Viral infection dramatically changes the levels of many types of RNA in a cell. In particular, certain oncogenic viruses activate expression of repetitive genes called retrotransposons, which are normally silenced due to their ability to copy and spread throughout the genome. Here, we established that infection with the gammaherpesvirus MHV68 leads to a dramatic induction of a class of noncoding retrotransposons called B2 SINEs in multiple cell types. We then explored how MHV68 activates B2 SINEs, revealing a role for the conserved herpesviral protein kinase ORF36. Both ORF36 kinase-dependent and kinase-independent functions contribute to B2 induction, perhaps through ORF36 targeting of proteins involved in controlling the accessibility of chromatin surrounding SINE loci. Understanding the features underlying induction of these elements following MHV68 infection should provide insight into core elements of SINE regulation, as well as disregulation of SINE elements associated with disease.
KEYWORDS: B2, MHV68, RNA polymerase III, SINE, herpesvirus, noncoding RNA
ABSTRACT
Short interspersed nuclear elements (SINEs) are RNA polymerase III (RNAPIII)-transcribed, retrotransposable noncoding RNA (ncRNA) elements ubiquitously spread throughout mammalian genomes. While normally silenced in healthy somatic tissue, SINEs can be induced during infection with DNA viruses, including the model murine gammaherpesvirus 68 (MHV68). Here, we explored the mechanisms underlying MHV68 activation of SINE ncRNAs. We demonstrate that lytic MHV68 infection of B cells, macrophages, and fibroblasts leads to robust activation of the B2 family of SINEs in a cell-autonomous manner. B2 ncRNA induction requires neither host innate immune signaling factors nor involvement of the RNAPIII master regulator Maf1. However, we identified MHV68 ORF36, the conserved herpesviral kinase, as playing a key role in B2 induction during lytic infection. SINE activation is linked to ORF36 kinase activity and can also be induced by inhibition of histone deacetylases 1 and 2 (HCAC 1/2), which is one of the known ORF36 functions. Collectively, our data suggest that ORF36-mediated changes in chromatin modification contribute to B2 activation during MHV68 infection and that this activity is conserved in other herpesviral protein kinase homologs.
IMPORTANCE Viral infection dramatically changes the levels of many types of RNA in a cell. In particular, certain oncogenic viruses activate expression of repetitive genes called retrotransposons, which are normally silenced due to their ability to copy and spread throughout the genome. Here, we established that infection with the gammaherpesvirus MHV68 leads to a dramatic induction of a class of noncoding retrotransposons called B2 SINEs in multiple cell types. We then explored how MHV68 activates B2 SINEs, revealing a role for the conserved herpesviral protein kinase ORF36. Both ORF36 kinase-dependent and kinase-independent functions contribute to B2 induction, perhaps through ORF36 targeting of proteins involved in controlling the accessibility of chromatin surrounding SINE loci. Understanding the features underlying induction of these elements following MHV68 infection should provide insight into core elements of SINE regulation, as well as disregulation of SINE elements associated with disease.
INTRODUCTION
A large fraction (40 to 45%) of mammalian genomes are composed of sequences derived from retrotransposable elements, which are capable of copying themselves (autonomous) or being copied (nonautonomous) and inserted semirandomly back into the genome. Retrotransposons are ubiquitously spread throughout the genome and are important components of genome architecture and chromatin remodeling (1–4). Among these, the short interspersed nuclear element (SINE) subfamily of retrotransposons makes up ∼12% of the genome and is transcribed by RNA polymerase III (RNAPIII) to produce short, ∼300-bp noncoding RNAs (ncRNAs). SINEs are evolutionarily derived from other common RNAPIII-transcribed genes, such as 7SL in the case of the human Alu SINE and tRNA in the case of the mouse B2 SINE. SINE ncRNAs are nonautonomous and co-opt the machinery encoded by the long interspersed nuclear elements (LINEs) for reverse transcription and reintegration. SINEs may act as functional enhancers and mobile RNA polymerase II promoters and are also present as “embedded elements” in many mRNA transcripts, where they can influence mRNA processing, localization, and decay (1, 5–7).
B2 SINE ncRNA transcription is RNAPIII dependent, requiring the transcription factor complexes TFIIIC and TFIIIB. TFIIIC binds to the internal A and B boxes present within type II RNAPIII promoters, such as those contained within B2 SINE and tRNA species. This is followed by recruitment of TFIIIB, comprising BDP1, BRF1, and TBP, which help position RNAPIII at the transcription start site. Absence of BRF1 abrogates transcription from type I and type II RNAPIII promoters but does not affect transcription from type III RNAPIII promoters, which utilize a Brf1 paralog, Brf2 (8). RNAPIII activity can be broadly controlled by its master repressor Maf1, a phosphoprotein that binds BRF1 and RNAPIII, thereby preventing TFIIIB assembly onto DNA and blocking the association of the polymerase with TFIIIB that is already assembled at transcription start sites (9). Phosphorylation of Maf1, for example, by mTORC1 (10), prevents Maf1-mediated repression of RNAPIII, thereby potentiating an increase in transcription.
SINE expression is normally repressed due to the maintenance of repressive tri-methylation of lysine 9 on histone H3 (H3K9me3) (11) and CpG methylation of DNA (12). However, SINEs become derepressed under conditions of cellular stress, such as chemical treatment and heat shock (13–15). SINEs from both humans and mice are also induced during infection with a variety of DNA viruses, including herpes simplex virus 1 (HSV-1), adenovirus, minute virus of mice, simian virus 40 (SV40), and murine gammaherpesvirus 68 (MHV68) (16–22). Several recent reports indicate that virus-induced SINEs and other RNAPIII-transcribed ncRNAs interface with innate immune pathways and thus may serve as signaling molecules during infection (23, 24). In particular, B2 ncRNAs induced upon MHV68 infection potentiate NF-κB signaling, in part through a pathway involving the mitochondrial antiviral signaling protein (MAVS), and also boost viral gene expression (21, 25). Aberrant accumulation of Alu RNAs contributes to age-related macular degeneration by inducing cytotoxic NLRP3 inflammasome activation (26–30) and can also induce epithelial-to-mesenchymal transition, a hallmark of progression of several cancers (31). Additionally, SINEs induced during heat shock can bind and inhibit RNA polymerase II transcription, indicating that these ncRNAs may have a variety of functions during stress (15, 32).
MHV68 is a model gammaherpesvirus related to Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) and has been widely used to dissect gammaherpesvirus biology and pathogenesis. A recent genome-wide mapping study revealed that MHV68 infection of murine fibroblasts leads to activation of ∼30,000 B2 SINE loci, although the mechanism of B2 induction is unknown (22). Here, we show that in addition to fibroblasts, B2 SINE induction occurs during MHV68 lytic infection of primary bone marrow-derived macrophages and during lytic reactivation of B cells, both physiologically relevant cell types for the virus. Induction is cell autonomous, occurs independently of innate immune signaling components, and does not involve RNAPIII regulation by the master repressor Maf1. Instead, a screen of MHV68 open reading frames (ORFs) revealed a role for the conserved herpesvirus protein kinase ORF36 in B2 SINE induction. Expression of wild-type (WT) ORF36 but not a kinase dead mutant was sufficient to activate B2 SINEs, and an MHV68 mutant lacking ORF36 displayed reduced SINE induction potential. ORF36 inhibits histone deacetylases 1 and 2 (33, 34), and we show that chromatin derepression contributes to B2 activation. Collectively, our results reveal a new function for the herpesviral protein kinase and provide insight into the mechanism of SINE activation during viral infection.
RESULTS
MHV68 infection induces B2 SINEs in physiologically relevant antigen-presenting cell types.
Our previous work established that MHV68 infection of murine fibroblasts results in robust activation of B2 SINEs (21). While fibroblasts are commonly used to study MHV68 infection in vitro, two of the most physiologically relevant cell types for the in vivo MHV68 life cycle and establishment of lifelong latency are B cells and macrophages (35). We therefore sought to determine whether B2 SINE induction is also a feature of MHV68 infection in these key cell types.
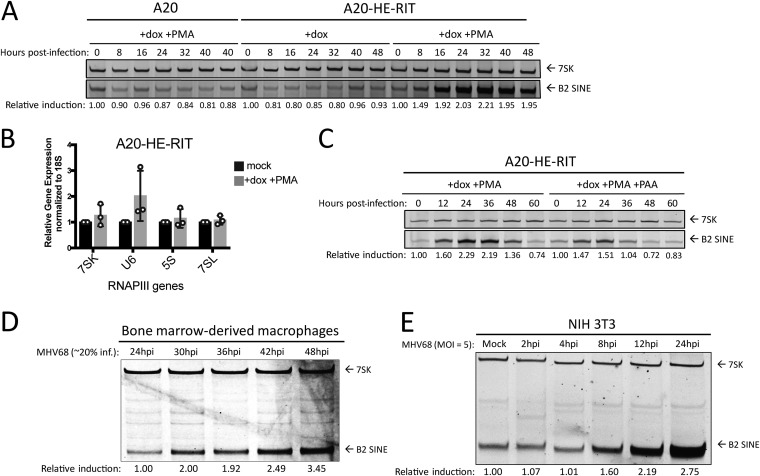
Although B cells are the main viral reservoir in vivo, they are highly resistant to de novo MHV68 infection in cell culture (36). The only latently infected B cell line isolated from an MHV68-infected mouse tumor, S11, reactivates to very low frequency, making study of lytic cell populations impractical (37). However, a B cell line has been generated (A20-HE-RIT) that is latently infected with MHV68 and contains a doxycycline (dox)-inducible version of the viral lytic transcriptional activator gene RTA. Treatment of these cells with Dox and phorbol ester (PMA) enables the switch from latency to lytic replication in approximately 80% of the cells (38, 39). Induction of the lytic cycle by dox and PMA treatment of the A20-HE-RIT cells caused a marked increase in B2 SINE levels as measured by primer extension, with levels peaking at 24 to 32 h poststimulation (Fig. 1A). Importantly, B2 RNA induction was not seen in the uninfected A20 parental cells subjected to the same dox and PMA treatment. Furthermore, the induction observed in infected cells was specific to B2 SINEs, as levels of another RNAPIII transcript, 7SK, remained unchanged. Further, reverse transcriptase quantitative PCR (RT-qPCR) analysis confirms that 7SK plus several other RNAPIII transcripts are not significantly elevated upon reactivation of A20-HE-RIT (Fig. 1B), similar to our previous findings during MHV68 lytic infection (21). Additionally, phosphonoacetic acid (PAA) treatment to block viral DNA replication did not prevent B2 SINE induction during reactivation in A20-HE-RIT cells, although the levels were modestly reduced (Fig. 1C). Thus, upon lytic reactivation of latently infected B cells, B2 SINEs are induced early in the viral lytic cycle and continue to accumulate as infection progresses.
