Abstract
Secreted and cell-surface proteases are major mediators of extracellular matrix (ECM) turnover, but their mechanisms and regulatory impact are poorly understood. We developed a mass spectrometry approach using a cell-free ECM produced in vitro to identify fibronectin (FN) as a novel substrate of the secreted metalloprotease ADAMTS16. ADAMTS16 cleaves FN between its (I)5 and (I)6 modules, releasing the N-terminal 30 kDa heparin-binding domain essential for FN self-assembly. ADAMTS16 impairs FN fibrillogenesis as well as fibrillin-1 and tenascin-C assembly, thus inhibiting formation of a mature ECM by cultured fibroblasts. Furthermore ADAMTS16 has a marked morphogenetic impact on spheroid formation by renal tubule-derived MDCKI cells. The N-terminal FN domain released by ADAMTS16 up-regulates MMP3, which cleaves the (I)5-(I)6 linker of FN similar to ADAMTS16, therefore creating a proteolytic feed-forward mechanism. Thus, FN proteolysis not only regulates FN turnover, but also FN assembly, with potential long-term consequences for ECM assembly and morphogenesis.
Keywords: Extracellular matrix*, Proteases*, Post-translational modifications*, Protein Degradation*, Proteolysis*, ADAMTS protease, Fibronectin, Metalloprotease
The extracellular matrix (ECM) 1 is a network of proteins, glycoproteins and complex carbohydrates that normally undergoes continuous remodeling via coupled proteolytic degradation and biosynthesis. Deposition of a provisional ECM during morphogenesis, tissue regeneration or ECM assembly by cultured cells is known to rely heavily on the assembly of FN, an ECM glycoprotein, which is a crucial cell adhesion molecule and pro-migratory substrate (1, 2). Plasma and cellular FN are secreted as disulfide-bonded dimers that bind to cell surface integrins. Subsequently, cell contractility and centripetal integrin translocation expose cryptic binding sites in the FN dimers that allow their association and promote FN fibril assembly (1, 3). A major FN domain mediating initial self-assembly and promoting fibrillogenesis is the N-terminal heparin-binding domain spanning the first five type I repeats (4–7). The FN network serves as a template for the assembly of fibrillins, collagens, tenascin-C, latent TGFβ-binding proteins and other molecules and therefore has a crucial role in the formation of mature ECM (8–11). Consistent with the observed significant impact on ECM ontogeny, cell adhesion and migration in vitro, inactivation of the Fn1 gene in mice leads to embryonic lethality by 8.5 days of gestation, with embryos showing impaired cardiovascular development and other morphogenetic defects (12).
Whereas initiation of FN fibril assembly has been extensively investigated, mechanisms regulating fibril turnover and maturation, are poorly understood (1, 13). Even though a number of proteases are known to cleave FN (4, 13–17) little is known about how they affect FN networks and so far, the relationship between FN proteolysis and assembly has not been specifically investigated. The ADAMTS (a disintegrin-like and metalloproteinase domain with thrombospondin type I motif) family includes 19 secreted proteases, most of which act on ECM and are crucial regulators of morphogenesis (18, 19). ADAMTS16 is a poorly characterized family member without a known substrate, although it was reported to play a role during renal and gonadal development. Homozygous Adamts16 knockout male rats are infertile, with reduced testicular size and cryptorchidism (20) and ADAMTS16 was linked to branching morphogenesis during renal development (21). In addition to its role during embryogenesis, ADAMTS16 was implicated in blood pressure regulation in rats and humans (22, 23) and linked to ovarian cancer and esophageal squamous cell carcinoma (24, 25). Thus, ADAMTS16 is well-connected to morphogenesis and human diseases, but without knowledge of its underlying mechanisms, i.e. its molecular characteristics, substrates, and the molecular pathways in which it participates.
Here, we identified FN as an ADAMTS16 substrate via a novel mass spectrometry (MS) approach, using a decellularized ECM produced in vitro by fibroblasts for substrate discovery. We show that ADAMTS16 cleaves FN near its N terminus with an immediate and long-term impact on ECM assembly. By upregulating MMP3 in the epithelial cell line MDCKI, ADAMTS16 creates an intriguing dual protease feed-forward loop that may serve to limit and fine-tune FN assembly and control tubular morphogenesis.
EXPERIMENTAL PROCEDURES
Antibodies
All antibodies and used dilutions are listed in supplemental Table S1.
Expression Plasmids
Mouse Adamts16 cDNA clone (with sequence NM_172053) was purchased from Creative Biogene (Shirley, NY). All constructs were generated by PCR using this clone as a template and HiFidelity Polymerase (Qiagen, Hombrechtikon, Switzerland) following the manufacturer's protocol. All constructs were C-terminally myc_His6 tagged and cloned into pCEP_pu (Invitrogen, Carlsbad, CA) for expression and verified by Sanger sequencing. ADAMTS16 mutant constructs were obtained by site directed mutagenesis PCR following a previously published protocol (26). For cloning and mutagenesis primer sequences see supplemental Table S2.
Cell Culture
HEK293-EBNA (CRL-10852, ATCC, Manassas, VA) cells were grown in DMEM supplemented with 10% FBS. To generate HEK293-EBNA cells stably expressing mouse ADAMTS16, mouse ADAMTS16-sh, mouse ADAMTS16-EA, or mouse ADAMTS16-sh-EA, cells were seeded on 0.1% gelatin-coated dishes (0.1% gelatin (Sigma Aldrich, St. Louis, MO) in H2O), transfected using jetPei® (Polyplus transfection, Illkirch, France) following manufacturer's instructions and selected with puromycin (2 μg/ml, Sigma Aldrich) for 14 days. Mouse BALB/c 3T3 fibroblasts (CCL-163, ATCC), human dermal fibroblasts (HDF, ATCC) and LN229 cells (CRL-2611, ATCC) were grown in DMEM supplemented with 10% FBS. MDCK strain I (MDCKI) cells, kindly provided by Dr. Martin Spiess (Basel, Switzerland), were grown in α-MEM supplemented with 10% FBS. To obtain MDCKI cells stably expressing mouse ADAMTS16, mouse ADAMTS16-sh, mouse ADAMTS16-EA, or mouse ADAMTS16-sh-EA, cells were transfected using FuGENE® 6 (Roche, Basel, Switzerland) and selected with puromycin for 14 days. All cell lines used in the experiments were tested for mycoplasma contamination by MycoAlertTM Mycoplasma Detection Kit (LT07–218, Lonza Basel, Switzerland).
Western Blotting
Cell extracts and conditioned media were prepared in Laemmli loading buffer with or without 2-mercaptoethanol and separated by SDS-PAGE. Proteins were transferred to PVDF membranes (Immobilon FL for fluorescence detection, EMD Millipore, Billerica, MA) and probed with primary antibody (supplemental Table S1) and detected by an enhanced fluorescence technique using an Odyssey CLx scanner (LI-COR Biosciences, Lincoln, NE).
Protein Purification
1 L of conditioned medium was filtered through a 0.22 μm filter (Corning, Corning, NY). After filtration the solution was applied to a pre-equilibrated 1 ml IMAC column (Ni-NTA agarose, Qiagen, Hombrechtikon, Switzerland) at 4 °C. Bound protein was eluted with 250 mm imidazole in 50 mm Tris-buffer (pH 7.5). For removal of imidazole, the protein solution was dialyzed against TBS (50 mm Tris-HCl, 150 mm NaCl, pH 7.5) supplemented with 5 μm ZnCl2. Protein concentration was determined with a NanoDrop ND-1000 spectrometer (ThermoFisher Scientific, Reinach, Switzerland). Protein absorption coefficients were calculated using the PDB ProtParam tool. The protein solution was stored in 50% (v/v) glycerol at −20 °C.
Immunofluorescence
HEK293-EBNA cells were seeded on 8-well glass chamber slides (Falcon Culture Slides, Thermo Fisher Scientific) and cultured in DMEM containing 10% FBS until they reached 80% confluence. Cells were transiently transfected with ADAMTS16-sh and ADAMTS16-CT, ADAMTS16 or empty vector only using Lipofectamine 3000 (Invitrogen/Life Technologies, CA). After 48 h under serum-free conditions the cells were fixed with 4% formaldehyde (PFA) for 20 min. The samples were incubated with anti-myc and anti-laminin for 1h at room temperature, followed by incubation with Alexa conjugated secondary antibody. Samples were mounted in ProLong Gold with DAPI (Life Technologies) and imaged with a Leica TCS5 SPII confocal microscope. Images comparing conditions were acquired under identical camera settings and analyzed using ImageJ software (U. S. National Institutes of Health, Bethesda, Maryland). LN229 cells were seeded on 8 well glass chamber slides and cultured in DMEM containing 10% FBS until they reached 90% confluence. Cells were transiently transfected using Lipofectamine 3000. Cells were maintained in DMEM with 10% FBS for another 48 h before fixation and staining.
Preparation of Cell-free ECM from HEK-EBNA Cells
HEK-EBNA cells were transiently transfected and cultured for 48 h in DMEM/10% FBS. Cells were removed using extraction buffer (0.5% Na-deoxycholate, 10 mm Tris-HCl, pH 7.5) supplemented with protease inhibitor (cOmplete™, Mini Protease Inhibitor Mixture, Sigma-Aldrich) for 30 min at 4 °C. ECM was washed with 2 mm Tris- HCl (pH 7.5). Total protein was analyzed by SDS-PAGE under reducing conditions.
Preparation of ECM Produced by Cultured BALB/c 3T3 Fibroblasts
BALB/c 3T3 fibroblasts were cultured in DMEM supplemented with 10% FBS for preparation of cell-free ECM as previously described (27). Briefly, 1 × 106 fibroblasts were seeded on a 6 cm culture dish (Corning) precoated with 0.1% gelatin and allowed to attach for 24 h. After 24 h the medium was replaced with fresh culture medium containing 10% FBS and 50 μg/ml ascorbic acid (Merck Millipore, Darmstadt, Germany). The medium was replaced every 24 h for 5 days. The cells were extracted from the matrix using PBS containing 0.5% Triton X-100 and NH4OH. PBS was added to dilute the cellular debris and the plate was stored overnight at 4 °C. Debris were removed, and ECM was washed with cold PBS. 5 × 105 HEK cells stably expressing mouse ADAMTS16, mouse ADAMTS16-sh, mouse ADAMTS16-sh-EA or transfected with empty vector as a control, were seeded on this matrix and cultured for 24 h in serum-free medium. ADAMTS16-sh-EA served as the control for both wtADAMTS16 and ADAMTS16-sh. The conditioned medium was analyzed by LC-MS/MS.
