Key Points
Virus entry into animal cells is initiated by attachment to receptors and is followed by important conformational changes of viral proteins, penetration through (non-enveloped viruses) or fusion with (enveloped viruses) cellular membranes. The process ends with transfer of viral genomes inside host cells.
Viral proteins mediating entry are very diverse, but many share common three-dimensional structural motifs.
Conformational changes in the viral proteins that drive entry are typically initiated by high-affinity interactions with receptors, or changes in pH after receptor binding and internalization. They include formation of coiled-coils in class I fusion proteins, dimer to trimer transitions in class II fusion proteins, movement of capsid proteins in non-enveloped viruses and exposure of membrane destabilizing sequences.
Fusion with, or penetration through, cell membranes might involve multimolecular protein complexes and requires structural rearrangements of membrane lipids.
Inhibitors of virus entry can prevent virus attachment, or bind to entry intermediates; small organic molecules, peptides, soluble receptors and antibodies are in clinical trials. Six virus-specific polyclonal human immunoglobulins, one monoclonal antibody and one peptide have been approved by the US Food and Drug Administration for clinical use.
Viral proteins involved in entry can induce immune responses and be used as vaccine immunogens.
Viral entry machineries could be beneficial for human physiology and retargeted for the treatment of cancer and other diseases.
Abstract
Viruses have evolved to enter cells from all three domains of life — Bacteria, Archaea and Eukaryotes. Of more than 3,600 known viruses, hundreds can infect human cells and most of those are associated with disease. To gain access to the cell interior, animal viruses attach to host-cell receptors. Advances in our understanding of how viral entry proteins interact with their host-cell receptors and undergo conformational changes that lead to entry offer unprecedented opportunities for the development of novel therapeutics and vaccines.
Main
Probably the first observation of specific attachment of a virus to a cell was made at the start of the twentieth century by d'Herelle1. He cultured Shigella and observed occasional clear spots — lysed bacteria — in a lawn of bacterial growth on a solid agar medium which he called plaques. The viruses that had lysed Shigella were named bacteriophages. Using co-sedimentation experiments, he showed that the attachment of the virus to the host cell is the first step in infection, and that attachment only occurred when the virus was mixed with bacteria that were susceptible to the virus. This early study showed that the host range of a virus was determined by the attachment step. A century later, we are beginning to understand the details of an increasing number of virus–receptor interactions at the atomic level. All viruses contain nucleic-acid genomes (either RNA or DNA), which are packaged with proteins that are encoded by the viral genome. Viruses can be divided into two main categories; enveloped viruses, which have a lipid membrane (envelope) that is derived from the host cell; and non-enveloped viruses, which lack a membrane. Viruses from 24 different families can cause, or are associated with, diseases in humans (Table 1), so it is crucial to understand how different viruses solve the problem of entry into cells, and how this process can be inhibited. This review summarizes recent advances in our understanding of virus entry mechanisms at the molecular level and options for therapeutic intervention of these processes.
Table 1.
Pathogenic human viruses
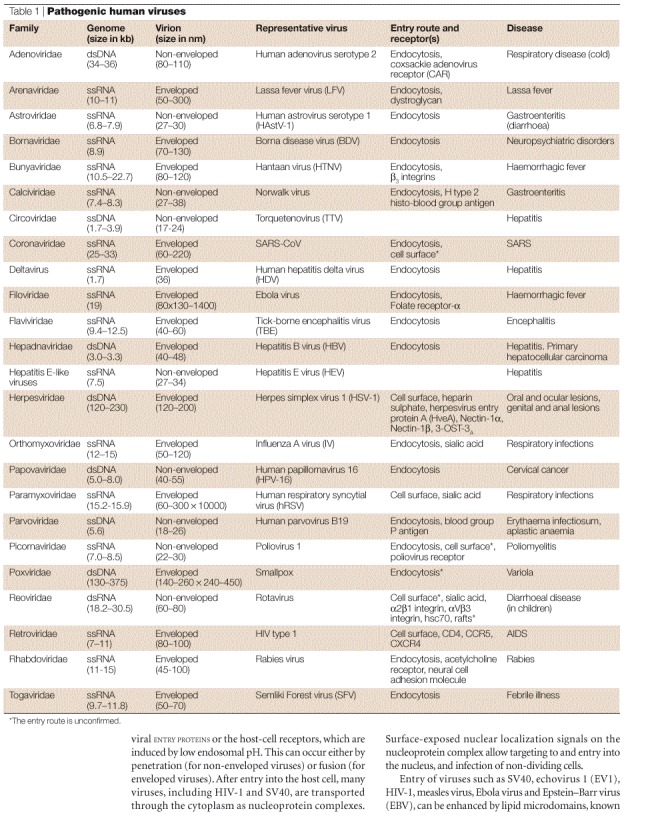
Modes of entry
Both non-enveloped and enveloped viruses share the same main steps and routes of virus entry — which begin with attachment to cell-surface receptors and end with the delivery of the viral genome to the cell cytoplasm (Fig. 1). After binding to receptors — which can be proteins, carbohydrates or lipids — viruses use two main routes to enter the cell — the endocytic and non-endocytic routes. The endocytic route is usually by transport in clathrin-coated vesicles or pits, but non-clathrin-coated pits, macropinocytosis or caveolae are also used2. Some viruses can induce internalization by endocytosis — for example, simian virus 40 (SV40), which induces local actin polymerization and dynamin recruitment at the site of entry3 (Fig. 1). The non-endocytic route of entry involves directly crossing the plasma membrane at neutral pH (Fig. 1). Viruses that use the non-endocytic route can also enter cells by the endocytic pathway — for example, human immunodeficiency virus type 1 (HIV-1). Membrane fusion — a basic cellular process that is essential for phagocytosis, pinocytosis and vesicular trafficking — is a basic mode of entry by enveloped viruses that use the endocytic or non-endocytic routes. The process is regulated and is mediated by membrane proteins once the membranes are in close proximity to each other. For both enveloped and non-enveloped viruses, entry into cells involves important conformational changes of the viral ENTRY PROTEINS or the host-cell receptors, which are induced by low endosomal pH. This can occur either by penetration (for non-enveloped viruses) or fusion (for enveloped viruses). After entry into the host cell, many viruses, including HIV-1 and SV40, are transported through the cytoplasm as nucleoprotein complexes. Surface-exposed nuclear localization signals on the nucleoprotein complex allow targeting to and entry into the nucleus, and infection of non-dividing cells.
Figure 1. Two main virus entry pathways.
a | Clathrin-mediated endocytosis, for example, adenovirus. Endocytosis by caveolae can also occur, for example, SV40. b | Fusion at the cell membrane, for example, HIV.Fusion can also occur from inside an endosome, for example, influenza.
Entry of viruses such as SV40, echovirus 1 (EV1), HIV-1, measles virus, Ebola virus and Epstein–Barr virus (EBV), can be enhanced by lipid microdomains, known as LIPID RAFTS4,5. Other viruses, including influenza, can use lipid rafts as a platform on which to concentrate a sufficient number of viral molecules into virions for efficient exit from one cell and entry into another cell6. However, fusion of Semliki Forest virus (SFV) or Sindbis virus (SIN) with LIPOSOMES does not require rafts, and the presence of rafts can be inhibitory to membrane fusion7. It remains to be established whether rafts are involved in the infectious cell entry of SFV and SIN8.
Some viruses enter cells through direct cell-to-cell contacts, using structures that are formed by the polarized cytoskeleton, adhesion molecules and viral proteins at the infected cell junction, which is known as the 'virological synapse'9. Direct cell-to-cell transmission of viruses by this process — for example, retroviruses (such as human T-cell lymphoma-leukaemia virus type 1 (HTLV-1) and HIV-1), herpesviruses (such as herpes simplex virus (HSV) and varicella-zoster virus) and poxviruses (such as vaccinia virus) — is poorly understood. In many cases it is not clear whether cell-to-cell transmission by this route involves membrane fusion or penetration, or direct transfer of the virus through cell junctions. Efficient and rapid cell-to-cell transmission of some viruses, such as HIV-1, could alternatively be mediated by virions that are budding or have just been released into the space between the closely apposed interacting cells or by cell-to-cell fusion. Cell-to-cell transmission might protect viruses from the actions of the immune system and could be an important route of transmission in vivo.
Viruses such as HIV-1 and poliovirus can enter and exit cells without crossing membranes by a process known as transcytosis10. Transcytosis — vesicular transport from one side of a cell to the other — is used by multicellular organisms to selectively move material (usually macromolecules) through cells between two environments without modifying it. Viruses have usurped this mechanism to cross the epithelial cell barrier and infect the underlying cells.
