Abstract
Abstract: Four members of the carcinoembryonic antigen family, CEACAM1, CEACAM8, CEACAM6 and CEACAM3, recognized by CD66a, CD66b, CD66c and CD66d monoclonal antibodies (mAb), respectively, are expressed on human neutrophils. CD66a, CD66b, CD66c and CD66d mAb binding to neutrophils triggers an activation signal that regulates the adhesive activity of CD11/CD18, resulting in an increase in neutrophil adhesion to human umbilical vein endothelial cells. Molecular modeling of CEACAM1 using IgG and CD4 as models has been performed, and three peptides from the N‐terminal domain were found to increase neutrophil adhesion to human umbilical vein endothelial cell monolayers. The peptides were 14 amino acids in length and were predicted to be present at loops and turns between β‐sheets. To better understand the amino acid sequences critical for this biological activity, in the present study we examined the other neutrophil CEACAMs and the highly homologous CEACAM, CEA. Molecular modeling of the N‐terminal domains of human CEACAM8, ‐6, ‐3 and CEA was performed. Twenty peptides, each 14 amino acids in length, that were homologous to the previously reported peptides from the N‐domains of CEACAM1, were synthesized and tested for their ability to alter neutrophil adhesion. Only one new peptide, from the N‐domain of CEA, was found to increase neutrophil adhesion, and this peptide differed from the corresponding CEACAM1 peptide by only a single conservative amino acid substitution. Importantly, minor amino acid differences between active and inactive homologous peptides suggest regions of these peptides that are critical for biological activity. The data suggest that the regions SMPF of peptide CD66a‐1, QLFG of peptide CD66a‐2 and NRQIV of peptide CD66a‐3 are critical for the activities of these peptides, and for the native CEACAMs.
Keywords: BGP, biliary glycoprotein, carcinoembryonic antigen, CEA, CD66, CEACAM, cell adhesion, inflammation, synthetic peptides
Abbreviations
- adhesion buffer
DMEM + 5% HIFBS
- BGP
biliary glycoprotein
- buffer B
PBS, pH 7.4, 0.2% BSA, 0.05% NaN3
- calcein labeling buffer
HBSS without Ca2+ or Mg2+ containing 0.02% BSA
- CEA
carcinoembryonic antigen
- endo wash buffer
HBSS + 4% HIFBS
- FMLP
N‐formyl‐Met‐Leu‐Phe
- HBSS
Hanks' balanced salt solution
- HIFBS
heat inactivated fetal bovine serum
- HUVECs
human umbilical vein endothelial cells
- NMS
normal mouse serum
- PBS
phosphate‐buffered saline, pH 7.4.
Carcinoembryonic antigen (CEA) family members appear to play a role in a wide variety of normal and pathological processes, including: cancer, embryonic development, bacterial infection, viral infection, inflammation, pregnancy and cell adhesion (1, 4). The CEACAM gene family belongs to the immunoglobulin (Ig) gene superfamily (for review see Refs 1, 3, 4, 5, 6). Structurally, each of the human CEACAMs contains one N‐terminal domain of 108–110 amino acid residues, homologous to Ig variable domains, followed by a differing number (0–6) of Ig C2‐type constant‐like domains. The structure of the N‐domain is predicted to be a stacked pair of β‐sheets with nine β‐strands (7). CD66 mAbs react with members of the CEACAM family (8, 9, 10, 11, 12, 13, 14, 15, 16, 17). In CD terminology, mAbs belonging to the CD66 cluster are classified according to their reactivity with each family member, as indicated by a lower case letter after ‘CD66’ as follows: CD66a, CEACAM1 or biliary glycoprotein; CD66b, CEACAM8; CD66c, CEACAM6; CD66d, CEACAM3; CD66e, CEA; and CD66f, pregnancy‐specific glycoprotein (PSG) (6, 13).
CEACAMs may function as adhesion molecules and can also transmit signals. CEACAM1, CEACAM6 and CEA are capable of homotypic and heterotypic adhesion, whereas CEACAM8 undergoes heterotypic adhesion with CEACAM6, as shown by use of recombinant CEACAM proteins (3, 4, 8, 9, 10, 11, 12, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34). CEACAM1, CEACAM8, CEACAM6 and CEACAM3, but not CEA, are expressed on human neutrophils, where they are ‘activation antigens’ in that their surface expression is increased following neutrophil activation by various stimuli. CD66a, CD66b, CD66c and CD66d mAb binding to the neutrophil surface trigger an activation signal that regulates the adhesive activity of CD11/CD18, and increases neutrophil adhesion to HUVECs (27, 28, 29, 30, 31, 32, 35). CEACAM1 is frequently downregulated in carcinomas (36, 37, 38, 39, 40), and may function as a tumor suppressor in some models of cancer (41, 42, 43, 44, 45, 46). CEACAMs are also important in bacterial infections, where pathogenic Neisseria meningitidis and N. gonorrhea interact with CEACAMs via Opa proteins. The binding site for these Opa proteins has been localized to the N‐domains of CEACAMs (47, 56). CEACAM1 also functions as a receptor for murine hepatitis virus (57, 58, 59, 60, 61).
The mechanism(s) by which CEACAMs transmit signals (e.g. activation in neutrophils, or growth regulating signals in epithelial cells and carcinomas) are unclear. However, CEACAM1 is phosphorylated on its cytoplasmic domain, largely on tyrosine with a lower level of phosphoserine, in neutrophils and colon cancer cells (12, 62, 63, 64, 65, 66). In addition, associated protein tyrosine kinase and phosphatase activities may be involved in CEACAM signaling (62, 64, 65).
Because of the adhesive and signaling properties of CEACAMs described above, we sought to identify functionally active domains of CEACAMs by use of synthetic peptides. In a previous study, we identified active sites on the extracellular domains of CEACAM1 by performing molecular modeling using IgG and CD4 as models. Three CEACAM1 peptides activated neutrophils, as determined by increasing neutrophil adhesion to human umbilical vein endothelial cell (HUVEC) monolayers and altering surface expression of CD11/CD18 and CD62L (67). These three peptides from the N‐domain of CEACAM1 were 14 amino acids in length and were predicted to represent regions connecting β‐sheets (67). It was concluded that at least three peptide motifs from the N‐terminal domain of CEACAM1 are involved in the interaction of CEACAM1 with other ligands, and can initiate signal transduction in neutrophils (67).
CEACAM8, CEACAM6 and CEACAM3 are also present on neutrophils and can transmit activation signals in neutrophils. CEACAM1, CEACAM8, CEACAM6, CEACAM3 and CEA are variably expressed in different tissues, undergo homotypic and/or heterotypic adhesion, and are homologous to each other. We therefore sought to identify other motifs of the N‐domains of CEACAM8, CEACAM6, CEACAM3 and CEA that might have biological activity. Molecular modeling was performed using IgG and CD4 as models as was done with CEACAM1, and 32 peptides of 14 amino acids in length that were predicted to contain regions connecting β‐sheets were identified. Twenty of these peptides that were nonidentical were synthesized and tested for their ability to alter neutrophil activation. Only one new peptide was found that activated neutrophils. This peptide was present in CEA; although CEA is not present on neutrophils it can bind to three of the neutrophil CEACAMs. Some peptides that had only minor amino acid differences from active peptides were inactive, suggesting that the amino acid residues at these positions are critical to biological activity.
Materials and Methods
Cell preparation
Normal peripheral blood neutrophils were prepared by a modification of the method described in Ref. 68, and were suspended at the indicated concentrations in Hanks' balanced salt solution (HBSS) (Gibco, Grand Island, NY, USA). Differential cell counts on Wright‐stained cells routinely revealed > 95% neutrophils. Viability as assessed by Trypan Blue dye exclusion was > 98%.
Antibodies and reagents
The PE‐labeled CD11b mAb (Leu−15) and the CD62L mAb (Leu−8) were obtained from Becton Dickinson (Mountain View, CA, USA). Monoclonal antibodies were diluted in PBS containing 1 mg/mL BSA as indicated. N‐Formyl‐met‐leu‐phe (FMLP) and normal mouse serum (NMS) were purchased from Sigma Chemical Co. (St Louis, MO, USA). Peptides were diluted in PBS containing 1 mg/mL BSA as indicated.
Fluorescence labeling of cells
Neutrophils were labeled with calcein AM (Molecular Probes, Eugene, OR, USA) (35, 69) by incubating 5 × 106/mL cells with 50 µg of calcein AM for 30 min at 37°C in 18 mL of calcein labeling buffer (HBSS without Ca2+ or Mg2+ containing 0.02% BSA). Cells were then washed twice with calcein labeling buffer at 23°C and resuspended in the desired media.
