Abstract
Human cytomegalovirus (HCMV) is responsible for life-threatening infections in immunocompromised individuals and can cause serious congenital malformations. Available antivirals target the viral polymerase but are subject to cross-resistance and toxicity. New antivirals targeting other replication steps and inducing fewer adverse effects are therefore needed. During HCMV replication, DNA maturation and packaging are performed by the terminase complex, which cleaves DNA to package the genome into the capsid. Identified in herpesviruses and bacteriophages, and with no counterpart in mammalian cells, these terminase proteins are ideal targets for highly specific antivirals. A new terminase inhibitor, letermovir, recently proved effective against HCMV in phase III clinical trials, but the mechanism of action is unclear. Letermovir has no significant activity against other herpesvirus or non-human CMV. This review focuses on the highly conserved mechanism of HCMV DNA-packaging and the potential of the terminase complex to serve as an antiviral target. We describe the intrinsic mechanism of DNA-packaging, highlighting the structure-function relationship of HCMV terminase complex components.
Keywords: cytomegalovirus, DNA packaging, terminase, letermovir
Human cytomegalovirus (HCMV) is responsible for life-threatening infections in immunocompromised individuals and can cause serious congenital malformations. Available antivirals target the viral polymerase but are subject to cross-resistance and toxicity. New antivirals targeting other replication steps are therefore needed. DNA-packaging are performed by the terminase complex. A new terminase inhibitor, letermovir, recently proved effective against HCMV in phase III clinical trials, but the mechanism of action is unclear. This review focuses on the highly conserved mechanism of HCMV DNA-packaging and the potential of the terminase complex to serve as an antiviral target.
INTRODUCTION
Human cytomegalovirus (HCMV) belongs to the betaherpesviruses. It has a double-stranded DNA genome of approximately 230 kb encoding over 200 proteins. Like other members of this subfamily, its main characteristics are high species specificity, various cellular targets and slow replication in cell culture. HCMV persists in a latent state after primary infection and is able to manipulate the immune system by expressing a large number of proteins. In healthy individuals, primary infection is usually asymptomatic or provokes only a self-limited febrile illness. Viremia is rapidly controlled by cell-mediated immunity, but HCMV establishes life-long latency in various cells. Viral reactivation can lead to life-threatening complications in immunocompromised individuals. Despite significant improvements in diagnostic and therapeutic management, cytomegalovirus (CMV) remains a significant problem in immunocompromised individuals, including solid-organ and hematopoietic stem cell transplant recipients and human immunodeficiency virus (HIV)-infected patients (Torres-Madriz and Boucher 2008). HCMV is also the most common infectious cause of congenital malformations, with developmental delay, sensorineural hearing loss and fetal death in 10%–15% of cases. The only drugs licensed for the treatment of HCMV infection and disease are ganciclovir (GCV, Cymevene®), valganciclovir (VGCV, Valcyte®), cidofovir (CDV, Vistide®) and foscarnet (FOS, Foscavir®), all of which target the viral polymerase pUL54. Acyclovir is approved for the prevention of HCMV infection in the European Union (EU). Limitations of these antivirals are their dose-limiting toxicity and resistances emergence. Resistance mutations occur in the UL97 kinase (GCV) or in the UL54 polymerase, leading to various levels of cross-resistance to all available antivirals (Lurain and Chou 2010; Chou 2015a; Chaer, Shah and Chemaly 2016). Recent attempts to develop new anti-HCMV compounds have focused mainly on novel targets such as the viral kinase UL97 (maribavir) and the viral terminase complex involved in viral DNA cleavage/packaging. Several molecules targeting terminase proteins have been discovered (2-bromo-5,6-dichloro-1-beta-D-ribofuranosyl benzimidazole (BDCRB), GW275175X and BAY 38-4766) (Underwood et al.1998; Reefschlaeger et al.2001; Williams et al.2003; Dittmer et al.2005) but none has reached phase 2 or 3 clinical development, mainly owing to poor bioavailability. Letermovir (AIC246) is derived from a new chemical class, the quinazolines (Lischka et al.2010), and acts via a novel, not fully understood mechanism involving the viral terminase protein pUL56 (Goldner et al.2011). Wildum, Zimmermann and Lischka observed no antagonistic effects during letermovir combination with current polymerase inhibitors or with anti-HIV drugs (Wildum, Zimmermann and Lischka 2015). Moreover, Wang and collaborators found that a hydroxypyridonecarboxylic acid compound, previously reported to inhibit HIV RNase H, also inhibited pUL89 at low concentrations (Wang et al.2016). pUL89 may thus present a potential drug target for HCMV. Here, we review the structure and function of HCMV terminase complex proteins, as new antiviral targets.
Genome cleavage/packaging, a highly conserved mechanism
Viruses generally use one of two main strategies for genome packaging: the virus either assembles the capsid around the genome (e.g. HIV) or packs the genome into a preformed procapsid. Double-stranded DNA viruses like tailed bacteriophages and herpesviruses use the latter strategy. Herpesviruses and most bacteriophages produce large concatemers during genome replication and then require a motor for cleavage/packaging. Knowledge of herpesvirus DNA-packaging is limited. However, many studies have highlighted similarities between bacteriophages and herpesviruses (Baker et al.2005). The herpes simplex virus 1 (HSV-1) DNA packaging ATPase is highly homologous to the large terminase subunit of bacteriophages (Przech, Yu and Weller 2003). Packaging mechanisms of bacteriophages such as λ, T3 and T7, which cut at specific sites along the concatemer, resemble those of herpesviruses and therefore present a good model for studying herpesvirus genome packaging. The following section describes current knowledge of the terminase complex of HCMV, partly extrapolated from bacteriophages and other herpesviruses.
