Significance
The development of an in vitro human bone marrow (BM) tissue appears essential to compile information on human hematopoiesis. Conventional systems fail at both capturing the complexity of the bone marrow niche while allowing the maintenance of functional hematopoietic stem cells (HSCs). Here, we report the development of a human 3D (BM) analogue in a perfusion-based bioreactor system, partially recapitulating structural, compositional, and organizational features of the native human osteoblastic niche environment. The engineered tissue supports the maintenance of some hematopoietic stem and progenitor cell (HSPC) properties. This provides an advanced technological platform of broad fundamental and translational relevance, including the study of human HSPC biology and interactions with their niche, the manipulation of functional human HSPCs, or the identification of factors influencing human hematopoiesis.
Keywords: hematopoiesis, bone marrow niche, 3D culture, tissue engineering, hematopoietic stem cell
Abstract
In adults, human hematopoietic stem and progenitor cells (HSPCs) reside in the bone marrow (BM) microenvironment. Our understanding of human hematopoiesis and the associated niche biology remains limited, due to human material accessibility and limits of existing in vitro culture models. The establishment of an in vitro BM system would offer an experimentally accessible and tunable platform to study human hematopoiesis. Here, we develop a 3D engineered human BM analog by recapitulating some of the hematopoietic niche elements. This includes a bone-like scaffold, functionalized by human stromal and osteoblastic cells and by the extracellular matrix they deposited during perfusion culture in bioreactors. The resulting tissue exhibited compositional and structural features of human BM while supporting the maintenance of HSPCs. This was associated with a compartmentalization of phenotypes in the bioreactor system, where committed blood cells are released into the liquid phase and HSPCs preferentially reside within the engineered BM tissue, establishing physical interactions with the stromal compartment. Finally, we demonstrate the possibility to perturb HSPCs’ behavior within our 3D niches by molecular customization or injury simulation. The developed system enables the design of advanced, tunable in vitro BM proxies for the study of human hematopoiesis.
The bone marrow (BM) microenvironment is responsible for the maintenance of hematopoietic stem cell (HSC) activity, enabling the lifelong production of mature blood cells (1, 2). The regulation of HSC self-renewal and differentiation is achieved by complex cellular (3), molecular (4, 5), structural (6), and physical (7, 8) cues defining the HSC niche (2, 9). The components of the human HSC niche, and how these elements interact to modulate stem cell fate, remain poorly understood.
The field is hampered by the limited possibilities to access and harness information from human specimens. Chimeric animal models (10) most closely recapitulate in vivo human physiology, but, in this setting, the niche has remained inaccessible to experimental manipulation and optical observation (11, 12). In addition, the interspecies-chimerism in both hematopoietic cells and their environment makes interpretation of experimental results difficult. The development of in vitro substitutes is a promising alternative with superior tunability and throughput (13, 14). Previous studies have described the combination of different stromal and hematopoietic progenitors using a variety of culture substrates (9–11), resulting in the phenotypic preservation of specific blood phenotypes. However, the recapitulation of the structural organization of BM, including essential cell–cell and cell–matrix interactions (15–19), and the associated functional preservation of HSCs (20) are still elusive.
The need for advanced culture systems of higher biological complexity has gained increasing recognition (21) to study the fundamental biology of stem cells. Similarly to the “organogenesis in a dish” proposed for complex organs [e.g., lung (22), breast (23), kidney (24), and liver (25)], the in vitro engineering of human BM environments (21, 26–28) capable to sustain HSCs (28, 29) would enable their study in xeno-free settings.
Here, we report an in vitro system supporting the development and maintenance of a human BM analog. Our approach consists in the use of porous hydroxyapatite scaffolds with structural and compositional features of bone (30), functionalized by human mesenchymal stromal cells (hMSCs) and the extracellular matrix (ECM) deposited during their progressive maturation into the osteoblastic lineage. The hMSC culture is performed under direct perfusion flow (31), offering efficient nutrient supply/waste removal, while mimicking interstitial flow and associated shear stress. The blood compartment was introduced into the resulting 3D stromal tissue by perfusion of human purified cord blood (CB)-derived CD34+ cells. This engineered organoid partially recapitulates structural and functional features of the human BM in defined and tunable settings.
Results
Three-Dimensional Microenvironments Can Be Engineered Within the Perfusion Bioreactor System.
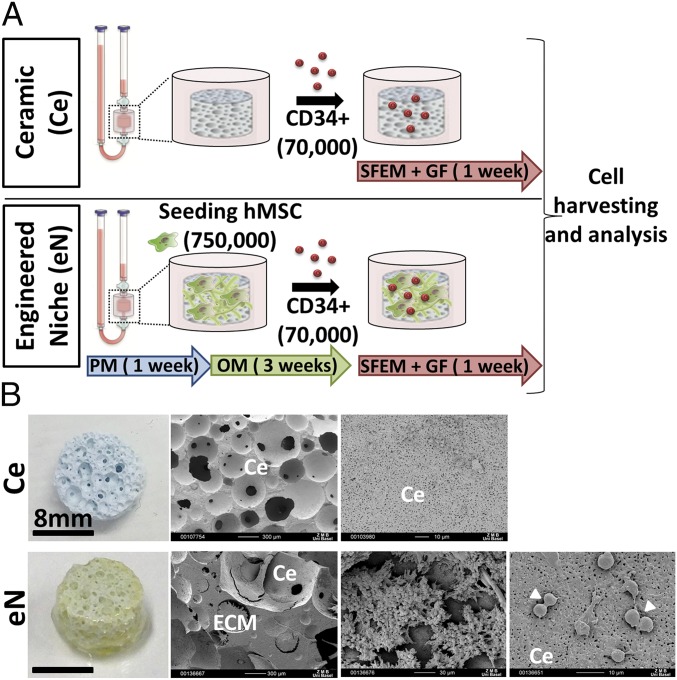
The generation of the 3D microenvironments was performed by differentiation of primary BM-derived hMSCs on ceramic materials within a perfusion bioreactor. Cells were first labeled with a VENUS transgene (>93%) (SI Appendix, Fig. S1A) to facilitate subsequent analysis. To achieve the engineering of an osteoblastic-like stroma, we adopted a protocol previously used for the generation of bone grafts (32) (Fig. 1A). hMSCs were first cultured 1 wk in proliferative medium (PM) to increase cell number and ensure scaffold colonization, followed by 3 wk of osteogenic medium (OM) supplementation, to promote cell differentiation while stimulating extracellular matrix (ECM) production (33). The resulting tissue was defined as “engineered niche” (eN) (Fig. 1A) while naked ceramic (Ce) (Fig. 1A), not containing hMSCs but loaded with CD34+ cells, was used as internal 3D culture control. The blood compartment was subsequently introduced by injecting CB-derived human CD34+ cells from single donors into the device tubing. The in vitro coculture was sustained for 1 wk in serum-free medium supplemented with a low concentration of hematopoietic cytokines [10 ng/mL thrombopoietin (TPO), stem cell factor (SCF), and Fms-related tyrosine kinase 3 ligand (Flt3-L)]. Upon retrieval from the bioreactor chamber, the Ce was devoid of apparent ECM structures (Ce, SI Appendix, Fig. S1B) while the eN exhibited features of an engineered tissue with thick gel-like structures homogenously covering the starting material (eN, Fig. 1B). Scanning electron microscopy confirmed the deposition of an ECM which embeds cells, presumably of both stromal (fibroblastic shape, Fig. 1B) and blood origins (round shape, Fig. 1B), including dividing cells (Fig. 1B).
