Abstract
Gap junctions establish direct pathways for cell-to-cell communication, through the assembly of twelve subunits (connexins) that form intercellular channels connecting neighboring cells. Co-assembly of different connexin isoforms produces channels with unique properties, and enables communication across cell-types. To gain access into the structural underpinnings of connexin co-assembly, we used single particle CryoEM to determine the structure of native lens gap junction channels, composed of connexin-46 and connexin-50 (Cx46/50). We provide the first comparative analysis to connexin-26 (Cx26), which together with computational studies elucidates key energetic features governing gap junction perm-selectivity. Cx46/50 adopts an open-state conformation that is unique from the Cx26 crystal structure, yet appears to be stabilized by a conserved set of hydrophobic anchoring residues. ‘Hot spots’ of genetic mutations linked to hereditary cataract formation map to the core structural-functional elements identified in Cx46/50, rationalizing many of the disease-causing effects.
Cell-to-cell communication directed by the gap junctions is essential to neuronal function, cardiac coupling, and for coordinating intercellular signaling and metabolic activity in most tissues (e.g. heart, skin, liver and eye lens)1. As such, genetic mutation or aberrant regulation is linked to a variety of pathological conditions, including cardiac arrhythmia, stroke, blindness, deafness, skin disease and cancers2–4.
Intercellular channel formation is achieved through an assembly of twelve subunits, known as connexins5. Within the plasma membrane, six connexins are organized into a hemi-channel structure. Hemi-channels from neighboring cells dock together to form complete cell-to-cell channels, which cluster to form large gap junction plaques. A remarkably large channel pore provides passage to an array of chemical information; including ions, metabolites, hormones, and other small signaling molecules less than ~1 kDa in size (e.g., K+, cyclic-AMP (cAMP), inositol triphosphate (IP3) and glucose). In this way, interconnected cells may exchange electrical and chemical information across an entire tissue or organ.
Humans express 21 connexin isoforms in a cell-type specific fashion6. Most cells express multiple isoforms and certain connexins display an ability to co-assemble, either by docking two hemi-channels composed of different isoforms (heterotypic), or through mixed isoform assembly within the same hemi-channel (heteromeric). This complexity is thought to allow cells to fine-tune the conductance of chemical information, and support coupling across different cell types7. However, our understanding of the physical basis for connexin isoform compatibility, conductance, substrate selectivity and channel gating remains limited8,9, as high-resolution structural information obtained by crystallographic analysis has so far been restricted to just a single model system, connexin-26 (Cx26)10,11.
To gain further insight into the mechanistic effects of gap junction isoform diversity and heteromeric assembly, we applied single particle imaging methods by electron cryo-microscopy (CryoEM) to elucidate the structure of native connexin-46 and −50 (Cx46/50) channels, isolated from the eye lens, where connexin-mediated communication is required for growth, differentiation and maintenance of lens transparency required to support vision12. Comparative molecular dynamics (MD) simulations reveal key features of ion permeation and selectivity, and suggest Cx46/50 adopts a more stable open-state conformation compared to the previously described Cx26 crystal structure10.
Structural overview of Cx46/50
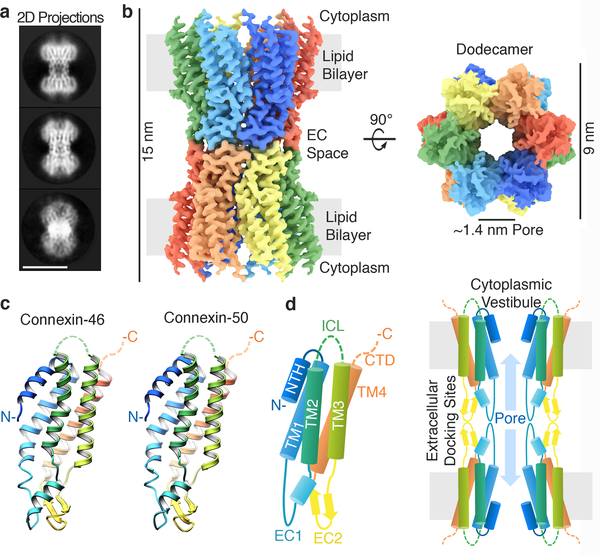
Cx46/50 form heteromeric/heterotypic intercellular channels in the mammalian lens13,14. We isolated native Cx46/50 intercellular channels from core lens tissue (sheep Cx44/49), and verified heteromeric co-assembly by biochemical analysis and chemical cross-linking mass-spectrometry (MS) (Extended Data Fig. 1). The structure of Cx46/50 intercellular channels was resolved by single particle CryoEM to near-atomic resolution (3.4 and 3.5 Å) (Fig. 1a–b, Extended Data Figs. 1–3). The resulting density maps revealed a 15 nm tall dodecameric (12-mer) channel with a girdled waist (~6–9 nm wide). When rotated 90°, a large unobstructed pore of ~1.4 nm diameter is visualized along the channel axis, consistent with the proposed open-state conformation (Fig. 1b).
Figure 1. Structure of connexin-46/50 intercellular channels.
(a) Representative projection averages of native Cx46/50 intercellular channels (selected from 165 classes). Scale bar = 10 nm. (b) 3D CryoEM reconstruction displaying the twelve subunits colored independently. Regions corresponding to the lipid bilayer, cytoplasm and extracellular (EC) space are indicated. (c) Atomic models of Cx46 (left) and Cx50 (right) monomers displayed in ribbon representation. (d) (left) Diagram of the connexin-fold and (right) cross-section of the assembled intercellular channel, with domains labeled: transmembrane domains 1–4 (TM1–4), extracellular domains 1–2 (EC1–2), and n-terminal helix domain (NTH). The intracellular loop (ICL) and c-terminal domains (CTD) were not visualized in the CryoEM density (dotted lines). EC1/2 form docking sites, establishing a continuous channel pore of ~1.4 nm diameter connecting the cytoplasm of neighboring cells.
Despite significant effort, we were unable to resolve a specific pattern of Cx46/50 heteromeric or heterotypic co-assembly using 3D-classification/refinement strategies (Methods and Extended Data Fig. 4,5). Nevertheless, high-resolution features corresponding to sidechain densities are observed throughout the reconstructions following 12-fold symmetry refinement (i.e., by averaging signal contributed by both Cx46 and Cx50). Thus, these two isoforms, which share ~80% core sequence identity (88% similarity), also share a highly similar 3D structure (Fig 1b-c, Extended Data Fig. 4–6), consistent with the ability of Cx46/50 to co-assemble in a variety of heteromeric/heterotypic states.
Atomic models for Cx46 and Cx50 were built into the averaged CryoEM density map, and various heteromeric/heterotypic channels were constructed for comparative analysis. While the structures display excellent validation statistics (Extended Data Fig. 2), local resolution assessment between the atomic models and experimental density identified features of both models that were less well defined by the density map, in particular at sites where the two isoforms differ in sequence (Extended Data Fig. 3–5 and Supplementary Table 2,3). We note, analysis of the models presented should be approached with caution due to intrinsic limitations of our heterogeneous dataset, which may go beyond local differences in primary sequence; for example, due to the possibility of one isoform being more well-ordered and potentially biasing interpretation.
The refined Cx46/50 structures comprise four alternating transmembrane α-helices (TM1–4), two extracellular domains (EC1/2) connecting TM1/2 and TM3/4 respectively, and an n-terminal helix (NTH) domain that folds into the channel vestibule and is connected to the pore-lining TM1 helix via a short linker (Fig. 1c,d). Density for Met1 is not observed in the CryoEM maps and was determined to be removed in both Cx46 and Cx50 by MS/MS, and the resulting n-terminal glycine (G2) is partially acetylated (Extended Data Fig. 1), as shown for the bovine isoforms15,16. The intracellular loop (ICL) connecting TM2/3 and cytoplasmic c-terminal domain (CTD) harboring the native cleavage sites of Cx46 and Cx50 are also not resolved16. The ICL and CTD were not observed in the crystallographic structures of Cx26 either10,11, presumably due to intrinsic disorder in these regulatory domains.
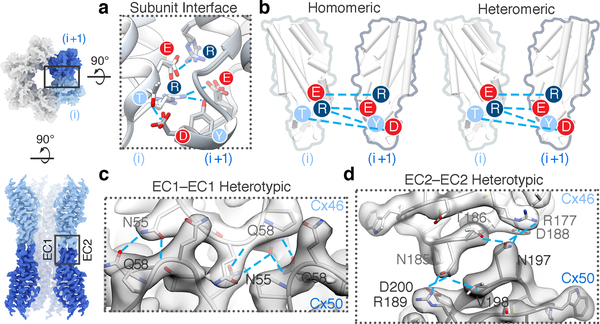
The close structural similarity of Cx46 and Cx50 results in highly similar interfacial interactions that include conserved regions of hydrophobic packing over the TM region and a strikingly similar hydrogen bond/ion-pair network between adjacent (heteromeric interface, Fig. 2a-b, Extended Data Fig. 6) and opposed subunits (heterotypic interface, Fig. 2c-d, Extended Data Fig. 6). Most of these stabilizing interactions are present in Cx2610,17, including the EC1 Q/N motif (Fig 2c) and EC2 paring involving the K/R-N-D motif (Fig 2d), a conserved element amongst Group I heterotypic compatible isoforms18. Although Cx46/50 and Cx26 are not classified as heteromeric compatible channels19, the conserved features at the heteromeric interface are congruent with the current understanding that heteromeric co-assembly of connexins is established during biogenesis in the ER/Golgi network19.
Figure 2. Heteromeric and heterotypic interactions between Cx46 and Cx50.
(a) Structural overlay and (b) illustration of the conserved electrostatic network between neighboring subunits (i and i+1) identified in homomeric Cx46 and Cx50 and heteromeric Cx46/50 models. Labels are colored according to amino acid charge characteristics (red – negative, dark blue – positive and light blue – polar). (c) Zoom-view of EC1-EC1 and (d) EC2-EC2 docking site interactions, with atomic models of Cx46 and Cx50 hemi-channels built into the CryoEM density in a heterotypic configuration. Conserved amino acids involved in hydrogen bond pairing (cyan lines) are labeled.
Overall, despite significant sequence differences, Cx46/50 (α-family connexins) display core structural features that are very similar to the β-family member, Cx2610 (pair-wise Cα root mean squared deviation (r.m.s.d.) = 2.18 Å and 2.14 Å versus Cx46 and Cx50, respectively). Thus, these different connexin family members share a conserved connexin-fold and gap junction channel architecture, as presented in Fig 1d, and are consistent with early low-resolution electron diffraction studies on connexin-43 (Cx43) obtained in a lipid bilayer20,21. Despite these general similarities; however, we uncovered significant differences between Cx46/50 and Cx26 localized to key functional sites that are expected to contribute to isoform-specific permeation and selectivity properties and provide insight into the interactions responsible for fully stabilizing the open-state conformation of these channels, detailed below.
Energetics of Ion Permeation/Selectivity
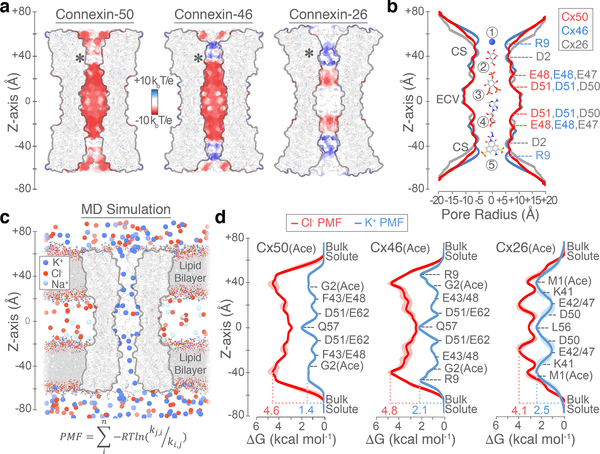
Comparisons between Cx50, Cx46 and Cx26 intercellular channels reveal unique electrostatic pore pathways, with shared regions of negative charge potential and steric constriction sites (CS) formed by the NTH domains that narrow the cytoplasmic vestibule (CV) to ~10–12 Å, at both ends of the channel (Fig. 3a,b). The pore diameters are within the range determined for other connexin channels22, and fitting with the general ability of gap junctions to permeate a variety of molecules less than ~1 kDa in size (e.g., Fig 3b, labeled 1–5). Remarkably; however, these channels can display a substantial level of isoform-specific selectivity for molecules beneath this size cut-off, even discriminating between small charged ions23.
Figure 3. Comparative pore profile and energetics of ion permeation.
