Summary
Aging is a complex process associated with a decline in functionality of adult stem cells affecting tissue homeostasis and regeneration. Calorie restriction (CR) is the only experimental manipulation known to extend lifespan and reduce the incidence of age-related disorders across numerous species. These benefits are likely mediated, at least in part, through the preservation of stem cell function. Here, we show that CR enhances the regenerative capacity of the intestinal epithelium through preservation of an injury-resistant reserve intestinal stem cell (ISC) pool. Cell-autonomous activity of mechanistic target of rapamycin complex 1 (mTORC1) governs the sensitivity of reserve ISCs to injury. CR inhibits mTORC1 in these cells, protecting them against DNA damage, while mTORC1 stimulation, either genetically or through nutrient sensing, sensitizes reserve ISCs to injury, thus compromising regeneration of the epithelium. These data delineate a critical role for mTORC1 in epithelial regeneration and inform clinical strategies based on nutrient modulation.
Keywords: calorie restriction, radiation injury, reserve intestinal stem cells, intestine, stem cells, mTORC1 signaling, radiosensitivity, regeneration
Graphical Abstract
Highlights
-
•
Calorie restriction increases the radioresistant intestinal stem cell (ISC) pool
-
•
Calorie restriction suppresses mTORC1 activity in reserve ISCs
-
•
Reserve ISC-autonomous mTORC1 activity governs ISC survival post injury
-
•
Calorie-restricted reserve ISCs have enhanced regenerative capacity post injury
In this study, Yousefi et al. identify reserve intestinal stem cell-autonomous suppression of mTORC1 activity in response to calorie restriction as the basis for enhanced regeneration of the intestinal epithelium after DNA-damaging injury. Conversely, the authors demonstrate that acute nutrient-based stimulation of mTORC1 prior to injury results in reserve stem cell apoptosis and intestinal regenerative failure.
Introduction
Calorie restriction (CR), i.e., receiving fewer calories while maintaining adequate essential nutrients, is one of the most well-established interventions that prolongs life span and retards aging across numerous species, possibly by preserving stem and progenitor cell function, which normally declines with age (Colman et al., 2009, Guarente, 2005, Houthoofd and Vanfleteren, 2006, McCay et al., 1989). Short-term CR in healthy young animals enhances the recovery from injury in several tissues including skeletal muscle and small intestine (Cerletti et al., 2012, Yilmaz et al., 2012).
In the intestinal epithelium, the most highly proliferative tissue in the body, the existence of both quiescent (residing outside the cell cycle in G0) and actively cycling intestinal stem cells (ISCs) in the crypts is becoming increasingly clear. Actively proliferating stem cells located at the crypt base (crypt base columnar stem cells [CBCs]) contribute robustly to tissue homeostasis under basal conditions, and exhibit high expression of canonical Wnt pathway target genes including Lgr5 (Lgr5high) (Barker et al., 2007). Extensive research on the effect of CR on Lgr5high CBCs has demonstrated that CR modestly increases the number of actively cycling Lgr5high CBCs in response to signals sent from adjacent Paneth cells that sense nutrient availability (Igarashi and Guarente, 2016, Yilmaz et al., 2012). However, high Wnt activity in cycling Lgr5high CBCs sensitizes them to DNA-damaging injury, and the functional contribution of CBCs to the enhanced regenerative response to injury after CR has never been tested (Tao et al., 2015, Tian et al., 2011). In addition, genetic ablation of Paneth cells has no effect on the regenerative capacity of the epithelium after high-dose radiation injury (Durand et al., 2012). Thus, the specific cell type, and by extension the underlying molecular mechanism, responsible for the enhanced regenerative capacity of the CR epithelium, remains unknown.
In addition to the Lgr5high CBCs, another population of more radioresistant, slower cycling ISCs has been described in the intestinal crypts, generally referred to as reserve ISCs. Reserve ISCs are located higher in the crypts outside of the WntHigh zone and are highly enriched in populations marked by a Hopx-CreER knockin allele, an mTERT-CreER transgene, and make up a significant fraction of the more heterogeneous population marked by Bmi1-CreER knockin allele (Li et al., 2014, Montgomery et al., 2011, Takeda et al., 2011, Tian et al., 2011). These cells are also likely represented in heterogeneous populations of cells marked by more broadly expressed reporter alleles (Asfaha et al., 2015, Li et al., 2016a, Powell et al., 2012). Reserve ISCs are more resistant to DNA damage than active CBCs, possibly due to their slower cycling rate, residence in G0, and lack of canonical Wnt pathway activity (Li et al., 2014, Li et al., 2016b, Yousefi et al., 2016, Tao et al., 2015). It is well established that these cells undergo a robust proliferative response and contribute broadly to regeneration of the intestinal epithelium following DNA damage, particularly high-dose (>10 Gy) ionizing radiation (Montgomery et al., 2011, Tao et al., 2015, Yan et al., 2012, Yousefi et al., 2016). Interestingly, these reserve ISCs appear to be a largely distinct population from non-cycling, label-retaining secretory progenitor cells, which can also possess stem cell activity (Buczacki et al., 2013, Li et al., 2016b).
We investigated the response of reserve ISCs to CR and subsequent DNA-damaging injury. The reserve ISC compartment expands in response to CR, contributes robustly to the CR-enhanced regenerative capacity of the epithelium, and is functionally important for optimal regeneration following radiation injury. We demonstrate that tight, cell-autonomous regulation of mechanistic target of rapamycin complex 1 (mTORC1) signaling in the reserve ISCs governs the regenerative response of the epithelium in response to DNA damage. These findings offer novel insight into the cell type specificity underlying the beneficial effects of CR, and have immediate implications for application of dietary modulation in patients exposed to DNA-damaging agents.
