Significance
Recently N6-methyladenosine (m6A) methylation has emerged as a biological process with significant impact on cellular functions. However, almost all the research regarding m6A methylation has been based on mRNAs. In our research, we focus on how m6A methylation affects microRNA (miRNA) biogenesis in Arabidopsis. In brief, we show that m6A methylation is necessary to maintain proper levels of mature miRNAs as well as their precursors. m6A mark affects pri-miRNA secondary structures and affects the recruitment of the Microprocessor to pri-miRNAs. We also demonstrate the interactions of MTA (m6A writer) with other proteins involved in miRNA biogenesis, namely RNA Polymerase II and TOUGH. Our study provides evidence of the role played by m6A in plant miRNA biogenesis.
Keywords: miRNA biogenesis, MTA, m6A methylation
Abstract
In Arabidopsis thaliana, the METTL3 homolog, mRNA adenosine methylase (MTA) introduces N6-methyladenosine (m6A) into various coding and noncoding RNAs of the plant transcriptome. Here, we show that an MTA-deficient mutant (mta) has decreased levels of microRNAs (miRNAs) but accumulates primary miRNA transcripts (pri-miRNAs). Moreover, pri-miRNAs are methylated by MTA, and RNA structure probing analysis reveals a decrease in secondary structure within stem–loop regions of these transcripts in mta mutant plants. We demonstrate interaction between MTA and both RNA Polymerase II and TOUGH (TGH), a plant protein needed for early steps of miRNA biogenesis. Both MTA and TGH are necessary for efficient colocalization of the Microprocessor components Dicer-like 1 (DCL1) and Hyponastic Leaves 1 (HYL1) with RNA Polymerase II. We propose that secondary structure of miRNA precursors induced by their MTA-dependent m6A methylation status, together with direct interactions between MTA and TGH, influence the recruitment of Microprocessor to plant pri-miRNAs. Therefore, the lack of MTA in mta mutant plants disturbs pri-miRNA processing and leads to the decrease in miRNA accumulation. Furthermore, our findings reveal that reduced miR393b levels likely contributes to the impaired auxin response phenotypes of mta mutant plants.
One of the most abundant mRNA modifications in eukaryotic cells, N6-methyladenosine (m6A), can regulate eukaryote gene expression at co- as well as posttranscriptional levels. m6A methylation in animal mRNAs is associated with several biological processes, ranging from cell development (1, 2) to carcinogenesis (3) and viral infections (4, 5), with the underpinning mechanisms including m6A regulated pre-mRNA splicing patterns, mRNA export, mRNA stability, and changes in translational efficiency (1). A group of proteins that collectively form the RNA methylation “writer” complex have been characterized and are well conserved between plants and animals. The mammalian m6A methyltransferase complex consists of methyltransferase-like 3 (METTL3) (2), methyltransferase-like 14 (METTL14) (6), Wilms’ tumor 1-associating protein (WTAP) (7), VIRMA (KIAA1429) (8), RNA-binding motif protein 15 (RBM15) (9), and zinc finger CCCH-type Containing 13 (ZC3H13) (10, 11). While METTL3 has been identified as the catalytic protein in this complex (2), auxiliary proteins provide specificity and help with proper localization of the complex (1). The m6A mark can be recognized by various “readers,” the best-characterized of which belong to the YT521-B homology (YTH) domain family (12–15). The modification can also be removed from transcripts by “erasers,” which in humans include the fat mass and obesity-associated protein (16) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5) (17).
In Arabidopsis thaliana, the presence of m6A was first reported in 2008 and was shown to be dependent upon the activity of mRNA adenosine methylase A (MTA; a homolog of human METTL3), the catalytic component of Arabidopsis m6A methyltransferase complex (18). FKBP12 interacting protein 37 kDa (FIP37, a homolog of WTAP) was the first identified methyltransferase-interacting complex member in eukaryotes (18). Later, FIP37 was further characterized and shown to be required for m6A formation in mRNA (19). Other protein components of the plant m6A methyltransferase complex that have been identified include: MTB (methytransferase B, a homolog of METTL14), VIR (Virilizer, a homolog of VIRMA), and AtHAKAI (a homolog of HAKAI) (20). In Arabidopsis, evolutionarily conserved C-terminal region proteins are the orthologs of the mammalian YTH family of proteins and are the only plant readers that have been identified so far (21–23). ALKBH10B and ALKBH9B represent the characterized Arabidopsis m6A erasers (24, 25).
Null mutants of MTA or any other members of the core writer complex (i.e., MTB, FIP37, and VIR) are embryo-lethal (18–20), indicating an essential function for this modification. However, hypomorphic knockdown mutants of these writers have been obtained and typically show an 80 to 90% reduction in m6A levels (19, 20, 26). In contrast, reader and eraser knockouts are viable (23–25). Studies utilizing these various mutant plant lines indicate a role of this modification in embryogenesis, proper plant development (trichome morphology, meristem maintenance, vascular development), flowering time and flower morphology, and pathogen response (18–20, 24–26).
Many aspects of plant development and metabolism are controlled by microRNAs (miRNAs), and the complex phenotypes of low methylation plants have aspects reminiscent of miRNA biogenesis pathway mutants (see ref. 27 for review) and might be, at least partially, explained by the influence of m6A on miRNA biogenesis. miRNAs are small endogenous noncoding RNAs that are ∼21 nt in length and are important players in regulating cellular metabolism. miRNAs are derived from hairpins present in primary miRNAs (pri-miRNAs) that are synthetized by RNA Polymerase II (RNA Pol II), and further processed by RNase III-type enzymes associated with and assisted by other proteins. In animals, where m6A is important for biogenesis of many miRNAs, m6A plays the role of a mark, identified by a reader protein (heterogeneous nuclear ribonucleoprotein A2/B1; HNRNPA2B1) that facilitates the recruitment of downstream enzymes and associated proteins to pri-miRNAs, thus facilitating their processing to miRNAs. Accordingly, it was shown that depletion of METTL3 leads to decreased accumulation of miRNAs and to an overaccumulation of pri-miRNAs due to their impaired processing (28, 29). While most of the findings related to m6A in plants concern mRNAs, its role in miRNA biogenesis in plants remains unknown.
In this study, we provide evidence that biogenesis of at least 25% of Arabidopsis miRNAs is affected by the absence of the m6A mark. We show that plant pri-miRNAs are m6A-methylated by MTA, and deficiency of MTA (and thus m6A) leads to accumulation of pri-miRNAs accompanied by lower miRNA levels. We also show that MTA interacts with RNA Pol II and TOUGH (TGH), suggesting that MTA acts at early stages of miRNA biogenesis. Lack of m6A leads to the stem–loop region of pri-miRNAs becoming less structured, which negatively impacts the binding of Hyponastic Leaves 1 (HYL1) to these precursors. Together, these results suggest that MTA affects Microprocessor assembly via its influence on secondary structure of methylated miRNA precursors, as well as direct interactions between MTA and a miRNA biogenesis protein TGH. Thus, the lack of m6A methylation in pri-miRNAs in the mta mutant plants results in the inefficient recruitment of Microprocessor components to plant pri-miRNAs, resulting in their less-efficient processing, and ultimately leading to the decrease in miRNA production. We also suggest that the impaired auxin response in plants with MTA deficiency is caused, at least partly, by decreased miR393b levels in the mta mutant plants.
Results
Lack of m6A Results in Impaired miRNA Biogenesis.
