Introduction
Pancreatic Cancer (PDA) is an aggressive malignancy, usually diagnosed at advanced stages and with a dismally low 5 year survival rate of less than 9%[1]. Early disease spread and local invasion combined with the high incidence of chemoresistance and lack of effective immunotherapy, makes PDA the 3rd leading cause of cancer-related death in the United States [2, 3].
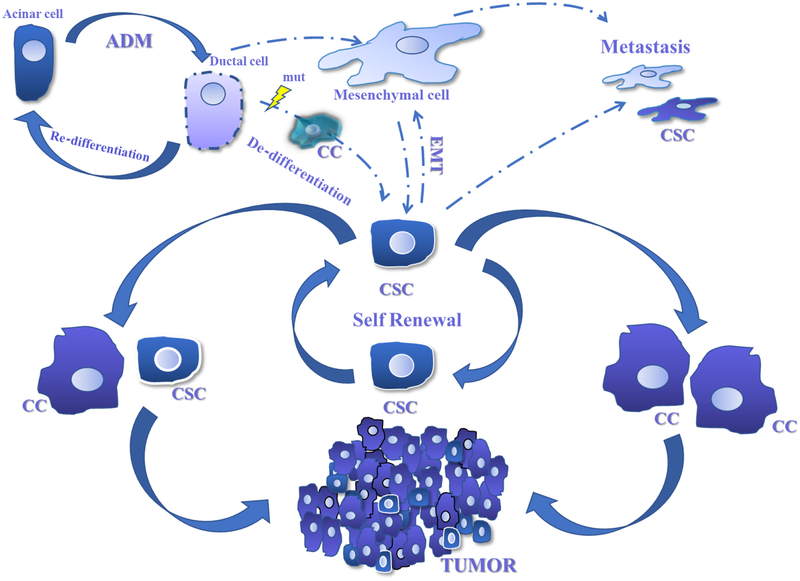
Cancer stem cells (CSC) are undifferentiated quiescent cells characterized by their ability to self-renew coupled with unique plasticity and metabolism [4]. In PDA, CSC are thought to comprise a small subpopulation within the tumor reported to be responsible for driving tumorigenesis and progression of the disease [5]. Emerging evidence suggests that CSC play an important role in disease recurrence and metastatic events and may represent the primary source of resistance to chemotherapy and radiation [6, 7]. The origin of pancreatic CSC remains unknown, but recent data cites local factors present in the tumor microenvironment as supportive of cellular persistence in a relatively undifferentiated state which may favor their development [8–15]. The classical CSC model proposes a hierarchical origin where the bulk of the tumor is constituted by non-CSC and a apical small portion is constituted by CSC that serve as a quiescent, self-renewing pool that can differentiate and replenish the non-CSC in the tumor ([16, 17] Figure 1).
Figure 1. Plasticity of Cancer Stem Cells in Pancreatic cancer.
Normal pancreatic acinar cells undergo trans-differentiation in response to injury and metabolic or inflammatory stress in a process known as Acinar-to-Ductal-Metaplasia (ADM). Oncogenic mutations can result in failure to re-differentiate leading to malignant degeneration and cancer formation. Cancer Stem Cells (CSC) are poised to originate from de-differentiation of genetically mutated epithelial cells or from non-cancer stem cells. Factors present in the PDA TME favor persistence of CSC and can promote a more malignant phenotype including metastatic potential. CSC can self-renew or produce differentiated progeny which comprise a majority of tumor cells.
Recent attention has focused on links between epigenetics and metabolism of CSC as potential mediators of their renewal capacity and chemoresistance suggesting possible new drug targets for these difficult-to-eradicate cells [18]. Because epigenetics represents a clear link connecting environmental effects and gene expression, this new arena of research could provide clues connecting known risk factors such as smoking and diet to the development of PDA [19, 20]. This review will focus on the metabolic and epigenetic features of pancreatic cancer CSC. A better understanding of this cellular subpopulation’s functional and phenotypic characteristics will allow us to develop more effective therapeutic strategies.
Cancer Stem Cell properties in PDA
Our understanding of the complexity of cancer at the cellular and molecular level has significantly expanded, yet the underlying mechanism aiding in evasion and resistance to therapy remains largely elusive. Advances such as the advent of single cell sequencing has revealed that tumors are composed not of single clones, but of a vast array of cellular subpopulations. The basis for the generation of the large cellular tumor heterogeneity remains, however, largely undetermined. It is believed that amongst the cancer cell population, a fraction of the cells hold stem-cell-like characteristics including capacity to not only replicate, but produce progeny that are unique from themselves [21, 22]. The CSC hypothesis has been postulated for decades, stating that a small portion of undifferentiated quiescent cancer cells with limited growth were the source of differentiated cancer cell progeny. Along with the ability to self-renew, this CSC population has unique plasticity and metabolism, as well as enhanced chemoresistance [23, 24].
