Significance
CDK8, the catalytic core of the Mediator kinase module, is activated by MED12. We describe the architecture of the ternary complex of CDK8, Cyclin C, and MED12, and we decipher the mechanism by which MED12 activates CDK8. We find the N-terminal segment of MED12 to wrap around the CDK8 molecule to place an “activation helix” in proximity of its T-loop. Exact placement of the helix, not MED12 binding alone, is crucial for CDK8 activation. Moreover, MED12 binding precludes binding of type II kinase inhibitors to the active site of CDK8. As many cancer patients carry mutations in the MED12 activation helix, our study invigorates the development of CDK8-specific compounds as well as offers functional insights into the Mediator kinase module.
Keywords: CDK8, MED12, Mediator kinase module, Mediator, kinase inhibitors
Abstract
The Mediator kinase module regulates eukaryotic transcription by phosphorylating transcription-related targets and by modulating the association of Mediator and RNA polymerase II. The activity of its catalytic core, cyclin-dependent kinase 8 (CDK8), is controlled by Cyclin C and regulatory subunit MED12, with its deregulation contributing to numerous malignancies. Here, we combine in vitro biochemistry, cross-linking coupled to mass spectrometry, and in vivo studies to describe the binding location of the N-terminal segment of MED12 on the CDK8/Cyclin C complex and to gain mechanistic insights into the activation of CDK8 by MED12. Our data demonstrate that the N-terminal portion of MED12 wraps around CDK8, whereby it positions an “activation helix” close to the T-loop of CDK8 for its activation. Intriguingly, mutations in the activation helix that are frequently found in cancers do not diminish the affinity of MED12 for CDK8, yet likely alter the exact positioning of the activation helix. Furthermore, we find the transcriptome-wide gene-expression changes in human cells that result from a mutation in the MED12 activation helix to correlate with deregulated genes in breast and colon cancer. Finally, functional assays in the presence of kinase inhibitors reveal that binding of MED12 remodels the active site of CDK8 and thereby precludes the inhibition of ternary CDK8 complexes by type II kinase inhibitors. Taken together, our results not only allow us to propose a revised model of how CDK8 activity is regulated by MED12, but also offer a path forward in developing small molecules that target CDK8 in its MED12-bound form.
Mediator is essential for all RNA polymerase II (Pol II)-mediated transcription in eukaryotes because it allows for the communication between enhancer-bound transcription factors and Pol II (1–4). In humans Mediator comprises 30 protein subunits that are subdivided into four modules: The head, the middle, the tail, and the kinase module. Whereas the first three modules form a complex that is often referred to as the Mediator complex, the kinase module only reversibly associates with the three-module Mediator complex (5–7). Structural information on the isolated Mediator complex and on the head and the middle module of Mediator, collectively termed core Mediator, bound to Pol II provided first insights into how Mediator is able to activate Pol II (8, 9). In contrast to the Mediator complex, the structure of the kinase module is unknown and its role during the transcription cycle is unclear. Despite the fact that, in yeast, the three-module Mediator complex is unable to simultaneously bind Pol II and the kinase module (10), deletion or inhibitor of CDK8 results only in minor gene-expression changes (11, 12). However, using human proteins in a mostly recombinant in vitro system, the kinase module was found to repress transcription by inhibiting Mediator coactivator function (13).
The Mediator kinase module consists of four subunits: Cyclin-dependent kinase 8 (CDK8), its corresponding cyclin, Cyclin C, and two large regulatory subunits, MED12 and MED13. Moreover, in metazoans all kinase module subunits, except for Cyclin C, possess paralogues that can be utilized for the assembly of kinase modules with altered subunit composition accompanied by a potentially altered activity and target specificity (14). CDK8 and its paralog CDK19 were found to phosphorylate a wide variety of substrates involved in transcription, DNA repair, and metabolic processes (11). This wealth of substrates likely lies at the heart of the context-dependent role of the Mediator kinase module in metazoan transcription. Moreover, the central role of CDK8 in transcription regulation explains why it is a potent oncogene that is involved in the progression of malignant cancers, like colorectal cancer and acute myeloid leukemia (15–17). Due to its oncogenic potential, the effective and selective inhibition of CDK8 is of great interest to drug development and therefore spurred significant efforts to develop CDK8-specific inhibitors. Despite the availability of structural information on CDK8 bound to Cyclin C (18), the development of CDK8-specific inhibitors is hampered by the to date unclear mechanism of CDK8 activation. In contrast to the common activation mechanism of cyclin-dependent kinases, CDK8 does not require phosphorylation of its T-loop for full activity. Instead, CDK8 T-loop phosphorylation seems to be functionally replaced by MED12 binding to CDK8, which results in its activation (5, 19). The current model for CDK8 activation proposes that the N-terminal segment of MED12 binds to a surface groove on Cyclin C, and thereby enhances CDK8 activity (19). However, how MED12 binding to this distant surface groove could be coupled to repositioning of the CDK8 T-loop remains unclear and the current model is therefore controversial.
To investigate how MED12 activates CDK8, we established a recombinant expression and purification scheme that resulted in monodisperse, highly pure ternary CDK8/Cyclin C/MED12 complexes. Using these complexes, we carried out chemical cross-linking coupled to mass spectrometry. Unexpectedly, the resulting lysine- and aspartate/glutamate-mediated cross-links suggest a substantially revised mechanism for how the N-terminal portion of MED12 binds and activates CDK8: MED12 makes extensive contacts to CDK8 with no crucial contacts to Cyclin C. Intriguingly, MED12 thereby places a critical “activation helix” in direct vicinity of the T-loop of CDK8, leading to its conformational stabilization by contacting its arginine triad. Using kinase assays and mutational profiling, we demonstrate that the N-terminal tip of the MED12 activation helix, in particular glutamate-33 (E33), is necessary for CDK8 activation. In addition, we find that disease-causing MED12 mutations likely lead to mis-positioning of the activation helix, and thereby abrogate CDK8 activity. In further support, we show that the transcriptome-wide gene expression changes in HCT116 cells that carry a MED12 E33Q mutation phenocopy multiple cancer-related transcriptome alterations. Finally, we demonstrate that, in contrast to type I inhibitors, type II kinase inhibitors lose much of their inhibitory potential when used against ternary MED12-bound CDK8/Cyclin C in vitro. Taken together, our results establish that an activation helix in MED12 functionally replaces T-loop phosphorylation of CDK8. Furthermore, MED12 binding likely remodels the CDK8 active site, thus preventing type II kinase inhibitors from efficiently targeting CDK8 bound by MED12 in vivo.
Results
MED12 Binding Stabilizes and Stimulates the Activity of the Binary CDK8/Cyclin C Complex.
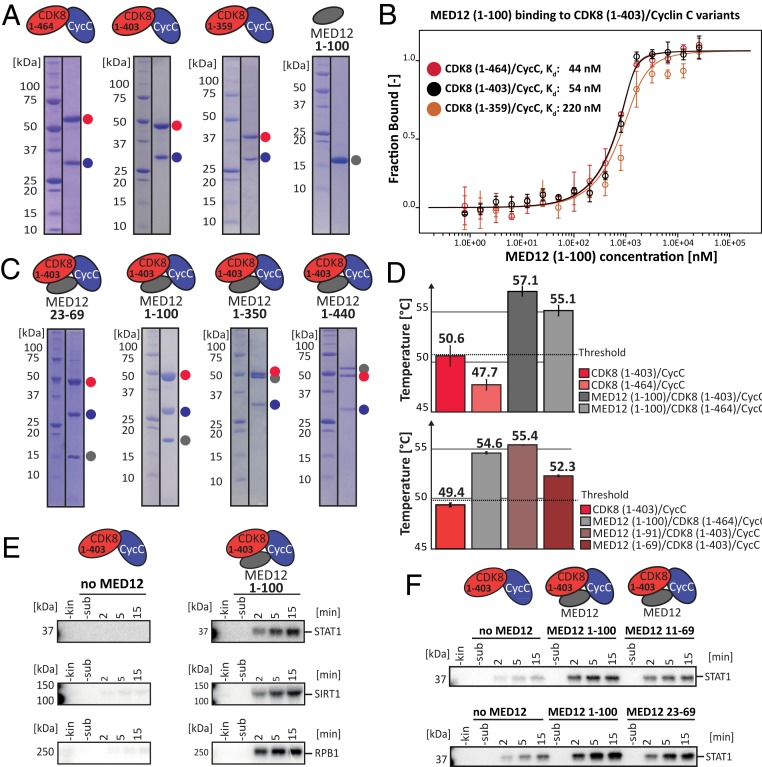
To study the affinity of MED12 for the binary CDK8/Cyclin C complex, we devised an expression and purification strategy for both components in insect cells (Fig. 1A and SI Appendix, Fig. S1A). As the N terminus of MED12 was reported to harbor the CDK8 activation domain (19), we measured the affinity of MED12 (1–100) to purified CDK8/Cyclin C complexes using microscale thermophoresis (MST). Using MST, we found a nanomolar affinity (Kd = 44 nM) of MED12 for full-length CDK8 (Fig. 1B). Shortening of the CDK8 C terminus reduced MED12 binding affinity by a factor of five for the shortest CDK8 variant (CDK8 [1–359], Kd = 220 nM). This suggests that the disordered C terminus of CDK8 only marginally influences MED12 binding. To cross-validate our MST measurements, we used isothermal titration calorimetry (ITC) to confirm an affinity of 72 nM of MED12 (1–100) for CDK8 (1–403)/Cyclin C (the Kd using MST was 54 nM) (Fig. 1B and SI Appendix, Fig. S1B). Having determined a nanomolar affinity of MED12 for binary CDK8/Cyclin C complexes, we aimed to develop expression and purification strategies for ternary CDK8/Cyclin C/MED12 complexes. Indeed, using MultiBacTurbo coexpression we were able to purify a variety of ternary complexes that comprised MED12 fragments with a length of up to 440 residues (Fig. 1C). Taking CDK8 (1–403)/Cyclin C/MED12 (1–100) as a representative of the ternary complexes investigated in this study, we validated its stoichiometry using static light scattering, which revealed a monodisperse complex with a 1:1:1 stoichiometry (SI Appendix, Fig. S1C). Furthermore, the minimal stable ternary complex could be formed with a MED12 fragment comprising residues 23 through 69 (Fig. 1C). To assess whether MED12 binding to different CDK8/Cyclin C complexes increases their stability, we measured their thermal melting behavior using differential scanning fluorimetry (nanoDSF). We found a significantly increased melting temperature (+7 °C) of ternary complexes, confirming that MED12 has a major stabilizing effect on CDK8/Cyclin C complexes (Fig. 1D). Note here that the stability of CDK complexes was reported to correlate with their activity (20). Moreover, our nanoDSF data uncover that the presence of CDK8’s likely unstructured C terminus destabilizes both binary and ternary complexes. We find that ternary complexes comprising a MED12 of fragments 1 to 91 or 1 to 100 result in the most stable complexes. In contrast, MED12 constructs shorter than 70 residues significantly destabilize the ternary complex, albeit still showing a stabilizing effect with respect to the binary complex (Fig. 1D).
