SUMMARY
Taxonomy: Lettuce mosaic virus (LMV) belongs to the genus Potyvirus (type species Potato virus Y) in the family Potyviridae.
Physical properties: The virion is filamentous, flexuous with a length of 750 nm and a width of 15 nm. The particles are made of a genomic RNA of 10 080 nucleotides, covalently linked to a viral‐encoded protein (the VPg) at the 5′ end and with a 3′ poly A tail, and encapsidated in a single type of capsid protein. The molecular weight of the capsid protein subunit has been estimated electrophoretically to be 34 kDa and estimated from the amino acid sequence to be 31 kDa.
Genome organization: The genome is expressed as a polyprotein of 3255 amino‐acid residues, processed by three virus‐specific proteinases into ten mature proteins.
Hosts: LMV has a worldwide distribution and a relatively broad host range among several families. Weeds and ornamentals can act as local reservoirs for lettuce crops. In particular, many species within the family Asteraceae are susceptible to LMV, including cultivated and ornamental species such as common (Lactuca sativa), prickly (L. serriola) or wild (L. virosa) lettuce, endive/escarole (Cichorium endiva), safflower (Carthamus tinctorius), starthistle (Centaurea solstitialis), Cape daisy (Osteospermum spp.) and gazania (Gazania rigens). In addition, several species within the families Brassicaceae, Cucurbitaceae, Fabaceae, Solanaceae and Chenopodiaceae are natural or experimental hosts of LMV.
Genetic control of resistance to LMV: The only resistance genes currently used to protect lettuce crops worldwide are the recessive genes mo1 1 and mo1 2 corresponding to mutant alleles of the gene encoding the translation initiation factor eIF4E in lettuce. It is believed that at least one intact copy of eIF4E must be present to ensure virus accumulation.
Transmission: LMV is transmitted in a non‐persistent manner by a high number of aphid species. Myzus persicae and Macrosiphum euphorbiae are particularly active in disseminating this virus in the fields. LMV is also seedborne in lettuce. The effectiveness of LMV transmission depends on the cultivar and the age of the seed carrier at the inoculation time.
Symptoms: The characteristic symptoms on susceptible lettuce cultivars are dwarfism, mosaic, distortion and yellowing of the leaves with sometimes a much reduced heart of lettuce (failure to form heads). The differences in virus strains, cultivars and the physiological stage of the host at the moment of the attack cause different symptom severity: from a very slight discoloration of the veins to severe necrosis leading to the death of the plant.
INTRODUCTION
Lettuce mosaic disease was first described in Florida (Jagger, 1921) and is now distributed worldwide, probably because seeds have been exchanged internationally over many years (Dinant and Lot, 1992). It occurs in all continents, including Europe, North and South America (Mexico, USA, Argentina, Brazil, Uruguay), the West Indies (Bermuda), Africa, the Middle East (Egypt, Israel, Jordan, Iraq, Iran, Turkey), Asia (China, Japan) and Oceania (Australia, Tasmania, New Zealand). Lettuce mosaic virus (LMV) is a major pathogen of commercial crops in lettuce‐growing areas of the world. Severe losses are recorded mainly in field crops, but the disease may be significant in the greenhouse when seedlings are not grown under insect‐proof conditions (Dinant and Lot, 1992). Elementary sanitary measures (keeping lettuce nurseries away from fields in which crops are grown etc.) may improve control of the disease. The detection of LMV in infected plants or in seed lots is routinely carried out using immunological techniques such as enzyme‐linked immunosorbent assay (Clark and Adams, 1977). Because of the prevalence of seedborne virus (Grogan et al., 1952; Newhall, 1923; Tomlinson, 1970), lettuce seeds can be tested by direct observation of lettuce seedlings. Indeed, seedlings with seedborne virus have misshapen cotyledons, the first true leaf is misshapen and has a dark green mottling appearance. Inoculation of a sensitive indexing host with sap extracted from the ground‐up seed, or a serological technique (Falk and Purcifull, 1983), can also ensure that each lettuce seed lot contains no infected seeds in a sample of 30 000 seeds (MT0, ‘Mosaic Tolerance Zero’ or zero infected seeds in 30 000). More recently, efforts have been made to develop other more sensitive techniques of detection of LMV, based on the polymerase chain reaction (Peypelut et al., 2004). It was shown that expression of a capsid protein transgene protects lettuce against LMV infection (Dinant et al., 1997). However, a more successful control measure is the incorporation of natural virus resistance into the principal lettuce types grown (Walkey et al., 1985).
The only resistance genes currently used to protect lettuce crops worldwide are the recessive allelic genes mo1 1 and mo1 2. The mo1 1 gene, formerly named g, was first identified in Argentina, in a Latin‐type cultivar named ‘Gallega de Invierno’ (Bannerot et al., 1969). In Europe, lettuce breeders used the Gallega source of resistance to incorporate the g gene in numerous varieties of lettuce, including butterhead, Batavia, cos and crisphead types (Pink et al., 1992a). The mo1 2 gene, identified in three Egyptian wild Lactuca sativa lines and named the recessive gene mo (Ryder, 1970), has been mostly used by North American breeders who introduced it into crisphead and cos types of lettuce (Pink et al., 1992a). Initially considered identical, these genes were later shown to have different specificities and to be either allelic or closely linked and therefore were renamed mo1 1 and mo1 2 (Dinant and Lot, 1992). These genes have been recently cloned and sequenced in our laboratory (Nicaise et al., 2003). The resistance alleles mo1 1 and mo1 2, as well as the susceptibility allele mo1 0, were found to code for forms of the eukaryotic translation initiation factor eIF4E in lettuce.
Depending on the viral isolate to which they are confronted, these genes can be considered as inducing resistance (no detectable virus accumulation) or tolerance (virus accumulation but failure to induce significant symptoms). Most of the field isolates of LMV are seedborne in susceptible lettuce cultivars, but not in resistant cultivars carrying mo1 1 or mo1 2, even in cultivars with low levels of resistance (Dinant and Lot, 1992; Pink et al., 1992a). Therefore, in addition to a reduction of viral infection or symptom expression, the mo1 1 and mo1 2 genes also provide a reduction in the dissemination of LMV through seed. For simplicity, only the term resistance will be used to refer the complex set of phenotypes associated with the mo1 1 and mo1 2 genes.
