Abstract
Microproteomic studies have improved our knowledge of cell biology. Yet, with mass spectrometry (MS) analysis, accuracy can be lost for protein identification and quantification when using heterogeneous samples. Laser capture microdissection (LCM) allows for the enrichment of specific subsets of cells to study their proteome; however, sample fixation is necessary. Unfortunately, fixation hampers MS results due to protein cross-linking. The aim of this study was to identify both a fixation protocol and an extraction method that returns the best yield of proteins for downstream MS analysis, while preserving cellular structures. We compared glutaraldehyde (GLU), a common fixative to preserve cells, to dithiobispropionimidate (DTBP), a cleavable cross-linker. Our DTBP fixation/extraction protocol greatly increased the protein recovery. In fact, while 1000 GLU fixed cells returned only 159 unique protein hits, from 1464 unique peptides of 1994 unique collected spectra, 1000 DTBP fixed cells resulted in 567 unique collected protein hits, from 7542 unique peptides, of 10,401 unique collected spectra. That is, a 3.57-fold increase in protein hits, 5.15-fold increase in unique peptides, and a 5.22-fold increase in unique collected spectra. Overall, the novel protocol introduced here allows for a very efficient protein recovery and good sample quality for MS after sample collection using LCM.
Keywords: dithiobispropionimidate, glutaraldehyde, laser capture microdissection, mass spectrometry
Proteomics, the large-scale study and identification of proteins in a given sample, has offered scientists a new and comprehensive way to look at the role proteins play in cell biology. Nevertheless, in order to discern relevant differences between samples, most proteomic studies require rather large sample sizes. While such requirements have recently been alleviated by microproteomic methods that allow for the identification and analysis of proteins from small sample sizes, different approaches are required to reproducibly identify proteins isolated from such limited samples.[1] For example, one method has sought to use microfluidic-based cell sorting in combination with mass spectrometry (MS) analysis to identify proteins from 100 and 1000 immune cells.[2] But while flow cytometry is fast and allows for the isolation of living cells, disadvantages such as the limited availability of relevant cell surface–specific antibodies,[3] the inability to use intracellular biomarkers, and the fact that cells cannot be sorted based on morphological features, still remain.[3] Solutions to these issues are critical since slight changes within a cell population may not be identifiable in small heterogeneous sample sizes. One way to bypass this limitation is to specifically isolate individual cells using laser capture microdissection (LCM). Indeed, the direct visualization of cells by microscopy allows for their selection and isolation based on cellular characteristics, such as cell morphology as well as intracellular and cell surface biomarkers. LCM, however, usually requires cell fixation during cell selection to maintain the cells in place and to avoid damage from the finely focused laser that cuts around cells.
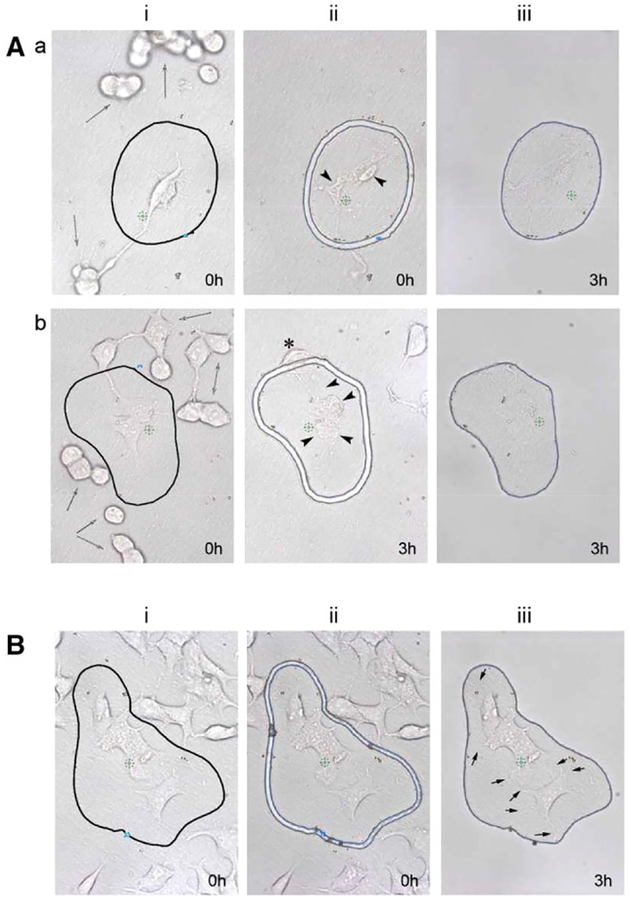
Thus, the first step was to determine whether living cells could be isolated by LCM. Some living cells were cut with the laser immediately after their selection (Figure 1A) or 3 h later (Figure 1Ab), the estimated period to select and isolate 2000 cells. As can be seen, live cells are loosely attached to the LCM membrane and many cells detached from the membrane over time (Figure 1A, i-dashed arrows) or move within the region of interest (ROI) after selection (Figure 1Ab, ii-star). In addition, cell blebbing and changes in overall morphology were observed (Figure 1A, ii-black arrowheads) and the live cells isolated appears very flat, with loss of structure and possibly cell content, as they became difficult to see on the LCM membranes (Figure 1A, iii). In comparison, GLU/PFA fixed cells showed no cell movements after 3 h (Figure 1B) and maintained the cells morphological structures, including small cellular protrusions (Figure 1B, iii-black arrows). Thus, cells can easily be distinguished and isolated based on any morphological feature, which was not possible with living cells or with the microfluidic isolation technique.[2]
Figure 1.