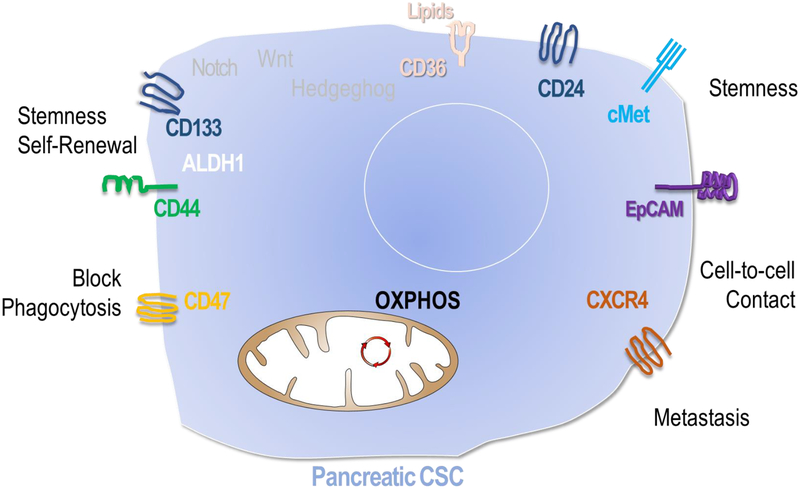
CSC in PDA were empirically defined by two major techniques, using tracing of genetic lineage markers [25] or by assessment of tumorigenesis in limiting dilution after transplantation into immunodeficient mice [26, 27]. CSCs were identified in other malignancies including lung, ovarian, and prostate cancer using similar methods [28–32]. The property that few or single cells from one subpopulation of tumor cells and not from others could reproduce an entire intact cancer is felt to reflect the stem-like potential of and therefore phenotypic core of CSCs but does not provide a reliable method to identify cells or follow their development in the tumor microenvironment (TME). [33–35]. In an effort to study CSCs in-situ, Driessens et al. used clonal analysis of squamous skin cancer in which single tumor cells were genetically labelled to follow their behavior within the TME [25]. They demonstrated that while most cells had a relatively limited proliferative capacity, a small population of cells persisted long-term and eventually their progeny constituted the majority of the tumor. Unfortunately no universal cell markers have been identified yet to unequivocally and reliably distinguish CSC populations in the TME. Cell surface markers including various combinations of CD34, CD44, CD133, ESA, ALDH1 and cMet have been shown to be associated with various CSC characteristics including limiting dilution transplantability [8, 28, 35, 36] (figure 2). Nevertheless, expression of these markers varies significantly among different cancer types and investigators [36, 37]. In PDA a small subset of tumor initiating cells were originally described by Li et al. using a xenograft mouse model transplanting subpopulations of cells from PDA patients into immunodeficient mice. They found that a small group of cells labelled as CD44+, CD24+, ESA+ could recapitulate features of the original tumor when injected into mice, while the larger population of cells could not. Although those cells constituted less than 1% of the original tumor they were a 100 times more tumorigenic than their non-CSCs counterparts [28]. In recent years additional PDA CSA specific cell markers have been characterized including CD133, CXCR4 and cMet [28, 38, 39]. Using these markers and various cell sorting techniques, researchers have begun to unfold some of the phenotypic characteristics unique to PDA CSC. Hermann et al. characterized CD133+ cells as CSC-like and gemcitabine-resistant and identified this population as essential for metastasis formation [39]. Isolation of cMet positive cells from human pancreas cancer specimens yielded cells with heightened tumorigenic potential as well as self-renewal capacity [38]. In addition to unique surface markers, PDA CSC were found to have high ALDH1 activity, a detoxifying enzyme which is essential for the early differentiation of stem cells [38]. Recent studies have shown that high levels of ALDH1 may provide protection against chemotherapy and represent a target in battling chemoresistance [40].
Figure 2. Pancreatic cancer stem cell characteristics.
CSC express unique surface and cytosolic proteins that aid in cell surface markers aid in macrophage evasion, self-renewal, stemness, and cell-to-cell in pancreatic cancer. They principally use OXPHOS but possess metabolic plasticity.
A subpopulation of Pancreatic cancer CSC, derived from parental PDA cell lines were demonstrated to have increased aggressive behavior. Bao et al. characterized the performance of triple positive CD44+, CD133+, EpCAM+ cells regarding migration and growth in addition to self-renewal [41]. More than 1500 genes were differentially expressed between triple positive and triple negative cells. In this study they used siRNA to demonstrate that the exacerbated aggressive behavior in this triple positive CSCs derived from MiaPaCa2 and L3.6pl cells was dependent on FoxQ1 expression. Additionally, xenograft tumors silenced for FoxQ1 in MiaPaCa2 cells displayed a clearly impaired ability to promote tumor development and growth [41].
Plasticity of CSC in PDA
Cellular plasticity is defined as the ability of the cells to alter their phenotypic state including differentiation status within a hierarchal order in response to environmental signals [4]. In the normal pancreas, acinar cells undergo trans-differentiation in response to injury, transcription factor activation, and/or metabolic or inflammatory stress, all of which promote conversion of cells to a more embryologic primitive duct-like state, a process known as ADM (Acinar-to-Ductal-Metaplasia) [42, 43]. This conversion is entirely reversible as these cells can re-differentiate back to their acinar origin when the promoting stimulus is removed, and the inhibition of ADM can prevent pancreatic regeneration (Figure 1 and [44, 45]). But if ADM happens in the presence of oncogenic KRAS mutations, the process becomes irreversible and promotes the formation of intraepithelial neoplastic regions in the pancreas, a prelude to malignant degeneration [46]. Some have proposed that persistence of cells in this trans-differentiated state, along with accumulation of further genetic mutations, is the origin of CSC in PDA [47]. The most common mutations found in PDA are aberrant activation in KRAS and inactivation of TP53, SMAD4, among others. [48]. Activation of some of these oncogenic signaling pathways influence metabolic reprogramming within the cell [49].
In PDA, a similar plasticity has been demonstrated for both CSC and non-CSC as shown by several studies where tumor cells undergo phenotypical changes in response to environmental signals [43]. CSC can change from a less differentiated stem-like phenotype to the highly active differentiated cells that constitute the bulk of the tumor. The resulting metaplastic cell can undergo further changes when exposed to sustained stimuli and become more mesenchymal and even potentially metastatic cells [41, 50] (figure 1).
Along with plasticity CSC exhibit relative quiescence which contributes to its therapeutic resistance. Quiescence is a non-proliferative state, defined as a reversible G-zero step of the cell cycle. These quiescent cells are thought to be in an actively preserved state from which the cells can escape and re-enter the cell cycle, which protects them from chemotherapeutics and often mediates their effects during cellular replication [51]. Entry and escape from the cell cycle are regulated both by epigenetic and metabolic cues from the TME and recent data has suggested that p53 is critical in supporting a stem cell quiescence state [52].
Metabolic plasticity in cancer cells and Cancer Stem Cells
Metabolic reprograming was recognized several decades ago as one of the hallmarks of cancer cells [53, 54]. Namely, rapidly proliferating tumor cells switch to the use of aerobic glycolysis to produce ATP displaying high consumption of glucose; features known as the “Warburg effect” [55]. Tumor cells are capable of having higher metabolic rates than their normal counterparts, utilizing glucose, glutamine, several amino acids and even fatty acids as substrates [56]. Thanks to their metabolic plasticity cancer cells quickly adapt to the challenges posed by the microenvironment reciprocally contributing to the heterogenous cellular metabolic landscape of the tumor niche.
