Abstract
Middle East respiratory syndrome‐coronavirus (MERS‐CoV) is a zoonotic virus transmitted between animals and human beings. It causes MERS with high mortality rate. However, no vaccine or specific treatment is currently available. Since antiviral activity of some flavonoids is known, we applied a flavonoid library to probe inhibitory compounds against MERS‐CoV 3C‐like protease (3CLpro). Herbacetin, isobavachalcone, quercetin 3‐β‐d‐glucoside and helichrysetin were found to block the enzymatic activity of MERS‐CoV 3CLpro. The binding of the four flavonoids was also confirmed independently using a tryptophan‐based fluorescence method. The systematic comparison of the binding affinity of flavonoids made it possible to infer their scaffolds and functional groups required to bind with MERS‐CoV 3CLpro. An induced‐fit docking analysis revealed that S1 and S2 sites play a role in interaction with flavonoids. The experimental and computational study showed that flavonol and chalcone are favourite scaffolds to bind with the catalytic site of MERS‐CoV 3CLpro. It was also deduced that some flavonoid derivatives with hydrophobic or carbohydrate attached to their core structures have a good inhibitory effect. Therefore, we suggest that flavonoids with these characteristics can be used as templates to develop potent MERS‐CoV 3CLpro inhibitors.
Keywords: flavonoid, FRET, inhibitory compounds, MERS‐CoV, MERS‐CoV 3CLpro
MERS‐CoV 3CLpro was assayed with a flavonoid library and some chalcone derivatives represented promising inhibitory activity. The docking study confirmed the experimental measurement of the better efficacy of helichrysetin than cardamonin. This study showed the antiviral effect of flavonoids in the atomic level.
1. INTRODUCTION
Coronaviruses (CoVs) are a positive sense, single‐stranded RNA virus coated with viral particles (Fehr & Perlman, 2015). Together with Arteriviridae and Roniviridae, CoVs belong to the Coronaviridae family in the order Nidovirales. These CoVs can infect a wide variety of hosts, including avian, swine and humans. Human coronaviruses (HCoVs) represent a major group of CoVs associated with various respiratory diseases from common cold to serious pneumonia and bronchitis (Mesel‐Lemoine et al., 2012). Today, HCoVs are recognized as one of the most rapidly evolving viruses originated from their characteristic high genomic nucleotide substitution rates and recombination. In human, their infection is known to cause approximately one‐third of common cold. Severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) CoVs are highly pathogenic and caused a fatal first major outbreak in 2003 and 2012, respectively (de Wit, Doremalen, Falzarano, & Munster, 2016). SARS‐ and MERS‐CoVs genomes contain two open reading frames ORF1a and ORF1b translated to two respective viral polyproteins pp1a and pp1ab by host ribosomes. ORF1a encodes two cysteine proteases, a papain‐like protease (PLpro) and a 3C‐like protease (3CLpro). While PLpro cuts the first three cleavage sites of its polyprotein, 3CLpro is responsible for cleavage of the remaining eleven locations resulting in release of a total of 16 non‐structural proteins (nsp) in both SARS‐ and MERS‐CoVs. The homodimeric form of 3CLpro is active in the presence of substrates. The crystal structure of 3CLpro showed that each monomer is composed of three structural domains: domains I and II form a chymotrypsin‐like architecture with catalytic cysteine and are connected to a third C‐terminal domain via a long loop (Neddle, Lountos, & Waugh, 2015). In the proteolytic site, all 3CLpros prefer glutamine at P1 position and leucine, basic residues, small hydrophobic residues at P2, P3 and P4 positions, respectively (Chuck, Chow, Wan, & Wong, 2011). At P1′ and P2′ positions, small residues are required. Since the autocleavage process is essential for viral propagation, 3CLpro is a good drug target for anti‐coronaviral infection. In this study, we used a proteolytic method to probe MERS‐CoV 3CLpro inhibitory compounds with a synthetic peptide labelled with the EDANS‐DABCYL FRET (Fluorescence resonance energy transfer) pair (Liu et al., 2005). Since emission wavelengths of EDANS are widely overlapped with absorbance wavelengths of DABCYL, the energy emitted from EDANS will be quenched by DABCYL in a close proximity (10–100 Å). Therefore, an increment of fluorescence can be a sign to judge whether a substrate is cleaved or not in this design. Hence from the fluorescence intensity change, the proteolytic activity of protease could be detected. With a synthetic peptide with the FRET pair, a flavonoid library was screened to search MERS‐CoV 3CLpro inhibitory compounds. Recent studies showed that some flavonoids have antiviral activity in some viruses (Frabasile et al., 2017; Jucá et al.,2018; Kiat et al., 2006; Yang, Lin, Zhou, Liu, & Zhu, 2017; Zakaryan, Arabyan, Oo, & Zandi, 2017). Therefore, we assayed flavonoids and tried to induce their structural property crucial to bind with MERS‐CoV 3CLpro.
