Abstract
Unprimed mice harbor a substantial population of "memory-phenotype" CD8+ T cells (CD8-MP cells) that exhibit hallmarks of activation and innate-like functional properties. Due to the lack of faithful markers to distinguish CD8-MP cells from bona fide CD8+ memory T cells, the developmental origins and antigen specificities of CD8-MP cells remain incompletely defined. Using deep T cell antigen receptor (TCR) sequencing, we found that the TCRs expressed by CD8-MP cells are highly recurrent and distinct from the TCRs expressed by naive-phenotype CD8+ T cells. CD8-MP clones exhibited reactivity to widely expressed self-ligands. T cell precursors expressing CD8-MP TCRs upregulated the transcription factor Eomes during maturation in the thymus, prior to induction of the full memory phenotype, suggestive of a unique program triggered by recognition of self-ligands. Moreover, CD8-MP cells infiltrate oncogene-driven prostate tumors and express high densities of PD-1, suggesting a potential role in anti-tumor immunity and response to immunotherapy.
INTRODUCTION
Classically, memory T cells arise after an immune response to a foreign pathogen in the periphery, and are poised to respond more rapidly upon repeated pathogen challenge. However, in conventionally housed mice and germ-free mice that have not been exposed to foreign pathogens, there exists a substantial population of CD8+ α T cells that exhibit a CD44hiCD122+ memory phenotype, suggestive of previous encounter with agonist ligands. This population, termed “memory-phenotype” CD8+ T cells (CD8-MP cells, also referred to as “virtual-memory”1, 2 or “innate memory”3 T cells), make up >5% of the CD8+ repertoire in adult mice, and exhibit numerous hallmarks of conventional memory CD8+ T cells reactive to foreign ligands. Although the existence of an analogous cell population has been suggested in humans4, 5, 6, the lack of validated markers has limited the ability to study CD8-MP cells in human samples. To date, diverse and dichotomous functions have been attributed to CD8-MP cells, including innate-like effector functions in the early stages of pathogen challenge2, 7, and roles in the maintenance of immune homeostasis at steady state8. However, it remains unclear whether these reflect broad functions of all CD8-MP cells, or distinct functions of heterogeneous T cell populations falling within the CD44hiCD122+ subset. Efforts to elucidate the mechanisms driving CD8-MP differentiation and the function of CD8-MP cells in the context of homeostasis, host defense, inflammation, and cancer have been hampered by the lack of available markers to directly identify CD8-MP cells and their precursors, especially in the context of immune activation. Thus, fundamental aspects of the biology of CD8-MP cells remain incompletely defined, including the nature of antigens recognized by these cells, the mechanisms driving their differentiation, and the functions of CD8-MP cells at steady state and in inflammatory contexts.
A long-standing question is whether CD8-MP differentiation is a T cell antigen receptor (TCR)-independent process driven by cytokines or accessory factors, or a TCR-instructed process triggered by the recognition of peptide/MHC-I ligands. CD8-MP cells exhibit slightly higher average densities of CD56, a surrogate marker of reactivity to positively selecting ligands. However, given that CD5 densities are thought to be “hard-wired” following positive selection in the thymus9, 10, 11, CD5 density cannot be used to assess the intensity of additional TCR signaling events occurring after positive selection. The finding that the phenotype and frequency of CD8-MP cells is not diminished in germ-free mice and germ-free mice fed an elemental diet1, 3 indicates that the absence of microbial and dietary antigens does not impact CD8-MP cells, and suggests that CD8-MP differentiation is either triggered by the recognition of endogenous self-ligands, or is driven by TCR-independent cues. In this regard, the observation that the endogenous repertoire harbors a small number of CD44hiCD122+ CD8+ T cells reactive to any foreign peptide/MHC-I complex2, 12 suggested that CD8-MP differentiation can occur in the absence of known agonist ligands, and that CD8-MP differentiation may be triggered by TCR-independent signals. However, a recent study identified two CD8-MP-biased TCRs, differing by one amino acid, that promoted CD8-MP differentiation when expressed in TCR retrogenic mice13 indicating that CD8-MP differentiation of this clonotype is TCR-directed. Thus, it is currently unknown whether TCR-instructed differentiation applies to the thousands of individual CD8-MP specificities within the endogenous T cell repertoire, and whether CD8-MP differentiation is a robust, orchestrated process that occurs reproducibly.
Regarding the developmental origins of CD8-MP cells, common thought suggests that CD8-MP differentiation occurs in the periphery. This notion is largely based on the fact that CD44hiCD122+ cells are first detected in the periphery of neonatal mice12 and are phenotypically similar to “lymphopenia-induced memory” (LIM) T cells, a population of CD44hiCD122+ cells that emerge in the periphery following intravenous transfer of naive-phenotype CD8+ T cells into lymphopenic mice1, 3. However, it is unclear whether peripherally induced LIM cells are representative of endogenous CD8-MP cells found in wild-type mice. Thus, there is a critical need for developmental studies in which the differentiation trajectories of individual CD8-MP clones can be tracked in the thymus and periphery.
Here, we address these key questions using an in-depth clonal approach, pairing complete TCR repertoire profiling with studies whereby the developmental arc of individual CD8-MP clones can be assessed at all stages of T cell maturation, and the reactivity of distinct CD8-MP clones can be directly assessed ex vivo. Our findings demonstrate that CD8-MP differentiation parallels the thymic differentiation of Foxp3+ regulatory T (Treg) cells in many respects, and reveal a previously unanticipated role for CD8-MP cells in the setting of cancer.
RESULTS
The CD8-MP TCR repertoire is distinct and recurrent
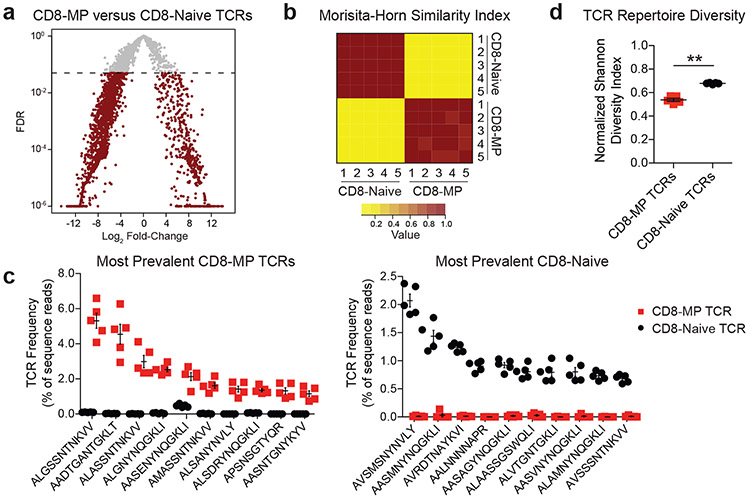
Despite comprising >5% of the CD8+ T cell repertoire in unprimed specific pathogen free (SPF) and germ-free (GF) mice (Extended Data 1), little is known about the array of TCRs expressed by CD8-MP cells. In particular, it is unknown whether the CD8-MP TCR repertoire has limited diversity, whether it varies stochastically from mouse to mouse, and whether it overlaps with the TCR repertoire expressed by naïve-phenotype CD8+ T cells. To comprehensively examine these questions, we sequenced the complete TCR repertoires of naïve-phenotype CD8+ T cells (CD8-Naïve cells, CD44loCD122−) and memory-phenotype CD8+ T cells (CD8-MP cells, CD44hiCD122+) using previously established methods14, 15. We focused our analysis on CD8-MP cells from C57BL/6 mice, in which interleukin 4 (IL-4)-induced CD8+ memory cells1, 3, 16 make minimal contributions to the peripheral CD8-MP pool12, 16. CD8-Naïve and CD8-MP cells from the pooled spleen and lymph nodes were purified by cell sorting from five different 9-week-old male mice expressing a fixed transgenic TCRβ chain on the C57BL/6J (B6) background. By fixing the TCRβ chain, a complete survey of the TCRαβ repertoire can be obtained by sequencing the endogenous Tcra chains using the iRepertoire platform14, 17. This approach yielded approximately 6.5 × 105 Tcra sequence reads per sample, providing a broad survey of TCR usage (Supplementary Table 1). Our data revealed the striking finding that the TCR repertoire of CD8-MP cells is largely distinct from that of CD8-Naïve cells, and was highly recurrent from mouse to mouse (Fig. 1). Comparative analysis of TCR frequency identified 493 recurrent Tcra chains that were significantly overrepresented in the CD8-MP subset relative to the CD8-Naïve subset (Fig. 1a, right arm of volcano plot). Cumulatively, these TCRs accounted for 65 +/− 1% of the CD8-MP TCR repertoire, demonstrating the broad extent of repertoire skewing. The recurrent nature of the CD8-MP repertoire was further illustrated using the Morisita-Horn (MH) similarity index, for which a value of 1 denotes identity and a value of 0 indicates complete dissimilarity. Pairwise comparison of the CD8-MP TCR repertoires from five different mice revealed a mean MH index of 0.92 +/− 0.03 (Fig. 1b), demonstrating that distinct T cell clones were reproducibly directed into the CD8-MP subset. By contrast, pairwise comparisons of CD8-Naïve vs. CD8-MP repertoires between the five mice revealed a mean MH index of 0.06 +/− 0.01, indicative of minimal overlap. The concepts that the CD8-MP TCR repertoires were highly recurrent and distinct from the CD8-Naïve TCR repertoire are also illustrated in Fig. 1c, which plots the frequency of the 10 most prevalent CD8-MP TCRs (left) and 10 most prevalent CD8-Naïve TCRs (right) within the CD8-Naïve and CD8-MP subsets in individual mice. Lastly, analysis of repertoire complexity using the Shannon diversity index demonstrated that the CD8-MP TCR repertoire was less diverse than the CD8-Naïve repertoire (Fig. 1d), with an average complexity of 6,979 TCR complementarity determining regions 3 (CDR3) segments for the CD8-MP subset and 24,443 CDR3s for the CD8-Naive subset. Analyses of CDR3α hydrophobicity and amino acid length revealed no consistent differences between the CD8-Naïve and CD8-MP TCRs (Extended Data 2). Collectively, these data suggest that CD8-MP differentiation is a robust TCR-directed process that occurs in a highly reproducible, orchestrated fashion.
Fig. 1. The TCR repertoire of memory-phenotype CD8+ T cells is recurrent and distinct from that of naïve-phenotype CD8+ T cells.
CD8-MP (CD8+ CD44hiCD122+) and CD8-Naïve (CD8+ CD44loCD122−) T cells were purified by FACS from the pooled spleen and lymph nodes of 9-week-old TCRβtg+ males and subjected to complete TCRα sequencing using the iRepertoire platform. N = 5 for CD8-MP and CD8-Naïve samples. TCRα chains were assessed solely based on their predicted CDR3 segment, regardless of V-region usage.
a, For the TCRα chain sequences, a volcano plot of false discovery rate (FDR) versus differential TCR representation (log2 fold-change) in the CD8-MP vs. CD8-Naïve subsets is shown for the 3,926 recurrently expressed TCRs. n = 5 for CD8-MP and CD8-Naïve samples. Comparisons were made using EdgeR and were adjusted for multiple comparisons. For the differential testing the GLM method was use (see Methods). Red dots denote TCRs with FDR < 0.05. The horizontal dashed line indicates FDR cutoff.
b, Heat map of the Morisita-Horn (MH) similarity index (see Methods), which is a measure of repertoire overlap, for the five CD8-MP and five CD8-Naïve samples. For the MH index, a value of 1 indicates identity and a value of 0 denotes complete dissimilarity.
c, For the 10 most prevalent TCRs recurrently expressed by CD8-MP (left plot) and CD8-Naïve (right plot) subsets, a summary plot of the frequency of these TCRs in individual mice is depicted, with the TCRα CDR3 sequences listed below. Red squares denote frequencies in CD8-MP samples, whereas black circles denote frequencies in CD8-Naïve samples. n = 5 for CD8-MP and CD8-Naïve samples. Mean ± SEM is indicated.
d, Summary plot of normalized Shannon diversity index for CD8-MP and CD8-Naïve subsets. The normalized index ranges from 0 (no diversity) to 1 (maximal diversity). Each symbol represents an individual mouse. n = 5 for CD8-MP and CD8-Naïve samples. Mean ± SEM is indicated. **p = 0.0079, two-tailed nonparametric Mann-Whitney test.
