SUMMARY
Noncoding RNAs (ncRNAs) play increasingly appreciated gene-regulatory roles. Here, we describe a regulatory network centered on four ncRNAs—a long ncRNA, a circular RNA, and two microRNAs—using gene editing in mice to probe the molecular consequences of disrupting key components of this network. The long ncRNA Cyrano uses an extensively paired site to miR-7 to trigger destruction of this microRNA. Cyrano-directed miR-7 degradation is much more effective than previously described examples of target-directed microRNA degradation, which come primarily from studies of artificial and viral RNAs. By reducing miR-7 levels, Cyrano prevents repression of miR-7–targeted mRNAs and enables accumulation of Cdr1as, a circular RNA known to regulate neuronal activity. Without Cyrano, excess miR-7 causes cytoplasmic destruction of Cdr1as in neurons, in part through enhanced slicing of Cdr1as by a second miRNA, miR-671. Thus, several types of ncRNAs can collaborate to establish a sophisticated regulatory network.
INTRODUCTION
MicroRNAs (miRNAs) are abundant ~22 nt RNAs that regulate gene expression by base-pairing to mRNAs to induce mRNA decay and inhibit translation (Bartel, 2018; Jonas and Izaurralde, 2015). Most miRNAs are extremely stable, with half-lives of days (Bail et al., 2010; Duffy et al., 2015; Gantier et al., 2011; Guo et al., 2015). This stability is attributed to their organization within the effector protein Argonaute, which protects them from exonucleases that degrade naked single-stranded RNAs in the cell. Nonetheless, some miRNAs are destabilized, either constitutively or in response to neuronal excitation or growth factors (Avraham et al., 2010; Hwang et al., 2007; Krol et al., 2010; Rissland et al., 2011).
One way that miRNAs are destabilized is through target RNA–directed miRNA degradation (TDMD). Artificial targets with extensive complementarity to the miRNA can trigger TDMD through a poorly understood process associated with tailing (addition of untemplated nucleotides) and trimming of the miRNA 3′ terminus (Ameres et al., 2010; Baccarini et al., 2011; de la Mata et al., 2015; Denzler et al., 2016; Xie et al., 2012). Natural TDMD triggers are found in some herpesviruses, which use this mechanism to promote degradation of specific host miRNAs (Cazalla et al., 2010; Libri et al., 2012; Marcinowski et al., 2012; Lee et al., 2013). In addition, a cellular transcript with TDMD activity limits miR-29b expression in cerebellar granule neurons and regulates behavior in mice and fish (Bitetti et al., 2018).
Long noncoding RNAs (lncRNAs) are transcribed and processed like mRNAs but do not code for functional proteins. Many lncRNAs have been annotated, but few have known functions (Kopp and Mendell, 2018). The lncRNA Cyrano is particularly intriguing because it is broadly conserved in vertebrates and contains a site with unusually high complementarity to the miR-7 miRNA (Ulitsky et al., 2011). Another class of abundant yet enigmatic RNAs is the circular RNAs (circRNAs), which are formed through back-splicing of mRNA or lncRNA exons (Ebbesen et al., 2017). Among the most extensively characterized circRNAs is Cdr1as, which is conserved in mammals and dampens neuronal activity in mice (Hansen et al., 2013; Memczak et al., 2013; Piwecka et al., 2017). Cdr1as has many sites to miR-7 (130 and 73 sites in mouse and human Cdr1as, respectively), the same miRNA that pairs to Cyrano, and Cdr1as and Cyrano are the two RNAs that most frequently cross-link to miR-7 in human and mouse brain (Piwecka et al., 2017). Here, we show that Cyrano promotes unusually efficient destruction of mature miR-7, which enables Cdr1as to accumulate in the brain.
RESULTS
Cyrano Promotes miR-7 Destruction
In zebrafish, Cyrano is expressed throughout the developing nervous system (Ulitsky et al., 2011). In the mouse embryo, Cyrano was also enriched in the early central and peripheral nervous systems (Figure S1A). In adult mice, Cyrano was broadly expressed with 5–10-fold higher expression in brain regions and muscle, a pattern largely conserved in human (Figure S1B-C). Within brain, Cyrano appeared more highly expressed in neurons than glia (Figure S1A, D), and within cells, Cyrano was mostly in cytoplasm (Figure S1E).
To investigate the function of Cyrano in mammals, we generated Cyrano-deficient mice. The first half of exon 3, which includes the most conserved region of Cyrano, was targeted (Figure 1A, S1F). RNA-seq confirmed complete loss of the targeted region as well as a >90% decrease in flanking Cyrano RNA, presumably due to destabilization caused by removal of the final splice site (Figure 1A). Cyrano-deficient mice were born at the expected Mendelian frequency and had no overt abnormalities (Table S1). Cyrano−/− adult mice had similar weights and one-year survival compared to sex-matched littermate controls, and Cyrano loss did not seem to affect performance in standard neurobehavioral tests (data not shown). In contrast, Cyrano knockdown in zebrafish appears to cause a neurodevelopmental phenotype (Ulitsky et al., 2011), a difference that might be due to the different experimental approaches used to query Cyrano loss of function, as reported for other genes (Kok et al., 2015). Despite their seemingly normal appearance, Cyrano−/− mice had striking molecular phenotypes.
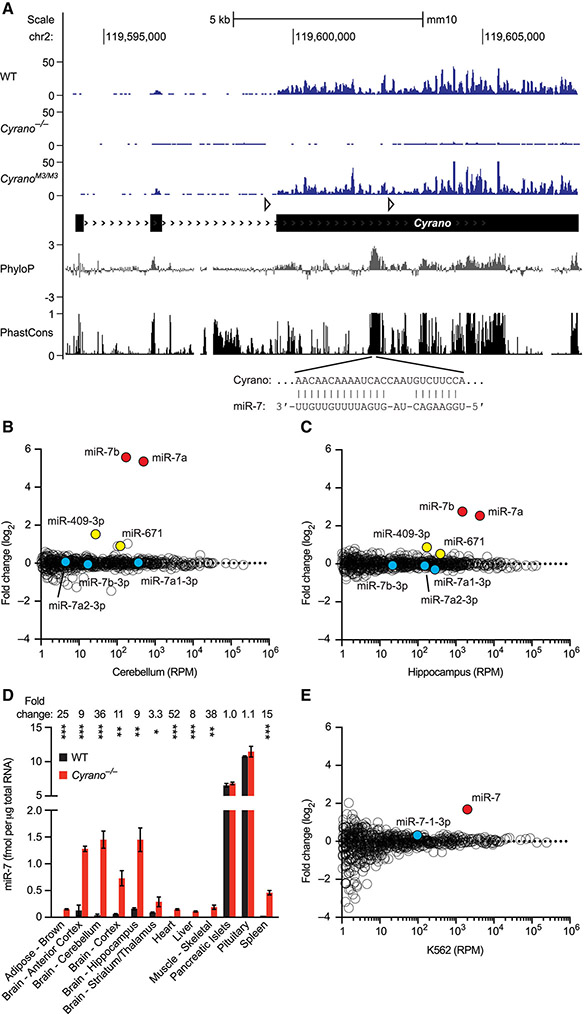
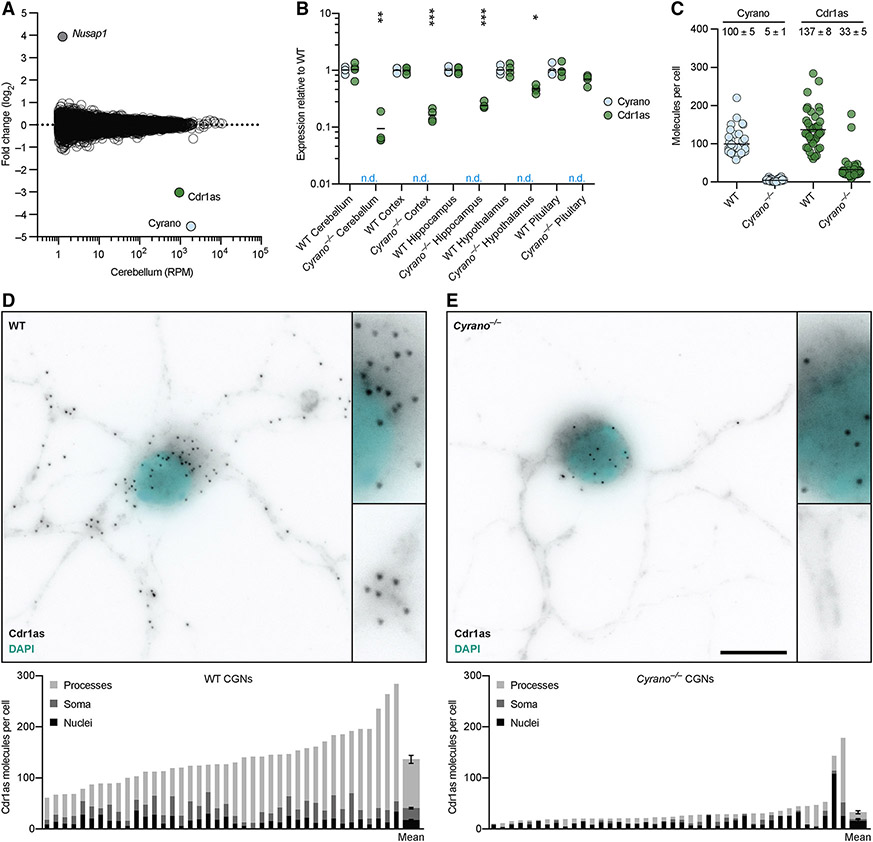
Figure 1. Cyrano Promotes miR-7 Destruction.
(A) Organization, conservation and expression of the murine Cyrano locus. Shown below the gene model (black boxes, exons; > > >, introns; open triangles, loxP sites), are conservation plots (PhyloP and PhastCons), which are based on a 40-genome placental mammal alignment generated relative to the mouse locus (Pollard et al., 2010; Siepel et al., 2005). Diagramed below that is the pairing of miR-7 to its highly complementary site, located within the most conserved region of Cyrano. Shown above the gene model are RNA-seq tracks for wild-type, Cyrano−/− and CyranoM3/M3 cerebellum (y-axis, reads) derived from libraries with similar sequencing depths.
(B) Influence of Cyrano on miRNA levels in cerebellum. Shown are log2-fold changes in mean miRNA levels observed between wild-type and Cyrano−/− cerebellum, as determined by small-RNA sequencing and plotted as a function of expression in wild-type cerebellum (n = 3 per genotype). Each circle represents one miRNA, showing results for all miRNAs expressed above 1 RPM (read per million miRNA reads) in wild-type cerebellum. Circles representing miR-7 paralogs, miR-7 passenger strands, and other indicated miRNAs are colored red, blue, and yellow respectively.
(C) Influence of Cyrano on miRNA levels in hippocampus; otherwise, as in (B).
(D) Influence of Cyrano on miR-7 levels in 12 mouse tissues. Plotted are mean levels of miR-7 per μg total RNA for wild-type (black) and Cyrano−/− (red) tissues, as determined by northern blots. For each tissue, 3–4 replicates of each genotype were analyzed alongside known quantities of miR-7 synthetic standards (error bars, s.d.). Statistically significant fold changes are indicated (*, p <0.05; **, p <0.01; ***, p <0.001, unpaired two-tailed t-test).
(E) Influence of Cyrano on miRNA levels in K562 cells; otherwise, as in (B).
See also Figures S1–S2 and Table S1.
The most conserved region of Cyrano contains an unusual site for miR-7 (Figure 1A). In addition to pairing to the miRNA extended seed region (nucleotides 2–8), a common feature of effective miRNA sites (Bartel, 2018), this site has extensive pairing to the remainder of the miRNA (Ulitsky et al., 2011), with only a short internal loop that disrupts pairing to miRNA nucleotides 9 and 10 and should prevent Argonaute2-catalyzed slicing of Cyrano (Elbashir et al., 2001). To determine whether Cyrano deficiency affected accumulation of miR-7 or other miRNAs, we sequenced small RNAs from both wild-type and Cyrano−/− brain samples. In humans, all three MIR7 loci express identical miRNAs, whereas in mice the three loci express two variants, miR-7a and miR-7b, which differ by a single nucleotide (A or U, respectively) at position 10. In Cyrano−/− mice, miR-7a and miR-7b increased 40 and 47 fold in cerebellum and 6 and 7 fold in hippocampus (Figure 1B–C). Only two other miRNAs, miR-671 and miR-409-3p, were differentially expressed in both tissues, changing 1.4–2.8 fold.
In principle, increased miR-7 could be caused by 1) increased transcription of Mir7 genes, 2) enhanced processing of primary or precursor miR-7 RNAs, and/or 3) decreased miR-7 degradation. To distinguish between these possibilities, we examined levels of intermediates in the miRNA-processing pathway. No changes were observed in either primary or precursor miR-7 RNAs in Cyrano−/− mice (Figure S2A-B). Most importantly, whereas possibilities 1 and 2 would each generate more miRNA duplex (a biogenesis intermediate in which the miRNA is paired to its passenger strand), our small-RNA sequencing results revealed no increase in miR-7 passenger strands (Figure 1B–C, species with 3p suffixes), which implied that Cyrano promotes degradation of mature miR-7 after it is loaded into Argonaute.
To further characterize the effect of Cyrano loss on miR-7 levels, we performed northern-blot analyses of samples from 12 adult tissues spanning the range of Cyrano expression (Figure 1D). In Cyrano−/− pituitary and pancreatic islets, two tissues with relatively low expression of Cyrano and the highest baseline levels of miR-7, the miR-7 fold changes were not statistically significant. However, in all other Cyrano-deficient tissues tested, miR-7 levels significantly increased, with magnitudes ranging from 3–52 fold and increases most prominent in brain tissues. Overall, miR-7 fold changes correlated with Cyrano expression (Figure S2C), as expected if Cyrano exerts greater effects in tissues in which it is more highly expressed. Increased levels of miR-7 were reliably observed in Cyrano−/− embryos as early as E13.5 (Figure S2D) and were also observed in primary cultures of Cyrano−/− neurons, where they increased 16–45 fold (Figure S2E).
To test whether regulation of miR-7 was conserved in humans, we used CRISPRi (Gilbert et al., 2013) to inhibit CYRANO transcription in human cells. CYRANO reduction in K562 cells led to a specific increase in miR-7, with no significant change in miR-7-3p (Figure 1E). Similar results were also observed in additional human and mouse cell lines, with the increase in miR-7 correlating with the degree of CYRANO knockdown (Figure S2F). These findings confirmed that the ability of Cyrano to destabilize miR-7 is conserved in humans and requires transcription of the CYRANO locus.
The miR-7 Site in Cyrano Directs Unusually Effective miR-7 Destruction
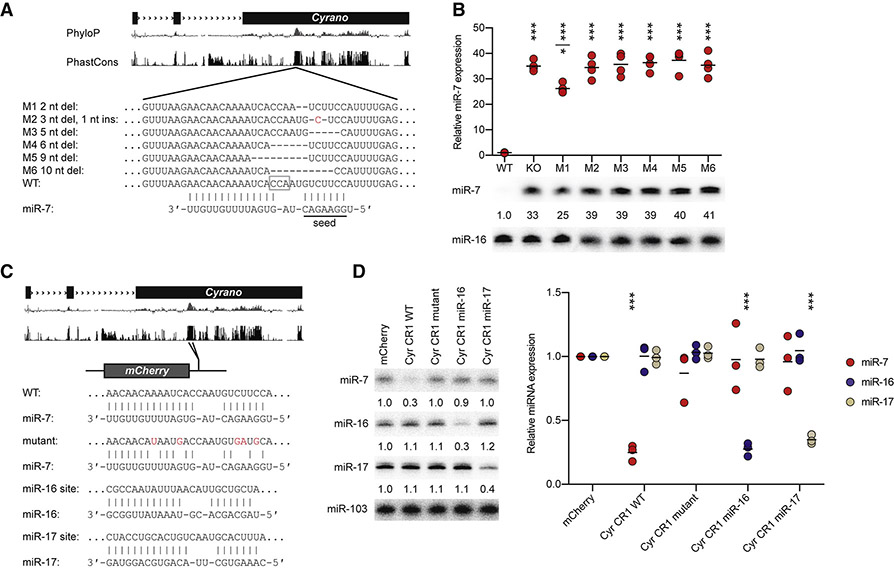
Based on previous studies of TDMD triggered by artificial and viral transcripts, we suspected that the miR-7 site within Cyrano was crucial for the ability of this endogenous transcript to direct miRNA degradation. To test this, we used Cas9 to generate six mouse lines (M1–M6) with perturbations in the site (Figure 2A). Although some of the mutations were expected to disrupt miR-7 binding (Bartel, 2018), none affected expression of Cyrano in the tissues tested (cerebellum and pituitary) (Figure S3). In contrast, all six disrupted the ability of Cyrano to induce miR-7 degradation (Figure 2B). The M2 and M3 mutations only perturbed miR-7 seed pairing, and mice harboring these mutations had miR-7 increases resembling those of Cyrano-deficient mice, indicating that, as expected, seed pairing is critical for this interaction. More surprisingly, the M1 allele, which had the smallest perturbation, a 2 nt deletion eliminating only a single Watson–Crick pair to the extended seed region and reducing the size of the internal loop from 4 to 3 nucleotides, was also largely unable to promote miR-7 destruction. Overall, the effects of perturbing the miR-7 site demonstrated that Cyrano acts through a form of TDMD reminiscent of that initially observed for artificial and viral transcripts.
