Abstract
Background
Cell‐free DNA (cfDNA) in the plasma of patients with both malignant and benign breast lesions was analyzed to determine whether the findings may have diagnostic and prognostic implications and to analyze the association between the levels of cfDNA and prognostic parameters.
Methods
Plasma samples were obtained from 99 subjects; 42 with breast cancer (BC), 30 with benign breast lesions, and 27 healthy women as normal controls. Circulatory cfDNA was extracted from the plasma samples and quantified using a real‐time quantitative PCR method. Immunohistochemistry was done on formalin‐fixed paraffin‐embedded sections to evaluate the status of hormonal receptors (estrogen receptor [ER] and progesterone receptor [PR]), and the protein expression of both Her2/neu and Topoisomerase IIα.
Results
The level of cfDNA in the BC group was significantly higher than in the benign lesions and control groups. cfDNA level was associated with malignant tumor size, lymph node involvement, stage, and grade as well as Her2/neu and Topoisomerase IIα expression, while it was not associated with ER or PR status.
Conclusions
The present study suggests that the level of cfDNA can be easily quantified using plasma samples. Thus, level of plasma cfDNA might constitute an important noninvasive diagnostic and prognostic valuable tool in cancer breast patients’ management.
Keywords: breast cancer, cell free plasma DNA, real‐time quantitative PCR
INTRODUCTION
Breast cancer (BC) is the most common cancer in the female population, representing a major health‐care challenge especially in developing countries as Egypt 1, 2 necessitating the development of new approaches that may facilitate better diagnosis and more effective treatment 3, 4.
The development of BC is associated with a number of genetic alterations involving the inactivation of tumor suppressor genes and the activation of oncogenes 5.
Human epithelial receptor type 2 (Her2/neu) (also known as c‐erb‐B2) is frequently amplified in BC and its overexpression is associated with poor clinical outcome. Amplification of Her2/neu can be detected in about 15–30% of invasive BCs and these carcinomas are often characterized by poor histological grade, high numbers of proliferating cells, DNA aneuploidy, and the lack of expression of estrogen (ER) and progesterone receptors (PR) 5.
Another biologically interesting gene is Topoisomerase IIα, which encodes for an important enzyme in DNA replication and in cell cycle progression. It has been suggested that amplification of Topoisomerase IIα and not overexpression of Her2/neu may, in fact, be the predictive marker for response to anthracyclines which are among the most effective chemotherapeutic agents in BC 6.
The presence of abnormally high levels of free circulating cell‐free DNA (cfDNA) in the plasma/serum of cancer patients was demonstrated in 1977 7. The presence of tumor DNA in blood is the result of different mechanisms such as apoptosis, necrosis, and circulating tumor cell lysis which produce DNA leakage to blood stream 8.
Recently, cfDNA in cancer has attracted attention and its possible use as a marker for diagnosis or prognosis has been investigated. Occurrence of mutations in cfDNA, as well as increase in the overall level of cfDNA, is not restricted to any particular tumor site, type, or grade 8.
However, there is tendency for significantly larger amounts of cfDNA in patients with late stage disease and metastasis. Thus, cfDNA may provide a very valuable source of genetic material as a surrogate for molecular analysis in cancer and precancer patients 9, 10.
The purpose of this study was to determine whether the amounts of free circulating DNA could discriminate between patients with benign or malignant breast diseases, and healthy individuals by using real‐time PCR‐based DNA quantification methodology and to analyze the association between the levels of cfDNA and prognostic parameters, including staging, grading, ERs, PRs, Her2/neu, and Topoisomerase IIα expression.
MATERIALS AND METHODS
The study approval was obtained from the ethics committee of the Faculty of Medicine, Alexandria University. All patients provided a written, signed consent to participate in this study.
A total of 99 females were enrolled in this study. Based on the results of the fine needle aspiration biopsy performed for all cases, 42 patients were diagnosed as BC, while 30 patients had benign breast diseases.
Twenty‐seven healthy volunteers were included as a control group. All blood samples were withdrawn before any surgical interventions or therapeutic treatments.
Pathological staging of cancer breast cases was done according to the TNM classification system developed by the American Joint Committee on Cancer (AJCC) 11, while grading was performed according to the Nottingham modification of the Scarff–Bloom–Richardson grading scheme (NSBR) 12.
Specimen Collection and DNA Isolation
About 8 ml of peripheral blood from each patient or control were centrifuged at 1,600 × g for 10 min. Plasma was transferred to new plain tubes and centrifuged again at maximum speed (16,000 × g) for 10 min 13.