CSC metabolic pathways have recently attracted a lot of interest since they pose an emerging source of new therapeutic targets in cancer. CSC’s distinct metabolic configuration is postulated to enhance tumorigenesis improving fitness of the cancer cells as a means of adaptation to nutrient or oxygen deprived conditions [57, 58]. As a relatively quiescent cell, CSCs also have vastly lower needs for energy and material to generate biomass as compared rapidly proliferating cells. Unsurprisingly, many 0differences exist between the metabolism of CSC and non-CSC in various tumor types [59]. As an example, CSCs appear to be more versatile than the non-CSC counterpart and able to use several energy source metabolic pathways [59]. Considerable controversy exists regarding the metabolic profile of CSC in many tumor types, as opposing reports in ovarian cancer for example, describe both oxidative phosphorylation (OXPHOS) and glycolysis dependence of the CSC metabolism [60, 61]. Additionally, extensive evidence suggests that metabolism of CSCs is tumor type specific with some relying on a glycolytic program such as nasopharyngeal and liver cancers [62, 63] while others use OXPHOS such as pancreatic [64], glioma [65] [60], lung [66] or colon cancer [67]. It is possible that CSCs have adapted a relatively plastic metabolism that can adjust to the settings in which the cells reside. If oxygen is available, the more energetically efficient OXPHOS is used rendering a higher number of ATP molecules per glucose molecule. In hypoxia or stress, the cell can revert to a glycolytic programing or even in some cases utilize mitochondrial fatty acid oxidation [68]. This likely renders downstream effectors of metabolism including epigenetic changes, which will be discussed later, context-dependent and variable-based on the current conditions of the specific TME.
Metabolism in Pancreatic Ductal Adenocarcinoma
Metabolic rewiring in cancer cells is largely influenced by oncogenic pathways which promote pro-growth programs such as glycolysis and activation of glycolytic enzymes [69, 70]. In PDA, this is nearly universally driven by activating mutations in the KRAS oncogene, which results in constitutive downstream signaling [71]. In a seminal paper by Ying et al., the authors used an inducible and reversible transgenic mutant KRAS mouse model to demonstrate that KRAS activation drove the initiation and maintance of PDA by its regulation of anabolic glucose metabolism [72]. In a follow-up study, Viale et al. demonstrated that turning off KRAS signaling led to initial tumor regression, followed by relapse from a small surviving cancer cell population. Metabolic and transcriptional characterization of these cells demonstrated that those cells had stem-like features and relied more heavily on OXPHOS rather than glycolysis for survival, suggesting a potentially important difference between CSC and non-CSC metabolism in PDA [72, 73].
In fact, efforts to target CSCs based on mitochondrial metabolism have been met with some success. A study by Sancho et al. observed that CSCs were highly dependent on mitochondrial OXPHOS and underwent rapid apoptotic death when treated with the mitochondrial inhibiting drug, metformin [74]. Interestingly, non-CSC cells which were predominately glycolytic exhibited cell cycle arrest, but little cell death. It was noted in this model of CSC enrichment that there was a relative lack of plasticity in CSC metabolism, demonstrating the potential vulnerability of CSC metabolism.
Along with differences in metabolic programing, CSCs rely on different metabolic substrates when compared to their differentiated counterparts for maintenance of cellular function. In culture, PDA is exquisitely reliant on the amino acid glutamine, which is necessary for tumor growth and persistence [75]. Unlike traditional cellular metabolism in which glutamine is used primarily as an anaplerotic substrate for the tricarboxylic acid cycle (TCA), glutamine carbon from the TCA cycle is shuttled from the mitochondria through the cytosolic malic enzyme pathways to maintain bioenergetics and cellular redox state. Perturbation of the metabolic enzymes involved led to suppression of PDA growth both in vitro and in vivo [75]. Further studies specifically addressing PDA CSC confirmed their reliance on glutamine for maintenance of a proper cellular redox state and found that when glutamine was unavailable, cells became more sensitive to radiation. Glutamine deprivation also negatively impacted other processes including their capacity for self-renewal and expression of stem-related genes, suggesting another potential therapeutic opportunity [76].
Proteomic and metabolomic profiling have shown additional metabolic alternatives used by CSCs, demonstrating reliance on both fatty acid and mevalonate pathways for CSC survival in PDA and other cancers [77, 78]. Using neutralizing antibodies against the fatty acid receptor CD36 in orthotopic models of human oral carcinoma, Pascual et al. demonstrated reduction of the metastatic potential of CSC [77]. Similarly, the proteomic comparison of cell-line-derived pancreatic CSCs with their parental cells revealed an increased dependence on fatty acid synthesis and mevalonate pathways. Inhibition of these mechanisms resulted in greatly reduced proliferation of these CSC compared to non-CSC [79]. Somewhat paradoxically, the CSC population had higher expression of proteins involved in glycolysis, again suggesting the possibility that their metabolism is context dependent.
Altogether these findings demonstrate important differences between the metabolomic profile and nutrient utilization of PDA CSC and PDA non-CSCs. Importantly, there also appears to be differences in metabolic weakness observed among difference CSCs suggesting a metabolic heterogeneity which may mirror that seen in PDA cell lines [80]. Furthermore, these studies shed some light on the ability of CSCs to survive in hostile environments. Improved understanding of their metabolomic plasticity is needed to further investigate therapeutic targets and explore impacts on downstream effectors including alterations in epigenetics.
Epigenetics and Pancreatic Cancer Stem Cells
Epigenetic changes within cells do not involve genetic sequence alterations but instead entail DNA and chromatin structural/chemical changes promoting or repressing DNA transcriptional accessibility. These changes occur as a result of both intrinsic and extrinsic signals which ultimately affect the overall phenotypic state of the cell and represent an important way in which cells interact with environmental factors [19].
PDA cells exhibit markedly altered epigenetic profiles and often have mutations within chromatin regulatory proteins [81]. Epigenetic context also controls the process of epithelial-mesenchymal transition (EMT), which is important in pancreatic cancer metastasis formation [82, 83]. Recent comparison of pancreatic metastases to the liver, lung, and peritoneum with primary tumors has revealed fundamental anatomic site-specific epigenetic signatures within the tumor cells [84]. Partly based on these ideas, many groups have begun to investigate whether inhibition of epigenetic regulatory processes could contribute to the development of new pancreatic cancer therapeutics [85, 86].
PDA CSC are thought to be related to the process of EMT through master transcription factors including Zeb1 acting within the proper epigenetic context [82, 83, 87]. Multiple groups have targeted different aspects of epigenetic and chromatin regulation, including DNA methylation, histone methylation, and acetylation and demonstrated concurrent inhibition of CSC function or CSC elimination [88–90].
