Abstract
The earliest autoantibodies in lupus are directed against the RNA binding autoantigen Ro60, but the triggers against this evolutionarily conserved antigen remain elusive. We identified Ro60 orthologs in a subset of human skin, oral, and gut commensal bacterial species and confirmed the presence of these orthologs in patients with lupus and healthy controls. Thus, we hypothesized that commensal Ro60 orthologs may trigger autoimmunity via cross- reactivity in genetically susceptible individuals. Sera from human anti-Ro60–positive lupus patients immunoprecipi-tated commensal Ro60 ribonucleoproteins. Human Ro60 autoantigen–specific CD4 memory T cell clones from lupus patients were activated by skin and mucosal Ro60-containing bacteria, supporting T cell cross-reactivity in humans. Further, germ-free mice spontaneously initiated anti-human Ro60 T and B cell responses and developed glomerular immune complex deposits after monocolonization with a Ro60 ortholog–containing gut commensal, linking anti- Ro60 commensal responses in vivo with the production of human Ro60 autoantibodies and signs of autoimmunity. Together, these data support that colonization with autoantigen ortholog-producing commensal species may initiate and sustain chronic autoimmunity in genetically predisposed individuals. The concept of commensal ortholog cross-reactivity may apply more broadly to autoimmune diseases and lead to novel treatment approaches aimed at defined commensal species.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a chronic, debilitating, multi-organ autoimmune disease with unclear etiology. Almost all patients with SLE have high titers of anti-nuclear autoantibodies, which can be detected years before the onset of symptoms (1). Anti-Ro antibodies are present in about 50% of patients with SLE, up to 90% of patients with subacute cutaneous lupus erythematosus (SCLE), >90% of newborns with neonatal lupus erythematosus (NLE) (2), and up to 80% of patients with Sjögren’s syndrome. Two distinct Ro antigens have been identified, Ro60 and Ro52, which differ substantially in structure and function. Although both antibodies can be found in patients with SLE, anti-Ro60 is the earliest and most common preclinical anti-nuclear antibody (1, 3). In addition, anti-Ro antibodies are pathogenic, as evidenced by the trans-placental spread of these antibodies in NLE, leading to potentially fatal cardiac conduction defects and cutaneous lesions similar to SCLE (4–7). Hence, identifying targetable triggers of anti-Ro60 antibodies would be beneficial for alleviating a variety of lupus manifestations and provide new insights into the pathogenesis of this disease. The fact that the incidence of SLE has tripled in the last 50 years underscores the need for fundamentally new approaches and also suggests that genetic factors alone may not be sufficient to explain disease pathology.
The Ro60 protein is a ring-shaped RNA binding protein that forms ribonucleoprotein (RNP) complexes with ~100-nucleotide noncoding RNAs (ncRNAs) called Y RNAs. Because Ro60 also binds certain mis-folded ncRNAs, it is proposed to function in ncRNA surveillance (5). A recent study suggests that endogenous Alu retroelements are also Ro60 targets in human cells (8). Most human anti-Ro60 autoantibodies bind epitopes that overlap with the RNA binding sites (6). Ro60 appears to have an important role in the environmental stress response; mammalian and bacterial cells lacking Ro60 are more sensitive to ultraviolet (UV) irradiation (9, 10), similar to the UV sensitivity seen in lupus patients. Immunization against human Ro60 (hRo60) protein in mice leads to intermolecular epitope spreading with subsequent production of antibodies against Ro52, La, Smith, and U1RNP (11), providing further evidence for the role of early anti-Ro60 auto-antibodies in disease initiation and progression in systemic autoimmunity. Mice lacking Ro60 develop a lupus-like syndrome with autoantibodies, nephritis, and photosensitivity (12), possibly due to the pathogenic accumulation of defective and excess ncRNAs and subsequent activation of Toll-like receptors (TLRs) (13). Thus, identifying targetable triggers of anti-Ro60 autoimmunity may improve our understanding of the pathogenesis of lupus, may serve as a paradigm for other systemic autoimmune diseases, and could be useful for developing therapies that prevent or alleviate lupus manifestations.
