Abstract
The HMMER webserver [http://www.ebi.ac.uk/Tools/hmmer] is a free-to-use service which provides fast searches against widely used sequence databases and profile hidden Markov model (HMM) libraries using the HMMER software suite (http://hmmer.org). The results of a sequence search may be summarized in a number of ways, allowing users to view and filter the significant hits by domain architecture or taxonomy. For large scale usage, we provide an application programmatic interface (API) which has been expanded in scope, such that all result presentations are available via both HTML and API. Furthermore, we have refactored our JavaScript visualization library to provide standalone components for different result representations. These consume the aforementioned API and can be integrated into third-party websites. The range of databases that can be searched against has been expanded, adding four sequence datasets (12 in total) and one profile HMM library (6 in total). To help users explore the biological context of their results, and to discover new data resources, search results are now supplemented with cross references to other EMBL-EBI databases.
INTRODUCTION
The use of profile hidden Markov models (HMM) for detecting sequence similarity is widespread. Their popularity stems from the fact that a few related and aligned sequences can be used to construct a profile HMM, which can then be used to search large sequence databases to find related sequences, even those distantly related (1). The sensitivity of profile HMMs is achieved by the position-specific probabilistic modelling of the alignment, which incorporates not only residue conservation, but also rates of insertions and deletions. The use of profile HMMs has been widely adopted by databases wishing to represent protein families, such as Pfam (2). Indeed, such databases represent some of the few biological data resources that have grown at linear rates. Until 2010, profile HMMs were somewhat confined to the niche of such protein family databases due to the computational expense of searching them. The accelerated profile HMM search algorithm in the third generation of the HMMER suite has significantly reduced this computation overhead (3). As such, it is possible to search a typical protein based profile HMM against 100 million protein sequences in a matter of ∼10 min on a single CPU. By scaling searches over multiple CPUs, this search time can be reduced to a matter of seconds. We have adopted this scaling approach to create the HMMER webserver (4), first launched in 2011, providing the ability to search a single sequence against profile HMM libraries or large sequence collection. Since then, this web service has increased significantly in popularity (as measured by the total number of searches and users). The interface has been described in detail (5,6). Herein, we describe the recent developments to the user interface, application program interface (API), portable JavaScript libraries and supported target databases.
WEB SERVER IMPROVEMENTS
Discoverability: links to other EMBL-EBI resources
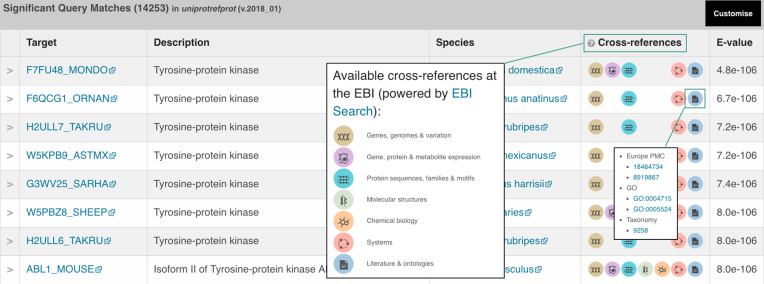
EBI-Search (7) is a scalable text search engine based on Apache Lucene [https://lucene.apache.org/core/] which collates and indexes data from across EMBL-EBI’s resources. A network of cross references provides navigation between the different resources, facilitating access to related information pertaining to different biological contexts or entities, for example a sequence is known to bind to a small chemical ligand. EBI-Search powers text searches from the EMBL-EBI website and many of the resources held at the institute via its API layer. The HMMER web server has similarly leveraged the EBI-Search API, adding extra value to matches (sequence or profile HMM) in the search results by showing connections to the match in related EMBL-EBI databases. Search results now contain an extra ‘Cross references’ column which shows these links (where they exist), grouped into seven broad categories: genes, genomes & variation; gene, protein & metabolite expression; protein sequences, families & motifs; molecular structures; chemical biology; systems; literature & ontologies. Hovering over each circular icon reveals a drop-down list of links grouped by resource (Figure 1). These links enable the user to discover new data sources and gain further insight by placing the results in a greater context of other biological resources.
