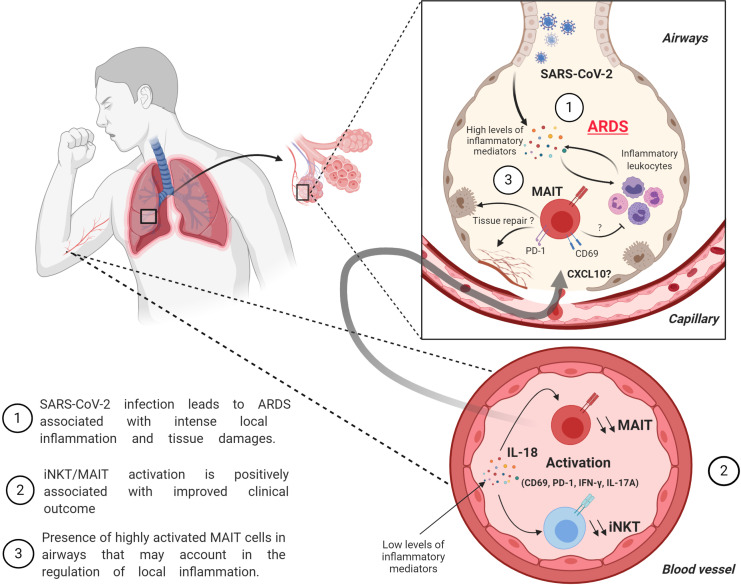
This study reveals profound alteration in biology of blood unconventional T cells (MAIT, iNKT, and γδT) in severe COVID-19. Highly activated MAIT cells are found in abundance in the airways of mechanically ventilated patients. Activation levels of MAIT/iNKT cells at admission positively correlate with clinical improvement.
Abstract
COVID-19 includes lung infection ranging from mild pneumonia to life-threatening acute respiratory distress syndrome (ARDS). Dysregulated host immune response in the lung is a key feature in ARDS pathophysiology. However, cellular actors involved in COVID-19–driven ARDS are poorly understood. Here, in blood and airways of severe COVID-19 patients, we serially analyzed unconventional T cells, a heterogeneous class of T lymphocytes (MAIT, γδT, and iNKT cells) with potent antimicrobial and regulatory functions. Circulating unconventional T cells of COVID-19 patients presented with a profound and persistent phenotypic alteration. In the airways, highly activated unconventional T cells were detected, suggesting a potential contribution in the regulation of local inflammation. Finally, expression of the CD69 activation marker on blood iNKT and MAIT cells of COVID-19 patients on admission was predictive of clinical course and disease severity. Thus, COVID-19 patients present with an altered unconventional T cell biology, and further investigations will be required to precisely assess their functions during SARS–CoV-2–driven ARDS.
Graphical Abstract
Introduction
In December 2019, in Wuhan, China, came the first reports of pneumonia cases due to a coronavirus, the severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2), a novel strain related to SARS-CoV and Middle East respiratory syndrome–CoV, responsible for previous outbreaks. Disease related to SARS-CoV-2 (i.e., coronavirus disease 2019 [COVID-19]) can vary from mild disease to life-threatening acute respiratory distress syndrome (ARDS). ARDS is caused by a sustained and dysregulated immune response triggered in the lung after an initial insult, resulting in alteration of alveolar–capillary membrane permeability and tissue repair (Thompson et al., 2017). This pathological process leads to interstitial and alveolar edema that strongly impairs gas exchange. The cellular and molecular factors that are responsible for this aberrant and persistent inflammatory response are poorly understood (Matthay et al., 2019).
During severe SARS-CoV-2 infection, elevated proinflammatory cytokine levels (e.g., IL-6 and TNF-α) were associated with more severe cases, supporting an inflammatory hypothesis (Chen et al., 2020; Mehta et al., 2020; Qin et al., 2020). In addition, T cell lymphopenia has been correlated with disease severity, suggesting a role for these cells in the pathophysiology of severe COVID-19 (Chen et al., 2020; Qin et al., 2020). Besides classic adaptive CD4+ and CD8+ T cells, the T cell compartment comprises several lineages of cells endowed with both innate and adaptive properties that are referred to as unconventional T (uT) cells (Godfrey et al., 2015). This heterogeneous class of T cells comprises three main lineages, including mucosa-associated invariant T (MAIT), γδT, and invariant natural killer T (iNKT) cells. These cells recognize nonpeptide antigens, are not restricted to classic MHC, and have emerged as key players in mucosal immunity and inflammatory response (Crosby and Kronenberg, 2018; McCarthy and Eberl, 2018; Trottein and Paget, 2018; Toubal et al., 2019). Given their versatile functions, uT cells could be important actors in the context of SARS-CoV-2–driven ARDS. First, uT cells mainly populate mucosal tissues, including the lung, and have the ability to promptly produce substantial amounts of inflammatory cytokines such as IFN-γ and IL-17A, two key cytokines in the antimicrobial response at barrier sites. Moreover, uT cells can fine-tune the intensity and quality of the host immune response, shaping the magnitude of the adaptive response. Hence, they have been shown to contribute in anti-infective responses to viruses (Déchanet et al., 1999; Paget et al., 2011; Loh et al., 2016; van Wilgenburg et al., 2016) and bacteria (Bonneville, O’Brien and Born, 2010; Le Bourhis et al., 2010; Crosby and Kronenberg, 2016), especially during pneumonia (Trottein and Paget, 2018). They can also participate in the process of the resolution of inflammation, including tissue repair and regeneration (Nielsen, Witherden and Havran, 2017; Hinks et al., 2019; Lamichhane et al., 2019; Leng et al., 2019; Paget and Trottein, 2019), a critical step that is impaired during ARDS. Despite that, the contribution of uT cells in the pathophysiological process of SARS-CoV-2–driven ARDS has never been explored.
Here, we dynamically assessed the relative frequencies and functions of uT cells in biological fluids of 30 patients with severe COVID-19 who were admitted to the intensive care unit (ICU). Our analysis indicates that uT cells from severe COVID-19 patients display a phenotype of activated cells associated with changes in their cytokine profile. Importantly, activated uT cells populated the airways of patients displaying strong local inflammation. In addition, the activation status of blood uT cells on admission was predictive of the level of hypoxemia during the course of infection. Thus, we show here that severe COVID-19 influences the phenotype and function of uT cells. This should encourage further investigation to assess the precise functions of uT cells and their associated activation mechanisms of uT cells during SARS-CoV-2–driven ARDS.
Results and discussion
Lymphopenia and compartmentalized lung inflammation are hallmarks of severe COVID-19 patients
30 patients admitted to the ICU for severe COVID-19 were included. Baseline characteristics of the patients are presented in Table 1. Median duration of symptoms before admission in ICU was 10 d, and ultimately, 24 patients (80%) required invasive mechanical ventilation (20 on admission). Among these mechanically ventilated patients, all presented with ARDS, 21 (70%) received neuromuscular blockade, 18 (60%) were placed in prone position, and 1 required extracorporeal membrane oxygenation. To better assess COVID-19 specificity in our study, two comparison groups were studied: 20 healthy volunteers (age- and sex-matched) and 17 critically ill patients with neither shock nor pneumonia, requiring invasive mechanical ventilation (referred to as “non–COVID-19 group”). Patients from the non–COVID-19 group (Table 1) were admitted to the ICU for cardiac arrest (n = 3), stroke or intracerebral hemorrhage (n = 9), or severe neuromuscular disease (n = 5).
