Abstract
mTORC2 plays critical roles in metabolism, cell survival, and actin cytoskeletal dynamics via phosphorylation of AKT. Despite its importance to biology and medicine, it is unclear how mTORC2-mediated AKT phosphorylation is controlled. Here, we identify an unforeseen principle by which a GDP-bound form of the conserved small G protein Rho GTPase directly activates mTORC2 in AKT phosphorylation in social amoebae Dictyostelium cells. Using biochemical reconstitution with purified proteins, we demonstrate that Rho-GDP promotes AKT phosphorylation by assembling the supercomplex with Ras-GTP and mTORC2. This supercomplex formation is controlled by chemoattractant-induced phosphorylation of Rho-GDP at serine 192 by GSK-3. Furthermore, Rho-GDP rescued defects in both mTORC2-mediated AKT phosphorylation and directed cell migration in Rho-null cells in a manner dependent on phosphorylation of serine 192. Thus, in contrast to the prevailing view that GDP-bound forms of G proteins are inactive, our study reveals that mTORC2-AKT signaling is activated by Rho-GDP.
Introduction
G proteins are molecular switches that control a wide range of biological processes, including signal transduction, protein and membrane trafficking, cytoskeletal and organelle remodeling, and cell growth and proliferation 1-6. The G protein family consists of two major groups: small monomeric GTPases and heterotrimeric G proteins. Both types of G proteins are regulated by a GDP/GTP cycle in which GTP binding activates them 1, 4, 7-9. The majority of G proteins are associated with GDP in cells and are considered to be in an inactive form.
G proteins control chemotaxis, directed cell migration toward chemoattractants. Upon ligand binding, chemoattractant receptors, such as G-protein coupled receptors (GPCRs) and receptor tyrosine kinases, activate small GTPase Ras by generating its GTP bound form. The activated Ras GTPase in turn leads to phosphorylation of a critical serine/threonine kinase, AKT (also known as protein kinase B), to regulate the actin cytoskeleton 10-15. This AKT phosphorylation is controlled by two evolutionarily-related kinases, Tor (target of rapamycin) and PI3K (phosphoinositide 3-kinase) 14. One of the TOR-containing serine/threonine kinase complexes, mTOR complex 2 (mTORC2), directly phosphorylates AKT 16-18.
This mTORC2-mediated AKT phosphorylation requires the recruitment of AKT to the plasma membrane. Upon chemoattractant stimulation, PI3K generates phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and PIP3 recruits AKT to the plasma membrane through its interaction with a PIP3-binding PH domain of AKT 19-21. Ras-GTP directly interacts with PI3K through its Ras binding domain and that this interaction stimulates PIP3 production by PI3K. It has been shown that Ras-GTP also interacts with mTORC2 and stimulates AKT phosphorylation when overexpressed in cells 22-24; however, how Ras regulates the enzymatic activity of mTORC2 against AKT is unknown. To address this critical question, it is essential to determine the function of Ras in mTORC2 activity by separating its function in the PI3K pathway.
In addition, PIP3 also stimulates the reorganization of the actin cytoskeleton, likely through activation of members of the Rho family GTPases, Rac and Rho 25-28. Rac controls actin polymerization and network formation at the front of migrating cells that extend pseudopods. Coordinating with pseudopod extension, Rho regulates actomyosin contraction at the rear end of the cell to move the cytoplasm forward 15. However, it is unknown whether Rac and Rho have a role in controlling mTORC2 in addition to their known roles downstream of mTORC2 and PI3K signaling
In the current study, we found that Rho forms a signaling supercomplex with Ras and mTORC2 and activates mTORC2-mediated AKT phosphorylation in vivo and in vitro. A critical regulatory step in this activation mechanism is not GTP binding to Rho, rather chemoattractant-stimulated serine phosphorylation of Rho-GDP by glycogen synthase kinase-3 (GSK-3). Our findings provide a principle that controls G proteins through a combination of GDP-binding and phosphorylation and identifies a direct activation mechanism of mTORC2 toward AKT.
Results
RacE-GDP promotes directed cell migration
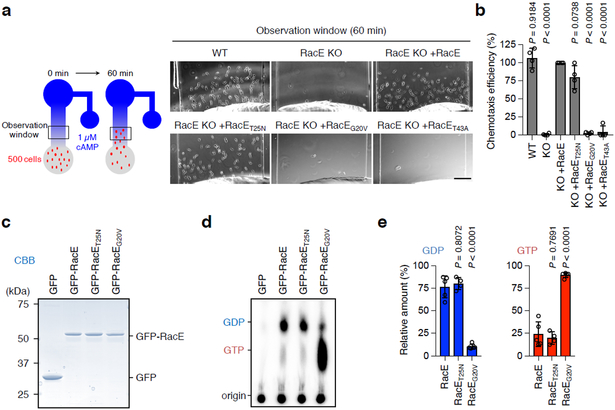
In Dictyostlium, all 20 members of the Rho family GTPases have been named Rac (Fig. S1a). We have previously shown that the closest homolog of human RhoA, RacE 29 (Fig. S1b), is required for directed cell migration toward the chemoattractant cAMP, but not for random cell migration, in Dictyostelium cells (Fig. 1a, 1b and S2) 30, 31. To investigate the mechanism by which RacE controls chemotaxis, we expressed GFP fused to WT RacE, GDP-bound RacET25N or GTP-bound RacEG20V in RacE-knockout (KO) cells, which normally grow on solid substrates 30, 31. We examined the chemotactic cell migration toward extracellular cAMP using a microfluidic chamber (Fig. 1a). Surprisingly, GDP-bound RacET25N almost completely rescued the migration defects in RacE-KO cells, similar to WT RacE (Fig. 1a and 1b), whereas GTP-bound RacEG20V or effector domain defective RacET43A failed to do so. Metabolic labeling using P32 showed that WT and RacET25N are mainly associated with GDP, while RacEG20V is predominantly bound to GTP (Fig. 1c-e). These data suggest that RacE-GDP mediates directed cell migration.
Figure 1. RacE-GDP functions in directed cell migration.
a-b, Cell migration toward the chemoattractant cAMP was analyzed in WT and RacE-KO Dictyostelium cells carrying WT RacE, GDP-bound RacET25N, GTP-bound RacEG20V or effector domain-defective RacET43A for 60 min in a microfluidic chamber in a. Bar, 100 μm. Chemotaxis efficiency was quantified by measuring the number of cells that moved toward a higher concentration of cAMP (observation window) in b. The chemotaxis efficiency in RacE-KO cells expressing WT RacE (KO+RacE) was set 100% (n = 4 independent experiments). Values are means ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison of KO+RacE with others. c-e, Guanine nucleotide binding to RacE. Differentiated Dictyostelium cells carrying GFP, GFP-RacE, GFP-RacET25N or GFP-RacEG20V were metabolically labeled using P32 for 1 h. GFP fusion proteins were immunopurified using GFP-Trap beads and analyzed using SDS-PAGE and CBB staining in c. Bound guanine nucleotides were analyzed by thin layer chromatography and phosphoimaging in d. Quantification of GDP and GTP (n=5 independent experiments for GFP-RacE and GFP-RacEG20V and n=4 independent experiments for GFP-RacET25N) in e. Values are means ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison of GFP-RacE with others.