2. METHODS AND MATERIALS
2.1. Protein expression and purification
The coding sequence of MERS‐CoV nsp5, a 3C‐like protease (NCBI Ref. seq. NC_019843.3) was synthesized chemically by Bioneer and cloned into a bacteriophage T7‐based expression vector. The plasmid DNA was transformed into E. coli BL21 (DE3) for protein expression. E. coli BL21 (DE3) cells were grown on Luria–Bertani (LB) agar plates containing 150 μg/ml ampicillin. Several colonies were picked and grown in capped test‐tubes with 10 ml LB broth containing 150 μg/ml ampicillin. A cell stock composed of 0.85 ml culture and 0.15 ml glycerol was prepared and frozen at 193 K for use in a large culture. The frozen cell stock was grown in 5 ml LB medium and diluted into 2,000 ml fresh LB medium. The culture was incubated at 310 K with shaking until an OD600 of 0.6–0.8 was reached. At this point, expression of MERS‐CoV 3CLpro was induced using isopropyl‐β‐d‐1‐thiogalactopyranoside (IPTG) at a final concentration of 1 mM. The culture was further grown at 310 K for 3 hr in a shaking incubator. Cells were harvested by centrifugation at 7,650 g (6,500 rev min−1) for 10 min in a high‐speed refrigerated centrifuge at 277 K. The cultured cell paste was resuspended in 25 ml of a buffer consisting of 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10 mM imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 10 μg/ml DNase I. The cell suspension was disrupted using an ultrasonic cell disruptor (Digital Sonifier 450; Branson). Cell debris was pelleted by centrifugation at 24,900 g (15,000 rev min−1) for 30 min in a high‐speed refrigerated ultra‐centrifuge at 277 K.
The protein was purified by affinity chromatography using a 5 ml Hi‐Trap Q column (GE Healthcare) followed by a 5 ml Hi‐Trap Blue column (GE Healthcare).
2.2. FRET protease assays with MERS‐CoV 3CLpro
The custom‐synthesized fluorogenic substrate, DABCYL‐KTSAVLQSGFRKME‐EDANS (ANYGEN), was used as a substrate for the proteolytic assay using MERS‐CoV 3CLpro (Kuo, Chi, Hsu, & Liang, 2004). This substrate contains the nsp4/nsp5 cleavage sequence, GVLQ↓SG (Wua et al., 2015), and works as a generic peptide substrate for many coronavirus including MERS‐CoV 3CLpro. The peptide was dissolved in distilled water and incubated with each protease. A SpectraMax i3x Multi‐mode microplate reader (Molecular Devices) was used to measure spectral‐based fluorescence. The proteolytic activity was determined at 310 K by following the increase in fluorescence (λexcitation = 340 nm, λemission = 490 nm, bandwidths = 9, 15 nm, respectively) of EDANS upon peptide hydrolysis as a function of time. Assays were conducted in black, 96‐well plates (Nunc) in 350 μl assay buffers containing protease and substrate as follows: for the MERS‐CoV 3CLpro assay, 1.84 μl of 0.19 mM protease containing 20 mM Tris pH 8.0 was incubated with 8.75 μl of 0.1 mM substrate at 310 K for 2 hr before measuring Relative Fluorescence Unit (RFU). Before the assay, the emission spectra of 40 flavonoids were surveyed after illuminating at 340 nm to avoid the overlapping with the emission spectrum of EDANS. Every compound was suitable to be tested. The final concentration of the protease, peptide and chemical used at the assay was 1, 2.5 and 20 μM each. At first, MERS‐CoV 3CLpro and chemical were mixed and preincubated at room temperature for 1 hr. The reaction was initiated by the addition of the substrate, and each well was incubated at 310 K for 2 hr. After 2 hr, we measured the fluorescence of the mixture on the black 96‐well plate using the end‐point mode of SpectraMax i3x where the excitation wavelength was fixed to 340 nm and the emission wavelength was set to 490 nm using 9, 15 nm bandwidth, respectively. All reactions were carried out in triplicate. Among the first forty flavonoids (Table S1), four of them were picked up to further assay at a concentration range of 2 μM–320 μM. IC50 value which is the value causing 50% inhibition of catalytic activity of MERS‐CoV 3CLpro was calculated by non‐linear regression analysis using graphpad prism 7.03 (GraphPad Software).
2.3. FRET protease assays with MERS‐CoV 3CLpro in the presence of Triton X‐100
The proteolytic assay using MERS‐CoV 3CLpro in the presence of Triton X‐100 has been performed to differentiate artificial inhibitory activity of chemicals through non‐specific binding with proteases by forming aggregate or complexation. The concentration used in this study was 0.01%.
