Summary
The brain of Down syndrome (DS) patients exhibits fewer interneurons in the cerebral cortex, but its underlying mechanism remains unknown. By morphometric analysis of cortical interneurons generated from DS and euploid induced pluripotent stem cells (iPSCs), we found that DS GABA neurons are smaller and with fewer neuronal processes. The proportion of calretinin over calbindin GABA neurons is reduced, and the neuronal migration capacity is decreased. Such phenotypes were replicated following transplantation of the DS GABAergic progenitors into the mouse medial septum. Gene expression profiling revealed altered cell migratory pathways, and correction of the PAK1 pathway mitigated the cell migration deficit in vitro. These results suggest that impaired migration of DS GABAergic neurons may contribute to the reduced number of interneurons in the cerebral cortex and hippocampus in DS patients.
Keywords: iPSC disease modeling, Down syndrome, differentiation, GABAergic neurons, migration
Highlights
-
•
DS iPSC-derived GABA interneurons show cellular deficits
-
•
DS GABA interneurons exhibit decreased migration in vitro and in vivo
-
•
RNA-seq reveals that expression of PAK1 is disrupted in the DS interneurons
-
•
Regulation of PAK1 pathway rescues the defects of migration
In this article, Liu and colleagues show that Down syndrome iPSC-derived GABAergic interneurons exhibit less complexity in morphology and impaired migration both in vitro and in vivo.
Introduction
Down syndrome (DS), resulting from an extra chromosome 21 (HSA21), or trisomy 21, is the most common genetic disorder of intellectual impairment (Epstein, 1986, Korenberg et al., 1990). Anatomical analysis from limited fetal brain tissues indicates a reduced brain size from gestation week 23 (Golden and Hyman, 1994). Careful analysis of brains from earlier stages (weeks 17–20) revealed a smaller hippocampus (Contestabile et al., 2007, Guidi et al., 2008, Sylvester, 1983) and decreased γ-aminobutyric acid (GABA) neurotransmitters (Whittle et al., 2007), suggesting an impaired neurogenesis and/or migration of interneurons, especially GABAergic interneurons (Rissman and Mobley, 2011, Ross et al., 1984). However, the relative inaccessibility of and inability to manipulate human fetal tissues impedes the dissection of cause-effect relationship between impaired production or migration of GABAergic neurons and reduced cortical interneurons.
Mouse models of DS have been used extensively to examine the effects of Ts21 and have led to significant insights into the mechanisms underlying DS. Analysis of the Ts65Dn DS mouse models show increased interneuron progenitor proliferation and increased production of interneurons (Chakrabarti et al., 2010). Although the mouse model data differ from human data that suggest fewer interneurons in human DS cortex, they highlight the fact that interneuron development is faulty in DS. Induced pluripotent stem cells (iPSCs) from DS patients provide an alternative but dynamic model system to investigate cellular, molecular, and functional changes that may contribute to DS pathogenesis. Progress has been made in revealing disease-relevant phenotypes in many neurological disorders using patient-derived iPSCs and some underlying mechanisms that are otherwise difficult to analyze (Brennand et al., 2011, Chen et al., 2014b, Kondo et al., 2013, Wen et al., 2014). Recent studies, including ours, revealed deficits in neurons and astrocytes by using DS iPSCs (Chen et al., 2014a, Shi et al., 2012, Weick et al., 2013). DS iPSC-derived cortical neurons showed decreased synapses (Weick et al., 2013) and presented Alzheimer’s disease-like pathologies at cellular levels (Shi et al., 2012). Nevertheless, none of these studies addressed why the cerebral cortex in DS patients has fewer interneurons.
Cortical interneurons are born in the medial ganglionic eminence (MGE) at gestation weeks 5–6 (Al-Jaberi et al., 2015, Zecevic et al., 2011) and migrate to the cerebral cortex during gestation weeks 8–15 (Ma et al., 2013). We hypothesize that the reduced cortical interneurons are attributed to the decreased production, proliferation, and/or migration of MGE progenitors. By comparing forebrain GABAergic interneurons that were differentiated from DS and euploid iPSCs, as well as wild-type iPSCs and human embryonic stem cells (hESCs) in vitro and after transplantation into the mouse brain, we found that the DS GABAergic interneurons showed altered subtypes with more somatostatin (SST), fewer calretinin (CR) neurons, and reduced soma size, branches, and neurite length in vitro and following transplantation into the medial septum in SCID mice. Importantly, there was a substantially reduced migration and axonal projection of DS GABAergic neurons to hippocampus and the olfactory bulb.
Results
DS GABAergic Interneurons Exhibit Less Complexity in Morphology In Vitro
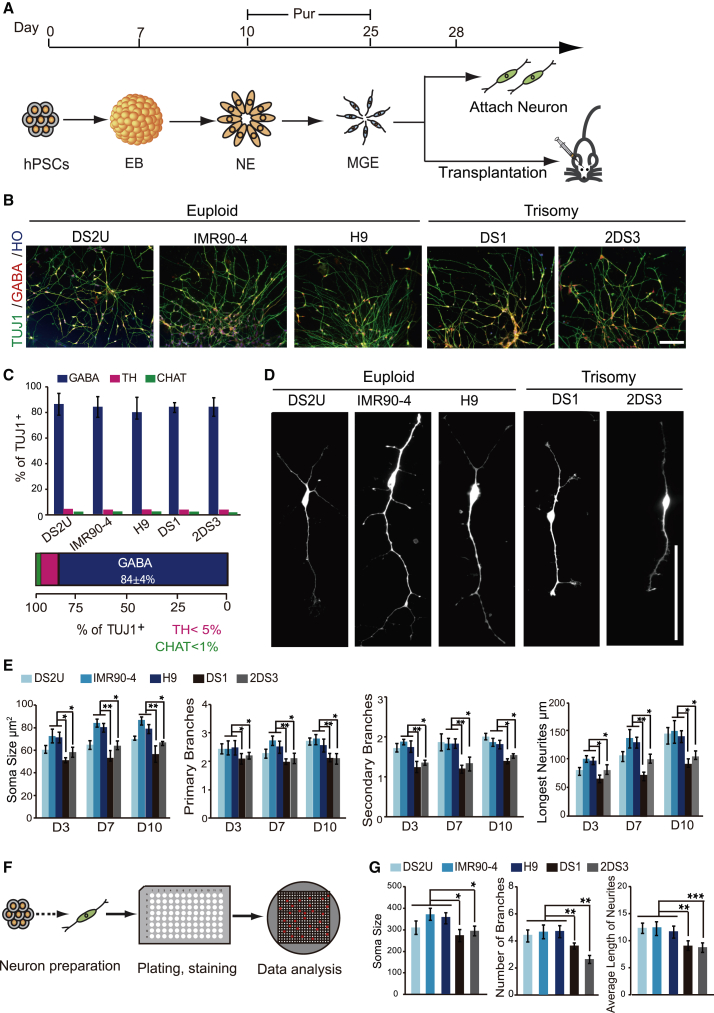
We first differentiated forebrain GABAergic interneurons from iPSCs derived from two DS patients (DS1 and 2DS3), euploid iPSCs (DS2U and IMR90-4), and euploid H9 hESCs (Figure 1A and Table S1). The euploid iPSCs (DS2U) were cloned from the same DS patient cells as DS1 in which about 10% of the cells exhibit the normal karyotype (Weick et al., 2013). Following our established protocol (Liu et al., 2013a), both the trisomy and euploid iPSCs differentiated to GABAergic neurons with a high efficiency (>80% of TUJ1+ neurons at day 35) (Figures 1B and 1C), in the presence of purmorphamine (Pur), a small-molecular sonic hedgehog (SHH) agonist. The remaining cells were positive for tyrosine hydroxylase (TH, <5%) and choline acetyltransferase (ChAT, <1%) (Figure 1C). The results suggest that trisomy iPSCs have a similar potential with euploid iPSCs to generate GABAergic interneurons.
Figure 1.
Trisomic iPSC-Derived GABAergic Interneurons Show Abnormal Gene Expression In Vitro
(A) Experimental outline. Euploid and trisomy iPSC- or hESC-derived MGE progenitors differentiated to GABAergic interneurons or transplanted to the SCID mouse brain.
(B) GABAergic interneurons were generated from euploid and trisomy iPSC lines at day 35. Scale bar, 50 μm.
(C) Proportion of GABA+, TH+, and ChAT+ neurons among total (TUJ1+) neurons derived from each cell line at day 35. At least 800 cells were counted from random selected field in each experiment, n ≥ 3 for independent experiments; bar graph presents the mean ± SEM.
