Significance
Viruses have coevolved with their hosts and developed strategies to dampen, evade, or subvert the host immune response to provide an advantage to the virus. We show that ectromelia virus (ECTV) encodes a TNF receptor (TNFR) homolog, which provides an advantage to the host and virus by dampening TNF levels and inflammation. Infection of ECTV-resistant mice with a mutant virus lacking viral TNFR (vTNFR) caused significant lung pathology and death due to secretion of excessive levels of TNF and other inflammatory cytokines. In vitro, recombinant vTNFR from ECTV and other orthopoxviruses bound to membrane-associated TNF and down-regulated inflammatory gene expression through reverse signaling. vTNFR benefits the host by enabling survival, potentially facilitating virus spread, which should advantage the virus.
Keywords: cytokine response modifier D, respiratory viral infection, lung pathology and pneumonia, cytokine storm, CrmD inhibits inflammation
Abstract
Ectromelia virus (ECTV) causes mousepox, a surrogate mouse model for smallpox caused by variola virus in humans. Both orthopoxviruses encode tumor necrosis factor receptor (TNFR) homologs or viral TNFR (vTNFR). These homologs are termed cytokine response modifier (Crm) proteins, containing a TNF-binding domain and a chemokine-binding domain called smallpox virus-encoded chemokine receptor (SECRET) domain. ECTV encodes one vTNFR known as CrmD. Infection of ECTV-resistant C57BL/6 mice with a CrmD deletion mutant virus resulted in uniform mortality due to excessive TNF secretion and dysregulated inflammatory cytokine production. CrmD dampened pathology, leukocyte recruitment, and inflammatory cytokine production in lungs including TNF, IL-6, IL-10, and IFN-γ. Blockade of TNF, IL-6, or IL-10R function with monoclonal antibodies reduced lung pathology and provided 60 to 100% protection from otherwise lethal infection. IFN-γ caused lung pathology only when both the TNF-binding and SECRET domains were absent. Presence of the SECRET domain alone induced significantly higher levels of IL-1β, IL-6, and IL-10, likely overcoming any protective effects that might have been afforded by anti–IFN-γ treatment. The use of TNF-deficient mice and those that express only membrane-associated but not secreted TNF revealed that CrmD is critically dependent on host TNF for its function. In vitro, recombinant Crm proteins from different orthopoxviruses bound to membrane-associated TNF and dampened inflammatory gene expression through reverse signaling. CrmD does not affect virus replication; however, it provides the host advantage by enabling survival. Host survival would facilitate virus spread, which would also provide an advantage to the virus.
Tumor necrosis factor (TNF) plays key roles in the maintenance of homeostasis, acute inflammation, and antimicrobial defense (1–6). This cytokine can also cause serious pathology during acute viral infections and chronic inflammatory diseases (7–9). TNF is produced early during the course of a viral infection following activation of the nuclear factor-κB (NF-κB) inflammatory pathway. It is expressed as a transmembrane protein (mTNF), which is cleaved by metalloproteinase enzymes to produce soluble TNF (sTNF) (10, 11). The two forms of TNF play important roles in the inflammatory response to viral infections and exert their activities by signaling through TNF receptor I (TNFRI) or TNFRII (12, 13). Whereas both forms of TNF are capable of activating the TNFRI signaling pathway, TNFRII is activated more efficiently by mTNF (14). TNFRI and TNFRII can exist as cell-associated and secreted proteins. Through binding to TNFR, sTNF and mTNF deliver a forward signal to the TNFR-bearing cell. In addition, mTNF can participate in reverse signaling, a process whereby the interaction of mTNF on one cell with TNFR (soluble or membrane-anchored on another cell) leads to the transmission of signals into the cell expressing mTNF (15, 16). Reverse signaling through mTNF has been shown to down-regulate LPS-induced TNF, IL-6, and IL-10 production (17), inhibition of the MEK–ERK pathway (18), and inhibition of NF-κB activation and suppression of IL-1β production (19).
Many viruses, including orthopoxviruses (OPV), encode proteins that can modulate the host TNF response (20–24). OPV-encoded homologs of mammalian TNF receptor (TNFR) can modulate inflammation and the immune response (22), a strategy that provides an advantage to the virus, allowing successful replication and transmission to other hosts. There are four types of viral TNFR (vTNFR), also known as cytokine response modifier B (CrmB), CrmC, CrmD, and CrmE (25, 26). CrmC and CrmE have specificity for TNF (26–28), whereas CrmB and CrmD bind TNF (29, 30) as well as lymphotoxin (LT)-α and -β (25, 31). All Crm proteins contain a TNF-binding domain at the N-terminal region, consisting of four cysteine-rich domains (CRDs), which are homologous to mammalian TNFR CRD (32, 33) and shown to bind to mTNF and inhibit its ability to activate host TNFRs (25). In addition, CrmB, encoded by variola virus (VARV) (agent of smallpox), and CrmD, the only vTNFR encoded by ectromelia virus (ECTV) (agent of mousepox), contain an extended C-terminal chemokine-binding domain known as the smallpox virus-encoded chemokine receptor (SECRET) domain (31).
ECTV is a strict mouse pathogen and causes mousepox, a disease similar to smallpox caused by VARV in humans. Mousepox has been utilized extensively as a small animal model to understand, among others, the pathogenetic basis for resistance and susceptibility of humans to smallpox. Among inbred strains of mice, resistance to mousepox requires robust inflammatory and polarized T helper 1-like immune responses, which are associated with the cytokines interleukin 2 (IL-2), IFN-γ and TNF, and robust natural killer (NK) cell, cytotoxic T lymphocyte (CTL), and neutralizing antibody responses (34–38). The C57BL/6 strain is resistant to ECTV infection, but deficiency in IFN (35, 39, 40) or TNF (41) makes it highly susceptible. ECTV-susceptible BALB/c mice generate weak inflammatory and immune responses, associated with suboptimal IFN-γ, TNF, NK cell, and CTL responses. However, infection of this strain with a CrmD deletion mutant virus (ECTVΔCrmD) augments inflammation, NK cell, and CTL activities, resulting in effective control of virus replication and survival (22). CrmD clearly provides an advantage to ECTV in the BALB/c strain, but the rapid death of a susceptible host would not assist in effective virus spread.
We report here the consequences of infecting ECTV-resistant C57BL/6 mice with ECTVΔCrmD or a virus that expresses only the SECRET domain (ECTVRev.SECRET). As the TNF-binding domain of CrmD (CRD) can also interact with LT-α and LT-β, we also determined the outcomes of infecting mice deficient in TNF production or those that only express mTNF. We show here that CrmD plays an essential role in dampening inflammation, involving reverse signaling through mTNF in C57BL/6 mice during a respiratory infection. Deficiency in CrmD substantially increased the susceptibility of C57BL/6 mice without increases in viral load but significantly augmented levels of several cytokines including TNF, IL-6, IL-10, and IFN-γ, which were responsible for lung pathology and death. IFN-γ causes pathology when the CRD and SECRET domain are deleted, whereas it is insufficient to cause pathology when only the SECRET domain is present.
Results
Absence of CrmD Increases Susceptibility of C57BL/6 Mice to Infection with ECTV.
A mutant virus lacking both copies of CrmD (ECTVΔCrmD) and wild-type ECTV (ECTVWT) were used. To dissect the contribution of the TNF- vs. the chemokine-binding domains, a virus expressing a mutant version of CrmD (point mutation in CRD) unable to bind TNF but retaining chemokine binding activity of the SECRET domain (ECTVRev.SECRET) was also used (22). The ability of CRD or the SECRET domain to bind TNF or chemokines, respectively, by each of these viruses is shown in SI Appendix, Table S1.
C57BL/6 wild-type (WT) mice, TNF-deficient (TNF−/−) mice, and triple-mutant (TM) mice, engineered to express only noncleavable mTNF but not TNFRI or TNFRII (mTNFΔ/Δ.TNFRI−/−.TNFRII−/−) were used. TNF−/− and TM mice were used to determine whether the presence or absence of vTNFR impacted on the pathogenesis of ECTV infection in the absence of TNF or presence of only mTNF, respectively. The TM mice are devoid of an endogenous TNF response but can respond to exogenous TNFR or CrmD. The characteristics of these mouse strains with respect to expression of sTNF, mTNF, TNFRI, or TNFRII are shown in SI Appendix, Table S2. The TM mice are marginally more susceptible than mTNFΔ/Δ mice, which express both TNFRI and TNFRII, to ECTVWT infection (SI Appendix, Fig. S1). We predicted that CrmD binding to sTNF in WT mice can reduce the bioavailability of the cytokine, whereas its binding to mTNF may potentially modulate the inflammatory response further through reverse signaling (15, 16) in ECTVWT-infected WT and TM mice but not in TNF−/− mice.
Mice were inoculated intranasally (i.n.) with virus and monitored for changes in body weight and disease signs, which are presented as clinical scores using a scoring system (SI Appendix, Table S3) (41).