In addition to the economic importance of LMV, the large biological diversity and the differences between the biological properties of isolates (symptoms, seed transmission, behaviour towards mo1 genes) make it a very good model to study plant–virus interactions, from both host and pathogen perspectives. Moreover, molecular tools available for LMV, including highly infectious cDNA copies of the LMV genome with the full‐length (FL) cDNA placed under the control of the enhanced CaMV 35S promoter and of the NOS terminator, have been constructed (Redondo et al., 2001; Yang et al., 1998). These infectious clones have provided a tool of tremendous importance to study the molecular genetics of LMV. This article aims to highlight advances made in understanding the lettuce–LMV interactions, at the population, individual and molecular levels.
LMV DIVERSITY: ‘LMV‐MOST ISOLATES’, A THREAT TO DURABLE RESISTANCE?
While LMV was considered appropriately controlled by the use of mo1 1 and mo1 2, occasional outbreaks of resistance‐breaking forms of LMV have been described for several decades (Dinant and Lot, 1992; Pink et al., 1992a). Usually, these resistance‐breaking isolates are not seedborne, which has suggested a link between the gain of virulence and the loss of seed transmission. However, since the beginning of the 1990s, mo1‐breaking isolates with high rates of seed‐transmissibility have been described (Dinant and Lot, 1992). In addition to the abilities of being seedborne and of infecting mo1 varieties, LMV isolates differ in the symptoms they induce, which can vary from barely detectable to strongly necrotic or even lethal for a same host (Fig. 1) (Krause‐Sakate et al., 2005; Kyriakopoulou, 1985).
Figure 1.
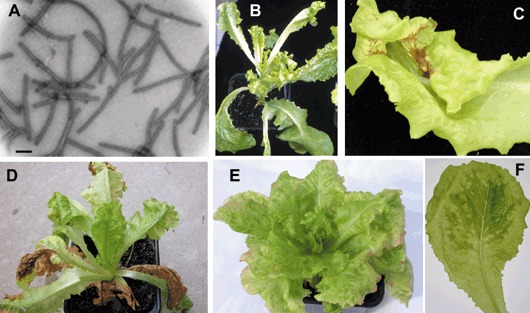
LMV particles and symptoms induced by LMV isolates on lettuce susceptible cultivar Trocadéro. (A) Electron micrograph of LMV virions (the bar represents 200 nm). (B–D) Severe symptoms induced by the LMV‐E isolate including dwarfing and necrosis (close‐up in C). (E) Mosaic symptoms induced by LMV‐AF199. (F) Leaf infected with LMV‐0 showing mosaic symptoms.
Attempts to link the key biological properties of LMV isolates with their sequence clustering have been carried out (Krause‐Sakate et al., 2002; Revers et al., 1997a). LMV isolates could be clustered in three main groups: a single isolate from Yemen (the upper branch in Fig. 2), a group from the Balkans (Greece and Croatia, named Gr) and a third group with very diverse geographical origins (including the Middle East and Greece, called ‘Rest of the World’ or RoW) (Fig. 2). No seedborne isolate was ever observed in the Yemen and Gr groups. Within the RoW group, two large subclusters of isolates contained all known seedborne LMV isolates: one with isolates unable to infect mo1 1 or mo1 2 plants, collectively named ‘LMV‐Common’, and the other with the isolates cumulating mo1 breaking and seed transmission, collectively named ‘LMV‐Most’ for mo1‐breaking, Seed Transmitted. Correlation of the sequence clustering of LMV isolates with their abilities to infect mo1 plants remains less clear than with their seed transmission properties. On the one hand, no mo1‐breaking isolate was ever observed in the LMV‐Common group, and all LMV‐Most isolates and LMV‐Gr assayed, as well as the Yemen isolate, were able to infect mo1 1 as well as mo1 2 plants. On the other hand, while the vast majority of LMV isolates outside the LMV‐Common group are able to overcome both mo1 alleles, variations in this respect occur sporadically in the dendrogram (Krause‐Sakate et al., 2002), the most remarkable occurrence being LMV‐1 (overcoming mo1 1 but not mo1 2), which clusters very close to LMV‐E (overcoming both mo1 alleles). Altogether, sequence clustering evidence suggests that the ability to infect mo1 plants did not evolve from isolates unable to do so, such as LMV‐Common.
Figure 2.
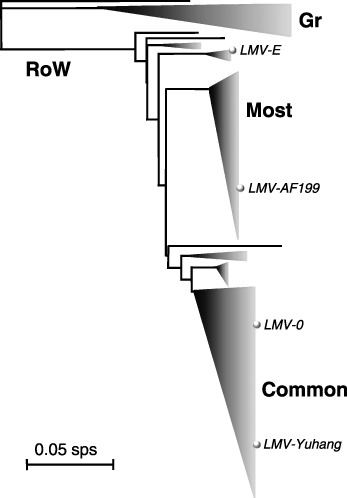
Dendrogram showing the relationships between LMV isolates. The dendrogram shows the levels of sequence divergence between LMV isolates using nucleotide sequences. It is derived from the Saitou & Nei distances calculated in an alignment of the variable nucleotide sequence of the NIB–CP junction (between nucleotide positions 8936 and 9151 of the LMV‐E genome). The bar represents 0.05 substitutions per site (sps). The upper line corresponds to the single isolate from Yemen. Remarkable clusters of isolates are represented by triangles. The cluster named Gr corresponds to the group of isolates from the Balkans. The cluster named RoW (Rest of the World) includes most of the LMV isolates, representing isolates from lettuce of various geographical origins (Europe, South America, North Africa, Middle East, China). Within this cluster, the Most and Common clusters have been named after Krause‐Sakate et al. (2002). The positions of the sequenced isolates LMV‐E, LMV‐0, LMV‐AF199 and LMV‐Yuhang are indicated by small spheres.
These studies therefore established a link between biological properties and sequence clustering, and provided the bases for strain‐specific detection of LMV, once the complete nucleotide sequence of an LMV‐Most isolate had been established (Krause‐Sakate et al., 2002; Peypelut et al., 2004). They also enabled a re‐writing of the scenario leading to local outbreaks of resistance‐breaking LMV: instead of the loss of seed transmission upon acquisition of the ability to overcome mo1 (Dinant and Lot, 1992), the available evidence more simply suggests that these outbreaks are caused by non‐Common‐non‐Most isolates occurring locally, perhaps in weed reservoirs, and which are primarily both unable to infect lettuce seed‐to‐seed and able to infect mo1 plants; in this scheme, LMV‐Most probably represents one of these local forms of LMV that occurred to be, or evolved to be, seedborne in lettuce and was therefore spread worldwide through seed trade.
