Abstract
In cells at steady state, two forms of cell compartmentalization coexist: membrane‐bound organelles and phase‐separated membraneless organelles that are present in both the nucleus and the cytoplasm. Strikingly, cellular stress is a strong inducer of the reversible membraneless compartments referred to as stress assemblies. Stress assemblies play key roles in survival during cell stress and in thriving of cells upon stress relief. The two best studied stress assemblies are the RNA‐based processing‐bodies (P‐bodies) and stress granules that form in response to oxidative, endoplasmic reticulum (ER), osmotic and nutrient stress as well as many others. Interestingly, P‐bodies and stress granules are heterogeneous with respect to both the pathways that lead to their formation and their protein and RNA content. Furthermore, in yeast and Drosophila, nutrient stress also leads to the formation of many other types of prosurvival cytoplasmic stress assemblies, such as metabolic enzymes foci, proteasome storage granules, EIF2B bodies, U‐bodies and Sec bodies, some of which are not RNA‐based. Nutrient stress leads to a drop in cytoplasmic pH, which combined with posttranslational modifications of granule contents, induces phase separation.
Keywords: membraneless organelles, metabolic enzyme foci, nutrient stress, P‐bodies, pH drop, prosurvival, Sec bodies, stress assemblies, stress granules
1. INTRODUCTION: MEMBRANE‐BOUND AND MEMBRANELESS ORGANELLES COEXIST IN INTERPHASE CELLS
Cells are highly compartmentalized to limit biochemical reactions in space. A large component of cell compartmentalization is provided by membrane‐bound organelles (Figure 1), that is, organelles which are surrounded by a sealed lipid bilayer. The membrane defines the boundary of the organelle, separates the lumen from the surrounding cytoplasm, and limits the biochemical/enzymatic reactions that are catalyzed by and within the organelle. Compartmentalization also allows for interactions with a specific pool of cytoplasmic proteins that are peripherally associated with the membrane. Together, these features define key aspects of organelle functional identity. The membrane defines the type of communication between membrane‐bound organelles as well with the other parts of the cell, mediated by small lipidic vesicle and tubule carriers as well as by membrane contact sites.1 Membrane‐bound organelles, their biogenesis, their maintenance, how they function and communicate—collectively referred to as membrane traffic—have been extensively studied in the last four decades and has yielded a Nobel Prize in Physiology and Medicine in 2013.2
Figure 1.
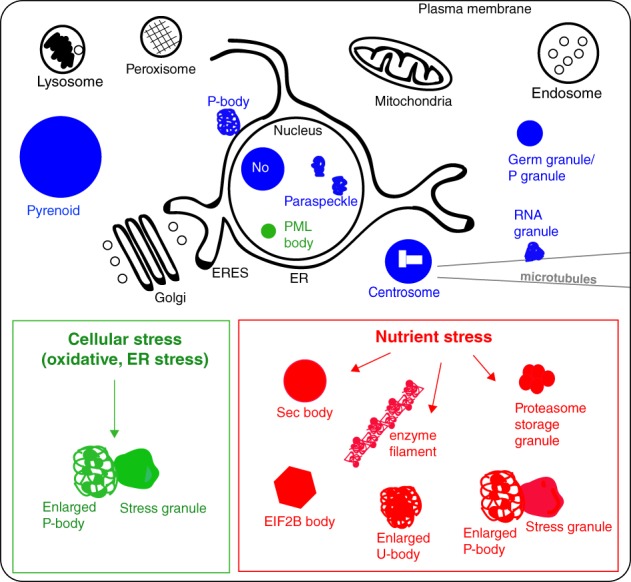
Schematic of cell compartmentalization. Membrane‐bound organelles are represented in black, the stable/basal membraneless organelles are in blue. The green box presents the formation of P‐bodies and stress granules upon many stresses, whereas the red box presents the stress assemblies formed upon nutrient stress
The second type of stable cellular compartments are membraneless organelles. Although first described nearly 200 years ago with the observation of the nucleolus,3, 4 membraneless organelles have recently (re‐)gained the attention of bio‐physicists for their unique mechanism of formation by phase separation and for their material properties.5 Like membrane‐bound organelles, membraneless organelles appear to support specific biochemistries with critical functions in cellular homeostasis and development. The differences and similarities in both types of cell compartmentalization have been described in 6
Membraneless organelles are present in both the nucleoplasm and the cytoplasm of most eukaryotes. Membraneless organelles in the nucleus include the nucleolus, Cajal bodies, nuclear stress bodies, nuclear speckles, interchromatin granule clusters, paraspeckles, Sam68 nuclear bodies, PML oncogenic domains, transcription histone locus bodies and Oct1/PTF/transcription domains (reviewed in References 5, 7. Membraneless organelles in the cytoplasm include the centrosome,5 processing‐bodies (P‐bodies) that are involved in mRNA decay, translational repression, microRNA‐induced RNA silencing and RNA storage (see below),8 posterior germ granules in Drosophila,9, 10 P‐granules in Caenorhabditis elegans 11, 12, 13 and neuronal granules transporting‐specific mRNAs,14, 15, 16 as well as the non‐RNA‐based Pyrenoid in photosynthetic organisms17, 18, 19, 20 (Figure 1).
Interestingly, although seemingly stable, most constitutive membraneless organelles are regulated according to the phase of the cell cycle.21 Indeed, it appears that the size and abundance of most membraneless organelles are reduced as the cell enters mitosis. Many components appear to become diffuse, as if to ensure optimal partitioning. This phenomenon is reminiscent of membrane‐bound organelles that fragment at the onset of mitosis,22 as extensively studied for the Golgi.23
While membraneless compartments have important functions in cell physiology, they can become pathological when formed upon the expression of mutated proteins. For instance, expression of amyloids, proteins with long poly‐glutamine tracts (PolyQ proteins; see review24), or mutated RNA‐binding proteins such as FUS25 or HnRNPA126, 27 can lead to the formation of irreversible membraneless compartments. Furthermore, the expression of the mutant form of C9Orf72 can modify the dynamics of membraneless compartments and make them pathological.28
Many reversible membraneless compartments are also strongly induced by cellular stress; we will refer to these as stress assemblies. After consideration of the general principles driving the formation of membraneless organelles in the first part of this review, we describe the formation of stress granules and enlarged P‐bodies upon different types of stress and describe their high level of heterogeneity (Figure 2). In the last part of this review, we focus on cytoplasmic stress assemblies that are induced by nutrient stress (Figure 3), a phenomenon that has mostly been described in yeast and Drosophila.
Figure 2.
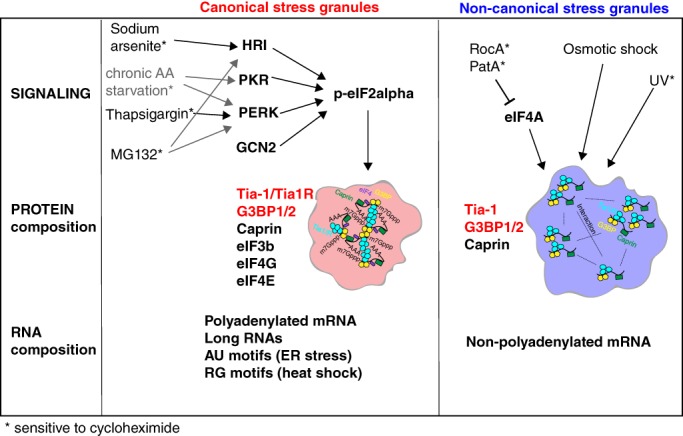
Heterogeneity in stress granule pathway formation and composition. Different cellular stress leads to the phosphorylation of eIF2a (p‐eIF2a) that in turn leads to the formation of canonical stress granules (in red) containing a number of RNA‐binding proteins and polyadenylated mRNA and RNAs with indicated features.96, 112, 113, 114, 115 Other stress mediate noncanonical stress granules (in blue) through the inhibition of eIF4A or modulation of unknown pathways. These stress granules appear to contain only a subset of RNA‐binding proteins and unpolyadenylated mRNAs
Figure 3.
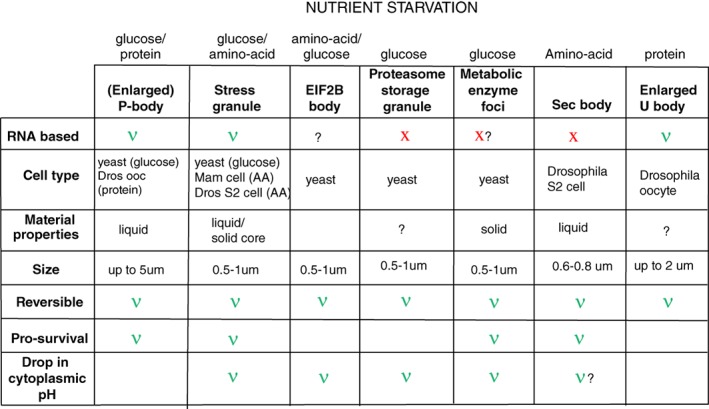
An overview of the features of membraneless stress assemblies triggered by nutrient starvation
2. AN OVERVIEW OF MECHANISMS DRIVING FORMATION OF MEMBRANELESS ORGANELLES
2.1. Membraneless compartments are formed by phase separation
The general consensus in the field is that membraneless organelles are formed by phase separation of their components from the surrounding nucleo‐ or cytoplasm.29, 30, 31, 32 Phase separation defines the behavior of a seemingly homogeneous solution of diffuse macromolecules that segregate into two distinct phases that then stably coexist.5, 12, 33 Phase separation can be either liquid‐liquid (leading to the formation of stable liquid droplets within another liquid) or liquid‐solid leading to gel‐like older stress granules,25 as occurs in vitro, as well as solid and crystalline structures.34, 35 The differences in the material properties of the separated phases can be distinguished by fluorescence recovery after photobleaching (FRAP). In the case of liquid droplets, bleaching half of the structure would result in a quick recovery through the efficient movement of the nonbleached molecules to the bleached area within the droplet. When the structure is solid and crystalline, the recovery does not occur as the molecules within the structure are immobile.