RacE-GDP interacts with mTORC2
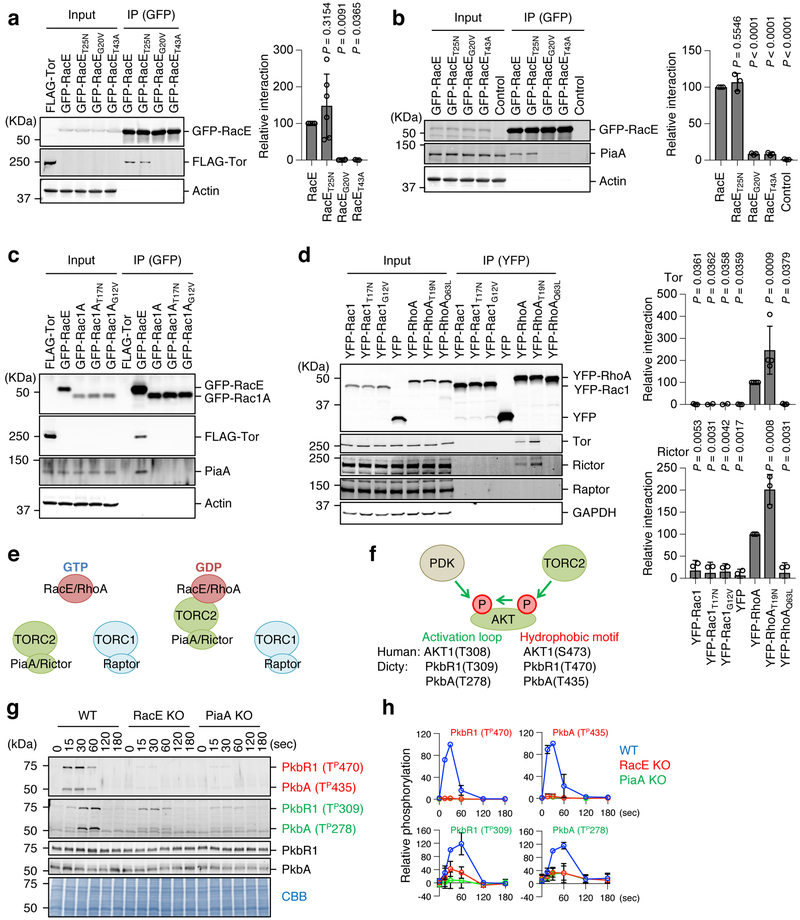
Chemoattractants stimulate signal transduction pathways involving mTORC2 and AKT via GPCRs in Dictyostelium cells and human neutrophils 15, 18, 32-35. Using proteomic analysis of RacE binding proteins, we identified two subunits of mTORC2, Tor (Tor homolog) and PiaA (rictor homolog) (Fig. S1c and S3, Table S1 and S2) 15, 16, 36, 37. Subsequent co-immunoprecipitation assays showed that both Tor and PiaA specifically co-precipitated WT RacE or GDP-bound RacET25N but not GTP-bound RacEG20V or effector domain defective RacET43A (Fig. 2a and 2b). As described above, WT RacE is primarily GDP-bound; therefore, RacE-GDP specifically interacts with mTORC2, and this interaction requires the effector domain. Unlike RacE, a homolog of human Rac1, Dictyostelium Rac1A (Fig. S1b), failed to interact with mTORC2 regardless of its guanine nucleotide binding status (Fig. 2c). To determine whether GDP-dependent interactions between Rho and mTORC2 are conserved in humans, we performed co-immunoprecipitation of YFP-RhoA and YFP-Rac1 with mTORC1 and 2 in human embryonic kidney (HEK) 293T cells. A GDP-bound RhoAT19N most strongly bound two subunits of mTORC2, Tor and Rictor, but not a subunit of mTORC1, Raptor, (Fig. 2d and 2e).
Figure 2. RacE-GDP specifically interacts with mTORC2.
a, Dictyostelium cell lysates carrying GFP fused to the indicated forms of RacE were incubated with cell lysates carrying FLAG-Tor and subjected to immunoprecipitation with GFP-Trap. Quantification of interaction is shown. The band intensity of FLAG-Tor in immunoprecipitates of cells expressing WT RacE was set 100% (n=6, 6, 6 and 3 independent experiments for RacE, RacET25N, RacEG20V and RacET43A, respectively). Values are average ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison between RacE and others. b, Dictyostelium cell lysates carrying the indicated GFP-RacE were subject to immunoprecipitation with GFP-Trap to analyze its association with endogenous PiaA. The band intensity of PiaA in immunoprecipitates of cells expressing WT RacE was set 100% (n = 3 independent experiments). Values are average ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison betwen RacE with others. c, Dictyostelium cell lysates carrying GFP fused to the indicated forms of Rac1A and RacE were incubated with cell lysates carrying FLAG-Tor and subjected to immunoprecipitation with GFP-Trap. Experiment was repeated independently three times with similar results. d, HEK293T cells were transfected with YFP fused to the indicated constructs of human Rac1 and RhoA and subjected to immunoprecipitation using GFP-Trap. The band intensities of Tor and rictor in immunoprecipitates of cells expressing WT RhoA was set 100% (n = 3 and n = 4 independent experiments for Tor and Rictor, respectively). Values are average ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison between YFP-RhoA and others. e, Summary of the data. f, AKTs are phosphorylated in the hydrophobic motif by mTORC2 and in the activation loop by PDK. g and h, WT, RacE-KO, and PiaA-KO Dictyostelium cells were stimulated with the chemoattractant cAMP (1 μM). (n = 3 independent experiments). The total amounts of two AKT homologs (PkbR1 and PkbA) and their phosphorylation (Red: hydrophobic motif, Green: activation loop) were analyzed by immunoblotting. PVDF membranes were stained with CBB as loading controls. The band intensity of phosphorylated AKTs was quantified in h: WT cells at 30 s were set at 100%. Values are average ± SD.
mTORC2-mediated AKT phosphorylation requires RacE-GDP in cells
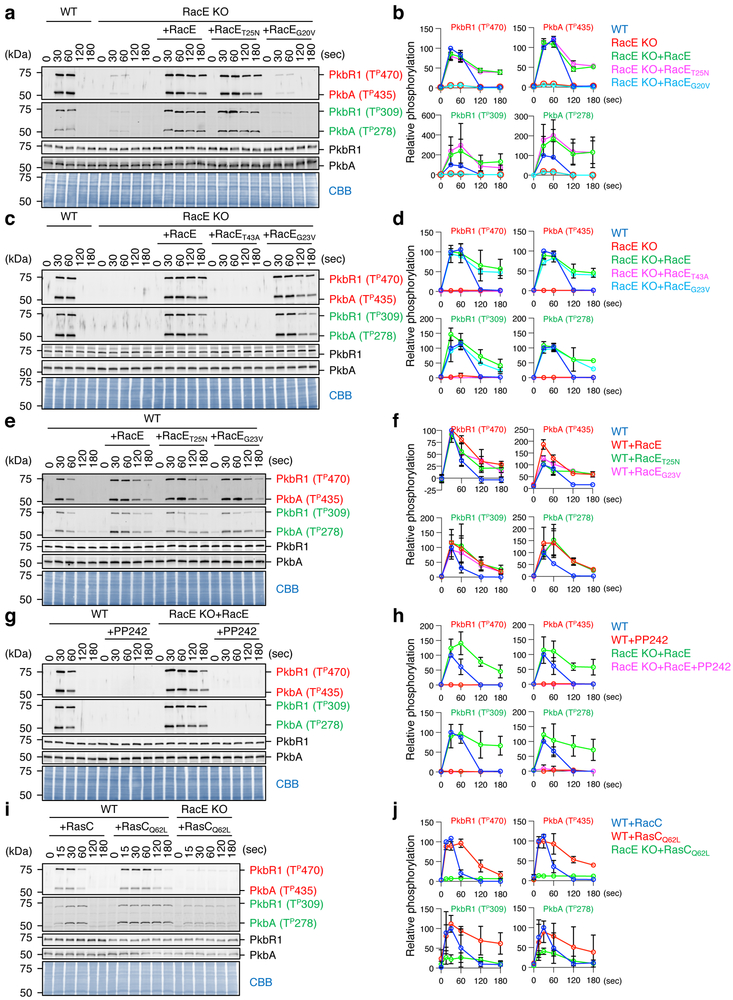
To determine the function of RacE in mTORC2 signaling in cells, we first examined the impact of RacE loss on the phosphorylation of AKT, a major substrate of mTORC2 15, 19. Two AKT homologs (PkbA and PkbR1) are phosphorylated by mTORC2 in the hydrophobic motif and by PDK in the activation loop (Fig. 2f and S4) 11, 19, 38. As previously reported, both sites were transiently phosphorylated in response to GPCR activation by the chemoattractant cAMP in WT cells, but not in cells lacking the Rictor homolog, PiaA (Fig. 2g and 2h) 39. Similar to PiaA-KO cells, chemoattractant-induced AKT phosphorylation was greatly decreased in RacE-KO cells (Fig. 2g and 2h). The total amounts of endogenous PkbR1 and PkbA were not changed in PiaA-KO or RacE-KO cells in the presence or absence of chemoattractant simulation (Fig. 2g). Second, we expressed WT RacE and various RacE mutants in RacE-KO cells and then stimulated these cells with cAMP. Intriguingly, WT RacE and GDP-bound RacET25N, but not GTP-bound RacEG20V or effector domain-defective RacET43A, restored mTORC2-mediated AKT phosphorylation in response to chemoattractants (Fig. 3a-d). It has been shown that a mutation (G17V) in human RhoA, which dissociates GTP from RhoA, is a prevalent driver mutation in T-cell lymphoma 40, 41; in cells carrying RhoAG17V, AKT phosphorylation is increased. To test whether an equivalent mutation promotes AKT phosphorylation in Dictyostelium cells, we expressed RacEG23V in WT and RacE-KO cells and found that RacEG23V increases the phosphorylation of PkbR1 and PkbA in both cell types (Fig. 3c-f). The expression levels of the GFP-RacE proteins were comparable in cells (Fig. S5). Third, the mTORC2 inhibitor PP242 completely blocked cAMP-induced AKT phosphorylation in WT cells and RacE-KO cells expressing WT RacE (Fig. 3g and 3h). Fourth, AKT phosphorylation requires RasC and is increased by ectopic expression of GTP-bound RasCQ62L 23. We found that RacE is necessary for this RasCQ62L-stimulated AKT phosphorylation (Fig. 3i and 3j). Fifth, consistent with the function of RacE-GDP and RasC-GTP in chemotactic signaling, metabolic labeling with P32 showed that GDP-binding to RacE and GTP-binding to RasC significantly increased in response to cAMP (Fig. S6).