Sample Preparation and Analysis of the Digested ECM by Mass Spectrometry
The proteins in the medium were precipitated using trichloroacetic acid (TCA), washed with HPLC grade ice-cold acetone and dissolved in 0.5 m Tris pH 8.6 containing 6 m guanidine hydrochloride and aliquoted into four fractions for treatment with the endopeptidase Lys-C (Wako, Neuss, Germany) or trypsin (sequencing grade modified, Promega, Dübendorf, Switzerland), Asp-N (Roche) and combinations thereof after reduction with TCEP and alkylation with iodoacetamide (both Fluka, Buchs, Switzerland) and further addition of digestion buffer (50 mm Tris/HCl pH 8.6, 5 mm CaCl2). Peptides were separated on an EASY n-LC 1000 liquid chromatography system equipped with a C18 Acclaim PepMap 100 trap-column (75 μm × 2 cm, 3 μm, 100 Å) and a C18 New Objective analytical column (75 μm × 25 cm, Reprosil, 3 μm) coupled to a Thermo Orbitrap Fusion mass spectrometer (Thermo Scientific) equipped with a New Objective Digital Pico View source. Data were collected over a 30 min linear gradient from 2% buffer B (0.1% formic acid in acetonitrile) to 25% buffer B, followed by a 5 min linear gradient from 25% to 40% buffer B. Buffer A contained 0.1% formic acid in water. Sample digests were acidified with trifluoroacetic acid (Pierce) to a final concentration of 0.1% TFA.
Analysis and Peptide Identification by LC-MS/MS
Peptides were identified by searching SwissProt restricted to mammalian proteins (version 2015–01, # of entries searched for: 16920 mouse proteins) using Mascot Distiller (version 2.5.1, Matrix Science) and Mascot (version 2.5.1, Matrix Science) allowing the following post-translational modifications; fixed modifications: Carbamidomethyl (C), variable modifications: acetylation at protein N termini, deamidation at asparagine and glutamine, oxidation at methionine and phosphorylation at serine/threonine and no enzyme specificity. The number of max. missed cleavages was set to 1, with a peptide mass tolerance ± 10 ppm and a fragment mass tolerance ± 0.6 Da. Data were compiled and evaluated with Scaffold (version Scaffold_4.4.1.1, Proteome Software), considering proteins identified with at least five peptides and having thresholds for protein and peptides of 90%, with peptide FDR 2.1% (Prophet) and protein FDR 0% (Prophet). The FDR was calculated using Scaffold software using the following parameters: Protein Grouping Strategy: Experiment-wide grouping with binary peptide-protein weights. Peptide Thresholds: 90.0% minimum and Protein Thresholds: 90.0% minimum and 5 peptides minimum. Scaffold software was used for a qualitative analysis of ADAMTS16 cleavage products. For a quantitative analysis of single peptides, the area under the curves of the MS1 spectra of single peptides was analyzed using a label free approach. For this, the corresponding data files were loaded into Progenesis QI for proteomics (version 2.0.5556.29015, Waters) and analyzed using the default settings of the program (medium sensitivity and peptide charge: 1–20). All the raw data files of the single runs, the scaffold file (.sf3) and the corresponding Mascot result files (file name: F077701–5.dat) as well as the Mascot peak list (mgf files) and the corresponding search files (xml files) for the Progenesis quantification are available via ProteomeXchange with identifier PXD007284.
Sample Preparation and LC-MS/MS Analysis of Human Fibronectin Cleavage Products
12-well tissue culture dishes were coated with purified human plasma fibronectin (FC010–10MG, Merck Millipore) in a final concentration of 30 μg/ml in PBS. HEK293-EBNA cells stably expressing various ADAMTS16 constructs were seeded on top of the FN under serum-free conditions and incubated for 24 h. The supernatant was analyzed by Western blotting or by LC-MS/MS for potential cleavage products. Prior to LC-MS/MS analysis the conditioned medium was TCA-precipitated and digested with LysC following the protocol described in the previous section. The generated peptides were acidified with 1 μl of 20% TFA and analyzed by capillary liquid chromatography tandem mass spectrometry with an EASY-nLC 1000 using a two-column set up (Thermo Scientific). The peptides were loaded with 0.1% formic acid, 2% acetonitrile in H2O onto a peptide trap (Acclaim PepMap 100, 75 μm × 2 cm, C18, 3 μm, 100 Å) at a constant pressure of 800 bar. Peptides were separated at a flow rate of 150 μl/min with a linear gradient of 2–6% buffer B in buffer A in 3 min followed by an linear increase from 6 to 22% in 40 min, 22–28% in 9 min, 28–36% in 8 min, 36–80% in 1 min and the column was finally washed for 14 min at 80% B (Buffer A: 0.1% formic acid, buffer B: 0.1% formic acid in acetonitrile) on a 50 μm x 15 cm ES801 C18, 2 μm, 100 Å column mounted on a DPV ion source (New Objective) connected to an Orbitrap Fusion mass spectrometer (Thermo Scientific). The data were acquired using 120,000 resolution for the peptide measurements in the Orbitrap and a top T (3s) method with HCD fragmentation for each precursor and fragment measurement in the LTQ according the recommendation of the manufacturer (Thermo Scientific).
Peptides were identified using Mascot (version 2.5.1, Matrix Science) and Mascot Distiller (version 2.5.1, Matrix Science) allowing the following post-translational modifications; fixed modification: Carbamidomethyl (C), variable modifications: acetylation at protein N termini, deamidation at asparagine and glutamine, oxidation at methionine and phosphorylation at serine/threonine. The number of max. missed cleavages was set to 1, with a peptide mass tolerance ± 5 ppm and a fragment mass tolerance ± 0.6 Da and quantified using Progenesis QI for proteomics (version 2.0.5556.29015, Waters). All raw data files of the single runs, a Scaffold file (.sf3) as well as the Mascot peak list (mgf files) and the corresponding search files (xml files) for the Progenesis QI quantification are available via ProteomeXchange with identifier PXD007284.
In Vitro Fibrillogenesis
HDF and HEK293 cells were co-cultured in a 3:5 (fibroblasts: HEK cells) ratio in DMEM containing 10% FBS for 24 h, 48 h, or 4 days respectively. For 8-day co-cultures the cells were seeded in a 4:1 (fibroblasts: HEK cells) ratio. Cells were fixed in 4% PFA, stained and imaged using a Leica TCS5 SPII confocal microscope. Images were analyzed using ImageJ software.
3D-culture Staining and Imaging
MDCKI cells were embedded in 3D collagen gels. Acid-solubilized collagen (Corning Collagen I) was mixed with 10X PBS in a 1:1 ratio, followed by pH adjustment to 7.5 with 0.1 m NaOH. The collagen concentration was adjusted to a final concentration of 2.5 mg/ml using α-MEM/10%FBS. 150 μl of collagen containing 1.5 × 104 cells was poured into each well of an 8-well glass chamber slide and allowed to solidify at 37 °C. After gelation α-MEM containing 10% FBS was added and replaced every 24 h.
After 4 days or 8 days of culture, the 3D gels were fixed using 4% PFA for 30 min and permeabilized 3X 15 min at room temperature using 0.3% Triton X-100 in PBS. Primary antibodies were diluted in PBST and applied overnight at 4 °C. Gels were washed with PBST and incubated with secondary antibodies in PBST for 2 h at room temperature. Images were taken with a Leica TCS5 SPII confocal microscope and analyzed with ImageJ software.
RNA Analysis by qRT-PCR
Total RNA was isolated using the RNeasy Plus Micro Kit (Quiagen). RNA was reversed transcribed and relative mRNA levels were detected as described (28). Relative mRNA levels for the genes, normalized to TBP, were measured using Platinum® SYBR® Green qPCR SuperMix-UDG with ROX (Invitrogen). Real-time PCR was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems, Rotkreuz, Switzerland) using a standard cycling profile. All samples were run in triplicates. Data were analyzed by the ΔΔCt method (29). RT-PCR primers are listed in supplemental Table S3.
Labeling of FN
0.5 mg of plasma FN (F0895, Sigma-Aldrich) was labeled using the Alexa Fluor 488 protein labeling kit (A10235, Thermo Fisher Scientific) following the manufacturer's instructions. 30 μg/ml of the labeled FN were plated on 8-well glass chamber slides. HEK-EBNA cells stably expressing wtADAMTS16, ADAMTS16-EA or empty vector were seeded under serum-free conditions and allowed to attach for 48 h prior to fixation and phalloidin staining.
FN Cleavage
Purified ADAMTS16-sh was incubated with plasma FN (Sigma-Aldrich) or 500 ng of the 70 kDa N-terminal FN fragment or the 4F1–2F2 produced in insect cells (30) for 4 h at 37 °C in TBS (pH 7.5) supplemented with 10 mm CaCl2. Cleavage products were analyzed by immunoblotting under reducing conditions.
Experimental Design and Statistical Rational
All grouped data are means ± S.E. The statistical analysis was performed using GraphPad InStat version 3.05. Differences between two groups were evaluated using a two-tailed Student t test for parametric data. Values of p < 0.05 were considered statistically significant. Empty vector and an inactive mutant of ADAMTS16 were used as control for all experiments. All qRT-PCR experiments were done in triplicates. Relative amount of FN was quantified using ImageJ software. Each condition was done in triplicate and a total of 12 images per condition were analyzed. For spheroid size quantification a total number of 50 spheroids per condition from three independent experiments together were evaluated. For lumen quantification a total of 16 spheroids per condition from 3 independent experiments were quantified.
