Significance
Membrane lipid homeostasis within cells requires a cooperation of vesicular transport and of lipid transfer proteins that mediate lipid exchange between membranes independent of bilayer fusion. Many such proteins also function as tethers connecting adjacent bilayers. For membranes that travel along the exo- and endocytic pathways, these tethers must be regulated to ensure their action selectively at specific stations of their journey. Here we identify a mechanism that recruits the endoplasmic reticulum (ER) to late endosomes/lysosomes and that may promote exchange of lipids between their membranes. These contacts rely on the binding of an ER-anchored lipid transfer protein to a key player in endosomal maturation, the small GTPase Rab7, thus explaining how their formation is regulated in time and space.
Keywords: SMP domain, lipid-transfer protein, membrane contact sites
Abstract
Contacts between the endoplasmic reticulum (ER) and other membranes are hot spots for protein-mediated lipid transport between the 2 adjacent bilayers. Compiling a molecular inventory of lipid transport proteins present at these sites is a premise to the elucidation of their function. Here we show that PDZD8, an intrinsic membrane protein of the ER with a lipid transport module of the SMP domain family, concentrates at contacts between the ER and late endosomes/lysosomes, where it interacts with GTP-Rab7. These findings suggest that PDZD8 may cooperate with other proteins that function at the ER–endo/lysosome interface in coordinating endocytic flow with lipid transport between endocytic membranes and the ER.
The endoplasmic reticulum (ER) is the compartment in which most membrane lipids are synthesized and to which lipid metabolites generated in other membranes are returned to be reused in biosynthetic reactions. Transport of lipids between the ER and other membranes occurs through vesicles that carry them in their bilayer or via lipid transport proteins (LTPs). LTPs contain modules that extract lipids from 1 bilayer, harbor them inside a hydrophobic cavity, and deliver them to other membranes bypassing vesicular transport (1–4). Many of them act at membrane contact sites (MCSs), thus enhancing the efficiency and specificity of transport (4–7).
One lipid harboring module found in LTPs localized at MCSs is the Synaptotagmin-like mitochondrial-lipid-binding (SMP) domain (8, 9). LTPs with this domain are expressed from unicellular organisms to humans. Although some of them are present in all phyla [e.g., the tricalbins/extended-synaptotagmins (10, 11)], others are restricted to a subset of living species (12). PDZD8 is an SMP domain containing ER protein anchored to the membrane by a N-terminal transmembrane region, which is followed, in sequence, by a PDZ domain, a C1 domain, and a coiled-coil region (12, 13). A putative split C2 domain has also been detected in its sequence (Fig. 1A) (14). PDZD8 was proposed to be the ortholog of the yeast protein Mmm1 because of the similarity of their SMP domains (13). Mmm1 is a subunit of the ERMES complex, which is found in fungi but not in metazoan cells, and which mediates lipid transport between the ER and mitochondria (15, 16). Supporting this possibility, defects in mitochondrial functions were identified in PDZD8 KD cells (13). However, a more recent analysis indicated that PDZD8 is a paralogue, not an ortholog, of Mmm1 (17). In addition, a BioID-based proximity labeling study identified PDZD8 as a neighbor of proteins of the endo-lysosomal system (18), prompting us to further examine its site of action. Here we show that PDZD8 tethers the ER to late endosomes and lysosomes via an interaction with Rab7.