Table 1. Clinical and laboratory information of the study groups at admission.
| Characteristic | COVID-19 (n = 30) | Non–COVID-19 (n = 17) |
|---|---|---|
| Age (yr) | 64 (57; 67) | 64 (56; 71) |
| Male/female | 2/1 | 1.1/1 |
| BMI (kg/m2) | 31 (28; 32) | 30 (24; 32) |
| SAPS2 | 32 (22; 36) | 47 (37; 55) |
| SOFA | 4 (2; 6) | 6 (4; 9) |
| Invasive mechanical ventilation at inclusion | 20 (66.7) | 17 (100) |
| Comorbidities | ||
| Hypertension | 12 (46.7) | 13 (76.5) |
| Type 2 diabetes | 9 (30) | 3 (17.6) |
| Active smoking | 0 | 5 (29.4) |
| Chronic respiratory failure | 0 | 0 |
| Chronic renal failure | 1 (3.3) | 0 |
| Cardiovascular disease | 3 (10) | 2 (11.8) |
| Immunosuppression | 1 (3.3) | 0 |
| Laboratory data | ||
| Lymphocytes (× 109/liter) | 0.715 (0.510; 0.998) | 1.16 (0.9; 1.3) |
| Neutrophils (× 109/liter) | 6.25 (4.3; 7.52) | 10 (7.8; 12.9) |
| C-reactive protein (mg/liter) | 187.7 (83.8; 258) | 123 (59.8; 178) |
Data are median (quartile 1; quartile 3) or n (%) unless noted otherwise.
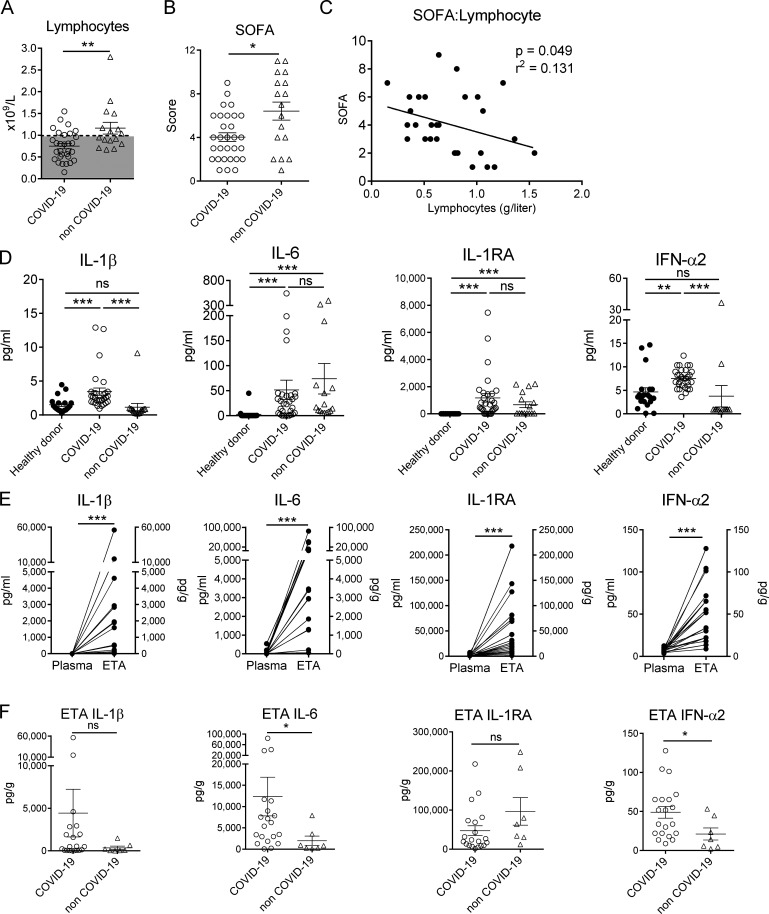
On admission, COVID-19 patients presented with more pronounced lymphopenia (0.75 × 109/liter ± 0.06) compared with non–COVID-19 patients (1.16 × 109/liter ± 0.13; Fig. 1 A) although the non–COVID-19 group displayed a higher sequential organ failure assessment (SOFA), indicating higher clinical severity (Fig. 1 B). As reported (Zhou et al., 2020), we observed a positive correlation between the degree of lymphopenia and SOFA score in severe COVID-19 patients (Fig. 1 C). Among the 30 COVID-19 patients, 1 died at day 2 after inclusion, and 14 (46.7%) were still in the ICU at day 15, including 9 still under invasive mechanical ventilation. The remaining 15 patients improved and were discharged to ward.
Figure 1.
Inflammatory status of study patients. (A) Lymphocyte count in whole blood from severe COVID-19 (n = 30) and non–COVID-19 patients (n = 16; the lymphocyte count of one patient was not determined) on day 1 after admission. Individuals and means ± SEM are depicted. Dashed line represents the threshold for lymphopenia. Mann–Whitney U test. (B) SOFA score of COVID-19 (n = 30) and non–COVID-19 (n = 17) patients. Individuals and means ± SEM are shown. Mann–Whitney U test. (C) Spearman’s rank correlation of SOFA and lymphocyte counts in COVID-19 patients. (D) Levels of IL-1β, IL-6, IL-1RA, and IFN-α2 in the plasma of healthy donors (n = 20) and COVID-19 (n = 30) and non–COVID-19 (n = 16) patients. Individuals and means ± SEM are shown. Kruskal–Wallis test with Dunn's multiple comparisons test. (E) Levels of IL-1β, IL-6, IL-1RA, and IFN-α2 in the plasma and ETA supernatants of matched COVID-19 patients (n = 20). Paired individual values are shown. Wilcoxon matched-pairs signed rank test. (F) Comparison of airway levels of IL-1β, IL-6, IL-1RA, and IFN-α2 in ETA supernatants of COVID-19 (n = 20) and non–COVID-19 (n = 7) patients. Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Although the circulating levels of the inflammatory mediators IL-1β and IL-6 were significantly higher in COVID-19 patients compared with control individuals (Fig. 1 D), the detected amounts were relatively low, as previously reported (Chen et al., 2020). The levels of plasma IL-1 receptor antagonist (IL-1RA) were high, suggesting an active anti-inflammatory process (Fig. 1 D). As judged by plasma levels of IFN-α2 in COVID-19 patients, the type I IFN response, a critical component of the antiviral response (Katze et al., 2002) was higher than in controls although the amount detected was fairly low (Fig. 1 D). To monitor the local inflammation, we analyzed the same mediators in the supernatants of endotracheal aspirates (ETAs) of matched COVID-19 patients. Strikingly, the amounts of IL-6, IL-1β, IL-1RA, and to a lesser extent IFN-α2 were much higher in the airways, suggesting intense local inflammation (Fig. 1 E). These inflammatory mediators were also detected in ETA supernatants of non–COVID-19 patients, albeit at lower levels (Fig. 1 F). Collectively, these data indicate compartmentalized and strong inflammation in the lungs of patients with severe COVID-19.
