Abstract
The chicken ovalbumin upstream promoter transcription factors (COUP-TFs) are members of the steroid/thyroid hormone receptor superfamily and function in transcriptional regulation of a wide variety of genes. The COUP-TFs purified from HeLa nuclear extract by COUP-affinity chromatography are composed of multiple Mr forms. The Low Mr COUP-TFs (43,000, 44,000, 46,000, and 47,000 Mr) produce a relatively fast migrating complex (Cl) with DNA in electrophoresis mobility shift assays, while the high Mr forms (66,000, 68,000, 72,000, and 74,000 Mr) produce a slower migrating (C2) complex. The high Mr COUP-TFs were purified by gel filtration chromatography and independently formed the C2 DNA complex, probably acting as dimers. The high Mr forms are indistinguishable from the low Mr COUP-TFs in DNA binding and in enhancement of in vitro transcription from the ovalbumin promoter. The finding of multiple COUP-TF forms led us to clone a second low Mr COUP-TF, “COUP-TF2.” The COUP-TF2 sequence has very strong homology with COUP-TF1. The N-termini of COUP-TF1 and COUP-TF2 are least similar, but both contain glutamine-rich and proline-rich motifs, putative activation domains.
The chicken ovalbumin upstream promoter transcription factor (COUP-TF) was first identified in HeLa and chick oviduct extracts by its binding to the distal promoter of the ovalbumin gene (Bagchi et al., 1987; Pastorcic et al., 1986; Wang et al., 1987). It was found to bind an element (COUP) between –90 and –70 on the ovalbumin promoter that is similar to thyroid and estrogen response elements (Pastorcic et al., 1986). The COUP-TF has also been shown to bind cis-elements involved in positive transcription regulation in the rat insulin II (Hwung et al., 1988; Hwung et al., 1988b), chicken VLDL II (Wijnholds et al., 1988), and human apolipoprotein AI and CIII genes (Ladias and Karathanasis, 1991). It was also reported to bind to negative regulatory elements in the proopiomelanocortin (Drouin et al., 1989a; Drouin et al., 1989b) and HIV-1 (Cooney et al., 1991) promoters. The COUP-TF, along with a non-DNA-binding transcription factor, S300-II, was essential for in vitro transcription of the ovalbumin gene (Sagami et al., 1986; Tsai et al., 1987). An interaction between COUP-TF and the S300-II factor was demonstrated in electro-phoretic mobility shift assays (EMSAs) by Tsai et al. (1987). The COUP-TFs were purified from HeLa cell nuclear extract by COUP-affinity chromatography (Wang et al., 1989). Antisera and labeled concatenated COUP elements were used to screen a HeLa cell mRNA library in lambda gtll and identify a COUP-TF1 cDNA (Wang et al., 1989). Sequence analysis showed that the COUPTF1 was a member of the steroid/thyroid hormone receptor superfamily (Beato, 1989; Evans, 1988; Miyajima et al., 1988). The 46 kDa translation product shares high levels of homology in the three regions (I, II, and III) with other members of the receptor superfamily (O’Malley, 1990; Wang et al., 1989). Thus, COUPTF1, identified initially as a transcription factor, joined the steroid/thyroid hormone receptor superfamily.
During the characterization of COUP-TF, the heterogeneity of the molecules became apparent. In EMSAs, two complexes formed on the COUP element (Wang et al., 1989). Additionally, SDS-PAGE analysis showed that several Mr species were present: a low Mr group (43,000, 44,000, 46,000, and 47,000 Mr), a 53,000 Mr band, and a high Mr group (around 68,000 Mr). The faster migrating complex in EMSAs (Cl) was shown to be the product of low Mr COUP-TFs. Also, the COUP-TF1 clone encoded five peptide fragments sequenced directly from low Mr COUP-TFs. The aims of the current work were to describe the proteins that bind to the COUP response element to form the slower (C2) EMSA complex, and to find new members of the COUP-TF family and determine their function. In this paper, we show by characterization of the high Mr forms that they are fully functional members of the COUP-TF family. Also, we describe the identification and sequence of another low Mr COUP-TF, “COUP-TF2.”
Materials and methods
Purification of the COUP-TFs
The COUP-TFs in nuclear extracts of HeLa cells (Invitron) were purified by three passes through a COUP-affinity column (Wang et al., 1989). High and low Mr COUP-TFs were separated by Superose 12 FPLC (Pharmacia) in buffer containing 20 mM HEPES, pH 7.9, 100 mM KCl, 10% (v/v) glycerol, 5 mM MgCl2, and 2 mM dithiothreitol. The flow rate (0.2 ml per minute) and fraction size (0.25 ml) were the same for the COUP-TF purification as they were for Mr standards (lactate dehydrogenase, bovine serum albumin, ovalbumin, and carbonic anhydrase; Boehringer Mannheim).
Electrophoretic mobility shift assays (EMSAs)
The COUP-TFs (1 μl of Superose fractions) bound a 32P-end-labeled fragment of the oval-bumin promoter (–269 to –44; see Sagami et al., 1986) and were analyzed on native 5% polyacrylamide gels. Competitor oligonucleotides contained the ovalbumin COUP sequence (–70 to –90) or a mutant sequence, with two base changes that abolish COUP-TF binding (Wang et al., 1987).
Renaturation of the individual high Mr COUP-TFs and amino acid sequencing of hERR1
Approximately 10 μg of affinity-purified COUP-TFs were separated on an SDS 10% polyacrylamide gel. After Coomassie blue staining, the 74,000, 72,000, 68,000, and 66,000 Mr bands were individually electroeluted and renatured (Wang et al., 1987) before testing in EMSAs.
The 53,000 and the 46,000–47,000 Mr bands on the SDS-PAGE gel, described above, were also electroeluted. After CNBr cleavage and HPLC separation, peptides with the largest yields were sequenced as previously described (Wang et al., 1987).