Figure 2. The miR-7 Site in Cyrano Specifies miR-7 Degradation.
(A) Schematic of wild-type and Cyrano mutant alleles that alter the extensively paired miR-7 site. Shown below the gene model and conservation tracks are the wild-type Cyrano miR-7 site and six mutant sites, M1–M6. The PAM motif used for mutagenesis is outlined (open gray box). Deleted nucleotides are indicated by dashes; an inserted nucleotide is colored red. The miR-7 extended seed is underlined.
(B) Importance of the miR-7 site for Cyrano function in cerebellum. At the bottom is a representative northern blot measuring miR-7 levels in cerebellum of the lines described in (A). In each lane, the level of miR-7 was normalized to that of miR-16 and reported relative to wild-type. Plotted above are relative miR-7 levels determined from all replicates (black bars, mean; n = 4 per genotype). Statistically significant changes compared to wild type are indicated (***, p <0.001, ANOVA with Tukey’s test). The miR-7 level in M1 cerebellum also differed from that in Cyrano−/− (KO) and M2–M6 cerebellum (*, p <0.05, ANOVA with Tukey’s test).
(C) Schematic of the wild-type CR1 (Cyrano conserved region) construct and constructs that changed its miR-7 site. The miR-7 site in the CR1 was mutated to either disrupt pairing to miR-7 or introduce an extensively complementary site to miR-16 or miR-17.
(D) Importance of the complementary site for specifying CR1 function. At the left is a representative northern blot measuring miR-7, miR-16, and miR-17 levels in HEK293T cells transfected with the constructs described in (C). Below each panel, levels of miRNA are reported relative to levels in the control mCherry transfection after normalizing to the level of miR-103. Plotted at the right are relative levels of miR-7 (red), miR-16 (blue), and miR-17 (tan) (black bars, mean; n = 3). Statistically significant changes compared to the mCherry control are indicated (***, p <0.001, ANOVA with Dunnett’s test).
One important difference between TDMD mediated by Cyrano and that described previously was its efficacy, as indicated by the decrease in miR-7 molecules per Cyrano molecule observed at steady state. In wild-type cerebellar granule neuron (CGN) cultures, Cyrano was present at 102 ± 16 molecules per cell, and miR-7 was present at 40 ± 7 molecules per cell, whereas in Cyrano-deficient CGNs, miR-7 levels increased 45 fold to 1800 molecules per cell (Figure S2E, Table S2). These values implied that Cyrano promoted TDMD with multiple turnover, with each Cyrano molecule responsible for destruction of an average of 17 miR-7 molecules [(1800 – 40) /102], especially when considering that the insensitivity of Cyrano to perturbation of either miR-7 pairing (Figure S3) or miR-7 abundance (described later) disfavored a model in which Cyrano degradation was coupled to miR-7 degradation. In contrast, similar calculations using values reported for previous examples of TDMD estimated the efficacy of viral HSUR1 to be ~0.1 miR-27 molecules per target (Cazalla et al., 2010; Lee et al., 1988) and that of artificial single-site transcripts to be either 0.1–0.8 miR-122 molecules per target (Denzler et al., 2016), or 0.3–1.0 miRNA molecules per target, with multiple turnover achieved only when artificially boosting the miRNA level (de la Mata et al., 2015). These estimates indicated that Cyrano-promoted TDMD is 17–170 times more effective than that of previously described examples.
To investigate further the specificity of this phenomenon, we examined whether the miR-7–destabilizing activity of Cyrano could be applied to other miRNAs. The 3′ UTR of a reporter gene was modified to include a 197 nt fragment of Cyrano, using versions of this region that contained either a wild-type miR-7 site, a mutant miR-7 site, or a site extensively complementary to either miR-16 or miR-17 (Figure 2C). Decreased miR-7 was observed only in cells expressing the wild-type Cyrano region, whereas decreased miR-16 or miR-17 was observed specifically in cells expressing the region with cognate sites (Figure 2D). These results resembled those of analogous experiments that start with viral or artificial TDMD-triggering transcripts, and like those previous experiments, the amount of degradation was modest, with only ~3-fold reductions of the cognate miRNA observed for our constructs in HEK293T cells—much less than the 45-fold reduction observed for endogenous Cyrano in CGN culture (Figure S2E).
Cyrano Induces Tailing and Trimming of miR-7, but Tailing Appears Dispensable
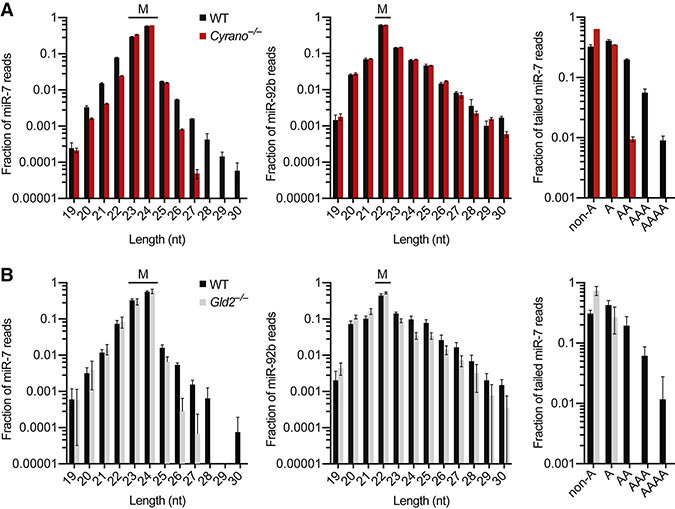
Tailing and trimming of the miRNA 3′ end are often associated with TDMD (Ameres et al., 2010; Baccarini et al., 2011; de la Mata et al., 2015; Denzler et al., 2016; Marcinowski et al., 2012; Xie et al., 2012). Likewise, we found that the fractions of tailed (≥25 nt) and trimmed (≤22 nt) miR-7 molecules were greater in wild-type than in Cyrano−/− hippocampus and cerebellum (Figure 3A, S4A). Such differences were not observed for any other miRNA with read coverage sufficient for analysis (Figure S4B-C), as exemplified by results for miR-92b (Figure 3A, S4A).
Figure 3. Cyrano Induces Tailing and Trimming of miR-7, but Tailing Appears Dispensable.
(A) Influence of Cyrano on miR-7 tailing and trimming in hippocampus. Plotted are length distributions of miR-7 reads (left) and miR-92b reads (middle) from wild-type (black) and Cyrano−/− (red) tissue, indicating the mature species for each miRNA (M) (error bars, s.d.; n = 3 per genotype). Also plotted are the fractions of tailed miR-7 reads that were mono- or oligoadenylylated (right) in wild-type and Cyrano−/− tissue.
(B) Influence of Gld2 on miR-7 tailing and trimming in hippocampus. Primary data from Mansur et al. (2016) were analyzed as in (A), with results for Gld2−/− tissue plotted in gray (n = 6 per genotype).
See also Figure S4.
In flies, tailing is primarily through addition of untemplated uridines (Ameres et al., 2010; Ameres et al., 2011), whereas in mammals, adenylylation appears more frequently than uridylylation, although both have been reported (Ameres et al., 2010; Baccarini et al., 2011; de la Mata et al., 2015; Marcinowski et al., 2012; Xie et al., 2012). We found that Cyrano mostly increased the fraction of mature miR-7 reads with two or more untemplated adenosines, whereas monoadenylylation and tailing with other nucleotides were less affected (Figure 3A, S4A). These results suggested that the cytoplasmic poly(A) polymerase Gld2 might be responsible for this activity. Indeed, re-analyses of small-RNA sequencing data from wild-type and Gld2−/− hippocampus (Mansur et al., 2016) revealed that Gld2 deletion substantially reduced the proportion of tailed miR-7 reads and eliminated oligoadenylylated miR-7 reads (Figure 3B). These results for Gld2 deletion mirrored those observed for Cyrano deletion, except the influence of Gld2 was not specific to miR-7 (Figure 3B, S4D). Although Gld2 appeared responsible for Cyrano-dependent oligoadenylylation of miR-7, unlike loss of Cyrano (Figure 3A, S4C), loss of Gld2 caused no decrease in the proportion of trimmed miR-7 reads (Figure 3B, S4E). Moreover, loss of Gld2 did not significantly affect levels of any miRNA, including miR-7 (Mansur et al., 2016) (Figure S4F). Likewise CRISPRi stable knockdown of PAPD4 (the human Gld2 homolog) by as much as 98% in K562 cells did not affect miR-7 levels (Figure S4G). These results showed that oligoadenylylation observed in the presence of Cyrano can be uncoupled from both trimming and degradation of miR-7. Results of additional experiments supported the idea that the miR-7 tailing observed in the presence of Cyrano is not on-pathway for Cyrano-directed miR-7 degradation (Figure S4H-I), which further implied that tailing might not be an obligate step of TDMD.
Loss of Cyrano Causes Increased miR-7 Target Repression
The change in miR-7 from very low to intermediate levels in many Cyrano-deficient tissues (Figure 1D) raised the question of whether miR-7 targets might be affected. To answer this question, we performed RNA-seq on wild-type and Cyrano−/− tissues and compared fold changes of predicted miR-7 targets to fold changes of mRNAs lacking a miR-7 site. The effects on all predicted targets were compared to the effects on both conserved predictions and the top ~80 predictions (Agarwal et al., 2015), expecting greater signal for subsets predicted with higher confidence (i.e., top targets > conserved targets > all targets).
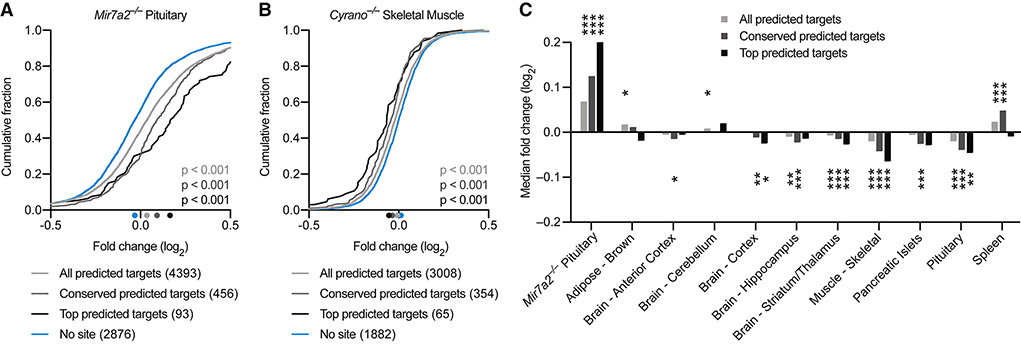
For context, we first processed RNA-seq data comparing wild-type to Mir7a2−/− pituitary (Ahmed et al., 2017). miR-7 is the most abundant miRNA in pituitary, and deletion of Mir7a2 leads to greatly reduced miR-7 levels in pituitary, an increase in predicted miR-7 targets, and infertility (Ahmed et al., 2017). Accordingly, we observed significant derepression of predicted miR-7 targets in Mir7a2−/− pituitary, with greater derepression for higher-confidence predictions (Figure 4A). When analyzing Cyrano−/− tissues, strong evidence for increased miR-7 target repression was observed in three of ten tissues tested—striatum/thalamus, skeletal muscle, and pituitary (Figure 4B, 4C). Detection of increased repression in pituitary suggested that miR-7 increased in pituitary, even though the increase did not pass our threshold for statistical significance when measured by northern blots (Figure 1D). Cyrano−/− spleen had an unanticipated increase in predicted miR-7 targets, although this effect did not persist when considering only top predictions. In tissues with increased miR-7 target repression, the amount of repression was substantially less than the effect of deleting miR-7 in pituitary (Figure 4), consistent with our results showing that absolute levels of miR-7 in Cyrano-deficient tissues were still ≥7-fold lower than that of wild-type pituitary (Figure 1D). Despite modest effect sizes, these results showed that by limiting miR-7 accumulation, Cyrano attenuates repression of miR-7 targets in some tissues.
Figure 4. Loss of Cyrano Can Cause Increased miR-7 Target Repression.
(A) Repression of predicted miR-7 targets by miR-7a-2 in pituitary. Plotted are cumulative distributions of mRNA fold changes observed after deleting Mir7a2 in pituitary (Ahmed et al., 2017), comparing the impact on all predicted miR-7 targets (light gray), conserved predicted miR-7 targets (dark gray), and top predicted miR-7 targets (black) to that of control mRNAs with no miR-7 site (blue). Median log2-fold changes for each set of mRNAs are indicated (colored dots below the x-axis). Statistical significance of differences between each set of predicted targets and control mRNAs was determined by the Mann–Whitney test.
(B) Influence of Cyrano on predicted targets of miR-7 in skeletal muscle. Plotted are cumulative distributions of mRNA fold changes observed in skeletal muscle after disrupting Cyrano; otherwise, as in (A).
(C) Influence of Cyrano on predicted targets of miR-7 in ten tissues. Plotted are median fold changes observed upon Cyrano disruption for the indicated sets of predicted miR-7 targets, normalized to the median fold change of control genes. For reference, effects of deleting Mir7a2 in pituitary, as evaluated in (A), are also shown (leftmost set of bars). For Cyrano-deficient tissues, the number of genes analyzed in each set ranged from 3008–4035 (all), 354–453 (conserved), 63–83 (top), and 1882–2531 (no site). Statistically significant changes from the control sets are indicated (*, p <0.05; **, p <0.01; ***, p <0.001, Mann–Whitney test).
Cyrano Enables the Cdr1as circRNA to Accumulate in the Brain
The statistically significant signal for increased miR-7 repression observed in some Cyrano-deficient tissues was attributable to many miR-7 targets being repressed—but each by only a small amount. Indeed, when searching for any genes that on their own were differentially expressed in Cyrano−/− cerebellum with adjusted p value <0.01, only three were identified (Figure 5A). One was Cyrano, as expected (Figure 1A), and another was the downstream gene Nusap1, which increased due to read-through transcription and aberrant splicing (Figure S5A). The third was the gene for Cdr1as, the circRNA with 130 sites to miR-7. Its expression decreased 10 fold in Cyrano−/− cerebellum.
Figure 5. Cyrano Enables Cdr1as to Accumulate in the Brain.
(A) Influence of Cyrano on RNA levels in cerebellum. Shown are fold changes in mean RNA levels observed between wild-type and Cyrano−/− tissue (n = 4 per genotype), as determined by RNA-seq and plotted as a function of expression in wild-type tissue. Each circle represents a unique mRNA or noncoding RNA, showing results for all RNAs expressed above 1 RPM in wild-type tissue. Circles representing Cyrano, Cdr1as, and Nusap1 are colored and labeled.
(B) Influence of Cyrano on Cdr1as expression in brain and pituitary. Levels of Cyrano (blue) and Cdr1as (green), as determined by RT-qPCR, were normalized to that of Actb and plotted relative to mean wild-type expression (black bars, mean; n = 4 per genotype; n.d., not detected). Statistically significant changes in Cdr1as levels in Cyrano−/− tissues compared to wild-type tissues are indicated (*, p <0.05; **, p <0.01; ***, p <0.001, unpaired two-tailed t-test).