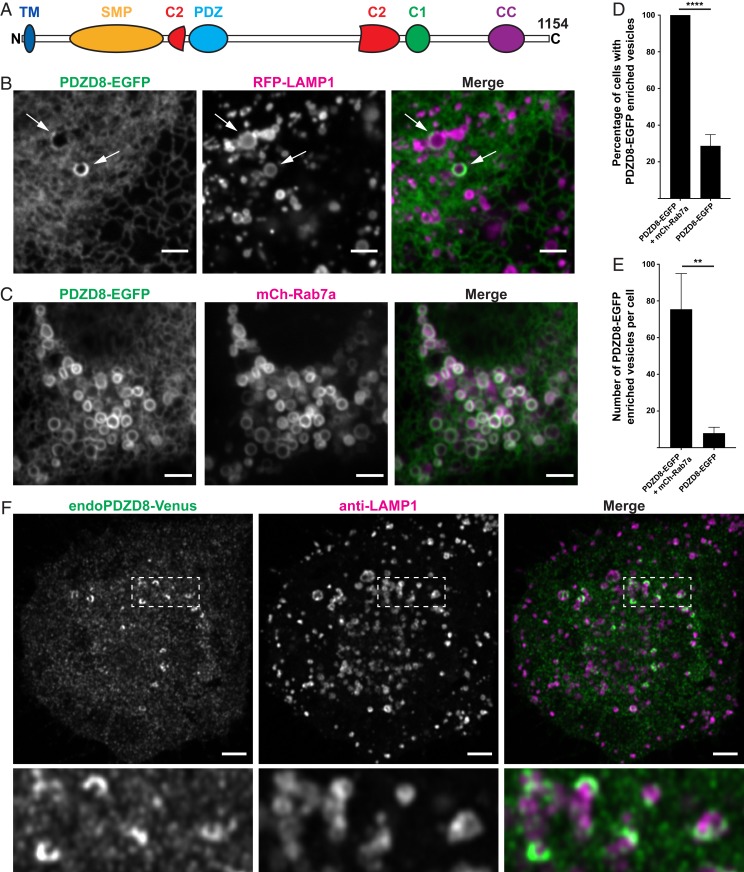
Fig. 1.
Localization of PDZD8 at MCS between the ER and late endosomes/lysosomes. (A) Domain organization of human PDZD8. (B) Confocal images of Cos7 cells showing that PDZD8-EGFP is localized throughout the ER but enriched at the ER surrounding a small subset of Lamp1-RFP–positive vesicles (arrows). (C) The expression of mCherry-Rab7a sequesters the bulk of PDZD8 on all mCherry-Rab7a–positive vesicles, producing a striking colocalization. (D) Percentage of cells with PDZD8-EGFP signal surrounding vesicles with and without mCherry-Rab7a coexpression. (E) Number of vesicles surrounded by PDZD8-EGFP per cell. Cells not expressing mCherry-Rab7a and without such vesicles were excluded from the count. Data for D and E represent mean ± SD, n = 3 experiments. **P = 0.004; ****P < 0.0001 (2-tailed, unpaired t test). (F) Anti-Venus immunofluorescence of Neuro2A cells showing concentration of endogenous PDZD8 (tagged with Venus) around Lamp1-positive vesicles and an additional diffuse punctate fluorescence as expected for an ER protein in fixed cells. (Scale bars, 3 µm.)
Results and Discussion
Expression of PDZD8-EGFP in Cos7 cells resulted in a reticular fluorescence, as expected for an ER protein. However, in a subset of cells, possibly reflecting a specific functional state, there was an additional intense signal on sparse vesicular structures, suggesting a concentration of PDZD8 on ER membranes that enwrap them (Fig. 1B). Based on the BioID-based study by Liu et al. (18), we investigated a potential proximity of PDZD8-EGFP to several fluorescently tagged proteins of the endolysosomal system. Colocalization was observed with a subpopulation of RFP-LAMP1–positive vesicles (Fig. 1B). Moreover, on cotransfection of mCherry-Rab7a, a late endosomal Rab (19), all cells positive for both constructs contained PDZD8-positive vesicles and the number of PDZD8-positive vesicles per cell dramatically increased, with a consistent striking colocalization of PDZD8-EGFP and mCherry-Rab7a (Fig. 1 C–E).
To exclude a role of PDZD8 overexpression on these findings, we analyzed previously described Neuro2A cells in which PDZD8 is tagged at the endogenous locus with a Venus fluorescent protein (13). Given the weak signal produced by the fluorescence of PDZD8-Venus, this protein was detected by anti-GFP antibodies, as performed in the previous study (13). Scattered PDZD8-Venus bright hot spots (often with a crescent shape) were visible in many of these cells in addition to lower-intensity widespread PDZD8-Venus fluorescence, reflecting its ER localization (Fig. 1F). Coimmunostaining for Lamp1 revealed that most hot spots were localized adjacent to clearly detectable Lamp1 vesicles, often partially surrounding them, although, as in Cos7 cells, there was high cell-to-cell variability in the number of Lamp1 vesicles positive for PDZD8. Only a subpopulation of LAMP1 vesicles located in the central region of the cells were positive for PDZ8-Venus, consistent with a localization on late endocytic compartments.