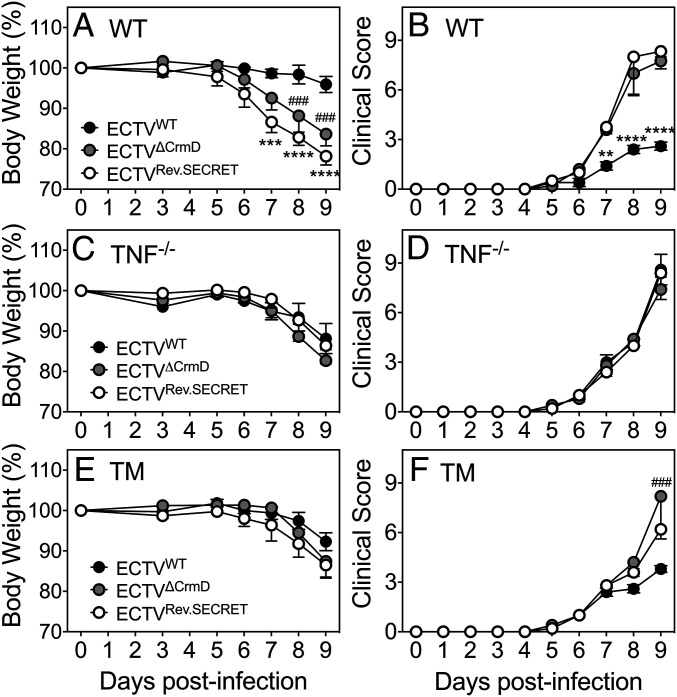
WT mice infected with the mutant viruses exhibited significant weight loss and higher clinical scores compared with ECTVWT-infected animals from day 7 postinfection (p.i.) (Fig. 1 A and B). Clinical scores between groups of WT mice infected with either of the mutant viruses were comparable. However, ECTVRev.SECRET-infected mice exhibited significantly higher weight loss at days 7 to 9 compared to ECTVWT-infected mice. TNF−/− mice fared poorly regardless of the type of virus used for infection, and the presence or absence of CrmD and/or SECRET did not influence weight loss or clinical scores (Fig. 1 C and D). TM mice infected with ECTVWT lost less weight than those infected with mutant viruses at days 8 and 9, but they were not statistically significant (Fig. 1E). The clinical scores in TM mice infected with any of the viruses were also similar until day 8, but by day 9, the clinical scores of ECTVWT-infected mice were significantly lower than ECTVΔCrmD-infected animals (Fig. 1 E and F). WT and TM mice infected with ECTVWT fared much better than all three strains infected with mutant viruses. Although no mortality was recorded in this experiment, all animals were killed at day 9 p.i. due to ethical and animal welfare reasons.
Fig. 1.
Increased susceptibility of WT and TM mice to ECTVΔCrmD and ECTVRev.SECRET. Groups of WT, TNF−/−, and TM female mice (five mice per group) were infected i.n. with 25 plaque-forming units (PFU) of ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. Weights (A, C, and E) and clinical scores (B, D, and F) were monitored during the course of infection. Mice were killed at day 9 p.i. Data for weights and clinical scores are expressed as means ± SEM. Statistical analysis was done using two way-ANOVA followed by Holm–Sidak’s posttests. Weight loss and clinical scores in mice infected with ECTVWT was compared to mice infected with the mutant viruses. *P < 0.05; **P < 0.01; ***P < 0.001; ###P < 0.001; ****P < 0.0001.
The results indicated that CrmD plays an important role in reducing weight loss and clinical symptoms during respiratory ECTV infection and that TNF is critical for CrmD to manifest its biological effects.
CrmD Deficiency Exacerbates Lung Pathology But Does Not Affect Viral Load in C57BL/6 Mice.
We investigated whether the significant morbidity among WT and TM animals infected with mutant viruses was due to exacerbated lung pathology, increased viral load, or both.
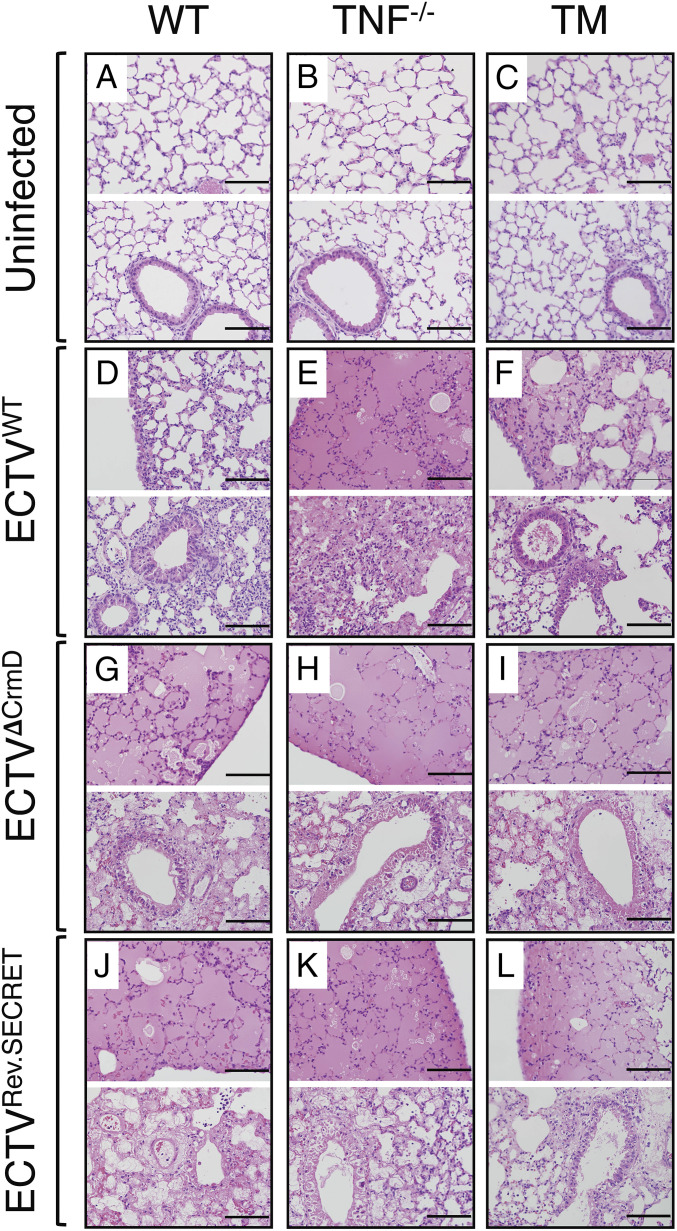
Fig. 2 shows hematoxylin and eosin (H&E)-stained lung sections from uninfected (Fig. 2 A–C) and virus-infected (Fig. 2 D–L) mice. WT mice infected with ECTVWT exhibited minimal parenchymal and perivascular edema with minor damage to the epithelial lining of the bronchioles, and minimal diffusion of cellular infiltrates (Fig. 2D). In contrast, ECTVWT-infected TNF−/− mice exhibited severe lung injury with total destruction of most alveoli due to fluid accumulation, diffuse to multifocal infiltration of leukocytes, and extensive damage to the entire epithelial lining of the bronchioles (Fig. 2E). Lung pathology in TM mice infected with ECTVWT was more severe than WT mice but less severe than TNF−/− mice (Fig. 2F).
Fig. 2.
Exacerbated lung pathology in mice infected with ECTVΔCrmD or ECTVRev.SECRET. Groups of WT, TNF−/−, and TM female mice (five mice per group) were either not infected or infected i.n. with 40 PFU ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. Representative lung histology sections from WT (A, D, G, and J), TNF−/− (B, E, H, and K), and TM (C, F, I, and L) mice at 9 d p.i. The Upper image in each panel shows lung sections with mainly alveoli, and the Lower image shows lung sections with alveoli, bronchioles, and bronchiolar epithelial cells. Slides were examined on all fields at 400× magnification. (Bars: 100 µm.)
ECTVΔCrmD infection exacerbated lung pathology in WT mice with massive fluid accumulation, destruction of most alveoli, diffuse to multifocal infiltration of leukocytes, and significant damage to the epithelial lining of the bronchioles (Fig. 2G). The absence of CrmD did not impact on lung pathology in TNF−/− (Fig. 2H) or TM (Fig. 2I) mice.
In ECTVRev.SECRET-infected WT (Fig. 2J), TNF−/− (Fig. 2K), and TM lungs (Fig. 2L), most of the bronchial walls exhibited complete or near-complete epithelial necrosis, interstitial infiltration of leukocytes, alveolar septal wall thickening, and collapse of alveolar septal walls.