Human FN digestion was done in biological triplicates and all triplicates were analyzed by LC-MS/MS to get statistically relevant quantifications of the cleavage peptides. The presence of the identified mouse FN peptide from decellularized ECM was verified in two individual runs using different working proteases and was further verified by a complete repetition of the experiment using the same experimental setup for ADAMTS16-sh, ADAMTS16-sh-EA and vector control.
RESULTS
ADAMTS16 is Secreted and Binds to ECM via its C-terminal Modules
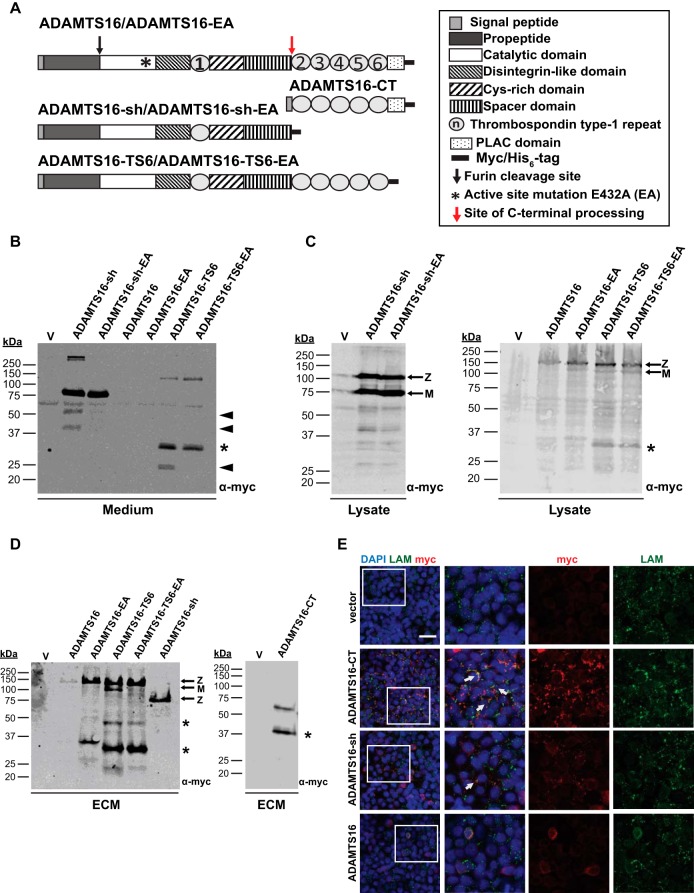
Constructs expressing wt myc-tagged ADAMTS16 and their corresponding active site mutants (E432A) (Fig. 1A) were transiently or stably expressed in HEK293-EBNA cells. Neither active full-length ADAMTS16 (wtADAMTS16) nor inactive ADAMTS16 (ADAMTS16-EA) were detectable in the conditioned medium of either stably or transiently transfected HEK-EBNA cells (Fig. 1B). In contrast, liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the medium of these cells detected an abundance of peptides spanning the catalytic domain to the spacer, but none derived from the C-terminal thrombospondin (TSR) repeats and the protease and lacunin (PLAC) domain (Fig. 1A, supplemental Fig. S1, supplemental Table S4). These findings suggested that wtADAMTS16 was C-terminally processed with release of a form containing the catalytic domain through the spacer module into the medium, whereas the remaining C-terminal modules were retained within the ECM and or the cell surface.
Fig. 1.
ADAMTS16 binds ECM and is C-terminally processed. A, Domain organization of ADAMTS16 constructs. B, Western blot analysis of conditioned medium from HEK293-EBNA cells transfected with the indicated constructs or empty vector (v) using anti-myc antibody and enhanced chemiluminescence. The black arrowheads and asterisk indicate autocatalytic and non-autocatalytic C-terminal fragments of ADAMTS16 respectively. C, Western blot analysis of HEK293-EBNA cell lysate transfected with the indicated constructs or empty vector (v). The asterisks indicate autocatalytic and non-autocatalytic C-terminal fragments of ADAMTS16. Z and M indicate the zymogen and mature forms arising from furin processing. D, Western blot analysis of cell-free ECM from HEK293-EBNA cells expressing various ADAMTS16 constructs or transfected with empty vector (v). The asterisks indicate C-terminal fragments. Z and M indicate the zymogen and mature forms arising from furin processing. E, HEK293-EBNA monolayers were stained for wtADAMTS16, ADAMTS16-CT and ADAMTS16-sh with anti-myc (red) and laminin (green) as ECM marker. The images show stronger localization of ADAMTS16-CT to the cell layer than ADAMTS16-sh. White arrows indicate co-localization of ADAMTS16 and laminin. The scale bar is 50 μm. The right-hand panels are 2X amplification of the boxed areas in the left-hand column.
To gain further insights into C-terminal processing of ADAMTS16, we designed a construct lacking the C-terminal TSRs and PLAC domain (ADAMTS16-sh) thereby mimicking wtADAMTS16 after C-terminal processing and release into the medium (Fig. 1A). In contrast to ADAMTS16, ADAMTS16-sh as well as its proteolytically inactive counterpart (ADAMTS16-sh-EA), were readily detectable in the conditioned medium using anti-myc (Fig. 1B). A reported isoform of ADAMTS16, lacking the C-terminal PLAC domain (ADAMTS16-TS6) (31) (Fig. 1A) was also detected in the conditioned medium of transfected HEK-EBNA cells (Fig. 1B), suggesting that the C-terminal modules, such as PLAC may regulate binding of ADAMTS16 to the cell surface and/or the ECM. Smaller than expected ADAMTS16-sh and ADAMTS16-TS6 molecular fragments in the medium are indicative of autocatalysis, in addition to proteolysis by ambient proteases within the TSR2- PLAC region (ancillary domain) (Fig. 1B, 1C). These findings support the assumption of C-terminal processing of ADAMTS16 between the spacer and TSR2, with retention of C-terminal modules in the cell layer and release of the remaining ADAMTS16 into the medium.
In addition, like other ADAMTS proteases, ADAMTS16 likely undergoes furin-processing at a consensus site, RHKR279, releasing the N-terminal propeptide. Consistent with this, the ADAMTS16 constructs observed in the medium by antibodies to the C-terminal myc tag were ∼30 kDa smaller than in the cell lysates (Fig. 1B, 1C). To determine if full-length ADAMTS16 binds to and retains in the ECM we performed a DOC insolubility assay using transiently transfected HEK-EBNA cells. All ADAMTS16 constructs could be detected within the cell-free ECM (Fig. 1D). In contrast to ADAMTS16-EA, ADAMTS16-TS6, ADAMTS16-TS6-EA, and ADAMTS16-sh, wtADAMTS16 was detected at markedly lower levels in the cell-free ECM of transfected HEK293-EBNA cells (Fig. 1D). This further confirms the finding that autoproteolysis likely had complex effects on full-length ADAMTS16 with increased lability compared with the other constructs. Immunostaining of ADAMTS16 transfected cells with laminin (a marker for ECM) showed that the C-terminal domain of ADAMTS16 (ADAMTS16-CT) was strongly localized to the ECM. ADAMTS16-sh was localized to the ECM as well, but at considerably reduced levels compared with ADAMTS16-CT (Fig. 1E). Together, these findings provide evidence that cell and ECM binding of ADAMTS16 is primarily mediated by its C-terminal ancillary domain, and that ADAMTS16 is released into the medium because of complex proteolytic processing, including autocatalysis (Fig. 1B–1E supplemental Fig. S1).
Mass Spectrometry Identifies FN as an ADAMTS16 Substrate
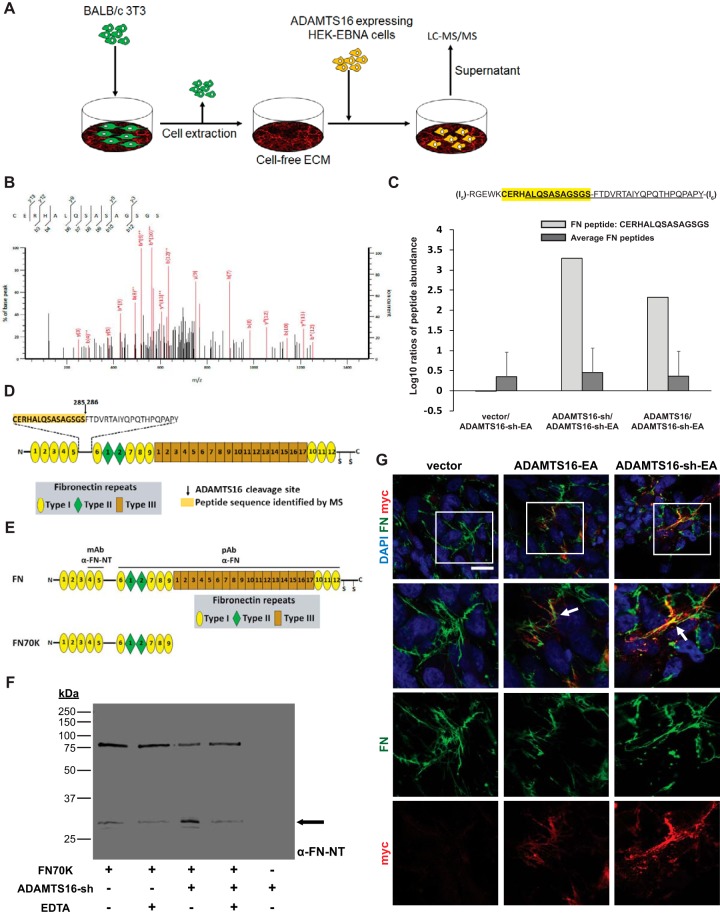
Because ADAMTS16 binds to the ECM and is extensively processed, purification of full-length catalytically active enzyme was not feasible. Therefore, to identify ADAMTS16 substrates, we designed a strategy in which a decellularized ECM, deposited by mouse BALB/c 3T3 fibroblasts in vitro, was digested by HEK293-EBNA cells expressing wtADAMTS16, ADAMTS16-sh or its catalytically inactive mutant ADAMTS16-sh-EA. The medium from these cultures was analyzed by LC-MS/MS to identify ADAMTS16 substrates (Fig. 2A, Table I, II) by searching for peptides with cleavage sites that did not arise from the proteases used to generate peptides for LC-MS/MS. This analysis identified a mouse FN peptide 270CERHALQSASAGSGS285 (GenBank ID NP_034363) exclusively in digests of ECM by the two active ADAMTS16 constructs but not the catalytically inactive mutant and/or the vector control (Fig. 2B, 2C, supplemental Fig. S2A). The presence of the FN peptide 270CERHALQSASAGSGS285 was found in both active samples (wtADAMTS16 and ADAMTS16-sh) using LysC alone as working protease or LysC in combination with AspN (Table I, supplemental Table S5–S7). The findings could be reproduced in a second independent experiment (data available in ProteomeXchange with identifier PXD007284). This finding suggests that ADAMTS16 cleaves FN within its N-terminal linker sequence between domain (I)5 and (I)6 (Fig. 2C). We identified additional peptides showing cleavage sites that did not arise from the sample preparation process (Table I). Because these peptides were also identified in the control samples, but at lower abundance, we conclude that they were not uniquely targeted by ADAMTS16 and were not pursued further (Table I, II, supplemental Table S5, S7, supplemental Fig. S2C, S2D). The similar results obtained for wtADAMTS16 and ADAMTS16-sh indicate that although C-terminal processing influences enzyme localization, it does not significantly influence FN recognition and proteolysis. Although a unique peptide identified FN as a potential substrate of ADAMTS16, the FN sequence coverage did not vary between the different samples, indicating the same overall amount of FN in the conditioned medium (Fig. 2C, supplemental Fig. S2C, supplemental Table S7). On the other hand, differential sequence coverage was noted for biglycan, plectin, and serpinH1, but no unique peptides arising from ADAMTS16 cleavage were identified (Table II, supplemental Table S7). These molecules could also be potential ADAMTS16 substrates where unique cleaved peptides were not detected by LC-MS/MS.
