Abstract
Adoptive cell therapy represents a new paradigm in cancer immunotherapy but can be limited by poor persistence and function of transferred T cells1. Here, through an in vivo pooled CRISPR-Cas9 mutagenesis screening, we demonstrate that CD8+ T cells are reprogrammed to long-lived effector cells with extensive accumulation, better persistence and robust effector function in tumors by targeting Regnase-1. Regnase-1-deficient CD8+ T cells show markedly improved therapeutic efficacy against mouse melanoma and leukemia. Through a secondary genome-scale CRISPR-Cas9 screening, we identify BATF as the key target of Regnase-1 and a rheostat in shaping antitumor responses. Loss of BATF suppresses the elevated accumulation and mitochondrial fitness of Regnase-1-deficient CD8+ T cells. Conversely, we reveal that targeting additional signaling factors including PTPN2 and SOCS1 improves the therapeutic efficacy of Regnase-1-deficient CD8+ T cells. Our findings suggest that T-cell persistence and effector function can be coordinated in tumor immunity and point to new avenues to improve the efficacy of adoptive cell therapy for cancer.
Adoptive cell therapy (ACT), including the use of T cells engineered to express chimeric antigen receptors (CARs), has produced unprecedented clinical outcomes for cancer immunotherapy. However, the therapeutic efficacy, especially in solid tumors, is often limited by poor in vivo accumulation, persistence and function of adoptively transferred T cells1. Paradoxically, terminal effector CD8+ T cells have been shown to have reduced antitumor efficacy and exhibit poor in vivo persistence2. How T-cell fate decision is regulated in the tumor microenvironment (TME) remains poorly understood.
Here through an in vivo pooled CRISPR-Cas9 mutagenesis screening of metabolism-associated factors, we identified Regnase-1 as a major negative regulator of antitumor responses. Regnase-1-deficient CD8+ T cells are reprogrammed in TME to long-lived effector cells by enhancing BATF function and mitochondrial metabolism, thereby improving ACT for cancer.
CRISPR screening for metabolic regulators of ACT
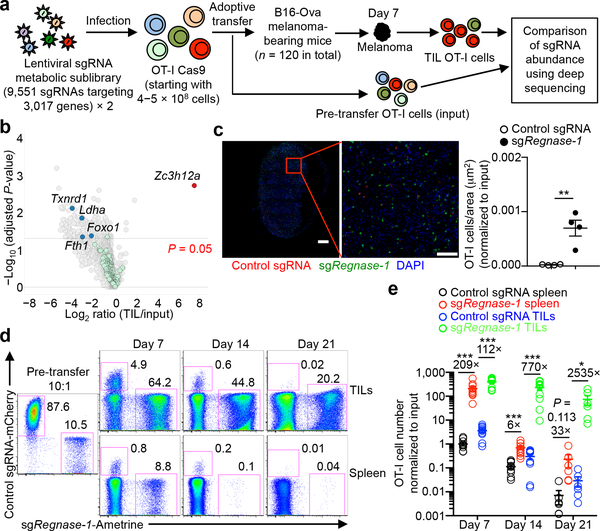
T-cell longevity and function in cancer immunotherapy have been proposed to closely correlate with cell metabolic fitness3, although the underlying molecular mechanisms are unclear. To systematically investigate the roles of metabolism-associated factors in T-cell−mediated antitumor immunity, we developed a pooled CRISPR mutagenesis screening approach in an ACT model (Fig. 1a), using CD8+ T cells expressing the OT-I T-cell receptor (TCR) and Cas9 and mice inoculated with B16 melanoma cells expressing the cognate antigen (B16-Ova). We developed two lentiviral sub-libraries of sgRNAs (6 sgRNAs per gene) targeting 3,017 metabolic enzymes, small molecule transporters, and metabolism-related transcriptional regulators (Supplementary Table 1). Seven days after adoptive transfer, sgRNA-transduced OT-I cells in tumor-infiltrating lymphocytes (TILs) were examined for library representation. A total of 218 genes were significantly depleted including Txnrd14, Ldha5, Fth16 and Foxo17, known regulators of cell survival and expansion (Fig. 1b and Supplementary Table 2). Strikingly, Zc3h12a (also known as Regnase-1, encodes Regnase-1) was the most highly enriched gene (Fig. 1b), suggesting that Regnase-1 could be a major negative regulator of antitumor responses. Regnase-1 has RNase activity and regulates immune cell activation8,9, but the function of Regnase-1 in tumor immunity remains unclear.
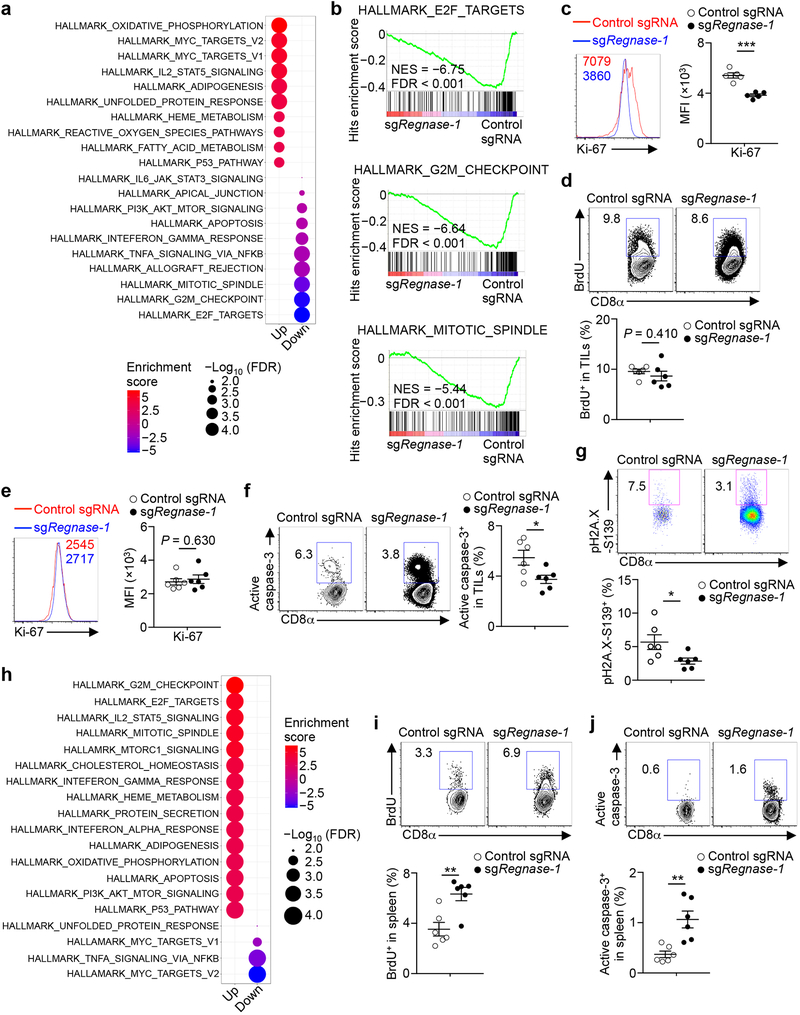
Figure 1. In vivo CRISPR screening identifies Regnase-1 as a major negative regulator of CD8+ T cell antitumor responses.
(a) Diagram of CRISPR screening for metabolic regulators of ACT. (b) Scatterplot of the enrichment of candidates (n = 6 sgRNAs per gene) with the most extensively enriched (red) and selective depleted (blue) genes, as well as ‘dummy’ genes (green; generated by random combinations of 6 out of 1,000 non-targeting control sgRNAs per ‘dummy’ gene) highlighted. (c) Representative images (left) and quantification of relative OT-I cell number per area (μm2) normalized to input (right) in the tumor section (n = 4). OT-I cells transduced with control sgRNA (red) and sgRegnase-1 (green) were mixed at a 10:1 ratio and transferred into tumor-bearing mice, and analyzed at day 7. Scale bars, 500 μm. (d, e) Control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed at a 10:1 ratio and transferred into tumor-bearing mice, followed by analyses of the proportion of OT-I cells in total CD8α+ cells (d), and quantification of normalized OT-I cell number relative to input (e) at days 7 (n = 10), 14 (n = 10) and 21 (n = 6). Cell number in the tumor indicates per gram tissue. Mean ± s.e.m. in c, e. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed paired Student’s t-test followed by Bonferroni correction in b, two-tailed unpaired Student’s t-test in c, e. Data are representative of two (c, d), or pooled from two (e) independent experiments.
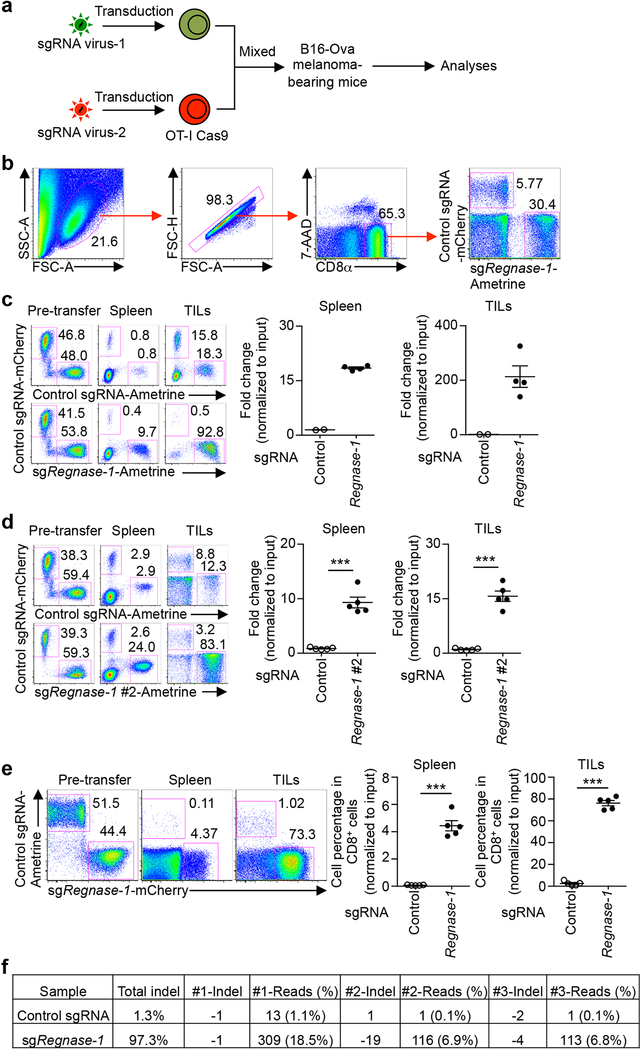
To validate our findings, we developed an in vivo dual transfer system to compare OT-I cells transduced with sgRNA vectors expressing distinct fluorescent proteins in the same tumor-bearing host (Extended Data Fig. 1a, b), without noticeable effects of different fluorescent proteins per se (Extended Data Fig. 1c, d, upper panels). We tested OT-I cells transduced with two different sgRNAs targeting Regnase-1 and found that the relative proportion of Regnase-1-null OT-I cells was drastically increased in both the spleen and tumor (Extended Data Fig. 1c–e). Imaging analysis identified significantly more Regnase-1-null OT-I cells within tumors than wild-type controls (Fig. 1c). Analyses of guide targeting efficacy revealed efficient disruption of Regnase-1 (Extended Data Fig. 1f). Next, we examined the persistence of Regnase-1-null OT-I cells at days 7, 14 and 21 after transfer. While wild-type OT-I cells declined over time, Regnase-1-null cells had markedly better persistence, especially in the tumor and at later time points (Fig. 1d, e). Therefore, loss of Regnase-1 endows tumor-specific CD8+ T cells with greatly improved accumulation and persistence, preferentially in the tumor.
Loss of Regnase-1 improves ACT efficacy
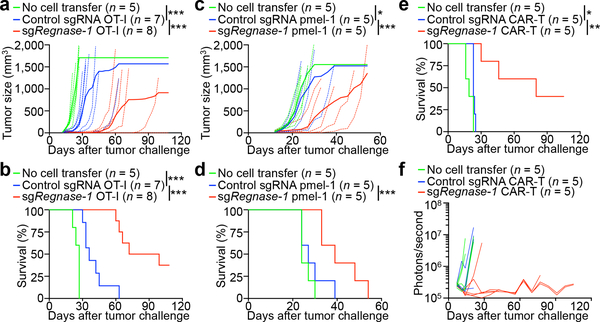
We assessed the efficacy of Regnase-1-null CD8+ T cells in ACT targeting different tumors. In the B16-Ova melanoma model, Regnase-1-null OT-I cells showed much stronger antitumor effects than wild-type cells, evidenced by markedly inhibited tumor growth and increased survival of melanoma-bearing mice (Fig. 2a, b). Similar results were observed in pmel-1 TCR-expressing T cells, which recognize the endogenous melanoma antigen gp100, transferred into B16-F10 melanoma-bearing mice (Fig. 2c, d). To assess the efficacy of CAR-T cells against leukemia, we used T cells expressing CARs targeting human CD19 (huCD19) and BCR-ABL1+ B progenitor acute lymphoblastic leukemia (Ph+ B-ALL) cells10 expressing huCD19 (huCD19-Ph+ B-ALL). Regnase-1-null CAR-T cells showed much stronger therapeutic efficacy than wild-type cells as indicated by mouse survival (Fig. 2e) and tumor burden analyses (Fig. 2f). Collectively, Regnase-1 deletion markedly enhances the efficacy of ACT against both solid and blood cancers.
Figure 2. Deletion of Regnase-1 enhances efficacy of ACT against solid and blood cancers.
OT-I (a, b), pmel-1 (c, d) or CD8+ CAR-T (e, f) cells (5 × 106) transduced with non-targeting control sgRNA or sgRegnase-1 were transferred into mice at day 12 after B16-Ova (a, b) or B16-F10 (c, d) melanoma engraftment, or at day 7 after Ph+ B-ALL cell engraftment (e, f), followed by analyses of tumor size (a, c), mouse survival (b, d, e) and tumor burden via Xenogen imaging of bioluminescent signal intensities (f). Non-treatment control mice received no T cell transfer. *P < 0.05; **P < 0.01; ***P < 0.001; two-way ANOVA in a, c, and Log-rank (Mantel-Cox) test in b, d, e. Data are representative of two (a, b, e, f) or four (c, d) independent experiments.