(D) Representative images of euploid and trisomy GABAergic interneurons generated from each cell lines in vitro. Scale bar, 100 μm.
(E) Quantification of soma size, branches, and longest neurite length of euploid and DS GABAergic interneurons at days 3, 7 and 10 after plating. At least 500 cells were counted from random selected field in each experiment, n ≥ 3 for independent experiments; bar graphs present the mean ± SD.
(F) High-content measurement workflow. After differentiation from DS iPSCs, GABAergic interneurons were plated on 96-well plates at a density of 2,000 cells per well.
(G) Quantification of soma size, branches, and neurite length of euploid and DS GABAergic interneurons at day 7 after plating. At least 2,500 cells were analyzed for each cell line, n ≥ 3 for independent experiments; bar graphs present the mean ± SD.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Morphometric analysis indicated that the DS GABAergic interneurons (DS1 and 2DS3) exhibited a reduced soma size in comparison with euploid cells (IMR90-4 and H9) and isogenic euploid control (DS2U) at days 3, 7, and 10 after plating day-28 MGE-like progenitors (Figures 1D and 1E). Measurement of neurite growth revealed that trisomy GABAergic interneurons (DS1 and 2DS3) displayed a shortened neurite length and diminished branch complexity as compared with euploid neurons (DS2U, IMR90-4, and H9) at day 3, 7, and10 after plating (Figure 1E). These cellular deficits were replicated by high-content measurement (Figure 1F) whereby neurons were fixed 7 days after plating and labeled with TUJ1 (Figures 1F and 1G). Thus, trisomy GABAergic interneurons were generated from iPSCs in a similar efficiency as their normal counterparts but displayed less complexity in morphology.
DS GABAergic Interneurons Exhibit Altered Subtypes and Impaired Migration In Vitro
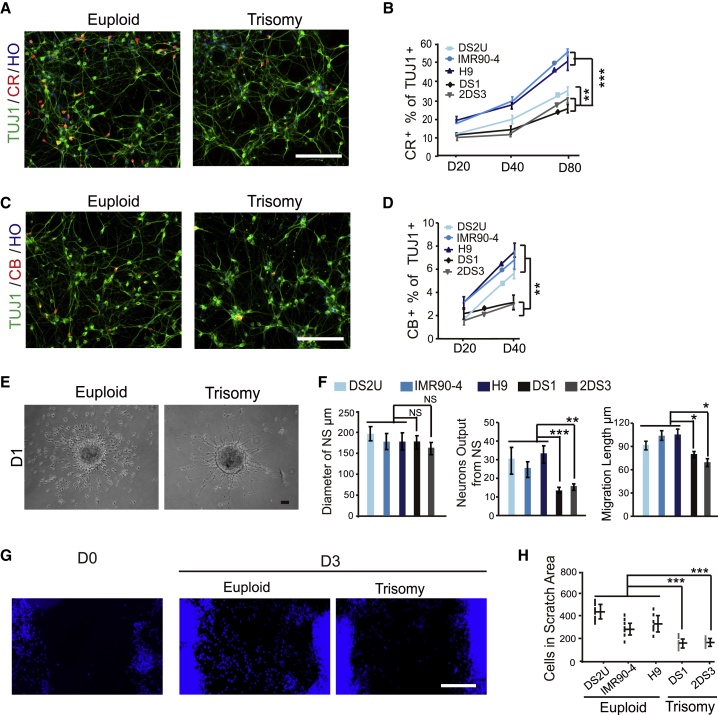
There are numerous types of GABAergic interneurons in the cerebral cortex. They may be subdivided into multiple subtypes according to the expression of calbindin (CB), SST, CR, and parvalbumin (PV) (Kawaguchi and Kubota, 1997, Wonders and Anderson, 2006). Dysfunction of any subtype may be associated with neurological disorders (Lawrence et al., 2010). The altered morphometric properties of DS neurons suggest a possibility of altered subtypes of GABAergic interneurons. Indeed, the percentage of CR+ interneurons in DS1 and 2DS3 was 23.1% ± 2.1% and 27.9% ± 1.7% at day 80, respectively, which is significantly lower than H9 (47.7% ± 4.7%) and DS2U (32.4% ± 2.1%) (Figures 2A and 2B). Similarly, the percentage of CB+ neurons in DS1 (2.2% ± 0.5%) and 2DS3 (2.0% ± 0.3%) is significantly lower than in H9 (7.4% ± 0.6%) and DS2U (5.6% ± 0.4%) at day 40 (Figures 2C and 2D). The SST+ or PV+ interneurons were rarely found at day 80 (less than 5 positive cells over one coverslip). Thus, the proportion of CR+ and CB+ subtype neurons in DS is reduced compared with the euploid neurons.
Figure 2.
Cellular and Migratory Analysis of GABAergic Interneurons In Vitro
(A) The expression of CR in euploid and trisomy iPSC-derived neurons at day 40. Scale bar, 100 μm.
(B) Quantification of CR at days 20, 40, and 80 in euploid and trisomy cell lines in vitro. At least 2,000 cells were counted from random selected field in each experiment, n ≥ 3 for independent experiments; bar graph presents the mean ± SEM.
(C) The euploid and trisomy iPSC-derived neurons expressed CB at day 40. Scale bar, 100 μm.
(D) Quantification of CB immunostaining analyses at days 20 and 40 in euploid and trisomy cell lines in vitro. At least 3,000 cells were counted from random selected field in each experiment, n ≥ 3 for independent experiments; bar graph presents the mean ± SEM.
(E) Representative phase image of euploid and trisomy GABAergic interneurons that migrated from neurospheres. Scale bar, 100 μm.
(F) Quantification of numbers and migration length of euploid and trisomy GABAergic neurons. Euploid groups include DS2U, IMR90-4, and H9. At least 12 neurospheres were plated, and 200 cells were counted in each experiment, n ≥ 3 for independent experiments; bar graphs present the mean ± SD.
(G) Representative images of scratch assay in euploid and trisomy cell lines. Scale bar, 100 μm.
(H) Quantification of the nuclei presented on the scratched area after 3 days for euploid and trisomy cell lines. n ≥ 3 for independent experiments; graph presents the mean ± SEM.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. NS, not significant.
During development, GABAergic interneurons originate from ganglionic eminences and tangentially migrate to the cortex (Ma et al., 2013). We asked whether the migration of GABAergic interneurons was affected in DS. The cell migration was assayed in two ways. First, GABAergic progenitor clusters with similar diameter (Figure 2E) (about 180 μm) were plated on glass coverslips for 24 hr and the number of cells and the distance of cell migrated away from the sphere were measured. There was a significant decrease in the number of cells and distance that cells migrated out of the sphere relative to euploid control (Figure 2F). Second, we used the scratch assay (Fattahi et al., 2016) and found that DS1 and 2DS3 GABAergic interneurons showed a notable migration deficit at day 3 after scratching (Figure 2G). There were 442 ± 63 cells that migrated to the scratched area after 3 days in H9, and 330 ± 80 cells in DS2U, compared with 150 ± 40 cells in DS1 and 160 ± 35 cells in 2DS3 (Figure 2H). Therefore, DS GABAergic interneurons exhibit decreased migration in vitro.