CD8-MP differentiation is a TCR-directed process
To study representative CD8-MP-biased specificities at the clonal level, we generated a series of monoclonal TCR “retrogenic” (TCRrg) mice expressing a single TCR of interest, and used T cells from these mice to perform both in vivo and ex vivo studies. In this study, we use the term “CD8-MP TCR” to define a TCR that is preferentially expressed by CD8-MP cells, and “CD8-Naïve TCR” for receptors skewed to the CD8-Naïve subset. For TCR nomenclature, we depict the amino acid sequence of the CDR3 segment of a given TCRα chain (Fig. 1c), and denote a TCR clone using a three-letter code reflecting the amino acids at positions 3-5 of the CDR3α. For example, TCRrg cells with CDR3α sequence AASMNYNQGKLI are denoted “SMNrg” cells. Our work focused on the study of four recurrent CD8-MP and four recurrent CD8-Naïve TCRs identified in Fig. 1, which were chosen because they span a range of frequencies and utilize distinct V and J-region segments (Supplementary Table 2). TCRrg mice were generated as previously described18. Briefly, bone marrow (BM) cells from Tcra−/− Cd4-Cre+ TCRβtg+ CD45.2/.2 mice were retrovirally transduced with a vector in which the genes encoding a TCRα chain of interest and an IRES-Thy1.1 reporter are preceded by a loxP-flanked stop cassette19. Transduced BM was engrafted along with wild-type filler BM cells into lethally irradiated 6-8-week-old CD45.1/.1 B6.SJL hosts, and mice were analyzed >6 weeks post-engraftment. Expression of Cd4-Cre at the CD4+CD8+ stage of thymic maturation induces TCRα expression by TCRrg cells, recapitulating the natural kinetics of TCR expression during development. Resulting TCRrg T cells were identified using the Thy1.1 and CD45.2 markers.
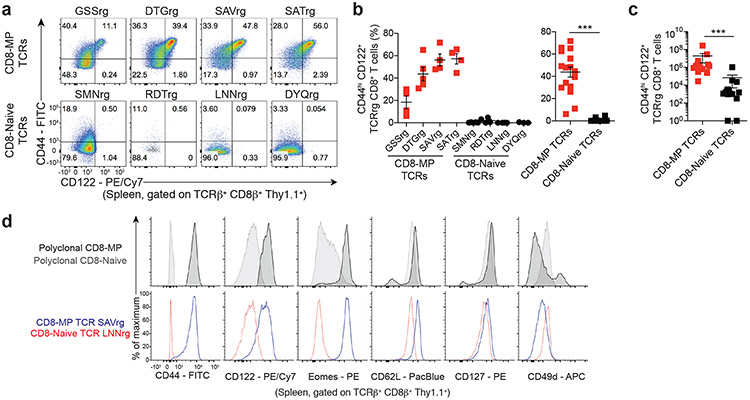
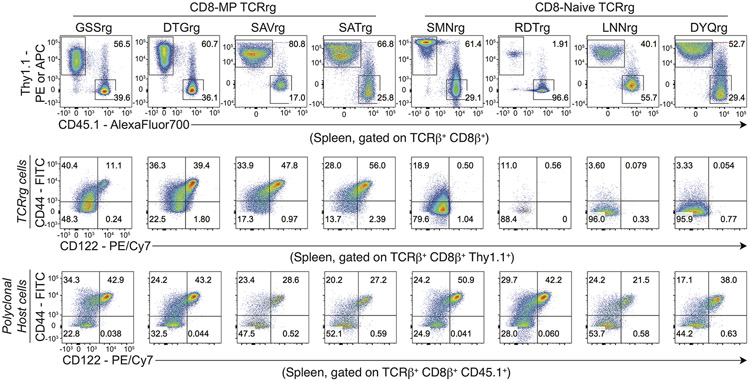
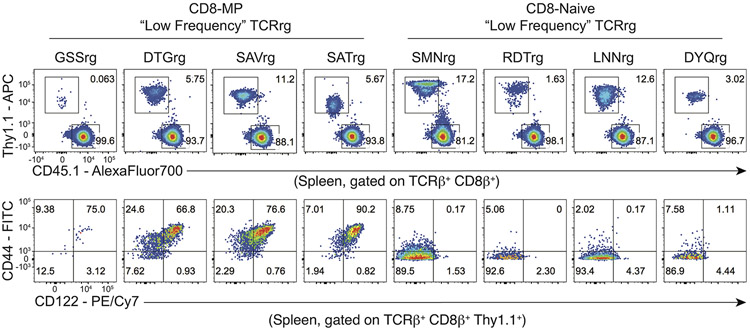
Using this approach, we found that a substantial fraction of T cells expressing CD8-MP TCRs adopted the CD44hiCD122+ memory phenotype in the periphery of TCRrg hosts, whereas very few TCRrg cells expressing CD8-Naïve TCRs acquired this phenotype (Fig. 2a-c, Extended Data 3). These findings validate the TCR sequencing and confirm the notion that CD8-MP differentiation is a TCR-instructed process. Notably, in TCRrg mice expressing CD8-MP TCRs, we found that not all peripheral TCRrg cells adopted a CD44hiCD122+ phenotype at the time of analysis, and that the fraction of TCRrg CD44hiCD122+ cells varied from mouse to mouse (Fig. 2a-c). A potential explanation for the lack of complete skewing to the CD8-MP phenotype is the existence of limited niches supporting CD8-MP differentiation, which may be overloaded in TCRrg mice harboring large numbers of monoclonal T cells. To define the phenotype and anatomical distribution of select CD8-MP clones, we generated TCRrg mice expressing CD8-MP TCRs at low clonal frequency (“low frequency” TCRrg mice; see Methods). In such mice, when TCRrg cells were present at frequencies of <15% of CD8+ T cells, the majority (>65%) of TCRrg cells expressing a CD8-MP TCR exhibited the CD44hiCD122+ phenotype (with nearly all cells shifted away from a naive phenotype), whereas TCRrg cells expressing CD8-Naive TCRs did not (Extended Data 4). These results are consistent with the existence of limited niches supporting the differentiation of CD8-MP clones. Furthermore, peripheral CD8-MP TCRrg cells from these mice adopted a central memory-like phenotype characteristic of polyclonal CD8-MP cells, including high expression of CD44, CD122, CD62L, CD127, and Eomes, and low expression of CD49d (Fig. 2d). These collective data demonstrate that the expression of CD8-MP TCRs in TCRrg hosts recapitulated differentiation into the CD8-MP subset, validating the TCRrg approach and providing further evidence that CD8-MP differentiation is a TCR-directed process.
Fig. 2. CD8-MP differentiation is a TCR-directed process.
Four representative CD8-MP (top) and four CD8-Naïve (bottom) TCRs defined in Fig. 1 were cloned and expressed in TCR retrogenic (TCRrg) mice (see Methods). 7-11 weeks after bone marrow reconstitution, the phenotype of TCRrg T cells was assessed.
a, Representative flow cytometric analysis of CD44 vs. CD122 expression by TCRrg cells from the spleen, gated on TCRβ + CD8β+ Thy1.1+ cells. The percentage of cells falling within the respective gates is indicated. Data are representative of six independent experiments.
b, Left: Summary plot of pooled data from (a) showing the frequency of TCRβ+ CD8β+ Thy1.1+ cells exhibiting a CD44hiCD122+ phenotype for the listed TCRs. Right: Data from the left plot were pooled from the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 17, CD8-MP TCRrg mice; n = 16, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from six independent experiments.
c, Summary plot of pooled data from (a) showing the absolute number of TCRβ+ CD8β+ Thy1.1+ cells exhibiting a CD44hiCD122+ phenotype for the CD8-MP TCRs and CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 17, CD8-MP TCRrg mice; n = 16, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from six independent experiments.
d, Representative flow cytometric analysis of CD44, CD122, Eomes, CD62L, CD127, and CD49d expression by splenic polyclonal TCRβ+ CD8β+ CD8-MP (CD44hiCD122+) and CD8-Naïve (CD44hiCD122−) cells from a 7-week-old C57BL/6 mouse (top panels) and monoclonal TCRβ+ CD8β+ Thy1.1+ SAVrg (CD8-MP TCR) and LNNrg (CD8-Naïve TCR) T cells 7 weeks after bone marrow reconstitution in “low frequency” TCRrg mice (bottom panels). Data are representative of three independent experiments.