(C) Influence of Cyrano on Cdr1as expression in DIV11 CGNs, as determined by single-molecule FISH. Plotted are the number of Cyrano and Cdr1as molecules per cell for wild-type and Cyrano−/− CGNs. Each circle represents mean molecules per cell from a 100x microscopy field, each field containing 1–3 cells (n = 40 fields per genotype; black bars, mean). The mean of all fields for each genotype (± s.e.m.) is also reported.
(D–E) Influence of Cyrano on accumulation of Cdr1as in neuronal cell bodies and processes. Shown is a representative image from single-molecule FISH experiments probing for Cdr1as in either wild-type (D) or Cyrano−/− (E) CGNs, with insets showing increased magnification of a portion of soma and processes (Cdr1as, black; nuclei, cyan; scale bar, 10 μm). Below each image is quantification from the same 40 fields quantified in (C), with each bar reporting mean molecules per cell, differentiating molecules located in nuclei (black), soma (excluding nuclei, dark gray), and processes (light gray), plotting at the far right of each panel the means ± s.e.m.
See also Figure S5.
Although our RNA-seq libraries were generated after poly(A) selection, which should not efficiently capture circRNAs, very few reads mapped beyond the portion of Cdr1as known to circularize (Figure S5B), suggesting that the RNA-seq was in fact capturing the circular isoform. Moreover, RT–qPCR analysis using primers that spanned the back-splice junction (and were thus specific to the circular isoform) found a similar 10-fold reduction in Cdr1as in Cyrano-deficient cerebellum (Figure 5B). In wild-type mice, Cdr1as is expressed in neurons throughout the brain, with highest expression in cerebellum (Hansen et al., 2013; Memczak et al., 2013; Piwecka et al., 2017) (Figure S5C). Cdr1as levels decreased throughout Cyrano-deficient brain, with the greatest effect observed in cerebellum, followed by cortex, hippocampus, and hypothalamus (Figure 5B), resembling the rank order of miR-7 fold changes in these regions (Figure 1D).
To determine the subcellular location of Cyrano activity, we performed single-molecule FISH (fluorescence in situ hybridization), probing for Cyrano and Cdr1as in cultured CGNs, the most abundant neuron in the cerebellum. An average of 100 molecules of Cyrano and 137 molecules of Cdr1as were observed in wild-type CGNs (Figure 5C), which agreed with our RT–qPCR estimates (Table S2). Cyrano was observed predominantly in cytoplasm (73%) with molecules distributed in both soma and processes (Figure S5D-E). As previously described in cortical neurons (Piwecka et al., 2017), Cdr1as was also predominantly localized to cytoplasm (86%), with most molecules found in processes (Figure 5D). In Cyrano−/− CGNs, a few Cyrano transcripts were detected (Figure S5D-E), consistent with our RNA-seq results (Figure 1C). The number of Cdr1as molecules also decreased in nearly all neurons (Figure 5C), and strikingly, this decrease was entirely attributable to depletion from soma and processes (Figure 5E). The preservation of Cdr1as levels in the nucleus suggested that the large reduction of Cdr1as in Cyrano−/− cells occurred through cytoplasmic destruction rather than reduced production or defective export. These results suggested a model in which high levels of miR-7 induce cytoplasmic destruction of Cdr1as, and Cyrano, by limiting miR-7, enables Cdr1as to accumulate in neuronal cell bodies and processes.
Cdr1as Is Regulated by miR-7 and miR-671
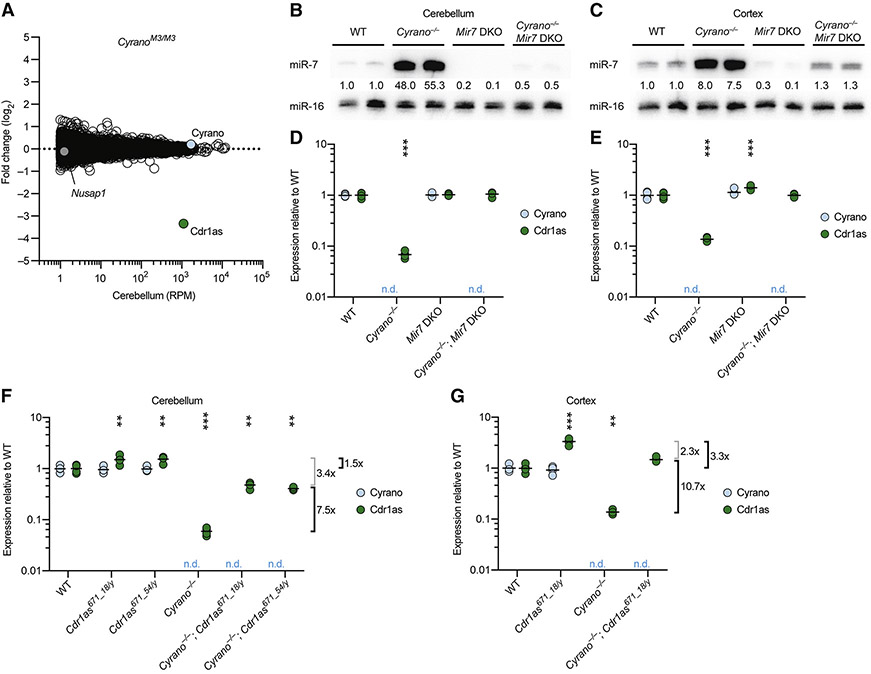
To explore further the idea that miR-7 links the fate of Cyrano to that of Cdr1as, we analyzed RNA-seq results from cerebellum of CyranoM3/M3 mice, which had a 5 nt deletion in the miR-7 site (Figure 2A). When compared to results from wild-type cerebellum, expression of Cyrano did not change (Figure 6A, 1C), as expected from our previous RT-qPCR analyses (Figure S3). Expression of the downstream gene Nusap1 also did not change, as expected in this mutant predicted to retain normal Cyrano splicing and 3′-end processing. Remarkably, Cdr1as was the only differentially expressed gene identified with adjusted p value <0.01, and its expression decreased 11 fold in CyranoM3/M3 cerebellum, similar to the decrease observed in Cyrano−/− cerebellum. This decrease in Cdr1as was corroborated by RT–qPCR analyses of cerebellar samples from all six lines with mutations in the miR-7 site of Cyrano (Figure S6A). These results showed that the miR-7 complementary site within Cyrano is required for this lncRNA to influence Cdr1as accumulation.
Figure 6. Cdr1as is Regulated by miR-7 and miR-671.
(A) Influence of the Cyrano miR-7 site on RNA levels in cerebellum. Shown are fold changes in mean RNA levels observed between wild-type and CyranoM3/M3 cerebellum (n = 3–4 per genotype); otherwise, as in Figure 5A.
(B–C) Substantially reduced miR-7 levels in brains of Mir7 DKO mice. Shown are northern blots measuring miR-7 levels in cerebellum (B) and cortex (C) of mice with the indicated genotypes. Levels of miR-7 were normalized to those of miR-16 and are reported relative to the mean wild-type level.
(D–E) A requirement of miR-7 for Cdr1as reductions observed in mice without functional Cyrano. Levels of Cyrano (blue) and Cdr1as circRNA (green) in cerebellum (D) and cortex (E) of mice with the indicated genotypes were determined by RT-qPCR, normalized to Actb, and plotted relative to mean wild-type expression (black bars, mean; n = 4 per genotype; n.d., not detected). Statistical significance of the differences in Cdr1as level in each mutant tissue compared to that in wild-type tissue is indicated (***, p <0.001, ANOVA with Tukey’s test).
(F–G) Influence of the miR-671 site within Cdr1as on Cdr1as levels both with and without functional Cyrano. Cyrano and Cdr1as in cerebellum (F) and cortex (G) of mice with the indicated genotypes were measured and plotted as in (D). Mean fold changes in Cdr1as attributable to the miR-671 site, either with or without functional Cyrano (black brackets), and not attributable to the miR-671 site (gray brackets) are indicated to the right of each plot.
To test whether miR-7 is required for destabilization of Cdr1as in the absence of Cyrano, we generated mice that lacked both miR-7a1 and miR-7b, the two miR-7 paralogs most highly expressed in brain (Figure S6B). These Mir7 double-knockout (DKO) mice were born at the expected Mendelian frequency and had no gross abnormalities, despite a report that mice expressing a miR-7 sponge have smaller brains (Pollock et al., 2014) (Table S1, Figure S6C).
Analysis of several brain regions showed that miR-7 levels substantially decreased throughout the brain of Mir7 DKO animals (Figure 6B–C, S6D). Global analyses of predicted miR-7 targets revealed no significant changes in cerebellum and cortex, which indicated that the absolute levels of miR-7 normally found in these tissues were too low to direct detectable target repression (Figure S6E). When combining the Cyrano deletion with the Mir7 DKO to generate triple-knockout mice (Cyrano−/−; Mir7 DKO), miR-7 levels contributed by the third paralog (Mir7a2) did not substantially exceed that of wild-type mice (Figure 6B–C), thus providing a context in which Cyrano loss was no longer tied to abnormally high miR-7 levels. In this context, Cdr1as was insensitive to Cyrano loss, demonstrating that increased miR-7 levels were required for Cdr1as destabilization in the absence of Cyrano (Figure 6D–E). Together, our results strongly supported a model in which Cyrano-directed miR-7 degradation protects Cdr1as from miR-7–mediated degradation, thereby allowing Cdr1as to accumulate in neuronal cell bodies and processes.
In the presence of Cyrano, Cdr1as levels were unaffected in Mir7 DKO cerebellum (Figure 6D, S6F), which indicated that in this region of the brain, Cyrano-mediated miR-7 degradation prevents miR-7 from detectably influencing Cdr1as levels. However, Cdr1as levels increased 1.4–2.3 fold in Mir7 DKO cortex, hippocampus, and hypothalamus (Figure 6E, S6F), indicating that in other regions, miR-7 reaches levels sufficient to influence Cdr1as accumulation even when Cyrano expression is intact.
In addition to many conserved miR-7 sites, Cdr1as also contains a single conserved miR-671 site, which pairs to the miRNA with complementarity sufficient to trigger Argonaute2-catalyzed slicing of the circRNA (Hansen et al., 2011). We hypothesized that miR-7-mediated degradation of Cdr1as might occur through enhanced miR-671–directed slicing. To be able to test this hypothesis, we generated mice with deletions (either 18 or 54 nt) that disrupted the miR-671 site (Figure S6G). Hemizygous males (Cdr1as is on the X chromosome) and homozygous females with either of these deletions were born at expected Mendelian frequencies, with no changes observed in miR-7 levels (Table S1; Figure S6H). Disruption of the miR-671 site within Cdr1as led to increased levels of Cdr1as in brain, up to 4 fold in cortex and hippocampus (Figure S6I). These results indicated that miR-671–directed slicing of Cdr1as, known to occur in cultured cells (Hansen et al., 2011), also occurs in the animal and is sufficiently robust to set Cdr1as levels in some brain regions.
With these Cdr1as671 strains in hand, we generated Cyrano−/−; Cdr1as671/y mice and, after confirming that Cyrano deletion in the Cdr1as671 background increased miR-7 levels (Figure S6J), we assessed Cdr1as expression. If enhanced miR-671–directed slicing was fully responsible for Cdr1as clearance in Cyrano−/− mice, Cdr1as expression in Cyrano−/−; Cdr1as671/y mice was expected to increase to the same level as that of Cdr1as671/y mice, whereas if enhanced slicing was not involved, Cdr1as was expected to increase relative to the Cyrano−/− by a fold change similar to that observed between wild-type and Cdr1as671/y mice (1.5 and 3.3 fold in cerebellum and cortex, respectively). Instead of either of these extremes, we observed that Cdr1as levels were partially restored. In cerebellum, Cdr1as increased 7.5 fold, implying a 5-fold (7.5 /1.5) increase in slicing at the miR-671 site in the absence of Cyrano activity (Figure 6F). The observation that preventing miR-671–directed slicing only partially restored Cdr1as levels indicated that at least one other miR-7–dependent mechanism acts to degrade Cdr1as in this tissue, with a presumed 3.4-fold effect. Likewise, in cortex, Cdr1as increased 10.7 fold, implying a 3.2-fold (10.7 / 3.3) increase in slicing at the miR-671 site, with another 2.3 fold attributed to another mechanism (Figure 6G). Taken together, our results showed that in the absence of Cyrano-directed miR-7 degradation, miR-7 levels in brain tissues increase such that miR-7 destroys Cdr1as through at least two mechanisms, one of which increases slicing at the miR-671 site.
DISCUSSION
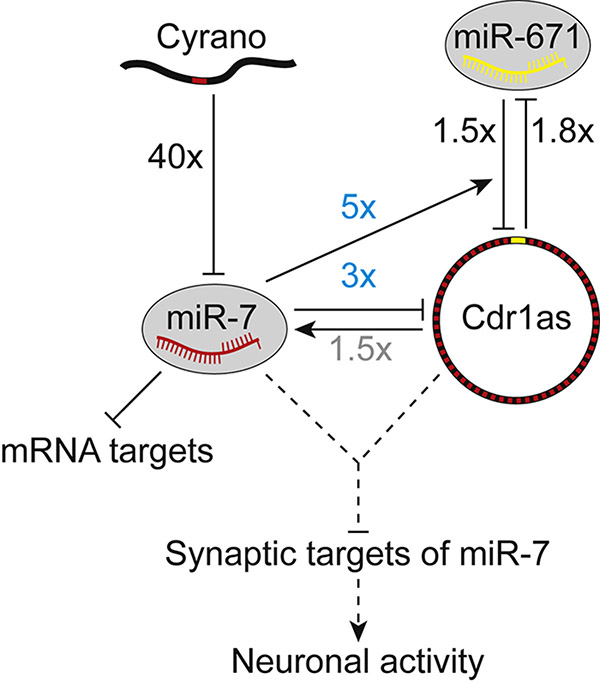
We characterize a posttranscriptional regulatory network centered on four noncoding RNAs—one lncRNA, one circRNA, and two miRNAs—using a panel of mouse knockouts to disrupt nodes and edges in the network and thereby learn their molecular consequences in the animal (Figure 7). The lncRNA Cyrano functions as a potent miR-7 assassin. In targeting miR-7 for degradation, Cyrano prevents miR-7 from repressing its mRNA targets in some tissues. Cyrano also prevents miR-7 from causing efficient cytoplasmic destruction of the circRNA Cdr1as in the brain, which occurs in part by enhancing miR-671–directed slicing of Cdr1as.
Figure 7. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain.
For each interaction, the effect was measured using a mouse-knockout model that disrupted either the relevant ncRNA (either Cyrano, miR-7, or Cdr1as) or miRNA-binding site (either the miR-7 site in Cyrano or the miR-671 site in Cdr1as). Fold changes are from the cerebellum, observed either in this study (black or blue) or previously (gray, Piwecka et al., 2017). The 1.8-fold increase in miR-671 observed in our mice with a disrupted Cdr1as miR-671 site resembled that observed in Cdr1as-knockout mice (Piwecka et al., 2017). Regulation that emerges with derepression of miR-7 in cerebellum (as opposed to deletion of miR-7) is indicated (blue fold changes). Repression of miR-7 targets is also depicted, but without a fold change because it was detected only in other tissues. Speculative interactions are shown as dashed lines.
Insights into TDMD
Cyrano-targeted miR-7 destruction is surprisingly potent compared to previously described TDMD examples. Although a highly complementary miRNA-binding site is required to induce TDMD (Ameres et al., 2010), additional sequences within the full-length RNA can augment the activity (Lee et al., 2013). Cyrano is substantially longer than viral transcripts that direct miRNA degradation and has deeply conserved blocks of sequence proximal to the miR-7 site and elsewhere within the transcript (Ulitsky et al., 2011) (Figure 1A). Consistent with these more distal conserved regions potentially enhancing Cyrano-directed miR-7 degradation, we observed only a 3-fold decrease in miR-7 after >100-fold overexpression of the 197 nt site-containing region of Cyrano (Figure 2D). This strategy of using multiple regions of a lncRNA to boost degradation efficacy would be suitable for an endogenous transcript, whereas the pressure to minimize genome size might favor the viral strategy of using a larger number of shorter, less effective transcripts.
