Abstract
Cajal bodies (CBs) are nuclear organelles concentrating two kinds of RNA–protein complexes (RNPs), spliceosomal small nuclear (sn), and small CB-specific (sca)RNPs. Whereas the CB marker protein coilin is responsible for retaining snRNPs, the tether for scaRNPs is not known. Here we show that Nopp140, an intrinsically disordered CB phosphoprotein, is required to recruit and retain all scaRNPs in CBs. Knockdown (KD) of Nopp140 releases all scaRNPs leading to an unprecedented reduction in size of CB granules, hallmarks of CB ultrastructure. The CB-localizing protein WDR79 (aka TCAB1), which is mutated in the inherited bone marrow failure syndrome dyskeratosis congenita, is a specific component of all scaRNPs, including telomerase. Whereas mislocalization of telomerase by mutation of WDR79 leads to critically shortened telomeres, mislocalization of telomerase by Nopp140 KD leads to gradual extension of telomeres. Our studies suggest that the dynamic distribution of telomerase between CBs and nucleoplasm uniquely impacts telomere length maintenance and identify Nopp140 as a novel player in telomere biology.
INTRODUCTION
The cell nucleus is devoid of physical borders such as membranes, yet contains several distinct organelles and domains recognized by light and electron microscopy. Many of these are compositionally characterized and harbor an abundance of ribonucleoproteins (RNPs). The most prevalent RNPs of the nucleus are preribosomes and spliceosomal small nuclear (sn)RNPs. The former localize to nucleoli, whereas the latter concentrate in speckles and Cajal bodies (CBs) (Staneˇk and Fox, 2017; Nozawa and Gilbert, 2019). Although CBs were identified more than 100 yr ago as accessory bodies of the nucleolus and ultrastructurally characterized as coiled bodies, their function is far from clear (Gall, 2003; Machyna et al., 2013). CBs concentrate two kinds of RNPs, snRNPs, and small CB-specific (sca)RNPs, forming the yin and yang of CBs (see Figure 6C later in this article). ScaRNPs site-specifically modify the snRNAs of snRNPs, apparently in CBs (Meier, 2016). Additionally, snRNPs may recycle in CBs during spliceosome regeneration (Staneˇk and Neugebauer, 2006). Although the CB marker protein coilin retains snRNPs in CBs (Tucker et al., 2001), the tether of scaRNPs including telomerase is unknown.
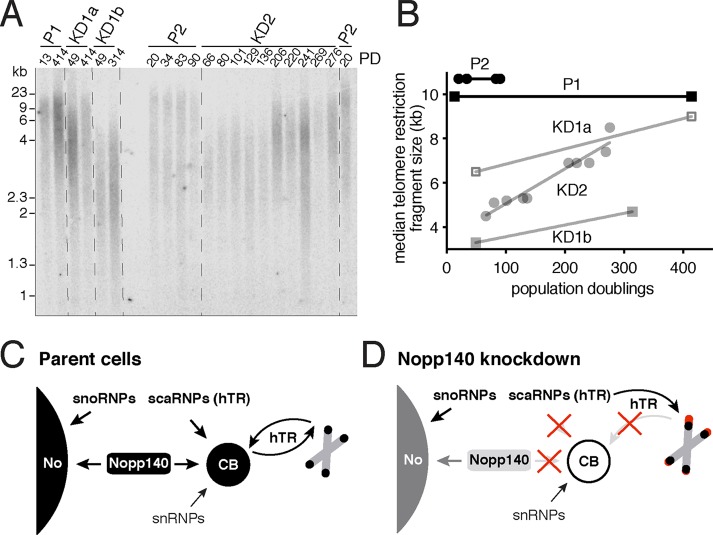
FIGURE 6:
Telomerase relocation from CBs to the nucleoplasm causes telomere lengthening. (A) Analysis of telomere restriction fragments isolated from the indicated cell lines (on top) at the indicated population doubling (PD), fractionated on agarose gel, denatured, and probed with a 32P-labeled TTAGGG probe. (B) Quantification of the median telomere restriction fragment sizes relative to PDs of the autoradiograph in A. Note that the parental cell telomeres (P1 and P2) retain their length over the entire PD range, whereas the telomeres of all three Nopp140 KD cell lines slowly extend (KD1a, KD1b, and KD2). (C) Schematic of Nopp140 and RNP trafficking in parental HeLa cells. Nopp140 distributes to nucleoli, together with snoRNPs, and to CBs, together with scaRNPs including telomerase (signified by hTR). Maturing snRNPs accumulate independently in CBs. During S phase of the cell cycle, telomerase extends some telomeres before returning to CBs. (D) Schematic of consequences of Nopp140 KD. Under limiting Nopp140 levels, residual Nopp140 partitions mostly to nucleoli allowing concentration of snoRNPs and rRNA biogenesis to continue. In contrast, residual Nopp140 fails to populate CBs restricting scaRNPs (but not snRNPs) to the nucleoplasm. As a consequence, telomerase access to telomeres is no longer restricted to S phase allowing gradual telomere lengthening throughout the cell cycle.
ScaRNPs are relatives of small nucleolar (sno)RNPs (see Figure 6C later in this article) and are part of two major snoRNP classes, the H/ACA and the C/D class. Their distinguishing features are short noncoding RNAs, which hybridize with target RNAs to specify a nucleotide for modification, H/ACA for guiding pseudouridylation, and C/D for guiding 2′-O-methylation (Kiss, 2001). A distinct set of four core proteins, including the pseudouridylase NAP57 (aka dyskerin and Cbf5) in H/ACA RNPs and the methylase fibrillarin in C/D RNPs, stabilizes the RNAs of each class (Meier, 2005; Kiss, 2006). ScaRNAs contain short sequence elements for CB localization, a CAB box in H/ACA RNAs, and a G•U/U•G wobble stem in C/D RNAs. These elements are recognized by the CB-specific protein WDR79 (aka TCAB1), encoded by the WRAP53 gene (Darzacq et al., 2002; Richard et al., 2003; Mahmoudi et al., 2009; Tycowski et al., 2009; Venteicher et al., 2009; Marnef et al., 2014). Unlike in nucleoli, where both H/ACA and C/D class RNPs are more or less equally represented, in CBs, H/ACA RNPs appear to outnumber the C/D class reflecting the larger number of pseudouridines over 2′-O-methylgroups in snRNAs (Meier, 2016). Although WDR79 is responsible for localizing scaRNPs including telomerase to CBs, what anchors them there is unknown. In addition to scaRNPs and snRNPs, the box C/D snoRNPs U3, U8, and U14 have also been reported in CBs (Wu et al., 1993; Samarsky et al., 1998; Narayanan et al., 1999; Verheggen et al., 2002). But it is not known what elements, in addition to the C/D box, are required for their CB localization.
Telomerase is a unique H/ACA scaRNP, which is required to counteract the replicative erosion of chromosome ends, telomeres (Schmidt and Cech, 2015). Human telomerase RNA (hTR), which templates the reverse transcription of telomeres, ends in an H/ACA domain that is stabilized by the H/ACA core protein complex (Mitchell et al., 1999; Nguyen et al., 2018). Additionally, hTR is endowed with a CAB box that is recognized by WDR79 localizing it to CBs (Tycowski et al., 2009; Venteicher et al., 2009). Although the purpose of CB localization of telomerase is not fully understood, it may be important for the association of the telomerase reverse transcriptase (TERT) with hTR. Indeed, live cell imaging of TERT demonstrates that among nuclear, CB, and telomeric fractions, CBs harbor by far the largest fraction of TERT with low diffusion coefficients (Schmidt et al., 2016). During the S-phase of the cell cycle when chromosomes replicate and telomeres are extended, the telomerase RNP visits some telomeres for that purpose (Jády et al., 2006; Tomlinson et al., 2006). Presumably, after maintaining those chromosome ends, telomerase returns to CBs preventing further extension (see Figure 6C later in this article). Mutation of the CAB box of hTR or of WDR79 itself abolishes CB accumulation of telomerase and causes precipitous telomere shortening (Jády et al., 2004; Cristofari et al., 2007; Venteicher et al., 2009). This is dramatically illustrated by mutations in WDR79 underlying the inherited bone marrow failure syndrome dyskeratosis congenita (DC) (Zhong et al., 2011). In fact, DC is further characterized by mutations in TERT and hTR and in the H/ACA core proteins NAP57, NOP10, and NHP2 (Savage, 2009). All DC patients suffer from extremely short telomeres in their peripheral blood lymphocytes below the first percentile (Mason and Bessler, 2011). Unfortunately, counteracting this pathological telomere erosion in DC patients remains a distant goal.
Nopp140 is an evolutionary conserved protein of nucleoli and CBs encoded by the gene NOLC1 (see Figure 6C later in this article). In coilin knockout cells, snRNPs are lost from CBs, but Nopp140 together with scaRNPs forms residual CBs (Tucker et al., 2001). The large intrinsically disordered central domain of Nopp140 consists of 10 repeats of alternating acidic and basic stretches (Meier and Blobel, 1992). Casein kinase 2 phosphorylates the serines of the acidic stretches with some 80 phosphates and Nopp140 associates with both H/ACA and C/D RNPs in a phosphorylation-dependent manner (Yang et al., 2000). The charged domains of Nopp140 present favorable interaction surfaces for multiple RNPs. Thus, Nopp140 may be the ideal molecule for allowing concentration of scaRNPs and snoRNPs in the confined spaces of CBs and the dense fibrillar component (DFC) of nucleoli, their respective homes. Cells are sensitive to exogenously increased concentrations of Nopp140, which causes unusual membrane proliferation in the normally membrane-free nucleoplasm and eventually cell death (Isaac et al., 1998, 2001). Although transient knockdown (KD) of Nopp140 has been studied (Watkins et al., 2004; Tsai et al., 2008; Renvoisé et al., 2009), the consequences for nucleoli and CBs in stable KD cell lines are unknown.
Nucleoli and CBs are liquid–liquid phase-separated organelles, which are often enriched in RNPs and multivalent proteins with intrinsically disordered domains (Zhu and Brangwynne, 2015). The highly disordered Nopp140 is the only protein that is not an integral part of RNPs, but is enriched in both, CBs and nucleoli (Meier and Blobel, 1994; Tantos et al., 2013; Na et al., 2018). Here we report on the generation of stable human Nopp140 KD cell lines and describe an unexpected role for Nopp140 in the dynamics and function of scaRNPs, including telomerase.