2.4. Absorption spectroscopic studies based on tryptophans of MERS‐CoV 3CLpro
To confirm the feasibility of the assay method independently, the fluorescence spectra from tryptophans of MERS‐CoV 3CLpro with candidate inhibitors were investigated (Lin, Lan, Guan, Sheng, & Zhang, 2009). The fluorescence measurements were recorded with a SpectraMax i3x Multi‐mode microplate reader (Molecular Devices) at excitation and emission wavelengths of 295 nm and 300–500 nm, respectively. The optimal excitation and emission wavelengths were determined by SoftMax Pro. Five tryptophans of MERS‐CoV 3CLpro showed a strong fluorescence emission with a peak at 340 nm at the excitation wavelength of 295 nm. In contrast, the flavonoids were almost non‐fluorescent under the same experiment condition. Each 40 μM chemical was incubated with 1 μM MERS‐CoV 3CLpro for 1 hr, and the fluorescence intensity of MERS‐CoV 3CLpro was measured.
2.5. Ligand preparation, ligand docking and target preparation
All the docking and scoring calculations were performed using the Schrödinger suite of software (maestro, version 11.5.011). The compounds were extracted from the PubChem database in sdf format and were combined in one file. The file was then imported into Maestro and prepared for docking using LigPrep. The atomic co‐ordinates of the crystal structure of MERS‐CoV 3CLpro (5WKJ) were retrieved from the Protein Data Bank and prepared by removing all solvent and adding hydrogens and minimal minimization in the presence of bound ligand using Protein Preparation Wizard. Ionizer was used to generate ionized state of all compounds at target pH 7.0 ± 2.0. This prepared low‐energy conformers of the ligand were taken as the input for docking analysis. Grids for molecular docking were built using Receptor Grid Generation. Compounds were docked using Ligand Docking mode with postdocking minimization including 5 poses per ligand. The docked poses were then refined using an XP (extra precision) option with the threshold value for rejecting minimized pose of 0.5 kcal/mol. The energies were calculated using the OPLS3 force field.
2.6. Induced‐fit docking
The induced‐fit docking protocol (Sherman, Day, Jacobson, Friesner, & Farid, 2006) was run from the graphical user interface accessible within Maestro 11.5.011. Receptor sampling and refinement were performed on residues within 5.0 Å of each ligand for each of the 20 ligand‐protein complexes. With Prime (Jacobson et al., 2004), a side‐chain sampling and prediction module, the side‐chains, as well as the backbone of the target protein, were energy minimized. A total of 20 induced‐fit receptor conformations were generated for each of the eight test ligands. Re‐docking the test ligands into their respective 20 structures within 30.0 kcal/mol of their lowest energy structure. Finally, the ligand poses were scored using a combination of Prime and GlideScore scoring functions (Friesner et al., 2006).
3. RESULTS
The amount of cell harvested for purification of MERS‐CoV 3CLpro was 3.01 g per 2,000 ml of E. coli culture. The protein was purified by ion chromatography using a 5 ml Hi‐Trap Q column (GE Healthcare). The column was equilibrated with a buffer consisting of 20 mM Bis‐Tris pH 7.0, and the pooled fractions were loaded. The column was eluted using a linear NaCl gradient to 1.0 M NaCl, and the protein was eluted at 0.16 M NaCl. The pooled fractions were loaded on a 5 ml Hi‐Trap Blue column (GE Healthcare) equilibrated with a buffer consisting of 10 mM sodium phosphate pH 7.0. The target protein was detected in unbound fractions. SDS–PAGE showed one band around 33(33,737.77Da) kDa, corresponding to the molecular weight of MERS‐CoV 3CLpro. The protein was concentrated to 31.14 mg/ml for protease assays in a buffer consisting of 10 mM sodium phosphate pH 7.0.
The purified MERS‐CoV 3CLpro was different from the previous method (Kumar et al., 2016) where the native form was obtained after removing of a His6‐tag wih Factor‐Xa. In our experiment, the enzyme activity was 10‐fold decreased when the His6‐tag protein was used (data not shown). That is due to the location of the His6‐tag connected to the N‐terminus of MERS‐CoV 3CLpro. The published crystal structure showed that the active site pocket might be easily hindered by the flexible N‐terminal His6‐tag.
A flavonoid library consisting of ten different scaffolds was also built (Figure 1). It contains three isoflavones, one isoflavane, six flavones, nine flavonols, four flavanols, five flavanones, one flavanonol, one prenylflavonoid, eight chalcones and two unclassified flavonoids (Table S1). We applied the library to assay MERS‐CoV 3CLpro. Since flavonoids are known to aggregate through a complexation and thus non‐specifically inhibit various proteases, the assay in the presence of Triton X‐100 was performed. Before assay, we tested the influence of Triton X‐100 on the catalytic activity of MERS‐CoV 3CLpro. As shown in the Figure S1, only a slight increment of the catalytic activity was observed even up to 0.1% Triton X‐100. Therefore, we performed the experiment at the concentration of 0.01% Triton X‐100 where no significant interference was detected.
Figure 1.
