Abstract
Actin is one of the most abundant and essential intracellular proteins that mediates nearly every form of cellular movement and underlies such key processes as embryogenesis, tissue integrity, cell division and contractility of all types of muscle and non-muscle cells. In mammals, actin is represented by six isoforms, which are encoded by different genes but produce proteins that are 95-99% identical to each other. The six actin genes have vastly different functions in vivo, and the small amino acid differences between the proteins they encode are rigorously maintained through evolution, but the underlying differences behind this distinction, as well as the importance of specific amino acid sequences for each actin isoform, are not well understood. This review summarizes different levels of actin isoform-specific regulation in cellular and developmental processes, starting with the nuclear actin’s role in transcription, and covering the gene-level, mRNA-level, and protein-level regulation, with a special focus on mammalian actins in non-muscle cells.
Introduction
Actin is one of the most abundant and essential intracellular proteins, highly conserved throughout the kingdoms of life. In mammals, actin is represented by six nearly identical isoforms that are encoded by different genes and are differentially expressed in tissues and organs throughout the body (Vandekerckhove and Weber, 1978). Globally, these six actins mediate nearly every form of cellular movement and underlie such key processes as embryogenesis, tissue integrity, cell division and contractility of all types of muscle and non-muscle cells.
Most of actin’s intracellular functions are tightly linked to its ability to self-associate into dynamic polymers, actin filaments, that form the structural basis of myofibrils in the muscle and the cytoskeleton in non-muscle cells. Through polymerization/depolymerization cycles, actin exists in a constant balance between monomeric (G-actin) and polymeric (F-actin) pool, tightly regulated by a variety of mechanisms to achieve different actin distribution and highly diverse functioning throughout the body (see, e.g., (Dominguez and Holmes, 2011; Pollard, 2016) for recent reviews). In addition to its function in the cytoplasm, actin is also found in the nucleus, where it can also undergo polymerization and depolymerization (recently reviewed in (Plessner and Grosse, 2019)) and has been shown to directly regulate transcription and participate in chromosome positioning, DNA rearrangements, and DNA repair (Kristo et al., 2016; Virtanen and Vartiainen, 2017).
One of the biggest mysteries around actin relates to the ways this highly conserved protein can incorporate into a variety of intracellular networks and simultaneously carry out a vast multitude of different functions. Some of this diversity relies on the tissue specificity of the actin genes (e.g., muscle versus non-muscle), however it is unclear how the highly similar proteins encoded by the actin genes (95-99% identical at the amino acid level), often present in comparable quantities in the same cell types, can preferentially incorporate into different actin structures (e.g., myofibrils versus cortical network) and mediate different cellular functions. Even more strikingly, a single actin isoform within the same cell type is believed to be able to simultaneously incorporate into different structures and networks and show distinct structural and functional segregation in response to different physiological stimuli. This differential regulation and the mechanisms behind actins isoform-specific functions have constituted an emerging area of interest in the past few years.
Many actin functions are believed to be mediated almost entirely by regulated interaction with hundreds of intracellular binding partners (Lee and Dominguez, 2010), often compounded by direct regulation of the properties of the actin monomer by a variety of posttranslational modifications (Terman and Kashina, 2013). It is assumed that these binding partners and modifying enzymes must be strictly compartmentalized within the cell to ensure precise coordination of the entire actin network, but this segregation itself is only partially characterized. Some key aspects of actin regulation arise at the mRNA, rather than the protein, level and are encoded by the nucleotide, rather than the amino acid sequence (Vedula et al., 2017; Zhang et al., 2010). This nucleotide-dependent regulation potentially extends into both coding and noncoding regions of the actin gene.
This review summarizes different levels of actin isoform regulation in cellular and developmental processes, with a special focus on mammalian actins in non-muscle cells.
Transcription regulation of cytoskeleton by the nuclear actin
For a very long time since its original discovery, actin has been considered a purely cytoplasmic protein, functionally restricted to the different cytoskeletal structures throughout cells and tissues. While a nuclear pool of actin has been initially observed over five decades ago (Ohnishi et al., 1963), this finding has been a highly debated topic for many years (reviewed in (Kelpsch and Tootle, 2018)). Subsequent studies found that actin is actively imported into the nucleus, and that its nuclear import/export can play dramatic roles in regulation of multiple cellular responses to global changes in the environment and cell’s physiological state (Dopie et al., 2012; Skarp et al., 2013; Skarp and Vartiainen, 2013; Viita and Vartiainen, 2017). Even more recently, it has been found that nuclear actin, similarly to the cytoplasmic actin, can form filaments, and changes in their assembly and disassembly have been proposed to underlie some of the actin’s nuclear roles (reviewed in (Plessner and Grosse, 2019)).
Nuclear actin’s regulatory roles in response to stimuli are tightly linked to its regulated nuclear import and export, which occurs with actin monomers and requires their sequestering by cofilin (for nuclear import) and profilin (for nuclear export). Cofilin provides a bipartite nuclear localization sequence (NLS), which is not present in actin itself, and the actin-cofilin complex interacts with importin 9 for nuclear entry (Viita and Vartiainen, 2017). This influx of actin monomers into the nucleus is counterbalanced by nuclear export via exportin 6 (Bohnsack et al., 2006; Dopie et al., 2012; Stuven et al., 2003), which is believed to involve the nuclear export sequences present in the actin itself (Wada et al., 1998), but also requires the presence of actin monomer binding to profilin. Thus, nuclear actin undergoes constant regulated exchange with the cytoplasmic actin pool ((Gieni and Hendzel, 2009; Skarp and Vartiainen, 2013)) that involves the activity of the monomer sequestering proteins (reviewed in (Hurst et al., 2019; Kelpsch and Tootle, 2018; Miyamoto and Gurdon, 2013)).
Inside the nucleus, actin is believed to play important roles in chromatin organization, DNA repair, RNA processing and export, chromatin remodeling and architecture, nuclear envelope assembly, and regulation of transcription. All of these functions are tightly linked to the balance between nuclear actin monomers and polymers in the nucleus (G- and F-actin, respectively), as well as – indirectly – to actin polymerization/depolymerization in the cytoplasm, which affect nuclear import/export. Thus, nuclear actin functions as an intricate part of the overall actin’s cellular homeostasis (reviewed in (Artman et al., 2014; Castano et al., 2010; Falahzadeh et al., 2015; Miyamoto and Gurdon, 2013; Percipalle, 2013; Wesolowska and Lenart, 2015)).
One of the best known examples of actin-dependent transcription regulation is its involvement in serum response, which directly links an increase in actin polymerization (and the resulting decrease in G-to-F actin ratio) to an increase in serum response factor (SRF)-mediated transcription. In this pathway, actin-dependent nuclear targeting of SRF1 coactivator, Myocardin-Related Transcription Factor A (MRTF-A) drives the expression of many cytoskeletal genes to facilitate the large-scale cytoskeletal rearrangements in response to changes in extracellular serum (Baarlink et al., 2013; Esnault et al., 2014; Olson and Nordheim, 2010; Vartiainen et al., 2007)). MRTF-A is actively transported into the nucleus when actin is highly polymerized, and can be prevented from nuclear import by binding to the cytosolic G-actin at an increase in actin disassembly, thus inhibiting serum response. On the nuclear side, this pathway is counteracted by mDia-dependent polymerization of nuclear actin, which can inhibit G-actin dependent MRTF-A nuclear export. Thus, actin in both nuclear and cytosolic compartments regulates transcriptional activation during serum response in a manner directly linked to its polymerization state (reviewed in (Virtanen and Vartiainen, 2017)).
Nuclear G-actin also regulates p53 and Hippo pathways by binding to JMY (reviewed in (Rajakyla and Vartiainen, 2014; Wesolowska and Lenart, 2015)), and nuclear F-actin, in addition to its role in balancing the G-actin concentration, can directly bind transcription regulatory complexes in response to retinoic acid (Ferrai et al., 2009). Intracellular levels of monomeric β–actin have been shown to regulate its own transcription (Lyubimova et al., 1997) through a signal located in the 3’ UTR of β– actin mRNA (Lyubimova et al., 1999). The specific pathway for this direct autoregulation, proposed to be involved in the intracellular maintenance of actin levels, has not been definitively characterized. Thus, nuclear actin can exert direct effects on its own expression (Salvany et al., 2014), as well as on the expression of many related cytoskeletal genes, and these effects are directly linked to its abundance and polymerized state in the cytosol.
Notably, most of the studies of nuclear actin suggests that this actin pool is predominantly or exclusively restricted to only one isoform, β-actin (Falahzadeh et al., 2015; Hofmann et al., 2004; Hu et al., 2004). A recent study reported that some amount of γ-cytoplasmic actin is also present in the nucleus (Migocka-Patrzalek et al., 2015), but likely in lower quantities. This nuclear actin isoform specificity is not well characterized, and it is unclear what specific determinant(s) are responsible for this apparent isoform specificity, and whether this prevalence of only one isoform in the nucleus is cell type-dependent. These questions require further investigation and will likely shed light on functional distinctions of actin isoforms in vivo.