DNase 1 footprinting
Purified low and high Mr COUP-TFs (15 μl of Superose fraction 50 or 40, respectively) were incubated with the EMSA probe described above. The protein-DNA complexes were digested with 3 μg/ml DNase 1 (DPFF, Worthington Diagnostics) for 1 minute at 20°C, and the DNA was purified and analyzed on an 8% polyacrylamide/urea sequencing gel (Pastorcic et al., 1986).
In vitro transcription
To construct the ovalbumin template for in vitro transcription with COUP-TFs, the ovalbumin promoter was excised by Cla I (at the –219 position, blunt-ended and ligated to EcoR I linkers) and Rsa I (at –1) from a 5′ deletion mutant ovalglobin plasmid (Knoll et al., 1983). The –219 to –1 ovalbumin promoter segment was cloned into the EcoR I and Sac I sites of the G-free cassette vector p(C2AT)19 (kindly provided by Dr. M. Sawadogo), after the latter site was blunt-ended. Transcription reactions were driven with crude fractions of HeLa nuclear extract (Sagami et al., 1986) lacking COUP-TFs unless provided by addition of purified high Mr COUP-TFs described above. In the presence of nucleotides (including 32P-UTP and 3-O-methyl GTP) and T1 RNase (Calbiochem), transcription from the ovalbumin promoter produced a 377-base product containing no G residues (Klein-Hitpass et al., 1990). An internal control template called AdML200, the generous gift of Dr. Uli Schibler, was added to each reaction and produced a 200-base transcript from the adenovirus major late (AdML) promoter. Transcripts from the ovalbumin and AdML promoters were analyzed on 7% acrylamide/urea sequencing gels by autoradiography. Competitor oligonucleotides contained COUP (Wang et al., 1989) or progesterone response elements (PREs; see Klein-Hitpass et al., 1990).
Identification and sequencing of COUP-TF2 cDNA
The EcoR 1-Fok I fragment (694 bp) of COUP-TF1 cDNA (Wang et al., 1989), which contains the N-terminal domain, the DNA binding domain, and part of the presumptive ligand binding domain, was labeled by nick translation and used as a probe to screen a HeLa cell cDNA library in lambda gt10, kindly provided by Dr. C. Hauser. Lambda plaques transferred to nylon membranes were incubated with probe and washed as previously described (Ritchie et al., 1990). Positives were subcloned into the EcoR I site of pGEM7 (Promega). One clone with a different Mbo II restriction map from the COUP-TF1 clone was sequenced by the chain termination method (Sanger et al., 1977) using Sequenase enzyme (United States Biochemicals). Sequencing was performed on cDNA fragments in M13mpl8 using the universal primer or specific oligonucleotide primers. Approximately 80% of the sequence was determined on both DNA strands (see Fig. 6).
Figure 6.
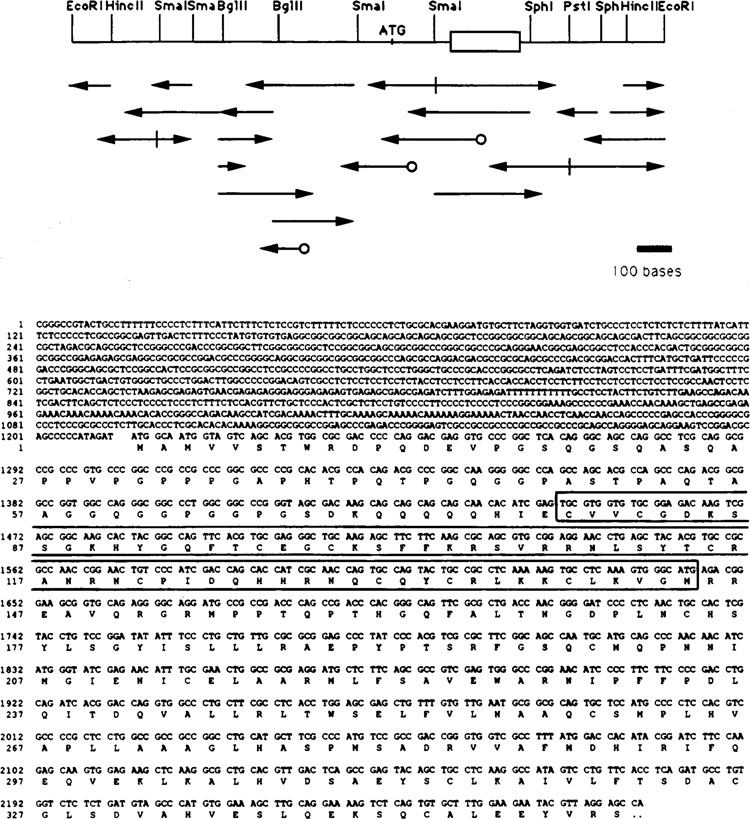
A second low Mr COUP-TF, COUP-TF2, has been cloned and sequenced. In the upper panel, the sequencing strategy is shown for the COUP-TF2 sequence. Sites for generating restriction fragments are pictured, along with the ATG translation start site and the DNA binding domain (boxed). Arrows denote sequencing information obtained, and circles indicate where specific oligonucleotide primers were used. A bar showing the relative size of 100 bases is at the lower right for reference. Shown below is the 2216-base sequence of a COUP-TF2 cDNA, identified by screening a lambda gt10 HeLa cell cDNA library with a COUP-TF1 probe. The inferred amino acid sequence is shown under the base sequence. The sequences are numbered at left for convenience. The DNA-binding domain, determined by homology to COUP-TF1, is boxed.