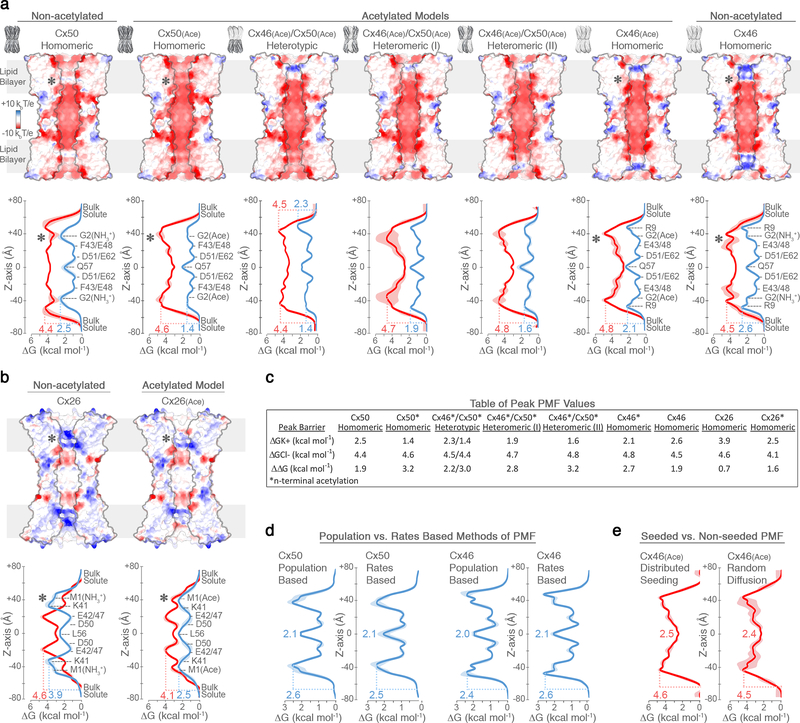
(a) Cut-away surface representation of Cx50 (left), Cx46 (center) and Cx26 (right) (PDB 2ZW310; residues 2–96 and 132–217), colored by coulombic potential (negative – red, neutral – white and positive – blue). Asterisk indicates the site of n-terminus. (b) Pore radius determined for experimental structures of Cx50 (red), Cx46 (blue) and Cx26 (grey, PDB 2ZW310). Locations of constriction sites (CS) and extracellular vestibule (ECV) are indicated. Structures of representative substrates are displayed to scale (1 – K+ ion, 2 – glucose, 3 – IP3, 4 – cAMP and 5 – Lucifer Yellow) (c) Snapshot of the Cx50 MD simulation, showing the membrane channel (white) embedded in two lipid bilayers and solvated in the presence of intracellular K+, extracellular Na+ and Cl− ions. Water molecules not shown. (d) Potential-of-mean-force (PMF) describing the free-energy landscape (ΔG, kcal mol−1) experienced by K+ ions (blue trace) and Cl− ions (red trace) permeating the channel pore. Symmetrized values are shown for acetylated models of Cx50(Ace) (left), Cx46(Ace) (center) and Cx26(Ace) (right), with non-symmetrized values in lighter shading. Amino acid positions are presented for correlation purposes, and do not represent deconvolution of the free-energy components.
To validate our structural models and gain insight into the mechanism of ion perm-selectivity, we conducted comparative all-atom molecular dynamics (MD) simulations and potential-of-mean-force (PMF) calculations to define the free-energy landscape of potassium (K+) and chloride (Cl−) permeation for Cx50, Cx46 and Cx26 (Fig. 3c-d, Extended Data Fig. 7,8). PMFs obtained for Cx26 should be interpreted cautiously due to significant dynamical behavior observed for the NTH domain during MD simulations (Extended Data Fig. 7), described in detail in the following section.
Cx50, Cx46 and Cx26 form high conductance ion channels, with preference for conductance of cations over anions. For MD simulation, the n-terminus of each of the models was acetylated (Cx50(Ace), Cx46(Ace) and Cx26(Ace)), as this form is expected to represent the predominant species in vivo24, and previous MD studies suggest this co-translational modification is required to obtain physiologically relevant charge selectivity of Cx2622. PMFs for K+, the major permeant ion, reveal peak energetic barriers within the CS regions, ranging from 1.4 kcal mol−1 for Cx50(Ace), to 2.1 kcal mol−1 in Cx46(Ace) and 2.5 kcal mol−1 in Cx26(Ace) (Fig. 3d). These relatively low barriers are similar to the peak energetic barrier determined for other high-conductance Na- and K-channels (~2–3 kcal mol−1)25–27, and are consistent with the range of experimental unitary conductance values of these channels, with Cx50 > Cx46 ≈ Cx26 (~220 pS for Cx5028 vs. ~140–135 pS for Cx4629 and Cx2630; in 130–140 mM CsCl).
The differences in K+ PMF correlate with isoform-specific differences in both steric and electrostatic environments. The CS of Cx50(Ace) displays the lowest barrier and is characterized by a nearly complete electronegative coulombic potential, due to neutralization of the n-terminus by acetylation (asterisk in Fig. 3a and Extended Data Fig. 8). The major K+ energy barrier of Cx46(Ace) correlates with the position of positively charged R9 residue (~50 Å z-axis) (Fig. 3d, Extended Data Fig. 8), which also constricts the CS of Cx46 to ~10 Å in our model (vs. ~12 Å for Cx50) (Fig. 3b). However, the CryoEM density map is not well defined at this site (Supplementary Table 2), likely due (at least in part) to the conformational flexibility of this residue, as dynamical behavior is observed during MD simulation. These dynamics of R9 effectively modulate the steric barrier of Cx46 (~10–12 Å). The CS K+ energy barrier of Cx26(Ace) correlates with the location of the basic residue K41 (~50 Å z-axis) (Fig. 3d), located on TM1 just below the NTH domain, as previously reported22,32.
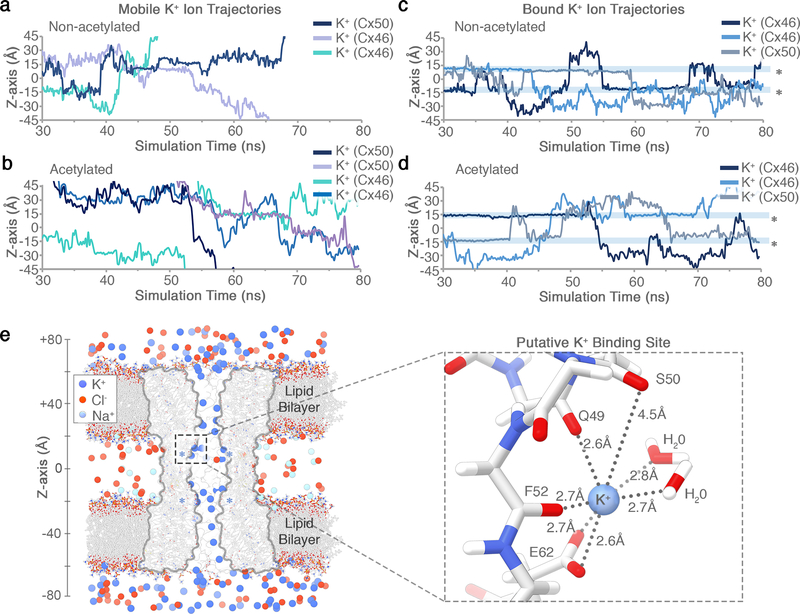
Free energy minima for K+ are localized within the extracellular vestibule (ECV; ~10 and ~30 Å, z-axis) of all three isoforms (Fig. 3d), supporting the role of EC1 in establishing charge selectivity and conductance31–33. In Cx46 and Cx50, several negatively charged residues (e.g., E48, D51 and E62) localize with regions of high K+ ion density (Fig. 3c-d, Extended Data Fig. 9). Notably, charge substitutions at D51 resulted in decreased unitary conductance in Cx46 hemi-channels34. The residues E48 and D51 are conserved in Cx26 (equivalent to E47 and D50) (Fig. 3d), and establish transient binding interactions with K+ ions during MD simulation. These sites have also been implicated in Ca2+ regulation in Cx26 by X-ray crystallography11, MD studies35,36, and by functional mutation studies of Cx4636. Therefore, competitive K+ binding at these sites may contribute to the mechanism of Ca2+ regulation/sensitivity. E62 (in Cx46/50) appears to form an additional cation binding site, through coordination by the carboxylate sidechain and nearby backbone carbonyl oxygens (Extended Data Fig. 9). E62 is not conserved in other human connexin isoforms (with the exception of Cx43), and therefore may constitute an isoform-specific regulatory site. Extracellular Ca2+ is involved in the mechanism of closing (or gating) connexin hemi-channels37, and competition by K+ binding at this putative site may contribute to the mechanism of potentiation of Cx50 and Cx46 hemi-channels by extracellular K+ ions38.
Cx50, Cx46 and Cx26 display an appreciable level of selectivity toward positively charged small ions, with permeability ratios of K+ to Cl− (PK+/Cl−) ranging between ~2.5 to ~1028,31,39–41. Hydrated K+ and Cl− ions (~7 Å diameter) would pass unobstructed through a 10–12 Å steric constriction site; yet, for all three isoforms the peak energy barriers to Cl− are considerably larger than for K+ (Fig. 3d, Extended Data Fig. 8). Peak Cl− barriers localize within the CS region of Cx46(Ace) and Cx50(Ace) (4.8 kcal mol−1, ~40 Å z-axis; and 4.6 kcal mol−1, ~38 Å z-axis, respectively), and slightly deeper into the channel pore for Cx26(Ace), near the CS-ECV border (4.1 kcal mol−1, ~20 Å z-axis). As a proxy for degree of PK+/Cl- selectivity, we assessed the difference in peak K+ and Cl− PMF barriers (ΔΔG = 3.2 kcal mol−1 for Cx50(Ace); 2.7 kcal mol−1 for Cx46(Ace); and 1.6 kcal mol−1 for Cx26(Ace)). These relatively small differences in free energy are in accordance with their moderate PK+/Cl- selectivity ratios, and on the order of those defined for bacterial sodium channels (~3.0–3.5 kcal mol−1)26,27, which also display modest ion selectivity (PNa+/K+ ~10–30). In contrast, voltage-gated K-channels display almost ideal selectivity for K+ over Na+ (PK+/Na+ ~1000), with energetic barrier differences to these ions reported to be ~6.6 kcal mol−1 for KcsA25.
Diffusion of Cl− ions across the CS energy barriers was relatively rare on the timescale of our equilibrium MD simulations, which necessitated enhanced sampling methods to construct robust Cl− PMF calculations (see Methods and Extended Data Fig. 8). Nevertheless, a few Cl− entry events were observed in our simulation data for Cx50 and Cx46, and in these cases, Cl− ions appear to co-migrate across the high energy barrier of the CS alongside a K+ counter ion. It is possible that similar mechanisms involving ionic charge-neutralization enable cation preferring gap junction channels to permit passage of negatively charged signaling molecules (e.g., cAMP and IP3). However, general insights into the mechanism of perm-selectivity are difficult, as ion conductance properties do not always correlate well with the conductance properties of larger molecules41,42.
The above analysis supports previous models proposing gap junction substrate selectivity and conductance properties are established by complex mechanisms involving both steric aperture and the unique pattern of electrostatic features contributed by isoform-specific amino acid composition42,43. In this way, Cx46/50 heteromeric/heterotypic channels confer unique conductance properties of potential functional significance. For example, rectification observed in Cx46/50 heterotypic channels can be explained by the resulting asymmetric free-energy landscape (Extended Data Fig. 8), induced by the uneven distribution of fixed charges30,40. Cx46/50 heteromeric assemblies also produced unique K+/Cl− PMF profiles, with peak barriers that were intermediate to their homomeric counterparts (Extended Data Fig. 8), supporting observations made by single-channel measurement29,44.
Additional fine-tuning of gap junction permeation properties may be achieved through co- and/or post-translational modification of pore-lining residues22. In our studies, n-terminal acetylation was found to enhance the cation-to-anion specificity of Cx50, Cx46 and Cx26 intercellular channels (Extended Data Fig. 8). Although n-terminal acetylation is irreversible, the effect of this co-translational modification illustrates how other dynamic and reversible charge-modifying post-translational modifications may serve to spatially and temporally modulate the behavior of inter-cellular communication.
Open-state stabilization of NTH domain
Despite general similarities, significant differences between the Cx46/50 CryoEM structures and the Cx26 crystal structure are localized to the NTH domain (Fig. 4, Extended Data Fig. 7). The connexin NTH domain contributes to ion selectivity and “fast” trans-junctional voltage gating that is common to all connexin isoforms45. The NTH folds into the cytoplasmic vestibule where it forms the CS, and is well positioned to function as a selectivity filter/gating domain (Fig. 1d and Fig. 4a).
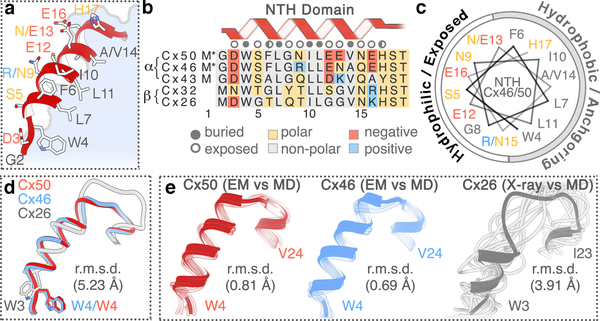
Figure 4. Open-state stabilization of the connexin-46/50 NTH domain.
(a) Zoom-view of the Cx46/50 NTH domain. (b) Sequence alignment of the NTH domain from representative α- and β-family connexins. Residues are annotated as buried, exposed, and partially exposed. Asterisk (∗) next to M1 indicates co-translational removal confirmed by MS/MS. (c) Helical wheel representation of the Cx46/50 NTH domain. In panels a–c, amino acids are labeled for Cx46/50 respectively, and colored by chemical properties (grey – hydrophobic, blue – positively charged, red – negatively charged and yellow – hydrophilic). (d) Overlay of NTH domains from experimental structures of Cx50 (red), Cx46 (blue) and Cx26 (grey; PDB 2ZW310), after super-positioning of TM1–4 and EC1–2 domains. (e) Superposition of NTH domains of each monomer captured from the MD production (faded tube), and aligned against the initial starting structures, Cx50(Ace) (left), Cx46(Ace) (center) and Cx26(Ace) (right) displayed as ribbons. Cα root mean squared deviation (r.m.s.d.) of the NTH domains are provided in panels d and e.