Cleavage/packaging and the HCMV terminase complex
As shown for bacteriophage λ, circularisation of the herpesvirus genome occurs early during infection and these circularised molecules act as templates for DNA replication. The viral DNA replicates according to an origin-dependent theta mechanism, in which circular templates are amplified. This step is followed by a rolling circle-based mode of replication that produces concatemers of the genome in head-to-tail fashion; these further act as substrates for the DNA-packaging process (McVoy and Adler 1994). A viral protein complex (terminase) then cleaves concatemeric HCMV DNA into unit-length genomes for DNA packaging. This involves site-specific cleavage at adenine or thymine (AT)-rich core sequences within pac motifs (‘cis-acting packaging signal’) located in the ‘a’ sequence of the terminal and internal repeat segments (Fig. 1). In general, terminase complexes are hetero-oligomers composed of two core proteins (pUL56 and pUL89 for HCMV), each carrying a different function required for the packaging process and associated with several essential cofactors of unknown function. HCMV packaging starts when a packaging signal called the pac sequence is recognised on concatemeric DNA by the viral terminase complex. The functional packaging holocomplex, a hetero-oligomer composed of proteins pUL56, pUL89 and pUL51, makes a first specific cut, thus generating a free end at which further packaging is initiated. The DNA/terminase complex then binds to an empty preformed procapsid at its unique portal vertex, across which the DNA is translocated. A second site-specific cleavage step terminates packaging when a unit length genome has been translocated. DNA packaging is followed by cleavage and expulsion of the scaffold protein and angularisation of the capsid. Knowledge of the DNA-packaging process in bacteriophages gave rise to a theoretical model of this process in HCMV (Bogner, Radsak and Stinski 1998). The DNA-packaging steps occur as follows (step numbers refer to respective numbers in Fig. 2): (i) after their translocation into the host cell nucleus, HCMV terminase proteins act to (ii) specifically bind the pac site on concatemeric DNA and recruit the empty capsid, (iii) cleave the duplex and (iv) exert ATPase activity to power the translocation of a unit-length DNA genome into the capsid. ATP depletion has been shown to inhibit HSV-1 packaging and lead to the accumulation of B capsid (scaffold containing capsid; Dasgupta and Wilson 1999). (v) The packaging process is completed by cutting off excess DNA at the portal region, leading to C capsids (viral DNA containing capsids). (vi) Finally, the DNA/terminase complex dissociates from the filled capsid and is ready for the next packaging step. Unlike the dsRNA motor, the HCMV terminase complex is a transient component of the viral particle and does not remain in the final virion. The small subunit, pUL89, seems to dissociate shortly after DNA cleavage, as it has never been detected together with pUL56 after this step, and is recycled for further cleavage/packaging (Scheffczik et al.2002).
Figure 1.
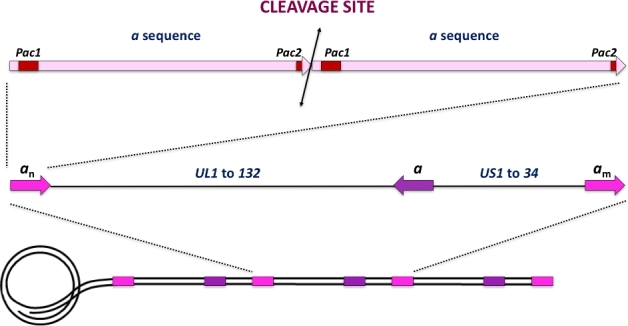
From full genome to cleavage site. Rolling circle replication results in the formation of head-to-tail concatamers that further act as substrates for the DNA-packaging process. The genome is organised as two regions. The unique long (UL) and the unique short (US) segments are flanked by repeated sequences that contain the ≪ a ≫ sequence. The pac1 and pac2 sequences are present in each ≪ a ≫ sequence.
Figure 2.
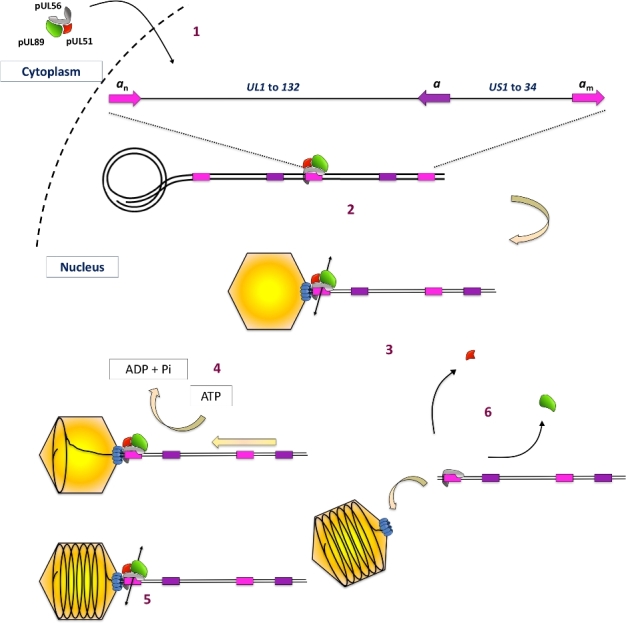
Genome cleavage/packaging and the HCMV terminase complexe adapted from Bogner, Radsak and Stinski (1998). (i) Translocation of the terminase complex into the nucleus, (ii) HCMV terminase specifically binds the pac site and recruits the empty capsid, (iii) cleaves the duplex, (iv) exerts its ATPase activity to power translocation of a unit-length DNA genome into the capsid and (v) completes the DNA-packaging process by cutting off excess DNA at the portal region. (vi) Finally, the DNA-terminase complex dissociates from the filled capsid and is ready for next DNA-packaging step.
Proteins involved in the DNA cleavage/packaging process
Like dsDNA bacteriophage terminases, the HCMV terminase complex includes a large (pUL56) and a small (pUL89) subunit encoding all the functions of ‘classical’ terminases, such as the processing of viral DNA concatemers. Electron microscopy revealed a toroidal architecture of both proteins, as is commonly the case of DNA-metabolizing proteins (Scheffczik et al.2002). Further studies suggested that multimers of pUL56 and pUL89 gather to form the oligomeric holoenzyme, but the exact stoichiometry of the complex remains unknown. Additionally, proteins pUL51, pUL52, pUL77 and pUL93 seem to be part of the terminase complex and/or to participate in the DNA cleavage/packaging process (Scheffczik et al.2002; Borst et al.2013; Borst et al.2016; Köppen-Rung, Dittmer and Bogner 2016; DeRussy and Tandon 2015; DeRussy, Boland and Tandon 2016).