Fig. 1.
Three-dimensional microenvironments can be engineered within the perfusion bioreactor system. (A) Experimental design for the generation of 3D niches in a perfusion bioreactor. OM, osteogenic medium; PM, proliferative medium; SFEM+GF, serum-free medium plus growth factors stem cell factor, thrombopoietin, and Flt3-ligand. (B) The engineered niche (eN, Left) exhibits thick gel-like structures homogenously covering the starting material (Ce). Scanning electron microscopy images of eN (Right) confirmed the deposition of an ECM which embeds cells, presumably of both stromal and blood origins. Arrowheads indicate the presence of dividing cells.
Engineered 3D Microenvironments Allow the Maintenance of Hematopoietic Stem and Progenitor Cells with Functional Properties.
To characterize their cellular composition, the 3D microenvironments were digested for cell retrieval (>92% retrieval efficiency, with an overall cell death below 1.5%) (SI Appendix, Fig. S1 B and C) and subsequent quantitative phenotypic analysis (SI Appendix, Fig. S1 D and E).
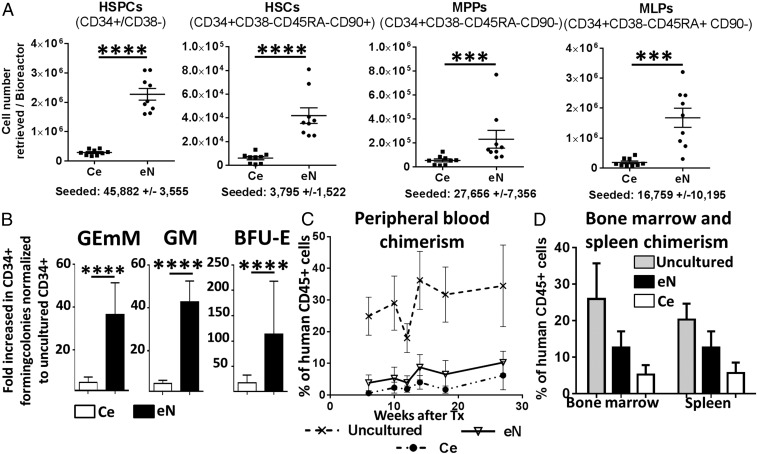
The eN was composed of over 4.7 × 106 blood cells (SI Appendix, Fig. S1F) per bioreactor system [61-fold increase (f.i.) over the initial 70,000 CD34+ cells] (SI Appendix, Fig. S1G). In contrast, in the absence of engineered stroma, blood cell expansion was limited to a 7.8 f.i. (SI Appendix, Fig. S1 F and G). With a ratio of HSPCs over committed cells (CD34−/CD38+) lower than the Ce (38 vs. 204) (SI Appendix, Fig. S1H), the eN was shown to sustain the proliferation of differentiated populations. The eN also promoted the maintenance of phenotypic hematopoietic stem and progenitor populations (HSPCs) (Fig. 2A), yielding systematically higher total numbers of HSPCs (46.2 f.i. vs. 6.4 for Ce), including HSCs (13.8 f.i. vs. 1.6), multipotent progenitors (MPPs) (8.5 f.i. vs. 2.3), and multipotent lymphoid progenitors (MLPs) (161 f.i. vs. 20) (SI Appendix, Fig. S2A).
Fig. 2.
Engineered 3D microenvironments allow the maintenance of HSPCs with functional properties. (A) The engineered niche (eN) supports the expansion of phenotypic hematopoietic stem and progenitor cells (HSPCs), hematopoietic stem cells (HSCs), multipotent progenitors (MPPs), and multipotent lymphoid progenitors (MLPs), as assessed by quantitative flow cytometry analysis post-3D culture. n ≥ 8 biological replicates. Ce, ceramic only. ***P < 0.001, ****P < 0.0001. (B) Improved maintenance of colony-forming potential of eN versus Ce cultured CD34+ cells. BFU-E, burst-forming unit-erythroid; GEmM, colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte; GM, colony-forming unit-granulocyte and macrophage. n ≥ 9 biological replicates. (C) The long-term repopulation capacity of eN and Ce cultured CD34+ cells. Reconstitution of the human blood compartment (percentage of human CD45+ cells in mononuclear cells) in NSG mice is shown by flow cytometry analysis of peripheral blood. Uncultured CD34+ cells served as positive control. n ≥ 4 biological replicates. Human CD34+ cells cultured on eN and Ce also robustly engrafted in the bone marrow and spleen (D) of transplanted mice, as assessed by flow cytometry 28 wk posttransplantation. n ≥ 4 biological replicates.
The capacity of hMSCs to support the proliferation of CD34+ cells was further confirmed in a 2D setup (SI Appendix, Fig. S2B). Using undifferentiated hMSCs as feeder layer (2D hMSCs) compared with a hMSC-free control (2D), we observed a significant increase of phenotypic HSPCs (100 f.i. vs. 15), HSCs (29 f.i. vs. 4), MPPs (43 f.i. vs. 16), and MLPs (279 f.i. vs. 21) (SI Appendix, Fig. S2B), in line with results derived from well-established Dexter cultures (34, 35). The capacity of the generated stroma to preserve the functionality of cultured blood cells was assessed both by in vitro and in vivo assays. The corresponding donor cells before in vitro culture (“uncultured”) were used as functional positive control. In vitro colony-forming unit assays demonstrated the maintenance of stem or progenitor cell capacities (with multilineage and proliferation potential) of cultured CD34+ cells, indicated by the generation of myeloid colonies with the same morphology as their uncultured counterparts (SI Appendix, Fig. S2C). Colony numbers were increased for cells retrieved from 3D cultures for colony-forming unit-granulocyte and macrophage (GM) (42.8 f.i vs. 4.8 for Ce), burst-forming unit-erythroid (BFU-E) (113 f.i vs. 18), and Colony-forming unit-Granulocyte (GEmM) (36.5 f.i vs. 4.8) colonies (Fig. 2B).