In the proposed open-state conformation of Cx46/50 described here, the NTH adopts a regular amphipathic α-helix and ordered loop connecting to TM1 (Fig. 4a-c). The hydrophobic face is established by a set of aromatic and hydrophobic residues that are conserved across various connexin isoforms (W4, L7, I10, L11, and V14 – in Cx46 and Cx50) (Fig. 4b). These anchoring sites pack against the pore lining helices (TM1/2), and along the interface of neighboring subunits. Despite sequence conservation at these sites, the NTH domain modeled in the crystal structure of Cx26 is in a distinctively different conformation and overall arrangement with respect to the TM domains compared to Cx46/50 (Cα r.m.s.d. = 5.2 Å, after alignment of the TM and EC domains) (Fig. 4d, Extended Data Fig. 7e). In Cx26, the NTH domain and loop connecting TM1 is less regular, and with the exception of W3 (Cx26 numbering) the conserved hydrophobic residues are modeled toward the solvent10.
We propose that the network of hydrophobic anchoring observed in the CryoEM structure of Cx46/50 supports a stabilized open-state conformation. Accordingly, analyses obtained from our MD simulations show the NTH domain for Cx50 and Cx46 is conformationally stable, in both acetylated and non-acetylated states, with only small amplitude backbone fluctuations (r.m.s.f. ~1.0–1.2 Å) (Fig. 4e, Extended Data Fig. 7). In contrast, the NTH of Cx26 (and Cx26(Ace)) becomes rapidly disordered (i.e., unfolded), and remains conformationally dynamic throughout the production phase of our MD simulations (PDB 2ZW310 as the starting structure; Fig. 4e, Extended Data Fig. 7). The dynamical behavior of the Cx26 NTH domain appears to be consistent with previous MD studies35,46,47. The functional significance of these differences in NTH domain structure and dynamic stability is currently unclear. Indeed, instability of the Cx26 NTH domain may be an intrinsic feature. In a more recent X-ray crystallographic study of Cx26 the NTH domain was completely unresolved, presumably due to local disorder11. However, potential effects of the conditions required for crystallization may not be ruled out.
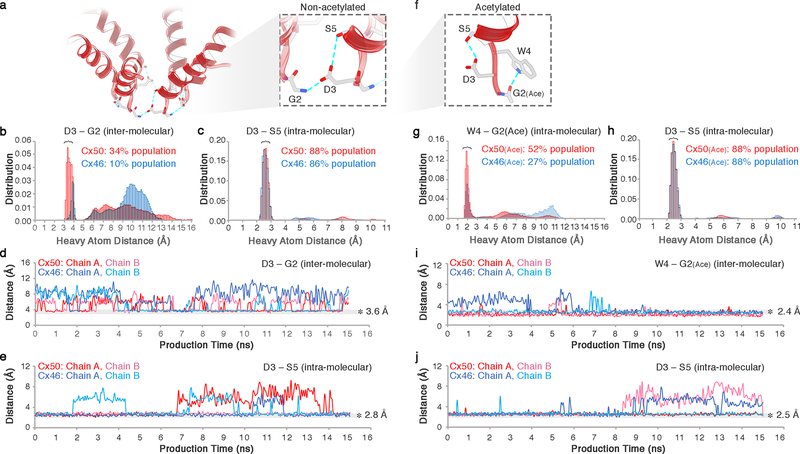
The amphipathic nature of the Cx46/50 NTH positions hydrophilic residues implicated in voltage sensing and ion selectivity at the solvent exposed face, forming the cytoplasmic vestibule45 (Fig. 4a-c). A network of h-bond interactions appears to contribute to the precise localization of some of these key residues, including the site of n-terminal acetylation. The carbonyl group at the acetylated-G2 site appears oriented, at least transiently, through h-bonding to the indole ring of W4 in the same subunit, while in the non-acetylated state G2 forms a transient inter-molecular ion-pair with D3 of a neighboring subunit (Extended Data Fig. 10). At the same time, the sidechain of D3 is oriented by a relatively stable intra-molecular h-bond with the hydroxyl of S5, in both acetylated and non-acetylated forms of both channels (Extended Data Fig. 10). D3 (in Cx46/50; D2 in Cx26) has been identified as a critical site for establishing polarity and/or sensitivity to trans-junctional voltage48–50. The precise spatial orientation imposed by these interactions may contribute to the strict conservation at this site, as replacement of D3 to a similarly negative charged residue, glutamate, results in significant perturbation to gating properties, conductance and free energy of the open-closed state in Cx50 gap junctions51.
Mutational Hot Spots Linked to Hereditary Cataracts
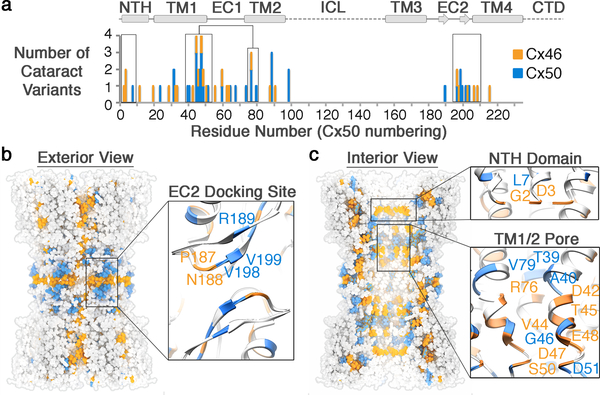
Cx46/50 gap junctions serve a critical role in maintaining the transparency of the eye lens by establishing a pathway for water, ion and nutrient circulation and removal of metabolic waste in this avascular organ12. Consequently, a variety of human genetic variations in Cx50 and Cx46 have been linked to hereditary cataract formation52. Age-related cataracts are currently incurable (except by surgery) and remain the leading cause of blindness in the world53. The more rare congenital forms of this disease have been linked to genetic mutation of various lens proteins, including Cx46/50 – offering critical insight into the mechanisms of maintaining lens transparency throughout life54. We mapped 46 mutation sites in Cx46/50 linked to congenital cataracts currently reported on the Cat-Map database54 (Fig. 5a). This analysis rationalizes many of the disease-causing effects induced by these polymorphisms, as identified mutational hot-spots localize to functionally important regions of the Cx46/50 gap junction structure. These include a cluster of residues localized within the EC2 docking site (e.g., Cx46 N188T/I and Cx50 R189Q/W), and at regions that deviate significantly from the Cx26 structure, such as the NTH gating/selectivity domain (e.g., Cx46 G2D and D3Y/H and Cx50 L7P) and several sites localized to the TM1/2 pore lining helix that form an interaction-network with the NTH domain, where mutation is expected to affect the permeation pathway or impact folding and stability within these regions of the protein (Fig. 5b,c). The localization of disease-causing mutations underscores the functional significance of these core structural/functional elements, and the importance of proper cell-to-cell communication through Cx46/50 gap junctions for the maintenance of lens transparency. The ability of CryoEM to provide high-resolution structural information on gap junctions may finally open the door to detailed mechanistic investigation of these disease-causing mutations in Cx46/50 and in other isoforms responsible for a diverse range of connexin-opathies.
Figure 5. Mutational hot-spots in connexin-46/50 linked to congenital cataracts.
(a) Histogram of connexin-46/50 genetic variants linked to hereditary (congenital) cataracts. Sites of Cx46 (orange) and Cx50 (blue) mutation are overlaid using Cx50 amino acid numbering, with secondary structure and domain elements indicated. Regions of high genetic variation “hot-spots” within the NTH, TM1/2 and EC2 domains are boxed. (b) Exterior view and (c) interior view of the Cx46/50 gap junction channel with cataract mutation sites mapped for Cx46 (orange) and Cx50 (blue), with zoom-views of the EC2 domain (panel b, inset), the NTH domain (panel c, top inset) and TM1/2 pore lining helices (panel c, bottom inset), with representative mutation sites labeled.
Methods
Cx46/50 purification and amphipol reconstitution.
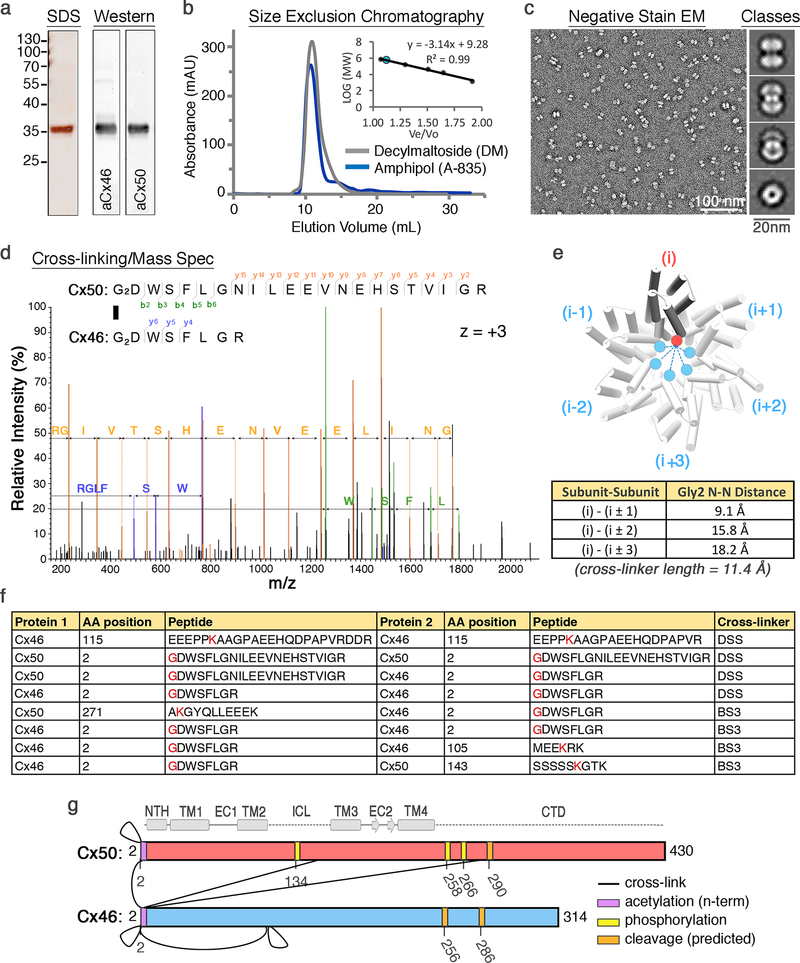
Lamb eyes were obtained from the Wolverine Packers slaughterhouse (Detroit, MI), and the lenses were removed using a surgical blade and stored at −86° C. Gap junction intercellular channels were isolated from the core lens fiber tissue, containing c-terminal truncation variants of Cx46 and Cx50 (a.k.a. MP38)16,55–57 (Extended Data Fig. 1). Details of the purification procedure are provided below.
Core lens fiber cell tissue was dissected from cortical tissue using a surgical blade, and stripped membranes were prepared as described58–60. Total protein concentration was determined by BCA (Pierce) and membranes were stored at −86° C suspended in storage buffer containing 10 mM Tris-8.0, 2 mM EDTA, 2 mM EGTA, at a total protein concentration of ~2 mg mL−1. Stripped membranes were thawed from −86° C and solubilized in 10 mM Tris-8.0, 2 mM EDTA, 2 mM EGTA, 1% (wt vol−1) n-Decyl-β-D-Maltoside (DM) for 30 minutes at 37° C. Unsolubilzed debris was cleared by ultra-centrifugation at 150,000 × g for 30 minutes at 4° C. The solubilized fraction was applied to an anion-exchange chromatography column (UnoQ, BioRad) equilibrated with buffer A, containing 10 mM Tris pH 8.0, 2 mM EDTA, 2 mM EGTA, 0.3% DM (wt vol−1). Protein was eluted with buffer B that additionally contained 500 mM NaCl. Elution peaks containing Cx46/50, as determined by SDS-PAGE, were pooled and applied to a size exclusion chromatography (SEC) column (ENC650, BioRad) equilibrated with SEC buffer, containing 20 mM HEPES, 150 mM NaCl, 2mM EDTA, 2mM EGTA and 0.3% DM (wt vol−1). Peak fractions containing purified Cx46/50 were pooled and protein concentration was determined by UV absorbance. All chromatography steps were performed at 4° C. The presence of both Cx46 and Cx50 was confirmed by western blot analysis using polyclonal antibodies directed against the n-terminal domain of Cx46 (AP11570PU-N, Acris) and c-terminal domain of Cx50 (sc-50432, Santa Cruz) (Extended Data Fig. 1a) and by mass-spectrometry analysis, described below.
Purified Cx46/50 was exchanged from DM to Amphipol A8–35 (Anatrace), as follows. Amphipol was added to freshly purified protein in an 5:1 amphipol:protein (wt vol−1) ratio using a stock solution prepared at 5% (wt vol−1) in water. This mixture was incubated for 2.5 hours at 4° C, while rotating. Detergent was then removed by application of SM-2 Biobeads (BioRad) at a ratio of 30:1 (wt wt−1) beads:detergent. Biobeads were incubated overnight at 4° C, while rotating. Biobeads were then removed by running samples over a polyprep column (BioRad) that had been washed with detergent-free SEC buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA). Samples were further cleared by ultra-centrifugation at 150,000 × g for 20 min at 4° C. The clarified sample was then applied to an SEC column (ENC650, BioRad) equilibrated with detergent-free SEC buffer to remove excess amphipol. Peak fractions corresponding to amphipol stabilized Cx46/50 were pooled and concentrated for single particle EM studies (Extended Data Fig. 1b). Final protein concentration was determined by UV absorbance at 280 nm.
Chemical cross-linking and mass-spectrometry.