Results
The COUP-TF family comprises high and low Mr members
Proteins from HeLa nuclear extracts were purified by affinity-chromatography on a column containing oligonucleotides of the ovalbumin COUP sequence. The COUP-TF preparation bound to a radiolabeled COUP element in two distinct complexes in an EMSA (Fig. 1, left panel.) The C2 complex migrated much more slowly than the Cl complex. Both Cl and C2 complexes were efficiently competed by a 25-fold molar excess of unlabeled COUP oligonucleotide, but were unaffected by a mutant COUP oligonucleotide at the same concentration (Cooney et al., 1991). The low Mr COUP-TFs, represented in the Cl complex, bound three to five times more probe than the high Mr COUP-TFs in C2, signifying their predominance in this cell type.
Figure 1.
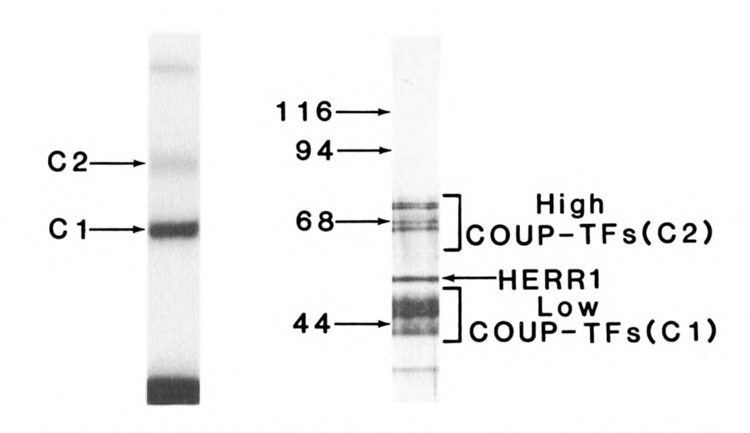
Purified COUP-TFs are composed of multiple Mr forms and produce complexes with COUP DNA of two distinct mobilities. Left panel: Electrophoretic mobility shift (EMSA) analysis of COUP-TFs. Approximately 30 ng of COUP-TFs, purified from HeLa nuclear extract by COUP affinity chromatography, bound a radiolabeled fragment of the ovalbumin promoter (−269 to −44). The two COUP-TF specific complexes, Cl and C2, are visible on the autoradiograph of the native 5% polyacrylamide gel, as is the free probe seen below. Right panel: Sodium dodecyl sulfate-PAGE analysis of purified COUP-TFs. Three hundred ng of purified COUP-TFs (above) were separated and silver-stained on a 10% polyacrylamide gel. The protein bands observed fall into three classes: low Mr COUP-TFs, responsible for the Cl complex in the left panel; high Mr COUP-TFs, responsible for the C2 complex; and hERRl, a relatively weak COUP-binding protein.
The Mr heterogeneity of the purified HeLa COUP-TFs is apparent from SDS-PAGE (Fig. 1, right panel). The low Mr COUP-TFs appear as two doublets, at 47,000 and 46,000, and 44,000 and 43,000 Mr. High Mr COUPTFs migrate in four major bands of 74,000, 72,000, 68,000, and 66,000 Mr. Other bands seen on the gel include human estrogen receptor-related protein (hERRl) at 53,000 Mr, which will be described later, and a minor low Mr band of unknown identity.
Gel filtration chromatography separates high and low Mr COUP-TFs and reveals that they exist as dimers
Since the low and high Mr COUP-TFs differed significantly in size on denaturing gels, separation of native forms was attempted by gel filtration chromatography. The high Mr forms eluted first, as seen in the EMSA analysis of fractions 40 to 44 (Fig. 2), and were fully capable of forming the C2 complex. The low Mr COUP-TFs eluted later, over a less well-defined peak (fractions 46 to 54), at least partly because of their high concentration in the starting material. The low Mr COUP-TFs formed the Cl complex in the EMSA.
Figure 2.
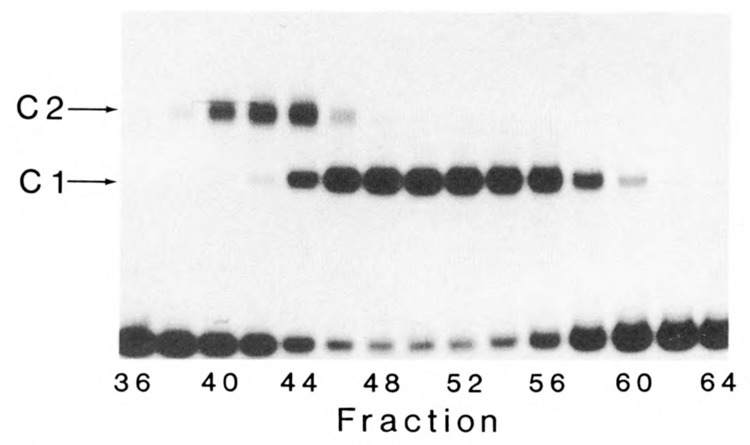
Dimers of high and low Mr COUP-TFs independently form C2 and Cl complexes, respectively. High and low Mr COUP-TFs were separated by gel filtration chromatography, and even-numbered fractions were analyzed by EMSA. The high Mr COUP-TFs formed the C2 complex on DNA, while the low Mr COUP-TFs formed Cl, as indicated. Elution of Mr standards from the column indicated that the native forms of high and low Mr COUP-TFs are approximately 130,000 and 90,000 Mr, respectively.
Since the low Mr COUP-TFs have been shown to be dimers by glycerol gradient centrifugation and UV-crosslinking on DNA (Sagami et al., 1986), we investigated whether high Mr forms exist as dimers. The native high Mr COUPTFs migrated as 130,000 M, moieties and the low Mr forms as approximately 90,000 Mr in gel filtration chromatography. These values are approximately double the respective Mrs of the denatured COUP-TFs on SDS-PAGE. Because only one EMSA complex is seen in the purified preparations of high and low Mr COUP-TFs (fractions 40 and 50, respectively), the high and low Mr forms appear to bind DNA independently to form C2 and Cl complexes. The simplest explanation of the data is that the high Mr COUP-TFs exist in solution as dimers with other members of their Mr class, as previously shown for the low Mr COUP-TFs.