Regnase-1 deletion reprograms T cells in TME
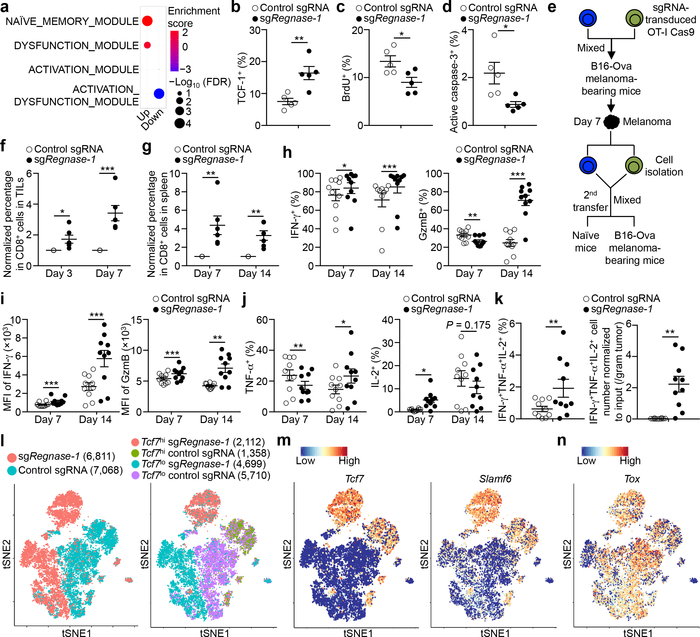
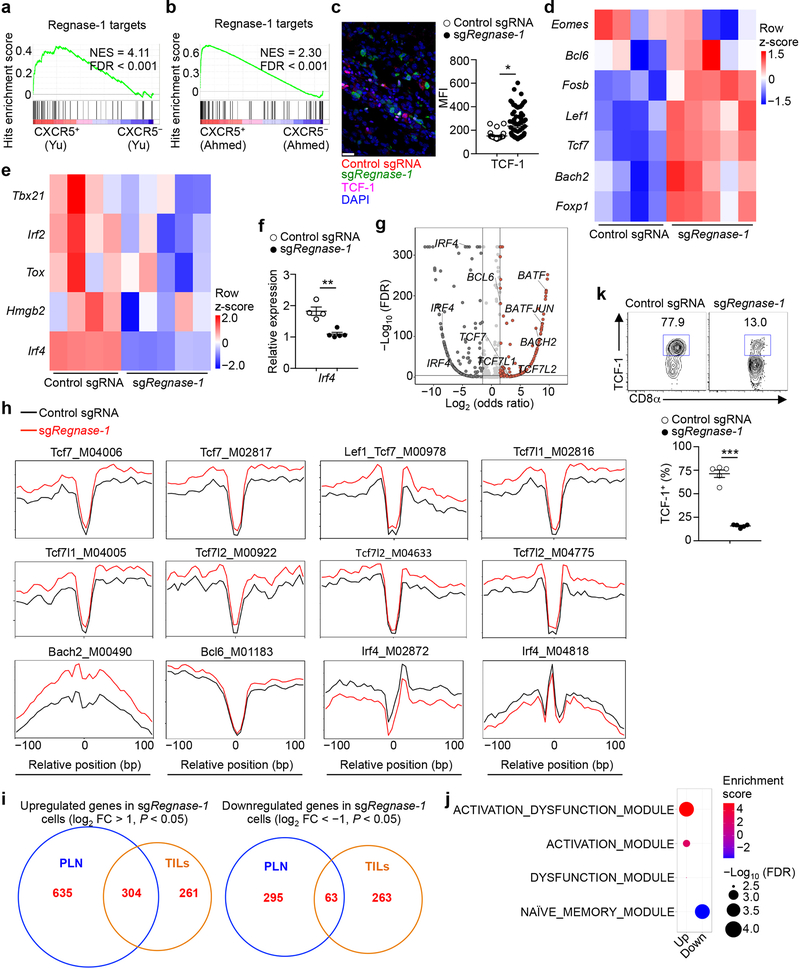
We performed RNA-Sequencing (RNA-Seq) of Regnase-1-null and wild-type OT-I cells isolated from the in vivo dual transfer system to address cell-intrinsic effects. Gene set enrichment analysis (GSEA) using gene modules associated with different functional states of tumor-infiltrating CD8+ T cells11 revealed that tumor-infiltrating Regnase-1-null cells were enriched with the naïve or memory module (Fig. 3a). Gene targets repressed by Regnase-1 were also enriched in memory-like CD8+ T cells in chronic infection12,13 (Extended Data Fig. 2a, b). Accordingly, tumor-infiltrating Regnase-1-null cells had increased expression of TCF-1, a transcription factor associated with naïve or memory T-cells14 (Fig. 3b, Extended Data Fig. 2c), as well as Lef1, Bach2, Tcf7 (encodes TCF-1), Foxp1, Bcl6, and Fosb14–17, but had lower expression of Irf2, Irf4 and Hmgb218,19 (Extended Data Fig. 2d–f). We next measured chromatin accessibility using ATAC-Seq20 of tumor-infiltrating Regnase-1-null and wild-type cells. Motif searches on accessible regions identified enrichment of TCF-1, Bach2 and Bcl6 but downregulated IRF4 motifs in Regnase-1-null cells (Extended Data Fig. 2g, h). Thus, Regnase-1-null CD8+ T cells are reprogrammed in TME with enhanced naïve/memory cell-associated gene expression programs.
Figure 3. Deletion of Regnase-1 reprograms tumor-infiltrating CD8+ T cells to long-lived effector cells.
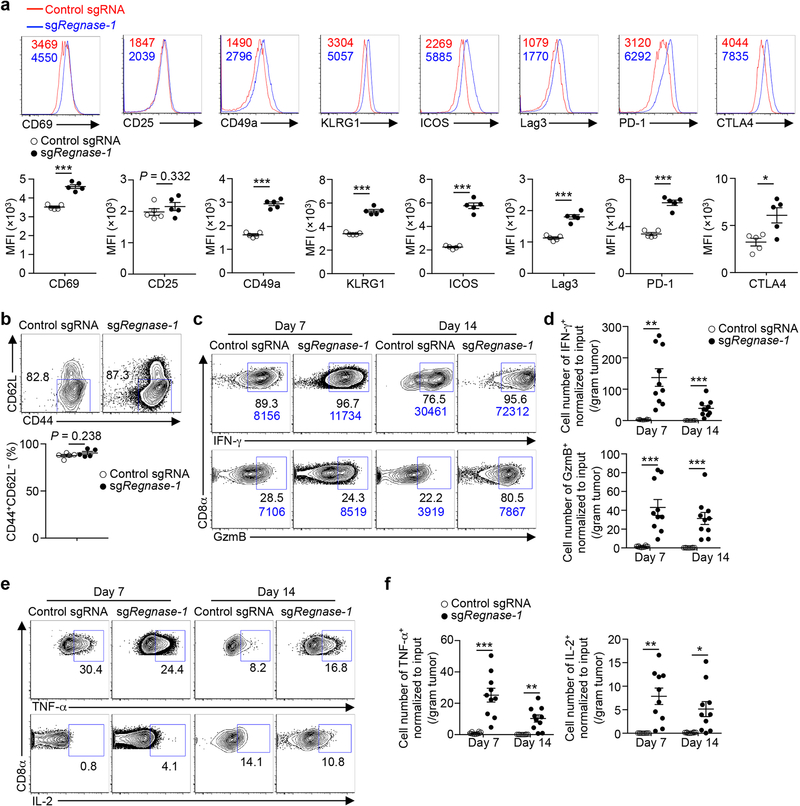
(a) GSEA enrichment plots of RNA-Seq from sgRegnase-1- (n = 5) versus non-targeting control sgRNA- (n = 4) transduced OT-I cells isolated from TILs from the dual transfer system, using gene sets of tumor-infiltrating CD8+ T cell activation states11. (b–d) Tumor-infiltrating sgRNA-transduced OT-I cells from the dual transfer system (n = 5) were analyzed at days 7 (b) and 14 (c, d) for the quantification of frequencies of TCF-1+ (b), BrdU+ (c) and active caspase-3+ (d) cells. (e–g) Diagram of in vivo persistence assay (e): sgRNA-transduced OT-I cells were isolated from TILs, mixed at a 1:1 ratio (1 × 105 each) and transferred into tumor-bearing hosts (f) or naïve mice (g). Quantification of normalized OT-I cell frequency in TILs of tumor-bearing hosts (n = 6) (f) or in the spleen of naïve hosts (n = 6) (g). (h–k) Tumor-infiltrating sgRNA-transduced OT-I cells from the dual transfer system were analyzed at days 7 (n = 10) and 14 (n = 10) for the quantification of frequencies of IFN-γ+ cells (h, left), GzmB+ cells (h, right), TNF-α+ cells (j, left), IL-2+ cells (j, right) and polyfunctional IFN-γ+TNF-α+IL-2+ cells (k, left) in OT-I cells, and mean fluorescence intensity (MFI) of IFN-γ and GzmB in IFN-γ+ and GzmB+ cells, respectively (i), and cell number (normalized to input) per gram tissue (k, right) of polyfunctional IFN-γ+TNF-α+IL-2+ OT-I cells. (l–n) scRNA-Seq analysis of tumor-infiltrating sgRNA-transduced OT-I cells isolated from the dual transfer system at day 7. tSNE visualization of OT-I cells indicating genotypes (l, left), Tcf7hi and Tcf7lo cells (l, right), and Tcf7 (m, left), Slamf6 (m, right) and Tox (n) gene expression in individual cells. Mean ± s.e.m. in b–d, f–k. *P < 0.05; **P < 0.01; ***P < 0.001; Kolmogorov-Smirnov test followed by Benjamini-Hochberg correction in a; two-tailed unpaired Student’s t-test in b–d, f, g; two-tailed paired Student’s t-test in h–k. Data are representative of three (b), or pooled from two (c, d, f–k) independent experiments.
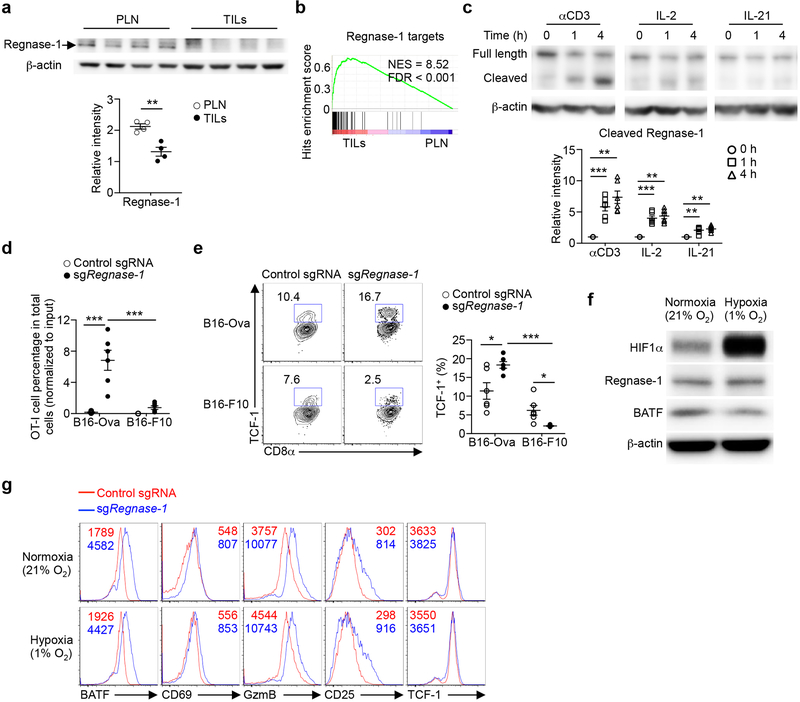
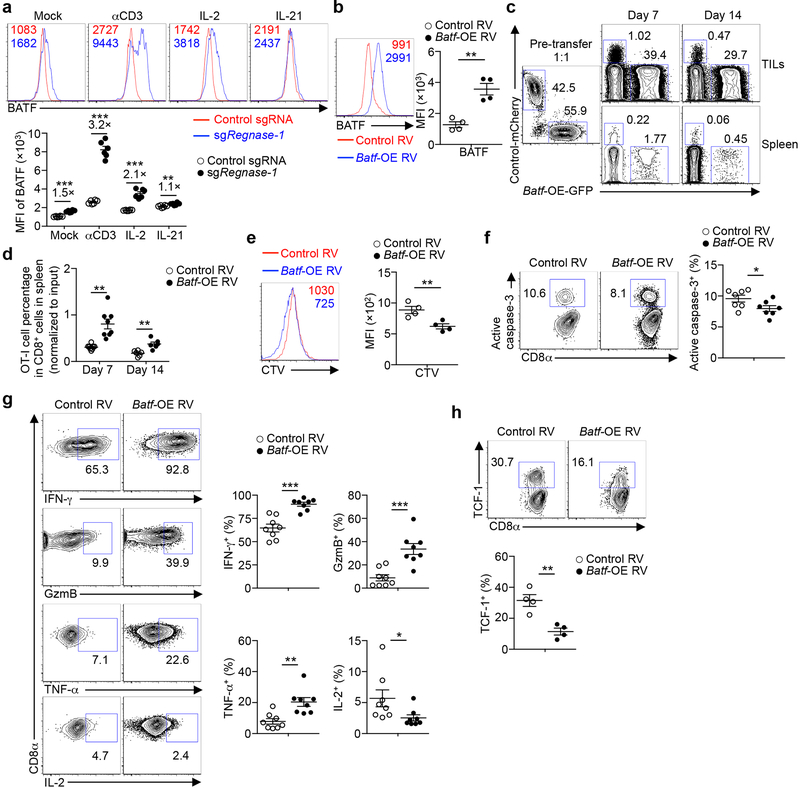
Transcriptional profiling revealed marked differences between tumor-infiltrating and peripheral Regnase-1-null cells (Extended Data Fig. 2i). Unlike the enrichment of naïve or memory module in Regnase-1-null cells in tumors (Fig. 3a) but in agreement with previous reports describing the negative role of Regnase-1 in T-cell activation under homeostasis8,9, peripheral Regnase-1-null cells were enriched with activation-associated but not naïve or memory module (Extended Data Fig. 2j), and had reduced expression of TCF-1 (Extended Data Fig. 2k). Given the TME-specific phenotypes of Regnase-1-null cells, we assessed regulation of Regnase-1 and found lower Regnase-1 expression in tumor-infiltrating than peripheral OT-I cells (Extended Data Fig. 3a). Additionally, gene targets repressed by Regnase-1 were elevated in tumor-infiltrating cells (Extended Data Fig. 3b), indicative of dampened Regnase-1 activity. Moreover, stimulation with TCR, and to a lesser extent, IL-2 or IL-21, induced Regnase-1 cleavage9 (Extended Data Fig. 3c). Antigen recognition was crucial in driving Regenase-1 deletion-induced CD8+ T-cell accumulation in TILs, as indicated by reduced Regnase-1-null OT-I cells in mice bearing B16-F10 (without cognate antigen) compared to B16-Ova melanoma (Extended Data Fig. 3d). Antigen stimulation was also required for Regnase-1-null cells to acquire increased TCF-1 expression (Extended Data Fig. 3e). In contrast, hypoxia did not alter expression of Regnase-1 or immune markers (Extended Data Fig. 3f, g). Thus, Regnase-1-null CD8+ T cells undergo specific reprogramming in TME in a process downstream of tumor antigen stimulation.