Cell‐free plasma DNA was extracted using the DNA Blood MiniKit (Qiagen, Hilden, Germany). DNA was stored at −20°C till further analysis.
Free DNA Quantification in Plasma
Free DNA was quantified in cell free plasma using a real‐time quantitative PCR method on Mx3000P™ Real‐Time PCR System (Stratagene, La Jolla, CA). Primers and probes amplified hTERTgene (Table 1) 13.
Table 1.
Sequences of the Primers and the Probe Used to Amplify hTERT Genes
| Primer/probe | Label |
|---|---|
| GGC ACA CGT GGC TTT TCG | None |
| GGT GAA CCT CGT AAG TTT ATG CAA | None |
| TCA GGA CGT CGA GTG GAC ACG GTG | VIC‐TAMRA |
Each sample was analyzed in duplicate and one negative control (no template control) was included in every run. For calculation of plasma cfDNA level, a standard curve was created using serial dilutions of TaqMan® Control Human Genomic DNA Standard (10 ng/μL) (Applied Biosystems, USA).
Amplification was carried out in a final volume of 25 μL containing 1x TaqMan universal PCR master mix No AmpErase UNG, containing AmpliTaq Gold DNA polymerase, 0.2 μM of each of the forward and reverse primers, and 0.1 μMTaqMan probe. Thermal cycling conditions included an initial hold at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec (denaturation step) and 60°C for 1 min (annealing/extension).
Operative Management
Excision of the lesion was done for all benign breast lesions. For early BC cases (stage I and II), lumpectomy with axillary clearance was done in 31% of patients. The rest of cases in early stages had multifocal lesions in mammogram, thus modified radical mastectomy was performed for them. As regards patients in late stages (stage III and IV), either simple mastectomy (in 23% of cases) or modified radical mastectomy with or without new adjuvant radio/chemotherapy was done (in 77% of cases).
Immunohistochemical Staining
Unstained slides from formalin‐fixed, paraffin‐embedded tumors were used for immunohistochemical procedures using primary monoclonal antibodies, ER, PR, Her2‐neu, and Topoisomerase IIα. Positive controls and negative controls were included in all the runs. The streptavidin–biotin–peroxidase complex method was used. This technique involves the sequential incubation of the specimens with an unconjugated primary antibody specific to the target antigen, a biotylinated secondary antibody that reacts with primary antibody, enzyme‐labeled streptavidin, and DAB substrate chromogen. The detection kit (Ultra Vision Detection System, Anti‐polyvalent, ready to use, HRP/DAB) (Lab Vision Corporation Fremont, CA) was used.
Interpretation of Immunohistochemical Results
Interpretation of ER and PR results was performed using Allred Method 14. ER and PR were defined as positive when 10% or more of tumor cells immunohistochemically stained positive.
For ER and PR hormonal receptors interpretations, the Allred score 14 which is semi‐quantitative system that takes into consideration the proportion of positive cells (scored on a scale of 0–5) and staining intensity (scored on a scale of 0–3) was applied. ER: (Clone SP1); PR: (Clone SP2) (Fremont).
The proportion and intensity were then summed to produce total scores of 0 or 2 through 8. A score of 0–2 was regarded as negative while 3–8 as positive 15.
For HER2 (Clone EP1045Y) testing, the American Society of Clinical Oncologists/College of American Pathologists guideline recommendations have clearly defined the positive (immunohistochemistry score 3), equivocal (immunohistochemistry score 2), and negative (immunohistochemistry score 0/1) categories. Among these categories, the equivocal or weakly positive 2 category creates confusion about trastuzumab treatment; therefore, it requires an additional FISH test 16.
TOP2A (clone Ki‐S1, monoclonal, IgG isotype, 1:100; DAKO, Tokyo, Japan) staining was defined as positive when 20% or more of tumor cells were stained positive 17.
Only nuclear staining was considered for Topoisomerase IIα immuno‐staining, frequency of the tumor cells was scored subjectively on a scale of 1–4 (1, 0–5% positive tumor cells; 2, 6–25%; 3, 26–75%; 4, more than 75%) 18.
Statistical Analysis
Data were analyzed using SPSS software package version 18.0 (SPSS, Chicago, IL). Nonparametric tests were used (Mann–Whitney U‐test and Kruskal–Wallis test). P < 0.05 was considered statistically significant. Cross‐validation using leave‐one‐out approach was used to assess robustness of the diagnostic evaluation of cfDNA plasma level.