Aberrant methylation of functionally relevant genes is a hallmark of pancreatic cancer [91]. This process is mediated by DNA methyltransferases (DNMT) which is frequently found upregulated in PDA [92, 93]. PDA CSC selected by flow cytometry label retention, nonadherent sphere growth conditions, and by CD133 expression have higher overall DNA methylation levels than the remaining cancer cells [94]. This correlates with higher expression levels of the DNA methyltransferase, DNMT1. When DNMT1 was pharmacologically inhibited or genetically deleted using a CRISPR-Cas9 approach, the CSCs demonstrated increased commitment and progression through the cell cycle along with epithelial-like differentiation [94]. At least part of this phenotypic switch is due to the hypomethylation of the miR-17–92 miRNA cluster after DNMT1 inhibition. In a separate study, Kwon et al. combined 5-aza-2-deoxycytidine (5-aza-dC), a DNA methylation inhibitor, with ionizing radiation in vitro and in vivo [95]. They observed dose-dependent reduction in the population of CSCs in the MiaPaCa-2 and Panc1 cells, reduced self-renewal markers, including Oct4, Nanog, and Sox2. These observations correlated with inhibition of their migration and tumorigenic properties [95]. Efforts to disrupt DNA methylation should form a part of our therapeutic approach to disrupt the pancreatic CSC compartment.
Gene expression is partly regulated by the combination of covalent histone modifications, including histone acetylation and methylation, that mark inactive heterochromatin or active euchromatin. Screening of compounds that inhibit ZEB1-mediated EMT identified the histone deacetylase (HDAC) inhibitor, mocetinostat, as a compound that synergizes with gemcitabine to inhibit in vivo pancreatic xenograft growth by disrupting EMT and the CSC phenotype [96]. Similarly, inhibition of HDACs with trichostatin A and vorinostat increased Panc-1 and MiaPaCa-2 cell death, promoted epithelial differentiation, disrupted tumorsphere formation, and subcutaneous tumor xenograft growth [97]. In a separate set of experiments, analysis of gemcitabine-resistant Panc1 cells showed dysregulation of multiple histone-modifying enzymes [98]. Among these, the histone methyltransferase (HMT) G9a played a key role in maintenance of the CSC subpopulation and chemotherapy resistance partly through production of the cytokine IL-8. Higher levels of G9a expression also correlated with poor survival in a cohort of pancreatic cancer patients [98]. Disruption of histone 3 lysine 27 (H3K27) methylation by enhancer of zeste homolog 2 (EZH2), a component of the Polycomb Repressor Complex 2 (PRC2), also disrupts pancreatic CSC tumorsphere growth and sensitizes the cells to gemcitabine by increasing the expression of cell surface nucleoside transporters [99, 100]. These preliminary studies in established pancreatic cancer cell lines suggest that interference with histone modifications can successfully inhibit the CSC state and potentially synergize with existing chemotherapy to stop pancreatic tumor growth. It will be important to replicate these findings in more high-fidelity models of pancreatic cancer including primary patient-derived xenografts and tumor organoids.
Metabolic and Epigenetic interactions in CSC
While the primary function of cellular metabolism is energy production, the sequalae of nutrient utilization and production of metabolites produce downstream effects that can additionally contribute to tumorigenesis by influencing gene expression and cell signaling [101]. This has been suggested as mechanism by which CSC maintain relative plasticity by up or down regulating genes under epigenetic control based on the environment in which they exist [19]. There are several substrates and cofactors for epigenetic enzymes that vary as a direct result of changes in OXPHOS or glycolysis which enhance or repress the enzymatic function [101]. Reciprocally, expression of specific metabolic genes could be the consequence of epigenetic dysregulation [102–105]. In PDA, several mechanisms in CSC and non-CSC appear to be involved in the crosstalk between the epigenetic and metabolic pathways ultimately contributing to cellular plasticity and enhanced tumorigenesis.
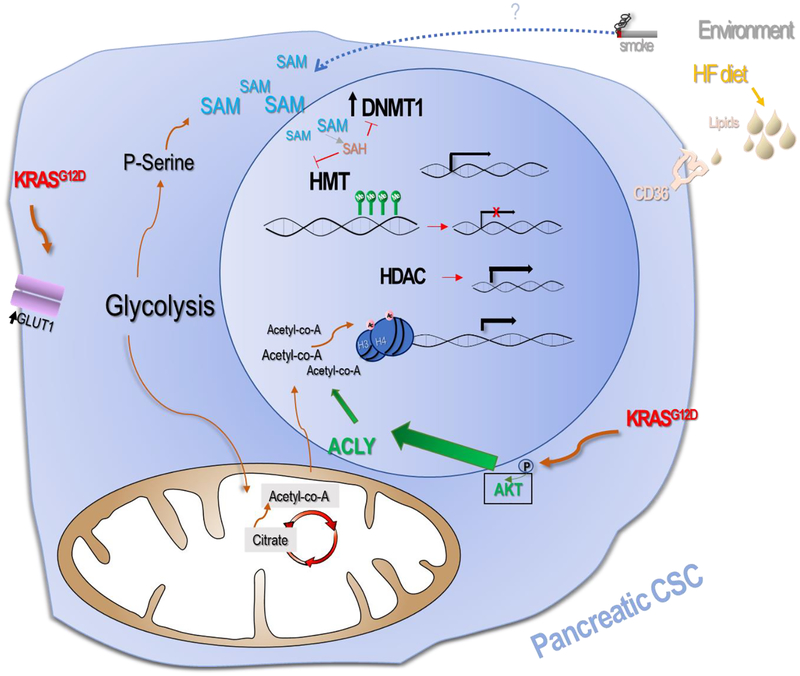
In murine PDA the metabolic reprograming which occurs in the presence of activated KRASG12D expression promotes increases in H3 and H4 acetylation, via acetyl-CoA nuclear accumulation after AKT-dependent signaling activation (Figure 3). Acetyl-CoA’s availability is influenced by oncogenic reprogramming of the cellular metabolism. There are two pools of Acetyl-CoA a mitochondrial and a nuclear-cytoplasmic pool. Several substrates contribute to the replenishment of this pool and histone acetylation correlates well with its availability. This histone acetylation is typically associated with active gene transcription and is dysregulated in tumors [106]. Glucose availability as well as acetate and glutamine represent Acetyl-CoA’s major contributors [107]. AKT phosphorylation and ACLY (ATP-citrate lyase) demonstrated to be linked to histone acetylation in tumors [106].
