Significance
N-terminal acetylation performed by N-terminal acetyltransferases (NATs) is a common protein modification in human cells. A unique NAT, NAA80, was recently found to control actin N-terminal acetylation and cytoskeletal dynamics. In this study, we developed potent and specific bisubstrate inhibitors against NAA80 and determined the crystal structure of NAA80 in complex with an inhibitor mimicking the β-actin N terminus, thus revealing molecular determinants for the substrate specificity and selective inhibition of NAA80. A yeast model uncovered how a cellular determinant, the NatB enzyme, acts to restrict the number of in vivo NAA80 substrates relative to the broader intrinsic capacity of NAA80. Our data provide a starting point for further development of inhibitors for the regulation of actin and cytoskeletal functions.
Keywords: NAA80, acetyltransferase, N-terminal acetylation, actin, inhibitor
Abstract
N-terminal (Nt) acetylation is a major protein modification catalyzed by N-terminal acetyltransferases (NATs). Methionine acidic N termini, including actin, are cotranslationally Nt acetylated by NatB in all eukaryotes, but animal actins containing acidic N termini, are additionally posttranslationally Nt acetylated by NAA80. Actin Nt acetylation was found to regulate cytoskeletal dynamics and motility, thus making NAA80 a potential target for cell migration regulation. In this work, we developed potent and selective bisubstrate inhibitors for NAA80 and determined the crystal structure of NAA80 in complex with such an inhibitor, revealing that NAA80 adopts a fold similar to other NAT enzymes but with a more open substrate binding region. Furthermore, in contrast to most other NATs, the substrate specificity of NAA80 is mainly derived through interactions between the enzyme and the acidic amino acids at positions 2 and 3 of the actin substrate and not residues 1 and 2. A yeast model revealed that ectopic expression of NAA80 in a strain lacking NatB activity partially restored Nt acetylation of NatB substrates, including yeast actin. Thus, NAA80 holds intrinsic capacity to posttranslationally Nt acetylate NatB-type substrates in vivo. In sum, the presence of a dominant cotranslational NatB in all eukaryotes, the specific posttranslational actin methionine removal in animals, and finally, the unique structural features of NAA80 leave only the processed actins as in vivo substrates of NAA80. Together, this study reveals the molecular and cellular basis of NAA80 Nt acetylation and provides a scaffold for development of inhibitors for the regulation of cytoskeletal properties.
N-terminal (Nt) acetylation is among the most abundant protein modifications in the eukaryotic cell, occurring on about 50–80% of soluble proteins (1, 2). The process is catalyzed by N-terminal acetyltransferases (NATs) and entails the transfer of an acetyl moiety from acetyl-CoA (Ac-CoA) to the polypeptide N terminus. To date, five cotranslational eukaryotic NAT enzymes have been described, NatA–NatE (3), as well as the Golgi membrane-associated NatF found in most multicellular eukaryotes (4), the plant chloroplast NatG (5), and the recently identified animal kingdom NAA80/NatH (6). NatA acetylates N termini where the initiator methionine (iMet) has been cleaved off and the new N-terminal amino acid carries a small side chain, NatB acetylates N-terminal methionine (Met) residues followed by a negatively charged residue, while NatC/E/F acetylates Met residues followed by hydrophobic/aromatic/aliphatic/positively charged residues. NatD is a specialized NAT with the histones H4 and H2A as the only known substrates (3), while NAA80/NatH has evolved in animals to posttranslationally Nt acetylate processed actins (6). The functional consequences of protein Nt acetylation are diverse, with roles in protein degradation (7), protein–protein interactions (8), and protein folding (9).
Actin is one of the most abundant proteins in the human cell, where it acts as one of the main constituents of the cytoskeleton and a regulator of cell motility (10). The N-terminal processing of animal kingdom actins is highly unusual and relatively complex compared with other cellular proteins (11). In human cells, there are two forms of cytosolic actin, β- and γ-actin, with MDDD- and MEEE- as their unprocessed N termini, respectively. The nascent actin N terminus is Nt acetylated by NatB at the ribosome (12) followed by removal of the acetylated Met residue by a still unidentified aminopeptidase. Finally, the neo-N termini (DDD-/EEE)- are acetylated by the recently identified NAT, NAA80 (6). In other eukaryotes, like budding yeast, the actin N terminus (MDSE-) is only processed by NatB-mediated cotranslational Nt acetylation to generate mature actin (12, 13).
In this investigation, we show that NAA80, controlling actin Nt acetylation and cytoskeletal dynamics (6), exhibits specific in vitro activity toward processed and unprocessed actin and other acidic N termini. We also develop a bisubstrate inhibitor mimicking the processed N terminus of β-actin and use it to determine a cocrystal structure with Drosophila melanogaster NAA80 (DmNAA80), thus uncovering the molecular basis of actin Nt acetylation and the unique structural features of this enzyme. A yeast model reveals the interplay between NatB and NAA80, highlighting that NatB is a major cellular factor restricting the in vivo substrates of NAA80 to processed actins.
Results
NAA80 Acetylates Acidic N Termini Using a Ternary Complex Mechanism.