Figure 3. RacE-GDP promotes chemoattractant-induced, mTORC2-mediated AKT phosphorylation in cells.
The indicated Dictyostelium cell lines were stimulated with the chemoattractant cAMP (1 μM). a-f, WT cells and RacE-KO cells expressing different GFP-RacE constructs were analyzed. g and h, WT cells and RacE-KO cells expressing GFP-RacE were pretreated with 0.5 μM of the mTORC2 inhibitor PP242 for 10 min and then stimulated with cAMP. i and j, WT and RacE-KO cells expressing FLAG-tagged RasC or GTP-bound RasCQ62L were analyzed. a-j, Total amounts of two AKT homologs (PKBR1 and PKBA) and their phosphorylation (Red: hydrophobic motif, Green: activation loop) were analyzed by immunoblotting. PVDF membranes were stained with CBB as loading controls in a, c, e, g, and i. The band intensity of phosphorylated AKTs was quantified in b, d, f, h, and j: WT cells at 30 s (b, d, f and h) and WT cells expressing RasC at 30 s (j) were set at 100%. Values are average ± SD (n = 3 independent experiments).
GSK-3 phosphorylates RacE at serine 192 in response to chemotactic stimulation
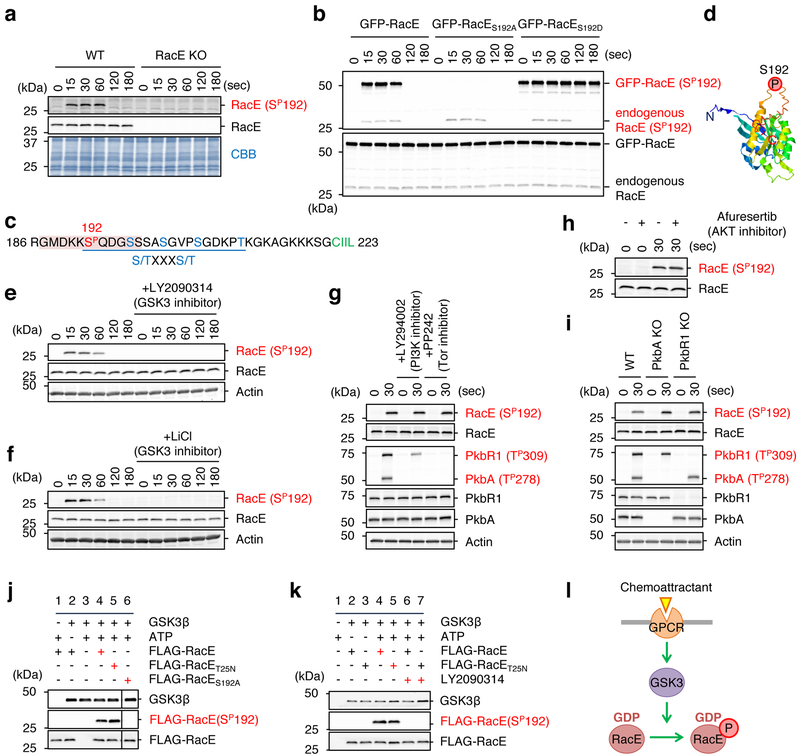
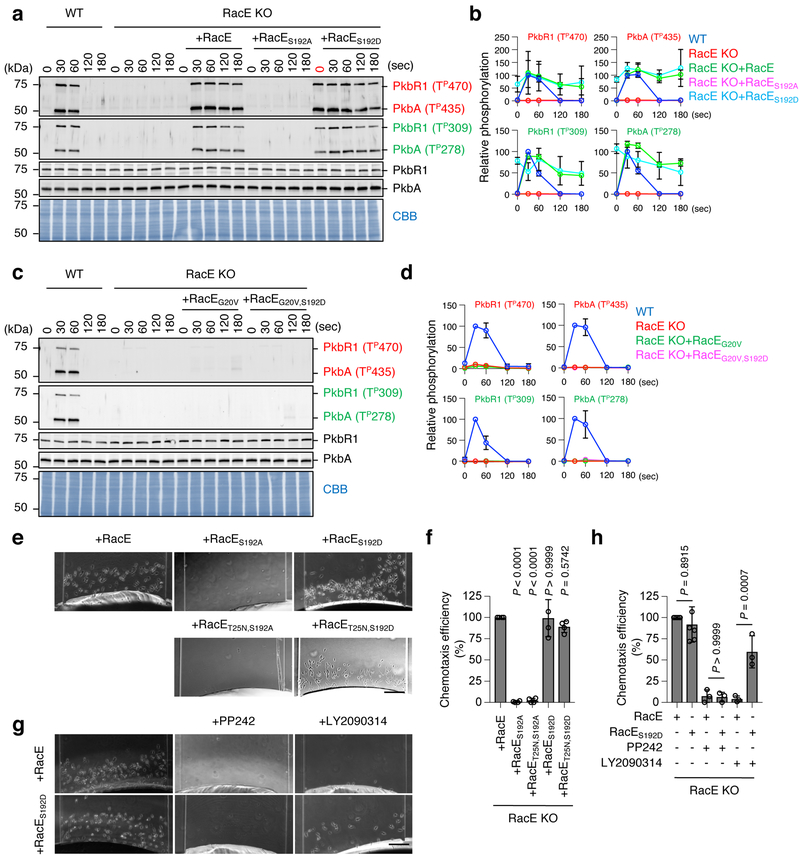
Previous phosphoproteomics has suggested that chemoattractant stimulation induces RacE phosphorylation at serine 192 (Ser192) in Dictyostelium cells 42. Raising antibodies to Ser192 phospho-RacE, we found that RacE indeed undergoes transient phosphorylation upon chemoattractant stimulation (Fig. 4a). To confirm the specificity of our Ser192 phospho-RacE antibodies, we expressed GFP-RacE, phospho-defective GFP-RacES192A or phospho-mimetic GFP-RacES192D in WT cells and stimulated these cells with the chemoattractant cAMP. We found that GFP-RacE was transiently phosphorylated at Ser192, similarly to endogenous RacE (Fig. 4b). In contrast, phospho-defective GFP-RacES192A was not detected by anti-phospho-RacE(Ser192) antibodies (Fig. 4b). As a positive control, GFP-RacES192D was detected by anti-phospho-RacE(Ser192) antibodies even in the absence of the chemoattractant stimulation.
Figure 4. GSK-3 phosphorylates RacE at Ser192 in response to the chemoattractant.
a, WT and RacE-KO cells were stimulated with the chemoattractant cAMP (1 μM). Total amounts of RacE and its phosphorylation at Ser192 were analyzed by immunoblotting with antibodies to RacE and phospho-RacE(Ser192). b, WT cells expressing GFP fused to WT RacE, phospho-defective RacES192A or phospho-mimetic RacES192D were stimulated with cAMP. Whole cell lysates prepared at the indicated time points were analyzed by immunoblotting with antibodies to RacE and phospho-RacE(S192). c, The amino acid sequence in the vicinity of the phosphorylation site (Ser192, red) of RacE. A consensus motif for GSK-3 phosphorylation — a cluster of (S/TXXXS/T) — is underlined. A phosphopeptide used to raise anti-phospho-RacE (S192P) is highlighted. d, The location of serine 192 in a modeled RacE 3-D structure. (e–h) WT cells were treated with inhibitors to GSK-3 (250 nM LY2090314 in e and 10 mM lithium in f), PI3K (250 μM LY294002 in g), mTORC2 (0.5 μM PP242 in g), or AKT (5 μM afuresertib in h) for 10 min. Cells were then stimulated by the chemoattractant cAMP for the indicated amounts of time. Whole cell lysates were analyzed by immunoblotting with antibodies to RacE and phospho-RacE(Ser192). i, WT and cells lacking AKT (PkbA-KO or PkbR1-KO) were stimulated with cAMP for 30 s. Whole cell lysates were analyzed by immunoblotting with the indicated antibodies. j, Purified human GSK-3β was incubated with purified WT, GDP-bound RacET25N or phospho-defective RacES192A for 15 min. Ser192 phosphorylation of RacE was tested by immunoblotting. k, WT or GDP-bound RacET25N was mixed with GSK-3β in the presence or absence of the GSK-3 inhibitor LY2090314 (10 nM) and examined for Ser192 phosphorylation using immunoblotting. l, Summary of the data. Experiments were repeated independently three times with similar results in a, b and e-k.