Stem and progenitor potential of cultured cells was further tested by intrafemoral transplantation of equal numbers of CD34+ cells from Ce or eN in irradiated NSG mice. The human blood compartment was successfully reconstituted in all groups [>1% human CD45 positive cells (hCD45) in peripheral blood] (Fig. 2C). Chimerism was detectable as early as week 6 and persisted over 28 wk posttransplantation, indicating the long-term repopulation capacity of transplanted cells. As anticipated, the highest chimerism was obtained by transplantation of uncultured CD34+ serving as positive control (27.9 ± 3.1%) (Fig. 2C). CD34+ cells derived from Ce and eN exhibited similar reconstitution levels, with an average of 2.7 ± 0.8% and 6.4 ± 1% of hCD45 respectively, detected in the peripheral blood (Fig. 2C). The chimerism was also assessed in the BM and the spleen of animals with human cells robustly engrafted in all conditions, with a trend for higher hCD45 frequency detected in the eN than in the Ce group, both in BM (6.4% ± 0.5 vs. 5.2% ± 2.5, Fig. 2D) and the spleen (12.6% ± 4.4 vs. 5.6% ± 2.8, Fig. 2D). Functionality of CD34+ cells was further assessed by multilineage reconstitution capacity. At week 18 posttransplantation, cells from all conditions successfully reconstituted the myeloid and lymphoid lineages (SI Appendix, Fig. S2D).
These data demonstrate that, compared with the Ce counterparts, the eN yields hematopoietic cells with similar reconstitution potential, but in higher numbers. This suggests that the generated tissue provides cues capable to enhance the maintenance/expansion of CFU-HSPCs with in vivo engraftment and multilineages reconstitution potential.
Molecular Characterization of the Engineered Niche Reveals the Establishment of an Osteoblastic-Like 3D Environment.
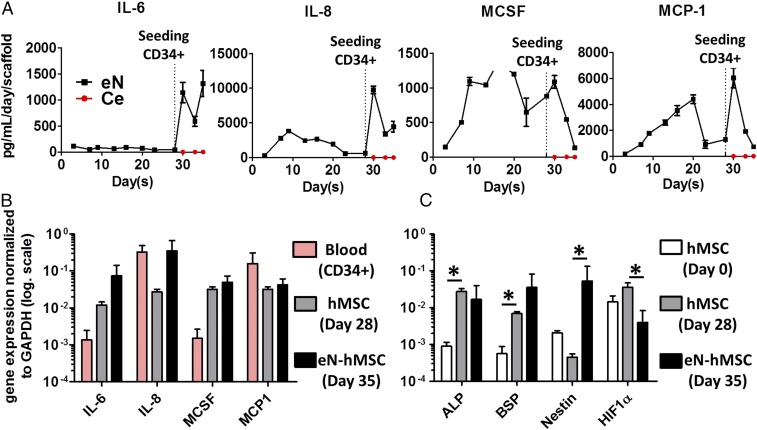
To identify factors associated with robust hematopoiesis development, the eN was further characterized. First, we monitored the secretion of key cytokines throughout culture times. Inflammatory factors [interleukin 6 (IL-6), interleukin 8 (IL-8), macrophage colony-stimulating factor (MCSF) (Fig. 3A), and monocyte chemoattractant protein-1 (MCP-1)] showed the highest differences and were found at high concentrations in eN. In particular, IL-6 and IL-8 production increased substantially upon HSPC addition (eN, Fig. 3A). Flt3-L, TPO, and SCF proteins were found at concentrations similar to those supplemented in the coculture medium (SI Appendix, Fig. S3A), suggesting that hMSCs secrete low levels of those HSPC supportive factors. Vascular endothelial growth factor α (VEGFα) and angiopoietin 1 (Ang-1) were also detected at significant levels (eN, SI Appendix, Fig. S3A) although Ang-1 decreased over time to remain stable after HSPC loading.
Fig. 3.
Molecular characterization of the engineered niche reveals the establishment of an osteoblastic-like 3D environment. (A) The engineered niche (eN) condition presents a higher concentration in inflammatory cytokines than the ceramic condition (Ce), based on Luminex analysis of 3D culture supernatants. IL-6, interleukin 6; IL-8, interleukin 8; MCP-1, monocyte chemotactic protein 1; MCSF, macrophage colony-stimulating factor. n ≥ 3 biological replicates. Addition of CD34+ cells to the niches only induces cytokine secretion in eN conditions. (B) hMSCs are principally responsible for the high levels of inflammatory cytokines detected in the eN. This was assessed by gene expression analysis of blood progenitor cells (CD34+), and hMSCs before (day 28) and after coculture with blood cells (day 35). n ≥ 3 biological replicates. (C) hMSCs acquire an osteoblastic-like niche genetic profile in culture. Gene expression analysis of hMSCs retrieved from the eN (eN hMSC). hMSCs (hMSC day 0) indicate the basic gene expression levels before their 3D culture. ALP, alkalyne phosphatase; BSP, bone sialoprotein; HIF1α, hypoxia-inducible factor 1α. n ≥ 3 biological replicates.
To obtain a more comprehensive understanding of the cellular compartments associated with factor secretion, we isolated both blood progenitors (CD34+) and mesenchymal populations before (defined as hMSC day 28) (SI Appendix, Fig. S3B) and after CD34+ coculture (defined as eN-hMSC day 35, SI Appendix, Fig. S3B). This confirmed the strong expression of inflammatory cytokines (IL-6, MCSF) by hMSCs during coculture with HSPCs, with levels markedly higher than those in blood progenitor cells (Fig. 3B). IL-8 and MCP-1 were expressed by both blood and mesenchymal cells (Fig. 3B). Interestingly, following coculture with CD34+ cells, hMSCs substantially increased their IL-6 and IL-8 expression (Fig. 3B).
To assess the role of IL-6 and IL-8 in the system, we investigated the effect of their addition in the Ce condition (SI Appendix, Fig. S4A). IL-6 and IL-8 at doses corresponding to those measured in the eN led to a significant increase in the number of HSPCs, committed progenitors (CD34+/CD38+), Granulocyte-monocytes progenitors (GMPs), and MLPs (SI Appendix, Fig. S4B). However, no effect was measured on HSCs and MPPs (SI Appendix, Fig. S4B). This suggests that these inflammatory cytokines mediate the proliferation of committed populations in the eN whereas the observed stem cell compartment expansion is driven by other hMSC factors.