The kinetics and efficiency of entry vary greatly between viruses from different families, between viruses within a family, between viruses within a genus and even between isolates of the same species. Some viruses, such as adeno-associated virus serotype 2 (AAV-2), SFV and influenza can cross the endosomal membranes very rapidly (within seconds), and the efficiency of entry can be as much, or more than, 50% (which means that 50% of attached viruses enter cells). Single AAV-2 virions can cross membranes in less than a second11 and individual influenza virions cross membranes in as little as one or more seconds12,13. Other viruses, such as HIV-1, take one or more minutes to enter cells, and the efficiency of entry is poor compared with AAV-2 — often as low as 0.1% (Refs 14,15). The kinetics and efficiency of virus entry might be related to the virus structure and it seems that the best kinetics and efficiency of entry are observed for viruses that use low pH as an entry trigger and have flattened structures — such as SFV. Cell-bound SFV can fuse in seconds with an efficiency of 80% (Ref. 16). Membrane lipid composition and structure also affect the kinetics and efficiency of virus entry.
Both non-enveloped and enveloped viruses can use the energy of METASTABLE STATES in viral entry proteins to expose hydrophobic sequences17,18 that can destabilize host-cell membranes. However, after this, the formation of different intermediates leads to the formation of membrane pores (in the case of non-enveloped viruses) or membrane fusion pores (in the case of enveloped viruses) (Fig. 1). Often, conformational changes in a single virus protein can mediate membrane fusion. Enveloped viruses can fuse with the plasma membrane or from inside an endosome. The penetration and entry of non-enveloped viruses might resemble the entry of toxins, such as anthrax toxin19. Entry of enveloped viruses has similarities with intracellular fusion processes, such as exocytosis20.
Virus structure and receptor recognition
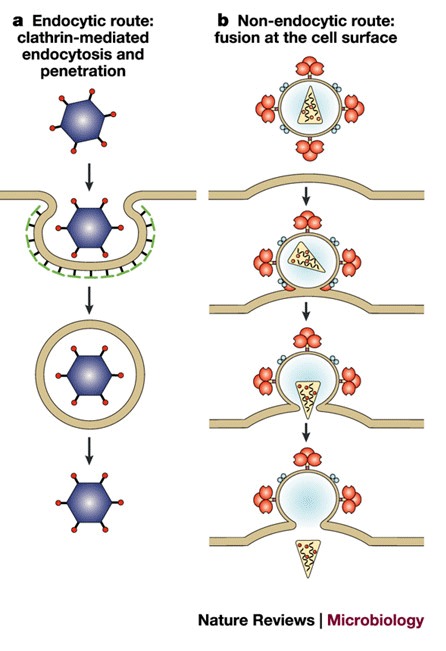
Virus evolution has resulted in several receptor-recognizing surface structures, which frequently have protrusions (spikes) about 10 nm or longer that are formed by the entry proteins — for example, coronaviruses (Fig. 2d) and AAVs (Fig. 2b) — or canyons — for example, the picornaviruses human rhinovirus 14 (HRV14)21 and poliovirus22 (Fig. 2a). Non-enveloped viruses are often small and stable, and can form crystals that diffract to good resolution, so the structures of a relatively large number of representatives from different virus families in this group have been solved by X-ray crystallography to high resolution — typically about 2 Å. By contrast, only a few structures of whole enveloped viruses — for example, SFV23 (Fig. 2c) and dengue virus24 — have been published, which have been solved to no more than 9-Å resolution by cryoelectron microscopy.
Figure 2. Structure of virus surfaces.
a | Structure of the 160S poliovirus particle. Reproduced with permission from Ref. 106 © (2000) American Society for Microbiology. b | Surface topology of adeno-associated virus 2. The protruding spikes are coloured white. Reproduced with permission from Ref. 105 © (2002) National Academies of Sciences, USA. c | Structure of Semliki Forest virus. The colour scheme reflects the radial distance from the centre of the virion, increasing from blue to red. Reproduced with permission from Ref. 23 © (2000) Elsevier Science. d | Structure of the severe acute respiratory syndrome (SARS) coronavirus. The coronavirus particle has club-shaped surface projections — known as spikes. Adeno-associated virus 2 and Semliki Forest virus particles (not to scale) are comparable in size to Poliovirus. Reproduced with permission from Ref. 107 © (2003) Massachusetts Medical Society.
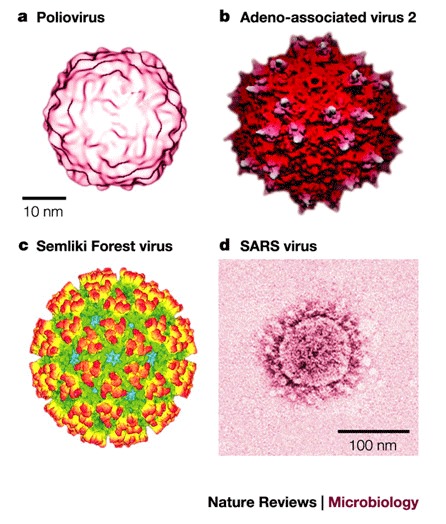
Structure of viral entry proteins. Virion-associated entry proteins are typically glycosylated oligomers — for example, homodimeric tick-borne encephalitis virus (TBE) E protein, homotrimeric adenovirus fibre and influenza haemagglutinin (HA) (Fig. 3). The ATTACHMENT PROTEINS of some viruses — for example, adenoviruses and retroviruses — are unlikely to significantly interact with each other, whereas the entry proteins of other viruses — including alphaviruses, such as SFV — form heterodimers with other proteins and interact with each other extensively to form a lattice of interacting proteins (Fig. 2c). The entire CAPSID of many non-enveloped viruses — for example the picornaviruses human rhinovirus 14 (HRV14)21 and poliovirus22 (Fig. 2a) — is formed by a network of interacting proteins that are involved in entry.
Figure 3. Structures of soluble fragments from virus entry proteins.
a | Ribbon tracing of reovirus attachment protein σ1. Reproduced with permission from Ref. 108 © (2003) Wiley. b | Ribbon tracing of adenovirus fibre. Reproduced with permission from Ref. 22 © (1985) American Association of Sciences. Both reovirus attachment protein σ1 (a) and adenovirus fibre (b) are homotrimers. The three monomers in each trimer are shown in red, orange and blue. Both proteins have head-and-tail morphology, with a triple β–spiral domain forming the tail and an eight-stranded-sandwich domain forming the head. c | Cleaved influenza haemagglutinin trimer26. Reproduced with permission from Ref. 38 © (2000) Annual Reviews. d | Model of the respiratory syncitial virus F protein structure (RSV-F), which is based on amino-acid sequence homology with the structure of the Newcastle disease virus F protein27. Reproduced with permission from Ref. 110 © (2003) Elsevier Science.
The topologies of viral entry proteins vary from those that form spikes and which comprise a stem and a globular head (Fig. 3) — for example reoviruses, adenoviruses, orthomyxoviruses and paramyxoviruses — to those that form relatively 'flat' structures, for example, the alphaviruses (family togaviridae) (Fig. 2c). The similarity between the topologies of the non-enveloped reovirus and adenovirus attachment proteins extends to their three-dimensional structures even though the amino-acid sequences of these two proteins share no sequence similarity25 (Fig. 3a,b). The similarity between the 3D structures, together with the conservation of function, indicates a common ancestor for these proteins, but does not preclude convergent evolution. The overall similarities between the topologies of the non-enveloped reovirus and adenovirus attachment proteins, and between the topologies of the ENVELOPE GLYCOPROTEINS (Envs) from enveloped viruses, such as influenza HA26 and the respiratory syncitial virus (RSV) fusion protein (which has been predicted based on sequence similarity with the homologous fusion protein of the Newcastle disease virus (NDV)27) is notable, particularly owing to the lack of amino-acid sequence identity between these proteins (Fig. 3). However, the stems of the reovirus and adenovirus attachment proteins are stabilized by triple β-spirals, whereas the stems of influenza HA and respiratory syncitial virus F protein are stabilised by α-helical coiled-coils, which indicates that these viruses have different ancestors. The structures of the enveloped flavivirus E proteins from TBE28 and dengue virus29 and the enveloped alphavirus SFV E1 protein30 are similar to each other, which, in combination with similar functions, might indicate the existence of a common ancestor for the entry proteins of flaviviruses and alphaviruses30. Virus entry proteins have been divided into two classes that are dependent on several criteria, including mechanism of action, whether the entry protein is cleaved and whether the entry protein is complexed with other viral proteins. Envs that contain coiled-coils, such as influenza HA, paramyxovirus fusion protein F and HIV glycoprotein 160 (gp160), have been designated class I FUSION PROTEINS and the Envs of alphaviruses and flaviviruses have been designated class II fusion proteins30,31.