Figure 3. Metabolic-Epigenetic crosstalk in PDA.
epigenetics and metabolic pathways ultimately contribute to tumorigenesis. In murine PDA the metabolic reprograming due to activated KRASG12D expression promotes increase in H3 and H4 acetylation, via acetyl-CoA nuclear accumulation after AKT-dependent activation. Histone acetylation is usually associated with active gene transcription. CSCs expression have higher overall DNA methylation levels. SAM might contribute to DNA hypermethylation and increase in SAM availability enhances gene silencing through promotion of DNA methylation. DNMTs are overexpressed in several cancers and promote silencing of tumor suppressor genes in PDA. A DNA methylation inhibitor promoted reduced self-renewal marker expression. Additionally, several observations suggest links between environmental factors such as smoke sun and diet with an increased risks of cancer development [119].
Another example of metabolic and epigenetic crosstalk involves Histone methylation and Histone methyltransferases (HMT) which catalyzes transfer of methyl groups to lysine and arginine residues on histones H3 and H4 changing the chromatin structure and gene transcription [108]. The cofactor and methyl source for both HMT enzyme as well as DNMT1, a methyltransferase frequently upregulated in PDA CSC (see epigenetics section) is S-adenosylmethionine (SAM). Surplus of SAM substrates in the cell promotes DNA hypermethylation in CpG islands and can result in gene silencing, particularly of tumor suppressor genes such as SOC2 [109]. When SAM is consumed it produces SAH (S-adenosyl-homocysteine) which in turn inhibits the HMT and DNMT enzymes. In other words, histone and DNA methylation are controlled by the resulting ratio between SAM and SAH within the cell. A recent study found that alterations in metabolism pathway that produce high levels of glycolysis and serine biosynthesis in PDA consequently led to generation of large amounts of SAM which in turn promotes hypermethylation of specific retrotransposon elements associated with transcriptional silencing [110]. Using an inducible transgenic murine model of PDA, Kottakis et al. proved the existence of a connection between the loss of the tumor suppressor LKB1 serine-threonine kinase and KRAS signaling activation, events that combined led to the induction of the serine pathway [110]. Specifically, epithelial primary duct cells were isolated from mice bearing KRASG12D/+; LKB1−/− cells; double and single mutants were injected in SCID mice and demonstrated increased tumorigenicity and proliferation compared to the single mutation counterparts. Loss of LKB1 improved glucose consumption of these subpopulations. The entire one carbon-Serine-Glycine pathway showed a strong enrichment by proteomic and sequencing of RNA analysis in the double positive cells. Furthermore, double mutant KRASG12D/+; LKB1−/− cells were dependent on the Serine pathway and had a clear CpG enrichment both of which were shown to strongly correlate with DNA methylation. Retrotransposons were found responsible of the observed gene expression modulatory effects. Moreover, that dependence was resolved when DNMT1 was chemically blocked.
The generation of chromatin modification substrates is an example of the interconnecting link between the epigenetic and metabolic stages within the tumor cell. In parallel to upregulation of DNA methyltransferases, SAM is ultimately generated and the lack of LKB1 finally translates into inhibition of the serine biosynthesis pathway and the DNA methylation mechanism [110]. These observations revealed the clear connection that exists in pancreatic cancer tumorigenesis between LKB1 expression, glycolysis and DNA methylation events.
Histone Acetylation, mediated by histone acetylases (HATs) is a crucial post translation modification that regulates histone activity and increases gene expression. HDACs have the opposite effect mainly promoting gene silencing by chromatin condensation [111]. HATs were shown to play a role in the self-renewal ability of embryonic stem cells [112–114]. The effect of HAT over PDA CSCs has yet to be determined but the effects of HATs over other CSCs suggests it could affect PDA CSC [113, 115, 116]. Additionally work by Zhao et al. reflects the direct effect of acetylation over the metabolic enzyme LDH-A in human PDA [117]. LDH-A is known to be frequently over-expressed in pancreatic cancer. Several studies have highlighted a role for HDAC in relation to CSCs as we previously mentioned [96] another example is in liver cancer where there was a significant decrease in stem markers expression upon specific HDAC3 inhibition [118].
While no studies directly address the role of metabolism in the epigenetic processes that control PDA CSCs, epigenetic activating and inhibiting enzymes require cofactors, substrates and donors that are generated and influenced by metabolic enzymes which exemplifies the interplay between those two processes. Additionally, it is likely that several of these influences will have a different outcome depending on whether they are acting over non-CSCs or CSCs respectively.
Accumulating observations suggest links between environmental factors such as smoking, sun and diet with an increased risks of cancer development [119]. The interplay between metabolism and epigenetics may constitute a major factor in many of the observed environment-related malignancies by enhancement of CSC persistence. For example, as previously stated, increase in SAM availability enhances gene silencing through promotion of DNA methylation [110]. On the other hand, recent human studies have established a clear linear association between circulating plasma concentration of SAM with BMI (body mass index) and fat mass, known risk factors for PDA [120, 121]. This suggests a potential molecular mechanism tying metabolism and metabolic changes associated with obesity and epigenetic reprograming which could consequently lead to silencing of tumor suppressor genes thus promoting pancreatic cancer formation. In other words, this provides a clear example of environmental factors promoting cancer development through interplays between metabolism and epigenetics of cancer cells. Another observation linking cancer, metabolism and stem-cells is the finding that the dietary fat palmitic acid, an abundant component of the western high-fat diet, has been linked to increased metastatic potential of CSC in squamous cancer cells [77]. The CSCs expressing CD36 receptor rely on lipids from the diet to increase metastasic events.
Concluding Remarks
Recent findings have expanded our understanding of CSC biology and, as the model of CSC continues to evolve, new therapeutic strategies are emerging aimed at targeting these difficult to eradicate cells. Because of their relative plasticity, it is likely that CSC exist within a spectrum of metabolic and epigenetic states making their study and treatment complex and likely context dependent. Metabolic-epigenetic cross-talk represents a mechanism by which CSC can maintain their plasticity and respond to changes in the local environment including availability of nutrients and oxygen. Strategies to target cellular and metabolic processes of CSC appear attractive, but their nimble nature makes development of resistance a likely result. One strategy developed specifically to target PDA CSC is currently in phase I/II trial and relies on a different approach, turning the body’s immune system against this cellular subpopulation [122]. Efforts to add metabolic and epigenetic manipulation to this strategy had also been proposed.