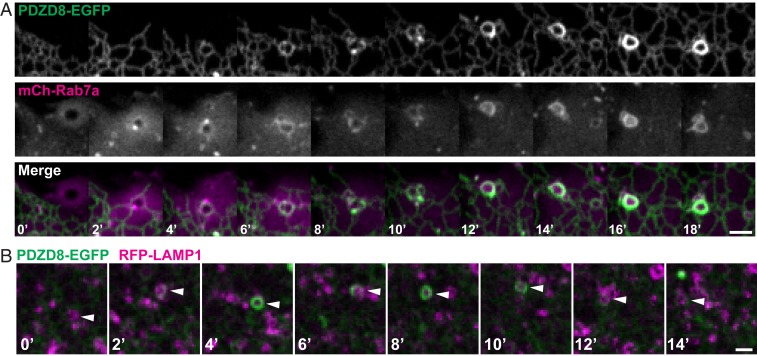
Time-lapse imaging of Cos7 cells revealed that appearance of mCherry-Rab7a on vesicles precisely correlated with an accumulation of PDZD8-EGFP signal around them, as expected if PDZD8 interacted with Rab7 (Fig. 2A and Movie S1). A dynamic association/dissociation was also observed between PDZD8-EGFP and RFP-LAMP1 in cells not overexpressing Rab7 (Fig. 2B and Movie S2).
Fig. 2.
PDZD8-mediated ER-wrapping of late endosomes/lysosomes is dynamic and correlates with presence of Rab7. (A) Confocal time-lapse images of PDZD8-EGFP and mCherry-Rab7a showing progressive ER wrapping and PDZD8 enrichment around the vesicles that acquire mCherry-Rab7a. (B) Confocal time-lapse images of Cos7 cells not overexpressing Rab7, showing reversible association of PDZD8-EGFP with a Lamp1-RFP–positive vesicle (arrowhead). (Scale bars, 3 µm.)
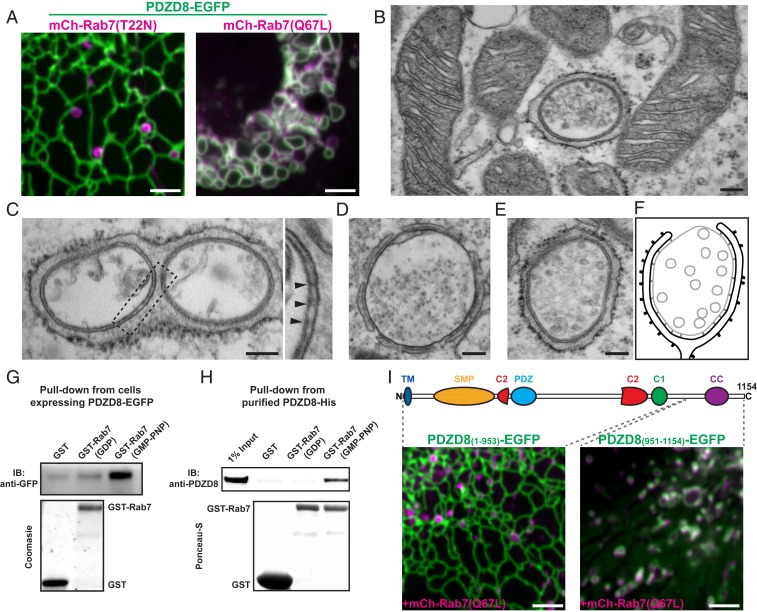
The strong impact of Rab7 on the localization of PDZD8 suggested that PDZD8 may be a Rab7 effector, prompting us to examine whether this action is regulated by Rab7’s GTPase activity. Upon coexpression of dominant negative Rab7 (mCherry-Rab7aT22N) in Cos7 cells, no colocalization with PDZD8-EGFP was observed (Fig. 3A). Conversely, transfection of the same cells with constitutively active Rab7 (mCherry-Rab7aQ67L) resulted in massive recruitment of PDZD8-EGFP to the large endosomes produced by the expression of this construct (Fig. 3A). These results were corroborated by electron microscopy, as inspection of sections of cells expressing PDZD8-EGFP and mCherry-Rab7aQ67L confirmed that this colocalization reflected massive enwrapping of vacuolar structures by ER (Fig. 3 B–F). Many such vacuoles contained heterogeneous material as expected for lysosomes (Fig. 3 B–D), whereas others contained homogenously sized vesicles, as expected for late endosomes/multivesicular bodies (Fig. 3 E and F). The narrow gap between the ER and other organelles was populated by electron-dense elements oriented perpendicular to the 2 adjacent membranes (Fig. 3 C, Right). Most likely, these densities represent tethers formed by PDZD8 and Rab7.