Decrease in blood uT cells in COVID-19 patients parallels their presence in airways
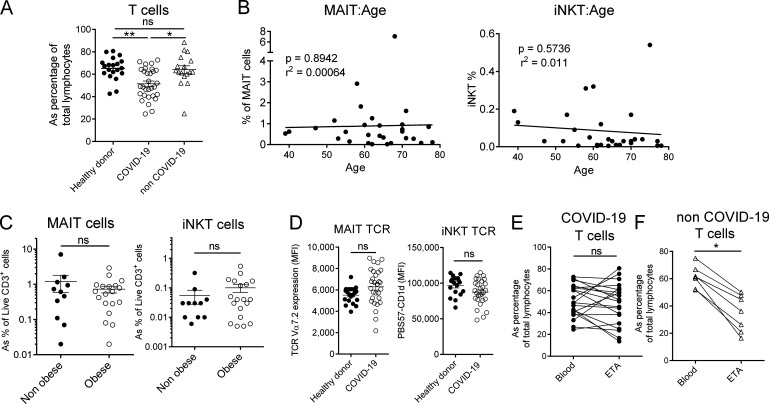
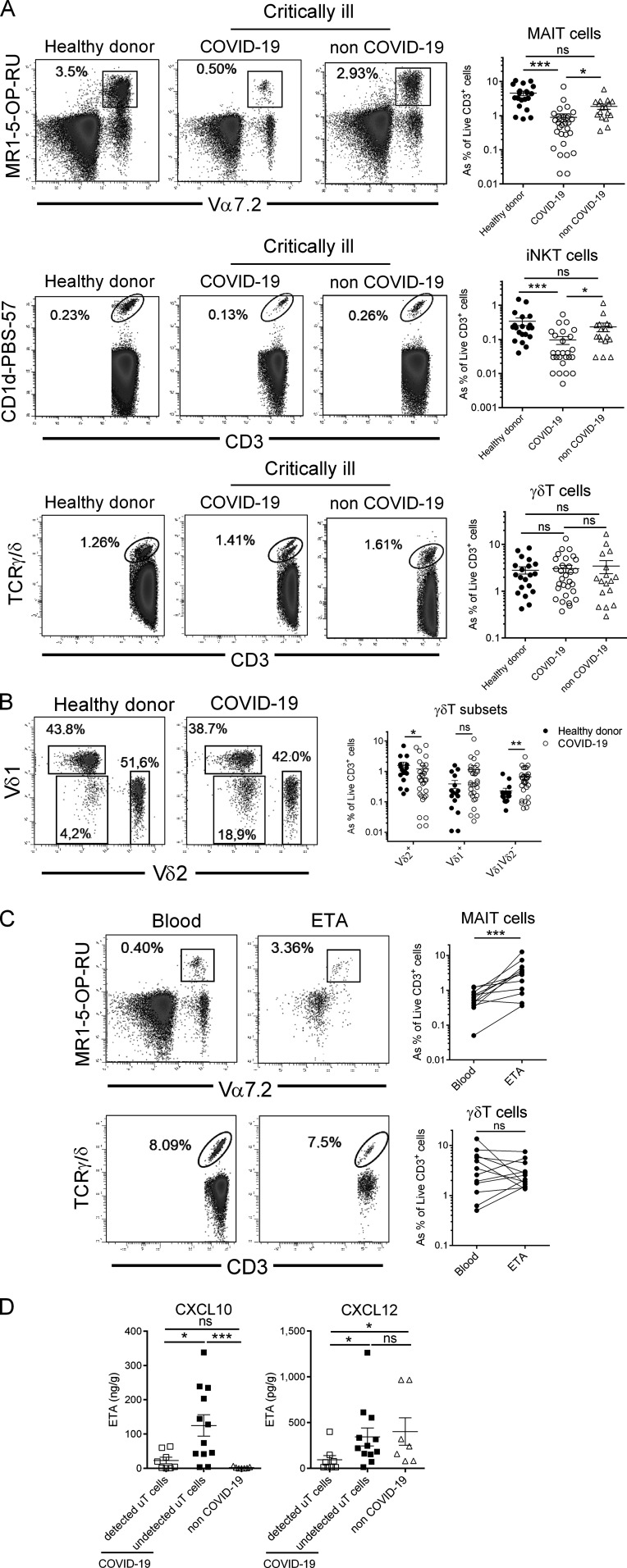
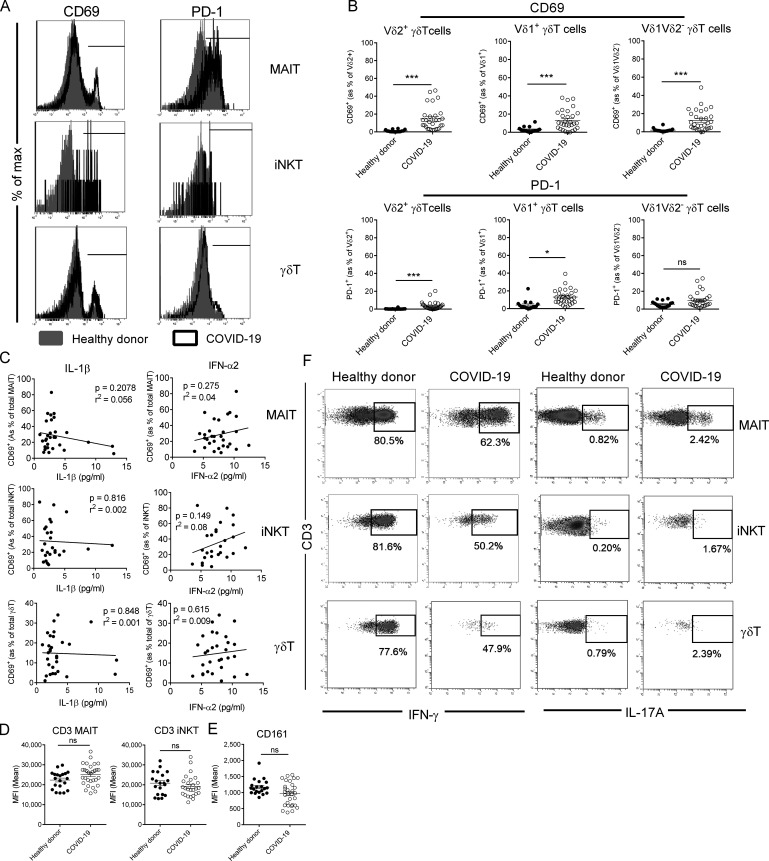
In line with a recent report (Chen et al., 2020), we observed T cell lymphopenia in severe COVID-19 patients compared with control individuals (Fig. S1 A). A detailed analysis of circulating uT cells indicated a decrease in MAIT and iNKT but not γδT cell relative proportions in severe COVID-19 patients (Fig. 2 A). To address potential confounding factors, MAIT and iNKT proportions were analyzed with respect to age and obesity status (Lynch et al., 2009; Magalhaes et al., 2015). No changes or correlations were noticed in relation to obesity (body mass index [BMI] ≥30) or age (Fig. S1, B and C). By focusing on γδT subsets, we observed distinct changes with a slight decrease for Vδ2+, moderate increase for Vδ1δ2−, and no change for the Vδ1+ subset (Fig. 2 B).
Figure S1.
Frequency, confounding effects, and TCR levels of T cell subsets in COVID-19 patients. (A) Frequency of circulating conventional T cells (within the lymphocyte gate) in healthy controls (n = 20) and COVID-19 (n = 30) and non–COVID-19 (n = 17) patients. Individuals and means ± SEM are shown. Kruskal–Wallis test with Dunn's multiple comparisons test. (B) Spearman’s rank correlation of age of COVID-19 patients and the frequency of blood MAIT (left panel) or iNKT (right panel) cells (n = 30). (C) Comparative analyses of MAIT and iNKT frequencies regarding the obese status (BMI > 30) of patients. Individuals and means ± SEM are shown. Mann–Whitney U test. (D) Mean intensity fluorescence (MFI) of TCR expression on MAIT and iNKT cells from controls (n = 20) or COVID-19 patients (n = 30) based on TCR Vα7.2 and PBS57-CD1d tetramer staining, respectively. Individuals and means ± SEM are shown. Mann–Whitney U test. (E and F) Comparative analysis of T cells in blood and ETA of COVID-19 (n = 20; E) or non–COVID-19 (n = 7; F) patients. Individuals and means ± SEM are shown. Wilcoxon matched-pairs signed rank test. ns, not significant; *, P < 0.05; **, P < 0.01.