It remains to be determined if the specific removal of CSC from a tumor, in addition to conventional chemotherapy against the bulk non-CSC compartment is the missing link to eradicating this difficult to treat disease. A lengthy history of failure to treat this population with traditional chemotherapy and radiation heightens the need to better understand its biology to define new avenues of attack.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors have read the manuscript and report no conflicts of interest.
REFERENCES
- 1.Hidalgo M, et al. , Addressing the challenges of pancreatic cancer: future directions for improving outcomes. Pancreatology, 2015. 15(1): p. 8–18. [DOI] [PubMed] [Google Scholar]
- 2.Mokdad AH, et al. , Trends and Patterns of Disparities in Cancer Mortality Among US Counties, 1980–2014. Jama, 2017. 317(4): p. 388–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel R, et al. , Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin, 2012. 62. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera MC, Hollingsworth RE, and Hurt EM, Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells, 2015. 7(1): p. 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abel EV and Simeone DM, Biology and clinical applications of pancreatic cancer stem cells. Gastroenterology, 2013. 144(6): p. 1241–8. [DOI] [PubMed] [Google Scholar]
- 6.Bao B, et al. , Overview of cancer stem cells (CSCs) and mechanisms of their regulation: implications for cancer therapy. Curr Protoc Pharmacol, 2013. Chapter 14: p. Unit 14.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gangemi R, et al. , Cancer stem cells: a new paradigm for understanding tumor growth and progression and drug resistance. Curr Med Chem, 2009. 16(14): p. 1688–703. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen L, et al. , Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol, 2010. 12(5): p. 468–76. [DOI] [PubMed] [Google Scholar]
- 9.Prasetyanti PR, et al. , Regulation of stem cell self-renewal and differentiation by Wnt and Notch are conserved throughout the adenoma-carcinoma sequence in the colon. Mol Cancer, 2013. 12(1): p. 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heddleston JM, et al. , Glioma stem cell maintenance: the role of the microenvironment. Curr Pharm Des, 2011. 17(23): p. 2386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takao S, Ding Q, and Matsubara S, Pancreatic cancer stem cells: regulatory networks in the tumor microenvironment and targeted therapy. J Hepatobiliary Pancreat Sci, 2012. 19(6): p. 614–20. [DOI] [PubMed] [Google Scholar]
- 12.Fessler E, et al. , Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett, 2013. 341(1): p. 97–104. [DOI] [PubMed] [Google Scholar]
- 13.Chen WJ, et al. , Cancer-associated fibroblasts regulate the plasticity of lung cancer stemness via paracrine signalling. Nat Commun, 2014. 5: p. 3472. [DOI] [PubMed] [Google Scholar]
- 14.Plaks V, Kong N, and Werb Z, The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell, 2015. 16(3): p. 225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albini A, et al. , Cancer stem cells and the tumor microenvironment: interplay in tumor heterogeneity. Connect Tissue Res, 2015. 56(5): p. 414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reya T, et al. , Stem cells, cancer, and cancer stem cells. Nature, 2001. 414(6859): p. 105–11. [DOI] [PubMed] [Google Scholar]
- 17.Dick JE, Stem cell concepts renew cancer research. Blood, 2008. 112(13): p. 4793–807. [DOI] [PubMed] [Google Scholar]
- 18.Wong TL, Che N, and Ma S, Reprogramming of central carbon metabolism in cancer stem cells. Biochim Biophys Acta, 2017. 1863(7): p. 1728–1738. [DOI] [PubMed] [Google Scholar]
- 19.Wainwright EN and Scaffidi P, Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer, 2017. 3(5): p. 372–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toh TB, Lim JJ, and Chow EK-H, Epigenetics in cancer stem cells. Molecular Cancer, 2017. 16(1): p. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, et al. , Cancer Stem Cells and Their Mechanism of Chemo-Radiation Resistance. International Journal of Stem Cells, 2009. 2(2): p. 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cioffi M, et al. , The miR-17–92 cluster counteracts quiescence and chemoresistance in a distinct subpopulation of pancreatic cancer stem cells. Gut, 2015. 64(12): p. 1936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clevers H, The cancer stem cell: premises, promises and challenges. Nat Med, 2011. 17(3): p. 313–9. [DOI] [PubMed] [Google Scholar]
- 24.Nowell PC, The clonal evolution of tumor cell populations. Science, 1976. 194(4260): p. 23–8. [DOI] [PubMed] [Google Scholar]
- 25.Driessens G, et al. , Defining the mode of tumour growth by clonal analysis. Nature, 2012. 488(7412): p. 527–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Facompre N, et al. , Stem-like cells and therapy resistance in squamous cell carcinomas. Adv Pharmacol, 2012. 65: p. 235–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Silva S, Frias-Aldeguer J, and Heeschen C, Stem cells & pancreatic cancer. Pancreatology, 2013. 13(2): p. 110–3. [DOI] [PubMed] [Google Scholar]
- 28.Li C, et al. , Identification of pancreatic cancer stem cells. Cancer Res, 2007. 67(3): p. 1030–7. [DOI] [PubMed] [Google Scholar]
- 29.Eramo A, Haas TL, and De Maria R, Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene, 2010. 29(33): p. 4625–35. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, et al. , Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res, 2008. 68(11): p. 4311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins AT, et al. , Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res, 2005. 65(23): p. 10946–51. [DOI] [PubMed] [Google Scholar]
- 32.Venkatesha VA, et al. , Sensitization of pancreatic cancer stem cells to gemcitabine by Chk1 inhibition. Neoplasia, 2012. 14(6): p. 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Hajj M, et al. , Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A, 2003. 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorico A and Rappa G, Phenotypic heterogeneity of breast cancer stem cells. J Oncol, 2011. 2011: p. 135039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricci-Vitiani L, et al. , Identification and expansion of human colon-cancer-initiating cells. Nature, 2007. 445(7123): p. 111–5. [DOI] [PubMed] [Google Scholar]
- 36.Miki J, et al. , Identification of Putative Stem Cell Markers, CD133 and CXCR4, in hTERT–Immortalized Primary Nonmalignant and Malignant Tumor-Derived Human Prostate Epithelial Cell Lines and in Prostate Cancer Specimens. Cancer Research, 2007. 67(7): p. 3153–3161. [DOI] [PubMed] [Google Scholar]
- 37.Hermann PC, Mueller MT, and Heeschen C, Pancreatic cancer stem cells--insights and perspectives. Expert Opin Biol Ther, 2009. 9(10): p. 1271–8. [DOI] [PubMed] [Google Scholar]
- 38.Li C, et al. , c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology, 2011. 141(6): p. 2218–2227. e5. [DOI] [PubMed] [Google Scholar]
- 39.Hermann PC, et al. , Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell, 2007. 1(3): p. 313–23. [DOI] [PubMed] [Google Scholar]