The large terminase subunit, pUL56
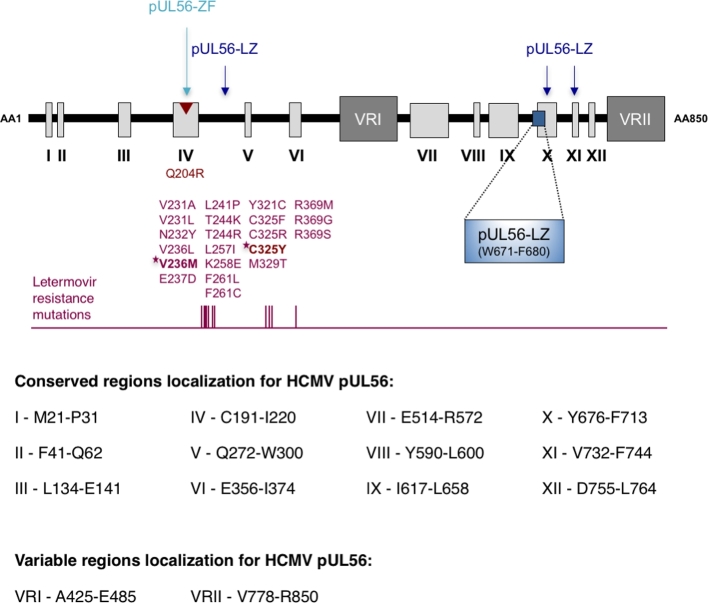
The large subunit of the HCMV terminase complex, pUL56, is composed of 12 conserved regions (I–XII; Champier et al.2008) and is encoded by ORF UL56 located on the unique long portion of the viral genome (Fig. 3). This highly conserved protein of approximately 130 kDa was identified and partially characterised through its homology with HSV-1 pUL28, which has an essential role in HSV-1 genome packaging (Bogner et al.1993; Addison, Rixon and Preston 1990). Three-dimensional reconstruction by electronic cryomicroscopy suggests that pUL56, when expressed alone, exists as a dimer formed by two ring-shaped structures connected to each other by a bridge to their base (Savva, Holzenburg and Bogner 2004). However, no crystal structure has so far been obtained, and its association with the other proteins of the complex has not been fully elucidated. Numerous in vitro studies confirmed the activity of pUL56. Using electrophoretic mobility shift assays, Bogner and colleagues demonstrated a sequence-specific interaction of pUL56 with pac motifs within ‘a’ sequences of the viral genome (Bogner, Radsak and Stinski 1998). A short 128-bp sequence containing regulatory cis elements in conjunction with pac motives is sufficient to mediate efficient HCMV genome maturation (Wang and McVoy 2011). Initiation of the DNA-packaging process takes place in nuclear structures known as replication centres, where pUL56 accumulates at late times post infection and co-localises with pUL112-113 and pUL44, three proteins involved in viral replication. Nuclear importation of pUL56 is mediated by an importin-dependent pathway through the interaction of hSRP1a with the C-terminal nuclear localisation signal (NLS) of pUL56 (amino acid residues 816–827). Alanine scanning identified arginine 822 and lysine 823 as the essential residues of the pUL56 NLS for nuclear translocation (Giesen, Radsak and Bogner 2000). Whereas pUL56 translocates into the nucleus when expressed alone, correct nuclear localisation of both pUL89 and pUL51 requires the concurrent presence of all three terminase subunits (Neuber et al.2017). pUL56 and pUL89 play a major role in driving the complete DNA cleavage and packaging process. Co-immunoprecipitation experiments performed by Hwang and Bogner detected a specific interaction between pUL56 C-terminal and pUL89, as confirmed by Thoma and colleagues (Thoma et al.2006). Recently, we have shown that a short sequence in the C-terminal region of pUL56 (671WMVVKYMGFF680) is essential for interaction with pUL89 (Ligat et al.2017). Furthermore, a recent study has shown a mutual interplay between terminase subunits pUL56, pUL89 and pUL51 as a prerequisite for terminase assembly and nuclear localisation (Neuber et al.2017). The first step during the cleavage of concatemeric DNA catalysed by pUL89 is an essential one and is followed by the binding of the terminase complex to a pac sequence. It is generally accepted that pUL56 acts as an ‘anchor’ for pUL89. Some studies indicate that pUL56 has ATP-independent endonuclease activity that seems to be pac specific (Bogner, Radsak and Stinski 1998). Moreover, pUL56 could enhance the endonuclease activity driven by pUL89 (Scheffczik et al.2002). pUL56 also interacts with the viral portal protein pUL104 during DNA-packaging via the C-terminal part of pUL56. This interaction is crucial during the DNA-packaging process: its prevention by the benzimidazole-D ribonucleosides BDCRB and 2,5,6-trichloro-1-beta-D-ribofuranosyl benzimidazole (TCRB) inhibits HCMV maturation (Krosky et al.1998; Dittmer et al.2005). More importantly, the viral DNA-packaging process is energy-dependent and requires terminase ATPase activity. In vitro studies of bacteriophages have shown that ATP hydrolysis, generating one ATP molecule, allows DNA-packaging of two base pairs (Guo, Peterson and Anderson 1987). In almost all bacteriophages, the large terminase subunit catalyses ATP-dependent translocation of genomic DNA to the proheads (Rao and Feiss 2015). Hwang and Bogner demonstrated that, in HCMV, the terminase ATPase activity is only associated with pUL56. pUL56 ATPase activity is enhanced by up to 30% when it is associated with pUL89 (Hwang and Bogner 2002). A similar mode of action has been documented for the phage T4 terminase complex, with the subunit gp16 enhancing the ATPase activity of the subunit gp17 (Leffers and Rao 2000). Site-directed mutagenesis indicates that the glycine and lysine at positions 714 and 715, respectively, of the putative ATP binding site 709YNETFGKQ716 are essential for ATP hydrolysis (Scholz et al.2003). The pUL56 Q204R substitution associated with BDCRB and TCRB resistance is located within a putative zinc finger, implicating this region in the benzimidazole d-ribonucleoside mechanism of action (Champier et al.2008) (Krosky et al.1998) while mutations conferring resistance to letermovir are located in a non-conserved region. Q204R does not confer resistance to letermovir (Lischka et al.2010; Chou 2015b; Fig. 3). This underlines the different mechanisms of action of these drugs and the need to further explore the possible functional roles of these domains.