Figure 1.
Results of a search in the ‘score’ view with the new ‘Cross references’ column, populated with links to the hit sequence in other EMBL-EBI databases. As the page is loaded, these symbols may disappear if no cross-references are found in this category.
Enriched application programming interface (API)
One of the value-added features that the HMMER website provides over the command line version is the ability to quickly filter sequence search results using taxonomy and/or domain architectures. While the website has provided this functionality for at least 5 years, the API has only recently been extended to enable similar querying and filtering of the search results. Thus, the ‘taxonomy’ and ‘domain architecture’ view data can now be accessed via the API. JSON and XML content types are enabled for all the result endpoints, with plain text being a third option for the ‘score’ view.
Having the complete repertoire of results available via the API has also presented other opportunities. We have just completed a process whereby the JavaScript tools for graphically representing taxonomy and domain graphics are able to consume the API endpoints. As well as being used within the HMMER website, they can also be integrated within third parties’ websites, facilitating the integration of the EMBL-EBI hosted search infrastructure. To effect this, the API now responds with appropriate cross-origin headers to allow not only scripts, but also other websites, to request search results directly from within their pages.
Standalone taxonomy viewer
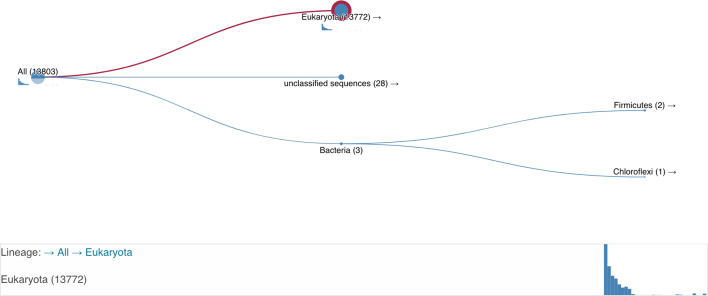
The existing taxonomy visualisation has been rewritten as a standalone library. The new visualisation library supports the same range of user interactions, but has been designed with the aim of providing a better user experience. Based on feedback, a key area of the redesign was to improve both the navigation of the tree and to show how the selection of nodes in the representative taxonomic tree filtered the results (Figure 2). As the user changes the selected node, additional subtle animations emphasise the impact on the number of results selected. Interaction with the taxonomy tree is now possible by both mouse and keyboard, allowing for precise node selections in densely branched trees. As well as the HMMER website, this library is also being used in the next revision of the InterPro (8) website. The library is available publicly on GitHub [github.com/ProteinsWebTeam/taxonomy-visualisation] and through NPM (JavaScript main package manager, see GitHub repository for more information) making distribution, extension and feedback from external implementers simpler through public channels.
Figure 2.
New taxonomy view. The tree is navigable via keyboard or mouse, with the red circle showing the currently selected node, and the size of the blue circles being proportional to the number of hits. Figure 3. As the user changes the selected node, the red line highlights the selected route to the node. The lineage is shown below the figure and the small barchart shows the distribution of E-values at the selected node.
Domain graphics
There has also been an effort to provide the domain graphics visualisation as a standalone library. The corresponding code has been extracted from the HMMER website, updated, and made available on GitHub and as a NPM package (see github.com/ProteinsWebTeam/domain-gfx). It keeps the same graphical style as the current visualisation but makes it easier to reuse.
Iterative searching with Jackhmmer
The jackhmmer search algorithm allows iterative searches against a sequence database, where subsequent queries are profile HMMs built from the aligned hit sequences of a preceding search. Unlike the command line version of HMMER, the set of sequences used for the alignment may be manually manipulated using checkboxes to add or remove sequences between successive iterations via the web interface. By default, all sequences above the selected significance threshold are pre-checked. However, if one wanted to select only a small number of sequences, the user would have to manually deselect all other sequences above the threshold: a number that could easily run into the thousands. In response to several users who have pointed out this drawback, there is now a choice of using either: (a) all sequences above the significance threshold, as before; or (b) have no sequences selected, manually adding only those that are required.