Cx46/50 was prepared for cross-linking and mass-spectrometry analysis in order to confirm heteromeric assembly of the two lens isoforms (Extended Data Fig. 1d-f). Crosslinking was carried out using the amine-to-amine cross-linking reagents bis(sulfosuccinimidyl)suberate (BS3; Thermo Fischer Scientific) and disuccinimidyl suberate (DSS H12:D12; Creative Molecules). For BS3 cross-linking, 0.5 μL of 12.5 mM BS3 dissolved in deuterium-depleted water (DDW) were added to 10 μL of purified 20 μM Cx46/50 complexes and incubated 2 hours on ice. For DSS cross-linking, 0.5 μL of a 1:1 mixture of 25 mM non-deuterated (d0) and deuterated (d12) DSS cross-linker dissolved in DMSO was added to 10 μL purified 20 μM Cx46/50 complexes. A control sample, in which only DMSO or DDW were added to the proteins was also done. DSS cross-linked samples were incubated at room temperature for 2 hours in a thermomixer at 300 rpm, while BS3 cross-linked samples were incubated for 2 hours on ice. The cross-linking reaction was quenched by adding Tris pH=7.4 at a final concentration of 100 mM for 15 minutes at room temperature in a thermomixer at 300 rpm.
The quenched reaction mixtures were separated on a NuPAGE gel (Thermo Fischer Scientific) and protein bands stained with InstantBlue (Expedeon). Cross-linked protein bands were excised and digested with trypsin (Promega) as described61. Peptides were re-suspended in 0.1% Formic Acid and separated on an Ultimate 3000 UHPLC system (Thermo Fischer Scientific) and electrosprayed directly into a QExactive mass-spectrometer (Thermo Fischer Scientific) through an EASY-Spray nano-electrospray ion source (Thermo Fischer Scientific). The peptides were trapped on a C18 PepMap100 pre-column (300 μm i.d. x 5 mm, 100 Å, Thermo Fisher Scientific) using solvent A (0.1% Formic Acid in water) at a pressure of 500 bar. The peptides were separated on an in-house packed analytical column (75 μm i.d. packed with ReproSil-Pur 120 C18-AQ, 1.9 μm, 120 Å, Dr.Maisch GmbH) using a gradient (length:50 minutes, 15% to 38% for 30 min followed by 38% to 58% solvent B (0.1% formic acid in acetonitrile, flow rate: 200 nL/min) for 15 min. The raw data was acquired on the mass-spectrometer in a data-dependent mode (DDA). Full scan MS spectra were acquired in the Orbitrap (scan range 350–2000 m/z, resolution 70000, AGC target 3e6, maximum injection time 50 ms). After the MS scans, the 10 most intense peaks were selected for HCD fragmentation at 30% of normalized collision energy. HCD spectra were also acquired in the Orbitrap (resolution 17500, AGC target 5e4, maximum injection time 120 ms) with first fixed mass at 180 m/z. Charge exclusion was selected for 1+ and 2+ ions. The dynamic exclusion set to 5 seconds. Cross-linking identification and analysis was done using pLink62 and Xcalibur 2.2 (Thermo Scientific). All peptides were manually validated.
For identification of proteins and post-translational modifications, protein bands were excised from the gel and processed as described above. Mass-spectrometry analysis was carried out similarly with a gradient of 15 – 38% for 30 min and the Orbitrap set to 350–1500m/Z. Charge exclusion was selected for 1+ and unassigned ions, dynamic exclusion was set to 5s. PTM identification was done using the MASCOT Daemon client program.
Negative stain electron microscopy.
Amphipol stabilized Cx46/50 was prepared for negative stain EM as described58,63. Briefly, a 3 μL drop of sample (~0.02 mg mL−1) was applied to a glow-discharged continuous carbon coated EM specimen grid (Ted Pella), blotted with filter paper and washed two times with detergent-free SEC buffer. The specimen was then stained with freshly prepared 0.75% (wt vol−1) uranyl formate (SPI-Chem).
Negatively stained specimens were visualized on a 120 kV TEM (iCorr, FEI) at a nominal magnification of 49,000× at the specimen level (Extended Data Fig. 1c). Digital micrographs were recorded on a 2K x 2K CCD camera (FEI Eagle) with a calibrated pixel size of 4.37 Å. A total of 75 micrographs were collected. CTF parameters were determined in EMAN264 and micrographs free of significant astigmatism and drift were selected based on Thon rings in the power spectra. A total of 5,330 particles were hand selected in EMAN2 and extracted with a box size of 84 × 84 pixels. Reference-free 2D class averages were generated using CTF-corrected (phase-flipped) images without applied symmetry (Extended Data Fig. 1c). A subset of 3,952 ‘good’ particles was selected following multiple rounds of 2D classification, and an initial model was generated de novo in EMAN2 using a subset of 12 class averages as input. This model was refined against the ‘good’ particle image dataset in EMAN2 with applied D6 symmetry to a final resolution of ~20 Å (Extended Data Fig. 2).
CryoEM data collection, image processing and 3D reconstruction.
Samples were prepared for CryoEM by applying 5 μL of amphipol-stabilized Cx46/50 (2.35 mg mL−1) to a glow-discharged holey carbon grid (Quantifoil R 1.2/1.3) for 10 seconds. The grid was blotted for 4.0 seconds and plunge frozen in liquid ethane using a Vitrobot (FEI) at 100% humidity and stored under liquid nitrogen.
CryoEM specimen grids were imaged on a Titan Krios (FEI) operated at 300kV. Image stacks were recorded using a K2 summit direct electron detector (Gatan) in counting mode with a super-resolution pixel size of 0.665 Å/pix. The dose rate was 3.2 electrons pixel−1 sec−1, with 4 frames per second collected for a total exposure time of 10 seconds. A Gatan energy filter with a slit width of 30 eV was used during data collection. An initial dataset of 1104 micrographs (Dataset 1) was obtained by automated data collected using SerialEM65, with nominal defocus values from 1.25 – 2.5 μm.
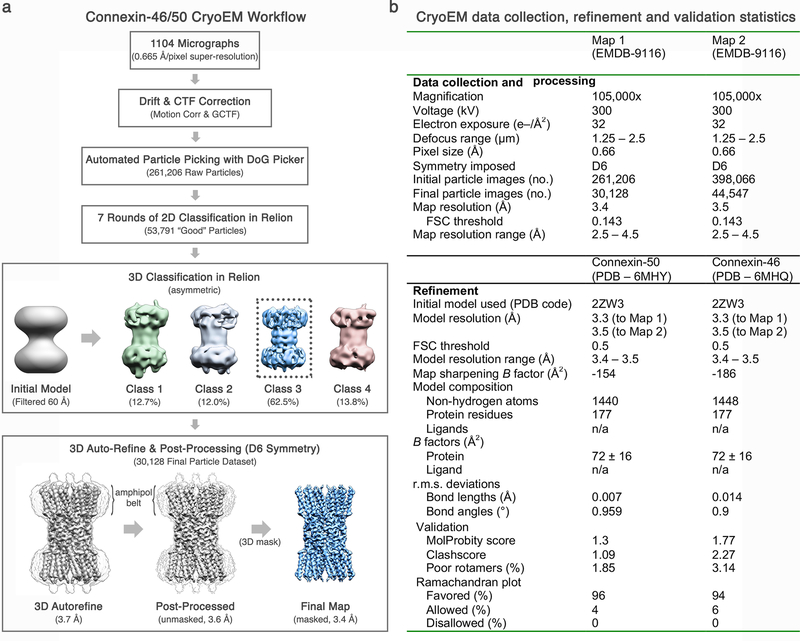
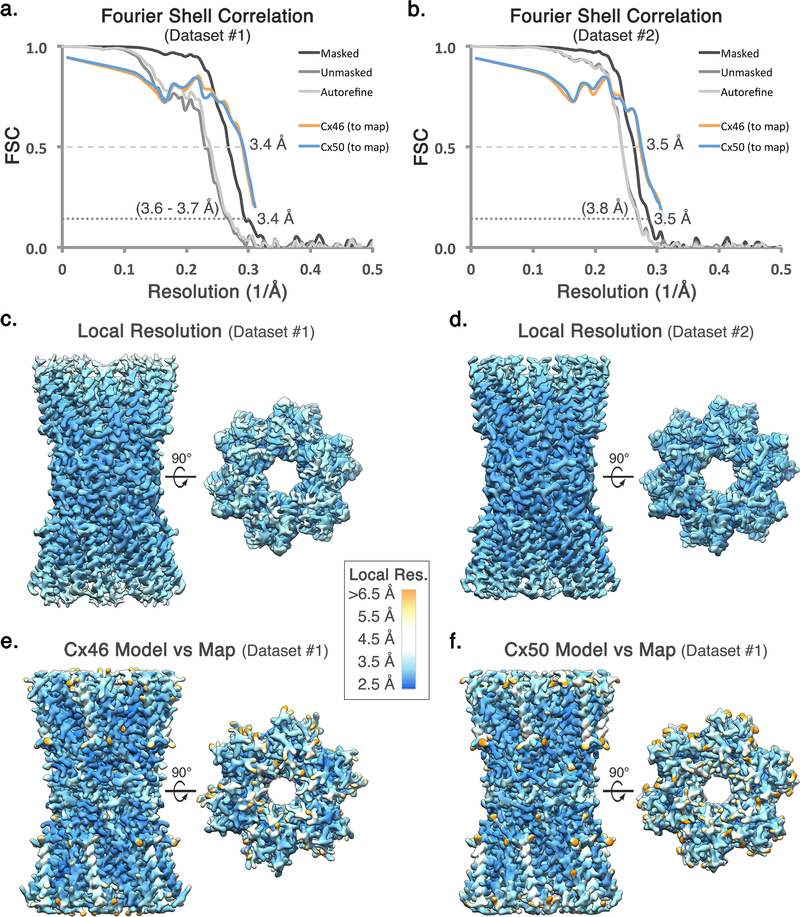
Drift correction and dose weighting was performed using MotionCor266 and CTF correction was performed using GCTF67. 261,206 particles were picked from Dataset 1 using DoGPicker68. Particles were extracted with 2× binning (resulting in a pixel size of 1.3 Å/pix). Five rounds of 2D classification in Relion 2.069 left 53,791 “good” particles. These particles were then subjected to 3D classification in Relion with four classes and no imposed symmetry. The most populated class contained 33,967 particles. These particles were unbinned and another round of 3D classification was performed, reducing the population of particles to 30,128. 3D auto-refinement was then performed on this set of particles with D6 symmetry imposed. After masking and post processing in Relion, the final map had a resolution of 3.4 Å by gold standard FSC (Extended Data Fig. 2 and Extended Data Fig. 3a).
An additional 1093 micrograph data set was collected and processed as above. Particles were picked from this set with DoGPicker, 2× binned and pooled with the original set of 261,206 particles for a total of 398,066 particles (Dataset 2). A set of 66,480 “good” particles was obtained after five rounds of 2D classification. These particles were subjected to 3D classification with four classes and no imposed symmetry. The most populated class contained 55,475 particles. This dataset was further culled by removing particles extracted from micrographs with Thon rings that did not extend beyond 3.5 Å, resulting in a final dataset of 44,547 particles. These particle images were subjected to 3D auto-refinement in Relion with D6 symmetry, resulting in an overall 3.5 Å resolution 3D reconstruction after post-processing as judged by gold-standard FSC (Extended Data Fig. 3b). Local resolution analysis using BlocRes70 showed the 3.4 Å map possessed exceptionally high resolution features within the central regions of the structure, while the 3.5 Å reconstruction contained more uniformly defined features throughout the density map, consistent with visual inspection (Extended Data Fig. 3c-d). An overview of CryoEM data collection and 3D refinement statistics is provided in Extended Data Fig. 2.
Cx46/50 symmetry analysis.
In an attempt to uncover a specific pattern(s) of Cx46/50 heteromeric/heterotypic co-assembly, 3D auto-refinement was also pursued in Relion using C1, C3, C6, and D3 symmetries, using the final 30,128-particle (Dataset 1) and 3.4 Å map (filtered to 15 Å) as input. These refinements converged to 4.1 Å (C1), 3.9 Å (C3), and 3.7 Å (D3 and C6). Examination of the resulting maps provided no indication that the Cx46/Cx50 subunits were being separately resolved (Extended Data Fig. 4,5). Further attempts were performed using 3D classification in Relion with C3, C6, and D3 symmetry, using the larger 55,475 particle set (which had already been subjected to one round of 3D classification with no imposed symmetry). The initial model was the 3.4 Å map filtered to 25 Å. No resolution limit was enforced, and classification was attempted with and without image alignment. Some classifications converged to a single class, while some maintained a more even distribution of particles throughout 3D classification. 3D auto-refine was attempted with the most populated class from each attempted symmetry group. C3 symmetry refined to 3.9 Å from a set of 47,074 particles; C6 symmetry refined to 3.8 Å from a set of 38,404 particles; D3 symmetry refined to 4.2 Å from a set of 16,520 particles. Inspection of the resulting maps provided no indication that isoform-specific features were being separately resolved into any specific symmetric arrangements (not shown). Finally, focused refinement strategies with signal subtraction were also explored using Relion, by masking a single hemi-channel or just a single subunit. However, these procedures did not produce isoform-specific features, or improved results compared to the D6 symmetrized maps.
As we were unable to identify a specific pattern of co-assembly for the Cx46/50 dodecameric channel, all further analysis and model building was performed using the 3D maps generated with imposed D6 symmetry. Both pre-processed and post-processed maps and associated masks generated from Datasets 1 and 2 have been deposited to the electron microscopy data bank (EMD-9116).
Atomic modeling, refinement and validation.