Figure 2.
Relative proportion of uT cells in PBMCs and ETA of COVID-19 patients. (A) Flow cytometry analyses of uT cells in the blood of healthy donors (n = 20) and COVID-19 (n = 30) and non–COVID-19 (n = 17) patients. Representative dot plots of MAIT, iNKT, and γδT cells of the three groups are shown in the left panel as percentage of CD3+ live cells. Individuals and means ± SEM are shown in the right panel. Of note, iNKT cells could not be detected in four COVID-19 patients and one non–COVID-19 patient. Kruskal–Wallis test followed by a Dunn's multiple comparisons test. (B) Proportion of γδT subsets in the blood of healthy donors (n = 20) and COVID-19 (n = 30) patients. Representative dot plots of γδT subsets are shown in the left panel. Individuals as percentage of CD3+ live cells and means ± SEM are shown in the right panel. Mann–Whitney U test. (C) Comparative analysis of MAIT and γδT cell subsets in blood and ETA of COVID-19 patients with detected uT cells in ETA. Representative dot plots are shown in the left panel. Individuals and means ± SEM are shown in the right panel. Wilcoxon matched-pairs signed rank test. (D) Levels of CXCL10 and CXCL12 in ETA supernatants of COVID-19 patients (n = 20), according to the presence (n = 12) or not (n = 8) of uT cells, and non–COVID-19 (n = 7) patients. Individuals and means ± SEM are shown. Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
The profound decrease in circulating MAIT and iNKT cells may have multiple causes, such as increased cell death, TCR down-modulation, or cell migration to the inflamed tissue. Mean fluorescence intensity for TCR expression indicated no modulation for MAIT and iNKT cells between COVID-19 patients and controls (Fig. S1 D), and thus does not support the hypothesis of an activation-induced TCR internalization. Another possibility is their active recruitment into the airways. To assess this hypothesis, uT cells were analyzed in ETA samples of intubated COVID-19 patients (n = 20; Table 1). While CD3+ cells were present in all samples (Fig. S1 E), MAIT and γδT cells were detected in ETA of only 12 patients (Fig. 2 C). In addition, airway iNKT cells were virtually undetectable. Importantly, the frequency of MAIT cells (but neither total T nor γδT cells) was higher in the airways compared with blood of matched patients (Fig. 2 C and Fig. S1 E). As a comparison, analysis of ETA samples from non–COVID-19 patients enabled detection of CD3+ cells (Fig. S1 F) but not uT cells. Of interest, the presence of uT cells in the airway fluids was associated with a higher level of T cell chemoattractants CXCL10 and CXCL12 (Mackay, 1996; Fig. 2 D). However, no change was observed for CXCL12 compared with non–COVID-19 patients, suggesting that CXCL10 could play a more specific role (Fig. 2 D).
Altogether, these data indicate a decrease in circulating MAIT and iNKT cells of COVID-19 patients. This is paralleled by a high frequency of MAIT cells in airways, which may be the result of non–mutually exclusive causes, such as their recruitment from the blood, the trafficking of lung-resident MAIT cells into the airways, or local expansion.
uT cells of severe COVID-19 patients display an altered functional profile
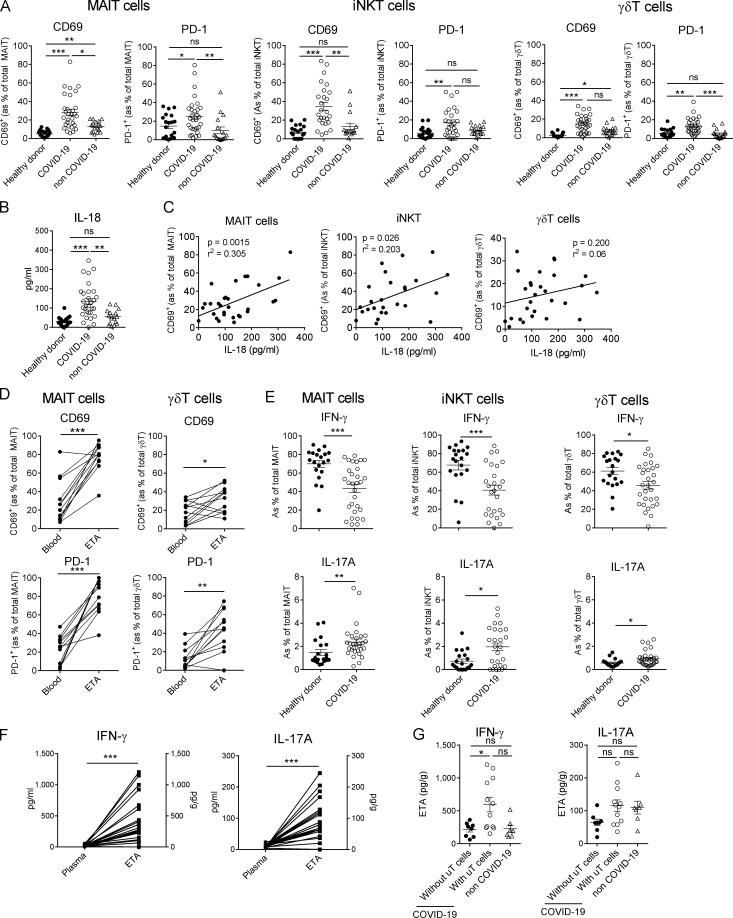
Phenotypic analysis of uT cells in blood of COVID-19 patients showed increased expression of the activation marker CD69 in all subsets compared with controls (Fig. 3 A and Fig. S2, A and B). Of note, levels of CD69 on γδT cells from COVID-19 patients were not significantly changed compared with non–COVID-19 critically ill controls, suggesting that this activation status could be a general reflection of a severe condition rather than a SARS-CoV-2 signature (Fig. 3 A). In parallel, we assessed the level of PD-1 on circulating uT cells, another activation marker routinely used to evaluate T cell exhaustion. The frequency of PD-1–expressing uT cells was increased in COVID-19 patients compared with controls (Fig. 3 A and Fig. S2, A and B).
Figure 3.
Functional analysis of uT cells during severe COVID-19. (A) Flow cytometry analyses of CD69 and PD-1 expression on MAIT, γδT, and iNKT cells in the blood of healthy donors (n = 20) and COVID-19 (n = 30) and non–COVID-19 (n = 17) patients. Individuals and means ± SEM are shown. Kruskal–Wallis test followed by a Dunn's multiple comparisons test. (B) Levels of IL-18 in the plasma of healthy donors (n = 20) and COVID-19 (n = 30) and non–COVID-19 (n = 17) patients. Individuals and means ± SEM are shown. Kruskal–Wallis test followed by a Dunn's multiple comparisons test. (C) Spearman’s rank correlation of CD69 expression on blood uT cells and levels of IL-18 in COVID-19 patients. (D) Relative proportions of CD69+ and PD-1+ MAIT and γδT cells in the blood and ETA of matched patients. Paired individual values are shown. Wilcoxon matched-pairs signed-rank test. (E) Intracellular staining for IFN-γ and IL-17A of PMA/ionomycin-activated PBMCs. Individuals and means ± SEM are shown. Mann–Whitney U test. (F) Levels of IFN-γ and IL-17A in the plasma and ETA supernatants of matched patients. Paired individual values are shown. Wilcoxon matched-pairs signed rank test. (G) Levels of IFN-γ and IL-17A in the ETA supernatants of COVID-19 patients, according to the presence (n = 12) or not (n = 8) of uT cells, and non–COVID-19 patients. Kruskal–Wallis test followed by a Dunn's multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
Figure S2.