We next determined cellular homeostasis of Regnase-1-null cells. GSEA revealed that cell cycling-associated hallmarks were the top downregulated pathways in tumor-infiltrating Regnase-1-null cells (Extended Data Fig. 4a, b). Accordingly, these cells had reduced BrdU and Ki-67 staining at day 14 after adoptive transfer (Fig. 3c, Extended Data Fig. 4), but not at day 7 (Extended Data Fig. 4d, e). Also, tumor-infiltrating Regnase-1-null cells had reduced levels of active caspase-3 (Fig. 3d, Extended Data Fig. 4f) and DNA damage biomarker (Extended Data Fig. 4g). Therefore, tumor-infiltrating Regnase-1-null cells are less proliferative after effector expansion and show better survival. In contrast but consistent with the increased activation signatures (Extended Data Fig. 2j), peripheral Regnase-1-null cells were enriched with cell cycling and apoptosis-associated signatures (Extended Data Fig. 4h), which was validated by increased BrdU and active caspase-3 staining (Extended Data Fig. 4i, j). These results further support the TME-specific phenotypes of Regnase-1-null CD8+ T cells. To test the effect on in vivo persistence, we isolated wild-type and Regnase-1-null OT-I cells from TILs and co-transferred them into tumor-bearing or naïve hosts (Fig. 3e). Regnase-1-null cells showed better accumulation in tumor sites (Fig. 3f) as well as the spleen of naïve recipients (Fig. 3g). These analyses collectively indicate that tumor-infiltrating Regnase-1-null CD8+ T cells are characterized by in vivo quiescence and survival, with better persistence in response to both antigenic and homeostatic signals.
Although Regnase-1-null OT-I cells acquired naïve/memory cell-associated programs in tumors, they had higher expression of many activation-associated markers (Extended Data Fig. 5a), retained an effector surface phenotype (CD44+CD62L–) (Extended Data Fig. 5b), and expressed more IFN-γ and granzyme B (GzmB) (Fig. 3h, i, Extended Data Fig. 5c, d). Additionally, these cells had similar or enhanced capacities to produce TNF-α and IL-2 (Fig. 3j, Extended Data Fig. 5e, f), and contained increased IFN-γ+TNF-α+IL-2+ polyfunctional cells (Fig. 3k). Thus, although tumor-infiltrating CD8+ T cells lacking Regnase-1 acquire better persistence and survival advantage, they retain potent effector function.
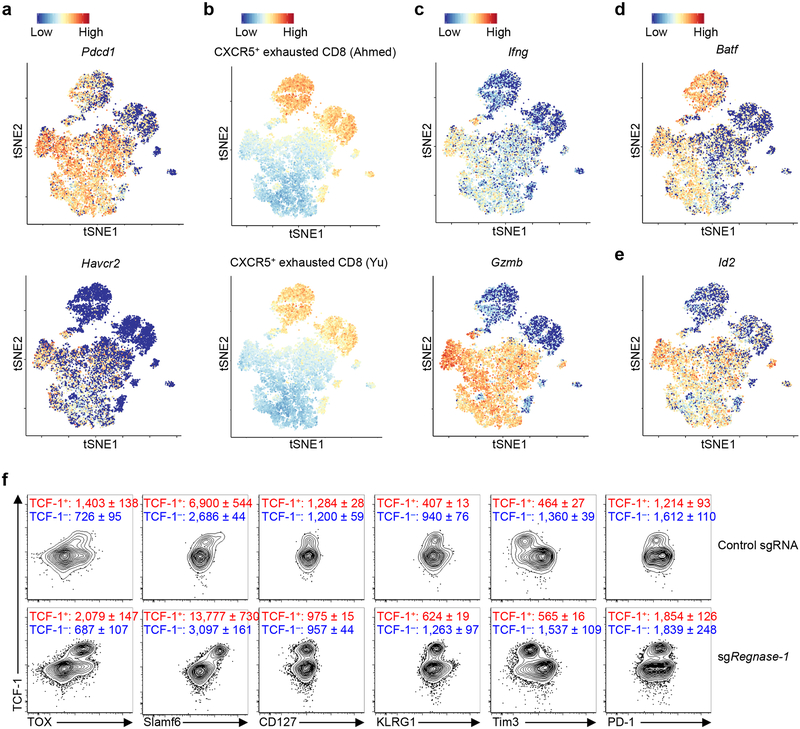
We used scRNA-Seq21 to dissect heterogeneity of TILs isolated from the in vivo dual transfer system. Regnase-1-null OT-I cells had distinct patterns from wild-type cells, including an increased proportion of Tcf7hi cells (Fig. 3l, m). Tcf7hi cells in both genotypes expressed Tox22,23 (Fig. 3n), and compared with Tcf7lo cells, had reduced expression of Pdcd1 (encodes PD-1) and Havcr2 (encodes Tim3) (Extended Data Fig. 6a). Wild-type Tcf7hi cells were enriched with TCF-1 target gene Slamf623,24 and memory-like gene signatures12,13, which were increased in Regnase-1-null cells (Fig. 3m, Extended Data Fig. 6b). In contrast, wild-type Tcf7hi cells had lower expression of Ifng and Gzmb than Tcf7lo cells, but in the absence of Regnase-1, Ifng and Gzmb were increased in both Tcf7hi and Tcf7lo cells (Extended Data Fig. 6c). Moreover, effector cell factor Batf25,26, but not Id227, was highly expressed by both Regnase-1-null Tcf7hi and Tcf7lo cells (Extended Data Fig. 6d, e). Flow cytometry validation revealed that TCF-1+ cells expressed TOX and Slamf6, with modestly higher levels observed in Regnase-1-null cells, but had low KLRG1 and Tim3 and intermediate PD-1 expression levels (Extended Data Fig. 6f). Collectively, these results establish the dual roles of Regnase-1 in coordinating T-cell effector function and memory-like features in antitumor immunity.
Regnase-1-BATF shapes effector responses
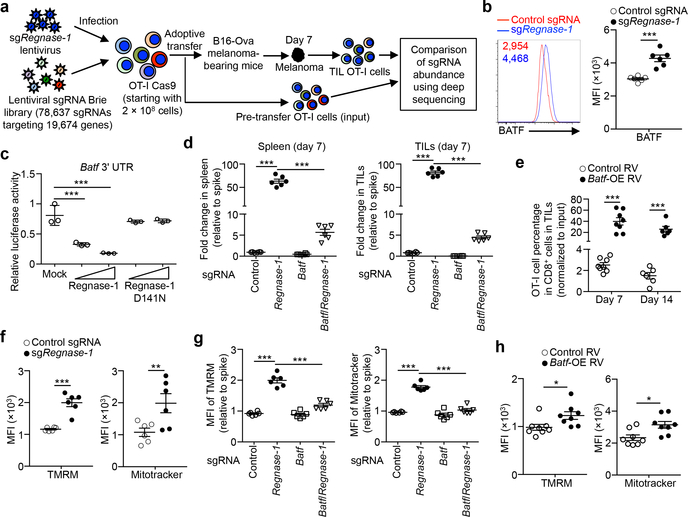
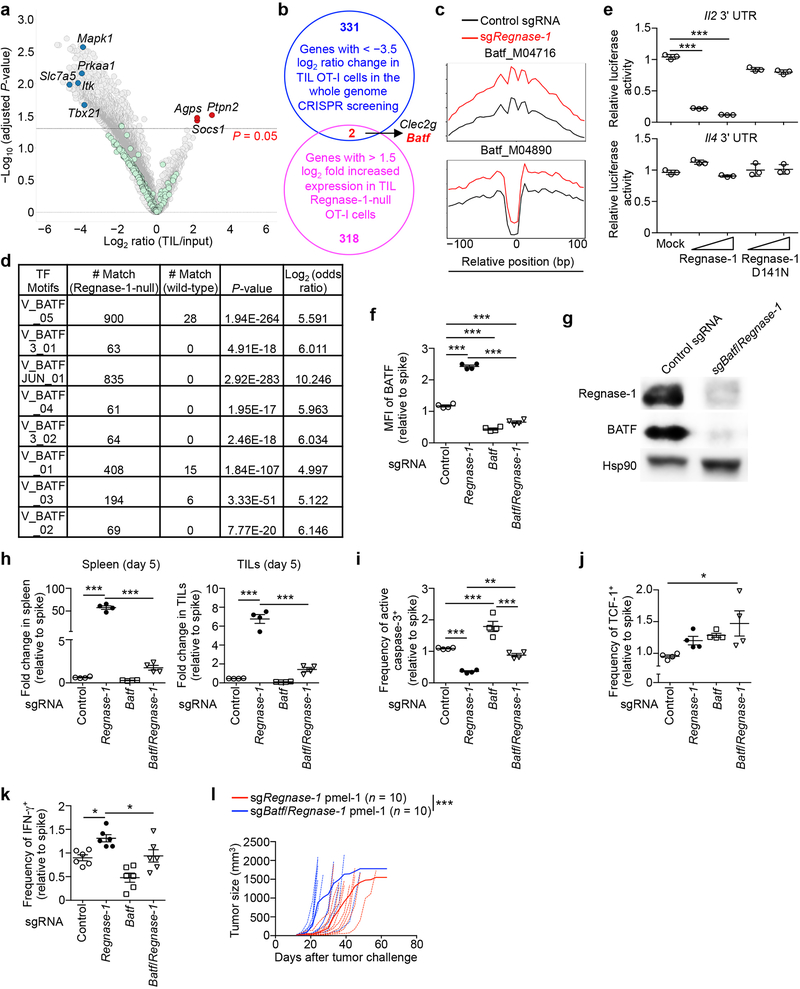
To identify mechanisms underlying Regnase-1 signaling, we took advantage of the extensive accumulation of tumor-infiltrating Regnase-1-null cells and performed a secondary in vivo genome-scale CRISPR screening by co-transducing OT-I cells with sgRegnase-1 and Brie lentiviral genome-scale sgRNA library28 (Fig. 4a). A total of 331 genes were strongly depleted in the screening, including Slc7a529, Itk30, Prkaa131, Mapk132 and Tbx2133 (Extended Data Fig. 7a). Given the role of Regnase-1 in inhibiting gene expression8,9, we applied two criteria to identify the functional targets of Regnase-1: such candidates should be upregulated in Regnase-1-null cells in RNA-Seq, but depleted in TILs in the genome-scale CRISPR screening. This analysis revealed two candidates including Batf25,26 (Extended Data Fig. 7b). Regnase-1-null cells showed increased BATF expression (Fig. 4b, Extended Data Fig. 6d) and enrichment of BATF binding motifs and gene targets26 (Extended Data Fig. 2g, 7c, d). We next determined whether Batf mRNA is regulated by Regnase-1, with the 3’ UTR of Il2 and Il4 genes as positive and negative controls, respectively9 (Extended Data Fig. 7e). The 3’ UTR of Batf gene was dose-dependently inhibited by Regnase-1, but not the nuclease-inactive Regnase-1 D141N (Fig. 4c), revealing BATF as a novel target. Importantly, co-deletion of BATF (Extended Data Fig. 7f, g) drastically reduced the accumulation of Regnase-1-null OT-I cells in both the periphery and tumor (Fig. 4d, Extended Data Fig. 7h), associated with elevated active caspase-3 (Extended Data Fig. 7i). In contrast, BATF/Regnase-1-null cells still had increased TCF-1 expression compared to wild-type cells (Extended Data Fig. 7j), suggesting a dispensable role of BATF in TCF-1 expression. Moreover, BATF co-deletion blocked increased IFN-γ production in Regnase-1-null cells, and dampened antitumor effects of Regnase-1-null cells in the pmel-1 ACT model (Extended Data Fig. 7k, l). Therefore, Regnase-1 targets BATF to impair the accumulation and effector function but not TCF-1 expression of tumor-specific T cells.
Figure 4. BATF is a key Regnase-1 functional target to mediate mitochondrial fitness and effector responses.
(a) Diagram of secondary genome-scale CRISPR screening. (b) Tumor-infiltrating sgRNA-transduced OT-I cells from the dual transfer system (n = 6) were analyzed at day 7 for BATF expression (left), and quantification of BATF MFI (right). (c) Luciferase activity of HEK293T cells after transfection with Batf mRNA 3’ UTR reporter, together with control (mock), wild-type or D141N Regnase-1-expressing plasmid (n = 3). (d, e) In vivo accumulation of sgRNA- and double sgRNA-transduced OT-I cells (d) or Batf-overexpressing retrovirus-transduced OT-I cells (e) in the dual transfer system (n = 6). OT-I cell percentage in CD8α+ cells was normalized to co-transferred non-targeting control sgRNA-transduced spike cells (d). (f–h) Tumor-infiltrating OT-I cells transduced with sgRNA (f; n = 6), double sgRNA (g; n = 6) and Batf-overexpressing retrovirus (h; n = 8) from the dual transfer system were analyzed at day 7 for the quantification of MFI of TMRM (left) and Mitotracker (right). MFI of TMRM and Mitotracker was normalized to those of co-transferred control sgRNA-transduced spike cells (g). Mean ± s.e.m. in b, d–h. Mean ± s.d. in c. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed paired Student’s t-test in f, two-tailed unpaired Student’s t-test in b, e, h, and one-way ANOVA in c, d, g. Data are representative of two (c), or pooled from two (b, d–h) independent experiments.
BATF expression was aberrantly induced in Regnase-1-null cells in response to TCR and to a lesser extent, IL-2, but not IL-21 (Extended Data Fig. 8a). To test whether BATF is a limiting factor in antitumor responses, we transduced wild-type OT-I cells with BATF (Extended Data Fig. 8b) and found that BATF overexpression improved cell accumulation in the spleen (Extended Data Fig. 8c, d) and even more profoundly in the tumor (Fig. 4e, Extended Data Fig. 8c). Accordingly, BATF-overexpressing OT-I cells in the tumor had increased cell proliferation and modestly reduced active caspase-3 (Extended Data Fig. 8e, f), and produced more IFN-γ, GzmB and TNF-α but not IL-2 (Extended Data Fig. 8g). In contrast but consistent with the role of BATF in promoting effector differentiation26, TCF-1 expression was reduced in BATF-overexpressing cells (Extended Data Fig. 8h). Therefore, BATF is regulated by Regnase-1 and immune signals and acts as an important rheostat in mediating antitumor effector responses.
To determine the contribution of aberrant BATF expression to the altered chromatin accessibility in Regnase-1-null cells, we performed ATAC-Seq analysis of wild-type, Regnase-1-null, BATF-null, and BATF/Regnase-1-null cells isolated from TILs. We identified 7,480 genes with significantly increased chromatin accessibility in Regnase-1-null cells as compared to wild-type cells (Extended Data Fig. 9a), and BATF co-deletion reversed the upregulation of a large proportion of these genes (5,052 in total) (Extended Data Fig. 9a). In addition, 2,527 among these 5,052 genes showed significantly downregulated chromatin accessibility in BATF-null cells as compared to wild-type cells (Extended Data Fig. 9a). Thus, a large majority of the genes with increased chromatin accessibility in Regnase-1-null cells are BATF dependent.