Fig. 2.
Identification and validation of FN as an ADAMTS16 substrate. A, Schematic of the proteomic strategy used to identify ADAMTS16 substrates. B, MS2 spectrum of the doubly charged FN peptide CERHALQSASAGSGS identified in the LysC-AspN digest of wtADAMTS16. C, Histogram of log10 ratios of relative abundance of the FN peptide CERHALQSASAGSGS identified by LC-MS/MS in the LysC/AspN digest showing its presence exclusively in matrix incubated with active ADAMTS16 and ADAMTS16-sh expressing cells, but not in the control samples. Log10 ratios compare wtADAMTS16 with ADAMTS16-sh-EA, ADAMTS16-sh with ADAMTS16-sh-EA and vector with ADAMTS16-sh-EA. Average of log10 ratios of all identified FN peptides, showed similar values between the samples, indicating equal amounts of FN in each sample. D, Domain structure of FN with expanded view of the amino acid sequence of the linker between domains (I)5 and (I)6 showing the peptide identified by LC-MS/MS (yellow highlight) and the potential ADAMTS16 cleavage site. E, The domain structure of FN and the N-terminal 70 kDa FN fragment (FN70K). Antibody binding sites of the monoclonal anti FN-NT (MAB1936) and the polyclonal anti-FN (Ruth Chiquet) are indicated above full-length FN. F, Western blot analysis of FN70K incubated with purified active ADAMTS16-sh in the presence or absence of EDTA. Anti-FN antibody (MAB1936) specific to the N-terminal heparin-binding domain of FN was used to identify release of the 30 kDa N-terminal heparin-binding domain (black arrow). G, Co-localization of ADAMTS16 with FN fibrils. Transiently transfected LN229 cells were cultured for 48 h and stained for ADAMTS16-EA and ADAMTS16-sh-EA with anti-myc (red) and FN (green). The images show co-localization of ADAMTS16-EA and ADAMTS16-sh-EA with FN fibers, indicating that the C terminus of ADAMTS16 is not essential for FN binding. White arrows show co-localization of ADAMTS16 with FN. Scale bar is 50 μm. Lower panels show 2× amplification of the boxed areas in the top row.
Table I. Peptides and peptide ratios obtained from ADAMTS16 digestion of BALB/c 3T3 extracellular matrix.
| Protein name | Preceding amino acid | Peptide seq. | Succeeding amino acid | Precursor charge | mass/charge (m/z) | Working protease | log10 wtADAMTS16/ADAMTS16-sh-EA | log10 ADAMTS16-sh/ADAMTS16-sh-EA | log10 vector/ADAMTS16-sh-EA |
|---|---|---|---|---|---|---|---|---|---|
| FN | K | 271CERHALQSASAGSGS285 | F | 2+ | 759.34 | LysC/AspN | 2.33 | 3.29 | −0.01 |
| FN | K | 271CERHALQSASAGSGS285 | F | 2+ | 759.34 | LysC | 3.51 | 4.50 | 1.66 |
| FN | K | 271CERHALQSASAGSGS285 | F | 3+ | 506.56 | LysC | 3.42 | 4.29 | 1.66 |
| FN | D | 557PIDQCQDSETR567 | T | 2+ | 674.79 | LysC/Trypsin | 0.65 | 0.37 | 0.15 |
| LTBP-1 | Y | 1581GRDALVDFSEQYGPETDPYFIQDR1604 | F | 3+ | 940.10 | LysC/Trypsin | 0.62 | 0.32 | 0.07 |
| POSTN | T | 604LLVNELK610 | S | 2+ | 415.25 | LysC | 0.2 | 0.14 | −0.08 |
| GELS | R | 450IEGSNKVPVDPATY463 | G | 2+ | 745.38 | AspN/Trypsin | 0.37 | −0.01 | 0.18 |
Comparison of peptides released from digests of decellularized BALB/c 3T3 ECM (mouse) with HEK-EBNA cells stably expressing wtADAMTS16, ADAMTS16-sh-EA, ADAMTS16-sh, and vector control as indicated in column headings. Peptides arising from cleavage by proteases different from the ones used for sample preparation (working proteases) are shown. No change in peptide abundance gives a ratio near 0. Increased peptide abundance in active samples gives a ratio >0. Ratios were calculated using Progenesis QI software. All peptides shown in the table are of murine (BALB/c 3T3) origin.
All Mascot peak lists and search files used to quantify peptide ratios and abundance are available via ProteomeXchange with identifier PXD007284.
Table II. Unique peptide count and sequence coverage of ECM proteins obtained from ADAMTS16 digestion of BALB/c 3T3 extracellular matrix.
| Protein name | Accession Nr. | Unique peptide count (wtADAMTS16) | Unique peptide count (ADAMTS16-sh) | Unique peptide count (ADAMTS16-sh-EA) | Unique peptide count (vector) | Sequence coverage % (wtADAMTS16) | Sequence coverage % (ADAMTS16-sh) | Sequence coverage % (ADAMTS16-sh-EA) | Sequence coverage % (vector) |
|---|---|---|---|---|---|---|---|---|---|
| FN | P11276 | 206 | 150 | 186 | 180 | 71 | 66 | 70 | 72 |
| LTBP1 | Q8CG19 | 41 | 27 | 21 | 25 | 28 | 20 | 17 | 21 |
| POSTN | Q62009 | 23 | 23 | 25 | 23 | 32 | 33 | 37 | 30 |
| GELS | P13020 | 46 | 32 | 46 | 40 | 57 | 45 | 58 | 50 |
| PGS1 | P28653 | 5 | 8 | 10 | 12 | 16 | 33 | 30 | 39 |
| SERPH | P19324 | 3 | 4 | 12 | 14 | 12 | 17 | 39 | 39 |
| PLEC | Q9QXS1 | 43 | 23 | 14 | 11 | 11 | 6.4 | 4.2 | 3.3 |
Differences in sequence coverage as well as in peptide counts were obtained for other ECM proteins. Sequence coverage and unique peptide counts were calculated using Scaffold software, the following posttranslational modifications were included in the Mascot search: acetylation at the protein N-terminus, oxidation, phosphorylation and deamidation. No enzyme specificity was applied to the search parameters to identify potential ADAMTS16 cleavage products. All ECM proteins shown in the table are of murine (BALB/c 3T3) origin. The original scaffold file and the corresponding Mascot search files are available via ProteomeXchange with identifier PXD007284.
For further validation of FN as substrate of ADAMTS16, cell culture dishes were coated with human plasma FN and cells stably expressing different ADAMTS16 constructs were seeded under serum-free conditions. ADAMTS16-mediated FN proteolysis was allowed for 24 h. The conditioned medium was analyzed for FN cleavage products by Western blotting using an antibody specific to the N-terminal domain of human FN. Samples incubated with ADAMTS16-sh showed a strong increase of the 30 kDa FN N-terminal heparin binding domain after 24 h of incubation in the conditioned medium compared with inactive mutant and vector control (supplemental Fig. S3A). To further validate FN as a substrate the conditioned medium was analyzed by mass spectrometry for specific FN cleavage peptides after 24 h of incubation with ADAMTS16-sh. The specific cleavage peptides 270CERHTSVQTTSSGSGPFTDVRAA292 from the (I)5-(I)6 linker sequence of FN was identified in very high abundance in the active ADAMTS16-sh sample but not in the inactive ADAMTS16-sh-EA and vector control samples, although overall FN peptide abundance was comparable supplemental Fig. S3B–S3D; supplemental Table S8, S9). This finding further suggests that ADAMTS16 cleaves FN within the N-terminal (I)5-(I)6 linker. We identified additional human FN cleavage peptides toward the C-terminal side of this cleavage indicating that ADAMTS16 might cleave FN elsewhere in addition to the characterized N-terminal cleavage site (supplemental Table S8).
