Abstract
Cu, Zn superoxide dismutase (SOD1) is one of the genes implicated in the devastating neurodegenerative disorder amyotrophic lateral sclerosis (ALS). Although the precise mechanisms of SOD1 mutant (SOD1mut)-induced motoneuron toxicity are still unclear, defects in SOD1 proteostasis are known to have a critical role in ALS pathogenesis. We previously reported that the SOD1mut adopts a conformation that exposes a Derlin-1–binding region (DBR) and that DBR-exposed SOD1 interacts with Derlin-1, leading to motoneuron death. We also found that an environmental change, i.e. zinc depletion, induces a conformational change in WT SOD1 (SOD1WT) to the DBR-exposed conformation, suggesting the presence of an equilibrium state between the DBR-masked and DBR-exposed states even with SOD1WT. Here, we conducted a high-throughput screening based on time-resolved FRET to further investigate the SOD1WT conformational change, and we used a genome-wide siRNA screen to search for regulators of SOD1 proteostasis. This screen yielded 30 candidate genes that maintained an absence of the DBR-exposed SOD1WT conformation. Among these genes was one encoding DDB1- and CUL4-associated factor 4 (DCAF4), a substrate receptor of the E3 ubiquitin–protein ligase complex. Of note, we found that DCAF4 mediates the ubiquitination of an ALS-associated protein and autophagy receptor, optineurin (OPTN), and facilitates autophagic degradation of DBR-exposed SOD1. In summary, our screen identifies DCAF4 as being required for proper proteostasis of DBR-exposed SOD1, which may have potential relevance for the development of therapies for managing ALS.
Keywords: amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), proteostasis, autophagy, protein misfolding, protein aggregation, DDB1 and CUL4 associated factor 4 (DCAF4), genome-wide siRNA screen, superoxide dismutase 1 (SOD1)
Introduction
Amyotrophic lateral sclerosis (ALS)2 is a late-onset and fatal motoneuron disease in which upper and lower motoneurons are specifically affected. More than 20 genes have been reported to cause ALS (1); however, the common molecular mechanism of ALS pathogenesis is still unclear, and there is no effective treatment. Cu, Zn superoxide dismutas (SOD1) is one of the ALS causative genes, and over 160 mutations in the SOD1 gene have been identified in ALS patients (2, 3). It is now widely accepted that mutant SOD1 (SOD1mut) exerts motoneuron toxicity through gain–of–toxic function mechanisms rather than changes in superoxide dismutase activity (4–9). Several hypotheses have been proposed for the toxicity of SOD1mut, including mitochondria abnormality, endoplasmic reticulum (ER) stress, and excitotoxicity (10).
We previously reported that more than 100 different versions of SOD1mut interact with Derlin-1, which is a component of the ER-associated degradation (ERAD) machinery (11–14). This interaction causes a defect in the ERAD system, resulting in the induction of ER stress and eventually motoneuron death (11). Moreover, inhibition of the SOD1–Derlin-1 interaction with a small-molecule compound ameliorated the ALS pathology in an in vitro model using patient-derived iPS motoneurons with SOD1 mutation and an in vivo model using ALS model mice expressing human SOD1mut (15). These data indicated the importance of the SOD1–Derlin-1 interaction in ALS pathology. We also revealed the molecular mechanism of the interaction between SOD1 and Derlin-1. WT SOD1 (SOD1WT) possesses a Derlin-1–binding region (DBR) in its N-terminal region, which is masked in the stationary state. Mutation in SOD1 causes a conformational change and exposure of the DBR, resulting in interaction of SOD1mut with Derlin-1 (12).
Several reports have indicated the involvement of SOD1WT in the pathogenesis of SOD1 mutation-negative ALS. Conformationally-disordered SOD1WT was observed in SOD1 mutation-negative sporadic ALS (SALS) patients (16). The noncell autonomous motoneuron toxicity of SOD1WT has also been shown in astrocytes or oligodendrocytes derived from SALS patients (17, 18). In addition, we previously reported that zinc deficiency induces a conformational change and DBR exposure even in SOD1WT through the loss of a coordinated zinc ion (19). These data suggest that the defect in SOD1WT proteostasis under certain conditions, including genetic and environmental factors, might contribute to ALS pathogenesis through disruption of SOD1WT proteostasis. However, the molecular mechanism by which the proteostasis of DBR-exposed (mutant-like) SOD1 is regulated is still unclear, and the factors required to sequester DBR-exposed SOD1 have not been identified. Thus, the elucidation of the regulatory mechanisms of SOD1 proteostasis that would lead to an understanding of the underlying molecular mechanism of ALS is a crucial issue.
In this study, we performed genome-wide small interfering RNA (siRNA) screens to identify the factors required to eliminate DBR-exposed SOD1. As a result, DCAF4, an assumed substrate receptor of the E3–ligase complex, was identified as an indirect but critical regulator of SOD1 proteostasis (20). We found that DCAF4 mediated the ubiquitination of OPTN, an ALS causative gene product, and facilitated autophagic degradation of DBR-exposed SOD1.
Results
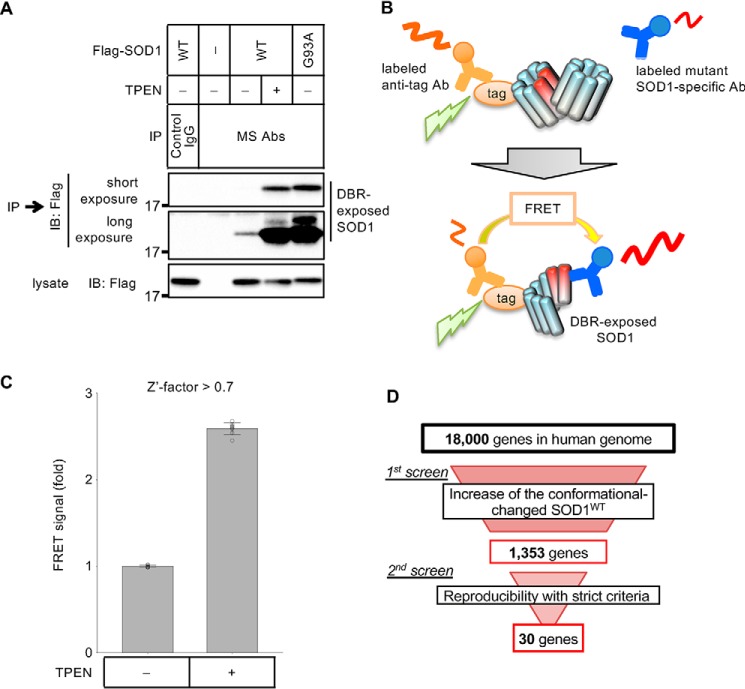
TR-FRET–based genome-wide siRNA screen for the regulators of SOD1 proteostasis
We have previously generated two antibodies that can specifically recognize DBR-exposed SOD1 in the immunoprecipitation assay (MS785 and MS27) (12, 21). During the analysis of the conformational change of SOD1WT with these antibodies (MS antibodies), we noticed that a portion of the SOD1WT population took the DBR-exposed conformation even in the absence of zinc deficiency (Fig. 1A, see long exposure panel). Previously, we have reported that even endogenous SOD1 adopted the DBR-exposed conformation during zinc deficiency, and several reports have indicated the involvement of conformationally-disordered SOD1WT in SOD1 mutation-negative sporadic ALS (SALS). Taken together, we assumed the presence of an equilibrium state between the DBR-masked and DBR-exposed conformation even in SOD1WT. Because SOD1WT mainly takes the DBR-masked conformation and unknown factors appear to be required for the zinc deficiency–dependent conformational change, there should be a regulator(s) of SOD1 proteostasis. To reveal the molecular mechanism of SOD1 proteostasis, we attempted to identify the factors that were required for sequestering DBR-exposed SOD1WT through a genome-wide siRNA screen.