Gene-level regulation of actin isoform tissue specificity.
The six mammalian actin isoforms, while encoding nearly identical proteins, are highly different from each other at the gene level (Ampe and Van Troys, 2017; Erba et al., 1986; Perrin and Ervasti, 2010). Originally these isoforms were differentiated by their prevalence in different tissues (e.g., cardiac, skeletal, or smooth muscle, as well as non-muscle cells) (Perrin and Ervasti, 2010; Simiczyjew et al., 2017; Tondeleir et al., 2009) – e.g., skeletal, cardiac, and smooth muscle tissues are dominated by α– and γ–-smooth muscle actin, respectively, while the non-muscle cells are prevalent in β– and γ–-non-muscle actin. However, subsequent studies found that this isoform specificity is far more complex. Most cells and tissues contain a combination of actin isoforms, expressed at specific ratios in every cell type (see, e.g., (Bachvarova et al., 1989; Barja et al., 1986; Buckingham, 1985; Gunning et al., 1997; Lloyd et al., 2004; Patrinostro et al., 2017), reviewed in (Simiczyjew et al., 2017)).
Recent explosion of high throughput data enabled a more detailed look at actin isoforms’ tissue specificity and revealed some unexpected trends in actin isoforms’ tissue expression. It has been assumed that differential abundance of actins in different tissues arises exclusively from tissue-specific changes in gene expression, predicted to result in the presence or absence of specific isoforms’ transcripts, or changes in their overall and relative levels. However, data available in the public depositories suggests that transcripts for all actin isoforms are prominently present in nearly every major tissue (Fig. 1). While some differences between the levels of these transcripts are seen, some actin isoforms’ mRNA are present at high levels even in tissues where they are believed to be a minority at the protein level – e.g., β- and γ-non-muscle actins in the skeletal muscle, which is heavily dominated by α-skeletal actin isoform. These data strongly suggest that, in addition to the promoter-level regulation, key determinants of actin isoform tissue specificity resides at post-transcriptional level through mechanisms that do not affect the abundance of their mRNA, likely via translation regulation.
Figure 1. mRNAs for the six mammalian actin isoforms are abundant in many mammalian tissues.
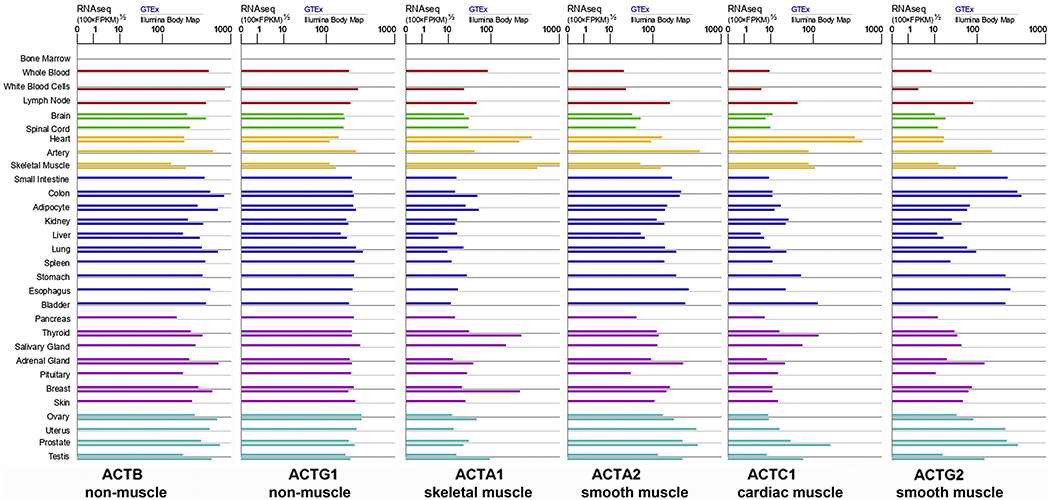
Data was adapted and simplified from the charts available at https://www.genecards.org/ for the human actin genes.
Gene knockout studies demonstrate dramatic differences between actin isoforms’ organismal functions (reviewed in (Perrin and Ervasti, 2010; Vedula and Kashina, 2018)). In mice, genetic ablation of one actin isoform tends to be accompanied by up-regulation of others, to compensate for the overall actin protein levels. This effect is believed to underlie the fact that most of these gene knockouts in mice lead to relatively mild phenotypes that affect specific organs and systems but do not perturb embryogenesis and generally lead to the birth of viable pups. The only exception is the non-muscle β-actin, the only single actin isoform absolutely essential for viability, which, if deleted, leads to early embryonic lethality (Bunnell et al., 2011; Perrin et al., 2010). This fact is especially striking, given that every cell expressing β-actin also contains comparable levels of γ–non-muscle actin, which differs from β–actin by only 4 N-terminal residues, and which is usually up-regulated to compensate for the loss of β–actin in mouse knockout models (Bunnell et al., 2011; Perrin et al., 2010).
Recent studies using CRISPR/Cas9 gene editing have definitively proven that this unique role of β-actin in organism’s survival is independent of its amino acid sequence: editing four N-terminal codons in the β-actin gene to produce γ– actin protein, without altering any other gene-level elements, does not affect mouse viability, despite complete absence of β-actin protein in these mice (Patrinostro et al., 2018; Vedula et al., 2017). Thus, it is the intact gene, not the protein, that defines β– actin’s essential role in organism’s survival and cell migration. While these studies have not further narrowed down which gene element(s) are critically involved in this effect, based on a number of considerations it has been proposed that mRNA coding sequence plays a primary role, by defining the rate of actin translation (Vedula et al., 2017). This hypothesis, however, still needs to be tested experimentally. Whether or not it proves to be correct, it appears likely that additional nucleotide elements also contribute to actin isoforms’ function. For example, it has been shown that intron III in the γ– actin gene can regulate cell morphology (Lloyd and Gunning, 1993). Both β– and γ– actin genes can generate alternatively spliced transcripts that exhibit distinct tissue specificity (in the case of β– actin, (Ghosh et al., 2008)) and are proposed to regulate actin’s abundance through mRNA decay (Drummond and Friderici, 2013). Other, as yet unidentified, gene elements may also contribute to the diverse organismal functions of actin isoforms. All these elements likely act in concert to achieve the complexity of actin-dependent processes in vivo.
Nucleotide-based regulation of actin translation.
In addition to high throughput data on actin isoforms’ mRNA abundance in different tissues (Fig. 1) recent proteomics studies have generated extensive data on the overall abundance of the corresponding proteins (Fig. 2). Comparison of these two data sets reveals another interesting trend. While the mRNA for all actin isoforms is generally present in all tissues, actin proteins exhibit marked tissue specificity, which generally agrees with the early studies on actin isoforms’ tissue assignments (Fig. 2). This observation strongly suggests that the determinants regulating actin’s presence at high levels in some tissues and near-absence in others (e.g., muscle versus non-muscle actins in the spleen, which contains comparable levels of all actin isoforms’ mRNA but only non-muscle actins at the protein level) is regulated independently of gene expression and mRNA abundance. This level of regulation, by default, excludes many of the known regulatory mechanisms that would ultimately affect the expression and decay of mRNA, leaving only two: mRNA translation activity and protein stability. Moreover, since actins are highly metabolically stable (Mayer et al., 1989; Rubinstein et al., 1976), the only likely mechanism that emerges as a major determinant regulating actin abundance and tissue specificity in vivo is differential translation. In support, ribosome profiling studies show that actin isoforms vastly differ from each other in ribosome density, suggesting vast differences in translation dynamics (Vedula and Kashina, 2018; Vedula et al., 2017).
Figure 2. Six mammalian actin isoforms show differential tissue abundance at the protein level.
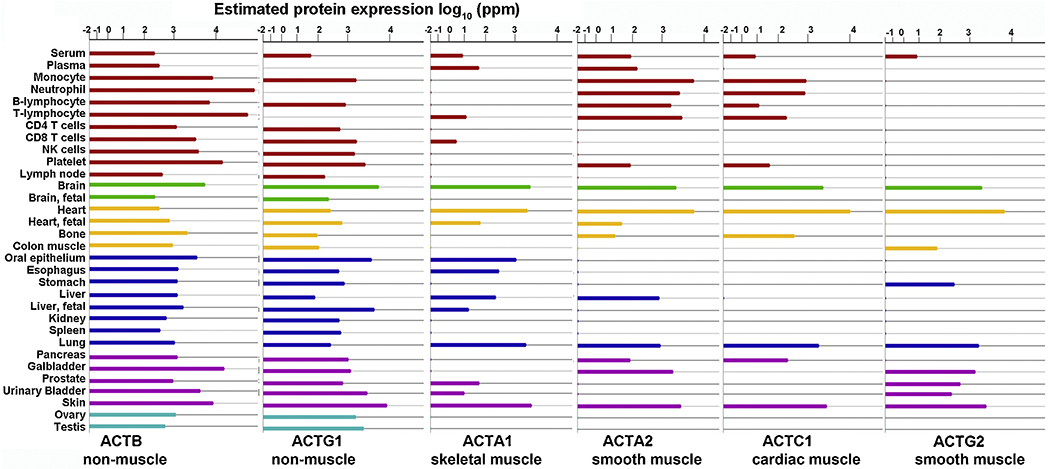
Data was adapted and simplified from the charts available at https://www.genecards.org/ for the human actin genes.