The high Mr COUP-TFs are indistinguishable from the low Mr COUP-TFs in binding to the ovalbumin COUP element
To define which high Mr COUP-TFs were active in binding to the COUP element, renaturation experiments were performed. Preparative SDS-PAGE was used to separate the high Mr COUP-TFs. Four bands, visualized by light Coomassie staining, were individually eluted, renatured, and tested by EMSA. All four produced diffuse complexes because of the harsh treatment of the proteins, but they migrated as C2 complexes like the ones shown in Fig. 3. The complexes were specific for COUP, as shown by competition with unlabeled COUP oligonucleotides. Mutant COUP oligonucleotides, to which COUP-TFs are unable to bind, did not affect the complexes.
Figure 3.
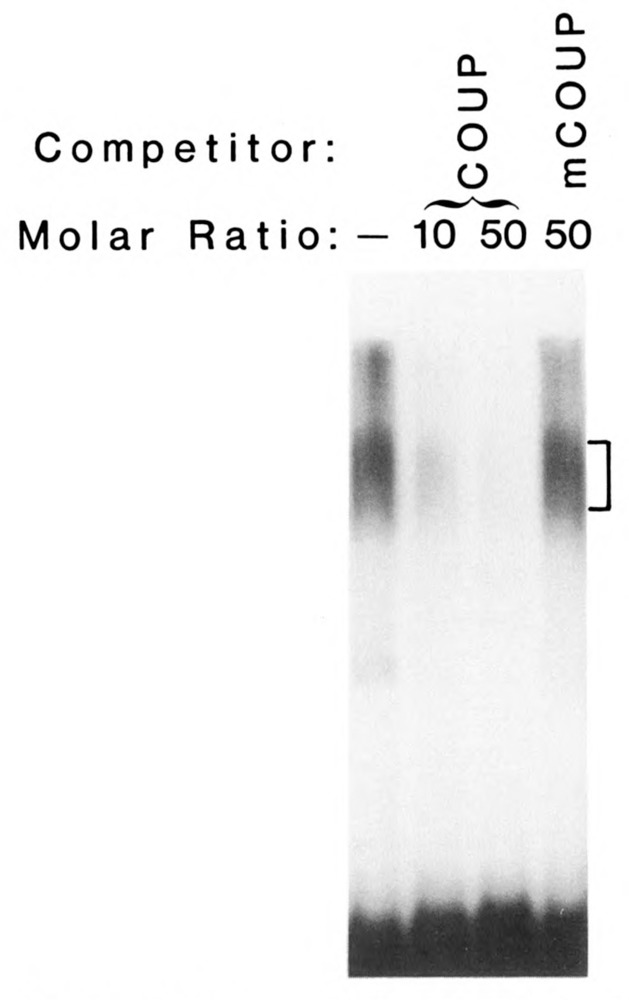
Individual forms of the high Mr COUP-TFs bind COUP specifically. Individual bands of the high Mr COUP-TFs were electroeluted from a preparative SDS-polyacrylamide gel, renatured, and tested in an EMSA. The results from only one high Mr COUP-TF are shown because all four were equivalent. The C2 complex (with brackets at right) was efficiently reduced by the addition of 10-fold and 50-fold molar excesses of unlabeled COUP oligonucleotide (COUP), but not by a 50-fold molar excess of mutant COUP oligonucleotide (mCOUP).
The binding characteristics of high Mr COUP-TFs were further examined to see if they resembled those of the low Mr COUP-TFs. High and low Mr COUP-TFs, separated by gel filtration chromatography, demonstrated identical DNase I footprints on a radiolabeled COUP sequence from −90 to −70 on the ovalbumin promoter (Fig. 4). Footprints from both high and low Mr COUP-TFs were eliminated with a 25-fold molar excess of unlabeled COUP oligonucleotides, but were unaffected by mutant COUP competitor (data not shown).
Figure 4.
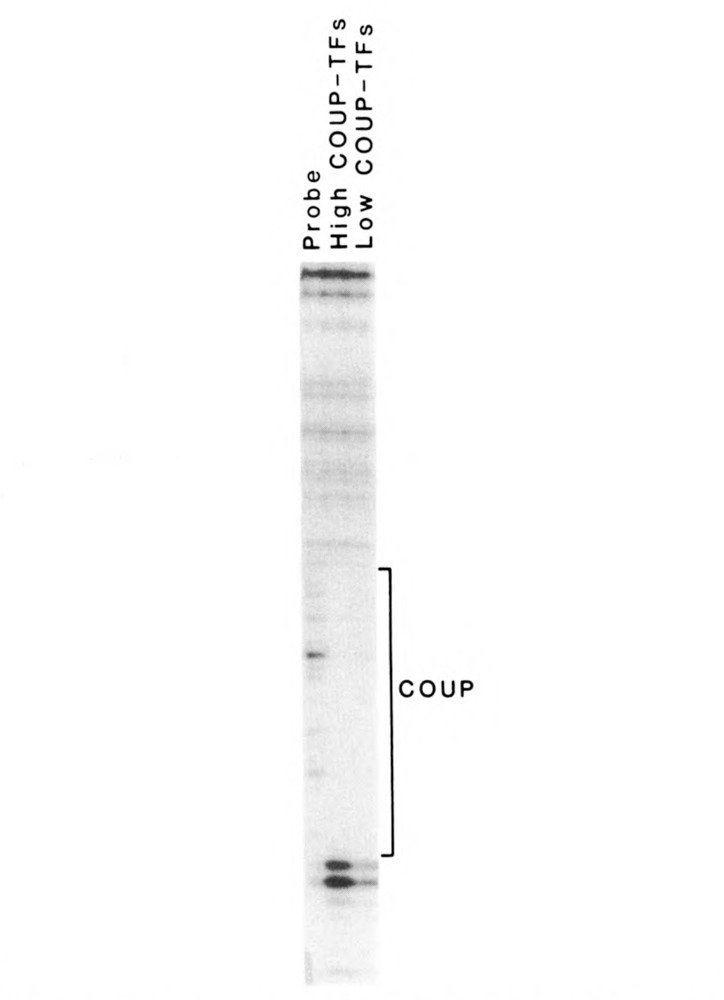
The high and low Mr COUP-TFs have identical DNase 1 footprints over the ovalbumin COUP element. Purified high and low Mr COUP-TFs were bound to the end-labeled −269 to −44 fragment of the ovalbumin promoter. During DNase 1 treatment, they protected identical areas on the coding strand of the ovalbumin promoter over the COUP element (−90 to −70; −70 is toward the bottom of the figure), as seen on this autoradiograph of a sequencing gel. The DNase 1 digestion of the probe in the absence of COUP-TFs is shown at left, for comparison.
