Abstract
Here we introduce fluorescence intensity fluctuation spectrometry for determining the identity, abundance, and stability of protein oligomers. This approach was tested on monomers and oligomers of known sizes and was used to uncover the oligomeric states of the epidermal growth factor receptor and the secretin receptor in the presence and absence of their agonist ligands. This method is fast and is scalable for high-throughput screening of drugs targeting protein-protein interactions.
The association of membrane proteins into oligomers is thought to regulate biological function. For example, the lateral association of receptor tyrosine kinases (RTKs) into oligomers controls RTK activation, as the proximity of two kinase domains in the oligomers is required for their cross-phosphorylation1. Likewise, the association of G protein-coupled receptors (GPCRs)2 into oligomers is believed to affect their interactions with G-proteins and other effector molecules3. Defects in the association of cell surface receptors have been linked to human diseases4,5. Yet, the discrimination of the oligomeric state of a membrane receptor in live cells remains a significant experimental challenge, with different studies often producing contradicting results6,7. Whether such contradictions stem from data over-interpretation or built-in structural and functional versatility of these receptors needs to be clarified in order to understand receptor function and thus facilitate the development of effective therapies targeting protein-protein interactions. Existing technologies either are laborious and slow or lack the bandwidth needed to discriminate between different oligomeric sizes.
For example, Förster Resonance Energy Transfer (FRET) is potent at extracting the geometry and stoichiometry of protein complexes8,9 but relies on sophisticated analyses that are not easily scalable to probing receptor oligomerization under different experimental conditions or are not fast enough for screening of ligands for their therapeutic potential.
By contrast, methods derived from analysis of fluorescence fluctuations10, such as Photon-Counting Histogram (PCH) analysis11 and Number and Brightness (N&B) analysis12, are comparatively simpler and faster but currently only provide average values of the oligomer size over mixtures of oligomers with different sizes. In addition, the number of species N in N&B is the average number of entities with different oligomerization status within the excitation voxel and not the more desirable concentration of molecules within that voxel. Moreover, fluctuation analysis based on time sampling is oblivious to the existence of comparatively less mobile oligomers that do not produce size-related fluctuations on the time scale of the measurements. In recognizing some of these difficulties, an enhanced N&B method (eN&B) has been proposed13 to extract brightness distributions from each image pixel. This method nevertheless does not solve the “immobile-fraction” problem and does not resolve the brightness distribution into distinct oligomer sizes. A related method, Spatial Intensity Distribution Analysis (SpIDA)14,15, captures oligomer size-related fluctuations from the immobile fraction by converting the problem of sampling the fluorescence in time to sampling it in space, but it does so at the expense of further reduced oligomer-size resolution. In addition, inhomogeneously distributed molecular concentrations may inadvertently appear as oligomer size distributions in SpIDA. Thus, a method that presents not only speed but also an ability to resolve fluorescence intensity distributions accurately into oligomer sizes and concentrations has not been described to date.
We have developed a method, termed one- or two-dimensional fluorescence intensity fluctuation spectrometry (i.e., FIF or 2D-FIF spectrometry, respectively), which can generate “spectrograms” of brightness distributions from intensity fluctuations across image pixels (i.e., from spatial distributions) for different concentrations of molecules, and unmixes them in order to determine the exact proportion of different oligomer sizes. FIF spectrometry extracts information on oligomer sizes and proportion from spatial intensity fluctuations within fluorescence images at the expense of single-cell resolution. We have implemented the 2D FIF analysis into a computer program (see Methods) that produces a complete set of data per type of sample within less than one day using a standard personal computer.
2D FIF spectrometry analysis starts from imaging cells expressing fluorescently labeled proteins of interest. Large regions of interest (ROI) are demarcated using an ROI-selection tool in a dedicated computer program which then generates small ROI segments (200-500 pixels in size) using available mathematical procedures described in the Methods section. The ROI segmentation procedure makes it possible to convert intensity fluctuations within the ROI into thousands of brightness and concentration data points from just hundreds of ROIs.
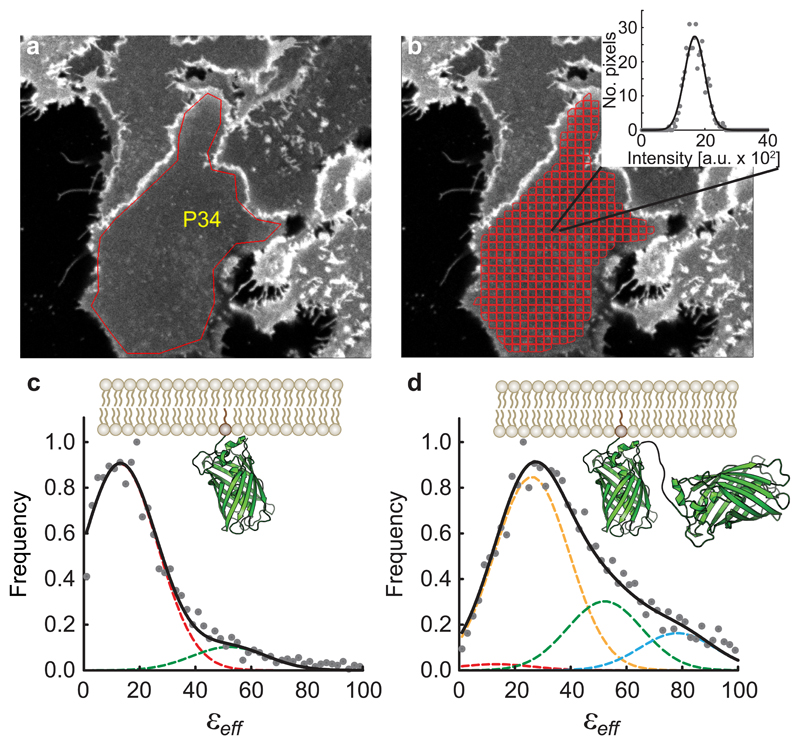
The ROI selection and segmentation process is illustrated in Fig. 1a,b using confocal images of the basolateral membrane of cells expressing a monomeric enhanced green fluorescent protein (mEGFP) construct associated to the plasma membrane via a lipidated peptide anchor (PM-1-mEGFP, see Methods). A fluorescence intensity distribution is generated for each ROI segment (Fig. 1b inset), from which the variance of the distribution and its average (i.e., position of its maximum) are computed. A one-dimensional brightness spectrogram is then computed for each ROI segment (Fig. 1c). Analysis of this spectrogram gives the effective molecular brightness of a monomer (or protomer, ). A similar analysis was performed for an equivalent tandem mEGFP construct, PM-2-mEGFP, in which two molecules of mEGFP are linked together (Fig. 1d). Here the effective brightness peak was located roughly at twice the effective brightness of PM-1-mEGFP. The results obtained using two-photon microscopy are shown in Supplementary Fig. 1.