Fig. 1.
MED12 both binds and activates binary CDK8/Cyclin C complexes. (A) SDS/PAGE analyses of purified binary CDK8/Cyclin C complexes and purified MED12 (1–100). Protein complexes are shown as cartoons above each gel, individual proteins are indicated next to the gels. CDK8 variants (1 to 464 [full-length], 1 to 403 and 1 to 359) are shown in red, Cyclin C is in blue, and MED12 is depicted in gray. (B) MST binding experiments using of MED12 (1–100) and different binary CDK8/Cyclin C complexes. Kd values are indicated. Error bars reflect the SD of four replicates. Please note that Kd cannot be read off directly due to the experimental necessity to use high protein concentrations. (C) SDS/PAGE analyses of purified ternary CDK8 (1–403)/Cyclin C/MED12 complexes. Protein complexes comprising MED12 constructs of different sizes are shown as cartoons above each gel, individual proteins are indicated next to the gels. Colors as in A. We note that MED12 (23–69) carries a C-terminal Strep-tag and hence runs higher than expected. (D) Thermal stability of binary CDK8/Cyclin C complexes and ternary CDK8/Cyclin C/MED12 complexes determined using nanoDSF. TM values are indicated. The SD of three experimental replicates is shown as error bars. (E) In vitro kinase assays using purified binary CDK8 (1–403)/Cyclin C and ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes. The 0.25 pmol kinase complex was incubated with 75 pmol GST-tagged STAT1 transactivation domain (TAD) in the presence of an excess of [γ-32P]-ATP. For SIRT1, a ratio of 0.5 pmol kinase and 50 pmol SIRT1 was used. For Pol II, 2 pmol of kinase complex was incubated in presence of 2 pmol Pol II. Please note that the Pol II C-terminal domain contains about 50 potential CDK8 phosphorylation sites, which made this 1:1 ratio necessary. Reactions were stopped at the indicated time points. (F) Same as in E; however, 7.5 pmol kinase complexes was incubated with 50 pmol STAT1 TAD.
MED12 binding to CDK8/Cyclin C was reported to activate the otherwise negligible kinase activity of CDK8 (19). We sought to recapitulate these findings using our purified binary CDK8/Cyclin C and ternary CDK8/Cyclin C/MED12 complexes. In addition, we asked which fragment of MED12 is sufficient to activate CDK8. To measure CDK8 activity, we established a radioactive kinase assay with the purified transactivation domain of STAT1 as a substrate (Fig. 1E). STAT1 is a known CDK8 target and is phosphorylated by CDK8 at Ser-727 (17, 21). Whereas we detected low kinase activity for binary CDK8/Cyclin C complexes using our in vitro system, the inclusion of MED12 in ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes resulted in a pronounced stimulation of kinase activity (Fig. 1E and SI Appendix, Fig. S1D). To extend our findings to more than a single CDK8 substrate, we also established Sirtuin 1 (SIRT1) and Pol II as known CDK8 targets in our in vitro assay system. Using these substrates, we observed similar MED12-dependant activation of CDK8 (Fig. 1E). This confirms earlier reports (5, 19), yet also demonstrates that the CDK8/Cyclin C complex possesses basal kinase activity whose extent varies with the ratio of kinase to substrate concentration (Fig. 1E and SI Appendix, Fig. S1D). Moreover, in addition to STAT1 phosphorylation (the substrate), we also observed signal for CDK8 phosphorylation, the extent of which varied dependent on the exact assay conditions and the used substrate (SI Appendix, Fig. S1D). Finally, using the aforementioned assay, we systematically truncated the MED12 N terminus. This allowed us to confine the minimal stable fragment sufficient for CDK8 activation to 46 amino acids at the MED12 N terminus (MED12 residues 23 to 69) (Fig. 1F and SI Appendix, Fig. S1E).
MED12 Makes Extensive Contacts to CDK8 without Substantially Contacting Cyclin C.
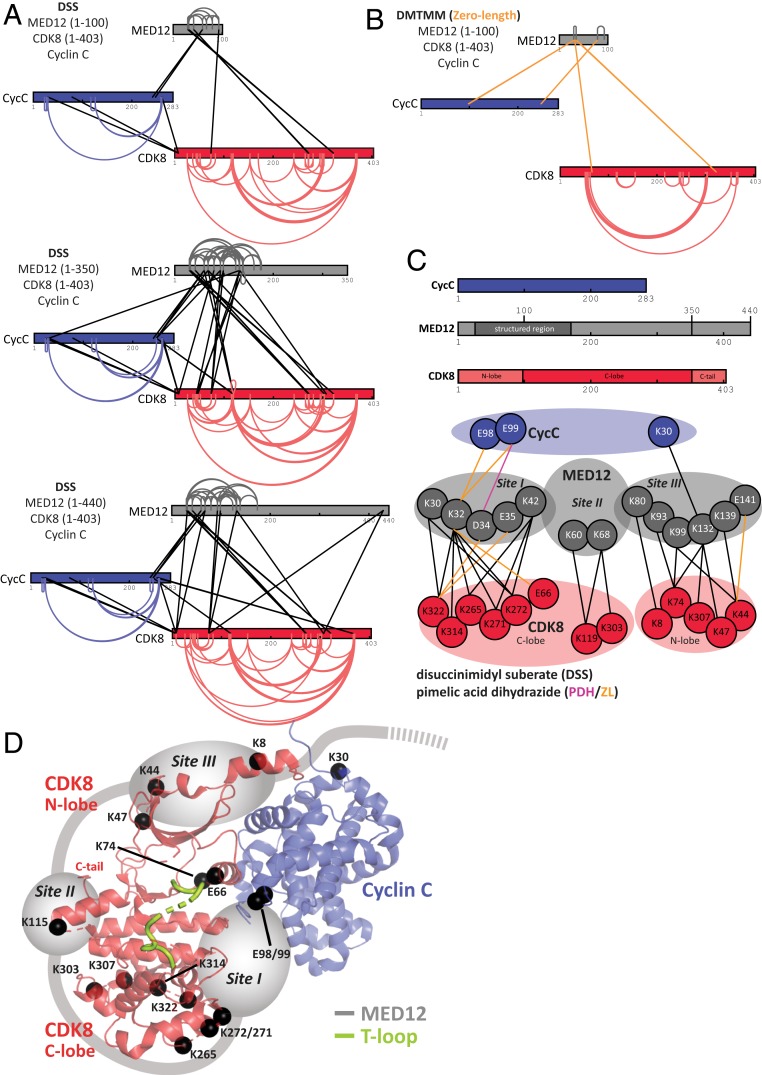
Prior work on the mechanism of CDK8 activation by MED12 identified a surface groove on Cyclin C that was suggested to constitute the MED12 binding site necessary for CDK8 activation (19, 20, 22). However, how such a distant MED12 binding site would activate CDK8 is controversial. Therefore, to determine the location of MED12 in ternary complexes, we carried out protein cross-linking coupled to mass spectrometry (XL-MS) (22). On the one hand, we utilized both 0.125 and 0.25 mM disuccinimidyl suberate (DSS) to form lysine–lysine cross-links in ternary CDK8 (1–403)/Cyclin C/MED12 complexes with varying lengths of MED12 (Fig. 2A). On the other hand, we obtained an independent set of distance restraints for CDK8 (1–403)/Cyclin C/MED12 (1–100) from carboxyl-specific and zero-length restraints introduced by the addition of 3.6 mg/mL pimelic acid dihydrazide (PDH) and 4.8 mg/mL coupling reagent DMTMM (4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride) (Fig. 2B and SI Appendix, Fig. S2C) (23). To properly assess the influence of MED12 binding on CDK8/Cyclin C, we also carried out DSS XL-MS for binary CDK8/Cyclin C complexes (SI Appendix, Fig. S2A).
Fig. 2.