Thus far, the miRNA-degradation factors responsible for TDMD have been elusive. As tailing and trimming of miRNA 3′ ends co-occur with TDMD, an attractive model has been that tailing of the 3′ end of the miRNA licenses the miRNA for degradation by a 3′-to-5′ exonuclease. This model is consistent with findings that 1) 2′-O-methylation, a modification that both prevents 3′ tailing and stabilizes small RNAs, is typically found on classes of small regulatory RNAs that have extensive pairing to most targets, yet is absent on metazoan miRNAs, which do not have extensive pairing to most targets (Ameres et al., 2010; Ji and Chen, 2012), 2) tailing of certain metazoan miRNAs and pre-miRNAs promotes their destruction (Boele et al., 2014; Chang et al., 2013; Katoh et al., 2015), and 3) nucleotidyltransferases and 3′-to-5′ exonucleases co-purify and cooperate in protein complexes (Kim and Richter, 2006; Reimao-Pinto et al., 2016). Despite the appeal of this model, our findings comparing Cyrano−/− and Gld2−/− mice indicated that the observed tailing is off-pathway for degradation, in which case trimming might also be an off-pathway event caused by transiently freeing the 3′ end of the miRNA from Argonaute. In this scenario, TDMD need not occur through a 3′-to-5′ exonuclease. Indeed, severe knockdown of candidate 3′-to-5′ exonucleases had no detectable effect on Cyrano-directed miR-7 degradation (Figure S4H–I). Although we have not ruled out redundancy or a role for other cytoplasmic exonucleases, perhaps the key factors recruited by Cyrano are not nucleases at all but instead act on Argonaute to either degrade the protein or help eject Cyrano-bound miR-7. Once removed from the protective grip of Argonaute, miR-7 could be destroyed by multiple cellular nucleases. Alternatively, Cyrano might induce a transient tailing of miR-7 that was not detected in our analyses because it was coupled to rapid, highly processive miRNA degradation. The discovery of the unusually effective TDMD mediated by Cyrano provides a promising system for dissection of this phenomenon.
Destruction of a circRNA
CircRNAs are much more stable than mRNAs, presumably due to the absence of free ends (Ebbesen et al., 2017). Cdr1as is unusual in that it has a built-in mechanism for destruction via miR-671–directed slicing. We found that this mechanism is active in the animal, controlling accumulation of Cdr1as in brain. In Cyrano−/− brains, miR-671-directed slicing of Cdr1as is further enhanced due to increased levels of miR-7. Perhaps miR-7, which has sites in close proximity to the miR-671 site, helps recruit or retain the miR-671 silencing complex through an undefined mechanism that also explains the cooperative action of closely spaced sites in mediating repression (Grimson et al., 2007; Saetrom et al., 2007). For example, the miR-7 and miR-671 silencing complexes might cooperatively bind to Cdr1as, or binding of miR-7 to Cdr1as might change the conformation of Cdr1as so as to increase accessibility of the miR-671 site.
Increased miR-671-directed slicing explains 3–5 fold of the Cdr1as degradation observed in Cyrano−/− cerebellum and cortex; at least one other miR-7–dependent mechanism is responsible for the remaining 2–3 fold. miR-7 is not expected to direct slicing of Cdr1as by Argonaute2 because Cdr1as has no site with extensive complementarity to miR-7. Perhaps Cdr1as studded with too many Argonautes is trafficked to a more degradative subcellular environment.
A Network of Noncoding RNAs Acting in Neurons
What might be the purpose of this baroque regulatory network centered on four noncoding RNAs? With its unusually high number of miR-7 sites, Cdr1as was initially proposed to act as a miR-7 inhibitor, titrating miR-7 away from mRNA targets while resisting deadenylation and decapping typically induced by stable Argonaute binding (Hansen et al., 2013; Memczak et al., 2013). Although this proposal has gained popularity, there is no evidence that endogenous Cdr1as acts as a sponge. Knocking out Cdr1as does not lead to the decreased levels of miR-7 targets expected if Cdr1as were a miR-7 sponge (Piwecka et al., 2017). Instead, the role of Cdr1as in dampening neuronal activity is partly attributed to the idea that it protects miR-7 from degradation thereby allowing increased repression of miR-7 targets (Piwecka et al., 2017).
One concern with this more recent idea is the small effect size observed for predicted miR-7 targets in Cdr1as-knockout tissues (Piwecka et al., 2017). Furthermore, the results of our experiments are difficult to reconcile with the notion that the ≤2 fold decrease in miR-7 observed in the Cdr1as knockout helps explain this knockout phenotype; when examining the effects of decreased miR-7 levels (>5 fold) in the Mir7 double knockout, we found no evidence for decreased repression of miR-7 targets in regions of the brain where Cdr1as is thought to be active (Figure S6E).
So how else might Cdr1as function to dampen neuronal activity? Although we do not favor the idea that its influence on miR-7 levels is at the heart of its function, the observation of its activity in tissues in which miR-7 levels seem too low to have any consequence on its general target repertoire imply that Cdr1as might potentiate miR-7 activity in some other way. We favor the proposal that Cdr1as acts to engineer spatial and perhaps temporal delivery of miR-7 in neurons (Piwecka et al., 2017). Controlled delivery of miR-7 to synapses might limit neuronal activity without detectable effects on most miR-7 targets. This idea is consistent with the observation that miR-7 has multiple conserved targets involved in vesicle trafficking (Latreille et al., 2014) and reports of miR-7 enrichment in neuronal processes (Siegel et al., 2009; Smalheiser et al., 2014). That Cdr1as contains sites for miR-7 instead of another lowly expressed neuronal miRNA suggests that what might matter is not merely the low expression of miR-7 but also its potent on–off switch in the form of Cyrano-directed degradation. Our insights regarding the crosstalk and molecular consequences of these noncoding RNAs, together with our knockout models disrupting nodes and edges of the network, help lay the foundation for experimental tests of these ideas.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David Bartel (dbartel@wi.mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse husbandry
Mice were group-housed in a 12 hr light/dark cycle (light between 07:00 and 19:00) in a temperature-controlled room (21.1 ± 1.1°C) at the Whitehead Institute for Biomedical Research with free access to water and food and maintained according to protocols approved by the Massachusetts Institute of Technology Committee on Animal Care. Euthanasia was performed by CO2 inhalation. The ages of mice are indicated in the figure legends or methods. Sex was not determined for embryos or neonatal pups. For analyses of older animals (≥P21), only male mice were used unless indicated otherwise.
Generation of Cyranofl/fl and Cyrano−/− mice
Mice carrying a conditional allele of the Cyrano gene (Cyranofl/fl mice) were generated by homologous recombination. The targeting vector was designed to insert a LoxP site in intron 2 and in a poorly conserved region of exon 3. This vector was based on the pPGKneoF2L2DTA vector (a gift from Philippe Soriano, Addgene #13445) and was constructed using genomic sequences amplified by PCR from a BAC clone (RP23–59M9; C57BL/6J; BAC-PAC Resources, Oakland, CA). The LoxP sites were flanked by 4.3 kb 5′ and 2.0 kb 3′ homology arms. NotI-linearized targeting construct was electroporated into hybrid 129S6;C57Bl/6J embryonic stem cells (ESCs). ESC colonies were screened for correct targeting by PCR and Southern blotting, and correctly targeted ESC clones were aggregated with albino CD-1 embryos and transferred into pseudopregnant recipient females. Chimeras were bred with FLPe transgenic mice (Cat #003946, Jackson Laboratories) to excise the neomycin resistance cassette. Cyranofl/+ mice were crossed to CMV-Cre transgenic mice (Cat #006054, Jackson Laboratories) to generate Cyrano+/− mice and then bred away from the FLPe and Cre transgenes. Cyranofl/+ and Cyrano+/− mice were each backcrossed ≥8 generations into C57Bl/6J (Cat #000664, Jackson Laboratories).
Generation of mutant mice using Cas9
Mice with mutations in the miR-7 site of Cyrano (CyranoM mice) were generated by injecting one-cell C57Bl/6J embryos with Cas9 RNA and an sgRNA designed to cut within the miR-7 site (Figure 2A, Table S3). F0 mice containing mutations in the miR-7 site were crossed to wild-type C57Bl/6J mice to generate heterozygotes and then heterozygotes with the same allele were bred to homozygosity. In total, we generated 6 lines, each with a unique mutant allele, M1–M6 (Figure 2A).
Mir7a1−/−; Mir7b−/− mice were generated by injecting one-cell C57Bl/6J embryos with Cas9 RNA and three pairs of sgRNAs—each pair of sgRNAs designed to excise the DNA encoding the pre-miRNA hairpin for a different miR-7 locus (Figure S6B, Table S3). Of 102 F0 mice, one had homozygous deletions of both Mir7a1 and Mir7b (Figure S6B), and several had heterozygous deletions of Mir7a1 and/or Mir7b. None had a deleted allele of Mir7a2. The double-knockout founder was crossed to C57Bl/6J, and then the double-heterozygous F1s were intercrossed to generate Mir7a1−/−; Mir7b−/− mice.
Mice with mutations in the miR-671 site of Cdr1as (Cdr1as671 mice) were generated by injecting one-cell C57Bl/6J embryos with Cas9 RNA and an sgRNA designed to cut within the miR-671 site (Figure S6G, Table S3). F0 mice containing deletions (18 and 54 nt, respectively) in the miR-671 site were bred directly to Cyrano+/− mice, and then F1 mice were intercrossed to generate lines with the desired homozygous mutations.
Genotyping
Genomic DNA was extracted from mouse earsnips using the HotSHOT method (Truett et al., 2000). For CyranoM mice, PCR was performed with KAPA HiFi HotStart ReadyMix (Roche), and amplicons were purified (QiaQuick PCR Purification Kit, Qiagen) and submitted for Sanger sequencing. For all other mutant mice, PCR was performed with KAPA 2G Fast Genotyping Mix (Roche), and amplicons were analyzed by agarose gel electrophoresis. PCR primers, PCR conditions, and expected amplicon sizes are indicated in Table S3.
Cell lines and cell culture
All cells were cultured at 37°C with 5% CO2. Human cell lines were cultured as follows: HEK293, HEK293T, HeLa, and MCF-7 in DMEM (Gibco) with 10% FBS (Clontech); SH-SY5Y and SK-N-SH in DMEM/F12 (Gibco) with 10% FBS; K562 in RPMI-1640 (Life Technologies) with 10% FBS. Mouse cell lines were cultured as follows: C2C12 in DMEM with 10% FBS; Neuro2a in 1:1 DMEM:Opti-MEM (Gibco) with 10% FBS. Reverse transfections were performed with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol. Transfection efficiencies, as estimated by microscopic evaluation of mCherry expression at 24 h, were >90%. Each of these cell lines is of female origin.
CRISPRi-based knockdown cell lines
Stable CRISPRi cell lines (MCF-7, SH-SY5Y, SK-N-SH, C2C12, HeLa) were generated by transduction with lentivirus carrying constitutively expressed dCas9-KRAB (Gilbert et al., 2013) (Addgene #46911). The CRISPRi K562 cell line expressed a doxycycline-inducible KRAB-dCas9 (Gilbert et al., 2014). Each stable CRISPRi cell line was transduced with lentivirus expressing an sgRNA, which was directed against either the TSS (transcription start site) region of the target gene, the TSS region of Cyrano, or no gene (a non-targeting negative-control sgRNA). sgRNA vectors were generated from the pU6-sgRNA EF1Alpha-puro-T2A-BFP plasmid (Addgene #60955) and prepared as described (Gilbert et al., 2014) using oligonucleotides (Table S3). After ~10 days of selection for sgRNA expression with puromycin (1 μg/mL in K562, 2 μg/mL in HeLa, 2.5 μg/mL in SH-SY5Y, and 3 μg/mL in MCF-7, SK-N-SH, and C2C12; Clontech), cells were harvested for RNA extraction. In the case of the inducible K562 cell line, doxycycline (Clontech) was added simultaneously with puromycin and adjusted daily to a concentration of 50 ng/mL, under the assumption of a 24 h half-life in culture (Gilbert et al., 2014).
Primary neuronal cultures
CGN cultures were prepared from male and female P5–P6 neonates as described (Lee et al., 2009) with some modifications. Neonates were decapitated with scissors and the cerebellum and midbrain were pinched off with forceps and transferred to a petri dish containing ice-cold HBSS-glucose (Hank’s Balanced Salt Solution, Life Technologies, 0.6% w/v glucose, 0.035% w/v sodium bicarbonate). Under a dissecting microscope, the cerebellum was detached from the meninges and midbrain and placed in an HBSS-glucose-containing 15 mL conical tube on ice. Additional cerebella were isolated, switching petri dishes every three neonates, and cerebella from the same genotype were stored together.
Tissue dissociation was performed using the Papain Dissociation System kit (Worthington). Cerebella were incubated in papain solution (Earle’s Balanced Salt Solution, 20 U/mL Papain, 100 U/mL DNase) at 37°C for 15–20 min, gently inverting the tube every 4 min. The tissue was triturated 10–12 strokes with an FBS-coated P1000 pipet tip and the supernatant was transferred to a new tube. Cells were pelleted at 200 g for 5 min, supernatant was removed, and cells were resuspended in resuspension solution (Earle’s Balanced Salt Solution, 10% albumin-ovomucoid inhibitor solution, ~100 U/mL DNase) using an FBS-coated pipet tip. The cell suspension was gently added to the top of a 15 mL conical tube containing 5 mL of albumin-ovomucoid inhibitor solution and centrifuged at 70 g for 6 min to separate intact cells from cell fragments and membranes. The supernatant was discarded and the pellet resuspended in 3–5 mL of serum-free media (Neurobasal medium, minus phenol red, supplemented with 1x GlutaMAX (Gibco), 100 U/mL penicillin-streptomycin (Gibco), 25 mM KCl, and 1x B-27 (Gibco)) then passed through a 70 μm cell strainer (BD Falcon). Cells were stained with Trypan Blue (Sigma Aldrich) and counted using a Countess cell counter (Thermo Fisher). Each cerebellum yielded 3–6 million cells with >90% viability.
Cells were plated either at ~50,000 cells/cm2 on #1 glass coverslips (Electron Microscopy Sciences) coated with poly-D-lysine (50 μg/mL; EMD Millipore) and laminin (2 μg/mL; Corning) and placed in 12-well plates or at ~90,000 cells/cm2 on tissue culture plates coated with poly-D-lysine (50 μg/mL). On DIV1, the media was exchanged for serum-free media containing 10 μM cytosine β-D-arabinofuranoside (AraC; Sigma Aldrich). On DIV3, the media was exchanged for serum-free media. Neurons were fed via half-media exchanges every 2–3 days thereafter. Culture viability was assessed by light microscopy. A subset of cultures were also examined by DAPI staining and immunofluorescence with anti-MAP2 (1:200; EMD Millipore), anti-Tau (1:500; Synaptic Systems), and/or anti-GFAP (1:500; Dako) antibodies to ascertain the fractions of neuronal and glial cells. These cultures typically were ~90% neuronal and consisted predominantly of smaller neurons with 2–5 processes, as expected for CGNs.
Hippocampal cultures were prepared as described (Beaudoin et al., 2012) with slight modifications. Hippocampi were dissected from male and female P0–P1 mouse pups in ice-cold dissection media, washed twice with ice-cold dissection media, trypsinized for 20 min at 37°C, and then treated with 0.1% DNase for 2 min at room temperature. Tissue was washed again in room-temperature dissection media and brought up to 10 mL in plating media. The tissue was then triturated in a 15 mL conical tube with an FBS-coated 5 mL serological pipet in 3 batches, performing no more than 10 triturations per batch and transferring the dissociated cells to a new 15 mL conical tube between each batch. Dissociated cells were passed through a 70 µm cell strainer, counted, and plated in plating media at ~100,000 cells/cm2 on tissue culture plates coated with poly-D-lysine (50 μg/mL). Plating media was exchanged for maintenance media between 2–6 h after plating. On DIV2, cells were treated with 5 µM AraC for 24 h and then maintenance media was fully exchanged. Neurons were fed via half-media exchanges every 3 days thereafter.