FIG 1.
B2 SINE transcription is upregulated in B cells, primary macrophages, and NIH 3T3 cells upon MHV68 infection. (A) MHV68 latently infected A20-HE-RIT B cells, or parental uninfected A20 B cells, were treated with doxycycline (dox) and phorbol ester (PMA) to induce lytic reactivation. Total RNA was isolated at the indicated time points postreactivation and subjected to primer extension for B2 SINEs or 7SK for loading. (B) RT-qPCR was performed to detect the indicated RNAPIII transcript levels using RNA extracted from mock or reactivated A20-HE-RITs. (C) A20-HE-RITs were reactivated in the presence or absence of phosphonoacetic acid (PAA) to block viral DNA replication, and total RNA was subjected to primer extension as described above. (D) BMDMs or (E) NIH 3T3 were either mock infected or infected with MHV68 for the indicated time periods, whereupon total RNA was isolated and subjected to primer extension as described above.
We next examined the potential for B2 SINE induction upon MHV68 infection of primary bone marrow-derived macrophages (BMDMs). Unlike fibroblasts, which are highly susceptible to MHV68, the highest level of infection we achieved in WT BMDMs was ∼20%, which occurred with a multiplicity of infection (MOI) of 20 and did not increase upon addition of more virus (A. M. Schaller, unpublished observation). Despite the lower infection efficiency, primer extension reactions demonstrated that in MHV68-infected primary BMDMs, B2 SINE induction began at 30 h postinfection (hpi) and reached their highest relative levels by 40 to 48 hpi (Fig. 1D). These induction kinetics were slower than what we observed in NIH 3T3 cells (Fig. 1E), likely due to overall slower replication kinetics of MHV68 in the BMDMs, as previously documented (40). In summary, B2 SINE RNA induction occurs during lytic MHV68 infection of multiple primary and immortalized cell types.
B2 SINE RNAs are not induced in uninfected cells by paracrine signaling.
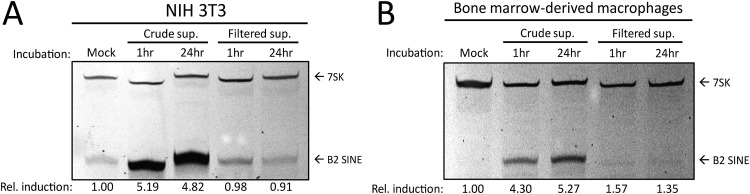
We were struck by the robust B2 upregulation in primary BMDMs, given that at most 20% of these cells were infected by MHV68. We therefore considered the possibility that infected cells produce paracrine signals that cause B2 upregulation in neighboring uninfected cells as well. We first tested this possibility using 3T3 cells, as their susceptibility to infection should yield a higher concentration of relevant paracrine signaling molecules. We performed a supernatant transfer assay, in which uninfected cells were incubated for 1 h or 24 h with cell supernatants from infected NIH 3T3 cells, either in crude form or after 0.1 μm filtration to remove viral particles. B2 SINE levels were then measured 24 h posttransfer using primer extension. We observed no B2 SINE induction in cells incubated in filtered supernatants, suggesting that paracrine signals derived from infected 3T3 cells are not sufficient to stimulate B2 induction in uninfected cells. In contrast, there was robust B2 SINE induction in cells incubated with crude supernatants, as expected since these supernatants contain MHV68 virions to initiate a de novo infection (Fig. 2A). This experiment was repeated in BMDMs, where filtered or crude supernatants were taken from infected 3T3 cells and incubated with plated BMDMs for 1 h or 24 h before removal. BMDMs were harvested 48 h after the beginning of incubation with 3T3 supernatants. These data were identical to those observed with 3T3s, in which paracrine signals contained within filtered supernatant were insufficient for B2 induction (Fig. 2B). While it is theoretically possible that some filterable component, and not viral infection, could be responsible for B2 SINE induction, these results strongly suggest that paracrine or cell-to-cell signaling through the supernatant is not sufficient to induce this phenotype.
FIG 2.
Paracrine signaling does not induce B2 SINE induction. (A) NIH 3T3 cells or (B) primary BMDMs were incubated with supernatants harvested from 24-h-infected NIH 3T3 cells, either in crude form or filtered to remove whole virus, for the indicated time period. Total RNA was isolated from cells at 24 h or 48 h postincubation and subjected to primer extension for B2 SINEs or 7SK, respectively.
B2 SINE induction is RNAPIII-dependent but does not involve the RNAPIII regulator Maf1.
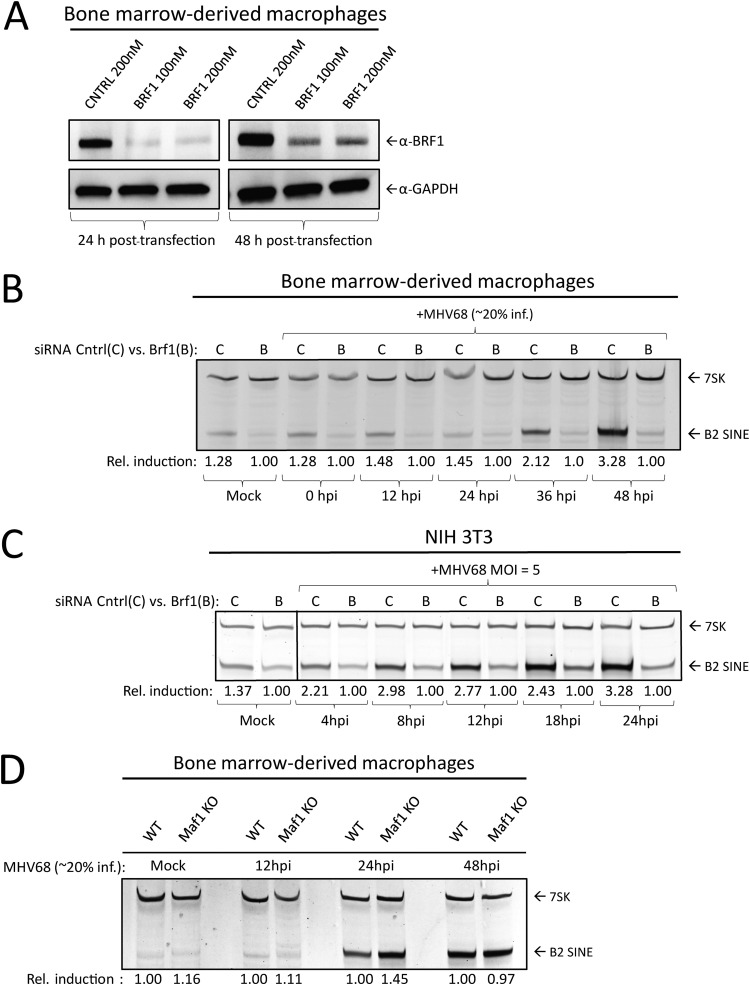
We previously showed that treatment of 3T3 cells with an RNAPIII inhibitor or B2-directed antisense oligonucleotides (ASOs) reduced the B2 RNA levels upon MHV68 infection, strongly suggesting that RNAPIII activity was required for their induction (21). However, given that small molecule inhibitors can have off-target effects and B2 ASOs will also target mRNAs containing embedded SINE elements, we sought to independently validate that the B2 SINE transcriptional induction is RNAPIII-dependent. We chose the strategy of depleting Brf1, a critical component of the TFIIIB transcription factor complex needed for RNAPIII transcription of type II (e.g., SINE) promoters using small interfering RNA (siRNA)-mediated knockdown (8). Knockdown of Brf1 was robust through 48 h posttransfection (Fig. 3A). In both BMDMs (Fig. 3B) and 3T3 cells (Fig. 3C), depletion of Brf1 completely abrogated B2 expression as measured by primer extension throughout the time course of infection. Notably, the levels of 7SK were not affected by Brf1 knockdown, as this RNAPIII transcript has a type III promoter that does not require Brf1 (41). Thus, these results confirm that RNAPIII is required for MHV68-induced B2 SINE activation.
FIG 3.
