Abstract
The DNA repair enzyme poly(ADP-ribose) polymerase 1 (PARP-1) is overexpressed in glioblastoma, with overall low expression in healthy brain tissue. Paired with the availability of specific small molecule inhibitors, PARP-1 is a near-ideal target to develop novel radiotherapeutics to induce DNA damage and apoptosis in cancer cells, while sparing healthy brain tissue. Methods: We synthesized an 131I-labeled PARP-1 therapeutic and investigated its pharmacology in vitro and in vivo. A subcutaneous tumor model was used to quantify retention times and therapeutic efficacy. A potential clinical scenario, intratumoral convection-enhanced delivery, was mimicked using an orthotopic glioblastoma model combined with an implanted osmotic pump system to study local administration of 131I-PARPi (PARPi is PARP inhibitor). Results: 131I-PARPi is a 1(2H)-phthalazinone, similar in structure to the Food and Drug Administration–approved PARP inhibitor AZD-2281. In vitro studies have shown that 131I-PARPi and AZD-2281 share similar pharmacologic profiles. 131I-PARPi delivered 134.1 cGy/MBq intratumoral injected activity. Doses to nontarget tissues, including liver and kidney, were significantly lower. Radiation damage and cell death in treated tumors were shown by p53 activation in U87-MG cells transfected with a p53-bioluminescent reporter. Treated mice showed significantly longer survival than mice receiving vehicle (29 vs. 22 d, P < 0.005) in a subcutaneous model. Convection-enhanced delivery demonstrated efficient retention of 131I-PARPi in orthotopic brain tumors, while quickly clearing from healthy brain tissue. Conclusion: Our results demonstrate 131I-PARPi’s high potential as a therapeutic and highlight PARP’s relevance as a target for radionuclide therapy. Radiation plays an integral role in brain tumor therapy, and radiolabeled PARP therapeutics could ultimately lead to improvements in the standard of care.
Keywords: PARP, radiotherapeutic, 131I-PARPi, 131I, convection enhanced delivery (CED)
Glioblastoma is the most common primary brain tumor in adults, with more than 12,000 diagnoses per year (1). Current standard treatment consists of maximal surgical resection followed by chemotherapy (temozolomide) and external beam radiation. This, however, only minimally extends median survival (2), and most patients develop recurrent tumors within months (3,4). The diffuse growth pattern is the fundamental reason why surgical resection and external beam radiotherapy are insufficient, because highly dispersed glioblastoma cells would require larger tumor margins to be treated and resected, which can result in unacceptably high healthy tissue loss and significant morbidities. A selective cellular-based approach would provide significant advantages over conventional therapy.
Recently, progress has been made toward the delivery of targeted therapeutics to central nervous system tumors, especially using convection enhanced delivery (CED) (5,6). This strategy uses catheters to infuse therapeutics directly into the affected brain tissue, where they are distributed by convective rather than diffusive properties. This approach offers considerable advantages for delivery and tissue penetrance of therapeutic antibodies, virus vectors, and cell-based therapeutics. Using this technology, even radiolabeled antibodies, which normally have limited tissue penetration, can be delivered (7), potentially improving therapeutic efficacy. Nevertheless, because of the heterogeneous nature of the disease (8), further enhancement of delivery and tissue penetration is necessary, which could be achieved through CED of targeted small-molecule radiotherapeutics.
One target for use in CED and targeted radiation therapy is poly(ADP-ribose) polymerase (PARP-1). PARP-1 is a fundamentally important member of the cellular DNA repair machinery that highly proliferative cancer cells rely on to maintain genomic integrity through an accelerated cell cycle (9). PARP-1 expression was reported in the nucleoli of neurons, oligodentritic cells, and astrocytes as well as the Purkinje cell layer in the cerebellum and the dentate gyrus (10). Nevertheless, it has been shown that malignant glial growths have elevated PARP-1 expression compared with healthy pediatric and adult brain tissue (11), forming an ideal foundation for CED-based therapeutics. Similarly important, PARP inhibitors not only quickly distribute and bind within PARP-expressing cells, but also simultaneously washout effectively from healthy tissues, resulting in high target-to-background contrast (10,12), potentially providing a large treatment window for CED therapy (7). Recently, advances have been made in PARP-targeted molecular imaging, and much of this work has focused on glioblastoma (13,14) and other central nervous system tumors (10). Intuitively, replacing a fluorescent or PET active imaging tag with a radiotherapeutic isotope would take advantage of the high specificity exhibited by the PARP-targeted imaging agents as well as the high PARP expression seen in glioblastoma. More information on the relevant chemistry and PARP inhibitors used for imaging are provided in comprehensive reviews (12,15).
In this study, we use 131I-PARPi, a small-molecule derived from an inhibitor screening library (14), and explore and develop its value as a CED agent for glioblastoma therapy. We found that the binding profile of 131I-PARPi matches that of other PARP inhibitors and imaging agents. In a mouse model of glioblastoma, we investigate the pharmacokinetics of 131I-PARPi and estimate the delivered radiotherapeutic doses. We show that p53 expression was activated by 131I-PARPi and that both single and fractionated doses extend the overall survival of tumor-bearing mice. In an orthotopic model of glioblastoma, we corroborate that high levels of 131I-PARPi accumulate via CED, whereas healthy brain tissue only retains the small molecule at low levels. Taken together, our results show that 131I-PARPi is a promising radiotherapeutic small molecule and could potentially improve glioblastoma therapy.
MATERIALS AND METHODS
General
Unless specified otherwise, all reagents were purchased from Sigma-Aldrich and used as received. 131I-Na in 0.1N NaOH with a specific activity more than 4,600 TBq/g was purchased from Nordion. 4-(4-fluoro-3-(4-(3-iodobenzoyl)piperazine-1-carbonyl)benzyl)phthalazin-1(2H)-one was synthesized as described previously (14). Olaparib (AZD-2281) was purchased from LC Laboratories. PARPi-FL was synthesized as previously described (16,17). Water (>18.2 MΩcm−1 at 25°C) was obtained from an Alpha-Q Ultrapure water system (Millipore) and acetonitrile (AcN) as well as ethanol (EtOH) were of high-performance liquid chromatography (HPLC) grade purity. Sterile 0.9% saline solution (Hospira) was used for all in vivo injections. HPLC purification and analysis were performed on a Shimadzu UFLC HPLC system equipped with a DGU-20A degasser, a SPD-M20A UV detector, a LC-20AB pump system, and a CBM-20A Communication BUS module. A LabLogic Scan-RAM radio–thin-layer chromatography/HPLC detector was used to detect activity. HPLC solvents (buffer A: water, buffer B: AcN) were filtered before use. HPLC analysis and purification were performed on a reversed-phase C18 Waters Atlantis T3 column (C18-RP, 5 μm, 6 mm, 250 mm). Purification of the iodinated benzoic acid was performed with method 1 (flow rate: 1 mL/min; gradient: 20 min 5%-95% B; 25 min 100% B; 26 min 100%-5% B); quality control analysis was performed with method 1. Purification of the final product was performed on a C6 Waters Spherisorb column (C6, 5 μm, 4.6 mm × 250 mm) with method 2 (flow rate: 1.5 mL/min; isocratic: 0–30 min 35% B). SPECT imaging was performed on a NanoSPECT/CT from Mediso Medical Imaging Systems. PET imaging experiments were conducted on a Focus 120 MicroPET (Concorde Microsystems). Digital phosphor autoradiography was obtained using a Typhoon FLA 7000 laser scanner from GE Healthcare. γ-counting and biodistributions were performed using a WIZARD2 automatic γ-counter (PerkinElmer).