Laser capture microdissection of living and fixed cells. For all of the experiments, a region of interest (ROI) was i) drawn around the chosen cells, ii) cut with the laser, and iii) isolated from the rest of the cells. A) Living cells were a) cut after delineation of the ROI with the laser immediately (0 h) and isolated 3 h later from the rest of the cells or b) cut 3 h after delineation of the ROI. As can be seen, numerous cell movements occur over time (i-dashed arrows) and the laser produced visible morphological alterations in the cells (black arrowheads). B) GLU/PFA fixed cells were iii) cut immediately after delineation of the ROI and isolated subsequently (3 h). Fixation maintained small structures such as cellular protrusions (black arrows) and no morphological changes were observed i) before or iii) after the isolation of the cells.
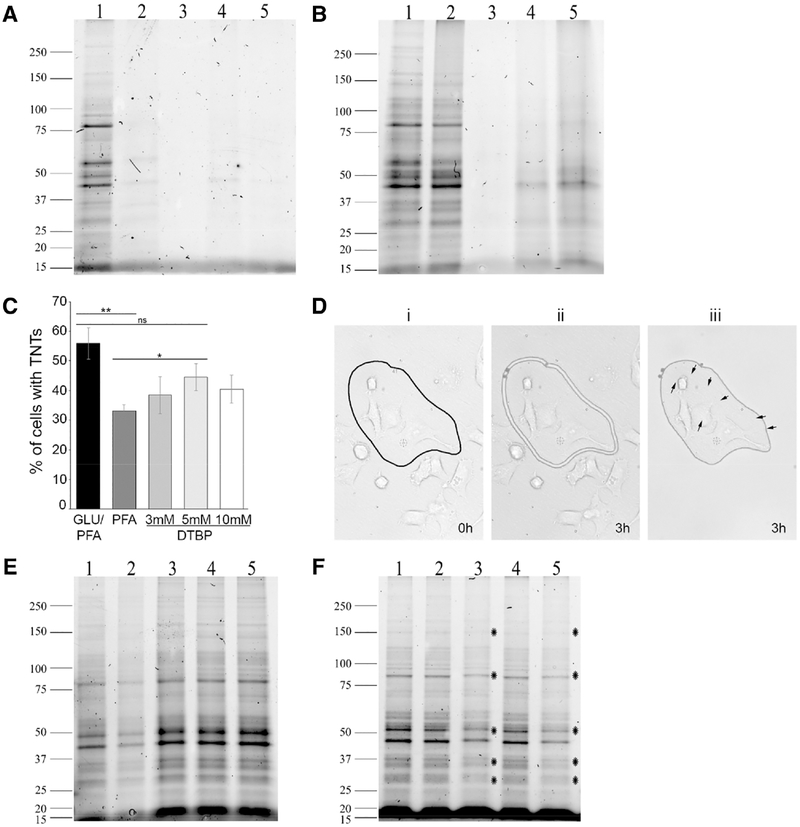
Next, we tested the effect of GLU/PFA fixation on the protein extraction recovery by gel electrophoresis. Lysis in RIPA buffer with 0.1% SDS at 4 °C resulted in little protein extraction from aldehyde-based fixed cell lysates (Figure 2A, lanes 2–5), compared to the control unfixed cell lysates, where numerous bands were observed (lane 1). In fact, even reducing the amount of GLU used fivefold (lane 4) or tenfold (lane 5) was not enough to reverse the cross-linking process impeding the detection of protein bands. We then tested a method, previously used to extract proteins from formalin-fixed tissue,[4] to determine if it would reverse paraformaldehyde (PFA) fixation by increasing the amount of SDS to 2% and using heat incubation (100 °C for 20 min followed by 60 °C for 2 h). Interestingly, this treatment reversed the PFA fixation (Figure 2B, lane 2) but it did not have any effect on the 0.05% GLU/PFA sample (Figure 2B, lane 3). Some proteins were extracted as the percentage of GLU was reduced fivefold to tenfold (lanes 4 and 5, respectively), but not up to satisfactory levels.