Viral entry proteins have diverse amino-acid sequences, but many of those for which the structure has been solved contain similar 3D structural motifs — although the overall topology can be different. This is especially notable for non-enveloped viruses; the viral proteins that are exposed to the environment and recognize receptor molecules from several plant, insect and mammalian viruses contain the same basic core structure, which consists of an 8-stranded β-barrel with a jelly-roll motif18. Although only a few Env structures have been solved, it seems that they might use similar types of structural motifs as non-enveloped viruses for the recognition of host cells. The globular head of the best-characterized viral Env that mediates entry — the HA of the orthomyxovirus influenza — contains a β-barrel-type structure26 (Fig. 3). The head of the NDV F protein, comprises an immunoglobulin-type β-sandwich domain and a highly twisted β-sheet domain32. Fragments of two other Envs — the HSV glycoprotein D33 and the receptor-binding domain of Friend murine leukaemia virus (Fr-MLV)34 — also contain β-barrel structures with similarity to the immunoglobulin-like fold. Analysis of this limited number of structures indicates that viruses use conserved frameworks of β-sheets joined by variable loops that can allow rapid adaptation to new receptors. The other three X-ray crystal structures of Envs with receptor-binding domains in an unbound state are soluble fragments of the main Envs of the flaviviruses TBE28 and dengue virus29, and a fragment of E1 from SFV30, which contain predominantly β-strands. Most of these β-strands are packed in sheets, including a six-stranded β-barrel in the second domain, and an immunoglobulin-like β-barrel fold in the third domain that could be important for binding to cellular receptors.
Recognition of virus receptors on host cells. Although VIRUS RECEPTORS have diverse sequences, structures and cellular functions35,36, there is a preference for molecules that are involved in cell adhesion and recognition by reversible, multivalent AVIDITY-determined interactions. Viruses might have evolved to bind to abundant cellular receptors, or to bind to cellular receptors that have relatively low affinity for their natural ligands with high affinity37 or to bind to receptors that have both of these characteristics (Table 2).
Table 2.
Human viruses and host cell receptors
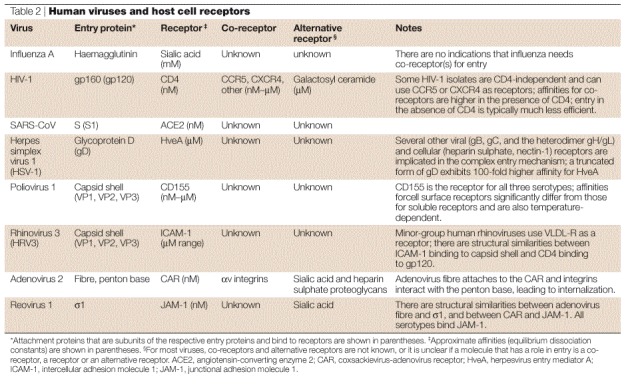
The receptor-recognition interactions of different viruses — and even different isolates of the same virus — can vary significantly. Typically, low-affinity (μM–mM) binding interactions between viral attachment proteins and their cognate receptors involve a small area of interaction between the viral and cellular receptors and do not lead to conformational changes in the entry proteins. For example, binding of influenza HA and SV40 to sialic acid38. High-affinity (nM–pM) interactions with virus receptors involve a large area of interaction (about 10 nm2) between the viral and cellular receptors, and often involve large conformational changes. For example, the binding of HIV-1 gp120 to CD4 and one of the CO-RECEPTORS CCR5 or CXCR4, and poliovirus to CD155, both result in conformational changes18,39. One exception is the high-affinity binding of the adenovirus fibre to the coxsackievirus-adenovirus receptor (CAR), which does not involve significant conformational changes (Fig. 4).
Figure 4. Receptor recognition.
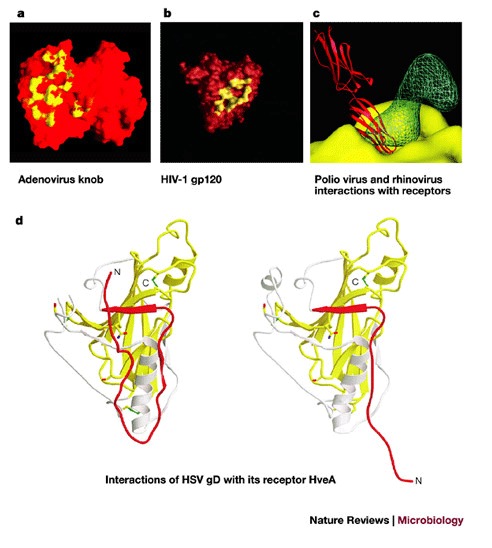
a | A molecular-surface representation of the interface between the adenovirus 12 (Ad12) knob and the coxsackievirus-adenovirus receptor D1 (CAR D1). The figure shows two adjacent Ad12 knob monomers, viewed at the interface between the Ad12 and CAR D1 molecules and coloured on a scale from yellow (contact with CAR D1) to red (no contact with CAR D1). Atoms in contact with CAR D1 are shared between the two Ad12 monomers. From Ref. 111 © (1999) American Association of Sciences. b | The HIV-1 gp120 and CD4 receptor contact surface. The gp120 surface is shown in red, with the surface that is 3.5 Å distant from the CD4 receptor (surface-to-atom-centre distance) shown in yellow. From Ref. 39 © (1998) Nature, Macmillan Magazines. c | Interaction of rhinovirus and poliovirus with their receptors. Comparison of poliovirus binding to its receptor CD155 and rhinovirus binding to its receptor ICAM-1. The figure shows the electron density of a poliovirus-bound CD155 molecule (green), the poliovirus molecular surface (yellow), including the canyon, and a rhinovirus-bound IC1 molecule (red). Structures of poliovirus and rhinovirus were superimposed to generate the figure. Reproduced with permission from Ref. 112 © (2000) Oxford University Press. d | The conformational change of soluble glycoprotein D (gD285) fragment. The left panel shows the gD285 fragment, in complex with herpesvirus entry mediator A (HveA). The amino-terminal portion of the HveA binding hairpin (red) can be seen interacting with residues 224–240 of an α-helix of gD285 (white). The right panel shows unliganded gD285. From Ref. 33 © (2003) American Society for Microbiology.
The high-affinity receptor-binding site can be located in a deep crevice (or canyon) on the viral protein, such as in the picornaviruses, or can contain loops, cavities and channels, such as the adenovirus knob and HIV gp120 (Fig. 4). Rather unusually, the HveA receptor-binding site on HSV-1 glycoprotein D is situated on an amino-terminal extension at one edge of the glycoprotein D molecule rather than being assembled from many parts of the glycoprotein D sequence, as for a typical binding surface or binding pocket. It undergoes conformational changes on binding to the receptor33. There is no correlation between the structure of the viral entry protein structure and the structure of the cellular receptor. Viruses from the same family, such as retroviruses, can bind to different cellular receptors, and the same cellular molecule, for example, sialic acid, can serve as a receptor for several different viruses.
The conformational changes that are induced by interactions with one receptor can be required to expose the binding site for another receptor, for example the interaction between CD4 and gp120 induces the exposure of a high-affinity binding site for a co-receptor (typically CCR5 or CXCR4) on HIV-1 gp120. In this case, CD4 serves as an 'attachment' receptor that ensures specific binding to CD4-expressing cells and the co-receptor serves as a 'fusion' receptor that induces conformational changes that lead to exposure of fusogenic sequences. In some strains of HIV-1, co-receptors can mediate both attachment and fusion in the absence of CD4. Entry can also be initiated by binding to a low-affinity receptor, such as heparin sulphate, followed by higher affinity interactions. The role of many receptors in entry remains unresolved or controversial. Entry into cells through interactions with more than one receptor seems to be widely used by viruses, especially for the infection of specific types of cells in vivo. In the case that a specific cell receptor is absent, ALTERNATIVE VIRUS RECEPTORS have also been identified for some viruses. For example, galactosyl ceramide40 and its sulphated derivative (sulphatide) can support low level HIV-1 infections in some CD4-negative cell lines, although the roles of such alternative receptors in vivo remain unknown.
Although the number of identified receptors for human viruses has increased rapidly during the past two decades41,42,43,44,45,46, most virus receptors remain uncharacterized, and there are only a few X-ray crystal or cryoelectron microscopy structures of entry proteins in complex with receptors (Fig. 4). Identification of new receptors is important for understanding virus tropism, pathogenicity and the mechanisms of entry. Recently, the receptor for the severe acute respiratory syndrome coronavirus (SARS-CoV) — the angiotensin-converting enzyme 2 (ACE2) — was identified, only months after the virus was discovered47, and the receptor-binding domain has been localized to amino-acid residues 303–537 of the SARS-CoV entry protein48.
Virus-receptor function is affected by membrane organization. Lipid rafts have been intensively studied to determine any possible role in virus entry5. Although rafts are well characterized, their role in virus entry is controversial. Studies on HIV-1 entry illustrate the controversies that still exist with regard to the role of rafts in virus entry. Together, depletion of cholesterol and inhibition of glycosphingolipid synthesis decrease the efficiency of HIV-1 Env-mediated membrane fusion — typically by about two-fold — which could indicate effects on membrane fusion owing to the disruption of raft integrity5. The raft component glycosphingolipids Gb3 and GM3 seem to interact with gp120 in the presence of CD4, which could also indicate that rafts are involved in HIV-1 entry49. However, recent data indicate that HIV-1 infection does not depend on the presence of CD4 and CCR5 in rafts and it has been proposed that cholesterol modulates HIV-1 entry by an independent mechanism, perhaps related to membrane merging or modulation of co-receptor binding50. Although the exact role of rafts in receptor-expressing host cells is controversial, recent experiments have clearly shown the importance of rafts for membrane fusion by clustering sufficient numbers of influenza HA molecules in rafts 200–280 nm in diameter6. So, it appears that in both receptor-expressing and Env-expressing cells the function of rafts is mainly to increase the local concentrations of molecules that are involved in entry. Glycosphingolipids might not only provide the structural basis for raft formation, but could also interact directly with viral entry proteins and cellular receptor molecules.