Before CD34+ loading, hMSCs in the engineered tissue predominantly consisted of not only osteoblastic-like cells, but also of a pool expressing progenitor markers (SI Appendix, Fig. S5). We further analyzed the transcription profile of eN-hMSCs as niche cells at the end of the culture (eN-hMSC day 35) and compared it to postexpanded hMSCs (hMSC Day 0) and hMSCs in the eN before CD34+ loading (hMSC day 28). This confirmed the osteoblastic profile of hMSC (day 35) at the end of the 3D culture, evidenced by the up-regulation of alkaline phosphatase (ALP) and bone sialoprotein (BSP) (17 and 869 f.i., respectively) (Fig. 3C). Interestingly, a marked increase in Nestin expression (117 f.i.) was acquired by hMSCs after coculture with blood cells (Fig. 3C), as well as a decrease in hypoxia-inducible factor 1-alpha (HIF1α, 8.9-fold) expression.
Thus, by combining protein and gene expression analyses, we describe an osteoblastic (36, 37) and niche-associated (1) molecular signature of hMSCs, associated with proinflammatory features, which is acquired following the coculture with the blood compartment. This suggests the establishment of an osteoblastic-like niche environment, mutually interacting with hematopoietic cells and capable of regulating HSPC activities, including proliferation and functional regulation.
The Engineered Niche Shares Structural and Compositional Features with Native Human BM.
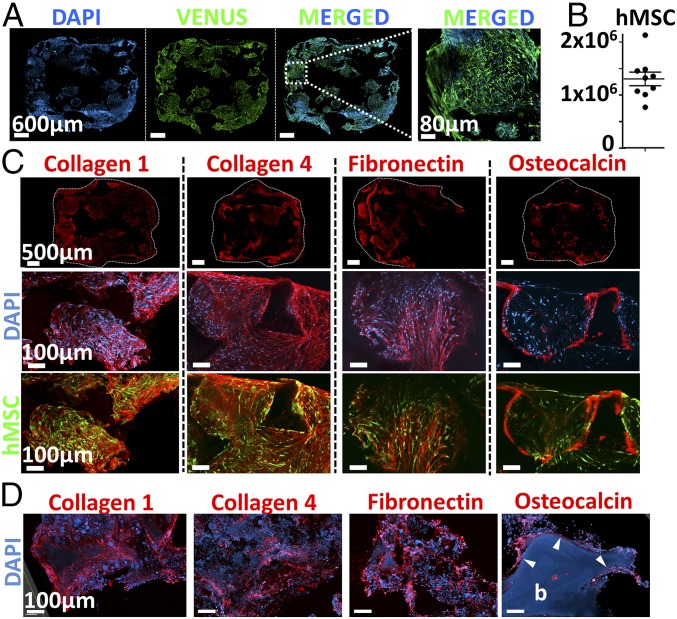
The role of hMSCs in the establishment of a supportive environment for the hematopoietic compartment was further investigated by studying the organization and composition of the eN. Immunofluorescence analysis of thick construct sections revealed a homogeneous network formed by hMSCs within the scaffold (VENUS signal, Fig. 4A). This observed mesenchymal fraction consisted in 1.3 × 106 hMSC-derived cells (Fig. 4B), as assessed by flow cytometry. The generated tissue resulting from the osteoblastic differentiation of hMSCs consisted in a dense human stroma filling the material pores and embedding mesenchymal cells (Fig. 4C). The composition of the deposited ECM included collagen type 1, collagen type 4, and fibronectin (Fig. 4C), reported as the main structural proteins of BM (38). Indeed, these were also abundantly found in healthy donor-derived BM specimens (human niche, Fig. 4D). The similarities between the human tissue and our engineered niche were not restricted to structural proteins. In human biopsies, osteocalcin staining revealed the presence of osteoblasts lining the bone surface (Fig. 4D). Remarkably, in the eN, a comparable pattern was observed by detection of osteocalcin in cells at the interface between the ceramic material and the cellular/ECM stroma (Fig. 4C).
Fig. 4.
The engineered niche shares structural and compositional features with native human BM. (A) hMSCs form a homogeneous cellular network distributed within the scaffolding material (confocal microscopy, representative images shown). (B) eN consists of 1.3 × 106 hMSC-derived cells at the end of the 3D culture (flow cytometry quantification). n = 9 biological replicates. Compositional and structural similarities of extracellular matrix in engineered niche (C) and human bone marrow samples (D), assessed by confocal microscopy. b, bone tissue (autofluorescence in the blue channel). Arrowheads indicate the presence of osteocalcin in osteoblasts lining at the bone surface.
Altogether, these results evidence the successful formation of a complex tissue under perfusion culture, associated with the development and support of human hematopoiesis (Fig. 2). The engineered environment was shown to share structural and compositional features typical of native human BM, suggesting the partial reconstitution of an osteoblastic-like niche environment.
The Bioreactor-Engineered Niche Displays a Functional Compartmentalization.
The developed culture system comprises two distinct phases: a liquid-fraction consisting in the supernatant (SN) SI Appendix, Fig. S6) and the stromal/ECM tissue confined within the scaffold chamber (ECM, SI Appendix, Fig. S6). This led us to further hypothesize that the two environments could differently impact the distribution of blood cell phenotypes. We thus separately analyzed HSPCs derived from these two phases by flow cytometry (SI Appendix, Fig. S6).