Synthesis of 127I-PARPi
4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazin-1(2H)-one (20 mg, 54.5 μmol) was dissolved in 1 mL of AcN and added to 10 mg (41.3 μmol) of 3-iodobenzoic acid, followed by 25 mg (66 μmol) of N,N,N′,N′-tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) and 20 μL (145.5 μmol) of Et3N. The reaction mixture was stirred for 30 min and purified by HPLC (method 2) to yield the title compound (13.4 mg, 22.3 μmol, 67%). 1H-NMR (CDCl3) δ = 10.48 (s, 1H), 8.40–8.39 (m, 1H), 7.74–7.66 (m, 5H), 7.27–7.26 (d, 2H), 7.09–7.07 (d, 2H), 4.22 (s, 2H), 3.73–3.14 (m, 8H). LC-ESI-MS (+) m/z = 597.1 [M+H+]+. HRMS-ESI [M-H+]−m/z calculated for [C27H22FIN4O3]−595.0642, found 595.0640.
Synthesis of 18F-PARPi
18F-PARPi was synthesized as described by Carney et al. (18). Briefly, a QMA cartridge containing cyclotron-produced 18F-fluoride (n.c.a.) was eluted with a solution containing 9 mg of Kryptofix [2.2.2] (4,7,13,16,21,24-hexaoxa-1,10-diazabicyclo[8.8.8]hexacosane), 0.08 mL of 0.15 M K2CO3, and 1.92 mL of AcN into a 5-mL reaction vial. Then, ethyl 4-nitrobenzoate (500 μg in 100 μL of dimthyl sulfoxide [DMSO]) was added to the azeotropically dried 18F− and heated to 150°C (15 min). After this time, 50 μL of 1M NaOH were added followed by HCl (1M, 50 μL) 1 min later. Then, 2 mg of 4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazin-1(2H)-one (in 100 μL of DMSO) were added followed by 10 mg of HBTU in 100 μL of DMSO. Finally, 20 μL of Et3N were added and the reaction mixture was further diluted with 400 μL of AcN and 1 mL of H2O. The crude mixture was purified by HPLC. The decay-corrected radiochemical yield for the final step was 38.4 ± 2.5% with a molar activity (MA) of 35.9 ± 15.2 GB/μmol. The purified final compound was formulated with 10% EtOH/0.9% NaCl 0.9%.
Synthesis of 131I-PARPi
131I-PARPi was obtained in a manner similar to synthetic procedures reported before (14). First, 131I-NHS-benzoate was obtained by adding N-succinimidyl-4-(tributylstannyl) benzoate (30 μg, 5.9 μmol) in 30 μL of AcN to a solution containing methanol (40 μL), chloramine T (6 μg, 30 nmol), acetic acid (2 μL), and 131I-NaI in NaOH 0.1 M (185–370 MBq [5–10 mCi]). After the reaction solution was driven for 15 min at room temperature, the reaction was purified by HLPC (method 1), and 131I-NHS-benzoate collected at 15.1 min. The collected purified fraction was concentrated to dryness in vacuum, reconstituted in a solution of 80 μL of AcN, and added to a solution of 4-(4-fluoro-3-(piperazine-1-carbonyl)benzyl)phthalazin-1(2H)-one (0.3 mg, 0.9 μmol) in 20 μL of DMSO and HBTU (0.3 mg, 0.8 μmol) in 20 μL of DMSO; 10 μL of 2,6-lutidine were further added and the reaction mixture stirred in an Eppendorf ThermoMixer overnight at 65°C (500 rpm). The following day, the reaction mixture was injected and purified by HPLC (method 2) and the final 131I-PARPi was collected at room temperature for 13.1 min (radiochemical yield > 70%; radiochemical purity > 99%) and concentrated to dryness under vacuum. 131I-PARPi was formulated with 30% PEG300/70% saline (0.9% NaCl) for both in vitro and in vivo assays (MA = 1.5 ± 0.3 GBq/μmol). Coelution with nonradioactive 127I-PARPi reference compound confirmed the identity of the radiotherapeutic.
PARP Inhibitor Binding Assays
Assay was performed by BPS Bioscience. DMSO solutions (100 μM) of each inhibitor were provided at a 100 nM, 50 nM, or 10 nM concentration. Measurements for each enzyme combined with each inhibitor were performed in triplicate in accordance with the BPS assay kit protocol (19). Luminescence was measured using a BioTek Synergy 2 microplate reader. Data were reported by BPS as percentage enzyme activity. Triplicates were combined to get a mean ± SD, which was then grouped and plotted onto a heat map using GraphPad Prism 7 (GraphPad Software).
Cell Culture
The human glioblastoma cell line U251 MG was kindly provided by the Laboratory of Dr. Ronald Blasberg (Memorial Sloan Kettering Cancer Center [MSKCC]). Cell lines were grown in Eagle’s minimal essential medium (MEM), 10% (vol/vol) heat-inactivated fetal bovine serum, penicillin 100 IU2, and streptomycin (100 μg/mL), purchased from the culture medium preparation facility at MSKCC.
Transduction Procedure of U87 Cell Line
U87 human glioblastoma cells were grown in MEM with 10% fetal calf serum. The retroviral transient producer cell line H29 was maintained in Dulbecco MEM supplemented with 10% fetal calf serum, puromycin (2 μg/mL), and tetracycline (1 μg/mL), transfected with Cis-p53/tdTomato-CBRluc-Neo and incubated for 48 h. The U87 cells were transduced with the Cis-p53/tdTomato-CBRluc-Neo later using H29-produced virus by incubating 30% confluent tumor cell culture with retroviral vector-containing medium for 48 h. Transduced U87-p53/tdTomato-CBRluc-Neo cells (short: U87-p53) were selected by culturing in MEM, supplemented with 10% fetal calf serum and G418 sulfate (0.5 mg/mL) (Corning, catalog no. 30-234-CR). Several fluorescence-activated cell sorting (FACS) sorting procedures were performed to enrich cells with a low background of p53 activity. Selected cells were additionally transduced with a second constitutively expressed SFG-Turquoise/rsRluc virus as described above.
Cell-Internalization, Targeting Specificity, and Binding Properties
U251 MG cells were adhered overnight in 6-well plates (1 million cells/well) and washed twice with phosphate-buffered saline (PBS) prior to addition of 131I-PARPi (222 kBq/well, 0.15 nmol) at 37°C for 0, 15, 30, 45, 60, 120, and 240 min in MEM to allow for binding. Binding specificity was challenged with a 50-fold excess of olaparib (3.26 μg, 7.5 nmol). Next, at each time point, cells were first washed twice with PBS 1× and then lysed by incubation with 1 M NaOH (10 min at 37°C). Finally, the resulting lysate in each well was collected and the radioactivity measured in an automated γ-counter. Targeting specificity was assessed by a competitive binding assay between 127I-PARPi and PARPi-FL. U251 MG cells (50,000 cells/well) were plated 48 h before the experiment. On the day of the experiment, binding affinity was determined by coincubating the plated cells (37°C, 40 min) with serial dilutions of 127I-PARPi, ranging from 0 to 10 μM and 0.5 μM PARPi-FL. After this time, cells were washed twice with PBS 1×, fixed, and counterstained with Hoechst DNA stain. PARPi-FL uptake and blocking effects were observed and measured by confocal microscopy.