2.2. Scaffold/drivers vs clients: Multivalency and low complexity sequences
Phase separation is driven by “driver/scaffold” proteins36, 37 that coalesce and attract “client” proteins or other macromolecules with which they normally interact. Drivers/scaffolds are proteins that are essential and sufficient to drive the formation of membraneless organelles.36, 38 When they are absent, these compartments are not formed or are not stable.
Drivers are proteins that engage in low affinity multivalent interactions (mutivalency).36 At the molecular level, drivers contain domains of low complexity that are often intrinsically disordered. These domains tend to have low amino acid diversity with repeating sequences that sometimes form prion‐like domains. Prions such as the N‐terminus of yeast Sup35 were first described to be enriched in glutamine and asparagine39, 40, 41 and to promote amyloid states. The development of the PrionW software (https://omictools.com/prionw-tool)42 allowed the discovery of many prion forming domains in human proteins, including several heterogeneous nuclear ribonucleoproteins (hnRNPs) that are dysregulated in neurodegenerative diseases, such as familial amyotrophic lateral sclerosis (fALS).43 Interestingly, a large number of proteins, which are either required for P‐body stability and formation, or RNA‐binding proteins known to coalesce, contains low complexity domains rich in glutamine and asparagine.44
Low complexity domains can also contain other amino‐acids, called “stickers,” such as arginine, histidine and tyrosine, that mediate Pi‐Pi interaction and cation‐Pi interactions.32, 45 Stickers have been well investigated in the RNA‐binding protein FUS, in which the low complexity PLD at its N‐terminus is necessary (although not sufficient) for spontaneous phase separation in vitro.25, 46, 47, 48 Phase separation is mediated by tyrosine residues within this prion‐like domain (QGSY) that interact strongly with the arginine residues of the RNA‐binding domain (RRM) via cation‐Pi interactions.48 Note that other models have been proposed to explain FUS coalescence, including the role of LARKS (low complexity aromatic‐rich kinked segments) that mediate interactions among pairs of closely aligned beta sheets with a kink,47 in accordance with the structures elucidated by Murray et al.46
Furthermore, stickers need to be separated from one another by spacers (typically composed of serine/threonine, glycine), which provide flexibility in the structure and allow for reversibility. However, spacers are not major determinants of the driving forces for phase separation.49
It is important to note that some of the clients of membraneless organelles also display properties similar to these of drivers (such as bearing low complexity repeats and multivalency) allowing them to engage in the low affinity interactions necessary for phase separation. This illustrates the challenge in understanding the formation of membraneless organelles (recently reviewed in Mittag and Parker50).
2.3. mRNAs can also drive phase separation
The presence of RNAs increases phase separation in vitro and in vivo for membraneless organelles that form from RNA‐binding proteins.25, 26, 27 This has been very well established using purified proteins such as FUS25 and HnRNPA1.26 However, until recently, it was not clear whether RNAs are drivers of phase separation (for instance, in scaffolding proteins) or simply increase interactions between the several components of membraneless organelles. This has been clarified through the finding that the long noncoding RNA NEAT1 (nuclear para‐speckle assembly transcript 1) drives the formation of nuclear paraspeckles51 by acting as a template for several proteins including FUS.52 A similar scaffolding role for rRNAs has been shown for the nucleolus.53
It has recently been shown that mRNA drives the formation of a phase separated compartment in Eremothecium gossypii. In this filamentous mold, the polyQ‐protein Whi3 induces conformational changes in specific RNA structures leading to oligomerization through RNA‐RNA interactions and phase separation into distinct droplets. Thus, the secondary structure/shape of mRNA can promote the formation and coexistence of a diverse array of RNA‐rich liquid compartments found in a single cell.54 RNA‐RNA interactions also contribute to the formation of other membraneless organelles, at least in vitro.55 Purified protein‐free total RNA from yeast was able to self‐assemble under conditions mimicking intracellular stress conditions. Interestingly, most of the RNAs found in these RNA‐RNA assemblies were long RNAs, similar to those found in stress granules (see below).55 This indicates that long mRNAs might be able to act as drivers for stress assemblies, especially when they contain repeat sequences capable of self‐base‐pairing.
3. CELLULAR STRESS STALLS TRANSLATION AND INDUCES THE FORMATION OF STRESS GRANULES AND P‐BODIES
As mentioned in Section 1, cellular stress leads to the formation of reversible membraneless compartments in the cytoplasm that we refer to as stress assemblies. Their formation appears to be part of a strategy for survival during stress (see below), and it is related to either inhibition of a given anabolic pathway and/or storage of key molecules. Steady state membraneless stress assemblies also form in the nucleus.56 For instance, stress appears to result in the abnormal segregation of some of the nucleolus components57 and/or inducible formation of anti‐apoptotic paraspeckles,58 and promyelocytic leukemia (PML) protein nuclear bodies form in response to virus infection59, 60 or oxidative stress.61 However, nuclear stress assemblies will not be discussed further as they have been recently reviewed.58, 62 Instead, we will focus this part of the review on the best studied stress assemblies, the P‐bodies and the stress granules.
3.1. Stress granules
In eukaryotic cells, many cellular stresses (the most canonical being oxidative stress through sodium arsenite treatment, and endoplasmic reticulum (ER) stress through heat shock or thapsigargin treatment) induce the inhibition of mRNA translation initiation and polysome disassembly. This leads to an accumulation of untranslated, 80S ribosome‐free mRNAs in the cytoplasm that can bind RNA‐binding proteins and coalesce into submicrometer large membraneless foci, the stress granules.63 Stress granules contain polyadenylated mRNAs, eukaryotic translation initiation factors eIF2A, eIF3, eIF4A/B, eIF4E and eIF4G, 40S ribosomes and the RNA‐binding proteins PAB1, Caprin, FMR1, TDP‐43, Tia1 and G3BP1/2.64, 65 Tia‐1 (and TiaR)66, 67 and G3BP1/265, 68 are the two best characterized drivers for stress granule formation in vivo. Overexpression of either leads to ectopic stress granule formation even in the absence of stress,68 and their depletion prevents stress granule formation in both mammalian and Drosophila cells.65, 68, 69, 70 A number of additional criteria establish that foci enriched in RNAs bound to RNA‐binding proteins are bona fide stress granules. Their formation is inhibited by cycloheximide, which locks the mRNA on the ribosomes and prevents its binding to RNA‐binding proteins. Conversely, their formation is stimulated by puromycin, which strips ribosomes from mRNAs. Furthermore, as mentioned above, they are membraneless, reversible upon stress relief and cytoprotective (see Section 6).
Stress granules have been shown to have liquid droplet properties,25, 26, 71 but appear, at least in yeast, to also contain a solid core.72, 73 This has been shown using the small organic alcohol 1,6‐hexanediol that differentiates between liquid‐like or solid‐like membraneless compartments.72 In the presence of this compound, liquids disperse but solids do not. In yeast, stress granules appear to be solid‐like amorphous aggregates that contain misfolded proteins and act as substrates for disaggregases and chaperones (eg, Hsp104).72 The presence of this solid core has been exploited to enrich the stress granules by centrifugation to determine their RNA and protein composition.73 In mammalian cells, the presence of a core is the subject of debate. 1,6‐hexanediol treatment of arsenite‐stressed‐HeLa cells triggered stress granule dispersion, suggesting that they are liquid.72 On the other hand, other studies have developed methods to isolate cores from mammalian stress granules.73, 74 The consensus, if any, is that stress granules contain a solid core surrounded by a liquid shell that allows exchange with the cytoplasm in both yeast, mammalian and Drosophila cells.75, 76
Stress granules have been proposed to act as triage centers for mRNAs77 that protect capped and polyadenylated mRNAs from degradation in P‐bodies (see below and Reference 71 and store them in such a way that they can be immediately translated upon stress relief.71, 78, 79, 80
3.2. P‐bodies
P‐bodies are dynamic cytoplasmic macromolecular assemblies composed of translationally inactive mRNAs and proteins involved in translation repression and mRNA turnover, such as 3′‐deadenylation, 5′‐decapping, 5′‐3′ exonuclease activity, nonsense‐mediated decay and miRNA‐targeted gene silencing.81 Mammalian P‐bodies are usually marked by the proteins AGO1/3, DCP2, XRN4, EDC3, EIF4E‐T, LSM1‐7, SMG7, HNRNPM and CPEB1,82, 83 whereas those in yeast are marked by Dcp1p, Dcp2p, Edc3p, Dhh1p, Pat1p, Lsm1p, Xrn1p, Ccr4p and Pop2p.84 At least in yeast, the deletion of any one of these genes does not impair P‐body integrity, suggesting that they are redundant and cooperative.84 In mammalian and Drosophila cells, P‐bodies are visible as microscopic entities even in the absence of stress, but stress triggers their enlargement.85 In yeast, they are only visible upon induction of stress86, 87 and they have liquid droplet properties as they are dissolved by 1,6‐hexanediol.
Given their concentration in RNA decay factors, P‐bodies have been proposed to be the sites of mRNA degradation and turnover. However, recent evidence shows that mRNA degradation might not occur in P‐bodies (at least not exclusively), and that P‐bodies are storage sites for repressed mRNAs that can be released and translated at the appropriate moment.82, 88 This was first shown by using a reporter called TREAT to visualize mRNA degradation in living HeLa cells. TREAT mRNAs were not degraded when present in P‐bodies, whether in nonstressed or in stressed cells.88 Second, purification of P‐bodies from human epithelial cells using a flow cytometric method for particle analysis (fluorescence‐activated particle sorting, FAPS) reveals that the thousands of mRNAs present in these structures are translationally repressed but not decayed.82 Therefore P‐bodies do not appear to be the sites of active mRNA turnover during growth and stress as was initially thought. This is in line with what was suggested in yeast where normal mRNAs could be targeted to P‐bodies but not degraded.81 The contradictory presence of intact mRNAs and RNA decay factors in P‐bodies is puzzling but may reflect protection of the mRNAs by specific RNA‐binding proteins and translational repressors that inhibit degradation.