CD8-MP T cell clones exhibit self-reactivity
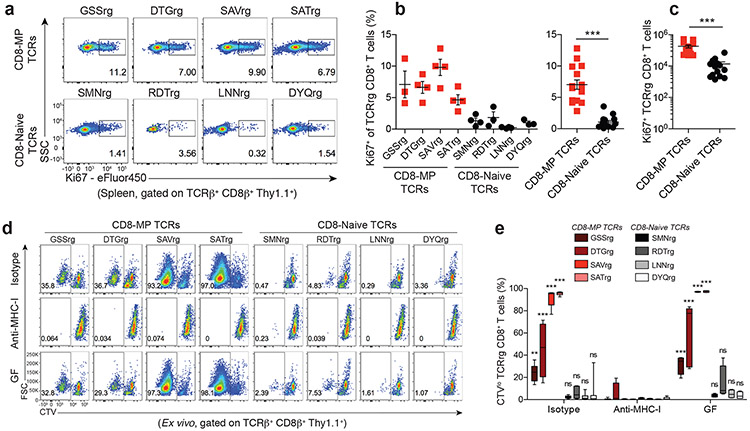
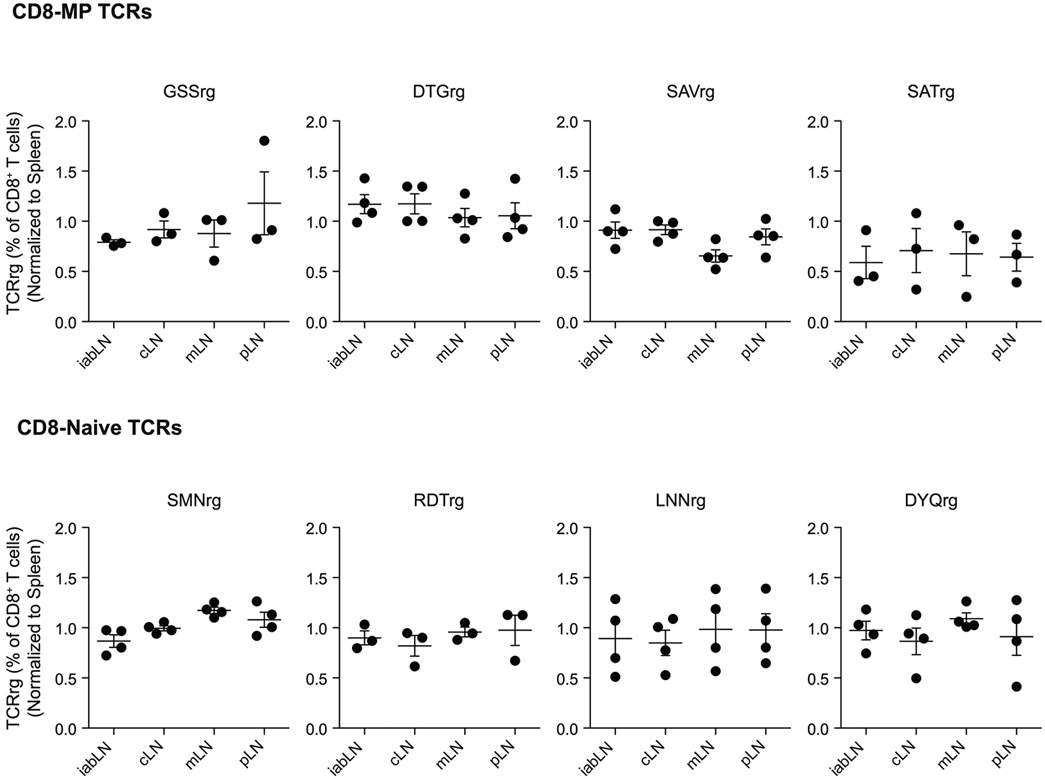
Our TCR repertoire and TCRrg mouse analyses suggest that CD8-MP differentiation is antigen driven. Since CD8-MP cells are abundant in naïve mice that have never been exposed to known foreign pathogens, we hypothesized that CD8-MP cells may be reactive to endogenous self-ligands. In TCRrg mice, we found that CD8-MP clones were equally distributed in all secondary lymphoid organs examined (Extended Data 5) and exhibited elevated percentages of proliferative cells as measured by Ki67 staining, suggestive of active sensing of ligands at steady state (Fig. 3a-c). To gain insight into the antigen specificity of CD8-MP clones, we assessed the ex vivo reactivity of purified naïve-phenotype (CD44loCD122−) CD8+ T cells isolated from TCRrg mice expressing CD8-MP or CD8-Naïve TCRs. We purified naïve-phenotype CD44loCD122− from TCRrg mice expressing each of the eight TCRs, and used these cells as a probe for antigen in in vitro stimulation assays. We found that the four CD8-MP clones underwent proliferation upon co-culture with splenic dendritic cells (DCs) and recombinant IL-2, whereas the four CD8-Naïve clones did not (Fig. 3d,e). This reactivity was abolished by the addition of anti-MHC-I blocking antibodies (Fig. 3d,e), indicative of reactivity to ligands displayed by classical MHC-I molecules. Notably, the reactivity of CD8-MP clones was not impaired using splenic DCs isolated from germ-free mice (Fig. 3d,e). Thus, our data provide direct evidence that four canonical CD8-MP TCRs examined confer overt reactivity to endogenous self-ligands presented by splenic DCs in the context of classical MHC-I molecules.
Fig. 3. CD8-MP T cell clones exhibit reactivity to MHC class-I restricted self-ligands presented by splenic dendritic cells.
a, Representative flow cytometric analysis of Ki67 expression by TCRβ+ CD8β+ Thy1.1+ cells from the spleens of TCRrg mice expressing the indicated CD8-MP TCRs (top) and CD8-Naïve TCRs (bottom), analyzed 6 weeks after bone marrow reconstitution. The percentage of cells in the indicated gates is denoted. Data are representative of five independent experiments.
b, Left: Summary plot of pooled data from (a) showing the frequency of TCRβ+ CD8β+ Thy1.1+ cells that are positive for Ki67 staining for the listed TCRs. Right: Data from the left panel were pooled from the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 15, CD8-MP TCRrg mice; n = 14, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from five independent experiments.
c, Summary plot of pooled data from (a) showing the absolute number of TCRβ+ CD8β+ Thy1.1+ cells that are positive for Ki67 staining for the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 15, CD8-MP TCRrg mice; n = 14, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from five independent experiments.
d, CD8+ naïve-phenotype (CD44loCD122neg) T cells were sorted from TCRrg mice expressing the indicated TCRs, labeled with CellTrace-Violet (CTV) and used as a probe for antigen. 1 × 104 TCRrg cells were cultured with 5 × 104 CD11c+ cells isolated from the spleens of SPF or GF C57BL/6 mice plus rmIL-2. Dilution of CTV was assessed by flow cytometry on day 5. Where indicated, cells were cultured with isotype control antibody or anti-MHC-I blocking antibody. The percentage of cells within the indicated gates is denoted. Data are representative of six independent experiments.
e, Summary plot of pooled data showing the frequency of divided cells in the indicated co-cultures from (d). Each sample represents an individual co-culture. The median, upper quartile, and lower quartile are indicated by the box plot and the whiskers indicate the minimum and maximum. GSSrg (n = 6 isotype, n = 6 anti-MHC-I, n = 4 GF), DTGrg (n = 4 isotype, n = 4 anti-MHC-I, n = 7 GF), SAVrg (n = 9 isotype, n = 9 anti-MHC-I, n = 4 GF), SATrg (n = 6 isotype, n = 6 anti-MHC-I, n = 4 GF), SMNrg (n = 6 isotype, n = 6 anti-MHC-I, n = 7 GF), RDTrg (n = 6 isotype, n = 6 anti-MHC-I, n = 4 GF), LNNrg (n = 9 isotype, n = 9 anti-MHC-I, n = 4 GF), DYQrg (n = 7 isotype, n = 7 anti-MHC-I, n = 4 GF). For each TCR, the frequency of divided cells in the isotype control condition (or GF condition) was compared to the proliferation in the anti-MHC-I condition using one-way ANOVA with Bonferroni post-test analysis, comparing all pairs of columns (ANOVA p < 0.0001, F = 103.1, df = 143). Adjusted p-values from the Bonferroni post-test are depicted: n.s., not significant, p > 0.9999; **p = 0.0014; ***p < 0.0001, Data are pooled from six independent experiments.
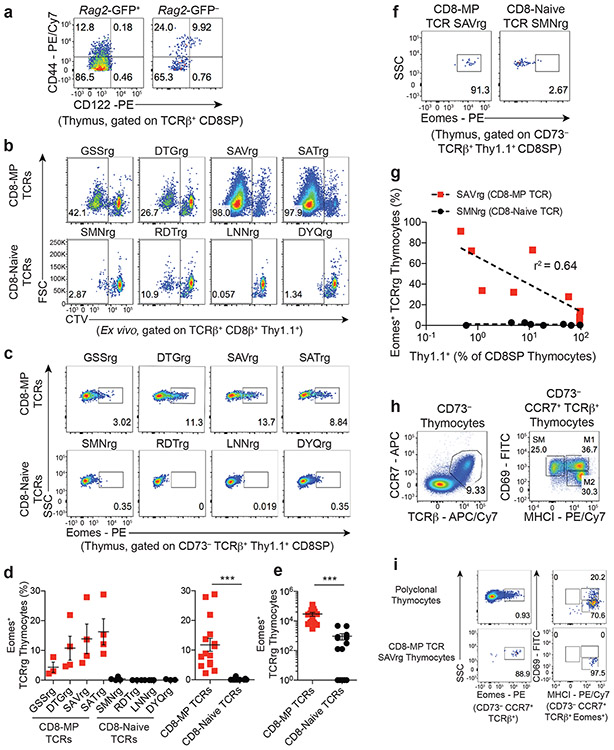
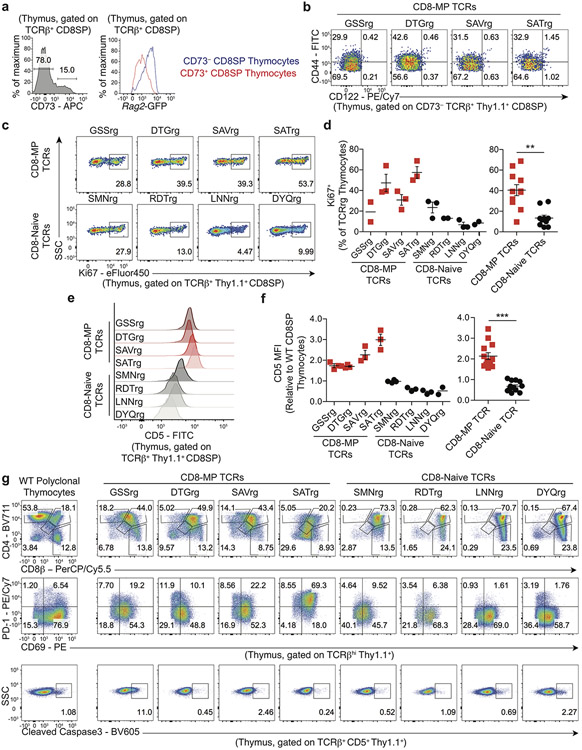
Our cumulative data suggest that CD8-MP differentiation is an orchestrated TCR-dependent process driven by reactivity to self-ligands. We next set out to define the stage at which CD8-MP differentiation is triggered. Previous studies report that CD44hiCD122+ CD8+ T cells first appear in the periphery of neonatal B6 mice12, suggesting that CD8-MP cells differentiate in the periphery1, 3. Congruent with this idea, we found that CD44hiCD122+ cells were not detected amongst polyclonal GFP+ CD8+CD4− (“CD8 single-positive”, hereafter referred to as “CD8SP”) thymocytes from adult Rag2-green fluorescent protein (GFP) reporter mice (Fig. 4a), in which GFP expression marks newly developing thymocytes that have recently undergone Rag2-dependent TCR rearrangement20.
Fig. 4. Thymocytes expressing CD8-MP TCRs upregulate Eomes during thymic maturation.
a, CD44hiCD122+ cells are not detected amongst newly developing thymocytes. Representative flow cytometric analysis of CD44 vs. CD122 expression by GFP+ (left) or GFP− (right) CD8+ T cells isolated from the thymus of a 7-week-old Rag2-GFP mouse. Rag2-GFP+ cells represent newly developing T cells that have recently rearranged their antigen receptors. Cells are gated on TCRβ+ CD8SP thymocytes. The percentage of cells within the indicated gates is denoted. Data are representative of five independent experiments.
b, CD8-MP clones exhibit reactivity to thymic dendritic cells. CD8+ naïve-phenotype (CD44loCD122−) T cells were sorted from TCRrg mice expressing the indicated TCRs, labeled with CellTrace-Violet (CTV) and used as a probe for antigen. 1 × 104 TCRrg cells were cultured with 5 × 104 CD11c+ cells isolated from the thymi of SPF C57BL/6 mice plus rmIL-2. Dilution of CTV was assessed by flow cytometry on day 5. At analysis, cells are gated on TCRβ+ CD8β+ Thy1.1+ TCRrg cells. The percentage of cells within the indicated gates is denoted. Data are representative of three independent experiments.
c, Representative flow cytometric analysis of Eomes expression by CD73− TCRβ+ Thy1.1+ CD8SP TCRrg thymocytes expressing the indicated TCRs, analyzed 6 weeks after bone marrow reconstitution. The percentage of cells within the indicated gates is denoted. Data are representative of five independent experiments.
d, Left: Summary plot of pooled data from (c) showing the frequency of TCRrg CD8SP thymocytes expressing Eomes for the indicated TCRs. Right: Data from the left panel were pooled from the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 15, CD8-MP TCRrg mice; n = 14, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from five independent experiments.
e, Summary plot of pooled data from (c) showing the absolute number of TCRrg CD8SP thymocytes expressing Eomes for the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 15, CD8-MP TCRrg mice; n = 14, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from five independent experiments.
f, Representative flow cytometric analysis of Eomes expression by CD73− TCRβ+ Thy1.1+ CD8SP TCRrg cells from "low frequency" TCRrg mice expressing the indicated TCRs, assessed 6 weeks after bone marrow reconstitution. The percentage of cells within the indicated gates is denoted. Data are representative of three independent experiments.
g, Summary plot of multiple TCRg mice from (f) demonstrating a “niche effect” of Eomes expression in developing TCRrg thymocytes. For the SAV and SMN TCRs, the percentage of TCRrg cells that express Eomes is plotted versus the frequency of Thy1.1+ TCRrg thymocytes as a percentage of all CD8SP thymocytes. Dashed lines indicate best-fit semilog curves. Data are pooled from nine independent experiments.
h, Representative flow cytometric plots of CD69 vs. MHCI expression for CD73− CCR7+ TCRβ+ thymocytes demonstrating the gating strategy to identify three subsets of medullary thymocytes, as described in 27. SM, semi-mature; M1, mature 1; M2, mature 2. The percentage of cells within the indicated gates is denoted. Data are representative of two independent experiments.
i, Left: Representative flow cytometric analysis of Eomes expression by CD73− CCR7+ TCRβ+ polyclonal (top) or monoclonal CD8-MP SAVrg (bottom) thymocytes. Right: Representative analysis of CD69 vs. MHCI expression by Eomes expressing cells. The percentage of cells within the indicated gates is denoted. Data are representative of two independent experiments.