Figure 3.
Terminase subunit pUL56 conserved regions adapted from Champier et al. (2008). pUL56 is composed of 12 conserved regions (I-XII). The conserved region IV represents the pUL56 zinc-finger domain. The central region of pUL56 and the C-terminus include two variable regions annotated VRI and VRII. The three putative leucine zippers, annotated pUL56-LZ, are indicated. The short sequence in the C-terminal region of pUL56 (671WMVVKYMGFF680), essential for interaction with pUL89 is highlighted. Positions of amino acids associated with in vitro resistance to letermovir are highlighted. Resistance mutations that have been identified in clinical studies are notified in bold with a star. The position of the Q204R benzimidazole resistance mutation is shown in red.
The small terminase subunit, pUL89
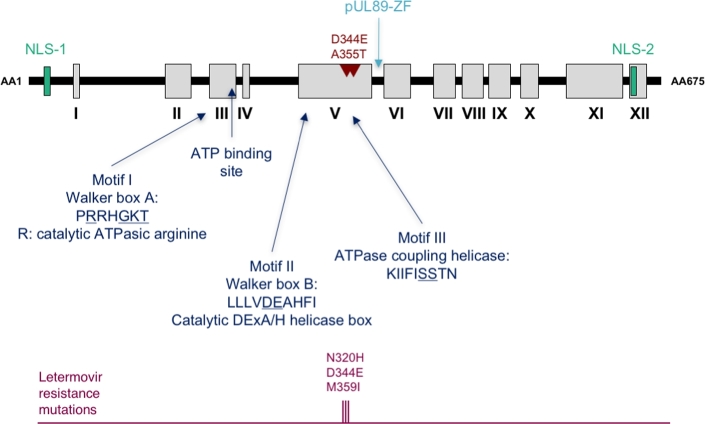
The small subunit of the HCMV terminase complex is encoded by ORF UL89, located on the long unit of the viral genome. In vitro translation and eukaryotic expression demonstrated that pUL89 is a 70- to 75-kDa protein in monomeric form, and 150 kDa when dimerised (Hwang and Bogner 2002; Thoma et al.2006). pUL89 is a homolog of HSV-1 protein pUL15 and was initially identified as a terminase subunit of the HCMV terminase complex by Hwang et al. A previous study by Davison had shown similarities in the amino acid sequence of HCMV pUL89 and the terminase subunit gp17 of phage T4. Because of the strong homology of part of pUL89 to the ATP binding motif of the bacteriophage T4 gp17 subunit, the possible role of pUL89 in HCMV DNA-packaging was investigated (Davison 1992; Hwang and Bogner 2002). Subsequently, our in silico study (Champier et al.2007) focusing on the amino acid sequence of pUL89 have highlighted the four motifs involved in the ATPase centre domains located in N-terminal part of pUL89: the adenine binding site (156EPFQ159 in HCMV), the Walker A box or motif I (213PRRHGKT219 motif in HCMV), the Walker B box or motif II (305LLLVDEAHFI314 in HCMV) and motif III (337SST339 in HCMV pUL89) (Champier et al.2007; Fig. 4). These motifs have also been identified by Mitchell et al in the terminase subunit of the bacteriophage T4 protein gp17 (Mitchell et al.2002). Despite its partial homology with the terminase subunit of T4 gp17, HCMV pUL89 did not exhibit enzymatic ATPase activity. However, it enhanced pUL56-associated ATPase activity described in the previous paragraph by about 30%, indicating a direct interaction of pUL89 with pUL56 (Hwang and Bogner 2002; Leffers and Rao 2000). In vitro binding assays using Glutathion-S-Transferase (GST) fusions, an HCMV-ΔUL89 mutant and BAC complementation experiments indicated that the 580–600 domain of pUL89 was necessary to bind with pUL56 (Thoma et al.2006). This short domain was then studied in silico and its structure was resolved as an alpha helix (Couvreux et al.2010). The crystal structure revealed that the pUL89 C-terminal region corresponds to an exposed helix with three fully conserved residues (Lys583, Ala586 and Asn595), probably forming the interaction domain between pUL56 and pUL89 (Nadal et al.2010). This interaction likely takes place in the cytoplasm, after which the terminase proteins are translocated to the nucleus. Two putative NLS have been proposed to catalyse the nuclear translocation of pUL89 (Champier et al.2007). Recent findings indicate that the pUL89 subunit translocates to the nucleus only in presence of pUL56 and pUL51, and otherwise remains exclusively in the cytoplasm (Wang et al.2012; Neuber et al.2017). Both HCMV terminase subunits were found to have random nuclease activity in vitro. Nevertheless, it has been suggested that pUL56 is unable to exert specific cleavage by itself and that, once again, synergy with pUL89 is necessary to complete the cleavage steps of the DNA-packaging process during HCMV replication (Scheffczik et al.2002). Structural data obtained by Nadal et al. indicate that the pUL89 C-terminal domain belongs to the RNase H-like superfamily of nuclease and polynucleotidyl transferases. Indeed, it has the characteristic fold of this superfamily, and three conserved acidic residues (Asp463, Glu534 and Asp651) coordinating two Mn2+ cations. The D344E and A355T substitutions in pUL89 are associated with BDCRB and TCRB resistance, implying that pUL89 is also involved in the mechanism of action of benzimidazole D-ribonucleosides (Krosky et al.1998; Underwood et al.1998).
