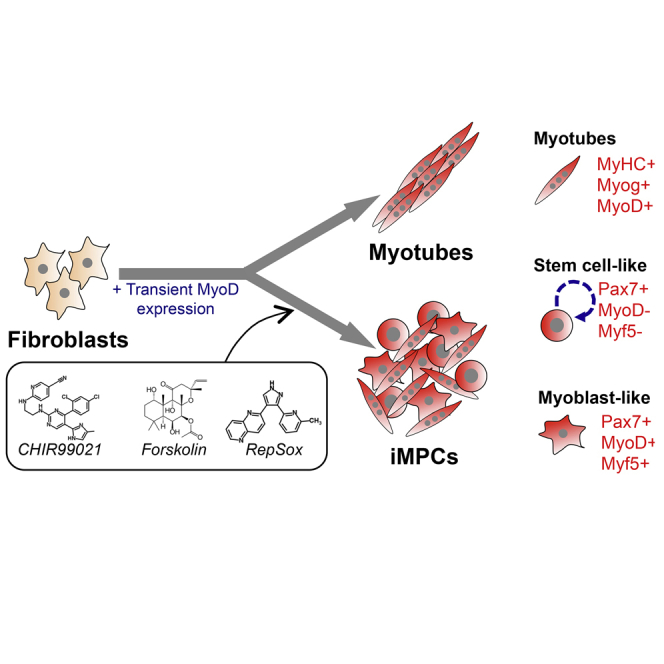
Summary
Skeletal muscle harbors quiescent stem cells termed satellite cells and proliferative progenitors termed myoblasts, which play pivotal roles during muscle regeneration. However, current technology does not allow permanent capture of these cell populations in vitro. Here, we show that ectopic expression of the myogenic transcription factor MyoD, combined with exposure to small molecules, reprograms mouse fibroblasts into expandable induced myogenic progenitor cells (iMPCs). iMPCs express key skeletal muscle stem and progenitor cell markers including Pax7 and Myf5 and give rise to dystrophin-expressing myofibers upon transplantation in vivo. Notably, a subset of transplanted iMPCs maintain Pax7 expression and sustain serial regenerative responses. Similar to satellite cells, iMPCs originate from Pax7+ cells and require Pax7 itself for maintenance. Finally, we show that myogenic progenitor cell lines can be established from muscle tissue following small-molecule exposure alone. This study thus reports on a robust approach to derive expandable myogenic stem/progenitor-like cells from multiple cell types.
Keywords: skeletal muscle, satellite cells, direct lineage reprogramming, induced muscle progenitor cells, MyoD, Pax7, small molecules, transplantation, muscular dystrophy
Graphical Abstract
Highlights
-
•
MyoD and small molecules reprogram fibroblasts to myogenic progenitors termed iMPCs
-
•
iMPCs self-renew and express key satellite cell and myoblast markers
-
•
iMPC growth is driven by Pax7+ cells and requires Pax7 gene function
-
•
Transplanted iMPCs engraft and sustain muscle regeneration in vivo
In this article, Hochedlinger and colleagues reprogrammed mouse fibroblasts into induced myogenic progenitors (iMPCs) by transient expression of MyoD and treatment with small molecules. iMPCs can be extensively propagated in vitro and exhibit skeletal muscle stem/progenitor cell characteristics, including the requirement for Pax7 function as well as the ability to sustain muscle regeneration upon repeated injury.
Introduction
Skeletal muscle is largely composed of differentiated, multinucleated myofibers responsible for contraction and, thus, movement (Almada and Wagers, 2016, Comai and Tajbakhsh, 2014, Yin et al., 2013). In addition, muscle tissue contains a quiescent population of mononucleated stem cells termed satellite cells, which are located between the myofibers' basal lamina and sarcolemma. Satellite cells are maintained in a quiescent state under homeostatic conditions but undergo activation following tissue injury. Once activated, satellite cells generate transit-amplifying progenitors termed uncommitted myoblasts, which then differentiate and fuse with one another or with residual myofibers to regenerate damaged tissue. Remarkably, individual satellite cells have the potential to produce myofibers and replenish the satellite cell niche when transplanted into damaged muscle, documenting their self-renewal and differentiation potential (Sacco et al., 2008). While satellite cells and myoblasts can be cultured and modestly expanded using growth factors or small molecules (Charville et al., 2015, Montarras et al., 2005, Parker et al., 2012), current protocols do not allow for long-term maintenance of primary stem/progenitor cells that retain myogenic potential in vitro and in vivo.
The different stages of adult myogenesis are distinguished by the expression of distinct transcription factors and surface markers (Almada and Wagers, 2016, Comai and Tajbakhsh, 2014, Yin et al., 2013). For example, quiescent satellite cells express the transcription factor Pax7 and the surface marker vascular cell adhesion molecule 1 (VCAM-1) (Fukada et al., 2007) but lack expression of the gene myogenic differentiation 1 (abbreviated hereafter as MyoD). By contrast, activated satellite cells co-express Pax7 and MyoD, whereas differentiating myoblasts and myotubes upregulate other myogenic factors such as myogenic regulatory factor 4 (MRF4 or Myf6) and myogenin (Myog) in addition to MyoD. Pax7 expression serves as a useful marker for quiescent and activated satellite cells and is often used to genetically mark or purify these immature cell populations using lineage-tracing alleles or fluorescent reporters (Bosnakovski et al., 2008, Murphy et al., 2011, Sambasivan et al., 2009, Rocheteau et al., 2012). Moreover, Pax7 expression is functionally required for the specification and maintenance of the adult satellite cell pool as well as for muscle repair (Gunther et al., 2013, Seale et al., 2000, von Maltzahn et al., 2013).
Given previous studies on transcriptional regulators important for the different stages of myogenesis, we reasoned that it should be feasible to induce muscle stem-like cells from heterologous somatic cell types using cellular reprogramming. Indeed, the generation of myotubes from fibroblasts upon ectopic expression of the transcription factor MyoD represents the first example of direct lineage conversion in a mammalian system (Davis et al., 1987). These seminal studies provided the intellectual framework for subsequent attempts to convert one mature cell type into another (e.g., murine embryonic fibroblasts [MEFs] to neurons, MEFs to cardiomyocytes, B cells to macrophages [Graf, 2011]). While these approaches have been important to dissect the mechanisms by which transcription factors control cell fate, they typically generate post-mitotic, non-expandable cells. This limitation is particularly problematic for potential clinical settings where millions to billions of mature cells may be required to achieve therapeutic benefit in patients. Although the transplantation of fibroblasts carrying a MyoD-inducible transgene has been proposed as a source of replacement muscle cells in vivo, this approach generates post-mitotic cells that cannot repopulate the satellite cell niche (Kimura et al., 2008). Induced pluripotent stem cells (iPSCs) may provide an alternative solution, as they can be expanded indefinitely and differentiated repeatedly into myogenic cells using recently developed protocols (Chal et al., 2015, Darabi et al., 2012, Tedesco et al., 2012). However, residual pluripotent cells sometimes form teratomas upon transplantation, complicating their therapeutic utility (Darabi et al., 2008).
In an attempt to overcome the limitations of previous approaches, we developed a system to generate expandable skeletal muscle stem/progenitor cells directly from fibroblasts by combining transient transcription factor expression and small-molecule treatment.
Results
MyoD and Small Molecules Induce Progenitor-like Program in Fibroblasts
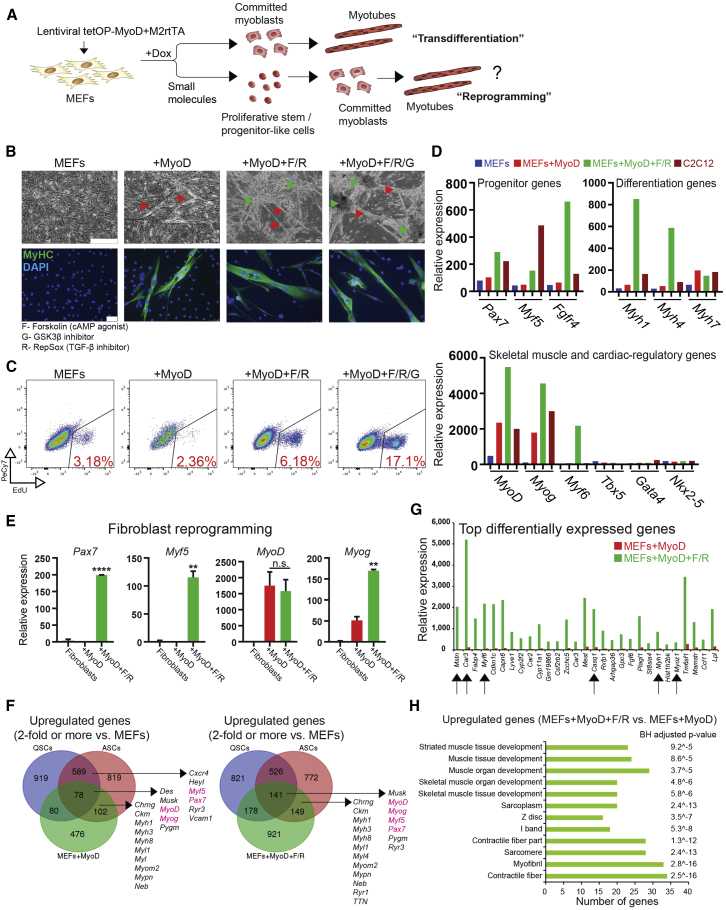
To induce conversion of fibroblasts into cells of the skeletal muscle lineage, we chose to overexpress MyoD, which has previously been shown to trigger transdifferentiation of different somatic cell types into post-mitotic myotubes (Davis et al., 1987) (Figure 1A, top row). To this end, we engineered a doxycycline (dox)-inducible lentiviral system (tetOP-MyoD), allowing for inducible and reversible activation of MyoD in target cells. MEFs transduced with lentiviral vectors co-expressing tetOP-MyoD and M2rtTA showed extensive nuclear staining for the MyoD protein after 24 hr of dox administration (Figure S1A) and gave rise to elongated myotubes after 24–96 hr of dox exposure, consistent with previous findings (Bergstrom et al., 2002) (Figure 1B, red arrowheads). We confirmed the myogenic identity of converted cells by immunofluorescence staining for the differentiation marker myosin heavy chain (MyHC), which was absent in uninfected MEFs (Figure 1B).