Because of the extensive post-secretion processing, purification of full-length ADAMTS16 was not feasible using the C-terminal tags, but ADAMTS16-sh was purified and its proteolytic activity was demonstrated by cleavage of the generic protease substrate α2-macroglobulin (supplemental Fig. S4A) as previously demonstrated (32). To obtain biochemical validation that fibronectin is a substrate of ADAMTS16 and to determine the requirements for proteolysis, we incubated purified ADAMTS16-sh with different FN constructs. A recombinant 70 kDa FN fragment extending from module (I)1 to module (I)9 was cleaved to generate a 30 kDa N-terminal fragment (Fig. 2E, 2F). Full-length plasma FN, whose globular structure potentially masks susceptible peptide bonds, was not cleaved by purified ADAMTS16-sh in vitro (supplemental Fig. S4B). ADAMTS16-sh failed to cleave the linker of a shorter recombinant FN fragment spanning the (I)4 to (II)2 modules suggesting that binding of ADAMTS16 to FN may occur via domains distant from the cleavage site (Fig. S4C, S4D). We also tested the ability of wtADAMTS16 to cleave FN using a cell-based approach. Seeding of HEK-EBNA cells expressing wtADAMTS16, but not ADAMTS16-EA on Alexa488 labeled FN showed a loss of FN fluorescence underlying the cells (supplemental Fig. S4E), suggesting that in contrast to plasma FN in solution, bound plasma FN could be cleaved by ADAMTS16-expressing cells. In transiently transfected LN-229 glioblastoma cells, ADAMTS16 co-localized with FN fibrils in vitro (Fig. 2G) and the FN network was reduced in cells expressing wtADAMTS16 (supplemental Fig. S4F). Together, these findings indicate that FN is directly cleaved by ADAMTS16, but that cleavage requires a distant binding region on FN and exposure of the scissile bond by traction provided by cell contraction.
ADAMTS16 Inhibits FN Assembly and Elaboration of a Mature FN-dependent ECM
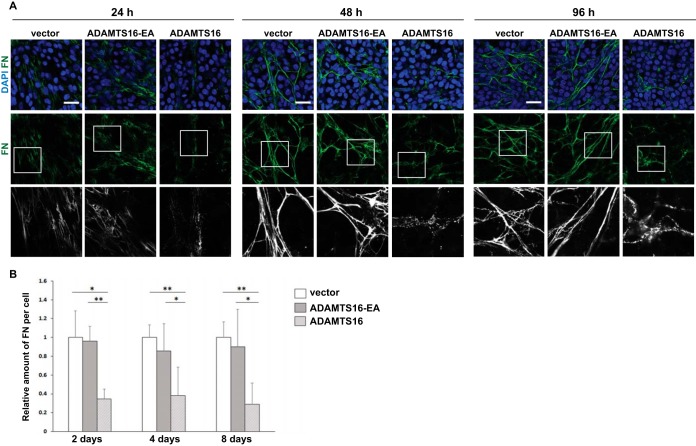
The 30 kDa N-terminal heparin binding domain released by wtADAMTS16 was previously shown to regulate FN self-association during fibrillogenesis (5, 6). To investigate whether ADAMTS16 had a direct effect on FN fibril networks, we investigated the temporal effect of ADAMTS16 on the FN network. Human dermal fibroblasts (HDF) co-cultured with HEK cells stably transfected with either vector or inactive ADAMTS16-EA established a robust FN network within 24 h in culture medium containing 10% FBS. In contrast, a profound reduction of FN fibrils was observed in the presence of wtADAMTS16 after 24 h despite comparable cell density (Fig. 3A). Indeed, in the presence of wtADAMTS16, FN was visible as punctate structures localized near the surface of fibroblasts, rather than in a well-defined fibrillar network. Upon continuing co-culture to 2, 4, and 8 days, a similar impact on the network was observed, indicative of a continuing impact on FN assembly, although digestion of preformed fibrils cannot be ruled out (Fig. 3A, 3B, Fig. 4). This temporal sequence in reduction of FN fibrillogenesis suggested that ADAMTS16 affected FN fibril maturation at the point of, or shortly after initial fibril assembly and therefore reduced further assembly and fibril maturation. FN1 mRNA levels were comparable in the various co-cultures (supplemental Fig. S5A), suggesting that reduction of fibrils in the presence of active ADAMTS16 did not have a transcriptional basis. The C-terminally truncated construct, ADAMTS16-sh had a similar effect on fibril maturation as full-length ADAMTS16, showing that the catalytic specificity for FN and inhibition of fibrillogenesis did not rely on the C terminus of the protease (supplemental Fig. S5B, S5C). Moreover, a similar impact was observed on FN networks formed by mouse BALB/c-3T3 fibroblasts, which were used for substrate identification, when co-cultured with wtADAMTS16 expressing HEK-EBNA cells (supplemental Fig. S5D). Impairment of FN assembly in LN-229 cells, HDF and 3T3 cells suggested that the effect of ADAMTS16 was independent of cell type.
Fig. 3.
ADAMTS16 affects FN fibril assembly by human dermal fibroblasts (HDF). A, Confocal analysis of FN networks after 24, 48 or 96 h of co-culture of HDF with ADAMTS16-expressing HEK293-EBNA cells, shows reduced FN fibril staining (green). Scale bars are 50 μm. Lower panels are 3X amplifications of the boxed areas in the upper row. B, The relative amount of fibrillar FN per cell was measured in wtADAMTS16, ADAMTS16-EA, and vector control co-cultures. Data are mean ± S.E., n = 12 images from 3 independent experiments. (*) p < 0.05 by Student t test, (**) p < 0.01, Student t test.
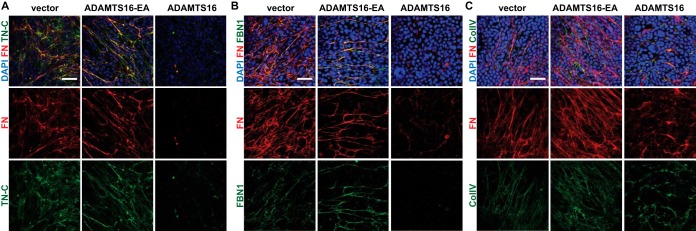
Fig. 4.
Reduction of FN fibrillogenesis in the presence of ADAMTS16 inhibits ECM maturation. A, 8 day co-culture of HDF with ADAMTS16-expressing HEK cells, shows reduction of the formation of FN (red, monoclonal FN-NT) and tenascin-C (green) networks. B, 8-day co-culture of HDF with ADAMTS16-expressing HEK cells, shows reduction of the formation of a FN (red) and fibrillin- 1 (green) network. C, 8 day co-culture of HDF with ADAMTS16-expressing HEK cells, shows thickening and shortening of collagen IV bundles. FN (red, monoclonal FN- NT (7D5)) and collagen IV (green). Scale bars are 50 μm.
Because fibrillin-1 (FBN1) and tenascin-C (TN-C) assembly requires a preformed FN network, we used them as reporters for the formation of a stable, mature ECM after formation of the primordial FN matrix. The 8-day co-culture was investigated for proper FBN1 and TN-C assembly, in addition to FN fibrillogenesis. Whereas the ADAMTS16 active-site mutant had no effect on the formation of FBN1 and TN-C fibrils, active ADAMTS16 strongly impaired the formation of FBN1 and TN-C networks (Fig. 4A, 4B). Whereas collagen IV formed a network with long thin fibers in the presence of ADAMTS16-EA, thicker and shorter collagen IV fibers accumulated in HDF cultures exposed to wtADAMTS16 (Fig. 4C). Therefore, we conclude that ADAMTS16 overexpression can influence the integrity of other ECM assemblies by interference with FN assembly.
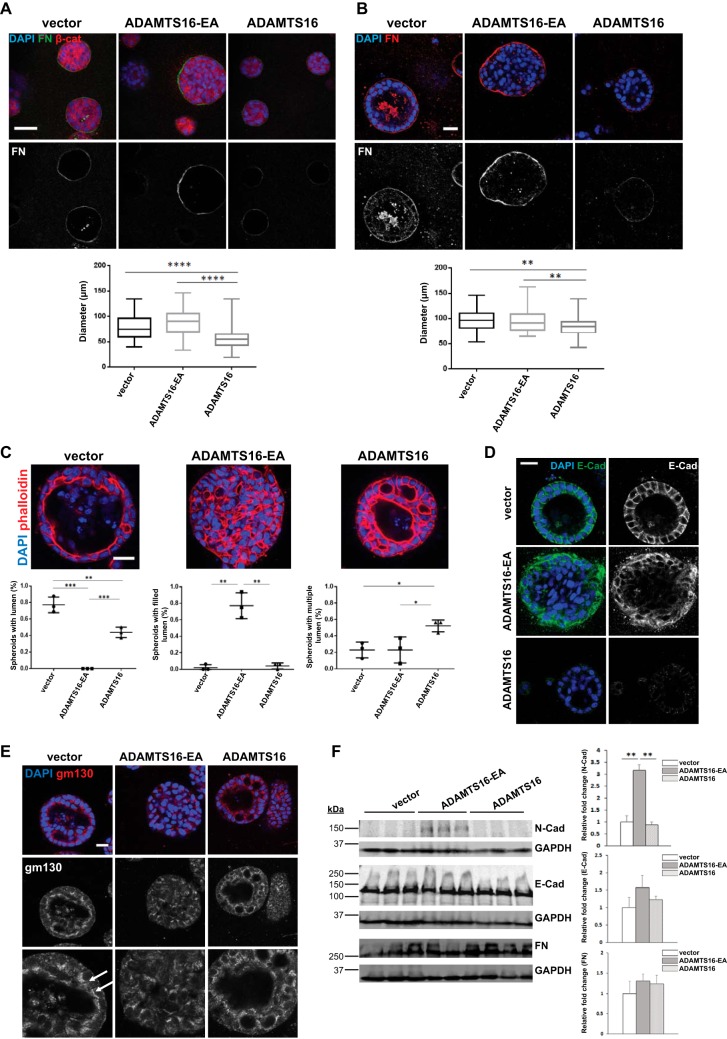
ADAMTS16 Impairs Spheroid Morphogenesis by MDCKI Cells in 3D Collagen Gels Via FN Proteolysis
Because of prior work indicating a role for ADAMTS16 in renal development (21, 22), renal tubule-derived MDCKI cells, which have very low levels of ADAMTS16 mRNA were used as a model to investigate the effect of ADAMTS16 on tubulogenesis. MDCKI cells stably transfected with ADAMTS16 constructs did not show any obvious morphological differences in monolayer culture (data not shown). When grown in 3-dimensional collagen gels, MDCKI cells form spheroids in which the cells are arranged with strict apical-basal polarity. In this spheroid-formation assay, wtADAMTS16 transfected cells showed less intense FN staining in the basement membrane whereas the ADAMTS16-EA expressing spheroids had stronger FN staining compared with vector control after 4 days and 8 days in 3D culture (Fig. 5A, 5B). This finding suggested increased FN digestion in the presence of ADAMTS16 and that ADAMTS16-EA may stabilize FN fibers in 3D cultures (Fig. 5A, 5B), or that binding to ADAMTS16-EA protects them from cleavage. Additionally we observed reduced laminin staining in spheroids expressing wtADAMTS16 after 8 days in culture compared with vector or ADAMTS16-EA expressing spheroids (supplemental Fig. S6A). Consistent with the reduction of FN, wtADAMTS16-expressing spheroids were significantly smaller after 4 days and 8 days compared with vector control and ADAMTS16-EA expressing cells (Fig. 5A, 5B). In addition to the obvious size difference, wtADAMTS16-expressing spheroids formed multiple lumina compared with spheroids made by MDCKI cells transfected with empty vector. Moreover, spheroids expressing ADAMTS16-EA failed to form a lumen (Fig. 5C). E-cadherin staining of spheroids expressing wtADAMTS16 showed weaker E-cadherin staining than spheroids expressing empty vector (Fig. 5D) although CDH1 mRNA was unaltered (supplemental Fig. S6B). In spheroids expressing ADAMTS16-EA, E-cadherin was localized to the membrane of cells attached to the basolateral FN matrix, whereas cells in the center of the spheroids showed no E-cadherin expression (Fig. 5D). To further investigate the polarity of these cells, we stained the spheroids for the Golgi marker gm130 after 8 days in culture. Whereas spheroids formed by vector-transfected cells showed a clear apical gm130 distribution, spheroids formed by cells expressing ADAMTS16-EA lacked polarity as evidenced by random distribution of gm130 (Fig. 5E). Spheroids formed by cells expressing wtADAMTS16 showed polarized gm130 staining in sections containing a single layer of cells, which was disrupted where multiple lumina were formed (Fig. 5E). Western blot analysis of the MDCKI monolayers showed similar E-cadherin levels when cultured in 2D and acquisition of N-cadherin expression in cells expressing ADAMTS16-EA (Fig. 5F). Together, these findings suggest that wtADAMTS16 overexpression reduces spheroid growth and promotes formation of multiluminal spheroids, whereas over-expression of ADAMTS16-EA promotes epithelial-mesenchymal transition in spheroids, evidenced by loss of cell polarity and acquisition of N-cadherin.