Ro60 is highly evolutionarily conserved, with orthologs found in taxa ranging from vertebrates to bacteria, and even bacteriophages (5, 14, 15). Relevant to human biology, multiple commensal bacteria identified on the skin, oral mucosa, and gut encode Ro60 orthologs with high sequence similarity to hRo60, including certain species of Corynebacterium, Propionibacterium, and Bacteroides (5, 14). Studies of the normal human gut have shown that the generation of commensal- reactive antibodies and T cell receptors (TCRs) is both abundant and physiologic (16, 17). Thus, the trillions of skin, oral, and gut bacteria that comprise the human microbiota provide a major source for antigenic variation that, to our knowledge, has never been explored for the possibility of ortholog cross-reactivity in autoimmunity. We hypothesized that autoimmune-predisposed individuals with the right combination of genetic susceptibility and environmental exposures who are chronically colonized by Ro60 commensals may develop antibodies against a bacterial Ro60 ortholog that leads to autoimmunity via cross- reactivity and epitope spreading. Moreover, the persistence of Ro60 orthologs on the skin or mucosa could sustain chronic autoantibody production by repetitively stimulating short-lived Ro60-specific B cell clones (18). Fluctuations of the load of Ro60 ortholog–containing bacteria in their respective niches could also be involved in disease flares. Although not live organisms, microbial peptides have previously been implicated as potential chronic triggers of lupus: the Epstein-Barr virus antigen EBNA-1 was reported to cross-react with the primary Ro60 B cell epitope (19), and cross-reactivity of murine T cell hybridomas was shown between hRo60 and commensal bacterial peptides (20), providing evidence that cross-reactive mechanisms may be at play in lupus pathogenesis.
Here, we show that Ro60 ortholog–containing commensal bacteria are common in the human skin, oral, and gut microbiota, and that lupus patients with Ro60 autoantibodies demonstrate T cell and B cell responses to commensal Ro60 bacterial orthologs in vitro. Moreover, monocolonization of the mouse gut with a common hRo60 ortholog–producing commensal leads to anti-Ro60 T and B cell responses in vivo. Together, these data suggest that Ro60 orthologs from human commensal bacteria may initiate and drive lupus pathogenesis.
RESULTS
Identification of human commensal Ro60 orthologs
GenBank was used to identify all known potential Ro60 orthologs in bacterial species. This list was cross-referenced with the Pathosystems Resource Integration Center (PATRIC) (21) and the Human Microbiome Project (22), resulting in several potential cross-reactive species within human microbiomes (table S1). Commensal bacterial genomes were searched for the presence of the most common bacterial Y RNA, YrlA (Y RNA–like A) RNA, as described (14), which was identified in most species (table S1). The full-length human and com-mensal Ro60 proteins were aligned (fig. S1), and a phylogenetic tree was constructed (Fig. 1A) to compare the sequence homology of commensals with each other and with mammalian Ro60. Bacterial Ro60 orthologs were identified from a wide variety of anatomical niches, including the gut, skin, oral, and nasopharyngeal cavities, airways, and urogenital tract (Fig. 1A).
Fig. 1. The human commensal microbiota contains multiple species with Ro60 orthologs.
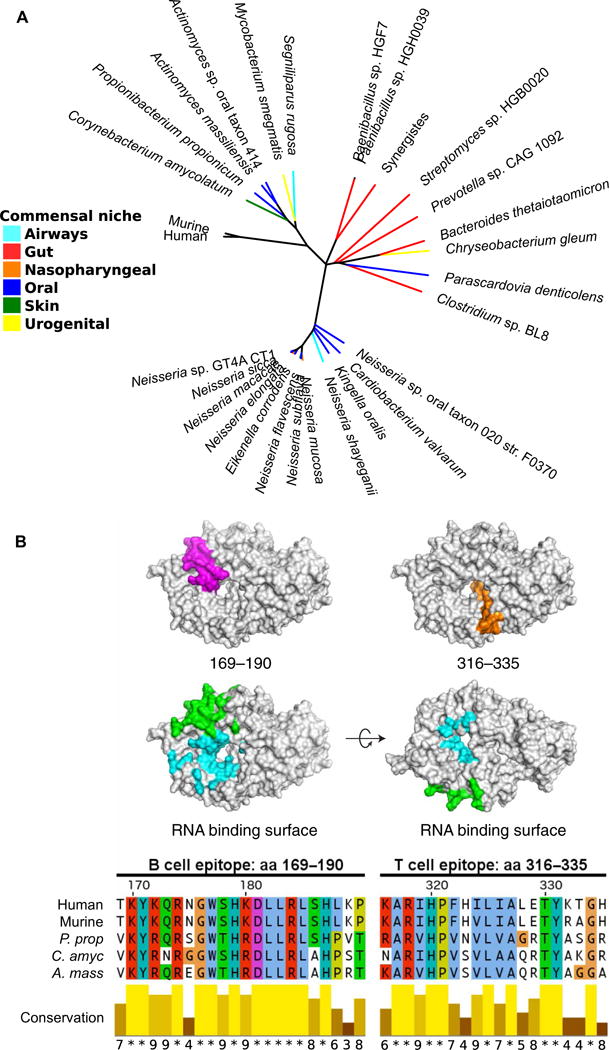
(A) Phylogenetic tree showing commensal bacterial ortholog Ro60 sequence homology to hRo60, with commensal niches identified by colors of each branch (see legend). (B) Structure of hRo60 with a major B cell epitope [amino acids (aa) 169 to 190] mapped in pink and T cell epitope (amino acids 316 to 335) mapped in orange, and the protein sequence alignments between human and commensal Ro60 at these epitopes compared below. Green color indicates residues that interact with Y RNA. Blue color indicates residues that interact with misfolded RNA. P. prop, P. propionicum; C. amyc, C. amycolatum; A. mass, A. massiliensis.