Figure 1.
Screens of the genes involved in SOD1 proteostasis. A, immunoprecipitation–immunoblotting (IP–IB) assay. HeLa cells transfected with the indicated constructs were treated with 15 μm TPEN for 5 h. The cell lysates were analyzed with an IP–IB assay using the indicated antibodies. The amount of DBR-exposed SOD1 was analyzed. MS Abs: mixture of MS785 antibody and MS27 antibody. B, model of the TR-FRET–based conformational change assay showing the case of the Eu-labeled anti-tag antibody and the d2-labeled MS785 antibody. C, FRET signals of HeLa cells stably expressing SOD1WT—Flag with Flag–Eu and MS785–d2 are shown as fold changes relative to nonstimulated cells. The cells were stimulated with 10 μm TPEN for 4 h. The data are shown as the mean ± S.D. (n = 8). D, flow chart of the screens.
The high-throughput assay system used to detect the SOD1 conformational change was established by utilizing time-resolved FRET (TR-FRET) technology (Fig. 1B). Here, an anti-tag antibody and anti-mutant SOD1-specific antibody (MS785) labeled with either a donor fluorophore (europium cryptate: Eu) or an acceptor fluorophore (d2) were used (12). We searched the SOD1WT construct that efficiently generates FRET signal in a zinc depletion-dependent manner. HEK293A cells were transfected with several SOD1WT constructs and stimulated with the cell-permeable zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN). Then, the cell lysates were mixed with the fluorophore-labeled antibodies (Fig. S1A). The combination of SOD1WT–Flag, an anti–Flag antibody labeled with Eu (Flag-Eu), and MS785 labeled with d2 (MS785-d2) efficiently generated a TPEN-dependent FRET signal (Fig. S1B).
Next, we established HeLa cells stably expressing SOD1WT–Flag and checked the induction of a FRET signal with TPEN stimulation. By using these HeLa cells, we could obtain a robust signal increase in a high-throughput manner, which was sufficient for siRNA screens (Z′-factor >0.7) (Fig. 1C). Using this assay system, we carried out a genome-wide siRNA screen to identify factors required for depleting DBR-exposed SOD1WT. In our screen, we explored genes whose suppression caused an increase in the FRET signal, indicating accumulation of DBR-exposed SOD1WT even in the absence of zinc deficiency. In the 1st screening, we selected the genes that showed a score higher than 1.96 either in the robust Z-score or B-score as positive genes (Fig. 1D and Fig. S1C). In the 2nd screen, we reassessed these positive genes in duplicate. To narrow down the candidate genes, genes that showed a Z-score higher than 2.58 in duplicate were regarded as positive (Fig. 2A and Fig. S1D). Finally, we obtained 30 candidate genes that may be regulators of SOD1 proteostasis (Fig. 1D and Table S1).
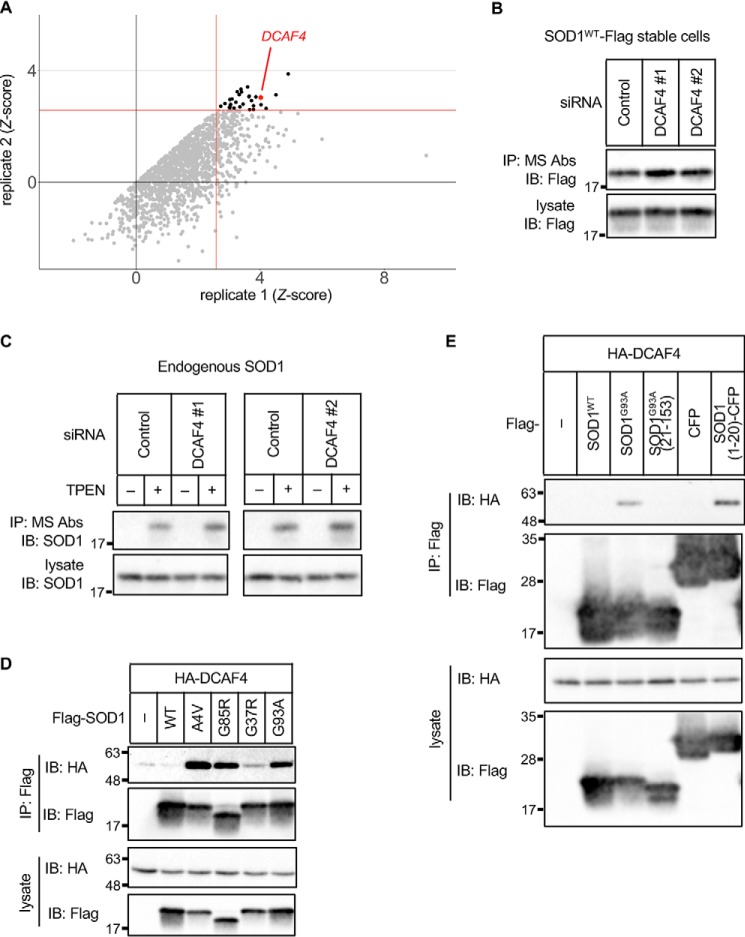
Figure 2.
DCAF4 specifically interacts with DBR-exposed SOD1 through the DBR. A, results of the 2nd screen. The dots indicate the Z-score of the duplicate. The replicates that showed a higher Z-score are described as replicate 1. Gray dots indicate negative genes; black dots indicate positive genes; red dot indicates DCAF4. Red lines indicate 2.58. B–E, IP–IB analysis of each cell lysate with the indicated antibodies is shown. B, HeLa cells stably expressing SOD1WT–Flag were transfected with the indicated siRNAs. The amount of DBR-exposed SOD1 was analyzed. C, HeLa cells were transfected with siRNAs and treated with 10 μm TPEN for 5 h. The amount of DBR-exposed endogenous SOD1 was analyzed. D, HEK293A cells were transfected with the indicated constructs. The interaction between SOD1 and DCAF4 was analyzed. E, HEK293A cells were transfected with the indicated constructs and treated with 5 μm MG132 to stabilize SOD1 fragments for 8 h. The interaction between SOD1 and DCAF4 was analyzed.