The molecular variability of LMV isolates was also revealed, using monoclonal antibodies directed against the coat protein, but these studies could not reveal any difference between LMV‐Common and LMV‐Most, probably owing to their identical amino‐acid sequence in the immunogenic N‐terminus of the coat protein (Candresse et al., 2007).
The occurrence of LMV‐Most is a concern for lettuce production worldwide, as these isolates are able to overcome the two major modes of control of LMV, namely genetic resistance and seed control. Therefore, specific effort must be made to avoid the spread of LMV‐Most in seeds, by promoting the propagation of lettuce seeds in LMV‐free environments and the dissemination of virus‐free seeds, developing specific detection tools (Peypelut et al., 2004), identifying potential reservoirs specific for LMV‐Most, and understanding the spread of LMV between host species and the molecular bases of the typical biological properties of LMV‐Most.
GENOMIC ORGANIZATION OF LMV
The genome of LMV consists of a single positive‐strand RNA of 10 080 nt in length. The genomic RNA of Potyviruses is covalently linked at its 5′ end to a virus‐encoded VPg protein (Murphy et al., 1991), and is polyadenylated at its 3′ end (Adams et al., 2005). There is a single open reading frame (ORF) flanked by two untranslated regions (UTRs, of 103 and 210 nt at the 5′ and 3′ ends, respectively) that is translated into a single, large polyprotein processed by three virus‐specific proteinases (Reichmann et al., 1992). To date, four full‐length genomic RNA sequences, corresponding to isolates of LMV differing in their biological properties, have been published: two mo1 resistance‐breaking isolates, LMV‐E and LMV‐AF199 (GenBank accession nos. X97705 and AJ278854 respectively) (Krause‐Sakate et al., 2002; Revers et al., 1997b), and two LMV‐common isolates, LMV‐0 from Europe (GenBank X97704) (Revers et al., 1997b) and LMV‐Yuhang from China (GenBank AJ306288) (Zheng et al., 2002).
We focus here on the three molecularly well‐characterized LMV isolates belonging to the same group of isolates: LMV‐E (isolated in Spain by H. Lot, INRA‐Avignon, France), a non‐seedborne resistance‐breaking isolate that provokes symptoms in cultivars carrying mo1 1 or mo1 2 genes (Pink et al., 1992b); LMV‐0, an LMV‐common isolate that provokes very mild or no symptoms on cultivars carrying the mo1 2 gene (tolerance) but does not invade systemically the cultivars carrying the mo1 1 gene (resistance) (Dinant and Lot, 1992); and LMV‐AF199 (LMV‐Most), which in addition to being seedborne, overcomes the mo1 1 and mo1 2 genes in lettuce (Krause‐Sakate et al., 2002).
Regardless of their large differences in pathogenicity, resistance‐breaking and seed‐transmission properties (Table 1), the entire genomic sequences differ from each other only by point mutations, with no deletion or insertions. The overall nucleotide sequence identities between LMV‐AF199 and LMV‐0, LMV‐AF199 and LMV‐E, and LMV‐E and LMV‐0 are 95.9, 93.9 and 94.0% respectively. At the amino acid sequence level, the identities are 98.0, 96.7 and 97.0%, respectively. Generally, the largest variability occurs in the P1 and the N‐terminal region of the coat‐protein (although more conserved between LMV‐0 and AF199) while the NIa protease domain, the NIb protein, the C‐terminus of the helper‐component protease (HcPro) and the 3′ non‐coding region are more conserved. A recombinant LMV isolate resulting from a natural exchange between LMV‐Most and LMV‐Common in a field where both strains occurred has been isolated in only one instance thus far. The putative recombination site was located within the P3 coding region (Krause‐Sakate et al., 2004).
Table 1.
Biological characteristics of the three LMV isolates of this study
| Symptoms on susceptible cultivar Trocadéro | mo1 1 breaking | mo1 2 breaking | Seed transmission | Phylogenetic group* | |
|---|---|---|---|---|---|
| LMV‐0 | Mosaic | No | No | Yes (2–9%) | LMV‐Common |
| LMV‐E | Severe mosaic with leaf deformation, general stunting, local necrosis | Yes | Yes | No | LMV‐RoW |
| LMV‐AF199 | Severe mosaic | Yes | Yes | Yes (5–10%) | LMV‐Most |
Major LMV phylogenetic group to which each isolate belongs (see Krause‐Sakate et al., 2002). LMV‐Common and LMV‐Most are subclusters of the LMV‐RoW main cluster. LMV‐E and LMV‐AF199 are able to accumulate and induce symptoms in the systemic infected leaves of both mo1 1 and mo1 2 lettuce cultivars.
mo1 1 confers high resistance to LMV‐0 (no systemic virus accumulation and no symptoms), mo1 2 confers lower resistance (no symptoms although reduced systemic virus accumulation). Seed transmission concerns susceptible cultivars.
PATTERN OF LMV INVASION IN SUSCEPTIBLE AND RESISTANT MO1 LETTUCE
In order to follow the viral invasion of susceptible and resistant lettuce, GFP and GUS markers have been used for LMV‐E and LMV‐0. When fused to the N‐terminus of the viral protein HcPro, both reporter genes affect the biological properties of recombinant LMV isolates in both susceptible and resistant lettuce varieties (German‐Retana et al., 2000). Upon addition of the NIa cleavage site between the reporter gene and HcPro, in such a way that a nearly wild‐type HcPro is produced upon action of the NIa proteinase, LMV‐0 and LMV‐E recombinant viruses recovered the behaviour of their wild‐type parent (symptoms, viral accumulation) in susceptible plants (German‐Retana et al., 2003) (Fig. 3A–G). In mo1 2 plants, the recombinant LMV‐E modified in this way recovered the breaking properties of its wild‐type counterpart. In mo1 2 plants, the LMV‐0‐derived recombinants showed a severe inhibition in systemic accumulation (3, 4), despite the fact that neither cell‐to‐cell movement nor phloem loading or unloading seemed to be severely affected in an mo1 2 genetic context (German‐Retana et al., 2003). Although infection foci are present in the LMV‐0‐GUS‐inoculated leaves of both quasi‐isogenic lettuce cultivars Salinas (susceptible) and Salinas 88 (mo1 2, resistant) (Fig. 4A,B), LMV‐0‐GUS systemic movement in the upper non‐inoculated systemic leaves is detected only in the Salinas cultivar, in contrast to Salinas 88 (Fig. 4C). This suggests a restriction in long‐distance movement and in systemic accumulation of the tagged LMV‐0 recombinants in mo1 2 lettuce (3, 4). An interesting observation is that the systemic movement to the upper non‐inoculated leaves appears to be more affected than the downward movement to the root system (3, 4). In general, viral invasion of the root system has been poorly studied and in most cases, when analysed, both upward and downward systemic movements were affected. However, similar to the situation reported here with LMV, Guerini and Murphy (1999) showed that in the resistant pepper (Capsicum annuum) variety Avelar, carrying the recessive gene pvr3, downward movement of Pepper mottle virus (PepMoV) to the roots still occurred while systemic movement to upper non‐inoculated leaves was completely blocked due to a block of entry into the internal phloem.