RESULTS
The study included 42 patients with histopathologically confirmed BC; all of them were of invasive ductal type. Females with benign breast lesions (n = 30) included 16 cases (53.4%) with fibroadenoma and 14 cases (46.6%) with fibrocystic disease of the breast. Twenty‐seven healthy females were included in the study as a control group. Histopathologic data of all cancer breast patients are presented in Table 2 and Fig. 1
Table 2.
Distribution of the Breast Cancer (BC) Cases According to the Different Histopathological Parameters
| No | Percentage | |
|---|---|---|
| Tumor size | ||
| <2 cm | 7 | 16.7 |
| 2–5 cm | 17 | 40.5 |
| >5 | 18 | 42.9 |
| Lymph node | ||
| Negative | 13 | 69 |
| Positive | 29 | 31 |
| Grade | ||
| Poorly differentiated | 6 | 14.3 |
| Moderately differentiated | 31 | 73.8 |
| Well differentiated | 5 | 11.9 |
| Stage | ||
| I | 7 | 16.7 |
| II | 22 | 52.4 |
| III | 10 | 23.8 |
| IV | 3 | 7.1 |
Data are presented as number and percentage.
Figure 1.
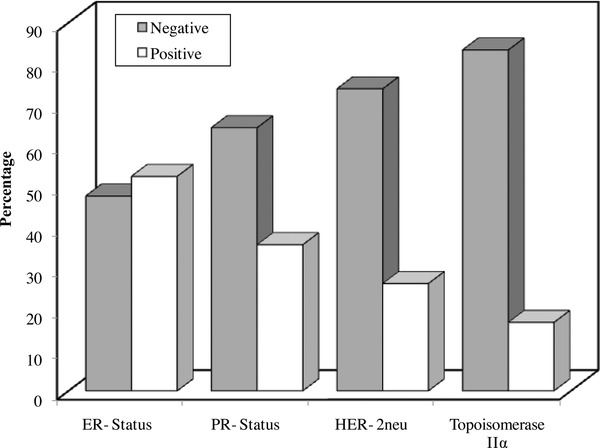
Distribution of the cancer breast cases according to the different biomarkers (ER, PR, Her‐2‐neu, and Topoisomerase IIα).
No statistically significant difference was detected between the three studied groups as regards age (P = 0.2).
A statistically significant difference was observed as regards plasma cfDNA level between the benign breast lesion group and the malignant lesion group (P < 0.001). In addition, a statistically significant difference in cfDNA level was observed between the control group and the malignant breast lesion group (P < 0.001), while no significant difference was found between the control group and the benign lesion group (P = 0.23) (Fig. 2) or between those complaining of fibroadenoma and those with fibrocystic disease of the breast (P = 0.19).
Figure 2.
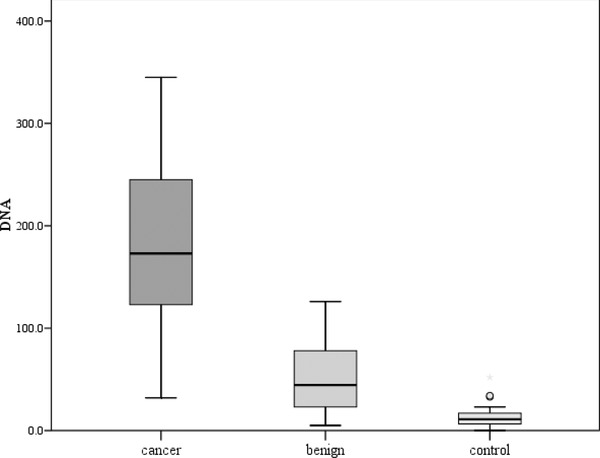
Comparison between the three studied groups as regards cfDNA plasma level.
To assess robustness of the diagnostic evaluation of cfDNA plasma level, cross‐validation using leave‐one‐out approach was used.
Using cfDNA serum level, 89.9% of the studied subjects were correctly classified (Table 3). This rate of correct classification tends to be slightly optimistic as it is based upon the cases originally used to evaluate cfDNA serum level. The cross‐validated section of the table attempts to correct this by classifying each case while leaving it out from the model calculations. Again, using leave‐one‐out cross validation, the rate of correctly classified subject remained the same (89.9%). This suggests that, overall, cfDNA serum level is correct about nine of ten times.
Table 3.