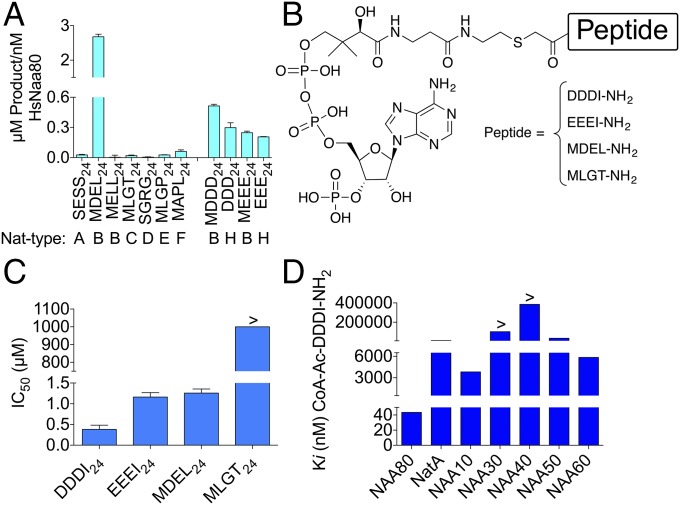
A unique NAT enzyme, NAA80, responsible for cellular Nt acetylation of processed β- and γ-actins was recently revealed (6). To investigate the in vitro activity of NAA80, an enzyme activity screening using a broad substrate library modified from Kuhn et al. (14) on purified enzyme was undertaken. Among all potential substrates tested, only the peptides with N-terminal sequences of MDEL24 (119 ± 5.61 μM), DDDI24 (50.5 ± 1.0 μM), and EEEI24 (42.4 ± 0.95 μM) (Table S1) were acetylated. Substrate specificity studies in the linear range of enzyme concentration revealed that the best substrate for NAA80 was MDEL24, with a fivefold higher product formation compared with the second best substrate, MDDD24, while its cellular substrate, processed β-actin, DDDI24 ranked third (Fig. 1A). Both MDEL24 (p65) and MDDD24 (unprocessed β-actin) represent cellular NatB substrates (12). Interestingly, another NatB substrate, MELL24, which as MDEL24, also contains a Met residue followed by an acidic residue, was not detectably acetylated. Thus, it seems that NAA80 strongly prefers peptide substrates with acidic amino acid residues both at positions 2 and 3, while it is more promiscuous regarding the nature of the amino acid at position 1.
Fig. 1.
NAA80 activity and inhibition. (A) Purified maltose-binding protein (MPB) tagged human NAA80 (MBP-HsNAA80) was tested for in vitro NAT activity toward a panel of potential substrates representing the different substrate classes (NatA–H). (B) Structures for the NAA80 bisubstrate inhibitors. (C) IC50 values for inhibitors measured in an Nt acetylation assay for NAA80. (D) Ki values for CoA-Ac-DDDI-NH2 for different human NATs measured in an Nt acetylation assay. All reactions were performed at least three times and in triplicate, and errors bars represent SD of each measurement.
To design inhibitors for NAA80, we investigated the enzymatic mechanism by bisubstrate kinetics and product inhibition experiments. In contrast to previous data suggesting a ping pong mechanism for NAA80/NAT6/Fus-2 (15), our data clearly supported a ternary complex mechanism (Fig. S1).
Selective and Potent Inhibitors of NAA80 Can Be Developed.
Since a ternary complex mechanism enables inhibition by bisubstrate analogs, we pursued NAA80 bisubstrate analog inhibitors (Fig. 1B) based on the results of the substrate screening and the sequences of the cellular substrates-processed β- and γ-actin (Table S1). We synthesized bisubstrate conjugates of CoA coupled to the tetrapeptides MDEL-NH2, DDDI-NH2, EEEI-NH2, and MLGT-NH2 via an acetamide linker (Fig. 1B). Inhibition studies revealed that CoA-Ac-DDDI-NH2 was the most potent NAA80 inhibitor, with an IC50 value of 0.38 μM: 3- and 3.3-fold more potent than CoA-Ac-EEEI-NH2 and CoA-Ac-MDEL-NH2, respectively (Fig. 1C). The negative control, CoA-Ac-MLGT-NH2 based on a NatC substrate (16), did not show detectable inhibition of NAA80 at the highest concentration tested of 1 mM. To determine the selectivity of the most potent NAA80 inhibitor, CoA-Ac-DDDI-NH2 was tested against a panel of human NATs (Fig. 1D). With a Ki value of 43 nM, the inhibitor was established to be both potent and selective. Interestingly, the monomeric NAA10, which has a known preference for acetylating acidic N termini in vitro (17), was the second most inhibited NAT enzyme. However, CoA-Ac-DDDI-NH2 showed a 88-fold reduced potency for NAA10 compared with NAA80.
Overall Structure of the Actin NAT DmNAA80 Reveals Unique Features.
Crystallization trials with Homo sapiens NAA80 (HsNAA80) did not produce well-ordered crystals. HsNAA80 has several predicted unstructured regions, particularly in the immediate N terminus, and a very long nonconserved β6–β7 loop that is not found in the other NATs that have been characterized (Fig. S2A). A sequence alignment of various model organisms revealed that Drosophila melanogaster NAA80 (DmNAA80) lacks both the long β6–β7 loop and the unstructured N-terminal region found in HsNAA80 (Fig. S2B). We, therefore, pursued crystallization trials using DmNAA80.