Mice
Female athymic nude CrTac:NCr-Fo mice were purchased from Taconic Laboratories at age 6–8 wk. Xenograft-bearing mice were used to determine all pharmacokinetics, the binding/imaging properties, and the treatment efficacy of the radiotherapeutic (ntotal = 98). During subcutaneous injections, mice were anesthetized using 2% isoflurane gas in 2 L/min medical air. During orthotopic injections, mice were anesthetized using a 150 mg/kg ketamine and 15 mg/kg xylazine cocktail (10 μL/g of body weight). For intravenous injections, the lateral tail vein was used. Mice were warmed with a heat lamp and placed in a restrainer, and the tail was sterilized with alcohol pads before injection. U251 MG or U87-p53 cells were implanted subcutaneously (5 × 106 cells in 150 μL of 1:1 PBS/Matrigel [BD Biosciences]) into the right shoulder and allowed to grow for approximately 2 wk until the tumors reached about 8 mm in diameter (100 ± 8 mm). For orthotopic injections, U251 MG or U87-p53 cells (5 × 105 cells in 2 μL of PBS) were injected 2 mm lateral and 1 mm anterior to the bregma using a Stoelting Digital New Standard Stereotaxic Device and a 5-μL Hamilton syringe and allowed to grow for approximately 3 wk. All mouse experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of MSKCC and followed National Institutes of Health guidelines for animal welfare.
SPECT/CT
SPECT/CT scans were obtained using a small-animal NanoSPECT/CT from Mediso Medical Imaging Systems. For subcutaneous U251 MG xenografts, 131I-PARPi (11.1 MBq, 1.5 GBq/μmol in 20 μL of 30% PEG300 in 0.9% sterile saline) was administered intratumorally (n = 3). At 0 and 6 h after injection, the mice were anesthetized with 1.5%–2.0% isoflurane (Baxter Healthcare) at 2 mL/min in oxygen, and SPECT/CT data were acquired for 20 min. For orthotopic U251 MG tumor–bearing mice, 131I-PARPi (1.9 MBq, 1.5 GBq/μmol in 5 μL of 30% PEG300 in 0.9% sterile saline) was injected intracranially using the same coordinates as for tumor cell injection (2 mm lateral and 1 mm anterior to the bregma using a Stoelting Digital New Standard Stereotaxic Device and a 5-μL Hamilton syringe). At 24 h after injection, the mice were anesthetized with 1.5%–2.0% isoflurane (Baxter Healthcare) at 2 mL/min in oxygen, and SPECT/CT data were collected for 20 min in the head region.
Autoradiography and Hematoxylin and Eosin Staining
Twenty-four hours after intratumoral injection of 131I-PARPi or olaparib/131I-PARPi in U251 MG subcutaneous xenograft–bearing mice, mice were sacrificed and tumors were harvested. The collected organs were embedded in Tissue-Tek O.C.T. compound (Sakura Finetek), flash-frozen in liquid nitrogen, and cut into 20-μm sections using a Vibratome UltraPro 5000 Cryostat (Vibratome). A storage phosphor autoradiography plate (BAS-MS2325; Fiji Photo Film) was exposed to the tissue slices overnight at −20°C and read the following day. Relative count intensity of the sections in each image was quantified using ImageJ 1.47u processing software (NIH). Tumor-to-muscle and brain-to-muscle ratios were calculated using Prism 6.0c (GraphPad Software). Sections were subsequently subjected to hematoxylin and eosin staining for morphologic evaluation of tissue pathology.
PET/CT Imaging
Small-animal PET imaging data were recorded on a Focus 120 MicroPET (Concorde Microsystems) and reconstructed using AsiPro VM MicroPET Analysis software (Siemens Medical Solutions). 18F-PARPi (5.55 MBq, 35.89 ± 15.17 GBq/μmol in 300 μL, 10% EtOH in 0.9% sterile saline) alone or following an intratumoral injection of 127I-PARPi (10 μg, 16.5 μmol, 20 μL, 30% PEG300/70% NaCl 0.9% 6 h before 18F-PARPi injection) was injected into U251 MG tumor–bearing mice (n = 3) via the tail vein. At 2 h after injection, the mice were anesthetized with 1.5%–2.0% isoflurane (Baxter Healthcare) at 2 mL/min in oxygen, and PET/CT imaging was performed over 15 min. Images were analyzed using AsiPro VM software (Concorde Microsystems). Then, mice were sacrificed and key organs harvested for quantification and autoradiography/hematoxylin and eosin staining.
Ex Vivo Biodistribution and Dosimetry (Subcutaneous Model)
Biodistribution studies were performed in subcutaneous U251 MG xenograft–bearing athymic nude mice (n = 44, 11 randomized cohorts, 4 mice per cohort). Mice were divided into 2 groups (blocked and unblocked), and 131I-PARPi (0.56 MBq, 1.5 GBq/μmol in 20 μL, 30% PEG300 in 0.9% sterile saline) was administered intratumorally. The blocked group was preinjected 30 min before 131I-PARPi with olaparib (8.15 μg, 18 nmol in 100 μL, 30% PEG300/70% NaCl 0.9%). Mice were sacrificed by CO2 asphyxiation at 1, 3, 6, 12, 24, 48, 72, and 96 h after injection (for 131I-PARPi–only groups) and at 1, 6, and 24 h after injections (for blocking experiments groups), and major organs were collected, weighed, and counted in a WIZARD2 automatic γ-counter (PerkinElmer). The 131I-PARPi uptake was expressed as a percentage of injected dose per gram (%ID/g) using the following formula: [(activity in the target organ/g of tissue)/injected dose] × 100% (Supplemental Figs. 5 and 6; supplemental materials are available at http://jnm.snmjournals.org). The average of the 4 organ specimens from each time point was fitted to a biexponential function. The best fit for most organs comprised a 2-phase exponential clearance model. For 2 organs (the thyroid and large intestine), there was measured uptake over the first 12 h, and therefore these organs were fitted to an uptake and clearance exponential model. Cumulative uptake was calculated from the areas under the tumor uptake curves. Absorbed doses to tumor and all normal organs were estimated assuming absorbed fractions of 1 for the β-emissions of 131I and 0 for the penetrating photon emissions and using an equilibrium dose constant of 0.111 g Gy/MBq h (0.405 g cGy/μCi h). The ratio of tumor to normal organ dose exceeded 20 for all organs except the thyroid (Supplemental Fig. 4). The high dose to the thyroid gland was a likely consequence of the deiodination followed by uptake and then slow clearance (evidence of trapping in the thyroid tissue) (Supplemental Fig. 7).