DCAF4 specifically interacts with DBR-exposed SOD1 through DBR
The Z score of the hit gene is relatively low in the screen, and we focused on the genes with high Z score (Table S1). There were genes related to the nucleotide metabolism, transcription, and cytoskeleton. However, we estimated that it is unlikely that these gene products directly regulate SOD1 proteostasis. The cullin–RING E3 ubiquitin ligases (CRLs) are the largest E3 ligase family and regulate many physiological processes, including proteostasis. Thus, among the candidate genes, we focused on damage-specific DNA-binding protein 1 (DDB1)–Cullin4 (CUL4)-associated factor 4 (DCAF4), which is speculated to function as a cellular substrate receptor of the CRL4 complex (Fig. 2A) (20). Suppression of DCAF4 with two independent siRNAs that differ from those used in the screens caused an accumulation of DBR-exposed SOD1WT even in the absence of zinc deficiency, confirming the result of the screens (Fig. 2B and Fig. S2, A and B). To evaluate the involvement of DCAF4 in endogenous SOD1 proteostasis, the amount of zinc deficiency–induced DBR-exposed endogenous SOD1 was investigated in DCAF4 knockdown cells (Fig. 2C and Fig. S2C). Suppression of DCAF4 induced an enhanced accumulation of DBR-exposed SOD1. These results demonstrate that DCAF4 is required to inhibit the accumulation of DBR-exposed SOD1. Because DCAF4 is predicted to be a substrate receptor of the CRL4 complex, we also investigated the involvement of CUL4A and CUL4B. Suppression of both CUL4A and CUL4B also induced accumulation of DBR-exposed SOD1WT, suggesting the involvement of the CRL4 complex (Fig. S2D).
To reveal the molecular mechanisms by which DCAF4 mediated the reduction in DBR-exposed SOD1, we first investigated their interaction. DCAF4 specifically interacts with SOD1mut but not with SOD1WT (Fig. 2D). This interaction was diminished with DBR-deleted SOD1G93A(21–153) (Fig. 2E). In addition, SOD1(1–20), which includes the DBR, was sufficient for interaction with DCAF4. Because SOD1WT possesses a masked DBR, which is exposed under zinc deficiency, we investigated the interaction between SOD1WT and DCAF4 during zinc depletion. As expected, zinc deficiency induced the association of DCAF4 with SOD1WT (Fig. S2E). These data indicate that DCAF4 specifically recognizes and interacts with DBR-exposed SOD1.
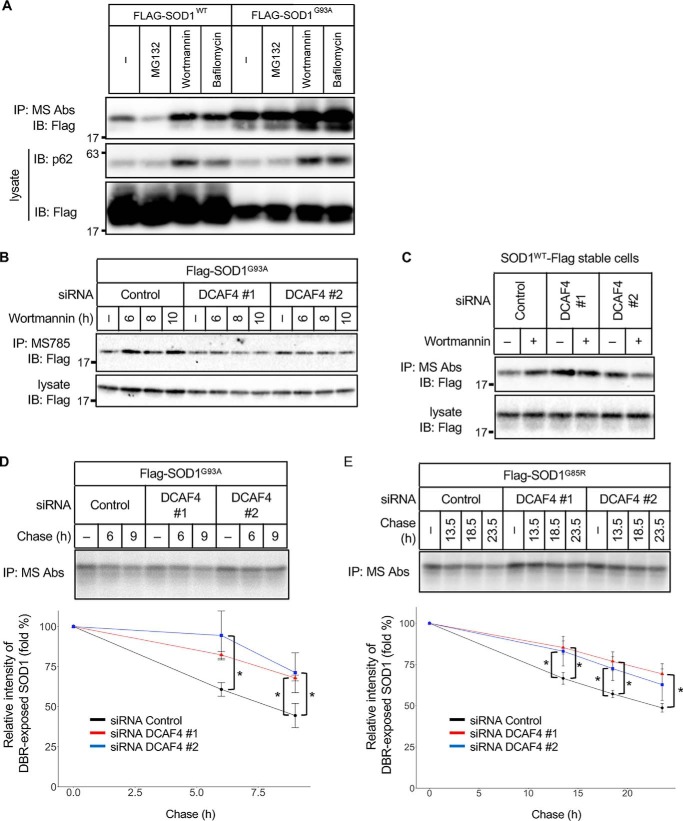
DCAF4 facilitates autophagic degradation of DBR-exposed SOD1
We next investigated the possibility that DCAF4 facilitates the degradation of DBR-exposed SOD1. To reveal the degradation pathway of DBR-exposed SOD1, we assessed the effect of MG132, a proteasome inhibitor, wortmannin and bafilomycin A1, autophagy inhibitors, on the amount of DBR-exposed SOD1. Treatment with wortmannin and bafilomycin A1 increased the amount of DBR-exposed SOD1 in both SOD1WT and SOD1G93A, although there was little effect with MG132 treatment (Fig. 3A and Fig. S3A). These data indicate that DBR-exposed SOD1 was predominantly degraded through the autophagic pathway, as suggested previously in studies using SOD1mut (22, 23).
Figure 3.
DCAF4 facilitates autophagic degradation of DBR-exposed SOD1. A–C, IP–IB analysis of each cell lysate with the indicated antibodies. A, HeLa cells were transfected with Flag–SOD1WT or Flag–SOD1G93A and treated with MG132 (5 μm, 9 h), wortmannin (1 μm, 9 h), or bafilomycin (50 nm, 3 h). The amount of DBR-exposed SOD1 and p62 was analyzed. B, HeLa cells were transfected with the indicated siRNAs and Flag–SOD1G93A and then treated with 1 μm wortmannin. The amount of DBR-exposed SOD1 was analyzed. C, HeLa cells stably expressing SOD1WT–Flag were transfected with the indicated siRNAs. Then, the cells were treated with 2 μm wortmannin for 9 h. The amount of DBR-exposed SOD1 was analyzed. D and E, pulse–chase assay of DBR-exposed SOD1. HeLa cells were transfected with the indicated siRNAs and Flag–SOD1mut and then were metabolically labeled with [35S]methionine and -cysteine and chased in HBSS (+). The cell lysates were immunoprecipitated with MS785 and MS27. The relative level of radioactivity in each sample was calculated. The decrease is shown as a percentage of the intensity observed at 0 h. The data are the mean ± S.D.; *, p < 0.05; n = 3 (ANOVA with a post hoc Dunnett's test).
To further evaluate the involvement of DCAF4 in autophagic degradation of DBR-exposed SOD1, we assessed the effect of autophagy inhibition in DCAF4 knockdown cells. Wortmannin treatment failed to increase the level of DBR-exposed SOD1G93A and DBR-exposed SOD1WT in DCAF4 knockdown cells compared with control cells (Fig. 3, B and C, and Fig. S3, B and C). To confirm that DCAF4 contributes to the degradation of DBR-exposed SOD1, the degradation rate of DBR-exposed SOD1 was assessed with a pulse–chase experiment. The DCAF4-suppressed cells showed an attenuated rate of DBR-exposed SOD1mut degradation compared with the control cells (Fig. 3, D and E). Collectively, these data indicate that DCAF4 contributes to autophagic degradation of DBR-exposed SOD1.