The Classification Pattern of Both the Original and Cross‐Validated Groups When Using Serum Level of cfDNA as a Classifier
| Predicted diagnosis | ||||
|---|---|---|---|---|
| Actual diagnosis | Benign | Malignant | Total | |
| Original | Benign | 27 (100.0) | 0 | 27 |
| Malignant | 7 (16.7) | 35 (83.3) | 42 | |
| Cross‐validateda | Benign | 27 (100.0) | 0 | 27 |
| Malignant | 7 (16.7) | 35 (83.3) | 42 | |
In cross‐validation, each case is classified by the functions derived from all cases other than that case.
Associations between levels of plasma cfDNA and variable clinical parameters, including tumor size, lymph node involvement, histological grading, stage, hormonal receptor status (ER, PR), Her2/neu, and Topoisomerase IIα expression were assessed (Table 4), Fig. 3.
Table 4.
Association Between cfDNA and Variable Histopathological Parameters
| cfDNA | ||
|---|---|---|
| Median (min–max) | P | |
| Tumor size | ||
| <2 cm | 58 (32–97) | |
| 2–5 cm | 160 (115–198) | <0.001* |
| >5 | 255 (210–345) | |
| Lymph node | ||
| − ve | 156 (32–231) | <0.001* |
| + ve | 289 (234–345) | |
| Grade | ||
| Poorly differentiated | 230 (234–345) | |
| Mod differentiated | 168 (32–312) | <0.001* |
| Well differentiated | 90 (43–178) | |
| Stage | ||
| I | (32–97) 58 | |
| II | (115–231) 167 | |
| III | 266 (234–320) | <0.001* |
| IV | (321–345) 324 | |
Data are presented as median (min–max) and were analyzed by Mann–Whitney test.
Statistical significance was set at P‐value < 0.05.
Statistically significant.
Figure 3.
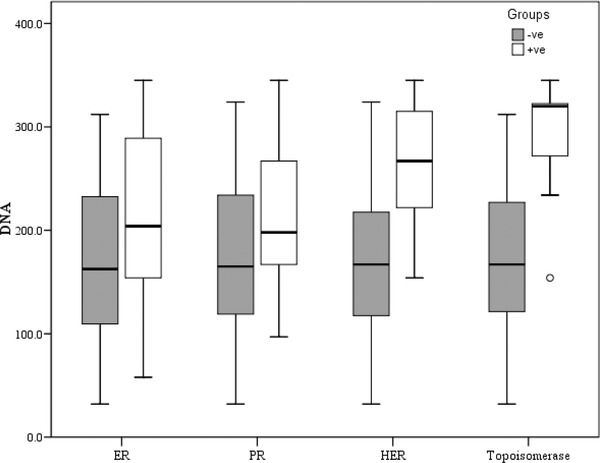
Association between cfDNA and different biomarkers (ER, PR, Her‐2‐neu, and Topoisomerase IIα).
Elevated levels of cfDNA were found to be associated with tumor size (P < 0.001), lymph node involvement (P < 0.001), histopathological grade (P < 0.001), and clinical staging (P < 0.001) with increasing cfDNA in tumors with larger sizes, or in those associated with positive lymph node involvement, also cfDNA plasma level increased significantly with advanced stage and higher grade of the tumor.
The level of cfDNA was not associated with the percentage of ER (P = 0.124) or PR (P = 0.176) but higher levels of cfDNA were associated with increased HER‐2neu (P = 0.002) and Topoisomerase IIα (P = 0.002) expression.
Using receiver operating characteristic (ROC) curve analysis, a cutoff level of 34 was set for cfDNA plasma level to distinguish cases with malignant breast lesion from the healthy control females. At a cutoff value of 34, the sensitivity of quantitative real‐time PCR in the detection of cancer breast was 97.6%, specificity 96.3%, positive predictive value 97.6% and negative predictive value 96.3%, and accuracy of 97.1%.
DISCUSSION
In Egypt, cancer of breast is still considered number one killing cancer in females despite increased general awareness of the disease, thus imposing a continuous search for a reliable, noninvasive diagnostic strategy 1.
In the present study, quantitative real‐time PCR was used for investigation of the role of plasma cfDNA extracted from a blood sample as a potential diagnostic and prognostic tool in cancer breast patients. It is anticipated that blood sampling if proved valuable, is considered an ideal, noninvasive simple technique in managing cancer breast patients.
Most cancer breast patients in the present study were presented in early stages (69.1% in early stages and 30.9% in late stages) reflecting relatively increased awareness among females as regards early cancer breast detection in Egypt.