METHOD DETAILS
RNA extraction
For cells, total RNA was extracted with TRI Reagent (Life Technologies) according to the manufacturer’s protocol. For adherent cells, TRI Reagent was added directly to the tissue culture plate, and cells were quickly detached by scraping. For suspension cells, cells were first pelleted by centrifugation. For mouse tissue, total RNA was extracted with TRI Reagent according to the manufacturer’s protocol with the following modifications. Mouse tissues were rapidly dissected after euthanasia (CO2) and flash frozen in Eppendorf tubes in liquid N2. Tissue was transferred to a 15 mL conical tube, 1–2 mL of TRI Reagent was added, and the tissue was homogenized with a TissueRuptor (Qiagen) and disposable probes. Samples were aliquots in Eppendorf tubes (1 mL per tube) and phase separation was achieved with either 200 μL chloroform (J.T. Baker Analytical) or 100 μL 1-Bromo-3-chloropropane (Sigma). For samples with high fat content, such as brain tissue, 2–3 volumes of 1-Bromo-3-chloropropane were necessary to fully separate the aqueous and organic layers. After isopropanol precipitation and two 70% ethanol washes, all RNA was resuspended in water.
Cytoplasmic/nuclear fractionation
Cytoplasmic and nuclear fractions of each cell line were isolated via mild lysis and centrifugation as follows. No more than 107 cells were harvested and pelleted by centrifuging at 300 g for 5 min. The plasma membrane was lysed by adding 175 μL of precooled RLN+EDTA buffer (50 mM Tris, pH 8.0, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 10 mM EDTA) freshly spiked with 1 μL/mL Superasin and 1 mM DTT and gently resuspended by pipetting, avoiding vigorous handling as this can cause partial nuclear lysis. Lysates were incubated on ice for 5 min then centrifuged at 500 g for 5 min to pellet nuclei. The supernatant was transferred to a new tube and spun again at 500 g for 1 min to remove any contaminating nuclei. The supernatant was transferred to a new tube containing 1 mL of TRI Reagent and vortexed vigorously. Meanwhile, the original nuclear pellet was washed once with 175 μL of precooled RLN+EDTA and spun at 500 g for 1 min. After aspirating the supernatant, the nuclei were gently resuspended in 175 μL of precooled RLN+EDTA. 1 μL was saved for microscopic confirmation of intact nuclei, and the remainder was added to a new tube containing 1 mL of TRI Reagent, vortexed vigorously, and passed through a 20-gauge needle at least 5 times. 25 pg of uncapped firefly luciferase RNA containing a 30-base poly(A) tail (Promega #L4561) was added to each fraction in TRI Reagent prior to RNA extraction.
Lentivirus production and transduction
HEK293T cells were used to package lentivirus. In a 6-well tissue culture plate, ~170,000 cells/cm2 were reverse-transfected with 1.4 μg transfer plasmid, 0.94 μg pCMV-dR8.91 packaging plasmid (a gift from Jonathan Weissman), and 0.47 μg pMD2.G envelope plasmid (a gift from Didier Trono; Addgene #12259) using Lipofectamine 2000 (Thermo Fisher). After 72 h, the media was collected and centrifuged at 500 g for 10 min to pellet debris. To infect cells, 250 μL of virus-containing supernatant (~1/7th of the total) was combined with 750 μL media and 8 μg/mL polybrene (Santa Cruz Biotechnology) and then added to one well of a 12-well plate (containing either 200,000 suspension cells or adherent cells plated 24 h before at 25,000/cm2). Plates were centrifuged at 12,000 g for 90 min at room temperature then returned to the 37°C incubator.
RNA-seq
Each poly(A)-selected RNA-seq library was prepared from 4 μg total RNA. For profiling predicted miR-7 targets in Cyrano−/− mice (Figure 4), libraries were prepared using the unstranded TruSeq kit (Illumina) with RNA from tissues from 4-month-old male wild-type and Cyrano−/− littermates on a mixed 129S6; C57Bl/6J background (1 replicate per library; n = 4–6 libraries per genotype for each tissue). For profiling mRNAs and noncoding RNAs (Figure 1A, 5A, 6A, S5A–B, S6E), libraries were prepared using the stranded NEXTflex kit (Bioo Scientific) with RNA from cerebella from 1-month-old male wild-type, Cyrano−/−, CyranoM3/M3 and Mir7a1−/−; Mir7b−/− mice on a C57Bl/6J background (1 replicate per library; n = 3–4 libraries per genotype). All libraries were sequenced on the Illumina HiSeq platform with 40 nt single-end reads.
Reads were aligned to the mouse genome (mm10) using STAR v2.4 (Dobin et al., 2013) with the parameters “--outFilterType BySJout --outFilterIntronMotifs RemoveNoncanonicalUnannotated --outSAMtype BAM SortedByCoordinate.” Aligned reads were assigned to genes using annotations from Ensembl (Mus_musculus.GRCm38.73.gtf; downloaded October 15, 2014) and htseq-count v0.6.1p1 (Anders et al., 2015) with the parameters “-m union -s no” for unstranded libraries and “-m union -s reverse” for stranded libraries. An annotation for Cdr1as (chrX: 61183248–61186174) was manually added to the gtf file. For counting pri-miR-7 species, Mir7 annotations were extended 200 nt on both the 5′ and 3′ ends, and the pre-miR-7 sequences were excluded from the annotation. Count files were merged to generate tables of counts for each tissue organized by genotype. Differential expression was determined using DESeq2 v1.10–v1.18 (Love et al., 2014) with the parameter “betaPrior = FALSE” and without the lfcShrink function. For RNA-seq plots (Figure 5D, 6A), only RNAs with a mean RPM >1 in wildtype samples and a CV <10 standard deviations above the median CV in both wildtype and mutant samples are shown. RNA-seq browser tracks were visualized in the UCSC genome browser and IGV v2.3.34 (Thorvaldsdottir et al., 2013).
The effects of miR-7 on target mRNA levels were determined by comparing fold changes for mRNAs containing miR-7 sites in their 3′ UTRs relative to those observed for control mRNAs. All predicted miR-7 targets were downloaded from TargetScanMouse v7.1 (Agarwal et al., 2015) and grouped by conservation of miR-7 targeting and by cumulative weighted context++ score to generate a set of conserved predicted targets and a set of top predicted targets, respectively. The set of no-site controls consisted of mRNAs lacking a perfect 6-nt seed match to miR-7 anywhere within the mature mRNA. For each tissue, only mRNAs with >50 mean counts (approximately equivalent to 5 RPM), as determined by DESeq2, were analyzed. Predicted targets generally have longer 3′ UTRs than no-site mRNAs; to minimize fold-change effects that correlate with 3′-UTR-length differences (rather than the presence of a miR-7 site), we normalized all fold changes based on mRNA 3′ UTR lengths (as annotated in Eichhorn et al., 2014). To do this, we first fit a linear function to the relationship between fold change and 3′-UTR length for no-site mRNAs and then used this function to normalize the fold changes for all mRNAs.
Small-RNA sequencing
Library preparation started with 5 μg of total RNA from either tissues of 1-month-old male mice on a C57Bl/6J background or stable CRISPRi K562 cell lines. From each total RNA sample, RNAs that co-migrated within the range of 18 and 32 nt radiolabeled internal standards were isolated on a 15% polyacrylamide urea gel. Purified RNA was ligated to a preadenylated 3′ adapter using T4 RNA Ligase 2 KQ mutant (NEB) in a reaction supplemented with 10% polyethylene glycol (PEG 8000, NEB). To reduce ligation biases, this 3′ adapter had four random-sequence positions at its 5′ end. After gel purification on a 10% polyacrylamide urea gel, RNA was ligated to a 5′ adapter using T4 RNA Ligase I (NEB) in a reaction supplemented with 10% PEG. To reduce ligation biases, this adapter had eight random-sequence positions at its 3′ end. After gel purification on a 8% polyacrylamide urea gel, RNA was reverse transcribed with SuperScript III (Invitrogen), and the cDNA was amplified 8–12 cycles with KAPA HiFi DNA polymerase (Kapa Biosystems). Amplified DNA was purified on an 8% polyacrylamide 90% formamide gel and submitted for sequencing. A step-by-step protocol for constructing libraries for small-RNA sequencing is available at http://bartellab.wi.mit.edu/protocols.html. Libraries were sequenced on the Illumina HiSeq platform with 50 nt single-end reads. Reads were trimmed of adaptor sequence using cutadapt (Martin, 2011) and filtered using fastq_quality_filter (FastX Toolkit; http://hannonlab.cshl.edu/fastx_toolkit/) with the parameters “-q 30 –p 100” to ensure that all bases had an accuracy of 99.9%.
To count the miRNAs in each library, the first 18 nt of each read were string-matched to a dictionary of miRNA sequences. Mouse and human miRNA dictionaries were based on annotations in miRbase_v20 and miRbase_v21, respectively. Matching to the first 18 nt had the advantage of combining all miRNA isoforms into a single count but also can result in ambiguous read assignments for mature miRNAs from distinct loci that are identical for the first 18 nt but differ at one or more positions closer to their 3′ ends (such as let-7a-5p and let-7c-5p). To avoid this ambiguity, these miRNAs (<5% of all annotations) were removed from the dictionary. Count files were merged to generate tables of counts for each tissue organized by genotype. Differential expression was determined using DESeq2 (Love et al., 2014) with the parameter “betaPrior = FALSE”. For small-RNA sequencing plots (Figure 2A–B, 2D, S6G–H), only miRNAs with a mean RPM >1 in wildtype samples and a CV <5 standard deviations above the median CV in both wildtype and mutant samples are shown.
For analyses of tailing and trimming, we used the same string-matching described above while also binning reads by lengths from 19–30 nt. For each miRNA with a mean RPM >100, we calculated the fraction of reads of each length. Any length with a fraction >14.5% was considered “mature” (this percentage was selected to include both the 23 and 24 nt isoforms of miR-7 in all samples). Trimmed reads were all reads shorter than the shortest mature length, and tailed reads were all reads longer than the longest mature length. In this fashion, we were able to determine the aggregate number of trimmed and tailed reads for each miRNA. To identify the nucleotide(s) added during tailing, we used custom scripts to count all homopolymers appended to mature miR-7 (both the 23 and 24 nt isoforms).
Northern blots
For each sample, 1–10 μg of total RNA was resolved on a denaturing polyacrylamide gel, transferred onto a Hybond-NX membrane (GE Healthcare) using a semi-dry transfer apparatus (Bio-Rad), and incubated at 60°C for 1 h with EDC (N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide; Thermo Scientific) diluted in 1-methylimidazole to chemically crosslink 5′ phosphates to the membrane. Blots were hybridized to radiolabeled DNA or LNA oligonucleotide probes (Table S3). Northern-blot results were analyzed with ImageQuant TL (v8.1.0.0). A step-by-step protocol is available at http://bartellab.wi.mit.edu/protocols.html. For some experiments, synthetic miR-7 standards were run in lanes adjacent to the experimental samples to generate a standard curve used for absolute quantification.
RT–qPCR
For mouse tissues, 1 μg total RNA was treated with RQ1 DNase (Promega) or Turbo DNase (Life Technologies), then reverse transcribed with Superscript III (Life Technologies) and either oligo(dT) primers (cytoplasm/nuclear fractionation experiments) or random hexamers (all other experiments). Expression of Cyrano and Cdr1as were measured by qPCR (primers listed in Table S3) using SYBR Green (Applied Biosystems) on a QuantStudio 6 system (Applied Biosystems) and quantified using the ∆∆CT method with either Actb expression or the geometric mean of Actb, U1, U2, and U6 expression as the internal normalization control.
For stable CRISPRi cell lines, 1 μg total RNA was reverse transcribed with QuantiTect RT Kit (Qiagen) followed by qPCR using SYBR Green. Expression of CYRANO and mRNAs were quantified (primers listed in Table S3) using the ∆∆CT method with the geometric mean of GAPDH, ACTN1, U1, U2, and U6 expression as the internal normalization control and the non-targeted sample as the inter-sample normalization control.
Absolute quantification of miR-7, Cyrano, and Cdr1as in CGNs and cell lines
To quantify the number of molecules of a given RNA per cell, we first determined the amount of total RNA per cell. For this, total RNA was extracted and quantified from a defined number of cells, with the assumption of negligible RNA loss. For human cell lines, cell number was determined using a Countess cell counter. For mouse DIV11 CGNs, cell number was determined from light micrographs of 6-well plates (10 20x images per sample) prior to RNA extraction. By this method, we estimated mean total RNA per cell as 7.7 pg per CGN, 19.9 pg per HEK293T cell, and 23.2 pg per HeLa cell (n = 3 samples per cell type; Table S2).
Next, we generated standards for miR-7, Cyrano, and Cdr1as. For miR-7, 10 pmol of a 23-mer RNA oligo (IDT) matching the human miR-7 and mouse miR-7a sequence was PNK-labeled with cold ATP, desalted on a P30 column (Bio-Rad) and then serially diluted in 200 ng/μL yeast tRNA (Life Technologies). For Cyrano, the first 3.5 kb of the mouse Cyrano cDNA and the first 3.7 kb of the human CYRANO cDNA were PCR amplified, using primers that added a T7-promotor sequence. After purification on an agarose gel (Qiaquick Gel Extraction, Qiagen), 1 pmol of each PCR product was in vitro transcribed with T7 MEGAscript (Ambion), according to manufacturer’s protocol, and cleaned up with Turbo DNase and MEGAclear (Ambion). Full-length RNA was mixed with deionized formamide (final concentration 60% v/v) and 6x loading dye, heated to 65°C for 5 min, snap-cooled on ice, and then size-selected on a native RNase-free 1% agarose gel run at 70V with RiboRuler HR ladder (Thermo Scientific). Size-selected RNA was purified with Zymoclean Gel RNA Recovery Kit (Zymo Research), and quantified on a NanoDrop spectrophotometer (Thermo Scientific). For Cdr1as, a 243 bp fragment of mouse Cdr1as and a 362 bp fragment of human CDR1AS, each containing the back-splice junction and each prepended with the T7-promotor sequence, were ordered from IDT as gBlocks (Table S3). Approximately 1 pmol of dsDNA was in vitro transcribed with T7 MEGAshortscript (Ambion) according to manufacturer’s protocol and then cleaned up with MEGAclear. The full-length RNAs were purified on a 4% polyacrylamide gel run with RNA Century markers (Ambion), eluted overnight at 4°C, purified with a Spin-X column (Corning), concentrated by EtOH precipitation, and quantified by NanoDrop. The molecular weight of each RNA standard was calculated based on its sequence (1,503,509.3 g/mol for mouse Cyrano; 1,147,517.4 g/mol for human Cyrano; 70,377.1 g/mol for mouse Cdr1as; 108,587.7 g/mol for human Cdr1as) and RNA standards were serially diluted, 1010 to 104 copies, in 0.2 μg/µL total RNA derived from either HeLa cells (for mouse standards) or 3T3 cells (for human standards).
To determine absolute amounts of miR-7, 5 μg total RNA from CGN and HEK293T and 15 μg total RNA from HeLa were analyzed by northern blot alongside a serial dilution of synthetic miR-7 standards. To determine absolute amounts of Cyrano and Cdr1as, RT-qPCR with Superscript III and gene-specific primers (Table S3) was performed on 1 μg total RNA from CGN, HEK293T, and HeLa alongside 5 µL of each serial dilution of in vitro transcribed standard. For each RNA of interest, knowing the absolute amount of this RNA per µg of total RNA as well as the amount of total RNA per cell enabled calculation of the number of molecules of this RNA per cell.
Single-molecule FISH
Single-molecule FISH (Batish et al., 2011) was performed on CGNs grown on glass coverslips according to the following protocol. Cells were washed twice in PBS (Ambion) and fixed for 10 min in freshly prepared 4% paraformaldehyde (Electron Microscopy Sciences) in PBS. After aspirating the fixation solution, cells were gently washed in PBS and then stored in 70% ethanol for ≥2 h (up to 2 weeks) at 4°C. Before probing, coverslips were equilibrated for ≥2 min in washing buffer (10% formamide, 2X SSC). Custom probes 3′-end–modified with Quasar 670 dye (Table S3) were diluted to 25 nM in hybridization solution (10% formamide, 2X SSC, 100 mg/mL dextran sulfate), and 50 μL of probe/hybridization mix was dotted on parafilm pressed to a glass plate, 1 dot per coverslip. Coverslips were removed from washing buffer with forceps and then placed on the probe/hybridization mix with the cell side facing the liquid. From this point forward, coverslips were protected from light as much as possible to minimize photobleaching of the 670 dye. After incubating overnight in a humidifying chamber at 37°C, the coverslips were washed 30 min in washing buffer to remove excess probe, 30 min in washing buffer containing 100 ng/mL DAPI, and 5 min in washing buffer to remove excess DAPI. Coverslips were mounted on glass slides with ProLong Gold (Invitrogen) and allowed to cure overnight before sealing with clear nail polish.
