HR2 was then dialyzed extensively against 10 mm NaPO4, pH 7.0, and concentrated by ultrafiltration (YM3; Amicon, Billerica, MA). The purity and identity of S2-HR2 were confirmed by SDS-PAGE and matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
NMR Spectroscopy—For the NMR experiments, the experimental conditions were 1 mm S2-HR2 in 10 mm NaPO4, pH 7.0, 30% TFE-d3 (Cambridge Isotope Laboratories, Andover, MA), and 10% 2H2O. NMR spectra were recorded at 25 °C on a Buker AVANCE 800 MHz spectrometer equipped with a cryogenic triple resonance probe. The spectra were processed by NmrPipe and visualized with NmrDraw (44). Backbone assignments were determined using a standard set of three-dimensional heteronuclear NMR experiments including: HNCO, CBCA-(CO)NH and CBCANH (45, 46). The side chain resonances were assigned by three-dimensional 15N-edited TOCSY-HSQC, threedimensional HCC(CO)NH, and three-dimensional CC(CO)NH experiments. A three-dimensional 15N-edited NOESY-HSQC (mixing time, 120 ms) was acquired for structural restraint information as well as confirmation of side chain assignments. An additional HNHA experiment was performed to add 3JHNα information (47). The intersubunit NOEs were identified by recording a three-dimensional F1-filtered, F2-edited 1H-13C NOESY-HSQC with a mixing time of 120 ms (48) on a 1 mm sample comprised of a mixture of 50% 14N/12C-labeled and 50% 15N/13C-labeled monomers. Exchangeable amide protons were identified by the CLEANEX-PM experiment with a mixing time of 100 ms (49). HNOE values were measured by a standard pulse sequence (reviewed in Ref. 50). The HNOE values for each residue were calculated as the intensity ratio (I/I 0)ofthe 15N-1H correlation peak in the presence (I) and absence (I 0) of proton saturation during the relaxation delay of 5 s. Spectral processing and analysis was performed using the program NMRPipe (44).
Structure Determination—NOEs were manually assigned and classified as: strong (1.8-2.7 or 1.8-2.9 Å for the NOE of NH), medium (1.8-3.3 or 1.8-3.5 Å for NOE of NH), weak (1.8-5.0 Å), or very weak (1.8-6.0 Å). For distances involving methyl protons, 0.5 Å was added to the upper limit to account for the higher apparent intensity of methyl protons. ψ and φ torsion angle restraints were derived from 3JHNA (51) and a data base analysis of chemical shifts (N, HN,Cα,Cβ,C′,Hα) using the program TALOS (52). Minimum error ranges for ψ and φ were set to ± 40 and ±20, respectively. H-bonds, which were identified by the 13C backbone chemical shifts, short range NOE patterns, and nonex-changeable protons, were incorporated for the helical region, residues 17-48, as two restraints per H-bond where rNH-O = 1.5-2.8 Å and rN-O = 2.4-3.5 Å. The structures were calculated by simulated annealing in torsion angle space (53) starting from three extended strands, followed by conventional simulated annealing (54), using the program CNS (55). The program was adapted to incorporate a conformational data base (56) and a pseudopotential term for noncrystallographic symmetry (17, 57). A family of the 30 lowest energy structures was chosen, and a minimized mean structure was calculated. The overall quality of the final structures was assessed using the program PRO-CHECK (58). The figures were generated using the program MOL-MOL (59).
RESULTS AND DISCUSSION
Optimization of Experimental Conditions—Previous analytical ultracentrifugation studies have demonstrated that SARS CoV S2-HR2 is a trimer (40). Accordingly, our initial NMR experiments on S2-HR2 were performed under varying conditions of pH and salt to optimize the solubility of the construct. However, at protein concentrations greater than 400 μm, which are typically necessary for NMR structural studies, S2-HR2 tends to form higher molecular mass species (larger than that of a trimer, 18 kDa) as evidenced by the presence of additional broad peaks in the 15N-edited HSQC (data not shown). The additional broad peaks of the higher molecular mass species are only observed at relatively high concentrations and therefore believed to be nonphysiologically relevant. The co-solvent TFE has previously been used to break up aggregates formed between coiled coil complexes without breaking up the true coiled coil quaternary structure (60). By the addition of 30% TFE, we were able to obtain NMR spectra with one set of resonance peaks at a concentration of 1 mm, which was suitable for high resolution structural studies.