A particularly striking example of this regulation is the differentiation of β– and γ– non-muscle actins. These two actins coexist in every cell type at comparable abundance (Fig. 1, 2), even though their ratios in different cells have been reported to vary 2-3 fold (Erba et al., 1988; Otey et al., 1987; Patrinostro et al., 2017; Skalli et al., 1987). Regardless of these cell and tissue-specific differences in relative protein levels, β– actin mRNA is nearly always far more abundant than that of γ– actin (reported to be 6-10 fold, in different studies focused on these specific isoforms). In addition, according to the ribosome profiling studies, ribosome densities on β– and γ– actin mRNA (often believed to directly reflect translation activity) can vary even more dramatically – in some cases, ribosome density on β– versus γ– actin differs by 100 fold or more, even when accounting for their different mRNA abundance (Vedula et al., 2017). Thus, the more abundant β– actin mRNA, which is also much more densely covered by the ribosomes, does not actually produce a lot more protein than the less abundant, less ribosome-covered γ– actin mRNA. This phenomenon suggests that β– actin mRNA spends most of the time in a translationally repressed state without undergoing active translation, and that its occasional translational de-repression is employed to precisely coordinate β– actin protein’s level in cells. In support, both predictions and experiments suggest that β– actin is capable of faster translation (Zhang et al., 2010), and has been proposed to undergo translational bursting (Strohl et al., 2017) however the mechanisms of its translational repression/derepression, as well as the upstream regulatory events, require further investigation.
Another interesting property of actin mRNA relates to its differential spatial distribution in cells (Hill and Gunning, 1993; Sundell and Singer, 1990). β–actin’s mRNA can be targeted to the cell leading edge via zipcode, present in the 3’UTR of β– actin, unlike any other actin isoform (Condeelis and Singer, 2005; Kislauskis et al., 1997; Oleynikov and Singer, 2003; Ross et al., 1997; Shestakova et al., 2001). This zipcode sequence mediates actin mRNA binding to the RNA-localizing zipcode-binding protein (ZPB1). ZBP1-mediated mRNA transport is estimated to localize approximately 10% of β-actin mRNA to the cell periphery, in complex with a number of other mRNAs (see (Rodriguez and Kashina, 2018) for a recent review).
Zipcode-mediated targeting of β– actin is essential for directional cell migration in non-muscle cells (Bassell et al., 1998; Kislauskis et al., 1997; Shestakova et al., 2001; Zhang et al., 1999), however the reasons for this requirement are unknown. It has been proposed that zipcode binding is at least one of the mechanisms that may cause β– actin’s translational repression, and that unpacking of β– actin mRNA at the cell leading edge mediates its translation on site to facilitate its rapid incorporation into the leading edge network through regulated translational bursts (Strohl et al., 2017). Curiously, the existence of these actin busts during cell migration, has been observed earlier on, in a study that proposed an existence of an active transport mechanism that can deliver localized bursts of actin monomers to the cell periphery (Zicha et al., 2003), even though this previous study did not propose this mechanism to involve translation on site. Overall, much debate exists about why this mechanism in case of actin makes a difference for directional migration, given the presence of highly abundant actin monomer pool in the cytosol and at the cell leading edge (Cramer et al., 2002; Kapustina et al., 2016; Koestler et al., 2009; Pollard et al., 2000, 2001; Raz-Ben Aroush et al., 2017). It is also unclear why only β– actin, and no other actin isoform, undergoes this targeting. Since β– actin is the most essential of all the six actin isoforms (Perrin and Ervasti, 2010), its ability for translational bursting, closely related to unique properties of its mRNA, may well prove to be the defining factor in β– actin’s function (Rodriguez and Kashina, 2018). However, the exact underlying mechanisms of this regulation remain to be determined.
Amino acid-based regulation of actin isoforms.
The six mammalian actins are very similar to each other in their amino acid sequences, but they are not identical (Fig. 3). It has been generally assumed in many prior studies that these amino acid differences constitute a key determinant of their in vivo function, even though the recent emergence of nucleotide-based actin regulation, summarized above, strongly argues that the nucleotide sequences of actin are at least equally important. Still, it is clear that amino acid differences in the actin isoforms play a substantial role in their functional distinctions in vivo. Indeed, vertebrate actin isoforms are 100% conserved in their amino acid sequence, which suggests a strong evolutionary pressure for specific amino acid residues in specific positions throughout the protein (Vedula and Kashina, 2018). This fact suggests that the six actin isoforms are uniquely adapted at the amino acid level to perform their diverse functions.
Figure 3. Six mammalian actin isoforms are highly conserved at the amino acid level.
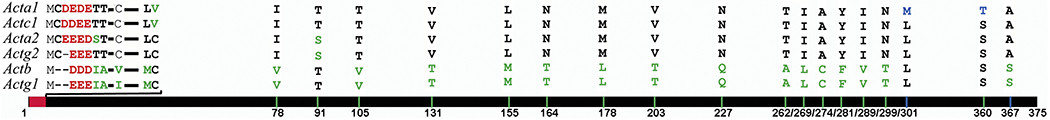
The solid line at the bottom represents the alignment of the six mammalian actins (following the gene symbols listed on the left). Red bar on the alignment represents the sequences unique to each actin isoform. Green bars represent the point amino acid substitutions that distinguish muscle from non-muscle actins. Blue represent the substitutions unique to only one actin isoform (a-skeletal actin). Letters on top list the amino acids in each of these positions for each isoforms (order corresponds to the gene list on the left). Dashes represent empty spaces. Numbers underneath represent the positions of these substitutions, counting from the initiator Met.
Biochemical studies of actin isoform-specific properties are in their infancy, due to the fact that the methods of obtaining pure and homogenous preparations of actin isoforms are still under development (reviewed in (Vedula and Kashina, 2018)). Physiologically active actin isoforms have been previously expressed in Dictyostelium discoideum (Noguchi et al., 2007), in Sf9 cells using baculovirus expression system (Anthony Akkari et al., 2003; Yates et al., 2007) and, more recently, in the yeast Pichia pastoris (Hatano et al., 2018), however technical issues related, e.g., to the potential contaminants of native actins from these expression systems, as well as to the potentially abnormal posttranslational processing, still pose challenge in these studies.
Muscle and non-muscle actins show distinct intracellular localization patterns, both in muscle cells (where non-muscle actins are present at the protein level but typically not found in the myfibrils), as well as in different types of non-muscle cells (Kaech et al., 1997; Mounier et al., 1997). Based on these data it is believed that in vivo muscle and non-muscle actins don’t incorporate into the same filaments in vivo, consistent with their higher amino acid divergence (Fig. 3). Different actin isoforms have been shown to co-polymerize in vitro (Bergeron et al., 2010; Muller et al., 2013), but even the highly similar non-muscle β- and γ-actin seem to have some differences in nucleotide and ion dependence as well as polymerization kinetics (Bergeron et al., 2010). Platelet and chicken gizzard actins appear to form non-overlapping filaments in vitro (Chen et al., 2017), suggesting that their predominant isoforms (~85% β- in platelets and ~75% γ-actin in chicken gizzard, likely dominated by the smooth muscle isoform) are potentially averse to copolymerization, β-actin shows preference for binding to myosin 2B, tropomyosin (Pathan-Chhatbar et al., 2018), and myosin 2C1 (Muller et al., 2013) and has been reported to have a specific capping protein betacap73 (Shuster et al., 1996; Welch et al., 2005) and a specific nucleator DIAPH3 (Chen et al., 2017). Depletion of intracellular cofilin preferentially affects β-actin polymerization in cells (Kapustina et al., 2016). γ-actin exhibits a preference toward myosin 7a (Muller et al., 2013) and has been recently shown to be specifically involved in nuclei localization in skeletal muscle syncytia (Roman et al., 2017). Non-muscle actin isoforms exhibit distinct preferences for binding to profilin (Ohshima et al., 1989), thymosin β4 (Weber et al., 1992), ezrin (Shuster and Herman, 1995; Yao et al., 1995), and plastin (Prassler et al., 1997). Some studies suggest that β- and γ-actin show different intracellular distribution (Dugina et al., 2009; Otey et al., 1988; Otey et al., 1986) and point to a number of functional differences between actin isoforms in vivo (Baranwal et al., 2012; Clement et al., 2005; Dugina et al., 2018; Dugina et al., 2009; Hinz et al., 2003), however some of these results, especially the differential intracellular distribution of β- and γ-actin, are not corroborated by other studies. It is possible that some, or all, of these effects are cell type specific, potentially dependent on the cells’ physiological state, and some of them may be mediated by nucleotide, rather than amino acid differences in actins.