RESULTS
CRISPR/Cas9 targeting of NOLC1 yields stable Nopp140 KD cell lines
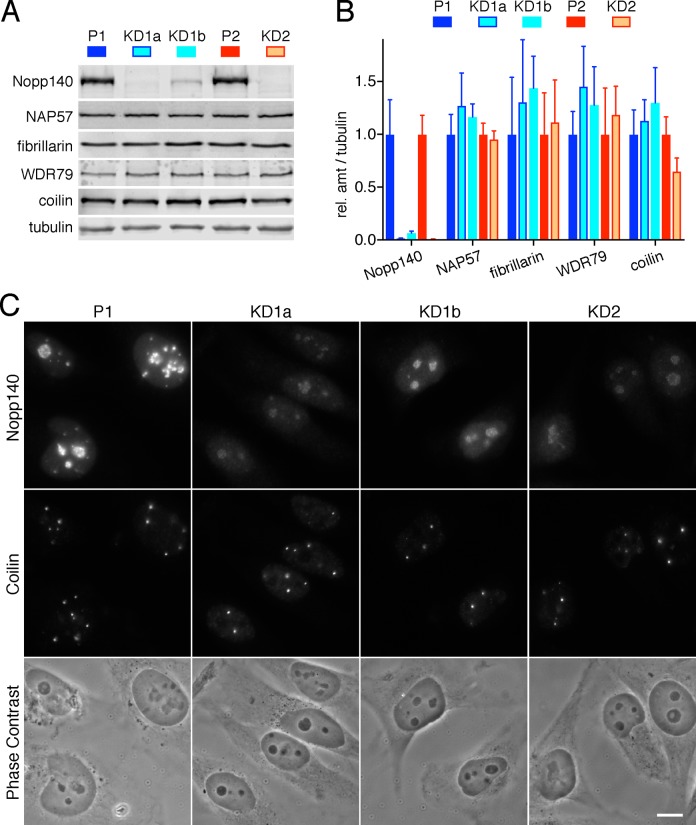
Nopp140 is the only protein that concentrates in both nucleoli and CBs without being an integral part of an RNP (see Figure 6C later in this article). Nevertheless, Nopp140 associates with both H/ACA and C/D RNPs. For functional analysis, we targeted the Nopp140 gene NOLC1 with CRISPR/Cas9 technology. Other studies using genomewide CRISPR screens identified Nopp140 as an essential core fitness gene in most cells, except HeLa, possibly due to high ploidy (Hart et al., 2015; Wang et al., 2015). Indeed, after induction of Cas9 expression, we cloned three viable Nopp140 KD cell lines from HeLa but not HEK-293T cells transduced with two single guide (sg)RNAs complementary to exon 1 or 3 of NOLC1. Parent 1 cells (P1) were transduced with sgRNA 1 giving rise to the clonal Nopp140 KD lines KD1a and KD1b, whereas parent 2 cells (P2) were transduced with sgRNA 2 yielding the KD2 line. Western blots (WBs) of whole cell extracts showed Nopp140 KD cells to express little Nopp140 (Figure 1A). The low Nopp140 amounts for each cell line remains stable for years. KD1a and KD2 expressed ∼1% of Nopp140 relative to their parent cells, whereas some 7% of Nopp140 remained in KD1b cells (Figure 1B). In contrast, expression of the enzymes of H/ACA and C/D RNPs, NAP57 and fibrillarin, and the scaRNP and Cajal body (CB) marker proteins, WDR79 and coilin, respectively, remained unaffected in the KD cells (Figure 1, A and B). By indirect immunofluorescence (IF), Nopp140 was barely detectable in nucleoli and not at all in CBs of KD cells (Figure 1C). Expression levels of Nopp140 are uniform for each cell line, whether parent or KD cells. Importantly, there is no significant growth difference between any of the cells, which have been dividing with the same frequency for at least 2 yr (Supplemental Figure S1). Further, there are no gross phenotypes (Figure 1C, phase contrast). In summary, we generated stable cell lines expressing minimal levels of the essential Nopp140.
FIGURE 1:
CRISPR/Cas9 targeting of two exons of the NOLC1 gene leads to KD of its gene product Nopp140 in three stable HeLa cell lines. (A) WBs on whole cell extracts of two parent cell lines (P1 and P2) and three corresponding Nopp140 KD cell lines (KD1a, KD1b, and KD2) cloned through two rounds of limited dilution. The blots were probed with the indicated antibodies and the signal of secondary antibodies detected by a near-infrared imaging system (Odyssey). (B) Quantification of the signal from three separate WBs as in A normalized to tubulin signal setting each parent as 1 (mean ± SD). Whereas only 1–5% of Nopp140 remain in KD cells, proteins representative of H/ACA RNPs (NAP57), C/D RNPs (fibrillarin), scaRNPs (WDR79), and CBs (coilin) are unaffected. (C) Indirect double IF of a parent and all KD cells show residual Nopp140 fluorescence in nucleoli but not CBs. Images were acquired with identical exposures and adjusted using identical parameters. However, no difference is detected between parent and KD cells by phase contrast imaging. Magnification is identical in all images, bar = 10 µm.
Nopp140 KD selectively displaces scaRNPs from CBs
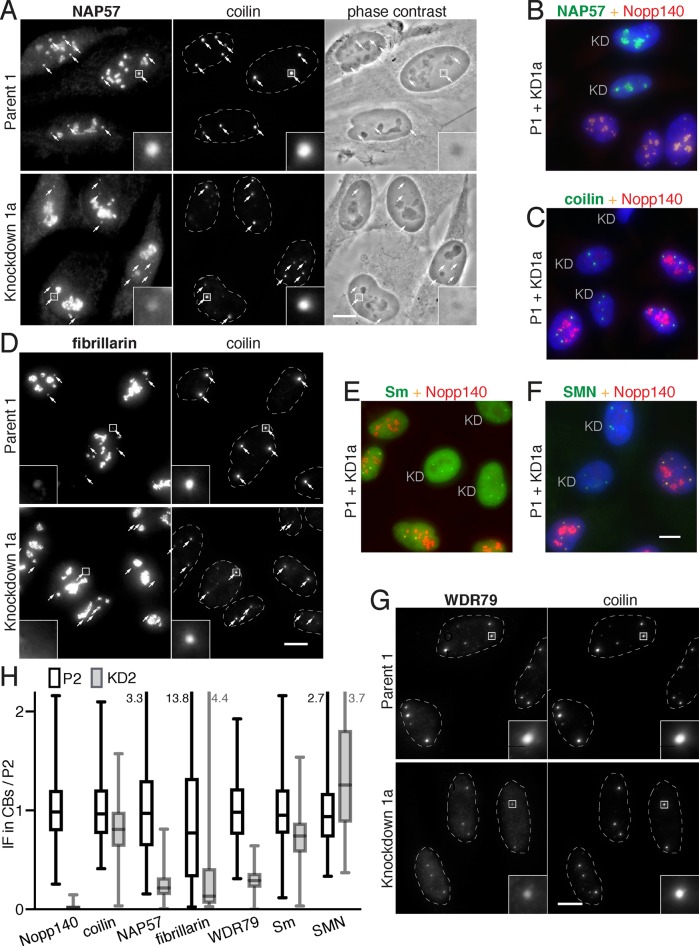
Although overall cellular levels of Nopp140-associated proteins are unaffected by Nopp140 KD (Figure 1A), we asked whether the KD impacted their subcellular distribution. We first tested the major core protein of H/ACA RNPs, NAP57, which was identified as a Nopp140-associated protein (Meier and Blobel, 1994). In parent cells, NAP57 fluorescence in CBs (small arrows) marked by coilin is equal or greater to that in nucleoli (Figure 2A, Parent 1). In contrast, in Nopp140 KD cells, NAP57 is visibly diminished in CBs (small arrows) while nucleolar fluorescence intensity remains unaffected if not increased (Figure 2A, Knockdown 1a). This is confirmed by quantification of the fluorescence signal in CBs for NAP57 and all other proteins (Figure 2H). No difference is detected between the phase dense CBs of P1 versus those of KD1a cells (Figure 2A, phase contrast) indicating the absence of gross effects. To ensure that the difference in staining between P1 and KD1a was not due to staining artifacts, we mixed P1 and KD1a cells 1:1 and costained them for NAP57 (Figure 2B, green) and Nopp140 (red). Whereas the overlapping red–yellow staining was seen in both nucleoli and CBs of P1 cells, KD1a cells only showed NAP57 staining in nucleoli confirming the results from independent staining (Figure 2, A and B). In contrast, coilin was unaffected by Nopp140 KD and equally present in CBs of P1 and KD1a cells (Figure 2C, see also Figures 2, A, D, and G, and 1C). The major box C/D core protein fibrillarin is not as concentrated in CBs as NAP57 because there are less C/D than H/ACA scaRNPs (Meier, 2016), but it was still reduced in Nopp140 KD cells (Figure 2D). To test whether components of the other major RNPs of CBs, the spliceosomal snRNPs, were impacted by Nopp140 KD, we tested for the presence of their Sm core ring antigens (Figure 2E) and their assembly factor SMN (Figure 2F). Both were equally present in CBs of cells with and without Nopp140. However, WDR79, the scaRNP-specific protein responsible for localization of H/ACA and C/D RNPs to CBs, was also reduced in CBs of Nopp140 KD cells (Figure 2G), as were the H/ACA core proteins GAR1 and NHP2 (unpublished data). We quantified the fluorescence of these antigens in CBs of Nopp140 KD cells relative to parent cells (Figure 2H). The large variability is due to the intra- and intercell differences in size and fluorescence signal of CBs. Nevertheless, the results show that, in addition to Nopp140, the overall amount of scaRNP proteins, NAP57, fibrillarin, and WDR79, was reduced by more than 50% in CBs, whereas that of coilin and the snRNP proteins Sm and SMN varied little (Figure 2H). We conclude that Nopp140 KD leads to a specific loss of scaRNP proteins from CBs (NAP57, fibrillarin, and WDR79) without affecting the localization of other CB components (SMN, Sm, and coilin). Meanwhile, nucleolar localization of snoRNP proteins remained unchanged.