Fig. 5.
ADAMTS16 impairs spheroid morphogenesis by MDCKI cells in 3D collagen gels. A, Confocal microscopy of spheroids formed by ADAMTS16-expressing MDCKI cells in 3D collagen gels after 4 days. Spheroids were stained for β-catenin (red), FN (green) and nuclei (DAPI, blue). Quantitative analysis of spheroid diameter in 3D collagen gels shows size reduction upon wtADAMTS16 overexpression compared with vector and ADAMTS16-EA control after 4 days. n = 50 spheroids from 3 independent experiments. (****) p < 0.0001, Student t test B, Confocal microscopy of spheroids formed by ADAMTS16-expressing MDCKI cells in 3D collagen gels after 8 days. Spheroids were stained for FN (red) and nuclei (DAPI, blue). Reduction of FN staining in spheroids expressing wtADAMTS16 was observed whereas spheroids expressing ADAMTS16-EA had increased FN staining. Quantitative analysis of spheroid diameter shows size reduction upon ADAMTS16 overexpression compared with vector and ADAMTS16-EA control after 8 days. n = 50 spheroids from 3 independent experiments. (**) p < 0.01, Student t test. C, Spheroids were stained for phalloidin (red) and DAPI (blue) after 8 days in culture. Quantification of lumen formation in spheroids after 8 days in culture. n = 16 per experiment and construct, experiments were repeated 3 times. (*) p < 0.05, (**) p < 0.01, (***) p < 0.001 D, Confocal microscopy of MDCKI spheroids in 3D collagen gels after 8 days showing perturbation of E-cadherin staining in spheroids expressing wtADAMTS16. Spheroids were stained for E-cadherin (green) and nuclei (DAPI, blue). E, Confocal microscopy of spheroids formed by ADAMTS16-expressing MDCKI cells in 3D collagen gels after 8 days. Spheres were stained for gm130 (red) and nuclei (DAPI, blue). White arrows show polarized gm130 staining. Lower images are 2× amplifications upper panel. F, Western blot analysis of MDCKI cells in monolayer culture. N-cadherin expression was strongly increased in cells expressing ADAMTS16-EA. No changes in E-cadherin or FN levels were observed. Scale bars are 50 μm in A, B and E and 25 μm in C, D. **, p < 0.01 by student t-test.
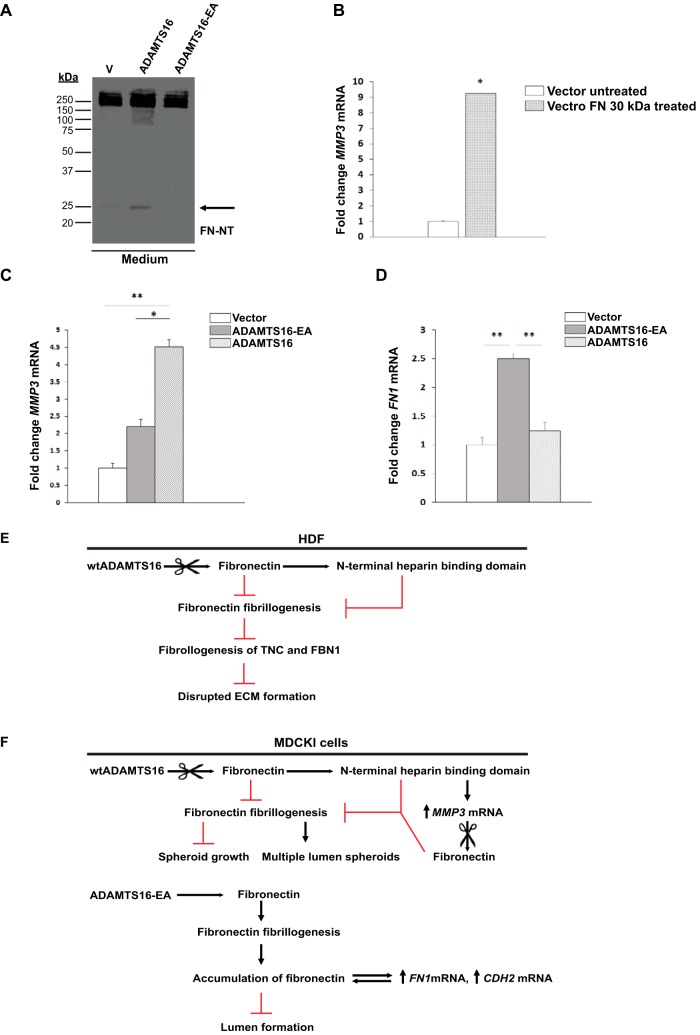
ADAMTS16 Overexpression Up-regulates MMP3 Expression by MDCKI Cells
Although Western blot analysis of the cell lysate of the MDCKI cells cultured in 2D did not show a significant differences in FN levels between cells expressing different ADAMTS16 constructs (Fig. 5F), the N-terminal 30 kDa FN fragment was evident in the concentrated medium of the wtADAMTS16- expressing cells (Fig. 6A). To ask whether this FN fragment induced MMP3 expression in MDCKI cells, as previously noted in chondrocytes (33), we added it to MDCKI monolayers and quantified MMP3 mRNA expression. We found a significant up-regulation of MMP3 mRNA after 24 h in treated MDCKI cells (Fig. 6B) and MMP3 mRNA levels were also significantly increased in cells expressing wtADAMTS16 for 6 days in 3D cultures (Fig. 6C). Because MMP3 cleaves FN within the (1)5-(I)6 linker i.e. generating the 30 kDa fragment similar to ADAMTS16 (34), it is likely that the observed MMP3 mRNA up-regulation leads to a feed-forward loop that may enhance FN proteolysis in MDCK I spheroids.
Fig. 6.
Release of the 30 kDa N-terminal heparin binding domain by ADAMTS16 up-regulates MMP3 mRNA in MDCKI cells to generate a proteolysis feed-forward loop. A, Western blot analysis of conditioned medium of MDCKI monolayers expressing wtADAMTS16, ADAMTS16-EA and empty vector control after 48 h under serum-free conditions, shows a 30 kDa FN fragment released by wtADAMTS16 activity (black arrow). B, MMP3 mRNA expression is significantly increased in MDCKI cells treated with recombinant 30 kDa N-terminal heparin binding domain (1F1–5F1). (*) p < 0.05 by Student t test. Experiments were done in duplicates. C, Quantitative RT-PCR analysis of MMP3 expression in MDCKI cells stably expressing ADAMTS16, ADAMTS16-EA and vector control after 6 days of culture in collagen gels. The expression of MMP3 is up-regulated in cells expressing active ADAMTS16. (*) p < 0.05, (**) p < 0.01 by Student t test.; n = 3 independent experiments per construct. D, Quantitative RT-PCR analysis of FN1 expression in MDCKI cells stably expressing ADAMTS16, ADAMTS16-EA and vector control after 6 days of culture in collagen gels. FN1 mRNA is up-regulated in cells expressing ADAMTS16-EA. (**) p < 0.01 by Student t test; n = 3 independent experiments per construct. E, F, Schematics depicting the proposed pathway and impact of ADAMTS16 in HDF and MDCKI cells respectively. ADAMTS16 is furin-processed and secreted into the extracellular space, where it binds to the ECM and is further C-terminally processed. ADAMTS16 cleaves FN at its N terminus releasing a 30 kDa N-terminal heparin-binding domain that inhibits FN fibrillogenesis and ECM maturation in fibroblast cultures (E). In MDCKI spheroids (F), the 30 kDa N-terminal heparin-binding domain of FN released by ADAMTS16 induces MMP3 expression, generating a feed-forward proteolytic loop. In contrast to active ADAMTS16, the inactive mutant ADAMTS16-EA binds to FN preventing its degradation. This enhances FN accumulation in the ECM, likely interfering with ECM stiffness and leading to increased FN1 and CDH2 expression.