Peptide selection, synthesis and purification
The N‐domains of CEACAM8, CEACAM6, CEACAM3 and CEA were modeled to conform to the IgV and Ig C2 domains of the heavy and light chains of Fab fragments of immunoglobulin and CD4 as we reported previously for CEACAM1 (67). Our rationale was to select sequences from the loop regions as these regions would potentially be more accessible to cells, and therefore may be more biologically active. The N‐domains were chosen, because in our earlier study only peptides from the N‐domain of CEACAM1 had biological activity in neutrophil activation (67). Hydropathy plots were also done using Kyte−Doolittle and Chou−Fassman analyses, and the results were consistent in that the chosen sequences coincided with hydrophilic regions. The sequences were then standardized to a length of 14 amino acid residues, because this length results in a relatively high yield of peptides during synthesis, and corresponds to peptides made in our earlier study of CEACAM1. In order to complete this standardization, amino acid residues were added to the amino or carboxyl ends of the sequences. We also attempted to avoid having cysteine residues appear in the final sequences, as peptide dimerization may then occur. Our final selection of peptides to be synthesized is shown in Table 1.
Table 1.
CEACAM N‐domain peptides
| Peptide | Amino acid sequence | Predicted loop | CEACAM |
|---|---|---|---|
| CD66a‐1 | SMPFNVAEGKEVL | AB | CEACAM1 |
| CD66a‐2 | LVHNLPQQLFGYSW | BC | CEACAM1 |
| CD66a‐3 | KGERVDGNRQIVGY | CC′ | CEACAM1 |
| CD66a‐4 | VGYAIGTQQATPG | C′C″ | CEACAM1 |
| CD66a‐5 | ATPGPANSGRETIY | C″D | CEACAM1 |
| CD66a‐6 | LLIQNVTQNDTGFY | EF | CEACAM1 |
| CD66a‐7 | VIKSDLVNEEATGQ | FG | CEACAM1 |
| CD66a‐8 | EATGQFHVYPELPK | link | CEACAM1 |
| CD66b‐1 | AVPSNAAEGKEVL | AB | CEACAM8 |
| CD66b‐2 | LVHNLPQDPRGYNW | BC | CEACAM8 |
| CD66b‐3 | KGETVDANRRIIGY | CC′ | CEACAM8 |
| CD66b‐4 | IGYVISNQQITPG | C′C″ | CEACAM8 |
| CD66b‐5 | ITPGPAYSNRETIY | C″D | CEACAM8 |
| CD66b‐6 | LLMRNVTKNDTGSY | EF | CEACAM8 |
| CD66b‐7 | VIKLNLMSEEVTGQ | FG | CEACAM8 |
| CD66b‐8 | EVTGQFSVHPETPK | link | CEACAM8 |
| CD66c‐1 | STPFNVAEGKEVL | AB | CEACAM6 |
| CD66c‐2 | LAHNLPQNRIGYSW | BC | CEACAM6 |
| CD66c‐3 | KGERVDGNSLIVGY | CC′ | CEACAM6 |
| CD66c‐4 | VGYVIGTQQATPG | C′C″ | CEACAM6 |
| CD66c‐5 | ATPGPAYSGRETIY | C″D | CEACAM6 |
| CD66c‐6 | LLIQNVTQNDTGFY | EF | CEACAM6 |
| CD66c‐7 | VIKSDLVNEEATGQ | FG | CEACAM6 |
| CD66c‐8 | EATGQFHVYPELPK | link | CEACAM6 |
| CD66d‐1 | STPFNVAEGKEVL | AB | CEACAM3 |
| CD66d‐2 | LVHNLPQHLFGYSW | BC | CEACAM3 |
| CD66d‐3 | KGERVDGNSLIVGY | CC′ | CEACAM3 |
| CD66d‐4 | VGYVIGTQQATPG | C′C″ | CEACAM3 |
| CD66d‐5 | ATPGAAYSGRETIY | C″D | CEACAM3 |
| CD66d‐6 | LLIHNVTQNDIGFY | EF | CEACAM3 |
| CD66d‐7 | VIKSDLVNEEATGQ | FG | CEACAM3 |
| CD66d‐8 | EATGQFHVY | link | CEACAM3 |
| CD66e‐1 | STPFNVAEGKEVL | AB | CEA |
| CD66e‐2 | LVHNLPQHLFGYSW | BC | CEA |
| CD66e‐3 | KGERVDGNRQIIGY | CC′ | CEA |
| CD66e‐4 | IGYVIGTQQATPG | C′C″ | CEA |
| CD66e‐5 | ATPGPAYSGREIIY | C″D | CEA |
| CD66e‐6 | LLIQNIIQNDTGFY | EF | CEA |
| CD66e‐7 | VIKSDLVNEEATGQ | FG | CEA |
| CD66e‐8 | EATGQFRVYPELPK | link | CEA |
Synthetic peptides of human CEACAMs as described in the text. Amino acids underlined and shown in bold represent amino acid residues that differ from the homologous CEACAM1 peptide.
Peptides were synthesized as amides by Fmoc solid‐phase methodology on a Gilson Automated Multiple Peptide Synthesizer AMS 422. Peptides were purified by preparative reverse phase‐HPLC on a Beckman System Gold equipped with a Regis Chemical ODS C18 column (10 µm particle size, 60 A pore size, 250 × 21.1 mm). The elution gradient was 12–50% B over 35 min at a flow rate of 5.0 mL/min, where A was water containing 0.1% trifluoroacetic acid, and B was acetonitrile containing 0.1% trifluoroacetic acid. Detection was at 235 nm. Peptides were analyzed for the correct amino acid composition by fast atom bombardment mass spectrometry, and all peptides were found to have the correct composition.
Endothelial cell adhesion assay
Neutrophil adhesion to HUVECs was determined as described previously (35, 69, 70, 71). Briefly, HUVECs (Clonetics Corp., San Diego, CA, USA) were passaged 1:5 in T‐25 flasks (Costar) no more than three times before plating in 96‐well microtiter plates at 3000 cells/well. HUVECs were grown to confluence in 96‐well microtiter plates in EGM media (Clonetics) and fed every 24 h. Using the adhesion assay described below, no difference in resting and stimulated neutrophil adhesion was observed, and, as expected (72), no difference in surface expression of CD54 (ICAM‐1) or CD62E (E selectin, ELAM‐1) in resting or TNF‐stimulated cells was noted using HUVECs passaged once compared with those passaged five times. In some experiments, the HUVECs were stimulated by culture for the indicated time with TNF‐α (Cetus, Emeryville, CA, USA). The wells were then washed four times with adhesion buffer (DMEM + 5% heat inactivated fetal bovine serum; HIFBS) and 25 µL of adhesion buffer containing the indicated peptide was added to each well, followed immediately by 100 µL of adhesion buffer containing 105 calcein‐labeled cells. Twenty‐five microliters of adhesion buffer containing the indicated concentration of FMLP was then added, and the plates were incubated at 37°C in 5% CO2 for 30 min. The wells were then aspirated and washed four times with endo wash buffer (HBSS + 4% HIFBS), and the fluorescence was quantitated with a Millipore fluorescence plate reader using an excitation wavelength of 485 nm and an emission wavelength of 530 nm. For each condition, quadruplicate wells were tested and values are reported as the mean±SD. Each experiment was performed at least three times using different HUVEC subcultures.
Statistical analyses
Effects of peptides on neutrophil adhesion to HUVECs was analyzed by the Student's t‐test when appropriate.
Analysis of CD11b and CD62L expression
Analyses of CD11b and CD62L expression were performed as previously described (67). For analysis of CD11b upregulation, purified neutrophils (105 in 100 µL HBSS + 0.02% BSA) were incubated with media containing the indicated peptide (167 µg/mL) or FMLP (10−7 m) for 15 min at 37°C. The cells were then cooled to 0°C for 10 min and 2 µg of the PE‐labeled CD11b mAb was added. The mixture was incubated at 0°C for 25 min, and 4 mL of buffer B (PBS, pH 7.4, 0.2% BSA, 0.05% NaN3, 0°C) was then added and the mixture was centrifuged at 400 g for 5 min at 4°C. The supernatant was removed and the cells were vortexed, and suspended in 1 mL of buffer B (0°C), and 250 µL of fixative (Coulter) (23°C) was then added. Three milliliters of buffer B (0°C) was then added, and the mixture centrifuged at 400 g at 4°C for 5 min. The cells were washed with 3 mL of buffer B as above, and resuspended in 200 µL of PBS containing 0.1% NaN3 (0°C) and stored at 4°C until analysis. Quantitative flow cytometric analysis of surface antigen expression was performed using a FACSTAR Plus (Becton Dickinson). Forward and right‐angle light scatter, as well as the peak fluorescence channel, were optimized with fluorescent beads. The cell population studied was determined by forward and right‐angle light scatter.