We next determined the functional pathways by which Regnase-1 regulates antitumor immunity. Functional enrichment of the top-ranking depleted genes of the genome-scale CRISPR screening revealed oxidative phosphorylation (OXPHOS) hallmark as the top-enriched pathway (Extended Data Fig. 9b). Also, OXPHOS was the top-ranking gene set enriched in tumor-infiltrating Regnase-1-null cells relative to wild-type cells (Extended Data Fig. 4a and 9c). While mitochondrial metabolism correlates with T-cell fitness and antitumor activity34,35, the negative signals involved, especially in the TME, remain elusive. Regnase-1-null cells showed increased mitochondrial fitness, as indicated by increased mitochondrial mass, membrane potential and volume (Fig. 4f, Extended Data Fig. 9d), as well as higher basal and maximal oxygen consumption rate (Extended Data Fig. 9e). Compared with Regnase-1-null cells, BATF/Regnase-1-null cells downregulated OXPHOS and cell cycling-associated hallmarks (Extended Data Fig. 9f, g). Moreover, BATF co-deletion largely blocked the increased mitochondrial mass and membrane potential in Regnase-1-null cells at days 5 and 7 after adoptive transfer (Fig. 4g, Extended Data Fig. 9h). Conversely, BATF overexpression was sufficient to upregulate mitochondrial mass and membrane potential (Fig. 4h). These results collectively reveal a role of BATF in linking Regnase-1 function and mitochondrial fitness.
To understand the molecular basis for Regnase-1 and BATF-mediated regulation of mitochondrial fitness, we mined our ATAC-Seq data for altered chromatin accessibility of mitochondrial genes. A total of 341 mitochondrial genes showed significantly upregulated chromatin accessibility in the absence of Regnase-1, and 214 of them were blocked by BATF co-deletion (Extended Data Fig. 9i). Moreover, 96 among these 214 genes showed downregulated chromatin accessibility in BATF-null cells as compared to wild-type cells (Extended Data Fig. 9i). These results further support a crucial contribution of BATF to the enhanced mitochondrial function in the absence of Regnase-1.
Targeting PTPN2 and SOCS1 for combined therapy
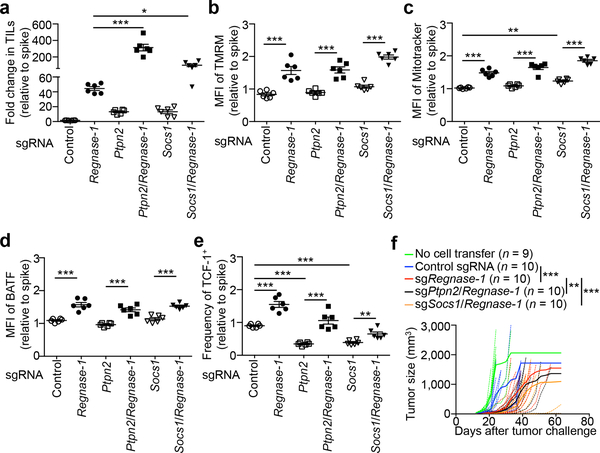
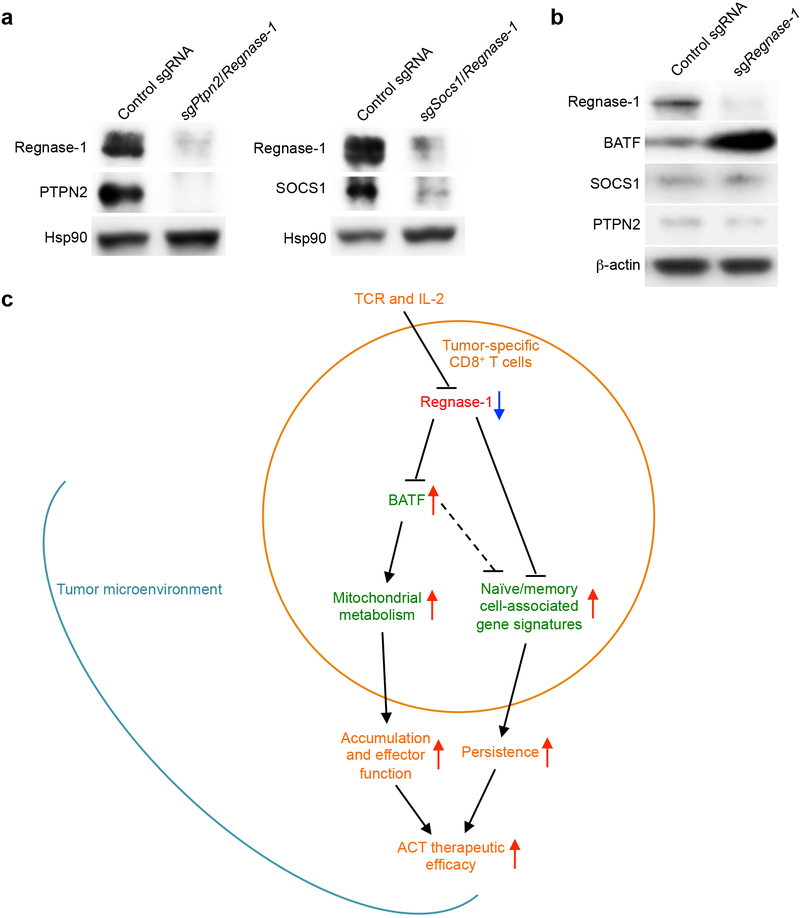
Combination therapy is key to clinical success of cancer immunotherapies36. To identify if the therapeutic potential of Regnase-1-null CD8+ T cells could be further potentiated, we focused on the top two genes enriched in TILs in our genome-scale CRISPR screening: Ptpn2 and Socs1. We validated the effects of co-deletion of these genes to enhance the accumulation of tumor-infiltrating Regnase-1-null cells (Fig. 5a, Extended Data Fig. 10a). Deletion of PTPN2 or SOCS1 alone resulted in a modestly increased accumulation of OT-I cells in the tumor (Fig. 5a). Of note, SOCS1 is identified to restrain human T-cell proliferation in vitro37, while PTPN2 deletion sensitizes cancer cells to immune checkpoint therapy38. Unlike BATF, PTPN2 and SOCS1 expression was not affected by Regnase-1 deletion (Extended Data Fig. 10b). Also, deletion of PTPN2 or SOCS1 alone did not affect or slightly increased mitochondrial mass and membrane potential (Fig. 5b, c), and co-deletion of Regnase-1 still elevated these mitochondrial profiles (Fig. 5b, c). Furthermore, deletion of PTPN2 or SOCS1 did not affect BATF expression (Fig. 5d), but significantly reduced TCF-1 expression (Fig. 5e), with Regnase-1 co-deletion still capable of upregulating TCF-1 expression (Fig. 5e). These comparative analyses reveal largely discrete mechanisms exerted by PTPN2 or SOCS1 in comparison to Regnase-1, including mitochondrial fitness and regulation of BATF and TCF-1 expression.
Figure 5. Genome-scale CRISPR screening identifies PTPN2 and SOCS1 as additional targets to enhance Regnase-1-null CD8+ T cell antitumor activity.
(a–e) OT-I cells transduced with non-targeting control sgRNA (spike) were mixed at a 1:1 ratio with cells transduced with non-targeting control sgRNA, sgRegnase-1, sgPtpn2, sgPtpn2/Regnase-1, sgSocs1 or sgSocs1/Regnase-1, and transferred into tumor-bearing hosts individually (n = 6). Tumor-infiltrating OT-I cells were analyzed at day 7 for quantification of relative OT-I cell percentage in CD8α+ cells normalized to spike (a), quantification of relative MFI of TMRM (b), Mitotracker (c) and BATF (d) normalized to spike, and quantification of relative frequency of TCF-1+ cells normalized to spike (e). (f) sgRNA- or double sgRNA-transduced pmel-1 cells (4 × 106) were transferred into mice at day 12 after B16-F10 melanoma engraftment, followed by analysis of tumor size. Non-treatment control mice received no T cell transfer. Mean ± s.e.m. in a–e. *P < 0.05; **P < 0.01; ***P < 0.001; one-way ANOVA in a–e, and two-way ANOVA in f. Data are pooled from two (a–f) independent experiments.
We assessed the therapeutic efficacy of pmel-1 T cells with PTPN2/Regnase-1 or SOCS1/Regnase-1 co-deletion. While Regnase-1 deletion alone greatly improved the therapeutic efficacy, PTPN2/Regnase-1-null and SOCS1/Regnase-1-null pmel-1 T cells exhibited additional effects to delay tumor growth (Fig. 5f). Altogether, our CRISPR screening identifies potential targets to combine with Regnase-1 deletion for cancer immunotherapy.
Discussion
There is a great need to understand how cell fate decisions occur in tumor-specific CD8+ T cells. Our work here reveals that tumor-specific CD8+ T cells can be reprogrammed in TME to acquire extensive accumulation and increased naïve/memory cell-associated features for long-term persistence, while retaining robust effector function (Extended Data Fig. 10c). Regnase-1 is a major regulator to be targeted to unleash this unique reprogramming in TME, with marked therapeutic efficacy against both solid and blood cancers in ACT. The specific transcriptional adaptation of Regnase-1-null CD8+ T cells in the TME highlights a previously unappreciated function of Regnase-1 after initial T-cell activation8,9, to enable precise temporal and spatial control of T-cell responses.
Despite recent emphasis on metabolic control of T-cell activation and differentiation, the metabolic reprogramming and the molecular events remain to be explored in antitumor T-cell responses3. Our results highlight that Regnase-1 restrains mitochondrial metabolism and effector responses through a key gene target BATF. BATF acts as a limiting factor for programming antitumor responses and mitochondrial metabolism, thereby advancing our understanding of context-dependent roles of pioneer factor BATF in adaptive immunity26,39. The genome-scale CRISPR screening also reveals PTPN2 and SOCS1 as potential targets to combine with Regnase-1 deletion to boost antitumor immunity. From a therapeutic perspective, our findings have identified novel targets for ACT against both solid and blood cancers and point to new avenues to reprogramming T-cell state and metabolism in cancer immunity and immunotherapy.
METHODS
Cell lines and mice
B16-F10 cell line was purchased from ATCC. B16-Ova cell line was kindly provided by Dario Vignali. huCD19-Ph+ B-ALL cell line was provided by Terrence Geiger. C57BL/6, OT-I, pmel-1 and Rosa26-Cas9 knockin mice40 were purchased from The Jackson Laboratory. CAR-T transgenic mice (T cells express CARs consisting of anti-human CD19 (huCD19) scFv fragments, CD8 transmembrane domain and 4–1BB-CD3ζ signaling tail) were provided by Terrence Geiger (to be described elsewhere). We crossed Rosa26-Cas9 knockin mice40 with OT-I41, pmel-142 or CAR-T transgenic mice to express Cas9 in antigen-specific CD8+ T cells. Gender-matched mice were used at 7–16 weeks old unless otherwise noted. All mice were kept in a specific pathogen-free facility in the Animal Resource Center at St. Jude Children’s Research Hospital. Experiments and procedures were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of St. Jude Children’s Research Hospital.
Cell purification and viral transduction
Naïve Cas9-expressing OT-I cells were isolated from the spleen and peripheral lymph nodes (PLNs) of Cas9-OT-I mice using naïve CD8α+ T cell isolation kit (Miltenyi Biotec 130–096-543) according to manufacturer’s instructions. Purified naïve OT-I cells were activated in vitro for 18 h with 10 μg/ml anti-CD3 (2C11; Bio X Cell), 5 μg/ml anti-CD28 (37.51; Bio X Cell) before viral transduction. Viral transduction was performed by spin-infection at 800 g at 25 °C for 3 h with 10 μg/ml polybrene (Sigma). Cells were continued to culture with human IL-2 (20 UI/ml; PeproTech), mouse IL-7 (25 ng/ml; PeproTech) and IL-15 (12.5 ng/ml; PeproTech) for 3−4 days. Transduced cells were sorted using a Reflection (i-Cyt) before adoptive transfer into recipients. sgRNAs were designed by using the online tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). sgRNAs used in this study were as follows: non-targeting control sgRNA: ATGACACTTACGGTACTCGT; sgRegnase-1: AAGGCAGTGGTTTCTTACGA; sgRegnase-1 #2: GGAGTGGAAACGCTTCATCG; sgBatf: AGAGATCAAACAGCTCACCG; sgBatf #2: AGGACTCATCTGATGATGTG (which gave similar results as sgBatf; data not shown); sgPtpn2: AAGAAGTTACATCTTAACAC; sgPtpn2 #2: CACTCTATGAGGATAGTCAT (which gave similar results as sgPtpn2; data not shown); sgSocs1: TGATGCGCCGGTAATCGGAG; sgSocs1 #2: TGGTGCGCGACAGTCGCCAA (which gave similar results as sgSocs1; data not shown). The coding sequence of Batf (Addgene # 34575) was subcloned into pMIG-II retroviral vector (Addgene # 52107), which was co-transfected into Plat-E cells with the helper plasmid pCL-Eco (Addgene # 12371) for the production of retrovirus.
Lentiviral sgRNA metabolic library CRISPR-Cas9 mutagenesis screening
Lentiviral and retroviral sgRNA vector design
The lentiviral sgRNA vector was generated from lentiGuide-puro vector by replacing the “EF-1α PuroR” fragment with a mouse PGK promoter-driven Ametrine (or GFP or mCherry) fluorescent protein. The retroviral sgRNA vector was generated from pLMPd-Amt vector43 by replacing the miR30 shRNA cassette with the U6 promoter driven gRNA cassette from the lentiGuide-puro vector.
Lentiviral sgRNA metabolic library construction
The gene list (3,017 genes) of mouse metabolic library was based on the reported human metabolic genes44. A total of 6 gRNAs were designed for each mouse metabolic gene according to our previously published selection criteria45 and were split into two sub-libraries (AAAQ05 and AAAR07, Supplementary Table 1), each containing 500 non-targeting controls. Oligonucleotides containing the guide sequence were synthesized (Custom Array), PCR amplified, and cloned into the recipient vector via a Golden Gate cloning procedure, including 5 μl Tango Buffer (ThermoFisher), 5 μl DTT (10 mM stock); 5 μl ATP (10 mM stock); 500 ng vector, pre-digested with Esp3I, gel-extracted, and isopropanol-precipitation purified; 100 ng insert PCR product; 1 μl Esp3I (ThermoFisher ER0452); 1 μl T7 ligase (Enzymatics, 3,000 Units/μl, L6020L); and water, up to 50 μl, and incubated in cycle (5 min at 37 °C and 5 min at 20 °C) for 100 times. The product was then purified by isopropanol precipitation and electroporated into STBL4 cells (Life Technologies 11635018). The distribution of the library was determined by Illumina sequencing.