To identify a protein kinase that phosphorylates RacE in response to chemoattractants, we first analyzed the amino acid sequence around Ser192 and found a GSK-3 substrate motif (Fig. 4c). A modeled structure of RacE based on human RhoA suggested that Ser192 is located in a C-terminal unstructured loop of RacE (Fig. 4d). To determine whether the Ser192 phosphorylation depends on GSK-3 in cells, we pre-treated Dictyostelium cells with LY2090314 and lithium, two structurally-distinct inhibitors specific to GSK-3, and stimulated these cells with the chemoattractant cAMP. Immunoblotting showed that chemoattractant-induced Ser192 phosphorylation of RacE was completely blocked (Fig. 4e and 4f). Conversely, other inhibitors against PI3K (LY294002) or mTORC2 (PP242) did not inhibit this RacE phosphorylation (Fig. 4g). We also found no inhibition of the RacE phosphorylation in WT cells treated with an AKT inhibitor (afuresertib) (Fig. 4h) or cells lacking PkbA or PkbR1 (Fig. 4i). To ask if GSK-3 directly phosphorylates RacE at Ser192, we incubated purified human GSK-3β with purified FLAG-RacE, FLAG-RacET25N (GDP-bound) or FLAG-RacES192A (phospho-defective). We found that GSK-3β phosphorylates FLAG-RacE and FLAG-RacET25N, but not FLAG-RacES192A, on Ser192 in vitro (Fig. 4j). This in vitro phosphorylation of RacE by purified GSK-3β was inhibited by the GSK-3 inhibitor LY2090314 (Fig. 4k). These data suggest that GSK-3 directly phosphorylates Ser192 in RacE in response to chemotactic stimulation (Fig. 4l).
Chemoattractant-induced phosphorylation of RacE-GDP at Ser192 controls mTORC2-mediated AKT phosphorylation and directed cell migration
When ectopically expressed in RacE-KO cells, phospho-defective RacES192A did not induce AKT phosphorylation in response to chemoattractant stimulation (Fig. 5a and 5b). Conversely, phospho-mimetic RacE192D increased AKT phosphorylation even without stimulation (0 s in Fig. 5a and 5b). Expression levels of these GFP-RacE proteins were comparable in cells (Fig. S5). GTP-bound RacEG20V remained inactive even when the phospho-mimetic S192D mutation was combined (Fig. 5c and 5d), suggesting that phosphorylation at Ser192 of RacE-GDP is crucial for the activation of mTORC2.
Figure 5. Ser192 phosphorylated RacE-GDP activates mTORC2.
a-d, WT cells and RacE-KO cells carrying different GFP-RacE constructs were stimulated by cAMP and analyzed for AKT phosphorylation. b and d, Quantification of band intensity (n = 3 independent experiments): WT cells at 30 s were set at 100%. e-h, Directed cell migration toward the chemoattractant cAMP was analyzed in RacE-KO cells carrying different GFP-RacE constructs in a microfluidic chamber in the presence or absence of inhibitors to mTORC2 (0.5 μM PP242) or GSK-3 (250 nM LY2090314). Bar, 100 μm. Chemotaxis efficiency was quantified in f and h for e and g, respectively. The chemotaxis efficiency in RacE-KO cells expressing WT RacE was set 100%. Values are average ± SD. Significance was calculated using ANOVA with post-hoc Tukey. p values are shown for comparison between RacE and others in f (n=4 independent experiments) and between untreated RacE and RacES129D, or between PP242-treated RacE and RacES129D, or LY2090314-treated RacE and RacES129D in f (n=6, 3, 3, 6, 3 and 3 independent experiments for untreated RacE, untreated RacES192D, PP242-treated RacE, PP242-treated RacES192D, LY2090314-treated RacE and LY2090314-treated RacES192D).
Both phospho-mimetic RacE192D and phospho-mimetic GDP-bound RacET25A,S192D rescued defects in directed cell migration toward the chemoattractant cAMP in RacE-KO cells (Fig. 5e and 5f). In contrast, the phospho-defective S192A mutation blocked the ability of WT RacE and GDP-bound RacET25N to reverse the cell migration defect (Fig. 5e and 5f). Furthermore, the mTORC2 inhibitor PP242 blocked the cell migration of RacE-KO cells expressing WT RacE or RacES192D while the GSK-3 inhibitor LY2090314 only blocked the migration of RacE-KO cells expressing WT RacE (Fig. 5g and 5h). Therefore, chemoattractant stimulation induces GSK-3-mediated S192 phosphorylation of RacE-GDP to activate mTORC2-mediated AKT phosphorylation in directed cell migration.
Reconstitution of GDP-bound RacE-regulated mTORC2 activation in vitro
To elucidate the molecular mechanism by which RacE-GDP regulates mTORC2, we reconstituted the chemoattractant-induced activation of mTORC2 using proteins purified from Dictyostelium cells (Fig. 6). To purify mTORC2 as an active kinase complex, we immunopurified FLAG-Tor under low-salt conditions in the presence of the detergent CHAPS, as described 43, 44 (Fig. S7a-c). We used inactive human AKT as a substrate and detected its phosphorylation using anti-phospho AKT antibodies (serine 473) 43, 44. AKT was robustly phosphoryalted when incubated with mTORC2, chemoattractant-stimulated RacE, and GTP-bound RasCQ62L (Fig. 6a, lane 3). For negative controls, we omitted ATP, RacE, RasC or mTOR2 or used kinase-dead TorD2146A (Fig. 6a, lanes 2, and 4–8).
Figure 6. Phosphorylated RacE-GDP activates mTORC2 in vitro.
mTORC2-mediated AKT phosphorylation was reconstituted using purified proteins. mTORC2 (FLAG-Tor, -PiaA or -Lst8), FLAG-RacE, and FLAG-RasC/G were purified from Dictyostelium cells. +cAMP indicates that cells were stimulated by the chemoattractant for 30 s before purification of FLAG-RacE. Purified proteins were mixed in the presence or absence of ATP and human unactive AKT for 5 min at room temperature. AKT phosphorylation was analyzed by immunoblotting using anti-phospho AKT (serine 473) antibodies. a, mTORC2 phosphorylates AKT in the presence of RacE and RasC. b and c, mTORC2 activation requires PiaA and Lst8, but not the mSIN1 homolog Rip3. FLAG-PiaA or FLAG-Lst8 was added to FLAG-Tor purified from the indicated KO cell lines in b. FLAG-PiaA and/or FLAG-Lst8 were incubated with high-salt washed FLAG-Tor in c. d, mTORC2 activation requires RacE-GDP. Purified RacE was incubated with EDTA (25 mM), GTPγS (0.5 mM) or GTPγS then GDP (2.5 mM) (GTPγS → GDP) before reconstitution. e, WT RacE or GDP-bound RacET25N, but not GTP-bound RacEG20V, activates mTORC2 after the chemoattractant stimulation. f, mTORC2 activation needs RasC-GTP but not RasG-GTP. g, RacE phosphorylation controls mTORC2 activation. Phospho-mimetic mutation S192D in GDP-bound RacET25N activates mTORC2 without chemoattractant stimulation, while the phospho-defective S192A mutation blocks it. h, mTORC2 activation requires RasC-GTP. Purified RacC was incubated with EDTA followed by either GTPγS or GDP before reconstitution. i, Summary of the data. j, RacEG23V-GDP activates mTORC2. Purified RacET25N and RacEG23V were incubated with EDTA followed by GTPγS or GDP prior to reconstitution. Experiments were repeated independently three times with similar results in a-h and j.
The Tor kinase forms mTORC2 with other subunits, including Rictor, mLst8, and mSin1 (Fig. S1c) 16, 24, 37. To define the minimum set of subunits for mTORC2 activation, we purified mTORC2 from WT or cells lacking each subunit (Rictor/PiaA-KO, Lst8-KO or mSin1/Rip3-KO) using FLAG-Tor under CHAPS-containing, low-salt conditions. While mTORC2 purified from WT cells phosphorylated AKT, mTORC2 purified from any of these KO cells failed to do so (Fig. 6b, lane 1, 2, 5 and 8). We then added high-salt washed, purified FLAG-Lst8 or FLAG-PiaA to the purified mTORC2 (Fig. 6b and S7f). FLAG-Lst8 restored AKT phosphorylation when added to mTORC2 purified from Lst8-KO cells, but not Rip3-KO or PiaA-KO cells (Fig. 6b, lanes 4, 7 and 10). Surprisingly, FLAG-PiaA restored AKT phosphorylation when added to mTORC2 purified from not just PiaA-KO cells but also Rip3-KO cells (Fig. 6b, lanes 3 and 6). PiaA was dissociated from mTORC2 in the absence of Rip3 since immunoblotting showed that endogenous PiaA did not co-purify with FLAG-Tor in Rip3-KO cells (Fig. 6b, lane 2). Whole cell lysates from WT and Rip3-KO cells contained similar amounts of PiaA (Fig. S7d). These data suggest that PiaA and Lst8 are essential subunits of mTORC2 for AKT phosphorylation while the mSIN1 homolog Rip3 stabilizes the interactions between PiaA and mTORC2.
To further test this notion, we purified FLAG-Tor under a high-salt condition, which failed to phosphorylate AKT (Fig. 6c, lane 2). We added back FLAG-PiaA and FLAG-Lst8 to the FLAG-Tor. AKT phosphorylation was only restored when both subunits were added together, but not individually (Fig. 6c, lanes 3–5). Therefore, Tor, PiaA, and Lst8 are sufficient components of mTORC2 to catalyze GDP-bound RacE-promoted AKT phosphorylation.