Bioluminescence
Bioluminescence studies were performed in subcutaneous U87-p53 xenograft–bearing mice (n = 8, 4 mice per cohort). Mice were divided into 2 groups (treated and untreated). For treatment, mice were administered 131I-PARPi (14.8 MBq, 1.5 ± 0.3 GBq/μmol in 20 μL, 30% PEG300 in 0.9% sterile saline) intratumorally. The untreated group received an intratumoral injection of a 30% PEG300 solution in 0.9% sterile saline (20 μL). Imaging was performed on an IVIS Spectrum preclinical in vivo imaging system from PerkinElmer the day before treatment (baseline) and on day 0, 1, and 2 after treatment. For detection of the constitutively expressed SFG-Turquoise/rsRluc reporter, mice were given a retroorbital injection of 10 μg of coelenterazine h ((2-(4-dehydroxy)coelenterazine from Biotium) dissolved in 5% ethanol in 0.9% sterile saline solution and imaged within 22 s of injections. For detection of p53 activation (Cis-p53/tdTomato-CBRluc-Neo), mice were imaged 20 min after an intraperitoneal injection of 2 mg of luciferin/100 μL of sterile 0.9% NaCl solution.
Therapeutic Efficacy
Therapeutic efficacy studies were performed in subcutaneous U87-p53 xenograft–bearing mice (n = 29, 3 randomized groups, 9–10 mice per cohort). Mice were divided into 3 cohorts (vehicle, nonradioactive/cold 127I-PARPi, and 3 × 14.8 MBq of 131I-PARPi). The cold group received mass equivalent doses (6 μg) of 127I-PARPi. Vehicle, 127I-PARPi, and 3 × 131I-PARPi were administered on day 0, 3, and 6 via intratumoral injections. Body weights were recorded every Monday, Wednesday, and Friday until tumor burden exceeded the endpoint of 800 mm.
Additional supporting tables and figures can be found in the supplemental materials. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee and followed the National Institutes of Health guidelines for animal welfare.
Orthotopic CED Model
Mice received intracranial injections of U87-p53 cells (5 × 104 cells in 1.5 μL of PBS). After 3 wk, bioluminescence imaging was used to determine presence of tumor. Then, we surgically implanted an ALZET Osmotic Pump into the mice, which slowly delivered an infusion into the brain using the same coordinates as for the tumor cell injection. Both healthy and diseased mice received an ALZET Osmotic Pump (Model 1003D) with Brain Infusion Kit 3 containing 131I-PARPi (40.7 ± 11.1 MBq in 101 ± 8 μL 30% PEG300/70% NaCl 0.9%). Pumps were weighed before and after filling to calculate added volume, as per the manufacturer’s instruction. After filling pumps and assembling catheter and cannula, infusion kits were incubated in 0.9 % saline at 37°C overnight to ensure flow was initiated (1.0 μL/h [±0.15 μL/h]) and air bubbles were removed. Immediately before implantation, pumps were measured in dose calibrators to measure activity in pumps at time of surgery. SPECT/CT images were acquired 1 h after surgery and again at 12 h. Mice were imaged every 24 h for 6 days. After the fifth day, the pumps were surgically removed, and the remaining dose was measured in dose calibrators to ensure payload had been delivered. The image volumes were segmented using a combination of manual and semiautomatic segmentation techniques in 3D Slicer, version 4.6 (www.slicer.org) for quantification of absolute activity in different organs at each time point. The presence of activity levels above background in the SPECT images was not observed except in the brain/tumor and the osmotic pump.
Dosimetry (CED)
Source/Target Definition
The popular Digimouse atlas (20) was used as a digital phantom for dosimetry calculations. The atlas was adapted in the open-source 3-dimensional modeling software Blender to incorporate models of the osmotic pump, tumor, and thyroid tissues. The adapted model was tetrahedralized via the Delaunay method in the mesh generation software Tetgen to construct a 3-dimensional finite element mesh suitable for implementation in the Particle and Heavy Ion Transport Simulation (PHITS) Monte Carlo code (21). Material attributes for each region were assigned as soft tissue, lung, or bone using standard International Commission on Radiological Protection reference data for atomic composition and density (www.physics.nist.gov).
Calculation of Injected Activity
The rate of injection of 131I activity into the tumor is:
| Eq. 1 |
where, , , , and . Given Ap = A0 at t = 0, the cumulative injected activity is obtained following solution of Equation 1 using an integrating factor or transform methods:
| Eq. 2 |
where, . A0 was determined by measuring the activity present in the pump in a dose calibrator immediately prior to implantation. Ap at different time points was determined from SPECT imaging volume-of-interest analysis. The resulting time–activity curve was fit as a 1-phase exponential decay; the fitted curve was then implemented for calculation of the injected activity via Equation 2 (Supplemental Fig. 10E).
Monte Carlo Simulation
PHITS was used to generate decay events in each source via rejection sampling; betas and photons were simulated using a spectrum of beta energies obtained from the RADAR database (www.doseinfo-radar.com) for 131I, gamma and x-ray spectra were obtained from the International Atomic Energy Agency Nuclear Data Center (www-nds.iaea.org), and a 3-keV energy cutoff was used. The activity in each source tissue is assumed to be distributed uniformly and is assigned a random initial direction vector. Total events (1–5 × 106) were simulated, resulting in less than 10% relative error in energy deposit in relevant target tissues. Absorbed doses were output either in units of Gy or normalized to the injected activity calculated using Equation 2 (Gy/uCi). Three-dimensional dose maps were rendered in Paraview (Sandia National Laboratory, Kitware Inc., Los Alamos National Laboratory).
RESULTS
Synthesis, Specificity, and In Vitro Performance of 131I-PARPi
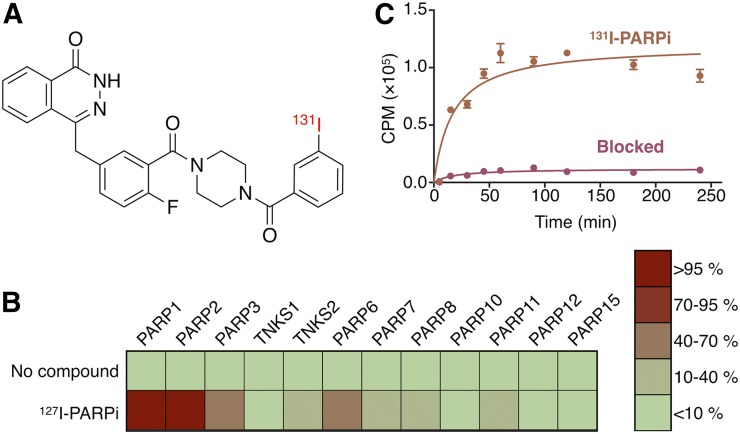
Several research groups have synthesized PARP-targeted radiotracers in the past; some of which were attached to radioiodine (14,22–24). We showed that one of these tracers, 131I-PARPi (14), has a high affinity with a concentration at which 50% of binding has been inhibited (IC50) in the nanomolar range (11 ± 3 nM) and a logPCHI of 2.3. 131I-PARPi was synthesized with a final molar activity of 1.5 GBq/μmol. Radiochemical purity was 99.1% ± 0.9% for all prepared compounds. Pharmacologically, 131I-PARPi behaves in a manner similar to the Food and Drug Administration–approved PARP inhibitor olaparib (Fig. 1; Supplemental Fig. 1; (25)). We tested the inhibitory activity of 127I-PARPi on a panel of 12 PARP enzymes, including tankyrase 1 and tankyrase 2 (Fig. 1B). We found near-perfect overlap with literature-known values for 18F-PARPi, PARPi-FL, and olaparib at 100 nM (12). 18F-PARPi is a PARP-targeted PET imaging agent with an IC50 of 3 nM (Supplemental Fig. 1) (18). PARPi-FL is a fluorescent PARP inhibitor with an IC50 value of 12.2 ± 1.1 (17). The 2 dominant targets for 127I-PARPi were found to be PARP-1 and PARP-2, just as for 18F-PARPi, PARPi-FL, and olaparib (26).