The N terminus of MED12 wraps around CDK8 and contacts its T-loop. (A) Schematic representation of all DSS-mediated cross-links obtained from ternary CDK8 (1–403)/Cyclin C/MED12 (1–100), CDK8 (1–403)/Cyclin C/MED12 (1–350), and CDK8 (1–403)/Cyclin C/MED12 (1–440) constructs using two different concentrations of DSS. Intramolecular cross-links are colored according to the individual protein subunits with CDK8 in red, Cyclin C in blue, and MED12 in gray. Intersubunit cross-links are shown as black lines. The endpoint of each line specifies a specific residue in the corresponding protein. All cross-links can be found in Dataset S1. Cross-linking data were displayed with help of xiVIEW (58). (B) Schematic representation of all cross-links detected after treatment of ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) with PDH and DMTMM. Intramolecular cross-links are colored as in A. Intersubunit cross-links are shown as orange lines. All cross-links can be found in Dataset S2. (C, Upper) Cyclin C, MED12 (1–440) and CDK8 (1–403) with different protein domains highlighted. (Lower) A cartoon depiction of all intersubunit cross-links involving MED12. The intersubunit cross-links were grouped in three interaction sites (sites I to III) according to their localization on the surface of the binary CDK8/Cyclin C complex. (D) Three-dimensional arrangement of the three major sites of cross-links between MED12 and CDK8 (1–403)/Cyclin C. Lysine and glutamate residues that were found cross-linked are represented as black spheres and plotted onto the structure of the CDK8 (1–403)/Cyclin C complex (PDB ID code 3RGF) (18). Please note that K115 is shown instead of the structurally unresolved, cross-linked residue K119.
DSS-mediated cross-linking of CDK8 (1–403)/Cyclin C/MED12 (1–100) resulted in the identification of 8 intersubunit cross-links and 45 intrasubunit cross-links using a DSS concentration of 0.125 mM (Fig. 2A and SI Appendix, Fig. S2B). A DSS concentration of 0.25 mM resulted in 6 intersubunit and 33 intrasubunit cross-links (Fig. 2A and Dataset S1). These numbers increased for ternary complexes encompassing MED12 (1–350) and MED12 (1–440) (SI Appendix, Fig. S2B). The average length for all DSS cross-links is 15.8 Å, falling well into the reported range for DSS cross-link distances (calculated using PDB ID code 5BNJ) (23, 24). The carboxyl-specific cross-linker was only successful for the CDK8 (1–403)/Cyclin C/MED12 (1–100) complex. It allowed for the identification of one intersubunit acidic cross-link and four intrasubunit acidic cross-links (SI Appendix, Fig. S2C and Dataset S2). Moreover, DMTMM catalyzed the formation of 6 intersubunit and 52 intrasubunit cross-links with zero-length distance (Fig. 2B). These cross-links are extremely valuable due to their high distance precision (23).
First and foremost, we detected 25 high-confidence cross-links of MED12 to CDK8, but only 4 cross-links of MED12 to Cyclin C (although Cyclin C contains many cross-linkable residues). This strongly suggests that MED12 primarily contacts CDK8, not Cyclin C. In addition, the number of intrasubunit cross-links is greatly diminished once MED12 binds to binary CDK8/Cyclin C complexes (Fig. 2A and SI Appendix, Fig. S2A). Next, we analyzed the distribution of all intersubunit cross-links involving MED12. This revealed three major sites of interaction between MED12 and CDK8/Cyclin C (Fig. 2C). The first site comprises MED12 residues 30 to 42, which interact with five residues in the C-lobe of CDK8 (K322, K314, K272, K271, K265). Interestingly, these five residues are located in an area of conserved protein–protein interactions across the CDK family (18, 25). Moreover, we also found this part of MED12 (30–42) to contact the αC helix of CDK8 (E66) and two acidic residues in Cyclin C (E98, E99). Interestingly, E99 in Cyclin C was hypothesized to be involved in CDK8 activation (18, 26). Taken together, these cross-links place residues 30 to 42 of MED12 between CDK8 and Cyclin C, in proximity to the activation segment of the T-loop of CDK8 whose structure was not resolved in the binary complex (18). The second site of MED12 interaction with CDK8 involves contacts between MED12 residues K60 and K68 and CDK8 residues K119 and K303 (Fig. 2C). This contact site is located close to the C terminus of CDK8, which was found to be disordered by X-ray crystallography (18). In support of this contact, we note that we found slightly higher affinities of MED12 (1–100) for full-length CDK8 than for CDK8 (1–403) (Fig. 1B). This indicates a weak contribution of the CDK8 C terminus to MED12 binding. A third site of intersubunit cross-links involving MED12 encompasses MED12 residues K80 through E141. Except for K30 on Cyclin C, all of these residues establish cross-links to the N-lobe of CDK8. Interestingly, all MED12 cross-links that are part of the third interaction site are situated on the surface of CDK8 that is oriented away from its active site, explaining how MED12 can wrap around CDK8 without obstructing substrate binding. To exclude that the C terminus of CDK8 alters the cross-linking patterns, we repeated the XL-MS experiments using DSS chemistry with binary and ternary complexes comprising full-length CDK8 [CDK8 (1–464)] (SI Appendix, Fig. S2D). Our results revealed unchanged cross-linking patterns, hence demonstrating that the CDK8 C terminus does not impact the structure of the CDK8/Cyclin C/MED12 complex. Lack of cross-linkable residues in the CDK8 C terminus did not allow to determine its exact position in the complex.
Taken together, our cross-linking data suggest a revised model of how MED12 binds the binary CDK8/Cyclin C complex (Fig. 2D). The N-terminal portion of MED12 wraps around the entire CDK8 molecule, making extensive contacts to both its C- and N-lobe (Fig. 2 C and D). Most interestingly, our data allow us to place a stretch of about 10 amino acids (MED12 residues 30 to 42) in immediate proximity of the T-loop of CDK8. This suggests that MED12 directly contacts the T-loop of CDK8, thereby resolving how MED12 is able to activate CDK8 in the absence of T-loop phosphorylation. The second and third site of interaction between MED12 and CDK8 illustrate that the N terminus of MED12 folds around the entire CDK8 molecule (Fig. 2 C and D). K139 through E141 are the last residues of MED12 for which we detected cross-links to the CDK8/Cyclin C complex. Furthermore, our cross-linking data of ternary complexes comprising MED12 constructs of various length suggest that the N terminus of MED12 is only structured until residue 174 (Fig. 2A). Beyond this residue, we neither detect intra-MED12 cross-links nor cross-links of MED12 to CDK8 or Cyclin C, hinting at the possibility that, beyond residue 174, MED12 is disordered in ternary complexes that lack MED13.
An Activation Helix in MED12 Is Essential for CDK8 Activation.
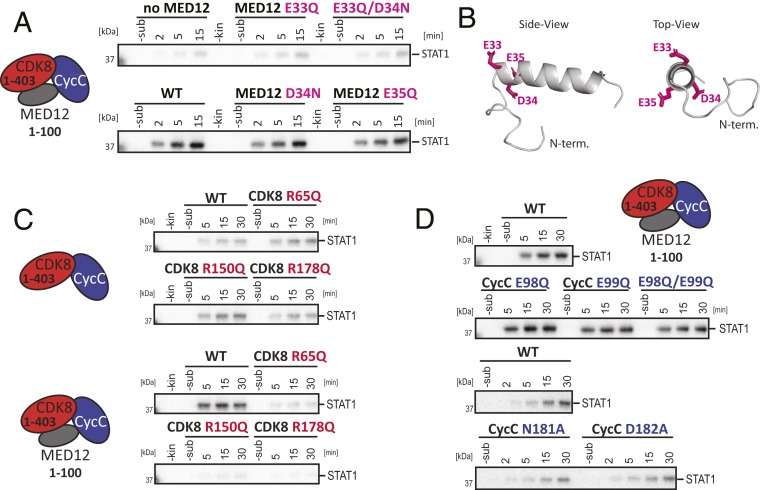
Our cross-linking data place MED12 residues 30 to 42 in vicinity of the T-loop of CDK8, suggesting that they are crucial for kinase activation. Interestingly, secondary structure prediction clearly hinted at the formation of an α helix for MED12 residues 32 through 44. To assess the importance of this α-helix for CDK8 activation, we systematically mutated residues that are part of the helix. In particular, we were intrigued by a cluster of three acidic residues (E33, D34, and E35) at the predicted N-terminal tip of the helix that stabilize the positive helical dipole at this position. Much to our surprise, an E33Q mutation completely abolished CDK8 activation by MED12 (Fig. 3 A and B). This was also the case for double-mutants involving E33. In contrast, neither the mutation of D34 nor of E35 had an effect on kinase activation. To exclude the direct involvement of other charged residues in CDK8 activation, we measured the effect of K30A, Q31A, and K32A mutations on kinase activation. We found none of them to influence CDK8 activation (SI Appendix, Fig. S3A). We thus conclude that MED12 E33 is essential to activate CDK8, prompting us to term the helix that harbors E33 at its tip “activation helix.” We note here that we were able to purify all ternary, mutation-containing complexes to homogeneity, demonstrating that MED12 binding to CDK8/Cyclin C and CDK8 activation can be experimentally separated.
Fig. 3.