The high Mr COUP-TFs function as positive regulators in transcription
To determine if the high Mr COUP-TFs were functional in transcriptional regulation, high Mr COUP-TFs devoid of low Mr forms, prepared as described above, were used in an in vitro transcription assay. The protein components driving transcription in the assay were reconstituted from partially purified fractions of HeLa nuclear extract that contained the S300-II factor and general transcription factors (e.g., TATA-binding factor), but no COUP-TFs. Cell-free transcription of the ovalbumin template was enhanced more than 10-fold by the addition of high Mr COUP-TFs (Fig. 5). The specificity of the response was proven by the inhibition of transcriptional enhancement with COUP oligonucleotides, but not with unrelated progesterone response element oligonucleotides. In contrast, the activity of the AdML promoter template, included as an internal control, was not affected by the oligonucleotide competitors. It should be noted that transcription from AdML is very low in the absence of the COUP-TF fraction (first lane) because it requires USF (Sawadogo et al., 1988), a minor contaminant present in the high Mr COUP-TF preparation. The differential effect of the oligonucleotide competition indicates that the high Mr COUP-TF indeed has positive transcriptional activity on the ovalbumin promoter.
Figure 5.
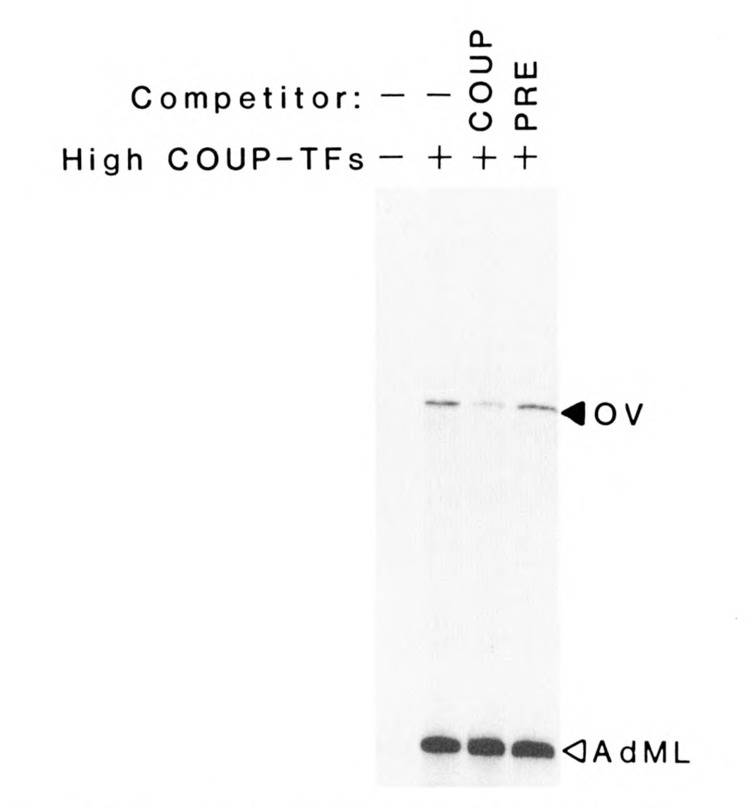
The high Mr COUP-TFs are activators of transcription from the ovalbumin promoter. The purified high Mr COUP-TFs activated transcription of a 377-base G-free cassette under the control of the ovalbumin promoter (−219 to −1). Each assay included fractions of HeLa nuclear extract containing RNA polymerase, its cofactors, and S300-II. Also added were nucleotides (including 32P.UTP), buffers, and the internal control template pAdML200, with the adenovirus major late promoter directing the formation of a 200-base G-less transcript. Specific transcripts from ovalbumin (OV) and adenovirus major late (AdML) promoters are seen at appropriate positions on this autoradiograph of a sequencing gel. The specificity of the COUP-TF enhancement of transcription was tested with 50-fold molar excesses of COUP oligonucleotide (COUP) or progesterone response element-containing oligonucleotides (PRE).
hERR1 copurifies with COUP-TFs
A 53,000 Mr protein was consistently copurified with COUP-TFs on COUP-affinity chromatography (Fig. 1). It appeared to be a moderately abundant protein with significant affinity for COUP and was present after three cycles of COUP-affinity chromatography in the presence of non-specific DNA competitor. To determine how this protein was related to the COUP-TFs, the 53,000 Mr polypeptide was electroeluted from a preparative gel, cleaved by CNBr, and two peptides separated by HPLC were sequenced:
(M)LKEGVRLDRVRGGRQKYKXRPEVDPLPFPG and
(M)EVLVLGVAQRSLPLQDELAFAEDL.