Figure 2.
Effect of fixations, protein extractions, and dehydration of samples on protein yield. GLU/PFA fixed samples (A) lysed with RIPA buffer (0.01% SDS) on ice or (B) lysed with RIPA buffer (2% SDS) and incubations at 100 °C/20 min and 60 °C/2 h: 1) Unfixed lysates, 2) 4% PFA alone, 3) GLU (0.05%)/PFA, 4) GLU (0.01%)/PFA, 5) GLU (0.005%)/PFA. C) Percent of cells with TNTs fixed with GLU/PFA, 4% PFA alone, PFA/3 mM DTBP, PFA/5 mM DTBP, or PFA/10 mM DTBP. Graph shows means (± SEM); **p = 0.00529; *p = 0.05507 and “ns” is not significant. D) PFA/5 mM DTBP fixed cells were plated on MMI dishes and ii) cut by LCM 3 h after i) delineation of the ROI and iii) isolated subsequently. No cellular changes were observed during the LCM isolation. E) PFA/DTBP fixed samples were lysed with RIPA buffer (2% SDS and 100 mM DTT) and incubations at 37 °C/30 min, 100 °C/20 min, and 60 °C/2 h; 1) Unfixed lysates, 2) 4% PFA alone, 3) PFA/3 mM DTBP, 4) PFA/5 mM DTBP, 5) PFA/10 mM DTBP. F) PFA/5 mM DTBP fixed samples extracted and loaded as in gel E; 1) cells were fixed and lysed the same day; 2) cells were fixed and kept in PBS for 24 h before lysing; 3) cells were fixed and kept dry for 24 h before lysing; 4) cells were fixed and kept in PBS for 72 h before lysing; 5) cells were fixed and kept dry for 72 h before lysing. Stars indicate protein bands with decreased intensity. Gels are representative of three independent experiments.
Thus, we searched for a fixative that would be strong enough to provide structural stability for morphological features, as observed by microscopy, yet not interfere with downstream MS analysis. Dithiobispropionimidate (DTBP) is a reversible cross-linker with an imidoester linkage that can easily be cleaved with a reducing agent like DTT, exposing the proteins for lysis and extraction.[5] Moreover, DTBP stood out since it was known in electron microscopy studies for its remarkable ability to preserve the fine filamentous or microtubular structures of the cell membrane.[6] Tunneling nanotubes (TNTs) are cellular extension notoriously fragile to fixation and therefore can be used to measure the cells’ structural integrity. While glutaraldehyde (GLU)-based fixation (GLU/PFA) is ideal to maintain TNTs[7] and therefore structural integrity (Figure 1B), it can hinder downstream MS analysis.[8] Thus, we first tested the ability of DTBP, compared to aldehyde-based fixations, to maintain the presence of fragile TNTs, as a measurement for the cells structural stability (Figure 2C). We compared GLU/PFA to PFA alone or three different PFA/DTBP concentrations, and counted the percent of cells with TNTs. As previously determined,[7] GLU/PFA allows for the best fixation, followed by PFA/DTBP (5 mm), with 80% of cells containing TNTs compared to GLU/PFA, and the worst fixative was PFA only with about 60% of cells containing TNTs compared to GLU/PFA. In addition, similar to GLU/PFA (Figure 1B), no cell movements were observed in PFA/DTBP fixed cells, numerous cell protrusions were visible (Figure 2D, black arrows), and the cells’ morphologies were identical before (i) or after (iii) LCM isolation.