In vivo screening
Lentivirus was produced by co-transfecting HEK293T cells with the lentiviral metabolic library plasmids, psPAX2 (Addgene plasmid # 12260) and pCAG4-Eco. At 48 h after transfection, virus was harvested and froze at −80 °C. Four hundred to five hundred million naïve Cas9-expressing OT-I cells were isolated from 8–14 Cas9-OT-I mice and transduced at a MOI of 0.3 to achieve ~20% transduction efficiency. After viral transduction, cells were cultured with human IL-2 (20 IU/ml; PeproTech), mouse IL-7 (25 ng/ml; PeproTech) and IL-15 (12.5 ng/ml; PeproTech) for 4 days to allow gene editing to occur. Transduced cells expressing Ametrine were sorted using a Reflection sorter (i-Cyt), and an aliquot of 5 × 106 transduced OT-I cells was saved as “input” (~500 × cell coverage per sgRNA). Transduced OT-I cells (5 × 106 cells per recipient) were i.v. transferred into mice at day 14 after B16-Ova melanoma engraftment. Sixty recipients were randomly divided into 3 groups as biological replicates in each sub-library screening. At 7 days after adoptive transfer, transferred Ametrine+ OT-I cells were recovered from the tumor pooled from 20 recipients per sample using a Reflection sorter (i-Cyt). On average, 5 × 105 OT-I cells per sample (~50 × cell coverage per sgRNA) were recovered and used for deep sequencing of sgRNA cassette, with the expectation that sgRNAs capable of improving ACT should be enriched in tumor-infiltrating OT-I cells.
Sequencing library preparation
Genomic DNA was extracted by using the DNeasy Blood & Tissue Kits (Qiagen 69506). Primary PCR was performed by using the KOD Hot Start DNA Polymerase (Millipore 71086) and the following pair of Nextera NGS primers (Nextera NGS-F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGttgtggaaaggacgaaacaccg; Nextera NGS-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGccactttttcaagttgataacgg). Primary PCR products were purified using the AMPure XP beads (Beckman A63881). A second PCR was performed to add adaptors and indexes to each sample. Hi-Seq 50-bp single-end sequencing (Illumina) was performed.
Data processing
For data analysis, FastQ files obtained after sequencing were demultiplexed using the HiSeq Analysis software (Illumina). Single-end reads were trimmed and quality-filtered using the CLC Genomics Workbench v11 (Qiagen) and matched against sgRNA sequences from the sgRNA metabolic library. Read counts for sgRNAs were normalized against total read counts across all samples. For each sgRNA, the fold change (log2 ratio) for enrichment was calculated between each of the biological replicates and the input experiment. After merging the quantification results from two sub-libraries, candidate genes were ranked based on the average enrichment of their 6 gene-specific sgRNAs in tumor-infiltrating OT-I cells relative to input (log2 ratio (TIL/input); adjusted P < 0.05). The gene level false discovery rate adjusted P-value was calculated among multiple sgRNAs (n = 6) of each gene, using a two-tailed paired Student’s t-test between log2 transformed average normalized read counts of tumor samples and those of input sample, and the P-value was further adjusted using Bonferroni correction with gene size.
Genome-scale sgRNA Brie library CRISPR-Cas9 mutagenesis screening
In vivo screening
Lentivirus was produced by co-transfecting HEK293T cells with lentiviral genome-scale Brie library plasmids with the puromycin resistant gene28, psPAX2 and pCAG4-Eco. At 48 h after transfection, virus was harvested and frozen at −80 °C. Two hundred million Cas9-expressing OT-I cells were isolated from 12 Cas9-OT-I mice and co-transduced with Brie sgRNA library and sgRegnase-1-Ametrine. After viral transduction, cells were cultured with human IL-2 (20 IU/ml; PeproTech), mouse IL-7 (25 ng/ml; PeproTech) and IL-15 (12.5 ng/ml; PeproTech) for 2 days. Brie sgRNA library-transduced cells were then selected by culture with 4 μg/ml puromycin in the presence of the abovementioned cytokines for another 3 days. Following puromycin selection, Ametrine+ cells were sorted using a Reflection sorter (i-Cyt) to select for cells co-transduced with sgRenase-1 and Brie library sgRNAs, and an aliquot of 10 × 106 transduced OT-I cells was saved as input (~120 × cells coverage per sgRNA). The majority of the co-transduced OT-I cells (5 × 106 cells per recipient) were then i.v. transferred into mice at day 14 after B16-Ova melanoma engraftment. Twenty recipients were randomly divided into 2 groups as biological replicates. At 7 days after adoptive transfer, transferred Ametrine+ OT-I cells were recovered from the tumor pooled from 10 recipients per sample using a Reflection sorter (i-Cyt). On average, 3 × 106 OT-I cells per sample (~40 × cell coverage per sgRNA) were recovered. DNA extraction and sequencing library preparation were as described above.
Data processing
For data analysis, FastQ files obtained after sequencing were demultiplexed using the HiSeq Analysis software (Illumina). sgRegnase-1 (GGAGTGGAAACGCTTCATCG) reads were removed, and single-end reads were trimmed and quality-filtered using the CLC Genomics Workbench v11 (Qiagen) and matched against sgRNA sequences from the genome-scale sgRNA Brie library. Read counts for sgRNAs were normalized against total read counts across all samples. For each sgRNA, the fold change (log2 ratio) for enrichment was calculated between each of the biological replicates and the input experiment. Gene ranking was based on the average enrichment (log2 ratio (TIL/input)) among replicates in representation of 4 individual corresponding sgRNAs in the genome-scale sgRNA Brie library. The gene level false discovery rate adjusted P-value was calculated among multiple sgRNAs (n = 4) of each gene, using a using a two-tailed paired Student’s t-test between log2 transformed average normalized read counts of tumor samples and those of input sample, and the P-value was further adjusted using Bonferroni correction with gene size.
Flow cytometry
For analysis of surface markers, cells were stained in PBS (Gibco) containing 2% (wt/vol) BSA (Sigma). Surface proteins were stained for 30 min on ice. Intracellular staining was performed with Foxp3/Transcription Factor Staining Buffer Set according to manufacturer’s instructions (eBioscience). Intracellular staining for cytokines was performed with fixation/permeabilization kit (BD Biosciences). Active caspase-3 staining was performed using instructions and reagents from the “Active Caspase-3 Apoptosis Kit” (BD Biosciences). BrdU staining (pulsed for 18 h) was performed using instructions and reagents from the “APC BrdU Flow Kit” (BD Biosciences). 7-AAD (Sigma) or Fixable viability dye (eBioscience) was used for dead cell exclusion. The following antibodies were used: anti-IFN-γ (XMG1.2), anti-TNFα (MAb11), anti-IL-2 (JES6–5H4), anti-CD69 (H1.2F3), anti-CD25 (PC61.5), anti-KLRG1 (2F1), anti-ICOS (7E.17G9), anti-Lag3 (C9B7W), anti-PD-1 (J43), anti-CTLA4 (1B8), anti-TOX (TXRX10) (all from eBioscience); anti-GzmB (QA16A02), anti-CD49a (HMα1), anti-CD44 (IM7), anti-Ki-67 (16A8), anti-CD127 (A7R34) (all from Biolegend); anti-BrdU (3D4), anti-active caspase-3 (C92–605), anti-pH2A.X-S139 (N1–431) (DNA damage biomarker, which measures phosphorylation of the histone variant H2A.X at Ser13946,47), anti-Slamf6 (13G3) (all from BD Biosciences); anti-BATF (D7C5), anti-Bim (C34C5), anti-TCF-1 (C63D9) (all from Cell Signaling Technology); anti-CD8α (53–6.7) (from SONY); anti-CD62L (MEL-14) (from TONBO Bioscience) and anti-Tim3 (215008) (from R&D). To monitor cell division, lymphocytes were labeled with CellTrace Violet (CTV; Life Technologies). For mitochondrial staining, lymphocytes were incubated for 30 min at 37 °C with 10 nM Mito Tracker Deep Red (Life Technologies) or 20 nM TMRM (tetramethylrhodamine, methyl ester; ImmunoChemistry Technologies) after staining surface markers. Flow cytometry data were analyzed using Flowjo 9.9.4 (Tree Star).
Adoptive T-cell transfer for tumor therapy
B16-Ova cells (2 × 105) or B16-F10 cells (2 × 105) were injected subcutaneously into female C57BL/6 mice (7–10 weeks age). At day 12, mice bearing tumors of similar size were randomly divided into 3 groups (5–8 mice per group), and sgRNA-transduced OT-I cells (5 × 106) (for the treatment of B16-Ova melanomas) or pmel-1 (5 × 106) (for the treatment of B16-F10 melanomas) were injected intravenously. Tumors were measured every three days with digital calipers and tumor volumes were calculated by the formula: Length × Width × [(Length × Width) ^ 0.5] × π/648. Death was defined as the point at which a progressively growing tumor reached 15 mm in the longest dimension. For the treatment of huCD19-Ph+ B-ALL, mice engrafted with huCD19-Ph+ B-ALL (1 × 106) were treated at day 7 with sgRNA-transduced CD8+ CAR-T cells (5 × 106). Mice were imaged using the Xenogen imaging system (Caliper Life Science).
Tumor-infiltrating lymphocyte (TIL) isolation
To isolate TILs, B16-Ova melanoma was excised, minced and digested with 0.5 mg/ml Collagenase IV (Roche) + 200 IU/ml DNase I (Sigma) for 1 h at 37 °C, and then passed through 70 μm filters to remove undigested tumor tissues. TILs were then isolated by density-gradient centrifugation over Percoll (Life Technologies).
Gene expression profiling and gene set enrichment analysis (GSEA)
OT-I cells transduced with control sgRNA (n = 4 biological replicates) and sgRegnase-1 (n = 5 biological replicates) were isolated from the tumors or PLN of the hosts of the in vivo dual color transfer assay, and analyzed with RNA-Seq. For RNA-Seq, RNA was quantified using the Quant-iT RiboGreen assay (Life Technologies) and quality checked by 2100 Bioanalyzer RNA 6000 Nano assay (Agilent) or LabChip RNA Pico Sensitivity assay (PerkinElmer) prior to library generation. Libraries were prepared from total RNA with the TruSeq Stranded Total RNA Library Prep Kit according to the manufacturer’s instructions (Illumina, PN 20020595). Libraries were analyzed for insert size distribution on a 2100 BioAnalyzer High Sensitivity kit (Agilent Technologies) or Caliper LabChip GX DNA High Sensitivity Reagent Kit (PerkinElmer.) Libraries were quantified using the Quant-iT PicoGreen dsDNA assay (Life Technologies) or low pass sequencing with a MiSeq nano kit (Illumina). Paired end 100 cycle sequencing was run on the HiSeq 4000 (Illumina). The raw reads were trimmed for adapter sequences using Trimmomatic v.0.36 using parameters ILLUMINACLIP:adapter.fa:2:30:10 LEADING:10 TRAILING:10 SLIDINGWINDOW:4:18 MINLEN:32, followed by mapping to mm9 reference genome downloaded from gencode release M1 (https://www.gencodegenes.org/mouse/releases.html) using star v.2.5.2b. with default parameters. Reads were summarized at gene level using python script htseq-count. Differential expression analysis was performed using R package DEseq2 v. 1.18.1. OT-I cells transduced with sgRegnase-1 (n = 3 biological replicates) and sgBatf/Regnase-1 (n = 3 biological replicates) were isolated from the tumors in the in vivo dual color transfer assay and used for microarray analysis (Affymetrix Mouse Clariom S Assay). For microarray, the expression signals were summarized robust multi-array average algorithm Affymetrix Expression Console v1.1, followed by differential expression analysis performed using R package limma v.3.34.9. All the plots were generated using R package ggplot2 v.2.2.1. Differentially expressed transcripts were identified by ANOVA (Partek Genomics Suite version 6.5), and the Benjamini-Hochberg method was used to estimate the false discovery rate (FDR) as described49. Differentially expressed (DE) genes were defined by |log2 FC| > 0.5; P < 0.05. GSEA was performed as described50 using the “Hallmark” database. For GSEA using manually curated gene signatures from public datasets, microarray dataset (GSE84105)12 was used for generating “CXCR5+ exhausted CD8 (Ahmed)” gene signatures (< 5% FDR); as total upregulated and downregulated genes were more than 200, we ranked genes by their log2 fold change of expression in CXCR5+ vs CXCR5− comparison and used top 200 upregulated genes as “CXCR5+ exhausted CD8 (Ahmed)”. RNA-Seq data (GSE76279)13 was processed using DEseq2 R package v 1.16.1 to generate “CXCR5+ exhausted CD8 (Yu)” using the similar strategy as the other signatures above.
ATAC-Seq and data analysis
Library preparation
To prepare ATAC-Seq library, tumor-infiltrating sgRNA-transduced OT-I cells were collected in the following two batches: (a) Control sgRNA- and sgRegnase-1-transduced OT-I cells (n = 4 biological replicates each group) were isolated from tumor-bearing mice using the in vivo dual color transfer assay; (b) Control sgRNA-, sgRegnase-1-, sgBatf- and sgBatf/Regnase-1-transduced TIL OT-I cells (n = 2–4 replicates each group) were isolated from the tumor-bearing mice that received the individual transfer of sgRNA-transduced OT-I cells. Sorted T cells were incubated in 50 μl ATAC-Seq lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) on ice for 10 min. Resulting nuclei were pelleted at 500 g for 10 min at 4 °C. Supernatant was carefully removed with a pipette and discarded. The pellet was resuspended in 50 μl transposase reaction mix (25 μl 2× TD buffer, 22.5 μl nuclease-free water, 2.5 μl Transposase) and incubated for 30 min at 37 °C. After the reaction, the DNA was cleaned up using the Qiagen MinElute kit. The barcoding reaction was run using the NEBNext HiFi kit based on manufacturer’s instructions and amplified for 5 cycles according to Buenrostro et al.20 using the same primers. Ideal cycle numbers were determined from 5 μl (of 50 μl) from the previous reaction mix using KAPA SYBRFast (Kapa Biosystems) and 20 cycle amplification on an Applied Biosystems 7900HT. Optimal cycles were determined from the linear part of the amplification curve and the remaining 45 μl of PCR reaction was amplified in the same reaction mix using the optimal cycle number.