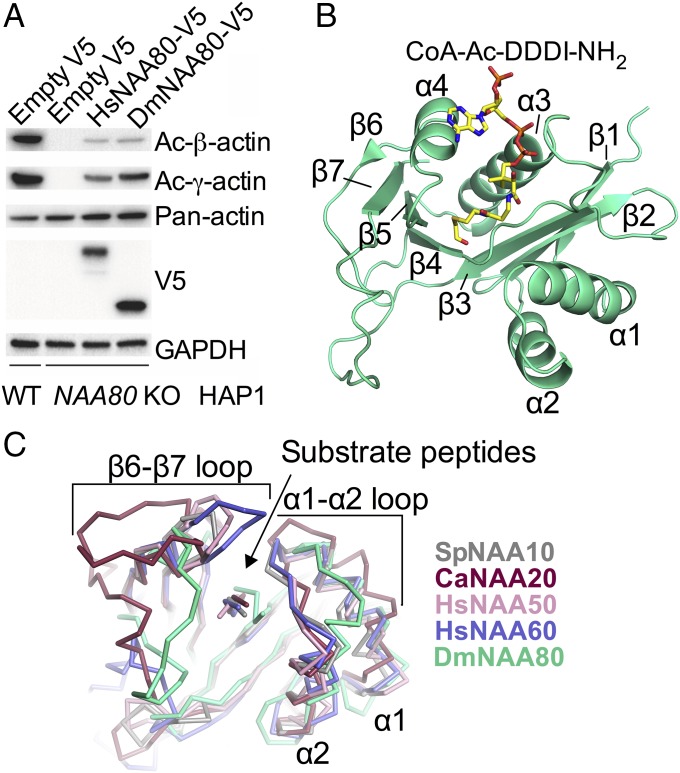
To validate that DmNAA80 had comparable in vitro activity with HsNAA80, activity assays using purified recombinant DmNAA80 against selected substrate peptides were conducted. The results showed a preference for the DDDI24 peptide (Fig. S2C), which matches well with the substrate preference of HsNAA80. Moreover, a rescue assay using NAA80 KO cells transfected with a control plasmid of empty V5, HsNAA80-V5, or DmNAA80-V5 verified the ability of DmNAA80 to Nt acetylate β- and γ-actin in human cells (Fig. 2A) (6). These in vitro and cellular data strongly support NAA80’s functional conservation as an actin-specific NAT in Drosophila.
Fig. 2.
Overall structure of the binary DmNAA80/CoA-Ac-DDDI-NH2 complex. (A) Cellular Nt acetylation of β- and γ-actin by DmNAA80. (B) Overall structure of DmNAA80 in green and bisubstrate inhibitor CoA-Ac-DDDI-NH2 (yellow). (C) Structural alignment of DmNAA80 and it substrate (green) with other NATs: SpNAA10 (PDB ID code 4KVM) and its substrate in gray (23), CaNAA20 (PDB ID code 5K18) and its substrate in raspberry (26), HsNAA50 (PDB ID code 3TFY) and its substrate in pink (24), and HsNAA60 (PDB ID code 5ICV) and its substrate in light blue (21). The shifted positioning of the DmNAA80 substrate is indicated with a black arrow. Also note the shifted α1–α2 loop of DmNAA80 compared with the other NATs.
We obtained crystals from an N-terminal deletion construct of DmNAA80, 9–168 (from here on only referred to as DmNAA80) bound to either Ac-CoA or the bisubstrate inhibitor CoA-Ac-DDDI-NH2 and determined their crystal structures to resolutions of 2.00 and 1.76 Å, respectively. The structures were determined using molecular replacement with the bacterial NAT RimI [Protein Data Bank (PDB) ID code 2CNT] (18) prepared with CHAINSAW to remove Rim1 side-chain residues as a search model. Refinement statistics can be found in Table S2.
The DmNAA80/Ac-CoA and DmNAA80/CoA-Ac-DDDI-NH2 structures are virtually identical, with an rms deviation of 0.33 Å for all shared atoms (Fig. S3). In the remainder of the text, the DmNAA80/CoA-Ac-DDDI-NH2 structure will be described unless otherwise indicated. DmNAA80 has a general Gcn5-related N-acetyltransferase fold (19, 20) with a central conserved Ac-CoA binding core region flanked by N- and C-terminal segments, similar to other NAT structures (21–26), with a total of four α-helices and seven β-strands (Fig. 2B). While the overall fold of DmNAA80 is similar to other NATs, the substrate binding loops differ noticeably (Fig. 2C) and help explain the substrate specificity of NAA80.
In the NAT family (excluding NAA40), the α1–α2 and/or β6–β7 loops play important roles in mediating N terminus-specific substrate recognition. In particular, these loop regions constrict the peptide binding site, such that polypeptides cannot sit across the protein to insert internal lysine substrates for acetylation, explaining why NATs are restricted to N-terminal peptide acetylation (27). Notably, both of these regions differ in NAA80 (Fig. 2C). The α1–α2 substrate binding region is more open in NAA80 than other NATs, as it shifts about 7 Å away from the β6–β7 loop compared with the other NATs. This is accompanied by a shift of the substrate peptide, which makes extensive contacts to conserved residues in the α1–α2 substrate binding region (Fig. 2C). The β6–β7 loop is also in an open conformation in the structure. Together, the α1–α2 and β6–β7 loops make the substrate binding groove wider compared with the other NATs (Fig. 2C) (21, 23, 24).
