Abstract
The blastocyst stage and the subsequent implantation are critical for a successful pregnancy, yet are challenging to study in vivo. In this issue of Stem Cell Reports, Kime et al. (2019) describe a novel way to generate blastocyst-like structures only from pluripotent stem cells. These structures mimic several aspects of the early embryo, offering a new promising tool to study this stage.
The blastocyst stage and the subsequent implantation are critical for a successful pregnancy, yet are challenging to study in vivo. In this issue of Stem Cell Reports, Kime et al. (2019) describe a novel way to generate blastocyst-like structures only from pluripotent stem cells. These structures mimic several aspects of the early embryo, offering a new promising tool to study this stage.
Main Text
Very little is known about the very first steps of human embryonic development, due to the small size of the embryo and limited accessibility in the womb. Several factors may affect implantation, and problems during implantation or shortly after it are the main reasons for pregnancy loss at early stages (Norwitz et al., 2001). Generating large numbers of isogenic, accessible embryos of the relevant stage is not possible through in vivo or IVF approaches, and an alternative approach may provide a valuable system for studying early pregnancy problems.
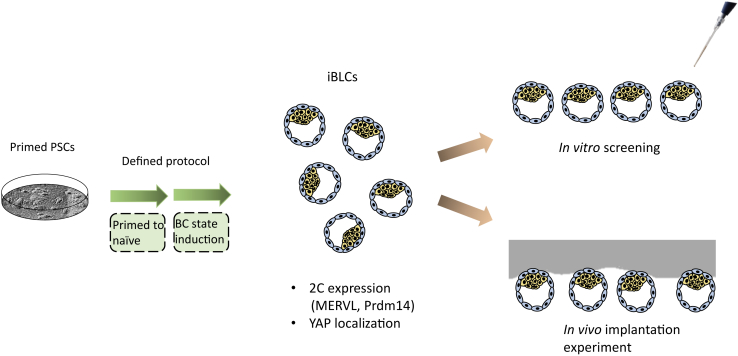
The relevant stage of embryonic development is the blastocyst (BC), a 32-cell stage shortly after the embryo loses its “ball of cells” shape and forms a cavity (called the blastocoel), in which a small number of inner cell mass (ICM) cells, which will form the embryo proper, are enveloped by trophectoderm (TE) cells, which will later contribute to extra-embryonic tissues (fetal placenta and membranes). A method to generate blastocyst-like structures from a combination of TE stem cells and ES cells has been recently introduced (Rivron et al., 2018). In this issue of Stem Cell Reports, Kime et al. (2019) present a novel system to generate blastocyst-like structures starting from just one type of cells—mouse PSCs (Figure 1). They demonstrate how these constructs (termed iBLCs) morphologically resemble blastocysts (BCs), including cavity formation and the separation to outer TE-like cells and inner ICM-like cells. Their iBLCs implant to a certain efficiency in pseudo-pregnant mice, inducing physiological changes in the uterus, though no further development of the embryo is demonstrated. Some molecular aspects, such as YAP subcellular localization and expression of totipotency markers, are demonstrated to resemble the BC.
Figure 1.
A Method for Deriving Isogenic BC-like Structures from Pluripotent Stem Cells
Embryonic stem cells are subjected to a two-step protocol that results in induced blastocyst-like structures (iBLCs). These can be later used both for studying implantation in vivo or further study of the BC stage in vitro.
The formation of iBLCs in this system goes through the generation of progenitor cells (termed iBLC-PCs), that express the 2C stage marker MERVL. MERVL is a retroviral transposable element that is active in vivo only at the 2C stage, and so its expression is indicative of the cells’ developmental stage (Macfarlan et al., 2012). Kime et al. (2016) had previously shown that such a naïve state can be driven from primed PSCs using a combination of ascorbic acid and lysophosphatidic acid. In conjunction with LIF, BMP, and inhibition of Activin, these conditions robustly drive primed to naïve conversion as the first step of the iBLC generation process. They had also noticed that blastocyst-like structures occasionally appear in these conditions, leading to the development of the current protocol. The protocol now leverages the natural tendency of PSC to spontaneously enter and exit the 2C state (as demonstrated in Macfarlan et al., 2012), as these cells have the potential to progress to either TE or ICM. The protocol provides these cells with suspension conditions that allow the formation of BC-like structures, with proper expression of some TE markers (such as Cdx2) and ICM markers (such as Oct4).
Interestingly, the transcription factor Prdm14 is found to be essential for the generation of iBLCs. Prdm14 is known to inhibit lineage-specifying genes (such as Fgfr1/2) and de novo DNMTs through recruitment of PRC2, thus promoting naïve pluripotency (Nakaki and Saitou, 2014). In vivo, it has been shown to be expressed asymmetrically at the 4-cell stage, directing cells at this stage toward the ICM fate (Burton et al., 2013). Understanding at exactly which stage Prdm14 knock-down hinders the generation of iBLCs may help establish its roles in the path to BC formation. This result also highlights a potential use for iBLCs as an in vitro tool for studying the role of different genes during early lineage allocation, such as the TE versus ICM decision.