Figure 3.
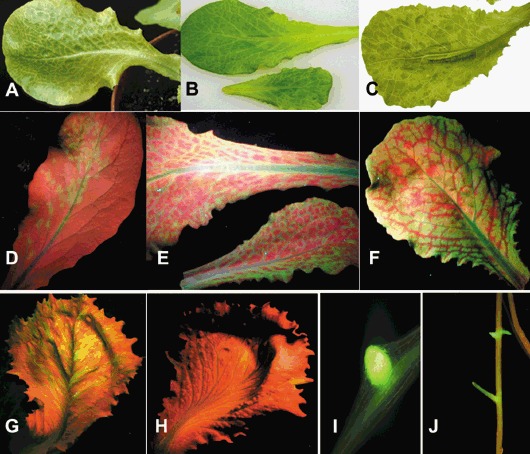
LMV‐GFP invasion in susceptible and resistant lettuce. (A–F) LMV‐GFP invasion in the susceptible lettuce cultivar Trocadéro. (A,B) Symptoms of vein clearing on the leaves located above the inoculated leaves at 10 dpi (days post inoculation). (C) Symptoms of mosaic on the upper non‐inoculated leaves at 20 dpi (photos taken under daylight). (D) GFP‐derived green fluorescence in the inoculated leaves showing infection foci at 7 dpi. (E) Vein clearing on the leaves located above the inoculated leaves at 10 dpi. (F) Mosaic on the upper non‐inoculated leaves at 20 dpi. (G) GFP‐derived green fluorescence in Salinas (susceptible cultivar) upper non‐inoculated leaves infected with LMV‐0‐GFP at 20 dpi. (H) Very sporadic GFP‐derived green fluorescence in Salinas 88 (resistant mo1 2) upper non‐inoculated leaves infected with LMV‐0‐GFP at 20 dpi. Detection of LMV‐0‐GFP in the roots of both Salinas (I) and Salinas 88 (J) cultivars at 20 dpi.
Figure 4.
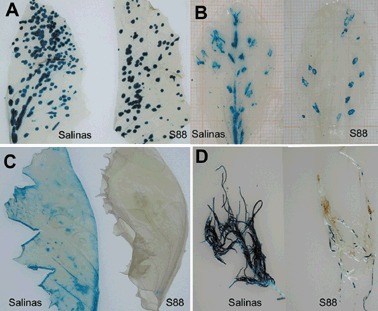
LMV‐GUS invasion in susceptible and resistant lettuce. Infection foci at 4 dpi (A) and 8 dpi (B) are present in inoculated leaves of both quasi‐isogenic lettuce cultivars: Salinas (left) and Salinas 88 (S88, right). The cell‐to‐cell movement of LMV‐0 is delayed in Salinas 88 compared with Salinas but is not abolished. (C) At 20 dpi, the LMV‐0‐GUS systemic movement in the upper non‐inoculated systemic leaves is detected in the Salinas cultivar, but not in Salinas 88. (D) At the same point‐time (20 dpi), LMV‐0‐GUS can be detected in the roots of both cultivars, although at a reduced rate in Salinas 88.
LMV‐GFP: TOOLS FOR LETTUCE BREEDERS
Beside their usefulness for studying viral invasion, both LMV‐0‐GFP and LMV‐E‐GFP can also be very useful tools to facilitate the screening of lettuce plants for LMV resistance, and identification of the resistance alleles present in a particular variety, both in vivo and in vitro (Candresse et al., 2002; Mazier et al., 2004). An evaluation of 101 cultivars of known status was carried out with these recombinant viruses and a 100% correlation was observed between LMV‐0‐GFP behaviour (whose systemic movement is abolished in resistant plants) and the mo1 resistance status. Similarly, the LMV‐E‐GFP (GFP fused to HcPro) allowed the identification of mo1 2 lines because its systemic movement was restricted in mo1 2 lines but not in susceptible or mo1 1 lines (Candresse et al., 2002). The use of these recombinant viruses can therefore greatly facilitate LMV resistance evaluation and speed up lettuce breeding programmes. Furthermore, the GFP LMV viruses constitute a simple and efficient tool for testing LMV resistance in in vitro cultivated lettuce, a method which reduces space requirements and improves environmental safety (Mazier et al., 2004).
LMV PATHOGENICITY DETERMINANTS IN SUSCEPTIBLE AND RESISTANT LETTUCE CULTIVARS MAP TO DIFFERENT REGIONS OF THE VIRAL GENOME
Severe symptoms on susceptible lettuce Trocadéro: role of HcPro
As with most viral diseases, the severity of symptoms induced by LMV isolates varies considerably depending on the host genotype, the stage of infection and the environmental conditions. In the susceptible cultivar Trocadéro, LMV isolates differ in their pathogenicity, namely in the severity of the symptoms they induce: while LMV‐0 and LMV‐AF199 induce relatively mild mosaic symptoms (Fig. 1E,F), LMV‐E induces severe mosaic symptoms accompanied by localized leaf necrosis, leaf deformation and general stunting of the infected plants (Fig. 1B–D) (Pink et al., 1992a). Analysis of the behaviour of recombinants constructed between LMV‐0 and LMV‐E determined that it is the HcPro protein of LMV‐E that causes the severe stunting and necrotic mosaic induced by this isolate in Trocadéro (Redondo et al., 2001). Involvement of HcPro in the determination of LMV symptom severity in Trocadéro was also demonstrated indirectly by analysis of the biological properties of GUS‐ or GFP‐tagged LMV‐E derivatives, in which the reporter gene was fused to the N terminus of HcPro, and their spontaneous deletion variants (German‐Retana et al., 2000). Indeed, the plants inoculated with the two tagged viruses or infected by the deletion mutants (lacking more than 100 amino acids in the N‐terminus of HcPro) failed to exhibit the severe stunting, leaf deformation or the necrotic reactions observed on LMV‐E‐inoculated plants.