Fig. 1. Illustration of the data reduction process in two-dimensional Fluorescence Fluctuation (2D-FIF) Spectrometry using single-photon excitation.
a, Typical fluorescence image of Flp-In™ T-REx™ 293 cells (out of 8 images containing 44 cells) expressing a plasma membrane-targeted mEGFP construct (PM-1-mEGFP). The overlaid polygon (P34) indicates a region of interest (ROI) comprising a patch of the basolateral membrane of a cell. b, Software-generated image segmentation of the ROI in (a) using the moving-squares method (see Methods). Inset, fluorescence intensity histogram (circles) of a single segment, together with its Gaussian curve fit (solid line). The mean and width of the Gaussian are used to calculate the brightness (εeff) and concentration for each segment (see Methods). c-d, Normalized frequency distribution assembled from (c) 3,582 and (d) 4,185 εeff values obtained from 8 images comprising several cells expressing monomeric (PM-1-mEGFP) or tandem (PM-2-mEGFP) mEGFP constructs was fit to a sum (solid black curves) of Gaussians (dashed lines with various colors), to find the brightness of single mEGFP protomers, Gaussian peak positions were set to where n is the number of protomers in an oligomer, and their widths were set equal to one another and determined from data fitting (13.4 a.u.). The εeff distribution for PM-1-mEGFP is primarily comprised of monomers (dashed, red Gaussian curve), with its peak positioned at while the PM-2-mEGFP spectrogram is mostly captured by a dimer model (dashed, yellow Gaussian curve), with its peak at
The in-cell measurement results were consistent with those obtained from purified monomeric and dimerizing EGFP constructs using a two-photon microscope (Supplementary Fig. 2a). We also analyzed (i) monomeric fluorescent proteins self-associating reversibly into dimers, (ii) covalently formed dimeric constructs self-associating into tetramers, and (iii) mixtures of covalently-formed dimers and tetramers self-associating reversibly to form tetramers and octamers, respectively. The analysis produced dissociation constants in agreement with the known number of binding sites for each oligomeric species (Supplementary Fig. 2b), and predicted oligomer sizes and concentrations in agreement with the known composition of the samples (Supplementary Fig. 2c).
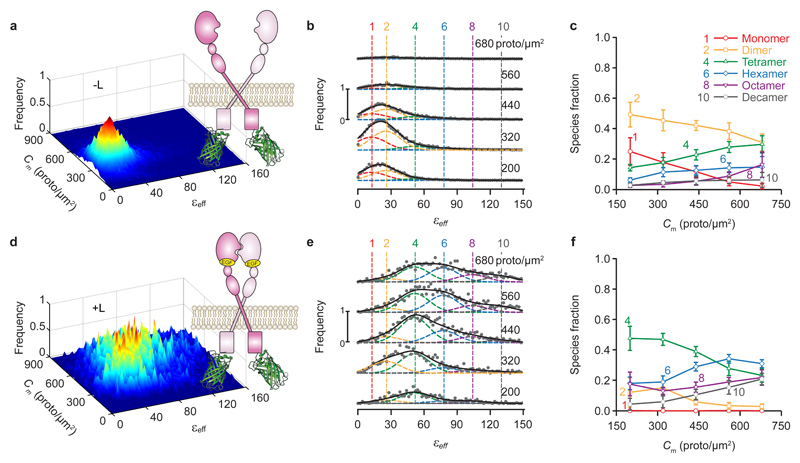
To test the 2D FIF method on biologically relevant receptors, we probed the self-association of the epidermal growth factor receptor (EGFR) fused to mEGFP. Wild-type EGFR presents ligand-dependent oligomerization1,16, while a mutant form containing a pair of domain II mutations (Tyr251Ala and Arg285Ser) is unable to oligomerize17, either in the presence or absence of ligand. Applying the same procedure as described in Fig. 1, we performed confocal imaging of cell membranes containing mEGFP-EGFR and obtained the brightness (εeff) and average intensity for each ROI segment. Dividing the average intensity by the brightness of the monomeric construct derived above, and the volume of the laser beam focal volume (see Methods), we obtained the concentration of EGFR protomers in each ROI segment. The frequency of occurrence of pairs consisting of effective brightness and concentration values was represented as two-dimensional surface plots in Fig. 2 for the wild-type EGFR, both in the presence and absence of its canonical ligand, the epidermal growth factor (EGF). For wild-type EGFR, the distribution of oligomer sizes was biased towards dimers in the absence of ligand (Fig. 2a,b,c), and it dramatically shifted towards larger oligomers in the presence of ligand (Fig. 2d,e,f) or as the concentration of receptor increased. By contrast, for the oligomerization-impaired EGFR mutant, the oligomer size remained unchanged after addition of the EGF ligand (Supplementary Fig. 3). All of these findings are in agreement with recent literature1,16.