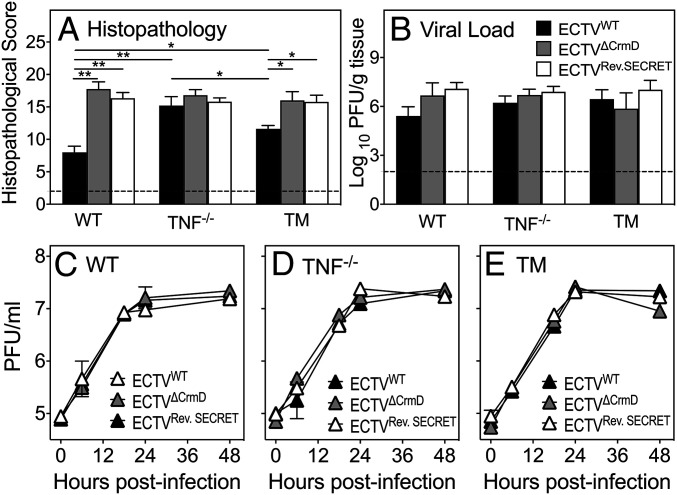
We semiquantified the histopathological changes in the lungs using a scoring system (SI Appendix, Table S4) (41). Five distinct observations were made from the analysis. First, WT lung sections had the lowest scores, TM lung sections had intermediate scores, and TNF−/− lung sections had the highest scores in response to ECTVWT infection (Fig. 3A). Second, WT mice infected with either mutant virus exhibited substantially greater lung histopathological scores. Third, in TNF−/− mice, all three viruses caused considerable levels of lung damage with no significant differences in the histopathological scores. Fourth, TM lung sections from mice infected with mutant either virus had significant increases in their histopathological scores compared to those infected with ECTVWT. Finally, the mutant viruses caused comparable levels of lung damage in all three strains of mice.
Fig. 3.
CrmD deficiency exacerbates lung pathology in WT and TM mice but has no effect on viral load. Groups of WT, TNF−/−, and TM mice (five mice per group) were infected i.n. with 39.5 PFU ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. (A) Histopathological scores of lung sections from mice 9 d p.i. Slides were examined on all fields at 400× magnification. Data are expressed as means ± SEM, and statistical analysis was done using ordinary one-way ANOVA test followed by Tukey’s multiple-comparisons tests, where *P < 0.05 and **P < 0.01. (B) Lung viral load at day 9 p.i. Data were log transformed and expressed as means ± SEM. Statistical analysis was undertaken using ordinary one-way ANOVA followed by Fisher’s least significant difference tests. The broken line corresponds to the limit of virus detection. In vitro virus replication in BMDMs from (C) WT, (D) TNF−/−, and (E) TM mice. BMDMs were infected at a multiplicity of infection of 1 with ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. Cells were harvested at 0, 6, 12, 24, and 48 h p.i., and viral load was determined by plaque assays. Data were log transformed and expressed as means ± SEM of duplicate samples for each time point. Statistical analysis was achieved using two-way ANOVA followed by Tukey’s posttests.
The lung viral load in all three strains of mice infected with ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET were comparable as the titers of viruses within each strain or across strains were not statistically different. This finding indicated that the TNF-binding or SECRET domains had no effect on viral replication in vivo (Fig. 3B). In vitro, the replication kinetics of ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET in bone marrow-derived macrophages (BMDMs) from WT (Fig. 3C), TNF−/− (Fig. 3D), or TM (Fig. 3E) mice over a 48-h period were comparable, consistent with the in vivo results.
CrmD Regulates Leukocyte Recruitment/Infiltration into Lungs of ECTV-Infected Mice.
We used flow cytometry of collagenase-digested lungs from uninfected and virus-infected mice to ascertain whether the TNF-binding domain and/or the SECRET domain affected leukocyte recruitment into the lungs, which might have contributed to pathology. We enumerated the total number of leukocytes and leukocyte subsets at early (day 4) and later (day 8) stages of infection.
At day 4 p.i., numbers of leukocytes or leukocyte subsets did not increase significantly in WT animals following infection with ECTVWT when compared with naive animals (Fig. 4 A–K). However, in the lungs of ECTVΔCrmD-infected animals, there were significant reductions in numbers of leukocytes (CD45.2+) (Fig. 4A) and leukocyte subsets [T cells (CD3+)] (Fig. 4B), T cell subsets (CD3+CD8+, CD3+CD4+, CD3+TCR γδ+) (Fig. 4 C–E), B cells (CD45R+) (Fig. 4F), NK cells (NK1.1+) (Fig. 4H), neutrophils (CD11bhiLy6clow) (Fig. 4J)], and inflammatory monocytes (CD11bhiLy6chi) (Fig. 4K) compared to those infected with ECTVWT or ECTVRev.SECRET. Although the absence of CrmD did not affect numbers of macrophages (F4/80+) (Fig. 4G) or dendritic cells (CD11c+) (Fig. 4I), the presence of only the SECRET domain in ECTVRev.SECRET-infected animals significantly increased numbers of these subsets. The SECRET domain appeared to be critical for leukocyte recruitment into the lungs of infected WT mice.
Fig. 4.
The SECRET domain of CrmD is important for leukocyte recruitment to the lungs early during the course of infection. Groups of WT, TNF−/−, and TM mice (five mice per group) were infected i.n. with 38 PFU ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. At day 4 p.i., lungs were collected from virus-infected mice and two naive animals from each group. Single-cell suspensions of digested lungs were stained with fluorochrome-conjugated mAb and analyzed by flow cytometry. Leukocytes were gated on (A) CD45+ cells (CD45.2+). Shown are numbers of (B) T cells (CD3+), (C) CD4+ T cells (CD3+ CD4+), (D) CD8+ T cells (CD3+ CD8+), (E) gamma delta T cells (CD3+ TCR γδ+), (F) B cells (CD45R+), (G) macrophages (F4/80+), (H) NK cells (NK1.1+), (I) dendritic cells (CD11c+), (J) inflammatory monocytes (CD11bhi Ly6chi), and (K) neutrophils (CD11bhi Ly6clow). Data were log transformed and expressed as means ± SEM. Statistical significance was obtained using the two-way ANOVA followed by Tukey’s multiple-comparisons tests, where *P < 0.05; **P < 0.01; ***P < 0.001.
In contrast to WT mice, TNF−/− animals did not exhibit any statistically significant increases or decreases in leukocyte subsets present in the lungs regardless of the type of infecting virus. This finding further indicated that TNF is critical for the function of CrmD and that any biological consequences of CrmD interacting with LT-α or LT-β had minimal impact on leukocyte recruitment.
In TM mice, the total infiltrating leukocytes (CD45.2+) (Fig. 4A) and most subsets examined were significantly higher in numbers in lungs of ECTVWT-infected animals compared to uninfected or mutant virus-infected animals. The absence of CrmD reduced leukocyte subset numbers in ECTVΔCrmD-infected mice, similar to the outcome in WT mice. However, unlike in WT mice, the presence of only the SECRET domain in ECTVRev.SECRET-infected mice did not contribute to any increases in numbers of leukocyte subsets. As TM mice do not produce sTNF, TNFRI, and TNFRII, the lack of any effect suggested that endogenous TNF signaling was likely necessary for SECRET to mediate its activity in relation to leukocyte recruitment into the lungs.
At day 8 p.i., leukocyte numbers in WT lungs were comparable irrespective of the infecting virus as was the case in TNF−/− mice (SI Appendix, Fig. S2). The one exception was a significant reduction in the number of B cells (CD45R+) in TNF−/− mice infected with ECTVRev.SECRET compared with those infected with ECTVΔCrmD or ECTVWT. TM mice were not included in the day 8 p.i. analysis of lung leukocytes as sufficient numbers of age- and sex-matched animals were unavailable. Results from the day 8 time point suggested that the exacerbated lung pathology seen at this time cannot be attributed to leukocyte numbers.
vTNFRs Dampen Inflammation In Vitro through Reverse Signaling.
The binding of TNFR to mTNF on a cell can induce reverse signaling in vitro and in vivo to dampen inflammatory gene expression (15, 16). We reasoned that CrmD and other OPV-encoded vTNFRs, which are known to bind mTNF (22), could also potentially reverse signal through mTNF and dampen inflammation in the lungs of infected hosts. Treatment with lipopolysaccharide (LPS), a potent inducer of TNF, stimulated the production of sTNF and mTNF in the murine macrophage cell line RAW264.7 and expectedly, it only induced mTNF in BMDMs from TM mice (SI Appendix, Fig. S3). To demonstrate reverse signaling, we used BMDMs from TM mice stimulated with LPS to induce mTNF and inflammatory gene expression, and subsequently treated the cells with vTNFR. We used the Mouse Signal Transduction Pathway Finder PCR array that screened for 84 genes (SI Appendix, Fig. S4 and Table S5; (42)). If vTNFRs dampened inflammation, we expected to see the down-regulation of mRNA transcripts for some LPS-induced inflammatory genes. We used recombinant CrmD from ECTV, CrmB, CrmC, and CrmD from cowpox virus (CPXV) and CrmB from VARV, expressed in the baculovirus system and purified as described (25).
