Abstract
Facultative heterochromatin that changes during cellular differentiation coordinates regulated gene expression, but its assembly is poorly understood. Here, we describe facultative heterochromatin islands in fission yeast and show that their formation at meiotic genes requires factors that eliminate meiotic messenger RNAs (mRNAs) during vegetative growth. Blocking production of meiotic mRNA or loss of RNA elimination factors, including Mmi1 and Red1 proteins, abolishes heterochromatin islands. RNA elimination machinery is enriched at meiotic loci and interacts with Clr4/SUV39h, a methyltransferase involved in heterochromatin assembly. Heterochromatin islands disassemble in response to nutritional signals that induce sexual differentiation. This process involves the antisilencing factor Epe1, the loss of which causes dramatic increase in heterochromatic loci. Our analyses uncover unexpected regulatory roles for mRNA-processing factors that assemble dynamic heterochromatin to modulate gene expression.
Heterochromatin assembly is critical for various chromosomal processes (1–3). In fission yeast (Schizosaccharomyces pombe), noncoding RNAs (ncRNAs) and RNA interference (RNAi) factors implicated in processing ncRNAs facilitate loading of Clr4/Suv39h to assemble constitutive heterochromatin domains (4, 5). Clr4 methylates histone H3 lysine 9 (H3K9me) to create binding sites for chromo-domain proteins, including the Chp1 subunit of the Ago1-containing RNA-induced transcriptional gene silencing (RITS) complex, as well as HP1 family proteins Swi6 and Chp2, which associate with chromatin modifiers, including Snf2–histone deacetylase repressor complex (SHREC) involved in transcriptional silencing (3).
Apart from constitutive heterochromatin domains at centromeres, subtelomeres, and mating-type locus, H3K9me and HP1 proteins can also be detected within additional genomic regions at discrete genes in the S. pombe genome (6). However, the assembly of heterochromatin targeting genes and its modifications in response to signals that modulate gene expression have not been explored. To address this, we mapped H3K9me across the genome (7).
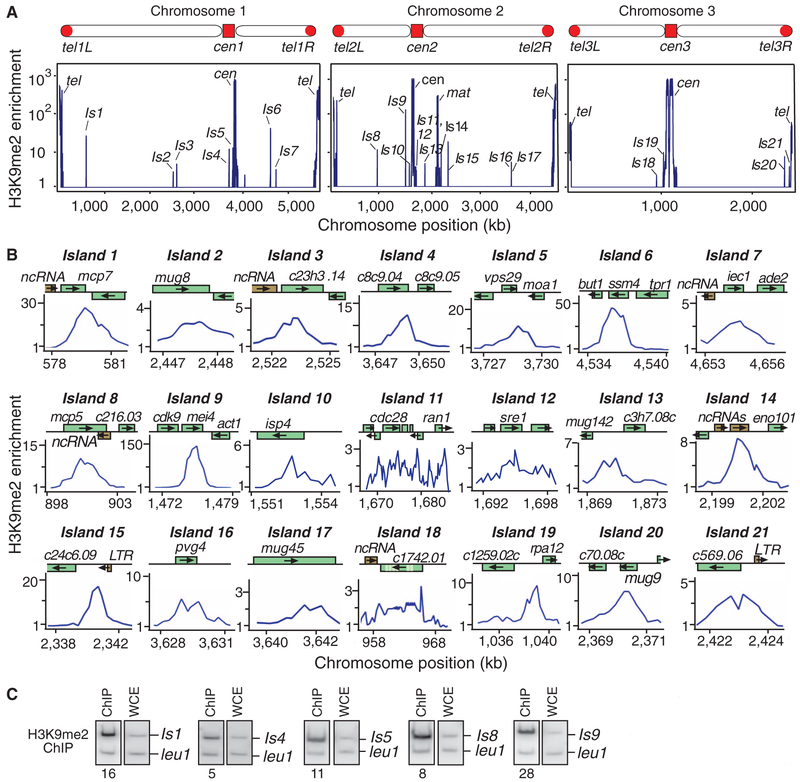
Besides previously reported heterochromatin loci, we reproducibly detected H3K9me at several additional sites (Fig. 1A). These heterochromatin islands encompass ~30 loci, which include genes located adjacent to ncRNAs and a few long terminal repeats (Fig. 1B). Overlapping transcription at convergent genes is believed to target H3K9me via RNAi (8), whereas heterochromatin islands correspond to both convergent and non-convergent loci (Fig. 1, B and C). A distinctive feature of heterochromatin islands is their preferential association with meiotic genes, which are silenced during vegetative growth (table S1). Most H3K9me enrichment corresponds to either open reading frames or 3′ ends of genes (Fig. 1B), consistent with transcription-coupled processes targeting H3K9me. Histone H3K4 methylation, a modification linked to RNA polymerase II transcription, could be detected at the 5′ ends of genes within heterochromatin islands (fig. S1).