Fig. 2. 2D-FIF results obtained from single-photon excitation of fixed cells expressing wild-type EGFR in the absence of ligand (-L) or after ten-minute treatment with 100 nM agonist ligand (+L).
a,d, Frequency of occurrence of effective brightness (εeff) for each protomer concentration using (a) 25,740 and (d) 6,812 total ROI segments to construct the distribution, extracted from 48 and 26 images, respectively (each of which contain several cells). Data were collected from at least two separate experiments. b,e, Cross sections through the surface plots in panels a,d, respectively, for different total concentration ranges; average concentration for each range (in protomer/μm2) is indicated above each plot. The vertical dashed colored lines indicate the peak positions for the brightness spectra of monomers, dimers, tetramers, etc., obtained from (or predicted based on) the simultaneous fitting of the PM1- and PM2-mEGFP spectrograms in Fig. 1, which were used as standards of brightness in the analysis. The εeff distribution for each concentration range was fitted with a sum of six Gaussians; the peak of each Gaussian was set to where n is the number of protomers in a given oligomer (e.g., 1, 2, 4, etc.), with the εeff and standard deviation obtained from measurements on cells expressing PM-1-mEGFP or PM-2-mEGFP (Fig. 1). Only the Gaussian amplitudes (An) were adjusted in the process of data fitting in b,e which gave the fraction of protomers (c,f) for each oligomeric species, i.e., niAi/∑n nAn. Number of Gaussians in each fit was chosen as discussed in the Methods section. c,f, Relative concentration of protomers in each oligomeric species vs. total protomer concentration, as derived by decomposing the spectrograms in column 2 into Gaussian components. Each data point and its error bar represent the mean ± standard deviation, respectively, of 1,500 different relative fraction values resulting from bootstrapping and refitting the original set of images as described in the Methods section.
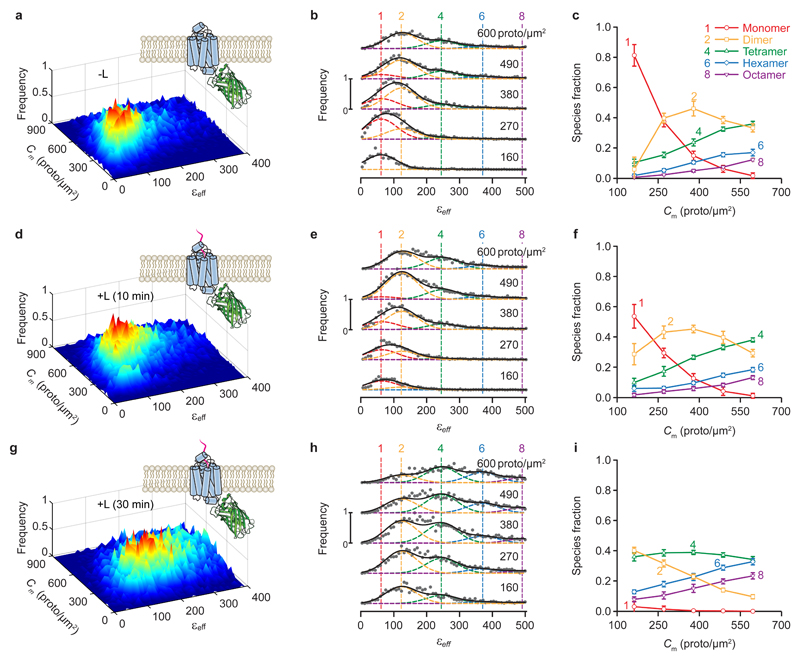
We next investigated the oligomerization of secretin receptor (SecR)18, a class B GPCR whose oligomerization behavior, like that of other GPCRs, is not fully understood. While several experiments have indicated that GPCRs form functional homo- or hetero-oligomeric complexes in vivo as well as in vitro7,19,20, there have been suggestions that not all GPCRs are multimeric or that oligomerization is not essential for function7. To help elucidate this question, we employed a two-photon microscope to image cells expressing SecR-mEGFP fusion proteins in their plasma membrane. The FIF spectrograms obtained from fixed cells were broader for higher expression levels of SecR (Fig. 3a,b) as well as for ligand-treated cells (Fig. 3d,e,g,h). Monomers were the dominant species at low expression levels, while higher order oligomers dominated at high expression levels. Prolonged treatment with ligand abolished the monomeric form of the receptor altogether, with dimers and tetramers dominating even at lower concentrations (Fig. 3i). Similar results were obtained when using a confocal microscope and fixed cells (Supplementary Fig. 4) as well as a two-photon microscope and live cells (Supplementary Fig. 5). For all untreated and 10-minute ligand-treated cells, the reduced fitting residuals (which ranged from 0.001 to 0.07) generally remained the same when Gaussian components corresponding to odd-number oligomer sizes were added to the fitting model, which did not justify the need to increase model complexity. By contrast, for the 30-minute treatment with ligand (Fig. 3h), the reduced fitting residuals decreased by 15% to 200% (!) after adding odd-number oligomer sizes to the model. We reasoned, nevertheless, that such an apparent improvement was caused by artefactual broadening of the intensity distribution of many of the ROI segments by ligand-induced accumulation of receptors in endocytic vesicles (Supplementary Fig. 6), which smeared the brightness spectrograms thereby prompting the need for including intermediate peaks in the data fitting model. Therefore, our final results shown in Fig. 3i were obtained with the simpler fitting model.