The earliest and most common Ro60 B cell epitope identified in lupus patients is the region between amino acids 169 and 190 (19, 23). A protein BLAST (Basic Local Alignment Search Tool) search of this peptide sequence against all bacterial genomes specifically identified the bacterial Ro60 orthologs as containing the most similar peptide sequences. Alignment of hRo60 peptide 169–190 with the commensal Ro60 orthologs showed very high amino acid sequence similarity at this epitope (Fig. 1B). Concordantly, alignment with the T cell epitope between amino acids 316 and 335, found in mice to initiate epitope spreading (11), also exhibits high sequence homology with commensal Ro60 orthologs (Fig. 1B). The conserved amino acids may act as anchor residues that allow for B and T cell cross-reactivity between human and bacterial Ro60.
Commensal Ro60 orthologs in human subjects
Subjects with SLE/SCLE and Ro60 autoantibodies (n = 8), SLE without Ro60 autoantibodies (n = 5), and healthy controls (n = 7) completed up to three longitudinal study visits, each 1 month apart, in which microbiome samples were collected from the buccal mucosa, skin over the manubrium, and stool. Blood samples were collected for serum, plasma, and peripheral blood mononuclear cells (PBMCs). A detailed health and medication history and 24-hour diet history was completed by all subjects at each visit (Table 1 and tables S2 and S3). The presence or absence of Ro60 autoantibodies was confirmed by enzyme-linked immunosorbent assay (ELISA) and by immunoprecipitation of Ro60–Y RNA complexes (fig. S2), which had concordant results. Human leukocyte antigen (HLA) typing was performed for the lupus risk alleles HLA-DR3 and HLA-DR15 (Table 1) (24).
Table 1. Human subject cohort and clinical characteristics.
SCLE02, SCLE03, SCLE05, and SCLE06 were de-identified serum or biopsy samples, so additional clinical information was unavailable. Ro60 column indicates the presence or absence of anti-Ro60 serum IgG. F, female; M, male; HCQ, hydroxychloroquine; NA, not available. Additional clinical information is available in tables S2 and S3.
| Subject | Diagnosis | Sex | Age (years) | HLA-DR3 | HLA-DR15 | Ro60 | Immunosuppressive medications | HCQ |
|---|---|---|---|---|---|---|---|---|
| SLE01 | SLE | F | 40 | − | − | + | Azathioprine | − |
| SLE02 | SLE | F | 47 | + | − | + | Rituximab Prednisone 5 mg daily | − |
| SLE03 | SLE | F | 49 | − | − | − | Mycophenolate mofetil Prednisone 7.5 mg daily | + |
| SLE04 | SLE | F | 29 | − | − | + | Discontinued mycophenolate mofetil and prednisone against medical advice 1 month prior | − |
| SLE05 | SLE | F | 49 | − | + | + | Prednisone 1 mg daily | + |
| SLE06 | SLE | F | 40 | − | + | − | Mycophenolate mofetil Prednisone 5 mg daily | + |
| SLE07 | SLE | F | 31 | − | − | − | — | + |
| SLE08 | SLE | F | 52 | − | − | + | — | + |
| SLE09 | SLE | F | 34 | + | − | − | — | + |
| SLE10 | SLE | F | 47 | + | + | + | — | + |
| SLE11 | SLE | F | 31 | + | + | − | Mycophenolic acid Prednisone 10 mg daily | + |
| SLE12 | DLE | F | 54 | + | − | − | — | + |
| SLE13 | SLE | F | 26 | + | − | + | Mycophenolic acid Methotrexate Prednisone 7.5 mg daily | + |
| SLE14 | SLE | F | 32 | − | − | − | — | − |
| SLE15 | SLE | F | 44 | − | + | − | — | − |
| SLE18 | SLE | F | 33 | − | − | + | NA | NA |
| SCLE01 | SCLE | F | 75 | + | − | + | Azathioprine | + |
| SCLE04 | SCLE | M | 72 | − | − | + | — | − |
High-throughput sequencing of the 16S ribosomal DNA (rDNA) V4 region was performed from the fecal, oral, and skin microbiomes of each study subject. Whereas principal coordinates analysis of weighted UniFrac distances showed expected differences in β-diversity between fecal, oral, and skin microbiomes, no clustering of anti- Ro60–positive versus anti-Ro60–negative patients was observed within different anatomic sites (Fig. 2A). The Shannon-Weiner diversity index was used to calculate α-diversity, and mean values were similar between anti-Ro60–negative and anti-Ro60–positive subjects within each site (fig. S3). Anti-Ro60–negative lupus subjects had slightly lower mean α-diversity values as compared to healthy controls in the fecal and oral samples, but no statistically significant differences in mean α-diversity were calculated between healthy and anti-Ro60–positive subjects. Thus, measures of intra- and inter-sample microbial diversity were similar between anti-Ro60–positive and anti-Ro60–negative patients.