For analysis of CD62L down regulation, purified neutrophils (105 in 100 µL HBSS + 0.02% BSA) were warmed to 37°C for 5 min and then incubated with media containing the indicated peptide (167 µg/mL) or FMLP (10−7 m) for 5 min at 37°C. The cells were then cooled to 0°C for 10 min and 5 µg of the PE‐labeled CD62L mAb was added. The cells were then incubated, washed and analyzed by flow cytometry as above.
Results
Synthesis of CEACAM peptides
As CEACAMs are members of the Ig superfamily, we modeled CEACAM8, CEACAM6, CEACAM3 and CEA using the known crystallographically determined structure of the IgV and Ig C2‐like domains of IgG and CD4. The amino acid sequences were also analyzed by a hydropathy plot using Kyte−Doolittle and Chou−Fassman analyses, and sequences predicted to be exposed on the surface of the molecules based on hydrophilicity were identified. There was good agreement in general between the peptides selected using these two methods. A series of 32 peptides of 14 amino acids in length was then identified that were predicted to contain turns and loops between β‐sheets (Table 1). Ten of the modeled peptides were identical to at least one other N‐domain peptide of CEACAMs 1, 8, 6, 3 or CEA. Twenty peptides were synthesized on a multipeptide synthesizer, purified and characterized as described in Materials and Methods. Synthesis of peptides CD66d‐6, CD66e‐6 and also CD66a‐7, which is identical to CD66c‐7, CD66d‐7 and CD66e‐7, was not successful. Peptide CD66a‐6, which is identical to CD66c‐6, was not soluble in our assay conditions and was therefore not studied further.
Effects of CEACAM peptides on neutrophil adhesion to endothelial cells
The CEACAM1 peptides CD66a‐1, CD66a‐2 and CD66a‐3 were previously reported to activate neutrophils and to increase neutrophil adhesion to HUVECs (67). As expected, when neutrophils were incubated for 30 min in the presence of media containing 167 µg/mL of peptide CD66a‐2 with HUVECs in the presence of 10−7 m FMLP and washed as described in Materials and Methods, peptide CD66a‐2 augmented neutrophil adhesion approximately 2‐fold compared with media (Fig. 1, solid bars). This effect was also seen when neutrophils were pretreated with TNF‐α (hatched bars). In contrast, none of the CEACAM8 peptides (CD66b‐1 to CD66b‐8), the CEACAM6 peptides (CD66c‐1 to CD66c‐5), or the CEACAM3 peptides (CD66d‐1 to CD66d‐5, and CD66d‐8) altered neutrophil adhesion compared with incubation in media alone (Fig. 1). Similar results were obtained in the absence of FMLP (not shown). Peptides CD66c‐6, CD66c‐7 and CD66c‐8 were not tested as they are identical to the CD66a‐6 (insoluble), CD66a‐7 (unsuccessful synthesis) and CD66a‐8 (inactive in this assay) peptides, respectively, previously reported (67). Peptide CD66d‐7 was not tested as it is identical to the CD66a‐7 peptide. We were not able to synthesize peptide CD66d‐6 in three separate attempts. Six peptides from CEA (CD66e‐1 to CD66e‐5 and CD66e‐8) were also tested for their ability to alter neutrophil adhesion to HUVECs (Fig. 1). Peptide CD66e‐3, but none of the other CEA peptides, altered neutrophil adhesion to HUVECs. Peptide CD66e‐7 was not tested as it is identical to CD66a‐7. We were not able to synthesize peptide CD66e‐6 in three separate attempts.
Figure 1.
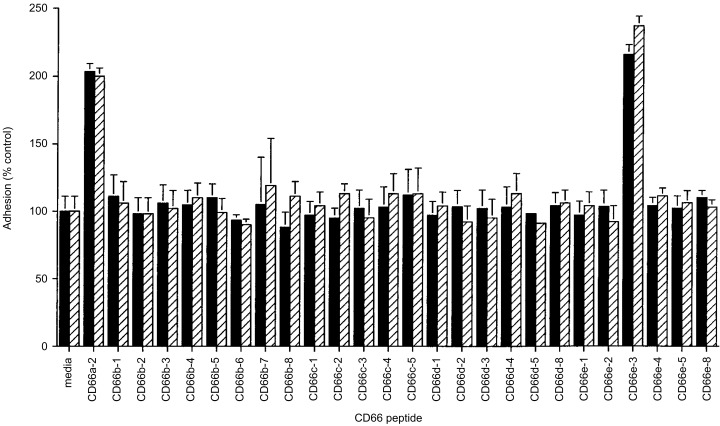
Effects of human CEACAM peptides on neutrophil adhesion to HUVECs. HUVECs were grown to confluence in 96‐well microtiter plates, and either used directly (solid bars) or stimulated by incubating in the presence of TNF‐α at 50 ng/mL final concentration for 4 h at 37°C (hatched bars). The wells were then washed and 25 µL of adhesion buffer containing the indicated human CEACAM peptide at 167 µg/mL (final concentration) was added. One hundred microliters of adhesion media containing 105 neutrophils was then added immediately, followed by 25 µL of adhesion buffer with 6 × 10−7 m FMLP, and the plates were incubated at 37°C for 30 min as described in the text. The wells were then washed and the number of adherent neutrophils determined with a fluorescence plate reader. Values are shown as the number of neutrophils remaining adherent to the HUVEC monolayers, expressed as a percent of those adherent in the presence of media without peptide, and represent the means±SD of four separate determinations. The adhesion observed in the presence of the active CEACAM1 peptide CD66a‐2 and the CEA peptide CD66e‐3 was statistically greater than that observed with the other CEACAM peptides or media alone (P < 0.05).
Concentration‐dependent effect of CD66e‐3 peptide
The CEA peptide CD66e‐3 was further tested for its effects on the adhesion of neutrophils to resting and TNF stimulated HUVECs in the presence of FMLP. Peptide CD66e‐3 increased neutrophil adhesion to HUVECs at concentrations as low as 75 µg/mL (≈ 50 µm) (Fig. 2).
Figure 2.
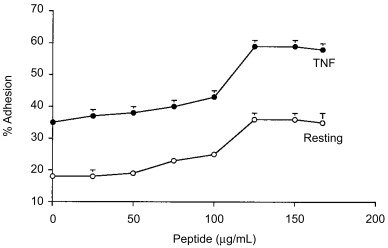
Effects of various concentrations of the CEA peptide CD66e‐3 on neutrophil adhesion to HUVECs. HUVECs monolayers were tested directly (○) or stimulated by incubating with TNF‐α (•), and neutrophil adhesion in the presence of the indicated final concentration of the CEA peptide CD66e‐3 and FMLP as in Fig. 1. Values are shown as the number of neutrophils remaining adherent to the HUVEC monolayers, expressed as a percent of those adherent in the presence of media without peptide, and represent the means±SD of four separate determinations. The adhesion observed in the presence of CEA peptide CD66e‐3 at 75 µg/mL was statistically greater than that observed with lower concentrations of peptides (P < 0.05).
Effect of CD66e‐3 peptide on CD11b expression
The effects of the CEA peptide CD66e‐3 on surface expression of CD11b on neutrophils was next examined. Although neutrophil adhesion to HUVECs is dependent on the functional activity of surface CD11/CD18, many adhesive stimuli also upregulate the surface expression of CD11/CD18, and this may play a role in regulating cell adhesion as well (73, 74, 75). To determine if an alteration in the surface expression of CD11/CD18 could contribute to the effect of the CD66e‐3 peptide on neutrophil adhesion, CD11b expression was analyzed by flow cytometry. Because CD11 and CD18 are translocated to the cell surface only when they are complexed with each other, the use of a directly labeled CD11b mAb was used to demonstrate upregulation of CD18 as well as CD11b. When neutrophils were incubated with HBSS for 15 min at 37°C and then reacted with a PE‐labeled CD11b mAb, CD11b expression was readily detected by flow cytometry (MCF = 584) (Fig. 3, left, upper panel). As expected, when neutrophils were incubated with FMLP (10−7 m) for 15 min, CD11b expression was increased (MCF = 709) (middle). When neutrophils were incubated with 167 µg/mL of the CEA peptide CD66e‐3 (MCF = 707) (lower), CD11b expression also increased, similar to that seen with incubation with 10−7 m FMLP.