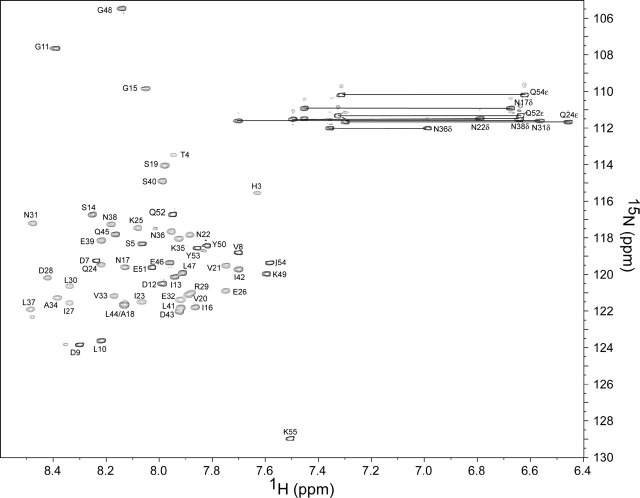
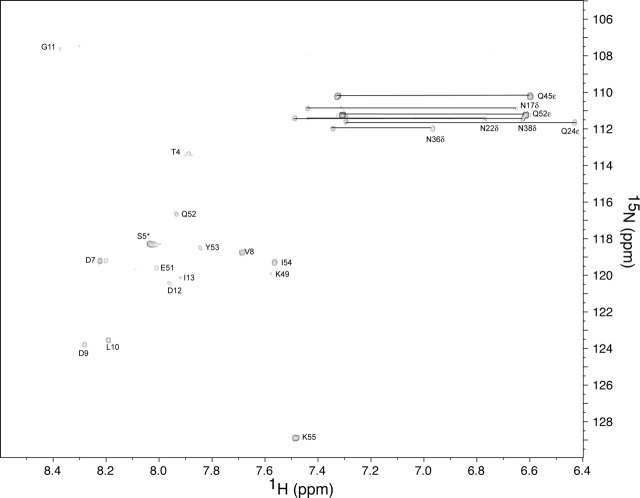
Resonance Assignment and Secondary Structure—A two-dimensional 15N-edited HSQC spectrum of S2-HR2 (buffer conditions 10 mm NaPO4, pH 7.0, 30% TFE-d3, 10% 2H2O) is shown in Fig. 1 . The backbone assignments were obtained using the standard set of three-dimensional triple resonance experiments as described under “Experimental Procedures” (the resonance assignments have been deposited in the Biological Magnetic Resonance Databank under entry 6969). The 15N-1H assignments are shown in Fig. 1. The 13C′ and 13Cα chemical shifts with respect to random coil values are shown in Fig. 2a . The secondary chemical shift suggests the presence of helix from residues 17 to 51. As shown by Fig. 2b, the HNOE profile further supports the presence of the helical region encompassing this region. For example, residues 3-16 and 52-55 clearly exhibit higher mobility with respect to the central region. The presence of exchangeable amide protons was next probed by CLEANEX-PM experiments (49). As shown in Fig. 3 , the amides of residues 7-13, 49, and 51-55 are in relatively fast exchange with solvent. Based on the 13C chemical shifts, HNOE, and amide exchange experiment, as well as the short range NOE patterns, S2-HR2 consists of a continuous helix from residues Asn17 to Leu47.
FIGURE 1.
15N-edited HSQC of SARS-CoV S2-HR2. The sample conditions were 1 mm S2-HR2 monomer, 10 mm NaPO4, pH 7.0, 30% TFE-d3 at 25 °C. Horizontal lines connect the side chain amide proton pairs of asparagine and glutamine residues.
FIGURE 2.