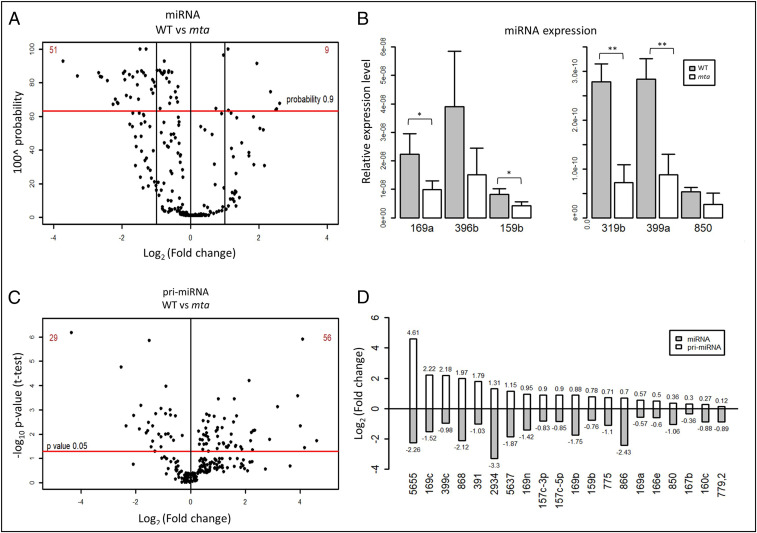
In order to assess possible effects of m6A on miRNA biogenesis, we performed small RNA (sRNA) sequencing on rosette leaves from 4-wk-old Arabidopsis plants of both WT Col-0 and a mutant line with severely reduced m6A levels (mta mutant). More specifically, the mta mutant line we used contains MTA cDNA under the ABI3 promoter in a homozygous MTA T-DNA insertion mutant background (mta ABI3:MTA). The ABI3 promoter drives strong embryo expression of MTA, enabling the plant embryo lethality to be bypassed. However, this promoter drives a very low level of expression postgermination, giving rise to plants with 80 to 90% less m6A modification compared to their WT counterparts (26). Our sequencing data showed a decrease in the overall abundance of mature miRNAs in mta mutant as compared to WT plants (SI Appendix, Fig. S1). Specifically, we identified 60 differentially expressed miRNAs that had high confidence scores (probability ≥ 0.9 or false-discovery rate ≤ 0.1), and 51 of these 60 miRNAs were found to be down-regulated, while only 9 miRNAs were up-regulated (Fig. 1A and SI Appendix, Table S1). This down-regulation of miRNAs in mta plants was confirmed by quantitative real-time PCR (RT-qPCR) for six arbitrarily selected miRNAs (miR159b, miR169a, miR319b, miR396b, mir399a, and miR850) (Fig. 1B).
Fig. 1.
miRNA biogenesis is impaired in mta mutant plants. (A) sRNA sequencing analysis of miRNAs in mta and WT plants. Each black dot represents one miRNA. The red horizontal bar represents the threshold (probability 0.9). (B) Relative abundance (as determined by TaqMan RT-qPCR) of miRNAs identified with altered abundance in WT vs. the mta mutant monitored by sRNA sequencing. *P < 0.05, **P < 0.005, and error bars represent SD (n = 3). (C) Levels of 230 pri-miRNAs as determined by RT-qPCR with separation of statistically significant pri-miRNAs (above the red horizontal bar). Each dot represents one pri-miRNA and the red horizontal bar represents P-value threshold (P value 0.05). (D) A set of cognate pairs of pri-miRNAs/miRNAs (selected from A and B) where pri-miRNA levels are up-regulated, while the levels of their cognate miRNA are down-regulated.
Next, we went on to investigate the levels of primary miRNAs (pri-miRNAs) in the low methylation plants (mta mutants) by using the mirEX2 platform (www.combio.pl/mirex2) developed by our group (30, 31). This platform utilizes a repository of 298 primer pairs, specifically for Arabidopsis pri-miRNA quantification by RT-qPCR. Of the 298 pri-miRNAs tested, 68 pri-miRNAs were at undetectable levels in our samples, leaving 230 for further analysis. The results of RT-qPCR revealed that 85 pri-miRNAs had statistically significant (P ≤ 0.05, n = 3) changes in their accumulation (WT vs. mta); 56 of these (∼66%) were found to be up-regulated, while 29 were down-regulated (Fig. 1C and SI Appendix, Table S1). Upon comparison of the RT-qPCR results with sRNA sequencing data, we identified 20 cognate pri-miRNA/miRNA pairs that showed higher accumulation of pri-miRNAs and lower levels of mature miRNAs (Fig. 1D). These results point toward an impaired processing of these pri-miRNAs in the absence of MTA.
Arabidopsis pri-miRNAs Are m6A-Methylated by MTA.
The general trend, showing an accumulation of pri-miRNAs and decreased levels of miRNAs in the conditions of reduced m6A methylation, raised the possibility that the presence of m6A is necessary for proper processing of some pri-miRNAs. Therefore, we tested the methylation status of the pri-miRNAs using m6A-RNA immunoprecipitation followed by sequencing (m6A-IP Seq). This method is not an equivalent of the commonly used MeRIPSeq (32), as we avoided the fragmentation of polyA RNA. We validated this protocol using spiked-in methylated and nonmethylated controls (SI Appendix, Fig. S2). We further tested the robustness of our data by comparing them to previously published data by Shen et al. (19) and Anderson et al. (33). In our dataset, we were able to identify 14,870 genes whose transcripts (15,562) are depleted in the mta mutant, indicating that these gene transcripts are methylated. Upon comparison of our data to the published datasets, we found an overlap of 2,868 (80%) and 3,100 genes (65%) between our data and data from Shen et al. (19) and Anderson et al. (33), respectively; 930 genes were common among all three datasets. A Venn diagram showing these overlaps is presented in SI Appendix, Fig. S3.
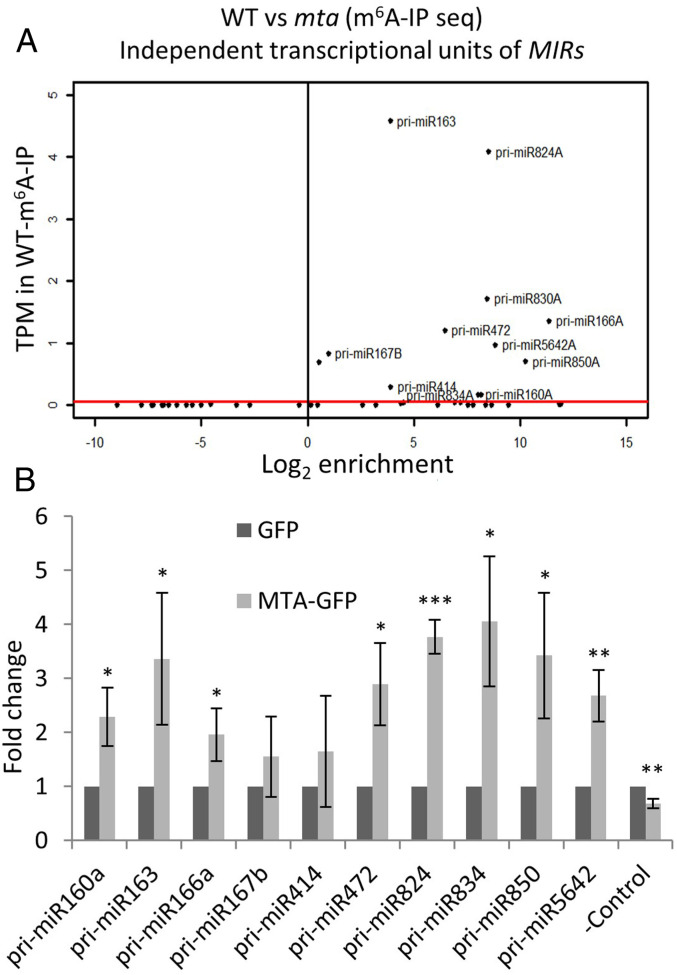
miRNAs can be encoded within intergenic regions as independent transcriptional units or can be embedded within protein-coding genes sharing transcriptional units with their host protein genes (34). For our analysis from the m6A-IP Seq data we selected transcripts of only those miRNA genes that are independent transcriptional units. Using this approach, we identified transcripts of 11 MIR genes that were enriched more than 1.5-fold in WT vs. mta, indicating that they are m6A-methylated in WT plants (hence can be immunoprecipitated with m6A antibody), and lack this methylation in the absence of MTA (Fig. 2A and SI Appendix, Fig. S4). All of the identified pri-miRNAs that passed the enrichment threshold were either absent or had significantly reduced abundance in the mta IP sample relative to WT IP.