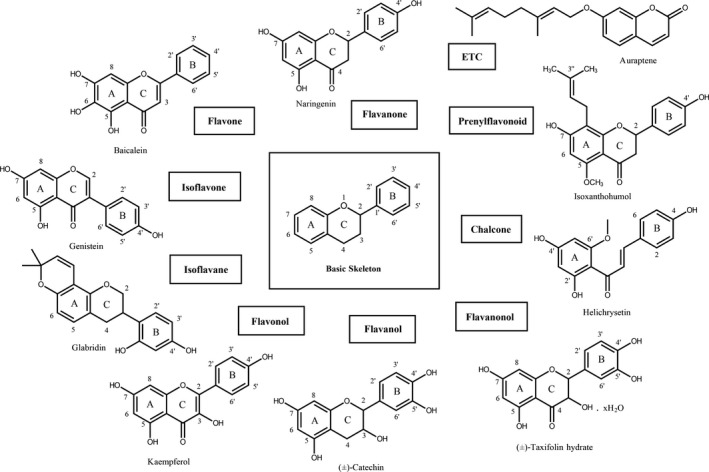
The basic skeleton structures of flavonoids and their scaffolds classified in this study. The basic structures of the most common flavonoids with a common numbering system
Using forty flavonoids, an inhibitory effect of each compound at 20 μM was tested. Among them, herbacetin (3,4′,5,7,8‐Pentahydroxyflavone), isobavachalcone (2′,4,4′‐Trihydroxy‐3′‐(3‐methyl‐2‐butenyl)chalcone), quercetin 3‐β‐d‐glucoside (3,3′,4′,5,7‐Pentahydroxyflavone 3‐β‐d‐glucoside) and helichrysetin (4,2′,4′‐trihydroxy‐6′‐methoxychalcone) were found to have prominent inhibitory activity (Figure 2). The four compounds showed the severely reduced fluorescent intensity and thus represented their MERS‐CoV 3CLpro inhibitory activity. The experimental data were plotted as log inhibitor concentration versus per cent fluorescence inhibition (Figure 2). IC50 values obtained from the dose‐dependent inhibitory curves of herbacetin, isobavachalcone, quercetin 3‐β‐d‐glucoside and helichrysetin are 40.59, 35.85, 37.03 and 67.04 μM, respectively.
Figure 2.
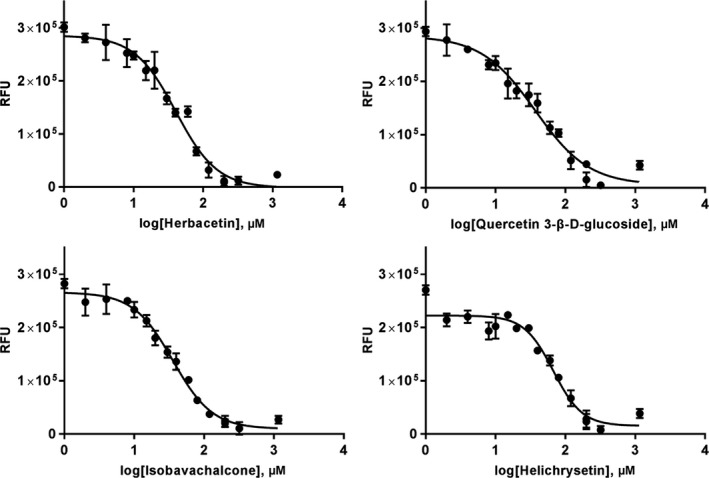
Results from the FRET method. Each data point represents the effect of each inhibitory compound against MERS‐CoV 3CLpro compared to the control. The RFU is plotted against the log‐concentration of inhibitory compounds. Each dot is expressed as the mean ± standard error of the mean (n = 3). RFU = Relative Fluorescence Units
In order to confirm the inhibitory activity of the flavonoids independently, a general tryptophan‐based assay method was employed. Tryptophan was well‐known to emit its own fluorescence. Therefore, if tryptophan is positioned adequately in proteins, the change in fluorescence intensity can reflect the binding state of chemicals and be used to judge interaction between proteins and chemicals. MERS‐CoV 3CLpro contains five tryptophan residues and thus displays a high fluorescence peak at 340 nm at the tryptophan excitation wavelength of 295 nm. We monitored the change in the fluorescence intensities of MERS‐CoV 3CLpro depending on the presence or absence of four chemicals. Since each compound in the flavonoid library was almost non‐fluorescent under the experiment condition, a change in fluorescence intensity reflects interactions between MERS‐CoV 3CLpro and chemicals. Only the four inhibitory compounds obviously reduced the fluorescence intensity of MERS‐CoV 3CLpro (Figure 3). The decreased emission intensity confirmed the complex formation between MERS‐CoV 3CLpro and inhibitory compounds.
Figure 3.