Interestingly, similar actin isoform diversity also exists in other, non-vertebrate species, including Drosophila, which also contains six actin isoforms encoded by different genes, including muscle and non-muscle isoforms. It has been recently shown that Drosophila muscle and non-muscle actins show specificity for different formins (Patel et al., 2018; Silkworth et al., 2018). It appears likely that isoform-specific mechanisms of actin regulation are likely universal across the tree of life.
Posttranslational regulation of actin
Studies accumulated throughout the years point to a large number of posttranslational modifications that directly target actin in different cell types during normal and disease-related processes (reviewed in (Terman and Kashina, 2013; Varland et al., 2019)). Actin has been shown to undergo tens of different covalent modifications (Fig. 4) that can potentially affect nearly every amino acid residue exposed on the surface of the folded actin monomer (Fig. 5). Relatively little, however, is known about the role of these modifications in modulating actin’s properties and cellular functions, or about their potential isoform specificity.
Figure 4. Actin undergoes a large variety of posttranslational modifications.
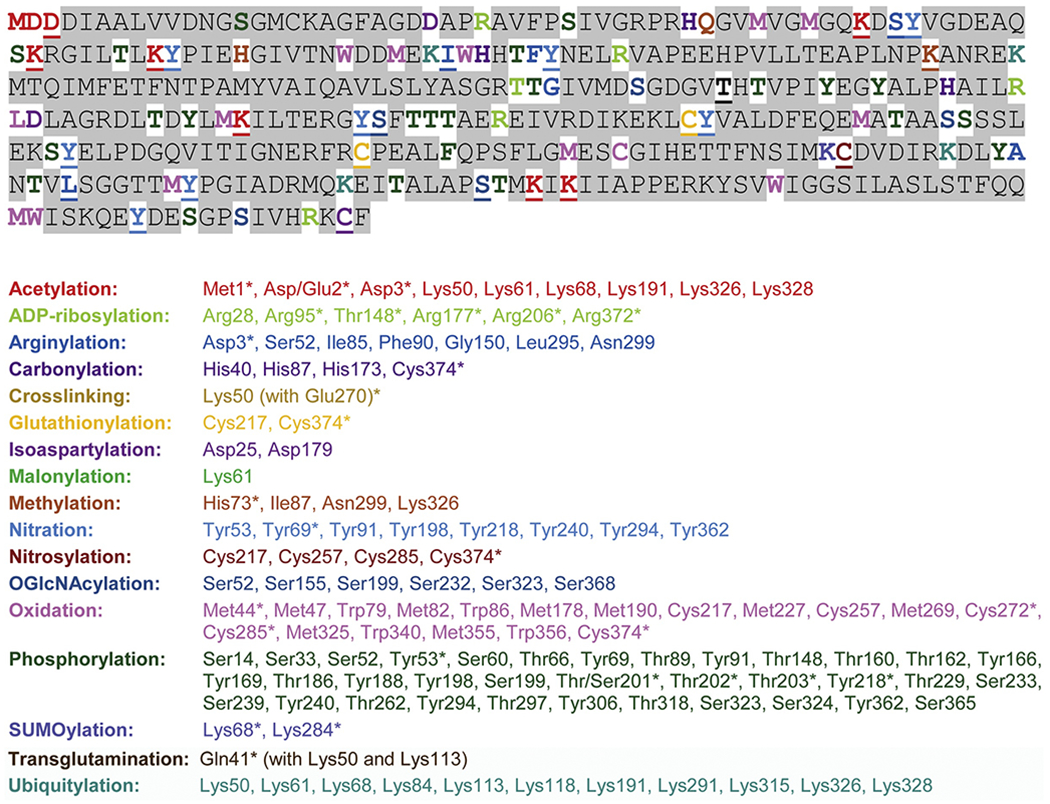
Top, amino acid sequence of mammalian β–actin, color-coded to depict sites of all modifications, listed alphabetically underneath. Underlined residues represent sites of multiple modifications, color-coded for the one found first in the list underneath. Areas shaded in gray represent the residues on which no posttranslational modifications have been described. Please note that amino acid positions in the list correspond to those reported in the published studies and are often counted not from the first Met but from the third residue, which appears as N-terminal after posttranslational processing in muscle actins. This list was adapted and updated from (Terman and Kashina, 2013), which lists many of the original publications that reported these arginylated sites.
Figure 5. Sites of actin’s posttranslational modifications mapped onto the folded actin monomer.
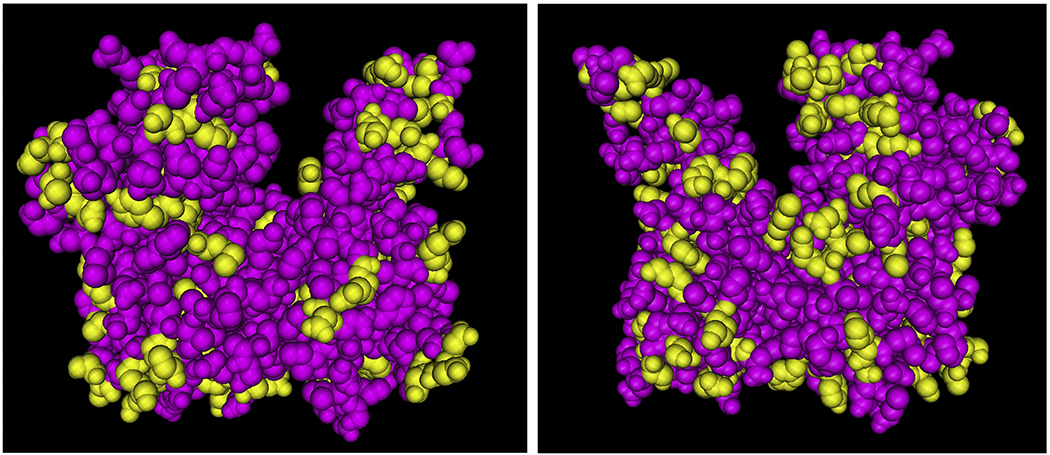
Yellowed residues represent the sites of possible posttranslational modifications, listed and mapped in Figure 3. Pink residues represent the sites where no posttranslational modifications have been described. The actin structure is based on the Cn3D view of b-actin (PDB identifier 1HLU).
Given actin’s high sequence conservation, presumably resulting from high evolutionary pressure applied on every residue in the actin’s amino acid sequence, it is expected that posttranslational modifications on any of these residues would potentially lead to drastic effects to actin’s ability to perform its functions in both monomeric and polymeric state. Compounding this is the fact that the majority of known posttranslational modifications are not known to be reversible, and actin is a very long-lived protein in the cell. Thus, many of the modified actin molecules are either selectively removed or expected to exist in the cell for a very long time, enabling the posttranslational modifications to continuously exert their effects on different actin molecules and intracellular structures. This regulation could potentially have profound effects on actin’s intracellular homeostasis.
Over 99% of intracellular actin is N-terminally acetylated (Drazic et al., 2018; Wiame et al., 2018). This acetylation is part of multi-enzyme processing that has been long known to be important for actin maturation (Rubenstein and Martin, 1983) and actin’s cytoskeleton function (Abe et al., 2000; Chaponnier et al., 1995). Interestingly, N-terminal acetylation affects every muscle and non-muscle actin isoform, even though the specific processing steps appear to be somewhat different between muscle and non-muscle actins and likely have some variability in affecting even the highly similar non-muscle actin isoforms. While deacetylating enzymes do exist in cells, no reversibility of actin’s N-terminal acetylation has been described (Ree et al., 2018).
Most of the actin’s posttranslational modifications have been identified by mass spectrometry. Due to high amino acid-level similarity of all actins, and the simultaneous presence of multiple actin isoforms in the same samples, it is typically impossible to distinguish posttranslational modifications on specific actin isoforms, with the exception of the N-terminus. It should be noted that, outside the N-terminus, all the currently known posttranslational modifications of actin have been found only on the residues that are identical between all actin isoforms (Fig. 3, 4). This suggests that most of actin’s posttranslational regulation, whether or not actin isoform-specific, has undergone stringent evolutionary pressure, and each of these modifications likely exerts universal effects on all actin isoforms in vivo.
The only currently known actin isoform-specific posttranslational modification is N-terminal arginylation – covalent addition of the arginine moiety – which affects specifically β–actin and has not been found in any other actin isoform (Karakozova et al., 2006; Kashina, 2006). A recent study shows that this arginylation is detected in a number of tissues and cell types and affects ~1% of endogenous β–actin (the portion of actin that is not normally N-terminally acetylated) (Chen and Kashina, 2019). Moreover, inhibition of N-terminal acetylation increases the intracellular β–actin arginylation level by multiple fold (Chen and Kashina, 2019). While biological effects of β–actin arginylation are unclear, interestingly, the differential arginylation of β– but not γ– actin is driven by differences in their mRNA coding sequence, rather than the amino acid sequence. Coding sequence differences confer faster translation to β– actin, and this faster translation facilitates preferential retention of arginylated β– actin and degradation of the incorrectly arginylated γ– actin (Zhang et al., 2010). Ultimately, these findings have driven our early understanding of the importance of the coding sequence, rather than the amino acid sequence, in actin’s function, and have led to the recent discovery that nucleotide-level differences underlie the unique role of β– actin in organism’s viability (Vedula et al., 2017). Thus, regardless of the specifics of β– actin’s regulation by N-terminal arginylation, its discovery has played a key role in uncovering a major mechanism of actin regulation in vivo.