FIGURE 2:
Proteins of sno- and scaRNPs, but not those of spliceosomal snRNPs, are specifically lost from CBs but not nucleoli in Nopp140 KD cells. (A) Comparison of indirect IF of NAP57 in parent and corresponding Nopp140 KD cells double labeled with coilin to identify the location of CBs (small arrows). A 5× magnification of one CB (framed) is included (inset). Note the reduced signal of NAP57 in CBs of Nopp140 KD vs. parent cells. No difference in coilin staining or CBs in phase contrast images is detected. (B) To ascertain identical immunolabeling, parent and KD cells were mixed 1:1 and double labeled for Nopp140 (red) and NAP57 (green). Nuclei were counterstained for DNA with DAPI (blue). In KD cells (KD, green only), NAP57 is in nucleoli but barely in CBs, whereas in parent cells (red/yellow), Nopp140 and NAP57 are present in nucleoli and CBs. (C) As in B but with coilin showing no difference between CBs of KD and parent cells. (D) As in A but with fibrillarin. Although the fibrillarin signal is low in CBs of even parent cells, it is completely gone from KD cells. (E) As in B without DAPI, but with Sm proteins, representative of spliceosomal snRNPs, showing no difference between CBs of KD and parent cells. (F) As in B but with SMN, a spliceosomal snRNP assembly factor, showing no difference between CBs of KD and parent cells. (G) As in D but with the scaRNP-specific WDR79 protein showing reduced staining in CBs of Nopp140 KD cells. (H) Quantification of fluorescence of the indicated proteins in CBs of P2 (black open boxes) and KD2 cells (gray filled boxes) normalized to the mean of P2. CB sample sizes (P2, KD2): Nopp140 (267, 364), coilin (482, 360), NAP57 (398, 335), fibrillarin (393, 192), WDR79 (482, 360), Sm (232, 247), SMN (267, 354). The bars indicate the median, the boxes the interquartile range, and the whiskers the minimum and maximum values (marked numerically where they go beyond the plot). Owing to the variability of intensity and size of CB signal, even within the same cell, the values cover a large range. Nevertheless, in addition to Nopp140 in the KD2 cells, only the signal of scaRNP proteins (NAP57, fibrillarin, and WDR79) is reduced compared with that of coilin and spliceosomal snRNPs (Sm and SMN). Magnification, bars = 10 µm; inset = 5× magnification of marked CB (square); B, C, and E are identical to F.
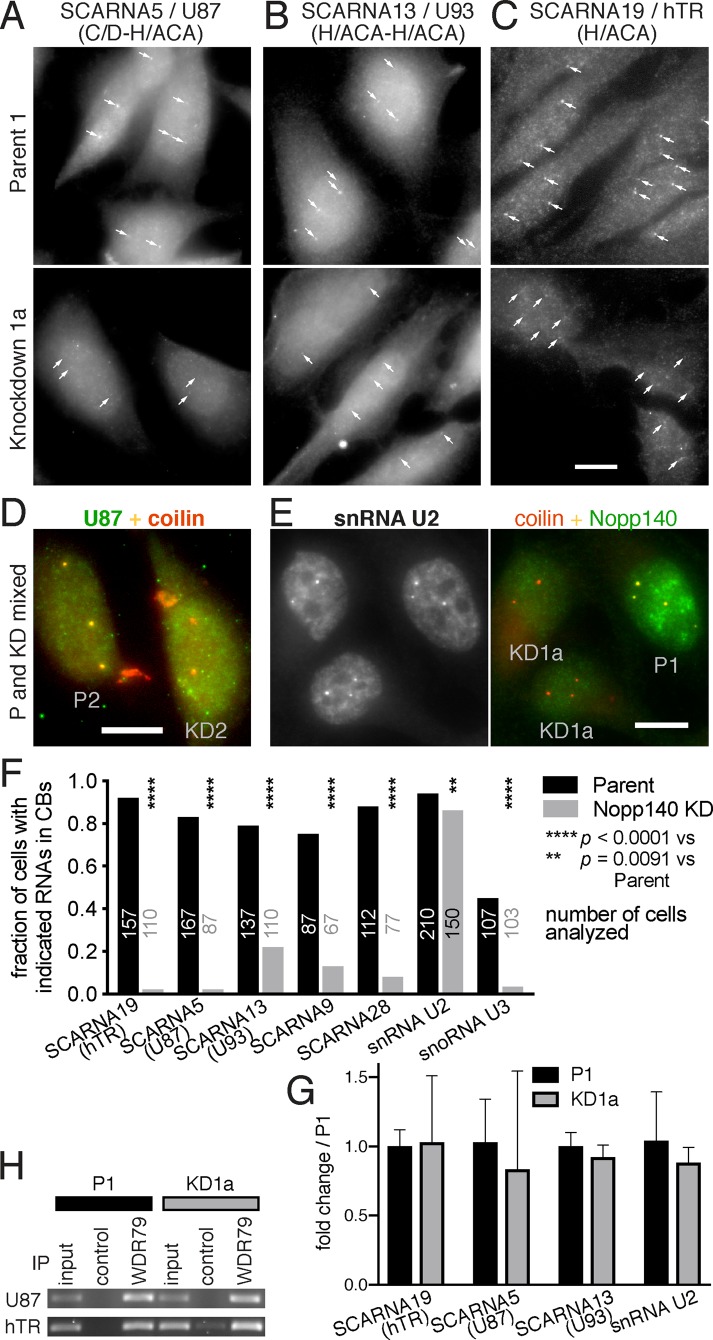
To test whether the loss of RNP proteins from CBs reflected that of intact protein–RNA complexes, we also interrogated the presence of scaRNAs and snRNAs in CBs using RNA fluorescence in situ hybridization (FISH) followed by indirect IF. The RNA FISH signal of various scaRNAs, hybrid C/D-H/ACA (Figure 3A), tandem H/ACA-H/ACA (Figure 3B), and the H/ACA telomerase RNA (Figure 3C), were reduced or lost from CBs (small arrows) of KD compared with those of parent cells. This was confirmed for SCARNA5 (U87, green) in CBs (coilin, red) of mixed P2 (yellow), and KD2 (red/orange) cells (Figure 3D). In contrast, U2 snRNA staining was indistinguishable in a mixed population of P1 and KD1a cells (Figure 3E, left panel) identified by Nopp140 (green) and coilin (red) presence (Figure 3E, right panel). The results for these and other scaRNAs were confirmed by quantification of hundreds of cells for the presence of the RNAs in CBs (Figure 3F). In all cases, KD1a cells exhibited significantly fewer cells with scaRNAs in CBs, whereas little difference was noted for the presence of the spliceosomal snRNA U2 (Figure 3F). Additionally, the C/D box snoRNA U3 was observed in significantly fewer CBs of KD2 compared with P2 cells (Figure 3F). Nevertheless, U3 was barely detectable in CBs and its CB levels were very low next to those in nucleoli and of bona fide scaRNAs (Supplemental Figure S2). The purpose for U3 transiting through CBs could be its need for cap hypermethylation by TGS1 and its pseudouridylation of at least two uridines (Reddy et al., 1979, 1980; Verheggen et al., 2002). Perhaps only U3 precursors make their way to CBs, explaining the low levels and detection in only about half of CBs. Regardless, the localization of U3 to nucleoli was unaffected by Nopp140 KD (Supplemental Figure S2).
FIGURE 3:
RNAs of scaRNPs, but not those of spliceosomal snRNPs, are lost from CBs in Nopp140 KD cells. (A) RNA FISH for the hybrid scaRNA5 combined with indirect IF for coilin (unpublished data but positions are indicated by small arrows). CB signal (small arrows) of SCARNA5 is lost from KD1a cells (bottom) compared with P1 cells (top). (B) As in A but with the tandem H/ACA SCARNA13. (C) As in A but with telomerase RNA (SCARNA19). (D) Same as A but on a mix of P2 and KD2 cells, identified by Nopp140 double IF (unpublished data). Note the yellow color of CBs in P2 cells due to the costaining of U87 (SCARNA5) and coilin (red), whereas in KD2 cells CBs are mostly orange/red due to absence of U87. (E) RNA FISH for the spliceosomal snRNA U2 (left, black and white) and indirect double IF for coilin (red) and Nopp140 (green) on a mix of P1 and KD1a cells. There is no difference in U2 staining between parent (P1, identified by nucleolar Nopp140) and KD cells (KD1a). (F) Quantification of FISH signal in P1 (black) and KD1a (gray) cells for all different kinds of scaRNAs, H/ACA SCARNA19 (hTR), hybrid SCARNA5, tandem H/ACA SCARNA13, tandem C/D SCARNA9, C/D SCARNA28, and spliceosomal U2 snRNA. All scaRNAs are significantly reduced in CBs but U2 snRNA barely. Additionally, the box C/D snoRNA U3 was significantly reduced in KD2 compared with P2 cells. The number of cells analyzed in each case is indicated. Statistical significance was assessed using Fisher’s exact test. (G) Quantification of total SCARNAs 19, 5, 13, and snRNA U2 using RT-qPCR in parent (black) and Nopp140 KD cells (gray). The mean ± SD are expressed relative to the parent cells (n = 3). Statistical significance was determined using the Holm-Sidak method (α = 0.05) without assuming a consistent SD. No significant change of total RNAs was observed (t test). (H) IP from whole cell extracts with antibodies to WDR79 or beads alone (control) followed by RT-PCR for associated RNAs. Note that scaRNAs U87 and hTR are precipitated equally from parent (P1) and KD cells (KD1a) indicating that the scaRNPs remain intact. Magnification, bars = 10 µm; A and B are identical to C.
The overall levels of scaRNAs did not change between P1 and KD1a cells as assessed by RT-qPCR (quantitative reverse transcription PCR) (Figure 3G), indicating that only their localization, but not their integrity, was affected by Nopp140 KD. Importantly, scaRNPs remained fully intact, that is, associated with WDR79, because immunoprecipitation (IP) with WDR79 antibodies followed by RT-PCR confirmed coprecipitation of equal amounts of SCARNA5/U87 and hTR from both parent and Nopp140 KD cells (Figure 3H). Taken together, our data document that Nopp140 KD displaces intact scaRNPs, but not snRNPs, from CBs.