B2 SINE upregulation is dependent on RNAPIII but independent of the RNAPIII master regulator Maf1. (A) BMDMs were transfected with the indicated concentrations of either control or Brf1 siRNA pools and harvested 24 to 48 h later. Then, 30 μg of total protein lysates was resolved by SDS-PAGE and Western blotting with antibodies against Brf1 or GAPDH (as a loading control). (B) Total RNA was harvested from mock- or MHV68-infected BMDMs and NIH 3T3 fibroblasts following control or Brf1 siRNA treatment at the indicated time points. Total RNA was subjected to primer extension using primers for B2 SINEs or 7SK (as a control). (C) WT or (D) Maf1–/– BMDMs were mock infected or infected with MHV68 for the indicated times, whereupon total RNA was harvested and subjected to primer extension as described in panel B.
We next considered the possibility that MHV68 infection alters the regulation of RNAPIII to increase its activity on B2 promoters. A master regulator of RNAPIII is Maf1, which acts by binding free RNAPIII at its clamp domain, thereby impairing RNAPIII binding to the TFIIIB-promoter complex and preventing RNAPIII transcription initiation (9, 42). To test the hypothesis that release of Maf1-mediated repression of RNAPIII transcription is responsible for B2 SINE induction, we derived primary BMDMs from Maf1–/– mice (43). Surprisingly, we observed no increase in B2 SINE RNA in uninfected Maf1–/– BMDMs compared to WT BMDMs, suggesting that Maf1 is not required for the normal silencing of B2 loci (Fig. 3D). We did observe somewhat more of an increase in B2 levels at 24 hpi with MHV68 in the Maf1–/– cells relative to WT cells, although this difference was not sustained at 48 hpi (Fig. 3D). We therefore conclude that the primary mechanism of B2 induction by MHV68 is not through interference with the RNAPIII repressor Maf1.
B2 SINE induction is independent of canonical innate immune signaling pathways.
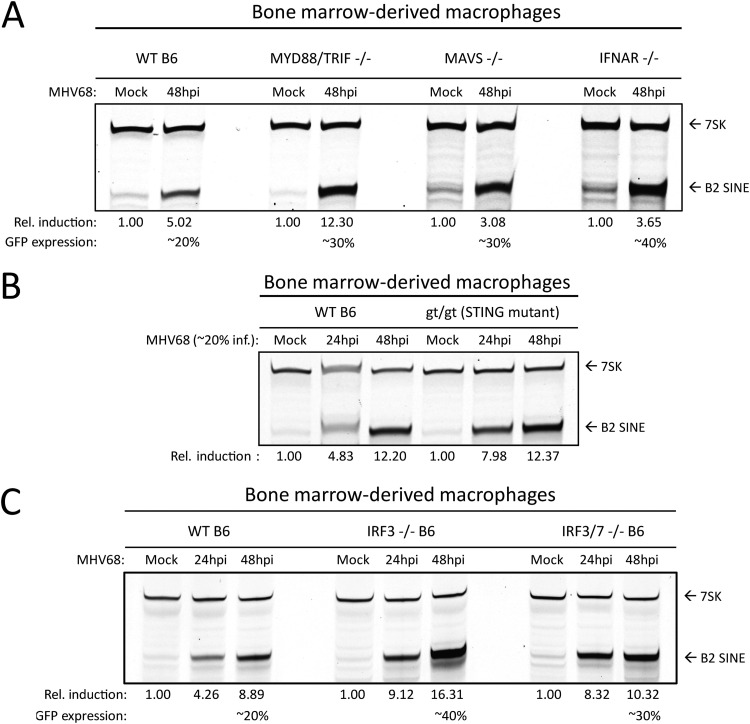
Due to their activation during herpesvirus infection and broadly acting signaling cascades, we considered that innate immune signaling may be involved upstream of B2 SINE induction. Pattern recognition receptors, namely, the toll-like receptors 2, 3, 7, and 9, RIG-I-like receptors, and AIM2, can become activated during lytic herpesvirus infection (44–47). To examine the possible upstream involvement of infection-induced innate immune signaling in the induction of B2 SINE transcription, we quantified B2 SINE levels in primary BMDMs derived from WT B6 mice versus mice lacking several canonical innate immune signaling pathways. These included mutants in Toll-like receptor signaling (MyD88/TRIF–/–), cytoplasmic RNA recognition signaling (MAVS–/–), or type I interferon (IFN) receptor-mediated signaling through the type I IFN receptor (IFNAR–/–) (Fig. 4A), as well as cGAS/STING-mediated DNA sensing using the golden ticket (gt/gt) mutant (48), which contains a missense mutation in exon 6 of the mouse STING gene, rendering STING inactive (Fig. 4B). In each case, primer extension experiments showed equivalent or greater B2 SINE RNA induction upon infection of the mutant BMDMs compared to the WT BMDMs. Thus, none of these innate immune components is individually required for SINE activation during MHV68 infection.
FIG 4.
B2 SINE induction occurs independently of innate immune signaling. (A to C) WT or the indicated innate immune factor knockout BMDMs were mock or MHV68 infected for 24 to 48 h. Total RNA was then harvested and subjected to primer extension using primers for B2 SINEs or 7SK (as a control).
To control for the possibility that multiple innate immune sensors could be activated in a redundant manner to induce B2 SINEs, we also tested primary BMDMs derived from mice lacking the downstream transcription factors interferon-regulatory factor 3 (IRF3) and interferon regulatory factor 7 (IRF7). All pattern recognition receptor signaling pathways converge on IRF3 and IRF7, which activate transcription of interferon-stimulated genes (ISGs) and inflammatory cytokines (49). In agreement with the data from BMDMs lacking the upstream innate immune sensors, MHV68 infection still caused robust B2 SINE induction in IRF3–/– and IRF3/7–/– BMDMs (Fig. 4C). Thus, innate immune signaling does not activate B2 SINE transcription during MHV68 infection.
We noted that the infection-induced B2 levels were even more pronounced in each of the single and double knockout BMDMs than in WT cells (Fig. 4A to C). We hypothesize that this is a result of increased MHV68 infection under conditions of impaired immune restriction, as we noted that the knockout BMDMs routinely achieved higher MHV68 infection rates (as measured by green fluorescent protein [GFP] positivity) than WT BMDMs (Schaller, unpublished).
The conserved herpesvirus kinase ORF36 is sufficient to induce B2 SINE transcriptional upregulation.
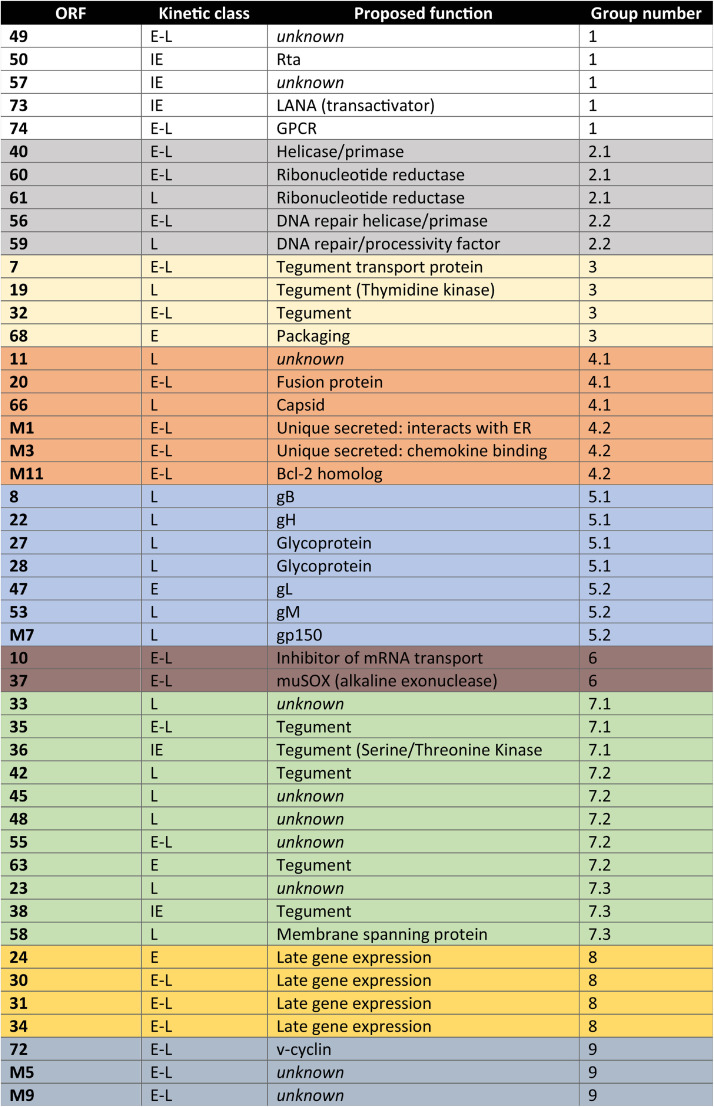
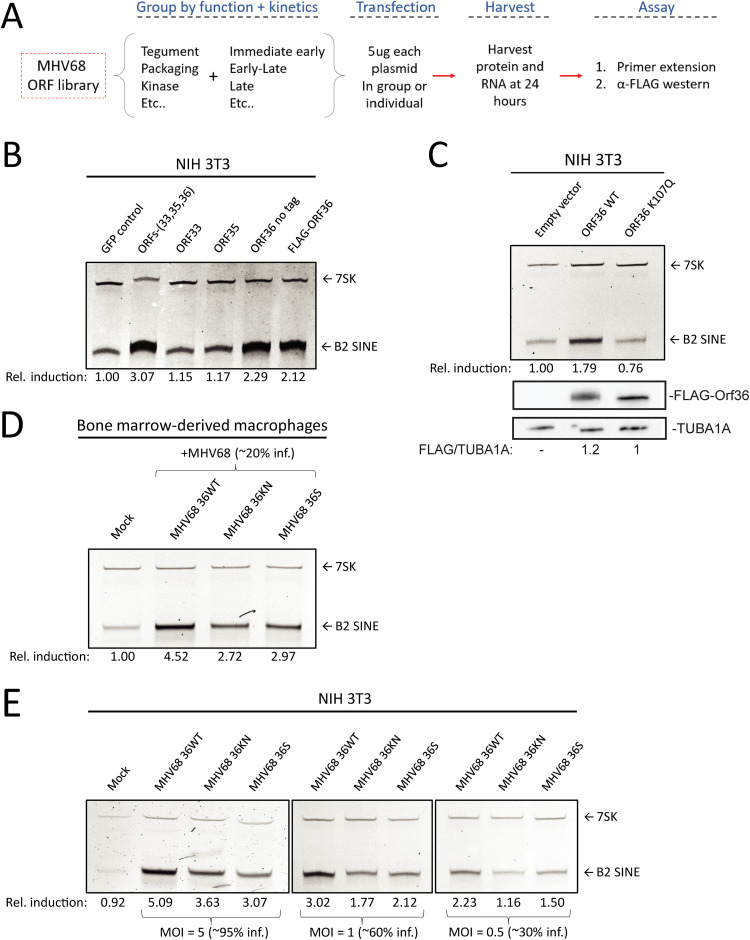
To search for viral factors involved in B2 SINE induction, we obtained and resequenced a partial MHV68 open reading frame (ORF) library previously generated by Ren Sun, which contained 47 full-length MHV68 ORF plasmids (50) (Table 1). The ORFs were first screened by cotransfection of 3T3 cells with 3 to 5 plasmids that were grouped based on similar temporal class and/or proposed or known function (Fig. 5A; Table 1) (51, 52). Only the group that contained ORFs 33, 35, and 36 showed B2 SINE induction above that of the control GFP-expressing plasmid as measured by primer extension (Fig. 5B). We then tested each of these ORFs individually for the ability to induce B2 SINEs, revealing that only MHV68 ORF36 expression was sufficient to upregulate B2 SINEs both as an untagged construct as well as with an N-terminal FLAG-tag (Fig. 5B).