MED12 utilizes an activation helix to stimulate the activity of CDK8. (A) In vitro kinase assays using ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes that harbor point-mutations within MED12. Kinase assays were carried out with the STAT1 TAD as described in Fig. 1F. WT denotes wild-type MED12. (B) Three-dimensional model of the MED12 activation helix comprising MED12 residues 19 to 50. The model was calculated using PEP-FOLD3 (59). The negatively charged triad of amino acids at the N-terminal tip of the activation helix (E33, D34, and E35) is shown as sticks in pink. (C) In vitro kinase assays using binary CDK8 (1–403)/Cyclin C and ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes. Each complex harbored a mutation in the CDK8 arginine triad. Kinase assays were carried out with the STAT1 TAD as a substrate as in Fig. 1F. (D) In vitro kinase assays using ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes that harbor point-mutations within Cyclin C. The activity of wild-type ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) is shown twice since the experiments were analyzed on separate gels. Kinase assays were carried out with the STAT1 TAD as a substrate as described in Fig. 1F.
We next asked how the activation helix is able to bind in the interface of CDK8 and Cyclin C. To that end we calculated the electrostatic surface potential of the binary CDK8/Cyclin C complex and of a model of the MED12 activation helix (SI Appendix, Fig. S3B). We noticed a basic patch at the interface of CDK8 and Cyclin C in the same region where we had detected cross-links of MED12 to CDK8. This warrants our speculation that the acidic triad E33-D34-E35 is responsible for positioning the MED12 activation helix properly for CDK8 activation.
Glutamate-33 Likely Positions the Arginine Triad of CDK8 and Thereby Activates the Kinase.
Having discovered a helix in MED12 that is crucial for its CDK8 activation potential, we next asked which CDK8 residues contact E33 in MED12. Our quest was guided by a comparison of CDK8 with a CDK homolog, the phosphate-dependent signaling complex Pho85/Pho80. In this complex, a salt bridge between R132 on the C lobe of Pho85 (the CDK) and D136 on Pho80 (the Cyclin) locks the activation loop of Pho85, thereby circumventing the requirement for T-loop phosphorylation (27). A similar phosphorylation-independent mechanism was observed for CDK5/p25. There, p25 tethers the unphosphorylated T-loop of CDK5 in an active conformation (28).
Just like all human CDKs, CDK8 possesses an arginine triad (R65, R150, and R178). However, as it lacks a phosphorylated T-loop residue that could coordinate these arginines, we asked whether one of the arginine residues instead contacts E33 in MED12. To that end, we prepared individual arginine mutants (R65Q, R150Q, and R178Q) of CDK8 and tested those in complex with Cyclin C and in ternary MED12-containing complexes (Fig. 3C). If one of the three arginine residues indeed contacts E33 in MED12, we expected to see no effect of this mutation on the basal kinase activity of the binary CDK8/Cyclin C complex. In contrast, we predicted the abrogation of MED12-dependent CDK8 activation by such mutation. This is exactly what we detected for all three arginine mutants (Fig. 3C). However, we were unable to detect significant and reproducible differences between the three arginine mutations, despite the fact that R65 is located in the αC helix of CDK8 and R150 and R178 are located in its T-loop. To exclude that the individual arginine mutants impair MED12 binding to CDK8/Cyclin C, we measured the affinity of MED12 (1–100) for binary CDK8 (1–403)/Cyclin C complexes carrying individual arginine mutations (R65Q, R150Q, or R178Q) by MST without detecting major changes in the affinity of MED12 for CDK8/Cyclin C (SI Appendix, Fig. S3C).
In summary, our data establish that both the CDK8 arginine triad and E33 of MED12 are essential for MED12-dependent CDK8 activation. Whether the active conformation of the CDK8 T-loop is induced by a direct salt bridge between E33 of MED12 and one of the members of the CDK8 arginine triad will require high-resolution structural information on the ternary CDK8/Cyclin C/MED12 complex.
Cyclin C Is Not Directly Involved in MED12-Dependent CDK8 Activation.
Cyclin binding triggers the repositioning of the αC helix in cyclin-dependent kinases, thereby allowing for the formation of their active site (29, 30). This mechanism also applies to binding of Cyclin C to CDK8 (18). In addition to this conserved role, residue E99 in Cyclin C and an unusually deep surface groove between its two cyclin boxes were also suggested to contribute to CDK8 activation (16, 17, 23, 26). We sought to confirm these results by using our in vitro kinase assay system. However, to our surprise neither an E98Q or E99Q mutation, nor the mutation of two surface groove residues (N181A, D182A) on Cyclin C that were suggested to bind MED12 (19, 31) had an impact on MED12-driven CDK8 activation (Fig. 3D). Using nanoDSF measurements, we are able to show that both Cyclin C surface mutants (N181A, D182A) destabilize ternary MED12-containing complexes (SI Appendix, Fig. S3D). Taken together, our functional and structural data suggest that the Cyclin C surface groove is not directly involved in the MED12-dependent activation of CDK8. Rather, upon mutation of residues in the surface groove, the entire complex becomes destabilized and thereby leads to reduced kinase activity under specific experimental conditions (19). Alternatively, the surface groove might contribute to substrate recognition or serve as a protein interaction site, as is the case for CDK2/Cyclin A (32, 33). In conclusion and despite the fact that we detected cross-links for Cyclin C E98 and E99 to the MED12 activation helix (Fig. 2C), Cyclin C seems only of minor importance for the MED12-driven activation of CDK8.
Malignant MED12 Mutations Retain Their Affinity for CDK8, yet Abolish MED12-Dependent CDK8 Activation.
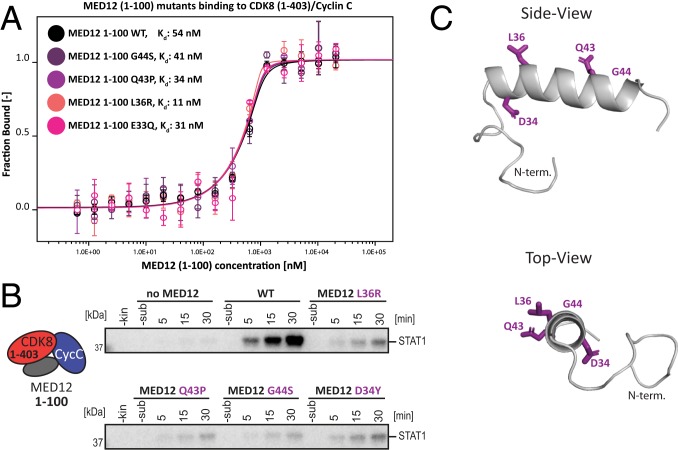
Frequent somatic MED12 mutations are associated with, among others, uterine leiomyomas, tumors of the breast, and chronic lymphocytic leukemia (34–37). Despite the fact that MED12 consists of 45 exons, the overwhelming majority of these mutations are found in MED12 exon 2 (amino acids 34 to 68) and to a lesser extent in exon 1 (amino acids 1 to 33) (34). Strikingly, when we mapped the mutations occurring in chronic lymphocytic leukemia (34) onto the sequence of MED12 (Fig. 2D), we found that the patient-derived mutations perfectly mirror the activation helix that we describe to be essential for CDK8 activation by MED12 (Figs. 3B and 4C). As the mutational analysis of the activation helix uncovered only a single amino acid to be critical for the CDK8-stimulatory function of MED12, we next examined the functional consequences of recurring disease mutations in MED12. First, we measured the affinity of MED12 (1–100) fragments carrying L36R, Q43P, or G44S mutations for binary CDK8 (1–403)/Cyclin C complexes. All three residues (L36, Q43, and G44) are mutational hotspots in uterine leiomyomas and were also found in chronic lymphocytic leukemia (34, 37). In contrast to previously published results, we did not detect significantly weakened binding of MED12 to binary complexes when compared to wild-type MED12 (1–100) (Fig. 4A) (19). The same holds true for MED12 carrying an E33 mutation (Fig. 4A). This raised the possibility that MED12 mutations that occur in different cancers might not influence MED12 binding to CDK8/Cyclin C, only its activation. To test this, we purified ternary complexes containing mutated MED12 variants (L36R, Q43P, and G44S) and measured their potential to activate the CDK8 kinase. As expected, for all complexes kinase activation was abolished or at least drastically reduced (Fig. 4B). Interestingly, the nature of the mutation has a profound impact on the activation potential of MED12. Whereas the mutation of Asp-34 to Tyr (D34Y), which is found in uterine leiomyomas, drastically reduces CDK8 kinase activity, a D34N mutation has no impact on kinase activity in our in vitro system (Figs. 3A and 4B). Taken together, we find that mutations in MED12 that are found in cancer patients lead to an abrogation of CDK8 kinase activation without altering the affinity of MED12 for CDK8/Cyclin C.
Fig. 4.
Cancer-associated mutations within the MED12 activation helix abolish CDK8 activation without altering MED12 affinity for CDK8/Cyclin C. (A) MST binding experiments using MED12 (1–100) variants carrying different cancer-associated mutations and binary CDK8/Cyclin C complexes. MED12 (1–100) E33Q serves as activation-dead control (Fig. 3A). Kd values are indicated. Error bars reflect the SD of four replicates. Please note that Kd cannot be read off directly due to the experimental necessity to use high-protein concentrations. (B) In vitro kinase assays using ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complexes harboring individual cancer-associated mutations within MED12. Kinase assays were carried out with the STAT1 TAD as a substrate as in Fig. 1F. (C) Three-dimensional model of the MED12 activation helix comprising MED12 residues 19 to 50. The model was calculated using PEP-FOLD3 (59). MED12 residues frequently mutated in cancer are shown as violet sticks (D34Y, L36R, Q43P, and G44S).
Type II Kinase Inhibitors Show Reduced Efficacy toward Ternary CDK8/Cyclin C/MED12 Complexes.