In the preimplantation embryo, YAP and its homolog TAZ are expressed uniformly in all cells but are distinct in their intracellular distribution. In the outer cells (fated to become TE) they are nuclear, while in inner cells (fated to become ICM) they are cytoplasmic. This differential localization is critical for the first fate decision in embryonic development and subsequently facilitates the activation of Cdx2 at the TE cells (Niwa et al., 2005). The current protocol produces a mixture of iBLC-PCs in terms of YAP localization: some iBLC-PCs homogenously express YAP, resembling an early embryo stage before compaction and polarization, while others show nuclear localization of YAP in outer cells and cytoplasmic localization in inner cells, resembling the polarized morula. In later iBLCs, YAP localization is differential as in BCs, with nuclear localization appearing only in the TE-like enveloping cells. It would be interesting to further quantify these localization dynamics, for example using a live YAP localization marker. This would allow researchers to test whether non-polarized iBLC-PCs later progress to a polarized pattern, and whether only polarized ones proceed to generate the correct BC-like YAP pattern as they mature into iBLCs.
A close comparison to similar systems (such as blastoids) may be instructive for future improvements. Currently, blastoids (formed by combining TS and ES cells) seem to develop further at the uterus and undergo implantation (Rivron et al., 2018). Can this point at partial differentiation of either the ICM or the TE part of the iBLC? Another pragmatic point is the throughput of this system. Here there is a manual selection step, which puts a limit on the throughput. Perhaps future improvements of the current protocol, that can be based on marker sorting, may automate these steps. In addition, several issues remain to be addressed. How are the iBLCs formed? Do different 2C stage cells turn into either a TE or an ICM path, and then spontaneously self-assemble at the right proportion? Or is it perhaps that the choice between these two fates is obtained only after aggregation, with paracrine signals affecting their segregation? Live imaging with TE/ICM markers at the formative stages (day 6–7 of the protocol) may answer these questions. How far can development after re-transfer be pushed? Currently, iBLCs fail into embryonic resorption after implantation. What is missing to progress further? A more comprehensive expression profile of iBLC and its constituent cells at different stages may help answer these questions.
One clear future use for iBLCs is their formation from iPS cells derived from individuals with repeated failures of early pregnancy. Such a system can be used to study infertility and genetic background effects on implantation and early development. In a broader perspective, the iBLC system joins a recently expanding collection of 3D in vitro models of early embryogenesis, alongside blastoids, ETS/ETX embryos, gastruloids, and embryoid bodies (Beccari et al., 2018, Boxman et al., 2016, Harrison et al., 2017, Rivron et al., 2018). These open exciting opportunities to study different developmental stages, with greater accessibility and amenability to genetic and external condition manipulation than their in vivo counterparts.
References
- Beccari L., Moris N., Girgin M., Turner D.A., Baillie-Johnson P., Cossy A.C., Lutolf M.P., Duboule D., Arias A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature. 2018;562:272–276. doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- Boxman J., Sagy N., Achanta S., Vadigepalli R., Nachman I. Integrated live imaging and molecular profiling of embryoid bodies reveals a synchronized progression of early differentiation. Sci. Rep. 2016;6:31623. doi: 10.1038/srep31623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A., Muller J., Tu S., Padilla-Longoria P., Guccione E., Torres-Padilla M.E. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013;5:687–701. doi: 10.1016/j.celrep.2013.09.044. [DOI] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:356. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Kime C., Sakaki-Yumoto M., Goodrich L., Hayashi Y., Sami S., Derynck R., Asahi M., Panning B., Yamanaka S., Tomoda K. Autotaxin-mediated lipid signaling intersects with LIF and BMP signaling to promote the naive pluripotency transcription factor program. Proc. Natl. Acad. Sci. USA. 2016;113:12478–12483. doi: 10.1073/pnas.1608564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime C., Kiyonari H., Ohtsuka S., Kohbayashi E., Asahi M., Yamanaka S., Takahashi M., Tomoda K. Induced 2C expression and implantation-competent blastocyst-like cysts from primed pluripotent stem cells. Stem Cell Reports. 2019;13:485–498. doi: 10.1016/j.stemcr.2019.07.011. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaki F., Saitou M. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem. Sci. 2014;39:289–298. doi: 10.1016/j.tibs.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Norwitz E.R., Schust D.J., Fisher S.J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.C., Korving J., Vivié J., Truckenmüller R.K., van Oudenaarden A., van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]