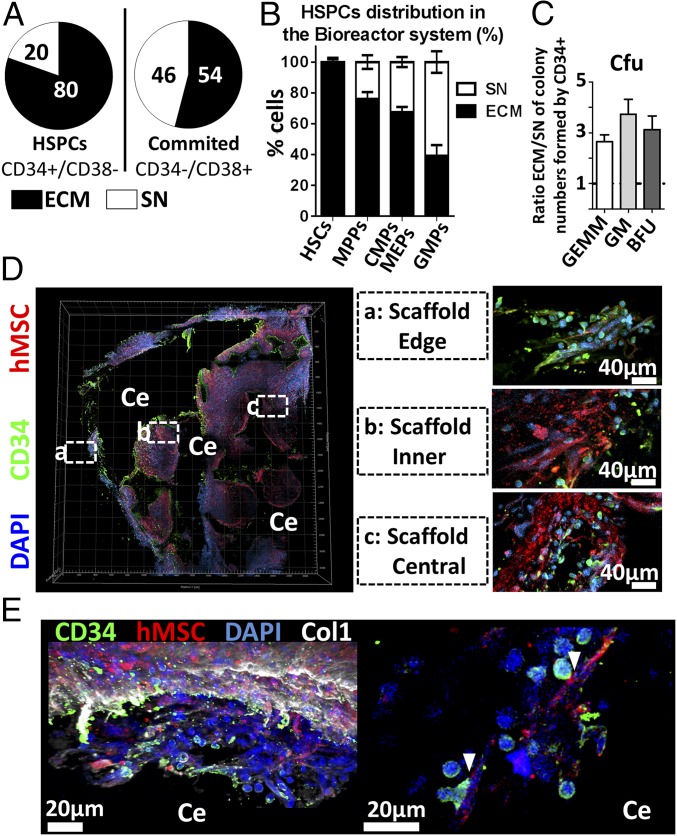
Despite the convection induced by perfusion flow, a specific cellular allocation was observed as 80% of the retrieved HSPC (CD34+/CD38−) populations resided in the ECM (Fig. 5A, Left). Conversely, more committed populations (CD34−/CD38+) exhibited a balanced distribution with 54% in ECM and 46% in SN (Fig. 5A, Right). A distinct pattern could be identified by further assessing the preferential localization of stem and progenitor populations, according to their progressive commitment (Fig. 5B). Remarkably, HSCs were found almost exclusively in the stroma (98%) (Fig. 5B) together with a vast majority of MPPs (76% in ECM) (Fig. 5B). This applied to a lower extent to common myeloid progenitors (CMPs) and megakaryocyte-erythroid progenitors (MEPs) (67% in ECM) while GMPs were more abundant in the SN (61%) (Fig. 5B). The localization was also shown to impact on the in vitro functionality of cells since CD34+ cells retrieved from ECM exhibited an increased capacity to form myeloid colonies compared with their SN counterpart (Fig. 5C). This includes the superior formation of GEmM, GM, and BFU colonies (2.6, 3.7, and 3.1 f.i., respectively).
Fig. 5.
The bioreactor-engineered niche displays a functional compartmentalization. (A) In the bioreactor system, the majority of HSPCs localized in the tissue (ECM) while committed cells are more equally distributed between the ECM and the supernatant (SN). n ≥ 8 biological repeats. (B) HSC, MPP, and CMP/MEP populations preferentially localize in the stroma (ECM), not in the liquid phase. n ≥ 8 biological repeats. (C) Superior myeloid colony formation potential of CD34+ cells retrieved from the ECM. BFU, burst-forming unit-erythroid; GEmM, colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte; GM, colony-forming unit-granulocyte and macrophage. n ≥ 12 biological replicates. (D) Confocal microscopy analysis of the eN demonstrates the presence of CD34+ cells at different depths throughout the tissue (Right, magnification). The CD34+ and hMSC fractions were found within an organized extracellular matrix (E, Left) in which they established physical interactions (E, Right). Ce, ceramic; Col1, collagen type 1.
The presence of human blood cells in the niche was confirmed by immunofluorescence, with human CD34+ cells abundantly detected within the ECM of the engineered tissue (Fig. 5D). Cells were predominantly located at the periphery of the scaffolds (Edge, Fig. 5D) but also largely infiltrated the tissue as shown by presence in the inner pores (Inner, Fig. 5D) and in the more central area (Central, Fig. 5D). Importantly, the mesenchymal and hematopoietic fractions in the engineered stroma were found within an organized ECM (Fig. 5E, Left) and closely interacted, as shown by explicit physical contacts (Fig. 5E, Right).
Altogether, this demonstrates the existence of a functional compartmentalization in the 3D perfusion system. The eN displays a hierarchical chemoattractant property on HSPC populations while commitment is progressively associated with a release into the liquid phase. The preferential stem cell confinement in the stroma, together with evidences of hMSC–CD34+ interactions, suggests an essential hMSC-triggered regulation of HSPC activities, resulting in a superior preservation of stem cell functionality.
BM Niches with Customized Molecular Signatures Can Be Genetically Engineered.
We next aimed at a proof of principle for the use of our system for the generation of customized hematopoietic niches with tailored molecular signature.
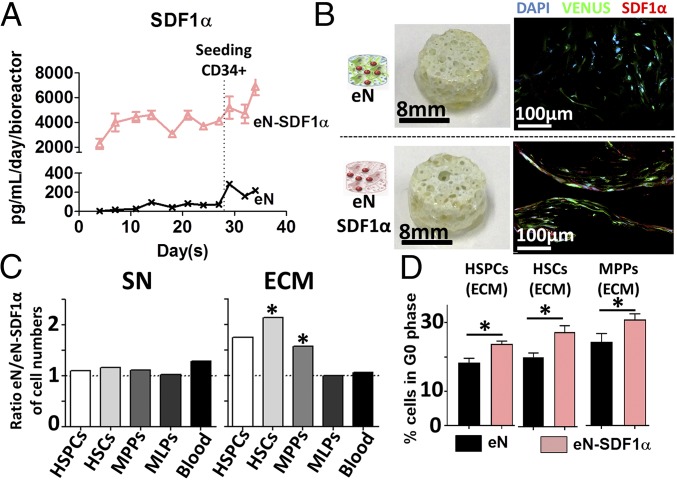
To this end, primary hMSCs were efficiently transduced using a VENUS-SDF1α lentivirus (>95%) (SI Appendix, Fig. S7 A and B), resulting in SDF1α overexpression (SI Appendix, Fig. S7C). SDF1α-engineered niches (eN-SDF1α) were subsequently produced, following the previously described protocol, and compared with eN without SDF1α enrichment. Throughout the culture period, substantial levels of SDF1α protein were continuously produced by the eN-SDF1α (>2,000 pg/mL/day/bioreactor) (Fig. 6A) compared with the eN (<300 pg/mL/day/bioreactor) (Fig. 6A).
Fig. 6.
Bone marrow niches with customized molecular signature can be genetically engineered. (A) The SDF1α-engineered niche (eN-SDF1α, red triangles) secretes superior levels of SDF1α than the control engineered niche (eN, black crosses) throughout the 3D culture period. n ≥ 3 biological replicates. (B) The eN and eN-SDF1α appeared macroscopically identical (Left), but confocal microscopy images revealed the increased presence of the SDF1α protein in the eN-SDF1α tissue. (C) Comparing the ratio of absolute number of hematopoietic populations from the eN and eN-SDF1α revealed an identical supernatant content (SN, Left) while the stroma of eN-SDF1α (ECM, Right) contained a reduced number of HSPCs, HSCs, and MPPs. n ≥ 8. (D) The SDF1α customization significantly increased the percentage of quiescent HSPCs, HSCs, and MPPs in the ECM of eN-SDF1α, as assessed by cell cycle analysis of corresponding populations. n = 5. HSCs, hematopoietic stem cells; HSPCs, hematopoietic stem and progenitor cells; MLPs, multilymphoid progenitors; MPPs, multipotent progenitors. *P < 0.05.