Conformational changes of entry proteins
After receptor recognition, viral entry proteins undergo marked conformational changes that drive the entry process to completion. One hypothesis is that entry proteins from many viruses, including poliovirus, influenza, HIV, TBE and SFV, are in a metastable high-energy state17,18. However, recent differential scanning-calorimetric measurements showed that the unfolding of influenza HA at neutral pH is an endothermic process, which might indicate that it is not in a metastable high-energy state51,52. According to the metastable-state hypothesis, receptor binding or a pH change (or possibly both events53,54, but this possibility is debated55) can provide the activation energy that is required to overcome energy barriers for the viral entry protein to reach a stable, energetically favourable state. The energy of the transition to this state is used to externalize sequences from internal parts of entry proteins that can destabilize membranes. The structural transition of the virus entry protein and subsequent action of the externalized membrane-destabilizing sequence induces the formation of membrane pores and membrane fusion pores.
Enveloped viruses: class I, class II and unclassified fusion proteins. The prototype class I fusion protein is influenza HA. Perhaps the best-characterized fusion intermediates are the helical coiled-coils that are formed by fragments from fusion proteins of representative orthomyxo-, paramyxo-, retro-, filo-, and coronaviruses38,56,57. These structures are thought to represent the lowest energy state, which is reached after a series of conformational changes that are induced by receptor binding or low pH. In addition to the formation of coiled-coils during fusion, the Envs from many of these viruses mature by proteolytic cleavage of precursor proteins to yield membrane-anchored subunits, which contain N-terminal fusion peptides, but some coronaviruses, including SARS-CoV, are not cleaved and remain trimeric throughout the fusion process30,31. A similar, but four-stranded, coiled-coil is involved in a number of intracellular fusion processes, which include synaptic vesicle fusion20. Although the formation of the coiled-coils is irreversible, it does involve some reversible steps — for example, in the case of influenza HA58. The molecular details of the pathway that leads to the formation of these structures are not fully understood, but include several intermediates that can be distinguished by kinetic measurements and structural analysis56,59 (Fig. 5). Molecular dynamics simulations of the conformational transitions of the influenza HA that are induced by a change from a neutral to a low pH might indicate that a complete dissociation of the globular domains of the HA proteins could expose the fusion peptides and reorientate the peptides towards the target membrane, which is consistent with a spring-loaded conformational-change hypothesis60.
Figure 5. Conformational changes of viral fusion proteins leading to membrane fusion.
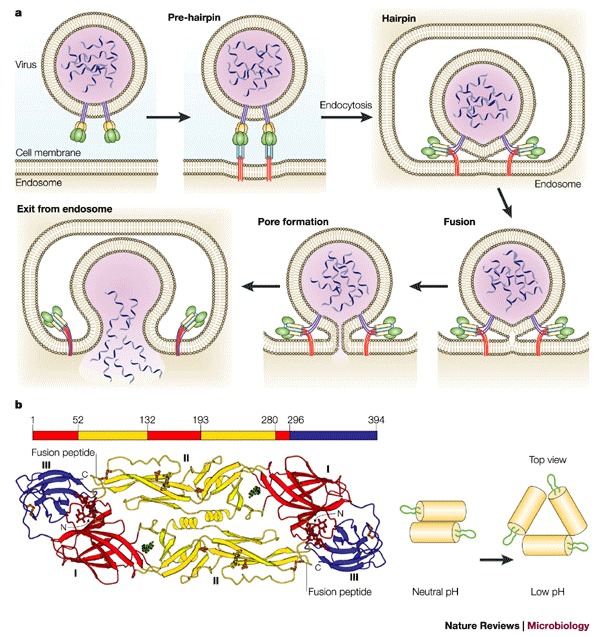
a | Schematic representation of a working model for viral membrane fusion mediated by class I fusion proteins. Influenza virus, which is internalized into an endosome, is shown as an example. In the native state of the fusion protein — which is a trimer — most of the surface subunit (green) is exposed. Part of the transmembrane subunit, including the fusion peptide, is not exposed. Following fusion-activating conditions, conformational changes occur to 'free' the fusion peptide (red) from its previously unexposed location. In the case of influenza HA, this occurs by a 'spring-loaded' mechanism. The 'pre-hairpin' intermediate spans two membranes — with the transmembrane domain positioned in the viral membrane and the fusion peptide inserted into the host-cell membrane. The pre-hairpin intermediate forms a trimer of hairpins, and membrane fusion occurs, which leads to pore formation and release of the viral genome into the cytoplasm. Modified with permission from Ref. 56 © (2001) Annual Reviews. b | Conformational changes of class II viral fusion proteins and entry. Structure of fragments of the class II fusion (E) glycoprotein from dengue virus29. The polypeptide chain begins in the central domain (domain I, red), which is an eight-stranded β-barrel with up-and-down topology. Two long insertions between strands in the central domain form the dimerization domain (domain II, yellow). The carboxy-terminal domain (domain III, blue) is an antiparallel β-barrel with an immunoglobulin-like topology, which is stabilized by three disulphide bridges. The domain definition is also highlighted on the peptide sequence (top). During entry, the oligomeric structure is reorganized. The configuration of dengue virus glycoproteins on the virion surface at neutral pH and the proposed configuration at low pH are shown. The E glycoproteins are shown as yellow cylinders and the fusion peptide is green. Reproduced from Refs 24,29 © (2002) Elsevier; (2003) National Academies of Sciences.
Class II fusion proteins are not proteolytically cleaved and have internal, rather than N-terminal fusion peptides30,31. They are synthesized as a complex with a second membrane glycoprotein, and the activation of the fusogenic potential of the class II fusion proteins involves the cleavage of this accessory protein. X-ray crystallography of three class II fusion proteins — the E proteins of TBE and dengue virus, and the E1 protein of SFV — has revealed a common fold for these proteins, which is structurally unrelated to the class I viral fusion proteins. Class II fusion proteins fold as heterodimers with the companion (chaperone) glycoprotein — for example, pE2 with pE1 in alphaviruses and prM with pE in flaviviruses — and form a protein network at the viral surface. The fusogenic conformational changes lead to a reversible dissociation of the dimers to release monomers, followed by irreversible reassociation into stable homotrimers in the presence of membranes (Fig. 5). Although the alphaviruses SFV and SIN require the membrane components cholesterol and sphingolipids for the binding and initiation of conformational changes, the flavivirus TBE does not require these membrane components — but binding to target membranes and trimerization of the TBE E protein might involve interactions with cholesterol61.
Unlike class I and class II fusion proteins, the conformational changes of the G proteins of rabies virus and the vesicular stomatitis virus (VSV) (family rhabdoviridae) that are induced by a low pH are reversible, which indicates that the low pH does not trigger a transition through a high-energy intermediate state62,63. However, interactions with membranes could induce irreversible conformational changes. VSV and rabies virus can both fuse with pure lipid vesicles, although neither virus requires any specific lipids for fusion. The transition between equilibrium states of the entry proteins could provide free energy to overcome the membrane fusion barrier, but formation of the fusion site might require many more trimers acting cooperatively than the 5–6 that have typically been estimated to be involved64. The G proteins of rhabdoviruses have several characteristics in common with class I fusion proteins (for example, they have an internal fusion peptide and are not complexed with other proteins at the virion surface) and class II fusion proteins (for example, they are not cleaved and do not have heptad sequences that are predictive of coiled-coils). The X-ray crystal structure of the rhabdovirus G protein has not been solved, and it remains to be seen whether it will become the founding member of a new class III fusion protein family.
Enveloped viruses: membrane fusion. An important role of the conformational changes that all classes of fusion proteins undergo is to overcome energy barriers to enable membrane destabilization and the formation of fusion pores. The energy barrier for the fusion of VSV with a cell is estimated to be about 42 kcal mol−1 (Ref. 65). For protein-free model lipid bilayers, the activation energies are comparable — estimated values of 37, 27 and 22 kcal mol−1 were reported for the formation of a reversible first intermediate, its conversion to a second, semi-stable intermediate and irreversible fusion-pore formation, respectively66, which indicates that the basic molecular mechanisms of viral fusion might involve similar lipid molecular rearrangements to those observed in the fusion of model lipid membranes.
How the energy that is released by conformational changes of the viral protein is used to overcome the membrane fusion energy barriers is unclear. For class I fusion proteins it was recently found that the formation of six-helix bundles stabilizes fusion-pore formation67. A spring-loaded conformational change68 (Fig. 5) is required, but might not be sufficient, for HA-mediated fusion, and the transition to the membrane fusion pore could depend, in part, on the subsequent action of the HA fusion peptide and transmembrane domain69. For class II fusion proteins it was proposed that the pH-triggered conformational changes result in insertion of class II protein β-barrels into the host membrane, which, in turn, leads to fusion-pore formation24. It seems that once the viral protein conformational changes have provided sufficient energy to enable close membrane apposition (less than 1 nm) and destabilization, formation of the fusion pore can proceed through intermediates, stalks and hemifusion diaphragms, which might be common to all known viral membrane fusion processes70,71. Data obtained for baculovirus gp64 are consistent with this hypothesis72.