Since PFA/DTBP was a good alternative to GLU/PFA for the maintenance of the cells’ structural integrity, we next determine the level of protein extractions from these fixed samples by electrophoresis. We have previously determined that the effects of PFA alone could be reversed by increasing SDS concentration and using heat incubation (Figure 2B), thus, we used a similar method, with the addition of 100 mm DTT in the RIPA buffer and a 37 °C incubation to cleave the disulfide bond in DTBP, prior to the high heat treatment. As expected, samples fixed with various concentrations of DTBP (Figure 2E, lanes 3–5) were easily de-crosslinked, independently of the DTBP concentration used. In fact, the protein pattern extracted from PFA/DTBP samples was close to that obtained from unfixed control samples (lane 1). Based on these microscopy and gel electrophoresis experiments (Figure 2C–E), PFA/DTBP (5 mm) was used for all subsequent experiments.
Fixed cells isolated by LCM can be cut in PBS (wet conditions) or without PBS (dry conditions). To determine if long periods of cutting in dry conditions could affect the quality of the extracted proteins during LCM isolation, we carried out an assay where cells were fixed with PFA/DTBP (5 mm) and kept dry or with PBS for 1 or 3 days (Figure 2F). We found that 1 day without PBS resulted in some protein degradation (lane 2 vs lane 3; stars indicate protein bands with reduced intensity). The protein degradation worsens with time, as band signals decreased when cells were fixed and kept dry for 3 days before extraction (lane 5; stars indicate protein bands with reduced intensity). In contrast, no change was observed when cells were maintained with PBS for 1 (lane 2) or 3 days (lane 4) after fixation. These data demonstrate that LCM isolation (i.e., selection and cutting) should be done in PBS in order to maintain optimal protein integrity for downstream analysis.
Since we drastically improved the LCM sample preparation using our new fixation/extraction protocol, we next analyzed its effect by MS.
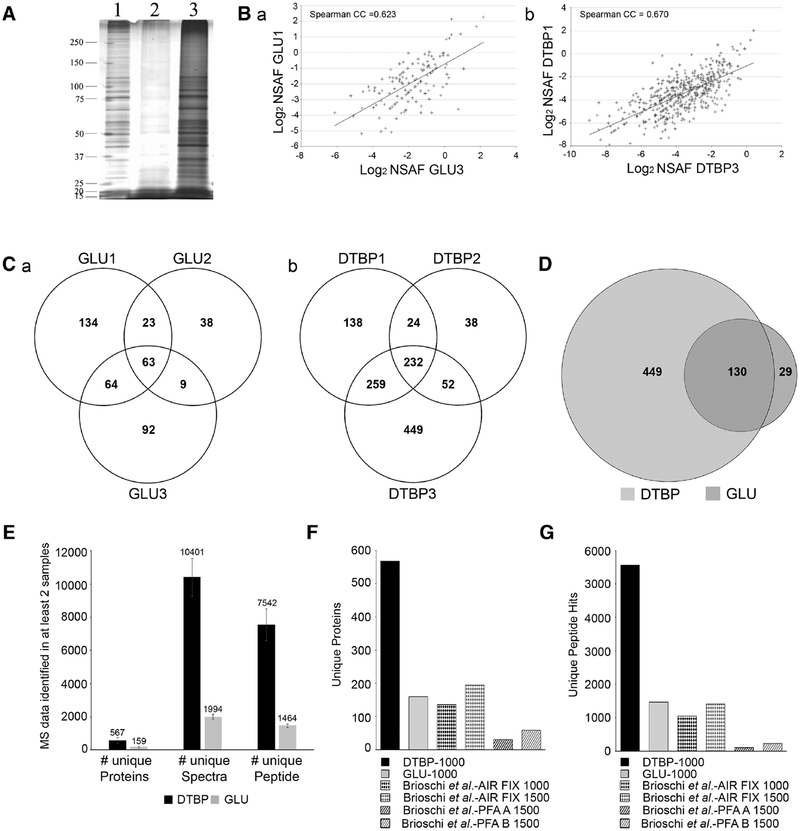
Thus, 1000 cells fixed with either GLU/PFA or PFA/DTBP were isolated by LCM in PBS, lysed, and extracted proteins were prepared for MS or ran on a silver-stained gel (Figure 3A). This gel confirmed that the extraction of 1000 GLU/PFA fixed cells yields a lower amount of proteins (lane 2) compared to 1000 PFA/DTBP fixed cells (lane 3).
Figure 3.