When retrieved from the bioreactor chambers, the different engineered tissues were macroscopically identical (Fig. 6B). Immunofluorescence analysis revealed the presence of hMSCs in both niches, but eN-SDF1α displayed an increased presence of SDF1α protein, confirming the secretion pattern previously observed and validating the molecular customization of the niche. The protein was particularly abundant intracellularly within hMSCs directly lining on the ceramic surface (eN-SDF1α, Fig. 6B and SI Appendix, Fig. S7D).
Flow cytometry quantitative phenotypic analysis was performed to assess the effect of the SDF1α enrichment in the system. The total number of blood cells pooled from both compartments (ECM and SN) was similar in the eN and eN-SDF1α groups (SI Appendix, Fig. S8A). However, a marked reduction in the number of HSPCs was observed in the eN-SDF1α, reflected by a significant fold increase in HSCs (1.9 f.i.) and MPPs (1.4 f.i.) in the eN systems compared with the eN-SDF1α. Importantly, the SDF1α effect appeared to be restricted to these populations since the MLPs and general blood contents were identical in both niches (SI Appendix, Fig. S8A). Interestingly, observed differences were correlated with the system compartmentalization. Indeed, the SN cellular content of both eN and eN-SDF1α was identical in composition (SN, Fig. 6C). Instead, the ECM compartment of the eN showed a higher number of HSPCs, including superior content in HSCs and MPPs (2.1. and 1.6 f.i., respectively) while, again, the MLPs and global blood numbers were not affected.
We investigated the cell cycle status of HSPC, HSC, and MPP populations retrieved from our 3D conditions. This revealed a quiescence-inducing effect of SDF1α in eN-SDF1α, with a significant increase of HSPCs, MPPs, and HSCs in G0 phase (Fig. 6D and SI Appendix, Fig. S8 B and C). In particular, in cells retrieved from the ECM, we measured an increase of 29.6%, 36.1%, and 26.9% in the number of quiescent HSPCs, HSCs, and MPPs, respectively (Fig. 6D). Thus, the observed lower proportions of HSCs and MPPs in the ECM of eN-SDF1α niches were due to the quiescence induction by SDF1α overexpression.
These findings exemplify the possibility to use the described model to engineer customized BM niches, capable to specifically impact on HSC and MPP distribution and behavior in the system.
Perturbation of HSC Behavior by Simulation of Injury in Engineered Niche.
We further assessed whether the blood cell distribution can be perturbed by simulating a niche injury (SI Appendix, Fig. S9A). Before CD34+ loading, the eN were exposed to bleomycin (eN-Bleo), as potent drug inducing DNA damage (39). The drug altered hMSC metabolism (30% reduction of activity) (SI Appendix, Fig. S9B) but not their viability in the system (SI Appendix, Fig. S9C).
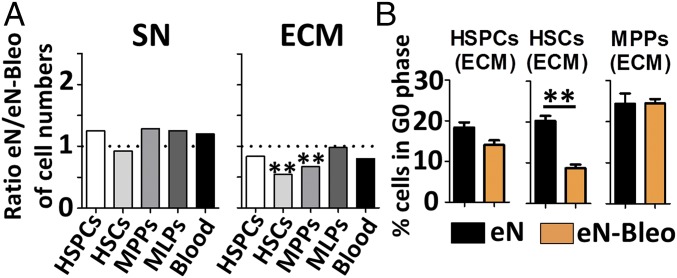
While both niches displayed similar HSPC content, the total number of HSCs was significantly higher in the eN-Bleo (SI Appendix, Fig. S9 D and E). The observed increase resulted from a higher HSC content in the ECM of eN-Bleo (Fig. 7A) while blood composition was identical in the SN of both eN and eN-Bleo (SI Appendix, Fig. S7A). Cells isolated from respective ECM niches showed a substantial decrease in the percentage of quiescent HSCs in eN-Bleo (8% vs. 20% in eN) (Fig. 7B). Interestingly, this was restricted to HSCs, and the cell cycle of HSPC and MPP populations was not affected.
Fig. 7.
Perturbation of HSC behavior by simulation of injury in engineered niche. (A) Engineered niches exposed to bleomycin (eN-Bleo) displayed a higher number of HSCs and MPPs in the ECM. In contrast, bleomycin injury did not affect the blood composition of the SN. n ≥ 3. (B) Injured niches (eN-Bleo) displayed an impaired capacity to maintain HSCs in a quiescent status, as assessed by cell cycle analysis. n ≥ 3. HSCs, hematopoietic stem cells; HSPCs, hematopoietic stem and progenitor cells; MLPs, multilymphoid progenitors; MPPs, multipotent progenitors. **P < 0.01.
These data indicate that the exposure to bleomycin impairs the capacity of hMSCs to maintain HSCs in a quiescent status, resulting in their increased proliferation. We thus validate the possibility to exploit our system for the study of human hematopoiesis in particular scenarios, like after injury.
Discussion
We report the successful in vitro engineering of BM-like tissues in a perfusion bioreactor system. The generated niches displayed high biological complexity, capturing structural, compositional, and organizational features of a native human osteoblastic environment, resulting in the support of HSPC functions. Moreover, using a proof-of-principle molecular customization of the 3D niche and through the design of specific injury scenarios, the system was validated as a BM engineering platform with tunable properties.
Tissue engineering offers new opportunities for stem cell research, enabling us to address fundamental biological questions that cannot be otherwise investigated using traditional culture plates. However, its application to the generation of viable BM environments in vitro has remained challenging, due to modeling constraints associated with the tissue complexity. This includes a precisely defined spatial organization, cellular diversity, and combined proliferation and maintenance of functionality of the blood compartment. Since existing models (14, 15) do not recapitulate all these features without bypassing the use of animals (40), we alternatively proposed the design of an “organ-like” tissue to support the development and maintenance of hematopoiesis.
Our system offers key advantages over existing approaches. First, unlike synthetic materials (41–43), the cell-deposited ECM more closely replicates native microenvironments. Despite substantial advances in the field, artificial matrices cannot recapitulate the distribution and diversity of signals existing in natural ECM nor offers their suitable and physiological presentation (44–46). Moreover, through hMSC genetic modifications and their tailored profile of secreted factors, we introduced the notion of modularity previously achieved by synthetic matrices (41–43). The biological delivery of defined cytokines by cells is a continuous process, as opposed to exogenous supplementation to culture medium, potentially associated with the issue of stability over time. This strategy is highly relevant when extended to putative niche factors, toward the identification of key cellular subsets/molecules that influence stem cell behavior (47). In this regard, the presence of a compartmentalization in our system can be exploited to address specific questions. These span from the possibility to study the chemoattractant effects of factors of interest to the investigation of mechanisms driving the release of stem cells outside of their niche, and the associated functional differences.