Phenotype and cytokine production of blood uT cells in COVID-19 patients. (A) Representative histogram plots of CD69 and PD-1 expression on uT cell subsets in healthy controls and COVID-19 patients. (B) Flow cytometry analyses of CD69 (upper panel) and PD-1 (lower panel) expression on γδT subsets in the blood of healthy donors (n = 20) and COVID-19 patients (n = 30). Individuals and means ± SEM are shown. Mann–Whitney U test. (C) Spearman’s rank correlation of CD69 expression on blood uT cells and levels of IL-1β or IFN-α2 in COVID-19 patients. (D) Mean intensity fluorescence (MFI) of CD3 expression on MAIT and iNKT cells from healthy controls (n = 20) and COVID-19 patients (n = 30). Individuals and means ± SEM are shown. Mann–Whitney U test. (E) Mean intensity fluorescence of CD161 expression on MAIT cells from healthy controls (n = 20) and COVID-19 patients (n = 30). Individuals and means ± SEM are shown. Mann–Whitney U test. (F) Representative dot plots of intracellular staining for IFN-γ (left panel) and IL-17A (right panel) of blood iT cells from healthy donors and COVID-19 patients following PMA/ionomycin activation. *, P < 0.05; ***, P < 0.001. ns, not significant.
The plasma levels of IL-18, a cytokine associated with uT cell activation during viral infections (Tyznik et al., 2014; Tsai et al., 2015; Loh et al., 2016; van Wilgenburg et al., 2016; Constantinides et al., 2019), were higher in COVID-19 patients (Fig. 3 B) and positively correlated with the level of CD69 expression on MAIT and iNKT but not γδT cells (Fig. 3 C). No correlation was observed with type I IFN (IFN-α2) and IL-1β, which are established uT cell–activating cytokines (Sutton et al., 2009; Doisne et al., 2011; Constantinides et al., 2019; Fig. S2 C). This may suggest a role for IL-18 in MAIT and iNKT cell activation during SARS-CoV-2 infection.
In the meantime, no down-modulation of CD3 was observed on uT cells of the two groups (Fig. S2 D), which does not support the involvement of a TCR ligand. Although CD161 expression on MAIT cells has been suggested to be modulated during certain viral infections (Leeansyah et al., 2013; Ussher et al., 2014), this was not observed in severe COVID-19 patients (Fig. S2 E). Levels of CD69 and PD-1–expressing MAIT and γδT cells were significantly higher in ETA compared with blood of matched patients (Fig. 3 D). As previously suggested, this could mirror the migration of activated blood uT cells into the lungs, even if the local inflammatory environment could also account for this hyperactivated phenotype.
Regarding cytokine production, circulating MAIT and iNKT cells and, to a lesser extent, γδT cells from COVID-19 patients produced less IFN-γ compared with cells from healthy donors (Fig. 3 E and Fig. S2 F). This decrease in IFN-γ production could be due to overinflammation-induced uT cell hyporesponsiveness and/or the migration of IFN-γ–producing cells into the lung tissue. Conversely, we observed an increased ability of uT cells from patients to produce IL-17A (Fig. 3 E and Fig. S2 F), although the levels detected were low. In line with the acute inflammation in the lungs of COVID-19 patients (Hadjadj et al., 2020), the levels of IFN-γ and IL-17A were more elevated in the supernatants of ETA compared with plasma of matched patients (Fig. 3 F). Notably, IFN-γ but not IL-17A levels were detected at higher levels in uT cell–containing ETA samples (Fig. 3 G). Collectively, our data indicate a functional alteration of uT cells in COVID-19 patients.
Alteration of uT cell behavior is persistent and correlates with disease severity
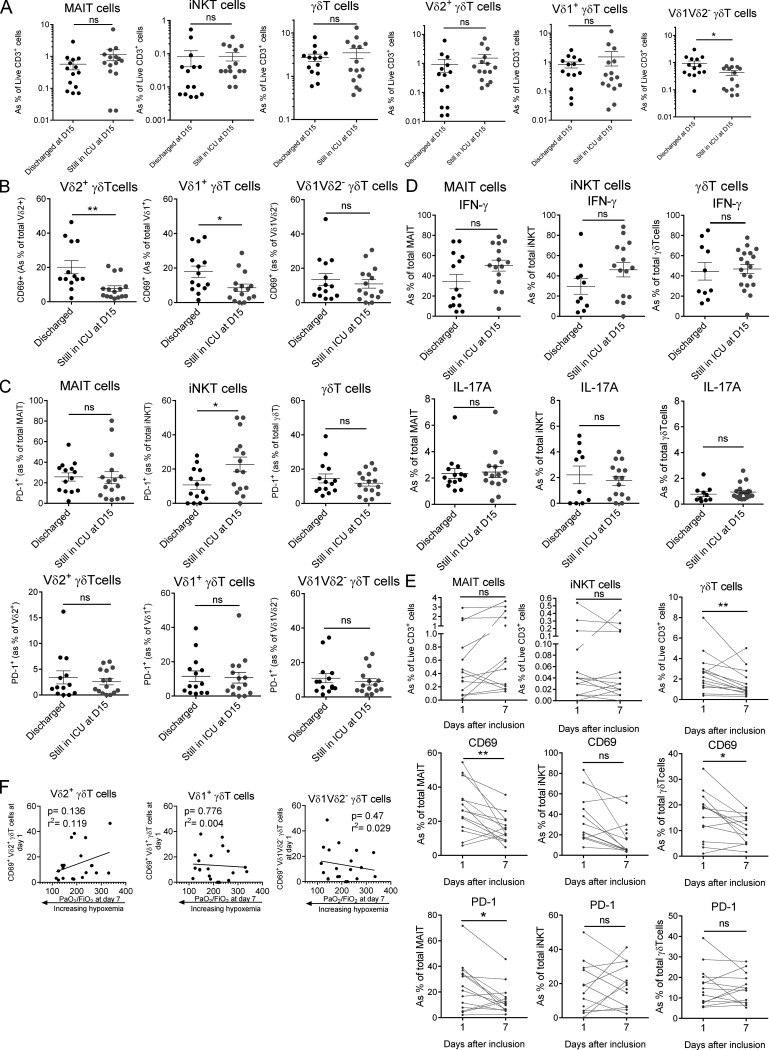
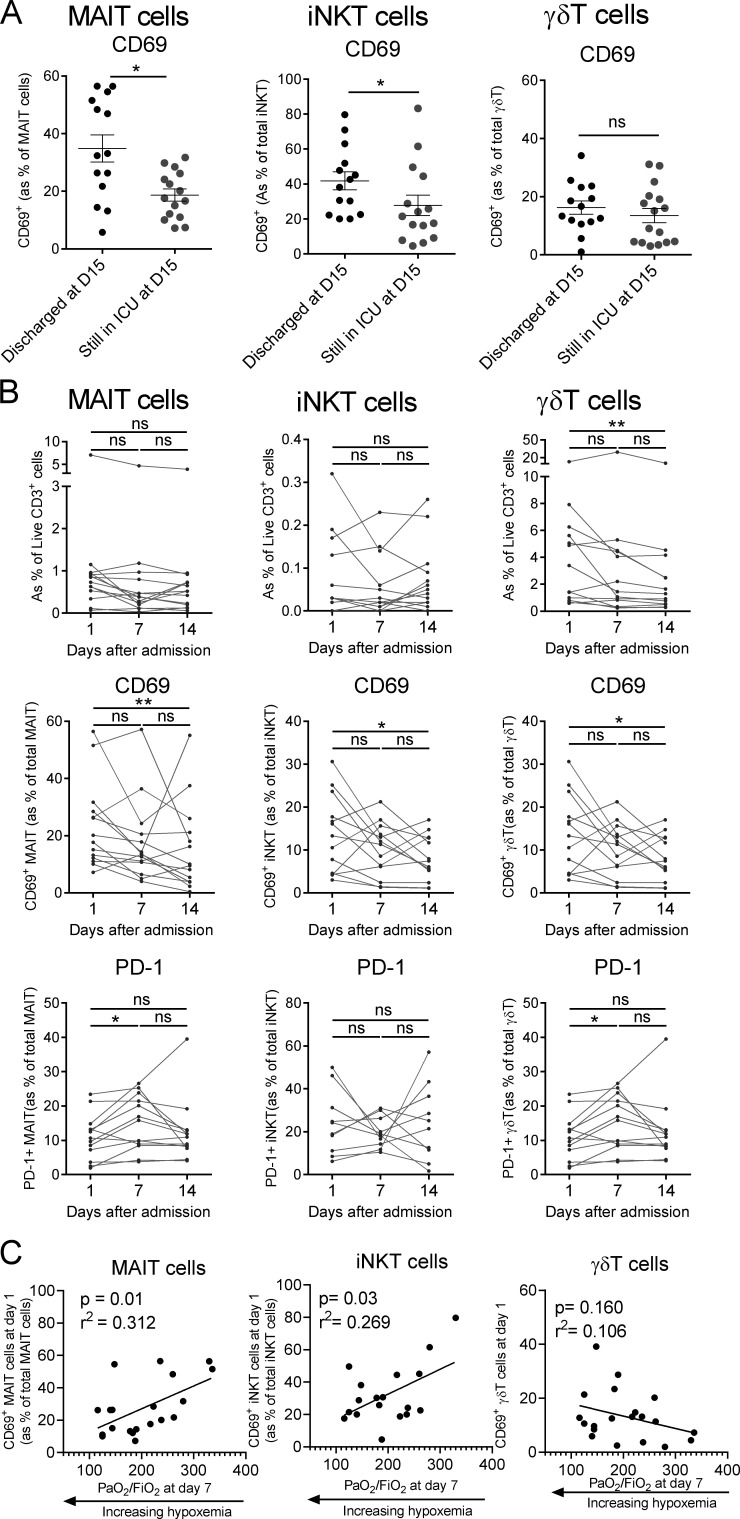
To gain insight into the potential links between uT cell biology and clinical evolution, we compared discharged patients (i.e., improved status) to those still in the ICU at day 15 (i.e., still in critical condition). Analysis of circulating uT cell frequency indicated no significant change between groups, with the exception of a slight decrease for the Vδ1Vδ2− γδT subset (Fig. S3 A). Interestingly, uT cell populations (except for Vδ1Vδ2− γδT) from discharged patients had a higher level of CD69 on admission (Fig. 4 A and Fig. S3 B). This was paralleled by a higher level of PD-1 on iNKT cells but not MAIT or γδT cells, from patients still in the ICU at day 15 (Fig. S3 C). No difference was noted regarding cytokine secretion (Fig. S3 D).
Figure S3.