LPS stimulation up-regulated the mRNA for 22 of the 84 genes that were screened (SI Appendix, Fig. S4). Treatment with Crm proteins down-regulated 18 of the 22 mRNA up-regulated by LPS (Table 1). Individual Crm proteins had varying effects on reducing mRNA levels, with CPXV CrmD downregulating the greatest number of mRNA transcripts induced by LPS. All of the Crm proteins significantly reduced IL-1α, CCL2, and IL-2Rα mRNA levels following reverse signaling (Table 1). The pathways that were most affected by reverse signaling such as NF-κB and JAK-STAT cytokine signaling pathways are involved in inflammatory processes. Crm proteins also modulated those genes that were down-regulated by LPS stimulation (SI Appendix, Table S6) and these are represented by several signaling pathways including stress, NF-κB, CREB, Wnt, Hedgehog, and PIA3K/AKT pathways.
Table 1.
Genes up-regulated by LPS stimulation and modulated following reverse signaling with vTNFRs
| Genes | Pathway | Fold change relative to unstimulated cells* | ||||
| ECTV crmD | CPXV crmB | CPXV crmC | CPXV crmD | VARV crmB | ||
| CXCL9 | JAK-STAT | 1.54 | 1.18 | 1.01 | −2.30 | −1.47 |
| IL1Α | NF-κB | −2.00 | −3.03 | −2.87 | −3.03 | −3.47 |
| PTGS2 | NF-κB | −1.47 | −2.24 | −3.11 | −5.29 | −1.83 |
| CCL2 | NF-κB | −2.30 | −2.64 | −2.79 | −2.87 | −4.95 |
| VCAM1 | NF-κB | −0.87 | −1.20 | −1.11 | −2.06 | −1.47 |
| NOS2 | NF-κB | −1.83 | −1.43 | −1.94 | −1.89 | −2.95 |
| IL2RΑ | JAK-STAT | −2.12 | −3.89 | −2.50 | −4.12 | −1.94 |
| SELP | Metabolism | −1.57 | −3.38 | −2.72 | −2.06 | −0.83 |
| FAS | NF-κB | −0.64 | −1.72 | −1.33 | −3.38 | −1.78 |
| ICAM1 | NF-κB | −0.87 | −1.11 | −1.67 | −3.47 | −1.67 |
| NFKBIA | NF-κB | −0.24 | −1.24 | −0.99 | −2.50 | −1.83 |
| GYS1 | Metabolism | 1 | 1.12 | −0.06 | 1.13 | −2.06 |
| IRF1 | JAK-STAT | −1.43 | −2.18 | −2.72 | −1.52 | −3.29 |
| CEBPB | Metabolism | −0.72 | −0.54 | −0.72 | −2.30 | −0.50 |
| TANK | NF-κB | −1.94 | −3.38 | −2.00 | −2.50 | −2.87 |
| HK2 | Metabolism | −0.15 | −0.18 | −0.12 | 1.01 | −2.06 |
| BIRC3 | NF-κB | −0.87 | −1.43 | −1.20 | −3.57 | −2.44 |
| MMP10 | JAK-STAT | −1.07 | −1.78 | −2.12 | −0.57 | −2.06 |
Values greater than twofold decrease in gene expression are in bold.
Absence of CrmD Results in Dysregulated Expression of Proinflammatory Mediators.
LPS stimulation of BMDMs followed by vTNFR treatment may not be a true reflection of changes in mRNA expression that can occur in the various cell types present in an organ as complex as the lung during viral infection, where many cell types produce TNF (41) and can thus be subjected to reverse signaling. We therefore determined changes in mRNA transcripts in lung tissue of WT, TNF−/−, and TM mice at day 9 p.i. using quantitative reverse transcription real-time PCR (qRT-PCR). We also measured the protein levels of some of the cytokines in separate groups of infected WT mice.
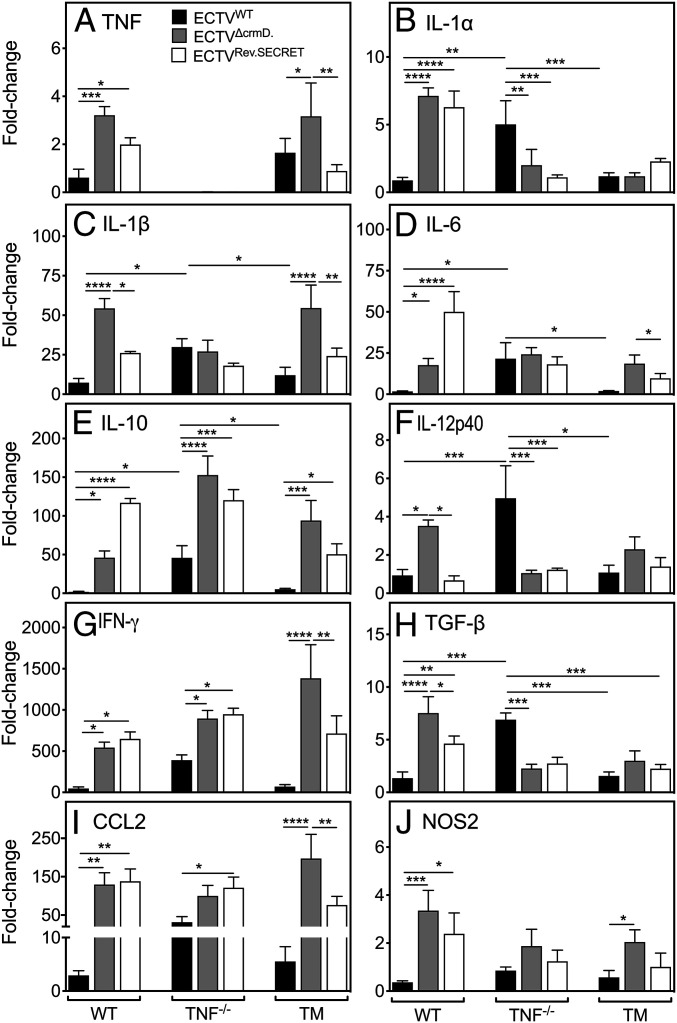
WT and TNF−/− mice do not express detectable levels of IFN-γ, IL-6, IL-10, TGF-β, and TNF (WT mice only) (41). The cytokine responses to ECTVWT infection in WT and TM lungs were similar, whereas the levels of most mRNA transcripts were significantly higher in TNF−/− lungs compared to WT or TM lungs (Fig. 5 A–J). The absence of CrmD in ECTVΔCrmD-infected WT mice led to significant increases in levels of mRNA transcripts for all mediators compared with ECTVWT infection. In the presence of the SECRET domain in ECTVRev.SECRET-infected WT mice, three types of responses were apparent when compared with ECTVΔCrmD infection. First, IL-1β, IL-12p40, and TGF-β mRNA levels were reduced. Second, mRNA levels of IL-6 and IL-10 were increased significantly. Third, mRNA levels of TNF, IL-1α, IFN-γ, CCL2, and NOS2 were similar but significantly higher than levels induced by ECTVWT.
Fig. 5.
Deletion of CrmD significantly up-regulates levels of mRNA transcripts for inflammatory mediators in virus-infected lungs. Groups of WT, TNF−/−, and TM mice (five mice per group) were infected i.n. with 25 PFU ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. At day 9 p.i., mice were killed, and lungs were collected. RNA was isolated from lungs and qRT-PCR was used to determine levels of mRNA transcripts for (A) TNF, (B) IL-1α, (C) IL-1β, (D) IL-6, (E) IL-10, (F) IL-12p40, (G) IFN-γ, (H) TGF-β, (I) CCL2, and (J) NOS2. UBC was used as the reference gene. The expression levels of each gene in infected mice are relative to the expression levels in naive mice. Data are expressed as means ± SEM. Statistical analysis was performed using two-way ANOVA followed by Tukey’s multiple-comparisons tests, where *P < 0.05; **P < 0.01; ***P < 0.001.
Expectedly, TNF−/− mice did not express mRNA for TNF, but infection with ECTVWT induced significantly higher levels of several inflammatory mediators compared to levels in WT or TM mice. The only exception was NOS2 mRNA, whose levels were similar to those in WT and TM mice. IL-1β, IL-6, and NOS2 mRNA levels in TNF−/− mice infected with any one of the three viruses were comparable. Some mRNA transcripts in TNF−/− mice infected with either of the mutant viruses changed significantly. IL-10, IFN-γ, and CCL2 levels increased, whereas IL-1α, IL-12p40, and TGF-β levels decreased regardless of whether both domains were absent or only the SECRET domain was present. Thus, any changes in mRNA expression in TNF−/− mice infected with ECTVΔCrmD or ECTVRev.SECRET were clearly not due to interactions of LT-α and/or LT-β with the CRD as the both the mutant viruses do not express the TNF-binding domain.
In TM mice, CrmD deficiency led to increases in levels of expression of a number of mediators, except IL-1α, IL-12p40, and TGF-β. TNF and IL-1β levels were significantly higher in the ECTVΔCrmD-infected group compared to ECTVWT- or ECTVRev.SECRET-infected mice. Similarly, IL-6 and IL-10 levels were higher in the absence of CrmD, reduced when only the SECRET domain was present, but were clearly higher than in ECTVWT-infected animals. NOS2 mRNA transcripts were only increased in the lungs of mice infected with ECTVΔCrmD.