Figure 3.
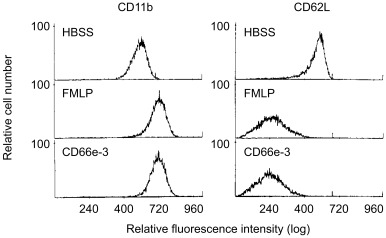
Representative flow cytometric histogram profiles of the effect of CD66e‐3 peptide on human neutrophil surface CD11b and CD62L expression. (Left) Purified neutrophils were incubated with HBSS (MCF = 584) (upper), 10−7 m FMLP (MCF = 709) (middle) or 167 µg/mL of the CEA peptide CD66e‐3 (MCF = 707) (lower), for 15 min at 37°C, and the binding of a PE‐labeled CD11b mAb was determined as described in the text. Vertical axis, relative cell number; horizontal axis, relative fluorescence intensity measured on a log scale. The MCFs represent the means of two determinations that agreed within 10%. (Right) Purified neutrophils were warmed to 37°C, incubated for 5 min with HBSS (MCF = 548) (upper), 10−7 m FMLP (MCF = 256) (middle), or 167 µg/mL of the CEA peptide CD66e‐3 (MCF = 239) (lower), and the binding of a PE‐labeled CD62L mAb was determined as described in the text. Vertical axis, relative cell number; horizontal axis, relative fluorescence intensity measured on a log scale.
Effect of CD66e‐3 peptide on CD62L expression
The effects of the CEA peptide CD66e‐3 on surface expression of CD62L on neutrophils was next examined. l‐Selectin, recognized by CD62L mAbs, also plays a role in neutrophil adhesion to endothelial cells, and its expression is altered by stimulation (73, 75). To determine if the surface expression of CD62L could be altered by the CEA peptide CD66e‐3, CD62L expression was analyzed by flow cytometry. When neutrophils were incubated with HBSS for 5 min at 37°C, and then reacted with a PE‐labeled CD62L mAb, CD62L expression was readily detected by flow cytometry (MCF = 548) (Fig. 3, left, upper). When neutrophils were incubated with 10−7 m FMLP, CD62L expression decreased as expected (MCF = 256) (middle). Similarly, when neutrophils were incubated with the CEA peptide CD66e‐3 (MCF = 239) (lower), CD62L expression also decreased.
Discussion
Twenty peptides were synthesized from regions of the N‐domains of human CEACAM8, CEACAM6, CEACAM3 and CEA, that we predict form loops and turns between regions of β‐sheets and may be exposed on the surface of the molecule. These peptides are homologous to a series of peptides from CEACAM1 that we described previously (67) that included three peptides that had activity in an assay examining stimulated neutrophil adhesion to HUVECs. These same three peptides also stimulated upregulation of CD11b/CD18 and downregulation of CD62L on the neutrophil surface. Thus, previous data suggested that peptide motifs from at least three regions of the N‐terminal domain of human CEACAM1 are involved in the interaction of CEACAM1 with other ligands and can initiate signal transduction in neutrophils. In the current study, although CEACAMs 1, 8, 6 and 3 can all transmit activation signals in neutrophils, only one of the 20 newly synthesized peptides (CD66e‐3) had biological activity in the neutrophil adhesion assay. Several other studies have proposed structural motifs of CD66a family proteins (7, 22, 54, 76). Although our peptides of 14 amino acid residues in length contain variable numbers of residues from proposed β‐sheet regions, our model is in general similar to other reported models. Although 19 of the 20 peptides tested in this study were not active in this assay, it is possible that they might have functional activity in a different assay.
We chose to focus our study on the N‐domains of CEACAMs 8, 6, 3 and CEA, for several reasons. First, these proteins are homologous to CEACAM1, second, CEACAMs 8, 6 and 3 can signal in neutrophils, and third, they can undergo homotypic and/or heterotypic adhesion with each other. Fourth, in our previous study, only peptides from the N‐domain of CEACAM1 had activity. In addition, studies of transfectants, recombinant proteins and domain‐specific mAbs, have suggested that the N‐terminal domain is critical for the homotypic and heterotypic adhesion activity of CEACAMs (16, 22, 24, 25, 26, 34, 77). Studies have also suggested that the A1, B1 or A2 domains may also be important in homotypic adhesion, and may interact with the N‐domain (16, 20, 21, 23, 24).
In addition to the finding that the CD66e‐3 peptide has biological activity, the observation that some other peptides homologous to the functionally active peptides CD66a‐1, CD66a‐2 and CD66a‐3 had no activity is also very informative. For example, CD66a‐1 is functionally active whereas peptides CD66c‐1, CD66d‐1 and CD66e‐1, which are identical, and differ from CD66a‐1 by a single amino acid substitution of M to T, are inactive (Table 2). These data suggest that the sequence of peptide CD66a‐1 in the SMPF region may be critical for activity. CD66b‐1 also differs in the SMPF region and is inactive. Similarly, although CD66a‐2 is functionally active, the homologous peptides CD66b‐2, CD66c‐2, CD66d‐2 and CD66e‐2, which differ from CD66a‐2 in the QLFG region, are inactive (Table 2). Thus, these data suggest that the amino acid sequence in this region is critical for activity. CD66d‐2 and CD66e‐2, which are identical, differ from CD66a‐2 by a single nonconservative Q to H substitution. CD66e‐3 differs from CD66a‐3 by a single conservative amino substitution at the end of the NRQIV sequence and retains activity (Table 2). In contrast, CD66c‐3 and CD66d‐3, which are identical, differ from CD66a‐3 by a RQ to SL substitution in the NRQIV sequence and are inactive. CD66b‐3 has a substitution in the NRQIV sequence of QIV to RII. Given the activity of the V to I substitution in CD66e‐3, this suggests that the loss of activity in CD66b‐3 was due to the nonconservative Q to R substitution, although two other amino acid substitutions are also present. These changes in primary amino acid sequence may evoke a change in secondary or tertiary structure reflected by biological activity.
Table 2.
CEACAM N‐domain peptides
| CEACAM | Peptide | Amino acid sequence | Activity | |
|---|---|---|---|---|
| CEACAM1 | CD66a‐1 | SMPFNVAEGKEVL | + | |
| CEACAM8 | CD66b‐1 | AVPSNAAEGKEVL | − | |
| CEACAM6 | CD66c‐1 | STPFNVAEGKEVL | − | |
| CEACAM3 | CD66d‐1 | STPFNVAEGKEVL | − | |
| CEA | CD66e‐1 | STPFNVAEGKEVL | − | |
| CEACAM1 | CD66a‐2 | LVHNLPQQLFGYSW | + | |
| CEACAM8 | CD66b‐2 | LVHNLPQDPRGYNW | − | |
| CEACAM6 | CD66c‐2 | LAHNLPQNRIGYSW | − | |
| CEACAM3 | CD66d‐2 | LVHNLPQHLFGYSW | − | |
| CEA | CD66e‐2 | LVHNLPQHLFGYSW | − | |
| * | ||||
| CEACAM1 | CD66a‐3 | KGERVDGNRQIVGY | + | |
| CEACAM8 | CD66b‐3 | KGETVDANRRIIGY | − | |
| CEACAM6 | CD66c‐3 | KGERVDGNSLIVGY | − | |
| CEACAM3 | CD66d‐3 | KGERVDGNSLIVGY | − | |
| CEA | CD66e‐3 | KGERVDGNRQIIGY | + | |
| # |
Comparison of active synthetic peptides of human CEACAM1 (CD66a‐1 to CD66a‐3), with homologous peptides of CEACAM8, CEACAM6, CEACAM3, and CEA as described in the text. Amino acids underlined and shown in bold represent amino acid residues that differ from the homologous CEACAM1 peptide. Activity in the neutrophil stimulation assay as described in the text is shown in the right column. * indicates the position in CD66a‐2, b‐2, c‐2, d‐2 and e‐2 of Tyr34 which has been shown by alanine mutagenesis studies to be important in binding to some Opa variants. # indicates the position in CD66a‐3, b‐3, c‐3, d‐3 and e‐3 of Val39 which has been shown by alanine mutagenesis studies to be important in binding to certain Opa variants and in CEACAM1 homotypic adhesion.