FIGURE 1.
(A) In vitro studies of PARP inhibitors. Structure of 131I-PARPi (14). (B) Heat map of binding characteristics of synthetic inhibitors (at 100 nM) to PARP family enzymes in percentage enzyme inhibition. (C) Cellular uptake of 131I-PARPi alone and blocked with mass excess dose of olaparib.
The in vitro binding specificity and internalization of 131I-PARPi was shown by incubating U251-MG cells with 131I-PARPi in the presence and absence of 150 μL of 6.67 μg/μL olaparib solution suspended in 25 μL of DMSO, 25 μL of polyethylene glycol (PEG), and 100 μL saline (Fig. 1C). In both cases, binding reached a plateau at approximately 50 min and was 10-fold higher in cells exposed to only 131I-PARPi than in cells coincubated with olaparib. Additionally, we showed that the radiotherapeutic binds to its intended target within the nucleus by incubating U251-MG cells with serial dilutions of its nonradioactive counterpart, 127I-PARPi, in the presence of 0.5 μM PARPi-FL. Overall, uptake of PARPi-FL retained in the nucleus at a concentration of 0.5 μM could be suppressed by addition of a 20-fold excess of 127I-PARPi (Supplemental Fig. 2B).
Intratumoral Injection and Tissue Perfusion of 131I-PARPi
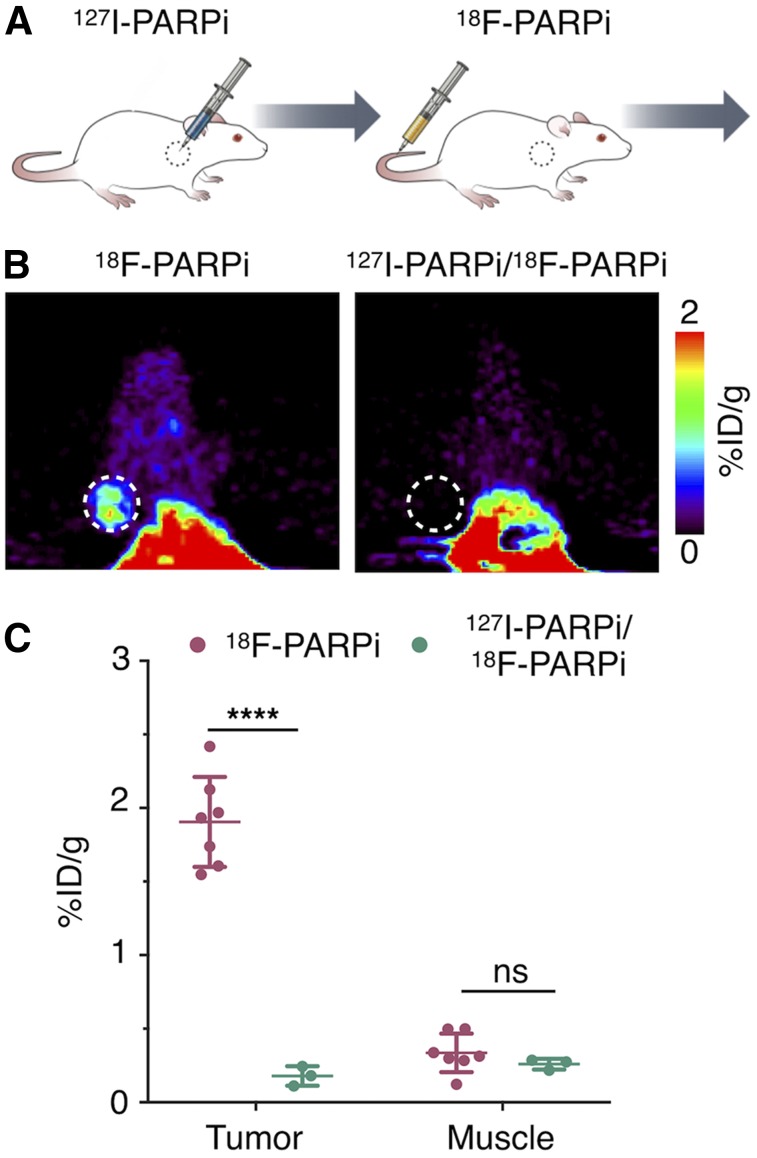
Intratumoral injections were implemented with a clinical CED model in mind, as well as for the favorable biodistribution (Fig. 2). To determine how intratumorally injected 131I-PARPi/127I-PARPi distributes throughout a tumor, we designed an in vivo competitive binding specificity experiment with 18F-PARPi, a previously described PET imaging agent for PARP-1. One cohort of animals was intratumorally injected with 127I-PARPi, followed by 18F-PARPi 30 min later. The second cohort was injected with 18F-PARPi only (Fig. 2A). We found that intratumoral injection of 127I-PARPi suppressed uptake of 18F-PARPi, indicating that the small molecule occupies 18F-PARPi binding sites (Figs. 2B and 2C), similar to what we showed in 18F-PARPi/olaparib blocking experiments previously (18). Ex vivo quantification at 1 h after injection of 18F-PARPi demonstrated that uptake dropped by 1.7 ± 0.1 %ID/g (1.9 ± 0.3 and 0.18 ± 0.07 %ID/g, P < 0.0001) between mice without and with intratumoral injection of 127I-PARPi (Fig. 2C). When replicated with olaparib and 131I-PARPi, brain uptake was low between mice without and with intravenous injection of olaparib (0.056 ± 0.020 and 0.061 ± 0.026 %ID/g, respectively) (Supplemental Figs. 3 and 4). Using autoradiography, we found heterogeneity in the intratumoral distribution of 131I-PARPi (Supplemental Fig. 5A). Overall quantification, however, corroborated the 18F-PARPi PET experiments (Fig. 2), and ratios between animals receiving only 131I-PARPi versus animals receiving a preinjection of olaparib were 7.8 (144.3 ± 32.7 AU and 18.5 ± 1.2 AU, P < 0.0001 for mice without and with preinjection of olaparib) (Supplemental Fig. 5B).
FIGURE 2.
131I-PARPi binding specificity. (A) Experimental outline of immuno-PET/CT imaging after injection of nonradioactive version of 131I-PARPi (127I-PARPi). (B) 18F-PARPi PET tracer injected intravenously 30 min before intratumoral injection of 127I-PARPi. PET scan of mice 2 h after receiving 18F-PARPi with and without 127I-PARPi before injection. (C) Ex vivo biodistribution of 18F-PARPi 1 h after injection in selected tissue. ****P < 0.0001. Error bars represent SD.