Fig. 3. 2D-FIF results obtained from two-photon excitation of fixed cells expressing wild-type secretin receptor in the absence of agonist ligand (-L) or after ten- or thirty-minute treatment with 100 nm ligand (+L).
a,d,g, Surface plots of the frequency of occurrence of εeff for each concentration of protomers using (a) 13,420, (d) 15,309 and (g) 12,979 total segments to construct the distribution, extracted from 82, 80, and 82 images, respectively (each of which contain several cells). b,e,h, Stacks of cross sections through the surface plots in panels (a), (d), and (g), respectively, i.e., frequency of occurrence vs. effective brightness for different concentration ranges; average concentration for each range (in protomer/μm2) is indicated above each plot. The vertical dashed lines indicate the peak positions for the brightness spectra of monomers, dimers, etc., obtained from (or predicted by) the simultaneous fitting of the PM-1- and PM-2-mEGFP spectrograms used as standards of brightness (Supplementary Fig. 1). Number of Gaussians in each fit was chosen as described in the Methods section. c,f,i, Relative concentration of protomers within each oligomeric species vs. total concentration of protomers, as derived from unmixing of the curves in (b), (e), and (h), respectively, into different Gaussian components. Samples were as follows: wild-type secretin receptor treated with vehicle (-L) (first row of graphs), secretin (+L) for 10 minutes (second row of graphs), or secretin (+L) for 30 minutes (third row of graphs). Each data point and its error bar represent the mean ± standard deviation, respectively, of 1,500 different relative fraction values resulting from bootstrapping and refitting the original set of images as described in the Methods section. Entire data analysis process followed the steps described in the caption to Fig. 2.
Overall, the above results suggested that SecR presents two different interfaces: one allowing monomers to dimerize and another one that permits association of dimers into higher-order oligomers8. The receptors switch between the associated and unassociated states, with the residence time in each state depending on the concentration (Fig. 3c). Ligand binding stabilizes the monomer-monomer interface to form dimers, thereby allowing dimers to associate into higher order oligomers via the second binding interface.
The reason why this wealth of information extracted with 2D FIF spectrometry is not accessible to other methods, such as SpIDA, lies in the observation that each ROI is characterized by broad distributions of molecular concentrations and oligomer sizes, which has two major adverse effects when using all the pixels within to create a single intensity histogram, as follows:
(i) Inhomogeneous distributions of oligomer sizes and concentrations across the ROI broaden the intensity histogram, which results in a larger apparent brightness (see equation 1 in the Methods section) than would be obtained for uniform intensity distributions. This effect is usually so large (Supplementary Fig. 7) that it obfuscates any dependence of the protein oligomer size on the concentration of molecules or treatment with ligand.
(ii) Although the oligomer sizes may fluctuate from place to place within the same ROI, only an average brightness value is obtained if the histogram is based on all the pixels in an entire ROI. Because of that, it is not the noise but rather the oligomer size distribution that is averaged out by using large ROIs, which is of course undesirable.
In conclusion, we demonstrated that 2D FIF spectrometry is able to distinguish between differing homo-oligomer sizes formed by a prototypical receptor tyrosine kinase and a class B GPCR. 2D FIF may be implemented on various fluorescence-based microscopes, including confocal and two-photon excitation microscopes, and should therefore be accessible to most research or pharmaceutical laboratories. 2D FIF is orders of magnitude faster than FRET spectrometry, which, nevertheless, also provides the oligomer geometry7, and as fast as N&B or SpIDA. It may be accelerated further by using automated image acquisition and analysis systems and is therefore scalable to high-throughput screening of ligands that can shift the monomer-dimer-oligomer equilibrium.
Methods
DNA constructs and cell lines
DNA constructs were made as previously described21,22. All Green Fluorescent Protein (GFP) variants incorporated the well-known A206K mutation to inhibit its self-association23. Monomeric A206K mEGFP construct (PM-1-mEGFP) or a tandem dimer of A206K mEGFP (PM-2-meGFP) were targeted to the plasma-membrane by adding the palmitoylation-myristoylation sequence, (Met)-Gly-Cys-Ile-Asn-Ser-Lys-Arg-Lys-Asp, at the amino terminus of the A206K mEGFP and the A206K mEGFP tandem.
Stable cell lines expressing receptors of interest were generated as described previously21,22. Wild-type and mutant EGFR as well as the plasma membrane targeted monomeric and dimeric constructs were expressed in Flp-In™ T-REx™ 293 cells (Invitrogen); these cells were maintained in DMEM (high glucose) supplemented with 10% (v/v) fetal bovine serum, 100 U ml−1 penicillin, 100 mg ml−1 streptomycin, 10 mg ml−1 blasticidin and 100 mg ml−1 zeocin. The wild-type secretin receptor constructs were stably expressed in Chinese hamster ovary (CHO) cells; stably transfected CHO cell lines were maintained in Hams F-12 Nutrient Mix (Invitrogen, Paisley, U.K.) supplemented with 5% (v/v) fetal bovine serum, 100 U ml−1 penicillin, 100 mg ml−1 streptomycin and 500 mg ml−1 zeocin. All cells were maintained in a humidified incubator with 95% air and 5% CO2 at 37°C.