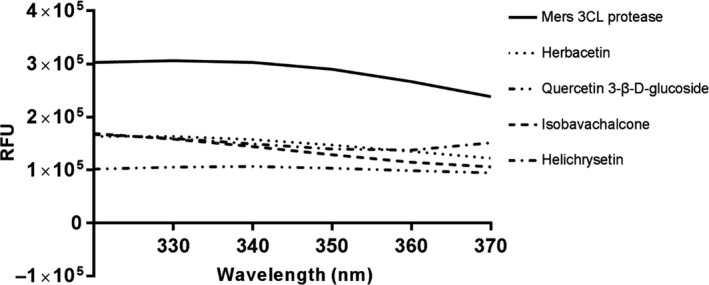
Fluorescence quenching spectra of MERS‐CoV 3CLpro. 1 μM MERS‐CoV 3CLpro shows a strong fluorescence emission (the solid line) with a peak at 340 nm at the excitation wavelength of 295 nm. Fluorescence quenching spectra of MERS‐CoV 3CLpro obtained in the presence of 40 μM each inhibitory compound, herbacetin (the dotted line), quercetin 3‐β‐d‐glucoside (one dashed two dotted line), isobavachalcone (the dashed line) and helichrysetin (one dashed one dotted line), into 1 μM MERS‐CoV 3CLpro are represented
In order to deduce the binding modes of the inhibitory flavonoids, an in‐depth theoretical investigation through an induced‐fit docking study was carried out. The interactions between MERS‐CoV 3CLpro and the inhibitory flavonoids were analysed to predict their binding affinities. Top ranked structures (according to the glide scores) from the induced‐fit docking results for herbacetin (−10.246), isobavachalcone (−9.364), quercetin 3‐β‐d‐glucoside (−9.751) and helichrysetin (−9.953) were selected and hypothesized to be biological complexes. In order to compare binding affinities to their closest homologues (Figure S2) based on the docking result, the poses of kaempferol (−10.159), 2,2′,4′‐trihydroxychalcone (−9.181), quercitrin (−8.891) and cardamonin (−7.393) were also calculated. The predicted complex structures and 2D schematic representations of them are illustrated in Figure S3. It is very obvious that the S1 site and hydrophobic S2 site of MERS‐CoV 3CLpro play a key role in predicted complexes.
4. DISCUSSION
Flavonoids are natural compounds with multiple pharmacological characteristics such as antioxidant, anti‐inflammatory, analgesic, anti‐carcinogenic, antibacterial infection, antifungal and antiviral properties (Frabasile et al., 2017). Naringenin has therapeutic effects on various neurological, cardiovascular, gastrointestinal, rheumatological, metabolic and malignant disorders (Rani et al., 2016). It also represents antiviral function (Zakaryan et al., 2017). Fisetin is a commercially available nutraceutical with anti‐inflammatory, antioxidant, anti‐tumorigenic, anti‐invasive, anti‐angiogenic, anti‐diabetic, neuroprotective and cardioprotective effects (Pal, Pearlman, & Afaq, 2016). Interestingly, fisetin has an anti‐noroviral activity by inducing cytokines (Seo & Choi, 2017). Although some of flavonoids show an antiviral effect, a molecular mechanism of their antiviral activity was rarely known.
In this study, we assayed the inhibitory activity of various flavonoids against MERS‐CoV 3CLpro. For the trial, a flavonoid library composed of nine different scaffolds plus one undefined group was constructed. They are classified based on a C6‐C3‐C6 skeleton and differ in the overall hydroxylation patterns and in the saturation of the heteroatomic ring C together with the position of the attached aromatic ring B (at the positions C‐2 or C‐3 of ring C) (Jucá et al., 2018). The flavonoid library was tested, and thus, a systematic analysis was possible. Among the ten groups, isoflavones, isoflavane, flavanols and flavanones did not show any inhibitory activity over MERS‐CoV 3CLpro. The other groups contains some flavonoids displaying moderate inhibitory activity. However, four flavonoids, herbacetin, isobavachalcone, quercetin 3‐β‐d‐glucoside and helichrysetin exhibited prominent inhibitory activity. The immediate inference of the primary scaffold required for binding with MERS‐CoV 3CLpro was as follows: first, the orientation of the aromatic ring B at the position C‐2 of ring C is essential as shown in isoflavones and isoflavane. Second, the saturated heteroatomic ring C is not preferred as shown in flavanols and flavanones. Third, chalcone and flavonol scaffolds show a promising binding property.