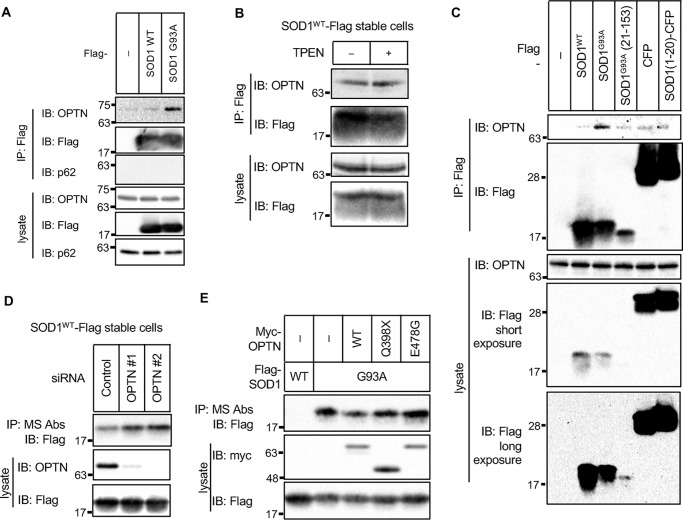
OPTN is involved in degradation of DBR-exposed SOD1
We speculated that DCAF4 mediates the ubiquitination of SOD1mut that leads to the autophagic degradation of DBR-exposed SOD1. However, the accumulation of ubiquitinated SOD1 could not be detected even in the presence of wortmannin (Fig. S4A). Thus, we assumed that the substrate of DCAF4 is not DBR-exposed SOD1 itself. One of the candidate substrates was the autophagic receptor. Among the autophagic receptors, OPTN and SQSTM1/p62 are reported to be ALS-causative genes (24, 25). Therefore, we investigated their potential involvement in the DBR-exposed SOD1 degradation.
First, we examined the interaction of these receptors with SOD1 and found that OPTN preferentially interacted with SOD1G93A and TPEN-stimulated SOD1WT rather than SOD1WT, whereas the interaction between SOD1mut and p62 could not be detected under the tested conditions (Fig. 4, A and B, and Fig. S4B). Next, we tested whether OPTN also recognizes and interacts with the exposed DBR. In contrast with DCAF4, DBR-deleted SOD1G93A(21–153) showed a weak but substantial interaction with OPTN even though its expression level was modest (Fig. 4C). In addition, the SOD1(1–20) fragment did not interact with OPTN (comparison of Flag–SOD1(1–20)–CFP with Flag–CFP). Thus, it is unlikely that OPTN interacts with the exposed DBR. OPTN may recognize the other region of conformationally disordered SOD1 (see “Discussion”).
Figure 4.
OPTN is involved in degradation of DBR-exposed SOD1. A–E, IP–IB analysis of each of the cell lysates with the indicated antibodies. A, HeLa cells were transfected with the indicated constructs. The interaction between SOD1 and endogenous OPTN or p62 was analyzed. B, HeLa cells stably expressing SOD1WT–Flag were stimulated with 10 μm TPEN for 5 h. The interaction with endogenous OPTN was analyzed. C, HeLa cells were transfected with the indicated constructs, and the interaction with endogenous OPTN was analyzed. D, HeLa cells stably expressing SOD1WT–Flag were transfected with the indicated siRNAs. The amount of DBR-exposed SOD1 was analyzed. E, HeLa cells were transfected with the indicated constructs. The amount of DBR-exposed SOD1 was analyzed.
To examine the contribution of OPTN to SOD1 proteostasis, we suppressed the expression of OPTN and assessed the amount of DBR-exposed SOD1WT. The suppression of OPTN expression induced accumulation of DBR-exposed SOD1WT, suggesting that OPTN is required for autophagic degradation of DBR-exposed SOD1 (Fig. 4D and Fig. S4C). Several OPTN mutants have been reported to be associated with ALS (26). We speculated that these mutants lost the ability to maintain SOD1 proteostasis. Although the interactions between SOD1G93A and two representative ALS-related OPTN mutants (OPTNQ398X and OPTNE478G) were preserved (Fig. S4D), they failed to reduce the amount of DBR-exposed SOD1G93A by co-expression, suggesting that they were inactive in terms of autophagic degradation of DBR-exposed SOD1 (Fig. 4E and Fig. S4E).
DCAF4-mediated ubiquitination of OPTN facilitates degradation of DBR-exposed SOD1
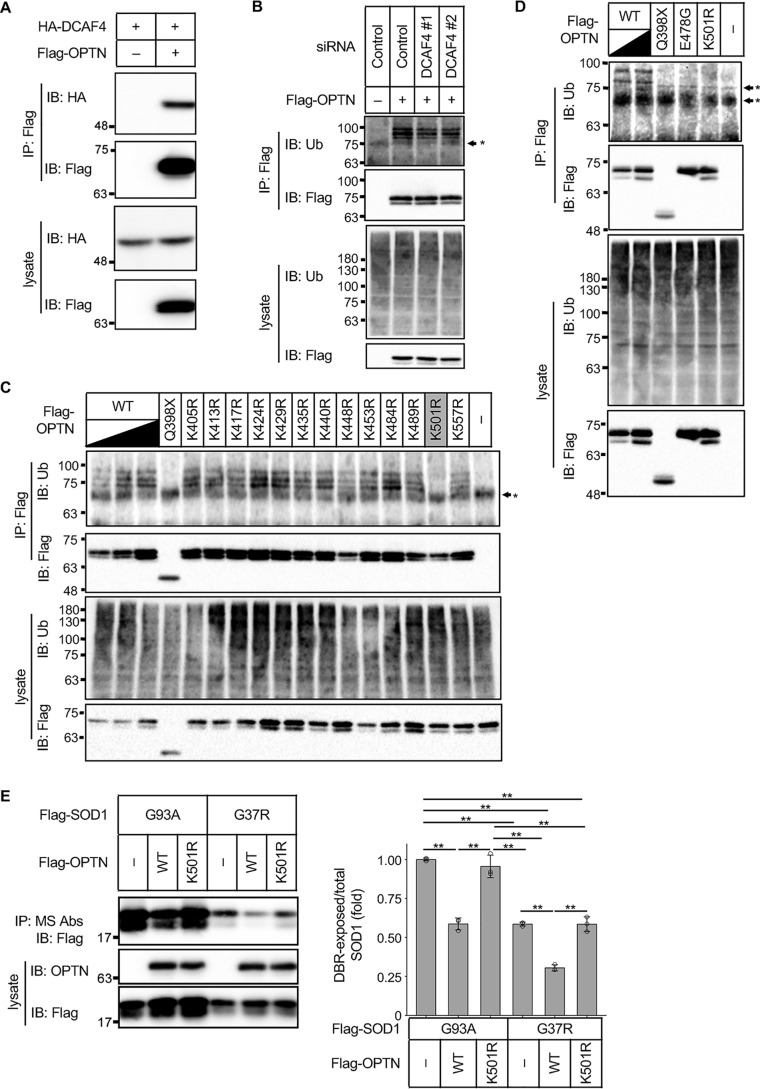
Because OPTN contributed to the degradation of DBR-exposed SOD1 and an interaction between DCAF4 and OPTN could be observed (Fig. 5A), we now assumed that DCAF4-mediated OPTN ubiquitination could be involved in the autophagic degradation of DBR-exposed SOD1. Thus, we first examined whether OPTN is ubiquitinated or not by the immunoprecipitation analysis of OPTN in a denatured condition to prevent a contamination of ubiquitinated substrates of OPTN followed by the detection of ubiquitinated proteins. Ubiquitinated OPTN was clearly observed (Fig. 5B). Then, the ubiquitination of OPTN was assessed in the DCAF4 knockdown cells. Suppression of DCAF4 partially but significantly attenuated the ubiquitination of OPTN, suggesting that the CRL4 complex, including DCAF4, ubiquitinates OPTN (Fig. 5B and Fig. S5). Next, we aimed to identify the ubiquitination site in OPTN. Because we found that the OPTNQ398X mutant was at least less ubiquitinated compared with the OPTNWT (Fig. 5C), all the lysine residues after 399 were mutated to arginine. Among these mutants, OPTNK501R showed resistance to ubiquitination (Fig. 5C). This is consistent with a previous report that identified the lysine 501 residue as a ubiquitination site in OPTN via MS (27). Notably, ubiquitination was also absent in the OPTNE478G mutant (Fig. 5D). These data demonstrate that DCAF4-including CRL4 complex mediates OPTN ubiquitination at lysine 501, and this ubiquitination is absent in the ALS-related OPTNmut.
Figure 5.
DCAF4-mediated ubiquitination of OPTN facilitates the degradation of DBR-exposed SOD1. A–E, IP–IB analysis of each of the cell lysates with the indicated antibodies. A, HeLa cells were transfected with the indicated constructs, and the interaction between OPTN and DCAF4 was analyzed. B, HeLa cells were transfected with the indicated siRNAs and Flag–OPTN. The ubiquitination of OPTN was evaluated. C and D, HeLa cells were transfected with the indicated Flag–OPTN constructs. The ubiquitination of OPTN was evaluated. E, left, HeLa cells were transfected with the indicated constructs. The amount of DBR-exposed SOD1 was analyzed. Right, DBR-exposed/total SOD1 ratios are shown as the mean ± S.D. (fold to nontreated SOD1G93A, n = 3). **, p < 0.01 (ANOVA with a post hoc Bonferroni). *, nonspecific band.