Cell preparation for imaging
Flp-In™ T-REx™ 293 cells expressing plasma-membrane targeted monomeric A206K mEGFP constructs (PM1-mEGFP) or dimeric A206K mEGFP constructs (PM2-mEGFP) were plated onto poly-D-lysine-coated 30-mm glass coverslips at a density of 2.5·105 cells/coverslip. These cells were induced using doxycycline at concentrations between 0.25 and 100 ng·ml-1, in order to achieve various expression levels. After induction, the cells were allowed to grow overnight, and the coverslips were then rinsed and resuspended in HEPES buffer (130mM NaCl, 5mM KCl, 1mM CaCl2, 1 mM MgCl2, 20mM HEPES, and 10mM D-glucose, pH 7.4), where they were then taken for imaging on a confocal microscope.
Secretin receptor-mEGFP, EGFR-mEGFP, and Tyr251Ala,Arg285Ser EGFR-mEGFP and cells were either grown on Lab-Tek 4-well-chambered cover glasses (Thermo Fisher Scientific, Paisley, U.K.) or 35-mm No. 1.5 coverglass bottom dishes containing a 14-mm diameter microwell (MatTek Corp.). Samples were either treated with ligand (EGF for EGFR expressing cells and secretin for secretin receptor expressing cells) or vehicle then fixed with 4% paraformaldehyde. After fixation, paraformaldehyde solution was removed from the chamber, rinsed multiple times in PBS, and the cells resuspended/imaged in PBS.
Imaging using single-photon confocal microscopy
Fluorescence images (1024 × 1024 pixels2) were acquired using a Zeiss LSM 510 PASCAL EXCITER laser scanning head coupled to a Zeiss Axiovert 200M inverted microscope (Carl Zeiss Microscopy) equipped with a 63× plan apochromat oil immersion lens with a numerical aperture of 1.4. The pixel dwell time was set to 12.8 μs/pixel. Detection of emitted fluorescence was accomplished using a photomultiplier tube (PMT) with settings: gain=850 V, offset=0, and amplifier gain=1. A long pass beam splitter with center wavelength 490 nm along with a long pass emission filter with a wavelength of 505 nm were chosen to efficiently collect the A206K mEGFP emission signal. All samples were excited using the 488-nm line of the 25-milliwatt multiargon laser. The 1/e2 laser beam waist was estimated by imaging sub-diffraction-sized 100-nm Tetraspeck fluorescent microspheres (Invitrogen, catalog no. T14792) and found to be ω0=0.266 μm for the radial direction.
Imaging using two-photon microscopy
Fluorescence images (800×480 pixels2) were acquired using a two-photon optical micro-spectroscope24,25 comprised of a Zeiss Axio Observer inverted microscope stand, an OptiMiS detection head and a line-scan module26 from Aurora Spectral Technologies. A mode-locked laser (MaiTai™, Spectra Physics), which generated 100 fs pulses, was used for fluorescence excitation at 960 nm. The excitation beam was focused in the plane of the sample using an infinity-corrected, C-Apochromat, water immersion objective (63×, NA=1.2; Carl Zeiss Microscopy). The optical scanning head (for laser beam scanning) was modified to incorporate a spatial light modulator (SLM) (P1920-1152-HDMI Nematic SLM System, Meadowlark Optics) for adaptive laser beam shaping. A multi-beam array was generated using the SLM and appropriate software for exciting 40 voxels in the sample simultaneously; the average power for each voxel was 3.9 mW. The OptiMiS detection head employed a non-descanned detection scheme, in which the emitted fluorescence was projected through a transmission grating onto a cooled electron-multiplying CCD (EMCCD) camera (iXon Ultra 897, Andor Technologies), allowing for the different wavelengths of light emitted by the sample to be separated into wavelength channels simultaneously (i.e. from a single exposure). The spectral bandwidth of the wavelength channels ranged from 450 nm to 600 nm with a spectral resolution of 22 nm. The 1/e2 laser beam waist was estimated by imaging sub-diffraction-sized 170-nm PS-Speck Microscope Point Source Kit fluorescent microspheres (Invitrogen, catalog no. P7220) and found to be ω0=0.429 μm for the radial direction. The spatial light modulator (for laser beam shaping), optical scanning head (for laser beam scanning), and EMCCD camera used for image acquisition were controlled by the same computer using in-house custom software written in C++. The pixel dwell time was set to 1.8 ms/pixel.
The intensity fluctuation analysis depends on the total average number of molecules present in the excitation volume. Future enhancements of 2D-FIF could include the use of STED microscopy for image acquisition, which features smaller excitation volumes compared to those of the usual confocal or two-photon microscopes, and would presumably enhance the fluorescence intensity fluctuations27.
Molecular brightness and receptor concentration determination
The essence of the fluorescence fluctuation spectroscopy methods is that the variance, σ2, of the distribution of measured intensities is dependent on both the number of photons emitted per second per molecule, i.e. the molecular brightness ε, and the average number of particles (or oligomers) within the observation volume, Noligo28,29. The assumption is that both the fluctuations in fluorescence and the detector shot noise follow Poisson statistics. When using analog detectors for signal collection, the molecular brightness, ε, can be extracted from the variance of the intensity distribution using the following relation30:
| (1) |
where εeff ≡ Gε and G is the analog gain in digital levels /photon, 〈IS〉 is the average background corrected measured intensity, γ is a shape factor which depends on the shape of the laser PSF as well as the geometry of the sample31, and is the variance arising due solely to the detector, which can be obtained from separate measurements on a constant intensity light source (see Supplementary Note 1 for a more detailed derivation).