The more detailed structural comparison also provided valuable structural information required for the binding affinity of each flavonoid. Helichrysetin is a chalcone derivative (Van Puyvelde et al., 1989). Therefore, we compared its inhibitory activity with a chalcone, cardamonin (2′,4′‐dihydroxy‐6′‐methoxychalcone) which is the structurally identical homologue except one hydroxyl group at the 4‐position of the benzyl moiety of chalcone. The inhibitory activity of cardamonin at the concentration of 40 μM was lower than that of helichrysetin (Figure 4). It implies that the 4‐hydroxyl group of helichrysetin is functionally important to bind with MERS‐CoV 3CLpro. The structural comparison of cardamonin with isobavachalcone also indicated the modification effect of acetophenone ring of chalcone: the hydrophobic modification at the 3′‐position of the acetophenone ring moiety of isobavachalcone improves its binding affinity to MERS‐CoV 3CLpro. Intriguingly, the docking study shows that the 4‐hydroxyl group of helichrysetin forms a hydrogen bond with the hydroxyl group of Tyr54 of MERS‐CoV 3CLpro. Since Tyr54 is located deep inside of the hydrophobic S2 site, helichrysetin is inserted into deeper than cardamonin (Figure S3a). As a result, helichrysetin seems to have a better affinity and thus represents a better inhibitory activity.
Figure 4.
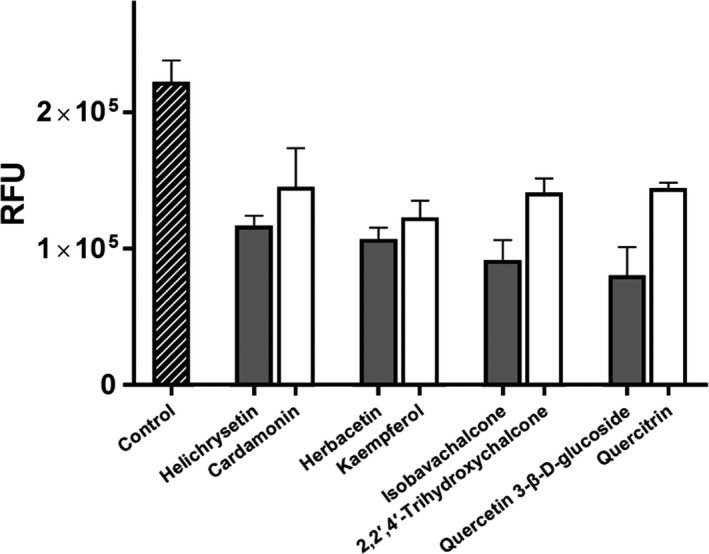
Comparison of inhibitory activity between homologue flavonoids. The two homologue flavonoids are compared side by side. Each of two bars represents the effect of two homologue inhibitory compounds at 40 μM against MERS‐CoV 3CLpro compared to the control. Each bar is expressed as the mean ± standard error of the mean (n = 3). RFU = Relative Fluorescence Units
In the flavonoid library, there are quite similar homologues of herbacetin, isobavachalcone and quercetin 3‐β‐d‐glucoside. They are kaempferol (3,4′,5,7‐Tetrahydroxyflavone), 2,2′,4′‐trihydroxychalcone and quercitrin (3,3′,4′,5,7‐Pentahydroxyflavone‐3‐L‐rhamnoside), respectively (Figure S2). Their MERS‐CoV 3CLpro inhibitory activity is lower than their corresponding compounds at 40 μM. Herbacetin is a kaempferol derivative where one more hydroxide is attached at 8‐position of kaempferol. The docking study indicates kaempferol derivatives occupy the S1 and S2 sites of MERS‐CoV 3CLpro (Figure S3b). The hydroxyl group at 7‐position looks important to bind at the S1 binding site. A bit better inhibitory activity of herbacetin indicates the hydroxide at 8‐position is favourable for its binding affinity to MERS‐CoV 3CLpro. In the predicted complex, the hydroxyl group at 8‐position of herbacetin induces a hydrogen bond with His41 at the S1 site.
The improved activity of isobavachalcone compared with 2,2′,4′‐trihydroxychalcone again points out the importance of its hydrophobic substituent. Coincided with the experimental result, the prenyl moiety of isobavachalcone makes hydrophobic interactions with Met25 and Leu49 at the hydrophobic S2 site (Figure S3c).
Quercetin 3‐β‐d‐glucoside is a homologue of quercitrin where rhamnoside is substituted for glucoside. The better inhibitory activity of quercetin 3‐β‐d‐glucoside means that hydroxymethyl is preferred to hydroxide in this position of the glucoside to interact with MERS‐CoV 3CLpro. The docking study represents that the hydroxymethyl group of the glucoside moiety makes a hydrogen bond with Glu169 and thus contributes its tighter binding to the S1 site than the rhamnoside moiety of quercitrin (Figure S3d).
Helichrysetin and isobavachalcone inhibiting the activity of MERS‐CoV 3CLpro are chalcone derivatives. Chalcone has immunomodulation, antibacterial, antifungal, antiviral, anti‐inflammatory, antioxidant, anticancer and anti‐diabetic activities (Yadav, Prasad, Sung, & Aggarwal, 2011). Recently, the non‐competitive inhibition of cardamonin against the dengue virus protease has been reported (Kiat et al., 2006). As discussed above, cardamonin is a strong homologue of helichrysetin. The analysis of the crystal structures of three viral proteases, MERS‐CoV 3CLpro, dengue virus NS2B/NS3 protease and norovirus 3CLpro were performed, and their active sites were compared (Figure S4) (Erbel et al., 2006; Nakamura et al., 2005). Despite their sequential divergence, their overall architecture resembles each other. Furthermore, the spatial positions of their catalytic residues were also highly conserved. The observation implies that chalcone and flavonol can be used as templates to develop a potential antiviral agent by targeting these viral proteases.