A number of models have been suggested for fusion-pore formation by lipid–protein complexes52,70. To ensure the transfer of the viral nucleoprotein complex, which is typically stable in solution, it seems that the internal lining of the fusion pore must be hydrophilic. Fusion-pore formation requires a minimal number (5–6) of oligomeric viral entry proteins to form a supramolecular lipid–protein complex. It is a dynamic process and small pores can open and close reversibly. Models of fusion-pore formation are mainly based on fusion mediated by influenza HA, but might prove generally valid. The enlargement of the fusion pore that allows transfer of the genome into the cell occurs by an unknown mechanism.
Non-enveloped viruses: membrane penetration. The mechanism of penetration of non-enveloped viruses remains poorly understood, although they are relatively small and simple in structure18. Fusion of the more complex enveloped viruses is better understood owing to the similarities between enveloped viruses and cells — both are surrounded by membranes and their fusion does not necessarily require a significant reorganization of the viral nucleoprotein complex. By contrast, either the whole non-enveloped virus must cross the membrane, or it must undergo important conformational changes and transfer the genome through the membrane. In the latter case, the energy of the metastable state must be used not only to destabilize the cell membrane, but also to reorganize the nucleoprotein complex so that the genome can be released through the destabilized membrane either through pores or other structures. Although the structures of several key intermediates in the conformational changes of viral entry proteins have now been solved to atomic resolution, it is clear that there is a great deal left to learn owing to the difficulties that are associated with studying the rapid conformational changes of only a few molecules of each virion against a background of large numbers of surface molecules on the virion that undergo no structural changes.
Entry proteins and pathogenesis
Viral proteins can destabilize not only plasma and endosomal membranes during entry, but can also destabilize other membranes inside cells to which the protein is in close proximity if there is a suitable trigger, such as a receptor, low pH or low calcium concentration. For example, such conditions arise during transportation of viral proteins after synthesis in the host cell in acidic Golgi vesicles. It seems that viruses have developed strategies to cope with this problem. For example, the G protein of rhabdoviruses, such as rabies and VSV, can exist in native, activated and inactive conformations. It was proposed that the inactive state helps to avoid membrane fusion during the transport of the G protein73. However, the HIV-1 Env can induce cell lysis after interaction with receptors — probably through disruption of important intracellular membranes — so methods to cope with destabilization of cell functions are not universal74. The significance of membrane fusion effects in human pathogenesis is unclear. Perhaps viruses that are well adapted to their hosts do not induce significant pathogenic effects that could lead to a reduction of virus production. However, viruses that infect a new host, such as HIV and SARS, might not be well adapted to this host and their entry proteins could cause CYTOPATHIC EFFECTS. Cell fusion — perhaps mediated by Envs — by endogenous retroviruses could contribute to cancer75.
Entry inhibitors, antibodies and vaccines
Entry is an attractive target for inhibition because the entry machinery is extracellular and it is therefore easier for drug molecules to reach than intracellular targets. Any step(s) of the entry process can be targeted by an entry inhibitor. Various types of molecules, such as proteins, peptides, carbohydrates, small organic molecules, nucleic acids and supramolecular structures, including liposomes and phage, have been found to inhibit entry. Yet, out of more than 30 antiviral drugs76, there are only two entry inhibitors — Synagis77 and T-20 (Ref. 78) — that have been approved by the US Food and Drug Administration (FDA) for clinical use (excluding human immune globulin for use against hepatitis A and measles, and virus-specific polyclonal human immune globulins for use against cytomegalovirus, hepatitis B, rabies, RSV, vaccinia and varicella-zoster79). Only T-20 (which is marketed as enfuvirtide) is used for the treatment of ongoing viral (HIV-1) infection. The humanized monoclonal antibody Synagis (which is also known as palivizumab) is used for the prevention of RSV infections in neonates and immunocompromised individuals. T-20 is not a small molecule — regarded as the 'gold standard' for a drug — but is a peptide that cannot be taken orally. A small organic molecule entry inhibitor (pleconaril) showed promising results for the treatment of infections caused by the picornaviruses that cause the common cold80, but was not approved by the FDA owing to concerns about potential interactions with other drugs — although different formulations are presently being evaluated for use in life-threatening disease. At present, a number of compounds are in clinical trials, including small organic molecules that bind to the HIV-1 co-receptor CCR5, and entry inhibitors are also being tested for efficacy as microbicides.
Challenges in the development of virus entry inhibitors. Major problems in the development of effective entry inhibitors include the generation of virus mutants that are resistant to such drugs, a low potency in vivo and toxic side effects. Although these problems are common to the development of other antiviral (and cancer) drugs and limit their efficacy, the development of entry inhibitors might also face specific challenges. Viruses have evolved to recognize cellular receptors and enter cells despite the presence of the host immune system. Antibodies that exhibit inhibitory effects on virus entry must bind viruses that have already developed a number of strategies for immune evasion, including the use of immunodominant variable loops and incomplete complementary binding surfaces, oligomeric occlusion, glycosylation81, conformational masking82 and multivalent interactions. Some problems that are inherent to the design of any inhibitor are especially challenging for those targeted to virus entry, such as the inhibition of high-affinity protein–protein interactions by small molecules. Another strategy used by viruses to counteract the action of antibodies and other entry inhibitors is the direct cell-to-cell transmission of viruses, which avoids or reduces the exposure of the virus to inhibitors that are not designed to penetrate through membranes or other barriers. This could be one important reason for the low efficacy that is observed in vivo of otherwise potent in vitro inhibitors. Low inhibitor potency combined with a high virus mutation rate is perhaps the most challenging problem in the development of entry inhibitors.
Although the rapid progress in our understanding of the structural mechanisms of virus entry promises new approaches that could 'outsmart' the virus, no entry inhibitors or treatment protocols in clinical use have so far been developed on the basis of predictions made by structural models, and the main source of new inhibitors is still from screening large libraries of small organic molecules, natural products, peptides and antibodies. However, structures of entry proteins have been invaluable for the development of our understanding of the mechanisms of inhibition and should allow further improvement of the inhibitors. It seems likely that sooner or later structure-based design will yield entry inhibitors that will be in clinical use.
Conserved entry intermediates as targets for inhibitors; multivalent inhibitors and other approaches. One direction of research that could hasten the arrival of entry inhibitors to the clinics is the development of compounds that interact with conserved intermediates of the entry process or with the protein structures that, on binding to receptors, trigger the conformational changes that lead to the formation of these entry intermediates. Typically, such intermediates are only transiently exposed, so viruses might not have evolved strategies to avoid inhibitors targeted to these structures. In addition, conserved intermediates are usually important for virus entry and presumably cannot easily be substituted by other structures after mutation.
An example of the successful design of an entry inhibitor that shows proof of the concept is the 5-Helix protein, which interferes with a conserved intermediate in the entry of HIV-1 (Ref. 83). A related example is a class of peptides that could have broad applications to several viruses containing class I fusion proteins. The peptides are derived from regions of fusion proteins (heptad repeats) that have a propensity to form coiled-coils and which serve as fusion intermediates and enable oligomerization of proteins. T-20 (also known as DP178) is derived from the carboxy-terminal heptad region of the HIV-1 gp41 and showed potent inhibitory activity in vivo78. Similar peptides from other viruses, including HTLV-1, RSV, measles virus, Nipah virus, Hendra virus, Ebola virus and SARS-CoV, are also promising entry inhibitor candidates. However, Ebola virus is inhibited only at very high concentrations, influenza HA-mediated fusion is not inhibited and SARS-CoV infection might not be inhibited either (K. Bossart and C. Broder, personal communication), perhaps owing to the endocytic entry route of these viruses. Although peptides have certain promising features, including a relatively small size that might ensure good penetration combined with high binding affinity, the lack of oral formulations, short half-life, possible toxicity and immunogenicity might limit their application.
Recently, a small molecule, BMS-378806, was identified that inhibits the entry of a broad range of HIV-1 isolates by a mechanism which was attributed to competition with the CD4 receptor for binding to gp120 (Ref. 84). However, this compound inhibited the entry of one isolate of HIV-1 (HIV-1JRFL) with an IC50 of 1.5 nM, and the binding of CD4 to gp120 from the same isolate with an IC50 of 100 nM. So, at inhibitory concentrations, BMS-378806 does not interfere with CD4 binding, indicating a different inhibitory mechanism. By analogy with inhibitors of picornavirus entry, such as pleconaril, BMS-378806 might inhibit the entry of HIV-1 by binding to conserved structures that are important for the conformational changes which gp120 must undergo for viral entry80. Potent virus-specific inhibitors of the viral-membrane-merging step have not been identified yet.