Both Trisomy and Euploid Progenitors Differentiate to GABAergic Interneurons in the Mouse Brain
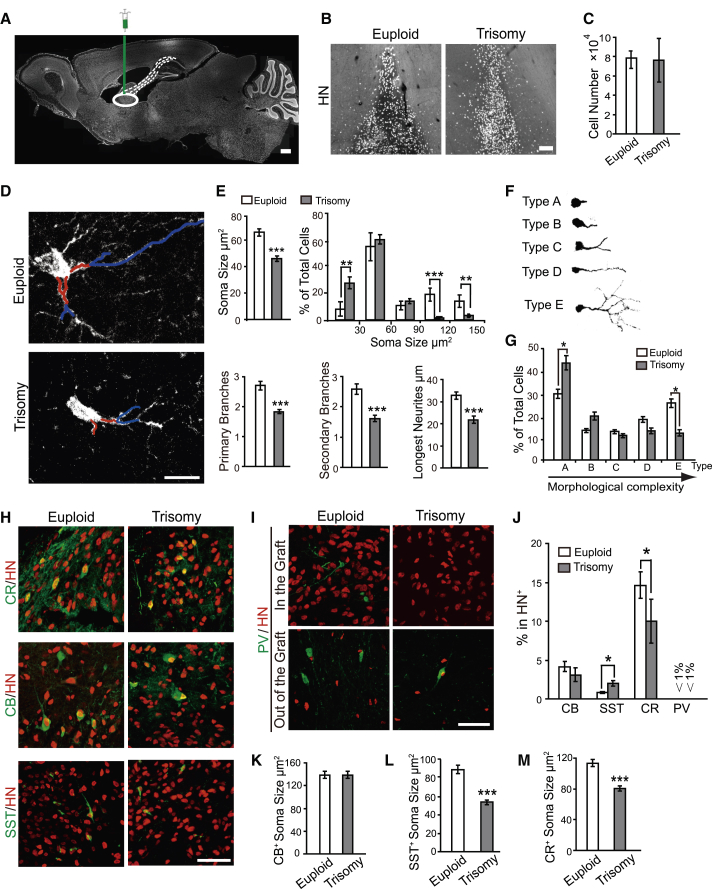
To determine whether the cellular phenotypes observed in vitro are intrinsic to DS GABAergic interneurons, we transplanted 50,000 7-week-old GABAergic progenitors, which were generated from trisomy and euploid control, into the medial septum (Figure 3A) in SCID mice (9 for DS1, 6 for 2DS3, 6 for H9, and 8 for DS2U). Transplanted human neural progenitors usually mature and form synaptic connections after 4–6 months (Liu et al., 2013b, Weick et al., 2011). When the grafts were analyzed by stereology 6 months after transplantation, we found that around 75,000 human nuclei (HN)-positive cells in the medial septum, and no obvious difference was discerned between the brains transplanted with trisomy and euploid cells (Figures 3B and 3C), suggesting that trisomy and euploid GABAergic progenitors survive in the brain in a similar manner.
Figure 3.
Survival and Differentiation of DS GABAergic Interneurons in the Mouse Brain
(A) GABAergic interneuron progenitors were injected into medial septum of SCID mice. The white dashed lines represent endogenous neuronal projections to the hippocampus. Scale bar, 500 μm.
(B) Grafted human (HN+) cells from both euploid and trisomy neurons survived in the medial septum 6 months after transplantation. Scale bar, 100 μm.
(C) Quantification of human cell numbers in the graft in euploid and trisomy groups show no significant difference for the survival grafted cells (9,522–15,055 and 5,758–20,617 HN+ cells were counted, n = 4; bar graph presents the mean ± SEM).
(D) Representative images of euploid and trisomy grafted human GABAergic interneurons in the mouse brain. The red lines illustrate the primary branches, and the blue lines illustrate the secondary branches. Scale bar, 20 μm.
(E) Quantification of soma size and its distribution, neurite arborization, and longest neurites of grafted euploid and trisomy GABAergic interneurons (n = 4; bar graphs present the mean ± SEM).
(F) Five representative neuronal types for the grafted human neurons.
(G) Distribution of the five types of grafted GABAergic neurons (n = 4; bar graph presents the mean ± SEM).
(H and I) Representative images of grafted human GABAergic interneuron subtypes, including calbindin (CB), calretinin (CR), somatostatin (SST), and parvalbumin (PV). The human PV+ neurons are observed out of the graft. Scale bars, 50 μm.
(J) Proportion of GABAergic interneuron subtypes, including CB, SST, CR, and PV, for euploid and trisomy groups (n = 4; bar graph presents the mean ± SEM).
(K) Quantification of CB+ neurons soma size in euploid and trisomy grafts. There was no significant difference between two groups (n = 3; bar graph presents the mean ± SEM).
(L and M) DS SST+ neuron (L) and CR+ neurons (M) show smaller soma size than euploid control. Euploid group refers to DS2U and Trisomy group refers to DS1 (n = 4; bar graphs present the mean ± SEM).
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Analysis of the grafted cells indicated that around 1% of the human cells were positive for NESTIN (Figure S1A) and hardly any were positive for Ki67 (Figure S1B), suggesting that the vast majority of the grafted cells become postmitotic. Indeed, 88.43% ± 5.34% of DS cells and 86.59% ± 2.64% of euploid cells expressed the neuronal marker TUJ1 (Figures S1D and S1E), 7.78% ± 5.48% of DS cells and 7.96% ± 0.91% of euploid cells were positive for an astrocyte marker glial fibrillary acidic protein (GFAP) (Figures S1D and S1E), and few were positive for an oligodendrocyte marker myelin basic protein (Figures S1C and S1E). In addition, about 40% HN+ cells expressed doublecortin (DCX) in both groups (Figures S1D and S1E). These results indicate that the majority of cells become postmitotic neurons and that some of them are migrating immature neurons.
Among the human TUJ1+ neurons in the medial septum, 77.34% ± 2.51% of the cells in DS and 78.35% ± 1.34% of those in euploid were positive for GABA, <2% for choline acetyltransferase (ChAT), and few (<1%) for TH (Figures S1F and S1G). No obvious difference in cell-type-specific marker expression was observed between DS and euploid cell transplant groups. Together, these results indicate that the transplanted trisomy and euploid cells generate GABAergic interneurons similarly in vivo.
DS GABAergic Interneurons Are Smaller with Shorter Neurites and Fewer Branches
We then asked whether the DS cells exhibit a similar phenotype in vivo. Morphometric analysis showed a wide range of soma size for the grafted GABAergic neurons. The average soma area for DS GABAergic interneurons was 38.76 ± 1.09 μm2, as compared with 52.18 ± 2.15 μm2 for euploid cells (Figures 3D and 3E). More DS GABAergic neurons were smaller than 30 μm2 than euploid cells. Less than 5% of the DS GABAergic neurons were larger than 90 μm2 compared with 30% for euploid cells (Figure 3E). Thus, more DS GABAergic neurons are in the range of smaller cell size.
Measurement of neurite branching revealed that DS GABAergic interneurons possessed fewer primary and secondary branches (Figures 3D and 3E), indicating reduced neurite arborization in DS neurons. Furthermore, neurite length, indicated by the longest constant neurites, was shorter in DS neurons compared with that in euploid cells (Figure 3E). These results were consistent with the in vitro properties.
Morphologically, the grafted neurons exhibited various shapes based on their soma size, neurite length, and branches, and we arbitrarily classified them into five categories (Figures 3F and 3G). The majority cells were type-A neurons, which exhibit short neurites and few branches. The DS groups had significantly more type-A neurons than the euploid group. In contrast, DS cells had fewer type-E neurons, those with rich branches and long neurites. Thus, the DS GABAergic interneurons are smaller with shorter neurites and fewer branches.
DS Progenitors Differentiate to More SST but Fewer CR GABAergic Neurons
Our in vitro analysis revealed altered subtypes in DS GABAergic neurons. We then asked whether this also happens in vivo. Immunostaining showed that the main types of GABAergic neurons were those positive for CR, CB, and SST (Figures 3H and 3J). Very few (<1%) were positive for PV, which were mostly on the edge or outside of the grafts (Figures 3I and 3J). The percentage and the soma size of CB+ neurons in DS were similar to those in euploid (Figures 3H, 3J, and 3K). However, the percentage of SST+ neurons increased but that of CR+ neurons decreased in DS (Figures 3H and 3J). Moreover, the soma size of SST+ and CR+ neurons in DS decreased compared with that in euploid (Figures 3L and 3M). Thus, the DS progenitors tend to generate more SST+ but fewer CR+ GABAergic interneurons at 6 months after transplantation.
DS GABAergic Neurons Exhibit Impaired Migration and Neurite Projection
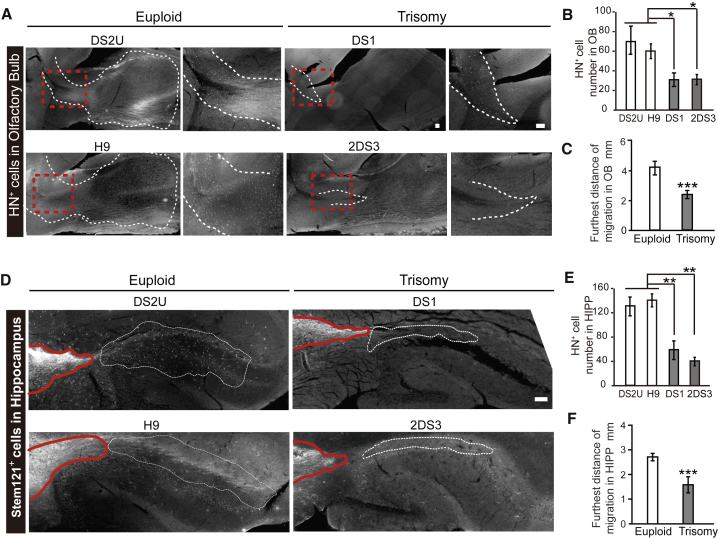
GABAergic neurons in the medial septum migrate to the olfactory bulb by the rostral migratory stream and project primarily to the hippocampus via the septohippocampal pathway (Bianchi et al., 2014, Merkle et al., 2017, Vega-Flores et al., 2014, Qin et al., 2017). We therefore asked whether human GABAergic interneurons follow the same pathways for migration/projection and whether the DS and euploid cells follow the same pattern. Immunostaining of sagittal brain sections with a human specific antibody, Stem121, showed that significantly fewer DS cells (DS1 and 2DS3) than euploid cells (DS2U and H9) were present in the olfactory bulb (Figures 4A and 4B). The distance of migration to olfactory bulb in euploid was up to 4.22 ± 0.43 mm, while in DS it was up to 2.38 ± 0.21 mm (Figure 4C).