Due to its prominent oncogenic role, considerable efforts were made to develop CDK8-specific inhibitors (17, 21, 37, 38). As we discovered that MED12 employs an activation helix to stimulate CDK8 activity, we asked whether the presence of MED12 alters the efficacy of commercially available inhibitors, all of which were developed against the binary CDK8/Cyclin C complex (38). We suspected that this might be the case because we found MED12 to contact the αC helix of CDK8 (Fig. 2). Moreover, MED12 likely positions the CDK8 T-loop in a catalytically competent conformation by contacting one or several members of its arginine triad (Fig. 3). In further support of a differential efficacy of commercial inhibitors against MED12-bound CDK8 complexes, we note that type II inhibitors, such as sorafenib, were previously reported not to translate their inhibitory efficacy into the cellular context (21).
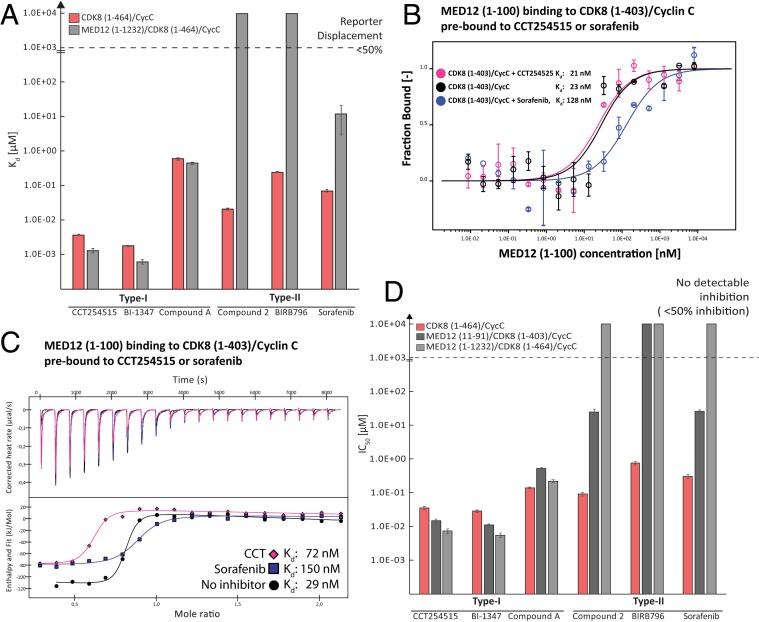
To test our hypothesis, we selected three type I kinase inhibitors: Compound A (PDB ID code 6T41) (SI Appendix, Fig. S4), CCT251545 (PDB ID code 5BNJ), and BI-1347 (BI-1347 was chosen as a type I inhibitor due to its chemical similarity to CCT251545 and its high in vivo potency [opnMe Boehringer Ingelheim Open Innovation Portal]). Moreover, we chose three known type II inhibitors: Sorafenib (PDB ID code 3RGF), compound 2 (PDB ID code 4F7L), and BIRB796 (39), and measured the dissociation constants and IC50 values of both classes of inhibitors toward a binary CDK8 (1–403)/Cyclin C and a ternary CDK8 (1–403)/Cyclin C/MED12 (1–100) complex (Fig. 5A and SI Appendix, Fig. S4A). To that end we used a reporter displacement assay that we had developed for CDK8/Cyclin C previously (40). In analogy to the reporter probe (SI Appendix, Fig. S4B), all type I inhibitors displayed similar affinities for both the binary and the ternary CDK8 complex (Fig. 5A and SI Appendix, Fig. S4A). However, strikingly, all type II inhibitors showed a significantly diminished affinity for the ternary CDK8 complex. Only sorafenib (a type II inhibitor) displaced the reporter probe by more than 50%, albeit with an affinity decreased by two orders-of-magnitude (Fig. 5A and SI Appendix, Fig. S4A). These results indicate that type II kinase inhibitors, which bind to the CDK8 hinge region and extend to the so-called deep pocket (18), are hindered from binding to CDK8 when MED12 is present. The deep pocket is only accessible when CDK8 is in its inactive state (DMG-out conformation of CDK8), suggesting that MED12 binding to CDK8 induces a DMG-in–like conformation that renders type I kinase inhibitors more potent once CDK8 is part of the entire Mediator kinase module.
Fig. 5.
Type II kinase inhibitors lose part of their inhibitory potential when used against ternary CDK8/Cyclin C/MED12 complexes. (A) Reporter displacement assay (RDA) with three type I kinase inhibitors (CCT251545, BI-1347, and compound A) and three type II inhibitors (compound 2, BIRB 976, and sorafenib). RDAs were measured using the full-length binary CDK8/Cyclin C complex and a ternary CDK8/Cyclin C/MED12 (1–1232) complex. Kd values of individual compounds were calculated using the Cheng–Prussoff equation as described previously (40). For clarity, Kd values of compound showing a probe displacement of less than 50% were set to 10 mM. (B) MST binding experiments using MED12 (1–100) and 2 nM binary CDK8 (1–403)/Cyclin C complexes that were either preloaded with 30 µM of the type I inhibitor CCT251545 or with 30 µM of the type II inhibitor sorafenib. Apo CDK8 (1–403)/Cyclin C was used as a control. Kd values are indicated. Error bars reflect the SD of two replicates. (C) Using ITC, the MST measurements from (B) were confirmed, Kd values are indicated. (Upper) The corrected heat rates; (Lower) The calculated enthalpies per injection are plotted against the molar ratio of ligand and target. Please note that CCT251545 addition resulted in some protein precipitate, which altered the observed molar ratio. (D) IC50 values for a binary CDK8 (1–464)/Cyclin C complex and two ternary complex variants [CDK8 (1–403)/CyclinC/MED12 (11–91), and CDK8 (1–464)/Cyclin C/MED12 (1–1232)] using the same set of inhibitors as in A and ADP-Glo data. IC50 values of individual compounds were calculated using a standard XLfit algorithm (SI Appendix, Fig. S6). For clarity, IC50 values of compounds showing an enzymatic inhibition of less than 50% were set to 10 mM.
To validate this conclusion, we next used MST to measure the affinity of the MED12 (1–100) fragment for binary CDK8 (1–403)/Cyclin C complexes in presence of saturating levels of a type I (CCT251545) or a type II inhibitor (sorafenib). The affinity of MED12 (1–100) for CDK8 (1–403)/Cyclin C prebound to CCT251545 was about 21 nM, just like for the apo CDK8 (1–403)/Cyclin C control. In contrast, a binary complex prebound to the type II inhibitor sorafenib showed a fivefold reduced affinity for MED12 with a Kd of about 130 nM (Fig. 5B). In further support of these results, we used ITC to, yet again, find a fivefold reduction in the affinity of MED12 (1–100) for CDK8 (1–403)/Cyclin C prebound to sorafenib as compared to prebinding of CCT251545, which only showed a minor reduction of MED12 affinity for the binary CDK8 complex (Fig. 5C). We note here that a fivefold reduction in MED12 affinity for the binary CDK8 complex is highly significant, as we do not expect the inhibitors to interfere with MED12 interaction sites II+III (Fig. 2D). Hence, we expected type II inhibitor prebinding to only reduce MED12 affinity for CDK8 (1–403)/Cyclin C, which is precisely what we observed (Fig. 5 B and C).
Finally, we sought to corroborate our binding data by comparing the enzymatic activity of binary full-length CDK8 (1–464)/Cyclin C with a minimal [CDK8 (1–403)/Cyclin C/MED12 (11–91)] and a longer ternary complex variant [CDK8 (1–464)/Cyclin C/MED12 (1–1232)]. To achieve this, we utilized the ADP-Glo technology (Promega) to measure CDK8 activity. We limited our measurements to CDK8 phosphorylation rates here, as the inclusion of substrates rendered the ADP-Glo data hard to interpret due to the presence of multiple CDK8 substrates. This strategy is valid as we had previously shown that CDK8 serves as an alternative, second substrate in kinase assays (SI Appendix, Fig. S1D). In analogy to the reporter displacement assay, the ADP-Glo data reveal that all tested type I kinase inhibitors abrogate CDK8 activity for both binary and ternary complexes with similar IC50 concentrations (Fig. 5D and SI Appendix, Fig. S6). In contrast, type II compounds only inhibited CDK8 when part of the binary complex, yet they showed significantly decreased inhibitory potential versus ternary MED12-bound CDK8 complexes (Fig. 5D). In more detail, for type II inhibitors used against ternary MED12-containing complexes, we observed incomplete IC50 curves with a maximal inhibition of 50% or significantly lower (Fig. 5D and SI Appendix, Fig. S7). Taken together, our data demonstrate that type II kinase inhibitors show drastically reduced inhibitory potential toward ternary MED12-bound CDK8 complexes. We did not observe this loss of efficacy for type I inhibitors, which prompts us to speculate that binding of MED12 induces a DMG-in conformation in the active site of CDK8. Thereby access to the deep pocket is blocked, precluding type II inhibitors from binding to CDK8. Moreover, the fact that we found similar enzymatic characteristics for ternary CDK8 complexes containing a small (11 to 91) or a longer MED12 variant (1 to 1232) (Fig. 5D) further supports the MED12-binding mode presented (Fig. 2D). Our model postulates that the CDK8 activation potential resides in the very N-terminal portion of MED12 (amino acids 23 to 69) (Fig. 1F) with only a marginal contribution of other parts of MED12.
Gene Expression Changes upon Mutation of MED12 E33 Phenocopy Transcriptome Changes in Human Cancers.