Figure 1.
MyoD and Small Molecules Induce Skeletal Muscle Progenitor-like Program in Fibroblasts
(A) Experimental outline. MEFs, murine embryonic fibroblasts.
(B) Representative bright-field images (scale bar, 500 μm) and immunofluorescence images for MyHC (green; scale bar, 50 μm) of MEFs induced with MyoD alone or MyoD in the presence of the indicated small molecules and medium containing fetal bovine serum (FBS), Serum Replacement (SR), and basic FGF (bFGF). Three-dimensional, proliferative, and contractile colonies were obtained only in the presence of the cyclic AMP agonist forskolin (F) and the TGF-β inhibitor RepSox (R), with or without GSK3β inhibitor (G). The experiment was validated using at least three different MEF lines. Green arrowheads indicate three-dimensional colonies; red arrowheads indicate multinucleated myofibers.
(C) Flow-cytometric analysis for EdU to measure proliferation in MEFs subjected to the indicated treatments.
(D) Expression of skeletal muscle- and cardiac-associated markers by microarray analysis in control MEFs, C2C12 myoblasts, and MEFs undergoing conventional transdifferentiation (MEF + MyoD) or reprogramming (MEFs + MyoD + F/R) for 14 days.
(E) qRT-PCR analysis for the indicated skeletal muscle genes during tail-tip fibroblast reprogramming. ∗∗p < 0.005, ∗∗∗∗p < 0.00005, n.s., not significant.
(F) Venn diagram showing the overlap of genes (>2-fold relative to MEFs) between quiescent satellite cells (QSCs), activated satellite cells (ASCs), and either MEFs expressing MyoD for 14 days or MEFs expressing MyoD and exposed to F/R for 14 days. Expression data for QSCs and ASCs were obtained from a previous publication (Liu et al., 2013).
(G) Graph showing the top differentially expressed genes by expression microarray analysis in MEFs expressing MyoD and exposed to F/R in comparison with MEFs expressing MyoD alone. Arrows highlight examples of mature muscle markers detected exclusively under reprogramming conditions (MyoD + F/R).
(H) Functional annotation analysis for upregulated genes (>2-fold) in MEFs + MyoD + F/R relative to MEFs + MyoD alone. Benjamini-Hochberg (BH) adjusted p values are presented. Top categories are shown together with the number of genes.
See also Figure S1.
We next exposed MEFs undergoing MyoD-induced transdifferentiation to various small molecules and cytokines in an attempt to induce reprogramming into a proliferative, myogenic progenitor-like cell state in addition to generating mature myotubes (Figure 1A, bottom row). Specifically, we used compounds that were previously shown to enhance reprogramming into iPSCs, including the GSK3β inhibitor CHIR99021 (Bar-Nur et al., 2014, Vidal et al., 2014) (abbreviated as “G”) and the transforming growth factor β1 (TGF-β1) receptor (ALK5) inhibitor RepSox (Ichida et al., 2009, Maherali and Hochedlinger, 2009) (abbreviated as “R”). In addition, we tested the effect of the cyclic AMP agonist forskolin (abbreviated as “F”), which reportedly facilitates the transient expansion of satellite cells in vitro (Xu et al., 2013). These compounds were added individually or combinatorially to MEFs expressing MyoD and cultured in KnockOut DMEM media containing 10% KnockOut Serum Replacement (KOSR) and 10% fetal bovine serum (FBS), and supplemented with 10 ng/mL basic fibroblast growth factor (bFGF), which promotes satellite cell and myoblast growth (Xu et al., 2013). We found that individual treatment of MyoD-expressing MEFs with F, G, or R as well as combined treatment with R/G or F/G did not noticeably enhance the formation of MyHC+, multinucleated myotubes (Figure S1B). By contrast, F/R treatment led to a marked increase in the number of proliferative cells, as confirmed by increased 5-ethynyl-2′-deoxyuridine (EdU) incorporation, and this effect was further enhanced by addition of G (i.e., F/R/G condition) (Figure 1C). Cultures treated with F/R or F/R/G typically assembled into three-dimensional colonies (Figure 1B, green arrowheads) with emanating myotubes and interspersed small, proliferative cells that we considered might be muscle progenitors. Strikingly, we also detected spontaneous and robust contractions within cultures treated with these small molecules, which we rarely detected in MEFs expressing MyoD alone (Video S1. MEF Control, Related to Figure 1, Video S2. MEFs Expressing MyoD, Related to Figure 1, Video S3. MEFs Expressing MyoD and Exposed to F/R, Related to Figure 1).
To assess whether concomitant expression of MyoD and exposure to our small molecules could activate a skeletal muscle-specific stem or progenitor cell program in fibroblasts, we performed microarray analysis of (1) untreated MEFs, or MEFs expressing (2) MyoD alone (MyoD condition) or (3) MyoD in combination with F/R (MyoD + F/R condition) for 14 days, or (4) the immortalized myoblast cell line C2C12. We found that the expression of genes associated with satellite cells and uncommitted myoblasts (e.g., Pax7, Myf5, Fgfr4) were significantly increased in the MyoD + F/R but not in the MyoD condition (Figure 1D, top left), as compared with MEFs alone. We further observed exclusive induction of genes associated with mature muscle tissue including Myh1, Myh4, and Myf6 in the MyoD + F/R condition, whereas markers associated with alternative lineages such as cardiomyocytes (e.g., Tbx5, Gata4, Nkx2-5) were not expressed at appreciable levels (Figure 1D, top right and bottom). We confirmed induction of Pax7 by qPCR and immunofluorescence in MEFs and tail-tip fibroblasts (TTFs) expressing MyoD and exposed to F/R or F/R/G (Figures 1E and S1B). In agreement with these results, we found that MEFs exposed to the MyoD + F/R condition but not to the MyoD condition share expression of other stem and progenitor cell-associated genes when compared with a previously published dataset of quiescent satellite cells (QSCs) and activated satellite cells (ASCs) (Figures 1F and S1C). A comparison of the most highly differentially expressed genes and associated gene ontology categories between MEFs expressing MyoD and those expressing MyoD and exposed to F/R corroborated the conclusion that the induced cell populations represent skeletal muscle and not cardiac muscle (Figures 1G and 1H). Together, these results demonstrate that the combined expression of exogenous MyoD and treatment with small molecules induces several characteristics of myogenic progenitors in MEFs including proliferation and transcriptional activation of key satellite cell and myoblast markers, as well as differentiation and contraction.