Results
Calorie Restriction Increases Reserve ISC Availability and Tissue Regeneration
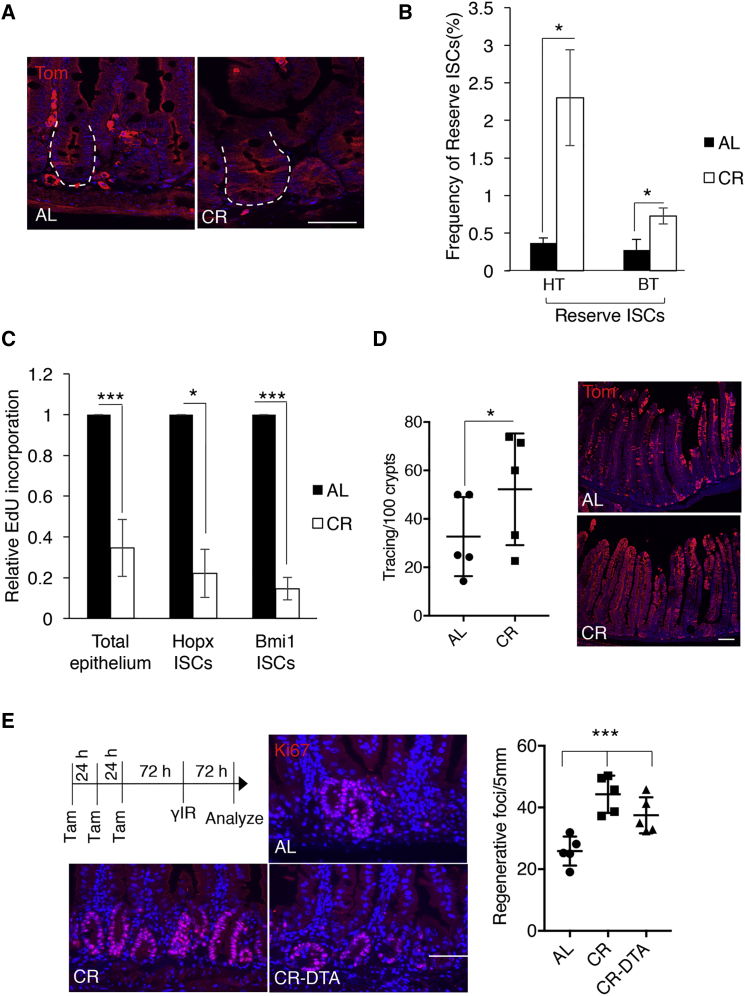
To assess the effects of CR on reserve ISCs, we reduced the caloric intake of mice harboring Hopx-CreER::Rosa26LSL-tdTomato reporter alleles (HT mice) by 40% for a period of 4–6 weeks starting at 2 months of age. Consistent with prior reports (Li et al., 2014, Takeda et al., 2011), we observed that 18 hr following induction of Hopx-CreER by tamoxifen injection in HT mice, single reserve ISCs were marked above the crypt base of ad libitum (AL)-fed mice. Interestingly, CR dramatically increased (514%) the number of cells marked by Hopx-CreER in HT mice (Figures 1A, 1B, and S1A). To investigate whether the increased number of tdTomato+ cells was a result of reserve ISC expansion or promiscuous activation of the Hopx locus, we examined the effect of CR on reserve ISCs marked with an independent allele, Bmi1-CreER::Rosa26LSL-tdTomato (BT) mice after tamoxifen induction. Bmi1-CreER and Hopx-CreER mark a largely overlapping (60%) population of reserve ISCs; however, Hopx-CreER marks a more homogeneous population relative to Bmi1-CreER (Li et al., 2014, Takeda et al., 2011, Tian et al., 2011, Yan et al., 2012). Consistent with this, we observed a robust but smaller increase (165%) in the number of cells marked in BT mice, indicating that CR expands the pool of reserve ISCs (Figure 1B). Next, we measured proliferation of reserve ISCs following CR using 5-ethynyl-2′-deoxyuridine (EdU) incorporation assays. Counterintuitively, we observed a reduction in proliferation of reserve ISCs, as well as the expected decreased proliferation of the bulk epithelium (Figure 1C). This suggests that the expansion observed in the reserve ISC pool may be relative, as CR decreases the overall cellular mass of the intestine (Yilmaz et al., 2012), or may also reflect a potential increase in self-renewal versus commitment to cell fate-determining decisions (Li et al., 2014) in these cells under CR conditions.
Figure 1.
Calorie Restriction Expands the Pool of Radioresistant Reserve Intestinal Stem Cells
(A) Immunofluorescence staining for tdTomato (red) on jejunal sections from Hopx-CreER::LSL-tdTomato ad libitum (AL)-fed and calorie-restricted (CR) mice 18 hr after a single tamoxifen injection. Epithelial crypts are outlined by a white dashed line (n = 4–6 mice). Scale bar, 50 μm.
(B) Analysis of reserve ISC frequency in CR and AL-fed Hopx-CreER::LSL-tdTomato and Bmi1-CreER::LSL-tdTomato mice using flow cytometry, 18 hr after tamoxifen injection (n = 4–6 mice, ∗p < 0.05). Details of gating strategy can be found in Figure S1A.
(C) Analysis of EdU incorporation in reserve ISCs and crypt cells of AL-fed and CR Hopx-CreER::LSL-tdTomato and Bmi1-CreER::LSL-tdTomato mice using flow cytometry, 18 hr after tamoxifen injection and 4 hr after EdU injection. The proliferation of CR groups is normalized to their control AL-fed counterparts (n = 4–6 mice, ∗p < 0.05 and ∗∗∗p < 0.0005).