3.3. Relationship between P‐bodies and stress granules
As mentioned above, P‐bodies and stress granules are functionally linked. They share approximately 10% to 25% of their protein components, including many RNA‐binding proteins (such as AGo1/2, Edc3/4, eIF4E, LSM1/3, PATL1 and XRN1) (reviewed in Reference 83, making them sometimes difficult to distinguish. Furthermore, electron microscopy of arsenite‐treated HeLa cells revealed that P‐bodies and stress granules closely appose each other,85 and in yeast they appear largely overlapping when observed by fluorescence microscopy.86 This close proximity appears to be instrumental for the exchange and triage of mRNAs.77, 89, 90 However, recent real‐time single‐molecule imaging revealed that mRNA movement between stress granules and P‐bodies is very marginal,91 suggesting that their proximity might serve a different yet unknown function. While both structures clearly store mRNAs, it is not known how different RNAs are targeted to each structure.
While P‐bodies and stress granules share some similarities, they are also different. As mentioned above, their material properties appear dissimilar, and the formation of P‐bodies and stress granules is triggered by different signaling pathways. In yeast, the formation of P‐bodies is regulated by the mitogen‐activated protein kinase (MAPK) upon arsenite92 or osmotic stress,93 and protein kinase A (PKA) is also involved.94, 95 Conversely, stress granules usually (but not always) form upon translation inhibition following phosphorylation of eIF2alpha (eIF2a) by either of four specific kinases (see Section 4.2).96 Furthermore, at least in yeast, PKA also appears to play a role in stress granule formation.97
Taken together, even though these two stress assemblies have been extensively studied for a long time, a lot remains to be discovered, especially regarding their function, the RNA sorting between them, and the manner by which some mRNAs escape degradation in P‐bodies. Furthermore, despite the unifying definition proposed above, stress granules (Figure 2) and P‐bodies are in fact diverse and heterogeneous.
4. STRESS GRANULES AND P‐BODIES ARE HETEROGENEOUS
4.1. Subcomparmentalization of stress granules and P‐bodies
Just like membrane‐bound organelles, membraneless organelles can be subcompartmentalized. The nucleolus3 and the paraspeckles52 have been shown for many years to contain several discrete parts. Posterior Drosophila embryonic germ granules comprise mRNAs that occupy distinct territories within the granules, whereas proteins appear to be more homogeneous (see reviews9, 10). P‐granules in C elegans also appear to display a MEG‐3‐containing shell surrounding a PGL‐3‐containing core both in vivo and in vitro.11
Evidence for subcompartmentalization of P‐bodies was first obtained by the Davis group in mid‐oogenesis Drosophila oocytes. The oocyte P‐bodies that are normally present at the dorsal anterior corner of the oocytes contain both gurken and bicoid mRNAs, and are required for the targeted localization of gurken in this very large cell. P‐bodies are meant to be translationally silent and indeed, they lack ribosomes and contain a number of translational repressors.98 Interestingly, the repressors were concentrated in the core of the P‐bodies, where bicoid mRNA was also present, consistent with the fact that bicoid mRNA is not translated until much later in oocyte development. However, gurken is translated during mid‐oogenesis into the protein Gurken, a ligand of the EGF receptor present in the adjacent follicle cells. In this regard, gurken mRNA is found enriched at the edge of the P‐bodies where the grk translational activator Orb (the Drosophila homolog of cytoplasmic polyadenylation element binding protein [CPEB]), is also enriched. Orb forms a complex with the poly(A) polymerase Wispy and is required for the hyper‐adenylation of grk transcript and for its translation.99 This led to the notion that P‐bodies are subcompartmentalized with a translationally silent core enriched in bicoid and a translationally active edge enriched in gurken.98 This subcompartmentalization is instrumental to oogenesis and strengthens the notion that P‐bodies store mRNAs instead of degrading them (see Section 3.3). Recently, single‐molecule live‐cell imaging analysis revealed similar RNA subcompartmentalization in mammalian P‐bodies.100, 101 lncRNAs, miRNAs and mRNAs are dynamically localized to P‐bodies in either the core or the periphery depending on whether they are used (periphery) or unused (core).
As mentioned above, stress granules also appear to be compartmentalized in yeast and mammalian cells with a solid‐core formed from prion‐like domains, surrounded by a more liquid edge.72, 73, 74 This has been revisited by the Drummond group who showed that heat shock‐induced stress granules in vitro are also heterogeneous.102 Indeed, in vitro the RNA‐binding protein Pab1 rapidly and efficiently phase separates into hydrogels upon higher temperatures. However, this is not driven by the Pab1 low complexity region and is inhibited by the presence of mRNAs. In fact, Pab1 compaction is proposed to exclude mRNAs encoding stress factors that would sense the stress of the increasing temperature and would promote stress granule formation. Stress granules would therefore be formed of a core of compacted Pab1 free of mRNAs surrounded by a shell of coalesced RNAs and other RNA‐binding proteins.
Altogether, it appears that many membraneless organelles, including P‐bodies and stress granules, contain different domains that potentially sustain specific functions. These functions remain to be elucidated in detail. In several cases, the membraneless organelles appear to present a core surrounded by an outer shell or edge. This suggests that they form in a step wise yet coordinated manner, deepening further the complexity of their formation as discussed above.
4.2. Stress‐specific differences in pathways leading to the assembly of stress granules
Stress granules appear to be heterogeneous not only in terms of subcompartmentalization, but also by the signaling pathways that drive their assembly and that vary depending on the inducing stress.
As described above, stress granule formation is triggered by many different types of stress that all lead to the accumulation of untranslated mRNAs via the inhibition of mRNA translation initiation by two mechanisms. The first mechanism is the inhibition of the RNA helicase eIF4a, which is necessary for the unwinding of RNA secondary structures in the 5′UTR of mRNAs to allow for efficient binding of the small ribosomal subunit.103 The second mechanism is the activation of kinases that phosphorylate eIF2alpha (eIF2a) on serine 51 (S51),104 thereby preventing the binding of tRNAiMet to the ribosome.105 Four eIF2a kinases are present in mammals: HRI (heme‐regulated initiation factor 2a kinase, eIF2a K1), PKR (protein kinase RNA‐activated, eIF2a K2), PERK (PKR‐like ER kinase, eIF2a K3) and GCN2 (general control nonderepressible 2, eIF2a K4).106 Although activation of any one of these kinases by a given stress is often coupled to stress granule formation, this is not always the case (Table 1).
Table 1.
elF2a phosphorylation, translation arrest and stress granule formation in mammalian cells upon different stresses (after [96])
| Conditions | Sodium arsenite | Heat shock | Thapsi gargin | MG132 | RocA | PatA | Osmotic Shock | UV | Chronic starvation | |
|---|---|---|---|---|---|---|---|---|---|---|
| HAP1 cells | Translation inhibition | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| SG formation | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| eIF2a Ser51>A HAP1 cells | Translation inhibition | No | No | No | No | Yes | Yes | Yes | Yes | No |
| SG formation | No | No | No | No | Yes | Yes | Yes | Yes | No | |
| ΔHRI | Translation inhibition | No | Yes | Yes | Yes | — | — | Yes | Yes | |
| SG formation | No | Yes | Yes | Reduced | — | — | Yes | — | Yes | |
| ΔPKR | Translation inhibition | Yes | Yes | Yes | Yes | — | — | — | Yes | Reduced |
| SG formation | Yes | Yes | Yes | Yes | — | — | Yes | — | Few | |
| ΔPERK | Translation inhibition | Yes | Yes | No | Yes | — | — | — | Yes | Reduced |
| SG formation | Yes | Yes | No | Reduced | — | — | Yes | — | Reduced | |
| ΔGCN2 | Translation inhibition | Yes | Yes | Yes | Yes | — | — | — | No | Yes |
| SG formation | Yes | Yes | Yes | Yes | — | — | Yes | — | Yes |
The experimental stresses most often used to induce stress granule formation are oxidative stress induction by sodium arsenite,107 ER stress through heat shock (44°C for mammalian cells108) or through Thapsigargin treatment73 (to deplete ER calcium stores), and proteotoxic stress through proteasome inhibition by MG132.109 The mechanism by which these stressors activate stress granule formation has been reinvestigated using a haploid mammalian cell line (HAP1) harboring either a wild type form of eIF2a or eIF2a carrying the nonphosphorylatable S51>A mutation.96 As expected all the four stresses mentioned above lead to eIF2a phosphorylation and stress granule formation in HAP1 cells expressing wild‐type eIF2a, whereas the cells harboring the Ser51>A mutant fail to form stress granules (Table 1). Of note, Drosophila and yeast cells can form stress granules upon heat shock independently of eIF2a phosphorylation, while in mammalian cells stress granule formation is eIF2a‐p dependent.110, 111 Interestingly, other stresses (such as hyperosmotic stress with NaCl, UV and eIF4A inhibition by RocA and PatA) also lead to stress granule formation, but this occurs through pathways that are independent of eIF2a phosphorylation.96 Accordingly, stress granules form in response to these stresses in both the wild type‐ and eIF2a Ser51>A mutant‐expressing HAP1 cells (Table 1).96
Differences were further unveiled with regard to the requirement of specific eIF2a kinases. HAP1 cells lacking either of the four eIF2a kinases were treated by the eight types of stresses mentioned above, and both translation arrest and stress granule formation were monitored (Table 1). Interestingly, it appears that stress granule formation in response to each type of stress requires a specific eIF2a kinase. For instance, stress granule induction by sodium arsenite needs HRI‐mediated eIF2a phosphorylation,107 induction by Thapsigargin needs PERK, and induction by heat shock, MG132 or chronic starvation seem to require two or more kinases.96, 112 Surprisingly, stress granule formation by UV stress requires the kinase GCN2 to inhibit translation, but is not associated with eIF2a phosphorylation on Ser51. This indicates that GCN2 may either phosphorylate eIF2a on another serine or phosphorylate another protein. Another possibility is that GCN2 inactivates the phosphatase that dephosphorylates p‐eIF2a. Taken together, this indicates that the formation of stress granules is not triggered via one uniform pathway, but that each type of stress can activate different kinases and pathways.