The downstream effects of β– actin arginylation are still poorly understood. Much debate revolves around its intracellular abundance. While earlier studies based on crude estimates by gel shifts proposed that 20-40% of total intracellular actin is arginylated, this estimate is likely incorrect, based on the difficulty of detecting arginylated actin in vivo. The real abundance of this modification, at least at the stationary level, is likely closer to single percent quantities, suggesting that its effects are either highly localized (e.g., to the cell leading edge), or are related to overall regulation rather than exerting direct influence on the actin network architecture. Deletion of arginyltransferase leads to a disruption of the cell leading edge morphology and directional migration (Karakozova et al., 2006), and an overall decrease in intracellular actin polymer levels (Saha et al., 2010). Transfection of cells with arginylated actin constructs rescues the leading edge morphology and cell migration (Karakozova et al., 2006). Moreover, antibody-based studies demonstrate localization of arginylated β– actin at the cell leading edge in different cell types (Pavlyk et al., 2018; Wang et al., 2017).While these data argue for a prevailing and specific role of arginylated actin at the cell leading edge, the exact effect of N-terminal arginylation of actin is still being debated, and requires further investigation.
Notably, additional sites within the actin molecule, as well as additional components of the actin cytoskeleton can also be arginylated (Cornachione et al., 2014; Kurosaka et al., 2012; Rai et al., 2008; Wong et al., 2007). It is possible that arginylation of these components, rather than actin itself, underlies some of the reported effects of arginylation on the actin cytoskeleton, which extend all the way to the protozoans (Batsios et al., 2019). A recent study, for example, found a direct link between arginyltransferase ATE1 and the transcriptional activity of MRTF-A, the component of the serum response pathway directly regulated by nuclear actin and expected to exert substantial effects on the actin cytoskeleton (Eisenach et al., 2014).
Given the wealth of posttranslational processes that can potentially target actin, it appears likely that in vivo only a fraction of these modifications would be present simultaneously on each actin monomer. It is also likely that these posttranslational modifications are hierarchically related and some of them may be mutually exclusive. For example, N-terminal acetylation would likely preclude arginylation, and N-terminal arginylation would definitely interfere with acetylation, which depends on the actin’s N-terminal stretch of negatively charged amino acid residues (Arnesen et al., 2018; Drazic et al., 2018). It is possible that development of new methods of isoform-specific actin isolation can address this constraint and enable new discoveries of isoform-specific actin regulation. One of such approaches could potentially involve editing of actin genes to create uni-actin isoform cells and model organisms, which could be individually analyzed by mass spectrometry to enable further insights into actin isoforms function.
Given the vast variety of proteins that can interact with actin, it is clear that every posttranslational modification, whether on monomer or polymer or both, would likely affect at least some of these interactions and modulate the binding of key actin regulator(s), which could in turn lead to strong downstream effects (Abe et al., 2000). Some of the modifications also affect the properties of the actin subunits themselves, such as Mical-mediated actin oxidation at Met 44 (Hung et al., 2011), shown to rapidly disassemble F-actin and prevent its polymerization, thus inhibiting tissue remodeling in vivo. This mechanism has recently emerged as a major regulator of attractive and repulsive signaling in cells and tissues, tightly linked to actin oxidation (Fremont et al., 2017; Grintsevich et al., 2017; Grintsevich et al., 2016; Yoon et al., 2017; Yoon and Terman, 2018). Direct roles of O-GlcNAc-ylation and phosphorylation of actin have been proposed to occur, an likely play differential roles, in diabetic nephropathy (Akimoto et al., 2019). A recent study showed that reversible actin mid-chain acetylation modulates the activity of inverted formin 2 and thus directly affects intracellular actin assembly (A et al., 2019). An actin-specific methyltransferase SETD3, which specifically methylates actin at His 73 – another posttranslational modifications that affects the majority of intracellular actin -- has recently been identified and shown to regulating actin’s organismal roles in female fertility and smooth muscle contraction (Dai et al., 2019; Guo et al., 2019; Kwiatkowski et al., 2018; Wilkinson et al., 2019). Overall, actin posttranslational regulation is rapidly coming into focus, and it can be anticipated in the future years to take center stage in actin research.
Conclusions.
Actin is one of the most abundant and essential intracellular proteins. Despite decades of study, many open questions remain in our understanding of actin’s intracellular regulation. Among these questions, actin isoform specificity and the mechanisms that control actin function have been recently coming into focus due to the development of new methods and genetic and biochemical models.
Intracellular actin homeostasis involves a complex interplay between many of its various functions, which are all tightly linked to the dynamic assembly and disassembly of actin filaments in the cells. This dynamics can regulate cells on many levels, starting with gene expression and chromatin organization, and including gene expression, mRNA transport, translation dynamics, and posttranslational regulation (Fig. 6). On top of this complexity, which is well studied but still not fully understood, major open questions in the field include regulation of actin isoform specificity in different tissues and intracellular structures, as well as the specificity and hierarchy of actin’s posttranslational modifications.
Figure 6. Levels of intracellular actin regulation.
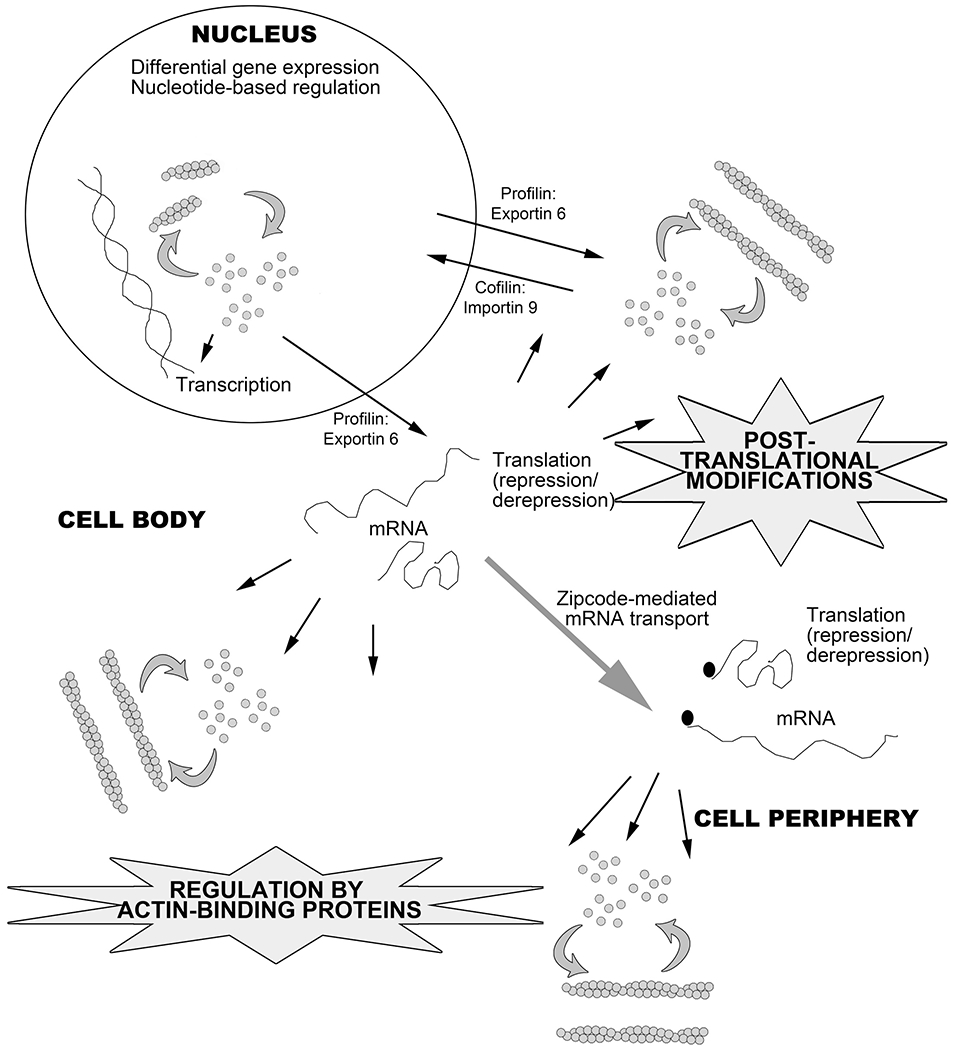
Actin is regulated at the gene level, through mRNA stability and accessibility to translation, as well as through interactions with multiple binding partners and direct targeting by posttranslational modifications. Interplay of this regulation dynamically controls the diversity of actin functions.
Acknowledgements.