- 40.Duong HQ, et al. , Aldehyde dehydrogenase 1A1 confers intrinsic and acquired resistance to gemcitabine in human pancreatic adenocarcinoma MIA PaCa-2 cells. Int J Oncol, 2012. 41(3): p. 855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bao B, et al. , Pancreatic cancer stem-like cells display aggressive behavior mediated via activation of FoxQ1. J Biol Chem, 2014. 289(21): p. 14520–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Husain S and Thrower E, Molecular and cellular regulation of pancreatic acinar cell function. Current opinion in gastroenterology, 2009. 25(5): p. 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storz P, Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nature Reviews Gastroenterology &Amp; Hepatology, 2017. 14: p. 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan M-R, et al. , The histone methyltransferase G9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget, 2016. 7(38): p. 61136–61151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halbrook CJ, et al. , Mitogen-activated Protein Kinase Kinase Activity Maintains Acinar-to-Ductal Metaplasia and Is Required for Organ Regeneration in Pancreatitis. Cell Mol Gastroenterol Hepatol, 2017. 3(1): p. 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopp JL, et al. , Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell, 2012. 22(6): p. 737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puri S, Alexandra E Folias, and M. Hebrok, Plasticity and Dedifferentiation within the Pancreas: Development, Homeostasis, and Disease. Cell Stem Cell, 2015. 16(1): p. 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernandez-Munoz I, et al. , Pancreatic ductal adenocarcinoma: cellular origin, signaling pathways and stroma contribution. Pancreatology, 2008. 8(4–5): p. 462–9. [DOI] [PubMed] [Google Scholar]
- 49.Tarrado-Castellarnau M, de Atauri P, and Cascante M, Oncogenic regulation of tumor metabolic reprogramming. Oncotarget, 2016. 7(38): p. 62726–62753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto Y, et al. , Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell, 2003. 3(6): p. 565–76. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, et al. , Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. 2016. 2016: p. 1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung TH and Rando TA, Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol, 2013. 14(6): p. 329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deberardinis RJ, et al. , Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev, 2008. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeBerardinis RJ and Chandel NS, Fundamentals of cancer metabolism. Sci Adv, 2016. 2(5): p. e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warburg O, über den Stoffwechsel der Carcinomzelle. Klinische Wochenschrift, 1925. 4(12): p. 534–536. [Google Scholar]
- 56.Martinez-Outschoorn UE, et al. , Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol, 2017. 14(1): p. 11–31. [DOI] [PubMed] [Google Scholar]
- 57.Pecqueur C, Oliver L, Oizel K, Lalier L, Vallette FM. Targeting metabolism to induce cell death in cancer cells and cancer stem cells. Int J Cell Biol. 2013;2013:805975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlashi E, et al. , Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci U S A, 2011. 108(38): p. 16062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peiris-Pages M, et al. , Cancer stem cell metabolism. Breast Cancer Res, 2016. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasto A, et al. , Cancer stem cells from epithelial ovarian cancer patients privilege oxidative phosphorylation, and resist glucose deprivation. Oncotarget, 2014. 5(12): p. 4305–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson AS, et al. , Ovarian tumor-initiating cells display a flexible metabolism. Exp Cell Res, 2014. 328(1): p. 44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen YA, et al. , Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle, 2015. 14(1): p. 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen CL, et al. , NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab, 2016. 23(1): p. 206–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sancho P, et al. , MYC/PGC-1alpha balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab, 2015. 22. [DOI] [PubMed] [Google Scholar]
- 65.Oliva CR, et al. , Identification of Small Molecule Inhibitors of Human Cytochrome c Oxidase That Target Chemoresistant Glioma Cells. J Biol Chem, 2016. 291(46): p. 24188–24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye XQ, et al. , Mitochondrial and energy metabolism-related properties as novel indicators of lung cancer stem cells. Int J Cancer, 2011. 129. [DOI] [PubMed] [Google Scholar]
- 67.Song IS, Jeong YJ, and Han J, Mitochondrial metabolism in cancer stem cells: a therapeutic target for colon cancer. BMB Rep, 2015. 48(10): p. 539–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sancho P, Barneda D, and Heeschen C, Hallmarks of cancer stem cell metabolism. Br J Cancer, 2016. 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barthel A, et al. , Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J Biol Chem, 1999. 274(29): p. 20281–6. [DOI] [PubMed] [Google Scholar]
- 70.Yun J, et al. , Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science, 2009. 325(5947): p. 1555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhu H, et al. , Upregulation of autophagy by hypoxia-inducible factor-1alpha promotes EMT and metastatic ability of CD133+ pancreatic cancer stem-like cells during intermittent hypoxia. Oncol Rep, 2014. 32(3): p. 935–42. [DOI] [PubMed] [Google Scholar]
- 72.Ying H, et al. , Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell, 2012. 149(3): p. 656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viale A, et al. , Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature, 2014. 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sancho P, et al. , MYC/PGC-1alpha Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab, 2015. 22(4): p. 590–605. [DOI] [PubMed] [Google Scholar]
- 75.Son J, et al. , Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature, 2013. 496: p. 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li D, et al. , Inhibition of glutamine metabolism counteracts pancreatic cancer stem cell features and sensitizes cells to radiotherapy. Oncotarget, 2015. 6(31): p. 31151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pascual G, et al. , Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature, 2017. 541(7635): p. 41–45. [DOI] [PubMed] [Google Scholar]
- 78.Brandi J, et al. , Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. Journal of Proteomics, 2017. 150: p. 310–322. [DOI] [PubMed] [Google Scholar]
- 79.Brandi J, et al. , Proteomic analysis of pancreatic cancer stem cells: Functional role of fatty acid synthesis and mevalonate pathways. J Proteomics, 2017. 150: p. 310–322. [DOI] [PubMed] [Google Scholar]
- 80.Daemen A, et al. , Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 2015. 112(32): p. E4410–E4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell, 2017. 32(2): p. 185–203. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krebs AM, et al. , The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol, 2017. 19(5): p. 518–529. [DOI] [PubMed] [Google Scholar]