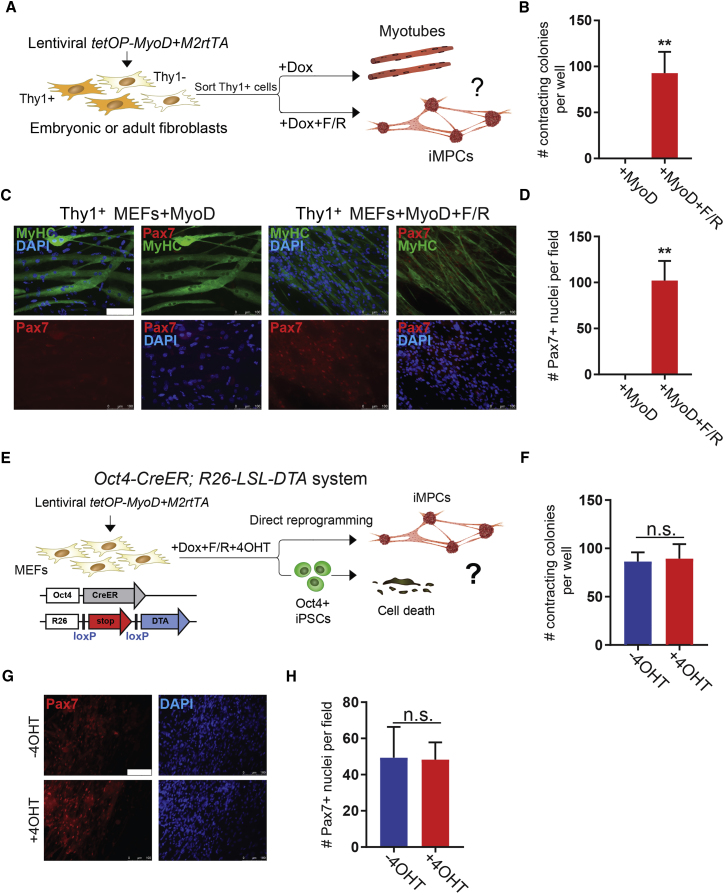
Transgene-Independent Progenitors Clonally Self-Renew and Express Key Satellite Cell and Myoblast Markers
We determined the temporal requirement for exogenous MyoD expression and small molecules during the establishment and maintenance of our progenitor-like cell population. To this end, we treated MyoD-transduced MEFs with dox for 0, 2, 4, 6, 8, or 12 days in the presence of F/R, followed by 7 days of dox withdrawal in the continuous presence of F/R, before scoring for contractile colonies containing Pax7+ cells (Figure S1D). We found that as little as 2 days of dox induction (i.e., exogenous MyoD expression) followed by 17 days of dox-independent growth was sufficient to generate contractile colonies with associated Pax7+ cells (Figures S1D–S1F). Individually picked colonies gave rise to stable cultures that could be propagated for at least 6 months or 20–24 passages in the absence of dox and containing both mononucleated cells and contracting myotubes (Figures S1D–S1F). However, continuous exposure of dox-withdrawn cultures to F/R was required for their maintenance as small-molecule removal led to their collapse (data not shown). We further derived transgene-independent myogenic progenitor cultures from subsets of MEFs or TTFs that were pre-sorted based on expression of the fibroblast-associated surface marker Thy1, confirming their origin from fibroblasts rather than a contaminating myogenic cell type (Figures 2A–2D). We also used an Oct4 lineage ablation system (Oct4-CreER;R26-LSL-DTA) (Bar-Nur et al., 2015) to rule out the possibility that our myogenic cells are generated indirectly via pluripotent intermediate cells, which subsequently differentiated (Bar-Nur et al., 2015, Maza et al., 2015) (Figures 2E–2H). Thus, we conclude that small molecules and growth factors collaborate with transient MyoD expression to endow MEFs and TTFs with a stable myogenic progenitor-like cell state. Once established, these cells no longer require exogenous MyoD expression and can be propagated long-term in the continuous presence of small molecules, demonstrating that they have acquired a transgene-independent self-renewing state. Based on these characterizations, we provisionally termed these cells “induced myogenic progenitor cells” (iMPCs).
Figure 2.
iMPCs Originate from Fibroblasts and Do Not Pass through a Pluripotent State
(A) Experimental design to confirm origin of iMPCs from Thy1+ fibroblasts.
(B) Quantification of contracting colonies obtained from sorted Thy1+ fibroblasts exposed to MyoD or MyoD + F/R (n = 3 biological replicates, 2 MEF lines and 1 tail-tip fibroblast [TTF] line were used; error bars denote SD; ∗∗p < 0.005).
(C) Representative immunofluorescence images for MyHC (green) and Pax7 (red) expression in sorted Thy1+ fibroblasts exposed to either MyoD or MyoD + F/R for 14 days. Note lack of Pax7 positivity in cells expressing MyoD alone. Scale bar, 100 μm.
(D) Quantification of Pax7+ nuclei in three random fields taken from sorted Thy1+ cells exposed to MyoD or MyoD + F/R for 14 days (n = 3 independent replicates; error bars denote SD; ∗∗p < 0.005).
(E) Experimental design to assess if iMPC formation requires passage through an Oct4+ pluripotent intermediate state using a DTA (diphtheria toxin A) lineage ablation system.
(F) Quantification of contracting colonies generated with and without 4OHT administration from Oct4-CreER;Rosa26-LSL-DTA MEFs and exposed to MyoD + F/R (n = 3 independent replicates; error bars denote SD).
(G) Representative immunofluorescence images show staining for Pax7 in Oct4-CreER;Rosa26-LSL-DTA MEFs exposed to MyoD + F/R with and without 4OHT. Scale bar, 100 μm.
(H) Quantification of Pax7+ nuclei in three random fields taken from Oct4-CreER;Rosa26-LSL-DTA MEFs exposed to MyoD + F/R with and without 4OHT (n = 3 independent replicates; error bars denote SD).
See also Figure S1.
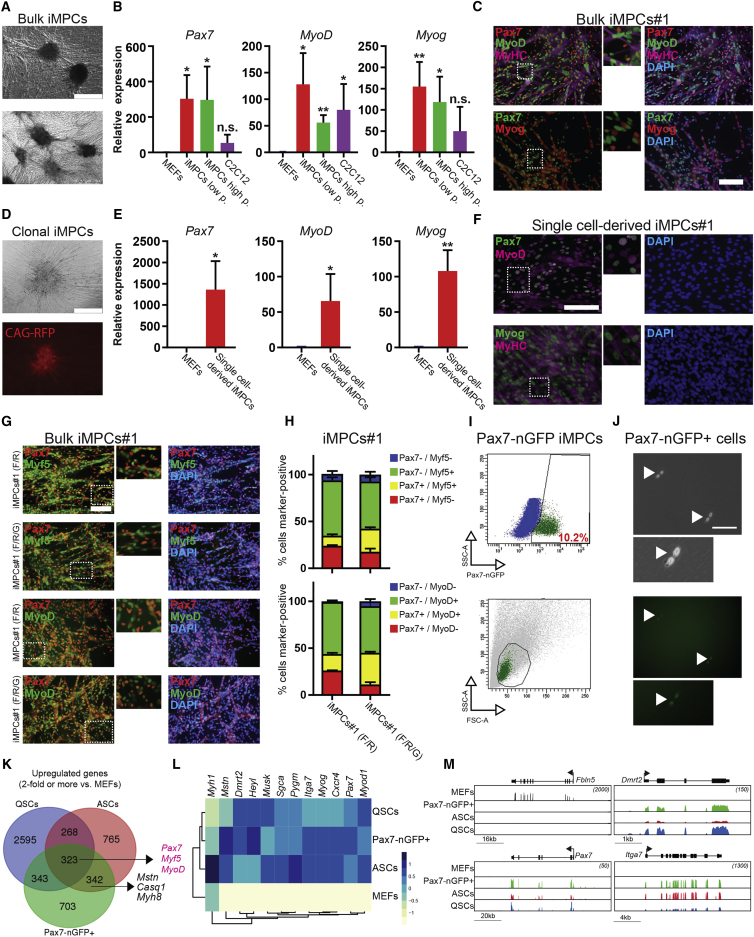
We next used qRT-PCR, microarray, and immunofluorescence analyses to assess whether transgene-independent iMPC cultures express markers associated with different stages of myogenesis (see Figure S2A for a scheme). Indeed, we found that bulk iMPC cultures at both low (∼P5) and high (∼P15) passage (Figures 3A–3C and S2B–S2D; Video S4) as well as clonal iMPC lines that were derived from singly plated mononucleated cells (Figures 3D–3F and S3A; Video S5) expressed the satellite cell marker Pax7 and the differentiation markers MyHC and Myog. Importantly, both bulk and clonal iMPC cultures activated the endogenous MyoD locus, confirming our earlier observation that dox exposure is only required for the initiation and not maintenance of iMPCs (Figures 3B, 3C, 3E, 3F, and S3A). We noted that Pax7+ cells lacked MyHC expression, supporting the notion that these cells resemble undifferentiated myogenic stem or progenitor cells (Figure 3C). Quantification of cells expressing these marker combinations within bulk iMPC cultures further revealed the co-existence of iMPC subsets that expressed Pax7 and Myf5 or MyoD as well as subsets that expressed Pax7 but neither Myf5 nor MyoD (Figures 3G, 3H, S3B, and S3C). This observation suggests that the Pax7+ population contains more primitive cells resembling QSC-like cells as well as more committed progeny resembling ASCs. Notably, clonal iMPC cultures also contained Pax7+/MyoD+ double-positive cells but lacked Pax7+/MyoD− cells, suggesting that the subcloning procedure may have selected for the outgrowth of more committed iMPC subsets (Figure S3D). This notion is consistent with the recent observation that differentiated myofibers and niche signals are critical for the prolonged maintenance of primitive subsets of Pax7+ satellite cells ex vivo (Quarta et al., 2016).
Figure 3.
iMPCs Self-Renew and Express Markers of Muscle Stem, Progenitor, and Mature Cells
(A) Representative images of dox-independent iMPC cultures containing spheroid structures, mononucleated cells, and multinucleated myotubes. Scale bar, 500 μm.
(B) qRT-PCR analysis for skeletal muscle-specific genes in low-passage (P < 5) and high-passage (P > 15) iMPC lines expanded from individual colonies. MEFs were used as negative control and C2C12 myoblasts as positive control (n = 3–4 biological replicates; error bars denote SD; ∗p < 0.05, ∗∗p < 0.005).
(C) Representative immunofluorescence images for the indicated muscle-specific proteins in a representative iMPC line. Scale bar, 100 μm.