4.3. Stress‐specific differences in protein composition of stress granules
The composition of stress granule proteins appears to be stress‐specific, and the molecular organization of stress granules is different between organisms and cell types.113
As mentioned above, proteins such as TIA‐1 and G3BP1/2 are essential for the formation of stress granules upon arsenite treatment. However, a subset of proteins is only present in stress granules formed upon a specific stress. For instance, the transcription initiation factors eIF3b and eIF4G are nearly absent from stress granules formed upon UV exposure, proteasome inhibition (MG132) and eIF4A inhibition (RocA), whereas they are present in stress granules formed in response to all other stresses.96
Stress granule protein content was further investigated in a high‐throughput immunofluorescence microscopy screen in HeLa cells using antibodies against 313 RNA‐binding proteins that were identified as potential stress granule content by proximity biotinylation using G3BP1 as prey.113 This comprehensive study confirmed the presence of stress‐specific proteins in stress granules formed upon either sodium arsenite or heat‐induced stress.113 Of the 313 RNA‐binding proteins tested, only 17% (52/313) actually localized to stress granules. Interestingly, 77% of these (such as UBAP2L) localized to stress granules formed in response to both stresses, and 23% (12/52) were stress‐specific. For instance, NOLC1 was specific to arsenite treatment whereas SF1 was specific to heat shock.113
Heterogeneity in stress granule protein composition has also been found in Drosophila S2 cells. Stress granules formed upon sodium arsenite treatment of Drosophila S2 cells requires the nonphosphorylated (S42) form of Rasputin (Drosophila G3BP), whereas the formation of stress granules by amino‐acid starvation requires its phosphorylated form and Sec16.69
Interestingly, the content of stress granules also appears to be cell type‐specific. A screen using three different mammalian cell lines (HepG2, HeLa and NPC) treated with sodium arsenite showed that approximately half of the RNA‐binding proteins (35/77) associated with stress granules exhibit a degree of cell‐type specificity.113
4.4. Stress‐specific differences in RNA composition of stress granules
The RNA content of stress granules also appears to be dictated by the type of stress to which the cells are exposed. For instance, poly‐adenylated mRNAs are a component of bona fide stress granules formed in mammalian cells by sodium arsenate treatment, but RocA‐ and UV‐induced stress granules do not contain them.96
For a while, it had been difficult to validate and extend these observations as the overall RNA composition of stress granules remained largely unknown. Recently, however, strategies have been developed to identify RNA molecules present in stress granules and to assess the stress‐specific subset.114, 115
Using centrifugation and immunoprecipitation, insoluble stress granule cores containing G3BP1‐green fluorescent protein (GFP) were isolated from mammalian U2OS cells exposed to sodium arsenite.114 Single molecule fluorescence in situ hybridization (smFISH) validated the localization of several transcripts (including AHNAK, DYNC1H1) in arsenite‐, heat shock‐, thapsigargin‐ and sorbitol‐induced stress granules.114 GAPDH was largely depleted and POLR2A was more enriched upon heat shock and sorbitol stress, while TFRC was only enriched upon heat shock. Sequencing of RNAs contained in these cores also revealed that 78% of them are mRNAs, most of them long and inefficiently translated.114 This might reflect the notion that longer mRNAs have potentially more binding sites for RNA‐binding proteins, and that poorly translated mRNAs are less engaged by ribosomes and thus have more opportunities to be recruited to stress granules.67, 89
The preferential recruitment of longer mRNA to stress granules has been confirmed in mammalian cells upon ER stress.115 The identification of these mRNAs has allowed the description of specific recruitment “motifs” for ER stress, such as adenylate‐uridylate (AU)‐rich elements (ARE). By contrast, stress granules formed upon heat shock appear to contain mRNAs with non‐ARE sequences, such as guanylate‐cytidylate (RG)‐rich motifs.115 This indicates that stress‐specific recruitment of RNA might be dependent on certain sequence motifs.
Recently, the RNA composition of HEK293 cells stress granules induced by heat shock (eIF2a‐p dependent) and by hippuristanol treatment (eIF2a‐p‐independent) was shown to be different using proximity‐biotinylation with the biotin ligase APEX2 fused to eIF4A1.116 Heat shock‐induced stress granules were enriched in longer mRNAs with lower translation efficiency (as above), whereas granules induce by hippuristanol treatment were not. This suggests that recruitment of longer and poorly translated mRNA is dependent on the type of stress. It may be possible that all eIF2a‐p dependent stress granules contain longer mRNAs, while eIF2a‐p independent stress granules do not.
4.5. P‐bodies are also heterogeneous in mRNA and proteins
Like stress granules, P‐bodies show many levels of heterogeneity. For example, RNAs in yeast P‐bodies that were induced by 10 minutes of glucose starvation or osmotic stress using high concentration of CaCl2 and NaCl were identified by in vivo crosslinking and affinity purification for epitope‐tagged Dcp2 or Scd6.87 A total of 1544 mRNAs were significantly present in P‐bodies upon glucose starvation and high Na+ and Ca2+ exposure, and 35% of them were specific for a given stress.87 Analysis of the RNA length revealed that P‐bodies induced by glucose starvation contained shorter RNAs when compared to the total pool of upregulated mRNAs under the respective stress conditions, whereas P‐bodies induced upon osmotic stress contained longer RNAs. This indicates that, as with stress granules, transcript length may be important for recruitment to P‐bodies. Interestingly, gene ontology analysis and smFISH combined with immunofluorescence microscopy analyses showed that P‐bodies formed upon glucose starvation were enriched for mRNAs encoding specific mitochondrial oxidative phosphorylation factors (ATP11, ILM1, MRPL38 and AIM2).87 By contrast, ATP11 was not found in P‐bodies induced by osmotic stresses.87 This enrichment is similar to that proposed for stress granules but much more specific and striking, as it reveals a clear link to the type of stress.
Taken together, these data reveal the extraordinary complexity of these two stress‐induced membraneless compartments that are related to both key cellular processes of RNA homeostasis and (at least for stress granules) to pathological situations. Digging further into their heterogeneity will unravel the multiple universal principles underlying how cells adapt to stress.
5. NUTRIENT STARVATION RESULTS IN THE FORMATION OF MANY CYTOPLASMIC STRESS ASSEMBLIES
As reviewed above in detail, many different types of stress lead to the formation of RNA‐based and heterogeneous P‐bodies and stress granules. Nutrient stress also inhibits translation initiation and induces a similar cellular response. For instance, glucose starvation of yeast induces the formation of P‐bodies87, 102, 117, 118 and of stress granules119 that appear to largely overlap with P‐bodies.35 Similarly, bona fide stress granule formation is induced by chronic112 or acute amino acid starvation in mammalian120 and Drosophila cells.70, 76 However, whereas many stresses appear to solely lead to P‐bodies and stress granule formation, starvation appears to be a strong stress that leads to the formation of many different cytoplasmic stress assemblies, not all of which are RNA‐based (Figure 3).
5.1. The EIF2B bodies in starved yeast
As mentioned above, translation initiation is suppressed during cellular stress and leads to the formation of stress granules and P‐bodies. It also triggers a third class of cytoplasmic assemblies called EIF2B bodies. EIF2B facilitates ternary complex formation and translation initiation through its guanine exchange activity on the eIF2 complex. However, when eIF2a is phosphorylated on Ser51 (see Section 4.2), it prevents ELF2B release from the elF2 complex resulting in blocking translation initiation. As a result, EIF2B appears to coalesce into EIF2B bodies, which are defined as either round or fibril‐like structures that contain subunits of the initiation complex eIF2.121
EIF2B bodies were first observed in growing yeast cells, but amino acid starvation increased their size and eIF2a content.121 Yeast EIF2B bodies can also be induced in 20% to 40% of cells by acute glucose deprivation.86, 119 Importantly, they also form in HeLa cells upon hypoxia and acidification.119 EIF2B bodies are rapidly and reversibly formed independently of stress granules that also form upon acute glucose deprivation.119 In fact, it appears that EIF2B bodies form more rapidly than stress granules but disassemble more slowly. Whether EIF2B bodies contain RNAs is not known.
5.2. Proteasomes assemblies in glucose‐starved yeast
Many more protein‐based assemblies form in starved yeast. The 26S proteasome, a 2.5‐MDa multi‐subunit protease, is the major nonmembrane‐based degradative machine that target proteins marked by ubiquitination for destruction. In growing and dividing yeast, proteasomes assemble in both the nucleus and the cytoplasm. In quiescent and glucose‐starved yeast, a large subset of proteasome subunits form large cytoplasmic assemblies called “Proteasome storage granules” that act as a reservoir for proteasome formation when cells are re‐fed.122, 123 The material properties of these assemblies have not been described but they appear to be non‐RNA‐based.
5.3. Metabolic enzyme foci in glucose‐starved yeast
In a more general manner, nutrient starvation/restriction as observed in stationary yeast growth phase leads to the formation of many large macroscopic protein complexes. A study screening a collection of 800 GFP‐tagged proteins for aggregation during stationary phase found that these complexes are made of primarily metabolic enzymes, with 180 proteins incorporated into macroscopic complexes.124, 125 Thirty‐three enzymes were further investigated biochemically and shown to function in purine metabolism, glycolysis, tRNA amino acylation and response to stress. Critically, these foci are reversible when nutrients are replenished.
The formation of these protein foci was reexamined using glutamine synthase (Gln1) as a model enzyme.34 The foci that form about 50 minutes after starvation are in fact solid filaments34, 126 and lack of glucose appears to be the critical factor inducing their formation. As for other stress assemblies, these filaments are rapidly reversible. A single point mutation in Gln1 prevents filament formation in starved yeast by interfering with back‐to‐back interactions of Gln1 dodecamers.
Enzyme foci formation in glucose‐starved yeast has been further investigated using the pyruvate kinase Cdc19 both in vivo and in vitro.35 Although the Cdc19 foci are also solid‐like and quickly reversible, the mechanism driving their formation appears different than that for Gln1. Cdc19 foci are driven by phosphorylation of monomeric CDc19, a modification that exposes the enzyme's low complexity domains. Furthermore, Cdc19 foci localize with Pab1 and Ecd3, two RNA‐binding proteins that partition to mixed stress granules/P‐bodies that are also induced by glucose starvation.35 Taken together, these data suggest that metabolic enzyme foci are not all homogenous. Indeed, the Gln1 foci that have not been reported to be incorporated into RNA‐based stress assemblies.