(D) Immunofluorescence staining for tdTomato (red) and quantification of lineage-tracing events (ribbons of tdTomato+ cells with contiguous tracing from crypts, through crypt-villus junction, and into villi) from Hopx-CreER+ ISCs 14 days after marking reserve ISCs with a single tamoxifen injection given to Hopx-CreER::LSL-tdTomato mice under AL and CR diets (n = 5–6 mice, ∗p < 0.05), Scale bar, 100 μm.
(E) Representative Ki67(red)-stained sections from jejunum of irradiated Hopx-CreER::LSL-DTA mice and their control counterparts (LSL-DTA) mice after 6 weeks of receiving CR or AL diet and quantification of the number of clonal proliferative crypt foci per unit length of small intestine. All groups of mice were given three daily consecutive doses of tamoxifen and were exposed to 12 Gy γ-IR 3 days after the final dose. Tissue was harvested 3 days after γ-IR (n = 5 mice, ∗∗∗p < 0.0005). Scale bar, 100 μm.
Error bars represent ±SD from the mean. See also Figures S1 and S2.
To assess stem cell activity within the expanded pool of reserve ISCs, we performed lineage-tracing experiments in HT mice. Under basal conditions in AL-fed mice, roughly 25% of reserve ISCs are in cycle at any given time, giving rise to all cell types in the intestinal epithelium, including CBCs, while the remainder reside in G0 (Li et al., 2014, Sangiorgi and Capecchi, 2008, Takeda et al., 2011, Tian et al., 2011). Considering the 514% increase in frequency of Hopx-CreER+ ISCs and the 75% decrease in their proliferation following CR, we would predict a roughly 1.5-fold increase in lineage-tracing events from these cells. Experimentally, we observed a 1.6-fold increase in tracing events emanating from reserve ISCs (defined as contiguous tdTomato+ ribbons emanating from crypts and passing through the crypt-villus junction into villi) in CR mice relative to AL-fed counterparts (Figure 1D), confirming stem cell activity of the expanded pool of Hopx-CreER+ cells. Performing analogous experiments in actively cycling CBCs marked by Lgr5-CreER-IRES-eGFP revealed no statistically significant differences in their frequency, proliferation, or stem cell activity in vivo (Figures S1B–S1D).
Next, we aimed to assess the functional contribution of the expanded reserve ISC pool to the enhanced epithelial regeneration observed upon CR. We bred the Rosa26LSL-DTA allele (Voehringer et al., 2008) into Hopx-CreER mice (HD mice). In HD mice, a floxed stop cassette prevents expression of the diphtheria toxin fragment A (DTA) such that tamoxifen injection results in selective ablation of Hopx-CreER+ cells. After 6 weeks of CR, we injected three doses of tamoxifen to HD mice and their control counterparts (lacking Hopx-CreER), then exposed them to 12 Gy of γ-irradiation (γ-IR) 3 days after the last dose of tamoxifen. We assessed regeneration efficiency by scoring the number of clonal regenerative foci per unit length of the intestine on histological sections. As previously reported (Yilmaz et al., 2012), we observed that CR enhanced epithelial regeneration following injury relative to AL-fed mice. Approximately 40% of this enhanced regenerative capacity was dependent on the presence Hopx-CreER+ reserve ISCs, emphasizing the functional importance of these cells in mediating beneficial effects of CR (Figures 1E and S2A). We further confirmed that ablation of these cells in the absence of injury has no discernible effects on tissue homeostasis and basal proliferation, as expected based on their low frequency (around 1% of the crypt epithelium) and residence in G0 in the absence of injury (Li et al., 2014) (Figure S2B). The lack of complete abrogation of CR-enhanced regeneration in HD mice may be due to contributions of additional cell populations, incomplete ablation of reserve ISCs due to inefficiencies of the tamoxifen-diphtheria toxin system, or a combination of these.