Fig. 2.
At least a set of pri-miRNAs are m6A-methylated by MTA. (A) MIR gene transcript identified by m6A-IP Seq. The red bar represents the threshold of transcripts per million (TPM) > 0.05. (B) Levels of pri-miRNAs in MTA-GFP samples after GFP trap-based RIP followed by RT-qPCR are presented as fold-changes as compared to the GFP control. *P < 0.05, **P< 0.005, ***P < 0.001. Error bars represent SD of three biological replicates. –Control = AT2G40000 gene.
As an orthogonal approach to verify that these pri-miRNAs were bona fide targets for MTA directed methylation, we carried out RNA IP (RIP) using an anti-GFP antibody and the GFP-tagged MTA (35S:MTA-GFP) plant line. As a control, the plants expressing only GFP in the WT Col-0 background were used. RIP was performed on the nuclear fraction and followed by RT-qPCR on oligo(dT) primed cDNA. Eighteen pri-miRNAs (10 from our m6A-IP Seq data and 8 randomly chosen) were selected for verification by RT-qPCR, and of these all but 4 showed a statistically significant enrichment (P < 0.05) in the MTA–GFP sample when compared to the GFP control (Fig. 2B and SI Appendix, Fig. S5). This indicates that pri-miRNAs are frequently bound by MTA. Taken together, these data suggest that MTA binds and methylates at least a set of pri-miRNAs, and that this binding promotes their processing to mature miRNAs.
Lack of m6A Affects Stem–Loop Structure of pri-miRNAs and Their Ability to Bind HYL1.
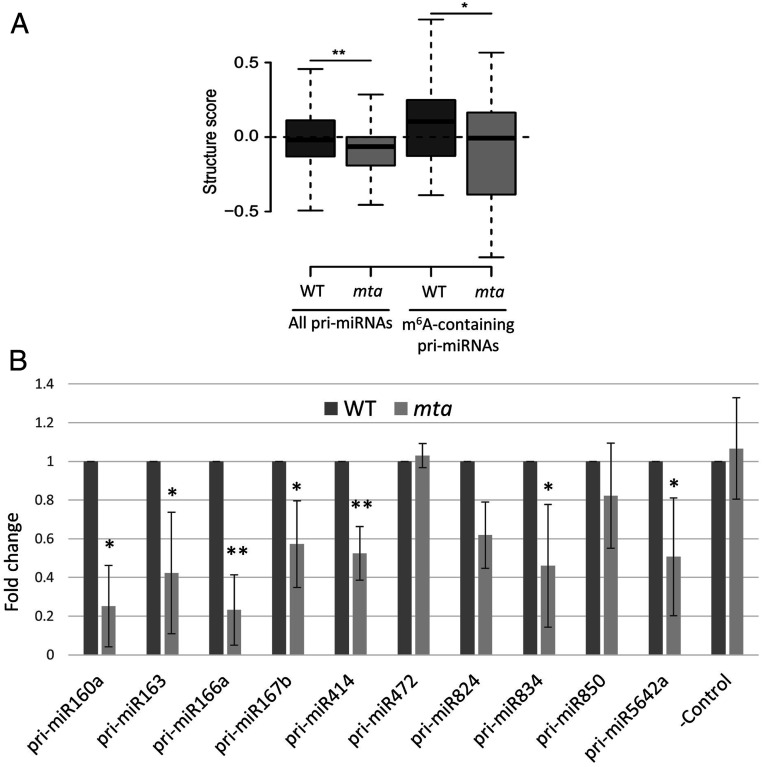
The effect of m6A on secondary structure of RNA has been well documented. Such alterations in RNA structure induced by m6A are known as “m6A switches” (35–37). These m6A switches influence the ability of RNA-binding proteins to recognize the RNA molecules, which further determines the substrate RNA fate. We used protein interaction profile sequencing (PIP-seq) in order to determine whether such structural changes could be observed in miRNA precursors and hence influence miRNA biogenesis. Using this approach, double-stranded RNAs and single-stranded RNAs can be detected after single- and double-stranded specific RNase treatment, respectively (38–40), and thus we were able to identify and distinguish between the structured and unstructured regions of the miRNA precursors. We noticed that the nucleotide accessibility for double- or single-stranded RNase(s) is altered in mta mutants. Our data show a significant reduction in the level of pri-miRNA stem–loop structure in the mta mutant as compared to WT plants (Fig. 3A, two box plots on the left). We observed even stronger differences when we focused only on those pri-miRNAs that we identified as m6A-methylated (Fig. 3A, two box plots on the right). By constraining a folding algorithm with PIP-seq–derived structure scores, we obtained 2D structural models for a few examples among the m6A-methylated pri-miRNAs (models for pri-miR160a, pri-miR163a, pri-miR166a, and pri-miR830 are presented in SI Appendix, Fig. S6). Presented models represent the most frequently interrogated structural conformation among the potentially multiple existing folding patterns for each RNA. These data-based structural models suggest that the pri-miRNA regions containing the miRNA/miRNA* stem regions tend to be in an unpaired conformation in the absence of the m6A mark (mta mutants) as compared to the methylated pri-miRNAs (WT plants) (SI Appendix, Fig. S6).
Fig. 3.
The structures of miRNA precursors and their binding to HYL1 is altered in absence of m6A. (A) Box plots show a decrease in the structure scores of miRNA precursors. All miRNA precursors include 235 precursors whose data are accessible from the PIP-seq analysis (two box plots on the left). m6A-containing precursors refer to the 11 pri-miRNA examples found as m6A-methylated in this study (two box plots on the right). (B) Levels of pri-miRNAs in mta mutants after HYL1-RIP RT-qPCR are presented as fold-changes as compared to WT control. Error bars represent SD of three biological replicates. *P < 0.05, **P < 0.005. –Control = WUSCHEL (WUS) gene (AT2G17950).
In light of these data, we reasoned that these changes in structure could in principle influence the binding of HYL1 to these precursors. HYL1 is a double-stranded RNA binding protein and is an important player in miRNA biogenesis (41, 42). Hence, we studied the binding of HYL1 to pri-miRNAs in WT and mta mutant plants using RIP (HYL1-RIP) followed by RT-qPCR. We found that 7 of the 10 pri-miRNAs tested were significantly less abundant in HYL1-IP samples from the mta mutants as compared to WT (Fig. 3B). A decrease in the binding efficiency of HYL1 that is a double-stranded RNA binding protein, to pri-miRNAs in the mta mutant is an additional indication that a double-stranded conformation of miRNA/miRNA* containing stem–loop regions of pri-miRNAs are formed less frequently in low methylation (mta) than in WT plants.
MTA Interacts with RNA Pol II In Situ.