Fig. 1.
Mapping H3K9me reveals heterochromatin islands. (A) Relative fold enrichment of dimethylated H3K9 (H3K9me2), as determined by ChIP-chip, is plotted. Besides centromere (cen), telomere (tel), and mating-type (mat) locus, H3K9me2 peaks distribute across the genome (islands 1 to 21). (B) H3K9me2 distribution at individual loci. Chromosome positions in (A) and (B) correspond to Sanger Center Pombe database 2004 assembly. LTR, long terminal repeat. (C) ChIP confirms H3K9me2 enrichment at selected loci. DNA isolated from immunoprecipitated chromatin (ChIP) and whole-cell crude extract (WCE) was used to perform multiplex polymerase chain reaction. Relative fold enrichment values are shown.
Factors that bind H3K9me and their associated effectors localize to heterochromatin islands: Chromatin immunoprecipitation (ChIP) detected the Clr4 complex (ClrC) subunit Raf2, Swi6/HP1, as well as posttranscriptional and transcriptional silencing activities, such as RNAi effector complex RITS (Chp1 and Ago1) and SHREC (Clr3 and Mit1), respectively, at heterochromatin islands (fig. S2). Additionally, Dcr1 localizes to three genes corresponding to islands (9). We also found the cohesin-loading factor Mis4, which interacts with Swi6/HP1 (10), and the cohesin sub unit Rad21 enriched at meiotic heterochromatin islands (fig. S2).
Our analysis identified the antisilencing factor Epe1 (11, 12) at heterochromatin islands (fig. S2). The loss of Epe1 caused the spread of H3K9me at most heterochromatin islands and at subtelomeric regions (fig. S3, A and B). Moreover, epe1Δ cells showed several H3K9me peaks that were not detected in wild-type cells (fig. S3C). More than 30 additional peaks mapped to ~100 convergent and nonconvergent loci. As in the case of the wild type, a major fraction of heterochromatin islands in epe1Δ mapped to meiotic genes. Thus, the S. pombe genome appears to harbor numerous heterochromatin nucleation sites, but heterochromatin assembly at many of these loci is suppressed by factors such as Epe1.
Given that heterochromatin islands map to transcribed regions, we wondered if RNAi targets heterochromatin to these loci. The loss of Dicer (Dcr1) or Argonaute (Ago1) caused only partial or no reduction in H3K9me at heterochromatin islands except island 5, which showed considerable reduction of H3K9me (fig. S4, A and B). Moreover, de novo targeting of H3K9me to ssm4 and mei4 occurred even in the absence of Ago1, albeit at levels lower than those of the wild type (fig. S4C), suggesting that additional RNAi-independent mechanism(s) target heterochromatin to meiotic loci. We also investigated the effects of histone deacetylases (HDACs), including Sir2 and SHREC (3), implicated in heterochromatin formation. Deletion of sir2 encoding a nicotinamide adenine dinucleotide–dependent HDAC (3) caused defective H3K9me at the majority of islands (fig. S5A), but SHREC subunits were dispensable (fig. S5B).