5.4. Sec bodies in amino‐acid‐starved Drosophila S2 cells
In addition to stress granules, Sec bodies are membraneless stress assemblies that are specifically formed upon 3 to 4 hours of amino‐acid starvation of Drosophila S2 cells.76 As stress granules are linked to the inhibition of translation initiation, Sec bodies are linked to the inhibition of secretory pathway function, especially at the level of ER exit. In growth conditions, the secretory pathway transports proteins (and lipids) from the ER to the Golgi from where they are dispatched to the plasma membrane, the extracellular environment or other intracellular membrane‐bound organelles.2, 127 One of the first steps of this pathway is the exit of newly synthesized proteins from the ER at specific ER exit sites, characterized by the concentration of COPII (coat protein II)‐coated buds and vesicles in which these proteins are packaged for transport to the Golgi. The COPII coat comprises six subunits, and the assembly of the coat is facilitated by the large scaffold protein, Sec16.128, 129
Upon amino‐acid starvation, protein transport in the secretory pathway is inhibited at the level of ER exit sites. The COPII subunits and Sec16 coalesce in large membraneless Sec bodies where they are stored and protected from degradation. Sec bodies are very quickly reversible upon refeeding. They act as the reservoir for ER exit site components, and upon stress relief, COPII subunits and Sec16 quickly recover their function even in the absence of protein synthesis.6, 76, 130, 131 Importantly, Sec bodies are the first example of a stress assembly that form from proteins normally associated with membrane traffic.
In contrast to stress granules, Sec bodies do not appear to be RNA‐based. However, like stress granules, Sec bodies have properties of liquid droplets,76 albeit with a high density slowly exchanging with the surrounding cytoplasm as detected by FRAP experiments. The drivers in Sec body phase separation have been shown to be Sec16 and Sec24AB, both of which are rich in low complexity domains.76 In Sec24AB, these domains are mostly located in the 400 N‐terminal residues. GFP fused to this N‐terminal region is efficiently recruited to Sec bodies, whereas a GFP‐Sec24AB fusion protein missing the low complexity sequences is not.76 In Sec16, the low complexity domains are spread throughout the protein (except for the conserved central region). However, overexpression of just a 44‐residue conserved domain (called SRDC) located at the C‐terminus of Sec16 is able to drive Sec body formation even in the absence of stress.69 Curiously, the SRDC itself is not incorporated into Sec bodies. This is reminiscent of a C elegans protein called SERF that has been identified to drive protein aggregation without being a component of the aggregates.132 Whether the SRDC functions as an intramolecular SERF remains to be elucidated.
5.5. Enlarged P‐bodies and U‐bodies in oocytes of starved Drosophila females
Female oogenesis is an energy demanding process, and organisms such as Drosophila modulate their oogenesis under conditions of protein starvation. Upon starvation, the late stage egg chambers that are present are removed, and early stages are stalled in their maturation. Importantly, key developmental mRNAs such as oskar—together with its known partner Yps—is trapped within both nurse cells133 and in the oocyte98, 133 in large cytoplasmic foci that also contain the decapping enzyme Dcp1, elF4E and the 5′‐3′ exoribonuclease Pacman.134 These foci likely correspond to enlarged P‐bodies.
In the oocytes, P‐bodies (marked by CUP and Otu) are in close proximity to and/or overlap with other membraneless structures known as U‐bodies. U‐bodies contain a fraction of cytoplasmic SMN (survival motor neurons) proteins that are also present in the nucleus.135 U‐bodies are thought to be responsible for the assembly and storage of uridine‐rich small nuclear ribonucleoproteins that are essential for pre‐mRNA splicing.
Several lines of evidence suggest that the association between U‐bodies and P‐bodies is functional and represents a specific pathway that may regulate multiple downstream events including nuclear organization. First, mutations in P‐body components affect the organization of U‐bodies. Second and conversely, SMN mutations affect both U‐body and P‐body organization. Third, SMN loss of function in the oocytes phenocopies P‐body component loss of function in causing nuclear disorganization.136 Last, U‐bodies and P‐bodies both grow during starvation showing that they are responsive to nutrition changes, presumably through the U‐body/P‐body pathway.137
Taken together, these examples illustrate the variety of membraneless stress assemblies that are induced by nutrient starvation. The described structures are likely only the tip of the iceberg as many more assemblies probably form to sustain and protect the components incorporated in these assemblies. As described below, some are also shown be prosurvival.
6. FORMATION AND ROLE OF NUTRIENT STRESS ASSEMBLIES
6.1. Phase separation upon nutrient starvation can be induced by a drop in the cytoplasmic pH
The current notion to explain the formation of stress assemblies is that stress assembly is driven by a slight change in the conformation of drivers that leads to their coalescence (see Section 2). In the case of stress assemblies, the conformational changes can be induced by modifying the biophysical properties of the cytoplasm and by protein posttranslational modifications.131
Glucose starvation has been shown to induce a drop of the cytoplasmic pH, and this is an important factor leading to the formation of stress assemblies at least in yeast. This drop is caused by the reduced ATP level normally provided by glycolysis. As ATP is needed to fuel the V‐ATPase proton pump that extrudes protons into endolysosomal organelles from the cytoplasm (thus ensuring that cytoplasmic pH remains neutral), a lower level of ATP leads to acidification of the cytoplasm.126 Inhibiting the proton pump in growing yeast leads to the same acidification. Interestingly, cytosolic pH acts as a cellular signal to activate Ras and TORC1 in response to glucose availability via Cdc19 (see above), thus linking foci formation to growth signaling.138 Importantly, cytoplasm acidification is the widely used mechanism that drives the formation of metabolic enzyme filaments. In this regard, a drop in cytoplasmic pH in growing yeast results in the same filament formation as in starved yeast.126 As a result, the cytoplasm of starved yeast exhibits a glass‐like material property, showing that many enzymes form filaments and foci.126 This is thought to be due to the fact that many enzymes “precipitate” when the pH drops below their pKa, which, for a large pool, is around 7.126 Interestingly, the screening for factors required for the formation of proteasome storage granules (see above) identified V‐ATPase as a critical factor. Direct depletion of this pump, like glucose starvation, leads to a similar drop of the cytoplasmic pH, leading to coalescence of the proteasome subunits.123 EIF2B bodies also form upon cytoplasm acidification upon glucose starvation.119
Based on these observations, the drop of cytoplasmic pH upon starvation has been proposed to be the signal for starvation‐induced stress assembly formation. Interestingly, preliminary results (Rabouille, unpublished) suggest that amino‐acid starvation of S2 cells would also lead to a drop in cytoplasmic pH, pointing to a possible convergent evolutionary mechanism for signaling by starvation. Overall, it appears that nutrient stress leads to a change in the biophysical properties of the cytoplasm (at least in term of pH) leading to the coalescence of many stress assemblies.
6.2. Posttranslational modifications are necessary for the formation of certain stress assemblies
The coalescence of metabolic enzymes and proteasome subunits in starved yeast appears to be mediated solely by the drop in the cytoplasmic pH seemingly without involving posttranslational protein modifications.131 However, cellular stress is known to activate signaling pathways leading to posttranslational modifications of key drivers.31 This would result in a slight modification of their conformation exposing their low complexity sequences, potentiating their multivalency, increasing their transient interactions and leading to their coalescence.139 For example, Cdc19 foci formation depends on its phosphorylation status.35 In this regard, phosphorylation (for instance of the serine of the PLD of FUS140), sumoylation,38, 141 arginine methylation142 and PARYation143 have been shown to be required for the formation of several stress assemblies. Furthermore, a role for mono‐ADP ribosylation (MARylation) catalyzed by Drosophila PARP16 has been established for Sec body formation. This enzyme is necessary for Sec body formation and appears to MARylate the small SRDC domain of Sec16 (mentioned above) upon amino‐acid starvation. This MARylation event is thought to be enough to drive the coalescence of Sec bodies.69
6.3. Stress assemblies are reversible upon stress relief
As mentioned above, in healthy cells, stress assemblies are reversible upon stress relief, and the coalesced components become diffuse again to adopt their nonstressed localization and function. This is the case for stress granules as well as for other nutrient stress assemblies mentioned above. For assemblies that form upon a fast change in the cytoplasm biophysical properties (such as acidification), the reversibility upon stress relief is quick. As mentioned above when yeast are supplemented with nutrients after starvation, the cytoplasmic pH raises and the enzymes are solubilized.
When posttranslational modifications are required for coalescence (such as stress granules and Sec bodies), the reversal necessitates an enzymatic activity. One category of proteins that can help dissolution is heat shock proteins (Hsp). Hsp104 has been clearly shown to help dissolution of irreversible protein aggregates144, 145 and interestingly, yeast stress granules need Hsp104 for their dissolution.72 Mammalian stress granules have also been shown to require the DYRK3 kinase for their dissolution, likely by phosphorylating multiple RNA‐binding proteins. This dissolution is proposed to release mTORC1 that appears sequestered in the granules, thus allowing protein synthesis to resume.146 Interestingly, DYRK3 activity appears to control the global cellular state of interphase (a largely phase separated state) vs mitotic (a largely “soluble” state) cells.21 Last, given that Sec bodies require MARylation for their formation, their reversal would be expected to require the Poly‐ADP‐ribosyse‐Glycolysase (PARG) or the related TARG147 for their dissolution, but this remains to be established.
6.4. Nutrient stress assemblies are prosurvival: Gel or die148
Despite the perception that protein coalescence and aggregation are deleterious and associated with pathologies, the formation and reversibility of stress assemblies largely provide cells with a means to survive during stress and a fitness advantage upon stress relief. As such, they confer cells with survival properties. In all the systems tested, when stress assembly formation is inhibited, cells die more quickly during stress and do not thrive as well as their control counterparts upon stress relief. For instance, yeast that form Gln1 filaments upon starvation thrive (growth and colony formation) better upon refeeding than yeast that do not form filaments (for instance those bearing the point mutation in Gln1 that prevents back‐to‐back packing).34 The fitness advantage of partitioning is also shown in a converse experiment in which yeast harboring a phosphorylated form of Cdc19 that partitions in foci more quickly than wild‐type Cdc19 upon glucose starvation (20 minutes instead of 240 minutes), and that does not revert upon glucose refeeding, fare poorly in response to stress.35 The premature foci formation impairs the resistance to stress as well as preventing the yeast to reenter the cell cycle upon stress relief. This is because Cdc19 modulates two pathways leading to ribosome biogenesis, and foci formation inhibits its activity.