The CRD and SECRET domain differentially regulated mRNA transcripts for a number of inflammatory genes in the three different strains of mice. In WT animals, the CRD was particularly important for dampening TNF, IL-1α, IL-1β, TGF-β, CCL2, IFN-γ, and NOS2, whereas the SECRET domain appeared critical for induction of IL-6 and IL-10. In TM mice, the presence of the SECRET domain alone reduced levels of CCL-2, IFN-γ, IL-1β, IL-6, IL-10, and TNF compared to levels in the absence of both the TNF-binding and SECRET domains.
We measured the levels of some cytokines by ELISA in lung homogenates of WT mice infected with ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET (SI Appendix, Fig. S5). The absence of CrmD in ECTVΔCrmD-infected mice increased levels of TNF, IL-10, and CCL2, whereas the presence of only the SECRET domain in ECTVRev.SECRET-infected mice significantly increased levels of IL-6 and IL-10, consistent with the gene expression data shown in Fig. 5. CrmD deficiency induced the highest levels of IL-1β and IL-12p40 mRNA transcripts; however, the SECRET domain alone was sufficient to induce significantly higher levels of these proteins. Finally, while the absence of CrmD significantly increased levels of mRNA transcripts for IFN-γ, the protein levels were comparable in mice infected with ECTVWT, ECTVΔCrmD, or ECTVRev.SECRET. Expression of mRNA transcripts and protein for some of the cytokines were incongruent (Fig. 5 and SI Appendix, Fig. S5). This may have been due to distinct expression kinetics, posttranscriptional regulation of cytokine gene expression, and/or the cytokine milieu in lungs of the different mouse strains infected with different viruses, which may have had an impact on protein translation.
Blockade of IFN-γ, IL-6, IL-10R or TNF Reduces Weight Loss and Disease and Increases Survival Rates.
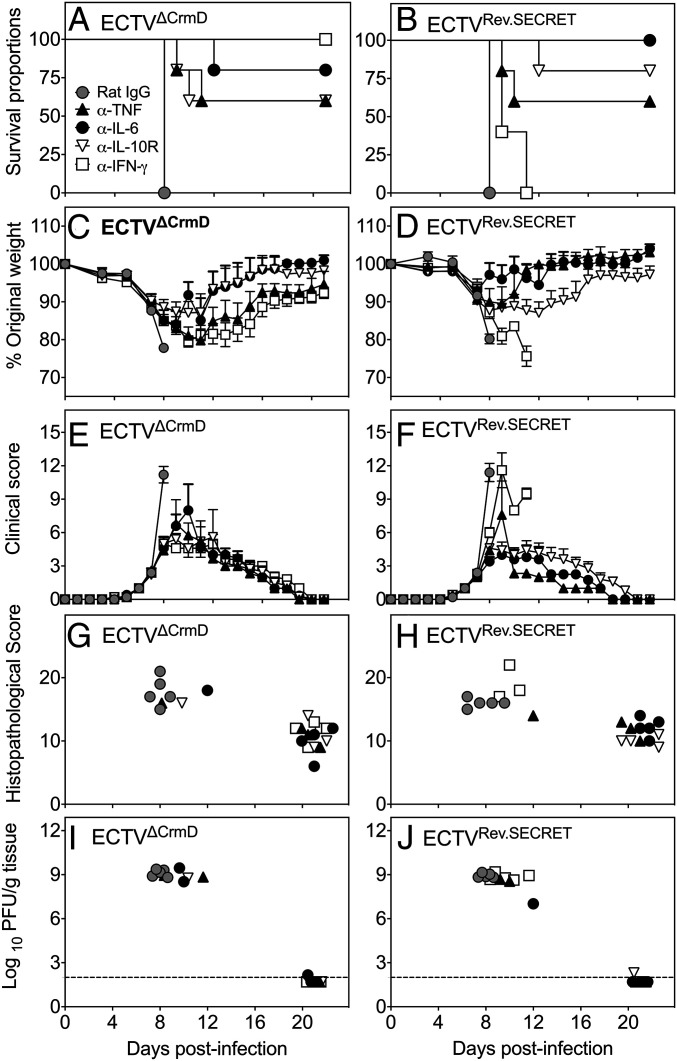
The increased production of a number of inflammatory cytokines in mice infected with mutant ECTV possibly contributed to the increased morbidity and mortality, as shown previously in ECTV-infected TNF−/− mice (41). We postulated that the in vivo blockade of some of these cytokines might reduce the severity of the clinical symptoms and lung pathology and possibly allow the recovery of mice infected with ECTVΔCrmD or ECTVRev.SECRET. Groups of WT mice infected with either mutant virus were treated with mAb against IFN-γ, IL-6, IL-10R, or TNF, beginning at day 7 p.i. and treatment continued every alternate day until day 20 p.i. and surviving animals were killed on day 21. We selected day 7 p.i. to start cytokine blockade therapy as this is the time when animals start losing weight, have relatively higher clinical scores, and lung pathology starts to develop. Control groups were treated with an isotype-matched rat IgG mAb, and the data are presented in Fig. 6.
Fig. 6.
Blockade of TNF, IFN-γ, IL-6, or IL-10R protects against mutant virus infection. Groups of WT female mice (five mice per group) were infected i.n. with 25 PFU of ECTVΔCrmD or ECTVRev.SECRET. On days 7, 9, 11, 13, 15, 17, 19, and 21 p.i., individual groups were treated with isotype control rat IgG mAb (control mAb) or specific mAb against TNF (α-TNF), IL-6 (α-IL-6), IL-10R (α-IL-10R), or IFN-γ (α-IFN-γ) at 500 µg, i.p. Mice were monitored daily and killed if they had lost ≥25% of their original weight or were severely moribund at any time during the course of infection. All surviving animals were killed on day 22 p.i. (A and B) Survival, (C and D) weight loss, (E and F) clinical scores, (G and H) histopathological scores, and (I and J) viral load in mice infected with ECTVΔCrmD or ECTVRev.SECRET. For A and B, statistical analysis was done using the log-rank (Mantel-Cox) test, P < 0.01, in survival proportions of mice infected with ECTVΔCrmD or ECTVRev.SECRET and treated with anti-cytokine mAb compared to mice treated with control mAb. For C–F, significance was obtained using two-way ANOVA followed by Dunnett’s posttests compared to the means of control mAb-treated groups at day 8 p.i. For C and D, in the ECTVΔCrmD-infected groups, P < 0.001 when control mAb-treated group is compared with α-IL-6– or α-IFN-γ–treated groups and P < 0.0001 when control mAb-treated group is compared with α-TNF– or α-IL-10R–treated groups. In the ECTVRev.SECRET-infected groups, P < 0.0001 when control mAb-treated group is compared with α-IL-6– or α-IFN-γ–treated groups and P < 0.01 when control mAb-treated group is compared with α-TNF– or α-IL-10R–treated groups. For E and F, in the ECTVΔCrmD- and ECTVRev.SECRET-infected groups, P < 0.0001 when control mAb-treated group is compared with α-IL-6–, α-IFN-γ–, α-TNF–, or α-IL-10R–treated groups. For G–J, datasets for individual mice are presented as the results come from different days for each of the groups. For that reason, statistical analysis was not done. Data shown are from one experiment.
In ECTVΔCrmD-infected WT mice, blockade of IFN-γ, IL-6, TNF, or IL-10R with specific mAb significantly increased survival rates compared to the rat IgG isotype control mAb-treated group (Fig. 6A). The control mAb-treated group exhibited 100% mortality by day 8 p.i., whereas 60% of animals treated with mAb against TNF or IL-10R were alive at day 21. Anti–IL-6 mAb treatment resulted in the survival of 80% of animals in the group, whereas IFN-γ blockade rescued 100% of the mice. Individually, IFN-γ, IL-6, TNF, and IL-10 were necessary and sufficient to cause significant morbidity and mortality due to lung pathology, but the blockade of just one of the cytokines was sufficient to protect 60 to 100% of the mice.
ECTVRev.SECRET-infected mice also succumbed to infection by day 8 p.i. (Fig. 6B). Survival rates significantly increased from 0 to 60% and from 0 to 80% after treatment with anti-TNF or anti–IL-10R mAb, respectively. There was 100% survival in the group treated with anti–IL-6 (Fig. 6B). Thus, blockade of IL-6, TNF, or IL-10R resulted in similar outcomes in animals infected with ECTVΔCrmD or ECTVRev.SECRET. The only exception was anti–IFN-γ treatment of ECTVRev.SECRET-infected mice, which had minimal effects on the outcome and all animals succumbed to infection. This finding is in complete contrast to ECTVΔCrmD-infected mice, which were fully protected from anti–IFN-γ treatment. Thus, IFN-γ, IL-6, TNF, and IL-10 are responsible for the pathology in the absence of the CRD and the SECRET domain in ECTVΔCrmD-infected mice. In contrast, in the presence of only the SECRET domain, IFN-γ was not sufficient to cause pathology.