I thank Dr. Pavan Vedula for helpful discussions and critical reading of the manuscript. This work was supported by NIG grants R35GM118017 and R01NS102435 to A.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A M, Fung TS, Kettenbach AN, Chakrabarti R, and Higgs HN (2019). A complex containing lysine-acetylated actin inhibits the formin INF2. Nature cell biology 21, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe A, Saeki K, Yasunaga T, and Wakabayashi T (2000). Acetylation at the N-terminus of actin strengthens weak interaction between actin and myosin. Biochemical and biophysical research communications 268, 14–19. [DOI] [PubMed] [Google Scholar]
- Akimoto Y, Yan K, Miura Y, Tsumoto H, Toda T, Fukutomi T, Sugahara D, Kudo A, Arai T, Chiba Y, et al. (2019). O-GlcNAcylation and Phosphorylation of beta-Actin Serine199 in Diabetic Nephropathy. American journal of physiology Renal physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampe C, and Van Troys M (2017). Mammalian Actins: Isoform-Specific Functions and Diseases. Handbook of experimental pharmacology 235, 1–37. [DOI] [PubMed] [Google Scholar]
- Anthony Akkari P, Nowak KJ, Beckman K, Walker KR, Schachat F, and Laing NG (2003). Production of human skeletal alpha-actin proteins by the baculovirus expression system. Biochemical and biophysical research communications 307, 74–79. [DOI] [PubMed] [Google Scholar]
- Arnesen T, Marmorstein R, and Dominguez R (2018). Actin’s N-terminal acetyltransferase uncovered. Cytoskeleton (Hoboken) 75, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artman L, Dormoy-Raclet V, von Roretz C, and Gallouzi IE (2014). Planning your every move: the role of beta-actin and its post-transcriptional regulation in cell motility. Seminars in cell & developmental biology 34, 33–43. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Wang H, and Grosse R (2013). Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340, 864–867. [DOI] [PubMed] [Google Scholar]
- Bachvarova R, Cohen EM, De Leon V, Tokunaga K, Sakiyama S, and Paynton BV (1989). Amounts and modulation of actin mRNAs in mouse oocytes and embryos. Development 106, 561–565. [DOI] [PubMed] [Google Scholar]
- Baranwal S, Naydenov NG, Harris G, Dugina V, Morgan KG, Chaponnier C, and Ivanov AI (2012). Nonredundant roles of cytoplasmic beta- and gamma-actin isoforms in regulation of epithelial apical junctions. Molecular biology of the cell 23, 3542–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barja F, Coughlin C, Belin D, and Gabbiani G (1986). Actin isoform synthesis and mRNA levels in quiescent and proliferating rat aortic smooth muscle cells in vivo and in vitro. Laboratory investigation; a journal of technical methods and pathology 55, 226–233. [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, and Kosik KS (1998). Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. The Journal of neuroscience : the official journal of the Society for Neuroscience 18, 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsios P, Ishikawa-Ankerhold HC, Roth H, Schleicher M, Wong CCL, and Muller-Taubenberger A (2019). Ate1-mediated posttranslational arginylation affects substrate adhesion and cell migration in Dictyostelium discoideum. Molecular biology of the cell 30, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron SE, Zhu M, Thiem SM, Friderici KH, and Rubenstein PA (2010). Ion-dependent polymerization differences between mammalian beta-and gamma-nonmuscle actin isoforms. The Journal of biological chemistry 285, 16087–16095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Stuven T, Kuhn C, Cordes VC, and Gorlich D (2006). A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nature cell biology 8, 257–263. [DOI] [PubMed] [Google Scholar]
- Buckingham ME (1985). Actin and myosin multigene families: their expression during the formation of skeletal muscle. Essays in biochemistry 20, 77–109. [PubMed] [Google Scholar]
- Bunnell TM, Burbach BJ, Shimizu Y, and Ervasti JM (2011). beta-Actin specifically controls cell growth, migration, and the G-actin pool. Molecular biology of the cell 22, 4047–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano E, Philimonenko VV, Kahle M, Fukalova J, Kalendova A, Yildirim S, Dzijak R, Dingova-Krasna H, and Hozak P (2010). Actin complexes in the cell nucleus: new stones in an old field. Histochemistry and cell biology 133, 607–626. [DOI] [PubMed] [Google Scholar]
- Chaponnier C, Goethals M, Janmey PA, Gabbiani F, Gabbiani G, and Vandekerckhove J (1995). The specific NH2-terminal sequence Ac-EEED of alpha-smooth muscle actin plays a role in polymerization in vitro and in vivo. The Journal of cell biology 130, 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Arora PD, McCulloch CA, and Wilde A (2017). Cytokinesis requires localized beta-actin filament production by an actin isoform specific nucleator. Nat Commun 8, 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Kashina A (2019). Quantification of intracellular N-terminal beta-actin arginylation. Scientific reports 9, 16669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement S, Hinz B, Dugina V, Gabbiani G, and Chaponnier C (2005). The N-terminal Ac-EEED sequence plays a role in alpha-smooth-muscle actin incorporation into stress fibers. Journal of cell science 118, 1395–1404. [DOI] [PubMed] [Google Scholar]
- Condeelis J, and Singer RH (2005). How and why does beta-actin mRNA target? Biology of the cell / under the auspices of the European Cell Biology Organization 97, 97–110. [DOI] [PubMed] [Google Scholar]
- Cornachione AS, Leite FS, Wang J, Leu NA, Kalganov A, Volgin D, Han X, Xu T, Cheng YS, Yates JR 3rd, et al. (2014). Arginylation of myosin heavy chain regulates skeletal muscle strength. Cell reports 8, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer LP, Briggs LJ, and Dawe HR (2002). Use of fluorescently labelled deoxyribonuclease I to spatially measure G-actin levels in migrating and non-migrating cells. Cell motility and the cytoskeleton 51, 27–38. [DOI] [PubMed] [Google Scholar]
- Dai S, Horton JR, Woodcock CB, Wilkinson AW, Zhang X, Gozani O, and Cheng X (2019). Structural basis for the target specificity of actin histidine methyltransferase SETD3. Nat Commun 10, 3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R, and Holmes KC (2011). Actin structure and function. Annual review of biophysics 40, 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, and Vartiainen MK (2012). Active maintenance of nuclear actin by importin 9 supports transcription. Proceedings of the National Academy of Sciences of the United States of America 109, E544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A, Aksnes H, Marie M, Boczkowska M, Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et al. (2018). NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proceedings of the National Academy of Sciences of the United States of America 115, 4399–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MC, and Friderici KH (2013). A novel actin mRNA splice variant regulates ACTG1 expression. PLoS genetics 9, e1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugina V, Shagieva G, Khromova N, and Kopnin P (2018). Divergent impact of actin isoforms on cell cycle regulation. Cell Cycle 17, 2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugina V, Zwaenepoel I, Gabbiani G, Clement S, and Chaponnier C (2009). Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. Journal of cell science 122, 2980–2988. [DOI] [PubMed] [Google Scholar]
- Eisenach PA, Schikora F, and Posern G (2014). Inhibition of arginyltransferase 1 induces transcriptional activity of myocardin-related transcription factor A (MRTF-A) and promotes directional migration. The Journal of biological chemistry 289, 35376–35387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erba HP, Eddy R, Shows T, Kedes L, and Gunning P (1988). Structure, chromosome location, and expression of the human gamma-actin gene: differential evolution, location, and expression of the cytoskeletal beta-and gamma-actin genes. Molecular and cellular biology 8, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erba HP, Gunning P, and Kedes L (1986). Nucleotide sequence of the human gamma cytoskeletal actin mRNA: anomalous evolution of vertebrate non-muscle actin genes. Nucleic acids research 14, 5275–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, and Treisman R (2014). Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes & development 28, 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahzadeh K, Banaei-Esfahani A, and Shahhoseini M (2015). The potential roles of actin in the nucleus. Cell journal 17, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrai C, Naum-Ongania G, Longobardi E, Palazzolo M, Disanza A, Diaz VM, Crippa MP, Scita G, and Blasi F (2009). Induction of HoxB transcription by retinoic acid requires actin polymerization. Molecular biology of the cell 20, 3543–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremont S, Romet-Lemonne G, Houdusse A, and Echard A (2017). Emerging roles of MICAL family proteins - from actin oxidation to membrane trafficking during cytokinesis. Journal of cell science 130, 1509–1517. [DOI] [PubMed] [Google Scholar]
- Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, and Pillai B (2008). MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene. Nucleic acids research 36, 6318–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieni RS, and Hendzel MJ (2009). Actin dynamics and functions in the interphase nucleus: moving toward an understanding of nuclear polymeric actin. Biochemistry and cell biology = Biochimie et biologie cellulaire 87, 283–306. [DOI] [PubMed] [Google Scholar]