Calorie Restriction Downregulates mTORC1 Signaling in Reserve ISCs
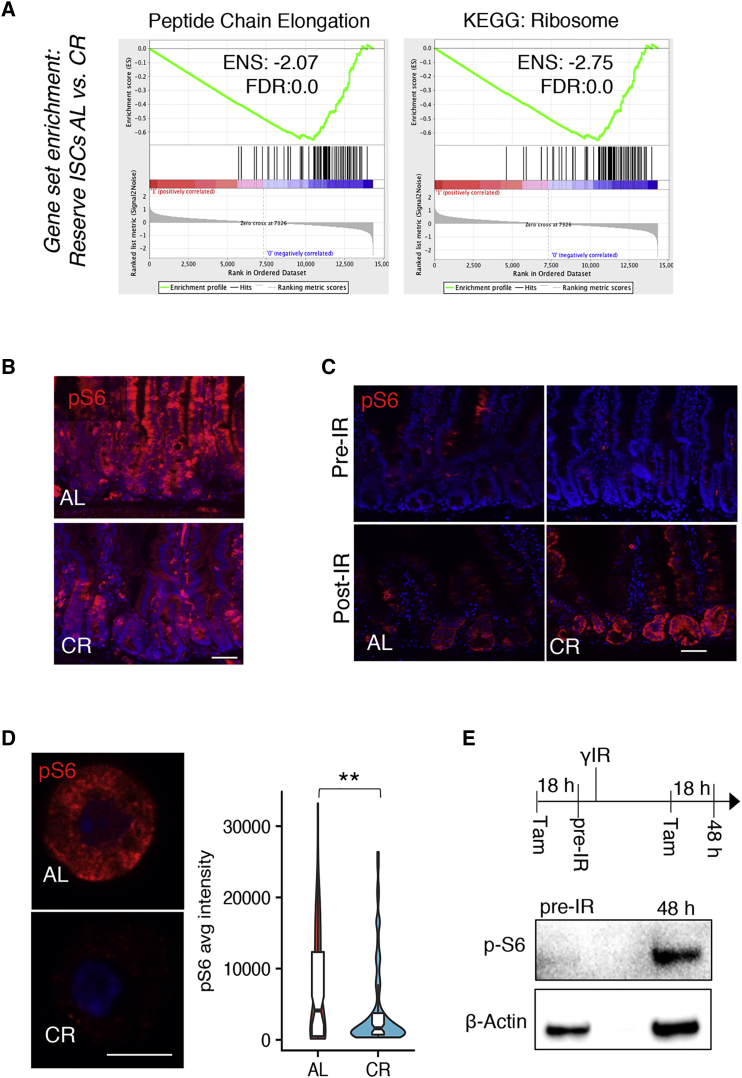
To understand the molecular mechanisms underlying enhanced tissue regeneration from reserve ISCs following CR, we performed RNA sequencing on FACS-purified populations of both active CBCs and reserve ISCs from control (AL) and CR mice (Table S1). We observed that the effects of CR on gene expression in these two cell types was largely distinct, with only four transcripts regulated similarly in both populations (Figure S3A and Table S1). Gene set enrichment analysis (GSEA) of reserve ISC transcriptome profiles revealed that genes encoding ribosomal proteins and proteins involved in translation were among most highly depleted gene sets in reserve ISCs of CR mice relative to AL-fed controls (Figure 2A). Ribosome biogenesis is a primary determinant of cellular translational capacity and accordingly has an essential role in the control of cell growth. mTORC1 is one of the major pathways coupling nutrient availability to the regulation of ribosome biogenesis and has been reported to regulate effects of CR in a number of somatic tissues (Dibble and Manning, 2013, Igarashi and Guarente, 2016, Powers and Walter, 1999, Yilmaz et al., 2012). Thus, we aimed to assess mTORC1 activity in reserve ISCs following CR.
Figure 2.
mTORC1 Activity Is Precisely Regulated during the Regenerative Response of the Intestinal Epithelium following Radiation Injury
(A) GSEA of the reserve ISC transcriptome isolated from Hopx-CreER::LSL-tdTomato mice under AL versus CR conditions.
(B and C) Immunofluorescence staining for pS6 (red) in the jejunum of CR and AL-fed mice under basal conditions (pre-IR) and 3 days after radiation injury. The exposure time in (B) is higher (5 s) than that in the top panels of (C) (1 s) in order to clearly highlight differences in pS6 levels under basal conditions (n = 5 mice). Scale bar, 50 μm.
(D) Immunofluorescence staining for pS6 and quantification of pS6 fluorescence intensity in single reserve ISCs from Hopx-CreER::LSL-tdTomato AL-fed and CR mice, 18 hr after tamoxifen injection (n = 3 mice, 30–40 ISCs scored per animal, ∗∗p < 0.005). Scale bar, 5 μm.
(E) Western blot analysis for pS6 in FACS-purified reserve stem cells from Hopx-CreER::LSL-tdTomato mice under basal conditions and 48 hr after irradiation injury. Both groups were given tamoxifen 18 hr before tissue harvest. In total, 21,000 cells were loaded per lane (representative blot from n = 3 mice).
We used phosphorylation of ribosomal protein S6 (pS6) by the mTORC1 target S6 kinase as a readout for mTORC1 activity (Morley and Traugh, 1990). CR repressed mTORC1 signaling under basal conditions (Figure 2B). Although it is well established that mTORC1 activity is dispensable for intestinal homeostasis, recent studies reported that mTORC1 activation is required during regeneration of the intestinal epithelium after DNA-damaging injury (Faller et al., 2015, Sampson et al., 2016). Interestingly, despite their lower mTORC1 activity under basal conditions, CR mice were fully capable of robust mTORC1 activation in regenerative foci in response to radiation injury to an extent greater than their AL-fed counterparts (Figure 2C). We confirmed that CR specifically represses mTORC1 in reserve ISCs by quantifying pS6 levels in fluorescence-activated cell sorting (FACS)-sorted Hopx-CreER+ ISCs from HT mice. This revealed that under AL-fed conditions, the majority of reserve ISCs exist in an mTORC1-low state (Figure 2D); however, a group mTORC1-high cells are clearly present, consistent with prior findings that roughly 25%–30% of reserve ISCs are active and in cycle during homeostasis (Li et al., 2016b, Yousefi et al., 2016). Upon CR, reserve ISCs shifted to a more inactive, mTORC1-low state, although few mTORC1-high cells remain present (Figure 2D). We next tested the ability of radiation damage to induce mTORC1 activity specifically in reserve ISCs and, consistent with immunostaining results, found low levels of pS6 in these cells in the basal state (Figure 2E). S6 phosphorylation was markedly induced in reserve ISCs 48 hr after radiation, a time when the first cell divisions are initiated in reserve ISCs (Figure 2E and Yan et al., 2012). These data led us to hypothesize that dynamic regulation and expanded response range of mTORC1 activity in reserve ISCs in CR mice is a major contributing factor to the enhanced tissue regeneration observed after caloric restriction.