In this study, we showed that the antiviral effects of some flavonoids may be directly from their inhibition of main viral proteases and thus nullify a process of virus peptides. Therefore, one of antiviral mechanisms of flavonoids may be originated from their direct binding capability to viral proteases. Consequently, flavonoid derivatives can be used as a template to design not only for broad‐spectrum but also for virus‐specific antiviral agents. A further study is going on to develop better inhibitory lead compounds derived from this study.
5. CONCLUSIONS
We built a flavonoid library to systematically search MERS‐CoV 3CLpro inhibitory compounds with a FRET method. We found four flavonoids effectively reducing the proteolytic activity of MERS‐CoV 3CLpro. The binding of the flavonoids was independently proven by a tryptophan‐based fluorescence method. The assay in the presence of Triton X‐100 also eliminated the chance of the false‐positive result from the aggregation of flavonoids. The analysis of the four compounds with their homologs using an induced‐fit docking study provided an insight of flavonoid scaffolds required to bind with MERS‐CoV 3CLpro. The prominent activity of helichrysetin and isobavachalcone indicates that the flexibility of the chalcone scaffold is good to bind with MERS‐CoV 3CLpro. In contrast, the comparable activity of flavones with a rigid γ‐pyrone ring indicates that their spatial rearrangement together with various functional groups may be an alternative strategy to interact with MERS‐CoV 3CLpro. Therefore, this study suggests that an antiviral effect of some flavonoids is directly from their structural characteristics binding to viral proteases.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Programs, 2018R1D1A1B07050781 to DHS and 2016R1A6A3A11935354 & 2018R1D1A1B07050942 to MK, funded by the National Research Foundation of Korea grant granted by the Ministry of Education, Science and Technology, Republic of Korea (MEST). S. Jo was supported by Brain Korea 21 (BK21) Project.
Jo S, Kim H, Kim S, Shin DH, Kim M‐S. Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chem Biol Drug Des. 2019;94:2023–2030. 10.1111/cbdd.13604
Contributor Information
Dong Hae Shin, Email: dhshin55@ewha.ac.kr.
Mi‐Sun Kim, Email: shfwk31@ewha.ac.kr.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Chuck, C. P. , Chow, H. F. , Wan, D. C. , & Wong, K. B. (2011). Profiling of substrate specificities of 3C‐like proteases from group 1, 2a, 2b, and 3 coronaviruses. PLoS ONE, 6(11), e27228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E. , van Doremalen, N. , Falzarano, D. , & Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14, 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel, P. , Schiering, N. , D'Arcy, A. , Renatus, M. , Kroemer, M. , Lim, S. P. , … Hommel, U. (2006). Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nature Structural & Molecular Biology, 13, 372–373. [DOI] [PubMed] [Google Scholar]
- Fehr, A. R. , & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology, 1282, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frabasile, S. , Koishi, A. C. , Kuczera, D. , Silveira, G. F. , Verri, W. A. Jr , Duarte Dos Santos, C. N. , & Bordignon, J. (2017). The citrus flavanone naringenin impairs dengue virus replication in human cells. Scientific Reports, 7, 41864 10.1038/srep41864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, R. A. , Murphy, R. B. , Repasky, M. P. , Frye, L. L. , Greenwood, J. R. , Halgren, T. A. , … Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein‐ligand complexes. Journal of Medicinal Chemistry, 49, 6177–6196. [DOI] [PubMed] [Google Scholar]
- Jacobson, M. P. , Pincus, D. L. , Rapp, C. S. , Day, T. J. , Honig, B. , Shaw, D. E. , & Friesner, R. A. (2004). A hierarchical approach to all‐atom protein loop prediction. Proteins, 55(2), 351–367. 10.1002/prot.10613 [DOI] [PubMed] [Google Scholar]
- Jucá, M. M. , Cysne Filho, F. M. S. , de Almeida, J. C. , Mesquita, D. D. S. , Barriga, J. R. M. , Dias, K. C. F. , Barbosa, T. M. , Vasconcelos, L. C. , Leal, L. K. A. M. , Ribeiro, J. E. , & Vasconcelos, S. M. M. (2018). Flavonoids: Biological activities and therapeutic potential. Natural Product Research, 16, 1–14. [DOI] [PubMed] [Google Scholar]