Ser192 phosphorylated RacE-GDP activates mTORC2-mediated AKT phosphorylation in vitro
Using this reconstitution system, we tested the effect of the guanine nucleotide binding status of RacE and RasC on mTORC2 activation in vitro. First, we removed guanine nucleotides from purified FLAG-RacE using EDTA and found that mTORC2 activation was completely blocked (Fig. 6d, lane 3). Second, no mTORC2 activation was observed when FLAG-RacE was loaded with a nonhydrolyzable GTP analog, GTPγS (Fig. 6d, lane 4). Subsequent replacement of GTPγS with GDP restored the ability of RacE to activate mTORC2 (Fig. 6d, lane 5). These guanine nucleotide switching experiments ruled out the possibility that other proteins, such as guanine nucleotide exchange factors that might associate with GDP-bound RacET25N, contribute to mTORC2 activation. Third, WT RacE and GDP-bound RacET25N, but not GTP-bound RacEG20V, activated mTORC2 (Fig. 6e). Fourth, regarding RasC, GTP-bound RasCQ62L, but not GDP-bound RasCS18N, activated mTORC2 (Fig. 6f, lanes 1, 2, 5 and 6). Another Ras homolog RacG 12, 22 did not activate mTORC2 regardless of its guanine nucleotide binding status (Fig. 6f, lane 3, 4, 7 and 8).
The chemoattractant stimulation of Dictyostelium cells prior to purification of FLAG-RacE was required for reconstitution of mTORC2 activation (Fig. 6d, lanes 1 and 2 and 6e, lanes 1–4). This chemoattractant stimulation was no longer necessary when phospho-mimetic GDP-bound RacET25N, S192D was used (Fig. 6g, lanes 1 and 5). Conversely, phospho-defective GDP-bound RacET25N, S192A failed to activate mTORC2-mediated AKT phosphorylation irrespective of the chemoattractant stimulation (Fig. 6g, lane 3 and 4). We also found that RacC must be in a GTP-bound form to stimulate mTORC2 in guanine nucleotide switching experiments (Fig. 6h). These data taken together show that chemoattractant-stimulated phosphorylation of RacE-GDP at Ser192, together with RasC-GTP, directly activates mTORC2-mediated AKT phosphorylation (Fig. 6i).
RacEG23V is equivalent to the cancer driver mutant RhoAG17V (Fig. 3c-f). The G17V mutation dissociates GTP from RhoA40, 41, 45, 46; however, it is unknown whether RhoAG17V is present in a guanine nucleotide-free or GDP-bound form. Using our reconstitution system, we tested the effect of the guanine nucleotide binding status of RacEG23V on mTORC2 activation. Similar to GDP-bound FLAG-RacET25N, purified FLAG-RacEG23V stimulated mTORC2-mediated AKT phosphorylation (Fig. 6j, lanes 1 and 2). When we treated FLAG-RacEG23V with EDTA, which removes potentially associated guanine nucleotides, this activity was lost (Fig. 6j, lane 4). Subsequent incubation of FLAG-RacEG23V with GDP, but not GTPγS, restored its ability to activate mTORC2 (Fig. 6j, lanes 6 and 8). Therefore, RacEG23V activates mTORC2 in a GDP-bound form.
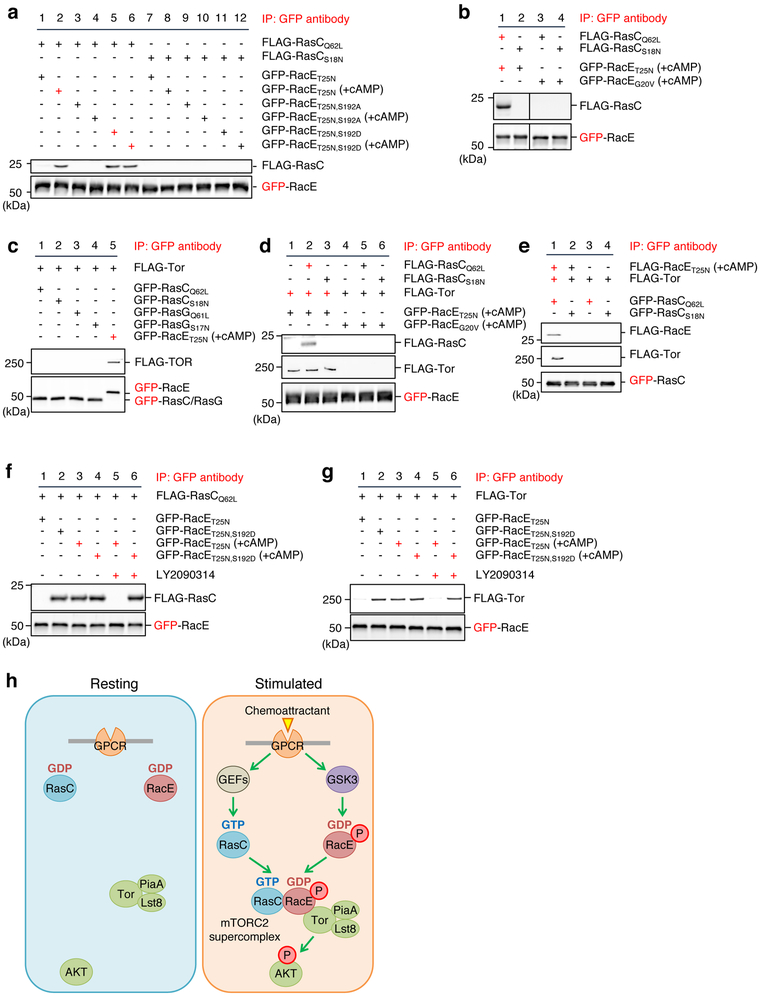
Ser192 phosphorylation of RacE-GDP assembles the RacE-RasC-mTORC2 supercomplex
To understand the biochemical basis of this phospho-mediated activation, we purified GDP-bound FLAG-RacET25N from Dictyostelium cells under high-salt conditions after chemoattractant stimulation and tested its interaction with purified FLAG-RasC proteins (Fig. 7a and S7e). The chemoattractant stimulation enabled GDP-bound RacET25N to interact specifically with GTP-bound RasCQ62L but not GDP-bound RasCS18N (Fig. 7a, lanes 2 and 8). The phospho-defective mutation S192A abolished this chemoattractant-induced interaction of RacE-GDP with RasC-GTP (Fig. 7a, lanes 2 and 4). Conversely, the phospho-mimetic mutation S192D bypassed the requirement for chemoattractant stimulation (Fig. 7a, lanes 1 and 5). GTP-bound RacEG20V did not interact with either RasC-GTP or RasC-GDP, even after chemoattractant stimulation (Fig. 7b, lanes 3 and 4). Therefore, the Ser192 phosphorylation of RacE-GDP enables its interaction with RasC-GTP.
Figure 7. Ser192 phosphorylated RacE-GDP forms a supercomplex with Tor and Ras-GTP.
a and b, The indicated GFP-RacE proteins were purified from Dictyostelium cells with or without 1 μM cAMP stimulation for 30 s. FLAG-RasC proteins were purified without cAMP stimulation. GFP-RacE was incubated with FLAG-RasC and pulled down using GFP-Trap. The pellet fraction was analyzed by immunoblotting using antibodies to GFP and FLAG. c, GFP-RacE, GFP-RasC or GFP-RasG was incubated with FLAG-Tor that was purified in a high-salt condition and pulled down with GFP-Trap. The pellet fraction was analyzed by immunoblotting using antibodies to GFP and FLAG. d, GFP fused to GDP-bound RacET25N or GTP-bound RacEG20V were purified from Dictyostelium cells under a high salt condition after stimulation with the chemoattractant cAMP. These GFP fusion proteins were incubated with high-salt washed FLAG-Tor and/or FLAG-RasC proteins. GFP-RacE was pulled down with GFP-Trap, and the pellet fractions were analyzed by immunoblotting. e, RacE forms a complex with Tor and RasC. The indicated proteins were purified under high-salt conditions and mixed for 15 min at room temperature. GFP-RasC proteins were pulled down with GFP-Trap, and the pellet fraction was analyzed by immunoblotting. f and g, Different GFP-RacE proteins were purified from Dictyostelium cells with or without 1 μM cAMP stimulation for 30 s in the presence or absence of the GSK-3 inhibitor LY2090314 (250 nM). GFP-RacE was incubated with FLAG-RasC in f or FLAG-Tor in g and pulled down using GFP-Trap. The pellet fraction was analyzed by immunoblotting using antibodies to GFP and FLAG. h, Model for GPCR-mediated mTORC2-AKT signaling. In response to GPCR activation by chemoattractant, Rho-GDP becomes phosphorylated by GSK-3 and assembles the super signaling complex with Ras-GTP and mTORC2 to promote AKT phosphorylation. Experiments were repeated independently three times with similar results in a-g.