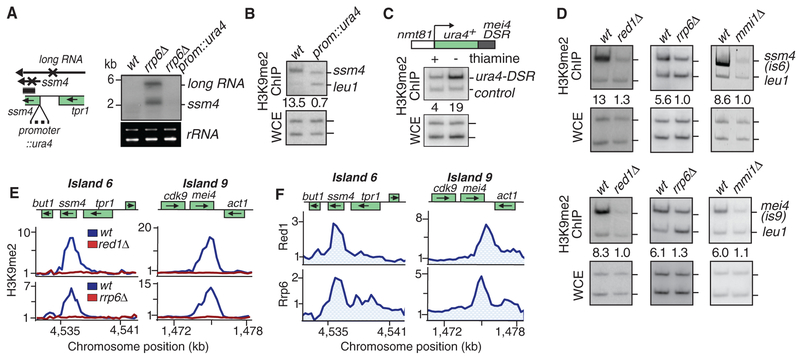
We next sought to investigate the mechanism of heterochromatin assembly at meiotic loci. Meiotic genes such as ssm4 are transcribed during vegetative growth, but their transcripts are processed by the exosome (13–16). The loss of Rrp6, an exosome-associated 3′-to-5′ exonuclease (17), led to the accumulation of ssm4 mRNA and a long RNA, which initiated from a promoter upstream of tpr1 (Fig. 2A and fig. S6). When production of ssm4 mRNA and the long RNA was disrupted by insertion of ura4 transcription terminator at the ssm4 promoter (Fig. 2A), the resulting strain showed the loss of H3K9me at the ssm4 (Fig. 2B). We also generated a strain in which ura4 terminator specifically blocks long RNA, without affecting ssm4 mRNA (fig. S7). ssm4 mRNA was sufficient to nucleate heterochromatin, as disruption of the long RNA did not abolish H3K9me (fig. S7). Based on these data, transcription is required for H3K9me at the ssm4 meiotic gene.
Fig. 2.
RNA elimination machinery affects heterochromatin assembly at meiotic genes. (A) Insertion of a mini-ura4 at the ssm4 promoter (prom::ura4) blocks the production of ssm4 mRNA and a long RNA, as determined by Northern blot analysis. rrp6Δis used to facilitate RNA detection. Ribosomal RNA (rRNA) serves as a loading control. wt, wild type. (B) Blocking ssm4 transcription abolishes H3K9me at this locus. H3K9me2 levels were assayed by ChIP. (C) mei4 DSR induces H3K9me at an ectopic site. DSR fused to ura4+ was expressed under the control of thiamine-repressible nmt1 promoter. Cells grown in the presence or absence of thiamine were used to detect H3K9me by ChIP. Endogenous ura4 serves as a control. (D and E) mmi1Δ, red1Δ, or rrp6Δ affect H3K9me at ssm4 and mei4. Results of ChIP (D) or ChIP-chip (E) are presented. (F) Red1-myc and Rrp6-myc distribution at ssm4 and mei4, as determined by ChIP-chip, are plotted.
The elimination of meiotic mRNAs in vegetative cells involves the YTH domain-containing protein Mmi1 (13). Mmi1 binds mRNAs containing determinant of selective removal (DSR) sequences and mediates their degradation by the exosome (13, 16). Meiotic mRNA suppression also requires the Red1 protein, which interacts with Mmi1, the exosome, and pre-mRNA 3′-processing factors (15). We wondered if the RNA elimination machinery targets H3K9me. Insertion of the mei4 DSR at the 3′ untranslated region of ura4 resulted in H3K9me at this site, especially when ura4-DSR was expressed (Fig. 2C), and ssm4 lacking its DSR failed to nucleate H3K9me (fig. S8). More importantly, the loss of Mmi1, Red1, or Rrp6, which caused accumulation of DSR-containing transcripts (fig. S9A) (13, 15), abolished H3K9me at both ssm4 and mei4, as determined by ChIP and ChIP-chip (Fig. 2, D and E). At other meiotic loci, we also observed defects in H3K9me in these mutants (Fig. 3A and fig. S10A), but we saw no change at heterochromatin islands targeting nonmeiotic loci, such as islands 14 and 15 (Fig. 3B). Based on these results, RNA elimination factors are required for the assembly of heterochromatin at meiotic genes.