These corresponded exactly to sequences of the human estrogen receptor-related protein 1 (hERRI), a molecule cloned and named for its resemblance to the estrogen receptor (Giguere et al., 1988), from amino acids 241 to 270 and 363 to 387, respectively. The renatured hERRI protein from SDS-PAGE does bind the COUP sequence and forms a complex of slightly less mobility than Cl, but only under conditions of low non-specific competitor DNA (Hwung et al., 1988a; Wang et al., 1987). Therefore, although hERRI is present in material purified by COUP-affinity chromatography, it interacts more weakly with the element than do the proteins responsible for the Cl and C2 complexes. The biological significance of this molecule in COUP-TF dependent gene regulation is unknown.
A second low Mr COUP-TF, “COUP-TF2,” is closely related to COUP-TF1
The heterogeneity of the Mr of the low COUP-TFs on SDS-PAGE was remarkable and indicated the possibility of multiple COUP-TF family members’ arising from different genes and/or splicing variants. Therefore, a cDNA probe from COUP-TF1 encoding the N-terminal domain and the conserved regions I and II was used to identify COUP-TF clones in a lambda gt10 cDNA library made from HeLa cell mRNA. One positive clone, sub-cloned into the plasmid pGEM 7, produced distinct MboII restriction fragments compared to COUP-TF1 clones. The clone, COUP-TF2, was sequenced and its translation product inferred, beginning with the AUG at base 1214 (Fig. 6). The purine (G) three bases upstream of this AUG is characteristic of a strong translational start site (Kozak, 1984). The 5′ untranslated region is very long and contains no other significant open reading frames. Sequencing of a CNBr-cleaved peptide from the 46,000 and 47,000 Mr COUP-TFs revealed the COUP-TF2 sequence from amino acid 155 to 167, confirming the identity of the COUP-TF2 clone. The cDNA for COUP-TF2 was incomplete at the 3′ terminus because of the absence of a stop sequence. The COUP-TF2 sequence was identified as the same as that of apolipoprotein regulatory protein-1 (ARP-1) that binds a COUP-like element (Ladias and Karathanasis, 1991). The ARP-1 sequence showed that our COUP-TF2 clone lacked 63 amino acid residues at the C-terminus.
In Figure 7, we compare the amino acid sequence of COUP-TF2, completed at the C-terminus by 63 amino acids of the ARP-1 sequence (Ladias and Karathanasis, 1991), with that of COUP-TF1. The inferred COUP-TF2 amino acid sequence was very similar to that of COUP-TF1, with 87% overall identity. The region containing the greatest mismatch of amino acids in the alignment (Fig. 7) was N-terminal to the DNA binding domain, with only 43 of 96 residues (45%) of COUP-TF2 identical to those of COUP-TF1. The remainder of the sequence was 96% identical, including the three domains conserved throughout the steroid/thyroid hormone receptor superfamily: region I (DNA-binding domain), 98% with a single conservative amino acid change (T to S); region II, 95%; and region III, 100%. The molecular mass predicted for COUP-TF2 (45.6 kDa) was just slightly below that for COUP-TF1 (46.2 kDa).
Figure 7.

The COUP-TF2 protein is highly homologous to COUP-TF1. The amino acid sequence of COUP-TF2 is aligned with that of COUP-TF1 to show the high level of positions with identical residues (indicated by a dash in the COUP-TF1 sequence). Spacing to optimize the alignment is designated by dots. The three conserved regions of the steroid/thyroid hormone receptor superfamily are boxed, with the most N-terminal being the DNA binding domain (region I), followed in order by regions II and III. Amino acid residues are numbered at right for convenience.
Two remarkable features of the amino acid sequence are proline-rich and glutamine-rich areas N-terminal to the DNA-binding domain. Regions such as these have been identified as transactivation motifs in other transcription factors (Mitchell and Tjian, 1989). The proline-rich region in the N-terminal area of COUP-TF2 has 15 out of 56 proline residues (27%), while COUP-TF1 has 11 out of 53 proline (21 %) over the aligned area. These levels of proline concentration are similar to the 25% levels seen in the transactivation domains of the members of the CTF/NF-1 transcription factor family (Mermod et al., 1989). The glutamine content of COUP-TF2 is more than 20% over a 64-amino acid stretch just N-terminal to the DNA binding domain, while that of COUP-TF1 over the aligned area is 15.5%. These levels are not quite as high as the 25% glutamine sequences of the Spl factor, proven to be transactivation domains (Courey et al., 1989; Courey and Tjian, 1988).
Also of note are consensus sites for phosphor-ylation, both by proline-directed kinase (-S/T-P-at amino acids 39, 42, 51, 278, and 299) and by protein kinase A (-K/R-X-S- at amino acids 87 and 106). These two kinases have been implicated in the phosphorylation and activation of the progesterone receptor (Denner et al., 1990a; Denner et al., 1990b). All of the noted sites are conserved between COUP-TF2 and COUP-TF1. Three of five possible sites for the proline-directed kinase are within 13 residues of each other in the middle of the N-terminal domain. The two possible protein kinase A sites are within the DNA binding domain, toward its N-terminus.