Analysis of uT cell biological parameters regarding the clinical evolution of COVID-19 patients. (A) Comparative analyses of the frequencies of blood MAIT, iNKT, and γδT subsets in COVID-19 patients who were discharged (n = 14) or not discharged (n = 15) from ICU at day 14. Individuals and means ± SEM are shown. Mann–Whitney U test. (B) Comparative analyses of the CD69 expression of blood γδT subsets in COVID-19 patients who were discharged (n = 14) or not discharged (n = 15) from ICU at day 14. Individuals and means ± SEM are shown. Mann–Whitney U test. (C) Comparative analyses of PD-1 expression on blood MAIT, iNKT, and γδT subsets in COVID-19 patients who were discharged (n = 14) or not discharged (n = 15) from ICU at day 14. Individuals and means ± SEM are shown. Mann–Whitney U test. (D) Comparative analysis of IFN-γ and IL-17A production by uT cells in COVID-19 patients who were discharged (n = 14) or not discharged (n = 15) from ICU at day 14. Individuals and means ± SEM are shown. Mann–Whitney U test. (E) Kinetics plots showing individual values of uT cell frequency and phenotype for each rapidly discharged (before day 15) patients (n = 14) at days 1 and 7 after admission. Wilcoxon matched-pairs signed rank test. (F) Spearman’s rank correlation of CD69 expression on blood γδT subsets on admission and hypoxemia levels at day 7 in COVID-19 patients (n = 20). *, P < 0.05; **, P < 0.01. ns, not significant.
Figure 4.
Predictive role and kinetics analysis of uT cell parameters in the clinical course of COVID-19. (A) Comparative analyses of CD69 expression on blood MAIT, γδT, and iNKT cells in COVID-19 patients who were discharged (n = 14) or not discharged (n = 15) from ICU at day 14. Individuals and means ± SEM are shown. Mann–Whitney U test. (B) Kinetics plots showing individual values of uT cell frequency and phenotype at days 1, 7, and 14 after admission for each patient still in critical condition at day 14 (n = 13). One-way ANOVA (Friedman test) with Dunn’s multiple comparison. (C) Spearman’s rank correlation of CD69 expression on blood uT cells on admission and hypoxemia levels at day 7 in COVID-19 patients (n = 20). *, P < 0.05; **, P < 0.01; ***, P < 0.001. ns, not significant.
In addition, the frequency and phenotype of blood uT cells were dynamically monitored in COVID-19 patients who were still in critical condition after 2 wk. We observed a persistent decrease in the frequency of uT cells at day 7 that was maintained to day 14 (Fig. 4 B). The levels of CD69 indicated a persistent activation status of uT cells during the course of the disease (Fig. 4 B), albeit a trend to decrease was noticed over time (Fig. 4 B). The proportion of PD-1–expressing uT cells remained largely stable (Fig. 4 B). Of note, CD69 and PD-1 expression on uT cells from rapidly discharged patients tended to decrease faster (Fig. S3 E).
Finally, we interrogated whether alterations in uT cell biology could be predictive of the level of hypoxemia, measured by the PaO2/FiO2 ratio, a routine clinical variable for ARDS management (Thompson et al., 2017). Thus, we compared multiple uT cell parameters on admission to the level of hypoxemia at day 7. Although no correlation could be observed for γδT cells (Fig. S3 F), we observed that CD69 expression on MAIT and iNKT cells was positively correlated with the PaO2/FiO2 ratio (Fig. 4 C). Notably, no additional correlation could be observed for MAIT and iNKT cells based on the other uT cell–related parameters, including PD-1 expression and cytokine production.
In summary, we show here that severe COVID-19 influences the phenotype and function of uT cells. It will be of interest to evaluate whether these parameters are influenced in different presentations of SARS-CoV-2 infection (i.e., mild and/or moderate). In addition, comparative analyses in pneumonia-induced ARDS patients of other microbiological origin will be informative to better gauge the SARS-CoV-2 specificity of our findings. In any case, our comparative analysis between the systemic (blood) and local (airways) compartment points toward localized inflammation and raises the possibility that activated MAIT cells may migrate into the inflamed tissue. As shown in other viral infections (Loh et al., 2016; van Wilgenburg et al., 2016), MAIT and iNKT cell activation during SARS-CoV-2 infection is likely antigen independent and may be driven by IL-18. Of importance, the levels of activation (as judged by CD69 expression) on MAIT and iNKT cells appear predictive of the clinical course. This could suggest a beneficial role for MAIT and iNKT cells during severe COVID-19, although their precise functions and associated mechanisms warrant further investigation. It is noteworthy that neither IFN-γ nor IL-17A secretion correlates with the positive evolution of clinical parameters, raising the possibility that uT cells could play a protective role through other functions. For instance, the role of MAIT and iNKT cells in tissue repair and regeneration has recently emerged (Trottein and Paget, 2018; Hinks et al., 2019; Lamichhane et al., 2019; Leng et al., 2019). This is particularly relevant in the context of severe COVID-19 with ARDS. Altogether, these findings should encourage further studies on MAIT and iNKT cells in pneumonia-induced ARDS to assess their potential as biomarkers and/or targets for immune intervention strategies.