Figure 4.
Terminase subunit pUL89 conserved regions adapted from Champier et al. (2007). pUL89 is composed of 12 conserved regions (I-XII) with several putative functional domains such as nuclear localisation site (NLS-1 and NLS-2), pUL89 zinc-finger domain annotated pUL89-ZF, adenine binding site (Walker A box, Walker B box and ATPase coupling helicase, ATP binding site. Underlined amino acids are residues involved in the activities of domains. Positions of amino acids associated with in vitro resistance to letermovir are highlighted. The position of the D344E and A355T benzimidazole resistance mutations are shown in red.
Other proteins of the terminase complex
DNA-packaging is probably more complex for herpesviruses genomes than for bacteriophages, and seems to involve more proteins than the terminase subunits pUL56 and pUL89. Based on data obtained with alphaherpesvirus mutants, it was suggested that, besides the genuine terminase subunits pUL56 and pUL89, at least four additional HCMV proteins, namely pUL51, pUL52, pUL77 and pUL93, contribute to this process. These proteins are homologous to the HSV-1 proteins pUL33, pUL32, pUL25 and UL17, respectively (Borst et al.2008; Borst et al.2013; Borst et al.2016; Köppen-Rung, Dittmer and Bogner 2016; DeRussy and Tandon 2015; DeRussy, Boland and Tandon 2016). Knowledge of these HSV-1 proteins is limited. HSV-1 pUL33 interacts with the HSV-1 terminase proteins pUL15 and pUL28 and with the portal protein pUL6 (Beard, Taus and Baines 2002). Although HSV-1 pUL32 is required for both DNA cleavage and packaging, HSV-1 pUL25 is not required for DNA cleavage but is necessary for efficient DNA-packaging (McNab et al.1998). Recent findings indicate that knockdown of HCMV pUL51 results in the absence of DNA-filled capsids (C capsids) in infected cells, suggesting a role of pUL51 in DNA-packaging. pUL51 interacts with the HCMV terminase subunits pUL56 and pUL89 and mediates their correct subnuclear localisation (Borst et al.2013). Moreover, Neuber et al. reported that only the fully assembled terminase complex consisting of pUL56, pUL89 and pUL51 is protected from proteasome turnover (Neuber et al.2017). In pUL52 deletion mutants, viral concatemers remain uncleaved, but pUL52 does not seem to be involved in the nuclear localisation of the pUL56 and pUL89 subunits. Furthermore, contrary to other packaging proteins, pUL52 is not detected in viral replication compartments. Thus, pUL52 might have a distinct function in HCMV DNA-packaging (Borst et al.2008). pUL93 interacts with pUL77 and components of the nuclear egress complex, namely pUL50, pUL53 and pUL97. Upon knockdown of pUL77 and pUL93, only B capsids are produced, indicating a putative role of these proteins during viral capsid formation (Köppen-Rung, Dittmer and Bogner 2016; DeRussy and Tandon 2015; DeRussy, Boland and Tandon 2016). These findings demonstrate an essential role of these proteins in HCMV DNA-packaging. However, their biochemical makeup and biological functions are poorly characterised. In addition, the three-dimensional structure of these proteins has not yet been resolved (Table 1).
Table 1.
Overview of terminases homologs of herpesviruses and phages.
| Homologs | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCMV | HSV | Phage λ | Phage T4 | Phage T3/T7 | |||||||||||
| Function | Name | Mass (kDa) | References | Name | Mass (kDa) | References | Name | Mass (kDa) | References | Name | Mass (kDa) | References | Name | Mass (kDa) | References |
| Large terminase subunit binds to DNA pac motifs | UL56 | 130 | (Scheffczik et al.2002), (Bogner et al.1993) | UL28 | 85 | (Addison, Rixon and Preston 1990), (Tengelsen et al.1993) | gpNu1 | 20 | (Becker and Murialdo 1990) | gp16 | 18 | (Bhattacharyya and Rao 1994) | gp18 | 20 | (Morita, Tasaka and Fujisawa 1995) |
| Small ATPase subunit of terminase | UL89 | 75 | (Champier et al.2007), (Couvreux et al.2010) | UL15 | 81 | (Yu and Weller 1998), (Przech, Yu and Weller 2003) | gpA | 73 | (Becker and Murialdo 1990) | gp17 | 70 | (Bhattacharyya and Rao 1994) | gp19 | 67 | (Morita, Tasaka and Fujisawa 1995) |
| DNA-packaging | UL51 | 17 | (Borst et al.2013) | UL33 | 36 | (Beard, Taus and Baines 2002) | NA | NA | NA | ||||||
| Interacts with terminase | UL52 | 76 | (Borst et al.2008) | UL32 | 150 | (Chang, Poon and Roizman 1996) | NA | NA | NA | ||||||
| Interacts with terminase | UL77 | 100 | (Köppen-Rung, Dittmer and Bogner 2016), (Borst et al.2016) | UL25 | 60 | (McNab et al.1998) | NA | NA | NA | ||||||
| Interacts with terminase | UL93 | 70 | (Köppen-Rung, Dittmer and Bogner 2016), (Borst et al.2016) | UL17 | 20 | (Toropova et al. 2011) | NA | NA | NA | ||||||
NA: not available
The HCMV terminase complex as a therapeutic target
Three major drugs, ganciclovir, cidofovir and foscarnet, all targeting the HCMV polymerase pUL54, are routinely used for the prevention and treatment of HCMV infection in the transplant setting. However, the emergence of CMV cross-resistance to available antiviral drugs, favoured by long-term exposure, use of low doses and prolonged immunosuppression, is a growing therapeutic challenge, along with the toxicity of these drugs. Because of their hematologic and nephrologic toxicity, these drugs are not recommended for use in pregnant women. The HCMV terminase complex is a critical component of the DNA-packaging process that translocates viral DNA into the empty capsid. The large subunits pUL56 and pUL89 have an essential role in this process, containing many of the functional sites required for DNA-packaging. Nevertheless, little is known of other proteins that belong to the terminase complex. Knowledge of the interactions between the HCMV terminase subunits can serve as a starting point for the generation of new