To evaluate the functional importance of this ubiquitination in SOD1 proteostasis, the ability of OPTNK501R to reduce the amount of DBR-exposed SOD1 was assessed. Exogenous expression of OPTNWT but not OPTNK501R reduced the amount of DBR-exposed SOD1G93A and SOD1G37R (Fig. 5E), demonstrating a requirement of the ubiquitination for facilitating the degradation of DBR-exposed SOD1.
Discussion
In this study, we carried out a genome-wide siRNA screen to elucidate the molecular mechanisms of SOD1 proteostasis. As a result, we obtained 30 candidate genes that were required to maintain the absence of DBR-exposed SOD1WT (Fig. 1D and Table S1). Among them, we focused on DCAF4, a substrate receptor of the CRL4 complex. DCAF4 was found to interact with DBR-exposed SOD1 through the DBR (Fig. 2, D and E) and was involved in autophagic degradation of DBR-exposed SOD1 (Fig. 3, A–E). OPTN, an ALS-causative gene product that is reported to act as an autophagy receptor, was identified as a ubiquitination substrate of DCAF4, and ubiquitination-resistant OPTN was unable to facilitate autophagic degradation (Fig. 5, B and E). These data indicate that DCAF4-mediated ubiquitination of OPTN is required for autophagic degradation of DBR-exposed SOD1 (Fig. S4).
Although the precise function is still unclear, some potential nucleotide metabolism proteins and transcription regulators (e.g. KTI12, CDKN2AIPL, SP1, and ZNF581) were included in the 30 candidate genes that were positive in the screen (Table S1). Because our screen could be affected by the expression level of SOD1WT, we estimated that it is unlikely that these gene products directly regulate SOD1 proteostasis. Of note, the mutation in one hit gene, DCNT1, was reported to be related to motor neuron disease and SALS (28). We are now investigating the molecular mechanisms of this gene in SOD1 proteostasis regulation. Further study may provide the molecular link between SOD1 and DCNT1.
Among the candidate genes identified in the screen, ATG family genes that are essential for autophagy were not included. Moreover, OPTN, CUL4A, and CUL4B were not included in the list. There could be several potential reasons for the absence of these genes. In our screen, siRNAs that exhibited toxicity became negative due to the reduction in DBR-exposed SOD1 as well as total SOD1WT. Redundancy also affects the results of the screen. Similar to CUL4A and CUL4B, which share high sequence similarity, single knockdown of redundant genes might not lead to a positive score. Another possibility is the limitation of our screen system. We used TPEN as the positive control in the present screen. Because chemically-induced accumulation of DBR-exposed SOD1 would be stronger than gene silencing–mediated accumulation, it is possible that the sensitivity of the screen was not sufficient to discover weakly-positive genes. Consistent with this, most of the positive genes had a relatively low Z-score in absolute value (<5) (Fig. 2A). Here, we identified DCAF4 and OPTN as critical genes for maintaining the absence of DBR-exposed SOD1WT. Further improvement of the screen system, with these genes as the positive controls, may lead to the discovery of all of the involved genes.
Here, we showed that DCAF4 and OPTN preferentially interact with SOD1mut compared with SOD1WT (Figs. 2E and 4C). Although DCAF4 interacted with SOD1 through the exposed DBR, OPTN appears to recognize other difference(s) in DBR-exposed SOD1. Of note, the UBAN domain is absent in OPTNQ398X, and its function was lost in OPTNE478G (29). Thus, the ubiquitination of SOD1 would not be required for the recognition of SOD1mut by OPTN (Fig. S3B). This is consistent with the observation that the wortmannin treatment failed to enhance the accumulation of ubiquitinated SOD1 (Fig. S3A) and the previous report that showed that OPTN recognizes mutant SOD1 in the ubiquitination-independent manner (30). Although the precise mechanism how OPTN recognizes SOD1mut is unclear, one of the candidate regions that OPTN bound is SOD1(32–40), which is adjacent to DBR and is also exposed in the SOD1mut (21). Because DCAF4 and OPTN interact with SOD1 through different sites, it is possible that SOD1 gathers DCAF4 and OPTN in close, enhancing the DCAF4-mediated ubiquitination of OPTN (Fig. S4). Notably, the phosphorylation of OPTN by TANK-binding kinase 1 (TBK1), another ALS-causative gene, has also been shown to facilitate the degradation of SOD1 aggregates (30, 31). Although OPTN interacts with soluble DBR-exposed SOD1 and the involvement of TBK1 is not evaluated in our system, the disturbance of SOD1 proteostasis could be the common feature of some ALS cases.