TABLE 1.
MHV68 ORFs tested in screena
The ORFs tested, their kinetic class, and their proposed function are listed. ORFs were grouped (last column) based on similarities of kinetic class and function.
FIG 5.
The MHV68 kinase ORF36 induces B2 SINE transcription. (A) Schematic representing the method for testing the MHV68 ORF library. (B) NIH 3T3 cells were transfected with plasmid(s) containing the indicated ORF(s) or a GFP control for 24 h, whereupon total RNA was extracted and subjected to primer extension using primers for B2 SINEs or 7SK (as a control). (C) NIH 3T3 cells were transfected with plasmids expressing either wild-type (WT) ORF36 or a kinase-null mutant (K107Q) for 24 h, and then total RNA was isolated and subjected to primer extension as described above. (D) BMDMs were infected with WT MHV68, kinase-null (KN), or ORF36 stop (S) virus. Total RNA was isolated at 48 hpi and subjected to primer extension as described in panel B. (E) NIH 3T3 cells were infected with WT MHV68, KN, or S virus at an MOI of 5. At 24 hpi, total RNA was isolated and subjected to primer extension as described in panel B.
MHV68 ORF36 is a conserved herpesvirus serine/threonine kinase with a variety of reported kinase-dependent and -independent roles relating to the DNA damage response, inhibition of histone deacetylation, and inhibiting IRF3-driven ISG production (33, 34, 53–55). To determine whether ORF36 kinase activity was required for B2 SINE upregulation, we compared the activity of WT ORF36 to an ORF36 kinase-null mutant (K107Q) (55). Primer extension of RNA from transfected 3T3 cells showed that only WT ORF36 but not K107Q induced B2 SINEs (Fig. 5C). To determine the contribution of ORF36 toward B2 induction in the context of infection, we obtained versions of MHV68 either lacking ORF36 (36S) or containing a kinase-null version of ORF36 (36KN) (54). Notably, infection of primary BMDMs with these viruses revealed a reduction in MHV68-induced B2 SINE RNA upon loss or kinase inactivation of ORF36 compared to infection with the repaired WT virus (Fig. 5D). We observed similar defects in B2 induction upon infection of 3T3 cells with 36S and KN viruses compared to WT, across a range of MOI (Fig. 5E). The fact that some residual B2 induction remained in BMDM and 3T3 cells infected with the ORF36 mutant viruses indicates that other viral factors also contribute to SINE induction. However, ORF36 expression is sufficient to activate B2 SINEs when expressed alone and is required for WT levels of B2 SINE induction in the context of MHV68 infection.
Induction of B2 SINE transcription is conserved among ORF36 CHPK homologs.
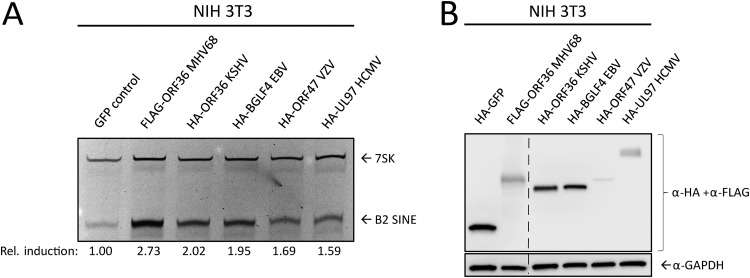
ORF36 homologs are found in all subfamilies of herpesviruses, where they are collectively referred to as the conserved herpesvirus protein kinases (CHPKs). Several examples exist of shared CHPK functions and shared substrate specificity (54, 56, 57). We therefore examined whether other CHPKs were able to induce B2 SINE RNA. We transfected NIH 3T3 cells with plasmids expressing HA- or FLAG-tagged CHPKs from KSHV (ORF36), varicella zoster virus (VZV) (ORF47), human cytomegalovirus (HCMV) (UL97), EBV (BGLF4), and MHV68 (ORF36) and measured B2 SINE RNA using primer extension (Fig. 6A). MHV68 ORF36 produced the most robust induction, followed by the other gammaherpesvirus CHPKs, KSHV ORF36, and EBV BGLF4. The alpha- and betaherpesvirus protein kinases, VZV ORF47 and HCMV UL97, induced B2 SINEs to a minimal degree, although they were expressed to similar (albeit low) levels as MHV68 ORF36 (Fig. 6B). Attempts at optimizing transfection conditions to achieve equivalent CHPK expression among homologs were unsuccessful, likely due to inherent properties of the expressed proteins in NIH 3T3s. However, these data suggest that B2 SINE induction is conserved among the CHPKs, with MHV68 ORF36 being highly potent for inducing B2 expression.
FIG 6.
Functional conservation of B2 SINE upregulation by several MHV68 ORF36 homologs. NIH 3T3 cells were transfected with plasmids containing FLAG-tagged MHV68 ORF36 or the indicated HA-tagged ORF36 homolog from Kaposi’s sarcoma-associated herpesvirus (KSHV ORF36), Epstein-Barr virus (EBV BGLF4), varicella zoster virus (VZV ORF47), or human cytomegalovirus (HCMV UL97). These cells were then harvested for total RNA for B2 and 7SK primer extension (A) or protein lysates, which were analyzed with Western blotting with antibodies against HA and FLAG, or GAPDH as a loading control (B). The dashed line indicates where an irrelevant lane was removed from the image.
Derepression of the chromatin landscape allows for B2 SINE induction.
Previous studies of features linked to SINE repression in uninfected cells indicated the importance of the repressive histone H3 lysine 9 tri-methylation (H3K9me3) mark and, to a lesser degree, DNA methylation at CpG sites (11, 12, 58, 59). These marks are deposited and maintained by the histone methyltransferases SU(VAR)3-9 and the DNMT family of DNA methyltransferases, respectively. Furthermore, ORF36 has been shown to inhibit histone deacetylases 1 and 2 (HDACs 1/2) (33), although whether HDACs are involved in repression of SINE loci is unknown.
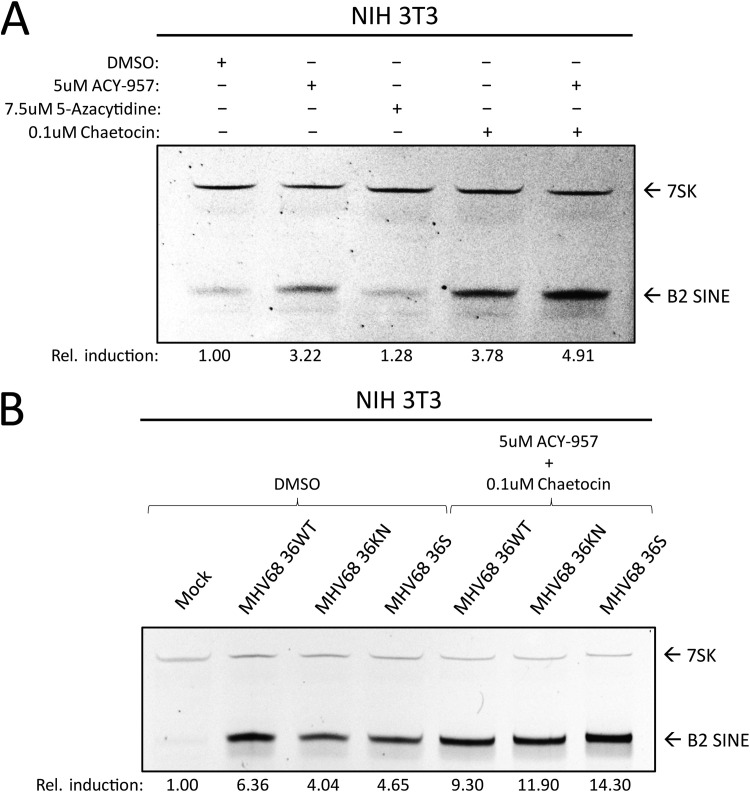
To test the role of each of these factors in B2 induction, we treated NIH 3T3 cells with inhibitors of HDACs 1/2 (ACY-957), DNMTs (5-azacytidine), and SU(VAR)3-9 (chaetocin) or a cocktail composed of ACY-957 and chaetocin together (Fig. 7A). We observed induction of B2 SINEs following treatment with ACY-957 and chaetocin and an additive effect when using both inhibitors together (Fig. 7A, lane 5). Treatment of cells with 5-azacytidine yielded no increase in levels of B2 RNA, in agreement with previous work (11).