Data analysis
ATAC-Seq analysis was performed as we described previously51. Briefly, 2 × 100 bp paired-end reads obtained from all samples were trimming for Nextera adapter by cutadapt (version 1.9, paired-end mode, default parameter with “ –m 6 –O 20 ”) and aligned to mouse genome mm9 (NCBIM37 from Sanger) by BWA (version 0.7.12-r1039, default parameter)52, duplicated reads were then marked with Picard (version 2.6.0-SNAPSHOT) and only non-duplicated proper paired reads have been kept by samtools (parameter “-q 1 -F 1804” version 1.2)53. After adjustment of Tn5 shift (reads were offset were offset by +4 bp for the sense strand and −5 bp for the antisense strand) we separated reads into nucleosome-free, mononucleosome, dinucleosome and trinucleosome as described20 by fragment size and generated bigwig files by using the center 80 bp of fragments and scale to 30 × 106 nucleosome-free reads. We observed reasonable nucleosome-free peaks and pattern of mono-, di-, tri-nucleosome on IGV (version 2.4.13)54 and all 8 samples have about 10 × 106 nucleosome-free reads so we conclude the data qualities are good. Next, we merged each 2 replicates to enhance peak calling on nucleosome-free reads by MACS2 (version 2.1.1.20160309 default parameters with “--extsize 200 --nomodel “)55. To assure the replicability, we first finalized nucleosome-free regions for each genotype and only retained a peak if it called with higher cutoff (macs2 -q 0.05) in one merged sample and at least called with lower cutoff (macs2 -q 0.5) in the other merged sample. The reproducible peaks were further merged between wild-type and Regnase-1-null samples and then we counted nucleosome-free reads from each of the 8 samples by bedtools (v2.24.0)56. To find the differentially accessible regions, we first normalized raw nucleosome-free reads counts used trimmed mean of M-values normalization method and applied Empirical Bayes Statistics test after linear fitting from voom package (R 3.23, edgeR 3.12.1, limma 3.26.9)57. FDR-correct P-value < 0.05 and fold change > 2 were used as cutoff for more accessible regions in Regnase-1-null samples (KO Larger) or less accessible regions in Regnase-1-null samples (KO Smaller). We annotated the differentially accessible regions in ATAC-Seq for the nearest genes, and also superimposed these genes with 1,158 mitochondrial genes defined in MitoCarta 2.0 database58. For motif analysis, we further selected regions < 0.05 fold change and P-value > 0.5 as control region. FIMO from MEME suite (version 4.11.3, “--thresh 1e-4 --motif-pseudo 0.0001”)59 have been used for scanning motif (TRANSFAC database, only included Vertebrata and not 3D structure-based) matches in the nucleosome-free regions and Fisher’s exact tests have been used to test whether a motif is significant enriched for differentially accessible regions compared to the control regions..
Transcription factor binding site footprinting
Footprinting was performed as described previously51. Briefly, we first generated bigwig files according to all tags of adjusted reads, and then normalized them according to the number of autosome reads to 2 × 108 reads (e.g. a sample with 1 × 108 autosome reads would be scaled so as to double the bigwig profile). We then generated average bigwig files from the mean of replicates at each bp for each sample, using motif matches within a nucleosome-free region for footprinting and taking the average profile across all motif matches at each bp from –100 bp from motif match centers to +100 bp. Finally, the footprinting profiles were smoothed with 10 bp bins and plot by deeptools (v2.5.7)60.
To identify the enrichment of BATF binding motifs, nucleosome-free differentially accessible regions were defined at |log2 FC| > 0.5; P < 0.05, and the peaks were further annotated as more or less accessible regions in Regnase-1-null OT-I cells compared to wild-type controls. For each group, differentially accessible peaks were overlapped with BATF ChIP-Seq peaks (downloaded from GSE5419126) to identify the common regions between ATAC-Seq peaks and BATF ChIP-Seq peaks using bedtools (version 2.25.0). Finally, FIMO61 from MEME suite (version 4.9.0) was used to scan the overlapping regions with TRANSFAC motifs associated with BATF to identify the number of motifs enriched in the differentially accessible regions in Regnase-1-null (shown as ‘# Match (Regnase-1-null)’ in Extended Data Fig. 7d) or wild-type control samples (shown as ‘# Match (wild-type)’), and Fisher’s exact test was used to test the significance of enrichment. This statistical bioinformatic method has been used successfully by us and others to circumvent cell number limitations51,62.
Imaging
B16-Ova melanomas were fixed in PBS containing 2% PFA, 0.3% Triton-100 and 1% DMSO for 24 h prior to cryoprotection in 30% sucrose. Cryosections were blocked with 1% BSA and 0.05% Tween-20 in TBS (20 mM Tris, pH 8.0, 100 mM NaCl) for 1 h at room temperature prior to overnight incubation in blocking buffer containing the following antibodies; anti-mCherry (Biorbyt orb11618), anti-GFP (Rockland Immuno 600–401-215), anti-TCF-7 (C63D9) (Cell Signaling Technology 2203), and anti-Tom20 (2F8.1) (Millipore MABT166). Slides were washed in TBS before application of AF488, Cy3 or AF647 secondary antibodies (Jackson Immuno) for 1 h at room temperature prior to mounting with Prolong Diamond hardset media containing DAPI (Thermofisher). Widefield fluorescence microscopy was performed using a motorized Nikon TiE inverted microscope equipped with a 20× Plan Apo 0.75NA objective, standard DAPI, FITC and TRITC filter sets, and an EMCCD camera (Andor). The entire tissue section was stitched based on the DAPI fluorescent signal and the subsequent large images were analyzed using NIS Elements software (Nikon Instruments). Images were segmented per channel, and further refined using a spot identification algorithm to identify single cells and positional information within the tumor. The number of cells per square area was determined following manual delineation of the tumor border. Analysis of transcription factor localization was performed using a Marianis spinning disk confocal microscope (Intelligent Imaging Innovations) equipped with a 100× 1.4NA objective and Prime 95B sCMOS camera, and analyzed using Slidebook software (Intelligent Imaging Innovations).
RNA isolation and real-time PCR
RNA was isolated using the RNeasy Micro Kit (Qiagen 74004) following the manufacturer’s instructions. RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher 4368813) according to manufacturer’s instructions. Real-time PCR was performed on the QuantStudio 7 Flex System (Applied Biosystems) using the PowerSYBR Green PCR Master Mix (ThermoFisher 4367659) and the following primers: Irf4-F: TCCGACAGTGGTTGATCGAC, Irf4-R: CCTCACGATTGTAGTCCTGCTT.
Protein extraction and immunoblot
Cells were lysed in RIPA buffer (ThermoFisher 89900), resolved in 4–12% Criterion™ XT Bis-Tris Protein Gel (Bio-Rad 3450124) and transferred to PVDF membrane (Bio-Rad 1620177). Membranes were blocked using 5% BSA for 1 h and then incubated for overnight with anti-MCPIP1 antibody (604421) (R&D), anti-BATF (D7C5) (Cell Signaling Technology), anti-PTPN2 (E-11) (Santa Cruz Biotechnology), anti-SOCS1 (E-9) (Santa Cruz Biotechnology), anti-Hsp90 (MAB3286) (R&D) and anti-β-actin (8H10D10) (Cell Signaling Technology). Membranes were washed 6 times with TBST and then incubated with 1:5,000 diluted HRP conjugated anti-mouse IgG (W4021) (from Promega) for 1 h. Following another 6 times of washes with TBST, the membranes were imaged using the ODYSSEY Fc Analyzer (LI-COR).
Luciferase assay
The full-length 3’ UTR constructs of Batf (MmiT031430-MT06), Il2 (MmiT092987-MT06) and Il4 (MmiT092992-MT06) mRNAs were purchased from GeneCopoeia, each containing two luciferase genes: firefly luciferase gene for 3’ UTR of the targeted gene, and Renilla luciferase gene as an internal control. The cDNA of wild-type Regnase-1 (Dharmacon MMM1013–202800061) was cloned into the pMIG-II vector. The D141N mutant Regnase-1 was generated by site-directed mutagenesis using the KOD Hot Start DNA Polymerase (Millipore 71086). HEK293T cells were transfected with 3’ UTR construct of interest together with wild-type or D141N mutant Regnase-1 expression plasmid or empty control plasmid. At 48 h after transfection, cells were lysed and luciferase activities in the lysates were determined using the Luc-Pair Duo-Luciferase Assay Kit (GeneCopoeia LF002) according to manufacturer’s instructions.
Seahorse metabolic assay
Oxygen consumption rates (OCR) were measured in XF media under basal conditions and in response to 1 μM oligomycin, 1.5 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP) and 500 nM rotenone using an XF96 Extracellular Flux Analyzer (EFA) (Seahorse Bioscience).
Single cell RNA-Sequencing (scRNA-Seq) and data analysis
Library preparation
Control sgRNA- and sgRegnase-1-transduced OT-I cells were sorted on an iCyt Reflection cell sorter from TILs pooled from the in vivo dual transfer hosts (6−8 mice per sample) at day 7 after adoptive transfer into tumor-bearing mice. The cells were counted and examined for viability using a Luna Dual Florescence Cell Counter (Logos Biosystems). All samples were spun down at 2,000 rpm for 5 min. The supernatant was removed, and cells were re-suspended in 100 μl of 1× PBS (Thermo Fisher Scientific) + 0.04% BSA (Amresco). The cells were then counted and examined for viability using a Luna Dual Florescence Cell Counter (Logos Biosystems). Cell counts were about 1 × 106 cells per milliliter and viability was above 98%. Single-cell suspensions were loaded onto the Chromium Controller according to their respective cell counts to generate 6,000 single cell GEMs (gel beads in emulsion) per sample. Each sample was loaded into a separate channel. Libraries were prepared using the Chromium Single Cell 3’ v2 Library and Gel Bead Kit (10X Genomics). The cDNA content of each sample after cDNA amplification of 12 cycles was quantified and quality checked using a High-Sensitivity DNA chip with a 2100 Bioanalyzer (Agilent Technologies) to determine the number of PCR amplification cycles to yield sufficient library for sequencing. After library quantification and quality check by DNA 1000 chip (Agilent Technologies), samples were diluted to 3.5 nM for loading onto the HiSeq 4000 (Illumina) with a 2 × 75 paired-end kit using the following read length: 26 bp Read1, 8 bp i7 Index, and 98 bp Read2. An average of 400,000,000 reads per sample was obtained (~approximately 80,000 reads per cell).
Alignment, barcode assignment and unique molecular identifier (UMI) counting
The Cell Ranger 1.3 Single-Cell software suite (10X Genomics) was implemented to process the raw sequencing data from the Illumina HiSeq run. This pipeline performed demultiplexing, alignment (using the mouse genome mm10 from ENSEMBL GRCm38), and barcode processing to generate gene-cell matrices used for downstream analysis. Specifically, data from two control sgRNA and two sgRegnase-1-transduced TIL OT-I cell samples were combined into one data set for consistent filtering, and UMIs mapped to genes encoding ribosomal proteins were removed. Cells with low UMI counts (potentially dead cells with broken membranes) or high UMI counts (potentially two or more cells in a single droplet) were filtered. A small fraction of outlier cells (888) was further removed because of their low transcriptome diversity (meaning that fewer genes were detected than in other cells with a comparable number of captured UMIs). A total of 13,879 cells (control sgRNA-transduced, 6,811; sgRegnase-1-transduced, 7,068) were captured, with an average of 11,040 messenger RNA molecules (UMIs, median: 9,391; range: 2,928–44,330). We normalized the expression level of each gene to 100,000 UMIs per cell and log-transformed them by adding 0.5 to the expression matrix.
Data visualization
Underlying cell variations derived from control sgRNA- and sgRegnase-1-transduced TIL OT-I cell single-cell gene expression were visualized in a two-dimensional projection by t-distributed stochastic neighbour embedding (tSNE). Expression of individual genes or pathway scores was color-coded (from low to high, blue–red) for each cell on tSNE plots. To visualize Tcf7-expressing cells, we defined Tcf7hi cells as cells with the highest third quantile of Tcf7 expression (with log2 gene expression intensity = 2.910317 as threshold) among all cells.
Statistical analysis for biological experiments
For biological experiment (non-omics) analyses, data were analyzed using Prism 6 software (GraphPad) by two-tailed paired Student’s t-test, two-tailed unpaired Student’s t-test, or one-way ANOVA with Newman-Keuls’s test. Two-way ANOVA was performed for comparing tumor growth curves. Log-rank (Mantel-Cox) test was performed for comparing mouse survival curves. P < 0.05 was considered significant. Data are presented as mean ± s.d. or mean ± s.e.m.
Extended Data
Extended Data Figure 1. Validation of the effect of Regnase-1 deletion on CD8+ T cell accumulation in tumor immunity using the in vivo dual transfer system.
(a) Diagram of the in vivo dual transfer system. OT-I cells transduced with sgRNA viral vectors expressing distinct fluorescent proteins were mixed and transferred into the same tumor-bearing hosts where further analyses were performed. (b) Gating strategy for sgRNA-transduced OT-I cell analysis. (c, d) OT-I cells transduced with non-targeting control sgRNA (mCherry+) were mixed at a 1:1 ratio with either cells transduced with control sgRNA (Ametrine+) (c (n = 2), d (n = 5), upper left) or two different sgRNAs targeting Regnase-1 (sgRegnase-1, Ametrine+, c (n = 4), lower left; or sgRegnase-1 #2, Ametrine+, d (n = 5), lower left), and transferred into tumor-bearing hosts. Mice were analyzed at 7 days after adoptive transfer for the proportion of OT-I cells in CD8α+ cells (c, d, left), and quantification of relative OT-I cell percentage in CD8α+ cells normalized to input in the spleen and TILs (c, d, right). Numbers in plots indicate frequencies of OT-I cells. (e) OT-I cells transduced with control sgRNA (Ametrine+) were mixed at a 1:1 ratio with cells transduced with sgRegnase-1 (mCherry+), and transferred into tumor-bearing hosts (n = 5). Mice were analyzed at 7 days after adoptive transfer for the proportion of OT-I cells in CD8α+ cells (left), and quantification of relative OT-I cell percentage in CD8α+ cells normalized to input in the spleen and TILs (right). Numbers in plots indicate frequencies of OT-I cells. (f) Indel mutations after CRISPR targeted disruption in OT-I cells transduced with either control sgRNA or sgRegnase-1, via deep sequencing analysis of indels generated at the exonic target site of Regnase-1 gene, including 97.3% indel events in sgRegnase-1-transduced cells isolated from tumors as compared to 1.3% in control sgRNA-transduced cells. Mean ± s.e.m. in c–e. ***P < 0.001; two-tailed unpaired Student’s t-test in d, e. Data are representative of two (e) independent experiments.
Extended Data Figure 2. Tumor-infiltrating and peripheral Regnase-1-null CD8+ T cells show distinct immune signatures.