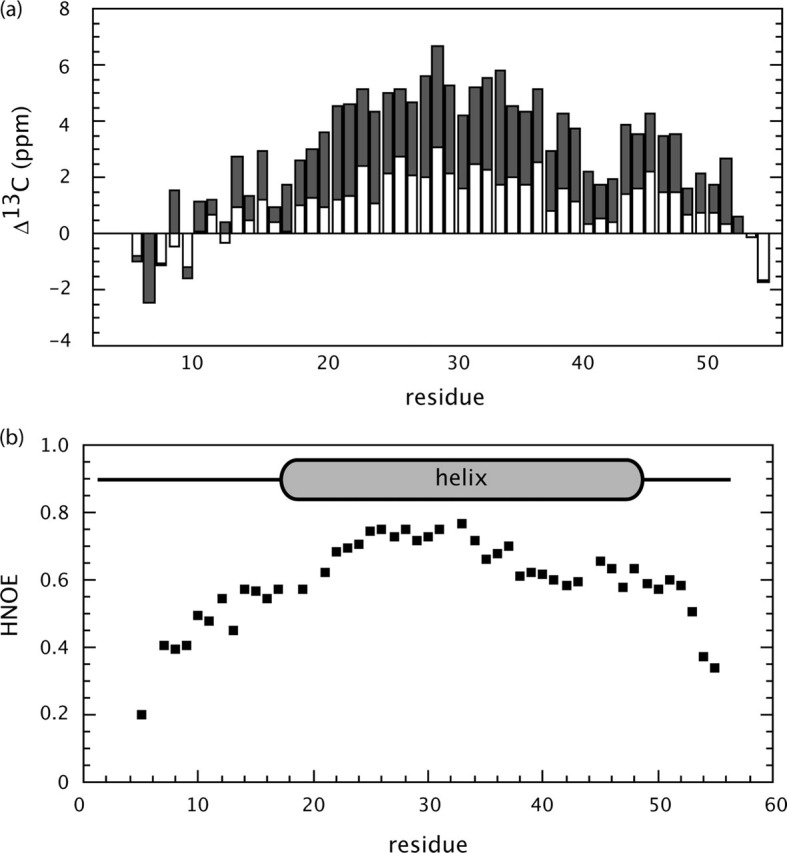
Secondary structure of SARS-CoV S2-HR2.a, secondary chemical shift of S2-HR2 13Cα (gray) and 13C′ (white) with respect to random coil values. Random coil values were taken from Wishart and Case (64). b, HNOE of S2-HR2.
FIGURE 3.
CLEANEX-PM 15N-edited HSQC. The sample conditions were 1 mm of S2-HR2 monomer, 10 mm NaPO4, pH 7.0, 30% TFE-d3 at 25 °C with a mixing time of 100 ms. Note that residues Thr4 and Ser5, which occur in an unstructured region at the N terminus, exhibit negative contours, presumably because of ROE effects and/or intermolecular NOEs with H2O (49). The absence of a correlations for residues 17-47 suggests that they are involved in H-bonds. The absence of correlations for residues 14-16 and 50 may suggest the presence of H-bonds. The other missing correlations include those of residues 1-3, which are also missing from the 15N-edited HSQC (residue 6 is a proline).
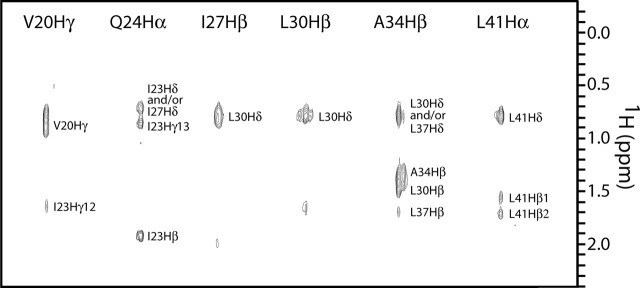
Structure Determination—The calculation of a symmetric trimer using NMR is a challenging task and requires the use of isotopic-filtered NOE experiments (reviewed in Ref. 61). In the present work, intermolecular NOEs were identified by a three-dimensional F1-filtered F2-edited 1H-13C NOESY-HSQC (48) experiment recorded on a 1:1 mixture of 13C/15N: 12C/14N-labeled S2-HR2. Based on previous analytical ultracentrifugation experiments, S2-HR2 is a trimer; hence one is not able to distinguish between intermolecular NOEs for example between subunit A and B or subunit A and C. We therefore assigned the NOEs in such a way that a proton on one subunit is allowed to interact with protons of the other subunits as (∑r-6)-1/6 sums (17). Examples of the quality of the filtered NOE spectrum are shown for selected residues in Fig. 4 . In total, all intermolecular NOEs were found between residues in the a, d, e, and g positions of the helical wheel, consistent with the interface of a trimeric coiled coil structure, to be discussed below. The solution structure of S2-HR2 was determined using information from a total of 344 NOEs (including 85 intermolecular NOEs), 30 H-bonds, 19 J-coupling constants, and 110 dihedral restraints/subunit (Table 1 ). The structure statistics of the family of the 30 lowest energy structures and the final minimized mean structure are summarized in Table 1. The superposition of the backbone of the final 30 lowest energy simulated annealing structures is shown in Fig. 5a . The final ensemble shows no NOE violations over 0.5 Å and no dihedral angles in the disallowed regions of the Ramachandran plot (Table 1). The root mean square deviation of residues Asn17-Leu47, which comprise the helical regions, relative to the mean is 0.46 Å for the backbone atoms and 1.04 Å for the heavy atoms.