Another promising direction is the development of multivalent inhibitors that can overcome problems caused by mutation of viral proteins to escape inhibition because multivalent inhibitors bind to several regions of the same (or different) protein(s) on the viral surface. One example of a multivalent inhibitor is the multimeric soluble receptors of influenza and HIV-1 (Ref. 82), which are potent inhibitors of entry in vitro and, to some extent, in vivo. The use of entry inhibitors in combination or as fusion proteins could also result in increased efficiency. Finally, improvement of current methods for structure-based design by accounting for protein flexibility and dynamics in binding to ligands85, and screening methods for inhibitors86,87 would certainly expand the range of possible inhibitors that can be tested.
Neutralizing antibodies and vaccine immunogen design. Neutralizing antibodies usually inhibit virus entry by preventing attachment of the virus to the cell or by binding to entry intermediates88,89,90. Human immunoglobulin composed of concentrated antibodies collected from pooled human plasma has been successfully used as a preventative treatment for virus infections, including rabies, hepatitis A and B, measles, mumps, varicella, cytomegalovirus and arenaviruses. Antibodies can completely prevent infection, but once infection is established they are a much less efficient treatment. The only monoclonal antibody in clinical use today to treat a viral disease — Synagis (MEDI-493) — is more potent than the polyclonal immunoglobulin that is presently in use, and is broadly active against numerous RSV type A and B clinical isolates91. It binds to the F protein of RSV with high affinity (3 nM) and inhibits virus entry and cell fusion in vitro with an IC50 of approximately 0.1 μg ml−1. It seems that the efficacy of Synagis in vivo is correlated with the high affinity of binding and potency of this antibody in vitro94. However, Synagis had no measurable clinical efficacy after administration as a single 15 mg kg−1 intravenous dose to infants that were hospitalized with established RSV infection, although it did significantly reduce RSV concentrations in tracheal aspirates92.
The X-ray crystal structures of rhinovirus21 and poliovirus22 indicate a possible mechanism by which picornaviruses can avoid neutralization by antibodies through the mutation of non-conserved amino acid residues surrounding the receptor-binding site — a 2 nm deep and 2 nm wide canyon (Fig. 4c). It was initially hypothesized that the conserved amino acid residues of the canyon are not accessible by antibodies; however, it was later shown that a strongly neutralizing antibody, Fab17, can penetrate deep within the receptor-binding canyon by undergoing a large conformational change without inducing conformational changes in the virus90,93. Unusually, not only the hypervariable residues but also residues from the framework region of Fab17 contact the canyon. Yet another remarkable mechanism of immune recognition of viruses is the recently discovered receptor mimicry by post-translational modification (tyrosine sulphation) of antibodies (Ref. 94; Huang, C. et al., manuscript in preparation). It seems that any accessible viral surface can be recognized by antibodies. Rapidly mutating viruses can escape neutralizing antibodies even if they bind to structures that are essential for virus replication, such as receptor-binding sites, unless they bind with energetically identical profiles95. Whether a virus will escape neutralization by antibodies depends on the interplay between the antibody affinity (avidity) and kinetics of binding, generation rate, concentration and the viral mutation rate and fitness. Mutations of immunodominant structural loops that form antibody-binding sites and mutations leading to changes in oligosaccharide attachment to viral entry proteins are common mechanisms by which viruses avoid neutralization90,96.
Mutations of conserved residues that have a role in the entry mechanism typically result in reduction or loss of infectivity. Antibodies or their derivatives that bind to epitopes where residues contribute most of the binding energy could have potential as entry inhibitors. Epitopes that are exposed after virus binding to receptors are typically well conserved — for example, the 17b39 and X5 (Ref. 97) epitopes on HIV-1 gp120. One potential problem with using antibodies as entry inhibitors in this case could be limited access to the post-receptor-binding state of the viral entry protein due to the relatively large size of the whole antibody. Solving this, and other problems, could lead to the development of potent broadly neutralizing antibodies which could limit the generation of resistant viruses, especially if these inhibitors are used in combination with other antibodies or inhibitory molecules.
Many viruses, especially RNA viruses such as HIV-1, exist as swarms of virions inside an infected individual, and might significantly differ in sequence between isolates. So, elicitation of potent, broadly neutralizing antibodies is an important goal for vaccine development. Potent, broadly neutralizing antibodies for HIV-1 Env do exist — for example, b12, 2G12, 447-52D, X5, 2F5 and 4E10/Z13. However, elicitation of these antibodies in vivo has not been successful. Identification of broadly neutralizing antibodies and the characterization of their epitopes could help to design vaccine immunogens that would be able to elicit these neutralizing antibodies in vivo — so-called retrovaccinology89. At present, all vaccines that elicit antibodies against entry proteins have been developed empirically using an antigen, rather than by designing an immunogen on the basis of the antibodies produced.
The important advantages of human antibodies as therapeutics are low or negligible toxicity combined with high potency and a long half-life. However, drawbacks include the generation of neutralization-resistant virus mutants, limited access of the large antibody molecules to the site of virus replication, lack of oral formulations and the high cost of production and storage.
Virus entry physiology and retargeting
Viruses are usually associated with disease. However, some viruses can be beneficial. Human endogenous retrovirus W (HERV-W) might be essential in human physiology — for the formation of the placenta. The HERV-W Env, known as syncytin, is fusogenic and has a role in human trophoblast cell fusion and differentiation98. Retroviral particles have been observed in the placenta, along with fused placental cells, which are morphologically reminiscent of virally induced syncytia. These studies led to the proposal that an ancient retroviral infection might have been a pivotal event in mammalian evolution99.
Viruses have long been used to transfer genes into cells. During the last decade, another important application has been the viral delivery of genes and drugs to treat cancer. A major challenge has been to develop virus entry proteins to deliver molecules to specific cells with high efficiency. To achieve this goal it is often desirable to engineer viruses that do not infect cells expressing the native receptor, but instead target a cell of choice. Engineering of entry proteins in this way is known as transductional retargeting100.
A conceptually simple approach to transductional retargeting is to incorporate the protein that determines cell tropism into the infecting virion of choice — known as virus 'pseudotyping'. This has been used in both retroviruses and adenoviruses, and does not require prior knowledge of specific virus–receptor interactions. In a related approach, viral entry proteins are used to produce drug and gene delivery vehicles, for example, the F protein of Sendai virus has been incorporated into liposomes to form virosomes101 and the L protein of hepatitis B has been incorporated into yeast-derived lipid vesicles102. Retargeting of retroviruses, adenoviruses and AAVs has been achieved by conjugation of entry proteins with molecular adaptors, such as bi-specific antibodies that have particular receptor-binding properties. Modification of the entry proteins so that the normal receptor-binding property is abolished, or a ligand for alternative receptor binding is incorporated has also been successful at redirecting adenovirus tropism in cell culture, but is unlikely to work for the entry of viruses that require receptor-induced conformational changes, such as retroviruses, unless detailed molecular mechanisms of those conformational changes are better understood. A related approach is based on screening libraries of chimaeric Envs from different strains of MLV103, or randomized peptides inserted at tolerant sites in viral proteins, such as VP3 of AAV104. This approach seems promising for the selection of specific retargeting vectors. Understanding the structure of AAV105 and other viruses could help to further improve the specificity and efficiency of retargeting. Retargeting viruses with complex entry mechanisms that involve several proteins, such as those of herpes viruses and poxviruses, remains challenging.
Challenges and perspectives
Elucidation of the molecular mechanisms and the dynamics of the conformational changes driving virus entry remains a significant challenge. It requires the development of new approaches to study the rapid conformational changes of a small number of membrane-interacting protein molecules that are surrounded by many more non-interacting molecules. A more realistic goal is the determination of the structures of proteins that mediate the entry of all human viruses and the identification of the cognate cellular receptors. If research continues at the present pace, this goal could be accomplished within the next decade. Identification of all the cellular receptors for human viruses would be an important contribution to our understanding of virus tropism and pathogenesis. The various, and in many cases unexpected, ways that entry proteins can affect pathogenesis could offer new opportunities for intervention. Rational structure/mechanism-based design of entry inhibitors and vaccine immunogens that are capable of eliciting potent, broadly neutralizing antibodies of known epitopes is expected to contribute towards the development of clinically useful therapeutics and vaccines. The development of panels of human monoclonal antibodies against every entry-related protein of all pathogenic human viruses could accelerate our understanding of entry mechanisms and help to fight viral diseases. Recent progress in virus retargeting also raises hopes for the possibility of designing entry machines that can deliver genes and other molecules to any cell of choice.
Acknowledgements
I thank R. Blumenthal, C. Broder, P. Kwong and the three reviewers for helpful comments, and members of my group, X. Xiao, M. Zhang, I. Sidorov and P. Prabakaran, for exciting discussions and help. I apologise to those colleagues whose research has not been highlighted owing to length constraints. The selected examples reflect my personal interests.
Glossary
- ENTRY PROTEINS
Proteins that mediate entry into cells. Entry proteins is a general term used here to denote attachment proteins and other proteins that are required for entry of non-enveloped and enveloped viruses into cells.