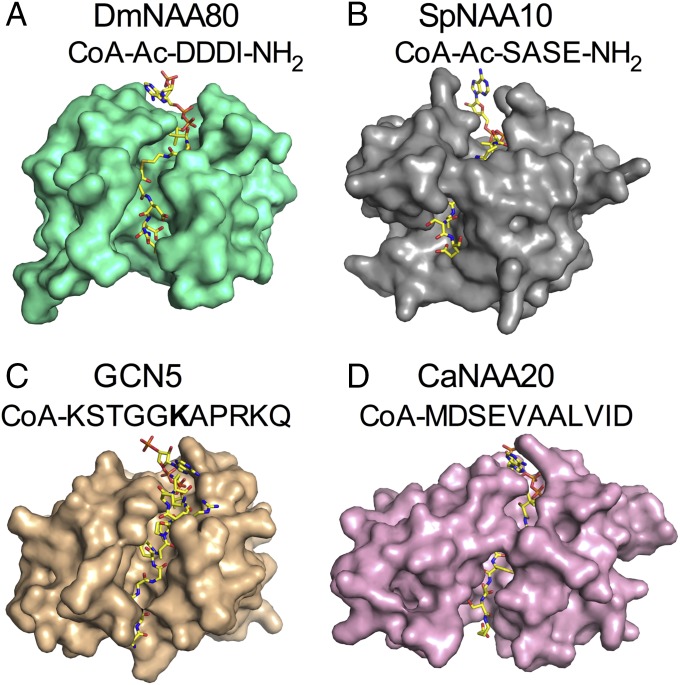
A surface representation of DmNAA80, Schizosaccharomyces pombe NAA10 (SpNAA10), and HsNAA20 (Fig. 3) highlights the wider substrate binding groove in DmNAA80. Interestingly, the wider substrate binding groove in DmNAA80 is reminiscent of the lysine acetyltransferase (KAT) Gcn5, which has a more open peptide substrate binding site (Fig. 3C), and could suggest that DmNAA80 could accommodate protein lysine substrates if not for the active site specificity for the DDD N-terminal sequence as will be described below. Also of note is the closed conformation of Candida albicans NAA20 (CaNAA20), which Nt acetylates unprocessed β- and γ-actin (Fig. 3D).
Fig. 3.
The substrate binding groove of DmNAA80. (A) DmNAA80 (green) bound to CoA-Ac-DDDI-NH2 (yellow) illustrating an open binding cleft. (B) SpNAA10 (PDB ID code 4KVM; gray) with CoA-Ac-SASE-NH2 (yellow) (23). (C) Lysine acetyltransferase GCN5 from Tetrahymena termophila (PDB ID code 1QSN; tan) with CoA-KSTGGKAPRKQ (yellow) (20); acetylated lysine is indicated in bold. (D) CaNAA20 (pink; PDB ID code 5K18) with CoA-Ac-MDSEVAALVID (yellow) (26).
DmNAA80 Substrate Binding Site Accommodates the Acidic Actin N Termini.
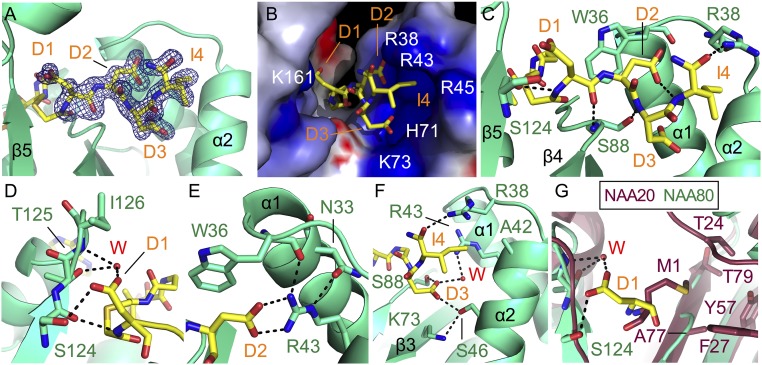
There was clear density for all four residues in the bisubstrate inhibitor (Fig. 4A). The DmNAA80 substrate binding site shows an electrostatic surface with many positively charged residues that would be able to accommodate the acidic β-actin N terminus (Fig. 4B), thus indicating substrate specificity toward acidic residues in the binding site. Both the backbone amides and the side chains make extensive interactions with the enzyme and ordered water molecules.
Fig. 4.
DmNAA80 active site and peptide binding. DmNAA80 is shown in green, and the bisubstrate inhibitor CoA-Ac-DDDI-NH2 is shown as yellow stick. Bonds are shown as dotted black lines. Key secondary structure elements are labeled. (A) Electron density map (|Fo| − |Fc| omit map) contoured at 3.0σ of the peptide moiety of the bisubstrate inhibitor. (B) Electrostatic surface of the DmNAA80 substrate binding site as calculated using the program Delphi (34). Blue indicates positive charges, and red indicates negative charges. Charged residues are indicated in white. (C) Close-up view of interactions between the inhibitor and the DmNAA80 substrate binding site. (D) Close-up view of D1 of the inhibitor in the active site. A water molecule is shown as a red sphere as well as three of its potential H bonds. (E) Close-up view of D2 of the inhibitor in the active site. (F) Close-up view of D3 of the inhibitor in the active site. A water molecule is shown as a red sphere as well as two of its potential H bonds. (G) C. albicans NAA20 (CaNAA20; PDB ID code 5K18; raspberry) (26) and DmNAA80 superimposed with the first residue of their substrates (M1 and D1, respectively) in their binding pockets. NAA20 residues are labeled in raspberry.