Phosphorylated RacE also binds Tor. GDP-bound RacET25N purified after chemotactic stimulation bound FLAG-Tor that was purified under high salt conditions (Fig. 7c, lane 5). Although it has been shown that recombinant GTP-loaded RasC binds the kinase domain of Tor purified from E. coli 22, RacC or RasG failed to bind Tor, regardless of their guanine nucleotide binding status, in our binding assay (Fig. 7c, lanes 1–4). Interactions of phosphorylated RacE with Tor require its GDP-binding but independent of RasC (Fig. 7d). Furthermore, chemoattractant-stimulated GDP-bound RacE bridges GTP-bound RasC and Tor (Fig. 7e). Finally, interactions between RacE and RacC or Tor were sensitive to the pre-treatment of chemoattractant-stimulated cells, before the purification of RacE, with the GSK-3 inhibitor LY2090314 (Fig. 7f and 7g); the phosphomimetic S192D mutation overcame this inhibition effect (Fig. 7f and 7g, lanes 5 and 6). These data suggest that chemoattractant-induced Ser192 phosphorylation of GDP-bound RacE assembles the RacE-RasC-mTORC2 supercomplex that phosphorylates AKT (Fig. 7h).
Discussion
In this study, we discovered an unforeseen mechanism by which phosphorylation of GDP-bound Rho GTPase activates mTORC2 signaling in GPCR-mediated directed cell migration. We propose that phosphorylated Rho-GDP forms the super signaling complex with Ras-GTP and mTORC2 (Fig. 7h). Our findings change the current view that switching guanine nucleotides from GDP to GTP activates G proteins 1, 4, 7-9. Because the majority of the G proteins are in a GDP-bound form, this mechanism for G protein regulation significantly broadens the understanding of intracellular signal transduction mediated by one of the largest protein families that control many biological processes. Our findings also demonstrate a role of Rho as an upstream component that connects GPCR activation to mTORC2 to initiate a signaling cascade. This role of Rho revises the prevailing model that Rho is a downstream regulator that couples signaling events to the remodeling of the actin cytoskeleton. In our previous studies, we identified a potential GTPase-activating protein (GflB) and guanine nucleotide exchange factor (GxcT) for RacE30, 47. Since both GflB and GxcT contribute to directed cell migration, it would be important to decipher how they regulate the function of phosphorylated RacE-GDP in mTORC2 activation in future studies.
We propose that phosphorylated RacE-GDP simultaneously binds both RasC-GTP and mTORC2 (Fig. 7h). In this supercomplex, RacE likely binds RasC and mTORC2 through distinct regions. We speculate that Tor binds the effector domain of RacE. On the other hand, RasC might interact with RacE in a manner similar to human KRas dimers 48-50. Our data show that the activation mechanism for mTORC2 by small GTPases is distinct from that for mTORC1. The localization of mTORC1 is regulated by small GTPases, Rags. A Rag dimer formed by RagA/B-GDP and RagC/D-GTP is anchored to the lysosomal membrane through the Regulator protein complex 16. In response to increased levels of amino acids, these GTPases switch guanine nucleotides and become a RagA/B-GTP and RagC/D-GDP dimer. This dimer associates with raptor and localizes mTORC1 to lysosomes. On the surface of lysosomes, the kinase activity of mTORC1 is activated by interactions with GTP-bound Rheb, a Ras-related GTPase. Similar to Rag dimers, RacE and RasC form a dimer in GDP- and GTP-bound forms, respectively. However, different from Rag dimers, RacE-RasC dimers directly stimulate the enzymatic activity of mTORC2.
It has been shown that GSK-3 regulates chemotaxis and differentiation in Dictyostelium cells51, 52; however, the role of GSK-3 in mTORC2-AKT signaling has been controversial: one group showed that Gsk3-KO cells increase this signaling in response to chemotactic stimulation 53, 54, whereas another group reported the opposite effect55. Significantly extending the latter findings, our current work reveals that GSK-3 directly phosphorylates the C-terminus of RacE at S192 upon cAMP stimulation to promote the assembly of the mTORC2 supercomplex that phosphorylates AKTs.
Our in vitro system identified a minimum set of proteins that reconstitute chemoattractant-stimulated mTORC2 activation. The kinase Tor, together with the Rictor homolog PiaA and LST8, are necessary and sufficient subunits of the mTORC2 in this system. The mSin1 homolog Rip3 is important for the stabilization of interactions between Tor and PiaA in cells, but is not essential for the enzymatic activity regulated by RacE and RasC. Supporting our findings, previous studies using mammalian cells have shown that mSin1 is important for the formation of mTORC2 24. Also, mSin1 contains a lipid-binding PH domain and is necessary for the recruitment of mTORC2 to the plasma membrane 24.
A mutation (G17V) that releases GTP from human RhoA is a prevalent driver mutation in T-cell lymphoma 40, 41. In cells carrying RhoAG17V or a GDP-bound RhoAT19N, AKT phosphorylation, the reorganization of the actin cytoskeleton, and cell migration are increased. These findings have been explained by the sequestration of guanine nucleotide exchange factors through stabilized interactions with RhoAG17V or RhoAT19N mutant proteins. In contrast to this model, we found that RacEG23V and RacET25N, which are Dictyostelium’s RhoAG17V and RhoAT19N counterparts, respectively, induce AKT phosphorylation in cells and directly activate mTORC2-mediated AKT phosphorylation in a GDP-bound form in vitro. In light of our findings in the Dictyostelium Rho GTPase, it would be important to test whether the unique cancer-associated RhoA mutation (G17V) is GDP-bound in cancer cells, activates the kinase activity of mTORC2 toward AKT, and thereby promotes cancer cell proliferation and metastasis.
Methods
Cells and plasmids
Disruption of the RacE, PiaA, Rip3, and Lst8 genes by homologous recombination using the blasticidin resistance cassette in Dictyostelium discoideum cells was done as previously described 30, 36, 38, 56. WT, PiaA-KO, Rip3-KO, and Lst8-KO Dictyostelium cells were cultured in HL5 medium (1% protease peptone, 1% glucose, 0.5% yeast extract, 2.5 mM Na2HPO4, 2.5 mM KH2PO4 [pH 6.5]) on a rotary shaker at 200 rpm and 22°C. RacE-KO cells were cultured in HL5 medium on Petri dishes and shaken for 1 day before being used for the experiments 30. Plasmids were introduced to Dictyostelium cells by electroporation, and stable cell lines carrying the plasmids were selected with G418 or hygromycin 30. To induce cell differentiation, exponentially growing cells were washed with development buffer (DB; 2 mM MgSO4, 0.2 mM CaCl2, 5 mM Na2HPO4, and 5 mM KH2PO4 [pH 6.5]), resuspended at 2×107 cells/ml, starved for 1 h, and shaken with 100 nM cAMP pulses at 6-min intervals for 4 h. To induce FLAG-RasC/G expression, 200 μg/ml doxycycline hyclate was included in the development buffer during differentiation. HEK293T cells were maintained in DMEM containing 10% FBS and were transiently transfected with a peYFP-C1 plasmid carrying WT or mutant versions of human RhoA or Rac1 using Lipofectamine 3000 (Life Technologies, L3000015) according to the manufacturer’s protocol. Transfected cells were cultured overnight before analysis. The plasmids are listed in Supplemental Table 3.
Directed cell migration assay
Chemotactic migration toward cAMP was analyzed using a microfluidic chamber, as reported 57 with some modifications. 1 μM cAMP, 500 μM SQ22536 (Sigma-Aldrich, S153), 2 mM MgSO4, 5 mM Na2HPO4, and 5 mM KH2PO4 (pH 6.5) was loaded into the chamber to generate a stable cAMP gradient. Next, differentiated Dictyostelium cells were resuspended at 1×105 cells/ml in 500 μM SQ22536, 2 mM MgSO4, 5 mM Na2HPO4, and 5 mM KH2PO4 (pH 6.5). Five-hundred cells were loaded into the opposite side of microfluidic chamber. Images of cells were recorded for 60 min using an Axio Vision inverted microscope (Zeiss) with a 10× objective. The number of cells that moved toward a higher concentration of cAMP was quantified.
Identification of RacE-binding proteins
Dictyostelium cells expressing GFP-RacE proteins were lysed in lysis buffer (150 mM NaCl, 0.5% Triton X-100, 1 mM NaF, 0.5 mM Na3VO4, 1 mM DTT, 10% glycerol, 25 mM Tris-HCl [pH 7.5] and protease-inhibitor cocktail [Roche, 4693159001]) and incubated with GFP-Trap magnetic beads (ChromoTek, gtma-200) for 4 h at 4°C. The beads were washed, and the bound fractions were analyzed using mass spectrometry at the Johns Hopkins Mass Spectrometry and Proteomics Facility (Supplementary Figure 3 and Supplementary Table 1 and 2).