As described above, the naturally occurring amino acid substitutions among the human CEACAMs provide insight into the identification of the amino acid residues that are critical for biological activity. Other studies of functionally active sites of CEACAMs have also identified regions critical to activity. The N‐terminal half of the N‐domain has been shown to have a critical Opa‐binding epitope (52). Studies of binding of Opa proteins of N. meningiditis and N. gonorrhoeae using alanine mutagenesis have suggested that the CFG face is important in Opa binding to N‐domains of CEACAMs (54). In particular, Tyr34 and Ile91 are critical to Opa binding (54). Ile‐91 is present in CD66a‐7, CD66b‐7, CD66c‐7, CD66d‐7 and CD66e‐7. Because we were not successful in synthesizing these peptides in a soluble form, we could not test their activities in our assay. These studies also found that Tyr34, present in CD66a‐2, CD66b‐2, CD66c‐2, CD66d‐2 and CD66e‐2, was also critical for some Opa variants, and Val39, present in CD66a‐3, CD66b‐3, CD66c‐3, CD66d‐3 and CD66e‐3, was critical for other Opa variants. Three of these peptides, CD66a‐2, CD66a‐3 and CD66e‐3, were active in our assay. Similar studies of CEACAM1 homotypic adhesion using alanine mutagenesis have also suggested that the CFG face is important in homotypic binding of CEACAM1, notably finding that Val39 in the CC′ loop, contained in peptides CD66a‐3, CD66b‐3, CD66c‐3, CD66d‐3 and CD66e‐3, is critical for homotypic adhesion (78). Recent mutagenesis studies have also suggested that the amino acid sequence NRQII, present in our peptide CD66e‐3, is important for homotypic adhesion of CEA (79). These studies also found that 1000 µg/mL of a cyclized version of NRQII could inhibit CEA homotypic adhesion.
Studies of murine hepatitis virus binding have identified the sequence YKGNTTAIDKEIARFVPNSN in the N‐domain to be critical for coronavirus binding to CEACAM1 (80). The regions of the human CEACAMs corresponding to this sequence are represented largely by peptides CD66a‐3, CD66b‐3, CD66c‐3, CD66d‐3 and CD66e‐3, and partly by peptides CD66a‐4, CD66b‐4, CD66c‐4, CD66d‐4 and CD66e‐4. Thus, two of these human peptides that correspond to the coronavirus binding region in murine CEACAM1 have biological activity in our study of neutrophil activation.
Site‐directed mutagenesis studies of rat CEACAM1 found that the integrity of Arg64 in the consensus ATPase domain (GPAYSGRET) was essential for homotypic aggregation (81). This arginine is highly conserved in Ig domains, being important in forming a salt bridge with a highly conserved aspartate within the same domain (7). The corresponding regions in our model are present in a loop/turn region comprising the sequences of peptides CD66a‐5, CD66b‐5, CD66c‐5, CD66d‐5 and CD66e‐5. Although none of these peptides had activity in our assay, the cells and assay systems utilized are quite different.
The finding that these short peptides can stimulate neutrophils, as can CD66 mAbs (27, 28, 29, 32, 35, 82) suggests that they have significant affinity for a surface structure, possibly native CEACAMs. If so, whether the activity derives from binding native CEACAMs and transducing a signal directly, or by another mechanism is unknown. The ability of the synthetic peptides described here to activate neutrophils could be mediated by alterations in CEACAM dimerization, for example by disrupting a pre‐existing association of CEACAM1 with other CEACAM members (including CEACAM1 itself in the form of dimers or oligomers already present on the cell surface) or by stimulating dimerization. It has been reported that CEACAM1 (83) and CEA (84) exist on the cell surface as dimers. Dimerization of CEACAMs could potentially occur via interactions between the extracellular domains of CEACAM molecules or via other mechanisms. CEACAM1 dimerization could be influenced by phosphorylation; CEACAM1 is phosphorylated on Thr453 in the calmodulin binding site by protein kinase C (2). CEACAM1 dimerization has been shown to be regulated by calmodulin and intracellular calcium in vitro (83, 85). Clearly, dimerization of CEACAMs could affect binding of other signal regulating molecules. The mechanisms by which CEACAM family members transmit signals (e.g. activation in neutrophils or growth regulating signals in epithelial cells and carcinomas) are unclear. In addition to the considerations described above, CEACAM1 is phosphorylated in neutrophils and colon cancer cells (12, 62, 63, 66), and protein kinase and phosphatase activity may associate with CEACAMs (62, 64, 65). CEACAM1 contains an immune tyrosine inhibitory motif (ITIM), as well as a motif similar to an immune tyrosine activating motif (ITAM) in its cytoplasmic domain (2, 62), and phosphorylation of ITAMs and ITIMs can result in binding of protein tyrosine kinases and protein tyrosine phosphatases, respectively, which can modify signal transduction (64, 65).
CEACAM family members appear to be involved in a wide variety of important biological processes, and their differential expression provides the possibility for diverse interactions. For example, CEACAM1, CEACAM8, CEACAM6 and CEACAM3, but not CEA, are expressed on neutrophils; CEA is expressed on many tumor cells but not leukocytes; CEACAM8 is expressed on neutrophils but not epithelial cells; CEACAM6 is expressed on both neutrophils and epithelial cells (reviewed in Refs 1, 2, 4, 13). In addition, the surface expression of these molecules in other cells may also be regulated; for example, CEACAM1 expression is induced on HUVECs by γ‐IFN (14). CEACAM members may play an important role in inflammation, as each of the CEACAM family members expressed on neutrophils are capable of transmitting activation signals, CEACAM1 is also present on T‐lymphocytes and a subset of NK cells (86), and CEA expression by tumor cells is correlated with resistance to NK/LAK cell mediated lysis (87, 88). It is possible that CEACAM family members could contribute to the immunosuppression often found in patients with cancer. Some bacteria bind to neutrophil CEACAMs (13, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 89, 90), and this interaction may result in signal transduction resulting in modification of neutrophil activity. It is possible that surface expression of CEACAM family members may modify the signal transduction capabilities of other cell surface CEACAM molecules.
In summary, the data suggest that three peptide motifs from the N‐terminal domain of CEACAM1 are involved in the interaction of CEACAM1 with other ligands, and can initiate signal transduction in neutrophils. One peptide motif from the N‐domain of CEA can also initiate signal transduction in neutrophils, and this may represent a region of CEA that can interact with CEACAM1, CEACAM6, or other ligands. Each of the four active CEACAM peptides has a unique amino acid sequence. The amino acid substitutions in the homologous peptides from other CEACAMs, compared with that found in the active peptides, that result in loss of activity provide further information on the critical amino acids generating the sequence necessary for biological activity.
Acknowledgements:
We thank Dr W. Gleason for a critical review of the manuscript.
To cite this article:
Skubitz, K. M., Campbell, K. D. & Skubitz, A. P. N. Synthetic peptides from the N‐domains of CEACAMs activate neutrophils.