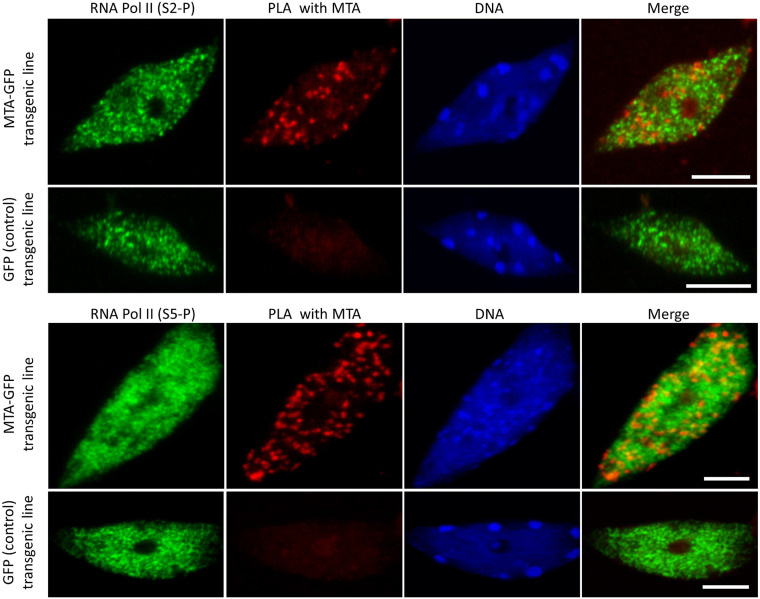
The strong tendency toward accumulation of pri-miRNAs and low abundance of miRNAs in mta mutants is reminiscent of various Arabidopsis mutants of genes encoding the proteins that participate in miRNA biogenesis, like Dicer-like 1 (DCL1), HYL1 (42–44), Serrate (SE) (45), and TGH (46). These proteins are involved in early stages of miRNA biogenesis and are needed for efficient processing of pri-miRNAs to miRNAs. These observations led us to suggest that MTA might act at early stages of miRNA biogenesis, probably cotranscriptionally. The deposition of m6A is usually assumed to be cotranscriptional; this assumption is supported by the fact that m6A can influence other processing events, such as selection of polyadenylation sites or pre-mRNA splicing, which are themselves cotranscriptional (47–49). However, a direct association between METTL3 and RNA Pol II has only been shown in mammalian cells when the rate of transcription was artificially slowed down (50). We confirmed colocalization of MTA and RNA Pol II in Arabidopsis using immunolocalization (SI Appendix, Fig. S7). Furthermore, we used the proximity ligation assay (PLA) to confirm in vivo interactions of MTA with RNA Pol II in plants under physiological conditions. Our results show direct interactions between MTA–GFP and RNA Pol II in cell nuclei with MTA–GFP expression but not in the GFP control (Fig. 4). These results were obtained for RNA Pol II phosphorylated at both Serine 5 and Serine 2. Thus, we show that MTA is associated with the RNA Pol II from an early stage of transcription and likely methylates pri-miRNAs (among other RNA Pol II transcripts) cotranscriptionally.
Fig. 4.
PLA shows the interaction between MTA and RNA Pol II phosphorylated at Serine 2 and Serine 5. Positive PLA signals (red spots in the second column) can be seen only in cells containing the MTA–GFP transgene, but not in control cells expressing GFP alone. RNA Pol II is represented in green. DNA is stained with HOECHST (blue). (Scale bars, 5 µm.)
MTA Acts at Early Stages of miRNA Biogenesis.
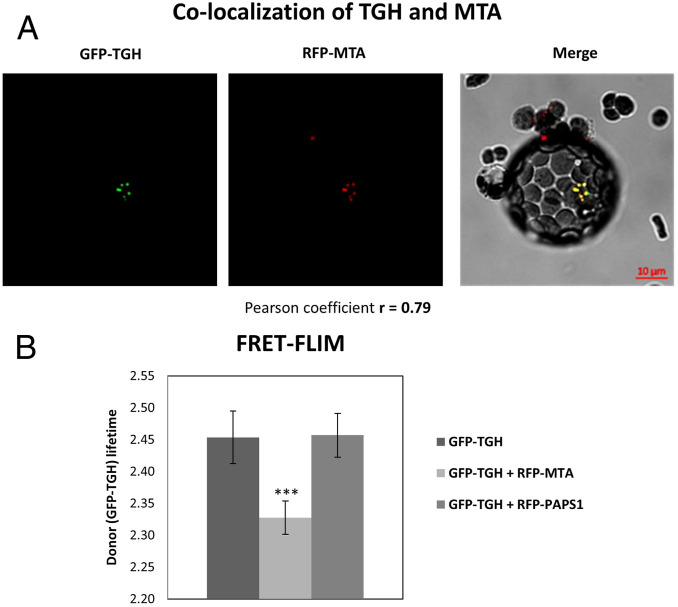
While MTA interacting with RNA Pol II is an indication of it methylating RNA Pol II transcripts cotranscriptionally in general, we investigated whether specific proteins involved in miRNA biogenesis could interact with MTA. To address this possibility, we performed a screen on possible MTA interactors involved in early steps of miRNA biogenesis, like HYL1, Cap binding protein 20 (CBP20), SE, TGH, and Dawdle (DDL1), using a microscopic approach. We performed FRET analyzed by fluorescence lifetime imaging microscopy (FLIM). As FRET can occur only when two proteins are within nanometers of each other, FRET–FLIM helps to quantify direct protein–protein interactions. This FRET–FLIM analysis confirmed the close association and interactions of MTA and TGH (Fig. 5). No such interactions could be seen in the case of any other tested proteins (SI Appendix, Fig. S8). Thus, we identified the miRNA biogenesis factor TGH as an MTA partner.
Fig. 5.
MTA interacts with the miRNA biogenesis related protein TGH. (A) Arabidopsis WT protoplasts cotransfected with GFP–TGH and RFP-MTA (Left and Center), showing colocalization of both proteins in the nucleus. (Right) A merged image. (B) FRET–-FLIM analysis in Arabidopsis protoplasts cotransfected with GFP–TGH and RFP–MTA, or GFP–TGH and RFP–PAPS1 [PAPS1 stands for Poly(A) Polymerase 1, used here as a control protein]. Reduction in a donor lifetime (GFP–TGH) was seen with RFP–MTA but not with the negative control (RFP–PAPS1); ***P < 0.001, n = 9.
Lack of m6A/MTA Impairs Microprocessor Complex Assembly.
Having identified TGH as an interactor of MTA, we focused on the m6A methylation status of pri-miRNAs in the tgh mutant. m6A-IP followed by RT-qPCR revealed that except for two pri-miRNAs, there is no overall significant reduction of m6A methylation in pri-miRNAs in the tgh mutant (SI Appendix, Fig. S9). This result indicates that TGH is not required for m6A methylation, and MTA acts upstream of TGH in miRNA biogenesis.