Metabolic labeling
Dictyostelium cells carrying different plasmids were differentiated in DB-MES (2 mM MgSO4, 0.2 mM CaCl2, 20 mM MES [pH 6.5]), washed in DB-MES, and resuspended at 5×107 cells/ml. Cells were incubated with 500 μCi/ml of P32 for 40 min at 22°C and then further incubated in the presence of 2 mM caffeine for 20 min 58. Cells were washed in ice-cold DB-MES twice, resuspended at 1×108 cells/ml, and lysed in an equal volume of ice-cold 2× lysis buffer (2% NP40, 300 mM NaCl, 20 mM MES [pH 6.5], protease inhibitor cocktail, and phosphatase inhibitors [Sigma, P5726]) on ice for 10 min. After clarification by centrifugation at 4°C, 1 ml of cell lysates were incubated with 20 μl of GFP-Trap agarose beads (ChromoTek, gta-200) or anti-FLAG agarose M2 beads (Sigma, A2220) at 4°C for 1 h with gentle agitation. After washing with 1× lysis buffer twice, the beads were incubated with 40 μl of elution solution (1 mM GTP, 0.2% SDS, 5 mM DTT, 2 mM EDTA [pH 8.0]) at 65°C for 5 min to elute guanine nucleotides. Samples were spotted on polyethylenimine cellulose TLC plates and developed in 0.75 M KH2PO4 (pH 3.4) 59. The TLC plates were exposed to phosphor-imaging screens and scanned using a Bio-Rad PharosFX Plus molecular imager; images were analyzed with NIH ImageJ. Proteins bound to the beads were eluted with SDS-PAGE sample buffer and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
Immunoprecipitation
Protein-protein interactions were assayed as previously performed 31 with some modifications. To prepare Dictyostelium cell lysates, cells carrying different plasmids were differentiated in DB and incubated with 2 mM caffeine at 22°C for 20 min. Cells were washed in DB, resuspended at 1×108 cells/ml, and lysed in an equal volume of ice-cold 2× lysis buffer (2% NP40, 300 mM NaCl, 20 mM sodium phosphate [pH 7.0], protease inhibitors, and phosphatase inhibitors) on ice for 10 min. Cell lysates were clarified by centrifugation at 4°C and incubated with GFP-Trap agarose beads to immunoprecipitate GFP-RacE proteins at 4°C for 2 h. After washing with 1× lysis buffer, bound fractions were eluted with 2× SDS-PAGE sample buffer and analyzed by SDS-PAGE and immunoblotting using appropriate antibodies. To analyze protein-protein interactions in HEK293T cells, HEK293T cells plated on a 6-cm dish were transfected with different YFP-RhoA or YFP-Rac1 plasmids, cultured overnight, and lysed in 0.25 ml 1× lysis buffer (150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM EDTA, 20 mM Tris [pH 7.5], protease inhibitors, and phosphatase inhibitors). YFP-RhoA or YFP-Rac1 proteins were immunoprecipitated using GFP-Trap agarose beads.
Chemoattractant-induced AKT phosphorylation in cells
Differentiated Dictyostelium cells were incubated in DB with 2 mM caffeine for 20 min at 22°C, washed twice with ice-cold DB, and resuspended at 5×107 cells/ml. Cells were shaken at 200 rpm at 22°C and stimulated with 1 μM cAMP 39. Aliquots were taken at different time points and lysed in SDS-PAGE sample buffer. Proteins were analyzed by immunoblotting with antibodies to PKBA and PKBR1. As extensively described in many studies 23, phosphorylation of PKBA and PKBR1 were detected using anti-phospho-RPS6KB1 antibodies (Cell Signaling, 9205) and anti-phospho-PKC antibodies (Cell Signaling, 2060) because of the similar amino acid sequences around the phosphorylation sites (Fig. S4).
Immunoblotting
Proteins were separated using SDS-PAGE and transferred onto PVDF membranes. The antibodies were GFP 31, 60, FLAG (Sigma, F7425 and F3165), PiaA 36, Tor (Cell Signaling, 2983), Rictor (Cell Signaling, 2114), Raptor (Cell Signaling, 2280), GAPDH (Thermo Fisher, MA5–15738), actin (Santa Cruz, Sc-1615), GSK-3 (Millipore, 05–412), phospho-RPS6KB1 (Cell Signaling, 9205), phospho-PKC (Cell Signaling, 2060), AKT (Cell Signaling, 9272) and phospho-AKT (serine 473) (Cell Signaling, 9271). Polyclonal rabbit antibodies to PKBA and PKBR1 were raised against the peptides, KNSDRKKRVNG and KKGNKNDETTP, respectively. Polyclonal rabbit antibodies to RacE and phospho-RacE (serine 192) were raised against the peptide REQQHPDPNSGKF and the phosphopeptide GMDKK(pS)QDGSS. Immunocomplexes were visualized using fluorescent-labeled secondary antibodies and detected using a Bio-Rad PharosFX Plus molecular imager. Images were analyzed using NIH ImageJ. The detailed information about the antibodies used in this study is provided in Supplementary Table 4.
Protein purification
Dictyostelium cells carrying the epitope-tagged proteins were differentiated, washed twice with ice-cold DB, and resuspended at 1×108 cells/ml. To purify mTORC2 using FLAG-Tor, cells were lysed in an equal volume of ice-cold 2× CHAPS lysis buffer (0.6% CHAPS, 250 mM NaCl, 2 mM EDTA, 100 mM HEPES [pH 7.4], protease inhibitors, and phosphatase inhibitors) for 10 min on ice. After clarification by centrifugation, 1 ml of cell lysates was incubated with 15 μl of anti-FLAG agarose beads to immunoprecipitate proteins for 2 h at 4°C with gentle agitation. The beads were washed using 1× CHAPS lysis buffer, snap frozen in liquid nitrogen, and stored at −80°C. In the experiments described in Figures 6b and c, in which individual subunits of mTORC2 were tested for AKT phosphorylation in vitro, FLAG-PiaA and FLAG-Lst8 were purified as described above and then washed twice in a high-salt condition (500 mM NaCl, 50 mM HEPES [pH 7.4]).
To purify FLAG- or GFP-RacE and FLAG- or GFP-RasC/G, cells resuspended at 1×108 cells/ml were lysed in an equal volume of ice-cold 2× NP40 lysis buffer (2% NP40, 250 mM NaCl, 2 mM EDTA, 10 mM sodium phosphate [pH 7.0], protease inhibitors, and phosphatase inhibitors) for 10 min on ice. After clarification, 1 ml of cell lysates was incubated with 15 μl of GFP-Trap or anti-FLAG agarose beads to immunoprecipitate proteins for 2 h at 4°C with gentle agitation. The beads were washed three times in 1× NP40 lysis buffer, twice in high salt wash buffer (500 mM NaCl, 10 mM sodium phosphate [pH 7.0]), and three times in 1× NP40 lysis buffer. The washed beads were snap-frozen in liquid nitrogen and stored at −80°C.
In vitro phosphorylation of RacE by GSK-3β
Purified 40 ng FLAG-RacE and purified 50 ng GSK-3β (Sigma, G4296) were mixed in 40 μl of kinase reaction buffer-1 (10 mM MgCl2, 0.1% 2-mercaptoethanol, 50 mM Tris-HCl, pH 7.5) in the presence or absence of 100 μM ATP for 15 min at room temperature. Phosphorylation of RacE at serine 192 was analyzed by immunoblotting with antibodies to RacE and phospho-RacE (serine 192).
Reconstitution of chemoattractant-stimulated mTORC2-mediated AKT phosphorylation
The kinase activity of mTORC2 was measured as described previously 43, 44, with some modifications. mTORC2, RacE, RasC, and RasG were eluted from anti-FLAG beads by incubation with 1 M arginine at room temperature for 5 min 61. Amounts of purified proteins were quantified on CBB-stained SDS-PAGE gels using ImageJ software with BSA as a standard. Purified mTORC2 (200 ng FLAG-Tor) was incubated with 40 ng RacE, 40 ng RasC or G, and 40 ng unactive human AKT1 (Sigma, 14–279) in 80 μl of kinase reaction buffer-2 (1 mM ATP, 1 mM MgCl2, 100 mM potassium acetate, and 25 mM HEPES [pH 7.4]) at room temperature for 5 min with gentle mixing. Reactions were stopped by adding 80 μl of 2× SDS-PAGE sample buffer. AKT phosphorylation was detected by immunoblotting with anti-phospho-AKT (serine 473) antibodies (Cell Signaling, 9271).
In vitro protein-protein interaction assay
To analyze interactions between RacE, RasC/G, and Tor, purified proteins were high-salt washed with 500 mM NaCl and 50 mM HEPES (pH 7.4). To examine the interactions of RacE with RasC/G and Tor, GFP-Trap carrying GFP-RacE (40 ng) was incubated with FLAG-RasC proteins (40 ng) or/and FLAG-Tor (200 ng) in kinase reaction buffer-2 without ATP at room temperature for 15 min. GFP-Trap beads were washed three times in the same buffer, and bound proteins were analyzed by immunoblotting. To examine interactions of RasC/G with RacE and Tor, GFP-Trap beads carrying Ras proteins (40 ng) were incubated with FLAG-tagged RacE (40 ng) and Tor (200 ng).
Statistics and reproducibility
Unpaired, two-tailed Student’s t-tests were performed using GraphPad Prism7 to determine statistical significance of the experiments described in Supplementary Fig. 2. For multiple group comparisons, one-way ANOVA analysis followed by Tukey’s test was performed using Prism7 in Fig. 1b, 1e, 2b, 2d, 5f and 5h. Statistical analysis and P value are described in each figure and figure legend. The number of independent experiments performed is described in each figure legend.