The total number of protomers within the beam volume, Nproto, can be written as a function of the measured intensity, as follows:
| (2) |
where is the molecular brightness of a single protomer, which must be determined from applying equation (1) to separate measurements of a calibration sample known to be monomeric. The concentration of protomers, C, can then be determined by dividing Nproto by the value for the observation volume:
| (3) |
Here PSF represents the laser distribution function of the focused laser beam and ∫∫∫ PSF2(x, y, z)dxdydz the volume of the laser beam comprising Nproto molecules (see Supplementary Note 2 for detailed derivation) and was numerically evaluated using a program written in Matlab™ (Mathworks Inc.). For measurements on the basal membranes of cells expressing membrane proteins, the concentration of molecules in the membrane, Cm, becomes Nproto per beam area, which is a particular case of equation (3), given by:
| (4) |
Description of the data analysis program
A graphical user interface was created that encompasses all the steps needed to quantify oligomeric actions from fluorescence fluctuation data. The software suite is separated into three modules: (1) region of interest (ROI) and segmentation generation, (2) brightness and concentration extraction, and (3) meta-analysis of brightness distributions. Each module is launched by a separate icon in the graphical user interface (GUI) toolbar.
In the first module, 2D fluorescence images are loaded as a stack, and the user selects ROIs using a polygon tool; multiple ROIs can be selected for each loaded image. A segmentation process is implemented which divides each ROI into smaller segments using either a moving square algorithm or a simple linear iterative clustering algorithm (SLIC) (for more details see Supplementary Note 3)32–34. The pixel locations for each segment are saved and paired with the corresponding source image. For a comparison of the results obtained with the two segmentation methods see Supplementary Fig. 8). The automatic ROI segmentation not only allowed the conversion into critical information of the inherent average intensity variations from segment to segment, but it also increased the number of data points using only a reasonably small number of manually selected ROIs of 100 or so. The number of pixels used in each segment ranged from 200-500, depending on the location of the segment within the cell. Pixel numbers larger than 500 introduced shifts in the effective brightness distributions (Supplementary Fig. 7), while pixel numbers lower than 200 are too few in number to reliably extract a brightness value.
Once all the ROIs are drawn and segments generated, the intensity histogram from each segment is fit with a single Gaussian function to determine the mean and the standard deviation for the intensity distribution of said segment; the algorithm executed in the fitting process incorporates the Nelder-Mead method 35,36. Using the mean and standard deviation obtained from the Gaussian fitting along with the corrections for the shot and background noise (see Supplementary Notes 1 and 4), the effective brightness, εeff, and concentration of the corresponding segment is found by applying equations (1) and (4), respectively. The entire procedure of fitting starting at intensity histogram calculation and ending in the calculation of a segments average effective brightness and concentration is performed after the click of a single button. Once εeff and concentrations have been found for each segment, multiple tools for visualization of the brightness distributions as a function of concentration are included. The first visualization tool creates a 2D surface plot of the concentration vs. εeff (Fig. 2a,d). The second visualization tool allows partitioning each 2D plot into one dimensional brightness spectrograms for several chosen concentration ranges and plots the histograms one on top of one another in order of increasing concentration, as is seen for instance in Fig. 2b,e.
The εeff distributions for various concentration ranges are further analyzed in the third module where the distributions are fit with a sum of multiple Gaussian functions this fitting, again, was performed using the Nelder-Mead method. The means of each Gaussian used in the fitting, are all linearly related and set equal to a multiple of the monomeric molecular brightness, The center of each Gaussian then corresponds to the expected peak of either monomers, dimers, or and various higher order oligomers, depending of the multiplication factor used, i.e. n = 1 for monomers, n = 2 for dimers, n = 4 for tetramers, n = 6 for hexamers, n = 8 for octamers, and n = 10 for decamers. The monomeric molecular brightness, can be found by applying the same software tools to images acquired from a standard monomeric sample and making a single bin of concentration values. For a given concentration range, the relative amplitudes of the ith Gaussian used in the fitting, niAi/∑n nAn, indicates the fraction of total protomers that the corresponding oligomeric species comprises. Plots of the species fraction values for each oligomer size, obtained from the relative amplitudes of the Gaussian fittings, are shown in, e.g., Fig. 2c,f (as well as panels in the third column of Fig. 3).
Statistical analysis
Statistical errors, indicated by error bars in Fig. 2c,f and Fig. 3c,f,i, showing the fractions of oligomeric species present in pixel-level mixture of oligomers, have been estimated using the bootstrapping method37. Briefly, we produced 500 “bootstrapped” sets of images by randomly selecting images from the original set of fluorescence images corresponding to a particular sample. Each of the datasets thus resampled was then ran through the whole gamut of analysis procedures, as described in the previous section. The resulting εeff distributions were then fit with three different values which were determined from: (i) the peak of the plasma membrane-targeted mEGFP monomeric construct εeff distribution, (ii) the peak of the dimeric construct εeff distribution, and (iii) a simultaneous fit of the monomeric and dimeric εeff distributions (Fig. 1c,d). Fitting 500 different datasets with Gaussians whose centers (i.e., means) were determined from three different values resulted in 1,500 relative fraction values for each oligomer size and protomer concentration. In other words, each data point in the plots of Fig. 2c,f and Fig. 3c,f,i was obtained by taking the mean of the 1,500 relative fraction values and the error bar for each data point represented ±1 standard deviation of the same set of values. The three values selected for the single photon excitation measurements were 15.6, 12.04, and 13.04, while the widths of the Gaussians were 13.78, 13.67, and 13.41, respectively. For the two-photon excitation measurements, the values were 72.6, 61.65, and 66, while the widths were 32.5, 48.02, and 48.02, respectively.