To address this question at the clonal level, we utilized our TCRrg approach to track the developmental trajectories of distinct CD8+ T cell clones that are destined to adopt a CD8-MP phenotype. For our thymic analyses of TCRrg mice, we used CD73-negativity as a surrogate marker for Rag2-GFP+ cells, as >97% of CD73− cells are Rag2-GFP+ in the thymus (21 and Extended Data 6a). Consistent with observations for polyclonal cells, we found that TCRrg cells expressing CD8-MP TCRs did not adopt a CD44hiCD122+ phenotype in the thymus (Extended Data 6b). However, CD8-MP TCRrg thymocytes exhibited elevated percentages of Ki67+ cells and increased densities of CD5 when compared to CD8-Naïve TCRrg thymocytes, suggestive of elevated TCR signaling in the thymus (Extended Data 6c-f). Consistent with this observation, we found that CD11c+ DCs isolated from the thymus stimulated the in vitro proliferation of TCRrg cells expressing CD8-MP, but not CD8-Naïve TCRs (Fig. 4b), indicating that ligands recognized by CD8-MP clones are displayed by DCs in the thymus. Comparative analysis of TCRrg thymocytes revealed that CD8-MP clones at the CD4+CD8+ stage displayed reduced densities of the CD4 and CD8 co-receptors (Extended Data 6g), a potential indicator of clonal deletion22. However, the findings that CD8-MP clones exhibited negligible staining for cleaved Caspase 3 (a marker of ongoing apoptosis, Extended Data 6g) and were readily detected within polyclonal and monoclonal repertoires suggests that CD8-MP clones are not substantially impacted by clonal deletion.
CD8-MP clones upregulate Eomes in the thymus
Collectively, the above results suggest that CD8-MP cells encounter endogenous self-ligands in the thymus. Thus, we considered the possibility that CD8-MP differentiation is triggered by the recognition of self-ligands in the thymus, and that the upregulation of the CD44hiCD122+ phenotype is delayed until cells emigrate to the periphery. To test this idea, we looked for early hallmarks of CD8-MP differentiation that lie upstream of CD122 upregulation. Our analysis focused on Eomes, a transcription factor that is highly expressed by peripheral CD8-MP cells, is required for the differentiation and/or survival of CD8-MP cells23, and promotes CD122 upregulation by direct binding to the Il2rb promoter24. Strikingly, in TCRrg mice expressing CD8-MP TCRs, a fraction of TCRrg CD8SP thymocytes upregulated expression of Eomes, whereas Eomes upregulation was not observed for TCRrg thymocytes expressing CD8-Naïve TCRs (Fig. 4c-e). These findings suggest that the differentiation of many CD8-MP clones is triggered during T cell maturation in the thymus, prior to upregulation of the CD44hiCD122+ phenotype in the periphery. To examine this phenomenon further, we determined whether there are saturable niches driving Eomes upregulation by select CD8-MP clones during thymic maturation, as has been described for Foxp3+ regulatory T cells25, 26. To do this, we generated a series of low-frequency TCRrg mice expressing the SAV CD8-MP TCR, and calculated the extent of Eomes upregulation at varying clonal frequencies. This approach demonstrated that the fraction of TCRrg thymocytes expressing Eomes increased with decreasing clonal frequency (peaking at >90%, Fig. 4f,g), suggesting the existence of a saturable niche supporting Eomes upregulation by the SAV CD8-MP clone. In contrast, we did not observe this phenomenon with the control SMN CD8-Naïve TCR (Fig. 4f,g), further highlighting the TCR-dependency of Eomes upregulation.
Next, we performed additional phenotypic analyses to examine hallmarks of antigen sensing at different stages of thymic maturation. To do this, we used a staging approach defined previously27, in which post-selection TCRβhiCCR7+ thymocytes are sub-divided into cells of progressing maturational stage, ranging from semi-mature (CD69+MHC-I−), mature-1 (CD69+MHC-I+), and mature-2 (CD69−MHC-I+) (Fig. 4h). Notably, we found that for developing polyclonal T cells and the CD8-MP SAVrg clone, Eomes+ cells primarily exhibited a CD69−MHC-I+ mature-2 phenotype (Fig. 4i), suggesting that Eomes expression is induced in the latest stages of thymic maturation, when cells are known to reside in the medulla.
Eomes identifies polyclonal thymic precursors of CD8-MP T cells
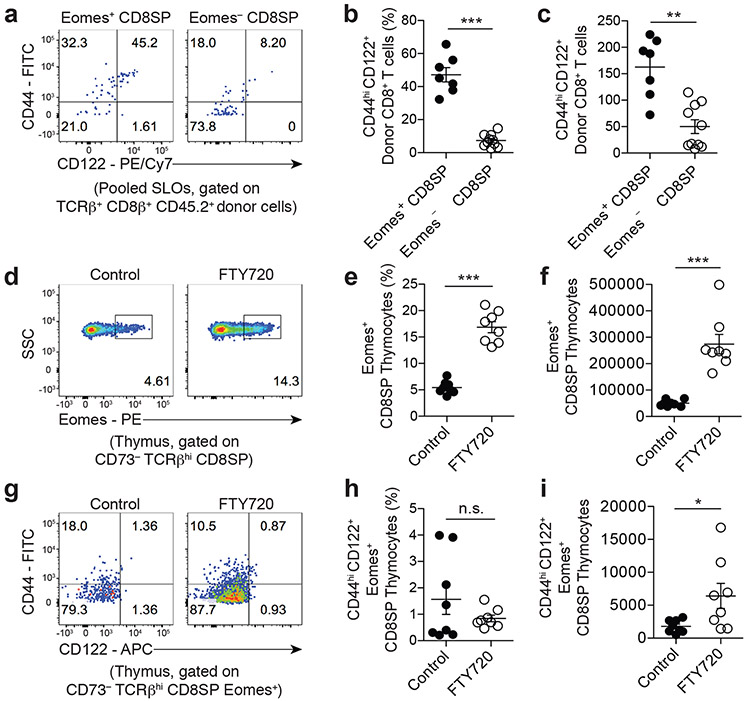
Next, we aimed to determine whether the principles observed using our clonal approach extended to polyclonal T cell populations in wild-type mice. Specifically, we hypothesized that Eomes-expressing CD8SP thymocytes represent thymic intermediates that are destined to upregulate the CD44hiCD122+ CD8-MP phenotype following emigration to the periphery. To test this hypothesis, we purified Eomes-GFP+ or Eomes-GFP− CD73− CD69− CD8SP thymocytes from 4-week-old Eomes-GFP reporter mice28, and transferred these cells separately into the periphery of congenically disparate wild-type hosts via intravenous injection. 3 weeks post-transfer, we found that ~47% of the Eomes-GFP+ donor cells recovered from recipient mice exhibited a CD44hiCD122+ phenotype, compared to ~7% of Eomes-GFP− donor cells (Fig. 5a-c), indicating that Eomes-expressing CD8SP thymocytes are enriched for thymic intermediates with the potential to upregulate the CD8-MP phenotype in the periphery. To determine whether Eomes+ CD8SP thymocytes upregulate CD44 and CD122 when thymic egress is pharmacologically delayed, we utilized FTY720 to block sphingosine 1-phosphate receptor-1 (S1PR1) and prevent thymic egress. We treated B6 mice with FTY720 or PBS control for 5 days and assessed the phenotype of CD8SP thymocytes at day 6. This treatment induced a marked increase in the percentage and number of Eomes-expressing CD8SP thymocytes (Fig. 5d-f), but did not induce an increase in the fraction of Eomes+ thymocytes that exhibit a CD44hiCD122+ phenotype (Fig. 5g-i). This finding is consistent with a model in which CD8-MP differentiation is triggered in the thymus, but requires a subsequent consolidation phase that can only be conferred in the periphery.
Fig. 5. Eomes identifies polyclonal thymic precursors of CD8-MP T cells.
a-c, 1 × 105 FACS-purified Eomes-GFP+ or Eomes-GFP− CD73− CD69− mature CD8SP thymocytes from the thymi of 4-week-old Eomes-GFP reporter mice were transferred intravenously into congenically disparate CD45.1+ recipients. 3-weeks post-transfer, the fate of the donor CD45.2+ cells was assessed in the pooled spleen and lymph nodes of recipient mice.
a, Representative flow cytometric analysis of CD44 vs. CD122 expression by donor CD45.2+ T cells recovered after transfer. The percentage of cells falling in the indicated gates is denoted. Data are representative of three independent experiments.
b, Summary plot of pooled data from (a) showing the frequency of recovered TCRβ+ CD8β+ CD45.2+ cells exhibiting a CD44hiCD122+ phenotype for the indicated donor cells. Each symbol represents an individual mouse. n = 7, Eomes-GFP+ CD8SP; n = 10, Eomes-GFP− CD8SP. Mean ± SEM is indicated. ***p = 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from three independent experiments.
c, Summary plot of pooled data from (a) showing the absolute number of recovered TCRβ+ CD8β+ CD45.2+ cells exhibiting a CD44hiCD122+ phenotype for the indicated donor cells. Each symbol represents an individual mouse. n = 7, Eomes-GFP+ CD8SP; n = 10, Eomes-GFP− CD8SP. Mean ± SEM is indicated. **p = 0.0020, two-tailed nonparametric Mann-Whitney test. Data are pooled from three independent experiments.
d-i, 5-week-old B6 mice were treated with 7 mg/ kg FTY720 or PBS control I.P. every other day for 5 days. On day 6 the thymocytes of the mice were assessed.
d, Representative flow cytometric analysis of Eomes expression by CD73− TCRβ+ CD8SP thymocytes in B6 mice receiving the indicated treatments. The percentage of cells falling in the indicated gates is denoted. Data are representative of two independent experiments.