Although the symptoms induced by LMV‐0 and LMV‐E are very different, the HcPro amino acid sequences of these two isolates are closely related, differing only in seven positions in the region identified as carrying the symptom determinant(s) for LMV (amino acids 35–286). These differences between LMV‐0 and LMV‐E HcPro proteins are scattered along this region and do not concern conserved motifs such as the FRNK block of amino acids implicated in symptom expression in Zucchini yellow mosaic virus (Gal‐On, 2000) nor the C‐terminus of HcPro involved in the necrosis response produced by strain PVYN of Potato virus Y in Nicotiana tabacum cv. Xanthi (Tribodet et al., 2005). Two‐dimensional crystals of LMV HcPro recombinant proteins revealed that HcPro of LMV is composed of two structural domains (domain 1 and 2) separated by a flexible constriction (the hinge domain) (Plisson et al., 2003). Amino acid region 35–286 of HcPro is associated with more than one structural domain of HcPro (domain 1 and the hinge domain). We hypothesize that domain 1 contains the active sites needed for various functions of HcPro and that the hinge domain regulates their accessibility by moving domain 2 to mask or expose domain 1. The movement of the hinge domain could be regulated by interactions with various hosts or viral proteins (Plisson et al., 2003).
VPg and other viral proteins play a role in overcoming mo1 1 and mo1 2 resistance
Although mo1 1 and mo1 2 resistance alleles of the mo1 gene are deployed worldwide and allow reasonably effective control of LMV disease, resistance‐breaking isolates such as LMV‐E and LMV‐AF199 may constitute a threat to the lettuce‐growing industry. In order to identify which region of the genome is responsible for the virulence of the resistance‐breaking isolates, recombinant isolates were constructed between LMV‐0 (common) and LMV‐E (resistance breaking). Using a reverse genetic approach, it was shown that the ability to overcome mo1 resistance and induce symptoms in the resistant cultivars was mapped to the 3′ half of the LMV‐E genome (Redondo et al., 2001), including the region encoding VPg. In any Potyviruses, the sequence of the central domain of VPg determines the ability to infect hosts harbouring recessive resistance genes from distinct plant families (Ayme et al., 2006, 2007; Borgstrom and Johansen, 2001; Keller et al., 1998; Moury et al., 2004; Nicolas et al., 1997; Rajamaki and Valkonen, 2002; Schaad et al., 1997). Although the identity of the viral genomic domain involved in the dialogue with recessive resistance (the central domain of the VPg) is remarkably conserved, the phenotypes associated with the corresponding resistance differ greatly, depending on the host and potyvirus partners considered: restriction of virus accumulation in single cells and inoculated leaves (Keller et al., 1998; Moury et al., 2004), restriction of long‐distance movement (Schaad et al., 1996; Schaad and Carrington, 1996). In lettuce, the mo1 1 and mo1 2 alleles are associated with a lack of symptoms or absence of systemic LMV accumulation depending on the virus isolate (Pink et al., 1992b; Revers et al., 1997a) and our results showed that neither the phloem loading not the phloem unloading was affected in resistant LMV‐0‐infected mo1 2 lettuce (German‐Retana et al., 2003). Recently we were able to narrow down the region carrying the LMV virulence to a portion of the LMV genome including the C‐terminal part of the CI protein as well as the 6K2 and VPg proteins (unpublished results). To date, the role of 6K2 in symptom induction and systemic movement has been described only for another potyvirus, Potato virus A (Spetz and Valkonen, 2004).
MOLECULAR DIALOGUE BETWEEN LMV AND LETTUCE: LOOKING FOR PLANT PARTNERS?
In the case of obligatory parasites such as viruses, absence or inadequacy of a single host factor may lead to the inability of the pathogen to multiply in the host or to invade it systemically (Ishikawa et al., 1997; Yamanaka et al., 2000). Such a mechanism implies that the dominant alleles of the host genes involved would be associated with susceptibility and the recessive alleles encoding non‐functional versions of this host factor with resistance.
Recessive resistance genes used to control Potyviruses agronomically have been estimated to represent about 40% of the known resistance genes (Provvidenti and Hampton, 1992). In the pathosystem lettuce/LMV, based on the observation that VPg was in the domain of the virus genome involved in mo1 breaking and that an interaction between VPg and the eukaryotic translation initiation factor 4E (eIF4E) had been described for two other potyvirus models (Schaad et al., 2000; Wittmann et al., 1997), we isolated three alleles of the lettuce eIF4E in their cDNA form, and obtained circumstantial and functional evidence that two of these alleles correspond to the recessive LMV resistance genes mo1 1 and mo1 2 (Nicaise et al., 2003). The immediate consequence of this conclusion is that mo1 1 and mo1 2 in lettuce are the mutant alleles of a unique mo1 gene encoding eIF4E.
During the last 5 years, it has been shown that natural mutations of components of the eukaryotic translation initiation complex that result in resistance to specific RNA viruses (especially Potyviruses) occur in a range of plant species (tomato, lettuce, pepper, pea, melon, barley, rice) (for reviews see Diaz‐Pendon et al., 2004; Maule et al., 2007; Robaglia and Caranta, 2006). However, how eIF4E is involved in the infection cycle in plants is currently not fully understood.
Several roles have been proposed for eIF4E in the potyvirus infection cycle based on its known biological and biochemical features. In particular, it was proposed that it could play a role during translation initiation through interaction with the genome‐linked protein VPg at the 5′ end of viral RNA (Lellis et al., 2002), which interacts with eIF4E in several plant–potyvirus systems (Beauchemin et al., 2007; Leonard et al., 2000; Roudet‐Tavert et al., 2007; Schaad et al., 2000; Wittmann et al., 1997). The in vitro interaction between the VPg of LMV and eIF4E from lettuce has been shown (Roudet‐Tavert et al., 2007) and characterized through spectroscopic studies (Michon et al., 2006). The central domain of the LMV VPg is involved in the interaction with the lettuce eIF4E. The VPg forms a ternary complex with both eIF4E and eIF4G, reducing eIF4E affinity for an mRNA cap analogue.
During mRNA translation, eIF4E provides the cap‐binding function and is associated with the protein eIF4G to form the eIF4F complex. Recently, susceptibility analyses of Arabidopsis mutants knocked‐out for At‐eIF4G genes showed that eIF4G factors are also indispensable for LMV infection, and that the eIF4G selective involvement parallels eIF4E recruitment, which suggests recruitment of the whole eIF4F for LMV infection in Arabidopsis (Nicaise et al., 2007).