The post-processed maps obtained with D6 symmetry were used to build and stereochemically refine atomic models for both Cx46 and Cx50, following similar procedures. An initial Cα model was generated using the available crystal structure of connexin-26 (PDB 2ZW310) and placed into the post-processed 3.4 Å density map using rigid-body fitting. Starting from this template, all atom models of Cx46 and Cx50 were built separately into the CryoEM density using COOT71. Disulfide bonds were modeled for Cx 50 (C54–C201, C61–C195 and C65–C190) and Cx46 (C54–C189, C61–C183 and C65–C178). Models were subjected to real space refinement in Phenix72 with non-crystallographic symmetry (D6-symmetry) and secondary structure restraints imposed. Successive rounds of modeling and refinement were conducted until refinement statistics converged, as judged by Molprobity73 (Extended Data Fig. 2). The FSC of the model versus map dropped below 0.5 at 3.4 Å (Dataset 1) and 3.5 Å (Dataset 2) for both Cx46 and Cx50, judged by the output of Phenix real space refine (Extended Data Fig. 3a-b). The NTH domain of Cx46 and Cx50 (residues 2 – 20) were further refined using the post-processed 3.5 Å density map (Dataset 2), as this region of the map was more well defined compared to the original 3.4 Å map. Over areas of the density maps where the sequence of Cx46 and Cx50 are identical or similar (80% identical and 8% similar) both models fit well into the D6 symmetrized map, and these regions tend to display well-resolved sidechain density. Over regions where the sequence of Cx46 and Cx50 differ, sidechain density is sometimes weaker. This observation is possibly due to the imposed D6 symmetry averaging the density of two different sidechains in these areas, or relative flexibility as many of these residues contain solvent exposed sidechains. In these areas of difference, where EM density is observed, both Cx46 and Cx50 can be fit into the density equally well (Extended Data Fig. 4,5). Fit of the models to the CryoEM density map were assessed quantitatively by local resolution analysis using BlocRes70, comparing the calculate maps of Cx50 and Cx46 atomic models to the 3.4 Å experimental CryoEM map (Extended Data Fig. 3). This analysis was tabulated by assigning each residue a range of resolution values corresponding to the output of this analysis, including the alpha carbon and extending to the end of the sidechain (Supplementary Table 2,3).
Completed models of the dodecameric Cx46 and Cx50 structures, corresponding to residues 2–97; 142–222 and 2–97; 154–234, respectively, have been deposited to the protein data bank (PDB ID’s 6MHQ and 6MHY, respectively). Additional density is observed for the region of TM2 that extends toward the cytoplasm; however, we did not model this region (corresponding to ~1–2 turns of an α-helix) due to the lack of identifiable sidechain density. Various heterotypic/heteromeric models of Cx46/50 were generated for analysis by applying appropriate symmetry operations to the monomeric subunits and combined to form a complete gap junction structure. Coulombic surface potentials were calculated and displayed using Chimera74.
Molecular dynamics simulations.
Visual Molecular Dynamics (VMD) v1.9.375 was used to build systems for Cx50, Cx46, Cx46/50 heteromeric and heterotypic models, and Cx26 (PDB 2ZW3)10. Representative Cx46/50 heteromeric models (heteromeric models I and II, with C3 or D3 point group symmetry, respectively) were constructed by applying the appropriate symmetry operations to the coordinates of the individual subunits. The Cx46/50 heteromeric and heterotypic channels were run through a steepest decent minimization routine using Phenix72 to ensure no clashes were introduced in the preparation of these models. Each system comprised the full dodecameric gap junction, and was prepared in explicit solvent and embedded in two lipid bilayers composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), mimicking a cell-cell junction. The Cx26 crystal structure was prepared for MD by completing the sidechains at residues Lys15, Ser17 and Ser19 and missing protons were added to all amino acids at standard positions. Sidechains were protonated according to neutral conditions, and the HSD model was used for all histidine residues. To facilitate comparison to the Cx46/50 models, the Cx26 model was constructed with a Met1 residue added, which was missing in the published crystal structure, but expected to be present in the protein based on proteomic analysis76, as previously described47. Disulfide bonds identified in the experimental structures were enforced for Cx50 and Cx46 (as described above), and for Cx2610 (C53–C180, C60–C174 and C64–C169). Amino acids corresponding to the intracellular loop (ICL) connecting TM2/TM3, and c-terminal domain (CTD) of Cx50, Cx46 and Cx26 and were not included for MD simulation, as experimental data describing the structure of these large domains (~50 residue ICL and ~200 residue CTD in Cx46/50) are missing. The introduced n- and c-terminal residues resulting from the missing ICL segment (Cx46 L97 and L142; Cx50 V97 and L154; and Cx26 G109 and K125) were neutralized. N-terminal acetylation sites were introduced in VMD through an all-atom acetylation patch in the automated PSF-Builder. A complete list of modeled residues for each system is provided in Supplementary Tables 1.
The prepared protein structures were submerged in a hydration shell using Solvate 1.0.177. Water was removed from sections of the channel corresponding to transmembrane domains, based on hydrophobic character and localization of amphipol observed in the experimental CryoEM data (~20–50 Å from the center of the channel). The VMD membrane-builder plugin was used to add two POPC bilayers, with dimensions of 152 × 152 Å for Cx46, Cx50 and Cx46/50 models, and 155 × 155 Å for Cx26 and lipids overlapping with protein were removed. The entire system was then placed in a water box with dimensions 150 × 150 × 180 Å for Cx46, Cx50 and Cx46/50 models, and 150 × 150 × 183 Å for Cx26, using VMD’s Solvate plugin. The system was neutralized using the Autoionize plugin, then 150 mM KCl and 150 mM NaCl were added to the solvent areas corresponding to intracellular and extracellular regions of the simulation box, respectively (see Fig. 3c). A summary of atoms counts for each system is provided in Supplementary Table 1.
GPU-accelerated nanoscale molecular dynamics (NAMD) 2.1278 was used for all classical MD simulations, using the CHARMM36 force-field79,80 for all atoms and TIP3P explicit model for water. Each system was prepared following the same minimization and equilibration protocol, as follows. An initial minimization of the lipid-tails, with all other atoms fixed, was performed for 1 ns with a 1 fs time-step, allowing the tails to ‘melt’. Next, the system, including lipids, solvent and ions were allowed to minimize around the protein, with the protein harmonically constrained for 1 ns. For the Cx46/50 heteromeric/heterotypic and acetylated models, a second minimization step was applied, where the system was free to minimize with a harmonic constraint on the protein backbone to ensure stable quaternary structure. The entire system was then released from restraints and subjected to all-atom equilibration runs employing Langevin thermostat, with a constant temperature of 310 K and constant pressure of 1 atm, with 1 or 2 fs time-steps and allowed to proceed for 30 ns (see Supplementary Table 1). Periodic boundary conditions were used to allow for the particle mesh Ewald (PME) calculation of electrostatics. Finally, all of the models were continued for a minimum of 50 ns of production. Root mean squared deviations (r.m.s.d.) and root mean squared fluctuations (r.m.s.f.) were calculated using VMD. All three gap junctions approached a steady r.m.s.d. within 20 ns of the equilibration phase (Extended Data Fig. 7a-b). All of these systems maintained an electro-chemical seal to extracellular sodium ions (Na+) during MD simulation (e.g., Fig. 3c), validating the stability of inter-molecular docking-site interactions and the various heteromeric/heterotypic models generated for analysis.
Calculation of the potential-of-mean-force (PMF) with respect to K+ and Cl− was performed using the fundamental principle of detailed balance via a one-dimensional Markov State Model (MSM). Configuration space was subdivided based on a natural coordinate, the channel pore (z-axis), and segmented into bins of 4 Å in length. Using a lag-time of 2 ps, a transition matrix was calculated from the trajectories of individual ions within the simulation. The i→j transition probability ki,j is computed via Equation 1:
| Equation (1) |
Where Ni,j is the count of transitions during the lag interval and Ni is the count of ions in bin i at the beginning of each lag interval. PMFs were constructed using the principle of detailed balance:
| Equation (2) |
| Equation (3) |
| Equation (4) |
Here, Peq are the equilibrium probabilities for an ion to occupy each bin respectively (Equation 2), ΔGi,i+1 is the free energy difference from bin i to i+1, R is the gas constant (1.986 Cal mol−1 K−1), and T is temperature (310 K) (Equation 3,4). Final PMF values were adjusted so that the values of the bulk solvent where zero. PMF curves in Fig. 3 and Extended Data Fig. 7 were derived by mapping z values to the corresponding bin index i and subsequently smoothed using MS Excel. The detailed-balance (rates-based) approach is justified by the high mobility of ions within the channel pore, and the time-scales used for analysis were validated by assessing the convergence of the unsymmetrized data to the symmetrized values presented in Fig. 3d and Extended Data Fig. 8. The results were shown to closely match PMFs constructed by taking the population profile, or average counts (<(Ni)>), of the K+ ions along the channel pore (z-axis) and solving: ΔG = −RTln(<(Ni)>)11,81,82 (Extended Data Fig. 8d).
Because the detailed-balance approach requires only local equilibrium sampling, we were able to apply a distributed seeding protocol to construct PMFs for Cl- ions. Initial analysis of Cl− trajectories revealed this ion to be poorly sampled inside the channel pore of the Cx50, Cx46 and Cx26 models, presumably due to an energetic barrier presented by significant regions of negative coulombic potential for each of these systems (see Fig. 3A). Therefore, a distributed seeding approach was employed where a single Cl− ion was randomly introduced (seeded) by replacing a K+ ion within the pore of the equilibrated channel. These coordinates were energy minimized and initial velocities were randomized before allowing the simulation to proceed for 10 ns. This procedure was repeated for each system 3–16 times until sufficient sampling was achieved, as determined by monitoring the resulting PMFs. Cl− PMFs were constructed based on the transition rates (as described above), which is not sensitive to the initial placement of the ion. This seeding approach was validated by showing the resulting PMF recapitulated the features of a Cl− PMF obtained for Cx46, where sufficient sampling had been achieved through random diffusion (Extended Data Fig. 8e). Trajectories from these distributed seeding simulations were combined with the production phase data and included in the MSM for calculation of final Cl− PMFs.
Analysis of hydrogen bonding within the NTH domains of Cx46 and Cx50 models was performed by recording the distance vs. time of potential donor-acceptor pairs. The three sets of interactions probed were potential inter-molecular hydrogen bonds between D3 and the neighboring n-terminal G2 residue, the intra-molecular hydrogen bonding between D3 and S5, and the intra-molecular hydrogen bonds between N-term acetyl and W4 for acetylated models of Cx46 and Cx50. To simplify the analysis of hydrogen bonding interactions involving equivalent rotameric donor-acceptor configurations, heavy atoms were selected for D3 (Cγ) and G2 (N) for analysis (Extended Data Fig. 10). For comparison, equivalent analysis for Cx26 was conducted (between D2 and M1 and T5 of the adjacent subunit). For Cx26, D2 appeared to form intra-molecular hydrogen bonding pairing with T5; however, stable inter-molecular pairing interactions with Met1 or T5 (as indicated in the crystal structure10) were not identified during the production phase of these simulations (data not shown), as previously reported35.
The NTH domain of Cx26 was found to be unstable (i.e., rapid unfolding) during MD simulation in either the acetylated or non-acetylated states (Extended Data Fig. 7 and Fig. 4e). The significance for this dynamical behavior is not clear; however, we attribute this feature to be a potential reason for the variation of our calculated PMFs of Cx26 compared to previous studies47. Notably, Kwon et al. reported the Cx26 channel to be anion selective in the absence of n-terminal acetylation47. These authors conducted an elegant set of experiments employing a GCMC/BD based approach for modeling ion conductance using a model of the Cx26 hemi-channel (by extracting a single hexamer of the Cx26 intercellular channel). However, a limitation of GCMC/BD method is the protein structure is held static, and as such the resulting PMFs obtained by this approach would be significantly influenced by the selected conformational state of the Cx26 NTH domain. These caveats should be considered when comparing results presented in this work.
Statistical analysis.
95% confidence intervals for comparison of Cα r.m.s.f. values were calculated using a two-tailed student t-test. No statistical methods were used to predetermine sample size for the CryoEM datasets. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Data Availability
CryoEM density maps have been deposited to the Electron Microscopy Data Bank (EMD-9116). Coordinates for Cx46 and Cx50 atomic models have been deposited to the Protein Data Bank (6MHQ and 6MHY). The original multi-frame micrographs have been deposited to EMPIAR (EMPIAR-10212).
Extended Data
Figure 1: Preliminary structural analysis of native lens Cx46/50 gap junction intercellular channels.