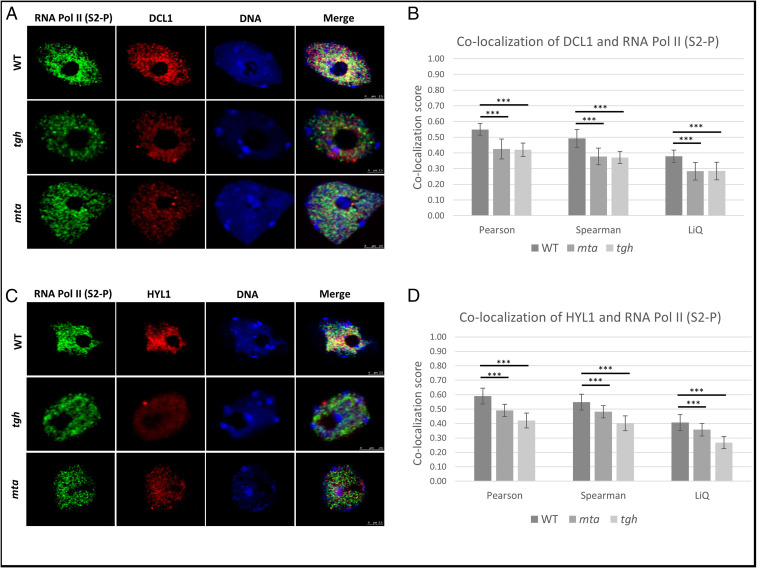
It is known that during miRNA biogenesis, TGH facilitates DCL1 action and pri-miRNA processing via promoting the pri-miRNA–HYL1 interaction (46). We then tested whether a lack of MTA, and hence loss of the MTA–TGH interaction, could also affect Microprocessor assembly. Using coimmunolocalization, we determined that in the tgh mutant DCL1 colocalization with RNA Pol II (phosphorylated at Serine 2) is significantly reduced. We found a similar level of reduction in DCL1 colocalization with RNA Pol II in the mta mutant (Fig. 6 A and B). Similarly, HYL1 colocalization with RNA Pol II is also reduced in mta mutant plants similar to the tgh mutant (Fig. 6 C and D). The observation that the lack of either TGH or MTA hampers colocalization of RNA Pol II with DCL1 and HYL1 suggests that MTA facilitates Microprocessor assembly either by attracting TGH via direct MTA–TGH interaction or with help of an additional m6A reader protein (unknown so far).
Fig. 6.
Microprocessor assembly is impaired in both mta and tgh mutants. (A) Localization of RNA Pol II (green), DCL1 (red), and DNA (blue) in the nuclei of WT plants as well as mta and tgh mutants. (B) Colocalization scores of DCL1 and RNA Pol II are significantly lower in mta and tgh mutants as calculated using three different approaches (Pearson, Spearman, and LiQ). (C) Localization of RNA Pol II (green), HYL1 (red), and DNA (blue) in the nuclei of WT plants as well as mta and tgh mutants. The merge column shows all three channels. (D) Colocalization scores of HYL1 with RNA Pol II are significantly lower in mta and tgh mutants as calculated by three different approaches (Pearson, Spearman, and LiQ). ***P < 0.001, n = 50.
MTA Regulates the Level of miR393b which Is Involved in Auxin Response.
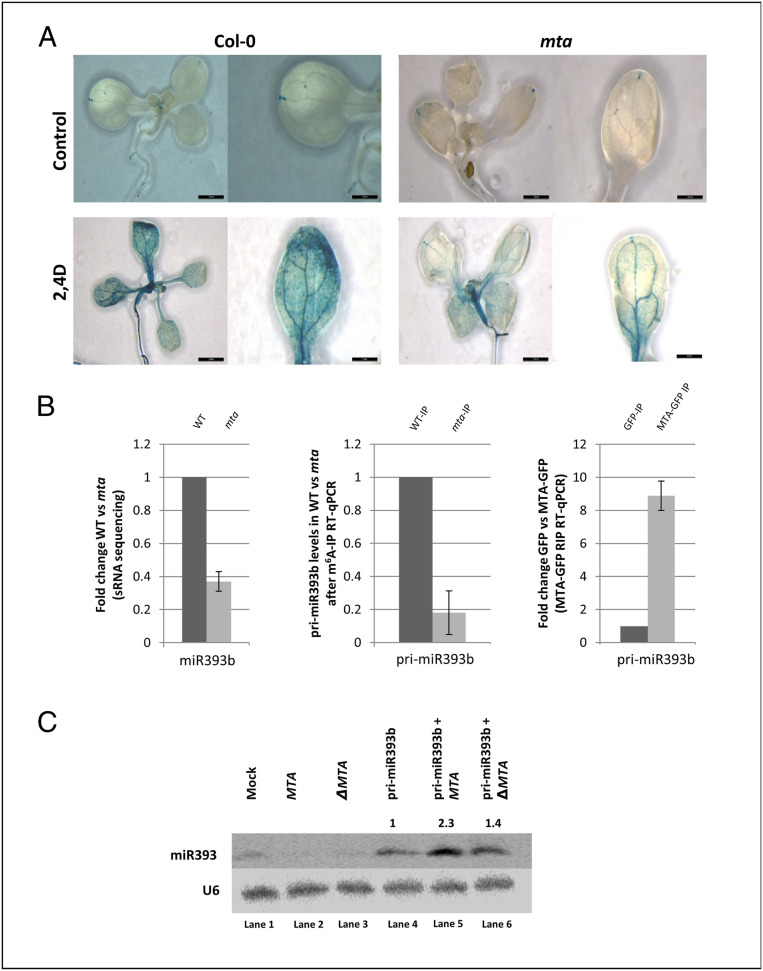
Auxin response defects have been reported for hypomorphic mutants of the known plant m6A methyltransferase complex (20). We found this to be the case for the auxin-responsive DR5pro:GUS reporter construct when introduced into both WT and the mta mutant background. Upon induction of 14-d-old seedlings with 2,4-dichlorophenoxyacetic acid (2,4-D), the mta plants showed much less GUS expression (Fig. 7A).
Fig. 7.
Auxin insensitivity in the mta mutant correlates with loss of methylation of pri-miR393b and the low level of mature miR393b. (A) Expression of auxin-responsive reporter DR5pro:GUS construct in the mta mutant background is reduced upon treatment with auxin (2,4-D). Strong inhibition of auxin response after induction is observed in the mta mutant. (Scale bars, 2 mm.) (B) The miR393b level is down-regulated in the mta mutant (sRNA sequencing data) as compared to WT plants (Left, n = 3), pri-miR393b carries m6A mark (m6A-IP followed by RT-qPCR, Center, n = 3), and pri-miR393b is bound by MTA (MTA-GFP RIP, Right, n = 3). Graphs represent fold-change values between WT and mta mutant plants (Left and Center) and GFP and MTA-GFP plants (Right). (C) N. benthamiana leaves were agroinfiltrated with one of following constructs: MTA, catalytically inactive MTA (ΔMTA), and pri-miR393b (lanes 2, 3, and 4, respectively) or with a combination of two constructs [encoding pri-miR393b + the construct encoding MTA (lane 5) or pri-miR393b + ΔMTA (lane 6)]. The levels of miR393b were monitored for each experimental variant using Northern blotting. Mock represents transfection with no plasmid control, and U6 serves as a loading control. Numbers above the last three lanes represent relative amounts of miR393b in the presence of MTA (2.3, penultimate lane) or ΔMTA (1.4, last lane) as compared to the expression of pri-miR393b alone (1, lane 4).
miR393b has been described to be involved in auxin response regulation, and in mutants lacking miR393b, auxin signaling is reduced (51). In our sRNA sequencing data, we found that miR393b is down-regulated in mta mutants, although we could not detect its precursor in our m6A-IP Seq. However, because of its recognized importance in regulating auxin responses, we further investigated the requirement of m6A for miR393b accumulation. We confirmed that pri-miR393b is m6A-methylated using m6A-IP followed by RT-qPCR. IP of RNA bound to MTA also confirmed MTA binding to pri-miR393b (Fig. 7B). Interestingly, our PIP-seq analysis revealed that pri-miR393b is less structured in mta than in WT plants. Moreover, the 2D model of this precursor revealed that its stem–loop region that contains mature miR393b sequence is formed more efficiently when pri-miR393b contains m6A (WT) in comparison to unmethylated precursors (mta mutant), similar to the other miRNAs showing underaccumulation in mta mutant plants in comparison to WT (SI Appendix, Fig. S10).