Data availability
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD014014 and provided in Supplementary Tables 1 and 2. Unprocessed images of all blots and gels are provided in Supplementary Figure 8. Source data for all graphical representations and statistical descriptions are provided in Supplementary Table 5, for Figures 1b, 1e, 2a, 2b, 2d, 2h, 3b, 3d, 3f, 3h, 3j, 5b, 5d, 5f, 5h, and Supplementary Figures S2 and S6c. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
We thank members of the Iijima and Sesaki labs for helpful discussions and technical assistance and Drs. P. Devreotes and T. Inoue for providing the plasmids and cell lines. This work was supported by grants to MI (NIH GM131768 and Allegheny Health Network-Sidney Kimmel Comprehensive Cancer Center), to H. Senoo (Japan Society for the Promotion of Science) and to H. Sesaki (NIH GM089853).
Abbreviations:
- mTORC2
mechanistic target of rapamycin complex 2
- GPCR
G-protein coupled receptor
- PI3K
phosphoinositide 3-kinase
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hodge RG & Ridley AJ Regulating Rho GTPases and their regulators. Nat Rev Mol Cell Biol 17, 496–510 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Burridge K & Wennerberg K Rho and Rac take center stage. Cell 116, 167–179 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Stenmark H Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Alberts B et al. Molecular Biology of the Cell, Sixth Edition Molecular Biology of the Cell, Sixth Edition, 1–1342 (2015). [Google Scholar]
- 5.Mayor R & Etienne-Manneville S The front and rear of collective cell migration. Nat Rev Mol Cell Biol 17, 97–109 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Weiner OD Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr Opin Cell Biol 14, 196–202 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langeberg LK & Scott JD Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol 16, 232–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon MA, Freed DM, Schlessinger J & Kiyatkin A The Dark Side of Cell Signaling: Positive Roles for Negative Regulators. Cell 164, 1172–1184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch CM, Elliott H, Danuser G & Hahn KM Imaging the coordination of multiple signalling activities in living cells. Nat Rev Mol Cell Biol 12, 749–756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang M et al. Disruption of PKB signaling restores polarity to cells lacking tumor suppressor PTEN. Molecular biology of the cell 22, 437–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamimura Y et al. PIP3-independent activation of TorC2 and PKB at the cell’s leading edge mediates chemotaxis. Curr Biol 18, 1034–1043 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Artemenko Y, Lampert TJ & Devreotes PN Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cellular and molecular life sciences : CMLS 71, 3711–3747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao Y et al. Altering the threshold of an excitable signal transduction network changes cell migratory modes. Nat Cell Biol 19, 329–340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pompura SL & Dominguez-Villar M The PI3K/AKT signaling pathway in regulatory T-cell development, stability, and function. J Leukoc Biol (2018). [DOI] [PubMed] [Google Scholar]
- 15.Devreotes P & Horwitz AR Signaling Networks that Regulate Cell Migration. Cold Spring Harbor perspectives in biology 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang K & Fingar DC Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol 36, 79–90 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Das S, Losert W & Parent CA mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 19, 845–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lien EC, Dibble CC & Toker A PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol 45, 62–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fruman DA et al. The PI3K Pathway in Human Disease. Cell 170, 605–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khanna A et al. The small GTPases Ras and Rap1 bind to and control TORC2 activity. Sci Rep 6, 25823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai H et al. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol 190, 233–245 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaubitz C, Prouteau M, Kusmider B & Loewith R TORC2 Structure and Function. Trends Biochem Sci 41, 532–545 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Berzat A & Hall A Cellular responses to extracellular guidance cues. The EMBO journal 29, 2734–2745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossman KL, Der CJ & Sondek J GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nature reviews. Molecular cell biology 6, 167–180 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Welch HC, Coadwell WJ, Stephens LR & Hawkins PT Phosphoinositide 3-kinase-dependent activation of Rac. FEBS letters 546, 93–97 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Iden S & Collard JG Crosstalk between small GTPases and polarity proteins in cell polarization. Nature reviews. Molecular cell biology 9, 846–859 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Larochelle DA, Vithalani KK & De Lozanne A Role of Dictyostelium racE in cytokinesis: mutational analysis and localization studies by use of green fluorescent protein. Mol Biol Cell 8, 935–944 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Senoo H, Sesaki H & Iijima M Rho GTPases orient directional sensing in chemotaxis. Proceedings of the National Academy of Sciences of the United States of America 110, E4723–4732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senoo H, Cai H, Wang Y, Sesaki H & Iijima M The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells. Mol Biol Cell 27, 1596–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Chen CL & Iijima M Signaling mechanisms for chemotaxis. Dev Growth Differ 53, 495–502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols JM, Veltman D & Kay RR Chemotaxis of a model organism: progress with Dictyostelium. Current opinion in cell biology 36, 7–12 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Kortholt A & van Haastert PJ Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal 20, 1415–1422 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Gerisch G, Schroth-Diez B, Muller-Taubenberger A & Ecke M PIP3 waves and PTEN dynamics in the emergence of cell polarity. Biophys J 103, 1170–1178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen MY, Long Y & Devreotes PN A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev 11, 3218–3231 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimobayashi M & Hall MN Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 15, 155–162 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Liao XH, Buggey J & Kimmel AR Chemotactic activation of Dictyostelium AGC-family kinases AKT and PKBR1 requires separate but coordinated functions of PDK1 and TORC2. J Cell Sci 123, 983–992 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamimura Y, Tang M & Devreotes P Assays for chemotaxis and chemoattractant-stimulated TorC2 activation and PKB substrate phosphorylation in Dictyostelium. Methods Mol Biol 571, 255–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palomero T et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 46, 166–170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo HY et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet 46, 371–375 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Charest PG et al. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Developmental cell 18, 737–749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J An in vitro assay for the kinase activity of mTOR complex 2. Methods Mol Biol 821, 75–86 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Sarbassov dos D, Bulgakova O, Bersimbaev RI & Shaiken T Isolation of the mTOR complexes by affinity purification. Methods Mol Biol 821, 59–74 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Gilbert-Ross M, Marcus AI & Zhou W RhoA, a novel tumor suppressor or oncogene as a therapeutic target? Genes Dis 2, 2–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dopeso H et al. Mechanisms of inactivation of the tumour suppressor gene RHOA in colorectal cancer. Br J Cancer 118, 106–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Senoo H, Cai H, Wang Y, Sesaki H & Iijima M The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells. Molecular biology of the cell 27, 1596–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ambrogio C et al. KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell 172, 857–868 e815 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Jang H, Muratcioglu S, Gursoy A, Keskin O & Nussinov R Membrane-associated Ras dimers are isoform-specific: K-Ras dimers differ from H-Ras dimers. Biochem J 473, 1719–1732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nan X et al. Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci U S A 112, 7996–8001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harwood AJ, Plyte SE, Woodgett J, Strutt H & Kay RR Glycogen synthase kinase 3 regulates cell fate in Dictyostelium. Cell 80, 139–148 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Kim L, Liu J & Kimmel AR The novel tyrosine kinase ZAK1 activates GSK3 to direct cell fate specification. Cell 99, 399–408 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Kolsch V et al. Daydreamer, a Ras effector and GSK-3 substrate, is important for directional sensing and cell motility. Mol Biol Cell 24, 100–114 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacal Romero J et al. The Dictyostelium GSK3 kinase GlkA coordinates signal relay and chemotaxis in response to growth conditions. Dev Biol 435, 56–72 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Teo R et al. Glycogen synthase kinase-3 is required for efficient Dictyostelium chemotaxis. Mol Biol Cell 21, 2788–2796 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S et al. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Molecular biology of the cell 16, 4572–4583 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakajima A, Ishida M, Fujimori T, Wakamoto Y & Sawai S The microfluidic lighthouse: an omnidirectional gradient generator. Lab Chip 16, 4382–4394 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Iijima M, Huang YE, Luo HR, Vazquez F & Devreotes PN Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem 279, 16606–16613 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Castro AF, Rebhun JF & Quilliam LA Measuring Ras-family GTP levels in vivo--running hot and cold. Methods 37, 190–196 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Iijima M & Devreotes P Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell 109, 599–610 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Futatsumori-Sugai M et al. Utilization of Arg-elution method for FLAG-tag based chromatography. Protein Expr Purif 67, 148–155 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited in ProteomeXchange with the primary accession code PXD014014 and provided in Supplementary Tables 1 and 2. Unprocessed images of all blots and gels are provided in Supplementary Figure 8. Source data for all graphical representations and statistical descriptions are provided in Supplementary Table 5, for Figures 1b, 1e, 2a, 2b, 2d, 2h, 3b, 3d, 3f, 3h, 3j, 5b, 5d, 5f, 5h, and Supplementary Figures S2 and S6c. All other data supporting the findings of this study are available from the corresponding author on reasonable request.