Nopp140 reexpression rescues the effects of Nopp140 KD
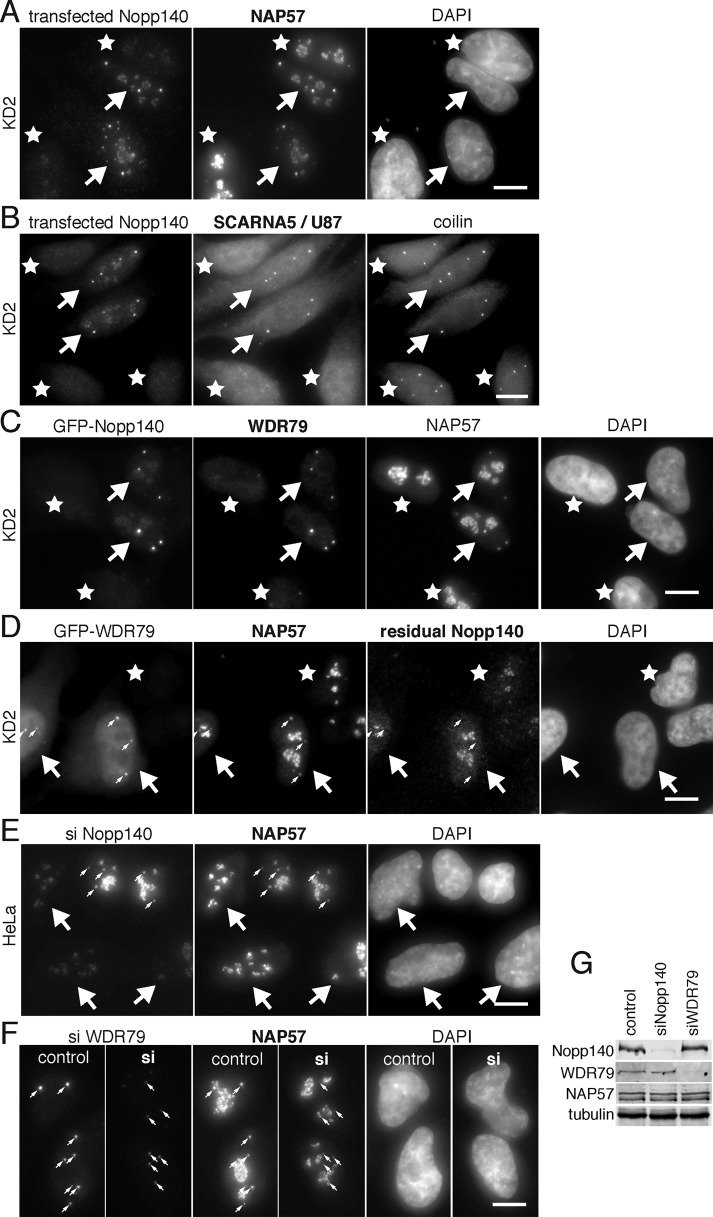
Although three CRISPR cell lines generated with two different sgRNAs targeting NOLC1 exon 1 or 3 produced identical results, we confirmed the specificity for Nopp140 KD by rescue of the KD cells through Nopp140 reexpression. Overexpression of Nopp140 generates intranuclear membrane tubules and is toxic to cells (Isaac et al., 1998, 2001; Chen et al., 1999). Therefore, we expressed human Nopp140 from the low-expressing UBC promoter. Over time, exogenously expressed Nopp140 first accumulated in CBs and subsequently in nucleoli of the Nopp140 KD cells (Figure 4, A–C, first panels). Unlike in untransfected cells (stars), endogenous NAP57 and SCARNA5/U87 followed the transfected Nopp140 back into CBs (Figure 4, A and B, arrows). Expression of GFP-tagged Nopp140 also restored CB localization of NAP57 and WDR79 (Figure 4C, arrows). Similar results were obtained with fibrillarin, NHP2, and GAR 1 (unpublished data) and with SCARNA19/hTR and U93 (Supplemental Figure S3). We conclude that Nopp140 reexpression in CBs restores scaRNPs to CBs and that their depletion from CBs is not due to CRISPR/Cas9 off-target effects but specific to Nopp140 loss.
FIGURE 4:
Displacement of scaRNPs from CBs in Nopp140 KD cells is not caused by CRISPR/Cas9 off-target effects. (A) Double IF demonstrates that reexpression of Nopp140 in KD cells (arrows) rescues NAP57 localization to CBs unlike in untransfected cells (asterisks). (B) Equally, Nopp140 transfection in KD cells (arrows) rescues SCARNA5/U87 localization (observed by RNA FISH) to CBs (identified by coilin double IF) unlike in untransfected cells (asterisks). (C) Transfected GFP-Nopp140 (fluorescence, arrows) rescues WDR79 and NAP57 localization to CBs unlike in untransfected cells (asterisks). (D) KD2 cells transfected with GFP-WDR79 (arrows) also rescue NAP57 localization to CBs (small arrows) but not that of residual Nopp140. (E) Transient Nopp140 KD in HeLa cells (siNopp140) shows a loss of NAP57 from CBs in siRNA transfected cells (arrows) but not from CBs of untransfected cells (small arrows). (F) Similarly, transient KD of WDR79 (si) reduced the WDR79 signal in CBs (small arrows) compared with untransfected cells (control), which was accompanied by a loss of NAP57 from CBs but not nucleoli, as previously noted (Zhong et al., 2011). (G) Transient KD of Nopp140 and WDR79, but not that of NAP57 and tubulin, was confirmed by WB on whole cell extracts. Magnification, bars = 10 µm.
ScaRNP localization to CBs depends on two factors, the CAB box of scaRNAs and the WDR79 which binds to it (Darzacq et al., 2002; Richard et al., 2003; Mahmoudi et al., 2009; Tycowski et al., 2009; Venteicher et al., 2009). This is illustrated by DC causing mutations in WDR79 that lead to scaRNP (including hTR) loss from CBs (Zhong et al., 2011). Therefore, we tested whether overexpression of WDR79 in the Nopp140 KD cells might also restore scaRNP localization to CBs. Indeed, expression of GFP-tagged WDR79 in Nopp140 KD cells (arrows), restored NAP57 (as a marker for scaRNPs) to CBs (Figure 4D, small arrows). In contrast, residual Nopp140 in the KD cells failed to reaccumulate in CBs (Figure 4D). Together with the fact that the Nopp140 loss from CBs is accompanied by a loss of WDR79 (Figure 2G), which is restored upon Nopp140 reexpression (Figure 4C), these data indicate that Nopp140 acts upstream of WDR79 in scaRNP localization to CBs.
While this work was in progress, TDP43 was shown to regulate the CB localization of a varying subset of four C/D scaRNAs (Izumikawa et al., 2019). Transient KD of TDP43 displaced some of these scaRNAs from CBs. A loss of TDP43 from CBs in the Nopp140 KD cells suggests that Nopp140 is also responsible for the TDP43-mediated localization of scaRNPs to CBs (Supplemental Figure S4). Finally, TGS1, the trimethyl transferase responsible for hypermethylating the monomethyl caps of snRNAs in the cytoplasm and hTR and snoRNA U3 possibly in CBs, is also concentrated in CBs (Verheggen et al., 2002) (Supplemental Figure S5). However, CB localization of TGS1 is reduced by about half in Nopp140 KD cells similar to the loss of hTR and U3 (Figure 3, C and F, and Supplemental Figure S5).
To further cement the role of Nopp140 in scaRNP localization to CBs, we replicated the effects of our stable Nopp140 KD cell lines by transient KD of Nopp140 using small interfering RNAs (siRNAs). Indeed, in regular HeLa cells transfected with Nopp140 siRNAs (Figure 4E, arrows), NAP57 was lost from CBs but not nucleoli and in contrast to CBs of untransfected cells (Figure 4E, small arrows). Similar results were observed in U2OS cells showing this to be a more general effect not unique to HeLa cells (unpublished data). Finally, we replicated the previously reported effect of transient WDR79 KD (Zhong et al., 2011) documenting a specific loss of NAP57 from CBs but not nucleoli in cells transfected with WDR79 siRNA (Figure 4F). In both cases, KD was confirmed by WB of whole cell extracts (Figure 4G). Overall, we conclude that Nopp140 is responsible for scaRNP accumulation in CBs upstream of WDR79. Consequently, we are left with CBs that still accumulate coilin, SMN, and snRNPs, but lack Nopp140 and scaRNPs.
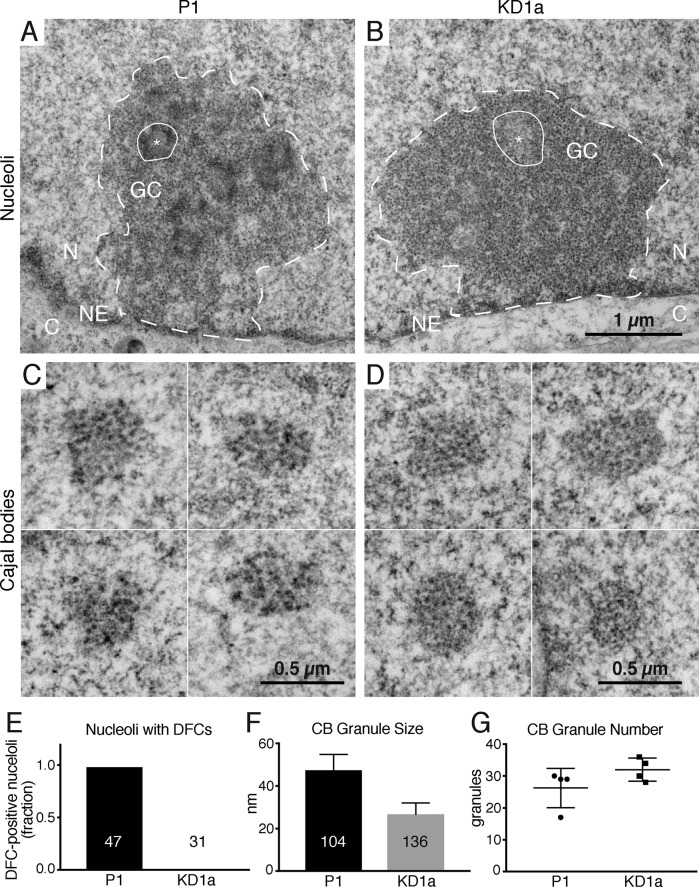
Nopp140 loss changes the ultrastructure of nucleoli and CBs
By light microscopy, Nopp140- and scaRNP-less CBs in KD cells look indistinguishable from their normal counterparts (Figures 1C and 2A). But might differences be detected by electron microscopy on an ultrastructural level? Nopp140 is equally concentrated in the DFC of nucleoli and in CBs (Meier and Blobel, 1992, 1994; Thiry et al., 2009). Transmission electron microscopic analysis of nucleoli in parent cells (Figure 5A, outlined) revealed the canonical tripartite structure of fibrillar centers (FCs, asterisk) surrounded by DFCs (dark, circled area) altogether embedded in the granular component (GC). KD1a cells showed the same nucleolar hallmarks, except for the dark DFCs (Figure 5B). In fact, whereas 46 of 47 nucleoli of P1 cells exhibited distinct DFCs, none of 31 KD1a nucleoli did (Figure 5E). We conclude that the loosening of DFCs is a direct consequence of low Nopp140 levels in the KD cells. CBs of parent cells can be identified by their characteristic coils of dark granules (Figure 5C). The only visible difference between CBs of KD1a to parent cells was a reduction in granule size by about half (Figure 5D, quantified in Figure 5F). In contrast, granule number did not vary significantly (Figure 5G) nor did the overall shape and size of CBs (compare Figure 5, C and D). The reduction of granule size is the only visible difference between KD and parent cells. This observation, together with the loss of Nopp140 and scaRNPs from CBs being the only physiological difference between the KD and parent cells, strongly implies that these granules are the CB residences of scaRNPs and, likely, their targets, snRNPs.