The amount of DCAF4 interacting with SOD1G37R was less compared with other mutants (A4V, G85R, and G93A) (Fig. 2D). One possibility is that because the ratio of DBR exposed to total SOD1 was smaller in SOD1G37R (Fig. 5E), a lesser amount of SOD1–DCAF4 complex was formed. It would also be possible that the DBR exposure is partial or the steric hindrance between DBR and DCAF4 exists in G37R mutant
Throughout this study, we revealed in part the molecular mechanism of SOD1 proteostasis. Although the accumulation of DBR-exposed SOD1 was not drastic here, the cumulation during the decades before the onset of ALS could lead to the motoneuron toxicity. Further studies would elucidate whether a defect in SOD1 proteostasis is involved in the pathogenesis of SOD1 mutation-negative ALS, including ALS caused by OPTN mutation.
Experimental procedures
Antibodies
Anti-tag antibodies labeled with either Eu or d2 (Fig. S1A) were purchased from Cisbio. The antibodies against Flag tag (Wako, 012-22384), HA tag (Roche Applied Science, 1867431), SOD1 (Enzo Life Science, ADI-SOD-100), ubiquitin (Santa Cruz Biotechnology, sc-8017), OPTN (Proteintech, 10837-1-AP), Cullin4 (Abcam, ab76470), and p62 (Progen Biotechnik, GP62-C) were purchased from the indicated suppliers. Control rat immunoglobulin G1 mAb was purchased from BioGenesis. MS785 and MS27 were prepared as described previously (12, 21). The purified MS785 was labeled with either Eu or d2 using a d2-labeling kit or europium cryptate labeling kit (Cisbio) according to the manufacturer's instructions.
Cell culture
HEK293A cells were purchased from Invitrogen and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 mg/ml glucose (Sigma, D5796), 10% FBS, and 100 units/ml penicillin (Meiji Seika, 6111400D2039). HeLa cells were purchased from ATCC and cultured in DMEM containing 1.0 mg/ml glucose (Sigma, D6046) supplemented with 10% FBS and 100 units/ml penicillin G. HeLa cells stably-expressing SOD1WT C-terminally tagged with Flag (SOD1WT–Flag stable HeLa cells) were previously generated (21) and cultured in DMEM containing 1.0 mg/ml glucose, 10% FBS, 100 units/ml penicillin, and 2 μg/ml puromycin (Gibco) in 5% CO2 at 37 °C. All cells were cultured in 5% CO2 at 37 °C and verified to be negative for mycoplasma.
Plasmids and transfection
The cDNAs encoding SOD1WT, OPTNWT, OPTNmut, and DCAF4 were cloned into a pcDNA3.0 plasmid (Invitrogen) using standard molecular biology techniques, and all constructs were verified by sequencing. The pcDNA3.0–Flag–SOD1WT, pcDNA3.0–Flag–SOD1mut, Flag–SOD1G93A(21–153), pcDNA3.0–Flag–CFP, and pcDNA3.0–Flag–SOD1(1–20)-CFP plasmids have been previously constructed (11, 12, 21).
Transfection was performed using polyethyleneimine-Max (Polysciences, 24765) according to the manufacturer's instructions. Transfections with siRNA were carried out by forward or reverse transfection using Lipofectamine RNAiMAX (Invitrogen, 133778-150) according to the manufacturer's instructions. When cells were transfected with both plasmid and siRNA, the transfection with siRNA was performed 12–24 h before the transfection with plasmid. The siRNAs used are listed in Table S2.
Immunoblotting analysis
Cells were lysed on ice in a lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10 mm EDTA, 1% sodium deoxycholate, and 1% Triton X-100) supplemented with 1 mm phenylmethylsulfonyl fluoride, and 5 g/ml leupeptin. After centrifugation, supernatants were sampled by adding 2× SDS sample buffer (80 mm Tris-HCl, pH 8.8, 80 μg/ml bromphenol blue, 28.8% glycerol, 4% SDS, and 10 mm DTT). The samples were resolved by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. After blocking with 5% skim milk in TBS-T (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 0.05% Tween 20), the membranes were probed with each primary antibody. After incubation with the appropriate secondary antibodies, the proteins were detected in an ECL system using a ChemiDoc imaging system and Image Lab software (Bio-Rad). All immunoblotting analyses were repeated at least three times, and almost the same results were obtained.
Immunoprecipitation analysis
The supernatants of centrifuged cell lysates were immunoprecipitated with anti-Flag antibody (Sigma, A2220, or Wako Pure Chemicals Industries, 016-22784), MS785, MS27, or control rat immunoglobulin G1 mAb (BioGenesis) using protein G–Sepharose (GE Healthcare, 17061802). The beads were washed with washing buffer 1 (20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 5 mm EGTA, and 1% Triton X-100) and washing buffer 2 (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 5 mm EGTA) or with lysis buffer. The proteins were eluted with 2× SDS sample buffer or 3× Flag peptide (Sigma) in lysis buffer.
Preparation of assay plate for siRNA screen
The genome-wide siRNA library used in the primary screen consisted of siRNAs targeting 18,023 human genes (Dharmacon siGENOME SMARTpool collections: human genome (G-005005-E2), human drug targets (G-004655-E2), and human druggable subsets (G-004675-E2)). The assay plates (Greiner, 784086) for the 1st screen were prepared as reported previously with minor modification in layout and the amount of siRNA by using Biomek FXP (Beckman Coulter), POD automation (LABCYTE), and Echo liquid handler (LABCYTE) (32). In the 2nd screen, the siRNAs targeting positive genes in the 1st screen were spotted onto a 384-well plate with POD automation and Echo liquid handler. The individual siRNAs targeting the same gene were placed on the same plate.