FIG 7.
Inhibitors of chromatin repression cause B2 SINE upregulation. (A) NIH 3T3 cells were treated with the indicated inhibitor(s) for 24 h, whereupon total RNA was isolated and subjected to primer extension for B2 SINEs or 7SK. (B) NIH 3T3 cells were subjected to pretreatment with DMSO or the indicated inhibitors for 1 h prior to infection with MHV68 WT, KN, or S virus for 24 h, whereupon total RNA was isolated and subjected to primer extension as described in panel A.
Given that the strongest effects on B2 induction were observed upon inhibition of histone methyltransferases combined with HDAC inhibition, we next tested whether treatment with these inhibitors during infection was sufficient to rescue B2 levels in ORF36 KN and S infection to ORF36 WT infection levels. We observed that, in the context of infection, treatment with ACY-957 and chaetocin restored the levels of B2 ncRNA in the ORF36 S- and KN-infected cells to those observed during WT MHV68 infection (Fig. 7B), showing that chromatin derepression induced B2 ncRNA accumulation in an additive manner. Taken together, these data show that keeping an actively repressed chromatin state, primarily through maintenance of H3K9me3, is important for preventing constitutive B2 SINE induction.
DISCUSSION
A growing body of literature indicates that RNAPIII transcripts are upregulated in herpesvirus-infected cells and can serve as substrates for innate immune recognition, although mechanisms underlying their induction remain largely unknown (23, 24, 60, 61). The most robustly induced class of such transcripts in MHV68-infected fibroblasts are the B2 SINE ncRNAs, whose transcription becomes activated across tens of thousands of loci (22). Here, we show that B2 SINEs are also strongly induced in an RNAPIII-dependent manner in reactivated B cells and primary bone marrow-derived macrophages, confirming that B2 activation is a prominent feature of MHV68 infection in physiologically relevant cell types. Induction of B2 SINEs occurs in a cell-autonomous manner, and they are not activated in uninfected cells via paracrine signaling. Furthermore, our data suggest that B2 induction is not a downstream product of antiviral signaling upon MHV68 infection, nor does it involve Maf1, a key negative regulator of RNAPIII activity. Instead, we link B2 activation to the conserved herpesviral serine/threonine protein kinase ORF36, which is sufficient to activate B2 RNA on its own and contributes to robust B2 accumulation during MHV68 infection. We hypothesize that changes in chromatin modification contribute to ORF36-mediated B2 activation and that this activity is at least partially conserved in other herpesviral protein kinase homologs. Altogether, ORF36-dependent B2 expression benefits the virus, as B2 RNAs help promote viral gene expression and replication (21).
Several immune sensing pathways can become activated during lytic herpesvirus infection, and B2 induction in uninfected cells has been linked to various types of cell stress. TLRs 2, 3, and 9, as well as the DNA-sensing AIM-2-like receptor family, the MAVS-dependent RNA-recognition receptors Mda5 and RIG-I, and the type I interferon signaling pathway have all been implicated in the sensing of herpesviral infection (45, 47, 61–63). However, our data from a variety of pattern recognition receptor and pathway knockout BMDMs indicate that engagement of these innate immune signaling components is not the mechanism by which MHV68 infection activates B2 SINEs. Indeed, B2 induction is even more robust in these infected knockout cells compared to WT BMDMs, likely reflecting enhanced replication of the virus in the absence of intact antiviral signaling. The innate immune independence of B2 activation is in agreement with the timing of B2 induction, which initiates with delayed early kinetics and continually increases late in infection.
RNAPIII transcription is broadly impacted by Maf1, which binds and negatively regulates polymerase activity (9). Thus, if B2 SINE induction were due to inactivation of Maf1, then we anticipated that Maf1–/– cells would have high baseline levels of B2 SINE RNA that would not further increase upon MHV68 infection. However, we did not observe any increase in B2 SINE levels in mock-infected Maf1–/– cells, and MHV68 infection of these cells resulted in B2 SINE activation that was comparable to that in WT cells. These findings indicate that regulation of Maf1 does not influence MHV68-mediated B2 SINE activation. Consistent with this, a recent chromatin immunoprecipitation-sequencing study of RNAPIII occupancy in wild-type mouse liver found relatively few B2 SINEs and identified only ∼30 of these elements with increased RNAPIII occupancy in Maf1–/– mice (64). We did observe a slight increase in B2 SINE levels at 24 hpi in Maf1–/– compared to WT cells, suggesting quicker RNAPIII transcription kinetics due to broad loss of Maf1-mediated repression (Fig. 3D).
A partial MHV68 ORF library screen revealed ORF36 to be a robust inducer of B2 SINE transcription. ORF36 is an early transcript (51, 52), which is consistent with the kinetics of B2 induction and with our current and prior observations that inhibition of viral DNA replication and late gene expression does not block B2 activation (21). Like other CHPKs, ORF36 displays homology to the host-encoded cyclin-dependent kinases but is thought to have broader substrate specificity (57). Indeed, it has been reported to phosphorylate many targets, including the retinoblastoma protein, H2AX, and lamin A/C (40, 56). Additionally, ORF36 has kinase-independent functions such as inhibition of HDACs 1/2 (34) and IRF-3 (55), both of which are beneficial for productive infection. Given our results showing that pharmacological inhibition of HDACs 1/2 and SU(VAR)3-9 stimulated B2 induction, we favor the hypothesis that ORF36 activities related to chromatin remodeling underlie its B2 induction phenotype. This would be in line with previous work in uninfected cells demonstrating that DNA CpG methylation and histone H3 trimethylation (H3K9me3) contribute to transcriptional repression of SINE loci (11, 12, 58, 59). The observation that the ORF36 kinase-null viral mutant was as defective as the ORF36 stop mutant for B2 induction indicates that while ORF36 modulation of HDACs 1/2 may contribute to such chromatin remodeling, this kinase-independent function of ORF36 is not the primary driver of B2 induction during infection. Instead, it may facilitate sustained B2 activation following a kinase-dependent initial activation event.
Whether ORF36 impacts SU(VAR)3-9 methyltransferases is unknown, although phospho-proteomics analysis of the EBV CHPK, BGLF4, suggests that SU(VAR)3-9h2 is phosphorylated in a BGLF4-dependent manner (65). An intriguing possibility is that ORF36 inhibits SU(VAR)3-9 function, through either direct phosphorylation of SU(VAR)3-9 or manipulation of an upstream regulator such as its repressor DBC1 (66). Additionally, recruitment of heterochromatin protein 1 (HP1) to H3K9me3 marks is dependent on HDAC activity (67), providing another link between these chromatin regulatory factors. Future experiments will be geared toward exploring epigenetic alterations to the host genome during MHV68 infection that could influence RNAPIII transcription.
The viral protein kinases are emerging as important players in gammaherpesvirus-associated lymphomagenesis, and an intriguing possibility is that its activation of Pol III retrotransposons—which are known to cause insertional mutagenesis (68–70)—may contribute to this phenotype. Indeed, prolonged expression of the ORF36 homolog in EBV (BGLF4) can contribute to genome instability leading to tumor formation, which has been linked to its phosphorylation of lamin A/C and topoisomerase-II (71, 72). KSHV ORF36 also displays functions associated with oncogenesis, including functional mimicry of the cellular ribosomal protein S6 kinase β-1 (S6KB1), which leads to enhanced protein synthesis, endothelial capillary tubule formation, and anchorage-independent growth (73). Notably, a recent study from the Damania lab showed that transgenic mice expressing KSHV ORF36 display increased B cell activation and develop high-grade B cell lymphomas that share many features of primary effusion lymphoma (74). In this regard, it is notable that among the vPK homologs, EBV BGLF4 and KSHV ORF36 showed the highest degree of B2 activation. The extent to which Pol III activation contributes to these oncogenic phenotypes, as well as whether MHV68 ORF36 also contributes to lymphomagenesis are important questions for the future.
MHV68 viral mutants lacking ORF36 or expressing a kinase-null version of the protein displayed a partial reduction in B2 RNA accumulation relative to WT virus. These results suggest that while ORF36 contributes to B2 induction during infection, one or more other viral activities may be involved. Our ORF screen encompassed a significant percentage of the annotated MHV68 genome (75); however, it should be noted that recent work from O’Grady et al. (76) shows pervasive alternate isoform usage overlapping ORF isoforms, suggesting that MHV68 encodes a more diverse proteome than previously anticipated. One or more of these untested proteins may also contribute to B2 induction, either via independent mechanisms or in cooperation with ORF36. Other MHV68-encoded ORFs involved in B2 SINE transcription and stabilization remain an open area of investigation.