Active site predictions
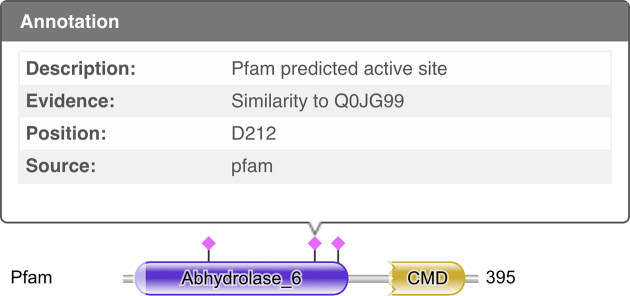
We have added the functionality to predict active sites using an approach developed by Mistry et al. (9) based on experimentally known data. These provide the likely sites of catalytically active residues. Unlike the original implementation which generated alignments between the query sequence and sequences with known active site residues using the Pfam profile HMMs, the sequences with known active site residues are now used to annotate the Pfam profile HMM, and the profile HMM match positions used to propagate the residue information. Conceptually, this is an equivalent process, but is significantly more computationally efficient. These are included with the domain graphic (‘Sequence Matches and Features’) which is automatically produced with every sequence search, with the sites represented as lollipops above the representation of the Pfam domain (Figure 3). Hovering the mouse cursor over the lollipops will present a description, evidence, position and source for the site.
Figure 3.
Annotation of the domain graphic with the catalytic active sites predicted by Pfam, with extra information about the middle site displayed in a mouse-activated pop-up.
CHANGES TO THE TARGET DATABASES
Over and above any algorithmic speed optimisations, a significant proportion of the speed of the HMMER web search relies on the databases being held in memory by the programs that execute the search itself. For profile HMM libraries, the files are typically only a few gigabytes in size and only need to be spread across a few CPUs to achieve search times in the range 1–200 milliseconds. Conversely, the sequence databases can be huge (millions of sequences and 10s of GBs in size). To provide the interactive searching of these resources (a few seconds), our searches are scaled over 16 compute nodes (128 CPUs, 2.3 GHz), using 1.2TB RAM in total (78GB RAM per node).
To support searching against multiple sequence databases in the current implementation of the HMMER software it is required that these all be merged into a single database (with each sequence in the combined dataset tagged to indicate the database(s) it occurs within). Whilst doing this, we take advantage of any redundancy by collapsing sequences having the same taxonomy across multiple databases into a single entity. This dramatically reduces the memory footprint compared with running the separate databases individually owing to the large overlap between them (for the January 2018 release 460 million sequences was reduced to a non-redundant set of 176 million). A further optimisation is to remove the sequence headers and store these in a separate database. These steps can only be followed if the sequence database is rebuilt in its entirety, which we perform at each release. Thus, the mapping of internal sequence identifiers to accessions changes from one release to another. Crucially, this means that a search performed just before a server update will no longer be valid afterwards. To try to mitigate the problem of invalid results discussed above we have added warnings to the site to give users advance notification of pending updates. These increase in severity as the update draws near and suggest approaches to the user: downloading results or deferring large batch jobs until after the update.
Target sequence database
Since the last update paper in 2015, our hmmpgmd specific sequence database has grown 3-fold to 48 Gb. As of January 2018, there are 168 million sequences in Ensembl Genomes and 106 million in UniProtKB. This rate of data growth has not been paralleled by growth in memory so the supported databases have been rationalised, with decisions based on scale, use and availability.
To enable us to scale, and given the substantial overlap between UniProtKB and the NCBI non-redundant (NR) and reference sequence (RefSeq) databases, we have temporarily stopped support for NR and RefSeq databases. While we understand that this may limit the utility of the service to some users, it is possible to convert UniProtKB accessions to the corresponding NCBI accessions by using the UniProtKB mapping tool (https://www.uniprot.org/uploadlists/). We are currently exploring a number of options to improve scalability to support multiple large sequence databases. The pfamseq database (Pfam's underlying sequence database) was not being searched against outside of EMBL-EBI, and due to the space constraints and low usage, is no longer supported as a target database.