TR-FRET–based siRNA screen
Lipofectamine RNAiMAX in Opti-MEM (Invitrogen) was added at 3 μl/well to the assay plates using a Multidrop Combi. After 15 min, 3.33 × 105 cells/ml of HeLa cells expressing SOD1WT–Flag were seeded at 15 μl/well using the Multidrop Combi, and the final siRNA concentration was set at 30 nm. After 48 h of incubation, DMSO or TPEN (final 10 μm in reference well) was overlaid using the Multidrop Combi and manually, respectively. After 4.5 h of stimulation, the medium was eliminated by centrifugation. Then, the fluorophore-labeled antibodies and MS27 antibody (for stabilization of the DBR-exposed SOD1 (21)) in FRET buffer (25 mm phosphate buffer, pH 7.0, 400 mm KF, 0.1% BSA, 0.5% Triton X-100, 10 mm EDTA, 5 μg/ml leupeptin, 1 mm phenylmethylsulfonyl fluoride) were added with the Multidrop Combi. After 1 h of incubation at room temperature, the FRET intensity (337-nm excitation filter/665-nm emission filter) and donor intensity (337-nm excitation filter/620-nm emission filter) were measured using PHERAstar (BMG Labtech), and the FRET ratio was calculated as FRET intensity/donor intensity. FRET signal was the FRET ratio of the samples divided by that of the DMSO-treated samples. The Z′-factor between the DMSO- and TPEN-treated reference groups in each plate was set at over 0.2 (1st screen) or 0.5 (2nd screen). Unsatisfactory plates were discarded and recreated. To compare between plates, the FRET ratio was normalized with a sample-based robust Z-score and B-score (1st screen) or a reference-based Z-score (2nd screen) (33–35).
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from cells using Isogen reagent (Wako) and reverse-transcribed using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). Primers were designed using the Universal Probe Library Assay Design Center (Roche Applied Science). Quantitative RT-PCR was carried out using SYBR Green PCR Master Mix and a LightCycler 96 (Roche Applied Science). The levels were normalized to the S18 mRNA level. The primer sequences are listed in Table S3.
Pulse–chase analysis
HeLa cells were labeled with l-[35S]Met and l-[35S]Cys (PerkinElmer Life Sciences) for 1 h in medium lacking methionine and cysteine. After washing with PBS, the cells were chased in HBSS(+). Cells were lysed on ice in lysis buffer, and the supernatants of centrifuged cell lysates were immunoprecipitated with MS785 and MS27. The beads were washed with washing buffer 1 and washing buffer 2. Immunoprecipitated samples were eluted with 2× SDS sample buffer, resolved by SDS-PAGE, and analyzed with an image analyzer (Storm 840, GE Healthcare). Quantification was performed using ImageJ software.
Ubiquitination assay
Cells were lysed in PBS containing 0.5% SDS, 5 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride. The lysates were diluted with PBS and sonicated. The supernatants of cell lysates were immunoprecipitated with anti–Flag antibody. The beads were washed with washing buffer 1 and washing buffer 2. The proteins were eluted with 2× SDS sample buffer or 3× Flag peptide in washing buffer 2.
Statistical analysis
Statistical analyses were performed via ANOVA with a post hoc Dunnett's test. The values are the mean ± S.D. calculated from at least three different experiments. Significance was assumed with *, p < 0.05.
Materials and data availability
The materials and data that support the findings of this study are available from the corresponding author on reasonable request.
Author contributions
K. H., I. N., and H. I. conceptualization; K. H., T. F., and H. I. funding acquisition; K. H., H. T., N. T., and T. F. investigation; K. H., H. T., N. T., I. N., T. F., and H. I. writing-original draft; K. H., I. N., T. F., and H. I. project administration.
Supplementary Material
Acknowledgments
We thank One-stop Sharing Facility Center for Future Drug Discoveries and Drug Discovery Initiative (University of Tokyo) for sharing the instruments. We are also grateful to H. Nishina (Tokyo Medical and Dental University) for assistance.
This work was supported by Grants-in-aid for Scientific Research from the Japanese Society for the Promotion of Science; MEXT Grants 25221302 and JP18H03995 (to H. I.), 16K18513 (to K. H.), JP18H02569 (to I. N.) and 18K15358 (to T. F.); Advanced Research for Medical Products Mining Program of the National Institute of Biomedical Innovation (to H. I.); Project for Elucidating and Controlling Mechanisms of Aging and Longevity from Japan Agency for Medical Research and Development (AMED) Grant JP17gm5010001 and JP19gm5010001 (to H. I.); the Kowa Life Science Foundation (to T. F.); the SERIKA Fund (to T. F.); and Grant-in-aid from the Tokyo Biochemical Research Foundation Grant 161000000310 (to K. H.). A portion of this study resulted from “Understanding of molecular and environmental bases for brain health” conducted under the Strategic Research Program for Brain Sciences by the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (to H. I.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Tables S1–S3.
- ALS
- amyotrophic lateral sclerosis
- DBR
- Derlin-1–binding region
- OPTN
- optineurin
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- SALS
- sporadic ALS
- TR-FRET
- time-resolved FRET
- TPEN
- N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- ANOVA
- analysis of variance
- IP–IB
- immunoprecipitation–immunoblotting
- CFP
- cyan fluorescent protein.
References
- 1. Nguyen H. P., Van Broeckhoven C., and van der Zee J. (2018) ALS genes in the genomic era and their implications for FTD. Trends Genet. 34, 404–423 10.1016/j.tig.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 2. Abel O., Powell J. F., Andersen P. M., and Al-Chalabi A. (2012) ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 33, 1345–1351 10.1002/humu.22157 [DOI] [PubMed] [Google Scholar]
- 3. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., and Deng H. X. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 4. Reaume A. G., Elliott J. L., Hoffman E. K., Kowall N. W., Ferrante R. J., Siwek D. F., Wilcox H. M., Flood D. G., Beal M. F., Brown R. H. Jr., Scott R. W., and Snider W. D. (1996) Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 13, 43–47 10.1038/ng0596-43 [DOI] [PubMed] [Google Scholar]
- 5. Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., and Deng H. X. (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 10.1126/science.8209258 [DOI] [PubMed] [Google Scholar]
- 6. Bruijn L. I., Becher M. W., Lee M. K., Anderson K. L., Jenkins N. A., Copeland N. G., Sisodia S. S., Rothstein J. D., Borchelt D. R., Price D. L., and Cleveland D. W. (1997) ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 18, 327–338 10.1016/S0896-6273(00)80272-X [DOI] [PubMed] [Google Scholar]
- 7. Bruijn L. I., Houseweart M. K., Kato S., Anderson K. L., Anderson S. D., Ohama E., Reaume A. G., Scott R. W., and Cleveland D. W. (1998) Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281, 1851–1854 10.1126/science.281.5384.1851 [DOI] [PubMed] [Google Scholar]