Figure 4.
Migration of Human GABAergic Interneurons
(A) Sagittal sections showing migration path and migrated cells (HN+) in olfactory bulb area. Scale bars, 100 μm.
(B) Quantification of migrated human cells in olfactory bulb (n = 3; bar graph presents the mean ± SEM).
(C) Quantification of furthest distance of cell migration to olfactory bulb from the center of the graft (n = 3; bar graph presents the mean ± SEM).
(D) Sagittal sections showing presence of Stem121+ human neurites (red outline) and cells (dotted outline) from medial septum to posterior hippocampus in euploid (DS2U and H9) and trisomy groups (DS2 and 2DS3). Scale bar, 100 μm.
(E) Quantification of migrated human cells in the hippocampus (n = 4; bar graph presents the mean ± SEM).
(F) Quantification of furthest distance of cell migration to hippocampus from the center of the graft (n = 4; bar graph presents the mean ± SEM).
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
The majority of the human cells in the olfactory bulb were DCX-positive cells (Figure S2A), reflecting the immature nature of the human cells. Some of the human cells in olfactory bulb were GFAP positive (Figure S2B). Few grafted cells were found in the cortex with the exception of areas around the needle track in both groups. Thus, the human GABAergic interneurons follow the endogenous pathways, albeit with differential migration capacities.
Besides the olfactory bulb, we also observed human cells in the hippocampus. Significantly fewer cells were present in the hippocampus from the DS groups (DS1 and 2DS3) than from the euploid groups (DS2U and H9) (Figures 4D and 4E). The distance of the DS GABAergic neurons migrated from the injection site to hippocampus was 1.58 ± 0.31 mm, whereas that for euploid GABAergic neurons was 2.73 ± 0.13 mm (Figure 4F). In both DS and euploid groups, most of those individual (migrating) cells in the hippocampus were GABA+ neurons, and less than 5% were GFAP+ (Figure S2C). Among the GABA+ neurons, very few were positive for SST, CR, CB, and PV. Instead, they were largely positive for DCX (Figure S2C), suggesting that they remain immature by 6 months post transplantation.
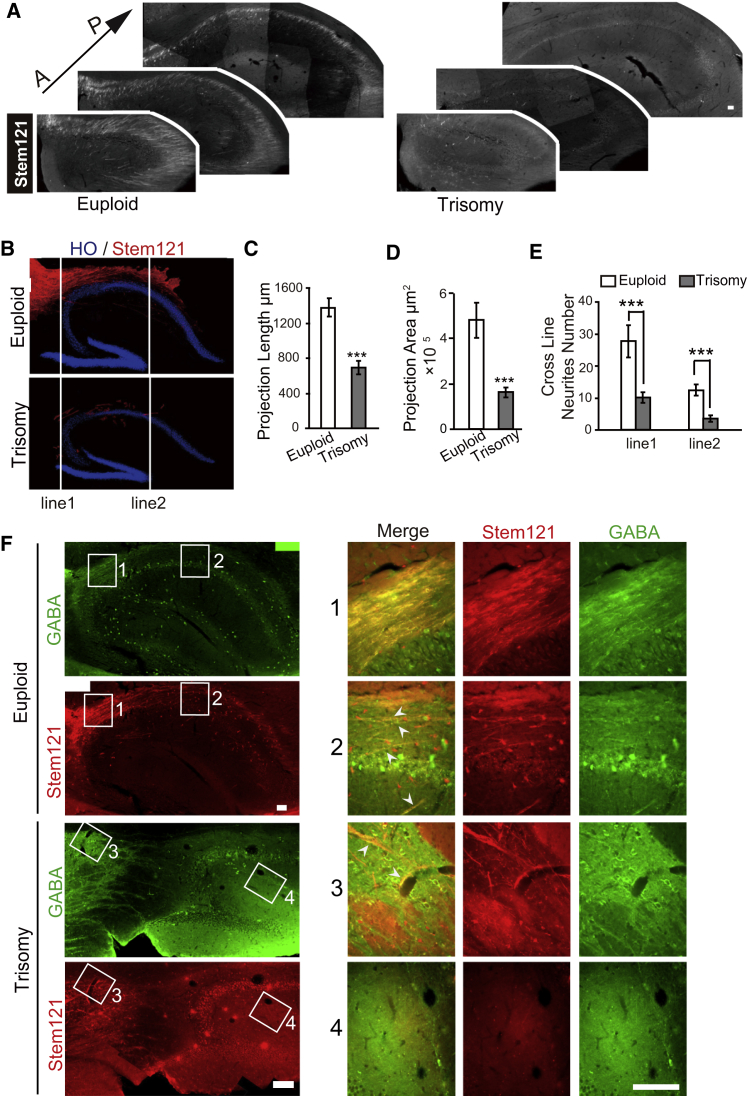
We also found that euploid cells presented a long projection trail along the septohippocampal pathway to the posterior part of hippocampus (Figures 4D and 5A). In contrast, fewer DS neurites were present in the hippocampus (Figures 5A and 5B). Quantitative measurement indicated a 50.02% reduction in neurite length from DS neurons compared with euploid cells (Figure 5C). Projection area by DS neurites was also significantly smaller than that by euploid neurites (Figure 5D). The reduced neurite projection was also demonstrated by counting the number of neurites that cross the two artificial lines according to the hippocampus structure (Figures 5B and 5E). In the euploid group, 28 ± 4.96 neurites crossed line 1, which is in the anterior hippocampus, and 12 ± 1.83 neurites traversed line 2 which is in the posterior hippocampus (Figure 5D). In contrast, approximately 10 ± 1.64 neurites crossed line 1 and only 4 ± 1.03 neurites traversed line 2 in the DS group (Figure 5E). These Stem121+ neurites coexpressed GABA (Figure 5F), indicating their GABAergic neuron identity. In addition, most of the GABA+ neurites were DCX+ with some positive for CR but not CB, SST, or PV (Figures S3A and S3B), suggesting that the majority of the neurites are from immature GABAergic neurons. Thus, DS GABAergic neurons exhibit a decreased axonal projection to the hippocampus.
Figure 5.
Axonal Projection of GABAergic Neurons toward Hippocampus
(A) Coronal sections showing presence of Stem121+ human neurites and cells in dorsal hippocampus from anterior to posterior in euploid and trisomy groups. Scale bar, 100 μm.
(B) Schematic images illustrating how the neurites in euploid and trisomy groups that cross line 1 (the line tangent to anterior CA3) and line 2 (the line tangent to posterior tip of dentate gyrus) are counted.
(C) Quantification of human neurite projection length in hippocampus in euploid and trisomy groups (n = 4; bar graph presents the mean ± SEM).
(D) Quantification of distribution areas with human neurites in hippocampus (n = 4; bar graph presents the mean ± SEM).
(E) Quantification of neurite numbers that cross line 1 and line 2 in euploid and trisomy groups (n = 4; bar graph presents the mean ± SEM).
(F) Colabeling of grafted Stem121+ neurites/cells with GABA (green) in hippocampus. Insets are shown at high magnification. Euploid group refers DS2U and Trisomy group refers DS1. The arrowheads represent the co-labeling of Stem121+ neurites/cells with GABA.
∗∗∗p < 0.001. Scale bars, 100 μm.