To further demonstrate that the low expression level of miR393b is in fact caused by the lack of MTA, we designed a transient expression assay in Nicotiana benthamiana. We used constructs designed to express pri-miR393b, MTA, or a catalytically inactive version of MTA, ΔMTA (D482A). ΔMTA was prepared by primer-induced point mutation resulting in the following change: Aspartic acid at position 482 to alanine (D482A) at the catalytic DPPW motif (52), making the protein catalytically inactive. These were introduced individually or in combination into Nicotiana leaves by agroinfiltration. We then assessed mature miR393b levels by Northern blot and found that the pri-miRNA393b transgene produces ∼2.3 times more miR393b when it is coexpressed with MTA, while this effect was abolished to a large extent when MTA was replaced by its catalytically inactive version (ΔMTA) (Fig. 7C). Thus, we show that the reduced auxin response in mta mutant plants is partially caused by the regulatory defects of miR393b biogenesis due to the lack of MTA activity.
Discussion
Here, we provide evidence that, in addition to influencing mRNA metabolism, m6A methylation is present in pri-miRNAs and affects miRNA biogenesis in A. thaliana. We show that the lower level of MTA (and hence m6A) leads to a reduction of miRNA levels in the case of at least 25% of miRNAs, whereas pri-miRNAs tend to overaccumulate in the mutant plants. This anticorrelation of pri-miRNAs with miRNA levels is reminiscent of observations from the Arabidopsis lines carrying mutations in genes encoding proteins involved in early stages of miRNA biogenesis: For example, hyl1 (42–44) and se (45). In contrast, mutants of HEN1 [a protein that is involved in methylation of miRNA/miRNA* duplexes at the 3′ ends (53), thus acting at the later step of miRNA biogenesis] do not show accumulation of pri-miRNAs, while down-regulation of miRNAs is observed in hen1 mutant plants (54). This suggests that MTA regulates miRNA production at early stages of their biogenesis.
Since MTA is a known mRNA methyltransferase and m6A is abundant in mRNA, we considered the possibility that the observed effects were indirect and can be a result of altered metabolism of other miRNA biogenesis proteins. We used two different and independent approaches to exclude this possibility. MTA–GFP-mediated RNA IP and m6A-IP experiments revealed that for at least a group of miRNAs, this effect is direct and is a result of MTA binding to pri-miRNAs and introducing m6A marks. Additionally, structural changes in the pri-miRNAs characterized by the loss of secondary structure and increase in single-stranded regions in the mta mutant background, specifically in the stem–loop regions of these miRNA precursors, also indicate the importance of m6A in miRNA biogenesis in plants. These structural analyses go in line with less efficient binding of HYL1, a double-stranded RNA binding protein, to unmethylated pri-miRNAs in the mta mutant.
Our studies also revealed that MTA can directly interact with TGH, a protein involved in early stages of miRNA biogenesis. As many other miRNA biogenesis mutants, tgh shows a decrease in accumulation of miRNAs and overaccumulation of many pri-miRNAs. Moreover, TGH has been described as a factor important for the recruitment of HYL1 to pri-miRNAs (46). Thus, MTA can influence miRNA biogenesis in two different ways: 1) by m6A methylation of miRNA precursors, and/or 2) by direct interactions with TGH. We show that the m6A marks introduced by MTA stabilize the secondary structure of the stem loop regions of pri-miRNAs, likely stimulating their recognition by HYL1, and afterward by the endonuclease DCL1 as well as the rest of the components of the Microprocessor complex. However, MTA may also stimulate Microprocessor assembly by its direct interactions with TGH, which in turn interacts with HYL1 and stimulates/stabilizes HYL1 binding to double-stranded regions of pri-miRNAs. Upon comparison of our results to the previously published data regarding miRNA levels in the tgh mutant, we found a 45% overlap within down-regulated miRNAs in both tgh and mta mutants, despite these two datasets originating from two different tissues (46) (SI Appendix, Fig. S11). These 23 miRNAs that are down-regulated in both mta and tgh mutants could be regulated by the direct MTA–TGH interactions. However, the mechanism of this MTA–TGH interaction-related alteration of miRNA biogenesis requires further studies.
Here, we also provide evidence that MTA interacts with RNA Pol II phosphorylated at both Serine 5 and Serine 2. This shows that MTA is associated with the RNA Pol II from the initiation of transcription and is present during the elongation step, modifying growing transcripts (see ref. 55 for review). In both mta and tgh mutants, colocalization of HYL1 and DCL1 with the elongation form of RNA Pol II (phosphorylated at Serine 2) is impaired, suggesting that both MTA/m6A as well as TGH contribute to cotranscriptional assembly of the Microprocessor.
In addition to the miRNAs that show decreased levels in the absence of m6A methylation, we also noticed nine miRNA species whose levels were up-regulated in mta hypomorphic plants. Interestingly, four precursors of these miRNAs adopt more structured conformations in low methylation plants (mta mutant) than in plants with the regular level of MTA (WT) (SI Appendix, Fig. S12). This may suggest that in some rare cases the absence of MTA or m6A methylation can have a positive effect on the proper conformation of pri-miRNAs. However, none of these precursors were shown to be m6A methylated, as demonstrated by our m6A-IP Seq. Moreover, they were also not found in any of the published lists of m6A-methylated Arabidopsis transcripts (19, 33). Thus, it is unlikely that the increased levels of the nine miRNAs in the mta mutant is connected to their pri-miRNA’s methylation or recognition by MTA, but rather it is an indirect effect of the lack of MTA activity, most probably via 1) pri-miRNA alternative splicing or polyA site selection or 2) up-regulation of transcription factors (either directly regulated by m6A mRNA methylation or regulated by miRNAs that are down-regulated in the mta mutant) stimulating their transcription. To answer which of these mechanisms are involved in the up-regulation of some miRNAs in mta plants, additional studies are required.
We also noticed that 29 pri-miRNAs were less abundant in mta in comparison to WT plants. Of these 29 pri-miRNAs, only 1 (pri-miR472) has been shown to be recognized and bound by MTA, and is m6A-modified. The additional 28 pri-miRNAs are most likely not m6A-methylated, so their lower accumulation cannot be directly explained by the presence of m6A marks, but rather by indirect effects. We show that HYL1 binds to pri-miR472 independently whether it is methylated (WT) or not (mta plants), and the level of mature miR472 in the mta mutant is similar to that observed in WT plants. This may suggest that the lack of MTA does not influence Microprocessor assembly on miR472 precursors. However, loss of MTA, hence m6A methylation of pri-miR472, affects the level of pri-miR472, which is decreased in low m6A methylation plants. It is worth noting that the Arabidopsis MIR gene encoding miR472 is very long (∼5 kbp) and contains four exons (31). Its transcription ends at several alternative polyadenylation sites and the transcripts undergo alternative splicing. Thus, it is also possible that m6A methylation of pri-miR472 changes maturation of MIR472 primary transcripts toward variants that are efficiently processed into mature miR472. This would explain the lower level of pri-miR472 and the constant level of miR472 in mta plants in comparison to WT. However, additional studies are needed to verify this interesting scenario.
As already mentioned above, while analyzing effects of m6A methylation on miRNA biogenesis, one should be aware that miRNA biogenesis is affected by a variety of processes like splicing, alternative polyadenylation, degradation, and so forth (56–60). The changes in structure caused by m6A can influence all of these processes by altering binding of proteins to MIR primary transcripts. The possible role of an m6A reader protein in the processing of pri-miRNAs should also not be overlooked. The overall expanse of m6A presence and the magnitude of its possible effects on miRNA biogenesis needs to be investigated and this work opens a new fascinating avenue for such future studies.