- 8. Cleveland D. W., Laing N., Hurse P. V., Brown R. H. (1995) Toxic mutants in Charcot's sclerosis. Nature 378, 342–343 10.1038/378342a0 [DOI] [PubMed] [Google Scholar]
- 9. Boillée S., Vande Velde C., and Cleveland D. W. (2006) ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron 52, 39–59 10.1016/j.neuron.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 10. Ilieva H., Polymenidou M., and Cleveland D. W. (2009) Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell Biol. 187, 761–772 10.1083/jcb.200908164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., and Ichijo H. (2008) ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 22, 1451–1464 10.1101/gad.1640108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujisawa T., Homma K., Yamaguchi N., Kadowaki H., Tsuburaya N., Naguro I., Matsuzawa A., Takeda K., Takahashi Y., Goto J., Tsuji S., Nishitoh H., and Ichijo H. (2012) A novel monoclonal antibody reveals a conformational alteration shared by amyotrophic lateral sclerosis-linked SOD1 mutants. Ann. Neurol. 72, 739–749 10.1002/ana.23668 [DOI] [PubMed] [Google Scholar]
- 13. Lilley B. N., and Ploegh H. L. (2004) A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429, 834–840 10.1038/nature02592 [DOI] [PubMed] [Google Scholar]
- 14. Ye Y., Shibata Y., Yun C., Ron D., and Rapoport T. A. (2004) A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429, 841–847 10.1038/nature02656 [DOI] [PubMed] [Google Scholar]
- 15. Tsuburaya N., Homma K., Higuchi T., Balia A., Yamakoshi H., Shibata N., Nakamura S., Nakagawa H., Ikeda S. I., Umezawa N., Kato N., Yokoshima S., Shibuya M., Shimonishi M., Kojima H., et al. (2018) A small-molecule inhibitor of SOD1–Derlin-1 interaction ameliorates pathology in an ALS mouse model. Nat. Commun. 9, 2668 10.1038/s41467-018-05127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., Frosch M. P., Agar J. N., Julien J. P., Brady S. T., and Brown R. H. (2010) Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 13, 1396–1403 10.1038/nn.2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haidet-Phillips A. M., Hester M. E., Miranda C. J., Meyer K., Braun L., Frakes A., Song S., Likhite S., Murtha M. J., Foust K. D., Rao M., Eagle A., Kammesheidt A., Christensen A., Mendell J. R., et al. (2011) Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 29, 824–828 10.1038/nbt.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferraiuolo L., Meyer K., Sherwood T. W., Vick J., Likhite S., Frakes A., Miranda C. J., Braun L., Heath P. R., Pineda R., Beattie C. E., Shaw P. J., Askwith C. C., McTigue D., and Kaspar B. K. (2016) Oligodendrocytes contribute to motor neuron death in ALS via SOD1-dependent mechanism. Proc. Natl. Acad. Sci. U.S.A. 113, E6496–E6505 10.1073/pnas.1607496113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Homma K., Fujisawa T., Tsuburaya N., Yamaguchi N., Kadowaki H., Takeda K., Nishitoh H., Matsuzawa A., Naguro I., and Ichijo H. (2013) SOD1 as a molecular switch for initiating the homeostatic ER stress response under zinc deficiency. Mol. Cell 52, 75–86 10.1016/j.molcel.2013.08.038 [DOI] [PubMed] [Google Scholar]
- 20. Jin J., Arias E. E., Chen J., Harper J. W., and Walter J. C. (2006) A family of diverse Cul4-Ddb1–interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- 21. Fujisawa T., Yamaguchi N., Kadowaki H., Tsukamoto Y., Tsuburaya N., Tsubota A., Takahashi H., Naguro I., Takahashi Y., Goto J., Tsuji S., Nishitoh H., Homma K., and Ichijo H. (2015) A systematic immunoprecipitation approach reinforces the concept of common conformational alterations in amyotrophic lateral sclerosis–linked SOD1 mutants. Neurobiol. Dis. 82, 478–486 10.1016/j.nbd.2015.08.010 [DOI] [PubMed] [Google Scholar]
- 22. Imamura K., Izumi Y., Watanabe A., Tsukita K., Woltjen K., Yamamoto T., Hotta A., Kondo T., Kitaoka S., Ohta A., Tanaka A., Watanabe D., Morita M., Takuma H., Tamaoka A., et al. (2017) The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 9, eaaf3962 10.1126/scitranslmed.aaf3962 [DOI] [PubMed] [Google Scholar]
- 23. Zhai J., Zhang L., Mojsilovic-Petrovic J., Jian X., Thomas J., Homma K., Schmitz A., Famulok M., Ichijo H., Argon Y., Randazzo P. A., and Kalb R. G. (2015) Inhibition of cytohesins protects against genetic models of motor neuron disease. J. Neurosci. 35, 9088–9105 10.1523/JNEUROSCI.5032-13.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H., Komure O., Matsuura S., Kobatake K., Morimoto N., Abe K., et al. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 10.1038/nature08971 [DOI] [PubMed] [Google Scholar]
- 25. Rea S. L., Majcher V., Searle M. S., and Layfield R. (2014) SQSTM1 mutations–bridging Paget disease of bone and ALS/FTLD. Exp. Cell Res. 325, 27–37 10.1016/j.yexcr.2014.01.020 [DOI] [PubMed] [Google Scholar]
- 26. Markovinovic A., Cimbro R., Ljutic T., Kriz J., Rogelj B., and Munitic I. (2017) Optineurin in amyotrophic lateral sclerosis: multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog. Neurobiol. 154, 1–20 10.1016/j.pneurobio.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 27. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikenaka K., Katsuno M., Kawai K., Ishigaki S., Tanaka F., and Sobue G. (2012) Disruption of axonal transport in motor neuron diseases. Int. J. Mol. Sci. 13, 1225–1238 10.3390/ijms13011225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wild P., Farhan H., McEwan D. G., Wagner S., Rogov V. V., Brady N. R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., and Dikic I. (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korac J., Schaeffer V., Kovacevic I., Clement A. M., Jungblut B., Behl C., Terzic J., and Dikic I. (2013) Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126, 580–592 10.1242/jcs.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cirulli E. T., Lasseigne B. N., Petrovski S., Sapp P. C., Dion P. A., Leblond C. S., Couthouis J., Lu Y. F., Wang Q., Krueger B. J., Ren Z., Keebler J., Han Y., Levy S. E., Boone B. E., et al. (2015) Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441 10.1126/science.aaa3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watanabe K., Umeda T., Niwa K., Naguro I., and Ichijo H. (2018) A PP6–ASK3 module coordinates the bidirectional cell volume regulation under osmotic stress. Cell Rep. 22, 2809–2817 10.1016/j.celrep.2018.02.045 [DOI] [PubMed] [Google Scholar]
- 33. Birmingham A., Selfors L. M., Forster T., Wrobel D., Kennedy C. J., Shanks E., Santoyo-Lopez J., Dunican D. J., Long A., Kelleher D., Smith Q., Beijersbergen R. L., Ghazal P., and Shamu C. E. (2009) Statistical methods for analysis of high-throughput RNA interference screens. Nat. Methods 6, 569–575 10.1038/nmeth.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malo N., Hanley J. A., Cerquozzi S., Pelletier J., and Nadon R. (2006) Statistical practice in high-throughput screening data analysis. Nat. Biotechnol. 24, 167–175 10.1038/nbt1186 [DOI] [PubMed] [Google Scholar]
- 35. Brideau C., Gunter B., Pikounis B., and Liaw A. (2003) Improved statistical methods for hit selection in high-throughput screening. J. Biomol. Screen. 8, 634–647 10.1177/1087057103258285 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.