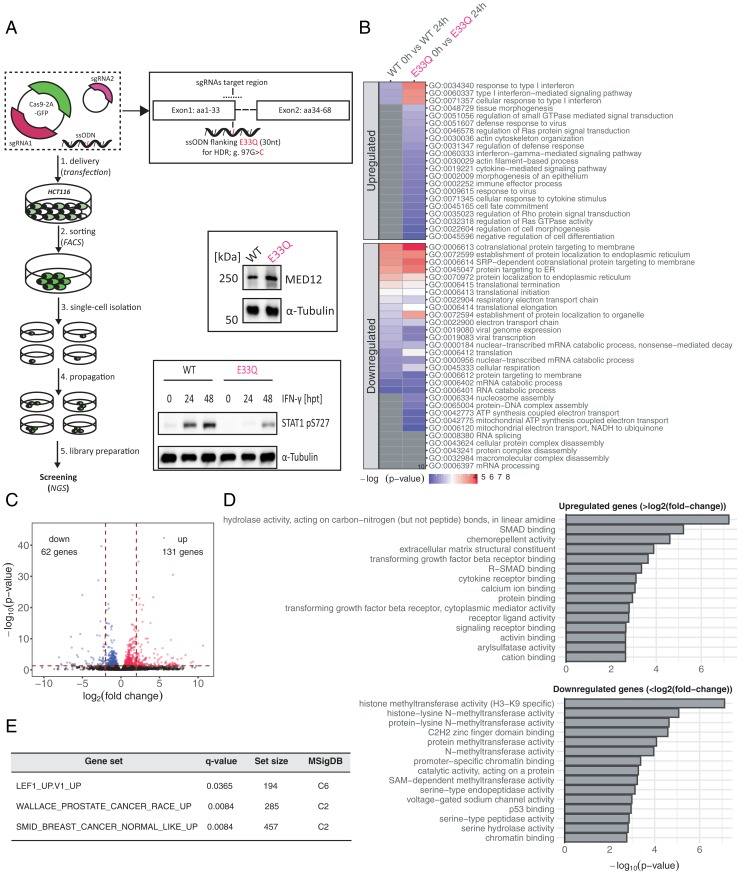
CDK8 was shown to be involved in Wnt/β-catenin–activated transcription, TGF-β signaling, and p53-signal transduction, all of which are pathways important for oncogenesis and cancer progression (15, 41, 42). However, none of these studies was able to address the functional share of MED12-dependent CDK8 activation. Therefore, given our discovery that a single amino acid (MED12 E33) is necessary for MED12-dependent CDK8 activation, we set out to generate a MED12 E33Q knockin in HCT116 colon cancer cells using the CRISPR/Cas9 system (Fig. 6A). We successfully obtained a stable HCT116 cell line carrying a MED12 E33Q mutation (SI Appendix, Fig. S7A). Besides the desired E33Q mutation, we also observed a K15N mutation in the generated cell line. However, this mutation neither altered MED12 expression levels nor its subcellular localization or its kinase activity (Fig. 6A and SI Appendix, Fig. S7 B and C). In order to enhance the magnitude of potential genome-wide gene expression changes, we decided to stimulate the HCT116 cells with IFN-γ prior to RNA isolation. We chose IFN-γ stimulation since its impact on expression changes of IFN-response genes relies on STAT1 phosphorylation by CDK8 (43, 44). In good agreement with our in vitro data, upon IFN-γ treatment we observed a reduction in STAT1 phosphorylation levels in the mutant (E33Q) cell lines as compared to HCT116 wild-type cells (Fig. 6A). Next, we prepared strand-specific libraries from HCT116 wild-type and E33Q cells in untreated condition and after 24 h of IFN-γ stimulation (Fig. 6B) (45). The subsequent transcriptome-wide gene-expression analysis uncovered the up-regulation of IFN response genes in both wild-type and E33Q cell lines upon IFN-γ stimulation (Fig. 6B). Moreover, when comparing E33Q cells to wild-type we found additional biological processes, such as tissue morphogenesis and innate immunity, to be affected by IFN-γ treatment. In support of this finding, >40% of all IFN-γ–responsive genes are known to be regulated by CDK8-mediated STAT1 phosphorylation, thus identifying CDK8 as an important regulator of antiviral responses (46). Furthermore, our data confirm that IFN-γ stimulation results in a significant down-regulation of translation and translation-related processes (Fig. 6B) (47).
Fig. 6.
Loss of MED12-dependent CDK8 activation resembles transcriptional profiles of human cancers. (A) Schematic representation of the employed CRISPR workflow to generate an HCT116 cell line carrying a MED12 E33Q mutation. The presence of MED12 in the MED12 E33Q mutant cell line was verified by Western blotting. The phosphorylation of Ser-727 in the STAT1 transactivation domain upon IFN-γ induction was also analyzed by Western blotting. (B) Heatmap showing coregulated (up-regulated and down-regulated) GO-terms in wild-type HCT116 (WT) and E33Q knockin (KI) HCT116 cells 24 h after IFN-γ treatment. (C) Volcano plot showing log2 fold changed up- and down-regulated genes 24 h after IFN-γ treatment. Wild-type HCT116 cells are compared to MED12 E33Q mutant cells. The dashed horizontal red line represents a P value threshold (P < 0.05). The two dashed vertical lines illustrate the threshold of log2 fold-changes >2 or <−2. (D) GO term enrichment analysis of up- (>log2 fold) and down-regulated (<log2 fold) genes. (E) GSEA of expression signatures that are up-regulated in MED12 E33Q knockin HCT116 cells. C2 is a collection of curated gene sets in MSigDB. C6 represents oncogenic signatures of cellular pathways as part of MSigDB.
Subsequently, we analyzed genes that were differentially expressed between wild-type and E33Q mutant cells (Dataset S3). We identified 830 such genes; 131 genes were significantly up-regulated (>log2 fold), whereas we found 62 to be down-regulated (<log2 fold) (Fig. 6C). Gene ontology (GO) term enrichment analysis revealed that down-regulated genes are involved in the modification of histones, chromatin binding, and p53 signaling, whereas up-regulated genes participate in TGF-β/SMAD signal transduction (Fig. 6D). Next, we performed a gene set enrichment analysis (GSEA) against curated gene sets (C2) and oncogenic gene sets (C6) that are part of the Molecular Signatures Database (MSigDB) (48, 49). To our surprise, we found that genes up-regulated in our MED12 E33Q cell line correlated with genes up-regulated in DLD1 colon carcinoma cells that undergo epithelial-mesenchymal transition (EMT) (Gene Set: LEF1_UP.V1_UP) (Fig. 6E and SI Appendix, Fig. S7D). Moreover, genes up-regulated in E33Q cells showed a significant overlap with genes associated with breast and prostate cancer (Fig. 6E and SI Appendix, Fig. S7D) (50, 51).
Altogether, our transcriptome-wide gene-expression data in HCT116 cells indicate that the reduction of CDK8 activity upon loss of the stimulatory activity of MED12 has profound effects on gene expression. It likely contributes to the oncogenic progression in patients carrying mutated alleles of MED12 and negatively impacts the innate immune response.
Discussion
A Revised Model of How MED12 Activates CDK8.
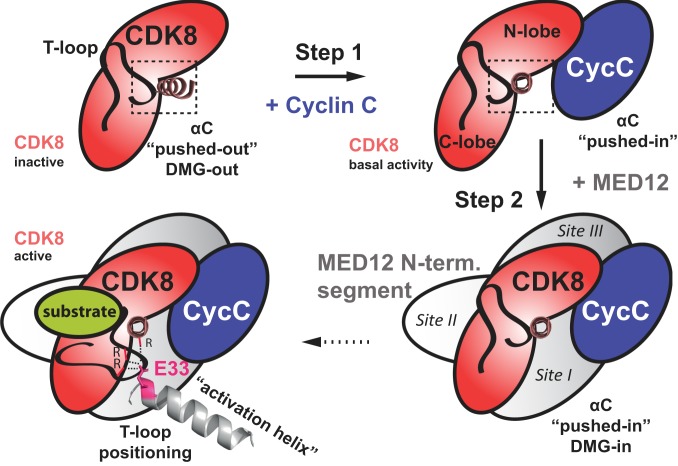
In taking together our in vitro and in vivo data, we propose a model of how MED12 activates CDK8 (Fig. 7). In a first step, CDK8 is bound by Cyclin C, which leads to a “pushed-in” conformation of the αC-helix of CDK8, generating the kinase active site (18). At this step CDK8 inhibitors were shown to trap the kinase in both a DFG-in conformation (type I) as required for catalysis (PDB ID code 5BNJ) (21) or in a DFG-out conformation (type II) that impedes catalysis and engages the deep pocket of the kinase (PDB ID code 3RGF) (18) (SI Appendix, Fig. S8). We envision that at this step the active site of the kinase can exhibit both a DFG-in, as well as a DFG-out conformation. Next, the N terminus of MED12 wraps around CDK8 and places its “activation helix” right next to its T-loop. Our mutational data indicate that the exact placement of the helix is crucial for activation of CDK8 by MED12. Binding alone is insufficient (Figs. 3 and 4). Moreover, our data suggest that MED12 wrapping around CDK8 is important for its activity. The minimal fragment of MED12, for which we were able to isolate a stable, active ternary complex, comprised MED12 residues 23 to 69 (Fig. 1C). This correlates with cross-linking sites I and II (Fig. 2 C and D) and suggests that MED12 has to at least bind to these two sites for a stable association with CDK8. In more detail, our data suggest a direct contact between the arginine triad of CDK8 and E33 in MED12 (Fig. 3 A and D). In support of this contact, we also detected a zero-length cross-link between K32 in MED12 and E66 in the αC-helix of CDK8 (Fig. 2C). K32 is right next to E33, the residue critical for MED12 function, and E66 is close to R65, one for the three members of the CDK8 arginine triad. Since E66 is pointing inside the CDK8 molecule in binary CDK8/Cyclin C complexes this suggest a further rearrangement of the αC-helix upon MED12 binding to CDK8 (SI Appendix, Fig. S8). Finally, we hypothesize that the contact between the arginine triad and the activation helix of MED12 leads to a stably “folded-away” conformation of the CDK8 T-loop, allowing unobstructed substrate binding to the active site.