Fig. 2. Ro60 commensal bacteria are common among lupus and healthy subjects without overt dysbiosis of the fecal, oral, or skin microbiome.
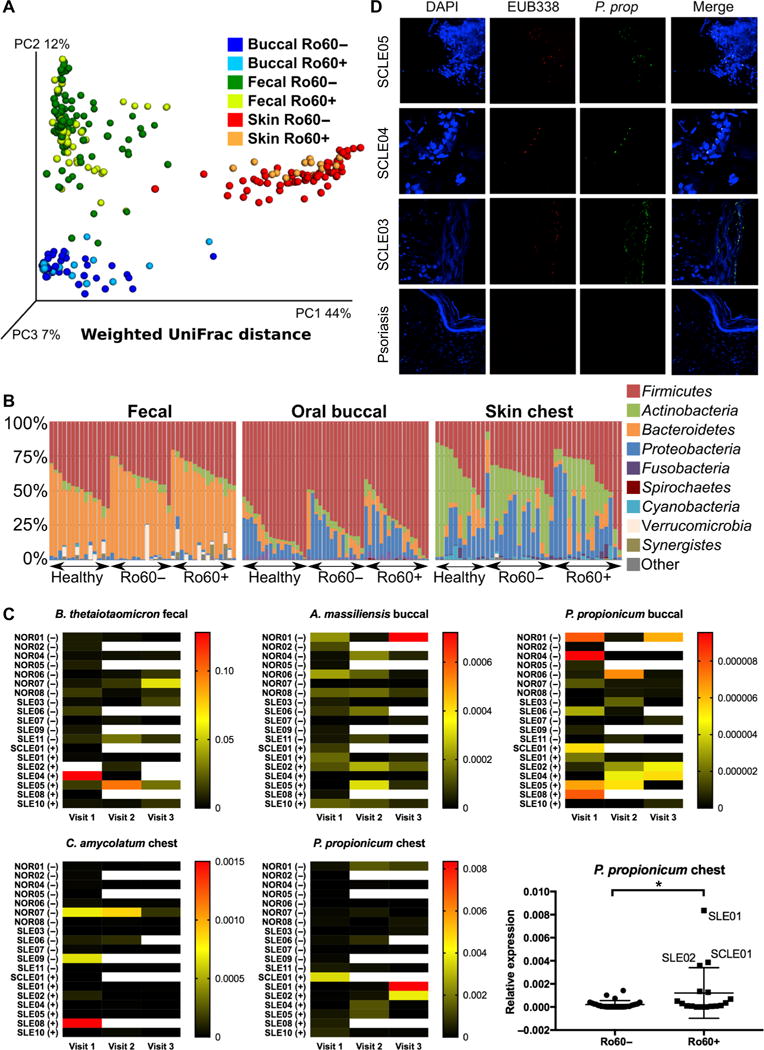
(A) 16S V4 sequencing was performed from the fecal, oral, and skin microbiomes of 15 subjects with lupus and 7 healthy controls. Principal coordinates analysis of weighted UniFrac distances represents β-diversity by body site but not by the presence of serum anti-Ro60 antibodies. (B) Vertical bars represent relative abundance of microbial phyla in individual fecal, oral, and skin microbiome samples. Legend indicates most abundant phyla. No significant differences in linear discriminant analysis effect size were found between groups. (C) Heat map of relative abundance of four Ro60 commensal bacteria from human microbiome samples measured by bacterial Ro60-specific qPCR. Each row represents a study subject: healthy (NOR), SLE, or SCLE. (+) or (−) indicates serum anti-Ro60 immunoglobulin G (IgG) autoantibodies. Subjects completed up to three longitudinal visits, shown by column. White space indicates no sample. Legend indicates color relative to ΔΔCt value. There was no significant difference in the mean abundance between Ro60(+) and Ro60(−) individuals except P. prop from the chest (two-sample t test, *P = 0.02) (plot shown at the bottom right). (D) SCLE patient cutaneous lesional biopsies stained with a P. prop 16S rDNA–specific FISH probe (green) as well as the eubacterial probe EUB338 (red). DAPI, 4′,6-diamidino-2-phenylindole.