With regard to backbone interactions, the N terminus of the peptide makes a hydrogen bond to a backbone carbonyl oxygen of S124. The backbone oxygen of D1 also makes a hydrogen bond to the backbone nitrogen of S88 in the β-bulge (Fig. 4C). W36 serves to orient the peptide substrate by acting as a hydrogen bond acceptor to the amide nitrogen of D2 (28) and through extensive van der Waals contacts. The amide nitrogen of D3 hydrogen bonds with the side chain of S88, and the backbone oxygen of I4 hydrogen bonds with the side chain of R38 (Fig. 4C). Unusually, the amide nitrogen of I4 makes an intramolecular hydrogen bond with the side chain of D2 (Fig. 4C). Most generalized NATs do not make a hydrogen bond with the amide nitrogen of the fourth residue in the substrate peptide; the hydrogen bonds stop at the third residue (23). Notably, NAA40 (the other specific NAT) makes a hydrogen bond with the fourth backbone amide as well (22). This could be another way that NAA80 mediates specificity for Asp or Glu in the second position.
The interactions with the side chains of the peptide portion of the CoA-Ac-DDDI-NH2 bisubstrate inhibitor help explain the substrate specificity of NAA80. D1 of CoA-Ac-DDDI-NH2 interacts with the substrate binding site via hydrogen bonding to the side chain of S124 and a water-mediated hydrogen bond to the backbone amide of I126 and the side chain of T125 (Fig. 4D). D2 of CoA-Ac-DDDI-NH2 forms a salt bridge with R43 of DmNAA80 (Fig. 4E), and R43 is additionally anchored in place by hydrogen bonds between the guanidino group, the backbone oxygen of W36, and the side chain of N33 (Fig. 4E). Hydrogen bonds are also made between D3 of CoA-Ac-DDDI-NH2 and the side chains of residues S46 and S88 (Fig. 4F). The side chain of S46 is hydrogen bonded to the side chain of K73, and the side chain of D3 makes a water-mediated contact with R43. These indirect contacts with positively charged residues may help mediate specificity for negatively charged residues in this position. Finally, the side chain of I4 makes van der Waals contacts with DmNAA80 A42 of the α1 helix (Fig. 4F). Combined, these observations in the structure suggest a substrate preference of DmNAA80 determined largely by residues 2 and 3 in the substrate peptide, where the D2 residue seems to be the anchoring residue of the inhibitor. Importantly, the residues that make contacts to the peptide substrate are highly conserved among NAA80 orthologs (Fig. S2B).
The majority of the peptide interactions between the CoA-Ac-DDDI-NH2 bisubstrate inhibitor and the enzyme are mediated by the α1–α2 region, with few contributions from the β6–β7 region, in contrast to the other more promiscuous NATs (excluding NAA40), where this is a major site for substrate recognition. In these NATs, three conserved tyrosines, one in the α1–α2 loop and two in the β6–β7 loop, hydrogen bond to the backbone amides and orient the peptide in place (21, 23, 24, 26). These tyrosines are not conserved in NAA80 (Fig. S2A). Indeed, the architecture of the peptide binding site in NAA80 causes the α1–α2 loop to shift away from its position relative to the other NATs (Fig. 2C), such that it can make the observed contacts with the substrate peptide. This difference in the structure helps account for the substrate preference of NAA80.
A structure alignment of CaNAA20 and DmNAA80 with the CoA-Ac-DDDI-NH2 bisubstrate inhibitor implicates why NAA20, which acetylates unprocessed actin, may not be able to also Nt acetylate processed actin. Specifically, a comparison of the CaNAA20 and DmNAA80 active sites reveals that CaNAA20 contains a hydrophobic pocket for M1 that would not accommodate an acidic residue at this position, which is accommodated by the more polar DmNAA80 site for residue 1 (Fig. 4G).
Kinetic and Mutational Analyses of DmNAA80 Are Consistent with Structural Observations.
We conducted a kinetic and mutational analysis of DmNAA80 against a variety of substrate peptides. We observed activity of DmNAA80 against peptides mimicking the N terminus of β- or γ-actin with or without the iMet as well as an MDEL24 peptide substrate. The catalytic efficiency of DmNAA80 was similar for the two iMet-starting peptides and the two processed actin peptides without the iMet (Table S3). Interestingly, the activity was higher against the peptides with iMet and much higher against the MDEL24 peptide. The major change in kinetic parameters between these peptides is the kcat, while the Km remains relatively unchanged (Table S3).
To further probe the residues important for substrate binding and catalysis, we prepared mutant forms of the enzyme that targeted conserved residues, which make contacts with the peptide. We carried out complete kinetic curves for the mutant enzymes against DDDI24, which represents the form of β-actin that NAA80 encounters in the cell (Table S3). Activity could not be detected for both the W36A and R43A mutants, corresponding to the residues that interact with D2. K73A and S88A, the residues that interact with D3, both had reduced catalytic efficiency compared with the WT enzyme, which was driven by a higher Km (Table S3).
The structure of DmNAA80 suggests that the only residue in a position to act as a general base is E87, a highly conserved residue located in β4 (Fig. S4). Although it is not in a position to directly deprotonate the N terminus of the peptide, it is hydrogen bonded through a network of two ordered water molecules, which could mediate the deprotonation of the peptide substrate amino group (Fig. S4). We, therefore, made the mutant proteins E87A and E87Q and tested their activity. Surprisingly, these mutants were active and showed the same pH dependence on activity as the WT enzyme, suggesting that this residue does not play a role as a general base (Fig. S4E).