Interestingly, the reduced auxin responsiveness of m6A writer mutant plants can be partially explained by the altered miR393b level. We show that pri-miR393b is m6A-methylated by MTA, and the level of miR393b is lower in the mta mutant in comparison to WT plants. miR393b is involved in homeostasis of AUX/IAA genes the expression of which is regulated by a complex feedback loop that involves regulation of these genes by the proteins they encode (51, 61, 62). miR393b has been shown to accumulate in leaves and is induced in response to auxin (52). We have found that expression of the auxin responsive DR5pro:GUS reporter is reduced in miR393b mutant plants (51) and a similar reduction is seen with the same reporter in the mta background. Thus, the altered miR393b biogenesis in the plants with the low level of m6A methylation of pri-miRNAs likely contributes to the reduced auxin response in the m6A writer mutant, linking this modification with miRNA biogenesis and, as a physiological effect, with one of the most crucial hormone response pathways.
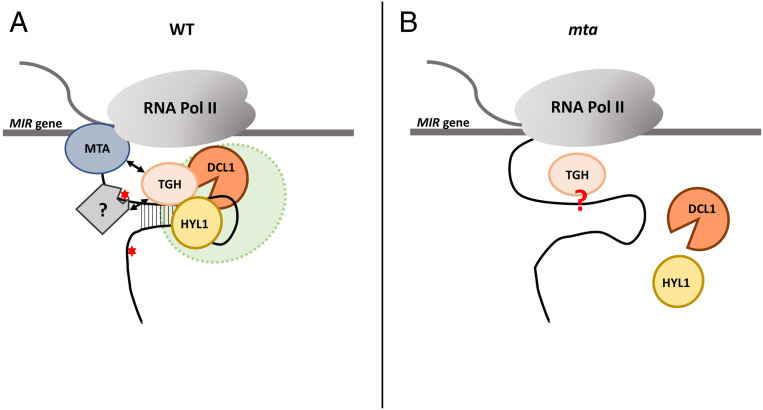
Based on our results, we propose a model showing the involvement of MTA/m6A in miRNA biogenesis in plants. m6A methylation of pri-miRNAs influences miRNA precursor secondary structure that in consequence stimulates the recognition of pri-miRNAs by HYL1. The direct interactions between MTA and the TGH protein and recognition of m6A marks by an unknown m6A reader that may also interact with TGH may also be involved in the HYL1 recruitment to the methylated miRNA precursor. This is strongly supported by the previously described involvement of TGH in the HYL1 recruitment to pri-miRNAs (46). Thus, the recognition of pri-miRNAs by HYL1, and in consequence assembly of the whole active Microprocessor, is controlled by pri-mRNA m6A methylation, which induces the proper secondary structure of the precursor as well as a specific MTA–TGH interaction that both support efficient assembly of the plant miRNA biogenesis machinery (Fig. 8).
Fig. 8.
A schematic representation of the role of m6A/MTA in miRNA biogenesis in plants. (A) In WT plants (Left), MTA methylates at least a set of pri-miRNAs which in turn leads to folding the proper secondary structure of a miRNA precursor. MTA and/or a putative reader of the m6A marks introduced by this enzyme (a gray square marked with “?”) facilitate the binding of TGH to the pre-miRNA. The properly folded miRNA precursor is recognized by HYL1. This eventually leads to efficient Microprocessor assembly and miRNA biogenesis at the appropriate level. (B) In the mta mutant, the lack of MTA, and thus m6A, leads to the loss of MTA–TGH interactions and inefficient formation of a stem–loop structure in the miRNA precursor, leading to reduced binding of HYL1 that in consequence disrupts Microprocessor assembly, and finally the down-regulation of the mature miRNA level is observed. Whether TGH can bind to the precursor in the mta mutant is unknown (marked in the scheme by a red question mark).
Materials and Methods
Plant Material.
A. thaliana WT Col-0, mta mutants (26), MTA-GFP (p35S:MTA-GFP), and GFP plants were sown on Jiffy pots and stratified for 2 d in dark at 4 °C. Thereafter, the plants were grown in plant growth chambers at 22 °C with 16-h light and 8-h dark cycles (70% humidity, 150- to 200-µmol m−2 s−1 photon flux density). Rosette leaves from 4-wk-old plants were harvested, immediately flash-frozen in liquid nitrogen, and used or stored at −80 °C until further use.
sRNA Sequencing and Analysis.
sRNA fraction from 4-wk-old A. thaliana WT Col-0 and mta mutant plants was used to prepare libraries that were sequenced at Fasteris on the HiSeq 4000 platform. A detailed description of the library preparation and data analysis can be found in SI Appendix, Supplementary Materials and Methods.
m6A-RNA IP of pri-miRNAs and Sequencing.
PolyA enriched RNA from 4-wk-old A. thaliana WT Col-0 and mta mutant plants was used to perform m6A IP. Libraries were prepared from RNA before IP (input) and after (IP). A detailed description of the library preparation and data analysis can be found in SI Appendix, Supplementary Materials and Methods.
Data Submission and Availability.
The data obtained in this study (sRNA and m6A-IP Seq) has been deposited under the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) accession no. GSE122528. The PIP-seq data used for the secondary structure analysis of miRNA transcripts can be found under NCBI GEO accession no. GSE108852.
RIP.
Transgenic Arabidopsis line p35S:MTA-GFP and mta mutants were grown along with the GFP and WT Arabidopsis lines. RIP was performed as described by Raczynska et al. (63). A detailed protocol for RIP and data analysis can be found in SI Appendix, Supplementary Materials and Methods.
PIP-Seq Analysis to Access RNA Structure Scores of miRNA Processors.
A total of two biological replicates of PIP-seq, including single-stranded RNA-seq and double-stranded RNA-seq libraries, for leaves five to nine from 4-wk-old WT and mta mutant plants were constructed as previously described (37, 39). The raw data has been deposited under the GEO accession no. GSE108852. Refer to SI Appendix, Supplementary Materials and Methods for the detailed protocol and data analysis pipeline.
Immunolocalization.
The immunolabeling experiments using the Duolink PLA fluorescence protocol were performed on isolated nuclei of 4-wk-old A. thaliana leaves (WT, plants with MTA-GFP expression or GFP expression, mta and tgh mutants). Before isolation, the leaves were fixed in 4% paraformaldehyde in PBS, pH 7.2, for 1 h, washed three times in PBS at room temperature, and then the nuclei isolation protocol according to the method of Pontvianne et al. (64) was followed.
Double Immunodetection of RNA Pol II with MTA, DCL1, and HYL1.
A detailed description of double-immunodetection protocol and the accompanying statistical analysis can be found in SI Appendix, Supplementary Materials and Methods.
PLA.
PLA was performed on isolated nuclei from leaves of MTA-GFP and GFP (control) lines. In situ PLA detection was carried out using the appropriate Duolink in situ Orange Kit Goat/Rabbit (Sigma-Aldrich) according to the protocol of the manufacturer. A detailed protocol for PLA can be found in SI Appendix, Supplementary Materials and Methods.
FRET–FLIM.
FRET–FLIM was performed using protoplasts were isolated from 3- to 4-wk-old WT Arabidopsis leaves using the method described in Knop et al. (65). For the detailed protocol, please refer to SI Appendix, Supplementary Materials and Methods.
GUS Staining for Auxin Response Analyses.