DISCUSSION
Identifying new substrates for proteases is a challenging yet crucial undertaking that is necessary for a better understanding of their function in diverse cellular and disease processes such as development, tissue repair, cancer, and inflammation. Because proteolysis is an irreversible post-translational modification, substrate identification can define pathways that are substantially modified by a protease with lasting consequences. Until now ADAMTS16 has been an orphan protease, i.e. one without known substrates. Although it has been linked to several human diseases, such as cancer and hypertension, its substrates and its mechanism of action were so far unknown. Our data shows that ADAMTS16 is a secreted metalloprotease that acts in a cell-proximate location. After secretion, ADAMTS16 is extensively cleaved within its C-terminal domains such that mature full-length ADAMTS16 is undetectable with antibodies to the C-terminal epitope tags. This results in release of a truncated form of ADAMTS16, which we modeled using the ADAMTS16-sh construct. ADAMTS16-sh had similar activity as ADAMTS16 in the assays utilized, suggesting that the TSR2-PLAC domain is primarily necessary for localizing ADAMTS16 to ECM through yet undetermined interactions, although ADAMTS16-sh clearly also retained some ECM-binding ability.
Here, the use of a cell-free ECM produced in vitro by BALB/c fibroblasts allowed substrate discovery using LC-MS/MS without prior purification of ADAMTS16. This approach is suitable for investigating other ECM-degrading proteases whose purification, like ADAMTS16, is challenging. The ECM used in this approach provided a physiological ECM containing a broad spectrum of potential substrates because fibroblasts typically produce several secreted molecules which are assembled into a well-organized ECM. The mass spectrometry strategy not only identified FN as a potential substrate of ADAMTS16, but also identified a cleavage site in the linker region between repeat (I)5 and (I)6 of FN. Release of the 30 kDa N-terminal heparin binding domain, likely affects FN assembly in two ways: FN dimers lacking this region may not assemble properly, and the 30 kDa fragment may interfere with the assembly of intact FN dimers by occupying cell-surface receptors or sites relevant for FN self-assembly (Fig. 6E). The temporal analysis of FN assembly in the presence of wild-type or catalytically inactive ADAMTS16, specifically at an early 24 h time point, suggests that ADAMTS16 primarily interferes with initiation and maturation of FN fibrils. However, release of the 30 kDa FN fragment, as observed by LC-MS/MS after digestion of a pre-formed ECM, indicates that ADAMTS16 is also capable of degrading FN fibrils. The conformation of FN dimers appears to be crucial for regulating proteolytic susceptibility, because ADAMTS16-sh did not cleave soluble plasma FN, which assumes a globular conformation (35) in which the (I)5-(I)6 linker region may be concealed. The cryptic site is likely exposed in the presence of cells, which apply tensile forces to FN and FN fibrils upon attachment (36). Other proteases, such as ADAMTS2 and ADAMTS3 (17), have been shown to cleave FN within its (I)5-(I)6 linker sequence, indicating its high susceptibility to proteolysis. The findings suggest that there is considerable redundancy in fibronectin processing by proteases. Thus, release of the 30 kDa fragment of FN by proteases is likely to be a widespread mechanism of regulation of FN assembly that has not been extensively studied.
The cell-proximate location of ADAMTS16 is relevant because FN assembly is initiated at the cell surface by integrin-binding to secreted dimers (37), and thus FN proteolysis by ADAMTS16 may occur pericellularly. WtADAMTS16 overexpression in MDCKI cells led to a significant reduction in the size of MDCK-derived spheroids associated with reduced FN staining and detection of the 30 kDa FN fragment. Additionally, we observed formation of multiluminal spheroids. These findings are consistent with a previously described reduction of spheroid size and multiple lumen formation in MDCKI cells upon FN knockdown (38, 39), suggesting that the observed impact is a result of reduction of FN in the spheroid ECM. Loss of FN in 3D cultures was also previously linked to loss of cell polarity (40). The 70 kDa N-terminal FN fragment, which blocks FN fibril assembly has been shown to impair angiogenesis in 3D cultures of vascular endothelium in collagen gels (40) and to promote loss of cell polarity during gastrulation (41). Interestingly, spheroids expressing the catalytically inactive ADAMTS16-EA had increased FN accumulation in their basement membrane. Most likely ADAMTS16-EA binds to FN without proteolysis of the protein and acts in a dominant negative manner to prevent other proteases from binding. The resulting accumulation of FN may change matrix stiffness leading to an up-regulation of FN1 expression, which further contributes to the extensive accumulation of FN. Overall these spheroids were larger and failed to form a lumen, in contrast to the vector or wtADAMTS16-expressing spheroids. The cells of ADAMTS16-EA expressing spheroids showed characteristics of epithelial-mesenchymal transition (EMT) upon FN accumulation as shown by the up-regulation of N-Cadherin (Fig. 5F). Previous studies showed that TGFβ overexpression in MDCK cells blocked lumen formation via induction of EMT (42, 43) and that the levels of laminin, fibronectin and collagen IV in the ECM of MDCK spheroids could influence their ability to form a proper lumen (42). FN turnover by ADAMTS16 can therefore potentially influence cell polarity, EMT and morphogenesis in 3D culture systems. The reduction of laminin staining in wtADAMTS16-expressing MDCKI spheroids, taken together with overlapping staining of ADAMTS16 and laminin in HEK293-EBNA cultures, suggests that laminins could also be potential ADAMTS16 substrates; however, BALB/c 3T3 cells produce very low levels of laminin and we did not identify unique laminin peptides in our LC-MS/MS screen.
The 30 kDa N-terminal heparin-binding domain of FN released by ADAMTS16 is not only functionally relevant to FN fibrillogenesis but was previously shown to activate MMP3 expression via integrin signaling in chondrocytes (33). MMP3 and its activation have been extensively studied during branching morphogenesis and cancer (44),(45, 46), but little is known about regulation of MMP3 expression by other proteases. Our data showed that the 30 kDa fragment of FN released by ADAMTS16 increased MMP3 expression in MDCKI cells. FN cleavage by MMP3 within the linker region between domain (I)5 and (I)6, like ADAMTS16, was previously shown to release the 30 kDa N-terminal heparin binding domain (33, 34). We therefore propose that induction of MMP3 upon ADAMTS16 overexpression creates a feed-forward loop augmenting FN proteolysis (Fig. 6F). Thus, ADAMTS16 and MMP3 might work synergistically in regulation of FN fibril maturation and ECM assembly (Fig. 6F). Although the data suggest that ADAMTS16 cleaves fibronectin directly, i.e. by showing it binds to fibronectin and that purified ADAMTS16 cleaves fibronectin, it is possible that ADAMTS16 may act not only as a terminal protease, but also via activation of an earlier step in a proteolytic pathway, such as by up-regulation of MMP3 or activation of other proteases.
In summary, we have demonstrated that FN turnover by ADAMTS16 has a profound effect on the formation of FN networks and therefore has the potential to modify cell behavior and influence ECM composition and integrity over extended time-scales. Additionally, we have shown that ADAMTS16 drives a feed-forward proteolytic loop, because the N-terminal FN fragment generated increases MMP3 expression in MDCKI cells. Although proteases are typically considered in the context of ECM turnover, the present work supports a major role for ADAMTS16 in regulating ECM assembly. These findings have broad ramifications for understanding cell behavior and extend the concept of the protease web (47, 48) to illustrate the complexity and interdependence of proteases in discrete cascades and circuits.
DATA AVAILABILITY
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files or are available from R.Sc upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD007284. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD007284; FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2018/04/PXD007284; PubMed ID: 29669734.
Supplementary Material
Acknowledgments
The authors express their appreciation of the many contributions of the late Ruth Chiquet-Ehrismann (deceased September 4, 2015) to whom this manuscript is dedicated. We thank Prof. Nancy E. Hynes for critical reading of the manuscript and members of the Chiquet-Ehrismann and Apte laboratory for constructive comments. We further thank Dr. Martin Spiess for the MDCKI cells and Dr. Jan Seebacher for input on MS analysis.
Footnotes
* This work was supported by grants from: Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (SNF, Project funding in biology and medicine (division III)) grant no. 31003A_156740 (to R. C-E), HHS/NIH awards HL107147 and EY024943 (to S.A.), and the Allen Distinguished Investigator Program, through support made by The Paul G. Allen Frontiers Group and the American Heart Association (to S.A.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ECM
- Extracellular Matrix
- FBN1
- Fibrillin1
- FN
- Fibronectin
- GELS
- Gelsolin
- LTBP1
- Latent-transforming growth factor beta-binding protein 1
- PG-S1
- Biglycan
- PLEC
- Plectin
- PLAC
- Protease and lacunin domain
- POSTN
- Periostin
- SERPH
- Serpin H1
- TNC
- Tenascin-C
- TSR
- Thrombospondin type 1 repeats.