Fig. 3.
The interactions of the C-terminal region of PDZD8 with GTP-bound Rab7 mediates ER tethering to late endosome/lysosomes. (A) Confocal images of Cos7 cells showing colocalization between PDZD8-EGFP and constitutively active Rab7 (mCherry-Rab7Q67L), but not with its dominant negative Rab7 (mCherry-Rab7T22N). (Scale bars, 3 µm.) (B–E) Representative electron micrographs of Cos7 cells overexpressing PDZD8-EGFP and mCherry-Rab7Q67L. (Scale bars, 150 nm.) Note a vesicle completely surrounded by the ER in the plane of the section in B, while there are no extensive contacts between the ER and mitochondria in the neighboring cell region. Lysosomes and a multivesicular body enwrapped by the ER are visible in C and D and in E, respectively. A portion of the ER-lysosome contact of field C is shown at higher magnification at right, where arrowheads point to protein densities that likely represent PDZD8-Rab7 tethers between the 2 organelles. (F) Schematic representation of the MCS shown in the images of (B–E). (G and H) GST pull-downs showing enrichment of PDZD8 on GMP-PMP-Rab7 relative to controls in cell lysates (G) and purified PDZD8 (H). (I) Confocal images showing no colocalization of mCherry-Rab7Q67L with PDZD8(1-953)-EGFP, but a striking colocalization with PDZD8(951-1154). (Scale bars, 3 µm.)
To confirm a GTP-dependent interaction of PDZD8 with Rab7, purified GST-tagged Rab7 preincubated with either GDP or GMP-PMP (nonhydrolyzable analog of GTP) was used in pull-downs from lysates of HEK293 cells overexpressing PDZD8-EGFP. The amount of PDZD8-EGFP retained by immobilized GST-Rab7a was much increased when Rab7 had been preincubated with GMP-PMP (Fig. 3G). A direct interaction was confirmed using His-tag purified PDZD8 as the bait in the pull-down (Fig. 3H), establishing that PDZD8 is a Rab7 effector. Removal of the C-terminal 200 residues of PDZD8, predicted to contain a coiled-coil domain, completely abolished its recruitment to vesicular structures even on coexpression with mCherry-Rab7aQ67L. Conversely, this C-terminal portion alone was robustly recruited to mCherry-Rab7aQ67L vesicles, indicating that the last 200 residues of PDZD8 are necessary and sufficient for its interaction with Rab7 (Fig. 3I).
Collectively, our findings support a model according to which PDZD8 and GTP-bound Rab7 drive the formation of MCS between the ER and either late endosomes or lysosomes. In agreement with our results, a recently published screen for Rab effectors identified PDZD8 as a potential interactor of Rab7 (20).
A previous study reported the presence of PDZD8 at ER-mitochondria MCS, and showed an effect of the loss of PDZD8 on mitochondrial Ca+2 dynamics (13). Based on our results, PDZD8 seems to be primarily enriched at MCSs between the ER and late endosomes/lysosomes, although an additional action at ER-mitochondria contacts remains possible. It is unclear to which extent the lack of PDZD8 at contacts between the ER and late compartments of the endo-lysosomal system could be responsible for such a pronounced defect in mitochondrial function (13). We note, however, that cross-talk between mitochondria and late endosomes/lysosomes was demonstrated by several studies in different organisms (21). Rab7 was recently implicated in the formation of contacts between lysosomes and mitochondria, and a Rab7 GAP was shown to be localized on mitochondria (22). Moreover, in yeast, a pathway for the flux of lipids between the ER and mitochondria via the vacuole is controlled by the GTPase Ypt7, the yeast equivalent of Rab7 (23, 24). Thus, indirect functional links between PDZD8 and mitochondria mediated by signaling via Rab7 may be plausible.