These results could be simply interpreted as the 5′ VPg of LMV RNA functionally playing a role equivalent to the 5′ cap of cellular mRNAs, as recently shown for an animal calicivirus (Goodfellow et al., 2005). Through its interaction with VPg and possibly other host and virus factors, eIF4E might be involved in the control of the successive fates encountered by the viral RNA, such as intracellular and cell‐to‐cell trafficking (Arroyo et al., 1996; Gao et al., 2004). Another possible implication of eIF4E in the virus cycle could be to allow RNA circularization by interaction of the 5′ VPg with the 3′ poly A, mediated by the same protein complex as in mRNA translation, namely eIF4E‐eIF4G‐PABP. Beside a role in translation, genome circularization may be required for virus RNA replication or other processes of the infection cycle. Indeed, genome circularization is an important feature of the replication of Picornaviruses (Herold and Andino, 2001), relatives of Potyviruses infecting animal hosts.
To date, however, the function of eIF4E and eIF4G during the infection process remains to be elucidated. Roles in the early events of infection are the main candidate hypotheses: viral RNA translation and/or replication, circularization of viral RNA, host protein sequestration, or virus movement from infected to uninfected cells.
CONCLUSIONS
In recent years, our knowledge concerning both LMV and host proteins involved in LMV/lettuce interactions has improved significantly.
LMV pathogenicity determinants in lettuce map to different regions. While the HcPro is involved in the determination of symptom severity in Trocadéro, the 3′ half of the genome including the VPg enables systemic infection and symptom induction on cultivars carrying the genes mo1 1 and mo1 2. These results indicate that the ability of LMV to induce severe symptoms and to overcome the protection in lettuce afforded by the recessive allelic genes mo1 1 and mo1 2 are independent phenomena. Furthermore, recent data have enabled us to narrow down this viral 3′ region and indicate which protein(s) other than VPg are actually involved in the virulence of LMV.
The identity of the resistance mo1 genes has been clarified since they were shown to encode natural variants of the cap‐binding protein eIF4E, which are unable, in the homozygous state, to provide the cellular and molecular background necessary for LMV accumulation or symptom induction (Nicaise et al., 2003). In order to develop targeted resistance to LMV in lettuce, efforts can focus on screening of a large collection of lettuce natural or artificial mutants in the candidate eIF4E gene using TILLING (targeting induced local lesions in genomes) (McCallum et al., 2000).
All these discoveries should lead to a better understanding of the interactions between Potyviruses and their hosts, a challenge that our laboratory is tackling with the objective of providing new and more sustainable sources of resistance to various Potyviruses in various crops.
ACKNOWLEDGEMENT
This work was supported by a grant from the French National Agency for Research (Poty4E, ANR‐05‐BLAN‐0302‐01).
REFERENCES
- Adams, M.J. , Antoniw, J.F. and Fauquet, C.M. (2005) Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. [DOI] [PubMed] [Google Scholar]
- Arroyo, R. , Soto, M.J. , Martinez‐Zapater, J.M. and Ponz, F. (1996) Impaired cell‐to‐cell movement of Potato Virus Y in pepper plants carrying the ya (pvr2 1) resistance gene. Mol. Plant–Microbe Interact. 9, 314–318. [Google Scholar]
- Ayme, V. , Petit‐Pierre, J. , Souche, S. , Palloix, A. and Moury, B. (2007) Molecular dissection of the potato virus Y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. [DOI] [PubMed] [Google Scholar]
- Ayme, V. , Souche, S. , Caranta, C. , Jacquemond, M. , Chadoeuf, J. , Palloix, A. and Moury, B. (2006) Different mutations in the genome‐linked protein VPg of potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant–Microbe Interact. 19, 557–563. [DOI] [PubMed] [Google Scholar]
- Bannerot, H. , Boulidard, L. , Marrou, J. and Duteil, M. (1969) Etude de la tolérance au virus de la mosaïque de la laitue chez la variété Gallega de Invierno. Ann. Phytopathol. 1, 219–226. [Google Scholar]
- Beauchemin, C. , Boutet, N. and Laliberte, J.F. (2007) Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in Planta. J. Virol. 81, 775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgstrom, B. and Johansen, I.E. (2001) Mutations in pea seedborne mosaic virus genome‐linked protein VPg after pathotype‐specific virulence in Pisum sativum . Mol. Plant–Microbe Interact. 14, 707–714. [DOI] [PubMed] [Google Scholar]
- Candresse, T. , Le Gall, O. , Maisonneuve, B. , German‐Retana, S. and Redondo, E. (2002) The use of green fluorescent protein‐tagged recombinant viruses to test lettuce mosaic virus resistance in lettuce. Phytopathology, 92, 169–176. [DOI] [PubMed] [Google Scholar]
- Candresse, T. , Lot, H. , German‐Retana, S. , Krause‐Sakate, R. , Thomas, J. , Souche, S. , Delaunay, T. , Lanneau, M. and Le Gall, O. (2007) Analysis of the serological variability of Lettuce mosaic virus using monoclonal antibodies and surface plasmon resonance technology. J. Gen. Virol. 88, 2605–2610. [DOI] [PubMed] [Google Scholar]
- Clark, M.F. and Adams, A.N. (1977) Characteristics of the microplate method of enzyme‐linked‐immunosorbent‐assay for the detection of plant viruses. J. Gen. Virol. 34, 475–483. [DOI] [PubMed] [Google Scholar]
- Diaz‐Pendon, J.A. , Truniger, V. , Nieto, C. , Garcia‐Mas, J. , Bendahmane, A. and Aranda, M.A. (2004) Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 5, 223–233. [DOI] [PubMed] [Google Scholar]
- Dinant, S. and Lot, H. (1992) Lettuce Mosaic Virus: a review. Plant Pathol. 41, 528–542. [Google Scholar]
- Dinant, S. , Maisonneuve, B. , Albouy, J. , Chupeau, Y. , Chupeau, M. , Bellec, Y. , Gaudefroy, F. , Kusiak, C. , Souche, S. , Robaglia, C. and Lot, H. (1997) Coat protein mediated protection in Lactuca sativa against lettuce mosaic virus strains. Mol. Breeding, 3, 75–86. [Google Scholar]