e, Summary plot of pooled data from (d) showing the frequency of CD73− TCRβ+ CD8SP thymocytes exhibiting a Eomes+ phenotype in B6 mice receiving the indicated treatments. Each symbol represents an individual mouse. n = 8, control; n = 8, FTY720. Mean ± SEM is indicated. ***p = 0.0002, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
f, Summary plot of pooled data from (d) showing the absolute number of CD73− TCRβ+ CD8SP thymocytes exhibiting a Eomes+ phenotype in B6 mice receiving the indicated treatments. Each symbol represents an individual mouse. n = 8, control; n = 8, FTY720. Mean ± SEM is indicated. ***p = 0.0002, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
g, Representative flow cytometric analysis of CD44 vs. CD122 expression by CD73− TCRβ+ CD8SP Eomes+ thymocytes in B6 mice receiving the indicated treatments. The percentage of cells falling in the indicated gates is denoted. Data are representative of two independent experiments
h, Summary plot of pooled data from (g) showing the frequency of CD73− TCRβ+ CD8SP Eomes+ thymocytes exhibiting a CD44hiCD122+ phenotype in B6 mice receiving the indicated treatments. Each symbol represents an individual mouse. n = 8, control; n = 8, FTY720. Mean ± SEM is indicated. n.s., not significant, p = 0.9591, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
i, Summary plot of pooled data from (g) showing the absolute number of CD73− TCRβ+ CD8SP Eomes+ thymocytes exhibiting a CD44hiCD122+ phenotype in B6 mice receiving the indicated treatments. Each symbol represents an individual mouse. n = 8, control; n = 8, FTY720. Mean ± SEM is indicated. *p = 0.0148, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
CD8-MP cells infiltrate murine prostate tumors
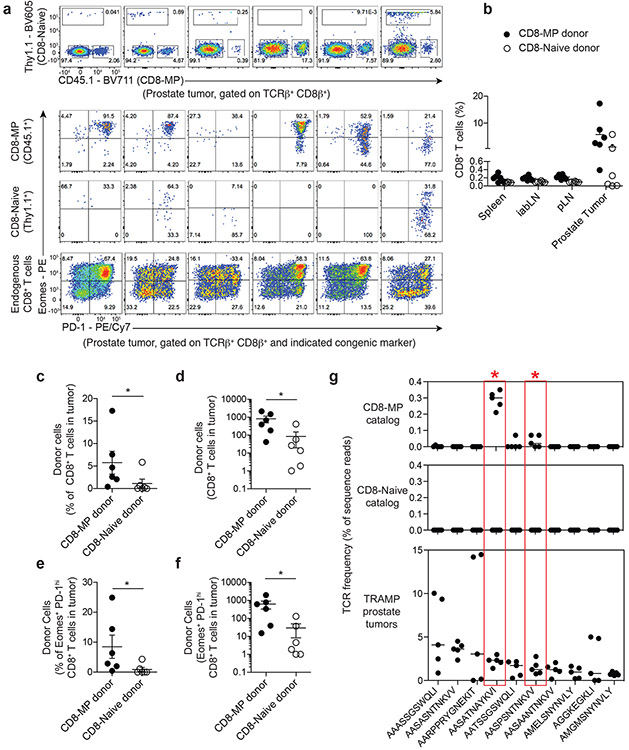
As introduced above, there remains a lack of available markers to distinguish CD8-MP cells from conventional CD8+ effector or memory T cells in the context of immune activation. Thus, the contribution of CD8-MP cells to the immune response to human and murine cancers is undefined. To examine this question from a unique perspective, we co-transferred congenically disparate polyclonal CD8-Naive and polyclonal CD8-MP cells into 2-month-old TRAMP males, in which transgenic expression of a model oncogene drives the development of prostatic adenocarcinoma with high penetrance29. Four months later, when the mice developed advanced prostate tumors, we analyzed the fate of the transferred cells. We found that donor CD8-MP cells constituted a substantial fraction of the tumor-infiltrating CD8+ T cells, ranging from 2-17% of intratumoral CD8+ T cells (Fig. 6a-f). Strikingly, the majority of donor CD8-MP cells expressed high densities of the inhibitory receptor PD-1 (Fig. 6a-f).
Fig. 6. CD8-MP cells infiltrate TRAMP prostate tumors and express high densities of PD-1.
a-f, 1 × 106 FACS-purified Thy1.1+ CD8-Naïve (CD44loCD122− Eomes-GFP−) and 1 × 106 CD45.1+ CD8-MP (CD44hiCD122+) CD8+ T cells from pooled spleen and lymph nodes of 2-month-old Thy1.1+ Eomes-GFP reporter mice or CD45.1+ B6.SJL mice were co-transferred intravenously into congenically disparate CD45.2+ TRAMP+/+ male recipients. 4 months later, the fate of the donor cells was assessed in the prostate tumors of recipient mice.
a, (Top) Flow cytometric analysis of congenic marker (Thy1.1 vs. CD45.1) expression by CD8+ T cells infiltrating the six prostate tumors. The percentage of cells falling in the indicated gates is denoted. (Bottom) Flow cytometric analysis of Eomes vs. PD-1 expression by TCRβ+ CD8+ T cells from indicated congenically marked populations infiltrating the six prostate tumors. The percentage of cells falling in the indicated gates is denoted. Data are pooled from two independent experiments.
b, Summary plot of pooled data of the frequency of CD8-MP (CD45.1+) and CD8-Naïve (Thy1.1+) TCRβ+ CD8+ T cells from indicated lymphoid sites of TRAMP+/+ recipients. Each symbol represents an individual mouse, n = 6. Mean ± SEM is indicated. Data are pooled from two independent experiments.
c, Summary plot of pooled data of the frequency of CD8-MP (CD45.1+) and CD8-Naïve (Thy1.1+) TCRβ+ CD8+ T cells infiltrating prostate tumors. Each symbol represents an individual mouse, n = 6. Mean ± SEM is indicated. *p = 0.0411, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
d, Summary plot of pooled data of the absolute number of CD8-MP (CD45.1+) and CD8-Naïve (Thy1.1+) TCRβ+ CD8+ T cells infiltrating prostate tumors. Each symbol represents an individual mouse, n = 6. Mean ± SEM is indicated. *p = 0.0260, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
e, Summary plot of pooled data of the frequency of CD8-MP (CD45.1+) and CD8-Naïve (Thy1.1+) amongst the Eomes+ PD-1+ TCRβ+ CD8+ T cells infiltrating prostate tumors. Each symbol represents an individual mouse, n = 6. Mean ± SEM is indicated. *p = 0.0295, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
f, Summary plot of pooled data of the absolute number of CD8-MP (CD45.1+) and CD8-Naïve (Thy1.1+) Eomes+ PD-1+ TCRβ+ CD8+ T cells infiltrating prostate tumors. Each symbol represents an individual mouse, n = 6. Mean ± SEM is indicated. *p = 0.0200, two-tailed nonparametric Mann-Whitney test. Data are pooled from two independent experiments.
g, (Bottom) CD8+ T cells were isolated from the prostate tumors of N = 5 27-week-old TRAMP+/− males expressing a fixed TCRβ chain (TCRβtg), and subjected to complete TCRα sequencing using the iRepertoire platform. Summary plots of the frequencies of the 10 most prevalent recurrent clones. The predicted CDR3α amino acid sequence of each clone is shown. (Top and middle) For the top 10 recurrent intratumoral clones listed at the bottom, the top and middle plots depict the frequencies of these clones in the CD8-MP and CD8-Naive TCR data sets derived from the secondary lymphoid organs of tumor-free mice (Fig. 1). Median is indicated. The asterisks and red boxes highlight the “SAT” and “SPS” clones, which represent clones that are significantly skewed to the CD8-MP subset in tumor-free mice (FDR < 0.05, defined in Fig. 1) and recurrently enriched in prostate tumors. Statistical analysis of differential representation in CD8-MP and CD8-Naive subsets was done using EdgeR-based methods (see Methods).
To further address the role of CD8-MP cells in anti-tumor immunity, we utilized our TCR sequencing approach to determine the extent to which CD8-MP-skewed clones contribute to the pool of tumor-infiltrating lymphocytes (TILs) in TRAMP mice. To do this, we isolated CD8+ T cells from the prostate tumors of five 27-week-old TRAMP+/− males expressing the fixed TCRβtg chain, and subjected these samples to deep Tcra sequencing (Supplementary Table 1). The survey identified numerous CD8+ T cell clones that are recurrently enriched in TRAMP prostate tumors (Fig. 6g, bottom). In addition, by examining the frequency of these intratumoral clones in the CD8-MP and CD8-Naive TCR data sets derived from the secondary lymphoid organs of tumor-free mice, we found that two of the ten most prevalent intratumoral clones were skewed to the CD8-MP subset (Fig. 6g, red boxes), and that the 8 remaining clones were rare within both the CD8-MP and CD8-Naive data sets. Notably, one of these CD8-MP-skewed clones was the “SAT” (AASATNAYKVI) clone examined elsewhere in this study. Thus, the use of comparative TCR profiling revealed that TRAMP prostate tumors drive the recurrent enrichment of self-specific “tumor-associated” CD8-MP clones that are uncommon in the periphery but are selectively enriched in prostate tumors. These collective findings demonstrate that recurrent CD8-MP clones make measurable contributions to the tumor-infiltrating T cell pool in TRAMP mice and express high densities of PD-1, suggesting that intratumoral CD8-MP cells may functionally impact anti-tumor immunity and may be directly impacted by anti-PD-1 or anti-PD-L1 checkpoint blockade antibodies.
Taken together, our findings demonstrate that CD8-MP differentiation is a robust TCR-directed process that is triggered by the recognition of self-ligands in the thymus, that Eomes identifies thymic precursors with a propensity to upregulate the CD8-MP phenotype in the periphery, and that CD8-MP cells make measurable contributions to the immune infiltrate of autochthonous prostate tumors.
DISCUSSION
Our cumulative findings reveal the unexpected finding that CD8-MP differentiation parallels the development of Foxp3+ Treg cells in several respects. Specifically, CD8-MP differentiation is a TCR-instructed process that is triggered by the recognition of self-ligands in the thymus, occurs optimally at low clonal frequencies25, 26, and involves a two-step process marked by an initial TCR-dependent triggering step followed by a second phase of consolidation30, 31. These findings challenge a common notion that CD8-MP cells represent "lymphopenia-induced memory" T cells that differentiate from naive CD8+ T cells in the periphery1, 3. The finding that hundreds of recurrent CD8+ T cell clones, constituting ~65% of the CD8-MP repertoire, strongly and reproducibly segregate to the CD8-MP subset demonstrates that TCR-dependent differentiation is a broad principle of most CD8-MP cells in B6 mice, and implies the existence of dedicated mechanisms coordinating the differentiation of these cells. Given our data showing that many CD8-MP clones exhibit overt reactivity to self-ligands, we hypothesize that the robust segregation of CD8-MP-biased clones to the CD8-MP subset is critical for removing overtly self-reactive CD8+ T cells from the naive compartment. Lastly, while TCR-independent factors such as IL-15 contribute to CD8-MP differentiation or survival6, 23, they are not sufficient to direct CD8-MP differentiation in unprimed mice, as only select CD8+ T cell clones will adopt the CD8-MP phenotype within the endogenous repertoire.
Our data demonstrate that some CD8-MP clones display overt reactivity to classical MHC-I-restricted ligands displayed by both thymic DCs and splenic DCs from germ-free mice. Despite this, CD8-MP clones readily populate the peripheral repertoire and are not substantially impacted by clonal deletion. This suggests either that the TCR/pMHC-I binding properties that trigger CD8-MP differentiation are distinct from the properties that drive clonal deletion, or that unique contextual cues (such as anatomical region, identity of the antigen presenting cell, or thymocyte maturational stage) trigger CD8-MP differentiation without inducing extensive deletion.