A statistically significant difference was observed as regards plasma cfDNA level between BC patients and patients with a benign breast tumor, also a statistically significant difference in plasma cfDNA was observed between the BC group and the benign breast lesion group.
Among many methods used in circulating plasma cfDNA assessment, real‐time PCR has the advantage of being quantitative. In the present study, ROC curve analysis revealed a cutoff of 34 to distinguish between those with BC and healthy control females.
Many studies were in accordance with the present results in investigating cfDNA as a tool in BC management. A study by Kohler et al. 19 used multiplex real‐time PCR to investigate the levels of nuclear cfDNA and mitochondrial DNA in plasma samples from patients with malignant and benign breast tumors, and from healthy controls aiming to evaluate cfDNA as a biomarker for distinguishing between the three studied groups, while no significant difference could be found in the level of cfDNA between the benign disease group and the healthy controls, the study showed significantly higher levels of cfDNA in the cancer group in comparison to the benign tumor group and the healthy control group. On using ROC curve analysis a cut‐off of 1,866 GE/ml was set to differentiate between BC cases and the healthy controls with a sensitivity of 81% and a specificity of 69%, thus, emphasizing on the role of cfDNA in the breast tumor management.
Catarino et al. 20 quantified circulating DNA using real‐time PCR revealing increased levels of circulating DNA in BC patients compared to control individuals. In addition, cfDNA levels were higher before than after breast surgery proving that cfDNA may be a good and simple tool for detection of BC with a potential to clinical applicability together with other current methods used for monitoring the disease.
In the present study, cfDNA was, in addition, compared against other traditional staging parameters, such as tumor size, lymph node involvement, extent of metastasis, and predictive markers, such as histological grades, receptor status, Her/2neu, and Topoisomerase IIα expression.
Elevated levels of cfDNA were found to be associated with tumor size, lymph node involvement, histopathological grade, and clinical staging.
In accordance with the present study results, Shao et al. 21 demonstrated that in BC patients, the level of plasma DNA also correlates with stage, lymph node metastasis, and tumor size.
Another study by Zhong et al. 22 analyzed cfDNA in the plasma of patients with both malignant and benign breast lesions by real‐time quantitative PCR to assess the role of cfDNA as a diagnostic and prognostic marker in these patients and found that cfDNA plasma level was associated with increased malignant tumor size, lymph node involvement, and distant metastasis, and concluded that cfDNA could have diagnostic as well as prognostic implications.
No association was detected between the levels of circulating cfDNA and the scoring of ER or PR, while it was associated with Her/2neu and Topoisomerase IIα expression.
Her2/neu have been linked with poor prognosis in BC either in the form of shorter disease‐free intervals, increased risk of metastasis, or resistance to many types of therapy. This was attributed to potentiation of tumor cell motility, protease secretion and invasion, and also modulation of cell‐cycle checkpoint function, DNA repair, and apoptotic responses. In addition, Her2/neu is expressed at low levels in normal adult tissues, thus, it is an ideal target for therapy 23.
Topoisomerase IIα expression has a key role in cell division by controlling and modifying the topological status of DNA 24. Furthermore, Topoisomerase IIα is the direct molecular target of TopoII inhibitors, which are among the most powerful cytostatic agents in the treatment of invasive BC, so immunohistochemical detection of Topoisomerase IIα is considered a predictive marker for treatment response.
In the present work, cfDNA was associated with Her2/neu and Topoisomerase IIα expression, thus cfDNA can be considered as a prognostic marker as well as a predictor of treatment response.
CONCLUSIONS
The level of cfDNA using quantitative real‐time PCR in patients with BC can be easily quantified using plasma samples and compared to normal individuals and patients with benign breast lesions.
The present study suggests that the level of cfDNA in the plasma is elevated in malignant BC and correlates with tumor size, lymph node status, stage, and grade as well as the expression of her2/neu and Topoisomerase IIα. Thus, level of plasma cfDNA might constitute an important noninvasive diagnostic and prognostic valuable tool in BC patients’ management and could provide a good platform for future studies.