Precise Control of mTORC1 Activity Is Required for Efficient Tissue Regeneration
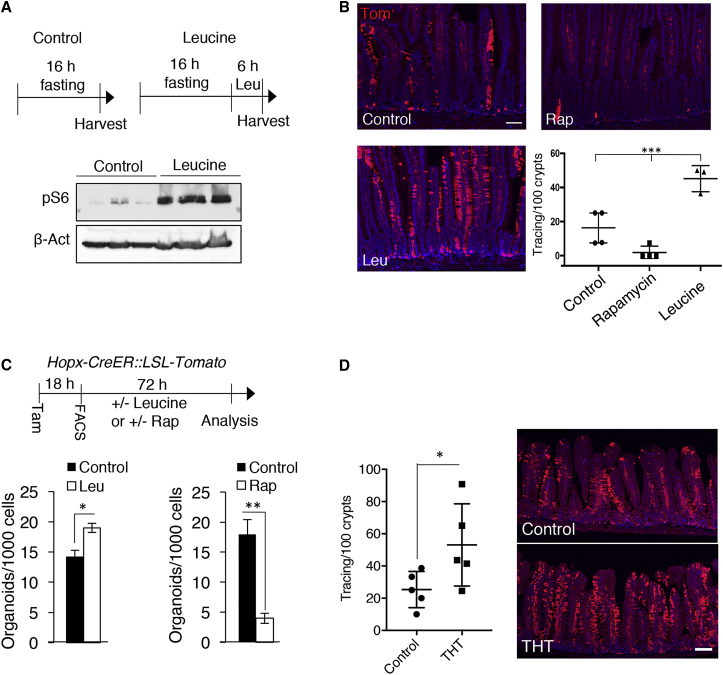
To further functionally validate our hypothesis, we tested the ability of leucine, a branched-chain amino acid known to be among the most potent nutrient agonists of the mTORC1 complex in vitro (Fox et al., 1998), to activate mTORC1 in vivo. Western blotting of crypt extracts after 1.5% leucine administration in drinking water confirmed the ability of this amino acid to potentiate mTORC1 activity in vivo (Figure 3A). Next, we assessed the effect of mTORC1 activation by leucine, as well as mTORC1 inhibition by rapamycin treatment, on lineage tracing from reserve ISCs 2 weeks after reporter activation in HT mice. Leucine stimulation dramatically increased the number of tracing events from reserve ISCs and rapamycin had the opposite effect, demonstrating both the necessity and sufficiency of mTORC1 signaling for activation of reserve ISCs in vivo (Figure 3B). To assess whether the effect of mTORC1 modulation on reserve ISC activation was cell autonomous, we FACS-purified reserve ISCs and tested the intestinal organoid-forming capacity of single cells in the presence of leucine or rapamycin in the culture media. Consistent with our in vivo results, in vitro addition of leucine increased organoid formation efficiency while rapamycin had the opposite effect, indicating that mTORC1 activity regulates reserve ISC activation in a cell-autonomous manner (Figures 3C and S3B). The repressive effects of rapamycin were markedly stronger than the inductive effects of leucine, possibly due to the presence of high levels of potent ISC mitogens and known mTORC1 agonists in the culture media, including Wnt, Notch, and epidermal growth factor ligands, as well as bone morphogenetic protein antagonists.
Figure 3.
mTORC1 Modulates Activation of Reserve ISCs in a Cell-Autonomous Manner
(A) Western blot analysis for pS6 in protein extracted from the intestinal epithelium of a cohort of mice that were fasted for 18 hr and then divided into two groups. One group was harvested after fasting and the other group received 1.5% leucine in the drinking water for 6 hr following fasting (n = 3 mice).
(B) Lineage tracing from Hopx-CreER+ ISCs using immunofluorescence staining for tdTomato in sections from Hopx-CreER::LSL-tdTomato and control mice 2 weeks after tamoxifen treatment. Rapamycin and leucine treatments started 3 days before tamoxifen injection and continued until time of harvest (n = 3–5 mice, ∗∗∗p < 0.0005). Scale bar, 100 μm.
(C) Organoid formation assays from single FACS-purified Hopx-CreER+ ISCs from Hopx-CreER::LSL-tdTomato mice that received one dose of tamoxifen 18 hr before tissue harvest. Sorted cells were treated with 10 μM leucine or rapamycin for 72 hr in culture (n = 3 replicate wells from each of two individual animals, ∗p < 0.05 and ∗∗p < 0.005). Images of organoid cultures are presented in Figure S3B.
(D) Lineage tracing from reserve ISCs in Hopx-CreER::Tsc1flox/flox::LSL-tdTomato (THT) mice visualized with immunofluorescence staining for tdTomato in sections from the jejunum 2 weeks after tamoxifen treatment (n = 5 mice, ∗p < 0.05). Scale bar, 100 μm.
Error bars represent ±SD from the mean. See also Figure S3.
To further confirm the cell-autonomous effect of mTORC1modulation on activation of reserve ISCs in vivo, we genetically activated mTORC1 specifically in Hopx-CreER+ reserve ISCs. Hopx-CreER::Rosa26LSL-tdTomato reporter alleles were bred into mice harboring floxed alleles of Tsc1 (Tsc1flox/flox::Hopx-CreER::Rosa26LSL-tdTomato, THT mice) (Kwiatkowski et al., 2002). Consistent with results using leucine, there were increased lineage-tracing events from reserve ISCs of THT mice 2 weeks after Hopx-CreER induction of reporter activity and Tsc1 deletion (Figure 3D). Conversely, inhibition of mTORC1 activity in reserve ISCs via Raptor inactivation in Raptorflox/flox::Hopx-CreER::Rosa26LSL-tdTomato (RHT) mice abrogated lineage-tracing events from reserve ISCs (Figure S3C), confirming the functional importance of cell-autonomous mTORC1-mediated regulation of reserve ISC activity.