Previous studies using pMHC-I tetramer enrichment assays demonstrated that in unprimed mice, a percentage of CD8+ T cells reactive to any given foreign pMHC-I complex display a CD44hiCD122+ phenotype2, 12 and exhibit innate-like effector functions in the early stages of pathogen challenge2, 7. Based on our findings presented here, we suggest that foreign pMHC-I-specific CD8-MP cells in unprimed mice likely represent CD8-MP clones that were selected on self-pMHC-I ligands, but cross-react with foreign pMHC-I ligands due to the plasticity inherent in TCR-pMHC ligand recognition32. Thus, we suggest that selection on endogenous self-ligands plays a critical role in poising CD8-MP cells for rapid recruitment into the early phases of an immune response2, 7.
Seminal studies of CD8+ T cell memory differentiation and survival examined the biology of lymphopenia-induced memory (LIM) cells, a population of CD44hiCD122+ T cells that emerge following the intravenous transfer of mature naive-phenotype CD8+ T cells into lymphopenic hosts33. Based on the fact that the phenotypic and functional properties of CD8-MP cells are similar to those of LIM cells, it has been tempting to equate these populations. However, two pieces of evidence presented here suggest that naturally occurring CD8-MP cells in B6 mice are distinct from LIM cells. First, we find that the differentiation of many CD8-MP clones is initiated in the thymus, suggesting that the forces driving LIM cell differentiation in the periphery may not be relevant for many naturally occurring CD8-MP clones. Second, our finding that CD8-MP differentiation is a TCR-instructed process, with hundreds of CD8-MP-biased clones exhibiting robust segregation to the CD8-MP subset, suggests that the naive-phenotype donor cells used in studies of LIM cell differentiation are likely depleted of the relevant TCRs that are naturally expressed by CD8-MP cells, implying that the antigen specificities of CD8-MP cells and LIM cells are inherently distinct.
Lastly, our transfer experiments and comparative TCR profiling approach revealed that CD8-MP cells are recurrently enriched in autochthonous prostate tumors in TRAMP mice and adopt a PD-1hi Eomes+ phenotype within tumors. Previously, the contribution of CD8-MP cells to the anti-tumor immune response in human or murine cancers had been largely undefined due to the lack of available markers or signatures to identify CD8-MP cells and distinguish them from bona fide tumor-specific CD8+ T cells. In this regard, an expanding body of evidence suggests that human tumors are infiltrated by a phenotypically diverse array of CD8+ T cells34, 35, 36, 37, 38, and that only a minor fraction of such cells are overtly reactive to tumor-expressed antigens39, 40. Given our findings presented here, we hypothesize that self-specific CD8-MP cells, selected on endogenous self-ligands in the thymus, make substantial contributions to the infiltrate of human and murine cancers and may be directly impacted by checkpoint blockade antibodies targeting the PD-1 axis. Elucidation of the functional role of CD8-MP cells in the tumor context will require the identification of unique markers that are exclusively expressed by tumor-associated CD8-MP cells, plus the development of loss-of-function approaches for the selective depletion of CD8-MP cells.
METHODS
Mice
The following mice were purchased from the Jackson Laboratory, and bred and maintained at the University of Chicago under specific-pathogen free conditions: C57BL/6J (B6) mice, CD45.1/.1 B6.SJL-Ptprca Pepcb/BoyJ mice, Rag1−/− B6.129S7-Rag1tm1Mom/J mice, Tcra−/− B6.129S2-Tcratm1Mom/J mice, CD4-Cre B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ mice and TRAMP C57BL/6-Tg(TRAMP)8247Ng/J mice. “TCRβtg” mice expressing a fixed TCRβ chain of sequence TRBV26-ASSLGSSYEQY were generated as described previously15. Rag2-GFP C57BL/6-Tg(Rag2-EGFP)1Mnz/J mice were received from the laboratory of M. Nussenzweig at Rockefeller University20. Eomes-GFP C57BL/6-Eomestm1.1Twa/Cnbc mice were received from the laboratory of T. Walzer at Inserm28. All mice were bred and maintained in accordance with the animal care and use regulations of the University of Chicago. Germ free C57BL/6 mice were housed at the University of Chicago gnotobiotic facility under strict germ-free conditions. Both male and female mice were used across individual experiments. Mice were not randomized for assignment to experimental group and experiments were not conducted in a blinded fashion.
Antibodies, Flow Cytometry, and FACs
All antibodies used were purchased from BioLegend or Fisher Scientific. Cells were stained with conjugated antibodies specific for the following proteins (with clone name in parentheses): Active caspase-3 (C92-605), CCR7 (4B12), CD4 (GK1.5, RM4-4, or RM4-5), CD5 (53-7.3), CD8β (YTS156.7.7), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD49d (R1-2), CD62L (MEL-14), CD69 (H1.2F3), CD73 (TY/11.8), CD122 (TM-β1), CD127 (A7R24), Eomes (Dan11mag), H-2Kb (AF6-88.5), Ki67 (SolA15), PD-1 (RMP1-30), TCRβ (H57-597), and Thy1.1 (OX-7). Cells were stained for 20 min on ice in staining buffer (phosphate-buffered-saline with 2% FCS, 0.1% NaN3, 5% normal rat serum, 5% normal mouse serum, 5% normal rabbit serum (all sera from Jackson Labs), and 10 μg/mL 2.4G2 antibody). Intracellular staining for Eomes, Ki67, and active Caspase3 was performed using fixation and permeabilization buffers from eBioscience. Cells being stained for CCR7 were incubated for 30 min at 23°C with surface stain antibodies in 2% FCS in PBS. Flow cytometry was performed on an LSR Fortessa (BD Biosciences) and data was analyzed using FlowJo software (Tree Star). FACS was performed using a FACSAria (BD Biosciences). An example of the gating strategy is shown in Extended Data 7.
TCR sequence analysis
CD8-Naïve (CD44loCD122−) and CD8-MP (CD44hiCD122+) T cells were purified by FACS from 9-week-old TCRβTg males. To identify recurrent, tumor-infiltrating CD8+ T cell clones, CD8+ T cells were purified by FACs from the prostate tumors of 27-week-old TRAMP+/− TCRβTg males. RNA was extracted from purified subsets and subjected to complete Tcra sequencing using the Amp2Seq service from iRepertoire, a platform based on semi-quantitative multiplex PCR coupled with Illumina sequencing. This approach allows analysis of the complete TCRα repertoire, regardless of variable-region usage. Typically, >6.5 × 105 TCR sequence reads were obtained per sample. TCRs were analyzed solely based on the predicted CDR3 sequence, regardless of V-region usage. We first filtered for TCRs with CDR3 segments between 7 and 17 amino acids in length. In order to focus on recurrent TCRs, we then removed those TCRs with counts that were less than ten in Ns-1 samples where Ns was the size of the smaller group. The differential analysis was done using R/Bioconductions edgeR package. The standard edgeR normalization function calcNormFactors was called. For the differential testing the GLM method was used. Since the data consisted of paired samples from each of several mice a paired test was done with the following design:
design ← model.matrix( ~ mouse + group )
where group is the “treatment” grouping variable and mouse is the mouse id. We then processed the data using the GLM version of the dispersion estimators:
d ← estimateGLMCommonDisp(d,design)
d ← estimateGLMTrendedDisp(d,design)
d ← estimateGLMTagwiseDisp(d,design)
and then for differential testing:
fit ← glmFit(d,design)
results ← glmLRT(fit,coef=group.cont)
where group.cont is the contrast that selects for comparisons between groups. For the purposes of computing average expression in a natural scale, the normalized counts were rescaled to the original size of the dataset. This was done by setting a scaling factor that is equal to the geometric mean of the total sample counts for the samples. Significant CDR3s were filtered to an FDR less than 0.05. To analyze TCR repertoire similarity, we also analyzed CDR3 elements using the Morisita-Horn similarity index41. The grand average of hydropathy (GRAVY) value (http://www.gravy-calculator.de) for CDR3 regions was calculated by taking the sum of hydropathy values of all amino acids divided by the protein length.
Retrovirus production, infection, and generation of TCRrg mice
TCRrg mice were generated as described previously18. Briefly, Tcra sequences encoding TCRα chains of interest were cloned into a modified retroviral construct18, 19. Plat-E cells42 were used to generate retrovirus. Tcra−/− CD4-Cre+ TCRβtg+ mice on a C57BL/6 background were injected with 5-fluorouracil (APP Pharmaceuticals) 3 days prior to bone marrow harvest. Bone marrow cells were cultured for 2 days in X-Vivo 10 (Lonza) containing 15% FCS, 1% penicillin/streptomycin, mouse SCF, mouse IL-3 and mouse IL-6 (BioLegend). Cells were infected with retrovirus by spinfection in the presence of 6 μg/mL polybrene (EMD Millipore) and cultured for an additional 24 h (TCRrg bone marrow). TCRrg bone marrow from one mouse was then mixed with 5 × 106 freshly harvested bone marrow "filler" cells from Rag1−/− mice and injected intravenously into one lethally irradiated (800 rad) CD45.1/.1 B6.SJL recipient mouse. "Low frequency" TCRrg mice were generated by mixing 12-30% cultured TCRrg bone marrow with 70-88% CD45.1/.11 B6.SJL bone marrow. 5 × 106 bone marrow cells were injected intravenously into one sub-lethally irradiated (500 rad) B6.SJL recipient mouse. TCRrg and “low frequency” TCRrg mice were analyzed 6-11 weeks after bone marrow reconstitution.
In vitro T cell stimulation assay
CD8+ T cells from pooled spleen and lymph nodes (axillary, brachial, cervical, inguinal, mesenteric, and periaortic) from TCRrg mice were enriched for CD8+ T cells by MACS (Miltenyi Biotech), and Thy1.1+ CD44loCD122− CD8+ T cells were purified by FACs. Thy1.1+ CD44loCD122− CD8+ TCRrg cells were CellTrace-Violet (ThermoFisher) labeled per manufacturer instructions with slight modification. Briefly, cells were pelleted, resuspended in CellTrace-Violet (CTV) at 1:1000 dilution and incubated for 20 min at 37°C. The reaction was quenched by the addition of 13 mL of complete culture media. To isolate dendritic cells, spleens were isolated from 6-week-old SPF or GF C57BL/6 mice and thymi were isolated from 3 to 4-week-old SPF C57BL/6 mice. Spleens or thymi were injected and digested with Liberase TL (400 μg/mL, Roche) and DNase (800 μg/mL, Roche) in RPMI for 30 min at 37°C. For thymic APC isolation, EDTA (10 mM) was added to digests and enriched by layering digested thymocytes on top of a discontinuous Percoll gradient (GE Healthcare) at 1.115 g/mL in PBS, followed by centrifugation at 1,350 × g for 30 min and isolation of cells settling at the Percoll interface43. APCs were enriched from spleens or thymi for CD11c+ cells by MACS-based (Miltenyi Biotech) positive selection. 1 × 104 CTV-labeled T cells were co-cultured with 5 × 104 CD11c+ APCs and 100 U/mL recombinant mouse interleukin-2 (IL-2). Where indicated, anti-MHC-I (H2) blocking antibody (clone M1/42.3.9.8, BioXCell) or rat IgG2a isotype control antibody (clone 2A3, BioXCell) was added to indicated cultures at a final concentration of 500 μg/mL. Cell cultures were set up in 384-well ultra-low attachment, round-bottom plates (Corning). Dilution of CTV was assessed by flow cytometry on day 5.