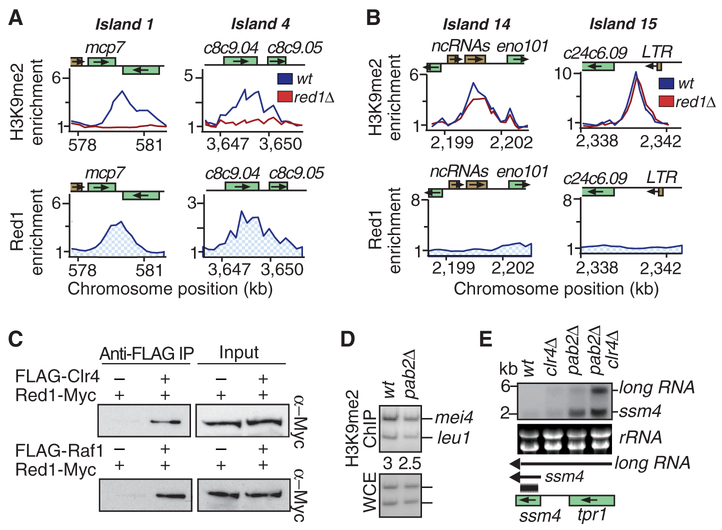
Fig. 3.
H3K9me at meiotic loci requires Clr4-interacting Red1 protein. (A and B) Relative enrichments of H3K9me2 and Red1-myc are plotted. (A) Red1 localized to islands 1 and 4 and is required for H3K9me2 at these loci. (B) red1Δ has no effect on H3K9me2 at islands 14 and 15, which lack detectable levels of Red1. (C) Clr4 and Raf1 interact with RNA processing factor Red1. Immuno-affinity purification of FLAG-Clr4 or FLAG-Raf1 was followed by Western blotting with myc antibody to detect Red1-myc. IP, immunoprecipitation. (D) pab2Δ causes only minor change in H3K9me at mei4, as determined by ChIP. (E) Clr4 and Pab2 act in parallel pathways to silence meiotic genes and to suppress long RNA. RNA from indicated strains was used to perform Northern blotting with an ssm4 probe (black bar).
Consistent with RNA elimination factors directly promoting H3K9me, we found Red1 and Rrp6 enriched at ssm4 and mei4 (Fig. 2F and fig. S9B). Red1 was also detected at heterochromatin islands at other meiotic genes (Fig. 3A). Localization of Red1 correlated with its requirement for H3K9me at individual loci (Fig. 3A and fig. S10A). Red1 could not be detected at islands that showed no reduction in H3K9me in red1Δ cells (Fig. 3B and fig. S10A). rec8 and spo5 genes, which encode DSR-containing transcripts degraded by a Mmi1-based mechanism (13, 16), lack detectable levels of H3K9me (fig. S10B). The lack of H3K9me enrichment correlated with the absence of Red1 binding at these loci (fig. S10B). The correlation between localization of RNA elimination factors and H3K9me prompted us to investigate interactions between these factors. Immunoprecipitation analyses detected RNA elimination factor Red1 interacting with Clr4 and another ClrC subunit, Raf1, and that these interactions were not sensitive to deoxyribonuclease I or ribonuclease A treatment (Fig. 3C). These data demonstrate that Red1 is a component of a protein network involved in the elimination of meiotic RNAs that recruits Clr4 to assemble heterochromatin islands.