FIGURE 5:
Nopp140 KD changes the ultrastructure of the DFC of nucleoli and that of CBs, its normal locations of residence. Transmission electron micrographs of a nucleolus (outlined) of parent (A) and Nopp140 KD (B) cells. One of several FCs is indicated (asterisk) surrounded by the DFC (encircled), which are altogether embedded in the GC. The nuclear envelope (NE), which separates the nucleoplasm (N) from the cytoplasm (C), is marked. The only difference between the P1 and KD1a nucleoli are the absence of a well-defined (dark) DFC, the natural habitat of Nopp140, from the KD cells, quantified in (E, P < 0.0001). C and D depict galleries of four CBs each from P1 (C) and KD1a (D) cells with their characteristic coils of granules. Whereas the overall size of all CBs is similar, the granule size of KD cell CBs is about half the size (F, P < 0.0001), but the overall granule number remains the same, that is, without any significant difference (G, P = 0.1605). All groups were compared using two-tailed unpaired t tests.
Telomerase relocation from CBs to the nucleoplasm causes telomere lengthening
One of the scaRNPs dislocated from CBs in the Nopp140 KD cells is telomerase (Figure 3, C and F), which is responsible for maintaining telomere length. If telomerase is lost from CBs due to mutation or KD of WDR79, as is observed in some cases of DC, telomeres shorten critically (Zhong et al., 2011; Vogan et al., 2016; Chen et al., 2018). Hence, we assessed the telomere length in our cell lines using telomere restriction fragment length analysis. Surprisingly, in all three Nopp140 KD clones, telomeres lengthened over time (Figure 6A). Note that the parental cells are a mixture of cells, whereas the KD cells are individual clones with shorter initial telomere lengths that vary according to clone. Importantly, the lengths of telomeres in all three KD clones gradually increase, whereas those of the parent cells remained stable for more than 1 yr (Figure 6B). This gradual increase is in stark contrast to telomere lengthening caused by hTR and telomerase overexpression, which proceeds significantly faster (Cristofari and Lingner, 2006).
DISCUSSION
We developed clonal cell lines expressing severely reduced levels of the snoRNP and scaRNP chaperone Nopp140. These cells document that normal Nopp140 levels are required to maintain scaRNPs in CBs but not snoRNPs in nucleoli (Figure 6C). Nopp140 KD cells provide an excellent tool to study the specific function of scaRNPs in CBs because localization of other RNPs and proteins in CBs remains undisturbed. One of the scaRNPs absent from CBs is the telomerase RNP required for telomere length maintenance. The role of WDR79 in increasing the presence of hTR at telomeres (Venteicher et al., 2009; Stern et al., 2012) emphasizes the importance of our Nopp140 KD cell lines, which now allows study of holo-telomerase without impacting its individual components. In stark contrast to preventing telomerase accumulation in CBs by manipulation of its CB localization elements in RNA or protein, which cause rapid telomere shortening (Darzacq et al., 2002; Richard et al., 2003; Cristofari et al., 2007; Zhong et al., 2011; Vogan et al., 2016; Chen et al., 2018), relocation from CBs of the intact holoenzyme leads to gradual telomere lengthening (Figure 6D). Thus, in the absence of other major phenotypes, Nopp140 silencing offers an avenue for therapy in DC patients who suffer from extremely short telomeres. Our studies reveal telomerase safeguarding as an unexpected function of CBs.
Nopp140 acts upstream of WDR79 in CB localization of scaRNPs
Our results show a clear dichotomy of RNP localization to CBs, that of scaRNPs and that of snRNPs, both accumulating independently. Indeed, KD of Nopp140 only affects localization of scaRNPs, whereas KD of coilin affects only that of snRNPs (Tucker et al., 2001). Therefore, and as previously suggested (Lemm et al., 2006), the congregation in CBs of the two types of RNPs, the scaRNPs and their modification targets, snRNPs, is handled independently by the cell. Localization of scaRNPs depends on their CB localizing elements, the CAB box and the GU/UG wobble stem (Darzacq et al., 2002; Richard et al., 2003; Marnef et al., 2014). Both elements are recognized by the protein WDR79, which is required for scaRNP accumulation in CBs (Tycowski et al., 2009; Venteicher et al., 2009). We now show that Nopp140 acts upstream of WDR79 in scaRNP localization to CBs because KD of Nopp140 prevents CB localization of WDR79, whereas overexpression of WDR79 fails to rescue CB localization of Nopp140 while rescuing that of scaRNPs. The latter may be achieved by excess binding of WDR79 to scaRNPs, for example, at two CAB boxes instead of one, or by weak binding to coilin (Mahmoudi et al., 2010; Enwerem et al., 2014). As it is, little is known about the mechanism of WDR79 association with scaRNPs, that is, are there assembly factors involved or are all sites on scaRNAs occupied? More importantly, how does Nopp140 concentrate in CBs in the first place? Liquid–liquid phase separation is the most likely answer (Zhu and Brangwynne, 2015). Nopp140 with its large central intrinsically disordered domain and its ability to bind both C/D and H/ACA scaRNPs, as well as coilin, is an ideal molecule to participate in the formation of CBs as phase-separated organelles in the membraneless nucleoplasm. Indeed, in the absence of coilin, Nopp140 together with scaRNPs forms residual CBs without snRNPs (Tucker et al., 2001). Hence Nopp140 is one of the core molecules underlying CB formation.
Safeguarding telomerase in CBs is required for telomere homeostasis
Telomerase is a specialized H/ACA scaRNP and concentrates with them in CBs. Although dependent on the CAB box of hTR and on WDR79, recruitment to CBs occurs through Nopp140. Mutation or ablation of the hTR CAB box or of WDR79 displaces telomerase from CBs and causes severe telomere shortening (Cristofari et al., 2007; Zhong et al., 2011; Vogan et al., 2016; Chen et al., 2018). However, displacement from CBs of the holo-telomerase scaRNP (with WDR79) leads to gradual telomere lengthening. How can this surprising and apparent contradiction be explained? We offer two possibilities.
First, CBs shelter telomerase during most of the cell cycle (Jády et al., 2004, 2006). However, during S-phase, telomerase shuttles from CBs to some telomeres to extend them (Jády et al., 2006; Cristofari et al., 2007). After extension, telomerase apparently returns to its harbor in CBs. We now show that Nopp140 is required as a landing pad or glue in CBs for all scaRNPs, including telomerase, which leads us to the following model. In the absence of its landing pad and CB shelter, telomerase concentration in the nucleoplasm slightly increases. Thus, telomerase may linger at telomeres for a prolonged period and slowly extend them (Figure 6D). There is an extreme precedence for such a mechanism when TERT and hTR are concomitantly overexpressed, telomeres extend severalfold beyond their normal length (Cristofari and Lingner, 2006). When telomerase is displaced from CBs, its nucleoplasmic concentration increases only slightly, explaining the observed slow but steady lengthening of telomeres.
Second, hTR may get modified in CBs, which would not happen or be reduced if it was displaced. For example, the monomethyl cap of hTR appears to be hypermethylated by TGS1 in CBs (Verheggen et al., 2002). Additionally, hTR is pseudouridylated attenuating the activity of telomerase while increasing its processivity (Kim et al., 2010). Thus, telomerase with unmodified hTR would exhibit heightened activity toward telomeres. If modification of hTR occurred in CBs, the absence of Nopp140 and of telomerase recruitment to CBs would result in telomerase with increased activity. This in turn could explain the observed slow telomere lengthening. Of course, both mechanisms could operate in parallel and additional explanations are possible. Regardless, the main difference between prior observations and our studies is that in our cells the holo-telomerase scaRNP remains intact; only its localization is affected.
Mechanism of scaRNP localization to CBs
Why is it particularly important to study effects on endogenous rather than exogenously expressed RNAs and proteins in the case of CBs? Recent evidence suggests CBs, like nucleoli, to be liquid–liquid phase-separated organelles (Zhu and Brangwynne, 2015). For example, expression of poly-arginine-glycine or -proline dipeptides disrupt nuclear phase-separated, membraneless organelles including CBs (Lee et al., 2016). Such organelles form in a manner that is exquisitely sensitive to the concentration of its constituents. Hence, exogenous expression of RNAs or proteins could change the partition of CB residents. Liquid–liquid phase separation is further stimulated by molecules that are intrinsically disordered and offer multiple binding sites, such as Nopp140 (Na et al., 2018). Nopp140, with its 10 alternating acidic and positively charged repeats, forms an ideal landing and neutralizing pad for sno- and scaRNPs with their acidic RNAs and positively charged proteins. Nopp140 can be considered as sca- and snoRNP glue. Why is it then that Nopp140 loss affects scaRNPs in CBs but not snoRNPs in nucleoli? Both organelles apparently form by liquid–liquid phase separation. Nucleoli nucleate around active clusters of rDNA transcription, whereas CBs may form spontaneously due to the high concentration of scaRNPs and snRNPs with the help of Nopp140 and coilin (Sawyer et al., 2019). There are several issues to be kept in mind when considering the differential effect of Nopp140 KD on nucleoli and CBs. First, these are KD and not knockout cells, so probably residual, if undetectable, levels of Nopp140 remain also in CBs. Second, these are clonal cells selected for survival, which depend on ribosome biogenesis in nucleoli but not on visible CBs, which are not essential and even undetectable in some cells. Third, the amounts of snoRNPs (and with it those of Nopp140) in the nucleolus is significantly higher than that of scaRNPs in CBs. Finally, the DFC of the nucleolus is not actually lost but its contrast in electron micrographs is reduced. In fact, the strong contrast of DFC and CBs may be partially caused by the extreme Nopp140 phosphorylation. Thus, even at reduced levels of Nopp140 in nucleoli (Figure 1C), evidenced by the loosened, less contrasted DFC in electron micrographs, RNA polymerase I transcription continues. Whether modification of rRNA is affected is currently being investigated and will be reported in a forthcoming publication. Regardless, the levels of Nopp140 are insufficient to keep scaRNPs focused in CBs. Interestingly, the Nopp140 organizing principle seems to be evolutionarily conserved as deletion of its yeast homologue Srp40p effects a loss of C/D RNAs from the nucleolar body, the closest relative to a CB in yeast (Meier, 1996; Verheggen et al., 2002).