Similar to previously reported microbial compositions in healthy individuals (22), microbiome samples in study subjects from the feces, oral mucosa, and skin were primarily composed of four microbial phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Fig. 2B). These four phyla represented on average 96.4% (±5.7%) of fecal samples, 98.4% (±1.6%) of oral samples, and 97.2% (±2.9%) of skin samples. The relative abundance of operational taxonomic units (OTUs) from anti-Ro60–positive and anti-Ro60–negative subjects was compared by linear discriminant analysis effect size using LEfSe (25), and P values were adjusted for the false discovery rate using the Benjamini-Hochberg procedure. This analysis revealed no significantly different bacterial OTUs in the fecal, oral, or skin microbiomes between anti-Ro60–positive and anti-Ro60–negative subjects. The same analysis was performed comparing healthy and lupus subjects, and no significantly different OTUs were found between groups at any of the three anatomic sites. Previous work showed a lower Firmicutes/Bacteroidetes ratio in the fecal microbiomes of subjects with lupus (26). This was also true in our study subjects. The fecal Firmicutes/Bacteroidetes ratio was 1.3 in healthy subjects and 0.75 in lupus subjects (P = 0.0039). No difference in this ratio was observed between anti-Ro60–positive and anti-Ro60–negative lupus subjects (0.70 versus 0.80, P = 0.47) or between lupus subjects with or without the HLA-DR3 or HLA-DR15 alleles. In summary, major dysbiosis of the oral, skin, and fecal microbiomes was not observed in anti-Ro60–positive lupus patients.
Given that 16S rDNA V4 region sequencing does not allow for discrimination at the species level, genetic strategies were developed to specifically detect commensal Ro60 orthologs. Because the prevalence, abundance, and role of most of the identified commensal species with Ro60 orthologs are poorly understood, three commensal species with the highest similarity to hRo60 and one common and abundant gut com-mensal were chosen for study and assessed by quantitative real-time polymerase chain reaction (qPCR) of genomic DNA extracted from human microbiota samples. Abundance of Propionibacterium propionicum (P. prop) Ro60 (on the buccal mucosa and skin), Corynebacterium amycolatum (C. amyc) Ro60 (on the skin), Actinomyces massiliensis (A. mass) Ro60 (on the buccal mucosa), and Bacteroides thetaiotaomicron (B. theta) Ro60 (in the feces) was compared with the amount of total 16S rDNA to calculate bacterial load of each species. These commensal bacteria with Ro60 orthologs were commonly identified in subjects with and without anti-Ro60 antibodies, including lupus subjects and healthy controls (Fig. 2C). The presence of the bacteria alone would not be expected to trigger autoimmunity in healthy individuals but may be a source of antigen in susceptible patients with genetic predispositions such as HLA polymorphisms that present the appropriate peptides. There was no difference in the mean bacterial load between anti-Ro60–positive and anti-Ro60–negative patients except in P. prop from chest swabs because of three Ro60-positive patients (SLE01, SLE02, and SCLE01) that were densely colonized with P. prop. Notably, the skin swab from patient SCLE01 was within an SCLE eruption on the chest (fig. S4) and contained relatively high levels of P. prop. Although these species represented a small component of the oral and skin micro-biota, all subjects tested had at least one of the four Ro60 ortholog–containing species and many subjects had all four species, indicating exposure to orthologous Ro60. Given that the absolute number of bacteria present in the human microbiota is similar to that of human cells, even species of low relative abundance could represent an ample source of orthologous Ro60. The skin surface swab method may also underrepresent anaerobes such as Propionibacteria that proliferate in the anaerobic environment found deep within the hair follicles (27).
To investigate whether P. prop is not only present in superficial skin swabs from lupus patients but also localized in situ within skin lesions, we performed bacterial fluorescence in situ hybridization (FISH). Skin biopsies from seven anti-Ro60–positive SCLE patients were stained with a eubacterial-specific (EUB338) (28) and a P. prop–specific 16S rDNA FISH probe (Fig. 2D) (29). There was high overlap between the signals for EUB338 and P. prop. Biopsies from five psoriasis patients were used as controls for inflammatory skin disease. Five of seven cutaneous SCLE biopsies stained positive for P. prop, whereas P. prop was not detected within psoriasis lesions, which may reflect the decreased cutaneous microbial diversity observed in psoriasis (30). We could detect these Ro60 commensals deeper than the surface of the epidermis, as was previously shown for the cutaneous microbiome (31). Overall, these data support the significance of Ro60 ortholog colonization within lesions of SCLE patients.