Heterochromatin and its associated factors such as RNAi components might promote meiotic gene silencing. Loss of Ago1 alone did not cause detectable changes in ssm4 and mei4 silencing (fig. S11). However, the effects of RNAi machinery might be masked by the activity of the exosome. Because the exosome is required for both mRNA processing and H3K9me, it was not possible to study the effects of heterochromatin factors in the rrp6Δ background. However, the degradation of meiotic mRNAs by the exosome requires the poly(A) binding protein Pab2 (16, 18), which does not interact with Red1 (15). pab2Δ impaired the degradation of transcripts but maintained considerable levels of H3K9me at meiotic loci (Fig. 3, D and E). When pab2Δ was combined with clr4Δ, the resultant double mutant showed cumulative accumulation of ssm4 mRNA and long RNA (Fig. 3E). These results suggest a role for Clr4 in silencing meiotic genes during vegetative growth. Clr4 might directly promote RNA processing (19) and/or facilitate loading of silencing effectors, such as chromatin modifiers and the RNAi machinery (fig. S2), via H3K9me-bound chromodomain proteins. Indeed, clr4Δ cells are defective in the localization of Swi6 and the RITS subunit Chp1 at ssm4 and mei4 loci (6).
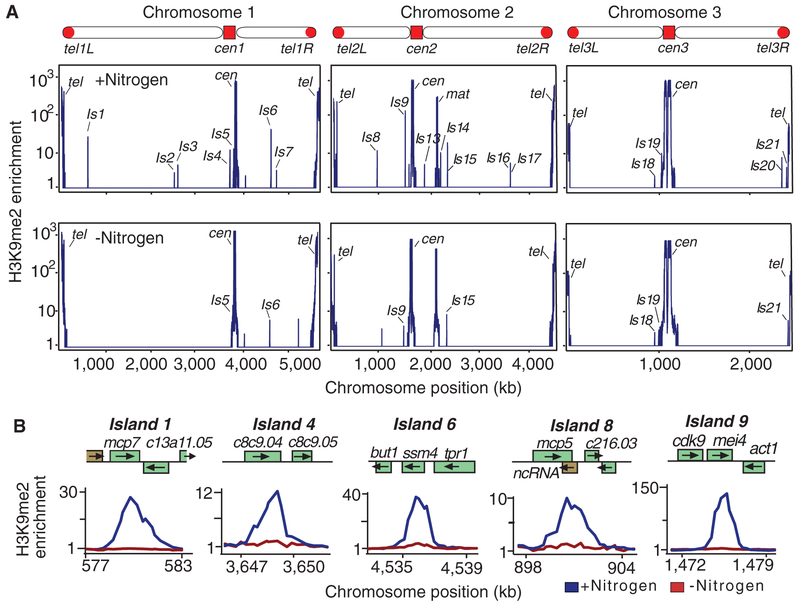
Meiotic gene expression is induced in response to sexual differentiation signals (20). Nitrogen starvation arrests cells in G1 phase of the cell cycle and triggers sexual differentiation. However, growth arrest can be reversed by the resupply of nitrogen. Nitrogen deprivation caused severe reduction in H3K9me at meiotic genes but had no effect on heterochromatin at centromeres, telomeres, and the mat locus (Fig. 4, A and B, and fig. S12A). The observed effect was specific to nitrogen starvation and was not observed when cells were arrested in G1 phase (fig. S12B). Appreciable reduction in H3K9me could be detected after 4 hours of nitrogen starvation (fig. S13A). The rapid loss of H3K9me involves Epe1, because disassembly of heterochromatin islands is delayed in epe1Δ mutant (fig. S13A). Resupplying nitrogen to starved cells restored heterochromatin to meiotic loci within 24 hours after the addition of nitrogen source to growth medium (fig. S13B). Thus, heterochromatin islands at meiotic loci are developmentally regulated.
Fig. 4.
Nutritional signals that induce meiosis trigger the loss of heterochromatin islands. (A and B) Nitrogen starvation causes reduction in H3K9me2 at meiotic heterochromatin islands. Levels of H3K9me2 measured by ChIP-chip performed using cells cultured in the presence or absence of nitrogen are plotted using a logarithmic (A) and linear (B) scale.