Images were acquired using a 100x oil-immersion objective with a numerical aperture of 1.4 on a wide-field fluorescence microscope (either a Zeiss AxioPlan2 with cooled CCD camera or a GE Healthcare Deltavision with sCMOS camera). In each field, 10–30 z-sections with 0.2 μm spacing were acquired. On the AxioPlan2, exposure times were 2 s Cy5, 0.5 s GFP, 0.5 s DAPI. On the Deltavision, exposure times were 0.5 s Cy5 (100% intensity), 0.3 s GFP (100%), 0.01 s DAPI (50%).
For quantification, hot pixels were removed from individual stacks using Remove Outliers (radius 1 pixel, threshold 10) and Despeckle functions in ImageJ. Paired Cy5 and GFP stacks were loaded into the MATLAB program ImageM (Lyubimova et al., 2013), z-projected, and the number of dots in each z-projection was enumerated using the Count Dots feature. After subtracting background dots, defined as those counted in both the Cy5 and GFP stacks, the number of dots was divided by the number of intact, non-pyknotic nuclei in each image to generate the average molecules per cell for each field. In total, 40 fields were analyzed for each probe and each genotype. To quantify the number of dots by subcellular location, the Segmentation feature in ImageM was used to outline either the soma using GFP autofluorescence or the nucleus using DAPI. In this fashion, the number of dots in the nucleus was determined directly, the number of dots in the cytoplasmic soma was calculated by subtracting nuclear dots from soma dots, and the number of dots in neuronal processes was calculated by subtracting soma dots from total dots per field. We note that because quantification was performed on z-projected images rather than individual sections in a z-stack, any dots above or below the nucleus would have been assigned to the nucleus. For generating images for figures, hot pixels were removed as described above, stacks were z-projected using the Max Intensity function and then combined using the Merge function in ImageJ. Merged files were converted to an RGB file and imported into Photoshop (Adobe) for cropping.
QUANTIFICATION AND STATISTICAL ANALYSIS
Graphs were generated in GraphPad Prism 7 and statistical analyses were performed using Excel or Graphpad Prism 7. Statistical parameters including the value of n, statistical test, and statistical significance (p value) are reported in the figures and their legends. For studies involving mouse tissues, replicates refer to samples derived from different mice. For studies involving cell culture, replicates refer to either transfections performed on different days or transductions performed on the same day with different viral supernatants derived from transfections performed in parallel. No statistical methods were used to predetermine sample size. Statistical tests were selected based on the desired comparison. Unpaired two-tailed t-tests (Excel) were used to assess significance when comparing expression of 1–2 genes (northern-blot analyses, RT-qPCR) between two genotypes. One-way ANOVA (Graphpad) was used to assess significance when comparing expression of 1–2 genes (northern-blot analyses, RT-qPCR) between ≥3 genotypes; significant ANOVA results were followed by post-hoc testing either comparing every mean with every other mean (Tukey’s multiple-comparison test) or comparing every mean to the wild-type mean (Dunnett’s multiple-comparison test). For differential expression of global measurements (RNA-seq, small-RNA sequencing), the DESeq2 software generated adjusted p values using the Wald test with the Benjamini-Hochberg procedure to correct for multiple-hypothesis testing. The Mann-Whitney test was used to compare cumulative distributions of mRNA fold changes between two gene sets.
DATA AND SOFTWARE AVAILABILITY
Sequencing datasets generated in this study have been deposited in the GEO under accession number GSE112635.
Supplementary Material
Figure S1. Cyrano Expression, Related to Figure 1
(A) Images of in situ hybridizations for Cyrano in mouse embryo (E14.5) and adult mouse brain (P56) obtained from GenePaint.org (Visel et al., 2004) and the Allen Brain Atlas (Lein et al., 2007), respectively. In both databases, Cyrano is annotated as the RIKEN cDNA 1700020I14Rik gene.
(B) Relative Cyrano expression in 25 tissues of 2-month-old C57Bl/6J mice. Shown are mean Cyrano levels, as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes and relative to cerebellum (error bars, s.d.; n = 3 for each tissue).
(C) CYRANO expression from RNA-seq of 32 post-mortem human tissues, provided by the GTEx Portal (01/15/2018). Expression values in TPM (transcripts per million) are shown as box plots (box, 25th, median, and 75th percentiles; whiskers, 1.5 times the interquartile range). In the GTEx Portal, CYRANO is annotated as the OIP5-AS1 gene.
(D) Cyrano expression from RNA-seq of purified cell populations from the mouse cortex (Dong et al., 2015). Mean expression values in FPKM (fragments per kb per million reads) are shown (error bars, range; n = 2 for each cell type).
(E) Percentage of CYRANO detected in the cytoplasm of mammalian cell lines. Ratios were determined by RT-qPCR, using RNA from cytoplasmic and nuclear fractions, normalizing to a luciferase spike-in transcript (error bars, range; n = 2 for HEK293, HeLa, and SH-SY5Y, n = 3 for K562 and Neuro2a). For comparison, results for GAPDH mRNA and the nuclear-localized XIST lncRNA are also shown (n.d., not detected above background in cytoplasm).
(F) Cyrano gene targeting strategy. Shown between the wild-type locus and the recombinant alleles is the vector used to target the first half of exon 3 (black boxes, exons; red line, miR-7 site with extensive pairing; open triangles, loxP sites; open diamonds, FRT sites; neo, neomycin-resistance cassette; DTA, diphtheria toxin A; AoH, arm of homology). Screening of ES cell clones was performed by long-range PCR as well as Southern blot of XbaI-digested genomic DNA with a 5′ probe upstream of exon 1.
Figure S2. Cyrano Promotes miR-7 Destruction, Related to Figure 1
(A) Influence of Cyrano on pri-miR-7 expression in the brain. The plot on the left shows mean pri-miR-7a1 and pri-miR-7b levels in wild-type (black) and Cyrano−/− (red) anterior cortex and cerebellum (key), as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes (error bars, s.d.; n = 3 per genotype for each tissue). The plot on the right shows mean expression values in RPM (reads per million) for all three pri-miR-7 transcripts in wild-type and Cyrano−/− anterior cortex, cerebellum, cortex, and pituitary, as determined by RNA-seq (error bars, s.d.; n = 4–5 per genotype for each tissue; n.d., not detected).
(B) Influence of Cyrano on pre-miR-7 expression. Shown is a representative northern blot measuring miR-7 levels in wild-type and mutant cortex. This blot is an uncropped version of the blot shown in Figure 6C, with the signal increased in order to detect the less-abundant pre-miR-7 species. Levels of pre-miR-7 species (identified by their absence in the Mir7 DKO) were unchanged in Cyrano−/− cortex.
(C) Relationship between miR-7 changes observed after disrupting Cyrano and the level of wild-type Cyrano expression. Shown are miR-7 fold changes from Figure 2C plotted as a function of Cyrano expression in wild-type tissues, as determined from RNA-seq data also used for the analyses of Figure 5C.
(D) Influence of Cyrano on miR-7 expression in developing mouse brain. Shown at the left is a representative northern blot measuring miR-7 levels in wild-type (WT) and Cyrano−/− (KO) brain at the indicated developmental time points. Plotted on the right are miR-7 levels in wild-type (black) and Cyrano−/− (red) embryonic brain normalized to miR-16 (error bars, s.d.; n = 3 per genotype). Statistically significant fold changes are indicated (*, p <0.05, unpaired two-tailed t-test).
(E) Influence of Cyrano on miR-7 expression in cultured primary neurons. Shown are northern blots for wild-type and Cyrano−/− DIV11 cerebellar neurons (left) and DIV12 hippocampal neurons (right). Levels of miR-7 (upper panels) were normalized to those of either miR-16 (lower panel, left) or U6 (lower panel, right), and reported relative to the mean wild-type level.
(F) Relationship between the magnitude of miR-7 fold change and extent of CYRANO knockdown. Shown are miR-7 fold changes, measured by northern blot, plotted as a function of CYRANO knockdown, as determined by RT-qPCR, for 35 mammalian CRISPRi cell lines stably expressing dCas9-KRAB and an sgRNA targeting CYRANO. Parental cell lines are indicated (key). For each cell line receiving a targeting sgRNA, levels of CYRANO and miR-7 were normalized to the geometric mean of the values for five housekeeping genes and miR-16, respectively, and shown relative to the non-targeting control.
Figure S3. Mutating the miR-7 Site in Cyrano Does Not Cause Detectable Changes in Cyrano Levels, Related to Figure 2
(A) Cyrano locus annotated with binding sites of 3 RT-qPCR primer pairs (#1, #2, #3). Otherwise, this panel is as in Figure 1A.
(B) Cyrano expression in wild-type and mutant cerebellum. Plotted are relative Cyrano levels in wild-type and Cyrano-mutant cerebellum, as determined by RT-qPCR with three primer sets pairing to sites mapped in (A). Cyrano levels, normalized to that of Actb, are shown as fold changes relative to the mean wild-type expression (n = 4 per genotype; n.d., not detected). No significant differences in Cyrano expression were detected (ANOVA with Dunnett’s test), indicating that either miR-7 does not influence Cyrano accumulation in this tissue, or abrogated repression at this site is offset by enhanced repression (due to increased levels of miR-7) at a second, more conventional site located at the beginning of exon 3.
(C) Cyrano expression in wild-type and Cyrano-mutant pituitary. Otherwise, this panel is as in (B).
Figure S4. Cyrano Induces Tailing and Trimming of miR-7, but Tailing Appears Dispensable, Related to Figure 3
(A) Influence of Cyrano on miR-7 tailing and trimming in cerebellum. Otherwise, this panel is as in Figure 3A. In both hippocampus and cerebellum, the trimmed miR-7 isoforms largely matched the genome-encoded miR-7 sequence, implying that tailing of trimmed species was either rare or destabilizing.
(B) Influence of Cyrano on global miRNA tailing in hippocampus (left) and cerebellum (right). Shown are aggregate tailing log2-fold changes for each expressed miRNA after disrupting Cyrano, plotting statistical significance of the change (based on p values determined by unpaired two-tailed t-tests, one per miRNA) as a function of the change in WT divided by Cyrano−/−. Each circle represents one miRNA, showing results for all miRNAs expressed above 100 RPM (read per million miRNA reads) in wild-type tissue. Circles representing miR-7 (miR-7a and miR-7b were combined for this analysis) and miR-92b are colored red and gray, respectively. Cyrano increased the aggregate tailed fractions 1.5 and 2.2 fold, respectively. Dotted lines indicate a p value of 0.05.
(C) Influence of Cyrano on global miRNA trimming in hippocampus (left) and cerebellum (right). Cyrano increased the aggregate trimmed fractions 3.2 and 1.6 fold, respectively. Otherwise, this panel is as in (B).
(D–E) Influence of Gld2 on global miRNA tailing (D) and trimming (E) in hippocampus, evaluated using data from Mansur et al. (2016). Otherwise, this panel is as in (B and C).
(F) Influence of Gld2 on miRNA levels in hippocampus. Shown are log2-fold changes in mean miRNA levels observed between wild-type and Gld2−/− hippocampus, as determined by small-RNA sequencing (Mansur et al., 2016) and plotted as a function of expression in wild-type hippocampus (n = 6 per genotype). Each circle represents one miRNA, showing results for all miRNAs expressed above 1 RPM (read per million miRNA reads) in wild-type hippocampus. Circles representing miR-7 paralogs are colored red.
(G) Influence of PAPD4 on miR-7 expression in stable PAPD4-knockdown K562 cells. Shown at the top is a northern blot monitoring miR-7 levels in stable cell lines expressing either a control sgRNA or an sgRNA targeting either CYRANO or PAPD4 for CRISPRi-mediated knockdown. The levels of miR-7 are reported relative to the miR-7 level in the control cell line after normalizing to the level of miR-16. Plotted at the bottom are CYRANO and PAPD4 levels, as determined by RT-qPCR, in stable lines cells expressing the indicated sgRNAs. CYRANO and PAPD4 levels were normalized to the geometric mean of the values for five housekeeping genes and plotted relative to levels in the control cell line. The level of PAPD4 in line #2 was <2% of that in the control.
(H) Influence of PARN on miR-7 expression. The levels of PARN in lines #1–3 were each <1% of that in the control. Otherwise, this panel is as in (G). One way to reconcile the prevailing idea that tailing is required for TDMD with our observation that miR-7 levels remained unchanged after loss of Gld2 would be to propose that Gld2 has opposing consequences that somehow offset each other. Indeed, for miR-122, monoadenylylation by Gld2 stabilizes the miRNA, whereas oligoadenylylation by Gld2 destabilizes it by making it a substrate for PARN-catalyzed exonucleolytic decay (Katoh et al., 2015; Katoh et al., 2009). Because monoadenylylation of miR-122 predominates, Gld2 deletion in mice leads to an overall decrease in miR-122 in liver (Katoh et al., 2009). Nevertheless, knockdown of PARN results in increased miR-122, revealing the destabilizing role for oligoadenylylation in this context (Katoh et al., 2015). However, the results of this panel, which show that stable knockdown of PARN by 99% had no effect on miR-7 levels, suggest that a similar phenomenon is not occurring for miR-7.
(I) Influence of DIS3L2 on miR-7 expression. The levels of DIS3L2 in lines #2–3 were <1% of that in the control. Otherwise, this panel is as in (G). Although DIS3L2 is thought to prefer oligouridylylated substrates, a recent biochemical effort to identify factors involved in viral TDMD enriched for Dis3l2 (Haas et al., 2016). However, the results of this panel indicate that CRISPRi knockdown of DIS3L2 by 98% had no discernable effect on miR-7 levels.
Figure S5. Cyrano Enables Cdr1as to Accumulate in Brain, Related to Figure 5
(A) Sashimi plot mapping splice junctions of Cyrano and Nusap1 transcripts in wild-type and Cyrano−/− cerebellum. Read density is shown on the y-axis with the range given by the numbers in brackets above each track. Each arc represents a splice junction, and the numbers associated with each arc are the number of reads mapping to that splice junction.
(B) Organization and expression of the extended murine Cdr1as locus. Shown are gene models (black boxes, exons; > > >, introns) for the circularized Cdr1as exon (Cdr1as) and adjacent RIKEN cDNA genes that might be misannotated fragments of the host gene for the Cdr1as circRNA (Barrett et al., 2017). Otherwise, this panel is as in Figure 1A.
(C) Relative Cdr1as expression in 25 tissues of 2-month-old C57Bl/6J mice. Shown are mean Cdr1as levels, as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes and relative to cerebellum (error bars, s.d.; n = 3 for each tissue).
(D) Representative images from single-molecule FISH experiments probing for Cyrano (black) in wild-type (upper) and Cyrano−/− (lower) CGNs, with insets showing increased magnification of portions of soma and processes. In Cyrano-deficient CGNs, a few Cyrano transcripts were detected, as expected for a probe set that extended beyond the deleted portion of Cyrano, but compared to wild type, the number of molecules detected was greatly reduced. Otherwise, this panel is as in Figure 5D–E.
(E) Quantitation of single-molecule FISH results obtained when probing for Cyrano in DIV11 wild-type (upper) and Cyrano−/− (lower) CGNs. Otherwise, this panel is as in Figure 5D–E.
Figure S6. Cdr1as is Regulated by miR-7 and miR-671, Related to Figure 6
(A) Importance of the miR-7 complementary site for the ability of Cyrano to influence Cdr1as levels in cerebellum. Cdr1as expression in wild-type and Cyrano mutant cerebellum (circle) was determined by RT-qPCR and plotted as in Figure 6D. For comparison, Cdr1as expression was also examined in pituitaries (diamond) of wild-type and some mutant mice (n.t., not tested). Statistical significance of the differences in Cdr1as level in each Cyrano mutant tissue compared to that in wild-type tissue is indicated (***, p <0.001, ANOVA with Dunnett’s test)..Otherwise, this panel is as in Figure 6D.