- LIPID RAFT
Areas of the plasma membrane that are rich in cholesterol, glycosphingolipids and glycosylphosphatidylinositol-anchored proteins. Also known as glycolipid-enriched microdomains (GEMs) and detergent-insoluble glycolipid-enriched membranes (DIGs).
- LIPOSOME
A lipid vesicle that is artificially formed by sonicating lipids in an aqueous solution.
- METASTABLE STATE
An energy state that is separated from one of lower energy by an energy barrier. Metastable states can exist for a long time if the height of the barrier is high and be undistinguishable from a truly stable state. The trigger leads to a decrease in the energy barrier and a transition to a more stable state of lower energy. The rate of transition is determined by the difference in the energies of the initial and final states, temperature and various parameters including molecular conformations and viscosity.
- ATTACHMENT PROTEINS
Proteins that mediate specific binding of viruses to their receptors. Attachment proteins are typically single proteins but in the case of many non-enveloped viruses, complexes of several proteins from the capsid bind receptor molecules and function as attachment proteins.
- CAPSID
Proteins that encapsulate viral genomes. Capsid shells of many non-enveloped viruses proteins bind receptor molecules and serve as attachment proteins. Capsid proteins also typically mediate membrane penetration by non-enveloped viruses.
- ENVELOPE GLYCOPROTEINS
(Envs). Viral proteins that are embedded in the envelope membranes of enveloped viruses. Many viruses, for example, rabies virus and HIV, have only one viral protein in their membranes that mediates viral entry into cells; others, for example, influenza have two or more, but some of the proteins might not be involved in the entry process. In this article, Envs is used to describe those entry proteins that mediate attachment and fusion of enveloped viruses.
- FUSION PROTEINS
Proteins that mediate the fusion of enveloped viruses. This term is frequently used to denote entry proteins that mediate the membrane fusion of enveloped viruses, for example, class I and class II fusion proteins. Typically, such proteins consist of an attachment protein (subunit or domain), sometimes denoted as surface protein, and a subunit mediating the actual membrane fusion event, sometimes denoted as transmembrane protein because it spans the viral membrane.
- VIRUS RECEPTORS
Host-cell molecules (usually membrane-associated) that bind virus-attachment proteins and are required for entry.
- AVIDITY
Effective affinity for multivalent interactions. Avidity is a complex function of the affinity for a monovalent interaction and the number of interactions, and can be many orders of magnitude higher than affinity.
- CO-RECEPTORS
Cell membrane-associated molecules that bind specifically to virus proteins and are required for entry (in addition to the primary receptor) typically to ensure continuation of the entry process after binding. By definition the term co-receptor implies a physical association with viral entry proteins after their binding to the primary receptor to distinguish them from molecules that are required for entry at later stages — for example, uncoating — or at unknown stages, and are denoted as entry cofactors.
- ALTERNATIVE VIRUS RECEPTORS
Receptors that viruses can use in the absence of the primary receptor, typically with a much lower efficiency for entry.
- CYTOPATHIC EFFECTS
(CPE). Effects caused by any agent, including viruses, that lead to deterioration of cellular functions and ultimately cell death.
Biography
Dimiter S. Dimitrov is a Senior Investigator at the National Cancer Institute, Frederick, USA. His research interests include mechanisms of virus entry and pathogenesis in humans, antibodies in viral diseases and cancer, and computational biology.
Related links
DATABASES
LocusLink
Protein Data Bank
FURTHER INFORMATION
Dimiter S. Dimitrov's laboratory
Competing interests
The author declares no competing financial interests.
References
- 1.d'Herelle F. The Bacteriophage and its Behavior. 1926. [Google Scholar]
- 2.Sieczkarski SB, Whittaker GR. Dissecting virus entry via endocytosis. J. Gen. Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- 3.Pelkmans L, Puntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- 4.Dimitrov DS. Cell biology of virus entry. Cell. 2000;101:697–702. doi: 10.1016/S0092-8674(00)80882-X. [DOI] [PubMed] [Google Scholar]
- 5.Rawat SS, et al. Modulation of entry of enveloped viruses by cholesterol and sphingolipids. Mol. Membr. Biol. 2003;20:243–254. doi: 10.1080/0968768031000104944. [DOI] [PubMed] [Google Scholar]
- 6.Takeda M, Leser GP, Russell CJ, Lamb RA. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl Acad. Sci. USA. 2003;100:14610–14617. doi: 10.1073/pnas.2235620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waarts BL, Bittman R, Wilschut J. Sphingolipid and cholesterol dependence of α-virus membrane fusion. Lack of correlation with lipid raft formation in target liposomes. J. Biol. Chem. 2002;277:38141–38147. doi: 10.1074/jbc.M206998200. [DOI] [PubMed] [Google Scholar]
- 8.Kielian M, Chatterjee PK, Gibbons DL, Lu YE. Specific roles for lipids in virus fusion and exit. Examples from the α-viruses. Subcell. Biochem. 2000;34:409–455. doi: 10.1007/0-306-46824-7_11. [DOI] [PubMed] [Google Scholar]
- 9.Igakura T, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299:1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 10.Bomsel M, Alfsen A. Entry of viruses through the epithelial barrier: pathogenic trickery. Nature Rev. Mol. Cell Biol. 2003;4:57–68. doi: 10.1038/nrm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seisenberger G, et al. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294:1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 12.Lowy RJ, Sarkar DP, Chen Y, Blumenthal R. Observation of single influenza virus-cell fusion and measurement by fluorescence video microscopy. Proc. Natl Acad. Sci. USA. 1990;87:1850–1854. doi: 10.1073/pnas.87.5.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc. Natl Acad. Sci. USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitrov DS, Willey R, Martin M, Blumenthal R. Kinetics of HIV-1 interactions with sCD4 and CD4+ cells: implications for inhibition of virus infection and initial steps of virus entry into cells. Virology. 1992;187:398–406. doi: 10.1016/0042-6822(92)90441-Q. [DOI] [PubMed] [Google Scholar]
- 15.Dimitrov DS, et al. Quantitation of HIV-1 infection kinetics. J. Virol. 1993;67:2182–2190. doi: 10.1128/jvi.67.4.2182-2190.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White J, Kartenbeck J, Helenius A. Fusion of Semliki forest virus with the plasma membrane can be induced by low pH. J. Cell. Biol. 1980;87:264–272. doi: 10.1083/jcb.87.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carr CM, Chaudhry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl Acad. Sci. USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogle JM. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stubbs MT. Anthrax X-rayed: new opportunities for biodefence. Trends Pharmacol. Sci. 2002;23:539–541. doi: 10.1016/S0165-6147(02)02127-2. [DOI] [PubMed] [Google Scholar]
- 20.Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nature Rev. Mol. Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 21.Rossmann MG, et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 22.Hogle JM, Chow M, Filman DJ. Three-dimensional structure of poliovirus at 2.9 A resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 23.Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell. 2000;5:255–266. doi: 10.1016/S1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn RJ, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/S0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chappell JD, Prota AE, Dermody TS, Stehle T. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 2002;21:1–11. doi: 10.1093/emboj/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/S0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 28.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 29.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lescar J, et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/S0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 31.Heinz FX, Allison SL. The machinery for flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 2001;4:450–455. doi: 10.1016/S1369-5274(00)00234-4. [DOI] [PubMed] [Google Scholar]
- 32.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nature Rev. Mol. Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 33.Carfi A, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell. 2001;8:169–179. doi: 10.1016/S1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 34.Fass D, et al. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 35.Wimmer, E. (ed.) Cellular receptors for animal viruses. 1–13 (Cold Spring Harbor Laboratory Press, New York,1994).