In summary, our results provide the first insights into how gammaherpesvirus infection induces SINE retrotransposons and identify a novel activity of the ORF36 protein kinase. Our work supports a model in which ORF36 functions to inhibit proteins involved in the maintenance of a repressive chromatin landscape. This may occur through both kinase-dependent and kinase-independent mechanisms, resulting in derepression of B2 SINE loci. How these activities selectively impact certain RNAPIII loci remains a key open question. Indeed, ongoing work to define how SINEs and other RNAPIII transcripts are activated during infection, as well as noncanonical functions of these ncRNAs, should provide insight into the emerging field of retrotransposon-linked cell signaling. Given the breadth of DNA viruses that activate these hyperabundant loci, viruses will continue to serve as unique tools to dissect the regulation of ncRNAs, as well as the mechanisms by which they influence the outcome of infection.
MATERIALS AND METHODS
Cells.
NIH 3T3 mouse fibroblasts were obtained from the UC Berkeley Cell Culture Facility and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal calf serum (FBS; Seradigm). A20 B cells were maintained in RPMI (Gibco), 10% fetal bovine serum (FBS; VWR), 2 mM l-glutamin, 100 U/ml penicillin,100 mg/ml streptomycin, and 50 mM β-mercaptoethanol (BME). A20-HE-RIT cell lines (kindly provided by Laurie Krug) (39) were maintained under the same conditions as A20 cells, with the addition of 300 μg/ml hygromycin B, 300 μg/ml G418 and 2 μg/ml puromycin. To reactivate A20-HE-RIT, cells were cultured in medium without antibiotic selection for 24 h and then seeded at a cell density of 1e6 cells/ml in the presence of 5 μg/ml doxycycline and 20 ng/μl phorbol myristate acetate (PMA) for the indicated time. To block viral DNA replication, PAA was used at a concentration of 200 μg/ml and was added at the start of reactivation. Bone marrow-derived macrophages (BMDMs) containing knockouts for innate immune pathway components (48, 77–79) were kindly provided by the lab of Gregory Barton (UC Berkeley, Department of Immunology). Wild-type and Maf1 knockout BMDMs were differentiated as follows: femurs and tibias from C57BL/6J (B6) mice (43) aged 3 to 6 months were flushed with bone marrow medium plus antibiotics (BMM+A; high glucose DMEM + 10% FBS, + 10% MCSF + 1% PenStrep) using a 3-ml syringe with attached 23-gauge needle. Cell-containing medium was filtered through a 70-μM filter to remove debris. Cells were pelleted at 280 × g in an Allegra X-15R Beckman Coulter centrifuge for 5 minutes. Supernatant was removed by aspiration, and cells resuspended in BMM+A. Cells were counted using a hemocytometer and plated in non-tissue culture (TC)-treated 15-cm petri dishes (ref. no. 351058; Falcon) at a concentration of 10e6 cells/25 ml BMM+A/plate. On day 3 of differentiation, 5 ml BMM+A was added to each plate to feed cells. On day 7 of differentiation, BMM+A was aspirated and replaced with 10 ml cold Dulbecco’s phosphate-buffered saline (DPBS; Invitrogen) per plate and placed at 4°C for 10 min. Cells were then lightly scraped from each plate and collected, pelleted as previously mentioned, and resuspended in bone marrow medium without antibiotics (BMM) containing 10% DMSO at a concentration of 10e6/ml. Then, 1.5-ml CryoTube vials containing 1 ml/10e6 BMDMs were frozen at –80°C for 24 h before being stored in liquid nitrogen for the duration. Subsequently, thawed vials of BMDMs were maintained in BMM except during infections. Experiments involving mice were performed under a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the Albert Einstein College of Medicine.
Transfections.
Transfection of BMDMs with Brf1 or control siRNA was completed as follows: BMDM cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal calf serum (FBS; Seradigm). Prior to siRNA transfection, BMDMs were grown for 3 days in 15-cm TC-treated plates to 90% confluence. Cells were removed and washed in DPBS twice by spinning at 475 × g for 5 minutes each time. Transfection of siRNA was done using the Neon transfection system (Thermo Fisher) as follows: for each condition, 2e6 cells were resuspended in buffer R at a concentration of 1e6 cells/100 μl. To this, 200 nM (assuming a final culture volume of 2 ml medium) siRNA was added from 100 μM stock or either control (ON-TARGETplus nontargeting control pool; Dharmacon) or Brf1 siRNA (SMARTpool; ON-TARGETplus mouse Brf1 siRNA; Dharmacon). Then, 100 μl Neon transfection system tips were used to transfect siRNA into cells as follows: parameters set for BMDMs were 1,680/20/1 (pulse/length/width). Cells were then quickly removed to 2 ml total medium and plated at 2e6 cells/plate in 60-mm TC-treated plates, before being placed at 37°C to incubate. Transfection of NIH 3T3 cells with MHV68 ORF library plasmids and ORF36 WT and mutant plasmids was completed as follows: NIH 3T3s were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% fetal calf serum (FBS; Seradigm). Prior to transfection, cells were maintained at parameters suggested in the PolyJet in vitro DNA transfection reagent protocol (http://signagen.com/DataSheet/SL100688.pdf). Guidelines for the advanced protocol for transfecting hard-to-transfect mammalian cells was followed strictly. Briefly, 2.7 × 106 cells were transfected with 5 μg of plasmid DNA for each sample. In cases of multiple ORF transfections, total plasmid DNA levels were not to exceed 25 μg.
Plasmids and cloning.
MHV68 ORF library plasmids were generously provided by the lab of Ren Sun (University of California Los Angeles), and their construction was previously described (50). For generation of the ORF36 kinase-null mutant, the K107Q mutation was introduced using QuikChange PCR with the following primers: 5′-GTGCTGTCAATTTTGGGATATACTGTATGCAGAGCGTGTCATCTGAT-3′ and 5′-ATCAGATGACACGCTCTGCATACAGTATATCCCAAAATTGACAGCAC-3′. Plasmids for conserved herpesvirus protein kinase homologs of ORF36 were purchased through Addgene from the laboratory of Robert Kalejta (https://www.addgene.org/Robert_Kalejta/) (56).
Virus preparation and infections.
MHV68 containing a stop mutation or kinase-null mutation in ORF36 as well as the corresponding mutant rescue virus were generously provided by Vera Tarakanova (Medical College of Wisconsin) (54). MHV68 was amplified in NIH 3T12 fibroblast cells, and the viral 50% tissue culture infective dose (TCID50) was measured on NIH 3T3 fibroblasts by limiting dilution. NIH 3T3 fibroblasts were infected at the indicated multiplicity of infection (MOI) by adding the required volume of virus to cells in 1 ml total volume (for each well of a 6-well plate), 2 ml total volume (for 6-cm plates), or 5 ml (for 10-cm plates). Infection was allowed to proceed for 45 min prior to removal of virus medium and replacement with DMEM plus 10% FBS. BMDMs were infected with the minimal volume of MHV68 required to achieve maximum infection (20% to 30%), as determined by titration experiments with GFP-marked MHV68 followed by flow cytometry for GFP. For infection of BMDMs, virus was added to cells in serum-free DMEM for 4 h in non-TC-treated plates. Virus-containing medium was then aspirated and replaced with macrophage medium without antibiotics.
RT-qPCR and primer extension.
Total RNA was extracted from cells using TRIzol reagent (Invitrogen). For RT-qPCR analysis, RNA was treated with Turbo DNase (Ambion) and reverse transcribed with avian myeloblastosis virus reverse transcriptase (AMV RT) (Promega) primed with random 9-mers. RT-qPCR was performed with iTaq universal SYBR green Supermix (Bio-Rad) using the following primers: (7SK_F: CCCCTGCTAGAACCTCCAAAC, 7SK_R: CACATGCAGCGCCTCATTT, U6_F: CGCTTCGGCAGCACATATAC, U6_R: AAAATATGGAACGCTTCACGA, 5S_F: TCTCGTCTGATCTCGGAAGC, 5S_R: AGCCTACAGCACCCGGTATT, 7SL_F: ATCGGGTGTCCGCACTAAGTT, 7SL_R: CAGCACGGGAGTTTTGACCT). The fold change was calculated using the ΔΔCT method. Primer extension was performed on 10 to 15 μg of total RNA using a 5′ fluorescein labeled oligo specific for B2 SINEs or 7SK. RNA was ethanol precipitated in 1 ml 100% EtOH, washed in 70% ethyl alcohol (EtOH), and pelleted at 21,130 × g and 4°C for 10 min. Pellets were resuspended in 9 μl annealing buffer (10 mM Tris-HCl, pH 7.5, 0.3 M KCl, 1 mM EDTA) containing 1 μl of (10 pmol/μl) 5′-fluorescein labeled primer (B2 SINE: TACACTGTAGCTGTCTTCAGACA, 7SK: GAGCTTGTTTGGAGGTTCT; Integrated DNA Technologies). Samples were heated briefly to 95°C for 2 min, followed by annealing for 1 h at 55°C. Then, 40 μl of extension buffer (10 mM Tris-HCl, pH 8.8, 5 mM MgCl2, 5 mM DTT, 1 mM deoxynucleoside triphosphate [dNTP]), and 1 μl AMV reverse transcriptase (Promega) were added, and extension was carried out for 1 h at 42°C. Samples were EtOH precipitated, and then pellets were briefly air dried and resuspended in 20 μl 1× RNA loading dye (47.5% formamide, 0.01% SDS, 0.01% bromophenol blue, 0.005% xylene cyanol, and 0.5 mM EDTA). Then, 10 μl of each sample was run on an 8% urea-PAGE gel for 1 h at 250 V. Gels were imaged on a Bio-Rad Chemidoc with fluoroscein imaging capability.