(a, b) GSEA enrichment plots of antigen-specific CXCR5+ and CXCR5– exhausted CD8+ T cells from chronic infection using gene targets repressed by Regnase-1 (i.e. top 100 upregulated genes in TIL sgRegnase-1- compared to control sgRNA-transduced OT-I cells as identified by RNA-Seq). (c) Representative images (left) and quantification of mean fluorescence intensity (MFI; right) of TCF-1 expression (pink) in control sgRNA- (mCherry+; red) and sgRegnase-1-transduced OT-I cells (Ametrine+; green) in the whole tumor section (n = 4 mice). Scale bars, 20 μm. (d, e) Gene expression heat maps normalized by row (z-score) for the naïve or memory T-cell−associated transcription factors (d) or effector or exhausted T-cell−associated transcription factors (e) in control sgRNA- (n = 4) and sgRegnase-1- (n = 5) transduced OT-I cells isolated from TILs. Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (f) Real-time PCR analysis of Irf4 mRNA expression in control sgRNA- (n = 4 samples) and sgRegnase-1- (n = 5 samples) transduced OT-I cells isolated from TILs. (g) Summary of ATAC-Seq motif enrichment data showing log2 (odds ratio) and –log10 (FDR) of cells from control sgRNA- and sgRegnase-1-transduced OT-I cells isolated from TILs (n = 4 samples each group). Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for ATAC-Seq analysis. (h) Tn5 insert sites from ATAC-Seq analysis were aligned to motifs for transcription factors from the TRANSFAC database, and the binding profiles of TCF-1, Bach2, Bcl6 and IRF4 are shown. (i) Venn diagram showing the overlap of significantly upregulated (left, sgRegnase-1- (n = 5 samples) versus control sgRNA-transduced OT-I cells (n = 4 samples)) or downregulated genes (right, sgRegnase-1- versus control sgRNA-transduced OT-I cells) by RNA-Seq profiling between TIL and peripheral lymph node (PLN) OT-I cells. Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (j) GSEA enrichment plots of PLN sgRegnase-1- (n = 5) versus control sgRNA- (n = 4) transduced OT-I cells using gene sets of four different tumor-infiltrating CD8 T-cell activation states11. Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and PLN OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (k) OT-I cells transduced with control sgRNA (mCherry+) and sgRegnase-1 (Ametrine+) were mixed and transferred into tumor-bearing mice (n = 5 mice), and OT-I cells in the spleen were analyzed at day 7 for expression of TCF-1 (upper), and quantification of frequency of TCF-1+ cells (lower). Numbers in graphs indicate frequencies of cells in gates. Mean ± s.e.m. in c, f, k. *P < 0.05; **P < 0.01; ***P < 0.001; Kolmogorov-Smirnov test followed by Benjamini-Hochberg correction in a, b, j, two-tailed unpaired Student’s t-test in c, f, k, two-sided Fisher’s exact test followed by Benjamini-Hochberg correction in g, and two-sided Fisher’s exact test in i. Data are representative of two (c, f, k) independent experiments.
Extended Data Figure 3. Upstream signals regulate Regnase-1 expression and Regnase-1-null cell phenotypes.
(a) Immunoblot analysis of Regnase-1 expression in control sgRNA-transduced OT-I cells isolated from PLN and TILs at 7 days after adoptive transfer (n = 4 samples each group) (upper). Quantification of relative intensity of Regnase-1 expression (lower). β-actin is loading control. (b) GSEA enrichment plots of PLN and TIL control sgRNA-transduced OT-I cells (n = 4) used in (a) by using gene targets repressed by Regnase-1 (i.e. top 100 upregulated genes in TIL sgRegnase-1- compared to control sgRNA-transduced cells as identified by RNA-Seq). (c) OT-I cells were stimulated with αCD3 and αCD28 for overnight before viral transduction, and then cultured in IL-7 and IL-15-containing medium for another 3 days in vitro. Pre-activated OT-I cells were then stimulated with αCD3, IL-2 or IL-21 for 0, 1 and 4 h (n = 5 samples each group) for immunoblot analysis of full length and cleaved Regnase-1 (upper), and quantification of relative intensity of cleaved Regnase-1 expression (lower). β-actin is loading control. (d, e) OT-I cells transduced with control sgRNA (mCherry+) and sgRegnase-1 (Ametrine+) were mixed at a 1:1 ratio and transferred into mice bearing B16-Ova (n = 6 mice) or B16-F10 (n = 6 mice) tumors. Mice were analyzed at day 7 after adoptive transfer for quantification of relative OT-I cell percentage in total cells normalized to input in the TILs (d), and expression of TCF-1 (e, left), and quantification of frequency of TCF-1+ cells (e, right) in tumor-infiltrating OT-I cells. (f, g) OT-I cells were stimulated with αCD3 and αCD28 for overnight before viral transduction, and then cultured in IL-2, IL-7 and IL-15-containing medium for another 3 days in vitro. Pre-activated OT-I cells were then continuously cultured in normoxia (21% O2) or hypoxia (1% O2) condition for 48 h for immunoblot analysis of expression of HIF1α, Regnase-1 and BATF (f), and for flow cytometry analysis of expression of BATF, CD69, GzmB, CD25 and TCF-1 (g). Numbers in graphs indicate MFI (g). β-actin is loading control. Mean ± s.e.m. in a, c–e. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test in a, Kolmogorov-Smirnov test followed by Benjamini-Hochberg correction in b, and one-way ANOVA in c–e. Data are representative of two (c, f, g) independent experiments, or pooled from two (d, e) independent experiments.
Extended Data Figure 4. Proliferation and survival analyses of Regnase-1-null CD8+ T cells in tumor immunity.
(a) List of the top 10 significantly (FDR < 0.05) upregulated and downregulated pathways in TIL sgRegnase-1-transduced OT-I cells, as revealed by performing GSEA using “Hallmark” gene sets. Specifically, control sgRNA- (n = 4) and sgRegnase-1- (n = 5) transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (b) GSEA enrichment plots of TIL sgRegnase-1-transduced OT-I cells using cell cycling-associated gene sets, including E2F targets (upper), G2M checkpoint (middle) and mitotic spindle (lower). (c–g) OT-I cells transduced with control sgRNA (mCherry+) and sgRegnase-1 (Ametrine+) were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were analyzed at day 7 (d–g) (n = 6 mice) and day 14 (c) (n = 5 mice) by flow cytometry for Ki-67 expression (c, left; e, left), BrdU incorporation (d, upper; pulse for 18 h), active caspase-3 expression (f, left), Ser139 phosphorylation of histone variant H2A.X (g, upper), and quantification of MFI of Ki-67 (c, right; e, right), frequency of BrdU+ cells (d, lower), frequency of active caspase-3+ cells (f, right) and frequency of Ser139 phosphorylated histone variant H2A.X+ cells (g, lower). Numbers in graphs indicate MFI of Ki-67 (c, right; e, left). Numbers in plots indicate frequencies of BrdU+ cells (d, upper), active caspase-3+ cells (f, left) and Ser139 phosphorylated histone variant H2A.X+ cells (g, upper). (h) List of the top 15 significantly (FDR < 0.05) upregulated and top 4 significantly downregulated pathways in PLN sgRegnase-1-transduced OT-I cells, as revealed by performing GSEA using “Hallmark” gene sets. Specifically, control sgRNA- (n = 4) and sgRegnase-1- (n = 5) transduced OT-I cells were mixed and transferred into tumor-bearing mice, and PLN OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (i, j) OT-I cells transduced with control sgRNA (mCherry+) and sgRegnase-1 (Ametrine+) were mixed and transferred into tumor-bearing mice, and OT-I cells in the spleen were analyzed at day 7 (i, j) (n = 6 mice) by flow cytometry for BrdU incorporation (i, upper; pulse for 18 h) and active caspase-3 expression (j, upper), and quantification of frequencies of BrdU+ cells (i, lower) and active caspase-3+ cells (j, lower). Numbers in plots indicate frequencies of BrdU+ cells (i, upper) and active caspase-3+ cells (j, upper). Mean ± s.e.m. in c–g, i, j. *P < 0.05; **P < 0.01; ***P < 0.001; Kolmogorov-Smirnov test followed by Benjamini-Hochberg correction in a, b, h, two-tailed unpaired Student’s t-test in c–g, i, j. Data are representative of two (c) independent experiments, or pooled from two (d–g, i, j) independent experiments.
Extended Data Figure 5. Effector molecule expression of tumor-infiltrating Regnase-1-null CD8+ T cells.
(a, b) OT-I cells transduced with control sgRNA (mCherry+) or sgRegnase-1 (Ametrine+) were mixed at a 5:1 ratio and transferred into tumor-bearing mice (n = 5 mice), and tumor-infiltrating OT-I cells were analyzed at day 7 for the expression of CD69, CD25, CD49a, KLRG1, ICOS, Lag3, PD-1 and CTLA4 (a, upper) and CD44 and CD62L (b, upper), and quantification of MFI of CD69, CD25, CD49a, KLRG1, ICOS, Lag3, PD-1 and CTLA4 (a, lower) and frequency of CD44+CD62L– cells (b, lower). Numbers in graphs indicate MFI (a, upper). Numbers in plots indicate frequency of CD44+CD62L– cells (b, upper). (c–f) OT-I cells transduced with control sgRNA (mCherry+) or sgRegnase-1 (Ametrine+) were mixed at a 5:1 ratio and transferred into tumor-bearing mice, and analyzed at days 7 (n = 10 mice) or 14 (n = 10 mice). Flow cytometry analysis of expression of IFN-γ (c, upper), GzmB (c, lower), TNF-α (e, upper) and IL-2 (e, lower) in TIL OT-I cells, and quantification of the numbers of IFN-γ+ cells (d, upper), GzmB+ cells (d, lower), TNF-α+ cells (f, left) and IL-2+ cells (f, right) normalized to input per gram tissue. Numbers adjacent to outlined areas indicate frequency of IFN-γ+ cells and MFI of IFN-γ in IFN-γ+ cells (c, upper), and frequency of GzmB+ cells and MFI of GzmB in GzmB+ cells (c, lower), and frequencies of TNF-α+ cells (e, upper), or IL-2+ cells (e, lower). Mean ± s.e.m. in a, b, d, f. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test in a, b, and two-tailed paired Student’s t-test in d, f. Data are representative of two (a–c, e) independent experiments, or pooled from two (d, f) independent experiments.
Extended Data Figure 6. scRNA-Seq and flow cytometry analyses of tumor-infiltrating Regnase-1-null OT-I cells.
(a–e) scRNA-Seq analysis of control sgRNA- and sgRegnase-1-transduced OT-I cells isolated from TILs. Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for transcriptional profiling by scRNA-Seq. tSNE visualization of Pdcd1 (a, upper), Havcr2 (a, lower), Ifng (c, upper), Gzmb (c, lower), Batf (d) and Id2 (e) gene expression, and “CXCR5+ exhausted CD8 (Ahmed)12” (b, upper) and “CXCR5+ exhausted CD8 (Yu)13” (b, lower) gene signatures in individual cells. (f) OT-I cells transduced with control sgRNA and sgRegnase-1 were mixed and transferred into tumor-bearing mice (n = 5 mice; data from one representative mouse were shown), and tumor-infiltrating OT-I cells were analyzed at day 7 for the expression of TOX, Slamf6, CD127, KLRG1, Tim3 and PD-1 in TCF-1+ and TCF-1– cells of control sgRNA- and sgRegnase-1-transduced OT-I cells. Numbers in graphs indicate mean ± s.e.m. of MFI of markers on the x-axis after gating on TCF-1+ or TCF-1– subsets. Data are representative of two (f) independent experiments.
Extended Data Figure 7. Genome-scale CRISPR screening identifies BATF as an important Regnase-1 functional target in tumor immunity.
(a) Scatterplot of the enrichment of each gene versus its adjusted P value in genome-scale CRISPR screening. Gene enrichment was calculated by averaging the enrichment of their sgRNAs (n = 4 for each gene) in tumor-infiltrating OT-I cells relative to input (log2 ratio (TIL/input)), with the most extensively enriched (red) and selectively depleted (blue) genes (adjusted P < 0.05), as well as ‘dummy’ genes (green; generated by random combinations of 4 out of 1,000 non-targeting control sgRNAs per ‘dummy’ gene). (b) Venn diagram showing the overlap of genes between top depleted genes in genome-scale CRISPR screening (by less than −3.5 log2 (TIL/input) fold change; adjusted P < 0.05) and top upregulated genes in TIL sgRegnase-1- versus control sgRNA-transduced OT-I cells as identified by RNA-Seq (by greater than 1.5 log2 fold change; P < 0.05). (c) Tn5 insert sites from ATAC-Seq analysis were aligned to motifs for transcription factors from the TRANSFAC database, and the binding profiles of BATF are shown. (d) Enrichment of BATF-binding motifs in the genomic regions with upregulated accessibility in Regnase-1-null cells. First, we analyzed common regions in our Regnase-1-null ATAC-Seq data and published BATF ChIP-Seq peaks (GSE5419126). Next, we scanned these common regions with TRANSFAC motifs for BATF, and numbers of motif matches and associated Fisher’s exact test P values and log2 (odds ratios) are shown (a positive log2 (odds ratio) value indicates that a motif is more likely to occur in Regnase-1-null cells than in wild-type samples; ‘E − x’ denotes ‘×10−x’). (e) Luciferase activity of HEK293T cells measured at 48 h after transfection with Il2 mRNA 3’ UTR (upper) or Il4 mRNA 3’ UTR (lower) luciferase reporter plasmid, together with control (mock), wild-type or D141N Regnase-1-expressing plasmid (n = 3 samples each group). (f) OT-I cells transduced with control sgRNA (mCherry+; spike) were mixed at a 1:1 ratio with cells transduced with control sgRNA (Ametrine+), sgRegnase-1 (Ametrine+), sgBatf (GFP+) or sgBatf/Regnase-1 (GFP+ and Ametrine+), and transferred into tumor-bearing hosts individually (n = 4 mice each group). Mice were analyzed at 5 days after adoptive transfer for quantification of relative OT-I cell percentage in CD8α+ cells normalized to spike in the spleen (f, left) and TILs (f, right), and quantification of relative MFI of BATF normalized to spike in the tumor-infiltrating OT-I cells (f). (g) Immunoblot analysis of Regnase-1 and BATF expression in in vitro cultured OT-I cells 3 days after transduction with control sgRNA or sgBatf/Regnase-1. Hsp90 is loading control. (h–k) The same transfer system as in (f) was used. Five days after adoptive transfer, mice were analyzed for quantification of relative OT-I cell percentage in CD8α+ cells normalized to spike in the spleen (h, left, n = 4) and TILs (h, right, n = 4). Tumor-infiltrating OT-I cells were analyzed at day 5 (n = 4 mice each group) for quantification of relative frequency of active caspase-3+ cells normalized to spike (i), and quantification of relative frequency of TCF-1+ cells normalized to spike (j), or at day 7 (n = 6 mice each group) for quantification of relative frequency of IFN-γ+ cells normalized to spike (k). (l) 4 × 106 pmel-1 cells transduced with sgRegnase-1 (Ametrine+) (n = 10 recipients) or sgBatf/Regnase-1 (GFP+ and Ametrine+) (n = 10 recipients) were transferred into mice at day 12 after B16-F10 melanoma engraftment, followed by analysis of tumor size. Mean ± s.e.m. in f, h–k. Mean ± s.d. in e. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s paired t-test followed by Bonferroni correction in a, two-sided Fisher’s exact test in d, one-way ANOVA in e, f, h–k, and two-way ANOVA in l. Data are representative of two (e) or three (h) independent experiments, or pooled from two (f, g, i–l) independent experiments.
Extended Data Figure 8. BATF overexpression markedly enhances CD8+ T cell antitumor responses.