Our analysis expands the repertoire of loci targeted by heterochromatin and uncovers a mechanism for heterochromatin assembly. Heterochromatin formation at meiotic loci requires transcription but can occur independent of RNAi or a specific gene orientation. We suggest that Mmi1 binding to DSR-containing RNAs engages Red1 and the exosome to nucleate heterochromatin by targeting Clr4/Suv39h (fig. S14). Because Red1-exosome degrades many RNAs and certain Mmi1 targets (e.g., rec8 and spo5) lack H3K9me, it is likely that additional factors, including specific features of RNAs, are important for loading heterochromatin. In this regard, Red1 and Mmi1-cooperate with pre-mRNA 3′-processing factors to promote hyperadenylation of target transcripts, which is critical for meiotic gene silencing (15, 16). Red1 and 3′-processing factors may be components of a mechanism that couples degradation of meiotic RNAs to heterochromatin assembly. Note that the Rik1 subunit of the Clr4 complex, which suppresses accumulation of aberrant RNAs (19), resembles cleavage and polyadenylation specificity factors involved in pre-mRNA 3′-processing (21). Similar to facultative heterochromatin in higher eukaryotes, heterochromatin islands are remodeled in response to cellular differentiation signals. These results suggest that signaling mechanisms exist that feed into heterochromatin pathways to facilitate coordinated changes in gene expression and reprogram the genome for other chromosomal events during meiotic induction. Similar pathways involving cotranscriptional RNA processing factors might promote RNA- and transcription-coupled epigenetic modification observed in other systems (22–25).
Supplementary Material
Acknowledgments:
We thank T. Sugiyama for constructions of red1Δ and tagged alleles during his time in the Grewal lab and M. Yamamoto and T. Sugiyama for their gift of strains. Microarray data are avaiable at the National Center for Biotechnology Information Gene Expression Omnibus repository under accession no. GSE33404. This work is supported by the Intramural Research Program of the NIH and National Cancer Institute.
Footnotes
Supporting Online Material
References and Notes
- 1.Henikoff S, Biochim. Biophys. Acta 1470, O1 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD, Science 293, 1074 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Grewal SI, Curr. Opin. Genet. Dev 20, 134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Mosch K, Fischle W, Grewal SI, Nat. Struct. Mol. Biol 15, 381 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Bayne EH et al. , Cell 140, 666 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cam HP et al. , Nat. Genet 37, 809 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Materials and methods are available as supporting material on Science Online.
- 8.Gullerova M, Proudfoot NJ, Cell 132, 983 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Woolcock KJ, Gaidatzis D, Punga T, Bühler M, Nat. Struct. Mol. Biol 18, 94 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Fischer T et al. , Proc. Natl. Acad. Sci. U.S.A 106, 8998 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isaac S et al. , Genetics 175, 1549 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zofall M, Grewal SI, Mol. Cell 22, 681 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Harigaya Y et al. , Nature 442, 45 (2006). [DOI] [PubMed] [Google Scholar]
- 14.McPheeters DS et al. , Nat. Struct. Mol. Biol 16, 255 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugiyama T, Sugioka-Sugiyama R, EMBO J. 30, 1027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M, EMBO J. 29, 2173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houseley J, LaCava J, Tollervey D, Nat. Rev. Mol. Cell Biol 7, 529 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Lemay JF et al. , Mol. Cell 37, 34 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Zhang K et al. , Science 331, 1624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata J, Lyne R, Burns G, Bähler J, Nat. Genet 32, 143 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Neuwald AF, Poleksic A, Nucleic Acids Res. 28, 3570 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S, Mol. Cell 29, 313 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Beisel C, Paro R, Nat. Rev. Genet 12, 123 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Tsai M-C et al. , Science 329, 689 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponting CP, Oliver PL, Reik W, Cell 136, 629 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.