antivirals that target the interaction between these key viral proteins. The terminase complex is highly CMV-specific, with no counterpart in mammalian cells, and thus represents a target of choice for new antivirals. This has been confirmed by the recent development of letermovir in the transplant setting (Lischka et al.2010). Preclinical data suggested that letermovir targets pUL56 (Lischka et al.2010). It is a potent antiviral with in vitro activity surpassing the current gold standard, GCV, by over 400-fold for the 50% effective concentration (EC50; 4.5 nM versus 2 μM) and over 2000-fold the EC90 (6.1 nM versus 14.5 μM), without significant cytotoxic effects (Marschall et al.2012; Goldner et al.2011; Lischka et al.2010). In phase II trials, letermovir effectively prevented HCMV infection in recipients of allogeneic hematopoietic cell transplants, with an acceptable safety profile (Chemaly et al.2014). The phase III trial started in 2014 (by Merck Sharpe and Dohme Corps) was completed at the end of 2016 (ClinicalTrials.gov Identifier: NCT02137772), and published in 2017 (Marty et al.2017). Letermovir significantly reduced the rate of clinically significant infection at week 24 post graft (end-organ disease or initiation of anti-HCMV pre-emptive therapy based on viremia). And was overall well tolerated. Under the name of Prevymis, letermovir has been recently approved (august 2017) by the US Food and Drug Administration as a new molecular entity for prophylaxis of cytomegalovirus infection and disease in adult CMV-seropositive recipients of an allogeneic hematopoietic stem cell transplant (www.accessdata.fda.gov, Reference ID 4179078). As transplant recipients receive antivirals coadministered with cyclosporine A (CsA) or tacrolimus (TAC) as immunosuppressants, clinical trials investigated the potential for letermovir–immunosuppressant interactions. Letermovir increased CsA and TAC exposure. Contrary to TAC, CsA altered letermovir pharmacokinetics (Kropeit et al.2017b). Hepatic and renal impairment also affect letermovir pharmacokinetics. Moderate hepatic impairment increases letermovir exposure less than 2-fold, and severe hepatic impairment 4-fold (Kropeit et al.2017a). Renal impairment also increases letermovir exposure (Kropeit et al.2017c) (www.accessdata.fda.gov, Reference ID 4179078).
UL89 and UL56 mutations are known to confer benzimidazole resistance. A large number of letermovir resistance mutations in UL56 that have been identified in vitro or in clinical studies, clustered at UL56 codons 231–369 (Fig. 3). Thus, these mutations were located outside the functional domains of pUL56 involved in DNA-packaging and do not impact viral replicative capacity (Goldner et al.2014; Chou 2015b); Lischka, Michel and Zimmermann 2016). Moreover, letermovir occasionally selected UL89 N320H, D344E and M359I mutations in vitro (Chou 2017a). After culture of HCMV laboratory strain under letermovir, P91S mutation in gene UL51 was observed in 2 experiments. P91S alone conferred a letermovir resistance and when combined, multiplied letermovir resistance conferred by UL56 mutations S229F or R369M (Chou 2017b). The cross-resistance mapping to 3 genes suggests that letermovir is targeting a mechanism that depends of the interaction of pUL56, pUL89 and pUL51. The impact of the UL56 V236M mutation, alone and combined with UL54 E756K, on drug susceptibility and the replicative capacity of recombinant HCMV was recently evaluated. The V236M and E756K double mutant exhibited borderline resistance to current antivirals and letermovir and replicated less efficiently than the wild-type virus in vitro (Piret, Goyette and Boivin 2017).
Emergence of resistance in the clinical studies has not been fully documented yet. Few patients experienced resistance during prophylaxis failure. In the phase II study only a short sequence including UL56 codons 231–369 has been sequenced. One out of 12 patients with prophylaxis failure had a pUL56 V236M substitution. In the phase III study, the entire UL56 and UL89 genes were sequenced. Three out of 28 patients who failed prophylaxis had pUL56 V236M, C325W and E237G substitutions during treatment (www.accessdata.fda.gov, Reference ID 4179078). However, the clinical significance of C325W and E237G substitutions is not known to date (www.accessdata.fda.gov, Reference ID 4179078).
Unlike other viral DNA-packaging inhibitors, letermovir is remarkably specific for HCMV (Table 2; Marschall et al.2012). Marschall et al. demonstrated potent in vitro activity of letermovir against 17 HCMV clinical isolates and no significant activity against any other herpesvirus. These findings point to a mechanism of action distinct from that of other DNA-packaging inhibitors, which are less specific and less efficient than letermovir. Although broad anti-herpesvirus activity would be a plus, the potential importance of letermovir, as a safe, specific and potent candidate antiviral, cannot be overstated, especially in view of the poor safety profile of drugs currently approved for prevention or treatment of HCMV infection (Griffiths and Emery 2014).
Table 2.
Antiviral activity of letermovir against alpha-, beta- and gammaherpesviruses adapted from Marshall et al. (2012).
| Virus (strain) | AIC246 EC50 ± SD (μM)a |
|---|---|
| Alphaherpesviruses | |
| VZV (Oka) | >10 |
| HHV_1 (166v VP22-GFP) | >10 |
| HHV_2 (01-6332) | >10 |
| Betaherpesviruses | |
| HCMV (AD169-GFP) | 0.0027 ± 0.0002 |
| MCMV (Smith) | 4.5 ± 2.0 |
| RCMV (Maastricht) | >10 |
| HHV_6 (A-GS) | >10 |
| Gammaherpesviruses | |
| EBV (B95-8) | >10 |
EC50 values were determined by specific cell culture-based antiviral test systems. Data are means of results from at least three independent experiments and are expressed with standard deviations.