- Kiat, T. S. , Pippen, R. , Yusof, R. , Ibrahim, H. , Khalid, N. , & Rahman, N. A. (2006). Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue‐2 virus NS3 protease. Bioorganic & Medicinal Chemistry Letters, 16(12), 3337–3340. 10.1016/j.bmcl.2005.12.075 [DOI] [PubMed] [Google Scholar]
- Kumar, V. , Tan, K. P. , Wang, Y. M. , Lin, S. W. , & Liang, P. H. (2016). Identification, synthesis and evaluation of SARS‐CoV and MERS‐CoV 3C‐like protease inhibitors. Bioorganic & Medicinal Chemistry, 24, 3035e3042 10.1016/j.bmc.2016.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, C.‐J. , Chi, Y.‐H. , Hsu, J.‐A. , & Liang, P.‐H. (2004). Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochemical and Biophysical Research Communications, 318, 862–867. 10.1016/j.bbrc.2004.04.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Lan, J. , Guan, M. , Sheng, F. , & Zhang, H. (2009). Spectroscopic investigation of interaction between mangiferin and bovine serum albumin. Spectrochimica Acta. Part A, Molecular and Biomolecular Spectroscopy, 73, 936–941. [DOI] [PubMed] [Google Scholar]
- Liu, Y. C. , Huang, V. , Chao, T. C. , Hsiao, C. D. , Lin, A. , Chang, M. F. , & Chow, L. P. (2005). Screening of drugs by FRET analysis identifies inhibitors of SARS‐CoV 3CL protease. Biochemical and Biophysical Research Communications, 333, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel‐Lemoine, M. , Millet, J. , Vidalain, P.‐O. , Law, H. , Vabret, A. , Lorin, V. , … Tangy, F. (2012). A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. Journal of Virology, 86(14), 7577–7587. 10.1128/JVI.00269-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K. , Someya, Y. , Kumasaka, T. , Ueno, G. , Yamamoto, M. , Sato, T. , … Tanaka, N. (2005). A norovirus protease structure provides insights into active and substrate binding site integrity. Journal of Virology, 79(21), 13685–13693. 10.1128/JVI.79.21.13685-13693.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neddle, D. , Lountos, G. T. , & Waugh, D. S. (2015). Structures of the Middle East respiratory syndrome coronavirus 3C‐like protease reveal insights into substrate specificity. Acta Cryst., D71, 1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, H. C. , Pearlman, R. L. , & Afaq, F. (2016). Fisetin and its role in chronic diseases. Advances in Experimental Medicine and Biology, 928, 213–244. [DOI] [PubMed] [Google Scholar]
- Rani, N. , Bharti, S. , Krishnamurthy, B. , Bhatia, J. , Sharma, C. , Amjad Kamal, M. , … Singh Arya, D. (2016). Pharmacological properties and therapeutic potential of naringenin: A citrus flavonoid of pharmaceutical promise. Current Pharmaceutical Design, 22(28), 4341–4359. 10.2174/1381612822666160530150936 [DOI] [PubMed] [Google Scholar]
- Seo, D. J. , & Choi, C. (2017). Inhibitory mechanism of five natural flavonoids against murine norovirus. Phytomedicine, 30, 59–66. 10.1016/j.phymed.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Sherman, W. , Day, T. , Jacobson, M. P. , Friesner, R. A. , & Farid, R. (2006). Novel procedure for modeling ligand/receptor induced fit effects. Journal of Medicinal Chemistry, 49(2), 534–553. 10.1021/jm050540c [DOI] [PubMed] [Google Scholar]
- Van Puyvelde, L. , De Kimpe, N. , Costa, J. , Munyjabo, V. , Nyirankuliza, S. , Hakizamungu, E. , & Schamp, N. (1989). Isolation of flavonoids and a chalcone from Helichrysum odoratissimum and synthesis of helichrysetin. Journal of Natural Products, 52(3), 629–633. 10.1021/np50063a025 [DOI] [PubMed] [Google Scholar]
- Wu, A. , Wang, Y. I. , Zeng, C. , Huang, X. , Xu, S. , Su, C. , … Guo, D. (2015). Prediction and biochemical analysis of putative cleavage sites of the 3C‐like protease of Middle East respiratory syndrome coronavirus. Virus Research, 208, 56–65. 10.1016/j.virusres.2015.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, V. R. , Prasad, S. , Sung, B. , & Aggarwal, B. B. (2011). The role of chalcones in suppression of NF‐κB‐mediated inflammation and cancer. International Immunopharmacology, 11(3), 295–309. 10.1016/j.intimp.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Lin, J. , Zhou, B. , Liu, Y. , & Zhu, B. (2017). Activity of compounds from Taxillus sutchuenensis as inhibitors of HCV NS3 serine protease. Natural Product Research, 31(4), 487–491. [DOI] [PubMed] [Google Scholar]
- Zakaryan, H. , Arabyan, E. , Oo, A. , & Zandi, K. (2017). Flavonoids: Promising natural compounds against viral infections. Archives of Virology, 162(9), 2539–2551. 10.1007/s00705-017-3417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