Previous studies of the NATs (23, 24) have described different residues important for catalysis for different members of the enzyme family. Some NATs use a general base to deprotonate the N terminus (21, 24). However, not all of the NATs use direct deprotonation of their substrates via a dedicated general base residue. The situation in NAA10 and NAA40 is ambiguous. Both of these enzymes have glutamate residues that are essential for catalysis yet are not directly positioned to deprotonate the N terminus based on the crystal structures (22, 23). Recently, the crystal structure of NatB was determined, and no residue was identified as a general base (26).
It is interesting that this is another NAT that does not use a general base for catalysis. This could be due to the fact that the pKa of protein N termini is generally 2–3 units lower than lysine side chains (29). Intriguingly, in many NATs, there is a water molecule positioned directly by the N terminus, and this water molecule may be responsible for the direct deprotonation of the N terminus and does not need the assistance of a base due to the relatively low pKa (Fig. S4).
The Narrow in Vivo Substrate Specificity of NAA80 Is Influenced by a Dominant Cotranslational NatB Activity.
A natural question arises of whether recognizing the second and third acidic residues of an N terminus is sufficient for limiting NAA80 to only acetylating β- and γ-actin in vivo. When searching for potential NAA80 substrates by only looking for N termini with acidic residues in the second and third positions (M-[D/E]-[D/E]), there are over 400 human proteins matching these criteria, which clearly cannot account for the specificity of this enzyme. Indeed, it was initially puzzling to us that the only substrates observed from cells were the processed acidic N termini of β- and γ-actin (6), yet NAA80 readily acetylates peptides beginning with methionine in vitro (Fig. 1A). In the cell, NatB (catalytic subunit NAA20) cotranslationally acetylates all proteins with an initial methionine followed by an acidic residue (12). Thus, to study the true in vivo capacity of NAA80, we made a yeast strain lacking NAA20/NAT3 (naa20Δ), and the strain was transformed with a plasmid carrying the human NAA80 gene. Fig. S5 shows tandem mass spectra of actin N-terminal peptides from the different yeast strains. As expected, no acetylated actin could be found in naa20Δ yeast, but surprisingly, the acetylation of the Met starting yeast actin was restored on expression of human NAA80. Furthermore, this pattern was observed for several other NatB-type N termini (MD, ME, MN, and MQ). The N-terminal peptides matching the NAA20 specificity, which were Nt acetylated in the WT strain and for which acetylation status could be determined in the NAA80 complemented strain, were assessed. Of the 11 unique NatB-type N-terminal peptides analyzed, 7 were acetylated in the naa20Δ strain expressing HsNAA80, while 4 were not (Fig. S5E). Although many of the substrates did not contain an acidic residue at both positions 2 and 3 of the N termini, all but one contained an acidic residue in at least one of these positions. In total, this supports the idea that NAA80 has the intrinsic capacity to acetylate a number of NatB-type substrates in vivo, but due to the cotranslational mode and high processivity of NatB, the only in vivo substrates of NAA80 are the processed actins.
Discussion
The data presented here have defined the catalytic properties and structure of a unique NAT, NAA80, responsible for Nt acetylation of processed β- and γ-actin. Our in vitro peptide screen verifies the enzymatic activity of NAA80 toward β- and γ-actins (6). Intriguingly, it also reveals an ability to acetylate Met-starting peptides, like MDEL24. In fact, HsNAA80 displays a preference for unprocessed actin isoforms (Met starting) compared with the processed isoform, where the iMet is removed (Fig. 1A). However, this does not necessarily imply that these N termini are acetylated by HsNAA80 in vivo as discussed below. We also wanted to establish the kinetic mechanism of NAA80 to design inhibitors. While an earlier study reported that NAA80 followed a ping pong mechanism (15), our initial velocity experiments indicate a ternary complex mechanism, where the acetyl group is transferred to the peptide without going through a covalent NAT–acetyl intermediate (Fig. S1), consistent with potent NAA80 inhibition by bisubstrate analogs.
We have developed a potent and selective bisubstrate NAA80 inhibitor based on Ac-CoA and its natural β-actin substrate (Fig. 1B), which together with the structure of NAA80, provide important molecular information and a structural scaffold for future development of NAA80 inhibitors. Actin is a major regulator of the cytoskeleton and numerous cellular activities (30), and the control of its functions by Nt acetylation provides an interesting potential therapeutic target. For instance, one might consider inhibiting NAA80 and actin Nt acetylation in cases where cell migration is desirable, such as nerve regeneration.
The DmNAA80 structure reveals similarities but also striking differences to other NAT structures, the most significant being the shifted substrate positioning in the substrate binding site accompanied by an altered positioning of the α1–α2 and β6–β7 loops (Fig. 2 B and C). In most of the other NATs, substrate recognition is mainly determined by the β6–β7 loop and to a certain extent, the α1–α2 loop; NAA80 recognizes its substrate almost exclusively using the α1–α2 loop. This loop contains highly conserved residues, which make critical contacts to the substrate peptide. These data along with kinetic data showing that NAA80 can acetylate peptides beginning with methionine or aspartic acid/glutamic acid indicate that the substrate specificity of DmNAA80 is driven largely by interactions with the second and third acidic residues in the peptide. This may also help explain why HsNAA80 has activity toward the MDEL24 peptide with no detectable activity toward MELL24, despite both starting with Met followed by an acidic amino acid (Fig. 1A).