(B) Schematic of the three murine Mir7 loci (black bars, pre-miRNAs; mature miRNAs and miRNA passenger strands are colored red and blue, respectively). PAM motifs used for Cas9-mediated mutagenesis are outlined (open gray boxes), and nucleotides deleted in Mir7a1 and Mir7b mutants are indicated by dashes.
(C) Relative brain weights in P0 wild-type, Mir7a1+/−; Mir7b+/−, and Mir7 DKO mice from three litters produced by crosses of double heterozygotes. Weights were mean-normalized for each litter. A significant difference in brain size between wild-type and DKO mice was not detected.
(D) miR-7 expression in wild-type and Mir7 DKO brain. Shown is a northern blot measuring miR-7 levels in the indicated brain regions of 2-month-old wild-type and Mir7 DKO female mice. Levels of miR-7 (upper panel) were normalized to those of U6 (lower panel) and reported relative to the wild-type level in each region. The residual miR-7 observed in DKO brain was presumably derived from the intact Mir7a2 locus.
(E) Influence of miR-7 on predicted targets of miR-7 in cerebellum and cortex. #1 and #2 refer to two independent experiments (n = 3–4 per genotype for each experiment). For Mir7 DKO tissues, the number of genes analyzed in each set ranged from 3809–3959 (all), 443–448 (conserved), 80–82 (top), and 2246–2376 (no site). Otherwise, this panel is as in Figure 4C.
(F) Influence of miR-7 on Cdr1as accumulation in brain and pituitary. Cdr1as levels were determined by RT-qPCR and plotted as in Figure 5B (green circles; black bars, mean; n = 4 per genotype). For comparison, Cyrano levels, determined in parallel, are also plotted (blue). Statistically significant changes in Cdr1as levels in Mir7 DKO tissues compared to wild-type tissues are indicated (*, p <0.05; **, p <0.01; ***, p <0.001, unpaired two-tailed t-test).
(G) Schematic of the murine Cdr1as locus. Shown below the gene model (black box, circularized exon), are conservation plots, as in Figure 1A, and lines indicating the location of either miR-7 sites (red) or the miR-671 site (yellow). Pairing of miR-671 to its slicing-competent site is diagramed below the conservation plots. The PAM motif used for Cas9-mediated mutagenesis is outlined (open gray box), and nucleotides deleted in the two different Cdr1as mutants are indicated by dashes.
(H) miRNA profiling in wild-type and Cdr1as671_18/y cerebellum. Shown are log2-fold changes in mean miRNA levels observed between wild-type and Cdr1as671_18/y cerebellum, as determined by small-RNA sequencing and plotted as a function of expression in wild-type cerebellum (n = 3 per genotype). Each circle represents one miRNA, showing results for all miRNAs expressed above 1 RPM (read per million miRNA reads) in wild-type cerebellum. Circles representing miR-7 paralogs and miR-671 are colored red and yellow, respectively. miR-671 increased 1.8 fold (adjusted p = 5.09 × 10−7, as determined by DESeq2), supporting the idea that the miR-671 complementary site in Cdr1as directs some degradation of miR-671 (Piwecka et al., 2017).
(I) Influence of the miR-671 site on Cdr1as accumulation in brain and pituitary. Otherwise, this panel is as in (F).
(J) miRNA profiling in wild-type and Cyrano−/−; Cdr1as671_18/y cerebellum. Otherwise, this panel is as in (H).
Table S1.
Table S2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-MAP2, clone ap20 | Millipore | RRID: AB_94856 |
| Guinea pig polyclonal anti-Tau | Synaptic Systems | RRID: AB_1547385 |
| Rabbit polyclonal anti-GFAP | Dako | RRID: AB_10013382 |
| Bacterial and Virus Strains | ||
| One Shot TOP10 chemically competent E. coli | Life Technologies | C404006 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| [γ-32P]ATP | PerkinElmer | NEG035C001MC |
| TRI Reagent Solution | Life Technologies | AM9738 |
| Chloroform | J.T. Baker Analytical | 9180-01 |
| Yeast tRNA | Life Technologies | 15401011 |
| Superase-In | Life Technologies | AM2696 |
| GE Healthcare Hybond-NX Membrane | VWR | 95038-412 |
| ULTRAhyb-Oligo hybridization buffer | Life Technologies | AM8663 |
| EDC (N-(3-dimethylaminopropyl)-N'-ethylcarbodiimide) | Thermo Scientific | 22891 |
| Spin-X Centrifuge Tube Filters | Corning | CLS8162 |
| Critical Commercial Assays | ||
| TruSeq unstranded RNA Library kit | Illumina | RS-122-2001 |
| NEXTflex stranded RNA-seq kit | Bioo Scientific | 5138-10 |
| Lipofectamine 2000 | Life Technologies | 11668019 |
| QuikChange Lightning Multi Site-Directed Mutagenesis | Agilent | 210515 |
| KAPA HiFi HotStart ReadyMix | Roche | KK2601 |
| KAPA 2G Fast Genotyping Mix | Roche | KK5121 |
| Papain Dissociation System | Worthington | LK003150 |
| Micro Bio-Spin P30 gel columns | Bio-Rad | 7326250 |
| QuantiTect RT kit | Qiagen | 205311 |
| Turbo DNase | Life Technologies | AM2239 |
| RQ1 Dnase | Promega | M6101 |
| T4 PNK | New England Biolabs | 101228-172 |
| SuperScript III Reverse Transcriptase | Life Technologies | 18080044 |
| SYBR Green PCR Master Mix | Applied Biosystems | 4309155 |
| Deposited Data | ||
| RNA-seq | This paper | GEO: GSE112635 |
| Small-RNA sequencing | This paper | GEO: GSE112635 |
| Mir7a2−/− pituitary RNA-seq | Ahmed et al., 2017 | ENA PRJEB12612 |
| Gld2−/− hippocampus RNA-seq | Mansur et al., 2016 | GEO: GSE114259 |
| Experimental Models: Cell Lines | ||
| Human: K562-dCas9-KRAB | Gilbert et al., 2014 | N/A |
| Human: HEK293T | ATCC | CRL-3216 |
| Human: HEK293 | ATCC | CRL-1573 |
| Human: HeLa | ATCC | CCL-2 |
| Human: SH-SY5Y | ATCC | CRL-2266 |
| Human: MCF7 | ATCC | HTB-22 |
| Human: SK-N-SH | ATCC | HTB-11 |
| Mouse: Neuro2a | ATCC | CCL-131 |
| Mouse: C2C12 | ATCC | CRL-1772 |
| Human: HeLa-dCas9-BFP-KRAB | This paper | N/A |
| Human: SH-SY5Y-dCas9-BFP-KRAB | This paper | N/A |
| Human: MCF7-dCas9-BFP-KRAB | This paper | N/A |
| Human: SK-N-SH-dCas9-BFP-KRAB | This paper | N/A |
| Mouse: C2C12-dCas9-BFP-KRAB | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Cyranofl/fl | This paper | N/A |
| Mouse: Cyrano−/− | This paper | N/A |
| Mouse: CyranoM1/M1 | This paper | N/A |
| Mouse: CyranoM2/M2 | This paper | N/A |
| Mouse: CyranoM3/M3 | This paper | N/A |
| Mouse: CyranoM4/M4 | This paper | N/A |
| Mouse: CyranoM5/M5 | This paper | N/A |
| Mouse: CyranoM6/M6 | This paper | N/A |
| Mouse: Mir7a1−/−; Mir7b−/− | This paper | N/A |
| Mouse: Cdr1as671_18/y | This paper | N/A |
| Mouse: Cdr1as671_54/y | This paper | N/A |
| Oligonucleotides | ||
| See Table S3 | This paper | N/A |
| Single-molecule FISH probes (also listed in Table S3) | Biosearch Technologies | SMF-1065-5 |
| miR-7 LNA northern probe | Exiqon | 38485-00 |
| Recombinant DNA | ||
| pPGKneoF2L2DTA | Hoch et al., 2006 | Addgene #13445 |
| pHR-SFFV-dCas9-BFP-KRAB | Gilbert et al., 2013 | Addgene #46911 |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP | Gilbert et al., 2014 | Addgene #60955 |
| Cyrano conditional knockout targeting vector | This paper | N/A |
| pcDNA3.1-mCherry | This paper | N/A |
| pcDNA3.1-mCherry-CyrCR1 | This paper | N/A |
| pcDNA3.1-mCherry-CyrCR1.mutant | This paper | N/A |
| pcDNA3.1-mCherry-CyrCR1.miR16 | This paper | N/A |
| pcDNA3.1-mCherry-CyrCR1.miR17 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.Control | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.CYRANO | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PAPD4-1 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PAPD4-2 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PAPD4-3 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PARN-1 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PARN-2 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.PARN-3 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.DIS3L2-1 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.DIS3L2-2 | This paper | N/A |
| pU6-sgRNA EF1Alpha-puro-T2A-BFP.DIS3L2-3 | This paper | N/A |
| Software and Algorithms | ||
| STAR RNA-seq Aligner | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Htseq-count | Anders et al., 2015 | https://htseq.readthedocs.io/en/release_0.9.1/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Targetscan v7.1 | Agarwal et al., 2015 | targetscan.org |
| ImageQuant TL (v8.1.0.0) | GE Healthcare | N/A |
| GraphPad Prism | GraphPad Software | N/A |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| ImageM (for MATLAB) | Lyubimova et al., 2013 | https://github.com/xuebingwu/ImageM |
| Photoshop CS5.1 | Adobe | N/A |
| Other | ||
| Small-RNA sequencing protocol, step-by-step | Fang and Bartel, 2015 | http://bartellab.wi.mit.edu/protocols.html |
| CRISPRi guide cloning protocol, step-by-step | Gilbert et al., 2014 | http://weissmanlab.ucsf.edu/CRISPR/CRISPR.html |
| Small-RNA blot protocol, step-by-step | Fang and Bartel, 2015 | http://bartellab.wi.mit.edu/protocols.html |
ACKNOWLEDGMENTS
We thank E. Kingston, A. Shkumatava, I. Ulitsky, other current and former members of the Bartel lab, M. Piwecka, N. Rajewsky, and A. Granger for helpful discussions; A. Liu for technical assistance; T. Eisen for hippocampal cultures; S. Eichhorn, J. Kwasnieski, K. Lin, J. Morgan, and X. Wu for computational advice; the Whitehead Institute Genome Technology Core for sequencing; W. Salmon and the Keck Imaging Facility for microscopy expertise; the Janelia Gene Targeting and Transgenic Facility, the MIT Transgenic Facility, and the Whitehead Institute Genetically Engineered Models Facility for generating mutant mice. This research was supported by NIH grants GM061835 and GM118135 (D.P.B), and T32CA009216 (B.K.). D.P.B is an investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agarwal V, Bell GW, Nam JW, and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, LaPierre MP, Gasser E, Denzler R, Yang Y, Rulicke T, Kero J, Latreille M, and Stoffel M (2017). Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. J. Clin. Invest 127, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, and Zamore PD (2010). Target RNA-directed trimming and tailing of small silencing RNAs. Science 328, 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Hung JH, Xu J, Weng Z, and Zamore PD (2011). Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA 17, 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham R, Sas-Chen A, Manor O, Steinfeld I, Shalgi R, Tarcic G, Bossel N, Zeisel A, Amit I, Zwang Y, et al. (2010). EGF decreases the abundance of microRNAs that restrain oncogenic transcription factors. Sci. Signal 3, ra43. [DOI] [PubMed] [Google Scholar]
- Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, and Brown BD (2011). Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol 21, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, and Kiledjian M (2010). Differential regulation of microRNA stability. RNA 16, 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, Raj A, and Tyagi S (2011). Single molecule imaging of RNA in situ. Methods in Molecular Biology (Clifton, NJ) 714, 3–13. [DOI] [PubMed] [Google Scholar]
- Beaudoin GM 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, and Arikkath J (2012). Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat. Protocols 7, 1741–1754. [DOI] [PubMed] [Google Scholar]
- Bitetti A, Mallory AC, Golini E, Carrieri C, Carreno Gutierrez H, Perlas E, Perez-Rico YA, Tocchini-Valentini GP, Enright AJ, Norton WHJ, et al. (2018). MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol 25, 244–251. [DOI] [PubMed] [Google Scholar]
- Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Sokilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, et al. (2014). PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 111, 11467–11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, and Steitz JA (2010). Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328, 1563–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, and Gregory RI (2013). A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Gaidatzis D, Vitanescu M, Stadler MB, Wentzel C, Scheiffele P, Filipowicz W, and Grosshans H (2015). Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 16, 500–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, and Stoffel M (2016). Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell 64, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Chen K, Cuevas-Diaz Duran R, You Y, Sloan SA, Zhang Y, Zong S, Cao Q, Barres BA, and Wu JQ (2015). Comprehensive Identification of Long Non-coding RNAs in Purified Cell Types from the Brain Reveals Functional LncRNA in OPC Fate Determination. PLoS Genet. 11, e1005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, and Simon MD (2015). Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol. Cell 59, 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbesen KK, Hansen TB, and Kjems J (2017). Insights into circular RNA biology. RNA Biol. 14, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu SH, Ghoshal K, Villen J, and Bartel DP (2014). mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 56, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, and Tuschl T (2001). Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. (2011). Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39, 5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, and Bartel DP (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Liu J, Elfenbein SJ, Ma Y, Zhong M, Qiu C, Ding Y, and Lu J (2015). Characterization of the mammalian miRNA turnover landscape. Nucleic Acids Res. 43, 2326–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, and Pfeffer S (2016). Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 44, 2873–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, and Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, and Kjems J (2011). miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, and Mendell JT (2007). A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100. [DOI] [PubMed] [Google Scholar]
- Ji L, and Chen X (2012). Regulation of small RNA stability: methylation and beyond. Cell Res. 22, 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, and Izaurralde E (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet 16, 421–433. [DOI] [PubMed] [Google Scholar]
- Katoh T, Hojo H, and Suzuki T (2015). Destabilization of microRNAs in human cells by 3' deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 43, 7521–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, and Suzuki T (2009). Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 23, 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, and Richter JD (2006). Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24, 173–183. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, and Mendell JT (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. (2010). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631. [DOI] [PubMed] [Google Scholar]
- Latreille M, Hausser J, Stutzer I, Zhang Q, Hastoy B, Gargani S, Kerr-Conte J, Pattou F, Zavolan M, Esguerra JL, et al. (2014). MicroRNA-7a regulates pancreatic beta cell function. J. Clin. Invest 124, 2722–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Greene LA, Mason CA, and Manzini MC (2009). Isolation and culture of postnatal mouse cerebellar granule neuron progenitor cells and neurons. J. Vis. Exp 23, e990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, and Ahn K (2013). Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe 13, 678–690. [DOI] [PubMed] [Google Scholar]
- Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, and Steitz JA (1988). Four novel U RNAs are encoded by a herpesvirus. Cell 54, 599–607. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, and Buck AH (2012). Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. USA 109, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimova A, Itzkovitz S, Junker JP, Fan ZP, Wu X, and van Oudenaarden A (2013). Single-molecule mRNA detection and counting in mammalian tissue. Nat. Protocols 8, 1743–1758. [DOI] [PubMed] [Google Scholar]
- Mansur F, Ivshina M, Gu W, Schaevitz L, Stackpole E, Gujja S, Edwards YJ, and Richter JD (2016). Gld2-catalyzed 3' monoadenylation of miRNAs in the hippocampus has no detectable effect on their stability or on animal behavior. RNA 22, 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinowski L, Tanguy M, Krmpotic A, Radle B, Lisnic VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, et al. (2012). Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Path. 8, e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, et al. (2017). Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, and Siepel A (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock A, Bian S, Zhang C, Chen Z, and Sun T (2014). Growth of the developing cerebral cortex is controlled by microRNA-7 through the p53 pathway. Cell Rep. 7, 1184–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimao-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, and Ameres SL (2016). Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J. 35, 2417–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, and Bartel DP (2011). MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell 43, 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saetrom P, Heale BS, Snove O Jr., Aagaard L, Alluin J, and Rossi JJ (2007). Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 35, 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol 11, 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Zhang H, Rizavi H, Cook EH, and Dwivedi Y (2014). Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PloS One 9, e86469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, and Warman ML (2000). Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). BioTechniques 29, 52, 54. [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, and Bartel DP (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Thaller C, and Eichele G (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. (2012). Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 9, 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Cyrano Expression, Related to Figure 1
(A) Images of in situ hybridizations for Cyrano in mouse embryo (E14.5) and adult mouse brain (P56) obtained from GenePaint.org (Visel et al., 2004) and the Allen Brain Atlas (Lein et al., 2007), respectively. In both databases, Cyrano is annotated as the RIKEN cDNA 1700020I14Rik gene.