Fig. 7.
Revised model of how MED12 activates CDK8. Step 1: Cyclin C binds to CDK8 and pushes the αC-helix of CDK8 into the “pushed-in” conformation. This binding event is crucial for the formation of the active site of CDK8 and results in basal kinase activity as demonstrated in Fig. 1E. Step 2: MED12 binding to CDK8/Cyclin C stabilizes and activates the entire ternary complex. In particular, an activation helix in MED12 contacts and stabilizes the T-loop of CDK8, thereby activating the kinase. Likely, this contact is established through an interaction of an acidic residue at the N-terminal tip of the MED12 activation helix (Glu-33) and the CDK8 arginine triad (Arg-65, Arg-150, and Arg-178). Moreover, MED12 binding favors the active site of CDK8 to adopt a DMG-in conformation to enhance its activity. The preference for a DMG-in conformation of the active site disfavors type II kinase inhibitors from binding and inhibiting CDK8 in ternary CDK8/Cyclin C/MED12 complexes.
A low-resolution cryoelectron microscopy map of the entire kinase module from Saccharomyces cerevisiae (52) was interpreted to show that MED12 contacts Cyclin C only, while our cross-linking, functional, and mutational data indicate the essential role of the MED12 N-terminal segment and the CDK8 αC-helix for contact formation. This apparent discrepancy may be due to the limited resolution (15 Å) of the cryoelectron microscopy map that cannot resolve the extended shape of the MED12 N-terminal segment of and its detailed interactions with the kinase.
Effective CDK8 Inhibitors Need to Be Developed Against MED12-Bound CDK8 Complexes.
Whereas class I kinase inhibitors inhibit both binary CDK8/Cyclin C and ternary CDK8/Cyclin C/MED12 complexes equally well, class II inhibitors lose a significant fraction of their inhibitory potential when utilized against the ternary CDK8/Cyclin C/MED12 complex (Fig. 5). This finding highlights a common problem in drug development: Whereas atomic resolution structures are needed for rational structure-based drug design, most drug targets are part of larger complexes, whose structures are unknown. Our study suggests that future drug development aimed at CDK8-specific drugs needs to focus on ternary, MED12-containing complexes. This is the case as our data suggest that MED12 induces—or at least favors—a DFG-in conformation of the active site of CDK8 (Fig. 7), thereby essentially precluding efficient binding of class II inhibitors. The fact that we found CCT251545 to be equally effective against both binary and ternary complexes is likely due to its discovery in cell-based SILAC (stable isotope labeling by/with amino acids in cell culture) pulldown assays (21). There, CDK8 is predominantly present in complex with MED12, which enabled the discovery of CCT251545 as potent inhibitor of ternary CDK8 complexes. Lastly, novel drugs intended at reducing CDK8 activity may also be developed by using small molecules that disrupt the interface between the MED12 activation helix and CDK8 (53). Such an approach would likely not impair the structural integrity of the Mediator kinase module and may only specifically target MED12’s role in activating CDK8.
Our data demonstrate that reducing CDK8 activity can have both positive and negative effects on target gene expression (Fig. 6). This is likely the result of the multitude of CDK8 targets and the differential functional outcome of target phosphorylation by CDK8 (11). For future drug development, it is therefore imperative to first decipher the exact role of CDK8 in the particular cancer under investigation and then to decide whether reducing CDK8 activity is the right route to take to impact the disease. This caution is warranted since MED12 was found to be involved in chemotherapy resistance without this finding having anything to do with its nuclear function as a CDK8 activator and part of the Mediator kinase module (54).
Materials and Methods
Expression and Purification of Binary CDK8/Cyclin C and Ternary CDK8/Cyclin C/MED12 Complexes.
To obtain binary CDK8/Cyclin C complexes, full-length Cyclin C (1–283) and different CDK8 variants (e.g., full-length [1 to 464] and C-terminally truncated CDK8 [1 to 403, 1 to 359]) were cloned into pAceBac1 and pUCDM vectors, respectively. To enable affinity purification, Cyclin C carried a C-terminal Strep-tag and a tobacco etch virus (TEV) cleavage site. Ternary complexes were constructed similarly. However, they contained the C-terminal Strep-tag on multiple N- or C-terminally truncated MED12 variants. Both binary CDK8/Cyclin C and ternary CDK8/Cyclin C/MED12 complexes were coexpressed in High5 insect cells by recombinant baculovirus infection via a titerless protocol. For large-scale production of desired complexes, 4 to 6 L High5 cells at a density of 1.25 × 106 cells/mL in serum-free SF-4 Baculo Express media (Bioconcept) were infected in Fernbach flasks. Flasks were incubated at 27 °C for 48 to 72 h on a rotary shaker before cells were collected by centrifugation and resuspended in buffer A (50 mM Hepes/NaOH [pH 7.4], 150 mM NaCl, 2 mM DTT) supplemented with protease inhibitors. Cells were lyzed by sonication and cell debris was removed by centrifugation at 35,000 rpm for 90 min in a Beckmann Ti-45 rotor. Supernatants were collected and bound to StrepTactin Superflow (IBA Lifesciences) resin followed by their elution with buffer A supplemented with 2.5 mM desthiobiotin. Cleavage of Strep-Tag fusions was performed overnight with TEV-protease. After cleavage, tag-free protein complexes were subjected to an anion-exchange column (ResourceQ, GE Healthcare). The flow-through after anion-exchange chromatography was subsequently loaded onto a cation-exchange column (Resource S, GE Healthcare) to recover the desired protein complexes. In a final step the protein complexes were polished by size-exclusion chromatography (SEC) using a Superdex 200 10/300 GL (GE Healthcare) or Superose6 10/300 GL (GE Healthcare) column in buffer B (50 mM Hepes/NaOH [pH 7.4], 100 mM NaCl, 2 mM DTT).
Chemical Cross-Linking and Sample Processing for Mass Spectrometry.
To optimize the cross-linking reaction conditions, pilot experiments were first carried out at small scale (5 μg protein) and monitored by SDS/PAGE. We selected conditions that resulted in an intensity reduction of the bands corresponding to monomeric subunits while at the same time preventing the formation of very high mass products. For the cross-linking reagent DSS, we chose two concentrations (125 and 250 μM). For the combination of PDH and the activating reagent DMTMM, we selected concentrations of 3.6 and 4.8 mg/mL, respectively. Final experiments for mass spectrometry were performed at a protein concentration of 1 mg/mL in cross-linking buffer (20 mM Hepes pH 7.4, 100 mM NaCl, 2 mM DTT/0.5 mM TCEP) and with 50 μg total protein. All cross-linking steps were carried out at 37 °C with mild shaking. DSS (a 1:1 mixture of d0- and d12-labeled forms, Creative Molecules) was added as a freshly prepared 25 mM stock solution in anhydrous dimethyl formamide. After incubation for 30 min, the reaction was stopped by adding ammonium bicarbonate to 50 mM. PDH (d0/d10) and DMTMM (both from Sigma-Aldrich) were added from freshly prepared stock solutions in sample buffer. After incubation for 45 min, the reaction was stopped by passing the sample solution through Zeba spin desalting columns (7 K molecular weight cutoff, ThermoFisher Scientific). For both chemistries, cross-linked samples were evaporated to dryness in a vacuum centrifuge. Further sample processing followed standard procedures for reduction of disulfide bonds (with TCEP), alkylation of free thiols (with iodoacetamide), and enzymatic digestion with endoproteinase Lys-C (Wako, 1:100, 37 °C for 2 to 3 h) and trypsin (Promega, 1:50, 37 °C overnight). Digests were purified by solid-phase extraction using SepPak tC18 cartridges (Waters), and purified peptide mixtures were enriched for cross-linked peptides using SEC fractionation on a Superdex Peptide PC 3.2/30 column (GE Healthcare), as described previously (55). Three to four SEC fractions were collected for LC-MS/MS analysis.
Liquid Chromatography-Tandem Mass Spectrometry.
LC-MS/MS was performed on an Orbitrap Fusion Lumos mass spectrometer coupled to an Easy nLC-1200 HPLC system via a Nanoflex electrospray source (all ThermoFisher Scientific). Peptides were separated by reversed-phase chromatography on an Acclaim PepMap RSLC C18 column (250 mm × 75 μm, ThermoFisher Scientific). Gradient elution was performed with the mobile phase A = water/acetonitrile/formic acid (98:2:0.15 [vol/vol/v]) and B = acetonitrile/water/formic acid (80:20:0.15 [vol/vol/v]) and a gradient from 11 to 40%B in 60 min at a flow rate of 300 nL/min. The mass spectrometer was operated in data-dependent acquisition mode using top-speed mode with a cycle time of 3 s. Full-scan mass spectra were acquired in the Orbitrap analyzer at 120,000 nominal resolution. Precursors with a charge state of +3 to +7 were selected for fragmentation with quadrupole isolation and an isolation window of 2.0 m/z. Fragmentation was performed using collision-induced dissociation in the linear ion trap and detected in the linear ion trap in rapid scan mode. Dynamic exclusion was activated for 30 s after one scan event.