131I-PARPi Pharmacokinetics and Dosimetry in Glioblastoma
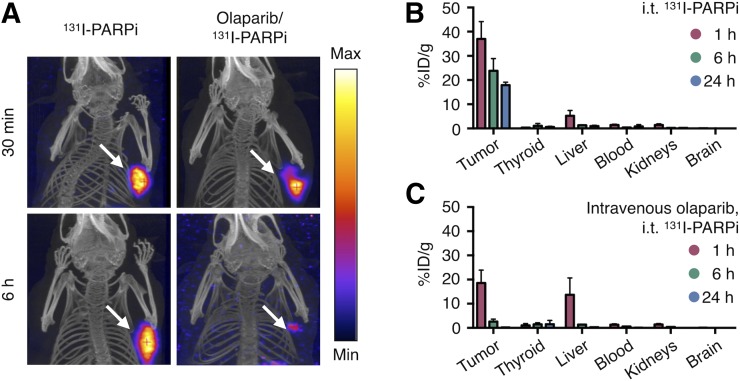
CED delivery of therapeutics to the brain relies on a drug’s fast perfusion, tissue permeability, and clearance from areas not expressing the targeted biomarker. To simulate this setting, we tested retention of 131I-PARPi in 2 cohorts of U251-MG subcutaneous xenografts. In 1 cohort, PARP was made unable to retain the radiotherapeutic through intravenous preinjection of olaparib. NanoSPECT/CT showed that at 6 h after intratumoral injection of 131I-PARPi (11.1 MBq, 20 μL, 30% PEG/0.9% NaCl solution), the radiotherapeutic was retained in tumors not previously injected with olaparib, whereas it had almost completely cleared from the blocked group (Fig. 3A). This observation was further corroborated by biodistribution experiments with endpoints (each time point n = 4) between 0 and 96 h after injection (Figs. 3B and 3C; Supplemental Figs. 3, 4, and 6). On the basis of these data, we calculated the activity and dose retained in different tissues (Supplemental Fig. 7). Intuitively, the highest dose was delivered to tumor tissue, with a calculated absorbed dose of 134.1 cGy/MBq. This was followed by the thyroid (68.6 cGy/MBq), which metabolizes and stores iodine (Supplemental Fig. 6; (27)). Besides PARP expression in the thyroid, storage of metabolized iodine is likely one reason for this comparatively high calculated dose. This is supported by the finding that in this setting, physical decay was observed as the only mode of clearance. However, moving forward, thyroid uptake can efficiently be blocked by administration of nonradioactive iodide (28).
FIGURE 3.
In vivo binding, biodistribution, and pharmacokinetics. (A) SPECT/CT images of subcutaneously U87-p53–xenografted mice intratumorally injected with (11.1 MBq) 131I-PARPi at time of injection and 6 h after injection. (B) Blocked mice received a systemic blocking dose of Olaparib 1 h before 131I-PARPi. 131I-PARPi (14.8 MBq) biodistribution in key organs after intratumoral injection at 1, 6, and 24 h (n = 4 per time point). (C) 131I-PARPi biodistribution in key organs at 1, 6, and 24 h postintratumoral injection after systemic blocking via intravenous injection of mass excess olaparib (n = 4 per time point).
Bioluminescence and p53 Response
We designed and transduced a reporter cell line responding to p53 activation, a marker of cell death and radiation damage. The cells coexpress p53/luciferase, together with constitutively expressing SFG-Turquoise/rsRluc. These bioluminescent reporters allow p53 expression to be imaged after injection of luciferase, and cell density to be imaged after injection of coelenterazine. Using this setup, we used bioluminescence imaging to determine p53 expression in relation to cell density (29). To use this experimental setup for testing whether 131I-PARPi retention is sufficient for inducing cell death, mice bearing subcutaneous transfected U87-p53 xenografts were injected with 16.3 ± 1.5 MBq of 131I-PARPi or vehicle (Supplemental Fig. 8). Dual reporter bioluminescence was measured 1 d before, on the day of, and 24 and 48 h after 131I-PARPi injection. For the control groups, no statistical differences were observed for any time points, suggesting that vehicle alone did not induce cell death (Supplemental Fig. 8). For mice injected with 131I-PARPi, however, a statistically significant increase in p53 signal was observed 24 h after administration of 131I-PARPi, which persisted at 48 h after injection (P < 0.005 after 24 h and P < 0.05 after 48 h when compared with day 0).
Therapeutic Efficacy of 131I-PARPi in Mouse Model of Glioblastoma
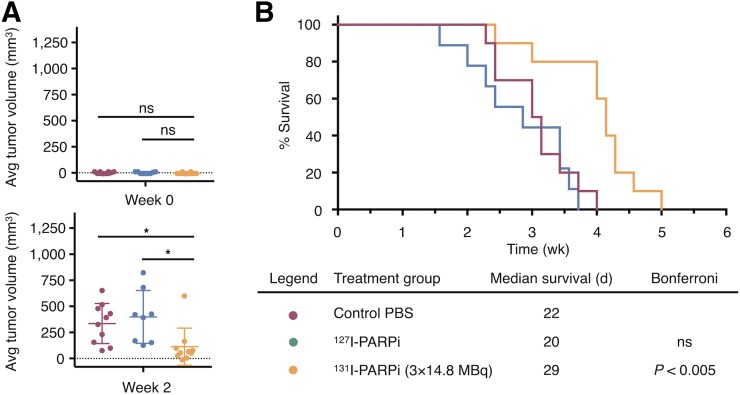
We next sought to determine the therapeutic efficacy of 131I-PARPi in a subcutaneous xenograft model of transduced U87-p53. For this purpose, tumor-bearing mice were randomly assigned to 3 different cohorts (n = 9–10 per cohort): vehicle (30% PEG-300/PBS), nonradioactive 127I-PARPi (5.9 μg, 9.9 nmol), or fractionated doses of 131I-PARPi (3 × 14.8 MBq, 3 × 9.9 nmol). For the fractionated-dose cohort, vehicle and cold 127I-PARPi intratumoral injections were performed on days 0, 3, and 6. Body weights and tumor volumes were recorded 3 times per week for the length of the study (Supplemental Fig. 9B); neither cohort experienced statistically significant weight loss, confirming that the administered activity did not lead to systemic toxicity (Supplemental Fig. 9B). We performed 2-sample t tests to compare tumor growth curves under dependent right censoring to account for sacrifices due to ethical guidelines and applied a correction for the multiple pairwise group comparisons (Supplemental Fig. 9D) (30). No statistical difference of tumor volume was observed between the vehicle cohort and the 127I-PARPi cohort. The fractionated-dose treatment led to a statistically significant reduction in tumor volume compared with the vehicle group (P < 0.005). This is also reflected in the tumor growth rates, which were not different at the beginning of the study (week 0) but showed reduced growth rates in the 2 radiotherapy groups at week 2 compared with the control group (Fig. 4A). When comparing survival of mice, statistical significance was found for the treatment group versus the vehicle (P < 0.005). No statistical difference was found between the vehicle group and cold 127I-PARPi. Mice receiving fractionated doses had the longest median survival (29 d). Median survival for the PBS group and cold 127I-PARPi group was 22 and 20 d, respectively (Fig. 4B).