(D) Representative images of single cell-derived iMPCs subcloned from CAG-RFP+ iMPCs. Scale bar, 250 μm.
(E) qRT-PCR analysis for skeletal muscle-specific genes in single cell-derived iMPCs. MEFs were used as a negative control (n = 3 biological replicates; error bars denote SD; ∗p < 0.05, ∗∗p < 0.005).
(F) Representative immunofluorescence images for the indicated muscle-specific proteins in a single cell-derived iMPC clone. Scale bar, 100 μm.
(G) Representative immunofluorescence images for Pax7 (red) and Myf5 (green) or Pax7 (red) and MyoD (green) expression in iMPCs#1 cultured in F/R or F/R/G conditions for at least five passages before analysis. Scale bar, 100 μm.
(H) Quantification of (G).
(I) Top: flow-cytometric analysis of GFP expression using low-passage iMPCs derived from Pax7-nGFP MEFs subjected to MyoD + F/R/G condition. Bottom: forward/side scatter flow-cytometric plot using Pax7-nGFP+ cells (green) compared with all mononucleated cells (gray).
(J) Representative images of sorted Pax7-nGFP+ cells using bright-field and GFP channels. Zoomed images show sorted Pax7-nGFP+ doublets. Arrowheads indicate Pax7-nGFP+ cell doublets. Scale bar, 100 μm.
(K) Venn diagram based on RNA sequencing data showing the overlap of upregulated genes (>2-fold, FDR < 0.05 relative to MEFs) between quiescent satellite cells (QSCs), activated satellite cells (ASCs), and sorted Pax7-nGFP+ cells purified from iMPCs. See Supplemental Experimental Procedures and main text for definition of QSCs and ASCs.
(L) Heatmap depicting expression of skeletal muscle stem and progenitor associated genes based on RNA sequencing data obtained from Pax-nGFP+ iMPCs, QSCs, ASCs, and MEFs.
(M) Integrative genomic viewer tracks for the indicated genes based on RNA sequencing data.
See also Figure S3.
To further explore the similarity of transgene-independent Pax7+ iMPC subsets to QSCs or ASCs, we derived iMPCs from MEFs carrying a satellite cell-specific Pax7-nGFP transgene (Sambasivan et al., 2009). We detected up to 10% Pax7-nGFP+ cells in dox-independent iMPCs, confirming robust activation of the endogenous Pax7 locus (Figure 3I). Differences in the frequency of recorded Pax7+ iMPCs using immunofluorescence analysis (Figure 3H) and the reporter system are likely due to method-specific detection limits. Like satellite cells, sorted Pax7-nGFP+ iMPCs were small by forward/side scatter and occasionally gave rise to adjoining cells in culture that exhibited equivalent GFP expression, suggesting active cell division (Figures 3I and 3J). By contrast, sorted Pax7-nGFP− cells were larger and morphologically resembled committed myoblasts (Figure S3E). We next extracted RNA from fluorescence-activated cell sorting-purified Pax7-nGFP+ iMPC cultures (referred to as “Pax7-nGFP+”) as well as from satellite cells that were harvested directly from mice (referred to as “QSCs”) or explanted on laminin-coated plates for 7 days in myoblast medium containing horse serum and FGF (Bosnakovski et al., 2008) to stimulate their proliferation (referred to as “ASCs”). We detected a similar number of genes that were upregulated in Pax7-nGFP+ iMPCs and either ASCs (342 genes) or QSCs (343 genes) in comparison with MEFs (Figure 3K). Representative examples of transcripts shared between Pax7-nGFP+ iMPCs, and ASCs include Itga7, Myog, and Mstn, and those shared between Pax7-nGFP+ iMPCs and QSCs include Dmrt2 and Heyl (Figures 3L, 3M, and S3F–S3H). Additionally, we noticed that Pax7-nGFP+ cells have silenced MEF-associated genes (e.g., Fbln5, Thy1), indicating effective extinction of the fibroblast program in iMPCs (Figures 3M and S3F–S3H). Together, these results show that purified Pax7-nGFP+ cells exhibit a gene expression profile that diverges from the parental MEFs and shares key molecular characteristics with skeletal muscle stem and progenitor cell populations.
iMPCs Differentiate into Dystrophin+ Myofibers in Dystrophic Recipient Mice
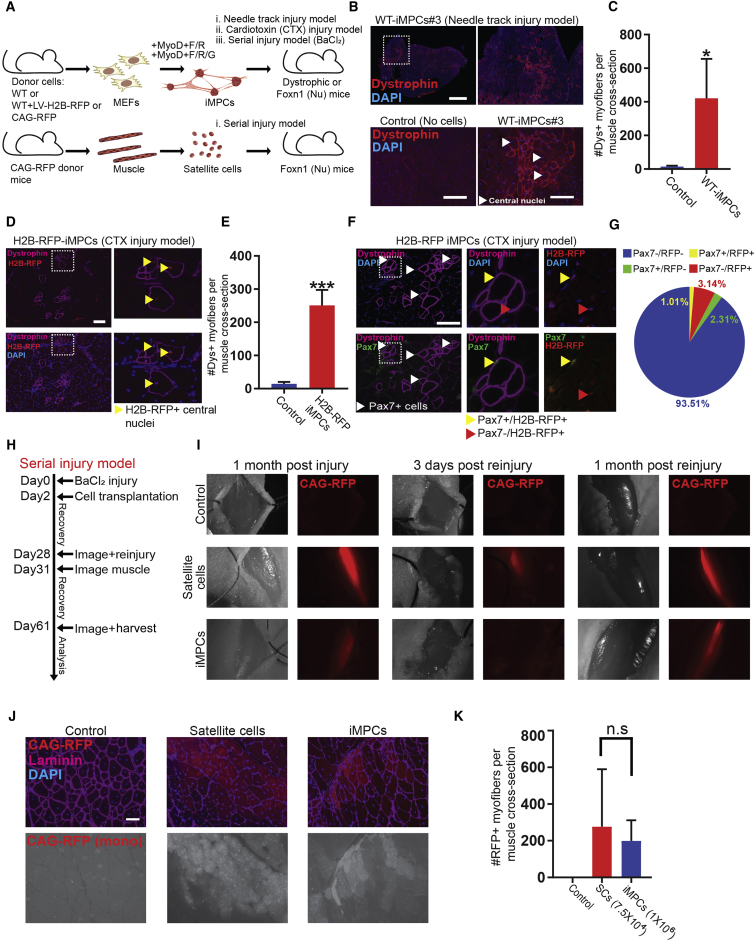
To assess whether iMPCs have the potential to contribute to muscle regeneration in vivo, we derived iMPC lines from MEFs using the aforementioned conditions and transplanted 1 × 106 cells into the tibialis anterior (TA) or gastrocnemius muscle of 12-week-old homozygous mdx dystrophic mice (Figure 4A). One month after cell injection, we consistently observed contiguous areas of dystrophin+ myofibers in mdx muscles transplanted with iMPCs, whereas we only detected rare dystrophin+ revertant myofibers in PBS-injected mdx control muscles (Figures 4B and S4A). iMPC-derived dystrophin+ myofibers had centrally located nuclei and varied in size, indicative of an active regenerative process involving both new fiber formation and repair of the damaged endogenous myofibers (Figures 4B, 4C, and S4A). We next transplanted 1 × 106 single cell-derived iMPCs into mdx recipients to test their in vivo regeneration potential. While these subclones also had the capacity to restore dystrophin expression, the dystrophin+ myofibers were reduced in size and smaller in number compared with bulk iMPC-derived grafts, which is consistent with our previous observation that these subclones exhibit a more differentiated phenotype (Figure S4B). Critically, we never observed tumor formation in muscles transplanted with either bulk or clonal iMPCs (data not shown). Altogether, these results demonstrate that iMPCs exhibit not only molecular but also key functional attributes of skeletal muscle progenitors.
Figure 4.
iMPCs Contribute to Skeletal Muscle Regeneration and the Satellite Cell Niche In Vivo
(A) Experimental design to assess the engraftment and differentiation potential of iMPCs in comparison with satellite cells.
(B) Immunofluorescence images for dystrophin (red) and DAPI (blue) for the indicated samples. One million cells were transplanted into the tibialis anterior or gastrocnemius of 12-week-old homozygous mdx dystrophic mice, and the muscles were isolated 1 month after transplantation for analysis. Non-transplanted or PBS-injected mdx muscle sections were used as negative controls. Rare dystrophin-positive myofibers, present in non-transplanted or PBS-injected control sections, are due to spontaneous reversion of the mdx mutation. Scale bars, 1,000 μm (top) and 100 μm (bottom).
(C) Quantification of (B) (n = 3 biological replicates; error bars denote SD; ∗p < 0.05).
(D) Immunofluorescence images of cardiotoxin (CTX)-injured SCID-mdx muscles transplanted with 1 × 106 iMPCs carrying a nuclear H2B-RFP reporter and analyzed 1 month after injury/transplantation. CTX injury was carried out 24 hr prior to transplantation. Successful engraftment was assessed by measuring dystrophin expression (purple) and H2B-RFP expression (red). Scale bar, 100 μm.
(E) Quantification of (D) (n = 3 biological replicates; error bars denote SD; ∗∗∗p < 0.0005).