J. Peptide Res., 2001, 58, 00−00.
References
- 1. Hammarstrom, S. (1999) The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 9, 67–81. [DOI] [PubMed] [Google Scholar]
- 2. Obrink, B. (1997) CEA adhesion molecules – multifunctional proteins with signal‐regulatory properties. Curr. Opin. Cell Biol. 95, 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shively, J.E. , Hinoda, Y. , Hefta, L.J.F. , Neumaier, M. , Hefta, S.A. , Shively, L. , Paxton, R.J. & Riggs, A.D. (1989) Molecular Cloning of Members of the Carcinoembryonic Antigen Gene Family. Elsevier Science, Amsterdam, pp. 97–110. [Google Scholar]
- 4. Thompson, J.A. , Grunert, F. & Zimmerman, W. (1991) Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J. Clin. Lab. Anal. 5, 344–366. [DOI] [PubMed] [Google Scholar]
- 5. Khan, W.N. , Frangsmyr, L. , Teglund, S. , Israelsson, A. , Bremer, K. & Hammarstrom, S. (1992) Identification of three new genes and estimation of the carcinoembryonic antigen family. Genomics 14, 384–390. [DOI] [PubMed] [Google Scholar]
- 6. Beauchemin, N. , Draber, P. , Dveksler, G. , et al. (1999) Redefined nomenclature or members of the carcinoembryonic antigen family. Exp. Cell Res. 252, 243–249. [DOI] [PubMed] [Google Scholar]
- 7. Bates, P.A. , Lou, J. & Sternberg, M.J.E. (1992) A predicted three‐dimensional structure for the carcinoembryonic antigen (CEA). FEBS Lett. 301, 207–214. [DOI] [PubMed] [Google Scholar]
- 8. Daniel, S. , Nagel, G. , Johnson, J.P. , Lobo, F.M. , Hirn, M. , Jantscheff, P. , Kuroki, M. , Von Kleist, S. & Grunert, F. (1993) Determination of the specificities of monoclonal antibodies recognizing members of the CEA family using a panel of transfectants. Int. J. Cancer 55, 303–310. [DOI] [PubMed] [Google Scholar]
- 9. Mayne, K.M. , Pulford, K. , Jones, M. , Micklem, K. , Nagel, G. & Van Der Schoot, E.C. (1993) Antibody By114 is selective for the 90 kD PI‐linked component of the CD66 antigen: a new reagent for the study of paroxysmal nocturnal haemoglobinuria. Br. J. Haematol. 83, 30–38. [DOI] [PubMed] [Google Scholar]
- 10. Nagel, G. , Grunert, F. , Kuijpers, T.W. , Watt, S.M. , Thompson, J. & Zimmerman, W. (1993) Genomic organization, splice variants and expression of CGM1, a CD66‐related member of the carcinoembryonic antigen gene family. FEBS Lett. 214, 27–35. [DOI] [PubMed] [Google Scholar]
- 11. Kuroki, M. , Matsuo, Y. , Kinugasa, T. & Matsuoka, Y. (1992) Three different NCA species, CGM6/CD67, NCA‐95, and NCA‐90, and comprised in the major 90–100‐KDa band of granulocyte NCA detectable upon SDS‐polyacrylamide gel electrophoresis. Biochem. Biophys. Res. Commun. 182, 501–506. [DOI] [PubMed] [Google Scholar]
- 12. Skubitz, K.M. , Ducker, T.P. & Goueli, S.A. (1992) CD66 monoclonal antibodies recognize a phosphotyrosine‐containing protein bearing a carcinoembryonic antigen cross‐reacting antigen on the surface of human neutrophils. J. Immunol. 148, 852–860. [PubMed] [Google Scholar]
- 13. Skubitz, K.M. , Grunert, F. , Jantscheff, P. , Kuroki, M. & Skubitz, A.P.N. (1997) Summary of the CD66 cluster workshop In Leukocyte Typing VI , Garland Publishing, New York, pp. 992–1000. [Google Scholar]
- 14. Skubitz, K.M. , Micklem, K. & Van Der Schoot, C.E. (1995) Summary of CD66 and CD67 Cluster Report, Oxford University Press, Oxford, pp. 889–899. [Google Scholar]
- 15. Stoffel, A. , Neumaier, M. , Gaida, F.‐J. , Fenger, U. , Drzeniek, Z. , Haubeck, H.‐D. & Wagener, C. (1993) Monoclonal, anti‐domain and anti‐peptide antibodies assign the molecular weight 160,000 granulocyte membrane antigen of the CD66 cluster to a mRNA species encoded by the biliary glycoprotein gene, a member of the carcinoembryonic antigen gene family. J. Immunol. 150, 4978–4984. [PubMed] [Google Scholar]
- 16. Watt, S.M. , Fawcett, J. , Murdoch, S.J. , Teixeira, A.M. , Gschmeissner, S.E. , Hajibagheri, N.M. & Simmons, D.L. (1994) CD66 identifies the biliary glycoprotein (BGP) adhesion molecule: cloning, expression and adhesion functions of the BGPc splice variant. Blood 84, 200–210. [PubMed] [Google Scholar]
- 17. Watt, S.M. , Sala‐Newby, G. , Hoang, T. , Gilmore, D.J. , Grunert, F. , Nagel, G. , Murdoch, S.J. , Tchilian, E. , Lennox, E.S. & Waldmann, H. (1991) CD66 identifies a neutrophil‐specific epitope within the hematopoietic system that is expressed by members of the carcinoembryonic antigen family of adhesion molecules. Blood 78, 63–74. [PubMed] [Google Scholar]
- 18. Oikawa, S. , Inuzuka, C. , Kuroki, M. , Matsuoka, Y. , Kosaki, G. & Nakazato, H. (1989) Cell adhesion activity of non‐specific cross reacting antigen (NCA) and carcinoembryonic antigen (CEA) expressed on CHO cell surface: hemophilic and heterophilic adhesion. Biochem. Biophys. Res. Commun. 164, 39–45. [DOI] [PubMed] [Google Scholar]
- 19. Benchimol, S. , Fuks, A. , Jothy, S. , Beauchemin, N. , Shirota, K. & Stanners, C.P. (1989) Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell 57, 327–334. [DOI] [PubMed] [Google Scholar]
- 20. Rojas, M. , Fuks, A. & Stanners, C.P. (1990) Biliary glycoprotein, a member of the immunoglobulin supergene family, functions in vitro as a Ca2+‐dependent intercellular adhesion molecule. Cell Growth Diff. 1, 527–533. [PubMed] [Google Scholar]
- 21. Pignatelli, M. , Durbin, H. & Bodmer, W.F. (1990) Carcinoembryonic antigen functions as an accessory adhesion molecule mediating colon epithelial cell–collagen interactions. Proc. Natl Acad. Sci. USA 87, 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oikawa, S. , Inuzuka, C. , Kuroki, M. , Arakawa, F. , Matsuoka, Y. , Kosaki, G. & Nakazato, H. (1991) A specific heterotypic cell adhesion activity between members of carcinoembryonic antigen family, W272 and NCA, is mediated by N‐domains. J. Biol. Chem. 266, 7995–8001. [PubMed] [Google Scholar]
- 23. Oikawa, S. , Kuroki, M. , Matsuoka, Y. , Kosaki, G. & Nakazato, H. (1992) Homotypic and heterotypic Ca++ ‐independent cell adhesion activities of biliary glycoprotein, a member of carcinoembryonic antigen family, expressed on CHO cell surface. Biochem. Biophys. Res. Commun. 186, 881–887. [DOI] [PubMed] [Google Scholar]
- 24. Zhou, H. , Fuks, A. , Alcaraz, G. , Bolling, T.J. & Stanners, C.P. (1993) Homophilic adhesion between Ig superfamily carcinoembryonic antigen molecules involves double reciprocal bonds. J. Cell Biol. 122, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou, H. , Stanners, C.P. & Fuks, A. (1993) Specificity of anti‐carcinoembryonic antigen monoclonal antibodies and their effects on CEA‐mediated adhesion. Cancer Res. 53, 3817–3822. [PubMed] [Google Scholar]
- 26. Teixeira, A.M. , Fawcett, J. , Simmons, D.L. & Watt, S.M. (1994) The N‐domain of the biliary glycoprotein (BGP) adhesion molecule mediates homotypic binding: domain interactions and epitope analysis of BGPc. Blood 84, 211–219. [PubMed] [Google Scholar]
- 27. Kuijpers, T. , Hoogerwerf, M. , Van Der Laan, L. , Nagel, G. , Van Der Schoot, C.E. , Grunert, F. & Roos, D. (1992) CD66 nonspecific cross‐reacting antigens are involved in neutrophil adherence to cytokine‐activated endothelial cells. J. Cell Biol. 118, 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuijpers, T.W. , Van Der Schoot, C.E. , Hoogerwerf, M. & Roos, D. (1993) Cross‐linking of the carcinoembryonic antigen‐like glycoproteins CD66 and CD67 induces neutrophil aggregation. J. Immunol. 151, 4934–4940. [PubMed] [Google Scholar]
- 29. Stocks, S.C. , Kerr, M.A. , Haslett, C. & Dransfield, I. (1995) CD66‐dependent neutrophil activation: a possible mechanism for vascular selectin‐mediated regulation of neutrophil adhesion. J. Leuk. Biol. 58, 40–48. [DOI] [PubMed] [Google Scholar]