(a) Denaturing protein gel and western-blot analysis of purified Cx46/50 isolated from lens core tissue (a.k.a. MP38). Protein bands corresponding to Cx46 and Cx50 co-migrate at a molecular weight of ~38 kDa, consistent with age-dependent proteolytic cleavage within the c-terminal domain of both isoforms16. Performed 3 times with the similar results. (b) Size exclusion chromatography elution profile of Cx46/50 gap junctions reconstituted in decyl-maltoside (DM, grey trace) or amphipol (A-835, blue trace), monitored by UV-absorbance. Performed >3 times with similar results. Inset, shows calibration curve (n = 3 runs) demonstrating Cx46/50 elutes at an apparent molecular weight of ~560 kDa, consistent with the size of a dodecameric protein complex (12 × ~38 kDa) and two micelles (2 × ~50kDa). (c) EM micrograph of negatively stained Cx46/50 gap junctions reconstituted into amphipol. Scale bar = 100 nm. (inset) Representative 2D class averages of negatively stained particles (selected from 25 classes). Scale bar = 20 nm. (d–g) Chemical crosslinking and mass spectrometry. (d) Representative MS/MS m/z spectrum, identifying inter-subunit cross-linking at the n-terminal Gly2 positions of Cx50 and Cx46. Identified peaks in the m/z spectrum and amino acid identities are indicated (Cx50 b-ions = green; Cx50 y-ions = yellow; Cx46 y-ions = blue). MS/MS data represent the consensus of 3 independent runs. (e) Structural analysis of cross-linking results, showing inter-subunit distances between the symmetrically related n-terminal Gly2 positions within the connexin hemi-channel, ranging from 9.1 Å (i to i±1), 15.8 Å (i to i±2), and 18.2 Å (i to i±3). The cross-linker spacer length is 11.4 Å, indicating an (i to i±1) arrangement of Cx50 and Cx46 within the same hemi-channel was most likely identified, although other arrangements may not be ruled out. (f) Overview of identified inter-subunit cross-links between Cx50 and Cx46 assembled gap junctions. Residues in red indicate the site of primary amines involved in the cross-linking reaction using either DSS or BS3. All detected inter-subunit crosslinks are between cytoplasmic domains. (g) Schematic showing sites of inter-subunit cross-linking between Cx46 and Cx50 (black lines) and post-translational modifications identified during proteomics analysis (yellow = phosphorylation; purple = n-terminal acetylation). Met1 was determined to be removed in both Cx46 and Cx50 and the resulting n-terminal Gly2 position was identified in both acetylated and non-acetylated forms for Cx46 and Cx50, consistent with the specificity of the NatA acetylation complex24. Also shown are the predicted c-terminal domain (CTD) cleavage sites in Cx46 and Cx50 (orange), based on previous analysis of bovine Cx46/50 isolated from lens core tissue16. Secondary structure and domain labels are indicated for the n-terminal helix (NTH), transmembrane helices (TM1–4) and extracellular domains (EC1–2) intracellular loop (ICL) and CTD.
Figure 2: Overview of CryoEM image processing and 3D reconstruction.
(a) A total of 1104 micrographs were collected in an automated fashion using SerialEM65 on a 300 kV Titan Krios (Dataset 1). Movie stacks were recorded using a K2 summit direct electron detector operated in super-resolution mode and acquired with an effective pixel size of 0.665 Å. Movie stacks were corrected for drift and contrast transfer function (CTF) using MotionCor266 and GCTF67, respectively. An initial dataset of 261,206 raw particles was obtained using unbiased autopicking procedures in DoG Picker68. A refined dataset of 53,791 ‘good’ particles was obtained following several rounds of 2D classification and removal of ‘bad’ particles (or ice contamination) in Relion69. Three-dimensional (3D) classification was seeded using an initial model obtained by negative stain EM, filtered to 60 Å. A majority of particles fell into a single 3D-class (~62.5% of the “good” particles). These 30,128 particles were used for final 3D auto-refinement and post processing, yielding a final map at 3.4 Å resolution by gold-standard FSC (Dataset 1). Dataset 2 was processed in a similar fashion from a total of 2,197 micrographs and 44,547 ‘good’ particles, resulting in a final map at 3.5 Å resolution by gold-standard FSC. (b) Summary of CryoEM data collection, refinement and model validation statistics. Dataset 1 was used to obtain the 3.4 Å resolution reconstruction (Map 1). Dataset 2 was used to obtain the 3.5 Å resolution reconstruction (Map 2). Pre-processed and post-processed maps and associated masks from both datasets have been deposited to the EM databank (EMD-9116). The original multi-frame micrographs have been deposited to EMPIAR (EMPIAR-10212). Coordinates for Cx50 and Cx46 atomic models have been deposited to the Protein Data Bank (6MHY and 6MHQ, respectively).
Figure 3: Global and local resolution analysis.
(a,b) Fourier Shell Correlation (FSC) analysis obtained from Dataset 1 and Dataset 2, respectively. Gold-standard FSC curves following auto-refinement (light grey), post-processing (grey), and masking (dark grey). The final masked maps display an overall resolution of ~3.4 Å (Dataset 1) and ~3.5 Å (Dataset 2), using a 0.143 cut-off. FSC curves comparing atomic models of Cx46 (orange) and Cx50 (blue) fit to the CryoEM maps display correlation at 0.5 cut-off to a resolution of 3.4 Å (Dataset 1) and 3.5 Å (Dataset 2). (c,d) Local resolution analysis using BlocRes70, obtained for the half-maps for Dataset 1 and 2, respectively. (e,f) Local resolution analysis comparing the experimental density map (Dataset 1) to the calculated maps of Cx46 and Cx50, respectively. Local resolution ranges in panels c-f are indicated by color (2.5 – 4.0 Å = blue – cyan; 4.0 – 5.0 Å = white; 5.0 – 6.5 Å yellow – orange). Tabulated values obtained for local resolution of Cx46 and Cx50 models compared to the experimental density map are provided (Supplementary Tables 2,3). Local resolution assessment comparing the density map to the two models indicates that the sites where the two isoforms differ in sequence were generally less well resolved as compared to equivalently exposed residues where Cx46 and Cx50 are conserved in sequence.
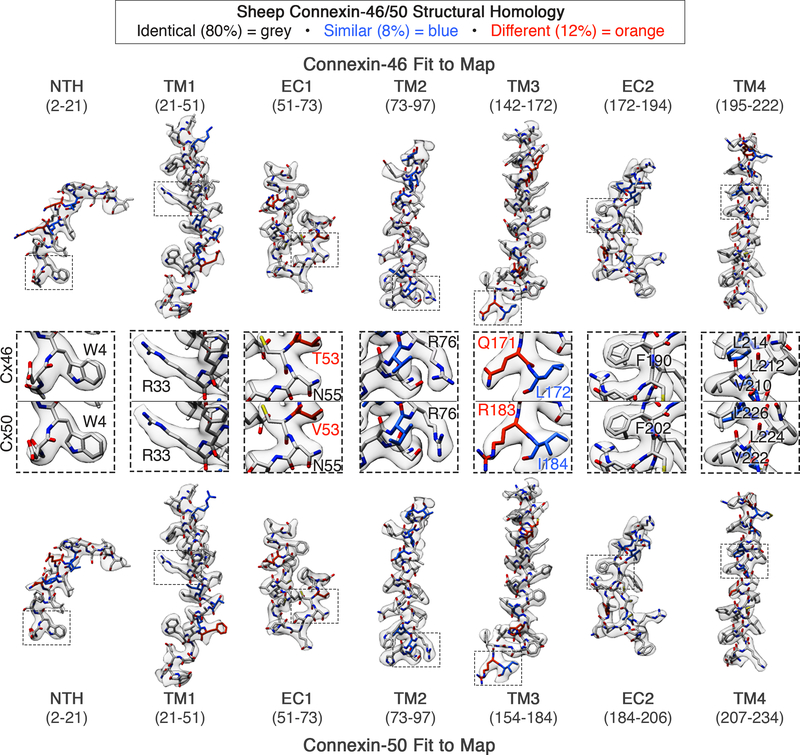
Figure 4: Connexin-46 and −50 atomic models fit to the CryoEM density maps.
Segmented CryoEM map with atomic models for sheep Connexin-46 (Cx46) and Connexin-50 (Cx50) fit to the experimental densities derived from Dataset 1 (3.4 Å, D6-symmetry), including regions for transmembrane domains 1–4 (TM1–4) and extracellular domains 1–2 (EC1–2). The n-terminal helix domain (NTH) is fit into the map from Dataset 2 (3.5 Å, D6-symmetry), which was more well defined in this region. Cx46 (top) and Cx50 (bottom) models are colored according to their pair-wise sequence homology, as being identical (grey, 80%), similar (blue, 8%) and different (orange, 12%). Windows show zoom-views corresponding to boxed regions of the segmented maps, highlighting representative sidechain densities and fit to the atomic models. Regions of identical or similar amino acids are fit equally well by both models (e.g., Cx46 L172 vs. Cx50 I184, blue labels). Over regions where the sequence of Cx46 and Cx50 differ, sidechain density is typically weaker (see also, Extended Data Fig. 5). This is possibly due to the imposed averaging of two different sidechains in these areas, or relative flexibility as many of these residues correspond to solvent exposed sidechains. In these areas of difference, and where EM density is present, both Cx46 and Cx50 models were typically fit equally well into the density map (e.g., Cx46 T53 vs. Cx50 V53 and Cx46 Q171 vs. Cx50 R183, orange labels).
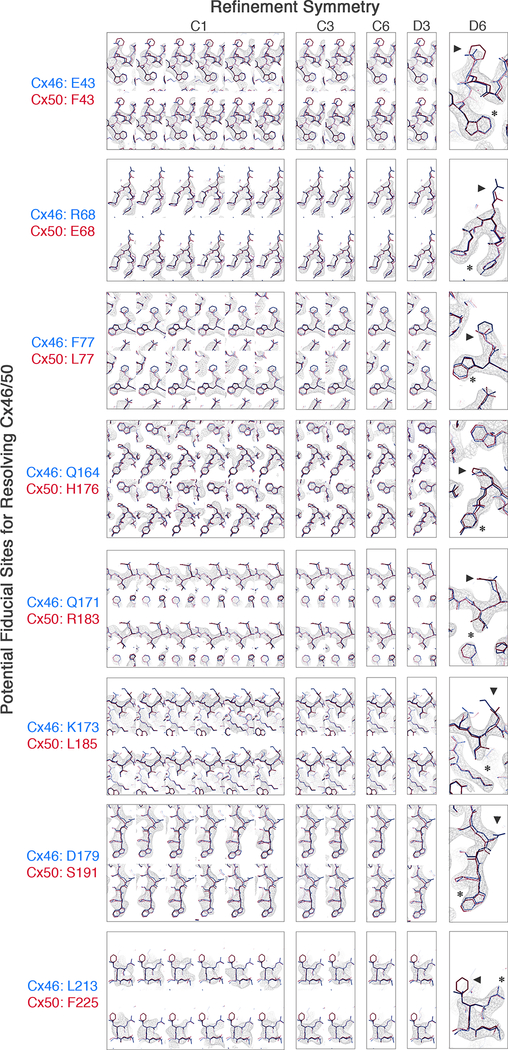
Figure 5: Analysis of different symmetry refinements on the ability to resolve differences between Cx46 and Cx50.
Eight sites of sequence differences involving bulky amino acids (labeled, and indicated by arrow head) were selected as potential fiducial markers for resolving the two different isoforms following 3D refinement with various applied symmetries (C1: 4.1 Å resolution; C3: 3.9 Å resolution, C6: 3.7 Å resolution, D3: 3.7 Å resolution; D6: 3.4 Å resolution). For the applied symmetries, views are presented for each unique asymmetric subunit (boxed). Despite the modest resolution of the asymmetric (C1) reconstruction, sidechain density for bulky amino acids were typically observed at sites where the two isoforms are conserved (asterisk). However, at the selected sites of sequence variation (arrow head) the sidechain densities are either not well resolved, or there was no systematic variation that indicated an ability to distinguish the two isoforms. The most resolved features at these sites of variation were obtained with D6 symmetry, and typically corresponded to regions where these different amino acids share similar structure (e.g., Cβ positions). Although the CryoEM density at these sites of variation were typically weak, the resolvable sidechain features throughout the rest of the map were generally enhanced when higher symmetry was applied during map refinement (indicated by asterisk), suggestion that regions of sequence similarity between Cx46/50 also share a high level of structural similarity.
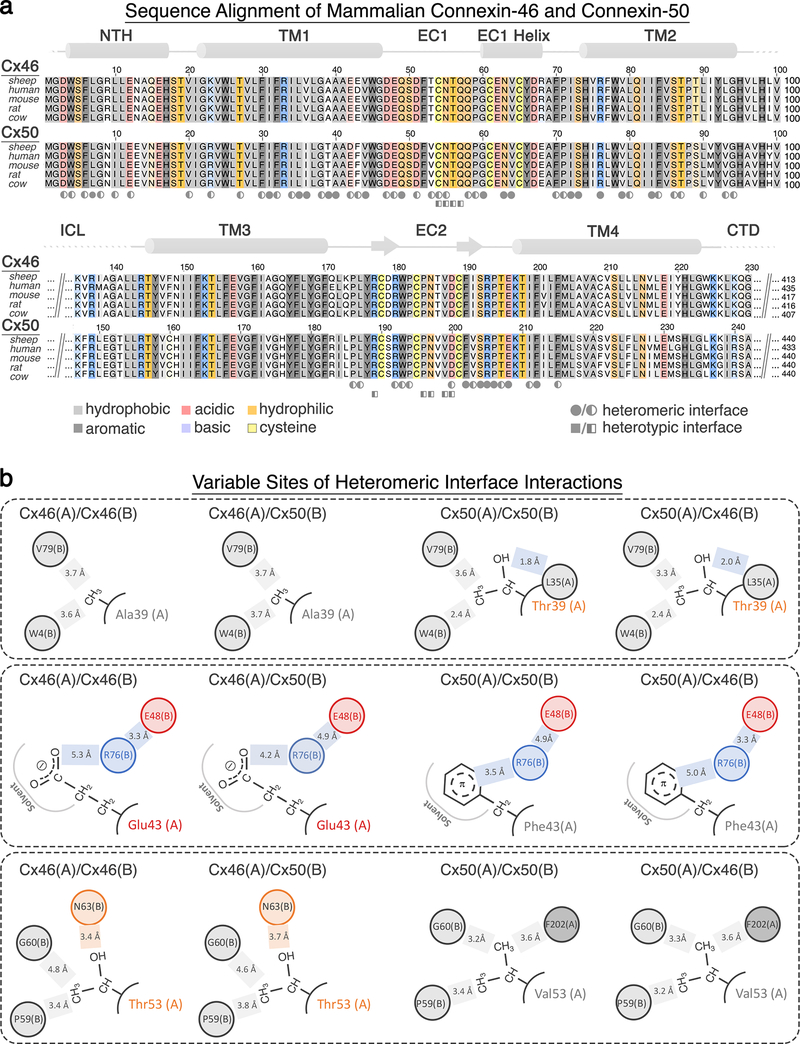
Figure 6: Sequence and structure conservation of Cx46 and Cx50 heteromeric/heterotypic interfaces.