(F) Immunofluorescence images for dystrophin (purple) and Pax7 (green) expression within an engrafted area. Insets indicate Pax7 nuclear staining co-localizing with the H2B-RFP reporter. Scale bar, 100 μm.
(G) Quantification of (F) (n = 3 biological replicates; error bars denote SD).
(H) Experimental outline for serial injury experiment.
(I) Representative images of tibialis anterior muscles transplanted with 1 × 106 iMPCs or 7.5 × 104 purified satellite cells carrying a CAG-RFP fluorescent reporter following BaCl2-induced muscle injury (48 hr prior to transplantation). Muscles transplanted with satellite cells or iMPCs were subjected to the same procedures as illustrated in (H). Note reduction of CAG-RFP signal 3 days after reinjury and restoration of CAG-RFP signal 1 month after reinjury, indicating successful regeneration by donor cells.
(J) Immunofluorescence images of a tibialis anterior muscle section transplanted with CAG-RFP iMPCs and analyzed 1 month after reinjury compared with a non-transplanted control. Scale bar, 100 μm.
(K) Quantification of serial injury procedure (n = 4 independent transplantation experiments per cell type). n.s., not significant.
See also Figure S4.
Transplanted iMPCs Populate the Satellite Cell Niche and Initiate Regeneration upon Serial, Acute Injury
We next determined whether transplanted iMPCs have the capacity to populate the satellite cell niche, which is a defining characteristic of bona fide skeletal muscle stem cells. Examination of the injection site of mice transplanted with bulk or clonal cell-derived iMPCs revealed numerous mononucleated Pax7+ cells within regenerating dystrophin+ myofibers (Figures S4C and S4D). However, our experimental setup did not allow us to exclude that these were host-derived satellite cells. To determine whether iMPCs can give rise to mononucleated Pax7+ cells in vivo, we infected iMPCs with a lentiviral vector driving constitutive expression of the nuclear H2B-RFP fusion gene, and transplanted 1 × 106 cells into mdx mice that were pre-injured with cardiotoxin (CTX) 24 hr prior to transplantation to enhance cell engraftment (Figure S4E). Consistent with our previous transplantation results, we observed robust contribution of iMPCs to mdx myofibers, assessed by restored dystrophin expression and centrally located nuclear H2B-RFP signal in a subset of these regenerating myofibers (Figures 4D and 4E). Importantly, we consistently found peripherally localized Pax7+ cells that were also H2B-RFP+ and DAPI+ within regions of engrafted, dystrophin+ myofibers (Figures 4F and 4G). This result suggests that at least a subset of transplanted iMPCs harbor the potential to populate the satellite cell niche in vivo.
To corroborate the conclusion that some of the transplanted iMPCs have acquired properties of muscle stem cells, we assessed their ability to initiate muscle regeneration using a serial, acute injury model (Hardy et al., 2016) (Figure 4H). In brief, we treated immunodeficient Foxn1nu (referred to as nude mice) with 50 μM (1.2%) barium chloride (BaCl2) to trigger muscle damage and enable engraftment of transplanted cells in non-dystrophic hosts (Figure S4E). Two days later, we transplanted either 1 × 106 bulk iMPCs or 7.5 × 104 satellite cells into the TA of recipient animals. Satellite cells were isolated using a previously reported protocol based on surface markers (Cerletti et al., 2008) (Figure S4F). We transplanted an excess of bulk iMPCs relative to purified satellite cells because of our prior observation that only 5%–10% of mononucleated iMPCs are Pax7+/MyoD− (Figure 3H) or Pax7-nGFP+ (Figure 3I). After 26 days, we imaged transplants to confirm successful engraftment before subjecting muscles to another round of BaCl2-induced injury (Figures 4I and S4G). Whereas analysis of individual transplants showed a noticeable loss or reduction of RFP signal 3 days after reinjury, we detected a robust RFP signal 31 days later, indicating successful regeneration by both iMPCs and satellite cells (n = 4 animals per cell type) (Figures 4I–4K, S4G, and S4H). Collectively, these results show that iMPCs have the potential to (1) engraft in dystrophic and non-dystrophic, injured muscles, (2) give rise to Pax7+ cells within transplants, and (3) contribute to muscle regeneration in a serial injury model.
iMPC Growth Is Driven by Pax7+ Muscle Stem-like Cells that Recapitulate Myogenesis In Vitro
To determine a possible lineage hierarchy within iMPC subsets in vitro, we first examined the expression of the surface marker VCAM-1, which has been associated with both QSCs and ASCs (Fukada et al., 2007). We noticed that the majority of mononucleated iMPCs were positive for VCAM-1 expression upon flow-cytometric analysis, which is consistent with a satellite cell or myoblast identity (Figure S5A). Consistently, immunofluorescence analysis showed that myofibers were negative while mononucleated cells were positive for VCAM-1 expression within heterogeneous iMPC cultures (Figure S5B). Critically, purified VCAM1+ iMPCs initially lacked MyHC expression by qRT-PCR, yet upregulated MyHC expression and gave rise to proliferative and contractile colonies upon culturing for 9 days (Figures S5C and S5D). However, VCAM-1− iMPCs did not give rise to proliferating cells or contractile myotubes, which indicates a differentiated phenotype (Figures S5C and S5D). These results suggest a differentiation hierarchy between undifferentiated VCAM-1+ stem/progenitor-like cells and VCAM-1− differentiated progeny akin to myogenic cells in vivo.
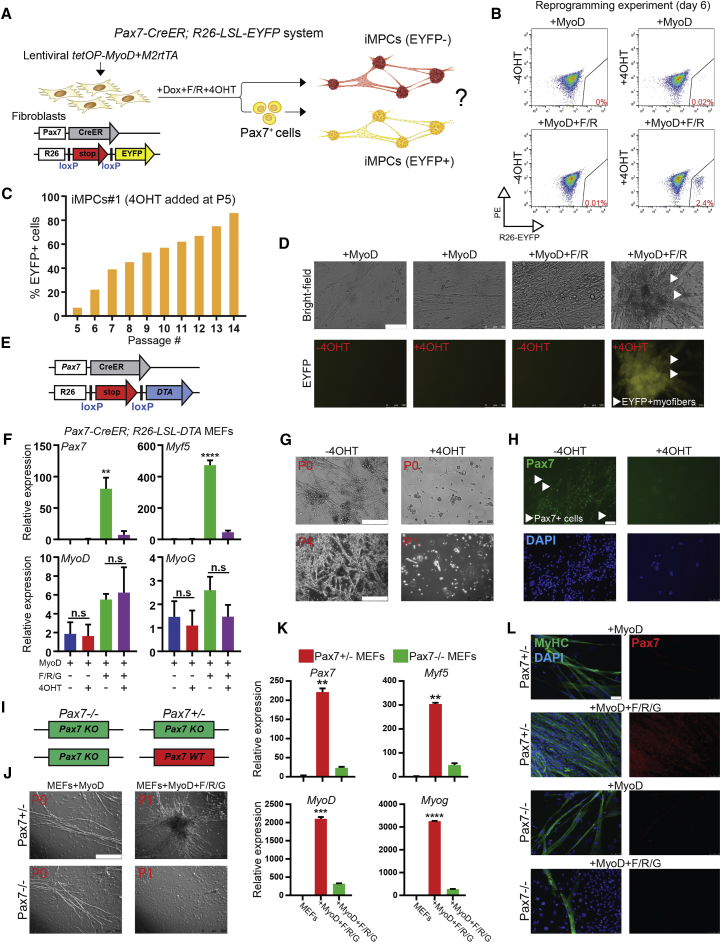
To further explore the hierarchical relationship among iMPC subsets and their possible resemblance to satellite cells, we generated iMPCs from MEFs carrying a satellite cell-specific Pax7-CreER allele (Murphy et al., 2011) as well as an ROSA26-LSL-EYFP reporter (Figure 5A). We failed to detect EYFP+ cells in MEFs exposed to 4OHT, ruling out the presence of contaminating Pax7+ myogenic or neural cells (Figure S5E). Moreover, expression of MyoD alone or in combination with single molecules was insufficient to activate the reporter in the presence of 4OHT in these MEFs, confirming our earlier observations (Figure S5F). By contrast, induction of MyoD in bulk MEFs, Thy1+ MEFs, or TTFs in the presence of bFGF and F/R or F/R/G activated the EYFP reporter in 3%–5% of cells after 6–10 days of MyoD induction and exposure to 4OHT (Figures 5B and S5G–S5J). Treatment of established, dox-independent bulk or clonal iMPC cultures with 4OHT for 2 days gave rise to a similar range of EYFP+ cells (1%–7%) (Figures S6A and S6B) and this fraction progressively increased to over 80% upon continuous passaging in the presence of 4OHT (Figures 5C, S6C, and S6D; Videos S6A and S6B). Importantly, we detected EYFP signal not only in contracting mononucleated cells but also in multinucleated myofibers 1–2 weeks after 4OHT treatment (Figure 5D). We further noticed that the vast majority of iMPCs that were EYFP+ after a 3-day 4OHT pulse were also VCAM-1+ (93%) (Figure S6E), which is consistent with our finding that VCAM-1 positivity corresponds to an immature myogenic state (Figures S5A–S5D). Together, these assays corroborate the conclusion that iMPC cultures contain undifferentiated Pax7+ stem-like cells, which self-renew and differentiate into mature, contracting myotubes over time, thus recapitulating aspects of normal myogenesis ex vivo.