Thymocyte adoptive transfer experiments and FTY720 treatment
Thymi were isolated from 4-week-old CD45.2+ Eomes-GFP+ mice and CD4+ thymocytes cells were depleted by incubating thymi with anti-mouse CD4 antibody conjugated to Biotin (clone RM4-5, BioLegend) for 10 min at 23°C, followed by incubation with Streptavidin beads (StemCell) for 3 min at 23°C. Cells were then placed on the EasySep magnet (StemCell) for 2 min and the supernatant was isolated. CD4− CD73− CD69− mature CD8β+ Eomes-GFP+ and Eomes-GFP− thymocytes cells were purified by FACs and 1 × 105 GFP+ or GFP− cell suspensions were transferred intravenously into CD45.1+ B6.SJL hosts. 3 weeks later, CD8+ T cells from pooled spleen and lymph nodes (axillary, brachial, cervical, inguinal, and periaortic) from host mice were enriched for CD8+ T cells by MACS (Miltenyi Biotech) and the phenotype of transferred cells was analyzed by flow cytometry. To prevent thymic egress of developing T cells, B6 mice were injected I.P. every other day for 5 days with 140 μg FTY720 (Sigma) in 50 μL diH20 or PBS control. The phenotype of thymocytes was analyzed by flow cytometry on day 6.
Lymphocyte isolation from TRAMP prostate tumors
Whole male genitourinary tracts were isolated and prostate lobes (anterior, dorsolateral, and ventral) were separated by microdissection from 24-week-old TRAMP+/+ or 27-week-old TRAMP+/− TCRβTg males. Prostate lobes were injected and digested with Liberase TL (10 mg/mL, Roche) and DNase (20 mg/mL, Roche) in RPMI for 30 min at 37°C. Digested tissue was mechanically disrupted with frosted microscope slides and viable lymphocytes were enriched using Histopaque 1119 (Sigma). CD8+ T cells were analyzed by flow cytometry or purified by FACs.
Statistical Analysis
Data were analyzed using Prism software (GraphPad). Significance testing was performed using the nonparametric Mann-Whitney test (two-tailed) or one-way ANOVA with Bonferroni’s multiple comparisons test (two-tailed). Statistical analysis of TCR sequencing data was performing with R (The R Project for Statistical Computing) using EdgeR-based methods (see “TCR sequence analysis” section for script details). Statistical testing on CDR3 length and GRAVY distributions was performed using R. Significance testing was performed using both the paired Wilcoxon signed-rank test (two-tailed), which tests for distribution shift, and the Kolmolgorov-Smirnov test, which tests for differences in distribution shape.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon request. The TCR sequence data are available at the Gene Expression Omnibus (GEO) repository under accession number GSE145365. The script used for TCR sequence analysis is available at https://github.com/soccin/MILLER_SAVAGE_CD8MP.
Extended Data
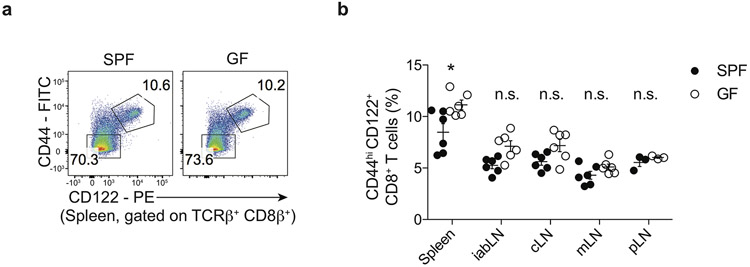
Extended Data 1. The frequency of CD8-MP cells is not diminished in germ-free mice.
a, Representative flow-cytometric analysis of CD44 vs. CD122 expression by CD8β+ T cells from the spleens of 16-week-old C57BL/6 specific pathogen free (SPF) and germ-free (GF) mice. The percentage of cells falling in the indicated gates is denoted. Data are representative of two independent experiments
b, Summary plot of the frequency of CD44hiCD122+ expressing TCRβ+ CD8β+ T cells from the indicated lymphoid sites in 6 to 16-week-old SPF or GF mice. iabLN: inguinal, axial, brachial lymph nodes; cLN: cervical lymph nodes; mLN: mesenteric lymph nodes; pLN: periaortic lymph nodes. Each symbol represents an individual mouse. Mean ± SEM is indicated. n = 6, Spleen, iabLN, cLN, mLN; n = 3, pLN. At each lymphoid site, the frequency of CD8-MP cells was compared between the SPF and GF mice using one-way ANOVA with Bonferroni post-test analysis, comparing all pairs of columns (ANOVA p < 0.0001, F = 17.43, df = 53). Adjusted p-values from the Bonferroni post-test are depicted: Spleen, *p = 0.0133; iabLN, n.s. p = 0.3669; cLN, n.s. p > 0.9999; mLN, n.s. p > 0.9999; pLN, n.s. p > 0.9999. Data are pooled from two independent experiments. (n.s., not significant)
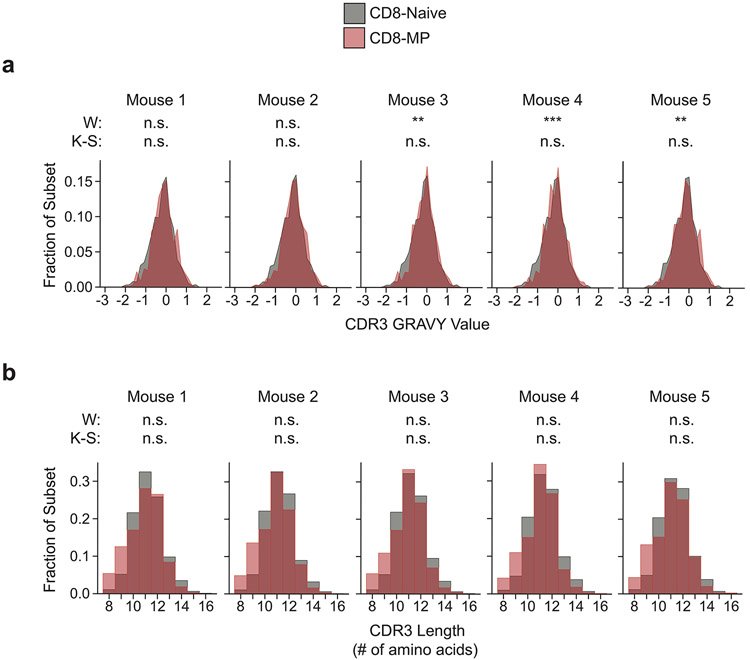
Extended Data 2. CD8-MP and CD8-Naïve CDR3α chain hydrophobicity and length analysis.
CD8-MP (CD8β+ CD44hiCD122+) and CD8-Naïve (CD8β+ CD44loCD122−) T cells were purified by FACS from the pooled spleen and lymph nodes of 9-week-old TCRβtg males and subjected to complete TCRα sequencing using the iRepertoire platform. N = 5 for CD8-MP and CD8-Naïve samples. TCRα chains were assessed solely based on their predicted CDR3 segment, regardless of V-region usage.
a, Histograms depicting grand average of hydropathy (GRAVY) values for CDR3 regions of the CD8-MP (red) and CD8-Naïve (black) subsets for each mouse, n = 5 mice. Significance testing was performed with the paired, two-tailed Wilcoxon signed-rank test (W) and the paired, two-tailed Kolmolgorov-Smirnov test (K-S). Mouse 1 (CD8-MP CDR3 n = 11999, CD8-Naïve CDR3 n = 29048), p = 0.2449 (W) and p = 1 (K-S); Mouse 2 (CD8-MP CDR3 n = 3327, CD8-Naïve CDR3 n = 14736), p = 0.0511 (W) and p = 1 (K-S); Mouse 3 (CD8-MP CDR3 n = 2159, CD8-Naïve CDR3 n = 26649), **p = 0.0017 (W) and p = 1 (K-S); Mouse 4 (CD8-MP CDR3 n = 2848, CD8-Naïve CDR3 n = 24501), ***p < 0.0001 (W) and p = 1 (K-S); Mouse 5 (CD8-MP CDR3 n = 13524, CD8-Naïve CDR3 n = 25367), **p = 0.0010 (W) and p = 1 (K-S). (n.s. not significant).
b, Histograms depicting CDR3 lengths of the CD8-MP (red) and CD8-Naïve (black) subsets for each mouse, n = 5 mice. Significance testing was performed with the paired, two-tailed Wilcoxon signed-rank test (W) and the paired, two-tailed Kolmolgorov-Smirnov test (K-S). Mouse 1 (CD8-MP CDR3 n = 11999, CD8-Naïve CDR3 n = 29048), p = 0.2031 (W) and p = 1 (K-S); Mouse 2 (CD8-MP CDR3 n = 3327, CD8-Naïve CDR3 n = 14736), p = 0.2031 (W) and p = 1 (K-S); Mouse 3 (CD8-MP CDR3 n = 2159, CD8-Naïve CDR3 n = 26649), p = 0.4258 (W) and p = 1 (K-S); Mouse 4 (CD8-MP CDR3 n = 2848, CD8-Naïve CDR3 n = 24501), p = 1 (W) and p = 1 (K-S); Mouse 5 (CD8-MP CDR3 n = 13524, CD8-Naïve CDR3 n = 25367), p = 0.9102 (W) and p = 1 (K-S). (n.s. not significant).
Extended Data 3. Phenotype of TCRrg and filler CD8+ T cell populations.
(Top) Representative flow cytometric analysis of Thy1.1 vs. CD45.1 expression by TCRβ + CD8β+ cells from the spleens of indicated TCRrg mice. The percentage of cells falling in the indicated gates is denoted.
(Bottom) Representative flow cytometric analysis of CD44 vs. CD122 expression by Thy1.1+ TCRrg or CD45.1+ “filler” TCRβ + CD8β+ cells from the spleens of indicated TCRrg mice. The percentage of cells falling in the indicated gates is denoted. Data are representative of six independent experiments.
Extended Data 4. A greater fraction of CD8-MP TCRrg cells adopt the CD44hiCD122+ phenotype at lower clonal frequencies.
(Top) Representative flow cytometric analysis of Thy1.1 vs. CD45.1 expression by TCRβ+ CD8+ cells and (Bottom) CD44 vs. CD122 expression by TCRβ+ CD8+ Thy1.1+ cells from "low frequency" TCRrg mice expressing the indicated TCRs, assessed 7 weeks after bone marrow reconstitution. It should be noted that the expression of the Thy1.1 reporter varies in different TCRrg mice, but the expression of TCRβ is uniform and comparable to that of endogenous cells (not shown). The percentage of cells falling in the indicated gates is denoted. Data are representative of four independent experiments.
Extended Data 5. CD8-MP TCRrg cells are broadly distributed across lymphoid sites.
Summary plots of the frequency of Thy1.1 expressing TCRrg cells of TCRβ+ CD8β+ T cells, normalized to the spleen from the indicated lymphoid sites 6-7 weeks after bone marrow reconstitution of the indicated “low frequency” TCRrg mice. Frequencies at different lymphoid sites were normalized to the spleen to control for varying engraftment of TCRrg bone marrow across different mice. iabLN: inguinal, axial, brachial lymph nodes; cLN: cervical lymph nodes; mLN: mesenteric lymph nodes; pLN: periaortic lymph nodes. Each symbol represents an individual TCRrg mouse. n = 3, GSSrg; n = 4, DTGrg; n = 4, SAVrg; n = 3, SATrg; n = 4, SMNrg; n = 3, RDTrg; n = 4, LNNrg; n = 4, DYQrg. Mean ± SEM is indicated. Data is pooled from four independent experiments.