Materials and methods
Clinical study design, patient population, and approval
Patients (>18 yr old) admitted to the ICU from March 18, 2020to April 17, 2020 with positive SARS-CoV-2 RT-PCR testing were prospectively included in this study. A control group of 17 critically ill patients admitted from March 14, 2018 to July 7, 2019, requiring mechanical ventilation but with no shock and no infectious illness, were also included. The study was conducted in one ICU from an academic hospital (Tours, France). All patients or their next of kin gave consent for participation in the study. This work was part of an ongoing study exploring immune response during community-acquired pneumonia (ClinicalTrial.gov identifier: NCT03379207). The study was approved by the ethics committee Comité de Protection de Personnes Ile-de-France 8 under agreement no. 2017-A01841-52, in accordance with national laws. Blood samples from healthy volunteers (age- and sex-matched; median age 56.5 yr (SD 6.8); male/female ratio 1.8/1) were obtained from the Etablissement Français du Sang.
Reagents and antibodies
Staining was performed using antibodies (Table S1) from BioLegend and Miltenyi Biotec. PBS-57 glycolipid-loaded and control CD1d tetramers (BV421-conjugated) as well as 5-OP-RU–loaded and control MR1 tetramers (BV421-conjugated) were obtained from the National Institute of Allergy and Infectious Diseases Tetramer Facility (Emory University, Atlanta, GA). Dead cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Thermo Fisher Scientific).
Human cell isolation
Peripheral blood mononuclear cells (PBMCs)
PBMCs were enriched by density gradient centrifugation using Histopaque-1077 solution (Sigma-Aldrich) according to the manufacturer’s instructions. Red blood cells were removed using a red blood cell lysis buffer (Sigma-Aldrich).
ETA
ETA was collected from intubated non–COVID-19 (n = 7) or COVID-19 (n = 20) patients who were under invasive mechanical ventilation. ETA samples were weighed and incubated in PBS (5 ml/g) with 1 mM dithiothreitol for 30 min at 4°C under gentle agitation. After centrifugation, supernatants were collected, and cell pellets were filtered through a 100-µm cell strainer. Red blood cells were removed using a red blood cell lysis buffer, and cells from ETA were passed through a 40-µm cell strainer before staining for flow cytometry.
Soluble mediator measurement
Inflammatory mediators were measured in sera and supernatants of ETA using the Bio-Plex Pro Human cytokines screening panel (Bio-Rad) in a multiplex fluorescent bead assay (Luminex), according to the manufacturer’s instructions. Inflammatory mediators in sera and ETA supernatants were recorded as picograms per milliliter and per gram, respectively. The ETA density ratio (g:ml) is 1:1.
Flow cytometry
Cells were stained with antibodies to surface epitopes and viability dye (LIVE/DEAD Fixable Aqua Dead Cell Stain). For cytokine profile analysis, cells were stimulated for 4 h in RPMI 1640 complete medium containing PMA (100 ng/ml) and ionomycin (1 µg/ml) in the presence of protein transport inhibitor cocktail (eBioscience) added 1 h after stimulation. For intracellular staining, cells were fixed and permeabilized using the Fixation/Permeabilization Solution Kit (BD Biosciences). Cells were stained with APC-conjugated mAb against IL-17A and PE-conjugated mAb against IFN-γ. Events were acquired on a MACS Quant cytometer (Miltenyi Biotec). Analyses were performed with VenturiOne software (Applied Cytometry).
Statistical analysis
All statistical analysis was performed with GraphPad Prism software. Statistical significance was evaluated by comparing biological values in each experimental group. In some cases, matched-pairs test was used. The tests used for each panel are indicated in the figure legends. Results with a P value of <0.05 were considered significant: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Correlation calculation between two parameters was performed using Spearman’s rho test.
Online supplemental material
Fig. S1 shows the absence of correlation regarding some potential confounding effects (age and obesity) as well as the modulation of TCR expression on uT cells in COVID-19 patients. Fig. S2 shows representative plots for activation markers and cytokine production by uT cells and absence of correlation between uT cell activation and some activating cytokines (IL-1β and IFN-α2). Fig. S3 shows a comparative analysis of multiple uT cell biological parameters with the clinical evolution of COVID-19 patients. Table S1 lists the monoclonal antibodies used in this study.
Supplementary Material
lists anti-human monoclonal antibodies.
Acknowledgments
We thank the National Institutes of Health tetramer core facility (Emory University, Atlanta, GA) for providing MR1 and CD1d tetramers. We acknowledge all health care coworkers involved in the ICU department at the Bretonneau Hospital, especially Aurélie Aubrey, Delphine Chartier, Véronique Siméon, and Julien Bontemps, for their excellent management of patient samples and clinical data. Annick Legras, Denis Garot, Emmanuelle Mercier, Charlotte Salmon-Gandonnière, Laetitia Bodet-Contentin, Marlène Morisseau, Stephan Mankikian, and Walid Darwiche are acknowledged for patient inclusions. We thank all patients and their families for their trust and confidence in our work. The graphical abstract was created using BioRender.com.
This work was supported by the Agence Nationale de la Recherche “JCJC program” (ANR-19-CE15-0032-01) and by the Fondation du Souffle, with the Fonds de Recherche en Santé Respiratoire. M. Si-Tahar and C. Paget are supported by the Institut national de la santé et de la recherche médicale. Y. Jouan, A. Guillon, Y. Perez, S. Ehrmann, and P-F. Dequin were supported by the Centre Hospitalier Régional Universitaire de Tours. M. Ferreira, T. Baranek, and L. Gonzalez are supported by the University of Tours.
Author contributions: Y. Jouan, A. Guillon, S. Ehrmann, B. François, P.-F. Dequin, M. Si-Tahar, T. Baranek, and C. Paget designed the research; L. Gonzalez, Y. Perez, C. Boisseau, M. Ferreira, T. Daix, R. Jeannet, T. Baranek, and C. Paget collected the data; Y. Jouan, L. Gonzalez, T. Baranek, and C. Paget analyzed the data; and Y. Jouan and C. Paget wrote the manuscript with the input of all authors.