Weight loss (Fig. 6 C and D) and clinical scores (Fig. 6 E and F) were also markedly lower after anti-cytokine treatment compared to the control mAb-treated groups. Mice treated with anti-cytokine mAb regained weight beginning at day 12 and those mice that recovered had similar clinical scores from this time. Improvements in weight loss and clinical scores correlated with the survival rates, as evidenced by body weight recovery and the absence of any clinical signs and symptoms in mice that were alive at day 21 p.i.
The histopathological scores (Fig. 6 G and H) generated from lung histology sections (SI Appendix, Fig. S6) were highest in those animals that had the highest clinical scores, lost significant body weights, and succumbed to mousepox compared to those animals that had survived. The viral load was very high in those animals that died early between days 8 and 12, but was at or below the limit of detection in animals that recovered and killed at day 21 (Fig. 6 I and J). Of particular interest is the outcome of IFN-γ blockade in mice infected with ECTVΔCrmD or ECTVRev.SECRET. In mice infected with ECTVΔCrmD and treated with anti–IFN-γ, viral load was below the limit of detection and the histopathological scores were substantially reduced. In contrast, in mice infected with ECTVRev.SECRET and treated with anti–IFN-γ, the viral load and histopathological scores were comparable with those of control mAb-treated mice. The results indicate that excessive IFN-γ production in mice infected with ECTVΔCrmD, when both CRD and SECRET domain are absent, results in significant lung pathology. In contrast, excessive IFN-γ production in mice infected with ECTVRev.SECRET, when only the SECRET domain is present, is not sufficient to cause pathology. It is possible that the significantly higher levels of IL-1β, IL-6, and IL-10 in ECTVRev.SECRET-infected mice, compared to levels induced by ECTVΔCrmD infection, could have overcome any protective effects that might have been afforded by IFN-γ blockade.
Discussion
OPV encode homologs of mammalian cytokine and chemokine receptors that can dampen, evade, or subvert the host immune response and influence the outcome of infection (20, 21, 23, 24). Such a strategy provides an advantage to the virus, allowing successful replication, transmission to other hosts, and even persistence within the host (43). As viruses have coevolved with their hosts and virus-encoded molecules can be host species specific, we utilized ECTV, a natural mouse pathogen, to address the function of CrmD in C57BL/6 mice that are normally resistant to infection with this virus. Both VARV and ECTV are related OPVs that cause respiratory infection and mousepox is a robust surrogate model for smallpox. The findings with mousepox will be relevant to smallpox and specifically to VARV-encoded CrmB, a vTNFR with TNF/LT binding properties for human TNF/LT and that also contains a SECRET domain that binds specific chemokines (25, 31). Respiratory infection of C57BL/6 mice with ECTVWT results in morbidity but animals completely recover, whereas infection with ECTVΔCrmD or ECTVRev.SECRET caused significant morbidity and 100% mortality. In the ECTV-resistant C57BL/6 mice, CrmD actually provided an advantage to both the host and virus.
The TNF-binding CRD not only reduces the bioavailability of host TNF, but it also dampens inflammatory cytokine production by reverse signaling through mTNF. The combined results from both in vivo experiments in ECTVWT-, ECTVΔCrmD-, or ECTVRev.SECRET -infected mice and in vitro results from BMDMs treated with Crm proteins from various OPVs revealed that the binding of CrmD to mTNF significantly reduces the levels of TNF production and some other cytokines that can mediate inflammatory or regulatory functions.
Previous studies on ECTV-encoded cytokine binding proteins (bp) such as IFN-γ-bp (44), IFN-α/β-bp (44, 45), and IL-18-bp (44) revealed that infection of ECTV-susceptible BALB/c mice with deletion mutant viruses was attenuated, some more than others. These mutant viruses were also attenuated in C57BL/6 mice, but the effects were far more dramatic in BALB/c mice. Indeed, some of us recently reported that infection of BALB/c mice with ECTVΔCrmD attenuated the infection to the extent that animals fully recovered from an otherwise lethal infection (22). When infected with ECTVWT, BALB/c mice respond with a weak inflammatory response and produce very little TNF or other inflammatory cytokines (35). However, when BALB/c mice were infected with ECTVΔCrmD, the inflammatory and cell-mediated immune responses were augmented resulting in effective virus control and survival (22). The CRD and SECRET domain were both found to be required for CrmD to mediate its function fully. The CRD neutralized TNF, whereas the SECRET domain reduced leukocyte recruitment, resulting in a reduction in the capacity of the host to generate an effective antiviral immune response (22).
In the present study, expression of CrmD protected the host from virus-induced immunopathology, instead of increasing virulence. Deletion of CrmD from ECTV significantly increased morbidity and mortality in ECTV-resistant C57BL/6 mice. In contrast to BALB/c mice, C57BL/6 mice infected with ECTVΔCrmD succumbed to mousepox due to significant lung inflammation and pathology. CrmD is critical for dampening lung inflammation in C57BL/6 mice, and its function requires host TNF. The increased susceptibility was not due to increased lung viral load as titers of WT and mutant viruses were comparable across the WT, TNF−/−, and TM mice. It is interesting to note that deletion of the secreted IL-1β receptor encoded by vaccinia virus also caused significant detrimental effects after i.n. infection of BALB/c mice, in this case with increased fever, accelerated weight loss, and mortality (4, 46).
C57BL/6 WT mice produce significantly higher levels of TNF compared to BALB/c mice (35). The absence of CrmD in WT and TM mice infected with ECTVΔCrmD resulted in significantly increased levels of TNF and other inflammatory cytokines including IL-6, IL-10, TGF-β, and IFN-γ. The dysregulated cytokine responses were contemporaneous with weight loss, higher clinical scores, exacerbated lung pathology, and 100% mortality in both WT and TM mice. WT mice in particular demonstrated clear increases in inflammatory cytokine responses but significantly reduced leukocyte recruitment into the lungs when both CRD and SECRET domain were absent. However, analysis of ECTVRev.SECRET-infected lungs established that it is the SECRET domain that contributed to leukocyte recruitment, at least at day 4 p.i. Nonetheless, the presence or absence of the SECRET domain when CRD was absent did not affect clinical scores, weight loss, lung pathology, or survival of WT and TM mice infected with either mutant virus. These findings are in sharp contrast to those reported for the BALB/c mice (22). As the SECRET domain has chemokine-binding activity and can bind to several chemokines (22), we had predicted that it would act in a similar manner in the C57BL/6 strain. It is likely that excessive production of a number of inflammatory cytokines in the absence of CRD in ECTVRev.SECRET-infected mice overcame the inhibitory effects of the SECRET domain on leukocyte recruitment to the lungs. It is also likely that virus inoculation through different routes, i.e., the i.n. route in C57BL/6 mice in the current study and through the s.c. route in BALB/c mice (22) may induce a distinct pattern of chemokine expression and influence leukocyte recruitment. In the current study, leukocyte numbers or phenotypes in lungs at days 4 or 8 p.i. did not correlate with pathology in mice infected with the mutant viruses, suggesting the possibility that dysregulated activation and production of excessive cytokines and chemokines by distinct leukocytes subsets present in lungs might be responsible.
The C57BL/6 TNF−/− strain is highly susceptible to respiratory ECTVWT infection and dies from serious lung pathology. Apart from binding to TNF, CRD can also interact with LT-α and LT-β and use of TNF−/− mice helped to dissociate the effects of CRD binding to TNF from those of binding to LT-α and/or -β. Unlike the significant effects seen in WT mice, infection of TNF−/− mice with ECTVΔCrmD or ECTVRev.SECRET did not make any difference to weight loss, clinical scores, leukocyte recruitment, viral load, lung pathology, or survival. These findings suggest that the major biological functions of CRD are mediated through binding to TNF. Nonetheless, mRNA transcripts for IL-1α, IL-12p40, and TGF-β were down-regulated, whereas IFN-γ and CCL-2 were up-regulated in mutant virus-infected TNF−/− mice, i.e., in the complete absence of CRD. Those changes are therefore not due to interactions of CRD with LT-α and/or LT-β.
The presence or absence of the SECRET domain when CRD was absent also did not affect clinical scores, weight loss, or survival of WT and TM mice. However, there were clear differences in the requirement for IFN-γ in causing pathology or controlling virus load in WT mice. IFN-γ has a well-established antiviral role in the mousepox model (39, 40), but excessive production can also cause lung pathology (41). In the current study, excessive IFN-γ production in mice infected with ECTVΔCrmD resulted in significant lung pathology, whereas it was not sufficient to cause pathology in mice infected with ECTVRev.SECRET. This is an unexpected but interesting finding that warrants further detailed investigation. The utilization of an additional mutant virus expressing only the CRD, in addition to the CrmD mutant and SECRET revertant viruses will help to delineate the distinct effects the two domains have on the immune response to ECTV infection. Nonetheless, some likely explanations include potential differences in the activation status of infiltrating leukocytes, which may have had an impact on the types and levels of cytokines/chemokines secreted in lungs of mice infected with ECTVΔCrmD or ECTVRev.SECRET. For example, there were distinct differences in levels of cytokines/chemokine in lungs of mice infected with the different mutant viruses. In particular, the levels of IL-1β, IL-6, and IL-10 were significantly higher in ECTVRev.SECRET-infected mice compared to ECTVΔCrmD-infected mice. Excessive levels of IL-6 or IL-10 production, as a consequence of TNF deficiency (41) or excessive TNF production (current study), during respiratory ECTV infection causes significant lung pathology. Consistent with our findings that high levels of IL-6 and IL-10 cause severe lung pathology is a recent report that showed that significantly higher levels of IL-6, IL-10, and TNF were detected in severe COVID-19 patients compared with those who had milder disease (47). IL-1β has been shown to drive chronic lung inflammation during influenza A virus infection in mice (48). It is conceivable that excessive levels of IL-1β, IL-6, and IL-10 in ECTVRev.SECRET-infected mice, treated with anti–IFN-γ mAb, could have overcome any protective effects that might have been otherwise observed.