- Falk, B.W. and Purcifull, D.E. (1983) Development and application of an enzyme‐linked immunosorbent assay (ELISA) test to index lettuce seeds for lettuce mosaic virus in Florida. Plant Dis. 67, 413–416. [Google Scholar]
- Gal‐On, A. (2000) A point mutation in the FRNK motif of the potyvirus helper component‐protease gene alters symptom expression in Cucurbits and elicits protection against the severe homologous virus. Phytopathology, 90, 467–473. [DOI] [PubMed] [Google Scholar]
- Gao, Z. , Johansen, E. , Eyers, S. , Thomas, C.L. , Noel Ellis, T.H. and Maule, A. J. (2004) The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell‐to‐cell trafficking. Plant J. 40, 376–385. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Candresse, T. , Alias, E. , Delbos, R. and Le Gall, O. (2000) Effects of GFP or GUS tagging on the accumulation and pathogenicity of a resistance breaking LMV isolate in susceptible and resistant lettuce cultivars. Mol. Plant–Microbe Interact. 13, 316–324. [DOI] [PubMed] [Google Scholar]
- German‐Retana, S. , Redondo, E. , Tavert‐Roudet, G. , Le Gall, O. and Candresse, T. (2003) Introduction of a NIa proteinase cleavage site between the reporter gene and HC‐Pro only partially restores the biological properties of GUS‐or GFP‐tagged LMV. Virus Res. 98, 151–162. [DOI] [PubMed] [Google Scholar]
- Goodfellow, I. , Chaudhry, Y. , Gioldasi, I. , Gerondopoulos, A. , Natoni, A. , Labrie, L. , Laliberte, J.F. and Roberts, L. (2005) Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 6, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan, R.G. , Welch, J.E. and Bardin, R. (1952) Common lettuce mosaic and its control by the use of mosaic‐free seeds. Phytopathology, 42, 573–578. [Google Scholar]
- Guerini, M.N. and Murphy, J.F. (1999) Resistance of Capsicum annuum‘Avelar’ to pepper mottle potyvirus and alleviation of this resistance by co‐infection with cucumber mosaic cucumovirus are associated with virus movement. J. Gen. Virol. 80, 2785–2792. [DOI] [PubMed] [Google Scholar]
- Herold, J. and Andino, R. (2001) Poliovirus RNA replication requires genome circularization through a protein‐protein bridge. Mol. Cell, 7, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, M. , Diez, J. , Restrepo‐Hartwig, M. and Ahlquist, P. (1997) Yeast mutations in multiple complementation groups inhibit brome mosaic virus RNA replication and transcription and perturb regulated expression of the viral polymerase‐like gene. Proc. Natl Acad. Sci. USA, 94, 13810–13815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger, I.C. (1921) A transmissible mosaic disease of lettuce. J. Agric. Res. 20, 737–741. [Google Scholar]
- Keller, K.E. , Johansen, I.E. , Martin, R.R. and Hampton, R.O. (1998) Potyvirus genome‐linked protein (VPg) determines pea seed‐borne mosaic virus pathotype‐specific virulence in Pisum sativum . Mol. Plant–Microbe Interact. 11, 124–130. [DOI] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Fakhfakh, H. , Peypelut, M. , Pavan, M.A. , Zerbini, F.M. , Marrakchi, M. , Candresse, T. and Le Gall, O. (2004) A naturally occurring recombinant isolate of Lettuce mosaic virus. Arch. Virol. 149, 191–197. [DOI] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Le Gall, O. , Fakhfakh, H. , Peypelut, M. , Marrakchi, M. , Varveri, C. , Pavan, M.A. , Souche, S. , Lot, H. , Zerbini, F.M. and Candresse, T. (2002) Molecular characterization of Lettuce mosaic virus field isolates reveals a distinct and widespread type of resistance‐breaking isolate: LMV‐Most. Phytopathology, 92, 563–572. [DOI] [PubMed] [Google Scholar]
- Krause‐Sakate, R. , Redondo, E. , Richard‐Forget, F. , Jadao, A.S. , Houvenaghel, M.C. , German‐Retana, S. , Pavan, M.A. , Candresse, T. , Zerbini, F.M. and Le Gall, O. (2005) Molecular mapping of the viral determinants of systemic wilting induced by a Lettuce mosaic virus (LMV) isolate in some lettuce cultivars. Virus Res. 109, 175–180. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou, P. (1985) A lethal strain of Lettuce mosaic virus in Greece. Phytoparasitica, 13, 271. [Google Scholar]
- Lellis, A.D. , Kasschau, K.D. , Whitham, S.A. and Carrington, J.C. (2002) Loss‐of‐susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Leonard, S. , Plante, D. , Wittmann, S. , Daigneault, N. , Fortin, M.G. and Laliberte, J.F. (2000) Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule, A.J. , Caranta, C. , and Boulton, M.I. (2007) Sources of natural resistance to plant viruses: status and prospects. Mol. Plant Pathol. 8, 223–231. [DOI] [PubMed] [Google Scholar]
- Mazier, M. , German‐Retana, S. , Flamain, F. , Dubois, V. , Botton, E. , Sarnette, V. , Le Gall, O. , Candresse, T. and Maisonneuve, B. (2004) A simple and efficient method for testing Lettuce mosaic virus resistance in in vitro cultivated lettuce. J. Virol. Methods, 116, 123–131. [DOI] [PubMed] [Google Scholar]
- McCallum, C.M. , Comai, L. , Greene, E.A. and Henikoff, S. (2000) Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michon, T. , Estevez, Y. , Walter, J. , German‐Retana, S. and Le Gall, O. (2006) The potyviral virus genome‐linked protein VPg forms a ternary complex with the eukaryotic initiation factors eIF4E and eIF4G and reduces eIF4E affinity for a mRNA cap analogue. Febs J. 273, 1312–1322. [DOI] [PubMed] [Google Scholar]
- Moury, B. , Morel, C. , Johansen, E. , Guilbaud, L. , Souche, S. , Ayme, V. , Caranta, C. , Palloix, A. and Jacquemond, M. (2004) Mutations in potato virus Y genome‐linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum . Mol. Plant–Microbe Interact. 17, 322–329. [DOI] [PubMed] [Google Scholar]
- Murphy, J.F. , Rychlik, W. , Rhoads, R.E. , Hunt, A.G. and Shaw, J.G. (1991) A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J. Virol. 65, 511–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhall, A.G. (1923) Seed transmission of lettuce mosaic. Phytopathology, 13, 104–106. [Google Scholar]