Concluding remarks
The HCMV terminase complex is highly herpesvirus specific and has no counterpart in mammalian cells. It thus represents a target of choice for antiviral drug development. A better understanding of the HCMV DNA-packaging process, together with the structure and function of the necessary components, will enable the development of antivirals with high specificity and low toxicity. Likewise, elucidation of the letermovir mechanism of action will hasten the development of new terminase inhibitors, not only in HCMV but also in other herpesviruses. Even if letermovir has no activity against herpesviruses other than HCMV, the conserved amino acid sequences of pUL56 and its homologues of other herpesvirus suggest that letermovir derivatives may be active against other herpesviruses. Finally, combining drugs such as letermovir with available pUL54 polymerase inhibitors could hold potential for the treatment of HCMV infection.
Acknowledgements
The authors thank MC. Ploy for her advice, E. Buelow and S. Demai for editorial assistance, L. Roland for his help with a figure and the reviewers for their constructive comments.
Conflict of interest. None declared.
REFERENCES
- Addison C, Rixon FJ, Preston VG. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol 1990;71:2377–84. [DOI] [PubMed] [Google Scholar]
- Baker ML, Jiang W, Rixon FJ et al. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol 2005;79:14967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard PM, Taus NS, Baines JD. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J Virol 2002;76:4785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol 1990;172:2819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SP, Rao VB. Structural analysis of DNA cleaved in vivo by bacteriophage T4 terminase. Gene 1994;146:67–72. [DOI] [PubMed] [Google Scholar]
- Bogner E, Radsak K, Stinski MF. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol 1998;72:2259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner E, Reschke M, Reis B et al. Identification of the gene product encoded by ORF UL56 of the human cytomegalovirus genome. Virology 1993;196:290–3. [DOI] [PubMed] [Google Scholar]
- Borst EM, Bauerfeind R, Binz A et al. The essential human cytomegalovirus proteins pUL77 and pUL93 are structural components necessary for viral genome encapsidation. J Virol 2016;90:5860–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst EM, Kleine-Albers J, Gabaev I et al. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J Virol 2013;87:1720–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst EM, Wagner K, Binz A et al. The essential human cytomegalovirus gene UL52 is required for cleavage-packaging of the viral genome. J Virol 2008;82:2065–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaer FEl, Shah DP, Chemaly RF. How I treat resistant cytomegalovirus infection in hematopoietic cell transplantation recipients. Blood 2016;128:2624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champier G, Couvreux A, Hantz S et al. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir Ther 2008;13:643–54. [PubMed] [Google Scholar]
- Champier G, Hantz S, Couvreux A et al. New functional domains of human cytomegalovirus pUL89 predicted by sequence analysis and three-dimensional modelling of the catalytic site DEXDc. Antivir Ther 2007;12:217–32. [PubMed] [Google Scholar]
- Chang YE, Poon AP, Roizman B. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J Virol 1996;70:3938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemaly RF, Ullmann AJ, Stoelben S et al. Letermovir for cytomegalovirus prophylaxis in hematopoietic-cell transplantation. N Engl J Med 2014;370:1781–9. [DOI] [PubMed] [Google Scholar]
- Chou S. Approach to drug-resistant cytomegalovirus in transplant recipients. Curr Opin Infect Dis 2015a;28:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Rapid in vitro evolution of human cytomegalovirus UL56 mutations that confer letermovir resistance. Antimicrob Agents Ch 2015b;59:6588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Comparison of cytomegalovirus terminase gene mutations selected after exposure to three distinct inhibitor compounds. Antimicrob Agents Ch 2017a;61:e01325–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. A third component of the human cytomegalovirus terminase complex is involved in letermovir resistance. Antiviral Res 2017b;148:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreux A, Hantz S, Marquant R et al. Insight into the structure of the pUL89 C-terminal domain of the human cytomegalovirus terminase complex. Proteins 2010;78:1520–30. [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Wilson DW. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J Virol 1999;73:2006–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison AJ. Channel catfish virus: a new type of herpesvirus. Virology 1992;186:9–14. [DOI] [PubMed] [Google Scholar]
- DeRussy BM, Boland MT, Tandon R. Human Cytomegalovirus pUL93 Links Nucleocapsid Maturation and Nuclear Egress. J Virol 2016;90:7109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRussy BM, Tandon R. Human cytomegalovirus pUL93 is required for viral genome cleavage and packaging. J Virol 2015;89:12221–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer A, Drach JC, Townsend LB et al. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-D-ribonucleosides. J Virol 2005;79:14660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesen K, Radsak K, Bogner E. The potential terminase subunit of human cytomegalovirus, pUL56, is translocated into the nucleus by its own nuclear localization signal and interacts with importin alpha. J Gen Virol 2000;81:2231–44. [DOI] [PubMed] [Google Scholar]
- Goldner T, Hempel C, Ruebsamen-Schaeff H et al. Geno- and phenotypic characterization of human cytomegalovirus mutants selected in vitro after letermovir (AIC246) exposure. Antimicrob Agents Ch 2014;58:610–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldner T, Hewlett G, Ettischer N et al. The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J Virol 2011;85:10884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths PD, Emery VC. Taming the transplantation troll by targeting terminase. N Engl J Med 2014;370:1844–6. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi 29. J Mol Biol 1987;197:229–36. [DOI] [PubMed] [Google Scholar]
- Hwang J-S, Bogner E. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J Biol Chem 2002;277:6943–8. [DOI] [PubMed] [Google Scholar]