Figure 5.
iMPC Maintenance Requires Pax7+ Cells and Pax7 Gene Function
(A) Schematic of lineage-tracing approach to determine lineage hierarchy among iMPC subsets.
(B) Flow-cytometric analysis of Pax7-CreER;Rosa26-LSL-EYFP MEFs exposed to indicated conditions for 6 days. The PE channel was used to control for autofluorescence.
(C) Quantification of flow-cytometric analysis of Pax7-CreER;Rosa26-LSL-EYFP iMPC line #1 continually treated with 4OHT from passages 5 to 14.
(D) Representative images of EYFP+ myotubes emerging from Pax7-CreER;Rosa26-LSL-EYFP MEFs after exposure to MyoD + F/R in the presence of 4OHT. Scale bar, 100 μm.
(E) Cell ablation system to determine if Pax7+ cells are required for the generation of iMPCs.
(F) qRT-PCR analysis for skeletal muscle-specific genes in Pax7-CreER;Rosa26-LSL-DTA MEFs undergoing reprogramming into iMPCs using the indicated conditions (n = 3 biological replicates; error bars denote SD; ∗∗p < 0.005, ∗∗∗∗p < 0.00005; n.s., not significant).
(G) Representative images of Pax7-CreER;Rosa26-LSL-DTA MEFs undergoing reprogramming into iMPCs using MyoD + F/R/G in the presence or absence of 4OHT. Scale bar, 250 μm.
(H) Representative immunofluorescence images for Pax7 expression (green) for the experiment depicted in (G). Scale bar, 50 μm.
(I) Schematic of Pax7 knockout (KO) alleles used to test whether iMPC generation requires Pax7 gene function.
(J) Bright-field images show myotubes derived from Pax7+/− and Pax7−/− MEFs upon MyoD overexpression (left panels), and an iMPC clone derived from Pax7+/− MEFs (top right) but not from Pax7−/− MEFs (bottom right). Scale bar, 500 μm.
(K) qRT-PCR analysis for indicated samples at day 21 of reprogramming. ∗∗p < 0.005, ∗∗∗p < 0.0005; ∗∗∗∗p < 0.00005.
(L) Representative immunofluorescence images show staining for MyHC (green) and Pax7 expression (red) in Pax7+/− and Pax7−/− MEFs exposed to MyoD or MyoD + F/R/G conditions followed by several passages without exogenous MyoD expression. Scale bar, 50 μm.
See also Figure S5.
To determine whether Pax7-expressing cells are crucial for the establishment of iMPCs, we generated MEFs carrying Pax7-CreER and ROSA26-LSL-DTA alleles and induced MyoD in the presence or absence of F/R/G and 4OHT (Figure 5E). Exposure of these cells to 4OHT is expected to ablate Pax7-expressing cells by inducing diphtheria toxin (DTA)-mediated apoptosis. MEFs expressing exogenous MyoD readily gave rise to myotubes that activated endogenous MyoD and Myog in the presence or absence of 4OHT (Figure 5F). By contrast, 4OHT treatment blunted the generation of Pax7 and Myf5 expressing iMPCs, yet did not affect the formation of MyoD/Myog-expressing myotubes, indicating that Pax7+ cells are indeed essential for the formation of iMPCs (Figures 5F–5H).
iMPC Maintenance Requires the Satellite Cell Master Regulator Pax7
We next tested whether the establishment or maintenance of iMPC cultures is dependent upon Pax7 itself. To this end, we intercrossed Pax7+/− mice (Mansouri et al., 1996) to obtain both Pax7−/− experimental and Pax7+/− control MEFs (Figure 5I). These cells were infected with lentiviral vectors expressing M2rtTA and tetOP-MyoD and subsequently exposed to either transdifferentiation (MyoD) or reprogramming (MyoD + F/R/G) conditions. MyoD expression alone yielded multinucleated myotubes from both Pax7+/− and Pax7−/− MEFs, indicating that Pax7 is dispensable for the direct conversion of fibroblasts to myotubes (Figure 5J, left). Moreover, we detected proliferative and contractile colonies in both Pax7+/− and Pax7−/− cultures upon overexpression of MyoD and treatment with dox and F/R/G, suggesting that Pax7 is also dispensable for the establishment of iMPC-like cells (data not shown). However, we were unable to maintain these Pax7−/− iMPC-like cultures despite the presence of F/R/G treatment (Figure 5J, bottom right). Specifically, Pax7−/− cultures ceased to proliferate and contract over time, leaving behind only post-mitotic myotubes that lacked Pax7 or Myf5 expression (Figures 5J–5L). We conclude that iMPC propagation in vitro relies on the same transcriptional regulator as satellite cell maintenance in vivo, providing further mechanistic evidence that these two cell states are related.
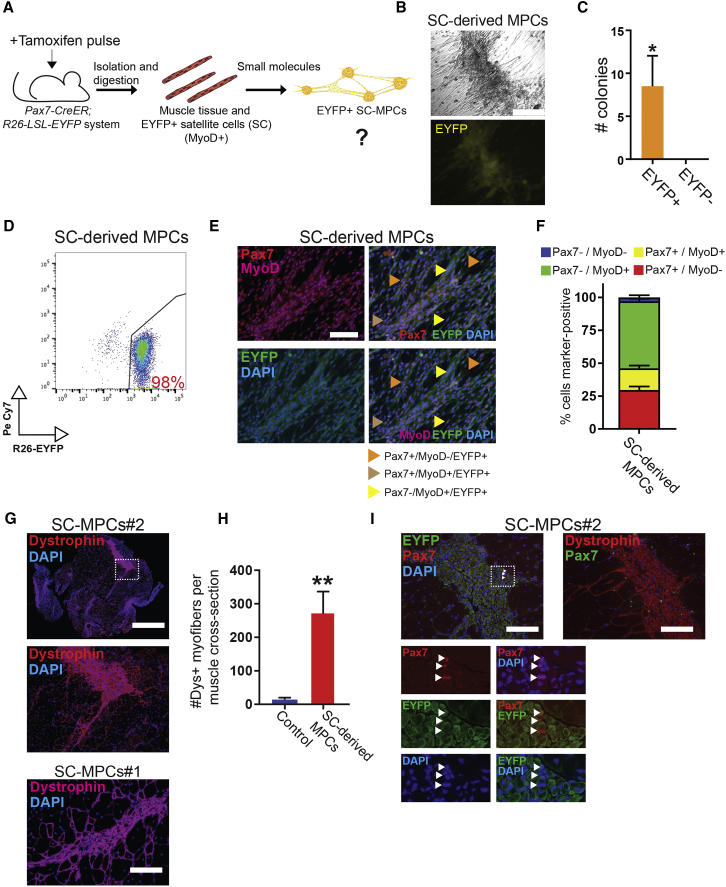
Derivation of MPCs from Muscle Tissue Using Small Molecules Alone
Our finding that concomitant MyoD expression and small-molecule treatment endows fibroblasts with a myogenic progenitor cell (MPC) state raises the possibility that small molecules alone are sufficient to capture an MPC-like state in primary muscle cells, which already express endogenous MyoD. To test this hypothesis, we explanted muscle tissue from Pax7-CreER;ROSA26-LSL-EYFP mice that had been treated with a pulse of tamoxifen to label satellite cells, isolated cells through mechanical and enzymatic digestion, and cultured these in iMPC medium (DMEM, KOSR, FBS, bFGF, F/R or F/R/G) without 4OHT (Figure 6A). We were indeed able to establish MPC-like colonies that could be propagated for several passages and were consistently EYFP+, demonstrating their origin from the satellite cell lineage (Figures 6B–6D; Videos S7A and S7B). Quantification of cells expressing Pax7 and MyoD within these satellite cell lineage-derived MPCs (SC-MPCs) revealed a similar proportion of Pax7+/MyoD+ and Pax7+/MyoD− subsets (Figures 6E and 6F) compared with MEF-derived iMPCs (Figure 3H). Moreover, SC-MPCs engrafted and differentiated into dystrophin+ myofibers with centrally located nuclei and EYFP fluorescence following transplantation into mdx mice, documenting their differentiation and regeneration potential in vivo (Figures 6G–6I). Notably, we detected rare EYFP+/Pax7+ cells within dystrophin+ areas, suggesting that transplanted SC-MPCs may also replenish the endogenous satellite cell pool (Figure 6I). However, we acknowledge that serial injury or serial transplantation experiments would be required to assess whether MPC-derived Pax7+ cells are similar to bona fide satellite cells. These results show that our small-molecule cocktail not only facilitates the reprogramming of MEFs expressing MyoD into iMPCs but also allows capture of MPCs from the satellite cell lineage (i.e., satellite cells or their progeny).
Figure 6.