- Grintsevich EE, Ge P, Sawaya MR, Yesilyurt HG, Terman JR, Zhou ZH, and Reisler E (2017). Catastrophic disassembly of actin filaments via Mical-mediated oxidation. Nat Commun 8, 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Yesilyurt HG, Rich SK, Hung RJ, Terman JR, and Reisler E (2016). F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nature cell biology 18, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, Weinberger R, and Jeffrey P (1997). Actin and tropomyosin isoforms in morphogenesis. Anatomy and embryology 195, 311–315. [DOI] [PubMed] [Google Scholar]
- Guo Q, Liao S, Kwiatkowski S, Tomaka W, Yu H, Wu G, Tu X, Min J, Drozak J, and Xu C (2019). Structural insights into SETD3-mediated histidine methylation on beta-actin. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Alioto S, Roscioli E, Palani S, Clarke ST, Kamnev A, Hernandez-Fernaud JR, Sivashanmugam L, Chapa YLB, Jones AME, et al. (2018). Rapid production of pure recombinant actin isoforms in Pichia pastoris. Journal of cell science 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MA, and Gunning P (1993). Beta and gamma actin mRNAs are differentially located within myoblasts. The Journal of cell biology 122, 825–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Dugina V, Ballestrem C, Wehrle-Haller B, and Chaponnier C (2003). Alpha-smooth muscle actin is crucial for focal adhesion maturation in myofibroblasts. Molecular biology of the cell 14, 2508–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. (2004). Actin is part of preinitiation complexes and is necessary for transcription by RNA polymerase II. Nature cell biology 6, 1094–1101. [DOI] [PubMed] [Google Scholar]
- Hu P, Wu S, and Hernandez N (2004). A role for beta-actin in RNA polymerase III transcription. Genes & development 18, 3010–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung RJ, Pak CW, and Terman JR (2011). Direct redox regulation of F-actin assembly and disassembly by Mical. Science 334, 1710–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst V, Shimada K, and Gasser SM (2019). Nuclear Actin and Actin-Binding Proteins in DNA Repair. Trends in cell biology 29, 462–476. [DOI] [PubMed] [Google Scholar]
- Kaech S, Fischer M, Doll T, and Matus A (1997). Isoform specificity in the relationship of actin to dendritic spines. The Journal of neuroscience : the official journal of the Society for Neuroscience 17, 9565–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapustina M, Read TA, and Vitriol EA (2016). Simultaneous quantification of actin monomer and filament dynamics with modeling-assisted analysis of photoactivation. Journal of cell science 129, 4633–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR 3rd, Mogilner A, Zebroski H, and Kashina A (2006). Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196. [DOI] [PubMed] [Google Scholar]
- Kashina AS (2006). Differential arginylation of actin isoforms: the mystery of the actin N-terminus. Trends in cell biology 16, 610–615. [DOI] [PubMed] [Google Scholar]
- Kelpsch DJ, and Tootle TL (2018). Nuclear Actin: From Discovery to Function. Anat Rec (Hoboken) 301, 1999–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, and Singer RH (1997). beta-Actin messenger RNA localization and protein synthesis augment cell motility. The Journal of cell biology 136, 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, and Small JV (2009). F- and G-actin concentrations in lamellipodia of moving cells. PloS one 4, e4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristo I, Bajusz I, Bajusz C, Borkuti P, and Vilmos P (2016). Actin, actin-binding proteins, and actin-related proteins in the nucleus. Histochemistry and cell biology 145, 373–388. [DOI] [PubMed] [Google Scholar]
- Kurosaka S, Leu NA, Pavlov I, Han X, Ribeiro PA, Xu T, Bunte R, Saha S, Wang J, Cornachione A, et al. (2012). Arginylation regulates myofibrils to maintain heart function and prevent dilated cardiomyopathy. Journal of molecular and cellular cardiology 53, 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski S, Seliga AK, Vertommen D, Terreri M, Ishikawa T, Grabowska I, Tiebe M, Teleman AA, Jagielski AK, Veiga-da-Cunha M, et al. (2018). SETD3 protein is the actin-specific histidine N-methyltransferase. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, and Dominguez R (2010). Regulation of actin cytoskeleton dynamics in cells. Molecules and cells 29, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C, and Gunning P (1993). Noncoding regions of the gamma-actin gene influence the impact of the gene on myoblast morphology. The Journal of cell biology 121, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM, Berendse M, Lloyd DG, Schevzov G, and Grounds MD (2004). A novel role for non-muscle gamma-actin in skeletal muscle sarcomere assembly. Experimental cell research 297, 82–96. [DOI] [PubMed] [Google Scholar]
- Lyubimova A, Bershadsky AD, and Ben-Ze’ev A (1997). Autoregulation of actin synthesis responds to monomeric actin levels. Journal of cellular biochemistry 65, 469–478. [PubMed] [Google Scholar]
- Lyubimova A, Bershadsky AD, and Ben-Ze’ev A (1999). Autoregulation of actin synthesis requires the 3’-UTR of actin mRNA and protects cells from actin overproduction. Journal of cellular biochemistry 76, 1–12. [DOI] [PubMed] [Google Scholar]
- Mayer A, Siegel NR, Schwartz AL, and Ciechanover A (1989). Degradation of proteins with acetylated amino termini by the ubiquitin system. Science 244, 1480–1483. [DOI] [PubMed] [Google Scholar]
- Migocka-Patrzalek M, Makowiecka A, Nowak D, Mazur AJ, Hofmann WA, and Malicka-Blaszkiewicz M (2015). beta- and gamma-Actins in the nucleus of human melanoma A375 cells. Histochemistry and cell biology 144, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, and Gurdon JB (2013). Transcriptional regulation and nuclear reprogramming: roles of nuclear actin and actin-binding proteins. Cellular and molecular life sciences : CMLS 70, 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier N, Perriard JC, Gabbiani G, and Chaponnier C (1997). Transfected muscle and non-muscle actins are differentially sorted by cultured smooth muscle and non-muscle cells. Journal of cell science 110 (Pt 7), 839–846. [DOI] [PubMed] [Google Scholar]
- Muller M, Diensthuber RP, Chizhov I, Claus P, Heissler SM, Preller M, Taft MH, and Manstein DJ (2013). Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PloS one 8, e70636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi TQ, Kanzaki N, Ueno H, Hirose K, and Uyeda TQ (2007). A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. The Journal of biological chemistry 282, 27721–27727. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Kawamura H, and Yamamoto T (1963). [Extraction of a Protein Resembling Actin from the Cell Nucleus of the Calf Thymus]. Journal of biochemistry 54, 298–300. [DOI] [PubMed] [Google Scholar]
- Ohshima S, Abe H, and Obinata T (1989). Isolation of profilin from embryonic chicken skeletal muscle and evaluation of its interaction with different actin isoforms. Journal of biochemistry 105, 855–857. [DOI] [PubMed] [Google Scholar]
- Oleynikov Y, and Singer RH (2003). Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Current biology : CB 13, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, and Nordheim A (2010). Linking actin dynamics and gene transcription to drive cellular motile functions. Nature reviews Molecular cell biology 11, 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, and Bulinski JC (1987). Identification and quantification of actin isoforms in vertebrate cells and tissues. Journal of cellular biochemistry 34, 113–124. [DOI] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, and Bulinski JC (1988). Immunolocalization of muscle and nonmuscle isoforms of actin in myogenic cells and adult skeletal muscle. Cell motility and the cytoskeleton 9, 337–348. [DOI] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, Lessard JL, and Bulinski JC (1986). Immunolocalization of the gamma isoform of nonmuscle actin in cultured cells. The Journal of cell biology 102, 1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Oztug Durer ZA, van Loon AP, Bremer KV, and Quinlan ME (2018). Drosophila and human FHOD family formin proteins nucleate actin filaments. The Journal of biological chemistry 293, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan-Chhatbar S, Taft MH, Reindl T, Hundt N, Latham SL, and Manstein DJ (2018). Three mammalian tropomyosin isoforms have different regulatory effects on nonmuscle myosin-2B and filamentous beta-actin in vitro. The Journal of biological chemistry 293, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinostro X, O’Rourke AR, Chamberlain CM, Moriarity BS, Perrin BJ, and Ervasti JM (2017). Relative importance of betacyto- and gammacyto-actin in primary mouse embryonic fibroblasts. Molecular biology of the cell 28, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrinostro X, Roy P, Lindsay A, Chamberlain CM, Sundby LJ, Starker CG, Voytas DF, Ervasti JM, and Perrin BJ (2018). Essential nucleotide-and protein-dependent functions of Actb/beta-actin. Proceedings of the National Academy of Sciences of the United States of America 115, 7973–7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlyk I, Leu NA, Vedula P, Kurosaka S, and Kashina A (2018). Rapid and dynamic arginylation of the leading edge beta-actin is required for cell migration. Traffic 19, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percipalle P (2013). Co-transcriptional nuclear actin dynamics. Nucleus 4, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, and Ervasti JM (2010). The actin gene family: function follows isoform. Cytoskeleton (Hoboken) 67, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Sonnemann KJ, and Ervasti JM (2010). beta-actin and gamma-actin are each dispensable for auditory hair cell development but required for Stereocilia maintenance. PLoS genetics 6, e1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M, and Grosse R (2019). Dynamizing nuclear actin filaments. Current opinion in cell biology 56, 1–6. [DOI] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and Actin-Binding Proteins. Cold Spring Harbor perspectives in biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, and Mullins RD (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annual review of biophysics and biomolecular structure 29, 545–576. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, and Mullins RD (2001). Actin dynamics. Journal of cell science 114, 3–4. [DOI] [PubMed] [Google Scholar]