Discussion
Two major classifications of COUP-TFs are proposed, based on their Mr in SDS-PAGE and the mobility of the complexes they form with COUP elements in EMSAs. The low Mr COUP-TFs (47,000, 46,000, 44,000, and 43,000 Mr) form the faster Cl complex, and the high Mr COUP-TFs (74,000, 72,000, 68,000, and 66,000 Mr) form the C2 complex. Separation of the four high Mr forms of COUP-TFs demonstrated that each could form C2 complexes independently with COUP elements in EMSAs. The low Mr COUP-TFs, similarly renatured from SDS-PAGE, could likewise form Cl complexes (Wang et al., 1989). The hERRl protein copurifies with the COUP-TFs and binds to COUP elements more weakly. Because the COUP-TFs appear in multiple bands in EMSA and SDS-PAGE analyses, we sought to clone novel members of the COUP TF family from a HeLa cell cDNA library. We cloned a second low Mr member, COUP-TF2, with a COUP-TF1 cDNA probe. The same sequence was recently cloned from a human pla-cental cDNA expression library by tracking a regulatory protein for the apolipoprotein Al gene promoter (Ladias and Karathanasis, 1991). Our COUP-TF cDNA, although lacking some 3’ coding region, contains 873 more nucleotides in the 5’ untranslated region. Only one nucleo-tide (T at position 1150) is different in the ARP-1 cDNA (C at 278). With the missing C-terminal amino acids supplied by the ARP-1 sequence, the COUP-TF2 sequence was compared to that of COUPTF1. These two COUP-TFs are highly homologous. The COUP-TF2 and COUPTF1 molecules differ primarily in their N-termini, where activation domains exist for the steroid/thyroid hormone receptors, raising the possibility that COUP-TFs may have different transactivation capabilities. Our laboratory has previously identified genomic clones for both COUP-TF1 and COUP-TF2 (Ritchie et al., 1990), and the genes are located on human chromosomes 5 (Miyajima et al., 1988) and 15 (Ladias and Karathanasis, 1991), respectively. Interestingly, the COUP-TF genes encode both Zn fingers of the DNA binding domain in the same exon (Ritchie et al., 1990). This feature, unique among the steroid/thyroid hormone receptors, suggests that the COUP-TFs are ancient and possibly ancestral members of the superfamily.
The first level of control of COUP-TF action is the expression of COUP-TF genes. Using COUP-TF1 cDNA probes, we detected no COUP-TF mRNA in Jurkat cells, where the C2 complex and high Mr COUP-TFs are predominant in EMSAs (A. Cooney, unpublished results). This suggests that the high Mr COUP-TFs may be produced from yet another gene. Although we have no sequence data to indicate structural relationships, we could not distinguish the high Mr COUP-TFs from the low Mr forms in function, both in in vitro transcription and in COUP binding. The COUP-TF family may be like that of the thyroid hormone receptors, comprising several members which differ in transcriptional activation properties and are differentially expressed in tissues during development (Glass et al., 1989; Hudson et al., 1990; Izumo and Mahdavi, 1988; Rentoumis et al., 1990).
Steroid and thyroid hormone receptors are modulated in their activity by binding their respective ligands (Beato, 1989; Evans, 1988). The presumptive ligand binding domains of COUP-TF1 and COUP-TF2, which align with the ligand binding domains of the other superfamily members, span the conserved regions II and III of the steroid/thyroid hormone receptor superfamily. The Drosophila “seven-up” homologue of the COUP-TFs, required for correct neuronal differentiation (Mlodzik et al., 1990), shares 92% homology with COUPTF1 and COUPTF2 in the putative ligand binding domain. Although no ligand has been identified, the conservation of these domains indicates that these may be important in the activity of COUP-TFs.
Many members of the steroid/thyroid hormone receptor superfamily act as dimers (Tsai et al., 1988). Thyroid hormone receptors can form dimers with themselves and heterodimers with retinoic acid receptors, which have various effects on transcription (Forman et al., 1989; Glass et al., 1989; Graupner et al., 1989). Low Mr COUP-TFs were shown to be dimers by glycerol gradient centrifugation and by UV-crosslinking on DNA in a previous study (Sagami et al., 1986). In this study, native COUP-TFs eluted from gel filtration chromatography at 130,000 and 90,000 Mr, approximately double the Mrs seen by SDS-PAGE. Also, the individual members of the high Mr COUP-TFs bind DNA, presumably as dimers, as do the low Mr COUP-TFs. There is no evidence for heterodimer formation between low and high Mr COUP-TFs, such as a complex of intermediate mobility to Cl and C2 in EMSAs. In vivo, dimerization between COUP-TFs may regulate their transacti-vation functions.
Another means of regulating transacting factor function involves protein-protein interactions with other transcription factors. The COUP-TFs interact with a non-DNA-binding transcription factor called S300-II (Tsai et al., 1987). Several other DNA-binding transcription factors have been found to require “mediators” (Kelleher et al., 1990), “adaptors” (Berger et al., 1990) or “coactivators” (Pugh and Tjian, 1990) for activity. Both transfected and endogenous steroid hormone receptors have been shown to compete with each other for a functionally limiting factor, so that each interfered with the transactivation of the other (Meyer et al., 1989). Limiting amounts of a factor, such as S300-II, could control the action of COUP-TFs on transcription.
The COUPTFs are unique members of the steroid/thyroid receptor superfamily: they are widespread, powerful DNA-binding transcription factors composed of multiple gene products. It is important to understand how the specific COUP-TFs, described here, regulate transcription of a variety of genes to affect cell growth and development.
Acknowledgments
The authors gratefully acknowledge the excellent help of Wanda Beattie, Maya Dajee, Shyun-Shyun Lee, Mark L. Wolfe, Craig Wilkinson, and Drs. Christina Chang and Richard Cook. This work was supported by Public Health Service grants HD08188 to BW.O. and HD17379 and Juvenile Diabetes Foundation #189123 to MJ.T.
The nucleotide sequence data of COUP-TF2 have been submitted to GenBank and have been assigned accession number M62760.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
Lee-Ho Wang is currently with the Division of Hematology and Oncology, Department of Internal Medicine, University of Texas Health Science Center, Houston, TX 77030.
References
- Bagchi M. K., Tsai S. Y., Tsai M.-J., and O’Malley B. W. (1987), Mol Cell Biology 7, 4151–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M. (1989), Cell 56, 335–344. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Cress W. D., Cress A., Triezenberg S. J., and Guarente L. (1990), Cell 61, 1199–1208. [DOI] [PubMed] [Google Scholar]
- Cooney A. J., Tsai S. Y., O’Malley B. W., and Tsai M.-J. (1991), J Virology (in press).