- 83.Tam WL and Weinberg RA, The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med, 2013. 19(11): p. 1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDonald OG, et al. , Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. 2017. 49(3): p. 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazur PK, et al. , Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. 2015. 21(10): p. 1163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Easwaran H, Tsai HC, and Baylin SB, Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell, 2014. 54(5): p. 716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Polireddy K, et al. , Targeting Epithelial-Mesenchymal Transition for Identification of Inhibitors for Pancreatic Cancer Cell Invasion and Tumor Spheres Formation. PLoS One, 2016. 11(10): p. e0164811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Momparler RL and Cote S, Targeting of cancer stem cells by inhibitors of DNA and histone methylation. Expert Opin Investig Drugs, 2015. 24(8): p. 1031–43. [DOI] [PubMed] [Google Scholar]
- 89.Glazer RI, et al. , 3-Deazaneplanocin: a new and potent inhibitor of S-adenosylhomocysteine hydrolase and its effects on human promyelocytic leukemia cell line HL-60. Biochem Biophys Res Commun, 1986. 135(2): p. 688–94. [DOI] [PubMed] [Google Scholar]
- 90.Momparler RL, et al. , Epigenetic therapy of acute myeloid leukemia using 5-aza-2’-deoxycytidine (decitabine) in combination with inhibitors of histone methylation and deacetylation. Clin Epigenetics, 2014. 6(1): p. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vincent A, et al. , Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res, 2011. 17(13): p. 4341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin R-K and Wang Y-C, Dysregulated transcriptional and post-translational control of DNA methyltransferases in cancer. Cell & Bioscience, 2014. 4: p. 46–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azizi M, et al. , MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther, 2014. 15(4): p. 419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zagorac S, et al. , DNMT1 Inhibition Reprograms Pancreatic Cancer Stem Cells via Upregulation of the miR-17–92 Cluster. Cancer Res, 2016. 76(15): p. 4546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kwon HM, et al. , Combinatorial effects of an epigenetic inhibitor and ionizing radiation contribute to targeted elimination of pancreatic cancer stem cell. Oncotarget, 2017. 8(51): p. 89005–89020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meidhof S, et al. , ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med, 2015. 7(6): p. 831–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cai MH, et al. , Depletion of HDAC1, 7 and 8 by Histone Deacetylase Inhibition Confers Elimination of Pancreatic Cancer Stem Cells in Combination with Gemcitabine. J Nanobiotechnology, 2018. 8(1): p. 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan MR, et al. , The histone methyltransferase G9a as a therapeutic target to override gemcitabine resistance in pancreatic cancer. Oncotarget, 2016. 7(38): p. 61136–61151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Avan A, et al. , Molecular mechanisms involved in the synergistic interaction of the EZH2 inhibitor 3-deazaneplanocin A with gemcitabine in pancreatic cancer cells. Mol Cancer Ther, 2012. 11(8): p. 1735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Vlerken LE, et al. , EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med, 2013. 2(1): p. 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaelin WG Jr. and McKnight SL, Influence of metabolism on epigenetics and disease. Cell, 2013. 153(1): p. 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang HB, et al. , Dysregulated metabolism contributes to oncogenesis. Nat Cell Biol, 2015. 35 Suppl: p. S129–s150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dong C, et al. , Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell, 2013. 23(3): p. 316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Menendez JA, The Metaboloepigenetic Dimension of Cancer Stem Cells: Evaluating the Market Potential for New Metabostemness-Targeting Oncology Drugs. Curr Pharm Des, 2015. 21(25): p. 3644–53. [DOI] [PubMed] [Google Scholar]
- 105.Myers SA, Panning B, and Burlingame AL, Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America, 2011. 108(23): p. 9490–9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee JV, et al. , Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab, 2014. 20(2): p. 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evertts AG, et al. , Quantitative dynamics of the link between cellular metabolism and histone acetylation. J Biol Chem, 2013. 288(17): p. 12142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trievel RC, Structure and function of histone methyltransferases. Crit Rev Eukaryot Gene Expr, 2004. 14(3): p. 147–69. [DOI] [PubMed] [Google Scholar]
- 109.Martinez-Chantar ML, et al. , Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology, 2008. 47(4): p. 1191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kottakis F, et al. , LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature, 2016. 539(7629): p. 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shahbazian MD and Grunstein M, Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem, 2007. 76: p. 75–100. [DOI] [PubMed] [Google Scholar]
- 112.Saraiva NZ, Oliveira CS, and Garcia JM, Histone acetylation and its role in embryonic stem cell differentiation. World Journal of Stem Cells, 2010. 2(6): p. 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dai X, et al. , Acetylation-dependent regulation of essential iPS-inducing factors: a regulatory crossroad for pluripotency and tumorigenesis. Cancer Medicine, 2014. 3(5): p. 1211–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, et al. , The Histone Acetyltransferase MOF is a Key Regulator of the Embryonic Stem Cell Core Transcriptional Network. Cell stem cell, 2012. 11(2): p. 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu N, et al. , Acetylation and deacetylation in cancer stem-like cells. Oncotarget, 2017. 8(51): p. 89315–89325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang XJ and Ullah M, MOZ and MORF, two large MYSTic HATs in normal and cancer stem cells. Oncogene, 2007. 26: p. 5408. [DOI] [PubMed] [Google Scholar]
- 117.Zhao D, et al. , Lysine-5 Acetylation Negatively Regulates Lactate Dehydrogenase A and Is Decreased in Pancreatic Cancer. Cancer cell, 2013. 23(4): p. 10.1016/j.ccr.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu C, et al. , Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Letters, 2013. 339(1): p. 60–69. [DOI] [PubMed] [Google Scholar]
- 119.Yadav D and Lowenfels AB, The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology, 2013. 144(6): p. 1252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elshorbagy AK, et al. , Serum S-adenosylmethionine, but not methionine, increases in response to overfeeding in humans. Nutrition & Diabetes, 2016. 6(1): p. e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Samocha-Bonet D, et al. , A family history of type 2 diabetes increases risk factors associated with overfeeding. Diabetologia, 2010. 53(8): p. 1700–8. [DOI] [PubMed] [Google Scholar]
- 122.Marcucci F, Rumio C, and Lefoulon F, Anti-Cancer Stem-like Cell Compounds in Clinical Development - An Overview and Critical Appraisal. Front Oncol, 2016. 6: p. 115. [DOI] [PMC free article] [PubMed] [Google Scholar]