FIGURE 4.
Intermolecular NOEs of SARS-CoV S2-HR2 showing intersubunit contacts involving the helix. Selected strips from the three-dimensional F1-filtered F2-edited 1H-13C NOESY-HSQC spectrum (mixing time of 120 ms) recorded on a 1:1 mixture of 12C/14N-and 13C/15N-labled S2-HR2.
TABLE 1.
Structural statistics
Shown are the structural statistics for the final 30 simulated annealing structures (<SA>) and the minimized mean (<SA>r). None of the structures exhibit distance violations greater than 0.5 Å, dihedral violations greater than 10°, or coupling constant violations greater than 2 Hz. The number of restraints/subunit is given in parentheses.
| <SA> | <SA>r | |
|---|---|---|
| Root mean square deviation from distance restraints | ||
| Total (344) | 0.014 ± 0.0020 | 0.012 |
| Intraresidue (57) | 0.004 ± 0.003 | 0.003 |
| Sequential () (153) | 0.010 ± 0.003 | 0.008 |
| Short range () (49) | 0.014 ± 0.003 | 0.014 |
| Intermolecular (85) | 0.024 ± 0.004 | 0.019 |
| Root mean square deviations from dihedral restraints (deg) (110)a | 0.19 ± 0.14 | 0.16 |
| Root mean square deviations from JHNα coupling (Hz) (19) | 0.449 ± 0.09 | 0.539 |
| Deviations from Idealized Covalent Geometry | ||
| Bonds (Å) | 0.0018 ± 0.00016 | 0.0016 |
| Angels (deg) | 0.389 ± 0.022 | 0.360 |
| Improper torsion (deg) | 0.339 ± 0.026 | 0.317 |
| Measures of Structure Qualityb | ||
| Residues in most favorable region (%) | 88.1 ± 3.2 | 89.8 |
| Residues in additionally allowed region (%) | 11.5 ± 3.5 | 8.2 |
| Residues in generously allowed region (%) | 0.4 ± 0.8 | 2.0 |
| Residues in disallowed region (%) | 0.0 ± 0.0 | 0.0 |
| Number of bad contacts/100 residues | 4.8 ± 3.3 | 6.0 |
| Coordinates precision (Å)c | ||
| Backbone residues 17-47 | 0.46 ± 0.17 | |
| Heavy residues 17-47 | 1.04 ± 0.15 |
Torsion angle restraints consisted of 51 φ, 51 ψ, 4 χ1, and 4 χ2 per subunit.
Overall quality of the structure as determined by the program PROCHECK (58).
Defined as average root mean square difference between the ensemble structures and the mean structure.
FIGURE 5.
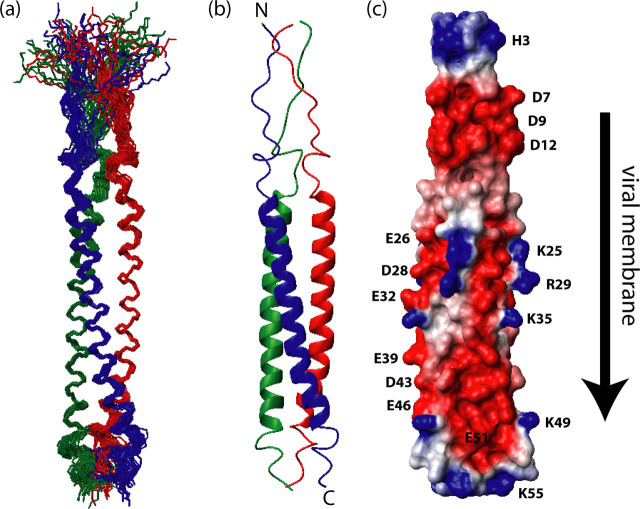
Solution structure of SARS-CoV S2-HR2.a, ensemble of 30 low energy structures of showing the superimposition of the backbone atoms. b, ribbon representation of the minimized mean structure. c, electrostatic map of the minimized mean structure. In a and b, subunits A, B, and C are colored red, green, and blue, respectively. The direction of the viral membrane is shown by an arrow.