Effect of type of fixation on protein yield and MS results from 1000 cells. A) Silver-stained representative gel from 1) unfixed cells (0.1 μg of protein); 2) 1000 GLU/PFA fixed cells; 3) 1000 PFA/DTBP fixed cells. B) Graphical representation of Spearman correlation between a representative replicate from the triplicate a) GLU/PFA or b) PFA/DTBP samples. The p-values <0.05 were considered statistically significant. Statistical analyses were done using StatPlus Pro. C) Venn diagram of identified proteins shared in the triplicate samples from a) 1000 GLU/PFA or b) 1000 PFA/DTBP fixed cells. D) Venn diagram showing the overlap of identified proteins between PFA/DTBP and GLU/PFA fixed samples. E) Average and SD of unique proteins, unique spectra, and unique peptides of samples from 1000 PFA/DTBP or GLU/PFA fixed cells. F) Comparison of unique proteins identified in 1000 PFA/DTBP fixed cells, 1000 or 1500 air fixed cells,[10] and 1500 PFA fixed cells extracted with two methods (A and B).[10] G) Comparison of unique peptides identified in 1000 PFA/DTBP fixed cells, 1000 or 1500 air fixed cells,[10] and 1500 PFA fixed cells extracted with two methods (A and B).[10]
For MS analysis, independently acquired triplicate samples from 1000 cells for both fixation conditions were run in limited gels, fixed and stored in 1% acetic acid until further MS processing. In order to gage the reproducibility of our triplicates, we used a spectral counting method known as normalized spectral abundance factor (NSAF).[9] This method is a label-free quantitation method used to determine the relative abundance of proteins in complex samples. NSAF quantification takes advantage of the fact that longer proteins have more spectra available to be identified. Thus, raw MS spectral data can be normalized by taking into account protein length. Here, we used the NSAF to analyze the abundance of proteins caught in our various samples and plotted correlation graphs (Figure 3B) to test the reproducibility between replicates of our GLU/PFA samples (a) or our PFA/DTBP samples (b). Two representative plots show that there is a significant high correlation between replicates in both groups. On average, we obtained a Spearman CC = 0.611; p < 0.0001 for the triplicate GLU/PFA samples and a Spearman CC = 0.674; p < 0.0001 for PFA/DTBP triplicate samples. This strong correlation is remarkable given the heterogeneity of our cells (Figure 1) and the small sample sizes used. These data demonstrate the precision of our LCM method in accurately isolating 1000 cells and shows that 1000 cells can give highly reproducible MS data.
Overall, we were able to identify a total of 1192 unique proteins from 1000 cells fixed with PFA/DTBP and 423 from 1000 cells fixed with GLU/PFA. Venn diagrams show the overlap of the triplicate GLU/PFA samples (Figure 3Ca) or PFA/DTBP (Figure 3Cb). Proteins identified in only one of the three samples were ignored. In total, 576 proteins were shared in at least two PFA/DTBP samples, and 159 proteins were shared in at least two GLU/PFA samples. To make sure that the differences between the PFA/DTBP and GLU/PFA samples were not sample collection related, we compared the two lists of identified proteins for their overlap. In total, 130 proteins were found to overlap (Figure 3D) which yields a 33-fold increase (p = 1.709E−185) over what would be expected from a random sample, meaning that while GLU/PFA yields less protein hits than PFA/DTBP, LCM sample collection is highly specific.
A comparison of the number of unique proteins, unique spectra, and unique peptides obtained from PFA/DTBP vs GLU/PFA samples shows a 3.57-, 5.22-, and 5.15-fold increase, respectively (Figure 3E). More importantly, our protein identification numbers obtained with PFA/DTBP fixed samples are very close to what was recently obtained with 1000 unfixed cells isolated by flow cytometry.[2] The authors did not report the number of unique proteins identified, but reported the identification of up to 911 total proteins from 1000 cells. In our study, the most unique proteins we obtained in one trial from 1000 cells fixed with PFA/DTBP was 992. This further confirms the efficiency of our DTBP fixation/extraction method observed by gel electrophoresis (Figure 2E) and shows the improvement achieved with this new method in extracting and identifying more proteins from a very limited sample compared to GLU/PFA.
What is more, our method greatly improves upon the LCM method introduced by Brioschi et al. in 2014,[10] which isolated 1000 and 1500 “air-fixed” cells for MS analysis. Our method using PFA/DTBP increases the number of proteins identified by 4.17- and 2.92-fold over their 1000 or 1500 “air-fixed” cells, respectively (Figure 3F) and the number of peptide hits by 5.35- and 3.96-fold, respectively (Figure 3G). Thus, our 1000 cells samples outperformed the 1500 cells samples from Brioschi et al.[10] (567 vs 194 for unique protein hits and 5558 vs 1403 for unique peptide hits) (Figure 3F,G), once again demonstrating the robustness of our new LCM/MS method.