There are likely also other factors mediating the specificity for processed actin that is observed in vivo. Of the major cotranslational NATs (NatA, -B, and -C), NatB is distinguished in acetylating nearly 100% of N termini starting with a Met followed by an acidic residue (3). Therefore, in the cell, nearly all of the M-[D/E]-[D/E] N termini are cotranslationally acetylated, thus removing them from the pool of potential NAA80 substrates, as NAA80 is found to act posttranslationally (6). However, the animal kingdom actins are distinguished in that they are further N-terminally processed, whereby unacetylated DDD or EEE N termini are exposed (11). Thus, due to the combination of the high activity of NatB, the unique N-terminal iMet processing of the animal actins, and the high concentration of actin in the cell (31), it is likely that most of the unacetylated N termini appearing in the cell posttranslationally with acidic second and third positions belong to actin.
In budding yeast, the actin N-terminus (MDSE-) is Nt acetylated by NatB to generate mature actin (12, 13), and we showed that, in naa20Δ yeast strains, Nt acetylation of actin and other NatB substrates was lost. Surprisingly, Nt acetylation of several NatB substrates could be restored on expression of HsNAA80. Interestingly, there was not a stringent need for acidic amino acids at both positions 2 and 3 of these Met-starting N termini, pointing to a broader intrinsic in vivo capacity at these residues. We cannot rule out the possibility that NAA80 acetylates other substrates, but based on the cellular proteomics survey and in vitro enzyme data (6), it is highly likely that the processed actins are the main targets of this enzyme. In human NAA80 KO cells, all NatB-type substrates, including several harboring MEE/MDD N termini, displayed a near-100% Nt acetylation status. Only the processed β- and γ-actins were less Nt acetylated in the NAA80 KO cells (0% Nt acetylation) compared with control cells (100% Nt acetylation). This narrow in vivo specificity is probably in large part caused by substrate availability, where a dominant NatB cotranslationally takes care of all cellular substrates while only the processed actins remain available to be Nt acetylated by NAA80. However, the intrinsic capacity of NAA80 to acetylate Met-starting NatB substrates might also imply that NAA80 may act as a backup system for NatB in circumstances where NatB is unable to fully acetylate its substrates, in particular unprocessed actins. Such a system would ensure that the unprocessed actins were always fully Nt acetylated in the animal kingdom.
Despite a major impact of NatB on the nature of the in vivo substrate pool of NAA80, if processed β- and γ-actins are, in fact, the sole substrates of NAA80, it argues for a highly specific substrate recognition by NAA80 similar to NAA40 (22), which is found to only Nt acetylate histones H4 and H2A (32). Similar to NAA80, the β6–β7 loop of NAA40 is flipped away from the substrate, but an extended α1–α2 loop in NAA40 is flipped toward the substrate (22). The substrate specificity of NAA40 has been shown to be dependent on at least the first four residues of the N terminus (22), in part explaining the selectivity of NAA40 compared with the other more promiscuous NAT family members that depend more on the first and second residues for substrate-specific acetylation. The altered binding pattern allows NAA80 and NAA40 to form contacts with the substrate side chains beyond the first two residues. This could not take place if the substrate binding was similar to the other NATs, thus indicating a potential common mechanism for selectivity among NAT enzymes with one or few substrates.
NAA80 also differs from the other NATs in that it has an open active site. For the other NATs, the restriction of the substrate binding site helps explain their preference for Nt acetylation over K acetylation (27). However, this does not necessarily mean that NAA80 can act as a KAT. In fact, an activity screen for potential NAA80 KAT activity indicated no such activity (Fig. S6). It is possible that the highly positively charged active site would repel a lysine from coming close enough to become acetylated. Since NAA80 targets such a negatively charged substrate, the selection of an N terminus over a lysine may come from the electrostatic nature of the active site and not steric blockage as in the other NATs.
Of interest is also the putative impact of early actin interactors on NAA80-mediated actin acetylation. For instance, TCP1 is a chaperonin that binds to newly synthesized actin and is required for actin folding and function in eukaryotes (33). A specific interaction between TCP1 and the actin methionine aminopeptidase (and potentially, NAA80) in animals might ensure specificity for this unique actin processing, since we know that all other proteins with an N terminus highly similar to actin (MEEE/MDDD) are not processed in this way. If a more classical posttranslational mode is followed, then it is likely that iMet cleavage and Nt acetylation occur after TCP1 action and rather, might involve an interaction with fully folded actin and possibly, its partners.
In conclusion, our study reveals the structural and cellular features allowing NAA80 to specifically acetylate processed actins in animals, and the selective bisubstrate analogs provide an interesting starting point for further development of inhibitors toward regulating actin and cytoskeletal functions. The discovery of a specific posttranslational NAT as well as its structure will form the basis for a broader understanding of the eukaryotic NAT machinery and its impact on cellular functions.