The metagenomics database, UniMes, is no longer supported by UniProt. However, there is a parallel HMMER service provided by the EBI Metagenomics team, which enables the searching of metagenomics sequences (10). This metagenomics database currently contains >334 million sequences retrieved from a diverse range of environments, but lacks some of the search functionality and results visualisations provided by this HMMER web server. For example, the metagenomics sequences do not have a known taxonomy and the iterative searches have not been enabled due to the size of the database resulting in alignments that could not be readily handled by the web servers.
Nevertheless, we have continued to update the existing databases and have increased the scope of the databases to reflect demand. The sequence databases are largely based on UniProtKB (11), with the option of searching either this database in its entirety, or one of the various subsets: UniProtKB Reference Proteomes, Representative Proteomes (12) or the curated sequences of UniProtKB/Swiss-Prot. A substantial addition has been the predicted peptides of Ensembl (13) and Ensembl Genomes (14). We have also added the sequences from the MEROPS (15) database of proteolytic enzymes, which allows the precise annotation of peptidase subunits on query sequences by querying against the MEROPS holotype sequence database. For the Ensembl database we provide a shortcut to the more commonly searched organisms (human, mouse and zebrafish). In the case of Ensembl Genomes, a search may be performed against the full database or one of the subsets: bacteria, fungi, protists, plants and metazoa. Ensembl Genomes has adopted the HMMER search engine into its own website, by providing a specific input form and wrapping the HMMER web result pages into its own website.
Profile HMM libraries
The lack of redundancy between profile HMMs and small size means that each protein family database is provisioned by a series of independent, small scale hmmpgmd arrangements. The existing profile HMM libraries (Pfam (2), CATH-Gene3D (16), TIGRFAMs (17), Superfamily (18) and PIRSF (19)) have been supplemented by those of the TreeFam (20) database of phylogenetic trees. The HMMER website now provides a single point of entry for those wishing to access the profile HMM libraries supported by EMBL-EBI. The profile HMM libraries have been updated as newer versions became available (Pfam to version 31.0 and CATH-Gene3D to 16.0.0). We have updated the different post-processing procedures that CATH-Gene3D and PIRSF perform on the raw HMMER output, to ensure that expected results are faithfully recreated. For searches against multiple databases only a single threshold may be applied and this will necessitate a compromise if the databases use conflicting approaches; for example, the curated gathering thresholds in Pfam versus the preset E-value thresholds of CATH-Gene3D. Finally, we anticipate the provision of the PANTHER database by summer 2018, but the size of the PANTHER 13.1 library that contains over 90 000 profile HMMs has presented new scaling challenges.
Release cycle
The UniProtKB sequence database and related derivatives thereof, represents the most widely queried database. To ensure the greatest consistency, we have synchronized to the monthly release cycle of UniProtKB to drive our own updates. This also strikes a good balance of the availability of new data against the disruption caused by expired search results. We consequently make a new release of the webserver once per month, on or immediately after a UniProt release date. PDB has a weekly release cycle which we cannot, at this time, fit into our current schedule, so is updated monthly. The two Ensembl resources change less frequently (approximately every three months): these are updated as and when the revised data sets are made public.
Documentation
To make our online documentation easier to use we have migrated it to ‘Read The Docs’ [hmmer-web-docs.readthedocs.io/en/latest]. This has enabled more media types to be used, facilitated better browsing/searching and allowed versioning of the documentation. Furthermore, this externally hosted service allows for text searches and downloads in a variety of formats and is backed by a publicly accessible GitHub repository [github.com/ProteinsWebTeam/HMMER-web-docs] which provides a simple mechanism for users to suggest improvements, as well as allow the whole team to contribute to the documentation.