(a) OT-I cells were stimulated with αCD3 and αCD28 for overnight before viral transduction, and then cultured in IL-7 and IL-15-containing medium for another 3 days in vitro. Control sgRNA- and sgRegnase-1-transduced OT-I cells were then stimulated with αCD3, IL-2 or IL-21 for overnight for flow cytometry analysis of BATF expression (upper), and quantification of the MFI of BATF (lower) (n = 6 samples each group). Numbers in graphs indicate MFI (upper) and fold change between comparisons (lower). (b–h) OT-I cells transduced with control retrovirus (mCherry+) were mixed at a 1:1 ratio with cells transduced with Batf-overexpressing retrovirus (GFP+), and transferred into tumor-bearing hosts. Mice were analyzed at day 4 (e) (n = 4 mice), day 5 (b, h) (n = 4 mice), day 7 (c, d, f, g) (n = 6–8 mice) or day 14 (c, d) (n = 6 mice) for the expression of BATF (b, left), active caspase-3 (f, left), IFN-γ, GzmB, TNF-α and IL-2 (g, left), and TCF-1 (h, upper) in TIL OT-I cells, and quantification of MFI of BATF in TIL OT-I cells (b, right), and quantification of frequencies of active caspase-3+ cells (f, right), IFN-γ+, GzmB+, TNF-α+ and IL-2+ cells (g, right), and TCF-1+ cells (h, lower) in TIL OT-I cells, and analysis of the proportion of donor-derived OT-I cells in total CD8α+ cells in TILs and spleen (c), and quantification of relative OT-I cell percentage in CD8α+ cells normalized to input in the spleen (d), and the dilution of CellTrace Violet (CTV) in TIL OT-I cells (e, left), and quantification of MFI of CTV in TIL OT-I cells (e, right). Numbers in graphs indicate MFI (b, left; e, left), frequencies of OT-I cells in gates (c), frequency of active caspase-3+ cells (f, left), frequencies of IFN-γ+, GzmB+, TNF-α+ or IL-2+ cells (g, left), and frequency of TCF-1+ cells (h, upper). Mean ± s.e.m. in a, b, d–h. *P < 0.05; **P < 0.01; ***P < 0.001; two-tailed unpaired Student’s t-test in a, b, d–h. Data are representative of two (a, c) independent experiments, or pooled from two (b, d–h) independent experiments.
Extended Data Figure 9. Genome-scale CRISPR screening identifies mitochondrial metabolism as an important downstream pathway of Regnase-1 and BATF.
(a) Chromatin accessibility heat maps normalized by row (z-score) for 7,480 genes with significantly increased chromatin accessibility (by |log2 FC| > 0.5; P < 0.05) in sgRegnase-1-transduced OT-I cells as compared to control sgRNA-transduced cells. Specifically, OT-I cells transduced with control sgRNA (mCherry+) (n = 4), sgRegnase-1 (Ametrine+) (n = 4), sgBatf (GFP+) (n = 2) or sgBatf/Regnase-1 (GFP+ and Ametrine+) (n = 4) were transferred into tumor-bearing hosts individually. OT-I cells were isolated from TILs at day 7 for ATAC-Seq analysis. We annotated the differential accessibility (DA) regions in ATAC-Seq for the nearest genes, and identified 7,480 genes with significantly increased chromatin accessibility in Regnase-1-null cells as compared to wild-type cells. BATF co-deletion reversed the upregulated chromatin accessibility for a large proportion of these genes (5,052 in total). Also, 2,527 among these 5,052 genes showed significantly downregulated chromatin accessibility in BATF-null cells as compared to wild-type cells. (b) Functional enrichment plots of the top 10 significantly (FDR < 0.05) enriched pathways in top-ranking depleted genes (n = 4 sgRNAs for each gene) identified in the genome-scale CRISPR screening (by less than −3.5 log2 (TIL/input) fold change; adjusted P < 0.05). (c) GSEA enrichment plots of TIL sgRegnase-1-transduced OT-I cells using the OXPHOS “Hallmark” gene set. Specifically, control sgRNA- and sgRegnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for transcriptional profiling by RNA-Seq. (d) Representative images (upper) and quantification of mitochondrial volume (stained with Tom20, white) per cell (lower) in control sgRNA- (mCherry+; red) and sgRegnase-1-transduced OT-I cells (Ametrine+; green) in tumors at 7 days after adoptive transfer (n = 4 mice). (e) Oxygen consumption rate (OCR) bioenergetic profiling of control sgRNA- and sgRegnase-1-transduced OT-I cells cultured in vitro for basal (left) and maximal (right) OCR (n = 9 samples each group). (f) List of the top 2 significantly (FDR < 0.05) upregulated and top 8 significantly downregulated pathways in TIL sgBatf/Regnase-1- (n = 3 samples) versus sgRegnase-1-transduced (n = 3 samples) OT-I cells isolated from TILs, as revealed by performing GSEA using “Hallmark” gene sets. Specifically, sgRegnase-1- and sgBatf/Regnase-1-transduced OT-I cells were mixed and transferred into tumor-bearing mice, and tumor-infiltrating OT-I cells were isolated at day 7 for transcriptional profiling by microarray. (g) GSEA enrichment plots of TIL sgBatf/Regnase-1- (n = 3 samples) versus sgRegnase-1-transduced OT-I cells (n = 3 samples) using OXPHOS gene set. (h) OT-I cells transduced with control sgRNA (mCherry+; spike) were mixed at a 1:1 ratio with cells transduced with control sgRNA (Ametrine+), sgRegnase-1 (Ametrine+), sgBatf (GFP+) or sgBatf/Regnase-1 (GFP+ and Ametrine+), and transferred into tumor-bearing hosts individually (n = 4 mice each group). Mice were analyzed at 5 days after adoptive transfer for quantification of relative MFI of TMRM (left) and Mitotracker (right) normalized to spike in tumor-infiltrating OT-I cells. (i) Chromatin accessibility heat maps normalized by row (z-score) for mitochondrial genes with significantly increased chromatin accessibility (by |log2 FC| > 0.5; P < 0.05) in sgRegnase-1-transduced OT-I cells as compared to control sgRNA-transduced cells as determined by ATAC-Seq as described in a. We annotated the DA regions in ATAC-Seq for the nearest genes, and superimposed these genes with 1,158 mitochondrial genes defined in MitoCarta 2.0 database. A total of 341 mitochondrial genes showed significantly upregulated chromatin accessibility in the absence of Regnase-1, 214 of which were blocked by BATF co-deletion in BATF/Regnase-1-null cells. Moreover, 96 among these 214 genes showed significantly downregulated chromatin accessibility in BATF-null cells as compared to wild-type cells. Mean ± s.e.m. in d, e, h. *P < 0.05; **P < 0.01; ***P < 0.001; two-sided Fisher’s exact test in a, i, right-tailed Fisher’s exact test in b, Kolmogorov-Smirnov test followed by Benjamini-Hochberg correction in c, f, g, two-tailed unpaired Student’s t-test in d, e, and one-way ANOVA in h. Data are representative of two (d, e) independent experiments, or pooled from two (h) independent experiments.
Extended Data Figure 10. PTPN2 and SOCS1 deletion efficiency and expression in Regnase-1-null cells, and model of Regnase-1 functions in tumor-specific CD8+ T cells.
(a) Immunoblot analysis of Regnase-1, PTPN2 and SOCS1 expression in in vitro cultured OT-I cells 3 days after transduction with control sgRNA, sgPtpn2/Regnase-1 (left), or sgSocs1/Regnase-1 (right). Hsp90 is loading control. (b) Immunoblot analysis of Regnase-1, BATF, SOCS1 and PTPN2 expression in control sgRNA- and sgRegnase-1-transduced OT-I cells cultured in vitro for 3 days after viral transduction. β-actin is loading control. (c) Regnase-1 is a major negative regulator of CD8+ T cell antitumor responses, and TCR and IL-2 inhibit its expression and activity. Deletion of Regnase-1 unleashes potent therapeutic efficacy of engineered tumor-specific CD8+ T cells against cancers by coordinating transcriptional and metabolic programs to achieve greatly improved cell accumulation and function. As a key functional target of Regnase-1, excessive BATF drives robust cell accumulation and effector function, in part through enhancing mitochondrial metabolism, in Regnase-1-null CD8+ T cells. Regnase-1 deletion also reprograms cells to acquire increased naïve/memory cell-associated gene signatures and gain survival advantage, which contribute to the improved persistence of Regnase-1-null effector CD8+ T cells. Targeting PTPN2 and SOCS1 (not depicted here) acts in coordination with Regnase-1 inhibition to promote CD8+ T cell antitumor responses. Data are representative of three (a, b) independent experiments.
Supplementary Material
Acknowledgements
The authors acknowledge Melissa Hendren for animal colony management, Chunliang Li for help with plasmids, Geoffrey Neale and Scott Olsen for assistance with sequencing, and St. Jude Immunology FACS core facility for cell sorting. This work was supported by NIH AI105887, AI131703, AI140761, CA176624 and CA221290 (to H.C.).
Footnotes
Competing interests
H.C. and J.W. are authors of a patent application related to Regnase-1 and BATF.
Data availability
Microarray, RNA-Seq, ATAC-Seq and scRNA-Seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number: GSE126072. All other relevant data are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Lim WA & June CH The Principles of Engineering Immune Cells to Treat Cancer. Cell 168, 724–740, doi: 10.1016/j.cell.2017.01.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gattinoni L et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J Clin Invest 115, 1616–1626, doi: 10.1172/JCI24480 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishton RJ, Sukumar M & Restifo NP Metabolic Regulation of T Cell Longevity and Function in Tumor Immunotherapy. Cell Metab 26, 94–109, doi: 10.1016/j.cmet.2017.06.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muri J et al. The thioredoxin-1 system is essential for fueling DNA synthesis during T-cell metabolic reprogramming and proliferation. Nat Commun 9, 1851, doi: 10.1038/s41467-018-04274-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng M et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 354, 481–484, doi: 10.1126/science.aaf6284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanoaica L et al. Conditional deletion of ferritin h in mice reduces B and T lymphocyte populations. PLoS One 9, e89270, doi: 10.1371/journal.pone.0089270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang W, Beckett O, Flavell RA & Li MO An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity 30, 358–371, doi: 10.1016/j.immuni.2009.02.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsushita K et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458, 1185–1190, doi: 10.1038/nature07924 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Uehata T et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 153, 1036–1049, doi: 10.1016/j.cell.2013.04.034 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Churchman ML et al. Synergism of FAK and tyrosine kinase inhibition in Ph(+) B-ALL. JCI Insight 1, doi: 10.1172/jci.insight.86082 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer M et al. A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 166, 1500–1511 e1509, doi: 10.1016/j.cell.2016.08.052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im SJ et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421, doi: 10.1038/nature19330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leong YA et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat Immunol 17, 1187–1196, doi: 10.1038/ni.3543 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Zhou X et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240, doi: 10.1016/j.immuni.2010.08.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurton LV et al. Tethered IL-15 augments antitumor activity and promotes a stem-cell memory subset in tumor-specific T cells. Proc Natl Acad Sci U S A 113, E7788–E7797, doi: 10.1073/pnas.1610544113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roychoudhuri R et al. BACH2 regulates CD8(+) T cell differentiation by controlling access of AP-1 factors to enhancers. Nat Immunol 17, 851–860, doi: 10.1038/ni.3441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ichii H, Sakamoto A, Kuroda Y & Tokuhisa T Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J Immunol 173, 883–891 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Man K et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141 e1125, doi: 10.1016/j.immuni.2017.11.021 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Sade-Feldman M et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020, doi: 10.1016/j.cell.2018.10.038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218, doi: 10.1038/nmeth.2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214, doi: 10.1016/j.cell.2015.05.002 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan O et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218, doi: 10.1038/s41586-019-1325-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336, doi: 10.1038/s41590-019-0312-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Utzschneider DT et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427, doi: 10.1016/j.immuni.2016.07.021 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Ciofani M et al. A validated regulatory network for Th17 cell specification. Cell 151, 289–303, doi: 10.1016/j.cell.2012.09.016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurachi M et al. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15, 373–383, doi: 10.1038/ni.2834 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CY et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol 12, 1221–1229, doi: 10.1038/ni.2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191, doi: 10.1038/nbt.3437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinclair LV et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol 14, 500–508, doi: 10.1038/ni.2556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atherly LO, Brehm MA, Welsh RM & Berg LJ Tec kinases Itk and Rlk are required for CD8+ T cell responses to virus infection independent of their role in CD4+ T cell help. J Immunol 176, 1571–1581 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Blagih J et al. The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54, doi: 10.1016/j.immuni.2014.12.030 (2015). [DOI] [PubMed] [Google Scholar]
- 32.D’Souza WN, Chang CF, Fischer AM, Li M & Hedrick SM The Erk2 MAPK regulates CD8 T cell proliferation and survival. J Immunol 181, 7617–7629 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan BM, Juedes A, Szabo SJ, von Herrath M & Glimcher LH Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A 100, 15818–15823, doi: 10.1073/pnas.2636938100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger R et al. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842 e813, doi: 10.1016/j.cell.2016.09.031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawalekar OU et al. Distinct Signaling of Coreceptors Regulates Specific Metabolism Pathways and Impacts Memory Development in CAR T Cells. Immunity 44, 380–390, doi: 10.1016/j.immuni.2016.01.021 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61, doi: 10.1126/science.aaa8172 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Shifrut E et al. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell 175, 1958–1971 e1915, doi: 10.1016/j.cell.2018.10.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manguso RT et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547, 413–418, doi: 10.1038/nature23270 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quigley M et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat Med 16, 1147–1151, doi: 10.1038/nm.2232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES
- 40.Platt RJ et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455, doi: 10.1016/j.cell.2014.09.014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogquist KA et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Overwijk WW et al. Tumor regression and autoimmunity after reversal of a functionally tolerant state of self-reactive CD8+ T cells. J Exp Med 198, 569–580, doi: 10.1084/jem.20030590 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41, 325–338, doi: 10.1016/j.immuni.2014.08.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birsoy K et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551, doi: 10.1016/j.cell.2015.07.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanson KR et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9, 5416, doi: 10.1038/s41467-018-07901-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sukumar M et al. Mitochondrial Membrane Potential Identifies Cells with Enhanced Stemness for Cellular Therapy. Cell Metab 23, 63–76, doi: 10.1016/j.cmet.2015.11.002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W et al. Effector T Cells Abrogate Stroma-Mediated Chemoresistance in Ovarian Cancer. Cell 165, 1092–1105, doi: 10.1016/j.cell.2016.04.009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei J et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol 17, 277–285, doi: 10.1038/ni.3365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng H et al. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490, doi: 10.1038/nature12297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karmaus PWF et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105, doi: 10.1038/s41586-018-0806-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137, doi: 10.1186/gb-2008-9-9-r137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842, doi: 10.1093/bioinformatics/btq033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Law CW, Chen Y, Shi W & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, doi: 10.1186/gb-2014-15-2-r29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calvo SE, Clauser KR & Mootha VK MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44, D1251–1257, doi: 10.1093/nar/gkv1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, doi: 10.1093/nar/gkp335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramirez F, Dundar F, Diehl S, Gruning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42, W187–191, doi: 10.1093/nar/gku365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuellar-Partida G et al. Epigenetic priors for identifying active transcription factor binding sites. Bioinformatics 28, 56–62, doi: 10.1093/bioinformatics/btr614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krishnamoorthy V et al. The IRF4 Gene Regulatory Module Functions as a Read-Write Integrator to Dynamically Coordinate T Helper Cell Fate. Immunity 47, 481–497 e487, doi: 10.1016/j.immuni.2017.09.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.