T cell Ro60 cross-reactivity in lupus subjects
To test the hypothesis of ortholog cross-reactivity in autoreactive human lupus CD4+ T cells, a T cell library assay developed by Sallusto and colleagues was used (32). T cells were isolated from blood of anti- Ro60–positive patients, sorted into T cell subsets, and expanded in vitro with phytohemagglutinin (PHA) and human recombinant interleukin-2 (IL-2) for 10 to 14 days. CD4+ CD25− T cell subsets were sorted into memory CCR6− and CCR6+ subsets; CCR6 was used as a marker to enrich for T helper 17 (TH17) cells frequently implicated in autoimmunity and inflammatory diseases (33). Memory T cells were expanded with IL-2/PHA, rested, and then stimulated with autologous, irradiated monocytes pulsed with whole, heat-killed P. prop. A stimulation index (SI) of 5 was used to define significant reactivity (Fig. 3, A and B) (32). T cell clones with SI ≥ 5 were collected, expanded, and restimulated with either recombinant hRo60 or an hRo60 T cell peptide that initiated epitope spreading in mice, amino acids 316 to 335 (11). Several P. prop–reactive T cell clones proliferated in response to hRo60 and the pathogenic Ro60 T cell peptide p316-335 (Fig. 3C). Similarly, expanded memory T cells reactive to recombinantly expressed Ro60 from the Ro60 ortholog–containing gut commensal B. theta (Fig. 3, D and E) also showed cross-reactivity to hRo60 or the p316-335 Ro60 peptide (Fig. 3F). The fact that several commensal- reactive or commensal Ro60–reactive clones proliferated in response to the hRo60 protein or peptide (Fig. 3, C and F) supports the idea that microbiota-reactive T cell clones cross-react with the major Ro60 autoantigen and could initiate Ro60 autoreactivity in vivo.
Fig. 3. Commensal-reactive T cell clones from lupus patients cross-react with hRo60 protein and a pathogenic Ro60 T cell peptide.
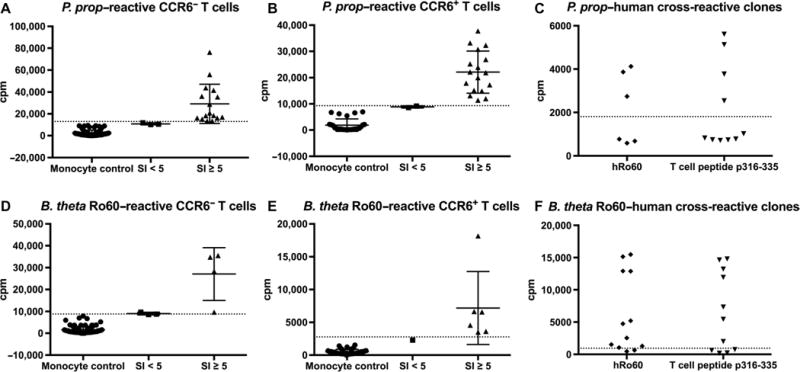
Human memory T cells from an anti-Ro60–positive SLE patient were sorted into (A) CCR6− and (B) CCR6+ subsets and stimulated with the Ro60 commensal P. prop. X axis indicates SI, and y axis indicates proliferation as counts per minute (cpm). Each point on the graph represents one clone. Dotted line indicates autologous, irradiated monocyte control. (C) Restimulation of CCR6+ clones with SI ≥ 5 with recombinant hRo60 or the pathogenic Ro60 T cell peptide p316-335. (D) Identical experiment as in (A) to (C), stimulating CD4+ memory CCR6− and (E) CCR6+ lupus T cells from another anti-Ro60–positive SLE patient with recombinant bacterial Ro60 from the Ro60-containing gut commensal B. theta. (F) Similar to (C), CCR6+ B. theta Ro60 (BtRo60)–reactive T cell clones were restimulated with recombinant hRo60 or the pathogenic Ro60 T cell peptide p316-335.
Next, we decided to test the reverse situation: the activation of hRo60-autoreactive T cells with Ro60 commensals. Freshly isolated anti-Ro60–positive patient memory T cells were first stimulated with recombinant mouse Ro60 or hRo60 protein (Fig. 4, A and B) and wells with SI ≥ 5 were subcloned as above to obtain Ro60 autoreactive T cell clones. After expansion, Ro60 clones were restimulated with heat-killed Ro60 orthologous commensal bacteria, identifying a CCR6+ T cell clone that proliferated in response to P. prop but not B. theta (Fig. 4C), supporting the concept of Ro60 ortholog epitope-specific cross-reactivity. TCR sequencing confirmed the presence of a single clonotype (TRBV9, CDR3 sequence CCAGCAGCCAAAGCCGCGGGAGATACGCAGTAT). Because CCR6 can be either a skin or mucosal marker, it is possible that the identified cross-reactive clone was derived from a skin-homing T cell as it reacted to a Ro60 epitope from the skin commensal P. prop but not the gut commensal B. theta. Differential reactivity may be due to subtle differences in sequence of the commensal Ro60 T cell epitope recognized by this clone or the Ro60 expression level in each commensal. Cytokine secretion studies to analyze the functional po-larization of the cross-reactive T cell clones revealed a diverse phenotype after restimulation with commensals (Fig. 4D), similar to previous studies of pathogen-specific memory T cell clones (34). The cross- reactive P. prop clone secreted proinflammatory cytokines interferon-γ (IFN-γ), granulocyte-macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor–α (TNFα), and IL-6, as well as IL-13, IL-9, and the T follicular helper cytokine IL-21.