Protein extraction and analysis.
Cells were washed with cold DPBS once before being lysed with RIPA lysis buffer (50 mM Tris HCl, 150 mM NaCl, 1.0% [vol/vol] NP-40, 0.5% [wt/vol] sodium deoxycholate, 1.0 mM EDTA, and 0.1% [wt/vol] SDS). Cell lysates were vortexed briefly, rotated at 4°C for 1 h, and then spun at 18,000 × g in a tabletop centrifuge at 4°C for 12 min to remove debris.
For Western blot analyses, 30 μg of whole-cell lysate was resolved with 4 to 15% mini-PROTEAN TGX gels (Bio-Rad). Transfers to polyvinylidene difluoride (PVDF) membranes were done with the Trans-Blot Turbo transfer system (Bio-Rad). Blots were incubated in 5% milk/TBS plus 0.1% Tween 20 (TBST) to block, followed by incubation with primary antibodies against FLAG (Sigma F1804, 1:1,000), Brf1 (Bethyl a301-228a, 1:1,000), hemagglutinin (HA) (Sigma H9658, 1:1,000), TUBA1A (abcam ab729, 1:1,000), or GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam ab8245, 1:1,000) in 5% milk/TBST. Washes were carried out in TBST. Blots were then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Southern Biotechnology, 1:5,000). Washed blots were incubated with Clarity Western ECL substrate (Rio-Rad) for 5 min and visualized with a Bio-Rad ChemiDoc imager.
Inhibitor treatment.
Cells were plated 12 h before inhibitor treatment to achieve 70% confluence at time of treatment. ACY-957 (MedChemExpress HY-104008), 5-azacytidine (Sigma A2385), and chaetocin (Cayman Chemicals 13156), were resuspended with DMSO prior to treatment. Inhibitors were diluted to working concentrations in warmed DMEM plus 10% FBS before addition to cells. Pretreatment of cells with inhibitor-containing medium preceded infection with MHV68 by 1 h. Upon removal of virus-containing medium, inhibitor-containing medium was replaced onto cells.
ACKNOWLEDGMENTS
We thank Ren Sun and Ting-Ting Wu (University of California Los Angeles) for providing their MHV68 ORF library, Vera Tarakanova (Medical College of Wisconsin) for providing ORF36.stop and kinase-null MHV68 and for helpful comments on the manuscript, and the labs of Greg Barton and Russell Vance (University of California Berkeley) for sharing immune factor-knockout macrophages.
This work was funded by National Institutes of Health grants AI147183, AI122528, and CA136367 to B.A.G. and GM120358 to I.W. A.M.S. was funded by a graduate research fellowship from the National Science Foundation (DGE 1752814). B.A.G. is an investigator at the Howard Hughes Medical Institute.
REFERENCES
- 1.Kramerov DA, Vassetzky NS. 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 107:487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kramerov DA, Vassetzky NS. 2011. SINEs. Wiley Interdiscip Rev RNA 2:772–786. doi: 10.1002/wrna.91. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su M, Han D, Boyd-Kirkup J, Yu X, Han J-DJ. 2014. Evolution of Alu elements toward enhancers. Cell Rep 7:376–385. doi: 10.1016/j.celrep.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Ferrigno O, Virolle T, Djabari Z, Ortonne JP, White RJ, Aberdam D. 2001. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet 28:77–81. doi: 10.1038/ng0501-77. [DOI] [PubMed] [Google Scholar]
- 7.Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 9.Willis IM, Moir RD. 2018. Signaling to and from the RNA polymerase III transcription and processing machinery. Annu Rev Biochem 87:75–100. doi: 10.1146/annurev-biochem-062917-012624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N. 2010. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. 2015. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun 6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu WM, Maraia RJ, Rubin CM, Schmid CW. 1994. Alu transcripts: cytoplasmic localisation and regulation by DNA methylation. Nucleic Acids Res 22:1087–1095. doi: 10.1093/nar/22.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu WM, Chu WM, Choudary PV, Schmid CW. 1995. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res 23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- 16.Singh K, Carey M, Saragosti S, Botchan M. 1985. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature 314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- 17.Williams WP, Tamburic L, Astell CR. 2004. Increased levels of B1 and B2 SINE transcripts in mouse fibroblast cells due to minute virus of mice infection. Virology 327:233–241. doi: 10.1016/j.virol.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 18.Jang KL, Latchman DS. 1989. HSV infection induces increased transcription of Alu repeated sequences by RNA polymerase III. FEBS Lett 258:255–258. doi: 10.1016/0014-5793(89)81667-9. [DOI] [PubMed] [Google Scholar]
- 19.Panning B, Smiley JR. 1995. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J Mol Biol 248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 20.Panning B, Smiley JR. 1994. Activation of RNA polymerase III transcription of human Alu elements by herpes simplex virus. Virology 202:408–417. doi: 10.1006/viro.1994.1357. [DOI] [PubMed] [Google Scholar]
- 21.Karijolich J, Abernathy E, Glaunsinger BA. 2015. Infection-induced retrotransposon-derived noncoding RNAs enhance herpesviral gene expression via the NF-κB pathway. PLoS Pathog 11:e1005260. doi: 10.1371/journal.ppat.1005260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karijolich J, Zhao Y, Alla R, Glaunsinger B. 2017. Genome-wide mapping of infection-induced SINE RNAs reveals a role in selective mRNA export. Nucleic Acids Res 45:6194–6208. doi: 10.1093/nar/gkx180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang JJ, Sparrer KMJ, van Gent M, Lässig C, Huang T, Osterrieder N, Hopfner K-P, Gack MU. 2018. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat Immunol 19:53–62. doi: 10.1038/s41590-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Y, Ye X, Dunker W, Song Y, Karijolich J. 2018. RIG-I like receptor sensing of host RNAs facilitates the cell-intrinsic immune response to KSHV infection. Nat Commun 9:4841. doi: 10.1038/s41467-018-07314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong X, Feng H, Sun Q, Li H, Wu T-T, Sun R, Tibbetts SA, Chen ZJ, Feng P. 2010. Murine gamma-herpesvirus 68 hijacks MAVS and IKKbeta to initiate lytic replication. PLoS Pathog 6:e1001001. doi: 10.1371/journal.ppat.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Karikó K, Yoo JW, Lee D-K, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJC, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MMW, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJC, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, Ogura Y, Yoo JW, Lee D-K, Provost P, Hinton DR, Núñez G, Baffi JZ, Kleinman ME, Ambati J. 2012. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright CB, Kim Y, Yasuma T, Li S, Fowler BJ, Kleinman ME, Ambati J, Gelfand BD. 2014. Enhanced Alu RNA stability due to iron-mediated DICER1 impairment causes NLRP3 inflammasome priming. Invest Ophthalmol Vis Sci 55:2187–2187. https://arvojournals.org/article.aspx?articleid=2267504. [Google Scholar]
- 29.Kim Y, Tarallo V, Kerur N, Yasuma T, Gelfand BD, Bastos-Carvalho A, Hirano Y, Yasuma R, Mizutani T, Fowler BJ, Li S, Kaneko H, Bogdanovich S, Ambati BK, Hinton DR, Hauswirth WW, Hakem R, Wright C, Ambati J. 2014. DICER1/Alu RNA dysmetabolism induces Caspase-8-mediated cell death in age-related macular degeneration. Proc Natl Acad Sci U S A 111:16082–16087. doi: 10.1073/pnas.1403814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida H, Matsushita T, Kimura E, Fujita Y, Keany R, Ikeda T, Toshimori M, Imanaka T, Nakamura M. 2019. Systemic expression of Alu RNA in patients with geographic atrophy secondary to age-related macular degeneration. PLoS One 14:e0220887. doi: 10.1371/journal.pone.0220887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Ruocco F, Basso V, Rivoire M, Mehlen P, Ambati J, De Falco S, Tarallo V. 2018. Alu RNA accumulation induces epithelial-to-mesenchymal transition by modulating miR-566 and is associated with cancer progression. Oncogene 37:627–637. doi: 10.1038/onc.2017.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espinoza CA, Goodrich JA, Kugel JF. 2007. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 13:583–596. doi: 10.1261/rna.310307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mounce BC, Mboko WP, Bigley TM, Terhune SS, Tarakanova VL. 2013. A conserved gammaherpesvirus protein kinase targets histone deacetylases 1 and 2 to facilitate viral replication in primary macrophages. J Virol 87:7314–7325. doi: 10.1128/JVI.02713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mounce BC, Mboko WP, Kanack AJ, Tarakanova VL. 2014. Primary macrophages rely on histone deacetylase 1 and 2 expression to induce type I interferon in response to gammaherpesvirus infection. J Virol 88:2268–2278. doi: 10.1128/JVI.03278-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flaño E, Kim I-J, Woodland DL, Blackman MA. 2002. Gamma-herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med 196:1363–1372. doi: 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarousse N, Chandran B, Coscoy L. 2008. Lack of heparan sulfate expression in B-cell lines: implications for Kaposi’s sarcoma-associated herpesvirus and murine gammaherpesvirus 68 infections. J Virol 82:12591–12597. doi: 10.1128/JVI.01167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usherwood EJ, Stewart JP, Nash AA. 1996. Characterization of tumor cell lines derived from murine gammaherpesvirus-68-infected mice. J Virol 70:6516–6518. doi: 10.1128/JVI.70.9.6516-6518.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forrest JC, Speck SH. 2008. Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade. J Virol 82:7688–7699. doi: 10.1128/JVI.02689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santana AL, Oldenburg DG, Kirillov V, Malik L, Dong Q, Sinayev R, Marcu KB, White DW, Krug LT. 2017. RTA occupancy of the origin of lytic replication during murine gammaherpesvirus 68 reactivation from B cell latency. Pathogens 6:9. doi: 10.3390/pathogens6010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mounce BC, Tsan FC, Droit L, Kohler S, Reitsma JM, Cirillo LA, Tarakanova VL. 2011. Gammaherpesvirus gene expression and DNA synthesis are facilitated by viral protein kinase and histone variant H2AX. Virology 420:73–81. doi: 10.1016/j.virol.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummins D, Doran TJ, Tyack S, Purcell D, Hammond J. 2008. Identification and characterisation of the porcine 7SK RNA polymerase III promoter for short hairpin RNA expression. J RNAi Gene Silencing 4:289–294. [PMC free article] [PubMed] [Google Scholar]