Despite being biologically inspired, our approach does not fully reflect the complexity of its in vivo counterpart (9). Important lacking components are, for instance, vascular and neuronal networks known to be regulators of HSC activity (48–50). This warrants the investigation of their integration into the system, though requiring the establishment of culture conditions sustaining the viability of multiple cell types (51). Nevertheless, the described model has reached a next step in complexity, illustrated by the observed self-organization of the mesenchymal and blood compartments that suggests the development of a BM proxy with organoid features.
As opposed to microfluidic approaches, we targeted the design of macroscale niches to combine reasonable throughput and multiple readouts. While microfluidic platforms consist in miniaturized systems with a high level of parallelization, the limited volume and cell numbers restrain multiplex analyses. Instead, the dimension of our system was compatible with several simultaneous assessments, including gene expression, flow cytometry, imaging, and necessary in vivo determination of stem and progenitor cell functionality.
We observed in our system an important increase in phenotypic HSC number. This was achieved in stringent xeno-free conditions, including low concentration of hematopoietic cytokines, and was validated with the use of single CB donors and multiple hMSC primary materials. However, HSC functionalities still need to be rigorously assessed by secondary transplantation and limiting-dilution assays before claiming a functional expansion of HSCs. Toward this objective, development and optimization of 3D parameters could be adopted to challenge gold-standard suspension cultures (52), currently requiring high levels of cytokine (53, 54) and agonist supplementation (52). Some of the parameters which may be tuned in the 3D-engineered culture platform include the types of niche cells, their specific stage of differentiation, the scaffolding material (e.g., structure/composition), or the physical factors driven by the perfusion system (e.g., shear stress, oxygen/hypoxia levels). All these act in concert with synergistic and competing effects and can be harnessed to modulate stem cell fate (55).
Finally, our system could be adapted to recapitulate pathological settings (56). The use of hMSCs and/or HSPCs harvested from patients suffering from blood disorders (e.g., leukemia) can offer the opportunity to model the disease in vitro, in an entirely human and ideally personalized setting. This could represent a powerful tool with a wide range of applications, from the identification of factors deregulating niche or blood functions to the screening of drugs to predict a patient-specific response to defined treatments.
Methods
Cord blood (CB) cells and human bone marrow aspirates were collected from healthy donors at the University Hospital Basel and the Universitäts-Kinderspital beider Basel (Ref.78/07), after obtaining informed consent. The study was approved by the ethics board of the canton Basel, Switzerland.
Osteoblastic niches were generated using hMSCs, following a preestablished protocol (32). Briefly, hydroxyapatite scaffolds were seeded directly in a perfusion bioreactor with 0.75 × 106 of hMSCs by overnight perfusion at a superficial velocity of 2,800 μL/s. After 24 h (cell-seeding phase), the superficial velocity was reduced to 280 μL/s for the perfusion culture of hMSCs. Cells were then cultured for 1 wk in proliferative medium (PM), followed by 3 wk of osteogenic medium (OM). Human CD34+ hematopoietic stem and progenitor cells (HSPCs) isolated from cord blood samples were seeded after 4 wk of hMSCs, by overnight perfusion at a superficial velocity of 2,800 μL/s with 7 × 105 HSPCs resuspended in serum-free expansion medium, supplemented with the following cytokines: SCF (10 ng/mL; cat. no. 130-093-991), FLT3-ligand (10 ng/mL; cat. no. 130-093-854); TPO (10 ng/mL; cat. no. 130-094-011), all from Miltenyi Biotec. After 24 h (cell-seeding phase), the superficial velocity was reduced to 280 μL/s for perfusion culture during 1 wk. Cells were then collected from the bioreactor system by collagenase and trypsinization treatments for subsequent analysis. A more detailed description of the methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (Systems-X program, 2014/266) (to R.S., T.S., and I.M.), as well as by the Freenovation program (Novartis AG) (to P.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805440115/-/DCSupplemental.
References
- 1.Méndez-Ferrer S, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kfoury Y, Scadden DT. Mesenchymal cell contributions to the stem cell niche. Cell Stem Cell. 2015;16:239–253. doi: 10.1016/j.stem.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
- 5.Zhang CC, Lodish HF. Cytokines regulating hematopoietic stem cell function. Curr Opin Hematol. 2008;15:307–311. doi: 10.1097/MOH.0b013e3283007db5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilak F, et al. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keung AJ, Healy KE, Kumar S, Schaffer DV. Biophysics and dynamics of natural and engineered stem cell microenvironments. Wiley Interdiscip Rev Syst Biol Med. 2010;2:49–64. doi: 10.1002/wsbm.46. [DOI] [PubMed] [Google Scholar]
- 8.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 9.Baryawno N, Severe N, Scadden DT. Hematopoiesis: Reconciling historic controversies about the niche. Cell Stem Cell. 2017;20:590–592. doi: 10.1016/j.stem.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 10.Rongvaux A, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: A human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Raic A, Rödling L, Kalbacher H, Lee-Thedieck C. Biomimetic macroporous PEG hydrogels as 3D scaffolds for the multiplication of human hematopoietic stem and progenitor cells. Biomaterials. 2014;35:929–940. doi: 10.1016/j.biomaterials.2013.10.038. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira MS, et al. Cord blood-hematopoietic stem cell expansion in 3D fibrin scaffolds with stromal support. Biomaterials. 2012;33:6987–6997, and erratum (2012) 33:9165. doi: 10.1016/j.biomaterials.2012.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Prewitz MC, et al. Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments. Nat Methods. 2013;10:788–794. doi: 10.1038/nmeth.2523. [DOI] [PubMed] [Google Scholar]
- 16.Leisten I, et al. 3D co-culture of hematopoietic stem and progenitor cells and mesenchymal stem cells in collagen scaffolds as a model of the hematopoietic niche. Biomaterials. 2012;33:1736–1747. doi: 10.1016/j.biomaterials.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 17.Jing D, et al. Hematopoietic stem cells in co-culture with mesenchymal stromal cells–Modeling the niche compartments in vitro. Haematologica. 2010;95:542–550. doi: 10.3324/haematol.2009.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Cuddihy MJ, Kotov NA. Three-dimensional cell culture matrices: State of the art. Tissue Eng Part B Rev. 2008;14:61–86. doi: 10.1089/teb.2007.0150. [DOI] [PubMed] [Google Scholar]