In Vitro Kinase Assays.
All in vitro kinase assays were carried out with purified binary or ternary complexes. Reactions contained 1 to 5 pmol of CDK8 and 50 to 200 pmol of substrate in kinase buffer (25 mM Tris/HCl [pH 8.0], 100 mM KCl, 10 mM MgCl2, 0.1 mM EGTA, and 2 mM DTT). Kinase reactions were started by the addition of 1 µCi γ-[32P]ATP and subsequent incubation at 37 °C. Reactions were separated by SDS/PAGE. Gels were dried and exposed to a phosphoimaging plate for 0.5 to 2 h. Images were recorded with a CR 35 Bio Phosphoimager (Dürr Medical). Data were quantified with ImageJ and fitted with GraphPad Prism.
Generation of a MED12 E33Q Knockin Mutant in HCT116 Cells.
The HCT116 cell line carrying a MED12 E33Q knockin was generated using the CRISPR-Cas9 system. The ssODN contained phosphorothioate modifications and encompassed the nucleotide to be mutated (g.97 G→C) (56). It was designed using the CRISPR Design Tool from Dharmacon. HCT116 cells were cotransfected in six-well plates at 80 to 90% confluency with either PX458-MED12-gRNA#1 or PX458-MED12-gRNA#2 plasmids and 5 µL ssODN (100 µM) using Lipofectamine 2000 according to the manufacturer’s instructions. Five hours posttransfection, cells were transfected again with 5 µL ssODN (100 µM) using Lipofectamine 2000. Forty-eight hours posttransfection, positive GFP-expressing cells were collected by using a FACSAria II cell sorter (BD Biosciences). Single cell-derived clones were subsequently cultivated and positive clones were confirmed by sequencing.
MED12 gRNA#1: AAGGAGGTGCGTTCGAAAAT
MED12 gRNA#2: CGAACGCACCTCCTTCTGTT
ssODN sequence (*: phosphorothioates):
G*A*TGTTTACCCTCAGGACCCCAAACAGAAGCAGGTGCGTTCGAAAATCGGGGCTCTGGAG*G*
Data Availability.
The mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (57) (dataset identifier PXD015394). The structure of Compound A in complex with CDK8 (1–403)/Cyclin C were deposited to the Protein Data Bank (PDB ID code 6T41). All sequencing data were deposited in the Gene Expression Omnibus (GEO), series GSE135458.
Supplementary Material
Acknowledgments
We thank Silke Spudeit, Sooruban Shanmugaratnam, Marc Heinrichmeyer, and Kathrin Oertwig for technical help; Norbert Eichner for performing sequencing analysis on CRISPR clones; Martin Humenik for static light-scattering experiments; Ramona Adolph and Dr. Clemens Steegborn for providing the SIRT1 expression plasmid; and Dr. Lars Neumann and Dr. Klaus Maskos from Proteros Biostructures GmbH for help with biochemical and biophysical experiments and their analysis. This work was supported by the Elite Network of Bavaria, the University of Bayreuth, and the Paul Ehrlich and Ludwig Darmstaedter Prize for Young Researchers (to C.-D.K.); and the Deutsche Forschungsgemeinschaft (SPP 1935) (to G.M.). Alexander Leitner thanks Ruedi Aebersold (Eidgenössiche Technische Hochschule Zürich) for access to instrumentation and infrastructure. The Orbitrap Fusion Lumos mass spectrometer was purchased using funding from the Eidgenössiche Technische Hochschule Scientific Equipment program and the European Union Grant ULTRA-DD FP7-JTI 115766 (to Ruedi Aebersold).
Footnotes
The authors declare no competing interest.
Data deposition: The mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (identifier PXD015394). The structure of Compound A in complex with CDK8 (1–403)/Cyclin C were deposited to the Protein Data Bank (PDB ID code 6T41). All sequencing data been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE135458).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1917635117/-/DCSupplemental.
References
- 1.Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W., The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Malik S., Roeder R. G., Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30, 256–263 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Soutourina J., Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19, 262–274 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Allen B. L., Taatjes D. J., The mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knuesel M. T., Meyer K. D., Donner A. J., Espinosa J. M., Taatjes D. J., The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 29, 650–661 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D., A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Hengartner C. J., et al. , Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9, 897–910 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Plaschka C., et al. , Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Tsai K.-L., et al. , Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmlund H., et al. , The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 103, 15788–15793 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poss Z. C., et al. , Identification of Mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 15, 436–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holstege F. C., et al. , Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Knuesel M. T., Meyer K. D., Bernecky C., Taatjes D. J., The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels L. D., et al. , Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-Mediator kinase module. J. Proteomics Bioinform. 1, 1–7 (2013). [Google Scholar]
- 15.Firestein R., et al. , CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455, 547–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark A. D., Oldenbroek M., Boyer T. G., Mediator kinase module and human tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 50, 393–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pelish H. E., et al. , Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526, 273–276 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider E. V., et al. , The structure of CDK8/CycC implicates specificity in the CDK/cyclin family and reveals interaction with a deep pocket binder. J. Mol. Biol. 412, 251–266 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Turunen M., et al. , Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep. 7, 654–660 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolen B., Taylor S., Ghosh G., Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Dale T., et al. , A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat. Chem. Biol. 11, 973–980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitner A., Faini M., Stengel F., Aebersold R., Crosslinking and mass spectrometry: An integrated technology to understand the structure and function of molecular machines. Trends Biochem. Sci. 41, 20–32 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Leitner A., et al. , Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9455–9460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitner A., et al. , Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics 9, 1634–1649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lolli G., Structural dissection of cyclin dependent kinases regulation and protein recognition properties. Cell Cycle 9, 1551–1561 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Hoeppner S., Baumli S., Cramer P., Structure of the mediator subunit cyclin C and its implications for CDK8 function. J. Mol. Biol. 350, 833–842 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Huang K., et al. , Structure of the Pho85-Pho80 CDK-cyclin complex of the phosphate-responsive signal transduction pathway. Mol. Cell 28, 614–623 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarricone C., et al. , Structure and regulation of the CDK5-p25(nck5a) complex. Mol. Cell 8, 657–669 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Jeffrey P. D., et al. , Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature 376, 313–320 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Pavletich N. P., Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287, 821–828 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Park M. J., et al. , Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 293, 4870–4882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulman B. A., Lindstrom D. L., Harlow E., Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. U.S.A. 95, 10453–10458 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo A. A., Jeffrey P. D., Patten A. K., Massagué J., Pavletich N. P., Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382, 325–331 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Kämpjärvi K., et al. , Somatic MED12 mutations are associated with poor prognosis markers in chronic lymphocytic leukemia. Oncotarget 6, 1884–1888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinonen H.-R., et al. , Multiple clinical characteristics separate MED12-mutation-positive and -negative uterine leiomyomas. Sci. Rep. 7, 1015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida M., et al. , Frequent MED12 mutations in phyllodes tumours of the breast. Br. J. Cancer 112, 1703–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mäkinen N., et al. , MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 334, 252–255 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Bergeron P., et al. , Design and development of a series of potent and selective type II inhibitors of CDK8. ACS Med. Chem. Lett. 7, 595–600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pargellis C., et al. , Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 9, 268–272 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Schneider E. V., Böttcher J., Huber R., Maskos K., Neumann L., Structure-kinetic relationship study of CDK8/CycC specific compounds. Proc. Natl. Acad. Sci. U.S.A. 110, 8081–8086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alarcón C., et al. , Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139, 757–769 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donner A. J., Szostek S., Hoover J. M., Espinosa J. M., CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27, 121–133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dannappel M. V., Sooraj D., Loh J. J., Firestein R., Molecular and in vivo functions of the CDK8 and CDK19 kinase modules. Front. Cell Dev. Biol. 6, 171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castro F., Cardoso A. P., Gonçalves R. M., Serre K., Oliveira M. J., Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 9, 847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Z., Theurkauf W. E., Weng Z., Zamore P. D., Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence 3, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bancerek J., et al. , CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 38, 250–262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kroczynska B., Mehrotra S., Arslan A. D., Kaur S., Platanias L. C., Regulation of interferon-dependent mRNA translation of target genes. J. Interferon Cytokine Res. 34, 289–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberzon A., et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallace T. A., et al. , Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 68, 927–936 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Smid M., et al. , Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68, 3108–3114 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Tsai K.-L., et al. , A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 20, 611–619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin L., Wang W., Fang G., Targeting protein-protein interaction by small molecules. Annu. Rev. Pharmacol. Toxicol. 54, 435–456 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Huang S., et al. , MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell 151, 937–950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leitner A., et al. , Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renaud J.-B., et al. , Improved genome editing efficiency and flexibility using modified oligonucleotides with TALEN and CRISPR-Cas9 nucleases. Cell Rep. 14, 2263–2272 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Perez-Riverol Y., et al. , The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Graham M. J., Combe C., Kolbowski L., Rappsilber J., xiView: A common platform for the downstream analysis of crosslinking mass spectrometry data. bioRxiv:10.1101/561829 (26 February 2019).
- 59.Lamiable A., et al. , PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 44, W449–W454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (57) (dataset identifier PXD015394). The structure of Compound A in complex with CDK8 (1–403)/Cyclin C were deposited to the Protein Data Bank (PDB ID code 6T41). All sequencing data were deposited in the Gene Expression Omnibus (GEO), series GSE135458.