(a) Multiple sequence alignment of mammalian Cx46 and Cx50 isoforms with residues contributing to heteromeric and heterotypic interfaces annotated (circle – heteromeric interface, square – heterotypic interface; filled = >70% buried and half-filled = 20–70% burial)83. Coloring corresponds to amino acid type (grey – hydrophobic; dark grey – aromatic; red – acidic; blue – basic; orange – hydrophilic; yellow – cysteine). Regions of sequence homology are indicated by the level of shading. Secondary structure and domain labels are indicated for the n-terminal helix (NTH), transmembrane helices (TM1–4) and extracellular domains (EC1–2). Regions lacking defined structure and of poor sequence homology within the intracellular loop (ICL) and c-terminal domain (CTD) have been omitted for clarity. Sheep and human Cx46 and Cx50 orthologs contain ~95% sequence identity (~98% similarity) over the structured regions of the protein. Numbering corresponds to the amino acid sequence of sheep Cx44 and Cx49 used in the main text. (b) Illustration of homomeric and heteromeric interface interactions involving the 3 sites lacking conservation between Cx44 and Cx49 at this interface (positions 39, 43 and 53). Despite these sequence differences, the interactions involving these residues are generally similar (hydrophobic – grey; h-bonding – orange; and ion-pairing – blue).
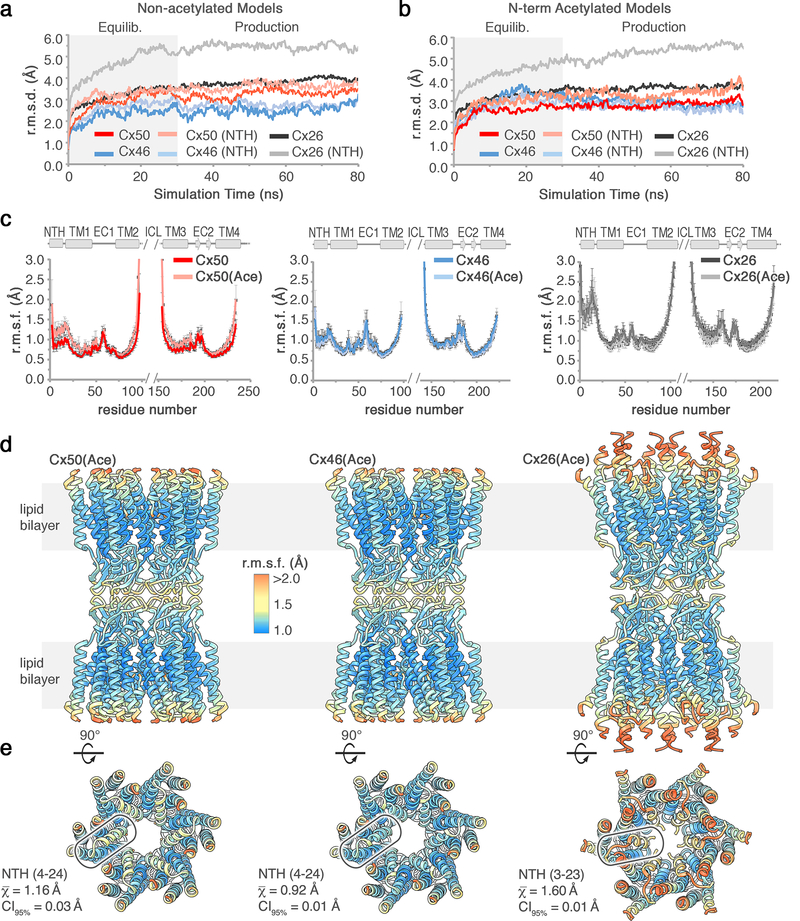
Figure 7: Analysis of protein backbone dynamics during MD equilibration and production.
(a) Cα root mean squared deviation (r.m.s.d.) analysis of equilibrium (0 – 30 ns) and production phases (30–80 ns) of the molecular dynamics (MD) simulations, calculated with respect to the experimental starting structure for non-acetylated models of Cx50 (red traces) and Cx46 (blue traces) and Cx26 (with Met1 added) (grey traces). Separate analysis for the n-terminal helix (NTH) domains are shown in lighter shades. (b) Same analysis as in panel a, for models with n-terminal acetylation added. The NTH domain of Cx26 (light grey traces) shows significantly higher r.m.s.d. values, for both non-acetylated and acetylated models. (c) Plot of average Cα root mean squared fluctuation (r.m.s.f.) during the production phase of the molecular dynamics (MD) simulations for Cx50 (left, red traces), Cx46 (center, blue traces) and Cx26 (right, grey traces). Data obtained for the n-terminal acetylated models are shown in lighter shades. Averages are determined for the 12 subunits composing the intercellular channel. Error bars represent 95% confidence intervals (n = 12 subunits). Secondary structure and domain labels are indicated for the n-terminal helix (NTH), transmembrane helices (TM1–4), extracellular domains (EC1–2) and intracellular loop (ICL; not modeled). (d,e) Average r.m.s.f. values of the acetylated models mapped to the experimental starting structures of Cx50 (left), Cx46 (center) and Cx26 (right). Colors correspond to r.m.s.f. amplitudes: 0 – 1.0 Å (cyan), 1.0 – 2.0 Å (yellow-orange), > 2.0 Å (red). In panel e, a single NTH domain is circled and the average r.m.s.f. values and 95% confidence intervals (student t-test) calculated over the NTH domain of each isoform are displayed (n = 12 subunits). The NTH domain of Cx26 shows significantly higher r.m.s.f. values, for both acetylated and non-acetylated models (P < 0.0001).
Figure 8: Modulation of coulombic surface potential and K+/Cl− PMFs resulting from n-terminal acetylation and Cx46/50 heterotypic/heteromeric assembly.
(a) Coulombic surface potential maps (top) and potential-of-mean-force (PMF, ΔG kcal mol−1) (bottom) obtained for a set of non-acetylated and acetylated Cx50/Cx50(Ace) and Cx46/Cx46(Ace) models characterized by MD simulation. Acetylated Cx50(Ace) and Cx46(Ace) monomers were used to construct a heterotypic channel and two different heteromeric channels (labeled, I and II). Icons at the top of each structure show the relative configurations of Cx50 (black) and Cx46 (white). The resulting coulombic surface potentials are colored as in Fig. 3a (negative – red, neutral – white and positive – blue). Only 8 subunits are shown to portray both the channel pore and subunit interfaces. An asterisk (panels a-b) indicates the site of n-terminal acetylation, which neutralizes the positively charged n-terminus. PMFs obtained for K+ (blue traces) and Cl− ions (red traces) are displayed directly beneath each model displayed in panel a. Free energy maxima are labeled and pore axis (z-axis) is indicated. Traces indicate symmetrized values, with unsymmetrized values in lighter shading. In the case of the asymmetric Cx46/50 heterotypic model (middle left), PMFs represent the average from the first and last ~60ns of simulation. (b) Coulombic surface potential (top) and PMFs (bottom) obtained for non-acetylated (left) and acetylated (right) Cx26/Cx26(Ace) are displayed as in panel a. (c) Table of peak free energy barriers for K+ (ΔGK+) and Cl− (ΔGCl-) and corresponding ΔΔG, reported as a proxy for charge selectivity. Asterisk indicates models with n-terminus acetylated. (d,e) Validation of methods used to construct the PMFs. (d) Comparison of K+ PMFs obtained for Cx50(Ace) and Cx46(Ace) using population states (left) or transition rates (right) (see Methods). Both methods yielded similar PMF profiles. All other PMFs were constructed using the transition rates method. (e) Comparison of Cl− PMFs obtained for Cx46(Ace) using transition rates of Cl− ions that diffused into the pore (left) and those that were randomly seeded within the pore (right). All other Cl− PMFs were constructed using the distributed seeding approach to enhance sampling (see Methods).
Figure 9: Analysis of K+ trajectories and putative binding site observed during MD simulation.
(a–d) K+ ion trajectories obtained for Cx50 and Cx46 along the channel pore (z-axis). (a,b) Representative traces of mobile ions transiting and exiting/entering the channel pore in both acetylated (panel a) and non-acetylated (panel b) models of Cx46 and Cx50. (c,d) K+ ions displaying long dwell times (~10–20 ns) localized at a putative binding site(s) within the channel pore (asterisk at ~14 Å, z-axis) in both models of Cx46 and Cx50. (a-d) Similar results were observed from 6 independent runs using non-acetylated models (1 × 80ns and 2 × 10ns runs for both Cx50 and Cx46) and 13 independent runs using the acetylated models (1 × 80 ns and 6 × 10ns runs for Cx50; and 1 × 80ns and 7 × 10ns runs for Cx46). (e) Representative snapshot showing a zoom view of the putative K+ binding site identified for Cx50 and Cx46, corresponding to the asterisked region in panels c and d. A single K+ ion is bound by a conserved set of amino acids (amongst Cx46/50 orthologs), coordinated by the sidechain carboxylate of Glu62 and backbone carbonyls of Gln49, Ser50 and Phe52 (identical in Cx46 and Cx50). Two transient water molecules observed coordinating the bond K+ ion are shown. A total of twelve binding sites are present within the dodecameric channel. Similar behavior was observed from simulations using both non-acetylated and acetylated models (a total of 19 independent simulations). A functional role for this putative binding site is not yet clear, but may represent a physiologically relevant cation binding site similar to the recently proposed Ca2+-binding site in Cx2611.
Figure 10: Dynamic hydrogen bond network within the NTH domain observed by MD simulation.
(a-e) Analysis of hydrogen bond interactions for non-acetylated models of Cx46 and Cx50 observed during MD simulation. (a) Inset, shows a zoom-view of D3 pairing with the positively charged n-terminal G2 position from a neighboring subunit (inter-molecular) and with the hydroxyl of S5 within the same subunit (intra-molecular). The D3-G2 interactions are dynamically formed and broken during MD simulation, while the intra-molecular D3-S5 h-bond is relatively stable (as shown in panels b–e). (b) Population statistics of inter-atomic distances involving D3 (Cγ) and G (N) of the neighboring chain and (c) Ser5 (Hγ) of the same chain, extracted from MD simulation production runs of Cx46 (blue histogram) and Cx50 (red histogram). For D3 and G2, heavy atoms were chosen as proxies to monitor hydrogen bonding interactions involving equivalent rotameric donor-acceptor configurations. The population centered at ~3.6 Å (panel b) and ~2.8 Å (panel c) are considered to be within hydrogen bond distance. (d) Trajectories extracted from MD simulation of Cx46 (blue traces) and Cx50 (red traces) showing the dynamical behavior of the D3-G2 inter-molecular charge paring and (e) D3-S5 intra-molecular h-bonding. The dwell times showing hydrogen bond pairing (~3.6 Å in panel d; and ~2.8 Å in panel e) are indicated with transparent grey shading. In the Cx26 crystal structure, the equivalent D2 site is modeled in hydrogen bond distance to a neighboring T5 site (Cx26 numbering)10, but this inter-molecular interaction is rapidly broken during MD simulations and does not appear to re-from within the time-scale of our MD experiments, and instead forms a stable intra-molecular interaction with T5, as described previously35 (data not shown). (f-j) Analysis of hydrogen bond interactions observed during MD simulation for Cx46(Ace) and Cx50(Ace) modeled with the n-terminal G2 position acetylated. (f) Inset, shows a zoom-view of acetylated G2(Ace) position h-bonded to the indole ring of W4 from the same subunit (intra-molecular) and the same intra-molecular D3-S5 h-bond interaction observed in the non-acetylated channel. (g) Population statistics of inter-atomic distances involving W4 (Nε) and G2(Ace) (acetyl-carbonyl) and (h) and D3 (Cγ) distance to Ser5 (Hγ) of the same chain, extracted from MD simulation production runs of Cx46(Ace) (blue histogram) and Cx50(Ace) (red histogram). (i) Trajectories extracted from MD simulation of Cx46(Ace) (blue traces) and Cx50(Ace) (red traces) showing the dynamical behavior of the W4-G2(Ace) h-bond paring and (j) D3-S5 intra-molecular h-bonding. The dwell times showing hydrogen bond pairing (~2.4 Å in panel i; and ~2.5 Å in panel j are indicated with transparent grey shading. For clarity, only the first 15 ns of the production period is shown (panels d-e and i-j). Similar results were observed from 6 independent runs using non-acetylated models (1 × 80ns and 2 × 10ns runs for both Cx50 and Cx46) and 13 independent runs using the acetylated models (1 × 80 ns and 6 × 10ns runs for Cx50; and 1 × 80ns and 7 × 10ns runs for Cx46).
Supplementary Material
Acknowledgements
We thank Tamir Gonen for early support of this work, and Tom White, Larry David, Upendra Adhikari and Barmak Mostofian for helpful discussions. We are grateful to the staff at the OHSU Multiscale Microscopy Core and Advanced Computing Center, and to William Garrick (PSU) for their assistance and training. C.V.R. and D.S.C. are supported by funding from the European Research Council (No. 695511-ENABLE). D.M.Z. and C.C.Y. are supported by the Center for Spatial Systems Biomedicine at OHSU. S.L.R. is supported by the Medical Research Foundation of Oregon and the National Institutes of Health (R35-GM124779).