- 19.Burdick JA, Vunjak-Novakovic G. Engineered microenvironments for controlled stem cell differentiation. Tissue Eng Part A. 2009;15:205–219. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazin T, Schaffer DV. Engineering strategies to emulate the stem cell niche. Trends Biotechnol. 2010;28:117–124. doi: 10.1016/j.tibtech.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Vunjak-Novakovic G, Scadden DT. Biomimetic platforms for human stem cell research. Cell Stem Cell. 2011;8:252–261. doi: 10.1016/j.stem.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson DC, et al. Development of a three-dimensional bioengineering technology to generate lung tissue for personalized disease modeling. Stem Cells Transl Med. 2017;6:622–633. doi: 10.5966/sctm.2016-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliopoulos N, Francois M, Boivin M-N, Martineau D, Galipeau J. Neo-organoid of marrow mesenchymal stromal cells secreting interleukin-12 for breast cancer therapy. Cancer Res. 2008;68:4810–4818. doi: 10.1158/0008-5472.CAN-08-0160. [DOI] [PubMed] [Google Scholar]
- 24.Takasato M, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt H, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2015;516:435–438. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 26.Di Maggio N, et al. Toward modeling the bone marrow niche using scaffold-based 3D culture systems. Biomaterials. 2011;32:321–329. doi: 10.1016/j.biomaterials.2010.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Dellatore SM, Garcia AS, Miller WM. Mimicking stem cell niches to increase stem cell expansion. Curr Opin Biotechnol. 2008;19:534–540. doi: 10.1016/j.copbio.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peerani R, Zandstra PW. Enabling stem cell therapies through synthetic stem cell-niche engineering. J Clin Invest. 2010;120:60–70. doi: 10.1172/JCI41158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores-Guzmán P, Fernández-Sánchez V, Mayani H. Concise review: Ex vivo expansion of cord blood-derived hematopoietic stem and progenitor cells: Basic principles, experimental approaches, and impact in regenerative medicine. Stem Cells Transl Med. 2013;2:830–838. doi: 10.5966/sctm.2013-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter JR, Ruckh TT, Popat KC. Bone tissue engineering: A review in bone biomimetics and drug delivery strategies. Biotechnol Prog. 2009;25:1539–1560. doi: 10.1002/btpr.246. [DOI] [PubMed] [Google Scholar]
- 31.Wendt D, Marsano A, Jakob M, Heberer M, Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol Bioeng. 2003;84:205–214. doi: 10.1002/bit.10759. [DOI] [PubMed] [Google Scholar]
- 32.Sadr N, et al. Enhancing the biological performance of synthetic polymeric materials by decoration with engineered, decellularized extracellular matrix. Biomaterials. 2012;33:5085–5093. doi: 10.1016/j.biomaterials.2012.03.082. [DOI] [PubMed] [Google Scholar]
- 33.Bourgine PE, et al. Engineered extracellular matrices as biomaterials of tunable composition and function. Adv Funct Mater. 2017;27:1605486. [Google Scholar]
- 34.Mayani H, et al. Kinetics of hematopoiesis in Dexter-type long-term cultures established from human umbilical cord blood cells. Stem Cells. 1998;16:127–135. doi: 10.1002/stem.160127. [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez-Rodríguez M, Reyes-Maldonado E, Mayani H. Characterization of the adherent cells developed in Dexter-type long-term cultures from human umbilical cord blood. Stem Cells. 2000;18:46–52. doi: 10.1634/stemcells.18-1-46. [DOI] [PubMed] [Google Scholar]
- 36.Pittenger MF. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 37.Frank O, et al. Real-time quantitative RT-PCR analysis of human bone marrow stromal cells during osteogenic differentiation in vitro. J Cell Biochem. 2002;85:737–746. doi: 10.1002/jcb.10174. [DOI] [PubMed] [Google Scholar]
- 38.Gattazzo F, Urciuolo A, Bonaldo P. Extracellular matrix: A dynamic microenvironment for stem cell niche. Biochim Biophys Acta. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolay NH, et al. Mesenchymal stem cells are sensitive to bleomycin treatment. Sci Rep. 2016;6:26645. doi: 10.1038/srep26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torisawa YS, et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods. 2014;11:663–669. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- 41.Gobaa S, et al. Artificial niche microarrays for probing single stem cell fate in high throughput. Nat Methods. 2011;8:949–955. doi: 10.1038/nmeth.1732. [DOI] [PubMed] [Google Scholar]
- 42.Gjorevski N, Ranga A, Lutolf MP. Bioengineering approaches to guide stem cell-based organogenesis. Development. 2014;141:1794–1804. doi: 10.1242/dev.101048. [DOI] [PubMed] [Google Scholar]
- 43.Perl A, Reinhoudt DN, Huskens J. Microcontact printing: Limitations and achievements. Adv Mater. 2009;21:2257–2268. [Google Scholar]
- 44.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilgus TA. Growth factor-extracellular matrix interactions regulate wound repair. Adv Wound Care (New Rochelle) 2012;1:249–254. doi: 10.1089/wound.2011.0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci. 2010;123:4195–4200. doi: 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kokkaliaris KD, et al. Identification of factors promoting ex vivo maintenance of mouse hematopoietic stem cells by long-term single-cell quantification. Blood. 2016;128:1181–1192. doi: 10.1182/blood-2016-03-705590. [DOI] [PubMed] [Google Scholar]
- 48.Katayama Y, et al. Signals from the sympathetic nervous system regulate hematopoietic stem cell egress from bone marrow. Cell. 2006;124:407–421. doi: 10.1016/j.cell.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura Y, et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood. 2010;116:1422–1432. doi: 10.1182/blood-2009-08-239194. [DOI] [PubMed] [Google Scholar]
- 50.Itkin T, et al. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532:323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourgine PE, Martin I, Schroeder T. Engineering human bone marrow proxies. Cell Stem Cell. 2018;22:298–301. doi: 10.1016/j.stem.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Boitano AEA, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda T, et al. Expansion of human NOD/SCID-repopulating cells by stem cell factor, Flk2/Flt3 ligand, thrombopoietin, IL-6, and soluble IL-6 receptor. J Clin Invest. 2000;105:1013–1021. doi: 10.1172/JCI8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuji K, Ueda T, Ebihara Y. Cytokine-mediated expansion of human NOD-SCID-repopulating cells. Methods Mol Biol. 2003;215:387–395. doi: 10.1385/1-59259-345-3:387. [DOI] [PubMed] [Google Scholar]
- 55.Lane SW, Williams DA, Watt FM. Modulating the stem cell niche for tissue regeneration. Nat Biotechnol. 2014;32:795–803. doi: 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sánchez-Aguilera A, Méndez-Ferrer S. The hematopoietic stem-cell niche in health and leukemia. Cell Mol Life Sci. 2017;74:579–590. doi: 10.1007/s00018-016-2306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.