In the same vein, the formation of proteasome storage granules also enhances resistance to genotoxic stress and confers fitness during aging,123 and Sec bodies increase cell survival during amino‐acid starvation and fitness upon stress relief. Depletion of Sec24AB—a driver for Sec body formation—under conditions that do not compromise protein export from the ER)—causes cells to die more during starvation and recover less upon refeeding compared to control cells.76 Furthermore, depletion of dPARP16—a key factor for Sec body formation—leads to the same phenotype.69
Stress granule formation is also prosurvival in response to many stresses. When key factors required for their formation are depleted or mutated, cell survival during stress is reduced. This is the case for Vgl during heat stress,149 FUS during hyperosmotic stress150 and 4E‐BP1 during selenite poisoning.151, 152 This is probably due to their roles in preserving nascent mRNA from degradation and in accumulating pro‐apoptotic kinases to prevent apoptosis.107, 153, 154, 155, 156 Simply preventing stress granule formation using cycloheximide or boosting it with puromycin (both inhibitors of protein synthesis) also resulted in modulating cell growth upon stress relief. Indeed, cells that were prevented from forming stress granules die 25% faster during heat shock than cells that did form stress granules.151, 152
This was reinvestigated recently in a study of the RNA‐binding protein, Pab1, that is integrated into stress granules. When yeast is stressed by heat or energy deprivation, Pab1 coalesces first into small droplets, and in doing so, releases the mRNAs that it had bound. These mRNA molecules are thought to encode key stress response factors (see above). Their release permits their translation, thus enabling cells to cope with stress. Hence, this Pab1 gelation/coalescence is proposed to be a sensor for stress. Importantly, prevention of Pab1 gelation (for instance by expressing a mutated or truncated form of Pab1) increased yeast cell death and decreased recovery relative to their control counterparts.102, 148
Taken together, the formation of stress assemblies upon nutrient stress shares common features that are summarized in Figure 2, including that they are prosurvival. The exact reason as to why impairment of their formation leads to cell death or compromises fitness is not always completely understood and needs to be investigated further.
6.5. Stress assemblies formed upon nutrient starvation are not substrates of autophagy
Nutrient starvation is also well known to inhibit mTORC1, the major amino‐acid sensor, and this inhibition leads to the induction of the catabolic pathway of autophagy that targets organelles and cytoplasmic regions marked for degradation upon fusion with lysosomes. The notion behind this process is to replenish the cell interior with essential nutrients derived from the degraded material. Importantly, in the few cases where this has been studied, the stress assemblies that are formed upon nutrient starvation, such as stress granules,70 Sec bodies76 and metabolic enzyme foci124 are not marked for degradation through the autophagic pathway.
However, older stress granules80 and pathological ones (irreversible, like those induced by mutated RNA‐binding proteins)157 appear to be removed by autophagy involving the AAA ATPases VCP/P97/CDc48. This suggests that these structures are likely marked by p62. Furthermore, the dissolution of stress granules formed upon oxidative stress and heat shock are VCP‐ and ULK1/2‐dependent, but autophagy‐independent. This is surprising because ULK1/2 is normally known to induce autophagy. Instead, here ULK1/2 appear to be recruited to stress granules where they phosphorylate and activate VCP, leading to stress granule disassembly.158
Taken together, nutrient stress appears to be a strong stress that leads to large reorganization of the cytoplasm, especially in yeast in which the formation of P‐bodies, stress granules, metabolic enzyme foci, proteasome storage granules and EIF2B bodies is stimulated by the drop in pH due to glucose starvation. Nutrient stress also induces phase separation of stress assemblies in Drosophila. It is quite striking that the formation of nutrient stress assemblies is largely not reported for mammalian cells. Is it because most well‐studied mammalian cells in culture are cancer‐derived and thus largely resistant to amino‐acid starvation? Or is it because removing glucose kills them due to metabolic impairment? Whether metabolic enzymes or ER exit site components coalescence in amino‐acid‐starved mammalian cells, and whether stress assembly in these cells is regulated similar to yeast and Drosophila cells will need to be addressed in the future.
7. CONCLUSION
In conclusion, cellular stress elicits the formation of many stress assemblies, among which stress granules and P‐bodies are the best studied. However, these two terms cover a large degree of heterogeneity that is only beginning to be unraveled. Signaling cues are the first source of heterogeneity. Canonical stresses lead to eIF2a phosphorylation via specific kinases, but other stresses use alternative pathways that are largely unknown. Second, the protein content of both stress granules varies by the inducing stress, and the specific recruitment/storage of proteins upon a given stress may be related to the fact that they are essential when the stress is relieved. Third, the RNA content of both stress granules and P‐bodies also varies with the inducing stress. For instance, glucose starvation induces the formation of P‐bodies that store mRNAs encoding proteins related to this particular stress. Stress granules induced by the canonical stresses store poly‐A mRNA, whereas those induced by UV and osmotic stress do not. However, the significance of these differences remains to be investigated.
Stress granules are even more complex than initially thought. For instance, they are directly linked to cyto‐nuclear transport through the nuclear pores159, 160, 161 and they appear to be dependent on pre‐mRNA splicing.162 They are also linked to several human neurodegenerative disorders such as Alzheimer, amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (reviewed in Reference 163. Thus, understanding the composition and mechanism of stress granules and the versatility of their dynamics may help in potential therapies to battle these diseases.
Conversely, nutrient stress leads to the formation of many prosurvival cytoplasmic stress assemblies, some of which are not RNA based. Whether these stress assemblies act solely as storage for key molecules or become crucibles of specific biochemical stress reactions, as is the case for stress granules, remains to be investigated.164 Furthermore, their propensity to become pathological upon expression of mutated proteins needs to be explored.
The fact that exogenous cellular stress leads to such a diversity of cytoplasmic (and nuclear) reorganization is intriguing as it opens the possibility that stress assemblies interact specifically with membrane‐bound‐organelles. They may act as 2D‐scaffolding platform like in the case of P‐bodies forming at the surface of the ER,165 or Sec bodies forming at the ER or ER exit sites that are entirely remodeled,76 but also interact biochemically to sustain specific functions. Overall, in the future, the cell biology of stress will need to take into account how membrane‐bound organelles react to and communicate with membraneless (stress) organelles.
ACKNOWLEDGMENT
We apologize to colleagues whose work was not discussed due to space constraints.
van Leeuwen W, Rabouille C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic. 2019;20:623–638. 10.1111/tra.12669
Peer Review The peer review history for this article is available at https://publons.com/publon/10.1111/tra.12669/
REFERENCES
- 1. Scorrano L, De Matteis MA, Emr S, et al. Coming together to define membrane contact sites. Nat Commun. 2019;10(1):1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonifacino JS. Vesicular transport earns a Nobel. Trends Cell Biol. 2014;24(1):3‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid‐like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA. 2011;108(11):4334‐4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3(3):1‐15. 10.001101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hyman AA, Weber CA, Julicher F. Liquid‐liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39‐58. [DOI] [PubMed] [Google Scholar]
- 6. Aguilera‐Gomez A, Rabouille C. Membrane‐bound organelles versus membrane‐less compartments and their control of anabolic pathways in Drosophila. Dev Biol. 2017;428(2):310‐317. [DOI] [PubMed] [Google Scholar]
- 7. Zaslavsky BY, Ferreira LA, Darling AL, Uversky VN. The solvent side of proteinaceous membrane‐less organelles in light of aqueous two‐phase systems. Int J Biol Macromol. 2018;117:1224‐1251. [DOI] [PubMed] [Google Scholar]
- 8. Luo Y, Na Z, Slavoff SA. P‐bodies: composition, properties, and functions. Biochemistry. 2018;57(17):2424‐2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 2015;6:7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Trcek T, Lehmann R. Germ granules in Drosophila. Traffic. 2019; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Putnam A, Cassani M, Smith JA, Seydoux G. A gel phase promotes condensation of liquid P granules in Caenorhabditis elegans embryos. Nat Struct Mol Biol. 2019;26(3):220‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brangwynne CP, Eckmann CR, Courson DS, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729‐1732. [DOI] [PubMed] [Google Scholar]
- 13. Marnik EA, Updike DL. Membraneless organelles: P granules in Caenorhabditis elegans . Traffic. 2019;20(6):373‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsang B, Arsenault J, Vernon RM, et al. Phosphoregulated FMRP phase separation models activity‐dependent translation through bidirectional control of mRNA granule formation. Proc Natl Acad Sci USA. 2019;116(10):4218‐4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Graeve F, Besse F. Neuronal RNP granules: from physiological to pathological assemblies. Biol Chem. 2018;399:623. [DOI] [PubMed] [Google Scholar]
- 16. Formicola N, Vijayakumar J, Besse F. Neuronal RNP granules: dynamic sensors of localized signals. Traffic. 2019; in press. [DOI] [PubMed] [Google Scholar]
- 17. Wunder T, Cheng SLH, Lai S‐K, Li H‐Y, Mueller‐Cajar O. The phase separation underlying the pyrenoid‐based microalgal Rubisco supercharger. Nat Commun. 2018;9(1):5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rochaix J‐D. The Pyrenoid: an overlooked organelle comes out of age. Cell. 2017;171(1):28‐29. [DOI] [PubMed] [Google Scholar]
- 19. Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, et al. The eukaryotic CO2‐concentrating organelle is liquid‐like and exhibits dynamic reorganization. Cell. 2017;171(1):148‐162.e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wunder T, Oh ZG, Mueller‐Cajar O. CO2‐fixing liquid droplets: towards a dissection of the microalgal pyrenoid. Traffic. 2019;20(6):380‐389. 10.1111/tra.12650 [DOI] [PubMed] [Google Scholar]
- 21. Rai AK, Chen J‐X, Selbach M, Pelkmans L. Kinase‐controlled phase transition of membraneless organelles in mitosis. Nature. 2018;559(7713):211‐216. [DOI] [PubMed] [Google Scholar]
- 22. Warren G, Wickner W. Organelle Inheritance. Cell. 1996;84(3):395‐400. [DOI] [PubMed] [Google Scholar]
- 23. Shorter J, Warren G. Golgi architecture and inheritance. Annu Rev Cell Dev Biol. 2002;18(1):379‐420. [DOI] [PubMed] [Google Scholar]
- 24. Spannl S, Tereshchenko M, Mastromarco GJ, Ihn SI, Lee HO. Membrane‐less organelles and disease. Traffic. 2019; Under consideration. [Google Scholar]