The present study expands the repertoire of MCS involving SMP domain containing proteins. It also adds information about mechanisms through which GTP-bound Rab7 controls interactions between late endosomes/lysosomes and the ER. Specifically, 2 other lipid transfer proteins localize to these contacts in a Rab7-dependent way: an ORD domain containing protein ORP1L (25) and a chorein domain containing protein VPS13C (26). In addition, another ER protein with no lipid transport modules, protrudin, is a Rab7 effector (27). The elucidation of the functional interplay between these proteins will be an important priority for future studies.
Materials and Methods
Cell Culture and Transfection.
Cos7 cells, HEK293 (ATCC) and Neuro2A cells (kind gift from F. Polleux, Columbia University, New York, NY) were cultured at 37 °C and 5% CO2 in DMEM containing 10% FBS, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 mg/mL streptomycin, and 2 mM l-glutamine. Plasmid transfections were performed with Lipofectamine 2000 (Life Technologies).
Antibodies and DNA Plasmids.
Primary antibodies were as follows: rat monoclonal anti-HA (3F10; Roche), rabbit monoclonal anti-GFP (ab290; abcam), rat monoclonal anti-LAMP1 (1D4B, DSHB), and rabbit polyclonal anti-PDZD8 (PA4-46771; Thermo Fisher). Sources of plasmids were as follows: mCherry-Rab7a (Addgene #68104) and RFP-LAMP1 (provided by W. Mothes, Yale University, New Haven, CT). PDZD8-EGFP was generated by PCR amplification of human PDZD8 cDNA (Dharmacon) and ligated into pEGFP-N1 (Addgene), using EcoRI and KpnI sites. PDZD8(1-953)-EGFP was generated by PCR amplification of the coding region from PDZD8-EGFP and ligated into pEGFP-N1 (Addgene), using EcoRI and KpnI sites. EGFP-PDZD8(951-1154) was generated by PCR amplification of the coding region from PDZD8-EGFP and ligated into pEGFP-C1 (Addgene), using EcoRI and XhoI sites. mCherry-Rab7 (Q67L) and mCherry-Rab7 (T22N) were generated using site-directed mutagenesis (QuikChange II XL; Agilent Technologies) of mCherry-Rab7. pGEX-Rab7 was generated by PCR amplification of Rab7’s coding sequence from mCherry-Rab7 and ligated into pGEX6p-1, using XhoI and EcoRI. His-PDZD8 was generated by PCR amplification of PDZD8 coding sequence (amino acids 91 to 1154) and ligated into pCMV6-AN-His using AscI and NotI.
Light Microscopy.
Live cell imaging.
Cos7 cells were seeded on glass-bottomed dishes (MatTek Corp.) at a concentration of 75 × 103 cells per dish, and transiently transfected after 24 h. Just before imaging, growth media was washed and cells were imaged in Live Cell Imaging solution (Life Technologies). All live cell imaging was carried out at 37 °C and 5% CO2. Spinning-disk confocal microscopy was performed using an Andor Dragonfly system equipped with a PlanApo objective (63×, 1.4NA, Oil) and a Zyla sCMOS camera.
Immunofluorescence.
Neuro2A cells were seeded in glass-bottomed dishes, fixed with 4% formaldehyde freshly prepared from PFA, permeabilized with 0.1% Triton X-100 and blocked with 5% BSA + 0.1% Triton X-100. anti-GFP and anti-Lamp1 were used as primary antibodies and donkey anti-rabbit and goat anti-rat, respectively, were used as secondaries. Cells were imaged using a Zeiss LSM880 with Airyscan system, equipped with a PlanApo objective (63×, 1.4NA, Oil).
Electron Microscopy.
Transfected Cos7 cells were fixed with Karnovsky fixative (28), postfixed in 1% OsO4, 1% K4Fe(CN)6 in 0.1 M cacodylate buffer for 1 h, contrasted in Kellenberger solution (veronal-acetate buffer in 0.1N HCl) for 1 h, dehydrated in ethanol and embedded in EMbed812. All solutions were acquired from Electron Microscopy Sciences. Ultrathin sections were then cut on a Leica Ultracut UCT (Leica Microsystems Inc) and stained with 1.5% uranyl acetate and lead citrate. Sections were analyzed in a Tecnei BioTwin electron microscope (FEI Company).
Pull-Down Assay.