It is striking that, under conditions of excessive TNF production (this study) or TNF deficiency (41) during respiratory ECTV infection, an overlapping set of cytokines and cytokine signaling pathways are dysregulated. Excessive TNF production involving NF-κB activation can induce the production of other inflammatory cytokines, including IL-6 and IL-10, which in turn can activate signal transducer and activator of transcription (STAT) 3 (49), which is downstream of the NF-κB signaling pathway. The two signaling pathways cross-regulate each other and regulate an overlapping group of target genes (50, 51). Therefore, perturbations in one pathway will disrupt or dysregulate the other pathway. Our results show that an overexuberant TNF response not only dysregulates the STAT3 signaling pathway (IL-6 and IL-10) but also the NLRP3 inflammasome (IL-1β), JAK/STAT (IFN-γ), NF-κB (IL-6), and TGF-β/Smad (TGF-β) signaling pathways. For this reason, blockading or dampening one inflammatory cytokine may be sufficient to ameliorate lung pathology, as we have shown in this study. However, if some of the inflammatory cytokines are produced at excessive levels, as in the case of ECTVRev.SECRET-infected mice, blockading a single inflammatory cytokine may not be sufficient to reduce lung pathology.
In conclusion, the outcome of infection of C57BL/6 vs. BALB/c mice with vTNFR mutant viruses would be determined not only by a balance between the host’s ability to produce TNF and the virus’ ability to neutralize its effects but also how CrmD signaling through mTNF affects production of other inflammatory cytokines. CrmD provides an advantage to ECTV in the BALB/c strain, but the rapid death of a susceptible host would not be conducive for effective virus spread. By enabling host survival in the C57BL/6 strain, CrmD benefits the host. Importantly, host survival would also facilitate virus spread and, as a consequence, provide an advantage to the virus. Based on the mousepox model, we would speculate that, in humans, VARV-encoded CrmB, expressing TNF and chemokine binding domains, may also protect against lung pathology in those individuals capable of generating potent inflammatory responses but otherwise contribute to increased susceptibility in those individuals not capable of generating an effective immune response.
Materials and Methods
Animal Studies.
Animal experiments were performed in accordance with protocols approved by the Animal Ethics and Experimentation Committee of the ANU (protocol numbers A2011/011 and A2014/018) and the Animal Ethics Committee of the University of Tasmania (protocol number A0016372). Six- to 12-wk-old female WT, Tnftm1Jods (TNF−/−) (2), Tnftm2Jods (mTNFΔ/Δ) (52), and Tnftm2JodsTnfrsf1atm1ImxTnfrsf1btm1Imx (designated triple mutant, TM) mice on a C57BL/6 background were bred under specific pathogen-free conditions at the Australian Phenomics Facility, Australian National University, Canberra, Australia. We generated the TM mice by crossing the mTNFΔ/Δ with TNFRI- and TNFRII-deficient mice (53). Animal experimental setup, virus inoculation, clinical scoring, weighing mice, and treatment with mAb were carried out as previously described (41) and outlined in SI Appendix. Details of sTNF, mTNF, TNFRI, and/or TNFRII expression by each mouse strain are presented in SI Appendix, Table S2.
Cell Lines and Viruses.
BS-C-1 (ATCC CCL-26) cells were cultured in Eagle’s minimum essential medium (EMEM) supplemented with 2 mM l-glutamine (Sigma-Aldrich), antibiotics (penicillin, 120 µg/mL; streptomycin, 200 µg/mL; and neomycin sulfate), 1 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), and 10% heat-inactivated fetal calf serum. This medium referred to as complete EMEM10. RAW264.7 (RAW 264.7; ATCC TIB-71) and NCTC clone 929 (L-929 cells; ATCC CCL-1) cells were cultured in complete DMEM (DMEM10) with additional supplements of 1 mM sodium pyruvate and 1 mM nonessential amino acids.
The Naval (22, 31) and Moscow (35) ECTV strains were propagated and quantified as described (54). The CrmD mutant viruses were generated from the Naval strain of ECTVWT (22, 31). Both strains of virus encode CrmD, with a single amino acid mutation (F263L) in a 320-amino acid polypeptide, have minor genetic differences (mainly due to different ways to annotate the viral genes) and exhibit similar degrees of virulence in BALB/c and C57BL/6 mice (55). A schematic overview of the three viruses with respect to their ability to bind TNF and/or chemokines is presented in SI Appendix, Table S1. For more information, see SI Appendix, Materials and Methods.
Generation of BMDMs and Virus Replication Kinetics.
BMDMs from WT, TNF−/− and TM mice were generated as described elsewhere (56) with some modifications and used to determine whether the CRD or SECRET domain influence ECTV replication in vitro. L929-cell conditioned medium was used as a source of macrophage-colony stimulating factor to generate BMDMs. For more details, see SI Appendix, Materials and Methods.
Viral Plaque Assay for Quantification of ECTV.
To enumerate the number of virus plaque-forming units (PFUs) present in samples, a viral plaque assay using BS-C-1 cells was used as previously described (54).
Reverse Signaling through mTNF in BMDMs from TM Mice.
A total of 5 × 106 mature BMDMs generated from TM mice was resuspended in 10 mL of complete DMEM10 and added to 25-cm2 tissue culture flasks. Cells were allowed to adhere for 2 h, prior to addition of LPS at 50 ng/mL, and incubated for 6 h. To induce reverse signaling through mTNF, recombinant Crm proteins were added to individual flasks at 10 μg/mL for 3 h. Cells were then harvested for isolation of RNA, generation of cDNA, and qRT-PCR analysis.
Histology and Microscopic Assessment of Lung Pathology.
Histological assessment of H&E-stained lung sections was undertaken as described previously (41). For more details, see SI Appendix, Materials and Methods.
Flow-Cytometric Analysis of Lung Leukocytes.
Lung tissue samples were digested with DNase I and collagenase, and flow cytometry of lung leukocytes was undertaken as described (41). Data were acquired on a BD LSR Fortessa flow cytometer with BD FACS Diva software and analyzed using FlowJo software, version 9.5 (Tree Star). For more details, see SI Appendix, Materials and Methods.
Detection of Cytokine Proteins in Lung Homogenates.
The levels of cytokines and chemokines in the lung homogenates were measured using capture ELISA kits (Biolegend), performed according to the manufacturer’s protocol. Optical density was measured at 450 nm with SOFTmax Pro software (Molecular Devices Corporation). For details on lung homogenate preparation, see SI Appendix, Materials and Methods.
RNA Extraction, cDNA Generation, and qRT-PCR.
RNA extraction, cDNA synthesis, and qRT-PCR were carried out as described (41). For more details, see SI Appendix, Materials and Methods.
Viral TNFR Expression and Purification.
Recombinant vTNFRs from ECTV (CrmD), CPXV (CrmB, CrmC, and CrmD), and VARV (CrmB) were expressed in the baculovirus system, fused to a C-terminal V5-6xHis tag as described elsewhere (25). Proteins were purified in metal chelate affinity columns from supernatants of infected Hi5 cell cultures. Permission from the World Health Organization was granted to hold VARV DNA encoding CrmB, and its manipulation was performed in accordance with the established rules.
Statistical Analysis.