- Nicaise, V. , Gallois, J.L. , Chafiai, F. , Allen, L.M. , Schurdi‐Levraud, V. , Browning, K.S. , Candresse, T. , Caranta, C. , Le Gall, O. and German‐Retana, S. (2007) Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana . FEBS Lett. 581, 1041–1046. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , German‐Retana, S. , Sanjuan, R. , Dubrana, M.‐P. , Mazier, M. , Maisonneuve, B. , Candresse, T. , Caranta, C. and LeGall, O. (2003) The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus lettuce mosaic virus. Plant Physiol. 132, 1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, O. , Dunnington, S.W. , Gotow, L.F. , Pirone, T.P. and Hellmann, G.M. (1997) Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology, 237, 452–459. [DOI] [PubMed] [Google Scholar]
- Peypelut, M. , Krause‐Sakate, R. , Guiraud, T. , Pavan, M.A. , Candresse, T. , Zerbini, F.M. and Le Gall, O. (2004) Specific detection of lettuce mosaic virus isolates belonging to the ‘Most’ type. J. Virol. Methods, 121, 119–124. [DOI] [PubMed] [Google Scholar]
- Pink, D.A.C. , Kostova, D. and Walkey, D.G.A. (1992a) Differentiation of pathotypes of lettuce mosaic virus. Plant Pathol. 41, 5–12. [Google Scholar]
- Pink, D.A.C. , Lot, H. and Johnson, R. (1992b) Novel pathotypes of lettuce mosaic virus—breakdown of a durable resistance? Euphytica, 63, 169–174. [Google Scholar]
- Plisson, C. , Drucker, M. , Blanc, S. , German‐Retana, S. , Le Gall, O. , Thomas, D. and Bron, P. (2003) Structural characterization of HC‐Pro, a plant virus multifunctional protein. J. Biol. Chem. 278, 23753–23761. [DOI] [PubMed] [Google Scholar]
- Provvidenti, R. and Hampton, R.O. (1992) Sources of resistance to viruses in the Potyviridae. Arch. Virol. Suppl. 5, 189–211. [DOI] [PubMed] [Google Scholar]
- Rajamaki, M.L. and Valkonen, J.P. (2002) Viral genome‐linked protein (VPg) controls accumulation and phloem‐loading of a potyvirus in inoculated potato leaves. Mol. Plant–Microbe Interact. 15, 138–149. [DOI] [PubMed] [Google Scholar]
- Redondo, E. , Krause‐Sakate, R. , Yang, S.J. , Lot, H. , Le Gall, O. and Candresse, T. (2001) Lettuce mosaic virus (LMV) pathogenicity determinants in susceptible and tolerant lettuce varieties map to different regions of the viral genome. Mol. Plant–Microbe Interact. 14, 804–810. [DOI] [PubMed] [Google Scholar]
- Reichmann, J.L. , Lain, S. and Garcia, J.A. (1992) Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 73, 1–16. [DOI] [PubMed] [Google Scholar]
- Revers, F. , Lot, H. , Souche, S. , Le Gall, O. , Candresse, T. and Dunez, J. (1997a) Biological and molecular variability of Lettuce mosaic virus isolates. Phytopathology, 87, 397–403. [DOI] [PubMed] [Google Scholar]
- Revers, F. , Yang, S.J. , Walter, J. , Souche, S. , Lot, H. , Le Gall, O. , Candresse, T. and Dunez, J. (1997b) Comparison of the complete nucleotide sequences of two isolates of Lettuce mosaic virus differing in their biological properties. Virus Res. 47, 167–177. [DOI] [PubMed] [Google Scholar]
- Robaglia, C. and Caranta, C. (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. [DOI] [PubMed] [Google Scholar]
- Roudet‐Tavert, G. , Michon, T. , Walter, J. , Delaunay, T. , Redondo, E. and Le Gall, O. (2007) Central domain of a potyvirus VPg is involved in the interaction with the host translation initiation factor eIF4E and the viral protein HcPro. J. Gen. Virol. 88, 1029–1033. [DOI] [PubMed] [Google Scholar]
- Ryder, E.J. (1970) Inheritance of resistance to common lettuce mosaic. J. Am. Soc. Hort. Sci. 95, 378–379. [Google Scholar]
- Schaad, M.C. and Carrington, J.C. (1996) Suppression of long‐distance movement of tobacco etch virus in a nonsusceptible host. J. Virol. 70, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C. , Anderberg, R.J. and Carrington, J.C. (2000) Strain‐specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two‐hybrid system. Virology, 273, 300–306. [DOI] [PubMed] [Google Scholar]
- Schaad, M.C. , Haldeman‐Cahill, R. , Cronin, S. and Carrington, J.C. (1996) Analysis of the VPg‐proteinase (NIa) encoded by tobacco etch potyvirus: effects of mutations on subcellular transport, proteolytic processing, and genome amplification. J. Virol. 70, 7039–7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaad, M.C. , Lellis, A.D. and Carrington, J.C. (1997) VPg of tobacco etch potyvirus is a host genotype‐specific determinant for long‐distance movement. J. Virol. 71, 8624–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spetz, C. and Valkonen, J.P. (2004) Potyviral 6K2 protein long‐distance movement and symptom‐induction functions are independent and host‐specific. Mol. Plant–Microbe Interact. 17, 502–510. [DOI] [PubMed] [Google Scholar]
- Tomlinson, J.A. (1970) Lettuce mosaic virus. CMI/AAB Descriptions of Plant Viruses, 9, Kew, UK, Commonwealth Mycological Institute/Association of Applied Biology. [Google Scholar]
- Tribodet, M. , Glais, L. , Kerlan, C. and Jacquot, E. (2005) Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 86, 2101–2105. [DOI] [PubMed] [Google Scholar]
- Walkey, D.G.A. , Ward, C.M. and Phelps, K. (1985) Studies on lettuce mosaic virus resistance in commercial lettuce cultivars. Plant Pathol. 34, 545–551. [Google Scholar]
- Wittmann, S. , Chatel, H. , Fortin, M.G. and Laliberté, J.F. (1997) Interaction of the viral protein genome linked of turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two‐hybrid system. Virology, 234, 84–92. [DOI] [PubMed] [Google Scholar]
- Yamanaka, T. , Ohta, T. , Takahashi, M. , Meshi, T. , Schmidt, R. , Dean, C. , Naito, S. and Ishikawa, M. (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl Acad. Sci. USA, 97, 10107–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.J. , Revers, F. , Souche, S. , Lot, H. , Le Gall, O. , Candresse, T. and Dunez, J. (1998) Construction of full‐length cDNA clones of Lettuce mosaic virus (LMV) and the effects of intron‐insertion on their viability in Escherichia coli and on their infectivity to plants. Arch. Virol. 143, 2443–2451. [DOI] [PubMed] [Google Scholar]
- Zheng, T. , Chen, J. and Chen, J. (2002) Complete sequence analysis of a Chinese isolate of lettuce mosaic virus. Chin. J. Virol. 18, 66–70. [Google Scholar]