- 25. Patel A, Lee HO, Jawerth L, et al. A liquid‐to‐solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066‐1077. [DOI] [PubMed] [Google Scholar]
- 26. Molliex A, Temirov J, Lee J, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell. 2015;60(2):208‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee K‐H, Zhang P, Kim HJ, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane‐less organelles. Cell. 2016;167(3):774‐788.e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li P, Banjade S, Cheng HC, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hyman AA, Brangwynne CP. Beyond stereospecificity: liquids and mesoscale organization of cytoplasm. Dev Cell. 2011;21(1):14‐16. [DOI] [PubMed] [Google Scholar]
- 31. Itakura AK, Futia RA, Jarosz DF. It pays to be in phase. Biochemistry. 2018;57(17):2520‐2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gomes E, Shorter J. The molecular language of membraneless organelles. J Biol Chem. 2018;294(18):7115‐7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brangwynne CP. Phase transitions and size scaling of membrane‐less organelles. J Cell Biol. 2013;203(6):875‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrovska I, Nuske E, Munder MC, et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. elife. 2014;3 10.7554/eLife.02409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, Dechant R. Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat Cell Biol. 2017;19:1202‐1213. [DOI] [PubMed] [Google Scholar]
- 36. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188‐1191. [DOI] [PubMed] [Google Scholar]
- 38. Banani SF, Rice AM, Peeples WB, et al. Compositional control of phase‐separated cellular bodies. Cell. 2016;166(3):651‐663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of Prionogenic proteins. Cell. 2009;137(1):146‐158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sabate R, Rousseau F, Schymkowitz J, Batlle C, Ventura S. Amyloids or prions? That is the question. Prion. 2015;9(3):200‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franzmann TM, Jahnel M, Pozniakovsky A, et al. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359(6371):pii: eaao5654. 10.1126/science.aao5654 [DOI] [PubMed] [Google Scholar]
- 42. Zambrano R, Conchillo‐Sole O, Iglesias V, et al. PrionW: a server to identify proteins containing glutamine/asparagine rich prion‐like domains and their amyloid cores. Nucleic Acids Res. 2015;43(W1):W331‐W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim HJ, Kim NC, Wang YD, et al. Mutations in prion‐like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. March ZM, King OD, Shorter J. Prion‐like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 1647;2016:9‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruff KM, Pappu RV, Holehouse AS. Conformational preferences and phase behavior of intrinsically disordered low complexity sequences: insights from multiscale simulations. Curr Opin Struct Biol. 2019;56:1‐10. [DOI] [PubMed] [Google Scholar]
- 46. Murray DT, Kato M, Lin Y, et al. Structure of FUS protein fibrils and its relevance to self‐assembly and phase separation of low‐complexity domains. Cell. 2017;171(3):615‐627.e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hughes MP, Sawaya MR, Boyer DR, et al. Atomic structures of low‐complexity protein segments reveal kinked β sheets that assemble networks. Science. 2018;359(6376):698‐701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang J, Choi J‐M, Holehouse AS, et al. A molecular grammar governing the driving forces for phase separation of prion‐like RNA binding proteins. Cell. 2018;174(3):688‐699.e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Semenow AN, Rubinstein M. Dynamics of strongly entangled polymer systems: activated reptation. Eur Phys J. 1998;B 1:87‐98. [Google Scholar]
- 50. Mittag T, Parker R. Multiple modes of protein–protein interactions promote RNP granule assembly. J Mol Biol. 2018;430(23):4636‐4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′‐end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012;31(20):4020‐4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. West JA, Mito M, Kurosaka S, et al. Structural, super‐resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol. 2016;214(7):817‐830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Picart‐Picolo A, Picault N, Pontvianne F. Ribosomal RNA genes shape chromatin domains associating with the nucleolus. Nucleus. 2019;10(1):67‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Langdon EM, Qiu Y, Ghanbari NA, et al. mRNA structure determines specificity of a polyQ‐driven phase separation. Science. 2018;360(6391):922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Treeck B, Parker R. Emerging roles for intermolecular RNA‐RNA interactions in RNP assemblies. Cell. 2018;174(4):791‐802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G. Stress‐induced nuclear bodies are sites of accumulation of pre‐mRNA processing factors. Mol Biol Cell. 2001;12(11):3502‐3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Boulon S, Westman BJ, Hutten S, Boisvert F‐M, Lamond A‐I. The nucleolus under stress. Mol Cell. 2010;40(2):216‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Staněk D, Fox AH. Nuclear bodies: news insights into structure and function. Curr Opin Cell Biol. 2017;46:94‐101. [DOI] [PubMed] [Google Scholar]
- 59. Shishido‐Hara Y, Ichinose S, Uchihara T. JC virus intranuclear inclusions associated with PML‐NBs: analysis by electron microscopy and structured illumination microscopy. Am J Pathol. 2012;180(3):1095‐1106. [DOI] [PubMed] [Google Scholar]
- 60. Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. 2016;90(13):5850‐5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lallemand‐Breitenbach V, de Thé H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154‐161. [DOI] [PubMed] [Google Scholar]
- 62. Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35(15):1603‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115(Pt 16):3227‐3234. [DOI] [PubMed] [Google Scholar]
- 64. Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803‐808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kedersha N, Panas MD, Achorn CA, et al. G3BP‐Caprin1‐USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212(7):845‐860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA‐binding proteins TIA‐1 and TIAR link the phosphorylation of eIF‐2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147(7):1431‐1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kedersha N, Cho MR, Li W, et al. Dynamic shuttling of TIA‐1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151(6):1257‐1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tourriere H, Chebli K, Zekri L, et al. The RasGAP‐associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160(6):823‐831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69. Aguilera‐Gomez A, van Oorschot MM, Veenendaal T, Rabouille C. In vivo vizualisation of mono‐ADP‐ribosylation by dPARP16 upon amino‐acid starvation. elife. 2016;5:pii: e21475. 10.7554/eLife.21475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Aguilera‐Gomez A, Zacharogianni M, van Oorschot MM, et al. Phospho‐Rasputin stabilization by Sec16 is required for stress granule formation upon amino acid starvation. Cell Rep. 2017;20(4):935‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668‐679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kroschwald S, Maharana S, Mateju D, et al. Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. elife. 2015;4:e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. elife. 2016;5:pii: e18413. 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase‐modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164(3):487‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mollet S, Cougot N, Wilczynska A, et al. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19(10):4469‐4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zacharogianni M, Aguilera‐Gomez A, Veenendaal T, Smout J, Rabouille C. A stress assembly that confers cell viability by preserving ERES components during amino‐acid starvation. elife. 2014;3 10.7554/eLife.04132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33(3):141‐150. [DOI] [PubMed] [Google Scholar]
- 78. Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19(10):R397‐R398. [DOI] [PubMed] [Google Scholar]
- 79. Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36(6):932‐941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461‐1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25(5):635‐646. [DOI] [PubMed] [Google Scholar]
- 82. Hubstenberger A, Courel M, Bénard M, et al. P‐body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017;68(1):144‐157.e145. [DOI] [PubMed] [Google Scholar]
- 83. Guzikowski AR, Chen YS, Zid BM. Stress‐induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip Rev RNA. 2019;10(3):e1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Teixeira D, Parker R. Analysis of P‐body assembly in Saccharomyces cerevisiae . Mol Biol Cell. 2007;18(6):2274‐2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P‐bodies in human cells. J Cell Sci. 2009;122(pt 20):3619‐3626. [DOI] [PubMed] [Google Scholar]
- 86. Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae . J Cell Biol. 2008;183(3):441‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, Spang A. Context‐dependent deposition and regulation of mRNAs in P‐bodies. elife. 2018;7:e29815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Horvathova I, Voigt F, Kotrys AV, et al. The dynamics of mRNA turnover revealed by single‐molecule imaging in single cells. Mol Cell. 2017;68(3):615‐625.e619. [DOI] [PubMed] [Google Scholar]
- 89. Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30(6):963‐969. [DOI] [PubMed] [Google Scholar]
- 90. Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871‐884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wilbertz JH, Voigt F, Horvathova I, Roth G, Zhan Y, Chao JA. Single‐molecule imaging of mRNA localization and regulation during the integrated stress response. Mol Cell. 2019;73(5):946‐958.e947. [DOI] [PubMed] [Google Scholar]
- 92. Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine‐387 facilitates its localization to processing bodies. Biochem J. 2008;413(3):429‐436. [DOI] [PubMed] [Google Scholar]
- 93. Romero‐Santacreu L, Moreno J, Pérez‐Ortín JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae . RNA. 2009;15(6):1110‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ramachandran V, Shah KH, Herman PK. The cAMP‐dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol Cell. 2011;43(6):973‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shah KH, Zhang B, Ramachandran V, Herman PK. Processing body and stress granule assembly occur by independent and differentially regulated pathways in Saccharomyces cerevisiae . Genetics. 2013;193(1):109‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Aulas A, Fay MM, Lyons SM, et al. Stress‐specific differences in assembly and composition of stress granules and related foci. J Cell Sci. 2017;130(5):927‐937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nilsson D, Sunnerhagen P. Cellular stress induces cytoplasmic RNA granules in fission yeast. RNA. 2011;17(1):120‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Weil TT, Parton RM, Herpers B, et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol. 2012;14(12):1305‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Norvell A, Wong J, Randolph K, Thompson L. Wispy and Orb cooperate in the cytoplasmic polyadenylation of localized gurken mRNA. Dev Dyn. 2015;244(10):1276‐1285. [DOI] [PubMed] [Google Scholar]
- 100. Pitchiaya S, Mourao MDA, Jalihal AP, et al. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol Cell. 2019;74(3):521‐533.e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chowdhury A, Yu M, Lemke EA. Phase separation comes of age: from phenomenology to single molecules. Mol Cell. 2019;74(3):413‐415. [DOI] [PubMed] [Google Scholar]