S2-HR2 Solution Structure—A ribbon representation of the minimized mean structure of S2-HR2 is shown in Fig. 5b. In this representation, subunits A, B, and C are colored red, green, and blue, respectively. S2-HR2 forms a symmetric trimeric coiled coil with an overall length of ∼96 Å and width of ∼13 Å. There are extensive contacts between the three subunits with ∼6150 Å2 buried upon complex formation. Interestingly, the side chains of the two conserved glycosylation sites of HR2, Asn17 and Asn38, are exposed to the surface as expected. A surface electrostatic representation of S2-HR2 shown in Fig. 5c reveals that the charge is evenly distributed along the entire helix. The main interface of the S2-HR2 coiled coil trimer was determined to include residues Asn17-Leu47.
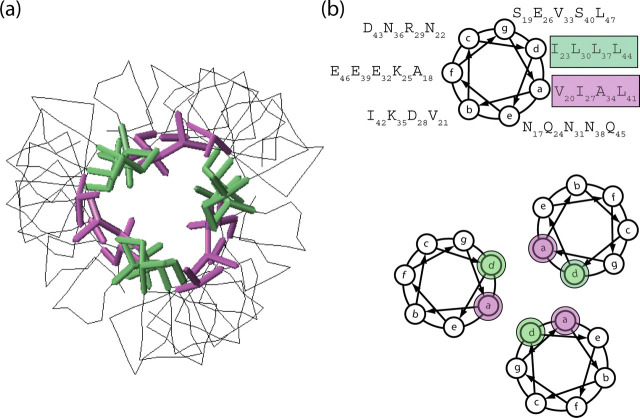
A coiled coil structure is a bundle of α-helices that interact with each other in such a way that they will form a left-handed superhelix. Residues in the a and d positions are often hydrophobic residues that will interact with each other to form the hydrophobic core. This packing is often referred to as knobs-into-holes and is the hallmark for a true coiled coil structure (62, 63). The packing of the amino acid side chains within S2-HR2 coiled coil is shown in Fig. 6 . The hydrophobic residues in the a position (Val20, Ile27, Ala34, and Leu41) are colored purple, and those in the d position (Ile23, Leu30, Leu37, and Leu44) are colored green to show the relative orientation and positioning. The stacking of these residues is arranged in a way that creates a tightly packed core within the S2-HR2 trimer with the hydrophobic interface buried. Positions e and g within a heptad repeat usually contain charged amino acids placed in such a way that the charges are complementary to each other to allow electrostatic interaction. In the case of S2-HR2, there is only one charged amino acid, in position g (Glu26), and therefore it is not likely that the e and g positions play important roles in the stability of the S2-HR2 trimer. On the other hand, positions b, c, and f are surface exposed in the coiled coil structure and generally encompass polar amino acids that contribute to the solubility of the protein. In the coiled coil region of S2-HR2, we note that there are two surface exposed hydrophobic residues, Val21 and Ile42, that are conserved among coronaviruses. Interestingly, in the fusion structure (25), post-Val and Ile form contacts with the HR1 that presumably stabilize the six-helix bundle conformation.
FIGURE 6.
Structural features of SARS-CoV S2-HR2.a, intermolecular contacts between the S2-HR helices. b, helical wheel representation of S2-HR2 residues Asn17-Leu47 looking down the helical axis, starting at the N-terminal end. There are seven residues/heptad where the individual positions of the seven residues are denoted by the letters a-f. The residues in the a and d positions make up the hydrophobic interface of the trimeric coiled coil of the S2-HR2 structure. The a positions are highlighted purple, and the d positions are highlighted green.