RNA Profiling Reveals Altered Gene Expression in DS GABAergic Interneurons
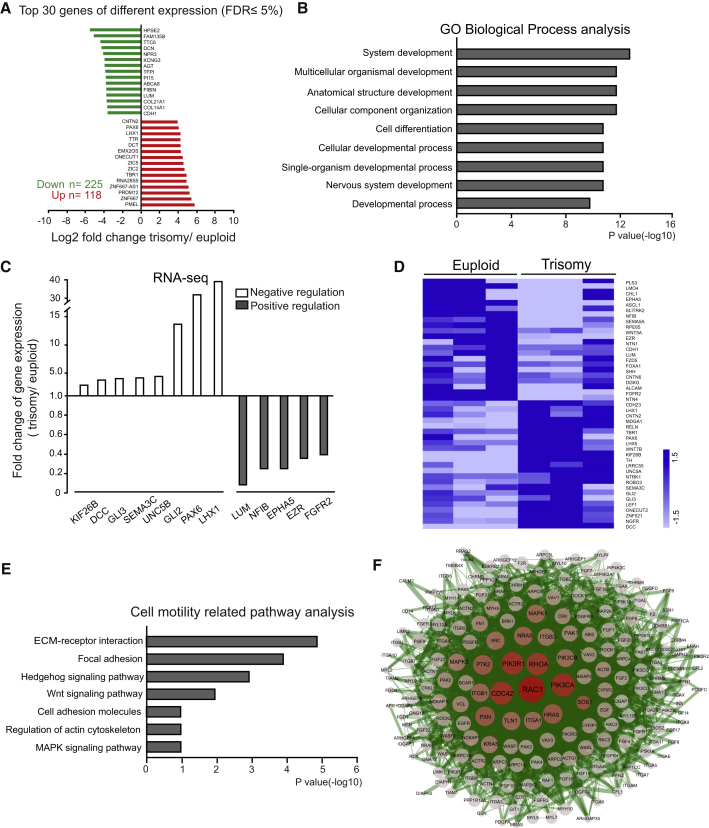
To gain insights into the molecular underpinning of DS GABAergic interneuron behaviors, we performed RNA sequencing (RNA-seq) using day-45 GABA interneurons that were differentiated from three independent Down syndrome patients (DS1, 2DS3, and DSP) and three euploid iPSCs (DS2U, IMR90-4, and H9). Of all 21,720 genes profiled, we found that 119 genes were significantly upregulated while 260 were downregulated in trisomic GABAergic interneurons compared with euploid control by using Cufflinks software (Figure 6A and Table S3), with a false discovery rate (FDR) ≤ 0.05 and |log2(fold change)| ≥ 1.
Figure 6.
Transcriptional Analysis of Trisomic and Euploid GABAergic Interneurons by RNA Sequencing
(A) Total number of genes and the top 30 fold-changed genes that were altered in trisomy GABAergic interneurons with an FDR of 5%.
(B) Top 9 altered biological processes in trisomy GABAergic interneurons in comparison with euploid controls by using gene ontology. p ≤ 0.05.
(C) The expression of representative genes involved in regulating cell morphology revealed by RNA-seq.
(D) Heatmap of differentiation-related gene expression in euploid and trisomy GABAergic interneurons.
(E) Cell motility-related pathways that were altered in trisomy GABAergic interneurons by using KEGG pathway enrichment.
(F) A network plot of “Regulation of actin cytoskeleton” pathway genes and their connections.
About 224 terms were significantly altered in DS GABAergic interneurons compared with the controls (p ≤ 0.05) by using gene ontology biological process (Table S4). Development-related terms were highly enriched in DS GABAergic interneurons, including system development, cell differentiation, and nervous system development, especially those relating to cell morphogenesis (Figure 6B). Lymphoid enhancer binding factor 1 (LEF1) and DCC netrin 1 receptor (DCC) were increased while lumican (LUM) and fibroblast growth factor receptor 2 (FGFR2) were decreased (Figure 6C and Table S5). Cell differentiation-related genes were also altered in DS GABAergic interneurons, such as achaete-scute family basic helix-loop-helix transcription factor 1 (ASCL1), Wnt family member 5A (WNT5A), nerve growth factor receptor (NGFR), and GLI family zinc finger 3 (GLI3) that are involved in basal forebrain development (Figure 6D and Table S6). This gene expression profile is consistent with our finding of impaired interneuron subtype differentiation in DS.
Pathway analysis using the DAVID Bioinformatics Resource revealed that146 pathways were significantly changed in DS GABAergic interneuron in comparison with controls (Table S7). Among them, cell motility-related pathways were prominent, including extracellular matrix (ECM)-receptor interaction, focal adhesion, hedgehog signaling pathways, cell adhesion molecules, and regulation of actin cytoskeleton (Figure 6E). Notably, the regulation of the actin cytoskeleton pathway is critical for neural migration (Solecki et al., 2009), hinting that the altered actin cytoskeleton pathway may play an important role in the defective migratory ability in DS. Protein-protein interaction network analysis revealed 212 genes that were involved in cytoskeleton pathway, including Rac family small guanosine triphosphatase 1 (Rac1), cell division cycle 42 (Cdc42), alkaline phosphatase (PhoA), and p21 activated kinase 1 (PAK1), which may contribute to the impaired migration of DS GABAergic interneurons (Figure 6F).
Correction of PAK1 Expression Rescues Migration Deficit of DS GABAergic Neurons
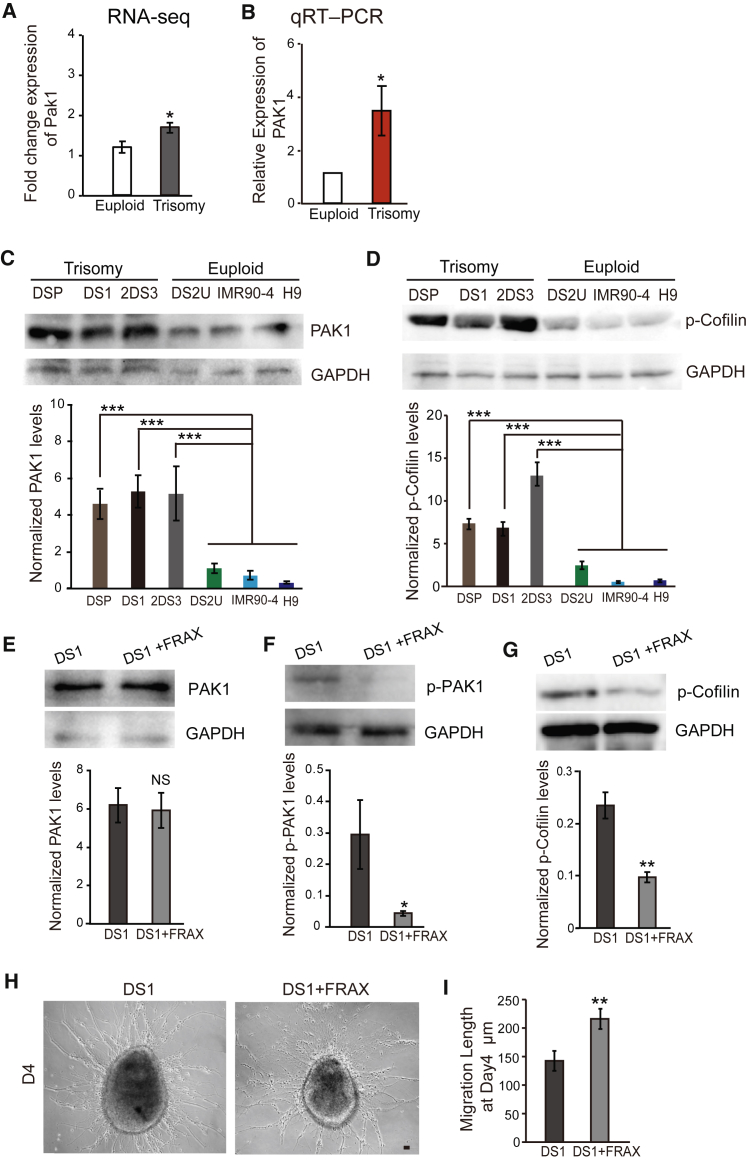
Our RNA-seq, confirmed by qPCR analysis, showed that the expression of PAK1 was increased in DS cells (Figures 7A and 7B). Western blotting revealed a substantial increase in PAK1 protein (Figure 7C). Its downstream gene cofilin was not altered. However, the phosphorylated cofilin (p-cofilin) was substantially increased in DS GABAergic neurons that were derived from DS1, 2DS3, and DSP iPSC lines compared with euploid controls (DS2U, IMR90-4, H9) (Figure 7D). PAK1 is important for neural migration during cortical genesis in mice (Causeret et al., 2008). Its downstream protein, cofilin, is an actin-binding protein that modulates cytoskeleton. We hypothesized that altered expression of PAK1 and cofilin is associated with the cell migration deficit seen in vitro and in vivo. The DS GABAergic progenitors were treated with FRAX486, an inhibitor of PAK1 with excellent kinase inhibition (Dolan et al., 2013), for 5 days. While the content of PAK1 was similar in the control and treated cells (Figure 7E), the expression of phosphorylated PAK1 (p-PAK1) and p-cofilin in DS GABAergic neurons was decreased after application of FRAX (Figures 7E–7G). Importantly, we found that there was a significant increase in migration distance of DS GABAergic interneurons with the treatment of FRAX486 (Figures 7H and 7I). These results suggest that an altered PAK1 pathway is at least partly responsible for DS interneuron migration defects.