Derivation of MPCs from Explanted Muscle Using Small-Molecule Treatment Alone
(A) Experimental design to assess whether prolonged exposure to small molecules of Pax7-CreER;Rosa26-LSL-EYFP hindlimb muscle-derived cells (top row) gives rise to EYFP+ myogenic progenitors in the absence of exogenous MyoD expression.
(B) Representative images of EYFP+ MPCs derived from explanted hindlimb muscles of Pax7-CreER;Rosa26-LSL-EYFP mice that were pulsed with tamoxifen prior to explantation. Scale bar, 250 μm.
(C) Quantification of EYFP expression in MPC colonies derived from explanted hindlimb muscles of tamoxifen-pulsed Pax7-CreER;Rosa26-LSL-EYFP mice. ∗p < 0.05.
(D) Flow-cytometric analysis of a satellite cell lineage-derived MPC line (SC-MPCs) carrying the Pax7-CreER;Rosa26-LSL-EYFP lineage label.
(E) Representative immunofluorescence images for indicated muscle-specific proteins in an EYFP+ SC-MPC clone. Scale bar, 100 μm.
(F) Quantification of (E).
(G) Immunofluorescence analysis for dystrophin expression (purple) 1 month after injection of 1 × 106 SC-MPCs derived from Pax7-CreER;Rosa26-LSL-EYFP muscles into the tibialis anterior of SCID-mdx recipients. Scale bars, 1,000 μm (top images) and 100 μm (bottom image).
(H) Quantification of (G); non-transplanted mdx muscle sections were used as negative control (error bars denote SD; ∗∗p < 0.005).
(I) Immunofluorescence images for dystrophin (red)/Pax7 (green) or EYFP (green)/Pax7 (red) expression in SC-derived MPC grafts. Scale bar, 100 μm.
Discussion
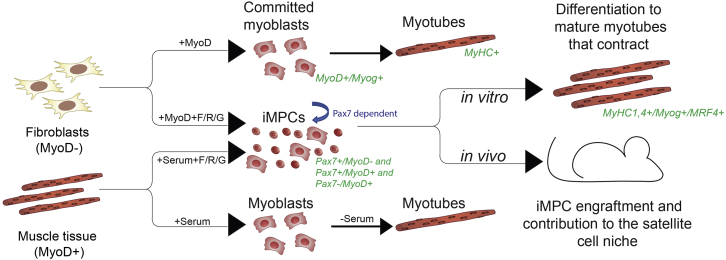
It has been notoriously difficult to culture primary myogenic cell populations for extended periods of time without losing proliferation and engraftment potential (Montarras et al., 2005). Similarly, previous attempts to directly reprogram fibroblasts to myogenic progenitors using transcription factors have had limited success, yielding cell populations that remain dependent on exogenous factors and fail to activate the endogenous Pax7 locus (Ito et al., 2017). Here, we provide evidence that transient MyoD induction in fibroblasts, combined with small-molecule treatment, readily induces an undifferentiated myogenic cell state, which shares characteristics with skeletal muscle stem cells. This includes (1) the activation of the endogenous Pax7 locus; (2) the requirement for Pax7 itself to self-renew; and (3) the potential to differentiate into functional myofibers in vitro and in vivo in the context of a serial injury model (Figure 7, top). Importantly, our culture conditions not only enable reprogramming of fibroblasts into iMPCs but also facilitate prolonged capture in culture of myogenic stem/progenitor cells from muscle tissue (Figure 7 bottom). Thus, our study reports on a long-term cell culture model of fibroblast-derived, non-transformed myogenic cells, which share key molecular and functional properties with muscle stem cells. Future work is certainly required to determine whether purified Pax7+/MyoD− iMPC subsets are transcriptionally and functionally equivalent to muscle-derived Pax7+/MyoD− satellite cells and whether specific iMPC subpopulations maintain a quiescent state in vitro akin to satellite cells in vivo. It will be equally interesting to understand why mononucleated iMPCs self-assemble into heterogeneous cultures containing both stem/progenitor cells and differentiated myotubes. One plausible explanation is that myotubes provide physical or chemical support for myogenic progenitors in vitro (Quarta et al., 2016). It may thus be possible to generate more homogeneous stem and progenitor cell cultures by supplementing our heterogeneous iMPC culture system with additional compounds that enhance satellite cell expansion (Charville et al., 2015).
Figure 7.
Schematic Summary of Results
MyoD is mostly known as a pro-differentiation factor in the context of myogenesis and direct lineage conversion from one mature cell type to another, while our data suggest that MyoD may also function as a dedifferentiation factor in the presence of appropriate signals, which may seem counterintuitive. However, our results are consistent with the prior observation that MyoD expression in fibroblasts induces a transient myoblast-like state before cells differentiate into post-mitotic myotubes (Bergstrom et al., 2002). The previous finding that MyoD promotes proliferation and suppresses differentiation of myoblasts in certain in vitro (Gillespie et al., 2009) and in vivo (Tedesco et al., 2011) contexts is also in agreement with our observations. We surmise that the concomitant expression of MyoD and exposure to small molecules enables the capture of a transient myoblast-like state and subsequently, the dedifferentiation of this cell type toward a Pax7+ stem-like state in a fraction of cells. We note that our data do not imply that Pax7 is a direct downstream target of MyoD. Rather, we hypothesize that the F/R/G treatment endows fibroblasts undergoing MyoD-induced cell fate conversion with a transcriptional and epigenetic state that facilitates downregulation of the fibroblast program and concomitant activation of muscle stem and progenitor cell genes such as Pax7 and Myf5. Consistent with this notion, TGF-β inhibition reportedly induces a mesenchymal-to-epithelial transition (Polo and Hochedlinger, 2010), whereas ascorbic acid, a key component of Serum Replacement (Stadtfeld et al., 2012), triggers epigenetic remodeling of targets via its role as a cofactor for TET enzymes and Jumonji-containing histone demethylases during the generation of iPSCs from fibroblasts (Chen et al., 2013). Once the endogenous MyoD, Myf5, and Pax7 loci have been activated in iMPCs, these small molecules may then be required to stabilize and maintain a self-renewing stem/progenitor cell state. Of note, these compounds also appear to promote terminal differentiation and maturation of myogenic stem/progenitor-like cells based on our finding that myofibers spontaneously contract and express markers associated with adult muscle (e.g., Myh1, Myh4, Myh6, Car3, Casq1, Mstn), which we rarely observed during transdifferentiation induced with MyoD alone.
In addition to providing mechanistic insights and a useful tool to study the role of transcription factors and external stimuli in cell fate control, our data may have therapeutic implications once translated to human cells. For example, patient-specific iMPCs might be useful to study myogenic disorders ex vivo as well as to screen for small molecules that could reverse disease phenotypes. Similarly, iMPCs derived from patients with Duchenne muscular dystrophy could in principle be used for cell therapy following restoration of dystrophin expression using CRISPR/Cas9 technology, as was recently shown in mouse satellite cells (Long et al., 2016, Nelson et al., 2016, Tabebordbar et al., 2016). Lastly, our observation that iMPC-derived myotubes express adult muscle markers and display vigorous contractions may provide a valuable source of material for tissue engineering purposes (Madden et al., 2015).
In conclusion, our study reports on a simple and robust approach to generate expandable, non-transformed myogenic cell populations with characteristics of muscle stem cells directly from fibroblasts and muscle tissue. Our results also raise the intriguing possibility that any other stem/progenitor populations of interest could be derived using similar principles, i.e., overexpression of differentiation-associated transcription factors and pharmacological suppression of pathways that resist reprogramming.
Experimental Procedures
Details on reprogramming conditions, cell culture and antibody reagents, vectors, expression analysis, animals, transplantation, and injury assays can be found in Supplemental Information. All procedures, including maintenance of animals, were performed in compliance with active IACUC protocols and according to institutional guidelines.
Author Contributions
O.B.-N., M.F.M.G., and K.H. conceived the experiments; O.B.-N. and M.F.M.G. conducted the majority of experiments with assistance from B.D.S., A.G., A.C., A.H., C.V., P.C., and D.P.-D.; A.E.A and A.J.W. helped with transplantation experiments; P.F. and M.A.R. provided Pax7+/− and Pax7−/− MEFs; S.P. and S.T. provided Pax7-nGFP MEFs; O.B.-N., B.D.S., A.A., and R.I.S. performed RNA sequencing and microarray analyses; H.C.O., A.J.W., M.A.R., and S.T. provided critical advice and contributed to discussion of results; O.B.-N., M.F.M.G., and K.H. wrote and revised the manuscript.
Acknowledgments
We thank members of the Hochedlinger lab for helpful comments and suggestions. We are further grateful to the MGH/CRM flow-cytometry core for assistance. O.B.-N. was supported by Gruss-Lipper Foundation and MGH ECOR postdoctoral fellowships; B.D.S. was supported by an EMBO long-term fellowship (#ALTF 1143–2015). K.H. was in part supported by the NIH (R01 HD058013, P01 GM099134) and MGH. M.A.R. holds the Canada Research Chair in Molecular Genetics. M.A.R. is supported with grants from the NIH (R01 AR044031), the Canadian Institutes for Health Research (FDN-148387), the Muscular Dystrophy Association, and the Stem Cell Network. A.J.W. was supported in part by the Glenn Foundation for Medical Research and NIH (R01 AG048917 and ES024935).