FIGURE 4.
In vivo therapeutic effect of 131I-PARPi therapy in subcutaneous mouse model. (A) Difference of tumor growth percentage at beginning of study and at 2 wk. (B) Kaplan–Meier plot and table of treatment groups with median survival. Nonparametric Student t test was used to calculate statistics for A. *P < 0.05. Error bars represent SD. P values were calculated by Mantel-Cox test for B, P = 0.0001.
Orthotopic Model
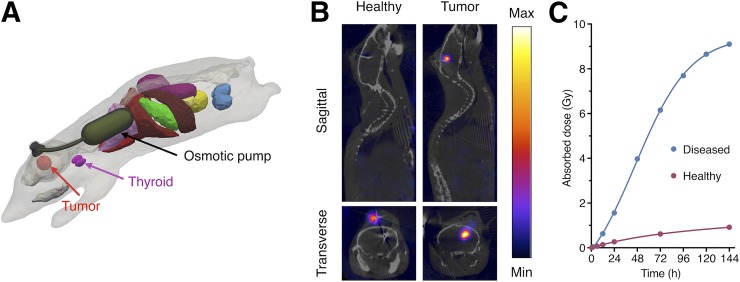
Simulating a potential clinical treatment scenario, we tested the delivery of 131I-PARPi in an orthotopic U87-p53 mouse model of glioblastoma paired with an implantable osmotic pump and cannula (Fig. 5A). The osmotic pump mimicked slow, CED-type delivery of our radiotherapeutic, and we compared the retention of 131I-PARPi in tumor-bearing versus healthy mice (Supplemental Fig. 10). On the basis of calculations of the administered dose from SPECT-derived pump activity concentrations versus time, it was shown that administration of 30.7 MBq (49.2 MBq initially implanted) resulted in the highest doses in the brain/tumor itself (30.1622 ± 0.1946 cGy/MBq ID), induced overwhelmingly by tumor-bound 131I-PARPi (27.4405 ± 0.03784 cGy/MBq ID) versus photon dose from the pump contents (2.0768 ± 0.100 cGy/MBq ID; Supplemental Fig. 5). Other organs, affected primarily by γ-radiation from the pump contents, received comparatively small doses (Fig. 5B). Only the left lung, which was in direct proximity to the osmotic pump activity reservoir, received a somewhat elevated dose (11.08 cGy/MBq injected dose). Experimental findings corroborated these models, as strong, durable retention of 131I-PARPi was observed in tumor-bearing mice, whereas healthy brain tissue retained the radiotherapeutic to a much lower degree (Fig. 5C); after 6 d, 9.1 Gy were delivered to the brains of tumor-bearing mice, whereas healthy mice retained less than 1 Gy.
FIGURE 5.
In vivo CED in orthotopic glioblastoma mouse model. (A) Three-dimensional model of orthotopic glioblastoma mouse model with CED-mimicking osmotic pump. (B) SPECT/CT of orthotopic U87-p53 tumors during osmotic pump treatment of 131I-PARPi at 72 h. (C) Calculated absorbed dose to brain* during treatment. *Brain and tumor have been considered together as a single organ in organ-level dose calculation.
DISCUSSION
There is a critical need for more and better therapeutic options for patients with glioblastoma. The past few decades have seen only modest improvements in treatment outcomes (31). Both PARP inhibition therapy and targeted radionuclide therapy are 2 new treatment approaches that have independently gained widespread attention; both types of therapy have led to clinical trials, which are currently ongoing (32). Here, we fuse these 2 approaches and use PARP as a target for a newly developed targeted radiotherapeutic. In this study, we sought to demonstrate a CED approach to delivering the radiotherapeutic molecule 131I-PARPi in orthotopic glioblastoma models.
In vitro and in vivo studies have shown that 131I-PARPi is a potent PARP inhibitor, and that introduction of radioiodine did not perturb binding specificity. This was also confirmed in a 12-member cross-family binding assay showing that 131I-PARPi possesses a binding profile akin to olaparib and other olaparib-based imaging agents (Fig. 1). PET imaging with 18F-PARPi demonstrated that intratumoral administration of the cold iodinated analog 127I-PARPi perfused efficiently throughout the tumor (Fig. 2), confirming that the tissue permeability of the small molecule is high, similar to what we found for PARPi-FL (33). We also measured the pharmacokinetics of the compound when administered intratumorally and observed strong retention of activity in the tumor if PARP is available, but only very little when PARP binding sites have been saturated with olaparib. Intuitively, the deposited dose is a function of the small molecule’s tissue half-life, and the difference between the 2 is a function of target selectivity (Fig. 3). These data were used in dosimetric analysis, which showed that the tumor received 134.1 cGy/MBq while the major clearance organs, the thyroid and liver, received doses of 68.4 and 6.2 cGy/MBq, respectively. Using a fluorescently transfected cell line capable of reporting p53 activation, we saw a statistically significant increase in p53 activation in subcutaneous mouse models treated with 131I-PARPi (Supplemental Fig. 7). This activation serves as a marker of radiation damage to cells, further verifying the therapeutic potential of 131I-PARPi. We then conducted a survival study with the subcutaneous mouse model that showed a significant reduction in tumor growth and improvement in median survival when 131I-PARPi was administered intratumorally as either a single dose of 14.8 MBq or as a fractionated dose of 3 × 14.8 MBq (Fig. 4). Low toxicity, as a function of the mouse’s body weight, was observed throughout this study.
131I-PARPi represents a theranostic pair in itself, insofar as it might be used for SPECT imaging when delivered systemically in addition to its therapeutic function when delivered intratumorally. However, in addition to SPECT, the 18F-labeled analog 18F-PARPi (18) can be used for PET imaging, which may be more suitable for patient stratification and treatment monitoring. Additionally, the fluorescently labeled analog PARPi-FL (16) might be used for fluorescence-guided resections.
A likely clinical scenario of 131I-PARPi treatment involves the radiotherapeutics’ administration as a CED agent, similar to what has been shown previously for antibodies (7). 131I-PARPi has structural similarity to olaparib, which features a cyclopropamide instead of the 131I-labeled meta-iodobenzamide. Olaparib is a substrate of phosphatidylglycerolphosphate synthase 1, a multidrug efflux transporter (34,35). PGP might therefore be important in mediating 131I-PARPi uptake in contrast-enhancing and non–contrast-enhancing regions of the brain, affecting both uptake and clearance of drug, but potentially also improving selective uptake. To mimic a potential clinical CED scenario, mice bearing orthotopic glioblastoma xenografts were treated with 131I-PARPi via implanted osmotic pumps (Fig. 5). Uptake and retention of 131I-PARPi in these mice were high, as determined via periodic SPECT/CT measurement for up to 6 d after implantation. Healthy mice showed no such retention. Dosimetry was calculated on the basis of the SPECT/CT data, indicating that 9.1 Gy were delivered to the whole brain of a tumor-bearing animal, whereas less than 1 Gy was delivered to a healthy animal. This shows that CED delivers its radiotherapeutic dose in such a way that PARP-overexpressing cells will retain the activity while the drug is cleared quickly from healthy brain cells (7).