Materials and Methods
Detailed descriptions of materials and methods, including purification and crystallization of NAA80 in various forms, acquisition of X-ray diffraction data, structural elucidation of DmNAA80/CoA-Ac-DDDI-NH2, and refinement as well as inhibitor assays, and the yeast model are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Kim Sharp (Department of Biochemistry and Biophysics, University of Pennsylvania) for his assistance with using Delphi to create an electrostatic surface shown in Fig. 4B. This work was supported by NIH Grants T32GM071339 (to student support for R.S.M.) and R35GM118090 (to R.M.), the Bergen Research Foundation (BFS) for support through the peptide synthesis laboratory at the Department of Chemistry at the University of Bergen (L.M.M., B.E.H., and T.A.), Norwegian Health Authorities of Western Norway Project 912176, the Meltzer Foundation (S.V.), Research Council of Norway Grants 230865 (to T.A.) and 249843 (to T.A.), and the Norwegian Cancer Society (T.A.). The Research Council of Norway is further acknowledged for support through Norwegian NMR Platform (NNP) 226244/F50.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates of the structures have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5WJD and 5WJE).
See Commentary on page 4314.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719251115/-/DCSupplemental.
References
- 1.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienvenut WV, et al. Comparative large scale characterization of plant versus mammal proteins reveals similar and Idiosyncratic N-α-Acetylation features. Mol Cell Proteomics. 2012;11:M111.015131. doi: 10.1074/mcp.M111.015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksnes H, Drazic A, Marie M, Arnesen T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem Sci. 2016;41:746–760. doi: 10.1016/j.tibs.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Aksnes H, et al. An organellar nα-acetyltransferase, naa60, acetylates cytosolic N termini of transmembrane proteins and maintains Golgi integrity. Cell Rep. 2015;10:1362–1374. doi: 10.1016/j.celrep.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 5.Dinh TV, et al. Molecular identification and functional characterization of the first Nα-acetyltransferase in plastids by global acetylome profiling. Proteomics. 2015;15:2426–2435. doi: 10.1002/pmic.201500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drazic A, et al. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA. 2018;115:4399–4404. doi: 10.1073/pnas.1718336115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun. 2014;5:4383. doi: 10.1038/ncomms5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubenstein PA, Martin DJ. NH2-terminal processing of actin in mouse L-cells in vivo. J Biol Chem. 1983;258:3961–3966. [PubMed] [Google Scholar]
- 12.Van Damme P, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci USA. 2012;109:12449–12454. doi: 10.1073/pnas.1210303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook RK, Sheff DR, Rubenstein PA. Unusual metabolism of the yeast actin amino terminus. J Biol Chem. 1991;266:16825–16833. [PubMed] [Google Scholar]
- 14.Kuhn ML, Majorek KA, Minor W, Anderson WF. Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N-acetyltransferases with unknown substrate specificity. Protein Sci. 2013;22:222–230. doi: 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zegerman P, Bannister AJ, Kouzarides T. The putative tumour suppressor Fus-2 is an N-acetyltransferase. Oncogene. 2000;19:161–163. doi: 10.1038/sj.onc.1203234. [DOI] [PubMed] [Google Scholar]
- 16.Starheim KK, et al. Knockdown of human N α-terminal acetyltransferase complex C leads to p53-dependent apoptosis and aberrant human Arl8b localization. Mol Cell Biol. 2009;29:3569–3581. doi: 10.1128/MCB.01909-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Damme P, et al. Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of N(α)-acetyltransferases and point to hNaa10p as the post-translational actin N(α)-acetyltransferase. Mol Cell Proteomics. 2011;10:004580. doi: 10.1074/mcp.M110.004580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vetting MW, Bareich DC, Yu M, Blanchard JS. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008;17:1781–1790. doi: 10.1110/ps.035899.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Rojas JR, et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 21.Støve SI, et al. Crystal structure of the Golgi-associated human Nα-acetyltransferase 60 reveals the molecular determinants for substrate-specific acetylation. Structure. 2016;24:1044–1056. doi: 10.1016/j.str.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magin RS, Liszczak GP, Marmorstein R. The molecular basis for histone H4- and H2A-specific amino-terminal acetylation by NatD. Structure. 2015;23:332–341. doi: 10.1016/j.str.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liszczak G, et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol. 2013;20:1098–1105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liszczak G, Arnesen T, Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J Biol Chem. 2011;286:37002–37010. doi: 10.1074/jbc.M111.282863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liszczak G, Marmorstein R. Implications for the evolution of eukaryotic amino-terminal acetyltransferase (NAT) enzymes from the structure of an archaeal ortholog. Proc Natl Acad Sci USA. 2013;110:14652–14657. doi: 10.1073/pnas.1310365110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong H, et al. Molecular basis of substrate specific acetylation by N-terminal acetyltransferase NatB. Structure. 2017;25:641–649.e3. doi: 10.1016/j.str.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Magin RS, March ZM, Marmorstein R. The N-terminal acetyltransferase Naa10/ARD1 does not acetylate lysine residues. J Biol Chem. 2016;291:5270–5277. doi: 10.1074/jbc.M115.709428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levitt M, Perutz MF. Aromatic rings act as hydrogen bond acceptors. J Mol Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 29.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard TD. What we know and do not know about actin. Handb Exp Pharmacol. 2016;235:331–347. doi: 10.1007/164_2016_44. [DOI] [PubMed] [Google Scholar]
- 32.Song OK, Wang X, Waterborg JH, Sternglanz R. An Nalpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J Biol Chem. 2003;278:38109–38112. doi: 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- 33.Sternlicht H, et al. The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci USA. 1993;90:9422–9426. doi: 10.1073/pnas.90.20.9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp KA. Electrostatic interactions in hirudin-thrombin binding. Biophys Chem. 1996;61:37–49. doi: 10.1016/0301-4622(96)00021-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.