Figure 7.
Deregulation of Cofilin Rescued Migration Deficit of DS GABAergic Neurons
(A and B) RNA-seq (A) and qRT-PCR analysis (B) of PAK1.
(C and D) Representative western blots for PAK1 (C) and p-cofilin (D) in euploid and trisomy GABAergic interneurons (n ≥ 3 for independent experiments; bar graphs present the mean ± SEM).
(E) Western blot analysis of PAK1 in trisomy GABAergic interneurons after treating with FRAX486 (FRAX = FRAX486, n ≥ 3 for independent experiments; bar graph presents the mean ± SEM).
(F and G) Western blot analysis of p-PAK1 (F) and p-cofilin (G) in trisomy GABAergic interneurons after treating with FRAX486 (n ≥ 3 for independent experiments; bar graphs present the mean ± SEM).
(H) Representative phase image of FRAX486-treated trisomy GABAergic interneuron that migrated from neurospheres. Scale bar, 100 μm.
(I) Quantification of migration length for (H). At least 10 neurospheres were plated and 400 cells were counted in each experiment, n ≥ 3 for independent experiments; bar graph presents the mean ± SD.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. NS, not significant.
Discussion
Our present study revealed that cortical inhibitory neural progenitors are generated from DS and euploid iPSCs similarly, but the differentiated DS GABAergic interneurons exhibit a reduced soma size and neurite complexity as well as a shift in GABAergic neuron subtypes with an increase in SST+ and reduction in CR+ cells. Such phenotypes are replicated following transplantation of the DS GABAergic neuronal progenitors into the mouse brain, strongly suggesting that these changes are directly associated with trisomy 21. More importantly, by transplanting the progenitors into the medial septum, which allows human cells to migrate along the same specific pathways as their endogenous counterparts in the adult brain, we discovered that the DS GABAergic neuron progenitors have a substantially reduced capacity to migrate and the differentiating GABAergic neurons to project axons to their targets. These results provide evidence of impaired development of cortical interneurons caused by trisomy, thus potentially explaining in part the reduced number of cortical interneurons in DS patients.
The observation that DS individuals have a smaller hippocampus from gestation weeks 19–20 (Contestabile et al., 2007) and overall reduced brain size from gestation week 23 (Golden and Hyman, 1994, Weitzdoerfer et al., 2001) suggests an impaired neurogenesis and/or migration of late-born cells, especially GABAergic interneurons. Consistent with this hypothesis, we have shown that specification of neuroectoderm cells and the generation of early-born cortical projection neurons from DS patient iPSCs are not altered (Weick et al., 2013). Our present study suggests that the generation of cortical GABAergic interneurons appears normal, although further work is needed to examine the differentiation propensity of GABAergic neuron progenitors from iPSCs under the condition that exogenous SHH does not override the effect of trisomy 21. Decreased neurogenesis and selective apoptosis of trisomic neurons (Rissman and Mobley, 2011) may become the underlying reason that causes delayed/impaired migration to a specific target. Nevertheless, our analysis did reveal smaller cell size and fewer processes of GABAergic interneurons in DS. Similar characteristics have been reported for fetus-derived trisomy 21 neurons (Bahn et al., 2002). Such a phenotype is not an artifact of culture conditions, as GABAergic interneurons differentiated from transplanted progenitors exhibit the same phenotype. Hence, even though the generation of GABAergic interneurons is not altered, the reduced cell size and fewer processes will at least partially contribute to a smaller brain observed in DS individuals.
Cortical GABAergic interneurons are born in the MGE at weeks 5–6 (Al-Jaberi et al., 2015, Zecevic et al., 2011) and migrate laterally and dorsally to the cerebral cortex and hippocampus over a long developmental period from week 8 to week 15 (Ma et al., 2013). Hence, impaired migration of the interneurons may account for the fewer interneurons in the DS cortex (Ross et al., 1984, Weitzdoerfer et al., 2001). Indeed, by transplanting the MGE progenitors to the medial septum, we have now clearly demonstrated that DS interneurons have migrated a shorter distance than isogenic cells along the septohippocampal pathway and the rostral migratory stream. Consequently, fewer cells have reached the target hippocampus or olfactory bulb even by 6 months after transplantation. As a note of coincidence, DS patients often present smell deficits (Bianchi et al., 2014, Murphy and Jinich, 1996, Nijjar and Murphy, 2002, Wetter and Murphy, 1999), and our study shows a defective migration of DS GABAergic neurons to the olfactory bulb. This is made possible by transplanting the progenitor cells into the medial septum where the grafted cells interact with multiple cell types and migrate following the physiological routes. Of course this is a xenotransplant paradigm, which may not completely recapitulate the cellular interactions. Nevertheless, this transplant paradigm validates our in vitro observation and reveals the cellular defects likely underlying the reduced interneurons in the DS cerebral cortex.
The molecular mechanism underlying the impaired migration is not known. Our RNA-seq results suggest the involvement of multiple pathways that affect migration, ranging from ECM-receptor interaction and cell adhesion molecules to regulation of actin cytoskeleton. The substantial increased expression of collagen genes such as COL6A1 prompted us to knock down COL6A1 as it inhibits cellular migration in trisomic fibroblasts, germ cells, and neural crest cells (Delom et al., 2009, Heuckeroth, 2015, Leffler et al., 1999, Roper et al., 2009). However, we did not see a significant increase in migration in DS GABAergic neurons. Interestingly, we found that the expression of PAK1, a critical gene related to the actin cytoskeleton pathway, was upregulated in DS iPSC-derived GABAergic interneurons. PAK1, located on chromosome 21, is a downstream gene of Down syndrome cell adhesion molecule (DSCAM) (Liu et al., 2009). Overexpression of DSCAM in primary mouse neurons deregulates the activity of pPAK1 (Pérez-Núñez et al., 2016). Strikingly, we observed altered (increased) expression of p-PAK1 and p-cofilin in DS GABAergic interneurons, suggesting activation of the PAK1 pathway in DS GABA interneurons. This may be at least partly responsible for the DS interneuron migration defects. Indeed, treatment of DS GABAergic interneurons by PAK1 inhibitor FRAX486 partially rescued the migration defect. Therefore, PAK1 is a potential therapeutic target for DS.
Together, our current analyses have revealed an impaired migratory capacity of the GABAergic interneurons, which explains the reduced number of interneurons in the cerebral cortex. Together with smaller cells and fewer processes, this might contribute to a smaller brain size and potentially impaired cognition.
Experimental Procedures
iPSC Culture and GABAergic Interneuron Differentiation
iPSC lines were generated from two Down syndrome patients as reported by Weick et al. (2013). DS iPSC lines were trisomy 21 (DS1, 2DS3, and DSP) and euploid iPSC line (DS2U, the isogenic control of DS1 with identical background) (Weick et al., 2013), and wild-type iPSC line (IMR90-4) and hESC line (H9) were used as additional controls. DSP was trisomy 21 and generated from a separate mosaic DS individual. For pluripotency maintenance, iPSCs/hESCs were maintained on a feeder layer of irradiated embryonic mouse fibroblast as previously described (Liu et al., 2013b).
For neural differentiation, iPSCs were detached with Dispase (1 U/mL, Gibco) to form embryoid bodies (EBs) in suspension in neural induction medium (NIM) consisting of DMEM/F12 (Gibco), N2 supplement (1:100; Gibco), and non-essential amino acids (NEAA) (1:100; Gibco) for 7 days. EBs were attached on day 7, and formed neural tube-like rosettes at days 10–16. To induce MGE-like progenitors, we treated the neuroepithelial cells with Pur (1.5 μM) or together with SHH at days 10–25 as previously described (Liu et al., 2013a). Rosette colonies were detached by gently blowing the neural colonies with a pipette and suspended to form neurospheres on day 16. At day 24, neurospheres were dissociated to single cells with TrypLE and plated on polyornithine-coated coverslips for relevant experiments.