Choosing the correct model for FIF spectrograms fitting
In searching for the correct number of Gaussian components to fit the brightness spectrograms presented in each figure, we tested several combinations of odd- and even-number oligomer sizes. Based on reducing the fitting residual values, two main models emerged: Model 1, which included monomers and even-numbered oligomer sizes (i.e., with peak positions at etc.); Model 2, which included odd as well as even number oligomer sizes (i.e., with peak positions at etc.). Using the 1,500 data sets obtained from bootstrapping as described in the previous paragraph, we computed averages and standard deviations for each experimental data point shown in the FIF spectrograms for each of the five concentration ranges shown in Figs. 2 and 3 in the main text. The standard deviations were used for weighting the fitting residuals obtained from simulating the experimental curves using each of the two models. Typical weighted residual values divided by the number of degrees of freedom ranged from 0.001 to 0.07 for all the samples investigated. Using Akaike’s information criterion, if the penalty for adding more fitting parameters was not compensated for by the improvement in the fitting, the simplest fitting model was retained, which was represented by Model 1. This may or may not be the case for other receptors investigated, and one would therefore have to perform a similar analysis for each case in part. More detailed analyses are beyond the scope of this work and will be provided in future publications.
Fluorescent protein constructs for testing the fluorescence fluctuation analysis
The following fluorescent protein constructs were engineered (Supplementary Note 5): monomeric EGFP (mEGFP), dimerization-prone EGFP, dimerizing YFP, and dimerizing tandem YFP dimers (yfp)2. Two Cysteins added to the C-terminus played a crucial role in our experiments on fluorescent protein solutions, in that they allowed slow end-to-end association via disulfide bridges of (yfp)2 dimers in the presence of small amounts of oxygen normally dissolved in the aqueous solvent. This led to about 20% of the dimers forming disulfide bridges, and hence irreversible tetramers (referred to as “(yfp)2ss(yfp)2” in Supplementary Fig. 2 and the main text). Using self-associating YFP domains expressed as dimers allowed us to induce formation of reversible tetramers via side-by-side interactions at two binding loci (i.e., represented by Alanine 206) on the barrel of each of the two YFP molecules in the (yfp)2ss(yfp)2 tandem dimers, as well as formation of octamers via four side-by-side interactions at four binding loci on the barrels of YFP in the (yfp)2ss (yfp)2 tetramers. Addition of 1 mM Dithiothreitol (DTT) led to complete dissociation of the disulfide bridges, so that only (yfp)2 and side-by-side tetramers were present in the (yfp)2 solution. These data (shown in Supplementary Fig. 2) were analyzed as described in Supplementary Note 6.
Step-by-step protocol
For further details regarding the sample preparation, measurements, and data analysis are provided in a Supplementary Protocol38.
Supplementary Material
Acknowledgments
We thank L. Miller, Mayo Clinic, Arizona, for provision of the Secretin receptor-mEGFP-expressing cells that were used to develop the current cell lines. We also thank M. McBride and A. Klug for assistance with data analysis. This work was partly funded by the National Science Foundation grant PHY-1626450 (awarded to V.R.), the Medical Research Council (U.K.) grant MR/L023806/1 (to G.M.), and the UWM Research Growth Initiative grants 101X333 (to V.R.) and 101X340 (to I.P.).
Footnotes
Code Availability
The compiled software used for data analysis described in this work has been deposited on the Figshare digital repository and is accessible from: https://figshare.com/s/acfd94b21b1105317f56. The computer code is available from the corresponding author upon request.
Data Availability
Fluorescence images and ROI files used to generate the FIF spectrograms in this study have been deposited on the Figshare digital repository and is accessible from: https://figshare.com/s/77b90d060901fa8b4cb3
Author contributions
M.R.S. prepared samples, performed two-photon microscopy measurements, designed and implemented algorithms, performed data analysis, and participated in manuscript writing. G.B. implemented data fitting algorithms, wrote the computer program for data reduction and analysis, and participated in manuscript writing. R.J.W. and J.D.P. designed the DNA constructs and cell lines, prepared the samples, and performed confocal microscopy measurements. D.B. participated in sample preparation and two-photon microscopy measurements. Annie Eis performed expression and purification of monomeric and multimeric fluorescent proteins. I.P. designed DNA constructs and supervised work on expression and purification of fusion fluorescent proteins. G.M. designed DNA constructs, supervised the work on development of cell lines and confocal microscopy imaging, and participated in manuscript writing. V.R. conceived and designed the study, generated algorithms, performed data analysis, supervised the project, and wrote the manuscript with contributions from M.R.S., G.B. and G.M.
Competing interests
M.R.S., G.B., and V.R. have submitted a provisional patent application covering aspects related to the generation, analysis and applications of one and two-dimensional brightness spectrograms.