mTORC1 Modulation Regulates the Intestinal Response to High-Dose γ-IR Injury
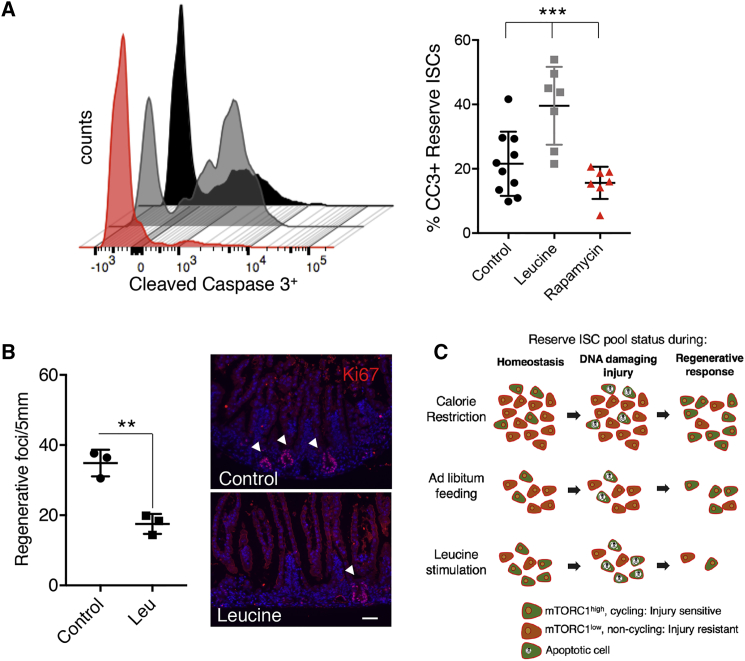
Ultimately, we tested whether inhibition of mTORC1 in reserve ISCs, as we observed in CR mice, has a radioprotective role at the time of injury. We assessed apoptotic cell death in response to radiation in reserve ISCs of mice that were treated with either leucine or rapamycin beginning 3 days prior to radiation exposure. Interestingly, inhibition of mTORC1 signaling prior to injury protected reserve ISCs against apoptosis, while stimulation of mTORC1 during this same period sensitized reserve ISCs to ionizing radiation (Figure 4A). We then assessed the effect of mTORC1 modulation prior to radiation exposure on the subsequent regenerative response. When mTORC1 was induced through administration of leucine in drinking water starting 3 days prior to radiation injury, the epithelial regenerative response was significantly impaired (Figure 4B). We observed similar regenerative failure using reserve ISC-specific genetic ablation of Tsc1 prior to ionizing radiation exposure (Figure S4A). Similarly, ISC-specific inhibition of mTORC1 did not affect tissue homeostasis under basal conditions but impaired regeneration following injury (Figures S4B and S4C).
Figure 4.
Premature Activation of mTORC1 Sensitizes Reserve ISCs to Radiation Injury and Results in Regeneration Failure
(A) Flow-cytometric analysis of apoptosis in reserve ISCs from Hopx-CreER::LSL-tdTomato mice by staining for cleaved caspase-3, 2 hr after 12 Gy γ-IR. Rapamycin and leucine treatments were started 3 days before the time of harvest, and all groups were given a single tamoxifen dose 18 hr prior to tissue harvest (n = 7–10 mice, ∗∗∗p < 0.0005).
(B) Ki67(red) staining and quantification of the regeneration efficiency after irradiation injury in mice receiving 1.5% leucine in their drinking water 3 days before γ-IR and control groups. Arrowheads point to regenerative crypt foci (n = 3 mice, ∗∗p < 0.005). Scale bar, 50 μm.
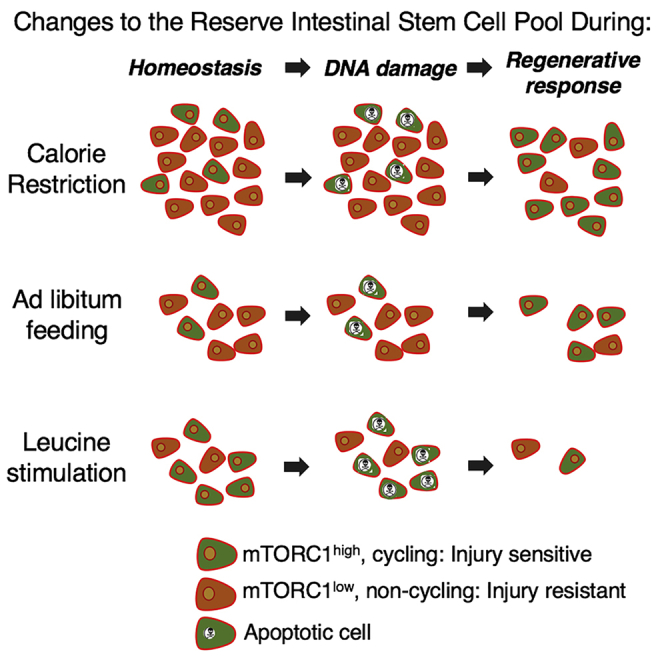
(C) Integrated model describing changes in the radioresistant reserve ISC pool in response to dietary modulation. CR increases the pool of reserve ISCs, while leucine stimulation drives activation of dormant ISCs (dormant: mTORC1-low, orange; active: mTORC1-high, green). Upon exposure to DNA-damaging injury, mTORC1-high ISCs undergo apoptosis. After injury, the surviving pool of ISCs is larger under CR, enabling a more robust regenerative response relative to AL-fed counterparts. In contrast, after leucine feeding, fewer ISCs survive the injury, resulting in regenerative failure.