REFERENCES
- 1. Seedhom A, Kamal N. Factors affecting survival of women diagnosed with breast cancer in El‐Minia governorate. Egypt Int J Prev Med 2011;2:131–138. [PMC free article] [PubMed] [Google Scholar]
- 2. Agarwal G, Ramakant P, Forgach E, et al. Breast cancer care in developing countries. World J Surg 2009;33:2069–2076. [DOI] [PubMed] [Google Scholar]
- 3. Pantel K, Muller V, Auer M, et al. Detection and clinical implications of early systemic tumour cell dissemination in breast cancer. Clin Cancer Res 2003;9:6326–6334. [PubMed] [Google Scholar]
- 4. Diel I, Solomayer E, Bastert G. Bisphosphonates and the prevention of metastasis: First evidences from preclinical and clinical studies. Cancer 2000;88:3080–3088. [DOI] [PubMed] [Google Scholar]
- 5. Kuzhan O, Ozet A, Ulutin C, et al. Impact of c‐erb2 status on survival after high dose chemotherapy in high‐risk breastcancer patients. Saudi Med J 2007;28:1374–1379. [PubMed] [Google Scholar]
- 6. Hannemann J, Kristel P, van Tinteren H, et al. Molecular subtypes of breast cancer and amplification of Topoisomerase IIa: Predictive role in dose intensive adjuvant chemotherapy. Br J Cancer 2006;95:1334–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leon S, Shapiro B, Sklaroff D, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646–650. [PubMed] [Google Scholar]
- 8. Gautschi O, Bigosch C, Huegli B, et al. Circulating deoxyribonucleic acid as prognostic marker in non‐small‐cell lung cancer patients undergoing chemotherapy. J Clin Oncol 2004;22:4157–4164. [DOI] [PubMed] [Google Scholar]
- 9. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–1665. [PubMed] [Google Scholar]
- 10. Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. Mut Res 2007;635:105–117. [DOI] [PubMed] [Google Scholar]
- 11. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. American Joint Committee on Cancer Staging Manual, seventh edition, New York: Springer; 2010. [Google Scholar]
- 12. Tavassoli F, Devilee P. World Health Organization Classification of Tumours: Tumors of the Breast and Female Genital Organs. Lyon: IARC Press, International Agency for Research on Cancer; 2003. p 18–19. [Google Scholar]
- 13. Catarino R, Ferreira M, Rodrigues H, et al. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol 2008;27:415–420. [DOI] [PubMed] [Google Scholar]
- 14. Onitilo A, Engel J, Greenlee R, Mukesh B. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin Med Res 2009;7:4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collins LC, Botero ML, Schnitt SJ. Bimodal frequency distribution of EstrogenReceptor immunohistochemical staining results in breast cancer. Am J ClinPathol 2005;123:16–20. [DOI] [PubMed] [Google Scholar]
- 16. Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology; College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists Guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Path Lab Med 2007;131:18–43. [DOI] [PubMed] [Google Scholar]
- 17. Yasojima H, Shimmura A, Naoi Y, et al. Association between c‐myc amplification and pathological complete response to neoadjuvant chemotherapy in breast cancer. Association between c‐myc amplification and pathological complete response to neoadjuvant chemotherapy in breast cancer. Eur J Cancer 2011;47:1779–1788. [DOI] [PubMed] [Google Scholar]
- 18. Sandri M, Hochhauser D, Ayton P, et al. Differential expression of the Topoisomerase II alpha and beta genes in human breast cancers. Br J Cancer 1996;73:1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohler C, Radpour R, Barekati Z, et al. Levels of plasma circulating cell free nuclear and mitochondrial DNA as potential biomarkers for breast tumors. Mol Cancer 2009;8:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Catarino R, Ferreira M, Rodrigues H, et al. Quantification of free circulating tumor DNA as a diagnostic marker for breast cancer. DNA Cell Biol 2008;27:415–421. [DOI] [PubMed] [Google Scholar]
- 21. Shao ZM, Wu J, Shen ZZ, Nguyen M. p53 mutationin plasma DNA and its prognostic value in breast cancer patients. Clin Cancer Res 2001;7: 2222–2227. [PubMed] [Google Scholar]
- 22. Zhong X, Ladewig A, Schmid S, et al. Elevated level of cell‐free plasma DNA is associated with breast cancer. Arch Gynecol Obstet 2007;276:327–331. [DOI] [PubMed] [Google Scholar]
- 23. Eccles S. The role of c‐erbB‐2/HER2/NEU/neu in breast cancer progression and metastasis. J Mammary Gland Biol Neoplasia 2002;6:393–406. [DOI] [PubMed] [Google Scholar]
- 24. Berger JM, Gamblin SJ, Harrison SC, et al. Structure and mechanism of DNA topoisomerase IIα. Nature 1996;379:225–232. [DOI] [PubMed] [Google Scholar]