- 36.Baranowski E, Ruiz-Jarabo CM, Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 37.Wang J. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci. 2002;27:122–126. doi: 10.1016/S0968-0004(01)02038-2. [DOI] [PubMed] [Google Scholar]
- 38.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 39.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harouse JM, et al. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science. 1991;253:320–323. doi: 10.1126/science.1857969. [DOI] [PubMed] [Google Scholar]
- 41.Fingeroth JD, et al. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl Acad. Sci. USA. 1984;81:4510–4514. doi: 10.1073/pnas.81.14.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 43.Klatzmann D, et al. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature. 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 44.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 46.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Entry of α-herpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 47.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammache D, Yahi N, Maresca M, Pieroni G, Fantini J. Human erythrocyte glycosphingolipids as alternative cofactors for human immunodeficiency virus type 1 (HIV-1) entry: evidence for CD4-induced interactions between HIV-1 gp120 and reconstituted membrane microdomains of glycosphingolipids (Gb3 and GM3) J. Virol. 1999;73:5244–5248. doi: 10.1128/jvi.73.6.5244-5248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Percherancier Y, et al. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 2003;278:3153–3161. doi: 10.1074/jbc.M207371200. [DOI] [PubMed] [Google Scholar]
- 51.Remeta DP, et al. Acid-induced changes in thermal stability and fusion activity of influenza hemagglutinin. Biochemistry. 2002;41:2044–2054. doi: 10.1021/bi015614a. [DOI] [PubMed] [Google Scholar]
- 52.Blumenthal R, Clague MJ, Durell SR, Epand RM. Membrane fusion. Chem. Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 53.Barnard RJ, Young JA. α-retrovirus envelope-receptor interactions. Curr. Top. Microbiol. Immunol. 2003;281:107–136. doi: 10.1007/978-3-642-19012-4_3. [DOI] [PubMed] [Google Scholar]
- 54.Mothes W, Boerger AL, Narayan S, Cunningham JM, Young JA. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell. 2000;103:679–689. doi: 10.1016/S0092-8674(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 55.Earp LJ, Delos SE, Netter RC, Bates P, White JM. The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH. J. Virol. 2003;77:3058–3066. doi: 10.1128/JVI.77.5.3058-3066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 57.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leikina E, Ramos C, Markovic I, Zimmerberg J, Chernomordik LV. Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin. EMBO J. 2002;21:5701–5710. doi: 10.1093/emboj/cdf559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim. Biophys. Acta. 2003;1614:36–50. doi: 10.1016/S0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 60.Madhusoodanan M, Lazaridis T. Investigation of pathways for the low-pH conformational transition in influenza hemagglutinin. Biophys. J. 2003;84:1926–1939. doi: 10.1016/S0006-3495(03)75001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stiasny K, Koessl C, Heinz FX. Involvement of lipids in different steps of the flavivirus fusion mechanism. J. Virol. 2003;77:7856–7862. doi: 10.1128/JVI.77.14.7856-7862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pak CC, Puri A, Blumenthal R. Conformational changes and fusion activity of vesicular stomatitis virus glycoprotein: [125I]iodonaphthyl azide photolabeling studies in biological membranes. Biochemistry. 1997;36:8890–8896. doi: 10.1021/bi9702851. [DOI] [PubMed] [Google Scholar]
- 63.Gaudin Y. Reversibility in fusion protein conformational changes. The intriguing case of rhabdovirus-induced membrane fusion. Subcell. Biochem. 2000;34:379–408. doi: 10.1007/0-306-46824-7_10. [DOI] [PubMed] [Google Scholar]
- 64.Roche S, Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 65.Clague MJ, Schoch C, Zech L, Blumenthal R. Gating kinetics of pH-activated membrane fusion of vesicular stomatitis virus with cells: stopped-flow measurements by dequenching of octadecylrhodamine fluorescence. Biochemistry. 1990;29:1303–1308. doi: 10.1021/bi00457a028. [DOI] [PubMed] [Google Scholar]
- 66.Lee J, Lentz BR. Secretory and viral fusion may share mechanistic events with fusion between curved lipid bilayers. Proc. Natl Acad. Sci. USA. 1998;95:9274–9279. doi: 10.1073/pnas.95.16.9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markosyan RM, Cohen FS, Melikyan GB. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol. Biol. Cell. 2003;14:926–938. doi: 10.1091/mbc.e02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carr CM, Kim PS. A spring-loaded mechanism for the conformational changes of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-W. [DOI] [PubMed] [Google Scholar]
- 69.Gruenke JA, Armstrong RT, Newcomb WW, Brown JC, White JM. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 2002;76:4456–4466. doi: 10.1128/JVI.76.9.4456-4466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chernomordik LV, Leikina E, Frolov V, Bronk P, Zimmerberg J. An early stage of membrane fusion mediated by the low pH conformation of influenza hemagglutinin depends upon membrane lipids. J. Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Melikyan GB, Chernomordik LV. Membrane rearrangements in fusion mediated by viral proteins. Trends. Microbiol. 1997;5:349–355. doi: 10.1016/S0966-842X(97)01107-4. [DOI] [PubMed] [Google Scholar]
- 72.Chernomordik L, Leikina E, Cho MS, Zimmerberg J. Control of baculovirus gp64-induced syncytium formation by membrane lipid composition. J. Virol. 1995;69:3049–3058. doi: 10.1128/jvi.69.5.3049-3058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaudin Y, Tuffereau C, Durrer P, Flamand A, Ruigrok RW. Biological function of the low-pH, fusion-inactive conformation of rabies virus glycoprotein (G): G is transported in a fusion-inactive state-like conformation. J. Virol. 1995;69:5528–5534. doi: 10.1128/jvi.69.9.5528-5534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.LaBonte JA, Madani N, Sodroski J. Cytolysis by CCR5-using human immunodeficiency virus type 1 envelope glycoproteins is dependent on membrane fusion and can be inhibited by high levels of CD4 expression. J. Virol. 2003;77:6645–6659. doi: 10.1128/JVI.77.12.6645-6659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–448. doi: 10.1016/S1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 76.De Clercq E. New anti-HIV agents and targets. Med. Res. Rev. 2002;22:531–565. doi: 10.1002/med.10021. [DOI] [PubMed] [Google Scholar]
- 77.Pollack P, Groothuis JR. Development and use of palivizumab (Synagis): a passive immunoprophylactic agent for RSV. J. Infect. Chemother. 2002;8:201–206. doi: 10.1007/s10156-002-0178-6. [DOI] [PubMed] [Google Scholar]
- 78.Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 2003;348:2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 79.Sawyer LA. Antibodies for the prevention and treatment of viral diseases. Antiviral Res. 2000;47:57–77. doi: 10.1016/S0166-3542(00)00111-X. [DOI] [PubMed] [Google Scholar]
- 80.Florea NR, Maglio D, Nicolau DP. Pleconaril, a novel antipicornaviral agent. Pharmacother. 2003;23:339–348. doi: 10.1592/phco.23.3.339.32099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 82.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 83.Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 84.Lin PF, et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl Acad. Sci. USA. 2003;100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Arkin MR, et al. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl Acad. Sci. USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 87.Braisted AC, et al. Discovery of a potent small molecule IL-2 inhibitor through fragment assembly. J. Am. Chem. Soc. 2003;125:3714–3715. doi: 10.1021/ja034247i. [DOI] [PubMed] [Google Scholar]
- 88.Dimmock NJ. Neutralization of animal viruses. Curr. Top. Microbiol. Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 89.Burton DR. Antibodies, viruses and vaccines. Nature Rev. Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 90.Smith TJ. Antibody interactions with rhinovirus: lessons for mechanisms of neutralization and the role of immunity in viral evolution. Curr. Top. Microbiol. Immunol. 2001;260:1–28. doi: 10.1007/978-3-662-05783-4_1. [DOI] [PubMed] [Google Scholar]
- 91.Johnson S, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- 92.Johnson S, et al. A direct comparison of the activities of two humanized respiratory syncytial virus monoclonal antibodies: MEDI-493 and RSHZl9. J. Infect. Dis. 1999;180:35–40. doi: 10.1086/314846. [DOI] [PubMed] [Google Scholar]
- 93.Smith TJ, Chase ES, Schmidt TJ, Olson NH, Baker TS. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature. 1996;383:350–354. doi: 10.1038/383350a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Choe H, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114:161–170. doi: 10.1016/S0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 95.Colman PM. Virus versus antibody. Structure. 1997;5:591–593. doi: 10.1016/S0969-2126(97)00214-1. [DOI] [PubMed] [Google Scholar]
- 96.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 97.Moulard M, et al. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120–CD4–CCR5 complexes. Proc. Natl Acad. Sci. USA. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frendo JL, et al. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Harris JR. Placental endogenous retrovirus (ERV): structural, functional, and evolutionary significance. Bioessays. 1998;20:307–316. doi: 10.1002/(SICI)1521-1878(199804)20:4<307::AID-BIES7>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 100.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 101.Sarkar DP, Ramani K, Tyagi SK. Targeted gene delivery by virosomes. Methods Mol. Biol. 2002;199:163–173. doi: 10.1385/1-59259-175-2:163. [DOI] [PubMed] [Google Scholar]
- 102.Yamada T, et al. Nanoparticles for the delivery of genes and drugs to human hepatocytes. Nature Biotechnol. 2003;21:885–890. doi: 10.1038/nbt843. [DOI] [PubMed] [Google Scholar]
- 103.Soong NW, et al. Molecular breeding of viruses. Nature Genet. 2000;25:436–439. doi: 10.1038/78132. [DOI] [PubMed] [Google Scholar]
- 104.Perabo L, et al. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol. Ther. 2003;8:151–157. doi: 10.1016/S1525-0016(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 105.Xie Q, et al. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl Acad. Sci. USA. 2002;99:10405–10410. doi: 10.1073/pnas.162250899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Belnap DM, et al. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 2000;74:1342–1354. doi: 10.1128/JVI.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 108.Stehle T, Dermody TS. Structural evidence for common functions and ancestry of the reovirus and adenovirus attachment proteins. Rev. Med. Virol. 2003;13:123–132. doi: 10.1002/rmv.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Weis W, et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 110.Morton CJ, et al. Structural characterization of respiratory syncytial virus fusion inhibitor escape mutants: homology model of the F protein and a syncytium formation assay. Virology. 2003;311:275–288. doi: 10.1016/S0042-6822(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 111.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 112.Xing L, et al. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 2000;19:1207–1216. doi: 10.1093/emboj/19.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]