Footnotes
Competing Interest
The authors declare no competing interests.
References
- 1.Goodenough DA & Paul DL Gap junctions. Cold Spring Harbor perspectives in biology 1, a002576, doi: 10.1101/cshperspect.a002576 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delmar M, Laird DW, Naus CC, Nielsen MS, Verselis VK, White TW Connexins and Disease. Cold Spring Harb Perspect Biol. September 4;10(9). pii: a029348 10.1101/cshperspect.a029348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia IE et al. Connexinopathies: a structural and functional glimpse. BMC Cell Biol 17 Suppl 1, 17, doi: 10.1186/s12860-016-0092-x (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aasen T, Mesnil M, Naus CC, Lampe PD & Laird DW Gap junctions and cancer: communicating for 50 years. Nat Rev Cancer 16, 775–788, doi: 10.1038/nrc.2016.105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosinsky GE & Nicholson BJ Structural organization of gap junction channels. Biochim Biophys Acta 1711, 99–125, doi: 10.1016/j.bbamem.2005.04.001 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Sohl G & Willecke K Gap junctions and the connexin protein family. Cardiovasc Res 62, 228–232, doi: 10.1016/j.cardiores.2003.11.013 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Cottrell GT & Burt JM Functional consequences of heterogeneous gap junction channel formation and its influence in health and disease. Biochim Biophys Acta 1711, 126–141, doi: 10.1016/j.bbamem.2004.11.013 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Beyer EC & Berthoud VM Gap junction structure: unraveled, but not fully revealed. F1000Res 6, 568, doi: 10.12688/f1000research.10490.1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosely R & Sorgen PL A history of gap junction structure: hexagonal arrays to atomic resolution. Cell Commun Adhes 20, 11–20, doi: 10.3109/15419061.2013.775256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda S et al. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458, 597–602, doi: 10.1038/nature07869 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Bennett BC et al. An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels. Nat Commun 7, 8770, doi: 10.1038/ncomms9770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathias RT, White TW & Gong X Lens gap junctions in growth, differentiation, and homeostasis. Physiol Rev 90, 179–206, doi: 10.1152/physrev.00034.2009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konig N & Zampighi GA Purification of bovine lens cell-to-cell channels composed of connexin44 and connexin50. J Cell Sci 108 (Pt 9), 3091–3098 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Jiang JX & Goodenough DA Heteromeric connexons in lens gap junction channels. Proc Natl Acad Sci U S A 93, 1287–1291 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shearer D, Ens W, Standing K & Valdimarsson G Posttranslational modifications in lens fiber connexins identified by off-line-HPLC MALDI-quadrupole time-of-flight mass spectrometry. Invest Ophthalmol Vis Sci 49, 1553–1562, doi: 10.1167/iovs.07-1193 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Wang Z & Schey KL Phosphorylation and truncation sites of bovine lens connexin 46 and connexin 50. Exp Eye Res 89, 898–904, doi: 10.1016/j.exer.2009.07.015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon T et al. Molecular dynamics simulations of the Cx26 hemichannel: insights into voltage-dependent loop-gating. Biophys J 102, 1341–1351, doi: 10.1016/j.bpj.2012.02.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai D Structural analysis of key gap junction domains--Lessons from genome data and disease-linked mutants. Semin Cell Dev Biol 50, 74–82, doi: 10.1016/j.semcdb.2015.11.015 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Koval M, Molina SA & Burt JM Mix and match: investigating heteromeric and heterotypic gap junction channels in model systems and native tissues. FEBS Lett 588, 1193–1204, doi: 10.1016/j.febslet.2014.02.025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unger VM, Kumar NM, Gilula NB & Yeager M Three-dimensional structure of a recombinant gap junction membrane channel. Science 283, 1176–1180 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Fleishman SJ, Unger VM, Yeager M & Ben-Tal N A Calpha model for the transmembrane alpha helices of gap junction intercellular channels. Mol Cell 15, 879–888, doi: 10.1016/j.molcel.2004.08.016 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Gong XQ & Nicholson BJ Size selectivity between gap junction channels composed of different connexins. Cell Commun Adhes 8, 187–192 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Goldberg GS, Valiunas V & Brink PR Selective permeability of gap junction channels. Biochim Biophys Acta 1662, 96–101, doi: 10.1016/j.bbamem.2003.11.022 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Varland S, Osberg C & Arnesen T N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401, doi: 10.1002/pmic.201400619 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berneche S & Roux B Energetics of ion conduction through the K+ channel. Nature 414, 73–77, doi: 10.1038/35102067 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Corry B & Thomas M Mechanism of ion permeation and selectivity in a voltage gated sodium channel. J Am Chem Soc 134, 1840–1846, doi: 10.1021/ja210020h (2012). [DOI] [PubMed] [Google Scholar]
- 27.Ulmschneider MB et al. Molecular dynamics of ion transport through the open conformation of a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A 110, 6364–6369, doi: 10.1073/pnas.1214667110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srinivas M et al. Voltage dependence of macroscopic and unitary currents of gap junction channels formed by mouse connexin50 expressed in rat neuroblastoma cells. J Physiol 517 ( Pt 3), 673–689 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopperstad MG, Srinivas M & Spray DC Properties of gap junction channels formed by Cx46 alone and in combination with Cx50. Biophys J 79, 1954–1966, doi: 10.1016/S0006-3495(00)76444-7 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh S, Rubin JB, Bennett MV, Verselis VK & Bargiello TA Molecular determinants of electrical rectification of single channel conductance in gap junctions formed by connexins 26 and 32. J Gen Physiol 114, 339–364 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trexler EB, Bukauskas FF, Kronengold J, Bargiello TA & Verselis VK The first extracellular loop domain is a major determinant of charge selectivity in connexin46 channels. Biophys J 79, 3036–3051, doi: 10.1016/S0006-3495(00)76539-8 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong X et al. The First Extracellular Domain Plays an Important Role in Unitary Channel Conductance of Cx50 Gap Junction Channels. PLoS One 10, e0143876, doi: 10.1371/journal.pone.0143876 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh S, Verselis VK & Bargiello TA Charges dispersed over the permeation pathway determine the charge selectivity and conductance of a Cx32 chimeric hemichannel. J Physiol 586, 2445–2461, doi: 10.1113/jphysiol.2008.150805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kronengold J, Trexler EB, Bukauskas FF, Bargiello TA & Verselis VK Pore-lining residues identified by single channel SCAM studies in Cx46 hemichannels. Cell Commun Adhes 10, 193–199 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zonta F, Polles G, Zanotti G & Mammano F Permeation pathway of homomeric connexin 26 and connexin 30 channels investigated by molecular dynamics. J Biomol Struct Dyn 29, 985–998, doi: 10.1080/073911012010525027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez W et al. Mechanism of gating by calcium in connexin hemichannels. Proc Natl Acad Sci U S A 113, E7986–E7995, doi: 10.1073/pnas.1609378113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris AL & Contreras JE Motifs in the permeation pathway of connexin channels mediate voltage and Ca (2+) sensing. Front Physiol 5, 113, doi: 10.3389/fphys.2014.00113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivas M, Calderon DP, Kronengold J & Verselis VK Regulation of connexin hemichannels by monovalent cations. J Gen Physiol 127, 67–75, doi: 10.1085/jgp.200509397 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trexler EB, Bennett MV, Bargiello TA & Verselis VK Voltage gating and permeation in a gap junction hemichannel. Proc Natl Acad Sci U S A 93, 5836–5841 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchyna TM et al. Different ionic selectivities for connexins 26 and 32 produce rectifying gap junction channels. Biophys J 77, 2968–2987, doi: 10.1016/S0006-3495(99)77129-8 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veenstra RD Size and selectivity of gap junction channels formed from different connexins. J Bioenerg Biomembr 28, 327–337 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Veenstra RD et al. Selectivity of connexin-specific gap junctions does not correlate with channel conductance. Circ Res 77, 1156–1165 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Nicholson BJ et al. The molecular basis of selective permeability of connexins is complex and includes both size and charge. Braz J Med Biol Res 33, 369–378 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Ebihara L, Xu X, Oberti C, Beyer EC & Berthoud VM Co-expression of lens fiber connexins modifies hemi-gap-junctional channel behavior. Biophys J 76, 198–206, doi: 10.1016/S0006-3495(99)77189-4 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xin L & Bai D Functional roles of the amino terminal domain in determining biophysical properties of Cx50 gap junction channels. Front Physiol 4, 373, doi: 10.3389/fphys.2013.00373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo Y, Rossi AR & Harris AL Computational Studies of Molecular Permeation through Connexin26 Channels. Biophys J 110, 584–599, doi: 10.1016/j.bpj.2015.11.3528 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon T, Harris AL, Rossi A & Bargiello TA Molecular dynamics simulations of the Cx26 hemichannel: evaluation of structural models with Brownian dynamics. J Gen Physiol 138, 475–493, doi: 10.1085/jgp.201110679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verselis VK, Ginter CS & Bargiello TA Opposite voltage gating polarities of two closely related connexins. Nature 368, 348–351, doi: 10.1038/368348a0 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Peracchia C & Peracchia LL Inversion of both gating polarity and CO2 sensitivity of voltage gating with D3N mutation of Cx50. Am J Physiol Cell Physiol 288, C1381–1389, doi: 10.1152/ajpcell.00348.2004 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Srinivas M, Kronengold J, Bukauskas FF, Bargiello TA & Verselis VK Correlative studies of gating in Cx46 and Cx50 hemichannels and gap junction channels. Biophys J 88, 1725–1739, doi: 10.1529/biophysj.104.054023 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xin L, Nakagawa S, Tsukihara T & Bai D Aspartic acid residue D3 critically determines Cx50 gap junction channel transjunctional voltage-dependent gating and unitary conductance. Biophys J 102, 1022–1031, doi: 10.1016/j.bpj.2012.02.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beyer EC, Ebihara L & Berthoud VM Connexin mutants and cataracts. Front Pharmacol 4, 43, doi: 10.3389/fphar.2013.00043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pascolini D & Mariotti SP Global estimates of visual impairment: 2010. Br J Ophthalmol 96, 614–618, doi: 10.1136/bjophthalmol-2011-300539 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Shiels A & Hejtmancik JF Mutations and mechanisms in congenital and age-related cataracts. Exp Eye Res 156, 95–102, doi: 10.1016/j.exer.2016.06.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kistler J, Christie D & Bullivant S Homologies between gap junction proteins in lens, heart and liver. Nature 331, 721–723, doi: 10.1038/331721a0 (1988). [DOI] [PubMed] [Google Scholar]
- 56.Kistler J, Schaller J & Sigrist H MP38 contains the membrane-embedded domain of the lens fiber gap junction protein MP70. The Journal of biological chemistry 265, 13357–13361 (1990). [PubMed] [Google Scholar]
- 57.White TW, Bruzzone R, Goodenough DA & Paul DL Mouse Cx50, a functional member of the connexin family of gap junction proteins, is the lens fiber protein MP70. Mol Biol Cell 3, 711–720 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reichow SL et al. Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat Struct Mol Biol 20, 1085–1092, doi: 10.1038/nsmb.2630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gold MG et al. AKAP2 anchors PKA with aquaporin-0 to support ocular lens transparency. EMBO Mol Med 4, 15–26, doi: 10.1002/emmm.201100184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichow SL & Gonen T Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure 16, 1389–1398, doi: 10.1016/j.str.2008.06.011 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shevchenko A, Tomas H, Havlis J, Olsen JV & Mann M In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1, 2856–2860, doi: 10.1038/nprot.2006.468 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Yang B et al. Identification of cross-linked peptides from complex samples. Nat Methods 9, 904–906, doi: 10.1038/nmeth.2099 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Myers JB et al. The CaMKII holoenzyme structure in activation-competent conformations. Nat Commun 8, 15742, doi: 10.1038/ncomms15742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46, doi: 10.1016/j.jsb.2006.05.009 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51, doi: 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voss NR, Yoshioka CK, Radermacher M, Potter CS & Carragher B DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol 166, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, doi: 10.1016/j.jsb.2012.09.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heymann JB & Belnap DM Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157, 3–18, doi: 10.1016/j.jsb.2006.06.006 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 72.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21, doi: 10.1107/S0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 75.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J Mol Graph 14, 33–38, 27–38 (1996). [DOI] [PubMed] [Google Scholar]
- 76.Locke D, Bian S, Li H & Harris AL Post-translational modifications of connexin26 revealed by mass spectrometry. Biochem J 424, 385–398, doi: 10.1042/BJ20091140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grubmuller H, Heymann B & Tavan P Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science 271, 997–999 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Phillips JC et al. Scalable molecular dynamics with NAMD. J Comput Chem 26, 1781–1802, doi: 10.1002/jcc.20289 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klauda JB et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B 114, 7830–7843, doi: 10.1021/jp101759q (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Best RB et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput 8, 3257–3273, doi: 10.1021/ct300400x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zuckerman DM Statistical Physics of Biomolecules: An Introduction. (CRC Press; 1 edition, 2010). [Google Scholar]
- 82.Im W, Seefeld S & Roux B A Grand Canonical Monte Carlo-Brownian dynamics algorithm for simulating ion channels. Biophys J 79, 788–801, doi: 10.1016/S0006-3495(00)76336-3 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krissinel E & Henrick K Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797, doi: 10.1016/j.jmb.2007.05.022 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CryoEM density maps have been deposited to the Electron Microscopy Data Bank (EMD-9116). Coordinates for Cx46 and Cx50 atomic models have been deposited to the Protein Data Bank (6MHQ and 6MHY). The original multi-frame micrographs have been deposited to EMPIAR (EMPIAR-10212).