Fig. 4. hRo60-reactive patient T cell clones generated by a CD4+ T cell library assay cross-react with the Ro60 commensal P. prop.
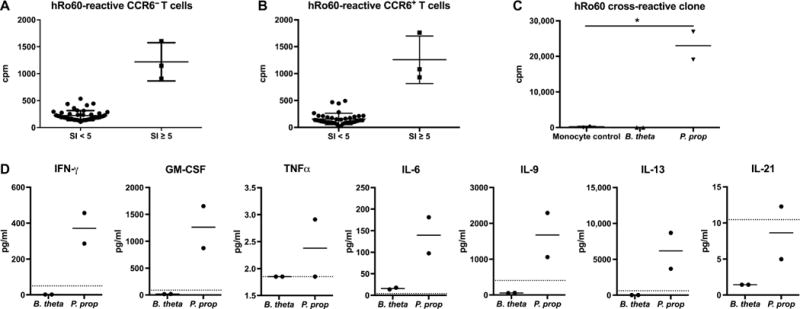
Human lupus memory T cells were sorted into (A) CCR6− and (B) CCR6+ subsets and stimulated with hRo60. X axis indicates SI, and y axis indicates proliferation as counts per minute (cpm). Each point on the graph represents one clone. (C) Three clones from each subset had SI ≥ 5 and were restimulated with the Ro60 ortholog–containing commensal bacteria B. theta and P. prop (two-sample t test, *P < 0.05). (D) Cytokine concentrations (in picograms per milliliter) from the supernatant of the cross-reactive clone 72 hours after stimulation. In (C) and (D), measurements were performed in duplicates, as indicated by the dots in each group. Dotted line indicates monocyte control.
As a negative control, CD4+ T cells from a healthy tetanus toxoid (TT)–vaccinated donor were stimulated with TT to obtain toxin-specific clones. After expansion, restimulation with hRo60 or whole, heat-killed Ro60 commensal bacteria (B. theta or P. prop) did not result in proliferation (fig. S5). Similarly, CD4+ T cells from an anti-Ro60–negative lupus patient did not proliferate when stimulated with hRo60 (fig. S6).
To determine whether a major T cell epitope was targeted by the autoreactive and P. prop cross-reactive T cell clone, we tested the previously identified KRFLAAVDVSASMNQ peptide 369–383 within mammalian Ro60 (20) and the corresponding Ro60 mimic region in P. prop (KRTMLALDVSGSMCS) and C. amyc (KRTLLSLDVSASMHW) (Fig. 5A). As above, human CD4+ lupus T cells were sorted into CCR6− and CCR6+ populations and hRo60-reactive T cell clones were identified using a T cell library assay (Fig. 5, B and C). hRo60-reactive clones were restimulated with the hRo60 peptide 369–383, bacterial mimic peptides, and whole, heat-killed bacterial lysates (Fig. 5D). The clones proliferated strongly in response to the commensal mimic peptides as well as to whole, heat-killed commensal bacteria including B. theta, likely due to high conservation of the amino acid sequence of this hRo60 T cell peptide. Cytokine secretion arrays showed a diverse cytokine profile after stimulation with commensal Ro60 mimic peptides (fig. S7), supporting a heterogeneous T cell phenotype, as described for human pathogen-specific memory T cell clones [see Fig. 4 and (34)].
Fig. 5. hRo60-reactive T cell clones cross-react with commensal Ro60-reactive mimic peptides and whole commensal bacteria.