Fourteen-day-old seedlings of WT and mta mutants with the DR5pro:GUS reporter gene construct were incubated in 1/2 MS medium containing 10 µM 2,4-D in ethanol or in ethanol alone as a control for 8 h. After GUS staining the pictures were taken using the Leica M60 stereo microscope.
pri-miR393b, MTA, and ΔMTA Constructs.
pri-miR393b (sequence from mirEX2, www.combio.pl/mirex2) and MTA cDNA (AT4G10760) were amplified and cloned into pENTR/D-TOPO vectors (Thermo Fischer Scientific). ΔMTA was prepared by a primer-induced point mutation resulting in the following change: Aspartic acid at position 482 was changed to alanine (D482A) at the catalytic DPPW motif. For transient expression in N. benthamiana, these sequences were then cloned into pMDC32 using the Gateway (Thermo Fisher Scientific) cloning system.
N. benthamiana Transient Expression.
pri-miR393b, MTA, and ΔMTA constructs were transformed into Agrobacterium tumefaciens (AGL1) using electroporation. After verification of constructs in Agrobacterium by sequencing, tobacco leaves were transformed as described by Bielewicz et al. (56). Leaves were transformed either with pri-miR393b, MTA, and ΔMTA alone or in combinations (pri-miR393b + MTA, pri-miR393b + ΔMTA); leaves were harvested after 72 h and RNA isolation (as described above) was followed by Northern blotting.
Northern Blotting.
Northern blotting was performed as described in Kruszka et al. (66). Briefly: 30 µg of RNA (per sample) isolated from transfected tobacco leaves was loaded on 8 M denaturing urea polyacrylamide gel (15%) in TBE buffer (0.089 M Tris, 0.089 M boric acid, and 0.002 M EDTA, pH 8.0). RNA was then transferred onto the Amersham Hybond-NX nitrocellulose membrane (GE Healthcare) using a Trans-Blot Electrophoretic Transfer Cell (Bio-Rad) and fixed using CL-1000 UV Crosslinker (UVP). Prehybridization and hybridization were performed in hybridization buffer (3.5% SDS, 0.375 M sodium phosphate dibasic, 0.125 M sodium phosphate monobasic) at 42 °C with DNA oligo probes (Sigma) labeled at their 5′ ends with γ32P ATP (Hartmann Analytic). U6 was used as a loading control. After washing, the blots were exposed for up to 3 d to a phosphorimaging screen (Fujifilm) and the results were visualized with the Fujifilm FLA5100 reader (Fujifilm) and quantified using Multi Gauge V2.2 (Fujifilm).
Supplementary Material
Acknowledgments
This work was funded by the Polish National Science Centre (Grants UMO-2019/32/T/NZ1/00122, UMO-2017/27/N/NZ1/00202, UMO-2016/23/B/NZ9/00862, UMO-2016/23/D/NZ1/00152, UMO-2013/10/A/NZ1/00557). The authors also received financial support from the Initiative of Excellence–Research University (05/IDUB/2019/94) at Adam Mickiewicz University, Poznan, Poland, and from the Poznan RNA Research Centre. Work in the R.G.F.’s laboratory was supported by the Biotechnology and Biological Sciences Research Council (Grant BB/M008606/1). Work in B.D.G.’s laboratory was supported by the National Science Foundation (Grants MCB-1623887 and IOS-1444490).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2003733117/-/DCSupplemental.
Data Availability.
The data reported in this paper have been deposited in the NCBI GEO database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE122528 and GSE108852).
References
- 1.Yang Y., Hsu P. J., Chen Y. S., Yang Y. G., Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokar J. A., Shambaugh M. E., Polayes D., Matera A. G., Rottman F. M., Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247 (1997). [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Y., Ma P., Liu Y., Li W., Shu Y., Multiple functions of m6A RNA methylation in cancer. J. Hematol. Oncol. 11, 48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brocard M., Ruggieri A., Locker N., m6A RNA methylation, a new hallmark in virus-host interactions. J. Gen. Virol. 98, 2207–2214 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Tan B., Gao S.-J., RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N6 -methyladenosine (m6 A). Rev. Med. Virol. 28, e1983 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J. et al., A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ping X.-L. et al., Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz S. et al., Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8, 284–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patil D. P. et al., m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen J. et al., Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knuckles P. et al., Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu C. et al., Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Wang X. et al., N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du H. et al., YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H. et al., YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia G. et al., N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng G. et al., ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S. et al., MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen L. et al., N(6)-Methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev. Cell 38, 186–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Růžička K. et al., Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei L.-H. et al., The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30, 968–985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arribas-Hernández L. et al., An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30, 952–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scutenaire J. et al., The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30, 986–1005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan H.C. et al., ALKBH10B is An RNA N6-methyladenosine demethylase affecting arabidopsis floral transition. Plant Cell 29, 2995–3011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez-Pérez M. et al., Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. U.S.A. 114, 10755–10760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bodi Z. et al., Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3, 48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., MicroRNA biogenesis and function in plants. FEBS Lett. 579, 5923–5931 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alarcón C. R., Lee H., Goodarzi H., Halberg N., Tavazoie S. F., N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcón C. R. et al., HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zielezinski A. et al., mirEX 2.0—An integrated environment for expression profiling of plant microRNAs. BMC Plant Biol. 15, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielewicz D. et al., mirEX: A platform for comparative exploration of plant pri-miRNA expression data. Nucleic Acids Res. 40, D191–D197 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominissini D. et al., Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Anderson S. J. et al., N6-Methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep. 25, 1146–1157.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Szarzynska B. et al., Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37, 3083–3093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu N. et al., N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roost C. et al., Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu N. et al., N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F. et al., Global analysis of RNA secondary structure in two metazoans. Cell Rep. 1, 69–82 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Gosai S. J. et al., Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol. Cell 57, 376–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson S. J., Willmann M. R., Gregory B. D., Protein interaction profile sequencing (PIP-seq) in plants. Curr. Protoc. Plant Biol. 1, 163–183 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Vazquez F., Gasciolli V., Crété P., Vaucheret H., The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Han M. H., Goud S., Song L., Fedoroff N., The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 1093–1098 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurihara Y., Takashi Y., Watanabe Y., The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song L., Han M.-H., Lesicka J., Fedoroff N., Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. U.S.A. 104, 5437–5442 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L., Liu Z., Lu F., Dong A., Huang H., SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Ren G. et al., Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 12817–12821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke S. et al., m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molinie B. et al., m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat. Methods 13, 692–698 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kasowitz S. D. et al., Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14, e1007412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slobodin B. et al., Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169, 326–337.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Windels D. et al., miR393 is required for production of proper auxin signalling outputs. PLoS One 9, e95972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang P., Doxtader K. A., Nam Y., Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Z., Ebright Y. W., Yu B., Chen X., HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park W., Li J., Song R., Messing J., Chen X., CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nechaev S., Adelman K., Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta 1809, 34–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bielewicz D. et al., Introns of plant pri-miRNAs enhance miRNA biogenesis. EMBO Rep. 14, 622–628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwab R., Speth C., Laubinger S., Voinnet O., Enhanced microRNA accumulation through stemloop-adjacent introns. EMBO Rep. 14, 615–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dolata J. et al., Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol. 172, 297–312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z. et al., SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Bajczyk M. et al., SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Res. 48, 6839–6854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z. H. et al., Regulation of auxin response by miR393-targeted transport inhibitor response protein 1 is involved in normal development in Arabidopsis. Plant Mol. Biol. 77, 619–629 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Si-Ammour A. et al., miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157, 683–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raczynska K. D. et al., The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 42, 1224–1244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pontvianne F. et al., Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Rep. 16, 1574–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knop K. et al., Active 5′ splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes. Nucleic Acids Res. 45, 2757–2775 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kruszka K. et al., Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics 14, 34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the NCBI GEO database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE122528 and GSE108852).