References
- 1.Needham SR, et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat Commun. 2016;7 doi: 10.1038/ncomms13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shivnaraine RV, et al. Allosteric modulation in monomers and oligomers of a G protein-coupled receptor. Elife. 2016;5 doi: 10.7554/eLife.11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh DR, Kanvinde P, King C, Pasquale EB, Hristova K. The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures. Communications Biology. 2018;1:15. doi: 10.1038/s42003-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panetta R, Greenwood MT. Physiological relevance of GPCR oligomerization and its impact on drug discovery. Drug Discov Today. 2008;13:1059–1066. doi: 10.1016/j.drudis.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 6.James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nature methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- 7.Raicu V, Schmidt WF. In: Recept Ser. Herrick-Davis K, Milligan G, Di Giovanni G, editors. Vol. 33. 2017. pp. 39–75. [Google Scholar]
- 8.Mishra AK, et al. Quaternary structures of opsin in live cells revealed by FRET spectrometry. Biochemical Journal. 2016;473:3819–3836. doi: 10.1042/BCJ20160422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellenkamp B, et al. Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study. Nature methods. 2018;15:669–676. doi: 10.1038/s41592-018-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian H, Elson EL. Distribution of Molecular Aggregation by Analysis of Fluctuation Moments. P Natl Acad Sci USA. 1990;87:5479–5483. doi: 10.1073/pnas.87.14.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojosnegros S, et al. Eph-ephrin signaling modulated by polymerization and condensation of receptors. P Natl Acad Sci USA. 2017;114:13188–13193. doi: 10.1073/pnas.1713564114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godin AG, et al. Revealing protein oligomerization and densities in situ using spatial intensity distribution analysis. P Natl Acad Sci USA. 2011;108:7010–7015. doi: 10.1073/pnas.1018658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pediani JD, Ward RJ, Marsango S, Milligan G. Spatial Intensity Distribution Analysis: Studies of G Protein-Coupled Receptor Oligomerisation. Trends Pharmacol Sci. 2018;39:175–186. doi: 10.1016/j.tips.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofman EG, et al. Ligand-induced EGF Receptor Oligomerization is Kinase-dependent and Enhances Internalization. Journal of Biological Chemistry. 2010;285:39481–39489. doi: 10.1074/jbc.M110.164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bessman NJ, Bagchi A, Ferguson KM, Lemmon MA. Complex Relationship between Ligand Binding and Dimerization in the Epidermal Growth Factor Receptor. Cell Rep. 2014;9:1306–1317. doi: 10.1016/j.celrep.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller LJ, Dong MQ, Harikumar KG. Ligand binding and activation of the secretin receptor, a prototypic family B G protein-coupled receptor. Brit J Pharmacol. 2012;166:18–26. doi: 10.1111/j.1476-5381.2011.01463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurel D, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: application to GPCR oligomerization. Nature methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferre S. The GPCR heterotetramer: challenging classical pharmacology. Trends Pharmacol Sci. 2015;36:145–152. doi: 10.1016/j.tips.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward RJ, Pediani JD, Godin AG, Milligan G. Regulation of oligomeric organization of the serotonin 5-hydroxytryptamine 2C (5-HT2C) receptor observed by spatial intensity distribution analysis. The Journal of biological chemistry. 2015;290:12844–12857. doi: 10.1074/jbc.M115.644724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward RJ, Pediani JD, Harikumar KG, Miller LJ, Milligan G. Spatial intensity distribution analysis quantifies the extent and regulation of homodimerization of the secretin receptor. Biochem J. 2017;474:1879–1895. doi: 10.1042/BCJ20170184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 24.Raicu V, et al. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat Photonics. 2009;3:107–113. [Google Scholar]
- 25.Stoneman MR, et al. Quaternary structure of the yeast pheromone receptor Ste2 in living cells. BBA-Biomembranes. 2017;1859:1456–1464. doi: 10.1016/j.bbamem.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Biener G, et al. Development and Experimental Testing of an Optical Micro-Spectroscopic Technique Incorporating True Line-Scan Excitation. International Journal of Molecular Sciences. 2014;15:261–276. doi: 10.3390/ijms15010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedde PN, et al. Stimulated emission depletion-based raster image correlation spectroscopy reveals biomolecular dynamics in live cells. Nat Commun. 2013;4 doi: 10.1038/ncomms3093. [DOI] [PubMed] [Google Scholar]
- 28.Qian H, Elson EL. On the Analysis of High-Order Moments of Fluorescence Fluctuations. Biophys J. 1990;57:375–380. doi: 10.1016/S0006-3495(90)82539-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Wei LN, Muller JD. Probing protein oligomerization in living cells with fluorescence fluctuation spectroscopy. P Natl Acad Sci USA. 2004;101:1792–1792. doi: 10.1073/pnas.2533045100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unruh JR, Gratton E. Analysis of molecular concentration and brightness from fluorescence fluctuation data with an electron multiplied CCD camera. Biophys J. 2008;95:5385–5398. doi: 10.1529/biophysj.108.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Muller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Achanta R, et al. SLIC Superpixels. EPFL Technical Report. 2010 149300. [Google Scholar]
- 33.Achanta R, et al. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012;34:2274–2281. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]
- 34.Zhang YX, Li XM, Gao XF, Zhang CM. A Simple Algorithm of Superpixel Segmentation With Boundary Constraint. Ieee T Circ Syst Vid. 2017;27:1502–1514. [Google Scholar]
- 35.Nelder JA, Mead R. A simplex method for function minimization. Computer Journal. 1965;7:308–313. [Google Scholar]
- 36.Lagarias JC, Reeds JA, Wright MH, Wright PE. Convergence properties of the Nelder-Mead simplex method in low dimensions. Siam J Optimiz. 1998;9:112–147. [Google Scholar]
- 37.Efron B. Bootstrap Methods: Another Look at the Jackknife. The Annals of Statistics. 1979;7:1–26. [Google Scholar]
- 38.Stoneman MR, et al. Quantifying membrane protein oligomerization using two-dimensional fluorescence intensity fluctuation spectrometry. Protocol Exchange. 2019 doi: 10.1038/protex.2019.1036. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.