REFERENCES
- 1. Schwarzbauer J. E., and DeSimone D. W. (2011) Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Prospect. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hynes R. O. (1990) Fibronectins. Springer- Verlag, New York [Google Scholar]
- 3. Grudzenko T., and Franz C. M. (2015) Studying early stages of fibronectin fibrillogenesis in living cells by atomic force microscopy. Mol. Biol. Cell 26, 3190–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pankov R., and Yamada K. M. (2002) Fibronectin at a glance. J. Cell Sci. 115, 3861–3863 [DOI] [PubMed] [Google Scholar]
- 5. Schwarzbauer J. E. (1991) Identification of the fibronectin sequences required for assembly of a fibrillar matrix. JCB 113, 1463–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McDonald J. A., Quade B. J., Broekelman T. J., LaChance R., Forsman K., Hasegawa E., and Akiyama S. (1987) Fibronectin's cell-adhesive domain and an amino-terminal matrix assembly domain participate in its assembly into fibroblast pericellular matrix. J. Biol. Chem. 262, 2957–2967 [PubMed] [Google Scholar]
- 7. McKEOWN-LONGO P. J., and MOSHER DF (1985) Interaction of the 70,000-mol-wt Amino-terminal Fragment of Fibronectin with the Matrix-assembly Receptor of Fibroblasts. J. Cell Biol. 100, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M. D., Annis D. S., Mosher D. F., and Reinhardt D. P. (2009) Fibrillin assembly requires fibronectin. Mol. Biol. Cell 20, 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiquet-Ehrismann R., Matsuoka Y., Hofer U., Spring J., Bernasconi C., and Chiquet M. (1991) Tenascin variants: differential binding to fibronectin and distinct distribution in cell cultures and tissues. Cell Regul. 11, 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kubow K. E., Vukmirovic R., Zhe L., Klotzsch E., Smith M. L., Gourdon D., Luna S., and Vogel V. (2015) Mechanical forces regulate the interactions of fibronectin and collagen I in extracellular matrix. Nat. Commun. 6, 8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dallas S. L., Sivakumar P., Jones C. J., Chen Q., Peters D. M., Mosher D. F., Humphries M. J., and Kielty C. M. (2005) Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J. Biol. Chem. 280, 18871–18880 [DOI] [PubMed] [Google Scholar]
- 12. George E. L., Georges-Labouesse E. N., Patel-King R. S., Rayburn H., and Hynes R. O. (1993) Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 119, 1079–1091 [DOI] [PubMed] [Google Scholar]
- 13. Shi F., and Sottile J. (2011) MT1-MMP regulates the turnover and endocytosis of extracellular matrix fibronectin. J. Cell Sci. 124, 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mosher D. F. (1989) Fibronectin. Academic Press. Inc. [Google Scholar]
- 15. Gronski T. J. J., Martin R. L., Kobayashi d, Walsh K. B. C., Holman M. C., Huber M., Wart H. E. V., and Shapiro S. D. (1997) Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J. Biol. Chem. 272, 12189–12194 [DOI] [PubMed] [Google Scholar]
- 16. Marchina E., and Barlati S. (1996) Degradation of human plasma and extracellular matrix fibronectin by tissue type plasminogen activator and urokinase. Int. J. Biochem. Cell Biol. 28, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 17. Bekhouche M., Leduc C., Dupont L., Janssen L., Delolme F., Goff S. V.-L., Smargiasso N., Baiwir D., Mazzucchelli G., Zanella-Cleon I., Dubail J., Pauw E. D., Nusgens B., Hulmes D. J., Moali C., and Colige A. (2016) Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-β signaling as primary targets. FASEB J. 30, 1741–1756 [DOI] [PubMed] [Google Scholar]
- 18. Kern C. B., Wessels A., McGarity J., Dixon L. J., Alston E., Argraves W. S., Geeting D., Nelson C. M., Menick D. R., and Apte S. S. (2010) Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 29, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelwick R., Desanlis I., Wheeler G. N., and Edwards D. R. (2015) The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 16, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abdul-Majeed S., Mell B., Nauli S. M., and Joe B. (2014) Cryptorchidism and infertility in rats with targeted disruption of the Adamts16 locus. PLoS ONE 9, e100967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jacobi C. L., Rudigier L. J., Scholz H., and Kirschner K. M. (2013) Transcriptional regulation by the Wilms tumor protein, Wt1, suggests a role of the metalloproteinase Adamts16 in murine genitourinary development. J. Biol. Chem. 288, 18811–18824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gopalakrishnan K., Kumarasamy S., Abdul-Majeed S., Kalinoski A. L., Morgan E. E., Gohara A. F., Nauli S. M., Filipiak W. E., Saunders T. L., and Joe B. (2012) Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc. Natl. Acad. Sci. U.S.A. 109, 20555–20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joe B., Saad Y., Dhindaw S., Lee N. H., Frank B. C., Achinike O. H., Luu T. V., Gopalakrishnan K., Toland E. J., Farms P., Yerga-Woolwine S., Manickavasagam E., Rapp J. P., Garrett M. R., Coe D., Apte S. S., Rankinen T., Pérusse L., Ehret G. B., Ganesh S. K., Cooper R. S., O'Connor A., Rice T., Weder A. B., Chakravarti A., Rao D. C., and Bouchard C. (2009) Positional identification of variants of Adamts16 linked to inherited hypertension. Hum. Mol. Genet. 18, 2825–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakamoto N., Oue N., Noguchi T., Sentani K., Anami K., Sanada Y., Yoshida K., and Yasui W. (2010) Serial analysis of gene expression of esophageal squamous cell carcinoma: ADAMTS16 is upregulated in esophageal squamous cell carcinoma. Cancer Sci. 101, 1028–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yasukawa M., Liu Y., Hu L., Cogdell D., Gharpure K. M., Pradeep S., Nagaraja A. S., Sood A. K., and Zhang W. (2016) ADAMTS16 mutations sensitize ovarian cancer cells to platinum-based chemotherapy. Oncotarget 8, 88410–88420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng L., Baumann U., and Reymond J. L. (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beacham D. A., Amatangelo M. D., and Cukierman E. (2007) Preparation of Extracellular Matrices Produced by Cultured and Primary Fibroblasts. Curr. Protoc. Cell Biol., 10.19.11–10.19.21 [DOI] [PubMed] [Google Scholar]
- 28. Asparuhova M. B., Ferralli J., Chiquet M., and Chiquet-Ehrismann R. (2011) The transcriptional regulator megakaryoblastic leukemia-1 mediates serum response factor- independent activation of tenascin-C transcription by mechanical stress. FASEB J. 25, 3477–3488 [DOI] [PubMed] [Google Scholar]
- 29. Schmittgen T. D., and Livak K. J. (2008) Analyzing real- time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 30. Maurer L. M., Tomasini-Johansson B. R., Ma W., Annis D. S., Eickstaedt N. L., Ensenberger M. G., Satyshur K. A., and Mosher D. F. (2010) Extended Binding Site on Fibronectin for the Functional Upstream Domain of Protein F1 of Streptococcus pyogenes. J. Biol. Chem. 285, 41087–41099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cal S., Obaya A. J., Llamazares M., Garabaya C., Quesada V., and López-Otín C. (2002) Cloning, expression analysis, and structural characterization of seven novel human ADAMTSs, a family of metalloproteinases with disintegrin and thrombospondin-1 domains. Gene 283, 49–62 [DOI] [PubMed] [Google Scholar]
- 32. Gao S., Geyter C. D., Kossowska K., and Zhang H. (2007) FSH stimulates the expression of the ADAMTS-16 protease in mature human ovarian follicles. Mol. Hum. Reprod. 13, 465–471 [DOI] [PubMed] [Google Scholar]
- 33. Ding L., Guo D., and Homandberg G. A. (2008) The cartilage chondrolytic mechanism of fibronectin fragments involves MAP kinases: comparison of three fragments and native fibronectin. Osteoarthritis Cartilage 16, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 34. Muir D., and Manthorpe M. (1992) Stromelysin generates a fibronectin fragment that inhibits Schwann cell proliferation. J. Cell Biol. 116, 177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maurer L. M., Ma W., and Mosher D. F. (2016) Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 51, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sivakumar P., Czirok A., Rongish B. J., Divakara V. P., Wang Y. P., and Dallas S. L. (2006) New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. J. Cell Sci. 119, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 37. Sechler J. L., Corbett S. A., and Schwarzbauer J. E. (1997) Modulatory roles for integrin activation and the synergy site of fibronectin during matrix assembly. Mol. Biol. Cell 8, 2563–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jiang S. T., Chiang H. C., Cheng M. H., Yang T. P., Chuang W. J., and Tang M. J. (1999) Role of fibronectin deposition in cystogenesis of Madin-Darby canine kidney cells. Kidney Int. 56, 92–103 [DOI] [PubMed] [Google Scholar]
- 39. Jiang S. T., Chuang W. J., and Tang M. J. (2000) Role of fibronectin deposition in branching morphogenesis of Madin-Darby canine kidney cells. Kidney Int. 57, 1860–1867 [DOI] [PubMed] [Google Scholar]
- 40. Zhou X., Rowe R. G., Hiraoka N., George J. P., Wirtz D., Mosher D. F., Virtanen I., Chernousov M. A., and Weiss S. J. (2008) Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev. 22, 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rozario T., Dzamba B., Weber G. F., Davidson L. A., and DeSimone D. W. (2009) The physical state of fibronectin matrix differentially regulates morphogenetic movements in vivo. Dev. Biol. 327, 386–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Santos O. F., and Nigam S. K. (1993) HGF-induced tubulogenesis and branching of epithelial cells is modulated by extracellular matrix and TGF-beta. Dev. Biol. 16, 293–302 [DOI] [PubMed] [Google Scholar]
- 43. Sakurai H., and Nigam S. K. (1997) Transforming growth factor-beta selectively inhibits branching morphogenesis but not tubulogenesis. Am. J. Physiol. 272, 139–146 [DOI] [PubMed] [Google Scholar]
- 44. Talhouk R. S., Chin J. R., Unemori E. N., Werb Z., and Bissell M. J. (1991) Proteinases of the mammary gland: developmental regulation in vivo and vectorial secretion in culture. Development 112, 439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagase H., Enghild J. J., Suzuki K., and Salvesen G. (1990) Stepwise activation mechanisms of the precursor of matrix metalloproteinase 3 (stromelysin) by proteinases and (4-aminophenyl)mercuric acetate. Biochemistry 29, 5783–5789 [DOI] [PubMed] [Google Scholar]
- 46. Overall C. M., and Kleifeld O. (2006) Tumour microenvironment—opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 47. Keller Ua. d., Doucet A., and CMO (2007) Protease research in the era of systems biology. Biol. Chem. 388, 1159–1162 [DOI] [PubMed] [Google Scholar]
- 48. Fortelny N., Cox J. H., Kappelhoff R., Starr A. E., Lange P. F., Pavlidis P., and Overall C. M. (2014) Network analyses reveal pervasive functional regulation between proteases in the human protease web. PLos Biol. 12, e1001869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its Supplementary Information files or are available from R.Sc upon request. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/archive/) with the dataset identifier PXD007284. Project Webpage: http://www.ebi.ac.uk/pride/archive/projects/PXD007284; FTP Download: ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2018/04/PXD007284; PubMed ID: 29669734.