Published: May 8, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and seven videos and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.04.009.
Accession Numbers
The accession numbers for the RNA sequencing and microarrays reported in this paper are GEO: GSE108543 and GSE92336.
Supplemental Information
Mouse embryonic fibroblasts infected with lentiviral vectors expressing M2rtTA and tetOP-MyoD and propagated without dox for 14 days.
Mouse embryonic fibroblasts infected with M2rtTA and tetOP-MyoD and propagated in the presence of dox for 14 days.
Mouse embryonic fibroblasts infected with lentiviral vectors expressing M2rtTA and tetOP-MyoD and propagated in the presence of dox, forskolin, and RepSox (F/R) for 14 days.
iMPCs at passage 11 cultured in F/R/G conditions.
Single cell-derived iMPC clone at passage 5.
Pax7-CreER;Rosa26-LSL-EYFP iMPCs (passage 16) cultured in F/R/G conditions and labeled with 4OHT between passages 5 and 16. Note EYFP expression in mononucleated and multinucleated cells.
Pax7-CreER;Rosa26-LSL-EYFP iMPCs derived from explanted muscles of tamoxifen-treated Pax7-CreER;Rosa26-LSL-EYFP mouse. Note EYFP expression in mononucleated and multinucleated cells.
References
- Almada A.E., Wagers A.J. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol. 2016;17:267–279. doi: 10.1038/nrm.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O., Brumbaugh J., Verheul C., Apostolou E., Pruteanu-Malinici I., Walsh R.M., Ramaswamy S., Hochedlinger K. Small molecules facilitate rapid and synchronous iPSC generation. Nat. Methods. 2014;11:1170–1176. doi: 10.1038/nmeth.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O., Verheul C., Sommer A.G., Brumbaugh J., Schwarz B.A., Lipchina I., Huebner A.J., Mostoslavsky G., Hochedlinger K. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 2015;33:761–768. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Bosnakovski D., Xu Z., Li W., Thet S., Cleaver O., Perlingeiro R.C., Kyba M. Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells. 2008;26:3194–3204. doi: 10.1634/stemcells.2007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M., Jurga S., Witczak C.A., Hirshman M.F., Shadrach J.L., Goodyear L.J., Wagers A.J. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chal J., Oginuma M., Al Tanoury Z., Gobert B., Sumara O., Hick A., Bousson F., Zidouni Y., Mursch C., Moncuquet P. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015;33:962–969. doi: 10.1038/nbt.3297. [DOI] [PubMed] [Google Scholar]
- Charville G.W., Cheung T.H., Yoo B., Santos P.J., Lee G.K., Shrager J.B., Rando T.A. Ex vivo expansion and in vivo self-renewal of human muscle stem cells. Stem Cell Reports. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Guo L., Zhang L., Wu H., Yang J., Liu H., Wang X., Hu X., Gu T., Zhou Z. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- Comai G., Tajbakhsh S. Molecular and cellular regulation of skeletal myogenesis. Curr. Top. Dev. Biol. 2014;110:1–73. doi: 10.1016/B978-0-12-405943-6.00001-4. [DOI] [PubMed] [Google Scholar]
- Darabi R., Gehlbach K., Bachoo R.M., Kamath S., Osawa M., Kamm K.E., Kyba M., Perlingeiro R.C. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat. Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- Darabi R., Arpke R.W., Irion S., Dimos J.T., Grskovic M., Kyba M., Perlingeiro R.C. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell. 2012;10:610–619. doi: 10.1016/j.stem.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Fukada S., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gillespie M.A., Le Grand F., Scime A., Kuang S., von Maltzahn J., Seale V., Cuenda A., Ranish J.A., Rudnicki M.A. p38-{gamma}-dependent gene silencing restricts entry into the myogenic differentiation program. J. Cell Biol. 2009;187:991–1005. doi: 10.1083/jcb.200907037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9:504–516. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Gunther S., Kim J., Kostin S., Lepper C., Fan C.M., Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy D., Besnard A., Latil M., Jouvion G., Briand D., Thepenier C., Pascal Q., Guguin A., Gayraud-Morel B., Cavaillon J.M. Comparative study of injury models for studying muscle regeneration in mice. PLoS One. 2016;11:e0147198. doi: 10.1371/journal.pone.0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., Son E.Y., Chung J.E., Egli D., Loh K.M., Carter A.C., Di Giorgio F.P., Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito N., Kii I., Shimizu N., Tanaka H., Shin'ichi T. Direct reprogramming of fibroblasts into skeletal muscle progenitor cells by transcription factors enriched in undifferentiated subpopulation of satellite cells. Sci. Rep. 2017;7:8097. doi: 10.1038/s41598-017-08232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura E., Han J.J., Li S., Fall B., Ra J., Haraguchi M., Tapscott S.J., Chamberlain J.S. Cell-lineage regulated myogenesis for dystrophin replacement: a novel therapeutic approach for treatment of muscular dystrophy. Hum. Mol. Genet. 2008;17:2507–2517. doi: 10.1093/hmg/ddn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Cheung T.H., Charville G.W., Hurgo B.M., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden L., Juhas M., Kraus W.E., Truskey G.A., Bursac N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. eLife. 2015;4:e04885. doi: 10.7554/eLife.04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A., Stoykova A., Torres M., Gruss P. Dysgenesis of cephalic neural crest derivatives in Pax7-/- mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- Maza I., Caspi I., Zviran A., Chomsky E., Rais Y., Viukov S., Geula S., Buenrostro J.D., Weinberger L., Krupalnik V. Transient acquisition of pluripotency during somatic cell transdifferentiation with iPSC reprogramming factors. Nat. Biotechnol. 2015;33:769–774. doi: 10.1038/nbt.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.H., Loretz C., Tyler A.E., Duddy W.J., Hall J.K., Olwin B.B., Bernstein I.D., Storb R., Tapscott S.J. Activation of Notch signaling during ex vivo expansion maintains donor muscle cell engraftment. Stem Cells. 2012;30:2212–2220. doi: 10.1002/stem.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7:5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- Quarta M., Brett J.O., DiMarco R., De Morree A., Boutet S.C., Chacon R., Gibbons M.C., Garcia V.A., Su J., Shrager J.B. An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 2016;34:752–759. doi: 10.1038/nbt.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M.A., Tajbakhsh S. A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell. 2012;148:112–125. doi: 10.1016/j.cell.2011.11.049. [DOI] [PubMed] [Google Scholar]
- Sacco A., Doyonnas R., Kraft P., Vitorovic S., Blau H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R.G., Tajbakhsh S. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell. 2009;16:810–821. doi: 10.1016/j.devcel.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102:777–786. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., Shioda T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S391–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M., Zhu K., Cheng J.K., Chew W.L., Widrick J.J., Yan W.X., Maesner C., Wu E.Y., Xiao R., Ran F.A. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco F.S., Gerli M.F., Perani L., Benedetti S., Ungaro F., Cassano M., Antonini S., Tagliafico E., Artusi V., Longa E. Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci. Transl. Med. 2012;4:140ra189. doi: 10.1126/scitranslmed.3003541. [DOI] [PubMed] [Google Scholar]
- Tedesco F.S., Hoshiya H., D'Antona G., Gerli M.F., Messina G., Antonini S., Tonlorenzi R., Benedetti S., Berghella L., Torrente Y. Stem cell-mediated transfer of a human artificial chromosome ameliorates muscular dystrophy. Sci. Transl. Med. 2011;3:96ra78. doi: 10.1126/scitranslmed.3002342. [DOI] [PubMed] [Google Scholar]
- Vidal S.E., Amlani B., Chen T., Tsirigos A., Stadtfeld M. Combinatorial modulation of signaling pathways reveals cell-type-specific requirements for highly efficient and synchronous iPSC reprogramming. Stem Cell Reports. 2014;3:574–584. doi: 10.1016/j.stemcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J., Jones A.E., Parks R.J., Rudnicki M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Tabebordbar M., Iovino S., Ciarlo C., Liu J., Castiglioni A., Price E., Liu M., Barton E.R., Kahn C.R. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Price F., Rudnicki M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse embryonic fibroblasts infected with lentiviral vectors expressing M2rtTA and tetOP-MyoD and propagated without dox for 14 days.
Mouse embryonic fibroblasts infected with M2rtTA and tetOP-MyoD and propagated in the presence of dox for 14 days.
Mouse embryonic fibroblasts infected with lentiviral vectors expressing M2rtTA and tetOP-MyoD and propagated in the presence of dox, forskolin, and RepSox (F/R) for 14 days.
iMPCs at passage 11 cultured in F/R/G conditions.
Single cell-derived iMPC clone at passage 5.
Pax7-CreER;Rosa26-LSL-EYFP iMPCs (passage 16) cultured in F/R/G conditions and labeled with 4OHT between passages 5 and 16. Note EYFP expression in mononucleated and multinucleated cells.
Pax7-CreER;Rosa26-LSL-EYFP iMPCs derived from explanted muscles of tamoxifen-treated Pax7-CreER;Rosa26-LSL-EYFP mouse. Note EYFP expression in mononucleated and multinucleated cells.