- Courey A. J., Holtzman D. A., Jackson S. P., and Tjian R. (1989), Cell 59, 827–836. [DOI] [PubMed] [Google Scholar]
- Courey A. J. and Tjian R. (1988), Cell 55, 887–898. [DOI] [PubMed] [Google Scholar]
- Denner L. A., Schrader W. T., O’Malley B. W., and Weigel N. L. (1990a), J Biol Chem 265, 16548–16555. [PubMed] [Google Scholar]
- Denner L. A., Weigel N. L., Maxwell B. L., Schrader W. T., and O’Malley B. W. (1990b), Science 1250, 1740–1743. [DOI] [PubMed] [Google Scholar]
- Drouin J., Nemer M., Charron J., Gagner J.-P., Sun Y. L., Therrien M., and Tremblay Y. (1989a), Genome 31, 510–519. [DOI] [PubMed] [Google Scholar]
- Drouin J., Sun Y. L., and Nemer M. (1989b), J Steroid Biochem 34, 63–69. [DOI] [PubMed] [Google Scholar]
- Evans R. M. (1988), Science 240, 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman B. M., Yang C., Au M., Casanova J., Ghysdael J., and Samuels H. H. (1989), Mol Endocrinol 3, 1610–1626. [DOI] [PubMed] [Google Scholar]
- Giguere V., Yang N., Segui P., and Evans R. M. (1988), Nature 331, 91–94. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Lipkin S. M., Devary O. V., and Rosenfeld M. G. (1989), Cell 59, 697–708. [DOI] [PubMed] [Google Scholar]
- Graupner G., Wills K. N., Tzukerman M., Zhang X., and Pfahl M. (1989), Nature 340, 653–656. [DOI] [PubMed] [Google Scholar]
- Hudson L. G., Santon J. B., Glass C. K., and Gill G. N. (1990), Cell 62, 1165–1175. [DOI] [PubMed] [Google Scholar]
- Hwung Y.-P., Crowe D. T., Wang L.-H., Tsai S. Y., and Tsai M.-J. (1988a), Mol Cell Biol 8, 2070–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwung Y.-P., Wang L.-H., Tsai S. Y., and Tsai M.-J. (1988b), J Biol Chem 263, 13470–13474. [PubMed] [Google Scholar]
- Izumo S. and Mahdavi V. (1988), Nature 334, 539–541. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J. III, Flanagan P. M., and Kornberg R. D. (1990), Cell 61, 1209–1215. [DOI] [PubMed] [Google Scholar]
- Klein-Hitpass L., Tsai S. Y., Weigel N. L., Allen G. F., Riley D., Rodriguez R., Schrader W. T., Tsai M.-J., and O’Malley B. W. (1990), Cell 60, 247–257. [DOI] [PubMed] [Google Scholar]
- Knoll B. J., Zarucki-Shultz T., Dean D. C., and O’Malley B. W. (1983), Nucl Acids Res 11, 6733–6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1984), Nucl Acid Res 12, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladias J. A. A. and Karathanasis S. K. (1991), Science 251, 561–565. [DOI] [PubMed] [Google Scholar]
- Mermod N., O’Neill E. A., Kelly T. J., and Tjian R. (1989), Cell 58, 741–753. [DOI] [PubMed] [Google Scholar]
- Meyer M.-E., Gronemeyer H., Turcotte B., Bocquel M.-T., Tasset D., and Chambon P. (1989), Cell 57, 433–442. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J. and Tjian R. (1989), Science 245, 371–378. [DOI] [PubMed] [Google Scholar]
- Miyajima K., Kadowaki Y., Fukushige S., Shimizu S., Semba K., Yamanashi Y., Matsubara K., Toyoshima K., and Yamamoto T. (1988), Nucl Acids Res 16, 11057–11074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Hiromi Y., Weber U., Goodman C. S., and Rubin G. M. (1990), Cell 60, 211–224. [DOI] [PubMed] [Google Scholar]
- O’Malley B. W. (1990), Mol Endocrinol 4, 363–369. [DOI] [PubMed] [Google Scholar]
- Pastorcic M., Wang H., Elbrecht A., Tsai S. Y., Tsai M.-J., and O’Malley B. W. (1986), Mol Cell Biol 6, 2784–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh B. F. and Tjian R. (1990), Cell 61, 1187–1197. [DOI] [PubMed] [Google Scholar]
- Rentoumis A., Chatterjee V. K. K., Madison L. D., Datta S., Gallagher G. D., Degroot L. J., and Jameson J. L. (1990), Mol Endocrinol 4, 1522–1531. [DOI] [PubMed] [Google Scholar]
- Ritchie H. H., Wang L.-H., Tsai S., O’Malley B. W., and Tsai M.-J. (1990), Nucl Acids Res 18, 6857–6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagami I., Tsai S. Y., Wang H., Tsai M.-J., and O’Malley B. W. (1986), Mol Cell Biol 6, 4259–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., and Coulson A. R. (1977), Proc Natl Acad Sci USA 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., VanDyke M. W., Gregor P. D., and Roeder R. G. (1988), J Biol Chem 263, 11985–11993. [PubMed] [Google Scholar]
- Tsai S. Y., Carlstedt-Duke J., Weigel N. L., Dahlman K., Gustafsson J.-A., Tsai M.-J, and O’Malley B. W. (1988), Cell 55, 361–369. [DOI] [PubMed] [Google Scholar]
- Tsai S. Y., Sagami I., Wang H., Tsai M.-J, and O’Malley B. W. (1987), Cell 50, 701–709. [DOI] [PubMed] [Google Scholar]
- Wijnholds J., Philipsen J. N. J., and AB G. (1988), EMBO J 7, 2757–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-H., Tsai S. Y., Cook R. G., Beattie W. G., Tsai M.-J., and O’Malley B. W. (1989), Nature 340, 163–166. [DOI] [PubMed] [Google Scholar]
- Wang L.-H., Tsai S. Y., Sagami I., Tsai M.-J., and O’Malley B. W. (1987), J Biol Chem 262, 16080–16086. [PubMed] [Google Scholar]