- 42.Boguta M. 2013. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta 1829:376–384. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 43.Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Marcelin G, Chua SC Jr, Martinez-Lopez N, Singh R, Moullan N, Auwerx J, Willemin G, Shah H, Hartil K, Vaitheesvaran B, Kurland I, Hernandez N, Willis IM. 2015. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev 29:934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uppal T, Sarkar R, Dhelaria R, Verma SC. 2018. Role of pattern recognition receptors in KSHV infection. Cancers 10:85. doi: 10.3390/cancers10030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bussey KA, Murthy S, Reimer E, Chan B, Hatesuer B, Schughart K, Glaunsinger B, Adler H, Brinkmann MM. 2018. Endosomal Toll-like receptors 7 and 9 cooperate in detection of murine gammaherpesvirus 68 infection. J Virol 93:e01173-18. doi: 10.1128/JVI.01173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Dittmer DP, Mieczkowski PA, Host KM, Fusco WG, Duncan JA, Damania B. 2018. RIG-I detects Kaposi’s sarcoma-associated herpesvirus transcripts in a RNA polymerase III-independent manner. mBio 9:e00823-18 doi: 10.1128/mBio.00823-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. 2011. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol 11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer J-D, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun 79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H-W, King K, Tu J, Sanchez M, Luster AD, Shresta S. 2013. The roles of IRF-3 and IRF-7 in innate antiviral immunity against dengue virus. J Immunol 191:4194–4201. doi: 10.4049/jimmunol.1300799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S, Salwinski L, Zhang C, Chu D, Sampankanpanich C, Reyes NA, Vangeloff A, Xing F, Li X, Wu T-T, Sahasrabudhe S, Deng H, LaCount DJ, Sun R. 2011. An integrated approach to elucidate the intra-viral and viral-cellular protein interaction networks of a gamma-herpesvirus. PLoS Pathog 7:e1002297. doi: 10.1371/journal.ppat.1002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, Nash AA. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J Gen Virol 84:99–109. doi: 10.1099/vir.0.18639-0. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Guzman D, Rickabaugh T, Wu T-T, Brown H, Cole S, Song MJ, Tong L, Sun R. 2003. Transcription program of murine gammaherpesvirus 68. J Virol 77:10488–10503. doi: 10.1128/jvi.77.19.10488-10503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romaker D, Schregel V, Maurer K, Auerochs S, Marzi A, Sticht H, Marschall M. 2006. Analysis of the structure-activity relationship of four herpesviral UL97 subfamily protein kinases reveals partial but not full functional conservation. J Med Chem 49:7044–7053. doi: 10.1021/jm060696s. [DOI] [PubMed] [Google Scholar]
- 54.Tarakanova VL, Leung-Pineda V, Hwang S, Yang C-W, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW. 2007. γ-Herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe 1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang S, Kim KS, Flano E, Wu T-T, Tong LM, Park AN, Song MJ, Sanchez DJ, O’Connell RM, Cheng G, Sun R. 2009. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe 5:166–178. doi: 10.1016/j.chom.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuny CV, Chinchilla K, Culbertson MR, Kalejta RF. 2010. Cyclin-dependent kinase-like function is shared by the beta- and gamma- subset of the conserved herpesvirus protein kinases. PLoS Pathog 6:e1001092. doi: 10.1371/journal.ppat.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacob T, Van den Broeke C, Favoreel HW. 2011. Viral serine/threonine protein kinases. J Virol 85:1158–1173. doi: 10.1128/JVI.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kondo Y, Issa J-PJ. 2003. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J Biol Chem 278:27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 59.Ichiyanagi K, Li Y, Li Y, Watanabe T, Ichiyanagi T, Fukuda K, Kitayama J, Yamamoto Y, Kuramochi-Miyagawa S, Nakano T, Yabuta Y, Seki Y, Saitou M, Sasaki H. 2011. Locus- and domain-dependent control of DNA methylation at mouse B1 retrotransposons during male germ cell development. Genome Res 21:2058–2066. doi: 10.1101/gr.123679.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vabret N, Najburg V, Solovyov A, Šulc P, Balan S. 2019. Y-RNAs lead an endogenous program of RIG-I agonism mobilized upon RNA virus infection and targeted by HIV. bioRxiv https://www.biorxiv.org/content/10.1101/773820v1.
- 61.Guggemoos S, Hangel D, Hamm S, Heit A, Bauer S, Adler H. 2008. TLR9 contributes to antiviral immunity during gammaherpesvirus infection. J Immunol 180:438–443. doi: 10.4049/jimmunol.180.1.438. [DOI] [PubMed] [Google Scholar]
- 62.Michaud F, Coulombe F, Gaudreault E, Kriz J, Gosselin J. 2010. Involvement of TLR2 in recognition of acute gammaherpesvirus-68 infection. PLoS One 5:e13742. doi: 10.1371/journal.pone.0013742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mboko WP, Mounce BC, Emmer J, Darrah E, Patel SB, Tarakanova VL. 2014. Interferon regulatory factor 1 restricts gammaherpesvirus replication in primary immune cells. J Virol 88:6993–7004. doi: 10.1128/JVI.00638-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bonhoure N, Praz V, Moir RD, Willemin G, Mange F. 2019. Chronic repression by MAF1 supports futile RNA cycling as a mechanism for obesity resistance. BioRxiv https://www.biorxiv.org/content/10.1101/775353v1.
- 65.Li R, Liao G, Nirujogi RS, Pinto SM, Shaw PG, Huang T-C, Wan J, Qian J, Gowda H, Wu X, Lv D-W, Zhang K, Manda SS, Pandey A, Hayward SD. 2015. Phosphoproteomic profiling reveals Epstein-Barr virus protein kinase integration of DNA damage response and mitotic signaling. PLoS Pathog 11:e1005346. doi: 10.1371/journal.ppat.1005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. 2009. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem 284:10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vaute O, Nicolas E, Vandel L, Trouche D. 2002. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res 30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cajuso T, Sulo P, Tanskanen T, Katainen R, Taira A, Hänninen UA, Kondelin J, Forsström L, Välimäki N, Aavikko M, Kaasinen E, Ristimäki A, Koskensalo S, Lepistö A, Renkonen-Sinisalo L, Seppälä T, Kuopio T, Böhm J, Mecklin J-P, Kilpivaara O, Pitkänen E, Palin K, Aaltonen LA. 2019. Retrotransposon insertions can initiate colorectal cancer and are associated with poor survival. Nat Commun 10:4022. doi: 10.1038/s41467-019-11770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anwar SL, Wulaningsih W, Lehmann U. 2017. Transposable elements in human cancer: causes and consequences of deregulation. Int J Mol Sci 10:E974. doi: 10.3390/ijms18050974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott EC, Devine SE. 2017. The role of somatic L1 retrotransposition in human cancers. Viruses 9:131. doi: 10.3390/v9060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang C-Y, Lee C-H, Wu C-C, Chang Y-T, Yu S-L, Chou S-P, Huang P-T, Chen C-L, Hou J-W, Chang Y, Tsai C-H, Takada K, Chen J-Y. 2009. Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int J Cancer 124:2016–2025. doi: 10.1002/ijc.24179. [DOI] [PubMed] [Google Scholar]
- 72.Negrini S, Gorgoulis VG, Halazonetis TD. 2010. Genomic instability: an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 73.Anders PM, Montgomery ND, Montgomery SA, Bhatt AP, Dittmer DP, Damania B. 2018. Human herpesvirus-encoded kinase induces B cell lymphomas in vivo. J Clin Invest 128:2519–2534. doi: 10.1172/JCI97053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhatt AP, Wong JP, Weinberg MS, Host KM, Giffin LC, Buijnink J, van Dijk E, Izumiya Y, Kung H-J, Temple BRS, Damania B. 2016. A viral kinase mimics S6 kinase to enhance cell proliferation. Proc Natl Acad Sci U S A 113:7876–7881. doi: 10.1073/pnas.1600587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Virgin HW 4th, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, Speck SH. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol 71:5894–5904. doi: 10.1128/JVI.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Grady T, Feswick A, Hoffman BA, Wang Y, Medina EM, Kara M, van Dyk LF, Flemington EK, Tibbetts SA. 2019. Genome-wide transcript structure resolution reveals abundant alternate isoform usage from murine gammaherpesvirus 68. Cell Rep 27:3988–4002.e5. doi: 10.1016/j.celrep.2019.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seth RB, Sun L, Ea C-K, Chen ZJ. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 78.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 79.Cai S, Batra S, Shen L, Wakamatsu N, Jeyaseelan S. 2009. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J Immunol 183:6629–6638. doi: 10.4049/jimmunol.0901033. [DOI] [PMC free article] [PubMed] [Google Scholar]