- Köppen-Rung P, Dittmer A, Bogner E. Intracellular distributions of capsid-associated pUL77 of HCMV and interactions with packaging proteins and pUL93. J Virol 2016; 90:5876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropeit D, McCormick D, Erb-Zohar K et al. Pharmacokinetics and safety of the anti-human cytomegalovirus drug letermovir in subjects with hepatic impairment. Brit J Clin Pharmaco 2017a;83:2678–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropeit D, von Richter O, Stobernack HP et al. Pharmacokinetics and Safety of Letermovir Coadministered With Cyclosporine A or Tacrolimus in Healthy Subjects. Clin Pharm Drug Dev 2017b;7:9–21. [DOI] [PubMed] [Google Scholar]
- Kropeit D, Scheuenpflug J, Erb-Zohar K et al. Pharmacokinetics and safety of letermovir, a novel anti-human cytomegalovirus drug, in patients with renal impairment. Brit J Clin Pharmaco 2017c83:1944–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosky PM, Underwood MR, Turk SR et al. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol 1998;72:4721–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem 2000;275:37127–36. [DOI] [PubMed] [Google Scholar]
- Ligat G, Jacquet C, Chou S et al. Identification of a short sequence in the HCMV terminase pUL56 essential for interaction with pUL89 subunit. Sci Rep 2017;7:8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Hewlett G, Wunberg T et al. In vitro and in vivo activities of the novel anticytomegalovirus compound AIC246. Antimicrob Agents Ch 2010;54:1290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Michel D, Zimmermann H. Characterization of cytomegalovirus breakthrough events in a phase 2 prophylaxis trial of letermovir (AIC246, MK 8228). J Infect Dis 2016;213:23–30. [DOI] [PubMed] [Google Scholar]
- Lurain NS, Chou S. Antiviral drug resistance of human cytomegalovirus. Clin Microbiol Rev 2010;23:689–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall M, Stamminger T, Urban A et al. In vitro evaluation of the activities of the novel anticytomegalovirus compound AIC246 (letermovir) against herpesviruses and other human pathogenic viruses. Antimicrob Agents Ch 2012;56:1135–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty FM, Ljungman P, Chemaly RF et al. Letermovir Prophylaxis for Cytomegalovirus in Hematopoietic-Cell Transplantation. N Engl J Med 2017;377:2433–2444. [DOI] [PubMed] [Google Scholar]
- McNab AR, Desai P, Person S et al. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol 1998;72:1060–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Adler SP. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol 1994;68:1040–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MS, Matsuzaki S, Imai S et al. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res 2002;30:4009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Tasaka M, Fujisawa H. Structural and functional domains of the large subunit of the bacteriophage T3 DNA packaging enzyme: importance of the C-terminal region in prohead binding. J Mol Biol 1995;245:635–44. [DOI] [PubMed] [Google Scholar]
- Nadal M, Mas PJ, Blanco AG et al. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. P Natl Acad Sci USA 2010;107:16078–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber S, Wagner K, Goldne T et al. Mutual Interplay between the Human Cytomegalovirus Terminase Subunits pUL51, pUL56, and pUL89 promotes Terminase Complex Formation. J Virol 2017;91:e02384–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J, Goyette N, Boivin G. Drug Susceptibility and Replicative Capacity of Multi-Drug Resistant Recombinant Human Cytomegalovirus Harboring Mutations in UL56 and UL54 Genes. Antimicrob Agents Chemother 2017;:e01044–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przech AJ, Yu D, Weller SK. Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging. J Virol 2003;77:9613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. Mechanisms of DNA packaging by large double-stranded DNA viruses. Ann Rev Virol 2015;2:351–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reefschlaeger J, Bender W, Hallenberger S et al. Novel non-nucleoside inhibitors of cytomegaloviruses (BAY 38–4766): in vitro and in vivo antiviral activity and mechanism of action. J Antimicrob Ch 2001;48:757–67. [DOI] [PubMed] [Google Scholar]
- Savva CG, Holzenburg A, Bogner E. Insights into the structure of human cytomegalovirus large terminase subunit pUL56. FEBS Lett 2004;563:135–40. [DOI] [PubMed] [Google Scholar]
- Scheffczik H, Savva CG, Holzenburg A et al. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res 2002;30:1695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz B, Rechter S, Drach JC et al. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res 2003;31:1426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengelsen LA, Pederson NE, Shaver PR et al. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol 1993;67:3470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma C, Borst E, Messerle M et al. Identification of the interaction domain of the small terminase subunit pUL89 with the large subunit pUL56 of human cytomegalovirus. Biochemistry 2006;45:8855–63. [DOI] [PubMed] [Google Scholar]
- Torres-Madriz G, Boucher HW. Immunocompromised hosts: perspectives in the treatment and prophylaxis of cytomegalovirus disease in solid-organ transplant recipients. Clin Infect Dis, 2008;47:702–11. [DOI] [PubMed] [Google Scholar]
- Underwood MR, Harvey RJ, Stanat SC et al. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J Virol 1998;72:717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, McVoy MA. A 128-base-pair sequence containing the pac1 and a presumed cryptic pac2 sequence includes cis elements sufficient to mediate efficient genome maturation of human cytomegalovirus. J Virol 2011;85:4432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Zhu Y, McVoy MA et al. Changes in subcellular localization reveal interactions between human cytomegalovirus terminase subunits. Virol J 2012;9:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mao L, Kankanala J et al. Inhibition of human cytomegalovirus pUL89 terminase subunit blocks virus replication and genome cleavage. J Virol 2016;91:e02152–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildum S, Zimmermann H, Lischka P. In vitro drug combination studies of Letermovir (AIC246, MK-8228) with approved anti-human cytomegalovirus (HCMV) and anti-HIV compounds in inhibition of HCMV and HIV replication. Antimicrob Agents Ch 2015;59:3140–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SL, Hartline CB, Kushner NL et al. In vitro activities of benzimidazole D- and L-ribonucleosides against herpesviruses. Antimicrob Agents Ch 2003;47:2186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Weller SK. Genetic Analysis of the UL15 Gene Locus for the Putative Terminase of Herpes Simplex Virus Type 1. Virology 1998;243:32–44. [DOI] [PubMed] [Google Scholar]