Immunostaining
Neurons were fixed with 4% paraformaldehyde for 30 min. Neurons were washed with PBS for 5 min three times, permeabilized in 0.2% Triton X-100 for 10 min, and blocked for 1 hr in 10% donkey serum before being incubated in the primary antibody in 5% serum and 0.1% Triton X-100 at 4°C overnight. Cells were subsequently washed and stained with Alexa Fluor-conjugated secondary antibodies and Hoechst in 5% donkey serum for 1 hr before being washed and mounted onto glass slides with Fluoromount-G mounting solution (SouthernBiotech). The primary antibodies are listed in Table S2. Images were taken using a Nikon 80i fluorescence microscope (Nikon Instruments).
High-Content Measurement
GABAergic progenitors (day 35) were dissociated and plated at a density of 2,000 neurons per well in 96-well flat-bottom plates that were pre-coated with Matrigel. Neurons were cultured with NIM as previously described. After 7 days of attachment, the neurons were immunostained for TUJ1. For high-content assays, image acquisition and analysis were performed using the Arrayscan VTI (Thermoscientifics, USA). The nuclei were detected based on the Hoechst staining, and soma size was detected based on TUJ1. A minimum of 500 cells per well were analyzed at a 20× objective, and at least five wells were analyzed for each iPSC-derived neuronal group. To quantify neurite outgrowth, we used the “neuronal profiling bioapplication” to track TUJ1-positive neurites. Each value was the average value of one well.
GABAergic Progenitor Migration Assay
Before assay, neurospheres were triturated by Pasteur pipette (Liu et al., 2013a). The narrow opening and bend in the pipette help to shear the clusters into smaller clusters with a roughly uniform size. Similar sizes of neurospheres were selected for the migration assay. Neurospheres were transferred carefully and plated dispersedly onto Matrigel-coated coverslips. After 24 hr, neurons migrated out from neurospheres and were assessed under a phase-contrast microscope (Nikon, TS100).
Scratch Assay
Neurospheres were dissociated into single cells and plated onto Matrigel-coated 96-well culture plates (450,000/cm2) in a neural differentiation medium containing DMEM/F12, N2 (1:100), NEAA (1:100), and B27 (1:50). After 3 days, the adherent culture was scratched using a 200-μL pipette tip. The neurons were fixed for imaging and migration analysis after 3 days. The scratched area was defined immediately after scratching. The cells that were present on the scratched area after a 3-day migration were counted.
Cell Transplantation
The animal experiments were carried out following the protocols approved by the Animal Care and Use Committee at the University of Wisconsin-Madison and Nanjing Medical University. The mice, aged 8–10 weeks, were anesthetized with 1% isoflurane mixed with oxygen. GABAergic progenitors were differentiated from DS1, 2DS3, DS2U, and H9 human PSCs for 7 weeks before transplantation. A week before transplantation, a small portion of GABAergic progenitors was dissociated and plated onto coverslips for the maturity and purity analysis. If more than 90% of the cells were NKX2.1-positive at day 30, and more than 50% of the cells were GABA-positive around day 35, the progenitors were “good” for transplantation. GABAergic progenitor clusters, around 30 μm of diameter, were injected into the medial septum with a glass pipette at a 15° angle toward the midline at the following stereotactic coordinates: anterior-posterior = +0.38 mm; left lateral = +1 mm; and dorsoventral = −4.12 mm. About 50,000 cells were injected into medial septum in 1 μL of artificial cerebrospinal fluid over a 5-min period.
Tissue Preparation and Immunohistochemistry
At 6 months after transplantation, animals were perfused transcardially with 4% paraformaldehyde. The brain was serially sectioned coronally and sagittally to 35 μm in thickness and subsequently stored in the preservation solution at −20°C.
Immunohistochemistry was performed as previously described (Liu et al., 2013b). Sections were washed with PBS for 5 min three times, and permeabilized and blocked for 1 hr in 10% donkey serum and 0.2% Triton X-100 before being incubated in the primary antibody in 5% serum and 0.2% Triton X-100 at 4°C overnight. Sections were subsequently washed and stained with Alexa Fluor (Life) secondary antibodies and Hoechst in 5% donkey serum for 1 hr before being washed and mounted onto glass slides with Fluoromount-G mounting solution (SouthernBiotech). The primary antibodies used in this study are listed in Table S2. Images were visualized using a Nikon 80i fluorescence microscope (Nikon Instruments) and a Zeiss confocal microscope (LSM700, Zeiss instruments).
Real-Time PCR
Total RNA was isolated using the TRIzol kit (Invitrogen, USA) according to the manufacturer's manual. One microgram of total RNA from each sample was reverse transcribed into cDNA and subjected to real-time PCR using SuperScript III First-Strand (Invitrogen). Primers for real-time PCR were: PAK1 forward primer CAG CCC CTC CGA TGA GAA ATA; PAK1 reverse primer CAA AAC CGA CAT GAA TTG TGT GT.
Western Blotting
For analysis of cofilin and p-cofilin protein, neurons were lysed in RIPA buffer with protease and protease inhibitor cocktail (Roche). Proteins was separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. Anti-PAK1 (1:500 dilution, #SC-166887, Santa Cruz Biotechnology), anti-Phospho-PAK1 (1:500 dilution, #2605P, Cell Signaling Technology), and anti-Phospho-cofilin (1:1,000 dilution, #3311, Cell Signaling) antibodies were used. Western blotting was performed using horseradish peroxidase-conjugated immunoglobulin G as a secondary antibody and the ECL system for detection.
Data Analysis
Data are shown as mean ± SEM. Statistical comparisons between two groups were performed by Student's t test. Statistical analyses were performed using one-way ANOVA (with Tukey’s or Dunnett’s test for multiple comparisons). p Values of <0.05 were considered statistically significant.
Author Contributions
Z.-Y.Q., H.-Q.H., S.-C.Z., and Y.L. designed the experiments; Z.-Y.Q., H.-Q.H., F.Y., L.M., L.Y., M.X., Y.H., and Y.L. performed the experiments; Z.-Y.Q., H.-Q.H., J.J., and Y.L. analyzed the data; H.-Q.H., A.B., S.-C.Z., and Y.L. wrote the paper.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (2016YFC1306703), the National Natural Science Foundation of China (grant 81471301), Jiangsu Outstanding Young Investigator Program (BK20160044 and BK20160047), Jiangsu Province's Innovation person, and the US NIH (R21HD085288, MH099587, and MH100031).
Published: March 8, 2018
Footnotes
Supplemental Information includes three figures and seven tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.02.001.
Contributor Information
Su-Chun Zhang, Email: zhang@waisman.wisc.edu.
Yan Liu, Email: yanliu@njmu.edu.cn.
Accession Numbers
The raw data have been deposited in the NCBI's Sequence Read Achieve (accession numbers SRR6517764, SRR6517763, SRR6517762, SRR6517761, SRR6517760, and SRR6517759).