It is worth noting that this study and the other two microproteomic studies cited above[2,10] have used different cell types. However, a study on 11 cell lines has previously shown that while the abundance of proteins within cell types can vary, the number of proteins identified by MS were remarkably similar between the various cell types.[11] In addition, Brioschi et al. used macrophages[10] with an average cell diameter of 20–80 μm,[12] while the cells we used are smaller with an average diameter of 15 μm,[13] suggesting that our data might be underrepresented compared to Brioschi et al.’s. Therefore, it suggests that the increase in the number of protein identified in our study results from the improvement in our fixation/extraction protocol rather than the cells used. Interestingly, Brioschi et al. used “air-fixed cells,”[10] and we show here that protein degradation can be affected by letting the samples air-dry even after fixation (Figure 2G). Thus, the use of “air-fixation” might explain their lower protein identification numbers. Our data were remarkably close to the data obtained in unfixed cells[2] and increasing the sample sizes to 1500[10] still resulted in a lower identification of unique proteins and peptide hits compared to our data collected from 1000 cells (Figure 3F,G). This suggests that the differences observed between these studies are the results of differences in the work flow, not the cell types used. Finally, the striking results obtained with PFA/DTBP samples compared to GLU/PFA samples that are within the range of those obtained by Brioschi et al. (Figure 3F,G) further highlight the improvement achieved with our new LCM/MS method.
Overall, we have described a greatly improved versatile methodology specifically suited for microproteomic analyses. It allows for the selection of specific cells within a larger cell population without restrictions. The identification can be based on cell morphology, or antibody labeling. Our new fixation and extraction protocols increase the sensitivity of our method, almost to the level of unfixed cells, without the constraints imposed by working with living cells[2] and allows for a greater protein recovery and identification by MS than any other currently available LCM/MS fixation methods.
Supplementary Material
Acknowledgements
A.G. and S.K.K. contributed equally to this work. This work was supported by a 2014 CSUPERB New Investigator Grant and the National Institute of General Medical Sciences of the National Institutes of Health under Award Number SC2GM111144 awarded to K.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (http://mass-spec.stanford.edu) for the MS analyses. Special thanks to Dr. Christopher M. Adams and Dr. Ryan Leib for their assistance with this project. A.G. was supported by the Fundación Alfonso Martín Escudero.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Gutstein HB, Morris JS, Annangudi SP, Sweedler JV, Mass Spectrom. Rev 2008, 27, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kasuga K, Katoh Y, Nagase K, Igarashi K, Proteomics 2017, 17, 10.1002/pmic.201600420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schütze K, Niyaz Y, Stich M, Buchstaller A, Methods Cell Biol 2007, 82, 649. [DOI] [PubMed] [Google Scholar]
- [4].Shi SR, Liu C, Balgley BM, Lee C, Taylor CR, J. Histochem Cytochem 2006, 54, 739. [DOI] [PubMed] [Google Scholar]
- [5].Charulatha V, Rajaram A, J. Biomed. Mater. Res 2001, 54, 122. [DOI] [PubMed] [Google Scholar]
- [6].Ono T, Yamamoto N, Yasuda K, Okajimas Folia Anat. Jpn 1976, 53, 199. [DOI] [PubMed] [Google Scholar]
- [7].Gousset K, Marzo L, Commere PH, Zurzolo C, J. Cell Sci 2013, 126, 4424. [DOI] [PubMed] [Google Scholar]
- [8].Scheler C, Lamer S, Pan Z, Li XP, Salnikow J, Jungblut P, Electrophoresis 1998, 19, 918. [DOI] [PubMed] [Google Scholar]
- [9].Arike L, Peil L, Methods Mol. Biol 2014, 1156, 213. [DOI] [PubMed] [Google Scholar]
- [10].Brioschi M, Eligini S, Crisci M, Fiorelli S, Tremoli E,Colli S, Banfi C, Anal. Bioanal. Chem 2014, 406, 2817. [DOI] [PubMed] [Google Scholar]
- [11].Geiger T, Wehner A, Schaab C, Cox J, Mann M, Mol. Cell Proteomics 2012, 11, M111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Freitas RA, Nanomedicine, Volume I: Basic Capabilities, Landes Bio-science, Austin, TX 1999.
- [13].Qi Y, Wang JK, McMillian M, Chikaraishi DM, J. Neurosci 1997, 17, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.