- Prassler J, Stocker S, Marriott G, Heidecker M, Kellermann J, and Gerisch G (1997). Interaction of a Dictyostelium member of the plastin/fimbrin family with actin filaments and actin-myosin complexes. Molecular biology of the cell 8, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Wong CC, Xu T, Leu NA, Dong DW, Guo C, McLaughlin KJ, Yates JR 3rd, and Kashina A (2008). Arginyltransferase regulates alpha cardiac actin function, myofibril formation and contractility during heart development. Development 135, 3881–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakyla EK, and Vartiainen MK (2014). Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 5, e27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz-Ben Aroush D, Ofer N, Abu-Shah E, Allard J, Krichevsky O, Mogilner A, and Keren K (2017). Actin Turnover in Lamellipodial Fragments. Current biology : CB 27, 2963–2973 e2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree R, Varland S, and Arnesen T (2018). Spotlight on protein N-terminal acetylation. Experimental & molecular medicine 50, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, and Kashina A (2018). Posttranscriptional and Posttranslational Regulation of Actin. Anat Rec (Hoboken) 301, 1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Martins JP, Carvalho FA, Voituriez R, Abella JVG, Santos NC, Cadot B, Way M, and Gomes ER (2017). Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nature cell biology 19, 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, and Singer RH (1997). Characterization of a beta-actin mRNA zipcode-binding protein. Molecular and cellular biology 17, 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein PA, and Martin DJ (1983). NH2-terminal processing of actin in mouse L-cells in vivo. The Journal of biological chemistry 258, 3961–3966. [PubMed] [Google Scholar]
- Rubinstein N, Chi J, and Holtzer H (1976). Coordinated synthesis and degradation of actin and myosin in a variety of myogenic and non-myogenic cells. Experimental cell research 97, 387–393. [DOI] [PubMed] [Google Scholar]
- Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, and Kashina A (2010). Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Molecular biology of the cell 21, 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvany L, Muller J, Guccione E, and Rorth P (2014). The core and conserved role of MAL is homeostatic regulation of actin levels. Genes & development 28, 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova EA, Singer RH, and Condeelis J (2001). The physiological significance of beta - actin mRNA localization in determining cell polarity and directional motility. Proceedings of the National Academy of Sciences of the United States of America 98, 7045–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, and Herman IM (1995). Indirect association of ezrin with F-actin: isoform specificity and calcium sensitivity. The Journal of cell biology 128, 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, Lin AY, Nayak R, and Herman IM (1996). Beta cap73: a novel beta actin-specific binding protein. Cell motility and the cytoskeleton 35, 175–187. [DOI] [PubMed] [Google Scholar]
- Silkworth WT, Kunes KL, Nickel GC, Phillips ML, Quinlan ME, and Vizcarra CL (2018). The neuron-specific formin Delphilin nucleates nonmuscle actin but does not enhance elongation. Molecular biology of the cell 29, 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simiczyjew A, Pietraszek-Gremplewicz K, Mazur AJ, and Nowak D (2017). Are non-muscle actin isoforms functionally equivalent? Histology and histopathology 32, 1125–1139. [DOI] [PubMed] [Google Scholar]
- Skalli O, Vandekerckhove J, and Gabbiani G (1987). Actin-isoform pattern as a marker of normal or pathological smooth-muscle and fibroblastic tissues. Differentiation; research in biological diversity 33, 232–238. [DOI] [PubMed] [Google Scholar]
- Skarp KP, Huet G, and Vartiainen MK (2013). Steady-state nuclear actin levels are determined by export competent actin pool. Cytoskeleton (Hoboken) 70, 623–634. [DOI] [PubMed] [Google Scholar]
- Skarp KP, and Vartiainen MK (2013). Actin as a model for the study of nucleocytoplasmic shuttling and nuclear dynamics. Methods Mol Biol 1042, 245–255. [DOI] [PubMed] [Google Scholar]
- Strohl F, Lin JQ, Laine RF, Wong HH, Urbancic V, Cagnetta R, Holt CE, and Kaminski CF (2017). Single Molecule Translation Imaging Visualizes the Dynamics of Local beta-Actin Synthesis in Retinal Axons. Scientific reports 7, 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, and Gorlich D (2003). Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. The EMBO journal 22, 5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundell CL, and Singer RH (1990). Actin mRNA localizes in the absence of protein synthesis. The Journal of cell biology 111, 2397–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman JR, and Kashina A (2013). Post-translational modification and regulation of actin. Current opinion in cell biology 25, 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondeleir D, Vandamme D, Vandekerckhove J, Ampe C, and Lambrechts A (2009). Actin isoform expression patterns during mammalian development and in pathology: insights from mouse models. Cell motility and the cytoskeleton 66, 798–815. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove J, and Weber K (1978). At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. Journal of molecular biology 126, 783–802. [DOI] [PubMed] [Google Scholar]
- Varland S, Vandekerckhove J, and Drazic A (2019). Actin Post-translational Modifications: The Cinderella of Cytoskeletal Control. Trends in biochemical sciences 44, 502–516. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, and Treisman R (2007). Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316, 1749–1752. [DOI] [PubMed] [Google Scholar]
- Vedula P, and Kashina A (2018). The makings of the ‘actin code’: regulation of actin’s biological function at the amino acid and nucleotide level. Journal of cell science 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula P, Kurosaka S, Leu NA, Wolf YI, Shabalina SA, Wang J, Sterling S, Dong DW, and Kashina A (2017). Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viita T, and Vartiainen MK (2017). From Cytoskeleton to Gene Expression: Actin in the Nucleus. Handbook of experimental pharmacology 235, 311–329. [DOI] [PubMed] [Google Scholar]
- Virtanen JA, and Vartiainen MK (2017). Diverse functions for different forms of nuclear actin. Current opinion in cell biology 46, 33–38. [DOI] [PubMed] [Google Scholar]
- Wada A, Fukuda M, Mishima M, and Nishida E (1998). Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. The EMBO journal 17, 1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Pavlyk I, Vedula P, Sterling S, Leu NA, Dong DW, and Kashina A (2017). Arginyltransferase ATE1 is targeted to the neuronal growth cones and regulates neurite outgrowth during brain development. Developmental biology 430, 41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Nachmias VT, Pennise CR, Pring M, and Safer D (1992). Interaction of thymosin beta 4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry 31, 6179–6185. [DOI] [PubMed] [Google Scholar]
- Welch AY, Riley KN, D’Souza-Schorey C, and Herman IM (2005). Arf6 modulates the beta-actin specific capping protein, betacap73. Methods in enzymology 404, 377–387. [DOI] [PubMed] [Google Scholar]
- Wesolowska N, and Lenart P (2015). Nuclear roles for actin. Chromosoma 124, 481–489. [DOI] [PubMed] [Google Scholar]
- Wiame E, Tahay G, Tyteca D, Vertommen D, Stroobant V, Bommer GT, and Van Schaftingen E (2018). NAT6 acetylates the N-terminus of different forms of actin. The FEBS journal 285, 3299–3316. [DOI] [PubMed] [Google Scholar]
- Wilkinson AW, Diep J, Dai S, Liu S, Ooi YS, Song D, Li TM, Horton JR, Zhang X, Liu C, et al. (2019). SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Xu T, Rai R, Bailey AO, Yates JR 3rd, Wolf YI, Zebroski H, and Kashina A (2007). Global analysis of posttranslational protein arginylation. PLoS biology 5, e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Chaponnier C, Gabbiani G, and Forte JG (1995). Polarized distribution of actin isoforms in gastric parietal cells. Molecular biology of the cell 6, 541–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates SP, Otley MD, and Dawson JF (2007). Overexpression of cardiac actin with baculovirus is promoter dependent. Archives of biochemistry and biophysics 466, 58–65. [DOI] [PubMed] [Google Scholar]
- Yoon J, Kim SB, Ahmed G, Shay JW, and Terman JR (2017). Amplification of F-Actin Disassembly and Cellular Repulsion by Growth Factor Signaling. Developmental cell 42, 117–129 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J, and Terman JR (2018). Common effects of attractive and repulsive signaling: Further analysis of Mical-mediated F-actin disassembly and regulation by Abl. Communicative & integrative biology 11, e1405197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Saha S, Shabalina SA, and Kashina A (2010). Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329, 1534–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, and Bassell GJ (1999). Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. The Journal of cell biology 147, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D, Dobbie IM, Holt MR, Monypenny J, Soong DY, Gray C, and Dunn GA (2003). Rapid actin transport during cell protrusion. Science 300, 142–145. [DOI] [PubMed] [Google Scholar]