Finally, the apparently well-tolerated consequences of Nopp140 KD on cell growth and morphology offer a reduction of Nopp140 levels as a therapeutic avenue to rescue telomere length in DC patients and aging.
MATERIALS AND METHODS
Cell culture and genome engineering
HeLa cells and different clones were cultured in DMEM and 10% fetal bovine serum at 37°C under 5% CO2 in air. Genome engineering was done using the CRISPR/Cas9 system. HeLa cells were transduced with Cas9 lentivirus, selected for 3 d with 1 µg/ml puromycin, then transduced with lentivirus containing two NOLC1-sgRNAs, sgRNA-Nopp140-Exon1, and sgRNA-Nopp140-Exon3 (Table 1) to obtain P1 and P2 cells, respectively, and selected for 7 d with 5 µg/ml blasticidin. Subsequently, Cas9 expression was induced for 7 d with 500 ng/ml doxycycline while maintaining puromycin and blasticidin selection. Single clones were obtained by limited dilution and tested by indirect IF and WB for Nopp140 loss. Clones with significantly decreased expression of Nopp140 underwent another round of limited dilution cloning and were used for further analysis. From P1, resulted the KD1a and KD1b clones, and from P2, the KD2 clone. For growth evaluation, 12,000 cells of each clone were plated on day 1 and expansion assessed on 4 subsequent days using a Countess II automated cell counter (Thermo Fisher Scientific). Correct targeting by the sgRNAs was confirmed by DNA amplification of the target sequences for each cell line (using primers U659 and U660 for KD1a/KD1b and primers U656 and U657 for KD2; Table 1) followed by Sanger sequencing (with primer U661 for KD1a/KD1b and U658 for KD2; Table 1). Proper targeting of the sgRNAs was also evident from RNA-Seq data of the cell lines (unpublished results). Even reinduction of Cas9 in our clones failed to reduce Nopp140 expression further. As in a prior study, the reasons for the residual expression are not entirely clear, but could be due to polyploidy (Hart et al., 2015). Perhaps one allele is located in a mostly silenced region of the chromatin preventing Cas9 access while allowing minimal expression. Obviously, single cell cloning would select for those cells that still exhibit some expression of an essential gene.
TABLE 1:
Primers and oligos.
| Target | Type of experiment | Primer | Sequence |
|---|---|---|---|
| GLUD2 | qPCR | U727 | TCGTGGAGGACAAGTTGGTG |
| U728 | TTGCAGGGCTTGATGATCCG | ||
| SCARNA19 (hTR) | qPCR and PCR | U590 | CCACCGCGAAGAGTTGGGCTC |
| U591 | GCATGTGTGAGCCGAGTCCTGG | ||
| SCARNA5 (U87) | qPCR and PCR | U717 | TCCCTTTGTTGCCCTCAACT |
| U718 | CCATGACTGCCACTCGTCAG | ||
| SCARNA13 (U93) | qPCR | U719 | ATCTGTAGTCTTGGAGCCGC |
| U720 | GTGGCAACAGTGACCAGAAAC | ||
| U2 | qPCR | U721 | ATCGCTTCTCGGCCTTTTG |
| U722 | CTATTCCATCTCCCTGCTCCA | ||
| Nopp140 Exon 1 | CRISPR/Cas9 | sgRNA-Nopp140-Exon1 | GCGCAGGAAGCCGAGCACGA |
| Nopp140 Exon 3 | CRISPR/Cas9 | sgRNA-Nopp140-Exon3 | GTTACAGGCAAATGGACCAG |
| Nopp140 intron 1 to exon 4 | PCR | U656 | ATGAAGATGTAACATTTATACCA |
| U657 | CTGCAAACCTGGACAGGCT | ||
| Nopp140 Exon 1 | Sanger sequencing | U658 | GTAATTATGTATGTAACAGGCCA |
| Nopp140 5′UTR to intron 1 | PCR | U659 | GAGCTCCGCCTCCACAAGCT |
| U660 | GCAACAAGATCTCAGCGATATCT | ||
| Nopp140 5′UTR | Sanger sequencing | U661 | GCTCCGCCCTTAACCAAGAT |
| UBC promoter | Cloning of UBC promoter | U662 | ATGCACATGTGAGCTCTTACGCGTAAAACTAGT |
| U663 | ATGCAGATCTAAGCTTCAAGTGACGATCAC | ||
| Linker STOP | Linker STOP insertion | U667 | GATCCTAACTAGGTGAGC |
| U668 | GGCCGCTCACCTAGTTAG | ||
| Telomeres | Telomere restriction fragment length analysis | 32P end-labeled telomere probe | TTAGGGTTAGGGTTAGGGTTAGGG |
Plasmids, siRNA, and transfection
Nopp140 plasmids used in this study are based on pNK65 (HA-hNopp140-mGFP under CMV promoter). HA-hNopp140 under the CMV promoter (pJB8) was generated by replacing the mGFP in pNK65 with a STOP codon using primers U667 and U668. HA-hNopp140-mGFP under the UBC promoter (pJB9) was generated by replacing the CMV promoter in pNK65 with the UBC promoter amplified off pGL3-UBC promoter-HSP70 using primers U662 and U663. HA-hNopp140 under the UBC promoter (pJB10) was generated by removal of mGFP from pJB9 like that from pNK65 to obtain pJB8.
Lipofectamine 3000 was used for plasmid and siRNA transfections following the manufacturer’s protocol. pJB9 and pJB10 plasmids were transfected for 24 h, pEGFP-WRAP53beta (WDR79; addgene.org) was transfected for 16 h, and Nopp140 and WDR79 siRNAs were transfected for 72 h before analysis.
Antibodies
Antibodies (dilutions in parentheses) for WB, IP, indirect IF, and FISH followed by IF (FISH-IF) were as follows: anti-Nopp140 rabbit serum (RS8 at 1:5000 for WB; 1:1000 for IF and FISH-IF) (Kittur et al., 2007); anti-NAP57 rabbit serum (RU10 at 1:50 for IP; 1:200 for WB and IF) (Darzacq et al., 2006); anti-NHP2 rabbit serum (p15 at 1:100 for IF) (Pogacic et al., 2000); anti-GAR1 rabbit serum (p16 at 1:100 for IF) (Dragon et al., 2000); anti-WDR79 rabbit serum (1:2000 for WB, 1:300 for IF, and 1:500 for IP; Novus Biologicals); anti-TDP43 rabbit serum (1:200 for IF; Proteintech); anti-TGS1 rabbit serum (1:500 for IF; ABclonal Science); anti-coilin rabbit serum (1:5000 for WB and 1:1000 for IF; Proteintech); anti-coilin mouse ascites fluid (5P10 at 1:1000 for IF and FISH-IF) (Almeida et al., 1998); mouse monoclonal anti-NAP57 immunoglobulin G (IgG) (H3 at 1:500 for IF; Santa Cruz Biotechnology); mouse monoclonal anti-fibrillarin culture supernatant (38F3 at 1:2000 for WB; EnCor Biotechnology); mouse monoclonal anti-Sm antibodies (Y12 at 1:100 for IF; Abcam); mouse anti-SMN ascites fluid (MANSMA1 at 1:100 for IF) (Young et al., 2000); mouse anti–γ-tubulin ascites fluid (GTU-88 at 1:5000 for WB; Sigma); DyLight488 goat anti-mouse IgG (1:500 for IF) and rhodamine (TRITC) goat anti-rabbit IgG (1:500 for IF; both Jackson Immuno Research); Alexa Fluor 647 goat anti-mouse IgG (1:500 for IF), Alexa Fluor 350 goat anti-mouse IgG (1:1500 for FISH-IF), Alexa Fluor 488 goat anti-rabbit IgG (1:1500 for FISH-IF), and Alexa Fluor 680 goat anti-rabbit IgG (1:10,000 for WB; all four from Thermo Fisher Scientific); IRDyeTM 800 goat anti-mouse IgG (1:10,000 for WB; Rockland Immunochemicals).
WBs
For each experiment, proteins from the same number of cells per condition were extracted into SDS-sample buffer (0.5 M Tris; pH6.8; 12% SDS; 0.05% bromophenol blue). The lysates were tip sonicated and total proteins were loaded (100,000 cell equivalents), separated on 9% SDS–PAGE, followed by transfer to nitrocellulose membrane. Transfer efficiency was confirmed by Ponceau red staining, and membranes were blocked in blocking buffer (Tris-buffered saline, 0.1% Tween, and 2.5% nonfat dry milk) for 30 min before incubation with primary antibodies diluted in blocking buffer overnight at 4°C. After extensive washes in blocking buffer, membranes were incubated with appropriate secondary antibodies diluted in blocking buffer for 1 h at room temperature in the dark. After extensive washes in blocking buffer, membranes were scanned on an Odyssey 9120 Imaging System (LI-COR Biosciences), and protein bands were quantified using Image Studio Lite (LI-COR Biosciences).