Extended Data 6. Thymocytes expressing CD8-MP-skewed TCRs exhibit hallmarks of elevated TCR signaling.
a, Left: Representative flow cytometric analysis of CD73 expression by TCRβ+ CD8SP thymocytes from a 7-week-old Rag2-GFP mouse. Right: Expression of Rag2-GFP on the CD73− and CD73+ CD8SP thymocyte populations. The percentage of cells falling in the indicated gates is denoted. Data are representative of two independent experiments.
b, Representative flow-cytometric analysis of CD44 and CD122 expression by indicated CD73− TCRβ+ Thy1.1+ CD8SP CD8-MP TCRrg thymocytes 6 weeks after bone marrow reconstitution. The percentage of cells falling in the indicated gates is denoted. Data are representative of five independent experiments.
c, Representative flow cytometric analysis of Ki67 expression by TCRβ+ CD8β+ Thy1.1+ cells from the thymi of TCRrg mice expressing the indicated CD8-MP TCRs (top) and CD8-Naïve TCRs (bottom), analyzed 6 weeks after bone marrow reconstitution. The percentage of cells in the indicated gates is denoted. Data are representative of five independent experiments.
d, Left: Summary plot of pooled data from (c) showing the frequency of TCRβ+ CD8β+ Thy1.1+ cells that are positive for Ki67 staining for the listed T cell clone. Right: Data from the left panel were pooled from the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 11, CD8-MP TCRrg mice; n = 10, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. **p = 0.0017, two-tailed nonparametric Mann-Whitney test. Data are pooled from five independent experiments.
e, Representative flow cytometric analysis of CD5 expression by TCRβ+ CD8β+ Thy1.1+ cells from the thymi of TCRrg mice expressing the indicated CD8-MP and CD8-Naïve TCRs, analyzed 6 weeks after bone marrow reconstitution. The percentage of cells in the indicated gates is denoted. Data are representative of six independent experiments.
f, Left: Summary plot of pooled data from (e) showing the normalized mean florescence intensity (MFI) of CD5 in TCRrg thymocytes compared to CD8SP thymocytes from a C57BL/6 thymus. Right: Data from the left panel were pooled from the CD8-MP TCRs and the CD8-Naïve TCRs. Each symbol represents an individual TCRrg mouse. n = 13, CD8-MP TCRrg mice; n = 12, CD8-Naïve TCRrg mice. Mean ± SEM is indicated. ***p < 0.0001, two-tailed nonparametric Mann-Whitney test. Data are pooled from six independent experiments.
g, Representative flow cytometric analysis of CD4 vs. CD8, PD-1 vs. CD69, and cleaved Caspase3 expression by TCRβ+ CD8β+ Thy1.1+ cells from the thymi of TCRrg mice expressing the indicated CD8-MP TCRs and CD8-Naïve TCRs, analyzed 6 weeks after bone marrow reconstitution. The percentage of cells in the indicated gates is denoted. Data are representative of three independent experiments.
Extended Data 7. Gating strategy to analyze TCRrg cells.
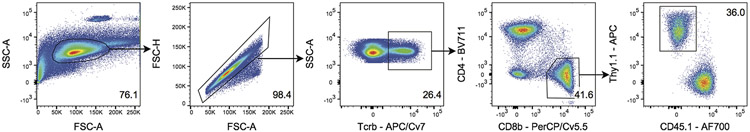
Lymphocytes were gated on forward and side scatter and doublets were removed by gating on FSC-H by FSC-A. TCRβ+ T cells were then gated for the expression of CD8β+ and CD4−. CD8β+ T cells were then gated for the expression of Thy1.1. This TCRrg cell population was used for subsequent phenotyping stains used throughout the paper. The percentage of cells in the indicated gates is denoted. Data are representative of six independent experiments.
Supplementary Material
Supplementary Table 1. Complete TCRα Sequencing Frequency Table.
For samples subjected to complete TCRα sequencing (see Methods), the frequencies of each TCR CDR3α segment, as a percentage of all TCRα sequence reads within a given sample, are listed. For this study, TCRs were cataloged solely based on predicted TCR CDR3α sequence, regardless of V-region usage. The table lists the Sample ID for the complete TCRα sequencing data set. For each biological sample, the total number of in-frame TCR sequence reads and the number of unique TCR sequence reads are also listed.
Supplementary Table 2. Selected CD8-MP and CD8-Naïve TCRs for retrogenic mouse generation.
TCRs were selected from the top 50 most prevalent, differentially expressed (FDR < 0.05) CD8-MP and CD8-Naive clones. From these lists, we sorted CDR3 sequences by TRAV usage. From each TRAV group, we selected the clone with the highest mean frequency. The selected TCR clones represent a range of frequencies (0.2 – 5.0% of the subset) and a diversity in TRAV and TRAJ usages, giving us broad representation of each CD8+ T cell repertoire. CDR3α, complementary determining region 3, alpha chain; FDR, false discovery rate; log2FC, log2 fold-change.
ACKNOWLEDGEMENTS
We thank A. Bendelac for critical reading of the manuscript. We thank S. Kasal, A. Bendelac, V. Leone, E. Chang, Z. Earley, B. Jabri, E. Hegermiller, and B. Kee for sharing reagents and resources. We thank M. Nussenzweig at Rockefeller University for Rag2-GFP mice. We thank T. Walzer at Inserm for Eomes-GFP mice. Flow cytometry and FACs were performed at the University of Chicago Cytometry and Antibody Technology Facility. This work was funded by R01-AI110507 (to P.A.S.). C.H.M. was supported by an NIH/NCI F30 predoctoral fellowship (F30-CA236061). V.L. was supported by an NIH/NCI F30 predoctoral fellowship (F30-CA217109). C.H.M and V.L. were supported by the Univeristy of Chicago Medical Scientist Training Program (T32-GM007281). D.K. was supported by T32-AI007090.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
REFERENCES
- 1.White JT, Cross EW & Kedl RM Antigen-inexperienced memory CD8(+) T cells: where they come from and why we need them. Nat Rev Immunol 17, 391–400 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haluszczak C et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med 206, 435–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jameson SC, Lee YJ & Hogquist KA Innate memory T cells. Adv Immunol 126, 173–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacomet F et al. Evidence for eomesodermin-expressing innate-like CD8(+) KIR/NKG2A(+) T cells in human adults and cord blood samples. Eur J Immunol 45, 1926–1933 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Warren HS et al. CD8 T cells expressing killer Ig-like receptors and NKG2A are present in cord blood and express a more naive phenotype than their counterparts in adult blood. J Leukoc Biol 79, 1252–1259 (2006). [DOI] [PubMed] [Google Scholar]
- 6.White JT et al. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat Commun 7, 11291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JY, Hamilton SE, Akue AD, Hogquist KA & Jameson SC Virtual memory CD8 T cells display unique functional properties. Proc Natl Acad Sci U S A 110, 13498–13503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rifa’i M, Kawamoto Y, Nakashima I & Suzuki H Essential roles of CD8+CD122+ regulatory T cells in the maintenance of T cell homeostasis. J Exp Med 200, 1123–1134 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzam HS et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med 188, 2301–2311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Persaud SP, Parker CR, Lo WL, Weber KS & Allen PM Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol 15, 266–274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong P, Barton GM, Forbush KA & Rudensky AY Dynamic tuning of T cell reactivity by self-peptide-major histocompatibility complex ligands. J Exp Med 193, 1179–1187 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akue AD, Lee JY & Jameson SC Derivation and maintenance of virtual memory CD8 T cells. J Immunol 188, 2516–2523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drobek A et al. Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD8 T cells. EMBO J 37 doi: 10.15252/embj.201798518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malchow S et al. Aire Enforces Immune Tolerance by Directing Autoreactive T Cells into the Regulatory T Cell Lineage. Immunity 44, 1102–1113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malchow S et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science 339, 1219–1224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YJ, Holzapfel KL, Zhu J, Jameson SC & Hogquist KA Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol 14, 1146–1154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leventhal DS et al. Dendritic Cells Coordinate the Development and Homeostasis of Organ-Specific Regulatory T Cells. Immunity 44, 847–859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald BD, Bunker JJ, Ishizuka IE, Jabri B & Bendelac A Elevated T cell receptor signaling identifies a thymic precursor to the TCRalphabeta(+)CD4(−)CD8beta(−) intraepithelial lymphocyte lineage. Immunity 41, 219–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner VM, Gardam S & Brink R Lineage-specific transgene expression in hematopoietic cells using a Cre-regulated retroviral vector. J Immunol Methods 360, 162–166 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Yu W et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature 400, 682–687 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Owen DL et al. Thymic regulatory T cells arise via two distinct developmental programs. Nat Immunol 20, 195–205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogquist KA Assays of thymic selection. Fetal thymus organ culture and in vitro thymocyte dulling assay. Methods Mol Biol 156, 219–232 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Sosinowski T et al. CD8alpha+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J Immunol 190, 1936–1947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Intlekofer AM et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Bautista JL et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol 10, 610–617 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung MW, Shen S & Lafaille JJ TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J Exp Med 206, 2121–2130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xing Y, Wang X, Jameson SC & Hogquist KA Late stages of T cell maturation in the thymus involve NF-kappaB and tonic type I interferon signaling. Nat Immunol 17, 565–573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daussy C et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med 211, 563–577 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenberg NM et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A 92, 3439–3443 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burchill MA et al. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity 28, 112–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lio CW & Hsieh CS A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia KC & Adams EJ How the T cell receptor sees antigen--a structural view. Cell 122, 333–336 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Sprent J & Surh CD Normal T cell homeostasis: the conversion of naive cells into memory-phenotype cells. Nat Immunol 12, 478–484 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savas P et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 24, 986–993 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Gide TN et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 35, 238–255 e236 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Guo X et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 24, 978–985 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Zheng C et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 169, 1342–1356 e1316 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Simoni Y et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 557, 575–579 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Scheper W et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat Med 25, 89–94 (2019). [DOI] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 41.Magurran. Ecological Diversity and Its Measurement. Princeton University Press: Princeton, NJ, 1988. [Google Scholar]
- 42.Morita S, Kojima T & Kitamura T Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene therapy 7, 1063–1066 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Aschenbrenner K et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol 8, 351–358 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Complete TCRα Sequencing Frequency Table.
For samples subjected to complete TCRα sequencing (see Methods), the frequencies of each TCR CDR3α segment, as a percentage of all TCRα sequence reads within a given sample, are listed. For this study, TCRs were cataloged solely based on predicted TCR CDR3α sequence, regardless of V-region usage. The table lists the Sample ID for the complete TCRα sequencing data set. For each biological sample, the total number of in-frame TCR sequence reads and the number of unique TCR sequence reads are also listed.
Supplementary Table 2. Selected CD8-MP and CD8-Naïve TCRs for retrogenic mouse generation.
TCRs were selected from the top 50 most prevalent, differentially expressed (FDR < 0.05) CD8-MP and CD8-Naive clones. From these lists, we sorted CDR3 sequences by TRAV usage. From each TRAV group, we selected the clone with the highest mean frequency. The selected TCR clones represent a range of frequencies (0.2 – 5.0% of the subset) and a diversity in TRAV and TRAJ usages, giving us broad representation of each CD8+ T cell repertoire. CDR3α, complementary determining region 3, alpha chain; FDR, false discovery rate; log2FC, log2 fold-change.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The TCR sequence data are available at the Gene Expression Omnibus (GEO) repository under accession number GSE145365. The script used for TCR sequence analysis is available at https://github.com/soccin/MILLER_SAVAGE_CD8MP.