Error bars represent ±SD from the mean. See also Figure S4.
Taken together, our findings demonstrate that reserve intestinal stem cell-autonomous activity of mTORC1 plays a significant functional role in governing the regenerative response of the epithelium to DNA-damaging injury and contributes to the beneficial effects of CR (Figure 4C).
Discussion
There is considerable interest in gaining a better understanding of the cellular basis underlying regeneration of the intestinal epithelium following DNA damage from radiation exposure, chemotherapy, microbiome dysbiosis, and ischemia-reperfusion injury, not only to enable strategies for targeting these cells to enhance regeneration but also because of the apparent parallels between intestinal regeneration and oncogenic transformation.
Previous studies suggested that CR improves regeneration of the intestinal epithelium following high-dose γ-IR through a non-cell-autonomous effect on actively cycling CBCs mediated by intercalating Paneth cells (Yilmaz et al., 2012). Given that WntHigh/Lgr5High CBCs are quantitatively ablated by high-dose γ-IR and that regeneration following radiation injury is driven by surviving WntLow/Off cells, the beneficial effects of CR are likely not mediated by active CBCs and likely rather reflect the activity of additional stem cell populations (Asfaha et al., 2015, Tao et al., 2015, Yan et al., 2012, Yousefi et al., 2016). This notion is further supported by recent demonstrations that Paneth cells, originally purported to be the niche for CBCs responsible for governing their activity, are dispensable for both CBC function and epithelial regeneration in response to radiation injury (Durand et al., 2012, Kabiri et al., 2014, Kim et al., 2012, San Roman et al., 2014).
Here, we show that radioresistant ISCs within Hopx-CreER-marked populations functionally contribute to the enhanced regenerative outcome following DNA-damaging injury in CR mice in an mTORC1-dependent manner. Diphtheria toxin-mediated ablation of these rare (roughly 1% of the crypt epithelium) radioresistant ISCs abrogates the beneficial effects of CR in response to DNA-damaging injury while having no discernible effects on the resting, homeostatic epithelium. These effects are cell autonomous and not an artifact of general tissue injury in response to diphtheria toxin activation, since genetic ablation of mTORC1 activity specifically in these cells results in similar regenerative failure only upon DNA-damaging injury.
We demonstrate that CR suppresses mTORC1 signaling in reserve ISCs and this, in turn, protects reserve ISCs against radiation injury. However, mTORC1 activation is required during regeneration following injury. Remarkably, despite the repression of mTORC1 in CR mice, their epithelium maintains the ability not only to activate mTORC1 in response to injury but does so to an extent greater than that seen in AL-fed animals. Curiously, a similar phenomenon has been observed in the context of acute fasting, whereby Pten inactivation through phosphorylation poises reserve ISCs to contribute to epithelial regeneration upon feeding (Richmond et al., 2015).
Ultimately, our findings emphasize the significance of precisely governed mTORC1 activity in protecting stem cells against injury and driving them to expand and repopulate the tissue when called upon. The discovery of the role of mTORC1 signaling in controlling reserve intestinal stem cell activity provides a framework for future studies aimed at the development of pharmacological or dietary interventions that delay ISC activation in an effort to protect patients against acute side effects of radiation therapy and radiation-induced acute gastrointestinal syndrome.
Experimental Procedures
Mouse Strains
We obtained Bmi1-CreER (JAX strain 010531), Tsc1 floxed (Jax mice strain 005680), Raptor floxed (Jax mice strain 013188), Lgr5-eGFP-IRES-CreERT2 (Jax mice strain 008875), and R26-CAG-LSL-tdTomato (JAX mice strain 007914) from the Jackson Laboratory. Hopx-CreER (JAX strain 017606) mice were generated at the Perelman School of Medicine, University of Pennsylvania. All mice were maintained on a C57/BL6N background.
Statistical Analysis
Shapiro-Wilk tests were used to assess normality of datasets. To compare differences in two groups, we calculated statistical significance using Student's t tests since all datasets passed the normality test. To compare differences in more than two groups, we used ANOVA for normal (Figures 1E and 4A) and a Kruskal-Wallis test (Figure 3A) for non-normal datasets. Error bars reflect SD. For animal studies, samples were only excluded from experiments if animals were considered unhealthy.
Study Approvals
All mouse protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania under protocol 803415, issued to Dr. Christopher Lengner.
Author Contributions
M.Y. and C.J.L. conceptualized the study, designed experiments, and wrote the manuscript. M.Y. conducted all experiments unless otherwise stated. A.N.D. performed and quantified experiments in Figure 3B and maintained all mouse strains used in this study. N.L. conducted organoid formation assays in Figures 3C and S3B. J.S. and R.J.C. assisted with mouse husbandry, feeding, and genotyping, as well as histological analysis. C.T.B. and K.P.S. performed transcriptome profiling analysis and graphical representation of quantitative data. Z.Q. contributed to manuscript editing and establishment of genetically modified mouse colonies. A.N.D., R.J.C., and C.T.B. contributed to editing of the manuscript.
Acknowledgments
We thank John Tobias at the Penn Genomic Analysis Core for assistance with analysis of transcriptome profiling data. M.Y. is a Howard Hughes Medical Institute International Student Research fellow (59107993). Z.Y. is supported by the National Natural Science Foundation of China no. 81772984 and SKLAB open grant 2018SKLAB-12. C.J.L. is supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK106309) and the National Cancer Institute (R01 CA168654). This work was supported in part by the Center for Molecular Studies in Digestive and Liver Diseases (NIH P30 DK050306) and its core facilities (Molecular Pathology and Imaging, Cell Culture, H-MARC, and Genetically Modified Mouse).