- 102. Riback JA, Katanski CD, Kear‐Scott JL, et al. Stress‐triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168(6):1028‐1040. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rogers GW, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274(18):12236‐12244. [DOI] [PubMed] [Google Scholar]
- 104. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljestrom P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell. 2005;16(8):3753‐3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kedersha N, Chen S, Gilks N, et al. Evidence that ternary complex (eIF2‐GTP‐tRNA(i)(Met))‐deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13(1):195‐210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci. 2013;70(19):3493‐3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. McEwen E, Kedersha N, Song B, et al. Heme‐regulated inhibitor kinase‐mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280(17):16925‐16933. [DOI] [PubMed] [Google Scholar]
- 108. Cuesta R, Laroia G, Schneider RJ. Chaperone Hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap‐initiation complexes. Genes Dev. 2000;14(12):1460‐1470. [PMC free article] [PubMed] [Google Scholar]
- 109. Mazroui R, Di Marco S, Kaufman RJ, Gallouzi I‐E. Inhibition of the ubiquitin‐proteasome system induces stress granule formation. Mol Biol Cell. 2007;18(7):2603‐2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P‐eIF2alpha‐dependent and ‐independent mechanisms. RNA. 2009;15(10):1814‐1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Groušl T, Ivanov P, Frydlová I, et al. Robust heat shock induces eIF2 ‐phosphorylation‐independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae . J Cell Sci. 2009;122(12):2078‐2088. [DOI] [PubMed] [Google Scholar]
- 112. Reineke LC, Cheema SA, Dubrulle J, Neilson JR. Chronic starvation induces noncanonical pro‐death stress granules. J Cell Sci. 2018;131(19):jcs220244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Markmiller S, Soltanieh S, Server KL, et al. Context‐dependent and disease‐specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590‐604.e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Khong A, Parker R. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction. J Cell Biol. 2018;217(12):4124‐4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Namkoong S, Ho A, Woo YM, Kwak H, Lee JH. Systematic characterization of stress‐induced RNA granulation. Mol Cell. 2018;70(1):175‐187.e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Padròn A, Iwasaki S, Ingolia NT. Proximity RNA labeling by APEX‐Seq reveals the organization of translation initiation complexes and repressive RNA granules. bioRxiv. 2018;454066 10.1101/454066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zid BM, O'Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014;514:117‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress‐dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P‐bodies. J Cell Biol. 2007;179(1):65‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Moon SL, Parker R. Analysis of eIF2B bodies and their relationships with stress granules and P‐bodies. Sci Rep. 2018;8(1):12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Damgaard CK, Lykke‐Andersen J. Translational coregulation of 5'TOP mRNAs by TIA‐1 and TIAR. Genes Dev. 2011;25(19):2057‐2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Campbell SG, Hoyle NP, Ashe MP. Dynamic cycling of eIF2 through a large eIF2B‐containing cytoplasmic body. J Cell Biol. 2005;170(6):925‐934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Laporte D, Salin B, Daignan‐Fornier B, Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol. 2008;181(5):737‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Peters LZ, Hazan R, Breker M, Schuldiner M, Ben‐Aroya S. Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J Cell Biol. 2013;201(5):663‐671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Narayanaswamy R, Levy M, Tsechansky M, et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc Natl Acad Sci U S A. 2009;106(25):10147‐10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. O'Connell JD, Zhao A, Ellington AD, Marcotte EM. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu Rev Cell Dev Biol. 2012;28:89‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Munder MC, Midtvedt D, Franzmann T, et al. A pH‐driven transition of the cytoplasm from a fluid‐ to a solid‐like state promotes entry into dormancy. elife. 2016;5:pii: e09347. 10.7554/eLife.09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Mellman I, Warren G. The road taken: past and future foundations of membrane traffic. Cell. 2000;100(1):99‐112. [DOI] [PubMed] [Google Scholar]
- 128. Gomez‐Navarro N, Miller E. Protein sorting at the ER‐Golgi interface. J Cell Biol. 2016;215(6):769‐778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sprangers J, Rabouille C. SEC16 in COPII coat dynamics at ER exit sites. Biochem Soc Trans. 2015;43(1):97‐103. [DOI] [PubMed] [Google Scholar]
- 130. Aguilera‐Gomez A, Rabouille C. Intra‐golgi transport In: Bradshaw Ralph A. and Stahl Philip D. (Editors‐in‐Chief), Encyclopedia of Cell Biology, Waltham, MA: Academic Press, 2016; Vol 2, 354–362. [Google Scholar]
- 131. Rabouille C, Alberti S. Cell adaptation upon stress: the emerging role of membrane‐less compartments. Curr Opin Cell Biol. 2017;47:34‐42. [DOI] [PubMed] [Google Scholar]
- 132. van Ham TJ, Holmberg MA, van der Goot AT, et al. Identification of MOAG‐4/SERF as a regulator of age‐related proteotoxicity. Cell. 2010;142(4):601‐612. [DOI] [PubMed] [Google Scholar]
- 133. Davidson A, Parton RM, Rabouille C, Weil TT, Davis I. Localized translation of gurken/TGF‐alpha mRNA during Axis specification is controlled by access to orb/CPEB on processing bodies. Cell Rep. 2016;14(10):2451‐2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Shimada Y, Burn KM, Niwa R, Cooley L. Reversible response of protein localization and microtubule organization to nutrient stress during Drosophila early oogenesis. Dev Biol. 2011;355(2):250‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Liu J‐L, Gall JG. U bodies are cytoplasmic structures that contain uridine‐rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci USA. 2007;104(28):11655‐11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lee L, Davies SE, Liu J‐L. The spinal muscular atrophy protein SMN affects Drosophila germline nuclear organization through the U body–P body pathway. Dev Biol. 2009;332(1):142‐155. [DOI] [PubMed] [Google Scholar]
- 137. Buckingham M, Liu J‐L. U bodies respond to nutrient stress in Drosophila. Exp Cell Res. 2011;317(20):2835‐2844. [DOI] [PubMed] [Google Scholar]
- 138. Dechant R, Saad S, Ibáñez A, Peter M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol Cell. 2014;55(3):409‐421. [DOI] [PubMed] [Google Scholar]
- 139. Bah A, Forman‐Kay JD. Modulation of intrinsically disordered protein function by post‐translational modifications. J Biol Chem. 2016;291(13):6696‐6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Monahan Z, Ryan VH, Janke AM, et al. Phosphorylation of the FUS low‐complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36(20):2951‐2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Jongjitwimol J, Baldock RA, Morley SJ, Watts FZ. Sumoylation of eIF4A2 affects stress granule formation. J Cell Sci. 2016;129(12):2407‐2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hofweber M, Dormann D. Friend or foe — post‐translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294(18):7137‐7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Leung AK. Poly(ADP‐ribose): an organizer of cellular architecture. J Cell Biol. 2014;205(5):613‐619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. DeSantis ME, Leung EH, Sweeny EA, et al. Operational plasticity enables Hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151(4):778‐793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Shorter J. Engineering therapeutic protein disaggregases. Mol Biol Cell. 2016;27(10):1556‐1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152(4):791‐805. [DOI] [PubMed] [Google Scholar]
- 147. Rack JGM, Ariza A, Drown BS, et al. (ADP‐ribosyl)hydrolases: structural basis for differential substrate recognition and inhibition. Cell Chem Biol. 2018;25(12):1533‐1546.e1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kroschwald S, Alberti S. Gel or die: phase separation as a survival strategy. Cell. 2017;168(6):947‐948. [DOI] [PubMed] [Google Scholar]
- 149. Wen WL, Stevenson AL, Wang CY, et al. Vgl1, a multi‐KH domain protein, is a novel component of the fission yeast stress granules required for cell survival under thermal stress. Nucleic Acids Res. 2010;38(19):6555‐6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Sama RR, Ward CL, Kaushansky LJ, et al. FUS/TLS assembles into stress granules and is a prosurvival factor during hyperosmolar stress. J Cell Physiol. 2013;228(11):2222‐2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Fujimura K, Sasaki AT, Anderson P. Selenite targets eIF4E‐binding protein‐1 to inhibit translation initiation and induce the assembly of non‐canonical stress granules. Nucleic Acids Res. 2012;40(16):8099‐8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Jevtov I, Zacharogianni M, van Oorschot MM, et al. TORC2 mediates the heat stress response in drosophila by promoting the formation of stress granules. J Cell Sci. 2015;128(14):2497‐2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Arimoto K, Fukuda H, Imajoh‐Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress‐responsive MAPK pathways. Nat Cell Biol. 2008;10(11):1324‐1332. [DOI] [PubMed] [Google Scholar]
- 154. Eisinger‐Mathason TS, Andrade J, Groehler AL, et al. Codependent functions of RSK2 and the apoptosis‐promoting factor TIA‐1 in stress granule assembly and cell survival. Mol Cell. 2008;31(5):722‐736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol. 2005;25(6):2450‐2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21(24):3381‐3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Marrone L, Poser I, Casci I, et al. Isogenic FUS‐eGFP iPSC reporter lines enable quantification of FUS stress granule pathology that is rescued by drugs inducing autophagy. Stem Cell Reports. 2018;10(2):375‐389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wang B, Maxwell BA, Joo JH, et al. ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol Cell. 2019;74:742‐757.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hofweber M, Hutten S, Bourgeois B, et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173(3):706‐719.e713. [DOI] [PubMed] [Google Scholar]
- 160. Guo L, Kim HJ, Wang H, et al. Nuclear‐import receptors reverse aberrant phase transitions of RNA‐binding proteins with prion‐like domains. Cell. 2018;173(3):677‐692.e620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yoshizawa T, Ali R, Jiou J, et al. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell. 2018;173(3):693‐705.e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Berchtold D, Battich N, Pelkmans L. A systems‐level study reveals regulators of membrane‐less organelles in human cells. Mol Cell. 2018;72(6):1035‐1049.e1035. [DOI] [PubMed] [Google Scholar]
- 163. Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416‐438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201(3):361‐372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Weidner J, Wang C, Prescianotto‐Baschong C, Estrada AF, Spang A. The polysome‐associated proteins Scp160 and Bfr1 prevent P body formation under normal growth conditions. J Cell Sci. 2014;127(pt 9):1992‐2004. [DOI] [PubMed] [Google Scholar]