GST and GST-Rab7 were expressed in bacteria and purified from the bacterial lysate using Glutathione Agarose resin (Thermo Scientific). His-PDZD8 was expressed in Expi293 cells and purified by a Ni-NTA column (Clontech), and further by gel filtration in buffer (25 mM Tris⋅HCl at pH 8.0, 150 mM NaCl, 0.5 mM TCEP). GST and GST-Rab7 were then used as a bait to pull down interactors from the lysate (PBS, 1% Triton X-100 and protease inhibitor mixture; Roche) of HEK293 cells expressing PDZD8-EGFP and to pull down purified His-PDZD8. Before being used for the pull-down, GST-Rab7 was incubated with 2.5 mM GDP (Sigma) or GMP-PNP (Sigma). The eluate from the pull-downs were subjected to SDS/PAGE and processed for Western blotting with anti-GFP and donkey anti-rabbit HRP.
Supplementary Material
Acknowledgments
We thank M. Hanna and M. Leonzino for discussion and S. A. Mentone for outstanding technical assistance. We are greatly indebted to Franck Polleux for his generous gift of N2A cells endogenously tagged with Venus at the PDZD8 locus. This work was supported in part by NIH grants NS036251 and DA018343 and the Kavli Foundation (to P.D.C.). A.G.-S. was supported by the Gruber Science Fellowship. X.B. was supported by a Human Frontier Science Program fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913509116/-/DCSupplemental.
References
- 1.Wirtz K. W., Phospholipid transfer proteins. Annu. Rev. Biochem. 60, 73–99 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Holthuis J. C. M., Menon A. K., Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Reinisch K. M., De Camilli P., SMP-domain proteins at membrane contact sites: Structure and function. Biochim. Biophys. Acta 1861, 924–927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong L. H., Gatta A. T., Levine T. P., Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 20, 85–101 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Prinz W. A., Bridging the gap: Membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205, 759–769 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saheki Y., De Camilli P., Endoplasmic reticulum—Plasma membrane contact sites. Annu. Rev. Biochem. 86, 659–684 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wu H., Carvalho P., Voeltz G. K., Here, there, and everywhere: The importance of ER membrane contact sites. Science 361, eaan5835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toulmay A., Prinz W. A., A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125, 49–58 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schauder C. M., et al. , Structure of a lipid-bound extended synaptotagmin indicates a role in lipid transfer. Nature 510, 552–555 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manford A. G., Stefan C. J., Yuan H. L., Macgurn J. A., Emr S. D., ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev. Cell 23, 1129–1140 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Giordano F., et al. , PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alva V., Lupas A. N., The TULIP superfamily of eukaryotic lipid-binding proteins as a mediator of lipid sensing and transport. Biochim. Biophys. Acta 1861, 913–923 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Hirabayashi Y., et al. , ER-mitochondria tethering by PDZD8 regulates Ca2+ dynamics in mammalian neurons. Science 358, 623–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong L. H., Levine T. P., Tubular lipid binding proteins (TULIPs) growing everywhere. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1439–1449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornmann B., et al. , An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang A. B., John Peter A. T., Walter P., Kornmann B., ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J. Cell Biol. 210, 883–890 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wideman J. G., Balacco D. L., Fieblinger T., Richards T. A., PDZD8 is not the ‘functional ortholog’ of Mmm1, it is a paralog. F1000 Res. 7, 1088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., et al. , An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 9, 1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavrier P., Parton R. G., Hauri H. P., Simons K., Zerial M., Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62, 317–329 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Gillingham A. K., Bertram J., Begum F., Munro S., In vivo identification of GTPase interactors by mitochondrial relocalization and proximity biotinylation. eLife 8, e45916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raimundo N., Fernández-Mosquera L., Yambire K. F., Diogo C. V., Mechanisms of communication between mitochondria and lysosomes. Int. J. Biochem. Cell Biol. 79, 345–349 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Wong Y. C., Ysselstein D., Krainc D., Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbaz-Alon Y., et al. , A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30, 95–102 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Hönscher C., et al. , Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 30, 86–94 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Ma X., et al. , A non-canonical GTPase interaction enables ORP1L-Rab7-RILP complex formation and late endosome positioning. J. Biol. Chem. 293, 14155–14164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar N., et al. , VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raiborg C., et al. , Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234–238 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Karnovsky M. J., A formaldehyde-glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27, 137A–138A (1965). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.