- 30. Stocks, S.C. & Kerr, M.A. (1992) Stimulation of neutrophil adhesion of antibodies recognizing CD15 (Lex(X)) and CD15‐expressing carcinoembryonic antigen‐related glycoprotein NCA‐160. Biochem. J. 288, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lund‐Johansen, F. , Olweus, J. , Symington, F.W. , Arli, A. , Thompson, J.S. , Vilella, R. , Skubitz, K.M. & Horejsi, V. (1993) Activation of human monocytes and granulocytes by monoclonal antibodies to glycosylphosphatidylinositol‐anchored antigens. Eur. J. Immunol. 23, 2782–2791. [DOI] [PubMed] [Google Scholar]
- 32. Stocks, S.C. , Ruchaud‐Sparagano, M.‐H. , Kerr, M.A. , Grunert, F. , Haslett, C. & Dransfield, I. (1996) CD66: role in the regulation of neutrophil effector function. Eur. J. Immunol. 26, 2924–2932. [DOI] [PubMed] [Google Scholar]
- 33. Yamanaka, T. , Kuroki, M. , Matsuo, Y. & Matsuoka, Y. (1996) Analysis of heterophilic cell adhesion mediated by CD66b and CD66c using their soluble recombinant proteins. Biochem. Biophys. Res. Commun. 219, 842–847.DOI: 10.1006/bbrc.1996.0320 [DOI] [PubMed] [Google Scholar]
- 34. Wikstrom, K. , Kjellstrom, G. & Obrink, B. (1996) Homophilic intercellular adhesion mediated by C‐CAM is due to a domain 1‐domain 1 reciprocal binding. Exp. Cell Res. 227, 360–366.DOI: 10.1006/excr.1996.0285 [DOI] [PubMed] [Google Scholar]
- 35. Skubitz, K.M. , Campbell, K.D. & Skubitz, A.P.N. (1996) CD66a, CD66b, CD66c, and CD66d each independently stimulate neutrophils. J. Leuk. Biol. 60, 106–117. [DOI] [PubMed] [Google Scholar]
- 36. Neumaier, M. , Paululat, S. , Chan, A. , Matthaes, P. & Wagener, C. (1993) Biliary glycoprotein, a potential human cell adhesion molecule, is down‐regulated in colorectal carcinomas. Proc. Natl Acad. Sci. USA 90, 10744–10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nollau, P. , Scheller, H. , Kona‐Horstmann, M. , Rohde, S. , Hagenmuller, F. , Wagener, C. & Neumaier, M. (1997) Expression of CD66a (human C‐CAM) and other members of the carcinoembryonic antigen gene family of adhesion molecules in human colorectal adenomas. Cancer Res. 57, 2354–2357. [PubMed] [Google Scholar]
- 38. Nollau, P. , Prall, F. , Helmchen, U. , Wagener, C. & Neumaier, M. (1997) Dysregulation of carcinoembryonic antigen group members CGM2, CD66a (biliary glycoprotein), and nonspecific cross‐reacting antigen in colorectal carcinomas. Am. J. Pathol. 151, 521–530. [PMC free article] [PubMed] [Google Scholar]
- 39. Riethdorf, L. , Lisboa, B.W. , Henkel, U. , Naumann, M. , Wagener, C. & Loning, T. (1997) Differential expression of CD66a (BGP), a cell adhesion molecule of the carcinoembryonic antigen family, in benign, premalignant, and malignant lesions of the human mammary gland. J. Histochem. Cytochem. 45, 957–963. [DOI] [PubMed] [Google Scholar]
- 40. Tanaka, K. , Hinoda, Y. , Takahashi, H. , Sakamoto, H. , Nakajima, Y. & Imai, K. (1997) Decreased expression of biliary glycoprotein in hepatocellular carcinomas. Int. J. Cancer 74, 15–19. [DOI] [PubMed] [Google Scholar]
- 41. Kunath, T. , Ordonez‐Garcia, C. , Turbide, C. & Beauchemin, N. (1995) Inhibition of colonic tumor cell growth by biliary glycoprotein. Oncogene 11, 2375–2382. [PubMed] [Google Scholar]
- 42. Kleinerman, D.I. , Troncosco, P. , Lin, S.‐H. , Pisters, L.L. , Sherwood, E.R. , Brooks, T. , Von Eschenbach, A.C. & Hsieh, J.‐T. (1995) Consistent expression of an epithelial cell adhesion molecule (C‐CAM) during human prostate development and loss of expression in prostate cancer: implication as a tumor suppressor. Cancer Res. 55, 1215–1220. [PubMed] [Google Scholar]
- 43. Kleinerman, D.I.C.P.N. , Dinney, W.‐W. , Zhang, S.‐H. , Van Lin, N.T. & Hsieh, J.‐T. (1996) Suppression of human bladder cancer growth by increased expression of C‐CAM1 gene in an orthotopic model. Cancer Res. 56, 3431–3435. [PubMed] [Google Scholar]
- 44. Luo, W. , Wood, C.G. , Earley, K. , Hung, M.‐C. & Lin, S.‐H. (1997) Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C‐CAM1): the adhesion and growth suppression are mediated by different domains. Oncogene 14, 1697–1704. [DOI] [PubMed] [Google Scholar]
- 45. Hsieh, J.‐R.W. , Luo, W. , Song, Y. , Wang, D.I. , Van Kleinerman, N.T. & Lin, S.‐H. (1995) Tumor suppressive role of an androgen‐regulated epithelial cell adhesion molecule (C‐CAM) in prostate carcinoma cell revealed by sense and antisense approaches. Cancer Res. 55, 190–197. [PubMed] [Google Scholar]
- 46. Luo, W. , Tapolsky, M. , Earley, K. , Wood, C.G. , Wilson, D.R. , Logothetis, C.J. & Lin, S.H. (1999) Tumor‐suppressive activity of CD66a in prostate cancer. Cancer Gene Ther. 6, 313–321. [DOI] [PubMed] [Google Scholar]
- 47. Virji, M. , Watt, S.M. , Barker, S. , Makepeace, K. & Doyonnas, R. (1996) The N‐domain of the human CD66a adhesion molecule is a target for Opa proteins of Neisseria meningitidis and Neisseria gonorrhoeae . Mol. Microbiol. 22, 929–939. [DOI] [PubMed] [Google Scholar]
- 48. Virji, M. , Makepeace, K. , Ferguson, D.J. & Watt, S.M. (1996) Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22, 941–950. [DOI] [PubMed] [Google Scholar]
- 49. Gray‐Owen, S. , Dehio, C. , Haude, A. , Grunert, F. & Meyer, T.F. (1997) CD66 carcinoembryonic antigens mediate interactions between Opa‐expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 16, 3435–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen, T. & Gotschlich, E.C. (1996) CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc. Natl Acad. Sci. USA 93, 14851–14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bos, M.P. , Grunert, F. & Belland, R.J. (1997) Differential recognition of members of the carcinoembryonic antigen family by Opa variants of Neisseria gonorrhoeae . Infect. Immun. 65, 2353–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bos, M.P. , Kuroki, M. , Krop‐Watorek, A. , Hogan, D. & Belland, J. (1998) CD66 receptor specificity exhibited by neisserial Opa variants is controlled by protein determinants in CD66 N‐domains. Proc. Natl Acad. Sci. USA 95, 9584–9589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bos, M.P. , Hogan, D. & Belland, R.J. (1999) Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J. Exp. Med. 190, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Virji, M. , Evans, D. , Hadfield, A. , Grunert, F. , Teixeira, A.M. & Watt, S.M. (1999) Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N‐domain of CD66 molecules. Mol. Microbiol. 34, 538–551. [DOI] [PubMed] [Google Scholar]
- 55. Virji, M. , Evans, D. , Griffith, J. , Hill, D. , Serino, L. , Hadfield, A. & Watt, S.M. (2000) Carcinoembryonic antigens are targeted by diverse strains of typable and non‐typable Haemophilus influenzae . Mol. Microbiol. 36, 784–795. [DOI] [PubMed] [Google Scholar]
- 56. Virji, M. (2000) The structural basis of CEACAM‐receptor targeting by neisserial opa proteins: response [In Process Citation]. Trends Microbiol. 8, 260–261. [DOI] [PubMed] [Google Scholar]
- 57. Dveksler, G.S. , Pensiero, M.N. , Cardellichio, C.B. , Williams, R.K. , Jiang, G.‐S. , Holmes, K.V. & Dieffenbach, C.W. (1991) Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65, 6881–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pensiero, M.N. , Dveksler, G.S. , Cardellichio, C.B. , Jiang, G.‐S. , Elia, P.E. , Dieffenbach, C.W. & Holmes, K.V. (1992) Binding of the coronavirus mouse hepatitis virus A59 to its receptor expressed from a recombinant vaccinia virus depends on posttranslational processing of the receptor glycoprotein. J. Virol. 66, 4028–4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Williams, R.K. , Jiang, G.‐S. & Holmes, K.V. (1991) Receptor for mouse hepatitis virus is a member of the carcijojembryonic antigen family of glycoproteins. Proc. Natl Acad. Sci. USA 88, 5533–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yokomori, K. & Lai, M.M.C. (1992) Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J. Virol. 66, 6194–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Holmes, K.V. , Dveksler, G. , Gagneten, S. , Yeager, C. , Lin, S.‐H. , Beauchemin, N. , Look, A.T. , Ashumn, R. & Dieffenbach, C. (1994) Coronavirus receptor specificity In Cornaviruses, Plenum Press, New York, pp. 261–266. [DOI] [PubMed] [Google Scholar]