Statistical analyses of experimental data, as indicated in the legend to each figure, were performed using appropriate tests to compare results using GraphPad Prism 8 (GraphPad Software). A value of P < 0.05 was taken to be significant: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Health and Medical Research Council of Australia to G.K. and G.C. (Grants 1007980 and 471426). The laboratory of A. Alcamí was funded by the Spanish Ministry of Science and Innovation and European Union (European Regional Development’s Funds) (Grants SAF2015-67485-R and RTI2018-097581-BI00). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank Prof. Jane Dahlstrom of the Australian National University Medical School and the Canberra Hospital, Australian Capital Territory, Australia, for helping us to develop the scoring system for the histopathology of lung sections. We also thank Dr. Jonathon D. Sedgwick, Boehringer Ingelheim Pharmaceuticals Inc. (Ridgefield, CT), for the gift of the TNF−/− and mTNFΔ/Δ mice. We acknowledge the assistance of staff at the Australian National University Phenomics Facility for animal breeding and the John Curtin School of Medical Research Microscopy and Flow Cytometry Research Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2004688117/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Arnett H. A.et al., TNF alpha promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 4, 1116–1122 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Körner H.et al., Distinct roles for lymphotoxin-alpha and tumor necrosis factor in organogenesis and spatial organization of lymphoid tissue. Eur. J. Immunol. 27, 2600–2609 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Papathanasiou S.et al., Tumor necrosis factor-α confers cardioprotection through ectopic expression of keratins K8 and K18. Nat. Med. 21, 1076–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcamí A., Smith G. L., A soluble receptor for interleukin-1 beta encoded by vaccinia virus: A novel mechanism of virus modulation of the host response to infection. Cell 71, 153–167 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Bean A. G., et al., Structural deficiencies in granuloma formation in TNF gene-targeted mice underlie the heightened susceptibility to aerosol Mycobacterium tuberculosis infection, which is not compensated for by lymphotoxin. J. Immunol. 162, 3504–3511 (1999). [PubMed] [Google Scholar]
- 6.Domm S., Cinatl J., Mrowietz U., The impact of treatment with tumour necrosis factor-alpha antagonists on the course of chronic viral infections: A review of the literature. Br. J. Dermatol. 159, 1217–1228 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Belisle S. E.et al., Genomic profiling of tumor necrosis factor alpha (TNF-alpha) receptor and interleukin-1 receptor knockout mice reveals a link between TNF-alpha signaling and increased severity of 1918 pandemic influenza virus infection. J. Virol. 84, 12576–12588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szretter K. J.et al., Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 81, 2736–2744 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tracey K. J.et al., Anti-cachectin/TNF monoclonal antibodies prevent septic shock during lethal bacteraemia. Nature 330, 662–664 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Black R. A.et al., A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385, 729–733 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Moss M. L.et al., Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-α. Nature 385, 733–736 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Beutler B., Cerami A., The biology of cachectin/TNF—a primary mediator of the host response. Annu. Rev. Immunol. 7, 625–655 (1989). [DOI] [PubMed] [Google Scholar]
- 13.Chu W. M., Tumor necrosis factor. Cancer Lett. 328, 222–225 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grell M.et al., The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell 83, 793–802 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Watts A. D.et al., A casein kinase I motif present in the cytoplasmic domain of members of the tumour necrosis factor ligand family is implicated in “reverse signalling.”. EMBO J. 18, 2119–2126 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juhász K., Buzás K., Duda E., Importance of reverse signaling of the TNF superfamily in immune regulation. Expert Rev. Clin. Immunol. 9, 335–348 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Eissner G., Kolch W., Scheurich P., Ligands working as receptors: Reverse signaling by members of the TNF superfamily enhance the plasticity of the immune system. Cytokine Growth Factor Rev. 15, 353–366 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Kirchner S.et al., LPS resistance in monocytic cells caused by reverse signaling through transmembrane TNF (mTNF) is mediated by the MAPK/ERK pathway. J. Leukoc. Biol. 75, 324–331 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Meusch U., Rossol M., Baerwald C., Hauschildt S., Wagner U., Outside-to-inside signaling through transmembrane tumor necrosis factor reverses pathologic interleukin-1beta production and deficient apoptosis of rheumatoid arthritis monocytes. Arthritis Rheum. 60, 2612–2621 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Alcami A., Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3, 36–50 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Alcami A., Koszinowski U. H., Viral mechanisms of immune evasion. Immunol. Today 21, 447–455 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alejo A.et al., Chemokines cooperate with TNF to provide protective anti-viral immunity and to enhance inflammation. Nat. Commun. 9, 1790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seet B. T.et al., Poxviruses and immune evasion. Annu. Rev. Immunol. 21, 377–423 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Stanford M. M., McFadden G., Karupiah G., Chaudhri G., Immunopathogenesis of poxvirus infections: Forecasting the impending storm. Immunol. Cell Biol. 85, 93–102 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Pontejo S. M., Alejo A., Alcami A., Comparative biochemical and functional analysis of viral and human secreted tumor necrosis factor (TNF) decoy receptors. J. Biol. Chem. 290, 15973–15984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith G. L., Virus proteins that bind cytokines, chemokines or interferons. Curr. Opin. Immunol. 8, 467–471 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Reading P. C., Khanna A., Smith G. L., Vaccinia virus CrmE encodes a soluble and cell surface tumor necrosis factor receptor that contributes to virus virulence. Virology 292, 285–298 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Saraiva M., Alcami A., Crm E., CrmE, a novel soluble tumor necrosis factor receptor encoded by poxviruses. J. Virol. 75, 226–233 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu F. Q., Smith C. A., Pickup D. J., Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology 204, 343–356 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Loparev V. N.et al., A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc. Natl. Acad. Sci. U.S.A. 95, 3786–3791 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alejo A.et al., A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc. Natl. Acad. Sci. U.S.A. 103, 5995–6000 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefkowitz E. J.et al., Poxvirus bioinformatics resource center: A comprehensive poxviridae informational and analytical resource. Nucleic Acids Res. 33, D311–D316 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ribas G., Rivera J., Saraiva M., Campbell R. D., Alcami A., Genetic variability of immunomodulatory genes in ectromelia virus isolates detected by denaturing high-performance liquid chromatography. J. Virol. 77, 10139–10146 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhri G., Panchanathan V., Bluethmann H., Karupiah G., Obligatory requirement for antibody in recovery from a primary poxvirus infection. J. Virol. 80, 6339–6344 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhri G.et al., Polarized type 1 cytokine response and cell-mediated immunity determine genetic resistance to mousepox. Proc. Natl. Acad. Sci. U.S.A. 101, 9057–9062 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang M., Sigal L. J., Antibodies and CD8+ T cells are complementary and essential for natural resistance to a highly lethal cytopathic virus. J. Immunol. 175, 6829–6836 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Karupiah G., Buller R. M., Van Rooijen N., Duarte C. J., Chen J., Different roles for CD4+ and CD8+ T lymphocytes and macrophage subsets in the control of a generalized virus infection. J. Virol. 70, 8301–8309 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker A. K., Parker S., Yokoyama W. M., Corbett J. A., Buller R. M. L., Induction of natural killer cell responses by ectromelia virus controls infection. J. Virol. 81, 4070–4079 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karupiah G., Fredrickson T. N., Holmes K. L., Khairallah L. H., Buller R. M., Importance of interferons in recovery from mousepox. J. Virol. 67, 4214–4226 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panchanathan V., Chaudhri G., Karupiah G., Interferon function is not required for recovery from a secondary poxvirus infection. Proc. Natl. Acad. Sci. U.S.A. 102, 12921–12926 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuazon Kels M. J.et al., TNF deficiency dysregulates inflammatory cytokine production, leading to lung pathology and death during respiratory poxvirus infection. Proc. Natl. Acad. Sci. U.S.A. 117, 15935–15946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Sayers E. W., GenBank. Nucleic Acids Res. 39, D32–D37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakala I. G.et al., Evidence for persistence of ectromelia virus in inbred mice, recrudescence following immunosuppression and transmission to naïve mice. PLoS Pathog. 11, e1005342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakala I. G., Chaudhri G., Eldi P., Buller R. M., Karupiah G., Deficiency in Th2 cytokine responses exacerbate orthopoxvirus infection. PLoS One 10, e0118685 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu R. H.et al., The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. J. Exp. Med. 205, 981–992 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alcamí A., Smith G. L., A mechanism for the inhibition of fever by a virus. Proc. Natl. Acad. Sci. U.S.A. 93, 11029–11034 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen G.et al., Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130, 2620–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sichelstiel A.et al., Targeting IL-1β and IL-17A driven inflammation during influenza-induced exacerbations of chronic lung inflammation. PLoS One 9, e98440 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong Z., Wen Z., Darnell J. E. Jr., Stat3: A STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994). [DOI] [PubMed] [Google Scholar]
- 50.Ma J. F.et al., STAT3 promotes IFNγ/TNFα-induced muscle wasting in an NF-κB-dependent and IL-6-independent manner. EMBO Mol. Med. 9, 622–637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oeckinghaus A., Hayden M. S., Ghosh S., Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Ruuls S. R.et al., Membrane-bound TNF supports secondary lymphoid organ structure but is subservient to secreted TNF in driving autoimmune inflammation. Immunity 15, 533–543 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Peschon J. J., et al., TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J. Immunol. 160, 943–952 (1998). [PubMed] [Google Scholar]
- 54.Chaudhri G., Kaladimou G., Pandey P., Karupiah G., Propagation and purification of ectromelia virus. Curr. Protoc. Microbiol. 51, e65 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Mavian C.et al., The genome sequence of ectromelia virus Naval and Cornell isolates from outbreaks in North America. Virology 462–463, 218–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weischenfeldt J., Porse B., Bone marrow-derived macrophages (BMM): Isolation and applications. Cold Spring Harbor Protoc. 2008, pdb.prot5080 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.