References
- Bonneville M., O’Brien R.L., and Born W.K.. 2010. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat. Rev. Immunol. 10:467–478. 10.1038/nri2781 [DOI] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., et al. 2020. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 130:2620–2629. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., Link V.M., Tamoutounour S., Wong A.C., Perez-Chaparro P.J., Han S.J., Chen Y.E., Li K., Farhat S., Weckel A., et al. 2019. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 366 eaax6624 10.1126/science.aax6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby C.M., and Kronenberg M.. 2016. Invariant natural killer T cells: front line fighters in the war against pathogenic microbes. Immunogenetics. 68:639–648. 10.1007/s00251-016-0933-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby C.M., and Kronenberg M.. 2018. Tissue-specific functions of invariant natural killer T cells. Nat. Rev. Immunol. 18:559–574. 10.1038/s41577-018-0034-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déchanet J., Merville P., Lim A., Retière C., Pitard V., Lafarge X., Michelson S., Méric C., Hallet M.M., Kourilsky P., et al. 1999. Implication of gammadelta T cells in the human immune response to cytomegalovirus. J. Clin. Invest. 103:1437–1449. 10.1172/JCI5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doisne J.-M., Soulard V., Bécourt C., Amniai L., Henrot P., Havenar-Daughton C., Blanchet C., Zitvogel L., Ryffel B., Cavaillon J.-M., et al. 2011. Cutting edge: crucial role of IL-1 and IL-23 in the innate IL-17 response of peripheral lymph node NK1.1- invariant NKT cells to bacteria. J. Immunol. 186:662–666. 10.4049/jimmunol.1002725 [DOI] [PubMed] [Google Scholar]
- Godfrey D.I., Uldrich A.P., McCluskey J., Rossjohn J., and Moody D.B.. 2015. The burgeoning family of unconventional T cells. Nat. Immunol. 16:1114–1123. 10.1038/ni.3298 [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. 2020. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 369:718–724. 10.1126/science.abc6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinks T.S.C., Marchi E., Jabeen M., Olshansky M., Kurioka A., Pediongco T.J., Meehan B.S., Kostenko L., Turner S.J., Corbett A.J., et al. 2019. Activation and In Vivo Evolution of the MAIT Cell Transcriptome in Mice and Humans Reveals Tissue Repair Functionality. Cell Rep. 28:3249–3262.e5. 10.1016/j.celrep.2019.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M.G., He Y., and Gale M. Jr.. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675–687. 10.1038/nri888 [DOI] [PubMed] [Google Scholar]
- Lamichhane R., Schneider M., de la Harpe S.M., Harrop T.W.R., Hannaway R.F., Dearden P.K., Kirman J.R., Tyndall J.D.A., Vernall A.J., and Ussher J.E.. 2019. TCR- or Cytokine-Activated CD8+ Mucosal-Associated Invariant T Cells Are Rapid Polyfunctional Effectors That Can Coordinate Immune Responses. Cell Rep. 28:3061–3076.e5. 10.1016/j.celrep.2019.08.054 [DOI] [PubMed] [Google Scholar]
- Le Bourhis L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11:701–708. 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- Leeansyah E., Ganesh A., Quigley M.F., Sönnerborg A., Andersson J., Hunt P.W., Somsouk M., Deeks S.G., Martin J.N., Moll M., et al. 2013. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood. 121:1124–1135. 10.1182/blood-2012-07-445429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng T., Akther H.D., Hackstein C.P., Powell K., King T., Friedrich M., Christoforidou Z., McCuaig S., Neyazi M., Arancibia-Cárcamo C.V., et al. ; Oxford IBD Investigators . 2019. TCR and Inflammatory Signals Tune Human MAIT Cells to Exert Specific Tissue Repair and Effector Functions. Cell Rep. 28:3077–3091.e5. 10.1016/j.celrep.2019.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh L., Wang Z., Sant S., Koutsakos M., Jegaskanda S., Corbett A.J., Liu L., Fairlie D.P., Crowe J., Rossjohn J., et al. 2016. Human mucosal-associated invariant T cells contribute to antiviral influenza immunity via IL-18-dependent activation. Proc. Natl. Acad. Sci. USA. 113:10133–10138. 10.1073/pnas.1610750113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch L., O’Shea D., Winter D.C., Geoghegan J., Doherty D.G., and O’Farrelly C.. 2009. Invariant NKT cells and CD1d(+) cells amass in human omentum and are depleted in patients with cancer and obesity. Eur. J. Immunol. 39:1893–1901. 10.1002/eji.200939349 [DOI] [PubMed] [Google Scholar]
- Mackay C.R. 1996. Chemokine receptors and T cell chemotaxis. J. Exp. Med. 184:799–802. 10.1084/jem.184.3.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes I., Pingris K., Poitou C., Bessoles S., Venteclef N., Kiaf B., Beaudoin L., Da Silva J., Allatif O., Rossjohn J., et al. 2015. Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J. Clin. Invest. 125:1752–1762. 10.1172/JCI78941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., and Calfee C.S.. 2019. Acute respiratory distress syndrome. Nat. Rev. Dis. Primers. 5:18 10.1038/s41572-019-0069-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N.E., and Eberl M.. 2018. Human γδ T-Cell Control of Mucosal Immunity and Inflammation. Front. Immunol. 9:985 10.3389/fimmu.2018.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., and Manson J.J.; HLH Across Speciality Collaboration, UK . 2020. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 395:1033–1034. 10.1016/S0140-6736(20)30628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M.M., Witherden D.A., and Havran W.L.. 2017. γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol. 17:733–745. 10.1038/nri.2017.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget C., and Trottein F.. 2019. Mechanisms of Bacterial Superinfection Post-influenza: A Role for Unconventional T Cells. Front. Immunol. 10:336 10.3389/fimmu.2019.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget C., Ivanov S., Fontaine J., Blanc F., Pichavant M., Renneson J., Bialecki E., Pothlichet J., Vendeville C., Barba-Spaeth G., et al. 2011. Potential role of invariant NKT cells in the control of pulmonary inflammation and CD8+ T cell response during acute influenza A virus H3N2 pneumonia. J. Immunol. 186:5590–5602. 10.4049/jimmunol.1002348 [DOI] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., et al. 2020. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 71:762–768. 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton C.E., Lalor S.J., Sweeney C.M., Brereton C.F., Lavelle E.C., and Mills K.H.. 2009. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 31:331–341. 10.1016/j.immuni.2009.08.001 [DOI] [PubMed] [Google Scholar]
- Thompson B.T., Chambers R.C., and Liu K.D.. 2017. Acute Respiratory Distress Syndrome. N. Engl. J. Med. 377:562–572. 10.1056/NEJMra1608077 [DOI] [PubMed] [Google Scholar]
- Toubal A., Nel I., Lotersztajn S., and Lehuen A.. 2019. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 19:643–657. 10.1038/s41577-019-0191-y [DOI] [PubMed] [Google Scholar]
- Trottein F., and Paget C.. 2018. Natural Killer T Cells and Mucosal-Associated Invariant T Cells in Lung Infections. Front. Immunol. 9:1750 10.3389/fimmu.2018.01750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-Y., Liong K.H., Gunalan M.G., Li N., Lim D.S.L., Fisher D.A., MacAry P.A., Leo Y.S., Wong S.-C., Puan K.J., et al. 2015. Type I IFNs and IL-18 regulate the antiviral response of primary human γδ T cells against dendritic cells infected with Dengue virus. J. Immunol. 194:3890–3900. 10.4049/jimmunol.1303343 [DOI] [PubMed] [Google Scholar]
- Tyznik A.J., Verma S., Wang Q., Kronenberg M., and Benedict C.A.. 2014. Distinct requirements for activation of NKT and NK cells during viral infection. J. Immunol. 192:3676–3685. 10.4049/jimmunol.1300837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussher J.E., Bilton M., Attwod E., Shadwell J., Richardson R., de Lara C., Mettke E., Kurioka A., Hansen T.H., Klenerman P., et al. 2014. CD161++ CD8+ T cells, including the MAIT cell subset, are specifically activated by IL-12+IL-18 in a TCR-independent manner. Eur. J. Immunol. 44:195–203. 10.1002/eji.201343509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wilgenburg B., Scherwitzl I., Hutchinson E.C., Leng T., Kurioka A., Kulicke C., de Lara C., Cole S., Vasanawathana S., Limpitikul W., et al. ; STOP-HCV consortium . 2016. MAIT cells are activated during human viral infections. Nat. Commun. 7:11653 10.1038/ncomms11653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 395:1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists anti-human monoclonal antibodies.