Indirect IF
Cells grown on coverslips were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 20 min, permeabilized with 1% Triton X-100 in PBS for 5 min, and blocked with 1% powdered milk in PBS (IF blocking buffer) for 15 min. The cells were then incubated for 2 h with primary antibodies in IF blocking buffer, washed, and incubated for 1 h with secondary antibodies in IF blocking buffer in the dark. This was followed by washing and nuclear staining with 4′,6-diamidino-2-phenylidone (DAPI; 1 µg/ml in PBS). Coverslips were mounted on glass slides using ProLong Diamond Antifade Mount (Thermo Fisher Scientific) and observed using a Zeiss Axio Observer Z1 fluorescence microscope (63× objective, NA 1.4) with filter sets 34-DAPI (Zeiss #000000-1031-334), 10-AF488 (Zeiss #488010-9901-000), 43HE-DsRED (Zeiss #489043-9901-000), and 50-Cy5 (Zeiss #488050-9901-000). Z-stack images in 200-nm steps were acquired with a Zeiss AxioCam MRm camera using Axiovision software (Zeiss). Maximum projections were generated using ImageJ (National Institutes of Health [NIH]). Quantification of protein signals in CBs was done using ImageJ with the help of macros (available upon request). Briefly, coilin images were used to locate the centroids of CBs around which masks were generated of circles with 8 pixel diameter (0.1024 µm/pixel). These masks were applied to the CB protein images to determine their signal intensity in CBs. Background was subtracted individually for each CB and was defined as the pixel with the lowest signal in a 2-pixel circumference of the mask using an ImageJ function. Images for figures were cropped and adjusted using Photoshop CC (Adobe). To compare parent and KD cell images, all images within the same panels and of the same antigens were acquired and adjusted identically. Methods to establish statistical significance are indicated in the figure legends and were calculated and plotted using Prism 8 (GraphPad Software).
FISH-IF
Cells on coverslips were washed with 5 mM MgCl2 in PBS (PBSM), fixed with 4% PFA in PBSM for 10 min, quenched with 50 mM Glycine in PBSM for 5 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with bovine serum albumin (BSA; 0.5 g/l) in PBSM (FISH-IF blocking buffer) for 15 min. Cells were then incubated with primary antibodies in FISH-IF blocking buffer for 1 h followed by secondary antibodies in FISH-IF blocking buffer for 1 h in the dark. After washes in FISH-IF blocking buffer, cells were incubated in 70% ethanol for 10 min followed by prehybridization buffer (2× SSC; 10% formamide; 0.5 g/l BSA; 1:1000 RNase OUT) for 30 min. Probes labeled with Quasar 570 on their 5′ and 3′ ends (Biosearch Technologies, each at 125 nM final; Table 2) were applied in hybridization buffer (2× SSC; 10% formamide; 0.5g/l BSA; 1 µg/µl yeast tRNA; 2 mM vanadyl ribonucleoside complex; 10% dextran sulfate; 1:100 RNase OUT) for 3 h at 37°C in the dark, then washed with wash buffer 1 (2× SSC; 10% formamide), followed by wash buffer 2 (2× SSC), and the coverslips were mounted as described for IF. The samples were observed using an Olympus BX61 epifluorescence microscope with PlanApo 60×, 1.4NA, oil-immersion objective (Olympus). An X-Cite 120 PC (Lumen Dynamics, Canada) light source was used for illumination, with filter sets DAPI-5060C-Zero (Semrock), Cy3-41007a (Chroma), and FITC-5050A-Zero (Semrock). Z-stack images in 200-nm steps were acquired with a CoolSNAP HQ camera (Photometrics) using MetaMorph (Molecular Devices) and processed as described for IF.
TABLE 2:
RNA-FISH probes.
| Target | RNA family | Probe name | Sequence |
|---|---|---|---|
| SCARNA19 (hTR) | H/ACA scaRNA | hTR-1 | GACAAAAAATGGCCACCACC |
| hTR-2 | GTCAGCGAGAAAAACAGCGC | ||
| hTR-3 | GCTCTAGAATGAACGGTGGA | ||
| hTR-4 | CAGCAGCTGACATTTTTTGT | ||
| hTR-5 | AGGCTTTTCCGCCCGCTGAA | ||
| hTR-6 | AAAGGCCTGAACCTCGCCCT | ||
| hTR-7 | CTCGCTCCGTTCCTCTTCCT | ||
| hTR-8 | ATGTGTGAGCCGAGTCCTGG | ||
| SCARNA5 (U87) | Hybrid scaRNA | U87-1 | GTGCAATCAGACCTTTTACC |
| U87-2 | AACAAAGGGACCGTGACATT | ||
| U87-3 | TACACATGACTTAAATGTGA | ||
| U87-4 | GAAACACACACTGACAGAAT | ||
| U87-5 | AGCTGTGAGCCGATCACTCT | ||
| U87-6 | CTGGTTTTGTTGGATACTCG | ||
| U87-7 | CACTCGTCAGTCTCCTGTGT | ||
| U87-8 | GGTCTCAGATTGAAAACTTG | ||
| SCARNA13 (U93) | Tandem H/ACA scaRNA | U93-1 | CGGCTCCAAGACTACAGATT |
| U93-2 | AACAGCTGGCTCTCGAGCAG | ||
| U93-3 | ATCAGAGGAAAATTGCACAT | ||
| U93-4 | GTGGCAACAGTGACCAGAAA | ||
| U93-5 | AATGACATAGCCCAGTCATT | ||
| U93-6 | CTCTTACTGTTGGCGGATAG | ||
| U93-7 | CAATATCTCGACTGCAAAGC | ||
| U93-8 | CTTGTGGCAGTACTTAGTGT | ||
| SCARNA9 | Tandem C/D scaRNA | SCARNA9-1 | TCCACTTCACTAAAAGCAGA |
| SCARNA9-2 | GATTTGGCTAGTTTCATCAT | ||
| SCARNA9-3 | CTGGAAAGACTTCTGATGCT | ||
| SCARNA9-4 | GTAGAGATCATGCATCAGGT | ||
| SCARNA9-5 | GTTTTATAGTTTTGCTTCTC | ||
| SCARNA9-6 | AGACATATGCCCTTATTGTT | ||
| SCARNA9-7 | CTACAGCTGTTATCTTTATA | ||
| SCARNA9-8 | CCACCCTCAATCTCATTCAT | ||
| SCARNA9-9 | ATAGACATACATACGCACCC | ||
| SCARNA9-10 | TTGCCCAGAAATGATTAGGC | ||
| SCARNA9-11 | AGCTCAGGTCAAGTGTAGAA | ||
| SCARNA28 | C/D scaRNA | SCARNA28-1 | CAGCCAGTATTACTCATCAC |
| SCARNA28-2 | CACAAACACACACACGTGCC | ||
| SCARNA28-3 | GCATGCACTGACAAGCATAT | ||
| SCARNA28-4 | CATTTGTACTCCCAGACATA | ||
| SCARNA28-5 | ACATAGCTAGTTCCCTACAA | ||
| SCARNA28-6 | GACTGTCATGGCCTAATAGA | ||
| SCARNA28-7 | AGCAATCAGATCTTATCAGT | ||
| U2 | snRNA | U2-1 | CCAAAAGGCCGAGAAGCGAT |
| U2-2 | ACAGATACTACACTTGATCT | ||
| U2-3 | CTCGGATAGAGGACGTATCA | ||
| U2-4 | CCTGCTCCAAAAATCCATTT | ||
| U2-5 | CGTGGAGTGGACGGAGCAAG | ||
| U2-6 | TGGAGGTACTGCAATACCAG |
Electron microscopy
Monolayers of cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide followed by 2% uranyl acetate, and dehydrated through a graded series of ethanol, and the cells were lifted from the monolayer with propylene oxide and embedded as a loose pellet in LX112 resin (LADD Research Industries, Burlington, VT) in Eppendorf tubes. Ultrathin sections were cut on a Leica Ultracut UC7, stained with uranyl acetate followed by lead citrate, and viewed on a JEOL 1400 Plus transmission electron microscope at 80 kv.
Total RNA extraction
RNA from the different cell lines was extracted using 500 µl TRIzol Reagent (Ambion) directly on 10-cm dishes (cell confluency ∼80%, ∼1,000,000 cells). Lysed cells were scraped into tubes, chloroform was extracted twice, and the RNA was precipitated with 0.7 vol isopropanol and 20 µg glycogen and resuspended in UltraPure distilled water. RNA concentration and quality were determined by Nanodrop (ratio 260/230 and 260/280 above 1.8). Total RNA was used as template for RT-qPCR analysis.
RT-qPCR
Residual DNA contamination was removed from 2 µg of RNA using DNase RQ1. 1 µg RNA (the other half was used as no RT control) was used for cDNA synthesis using random hexamers and SuperScript III reverse transcriptase (Thermo Fisher Scientific). cDNA corresponding to 5 ng equivalents of RNA templated the qPCR reaction with Power SYBR Green PCR master mix and amplified on a QuantStudio 6 Flex real-time PCR system (Applied Biosystems) The gene expression levels were calculated by comparative CT approach (ΔΔCT) and normalized for that of GLUD2 mRNA. Primers (Thermo Fisher Scientific) used for RT-qPCR are described in Table 1.
RNA-IP and RT-PCR
Cells (2,000,000 from two 10-cm dishes) were lysed in buffer A (20 mM Tris-HCl, pH 7.9; 150 mM NaCl; 1% Triton X-100; 0.2% SDS; 2 mM EDTA; protease inhibitor, complete EDTA-free cocktail tablet; Roche, 1 tablet/50 ml) and incubated on ice for 10 min. Lysates were cleared by centrifugation at 15,000 rpm (21.1 × g) for 10 min. A 6000 cell equivalent was kept as input control and 60,000 cell equivalents were incubated with anti-WDR79 serum at a dilution of 1:500 for 1 h at room temperature. Incubations were added to 25 µl of protein A sepharose beads (GE Healthcare) for 1 h at room temperature. The beads were washed once with buffer A, three times with buffer B (20 mM Tris-HCl, pH 7.4; 150 mM NaCl; 0.02% Triton X-100), and once with buffer C (20 mM Tris-HCl, pH 7.4; 150 mM NaCl) before RNA extraction using TRIzol (Ambion, following the manufacturer’s instructions). RNA concentrations of the inputs were determined using Nanodrop. Semiquantitative RT-PCR was performed using SuperScript III One-Step RT-PCR System (Thermo Fisher Scientific) on 200 ng of total input RNA and all RNA from the beads (input = 1/10 of precipitates). Primers (Thermo Fisher Scientific) used for RT-PCR are described in Table 1.