CONCLUSION
We have identified PARP-1 as a potential anchor for radiotherapeutic PARP inhibitors in glioblastoma. We explored and validated the pharmacokinetics and pharmacodynamics of 131I-PARPi and showed that animals treated with the radiotherapeutic have a survival benefit, and healthy brain cells do not retain the radiotherapeutic. Future studies will need to show whether 131I-PARPi demonstrates an improvement in therapeutic efficacy over the existing standard of care.
DISCLOSURE
This work was supported by National Institutes of Health grants 1 R01 CA204441 (Thomas Reiner) and P30 CA008748. The authors thank the Tow Foundation and Memorial Sloan Kettering Cancer Center’s Center for Molecular Imaging & Nanotechnology (Brandon Carney and Susanne Kossatz), the National Science Foundation Integrative Graduate Education and Research Traineeship (IGERT 0965983 at Hunter College; Brandon Carney), the American-Italian Cancer Foundation (Giuseppe Carlucci), and the Imaging and Radiation Sciences Program (Thomas Reiner) for financial support. No other potential conflict of interest relevant to this article was reported.
Supplementary Material
Acknowledgments
We thank the support of Memorial Sloan Kettering Cancer Center’s Animal Imaging Core Facility, Radiochemistry & Molecular Imaging Probes Core Facility, and Molecular Cytology Core Facility. In addition, we thank Dr. Ira Dunkel for helpful discussions and Leah Bassity for editing the manuscript.
REFERENCES
- 1.Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35:2402–2409. [DOI] [PubMed] [Google Scholar]
- 2.Bi WL, Beroukhim R. Beating the odds: extreme long-term survival with glioblastoma. Neuro-oncol. 2014;16:1159–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland EC. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA. 2000;97:6242–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park JK, Hodges T, Arko L, et al. Scale to predict survival after surgery for recurrent glioblastoma multiforme. J Clin Oncol. 2010;28:3838–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anjum K, Shagufta BI, Abbas SQ, et al. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: a review. Biomed Pharmacother. 2017;92:681–689. [DOI] [PubMed] [Google Scholar]
- 6.Vogelbaum MA, Aghi MK. Convection-enhanced delivery for the treatment of glioblastoma. Neuro-oncol. 2015;17(suppl 2):ii3–ii8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luther N, Zhou Z, Zanzonico P, et al. The potential of theragnostic 124I-8H9 convection-enhanced delivery in diffuse intrinsic pontine glioma. Neuro-oncol. 2014;16:800–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR. Molecular heterogeneity in glioblastoma: potential clinical implications. Front Oncol. 2015;5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. [DOI] [PubMed] [Google Scholar]
- 10.Kossatz S, Carney B, Schweitzer M, et al. Biomarker-based PET imaging of diffuse intrinsic pontine glioma in mouse models. Cancer Res. 2017;77:2112–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murnyák B, Kouhsari MC, Hershkovitch R, et al. PARP1 expression and its correlation with survival is tumour molecular subtype dependent in glioblastoma. Oncotarget. 2017;8:46348–46362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carney B, Kossatz S, Reiner T. Molecular imaging of PARP. J Nucl Med. 2017;58:1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlucci G, Carney B, Brand C, et al. Dual-modality optical/PET imaging of PARP1 in glioblastoma. Mol Imaging Biol. 2015;17:848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salinas B, Irwin CP, Kossatz S, et al. Radioiodinated PARP1 tracers for glioblastoma imaging. EJNMMI Res. 2015;5:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight JC, Koustoulidou S, Cornelissen B. Imaging the DNA damage response with PET and SPECT. Eur J Nucl Med Mol Imaging. 2017;44:1065–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin CP, Portorreal Y, Brand C, et al. PARPi-FL: a fluorescent PARP1 inhibitor for glioblastoma imaging. Neoplasia. 2014;16:432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiner T, Lacy J, Keliher EJ, et al. Imaging therapeutic PARP inhibition in vivo through bioorthogonally developed companion imaging agents. Neoplasia. 2012;14:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carney B, Carlucci G, Salinas B, et al. Non-invasive PET imaging of PARP1 expression in glioblastoma models. Mol Imaging Biol. 2016;18:386–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown JA, Marala RB. Development of a high-throughput screening-amenable assay for human poly(ADP-ribose) polymerase inhibitors. J Pharmacol Toxicol Methods. 2002;47:137–141. [DOI] [PubMed] [Google Scholar]
- 20.Dogdas B, Stout D, Chatziioannou AF, Leahy RM. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys Med Biol. 2007;52:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Niita K, Matsuda N, et al. Particle and heavy ion transport code system, PHITS, version 2.52. J Nucl Sci Technol. 2013;45:634–638. [Google Scholar]
- 22.Zmuda F, Malviya G, Blair A, et al. Synthesis and evaluation of a radioiodinated tracer with specificity for poly(ADP-ribose) polymerase-1 (PARP-1) in vivo. J Med Chem. 2015;58:8683–8693. [DOI] [PubMed] [Google Scholar]
- 23.Makvandi M, Xu K, Lieberman BP, et al. A radiotracer strategy to quantify PARP-1 expression in vivo provides a biomarker that can enable patient selection for PARP inhibitor therapy. Cancer Res. 2016;76:4516–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson RC, Makvandi M, Xu K, et al. Iodinated benzimidazole PARP radiotracer for evaluating PARP1/2 expression in vitro and in vivo. Nucl Med Biol. 2016;43:752–758. [DOI] [PubMed] [Google Scholar]
- 25.Menear KA, Adcock C, Boulter R, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin-1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–6591. [DOI] [PubMed] [Google Scholar]
- 26.Carney B, Lok BH, Schneeberger VE, et al. Target engagement imaging of PARP inhibitors in small cell lung cancer. Nat Commun. 2018;9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bianco AC, Kim BW. Deiodinases: implications of the local control of thyroid hormone action. J Clin Invest. 2006;116:2571–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brans B, Monsieurs M, Laureys G, Kaufman JM, Thierens H, Dierckx RA. Thyroidal uptake and radiation dose after repetitive I-131-MIBG treatments: influence of potassium iodide for thyroid blocking. Med Pediatr Oncol. 2002;38:41–46. [DOI] [PubMed] [Google Scholar]
- 29.Doubrovin M, Ponomarev V, Beresten T, et al. Imaging transcriptional regulation of p53-dependent genes with positron emission tomography in vivo. Proc Natl Acad Sci USA. 2001;98:9300–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vardi Y, Ying Z, Zhang C-H. Two-sample tests for growth curves under dependent right censoring. Biometrika. 2001;88:949–960. [Google Scholar]
- 31.Bianco J, Bastiancich C, Jankovski A, des Rieux A, Préat V, Danhier F. On glioblastoma and the search for a cure: where do we stand. Cell Mol Life Sci. 2017;74:2451–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11:239–253. [DOI] [PubMed] [Google Scholar]
- 33.Thurber GM, Yang KS, Reiner T, et al. Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo. Nat Commun. 2013;4:1504–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schinkel AH. P-glycoprotein, a gatekeeper in the blood-brain barrier. Adv Drug Deliv Rev. 1999;36:179–194. [DOI] [PubMed] [Google Scholar]
- 35.Vaidyanathan A, Sawers L, Gannon AL, et al. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br J Cancer. 2016;115:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.