Supplemental Information
References
- Al-Jaberi N., Lindsay S., Sarma S., Bayatti N., Clowry G.J. The early fetal development of human neocortical GABAergic interneurons. Cereb. Cortex. 2015;25:631–645. doi: 10.1093/cercor/bht254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S., Mimmack M., Ryan M., Caldwell M.A., Jauniaux E., Starkey M., Svendsen C.N., Emson P. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet. 2002;359:310–315. doi: 10.1016/S0140-6736(02)07497-4. [DOI] [PubMed] [Google Scholar]
- Bianchi P., Bettini S., Guidi S., Ciani E., Trazzi S., Stagni F., Ragazzi E., Franceschini V., Bartesaghi R. Age-related impairment of olfactory bulb neurogenesis in the Ts65Dn mouse model of Down syndrome. Exp. Neurol. 2014;251:1–11. doi: 10.1016/j.expneurol.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Brennand K.J., Simone A., Jou J., Gelboin-Burkhart C., Tran N., Sangar S., Li Y., Mu Y., Chen G., Yu D. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causeret F., Terao M., Jacobs T., Nishimura Y.V., Yanagawa Y., Obata K., Hoshino M., Nikolic M. The p21-activated kinase is required for neuronal migration in the cerebral cortex. Cereb. Cortex. 2008;19:861–875. doi: 10.1093/cercor/bhn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti L., Best T.K., Cramer N.P., Carney R.S., Isaac J.T., Galdzicki Z., Haydar T.F. Olig1 and Olig2 triplication causes developmental brain defects in Down syndrome. Nat. Neurosci. 2010;13:927–934. doi: 10.1038/nn.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jiang P., Xue H., Peterson S.E., Tran H.T., McCann A.E., Parast M.M., Li S., Pleasure D.E., Laurent L.C. Role of astroglia in Down's syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat. Commun. 2014;5:4430. doi: 10.1038/ncomms5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Qian K., Du Z.W., Cao J.Y., Petersen A., Liu H.S., Blackbourn L.W., Huang C.L., Errigo A., Yin Y.N. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contestabile A., Fila T., Ceccarelli C., Bonasoni P., Bonapace L., Santini D., Bartesaghi R., Ciani E. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus. 2007;17:665–678. doi: 10.1002/hipo.20308. [DOI] [PubMed] [Google Scholar]
- Delom F., Burt E., Hoischen A., Veltman J., Groet J., Cotter F.E., Nizetic D. Transchromosomic cell model of Down syndrome shows aberrant migration, adhesion and proteome response to extracellular matrix. Proteome Sci. 2009;7:31. doi: 10.1186/1477-5956-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan B.M., Duron S.G., Campbell D.A., Vollrath B., Shankaranarayana R.B.S., Ko H.Y., Lin G.G., Govindarajan A., Choi S.Y., Tonegawa S. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc. Natl. Acad. Sci. USA. 2013;110:5671–5676. doi: 10.1073/pnas.1219383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C.J. Developmental genetics. Experientia. 1986;42:1117–1128. doi: 10.1007/BF01941286. [DOI] [PubMed] [Google Scholar]
- Fattahi F., Steinbeck J.A., Kriks S., Tchieu J., Zimmer B., Kishinevsky S., Zeltner N., Mica Y., El-Nachef W., Zhao H. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. 2016;531:105–109. doi: 10.1038/nature16951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.A., Hyman B.T. Development of the superior temporal neocortex is anomalous in trisomy 21. J. Neuropathol. Exp. Neurol. 1994;53:513–520. doi: 10.1097/00005072-199409000-00011. [DOI] [PubMed] [Google Scholar]
- Guidi S., Bonasoni P., Ceccarelli C., Santini D., Gualtieri F., Ciani E., Bartesaghi R. Neurogenesis impairment and increased cell death reduce total neuron number in the hippocampal region of fetuses with Down syndrome. Brain Pathol. 2008;18:180–197. doi: 10.1111/j.1750-3639.2007.00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuckeroth R.O. Hirschsprung's disease, down syndrome, and missing heritability: too much collagen slows migration. J. Clin. Invest. 2015;125:4323–4326. doi: 10.1172/JCI85003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb. Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kondo T., Asai M., Tsukita K., Kutoku Y., Ohsawa Y., Sunada Y., Imamura K., Egawa N., Yahata N., Okita K. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Korenberg J.R., Kawashima H., Pulst S.M., Allen L., Magenis E., Epstein C.J. Down syndrome: toward a molecular definition of the phenotype. Am. J. Med. Genet. Suppl. 1990;7:91–97. doi: 10.1002/ajmg.1320370719. [DOI] [PubMed] [Google Scholar]
- Lawrence Y.A., Kemper T.L., Bauman M.L., Blatt G.J. Parvalbumin-, calbindin-, and calretinin-immunoreactive hippocampal interneuron density in autism. Acta Neurol. Scand. 2010;121:99–108. doi: 10.1111/j.1600-0404.2009.01234.x. [DOI] [PubMed] [Google Scholar]
- Leffler A., Ludwig M., Schmitt O., Busch L.C. Germ cell migration and early development of the gonads in the trisomy 16 mouse—an animal model for Down's syndrome. Ann. Anat. 1999;181:247–252. doi: 10.1016/S0940-9602(99)80039-9. [DOI] [PubMed] [Google Scholar]
- Liu G., Li W., Wang L., Kar A., Guan K.L., Rao Y., Wu J.Y. DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc. Natl. Acad. Sci. USA. 2009;106:2951–2956. doi: 10.1073/pnas.0811083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu H., Sauvey C., Yao L., Zarnowska E.D., Zhang S.C. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat. Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Weick J.P., Liu H., Krencik R., Zhang X., Ma L., Zhou G.M., Ayala M., Zhang S.C. Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat. Biotechnol. 2013;31:440–447. doi: 10.1038/nbt.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Wang C., Wang L., Zhou X., Tian M., Zhang Q., Zhang Y., Li J., Liu Z., Cai Y. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- Merkle F.T., Mirzadeh Z., Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2017;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Murphy C., Jinich S. Olfactory dysfunction in Down's syndrome. Neurobiol. Aging. 1996;17:631–637. doi: 10.1016/0197-4580(96)00008-5. [DOI] [PubMed] [Google Scholar]
- Nijjar R.K., Murphy C. Olfactory impairment increases as a function of age in persons with Down syndrome. Neurobiol. Aging. 2002;23:65–73. doi: 10.1016/s0197-4580(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Pérez-Núñez R., Barraza N., Gonzalez-Jamett A., Cárdenas A.M., Barnier J.V., Caviedes P. Overexpressed Down syndrome cell adhesion molecule (DSCAM) deregulates P21-activated kinase (PAK) activity in an in vitro neuronal model of Down syndrome: consequences on cell process formation and extension. Neurotox Res. 2016;30:76–87. doi: 10.1007/s12640-016-9613-9. [DOI] [PubMed] [Google Scholar]
- Qin S., Ware S.M., Waclaw R.R., Campbell K. Septal contributions to olfactory bulb interneuron diversity in the embryonic mouse telencephalon: role of the homeobox gene Gsx2. Neural Dev. 2017;12:13–26. doi: 10.1186/s13064-017-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman R.A., Mobley W.C. Implications for treatment: GABAA receptors in aging, Down syndrome and Alzheimer's disease. J. Neurochem. 2011;117:613–622. doi: 10.1111/j.1471-4159.2011.07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R.J., VanHorn J.F., Cain C.C., Reeves R.H. A neural crest deficit in Down syndrome mice is associated with deficient mitotic response to Sonic hedgehog. Mech. Dev. 2009;126:212–219. doi: 10.1016/j.mod.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross M.H., Galaburda A.M., Kemper T.L. Down's syndrome: is there a decreased population of neurons? Neurology. 1984;34:909–916. doi: 10.1212/wnl.34.7.909. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Smith J., MacLean G., Orkin S.H., Livesey F.J. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci. Transl. Med. 2012;4:124ra129. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki D.J., Trivedi N., Govek E.E., Kerekes R.A., Gleason S.S., Hatten M.E. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester P.E. The hippocampus in Down's syndrome. J. Ment. Deficiency Res. 1983;27(Pt 3):227–236. doi: 10.1111/j.1365-2788.1983.tb00294.x. [DOI] [PubMed] [Google Scholar]
- Vega-Flores G., Rubio S.E., Jurado-Parras M.T., Gomez-Climent M.A., Hampe C.S., Manto M., Soriano E., Pascual M., Gruart A., Delgado-Garcia J.M. The GABAergic septohippocampal pathway is directly involved in internal processes related to operant reward learning. Cereb. Cortex. 2014;24:2093–2107. doi: 10.1093/cercor/bht060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick J.P., Liu Y., Zhang S.C. Human embryonic stem cell-derived neurons adopt and regulate the activity of an established neural network. Proc. Natl. Acad. Sci. USA. 2011;108:20189–20194. doi: 10.1073/pnas.1108487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick J.P., Held D.L., Bonadurer G.F., 3rd, Doers M.E., Liu Y., Maguire C., Clark A., Knackert J.A., Molinarolo K., Musser M. Deficits in human trisomy 21 iPSCs and neurons. Proc. Natl. Acad. Sci. USA. 2013;110:9962–9967. doi: 10.1073/pnas.1216575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzdoerfer R., Dierssen M., Fountoulakis M., Lubec G. Fetal life in Down syndrome starts with normal neuronal density but impaired dendritic spines and synaptosomal structure. J. Neural Transm. Suppl. 2001:59–70. doi: 10.1007/978-3-7091-6262-0_5. [DOI] [PubMed] [Google Scholar]
- Wen Z., Nguyen H.N., Guo Z., Lalli M.A., Wang X., Su Y., Kim N.S., Yoon K.J., Shin J., Zhang C. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetter S., Murphy C. Individuals with Down's syndrome demonstrate abnormal olfactory event-related potentials. Clin. Neurophysiol. 1999;110:1563–1569. doi: 10.1016/s1388-2457(99)00086-3. [DOI] [PubMed] [Google Scholar]
- Whittle N., Sartori S.B., Dierssen M., Lubec G., Singewald N. Fetal Down syndrome brains exhibit aberrant levels of neurotransmitters critical for normal brain development. Pediatrics. 2007;120:e1465–1471. doi: 10.1542/peds.2006-3448. [DOI] [PubMed] [Google Scholar]
- Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Zecevic N., Hu F., Jakovcevski I. Interneurons in the developing human neocortex. Dev. Neurobiol. 2011;71:18–33. doi: 10.1002/dneu.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.