- 62. Skubitz, K.M. , Campbell, K.D. , Ahmed, K. & Skubitz, A.P.N. (1995) CD66 family members are associated with tyrosine kinase activity in human neutrophils. J. Immunol. 155, 5382–5390. [PubMed] [Google Scholar]
- 63. Skubitz, K.M. , Ducker, T.P. , Skubitz, A.P.N. & Goueli, S.A. (1993) Anti‐serum to carcinoembryonic antigen recognizes a phosphotyrosine‐containing protein in human colon cancer cell lines. FEBS Lett. 318, 200–204. [DOI] [PubMed] [Google Scholar]
- 64. Brummer, J. , Neumaier, M. , Gopfert, C. & Wagener, C. (1995) Association of p60c–src with biliary glycoprotein (CD66a), an adhesion molecule of the carcinoembryonic antigen family downregulated in colorectal carcinomas. Oncogene 11, 1649–1655. [PubMed] [Google Scholar]
- 65. Beauchemin, N. , Kunath, T. , Robitaille, J. , Chow, B. , Turbide, C. , Daniels, E. & Veillette, A. (1997) Association of biliary glycoprotein with protein tyrosine phosphatase SHP‐1 in malignant colon epithelial cells. Oncogene 14, 783–790. [DOI] [PubMed] [Google Scholar]
- 66. Afar, D.E. , Stanners, C.P. & Bell, J.C. (1992) Tyrosine phosphorylation of biliary glycoprotein, a cell adhesion molecule related to carcinoembryonic antigen. Biochim. Biophys. Acta 1134, 46–52. [DOI] [PubMed] [Google Scholar]
- 67. Skubitz, K.M. , Campbell, K.D. & Skubitz, A.P. (2000) Synthetic peptides of CD66a stimulate neutrophil adhesion to endothelial cells. J. Immunol. 164, 4257–4264. [DOI] [PubMed] [Google Scholar]
- 68. Skubitz, K.M. & Snook, R.W. , I.I. (1987) Monoclonal antibodies that recognize lacto‐N‐fucopenatose III (CD15) react with adhesion‐promoting glycoprotein family (LFA‐1/HMAC‐1/GP 150,95) and CR1 on human neutrophils. J. Immunol. 139, 1631–1639. [PubMed] [Google Scholar]
- 69. Vaporciyan, A.A. , Jones, M.L. & Ward, P.A. (1993) Rapid analysis of leukocyte‐endothelial adhesion. J. Immunol. Methods 159, 93–100. [DOI] [PubMed] [Google Scholar]
- 70. Skubitz, K.M. , Cambell, K.D. , Iida, J. & Skubitz, A.P.N. (1996) CD63 associates with tyrosine kinase activity and CD11/CD18, and transmits an activation signal in neutrophils. J. Immunol. 157, 3617–3626. [PubMed] [Google Scholar]
- 71. Skubitz, K.M. , Campbell, K.D. & Skubitz, A.P.N. (1997) CD50 monoclonal antibodies inhibit neutrophil activation. J. Immunol. 159, 820–828. [PubMed] [Google Scholar]
- 72. Wertheimer, A.J. , Myers, C.L. , Wallace, R.W. & Parks, T.P. (1992) Intercellular adhesion molecule‐1 gene expression in human endothelial cells. J. Biol. Chem. 267, 12030–12035. [PubMed] [Google Scholar]
- 73. Springer, T.A. (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76, 301–314. [DOI] [PubMed] [Google Scholar]
- 74. Wright, S.D. & Meyer, B.C. (1986) Phorbol esters cause sequential activation and deactivation of complement receptors on polymorphonuclear leukocytes. J. Immunol. 136, 1759–1764. [PubMed] [Google Scholar]
- 75. Carlos, T.M. & Haran, J.M. (1994) Leukocyte‐endothelial adhesion molecules. Blood 84, 2068–2101. [PubMed] [Google Scholar]
- 76. Boehm, M.K. , Mayans, M.O. , Thornton, J.D. , Begent, R.H.J. , Keep, P.A. & Perkens, S.J. (1996) Extended glycoprotein structure of the seven domains in human carcinoembryonic antigen by X‐ray and neutron solution scattering and an automated curve fitting procedure: implications for cellular adhesion. J. Mol. Biol. 259, 718–736. [DOI] [PubMed] [Google Scholar]
- 77. Oikawa, S. , Sugiyama, M. , Kuroki, M. & Nakazato, H. (2000) Extracellular N‐domain alone can mediate specific heterophilic adhesion between members of the carcinoembryonic antigen family, CEACAM6 and CEACAM8. Biochem. Biophys. Res. Commun. 278, 564–568. [DOI] [PubMed] [Google Scholar]
- 78. Watt, S.M. , Teixeira, A.M. , Doyonnas, R. , Fritz, G. , Blumberg, R.S. , Kuroki, M. , Skubitz, K.M. & Bates, P.A. (2000) The Identification of Critical Adhesiotopes on the N‐Domain of Human CEACAM1 Required for Homophilic Interactions, Garland, New York. [Google Scholar]
- 79. Taheri, M. , Saragovi, U. , Fuks, A. , Makkerh, J. , Mort, J. & Stanners, C.P. (2000) Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J. Biol. Chem. 275, 26935–26943. [DOI] [PubMed] [Google Scholar]
- 80. Wessner, D.R. , Shick, P.C. , Lu, J.H. , Cardellichio, C.B. , Gagneten, S.E. , Beauchemin, N. , Holmes, K.V. & Dveksler, G.S. (1998) Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain A59. J. Virol. 72, 1941–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sippel, C.J. , Shen, T. & Perlmutter, D.H. (1996) Site‐directed mutagenesis within an ectoplasmic ATPase consensus sequence abrogates the cell aggregating properties of the rat liver canalicular bile acid transporter/ecto‐ATPase/cell CAM 105 and carcinoembryonic antigen. J. Biol. Chem. 271, 33095–33104. [DOI] [PubMed] [Google Scholar]
- 82. Jantscheff, P. , Nagel, G. , Thompson, J. , Kleist, S.V. , Embleton, M.J. , Price, M.R. & Grunert, F. (1996) A CD66a‐specific, activation‐dependent epitope detected by recombinant human signal chain fragments (scFvs) on CHO transfectants and activated granulocytes. J. Leuk. Biol. 59, 891–901. [DOI] [PubMed] [Google Scholar]
- 83. Hunter, I. , Sawa, H. , Edlund, M. & Obrink, B. (1996) Evidence for regulated dimerization of cell−cell adhesion molecule (C‐CAM) in epithelial cells. Biochem. J. 320, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lisowska, E. , Krop‐Watorek, A. & Sedlaczek, P. (1983) The dimeric structure of carcinoembryonic antigen (CEA). Biochem. Biophys. Res. Commun. 115, 206–211. [DOI] [PubMed] [Google Scholar]
- 85. Edlund, M. , Blikstad, I. & Obrink, B. (1996) Calmodulin binds to specific sequences in the cytoplasmic domain of C‐CAM and down‐regulates C‐CAM self‐association. J. Biol. Chem. 271, 1393–1399. [DOI] [PubMed] [Google Scholar]
- 86. Moller, M.J. , Kammerer, R. , Grunert, F. & Von Kleist, S. (1996) Biliary glycoprotein (BGP) expression on T cells and on a natural‐killer‐cell sub‐population. Int. J. Cancer 65, 740–745. [DOI] [PubMed] [Google Scholar]
- 87. Prado, I.B. , Laudanna, A.A. & Carneiro, C.R.W. (1995) Susceptibility of colorectal carcinoma cells to natural‐killer‐mediated lysis: relationship to CEA expression and degree of differentiation. Int. J. Cancer 61, 854–860. [DOI] [PubMed] [Google Scholar]
- 88. Kammerer, R. & Von Kleist, S. (1994) CEA expression of colorectal adenocarcinomas is correlated with their resistance against LAK‐cell lysis. Int. J. Cancer 57, 341–347. [DOI] [PubMed] [Google Scholar]
- 89. Leusch, H.G. , Drezeniek, Z. , Markos‐Pusztai, Z. & Wagener, C. (1991) Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic α‐glycosides of mannose. Infect. Immun. 59, 2051–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sauter, S.L. , Rutherfurd, S.M. , Wagener, C. , Shively, J.E. & Hefta, S.A. (1993) Identification of the specific oligosaccharide sites recognized by type 1 fimbriae from Escherichia coli on nonspecific cross‐reacting antigen, a CD66 cluster granulocyte glycoprotein. J. Biol. Chem. 268, 15510–15516. [PubMed] [Google Scholar]