Published: February 22, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.026.
Accession Numbers
RNA sequencing data are available on the Gene Expression Omnibus (GEO) under accession number GEO: GSE109002.
Supplemental Information
References
- Asfaha S., Hayakawa Y., Muley A., Stokes S., Graham T.A., Ericksen R.E., Westphalen C.B., von Burstin J., Mastracci T.L., Worthley D.L. Krt19(+)/Lgr5(-) cells are radioresistant cancer-initiating stem cells in the colon and intestine. Cell Stem Cell. 2015;16:627–638. doi: 10.1016/j.stem.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Cerletti M., Jang Y.C., Finley L.W., Haigis M.C., Wagers A.J. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman R.J., Anderson R.M., Johnson S.C., Kastman E.K., Kosmatka K.J., Beasley T.M., Allison D.B., Cruzen C., Simmons H.A., Kemnitz J.W. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C.C., Manning B.D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand A., Donahue B., Peignon G., Letourneur F., Cagnard N., Slomianny C., Perret C., Shroyer N.F., Romagnolo B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc. Natl. Acad. Sci. USA. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller W.J., Jackson T.J., Knight J.R., Ridgway R.A., Jamieson T., Karim S.A., Jones C., Radulescu S., Huels D.J., Myant K.B. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox H.L., Pham P.T., Kimball S.R., Jefferson L.S., Lynch C.J. Amino acid effects on translational repressor 4E-BP1 are mediated primarily by L-leucine in isolated adipocytes. Am. J. Physiol. 1998;275:C1232–C1238. doi: 10.1152/ajpcell.1998.275.5.C1232. [DOI] [PubMed] [Google Scholar]
- Guarente L. Calorie restriction and SIR2 genes—towards a mechanism. Mech. Ageing Dev. 2005;126:923–928. doi: 10.1016/j.mad.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Houthoofd K., Vanfleteren J.R. The longevity effect of dietary restriction in Caenorhabditis elegans. Exp. Gerontol. 2006;41:1026–1031. doi: 10.1016/j.exger.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Igarashi M., Guarente L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- Kabiri Z., Greicius G., Madan B., Biechele S., Zhong Z., Zaribafzadeh H., Edison, Aliyev J., Wu Y., Bunte R. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141:2206–2215. doi: 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- Kim T.-H., Escudero S., Shivdasani R.A. Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. USA. 2012;109:3932–3937. doi: 10.1073/pnas.1113890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D.J., Zhang H., Bandura J.L., Heiberger K.M., Glogauer M., el-Hashemite N., Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Li N., Yousefi M., Nakauka-Ddamba A., Jain R., Tobias J., Epstein J.A., Jensen S.T., Lengner C.J. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 2014;3:876–891. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Yousefi M., Nakauka-Ddamba A., Tobias J.W., Jensen S.T., Morrisey E.E., Lengner C.J. Heterogeneity in readouts of canonical wnt pathway activity within intestinal crypts. Dev. Dyn. 2016;245:822–833. doi: 10.1002/dvdy.24423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Nakauka-Ddamba A., Tobias J., Jensen S.T., Lengner C.J. Mouse label-retaining cells are molecularly and functionally distinct from reserve intestinal stem cells. Gastroenterology. 2016;151:298–310.e7. doi: 10.1053/j.gastro.2016.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay C.M., Crowell M.F., Maynard L.A. The effect of retarded growth upon the length of life span and upon the ultimate body size. Nutrition. 1989;5:155–171. (January 1935) discussion 172. [PubMed] [Google Scholar]
- Montgomery R.K., Carlone D.L., Richmond C.A., Farilla L., Kranendonk M.E., Henderson D.E., Baffour-Awuah N.Y., Ambruzs D.M., Fogli L.K., Algra S. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley S.J., Traugh J.A. Differential stimulation of phosphorylation of initiation factors eIF-4F, eIF-4B, eIF-3, and ribosomal protein S6 by insulin and phorbol esters. J. Biol. Chem. 1990;265:10611–10616. [PubMed] [Google Scholar]
- Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C.A., Shah M.S., Deary L.T., Trotier D.C., Thomas H., Ambruzs D.M., Jiang L., Whiles B.B., Rickner H.D., Montgomery R.K. Dormant intestinal stem cells are regulated by pten and nutritional status. Cell Rep. 2015;13:2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson L.L., Davis A.K., Grogg M.W., Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Roman A.K., Jayewickreme C.D., Murtaugh L.C., Shivdasani R.A. Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Reports. 2014;2:127–134. doi: 10.1016/j.stemcr.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S., Tang D., Morita Y., Sperka T., Omrani O., Lechel A., Sakk V., Kraus J., Kestler H.A., Kühl M. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2015;34:624–640. doi: 10.15252/embj.201490700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Biehs B., Warming S., Leong K.G., Rangell L., Klein O.D., de Sauvage F.J. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D., Liang H.E., Locksley R.M. Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J. Immunol. 2008;180:4742–4753. doi: 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Ö.H., Katajisto P., Lamming D.W., Gültekin Y., Bauer-Rowe K.E., Sengupta S., Birsoy K., Dursun A., Yilmaz V.O., Selig M. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi M., Li N., Nakauka-Ddamba A., Wang S., Davidow K., Schoenberger J., Yu Z., Jensen S.T., Kharas M.G., Lengner C.J. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J. Cell Biol. 2016;215:401–413. doi: 10.1083/jcb.201604119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.