DISCUSSION
Over the past two years (Jan 2016 - Dec 2017), >23 million jobs have been submitted to the web server, corresponding to over 28 million individual searches (a single batch or jackhmmer job can comprise multiple searches). As described above, the functionality of the site has been enriched to both facilitate the searching of a wide range of target databases (both sequence and profile HMM) and support both interactive usage and programmatic access. By design, the URL namespaces between the API and website overlap, allowing job unique identifiers to be used interchangeably. Consequently, these usage figures include interactive web searches, queries via the API and batch submissions. The statistics also include a number of ‘third party’ searches, for examples those redirected from the Ensembl Genomes (14) browser and the Pfam website, which uses HMMER for its batch searches.
The reusability of our JavaScript widgets, coupled with the associated API, has already facilitated the integration of HMMER search and visualization capabilities into other EMBL-EBI based resources, which increases the sustainability of these services. They also enable users outside of EMBL-EBI to build their own (potentially lightweight) tools and clients which can access the fast, efficient search infrastructure we maintain, without having the overhead of setting up and maintaining this infrastructure. To ensure longer term support and scalability we are working on multiple approaches to provide solutions to deliver better horizontal scaling of resources and balance between interactive use and API use. While we want to provide faster searches for the manual user, total throughput is more important than ultimate speed of each search for users of the API.
FUNDING
EMBL core funds; Howard Hughes Medical Institute; NIH [R01 HG009116 to S.R.E.]. Funding for open access charge: EMBL core funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998; 14:755–763. [DOI] [PubMed] [Google Scholar]
- 2. Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016; 44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Finn R.D., Clements J., Eddy S.R.. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R.. HMMER web server: 2015 update. Nucleic Acids Res. 2015; 43:W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prakash A., Jeffryes M., Bateman A., Finn R.D.. The HMMER Web server for protein sequence similarity search. Curr. Protoc. Bioinformatics. 2017; 60:3.15.1–3.15.23. [DOI] [PubMed] [Google Scholar]
- 7. Park Y.M., Squizzato S., Buso N., Gur T., Lopez R.. The EBI search engine: EBI search as a service-making biological data accessible for all. Nucleic Acids Res. 2017; 45:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M. et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mistry J., Bateman A., Finn R.D.. Predicting active site residue annotations in the Pfam database. BMC Bioinformatics. 2007; 8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitchell A.L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., Salazar G.A., Pesseat S., Boland M.A., Hunter F.M.I. et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018; 46:D726–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018; 46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen C., Natale D.A., Finn R.D., Huang H., Zhang J., Wu C.H., Mazumder R.. Representative proteomes: a stable, scalable and unbiased proteome set for sequence analysis and functional annotation. PLoS ONE. 2011; 6:e18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al. Ensembl 2018. Nucleic Acids Res. 2018; 46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kersey P.J., Allen J.E., Allot A., Barba M., Boddu S., Bolt B.J., Carvalho-Silva D., Christensen M., Davis P., Grabmueller C. et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018; 46:D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D.. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018; 46:D624–D632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dawson N.L., Lewis T.E., Das S., Lees J.G., Lee D., Ashford P., Orengo C.A., Sillitoe I.. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017; 45:D289–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haft D.H., Selengut J.D., Richter R.A., Harkins D., Basu M.K., Beck E.. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013; 41:D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oates M.E., Stahlhacke J., Vavoulis D.V., Smithers B., Rackham O.J.L., Sardar A.J., Zaucha J., Thurlby N., Fang H., Gough J.. The SUPERFAMILY 1.75 database in 2014: a doubling of data. Nucleic Acids Res. 2015; 43:D227–D233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu C.H., Nikolskaya A., Huang H., Yeh L.-S.L., Natale D.A., Vinayaka C.R., Hu Z.-Z., Mazumder R., Kumar S., Kourtesis P. et al. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res. 2004; 32:D112–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schreiber F., Patricio M., Muffato M., Pignatelli M., Bateman A.. TreeFam v9: a new website, more species and orthology-on-the-fly. Nucleic Acids Res. 2014; 42:D922–D925. [DOI] [PMC free article] [PubMed] [Google Scholar]