Telomere restriction fragment length analysis
DNA was isolated from the different cell lines at different population doublings on 15-cm plates (confluency roughly 80%) and treated overnight with proteinase K (in 10 mM Tris, pH 7.4; 100 mM NaCl; 10 mM EDTA; 1% SDS; 100 µg/ml proteinase K). DNA was extracted with phenol/chloroform and digested overnight with HinfI, Alu1, MboI, and RsaI (NEB enzymes in NEB buffer 2). Approximately 3 µg of the digested DNA was fractionated by electrophoresis on 1% agarose gels. Gels were dried and DNA was denatured in gel for 30 min with 0.5 M NaOH and 1.5 M NaCl. Telomeres were detected by hybridization at 55°C overnight with a 32P end-labeled oligonucleotide probe (TTAGGG)4. Probe (2 µg) was prepared with γ-32P-ATP using T4 Kinase (Thermo Fisher Scientific) in Church buffer (0.5 M sodium phosphate, pH 7.2; 1 mM EDTA; 7% SDS; 10 g/l BSA). Gels were scanned on a Typhoon PhosphorImager (GE Healthcare). Lanes were quantified using ImageJ (NIH) and median telomere length was determined in Excel (Microsoft).
Supplementary Material
Acknowledgments
We are grateful to John Reidhaar-Olson of the Einstein Gene Modification (formerly shRNA) Facility for generation and transduction of the Cas9 and NOLC1-sgRNA lentiviruses. We acknowledge for the referenced antibodies Maria Carmo-Fonseca, Phil Young, Francois Dragon, and Witek Filipowicz and Maria Vera for pGL3-UBC. The microscopes used are maintained by the Einstein Analytical Imaging Facility (AIF), which is partially supported by the NIH-funded Einstein Cancer Center (P30CA013330). We thank Maria Vera for her assistance with RNA-FISH and Rob Singer for use of his microscope. Leslie Gunther-Cummins (AIF) prepared and imaged the samples on an electron microscope supported by an NIH-funded Shared Instrumentation Grant (S10OD016214).
This work was supported by grants from the National Institutes of Health, HL136662 (to U.T.M.) and CA116352 (to S.S.).
Abbreviations used:
- BSA
bovine serum albumin
- CB
Cajal body
- DAPI
4′,6-diamidino-2-phenylidone
- DC
dyskeratosis congenita
- DFC
dense fibrillar component
- FC
fibrillar center
- FISH
fluorescence in situ hybridization
- GC
granular component
- hTR
human telomerase RNA
- IF
immunofluorescence
- IP
immunoprecipitation
- KD
knockdown
- NIH
National Institutes of Health
- P1
parent 1
- P2
parent 2
- PBS
phosphate-buffered saline
- PBSM
5 mM MgCl 2 in phosphate-buffered saline
- PFA
paraformaldehyde
- RNP
ribonucleoprotein
- sca
small Cajal body–specific RNP
- sg
single guide
- sn
small nuclear
- sno
small nucleolar
- TERT
telomerase reverse transcriptase
- WB
Western blot
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-08-0429) on October 30, 2019.
REFERENCES
- Almeida F, Saffrich R, Ansorge W, Carmo-Fonseca M. (1998). Microinjection of anti-coilin antibodies affects the structure of coiled Bodies. J Cell Biol , 899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Pai C, Huang J, Yeh N. (1999). Human Nopp140, which interacts with RNA polymerase I: implications for rRNA gene transcription and nucleolar structural organization. Mol Cell Biol , 8536–8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Roake CM, Freund A, Batista PJ, Tian S, Yin YA, Gajera CR, Lin S, Lee B, Pech MF, et al (2018). An activity switch in human telomerase based on RNA conformation and shaped by TCAB1. Cell , 218–230.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. (2007). Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell , 882–889. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. (2006). Telomere length homeostasis requires that telomerase levels are limiting. EMBO J , 565 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. (2002). Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J , 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. (2006). Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol , 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Pogacic´ V, Filipowicz W. (2000). In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol Cell Biol , 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enwerem II, Velma V, Broome HJ, Kuna M, Begum RA, Hebert MD. (2014). Coilin association with Box C/D scaRNA suggests a direct role for the Cajal body marker protein in scaRNP biogenesis. Biol Open , 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. (2003). The centennial of the Cajal body. Nat Rev Mol Cell Bio , 975–980. [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al (2015). High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell , 1515–1526. [DOI] [PubMed] [Google Scholar]
- Isaac C, Pollard J, Meier UT. (2001). Intranuclear endoplasmic reticulum induced by Nopp140 mimics the nucleolar channel system of human endometrium. J Cell Sci , 4253–4264. [DOI] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. (1998). Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol , 319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa K, Nobe Y, Ishikawa H, Yamauchi Y, Taoka M, Sato K, Nakayama H, Simpson RJ, Isobe T, Takahashi N. (2019). TDP-43 regulates site-specific 2′-O-methylation of U1 and U2 snRNAs via controlling the Cajal body localization of a subset of C/D scaRNAs. Nucleic Acids Res , 2487–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T. (2004). Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol , 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Richard P, Bertrand E, Kiss T. (2006). Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell , 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N-K, Theimer CA, Mitchell JR, Collins K, Feigon J. (2010). Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res , 6746–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2001). Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J , 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. (2006). SnoRNP biogenesis meets Pre-mRNA splicing. Mol Cell , 775–776. [DOI] [PubMed] [Google Scholar]
- Kittur N, Zapantis G, Aubuchon M, Santoro N, Bazett-Jones DP, Meier UT. (2007). The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol Biol Cell , 2296–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell , 774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R. (2006). Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell , 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. (2013). Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA , 17–34. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Corcoran M, Méndez-Vidal C, Wiman KG, Farnebo M. (2009). Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol Cell , 462–471. [DOI] [PubMed] [Google Scholar]
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Söderberg O, Strömblad S, Wiman KG, Farnebo M. (2010). WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol , e1000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Richard P, Pinzón N, Kiss T. (2014). Targeting vertebrate intron-encoded box C/D 2′-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res , 6616–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PJ, Bessler M. (2011). The genetics of dyskeratosis congenita. Cancer Genet , 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. (1996). Comparison of the rat nucleolar protein Nopp140 with its yeast homolog SRP40 differential phosphorylation in vertebrates and yeast. J Biol Chem , 19376–19384. [PubMed] [Google Scholar]
- Meier UT. (2005). The many facets of H/ACA ribonucleoproteins. Chromosoma , 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT. (2016). RNA modification in Cajal bodies. RNA Biol , 693-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT, Blobel G. (1992). Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell , 127–138. [DOI] [PubMed] [Google Scholar]
- Meier UT, Blobel G. (1994). NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol , 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. (1999). A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol , 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na J-H, Lee W-K, Yu Y. (2018). How do we study the dynamic structure of unstructured proteins: a case study on Nopp140 as an example of a large, intrinsically disordered protein. Int J Mol Sci , 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Speckmann W, Terns R, Terns MP. (1999). Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell , 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. (2018). Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature , 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa R-S, Gilbert N. (2019). RNA: nuclear glue for folding the genome. Trends Cell Biol , 201–211. [DOI] [PubMed] [Google Scholar]
- Pogacic V, Dragon F, Filipowicz W. (2000). Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol , 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Henning D, Busch H. (1979). Nucleotide sequence of nucleolar U3B RNA. J Biol Chem , 11097–11105. [PubMed] [Google Scholar]
- Reddy R, Henning D, Busch H. (1980). Substitutions, insertions, and deletions in two highly conserved U3 RNA species. J Biol Chem , 7029–7033. [PubMed] [Google Scholar]
- Renvoisé B, Colasse S, Burlet P, Viollet L, Meier UT, Lefebvre S. (2009). The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy. Hum Mol Genet , 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. (2003). A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J , 4283–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ, Singer RH, Bertrand E. (1998). The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J , 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA. (2009 [updated 2019]). Dyskeratosis congenita. GeneReviews [Internet].
- Sawyer IA, Sturgill D, Dundr M. (2019). Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip Rev RNA , e1514. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Cech TR. (2015). Human telomerase: biogenesis, trafficking, recruitment, and activation. Gene Dev , 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Zaug AJ, Cech TR. (2016). Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell , 1188–1197.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneˇk D, Fox AH. (2017). Nuclear bodies: news insights into structure and function. Curr Opin Cell Biol , 94–101. [DOI] [PubMed] [Google Scholar]
- Staneˇk D, Neugebauer KM. (2006). The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma , 343–354. [DOI] [PubMed] [Google Scholar]
- Stern LJ, Zyner KG, Pickett HA, Cohen SB, Bryan TM. (2012). Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol , 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantos A, Szrnka K, Szabo B, Bokor M, Kamasa P, Matus P, Bekesi A, Tompa K, Han K-H, Tompa P. (2013). Structural disorder and local order of hNopp140. Biochim Biophys Acta , 342–350. [DOI] [PubMed] [Google Scholar]
- Thiry M, Cheutin T, Lamaye F, Thelen N, Meier UT, O’Donohue M-F, Ploton D. (2009). Localization of Nopp140 within mammalian cells during interphase and mitosis. Histochem Cell Biol , 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. (2006). Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell , 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y-T, Lin C-I, Chen H-K, Lee K-M, Hsu C-Y, Yang S-J, Yeh N-H. (2008). Chromatin tethering effects of hNopp140 are involved in the spatial organization of nucleolus and the rRNA gene transcription. J Biomed Sci , 471–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KE, Berciano M, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan E, Lafarga M, Conlon RA, Matera GA. (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol , 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu M-D, Kukoyi A, Steitz JA. (2009). A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell , 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Reu E, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science , 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine D, Samarsky D, Mouaikel J, Blanchard J, Bordonné R, Bertrand E. (2002). Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J , 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, Hockemeyer D, Collins K. (2016). Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. Elife , e18221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. (2015). Identification and characterization of essential genes in the human genome. Science , 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Lührmann R. (2004). Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell , 789–798. [DOI] [PubMed] [Google Scholar]
- Wu Z, Murphy C, Wu C, Tsvetkov A, Gall J. (1993). Snurposomes and coiled bodies. Cold Spring Harb Sym Quant Biol , 747–754. [DOI] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT. (2000). Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell , 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PJ, Le TT, thi Man N, Burghes A, Morris GE. (2000). The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res , 365–374. [DOI] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. (2011). Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Gene Dev , 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Brangwynne CP. (2015). Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol , 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.