Conclusions—In conclusion SARS-CoV S2-HR2 forms a coiled coil structure in solution consisting of three helices coming together to form a parallel trimer. As mentioned in the introduction, SARS-CoV S2 is thought to exist in two conformations: a prefusion conformation in which HR1 and HR2 do not interact with each other and a fusion conformation in which HR2 interacts with the trimeric coiled coil of HR1 in an antiparallel fashion to form the six-helix bundle, which was also observed for HIV and simian immunodeficiency virus (SIV) gp41 and Ebola glycoprotein. In light of our results, it is enticing to speculate that the present structure represents a prefusion intermediate of the S2-HR2 region, which has never before been observed for coroniviridae. Importantly, the information provided in this study may be exploited for the design of novel antivirals. For example, the relatively poor antiviral activity of SARS-CoV S2-HR2 may be a consequence of its tendency to form the trimeric coiled coil, and thus one potential strategy would be to mutate residues that are important to the formation of a HR2 trimer while at the same time maintaining the interactions necessary for the HR2-HR1 interaction.
Acknowledgments
We thank the members of the Brian Volkman laboratory (Uninversity of Wisconsin Medical College) for help with the intermolecular NOE experiment.
Footnotes
The atomic coordinates and structure factors (code 2FXP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The abbreviations used are: SARS, severe acute respiratory syndrome; HR, heptad repeat; CoV, coronavirus; TFE, trifluoroethanol; HA, hemagglutinin; HIV, human immunodeficiency virus; NOE, nuclear Overhauser effect; HSQC, heteronuclear single quantum correlation; NOESY, NOE spectroscopy; HNOE, heteronuclear NOE.
References
- 1.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., Rappuoli R. Nat. Rev. Microbiol. 2003;1:209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Preiser W., Gunther S., Schmitz H., Doerr H.W. Trends Mol. Med. 2003;9:325–327. doi: 10.1016/S1471-4914(03)00133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez L.D., Hoffman L.R., Wolfsberg T.G., White J.M. Annu. Rev. Cell Dev. Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 7.Eckert D.M., Kim P.S. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 8.Lamb R.A., Paterson R.G., Jardetzky T.S. Virology. 2006;344:30–37. doi: 10.1016/j.virol.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher T.M., Buchmeier M.J. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo S.A., Finnegan C.M., Viard M., Raviv Y., Dimitrov A., Rawat S.S., Puri A., Durell S., Blumenthal R. Biochim. Biophys. Acta. 2003;1614:36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann H., Pohlmann S. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn J.H., Li W., Choe H., Farzan M. Cell. Mol. Life Sci. 2004;61:2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan D.C., Fass D., Berger J.M., Kim P.S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 15.Tan K., Liu J., Wang J., Shen S., Lu M. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 17.Caffrey M., Cai M., Kaufman J., Stahl S.J., Wingfield P.T., Covell D.G., Gronenborn A.M., Clore G.M. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malashkevich V.N., Chan D.C., Chutkowski C.T., Kim P.S. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9134–9139. doi: 10.1073/pnas.95.16.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Z.N., Mueser T.C., Kaufman J., Stahl S.J., Wingfield P.T., Hyde C.C. J. Struct. Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 20.Wilson I.A., Skehel J.J., Wiley D.C. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 21.Weissenhorn W., Carfi A., Lee K.H., Skehel J.J., Wiley D.C. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 22.Malashkevich V.N., Schneider B.J., McNally M.L., Milhollen M.A., Pang J.X., Kim P.S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y., Liu Y., Lou Z., Qin L., Li X., Bai Z., Pang H., Tien P., Gao G.F., Rao Z. J. Biol. Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F., Rao Z. J. Biol. Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supekar V.M., Bruckmann C., Ingallinella P., Bianchi E., Pessi A., Carfi A. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin H.S., Paterson R.G., Wen X., Lamb R.A., Jardetzky T.S. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carr C.M., Kim P.S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 28.Bullough P.A., Hughson F.M., Skehel J.J., Wiley D.C. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 29.Yin H.S., Wen X., Paterson R.G., Lamb R.A., Jardetzky T.S. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wild C., Oas T., McDanal C., Bolognesi D., Matthews T. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wild C.T., Shugars D.C., Greenwell T.K., McDanal C.B., Matthews T.J. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuta R.A., Wild C.T., Weng Y., Weiss C.D. Nat. Struct. Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 33.Russell C.J., Jardetzky T.S., Lamb R.A. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilby J.M., Hopkins S., Venetta T.M., DiMassimo B., Cloud G.A., Lee J.Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M.R., Nowak M.A., Shaw G.M., Saag M.S. Nat. Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 35.Kilby J.M., Lalezari J.P., Eron J.J., Carlson M., Cohen C., Arduino R.C., Goodgame J.C., Gallant J.E., Volberding P., Murphy R.L., Valentine F., Saag M.S., Nelson E.L., Sista P.R., Dusek A. AIDS Res. Hum. Retroviruses. 2002;18:685–693. doi: 10.1089/088922202760072294. [DOI] [PubMed] [Google Scholar]
- 36.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosch B.J., Martina B.E., Van Der Zee R., Lepault J., Haijema B.J., Versluis C., Heck A.J., De Groot R., Osterhaus A.D., Rottier P.J. Proc. Natl. Acad. Sci. U. S. A. 2004;101:8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C.R., Xiong H., Farmar J., Debnath A.K., Tien P., Jiang S. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y., Yan H., Cole D.K., Bell J.I., Rao Z., Tien P., Gao G.F. Biochem. Biophys. Res. Commun. 2004;319:283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tripet B., Howard M.W., Jobling M., Holmes R.K., Holmes K.V., Hodges R.S. J. Biol. Chem. 2004;279:20836–20849. doi: 10.1074/jbc.M400759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huth J.R., Bewley C.A., Jackson B.M., Hinnebusch A.G., Clore G.M., Gronenborn A.M. Protein Sci. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dougherty W.G., Parks T.D., Cary S.M., Bazan J.F., Fletterick R.J. Virology. 1989;172:302–310. doi: 10.1016/0042-6822(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 43.Marley J., Lu M., Bracken C. J. Biomol. NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Caffrey M., Cai M., Kaufman J., Stahl S.J., Wingfield P.T., Gronenborn A.M., Clore G.M. J. Mol. Biol. 1997;271:819–826. doi: 10.1006/jmbi.1997.1217. [DOI] [PubMed] [Google Scholar]
- 46.Jiang S., Caffrey M. J. Biomol. NMR. 2002;24:365–366. doi: 10.1023/a:1021641814058. [DOI] [PubMed] [Google Scholar]
- 47.Kuboniwa H., Grzesiek S., Delaglio F., Bax A. J. Biomol. NMR. 1994;4:871–878. doi: 10.1007/BF00398416. [DOI] [PubMed] [Google Scholar]
- 48.Stuart A.C., Borzilleri K.A., Withka J.M., Palmer A.G. J. Am. Chem. Soc. 1999;121:5346–5347. [Google Scholar]
- 49.Hwang T.L., van Zijl P.C., Mori S. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 50.Cavanagh J., Fairbrother W., Palmer A., Skeleton N. Academic Press; San Diego: 1996. Protein NMR Spectroscopy Principles and Practice. [Google Scholar]
- 51.Bax A., Vuister G.W., Grzesiek S., Delaglio F., Wang A.C., Tschudin R., Zhu G. Methods Enzymol. 1994;239:79–105. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 52.Cornilescu G., Delaglio F., Bax A. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 53.Stein E.G., Rice L.M., Brunger A.T. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- 54.Nilges M., Clore G.M., Gronenborn A.M. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 55.Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Kuszewski J., Gronenborn A.M., Clore G.M. Protein Sci. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caffrey M. Biochim. Biophys. Acta. 2001;1536:116–122. doi: 10.1016/s0925-4439(01)00042-4. [DOI] [PubMed] [Google Scholar]
- 58.Laskowski R.A., MacArther M.W., Moss D.S., Thornton J.M. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 59.Koradi R., Billeter M., Wuthrich K. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 60.Lindhout D.A., Litowski J.R., Mercier P., Hodges R.S., Sykes B.D. Biopolymers. 2004;75:367–375. doi: 10.1002/bip.20150. [DOI] [PubMed] [Google Scholar]
- 61.Otting G., Wuthrich K. Q. Rev. Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 62.Crick F.H.C. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 63.Lupas A. Trends Biochem. Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 64.Wishart D.S., Case D.A. Methods Enzymol. 2001;338:3–34. doi: 10.1016/s0076-6879(02)38214-4. [DOI] [PubMed] [Google Scholar]