(B) Relative Cyrano expression in 25 tissues of 2-month-old C57Bl/6J mice. Shown are mean Cyrano levels, as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes and relative to cerebellum (error bars, s.d.; n = 3 for each tissue).
(C) CYRANO expression from RNA-seq of 32 post-mortem human tissues, provided by the GTEx Portal (01/15/2018). Expression values in TPM (transcripts per million) are shown as box plots (box, 25th, median, and 75th percentiles; whiskers, 1.5 times the interquartile range). In the GTEx Portal, CYRANO is annotated as the OIP5-AS1 gene.
(D) Cyrano expression from RNA-seq of purified cell populations from the mouse cortex (Dong et al., 2015). Mean expression values in FPKM (fragments per kb per million reads) are shown (error bars, range; n = 2 for each cell type).
(E) Percentage of CYRANO detected in the cytoplasm of mammalian cell lines. Ratios were determined by RT-qPCR, using RNA from cytoplasmic and nuclear fractions, normalizing to a luciferase spike-in transcript (error bars, range; n = 2 for HEK293, HeLa, and SH-SY5Y, n = 3 for K562 and Neuro2a). For comparison, results for GAPDH mRNA and the nuclear-localized XIST lncRNA are also shown (n.d., not detected above background in cytoplasm).
(F) Cyrano gene targeting strategy. Shown between the wild-type locus and the recombinant alleles is the vector used to target the first half of exon 3 (black boxes, exons; red line, miR-7 site with extensive pairing; open triangles, loxP sites; open diamonds, FRT sites; neo, neomycin-resistance cassette; DTA, diphtheria toxin A; AoH, arm of homology). Screening of ES cell clones was performed by long-range PCR as well as Southern blot of XbaI-digested genomic DNA with a 5′ probe upstream of exon 1.
Figure S2. Cyrano Promotes miR-7 Destruction, Related to Figure 1
(A) Influence of Cyrano on pri-miR-7 expression in the brain. The plot on the left shows mean pri-miR-7a1 and pri-miR-7b levels in wild-type (black) and Cyrano−/− (red) anterior cortex and cerebellum (key), as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes (error bars, s.d.; n = 3 per genotype for each tissue). The plot on the right shows mean expression values in RPM (reads per million) for all three pri-miR-7 transcripts in wild-type and Cyrano−/− anterior cortex, cerebellum, cortex, and pituitary, as determined by RNA-seq (error bars, s.d.; n = 4–5 per genotype for each tissue; n.d., not detected).
(B) Influence of Cyrano on pre-miR-7 expression. Shown is a representative northern blot measuring miR-7 levels in wild-type and mutant cortex. This blot is an uncropped version of the blot shown in Figure 6C, with the signal increased in order to detect the less-abundant pre-miR-7 species. Levels of pre-miR-7 species (identified by their absence in the Mir7 DKO) were unchanged in Cyrano−/− cortex.
(C) Relationship between miR-7 changes observed after disrupting Cyrano and the level of wild-type Cyrano expression. Shown are miR-7 fold changes from Figure 2C plotted as a function of Cyrano expression in wild-type tissues, as determined from RNA-seq data also used for the analyses of Figure 5C.
(D) Influence of Cyrano on miR-7 expression in developing mouse brain. Shown at the left is a representative northern blot measuring miR-7 levels in wild-type (WT) and Cyrano−/− (KO) brain at the indicated developmental time points. Plotted on the right are miR-7 levels in wild-type (black) and Cyrano−/− (red) embryonic brain normalized to miR-16 (error bars, s.d.; n = 3 per genotype). Statistically significant fold changes are indicated (*, p <0.05, unpaired two-tailed t-test).
(E) Influence of Cyrano on miR-7 expression in cultured primary neurons. Shown are northern blots for wild-type and Cyrano−/− DIV11 cerebellar neurons (left) and DIV12 hippocampal neurons (right). Levels of miR-7 (upper panels) were normalized to those of either miR-16 (lower panel, left) or U6 (lower panel, right), and reported relative to the mean wild-type level.
(F) Relationship between the magnitude of miR-7 fold change and extent of CYRANO knockdown. Shown are miR-7 fold changes, measured by northern blot, plotted as a function of CYRANO knockdown, as determined by RT-qPCR, for 35 mammalian CRISPRi cell lines stably expressing dCas9-KRAB and an sgRNA targeting CYRANO. Parental cell lines are indicated (key). For each cell line receiving a targeting sgRNA, levels of CYRANO and miR-7 were normalized to the geometric mean of the values for five housekeeping genes and miR-16, respectively, and shown relative to the non-targeting control.
Figure S3. Mutating the miR-7 Site in Cyrano Does Not Cause Detectable Changes in Cyrano Levels, Related to Figure 2
(A) Cyrano locus annotated with binding sites of 3 RT-qPCR primer pairs (#1, #2, #3). Otherwise, this panel is as in Figure 1A.
(B) Cyrano expression in wild-type and mutant cerebellum. Plotted are relative Cyrano levels in wild-type and Cyrano-mutant cerebellum, as determined by RT-qPCR with three primer sets pairing to sites mapped in (A). Cyrano levels, normalized to that of Actb, are shown as fold changes relative to the mean wild-type expression (n = 4 per genotype; n.d., not detected). No significant differences in Cyrano expression were detected (ANOVA with Dunnett’s test), indicating that either miR-7 does not influence Cyrano accumulation in this tissue, or abrogated repression at this site is offset by enhanced repression (due to increased levels of miR-7) at a second, more conventional site located at the beginning of exon 3.
(C) Cyrano expression in wild-type and Cyrano-mutant pituitary. Otherwise, this panel is as in (B).
Figure S4. Cyrano Induces Tailing and Trimming of miR-7, but Tailing Appears Dispensable, Related to Figure 3
(A) Influence of Cyrano on miR-7 tailing and trimming in cerebellum. Otherwise, this panel is as in Figure 3A. In both hippocampus and cerebellum, the trimmed miR-7 isoforms largely matched the genome-encoded miR-7 sequence, implying that tailing of trimmed species was either rare or destabilizing.
(B) Influence of Cyrano on global miRNA tailing in hippocampus (left) and cerebellum (right). Shown are aggregate tailing log2-fold changes for each expressed miRNA after disrupting Cyrano, plotting statistical significance of the change (based on p values determined by unpaired two-tailed t-tests, one per miRNA) as a function of the change in WT divided by Cyrano−/−. Each circle represents one miRNA, showing results for all miRNAs expressed above 100 RPM (read per million miRNA reads) in wild-type tissue. Circles representing miR-7 (miR-7a and miR-7b were combined for this analysis) and miR-92b are colored red and gray, respectively. Cyrano increased the aggregate tailed fractions 1.5 and 2.2 fold, respectively. Dotted lines indicate a p value of 0.05.
(C) Influence of Cyrano on global miRNA trimming in hippocampus (left) and cerebellum (right). Cyrano increased the aggregate trimmed fractions 3.2 and 1.6 fold, respectively. Otherwise, this panel is as in (B).
(D–E) Influence of Gld2 on global miRNA tailing (D) and trimming (E) in hippocampus, evaluated using data from Mansur et al. (2016). Otherwise, this panel is as in (B and C).
(F) Influence of Gld2 on miRNA levels in hippocampus. Shown are log2-fold changes in mean miRNA levels observed between wild-type and Gld2−/− hippocampus, as determined by small-RNA sequencing (Mansur et al., 2016) and plotted as a function of expression in wild-type hippocampus (n = 6 per genotype). Each circle represents one miRNA, showing results for all miRNAs expressed above 1 RPM (read per million miRNA reads) in wild-type hippocampus. Circles representing miR-7 paralogs are colored red.
(G) Influence of PAPD4 on miR-7 expression in stable PAPD4-knockdown K562 cells. Shown at the top is a northern blot monitoring miR-7 levels in stable cell lines expressing either a control sgRNA or an sgRNA targeting either CYRANO or PAPD4 for CRISPRi-mediated knockdown. The levels of miR-7 are reported relative to the miR-7 level in the control cell line after normalizing to the level of miR-16. Plotted at the bottom are CYRANO and PAPD4 levels, as determined by RT-qPCR, in stable lines cells expressing the indicated sgRNAs. CYRANO and PAPD4 levels were normalized to the geometric mean of the values for five housekeeping genes and plotted relative to levels in the control cell line. The level of PAPD4 in line #2 was <2% of that in the control.
(H) Influence of PARN on miR-7 expression. The levels of PARN in lines #1–3 were each <1% of that in the control. Otherwise, this panel is as in (G). One way to reconcile the prevailing idea that tailing is required for TDMD with our observation that miR-7 levels remained unchanged after loss of Gld2 would be to propose that Gld2 has opposing consequences that somehow offset each other. Indeed, for miR-122, monoadenylylation by Gld2 stabilizes the miRNA, whereas oligoadenylylation by Gld2 destabilizes it by making it a substrate for PARN-catalyzed exonucleolytic decay (Katoh et al., 2015; Katoh et al., 2009). Because monoadenylylation of miR-122 predominates, Gld2 deletion in mice leads to an overall decrease in miR-122 in liver (Katoh et al., 2009). Nevertheless, knockdown of PARN results in increased miR-122, revealing the destabilizing role for oligoadenylylation in this context (Katoh et al., 2015). However, the results of this panel, which show that stable knockdown of PARN by 99% had no effect on miR-7 levels, suggest that a similar phenomenon is not occurring for miR-7.
(I) Influence of DIS3L2 on miR-7 expression. The levels of DIS3L2 in lines #2–3 were <1% of that in the control. Otherwise, this panel is as in (G). Although DIS3L2 is thought to prefer oligouridylylated substrates, a recent biochemical effort to identify factors involved in viral TDMD enriched for Dis3l2 (Haas et al., 2016). However, the results of this panel indicate that CRISPRi knockdown of DIS3L2 by 98% had no discernable effect on miR-7 levels.
Figure S5. Cyrano Enables Cdr1as to Accumulate in Brain, Related to Figure 5
(A) Sashimi plot mapping splice junctions of Cyrano and Nusap1 transcripts in wild-type and Cyrano−/− cerebellum. Read density is shown on the y-axis with the range given by the numbers in brackets above each track. Each arc represents a splice junction, and the numbers associated with each arc are the number of reads mapping to that splice junction.
(B) Organization and expression of the extended murine Cdr1as locus. Shown are gene models (black boxes, exons; > > >, introns) for the circularized Cdr1as exon (Cdr1as) and adjacent RIKEN cDNA genes that might be misannotated fragments of the host gene for the Cdr1as circRNA (Barrett et al., 2017). Otherwise, this panel is as in Figure 1A.
(C) Relative Cdr1as expression in 25 tissues of 2-month-old C57Bl/6J mice. Shown are mean Cdr1as levels, as determined by RT-qPCR, normalized to the geometric mean of the values for four housekeeping genes and relative to cerebellum (error bars, s.d.; n = 3 for each tissue).
(D) Representative images from single-molecule FISH experiments probing for Cyrano (black) in wild-type (upper) and Cyrano−/− (lower) CGNs, with insets showing increased magnification of portions of soma and processes. In Cyrano-deficient CGNs, a few Cyrano transcripts were detected, as expected for a probe set that extended beyond the deleted portion of Cyrano, but compared to wild type, the number of molecules detected was greatly reduced. Otherwise, this panel is as in Figure 5D–E.
(E) Quantitation of single-molecule FISH results obtained when probing for Cyrano in DIV11 wild-type (upper) and Cyrano−/− (lower) CGNs. Otherwise, this panel is as in Figure 5D–E.
Figure S6. Cdr1as is Regulated by miR-7 and miR-671, Related to Figure 6
(A) Importance of the miR-7 complementary site for the ability of Cyrano to influence Cdr1as levels in cerebellum. Cdr1as expression in wild-type and Cyrano mutant cerebellum (circle) was determined by RT-qPCR and plotted as in Figure 6D. For comparison, Cdr1as expression was also examined in pituitaries (diamond) of wild-type and some mutant mice (n.t., not tested). Statistical significance of the differences in Cdr1as level in each Cyrano mutant tissue compared to that in wild-type tissue is indicated (***, p <0.001, ANOVA with Dunnett’s test)..Otherwise, this panel is as in Figure 6D.
(B) Schematic of the three murine Mir7 loci (black bars, pre-miRNAs; mature miRNAs and miRNA passenger strands are colored red and blue, respectively). PAM motifs used for Cas9-mediated mutagenesis are outlined (open gray boxes), and nucleotides deleted in Mir7a1 and Mir7b mutants are indicated by dashes.
(C) Relative brain weights in P0 wild-type, Mir7a1+/−; Mir7b+/−, and Mir7 DKO mice from three litters produced by crosses of double heterozygotes. Weights were mean-normalized for each litter. A significant difference in brain size between wild-type and DKO mice was not detected.
(D) miR-7 expression in wild-type and Mir7 DKO brain. Shown is a northern blot measuring miR-7 levels in the indicated brain regions of 2-month-old wild-type and Mir7 DKO female mice. Levels of miR-7 (upper panel) were normalized to those of U6 (lower panel) and reported relative to the wild-type level in each region. The residual miR-7 observed in DKO brain was presumably derived from the intact Mir7a2 locus.
(E) Influence of miR-7 on predicted targets of miR-7 in cerebellum and cortex. #1 and #2 refer to two independent experiments (n = 3–4 per genotype for each experiment). For Mir7 DKO tissues, the number of genes analyzed in each set ranged from 3809–3959 (all), 443–448 (conserved), 80–82 (top), and 2246–2376 (no site). Otherwise, this panel is as in Figure 4C.
(F) Influence of miR-7 on Cdr1as accumulation in brain and pituitary. Cdr1as levels were determined by RT-qPCR and plotted as in Figure 5B (green circles; black bars, mean; n = 4 per genotype). For comparison, Cyrano levels, determined in parallel, are also plotted (blue). Statistically significant changes in Cdr1as levels in Mir7 DKO tissues compared to wild-type tissues are indicated (*, p <0.05; **, p <0.01; ***, p <0.001, unpaired two-tailed t-test).
(G) Schematic of the murine Cdr1as locus. Shown below the gene model (black box, circularized exon), are conservation plots, as in Figure 1A, and lines indicating the location of either miR-7 sites (red) or the miR-671 site (yellow). Pairing of miR-671 to its slicing-competent site is diagramed below the conservation plots. The PAM motif used for Cas9-mediated mutagenesis is outlined (open gray box), and nucleotides deleted in the two different Cdr1as mutants are indicated by dashes.
(H) miRNA profiling in wild-type and Cdr1as671_18/y cerebellum. Shown are log2-fold changes in mean miRNA levels observed between wild-type and Cdr1as671_18/y cerebellum, as determined by small-RNA sequencing and plotted as a function of expression in wild-type cerebellum (n = 3 per genotype). Each circle represents one miRNA, showing results for all miRNAs expressed above 1 RPM (read per million miRNA reads) in wild-type cerebellum. Circles representing miR-7 paralogs and miR-671 are colored red and yellow, respectively. miR-671 increased 1.8 fold (adjusted p = 5.09 × 10−7, as determined by DESeq2), supporting the idea that the miR-671 complementary site in Cdr1as directs some degradation of miR-671 (Piwecka et al., 2017).
(I) Influence of the miR-671 site on Cdr1as accumulation in brain and pituitary. Otherwise, this panel is as in (F).
(J) miRNA profiling in wild-type and Cyrano−/−; Cdr1as671_18/y cerebellum. Otherwise, this panel is as in (H).
Table S1.
Table S2.
Data Availability Statement
Sequencing datasets generated in this study have been deposited in the GEO under accession number GSE112635.