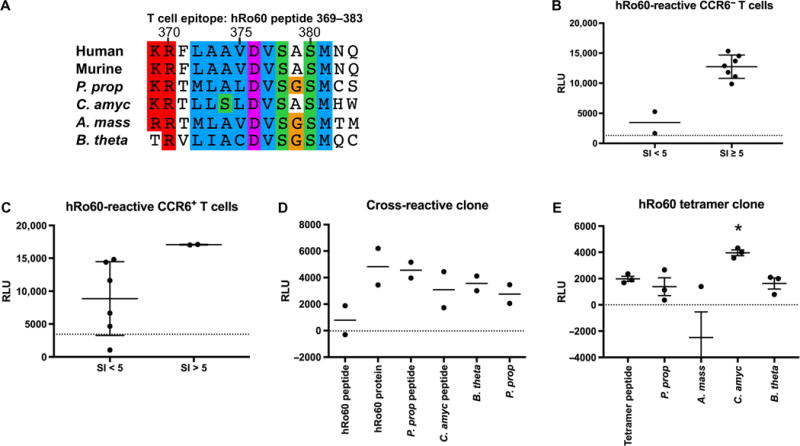
(A) Alignment of hRo60 T cell autoepi-tope peptide 369–383 with the corresponding amino acid sequences in commensal Ro60 orthologs of P. prop., C. amyc, and B. theta. CCR6− (B) and CCR6+ (C) memory CD4+ T cell subsets of an anti-Ro60–positive SLE patient stimulated with hRo60 protein. Y axis indicates proliferation as reactive light units (RLU) using a nonradioactive adenosine 5′-triphosphate (ATP) release assay. Dotted lines indicate monocyte control, which was set as baseline to zero for restimulation assays. (D) hRo60-reactive T cell clones of the same patient proliferated to hRo60 peptide 369–383, commensal ortholog mimic peptides, and whole commensal bacteria. (E) CD4+ T cell clone isolated from the peripheral blood of an anti-Ro60–positive SLE patient using a hRo60 peptide–specific tetramer cross-reacts with the Ro60 commensal C. amyc (*P < 0.05, two-sample t test).
Because the frequency of autoantigen-specific T cells in the peripheral blood is low, and to confirm cross-reactivity of T cell clones with another method, major histocompatibility complex (MHC) class II tetramers were synthesized for isolation of hRo60 peptide–reactive memory CD4+ T cells ex vivo from a Ro60-positive patient. The tetramer was loaded with the hRo60 peptide KRFLAAVDVSASMNQ (peptide 369–383, as used in Fig. 5) (20). CD45RA−CD45RO+CD25−tetramer+ single T cells were expanded with IL-2 and PHA and then restimulated with the hRo60 tetramer peptide and with Ro60 commensal bacteria (Fig. 5E). The tetramer-positive clone proliferated significantly (P < 0.05) in response to the Ro60 ortholog commensal C. amyc, and to a lesser degree also to B. theta and P. prop, which contain a slightly different Ro60 similarity to the hRo60 peptide from C. amyc (Fig. 5A). Overall, these data support commensal Ro60 T cell cross-reactivity using both T cell library assays and tetramer-sorted memory CD4+ T cells.
Commensal Ro60 bound by human lupus serum antibodies
In addition to T helper cell cross-reactivity, antibody cross-reactivity with commensal Ro60 orthologs could also drive human anti-Ro60 responses in lupus patients. To assess for the presence of antibodies reactive with bacterial Ro60 RNPs in patients’ sera, immunoprecipitations were performed with human sera and lysates from cultured P. prop and B. theta, because a putative ortholog of the most conserved bacterial Y RNA, YrlA, had been identified in each of these species (14). RNA was extracted from immunoprecipitated complexes and subjected to Northern blotting to detect YrlA. Human sera from anti-Ro60–positive subjects with SLE and SCLE immunoprecipitated RNPs containing the P. prop YrlA RNA (n = 4 of 6), whereas anti- Ro60–negative SLE subjects and healthy controls did not (n = 3 of 3) (Fig. 6A), suggesting the presence of cross-reactive Ro60 autoanti-bodies in most lupus patients. Oligonucleotide probes to the YrlA RNA 5′ stem region, 3′ stem region, and loop region all bound P. prop YrlA RNA by Northern blot, confirming the predicted bacterial YrlA RNA sequence (Fig. 6B) (14). Some lupus subjects with Ro60 autoantibodies did not immunoprecipitate P. prop Ro60 RNPs containing YrlA RNA (n = 2 of 6), highlighting the heterogeneity among SLE patients and the possibility that other bacterial Ro60 orthologs with different conservation of hRo60 B cell epitopes than P. prop could be cross-reactive with these sera. Because bacterial YrlA RNAs contain a domain that is structurally similar to transfer RNA (tRNA) (14), the Northern blots were reprobed for the far more abundant P. prop tRNA-proline- GGG as a negative control. None of the human sera immunoprecipitates contained this tRNA, supporting specificity of the assay (Fig. 6C). To confirm the presence of cross-reactive antibodies in anti-Ro60– positive patients, sera from a larger cohort of 63 well-characterized SLE patients from Brigham and Women’s Hospital (called Harvard sera thereafter) were obtained as a validation cohort. Anti-Ro60 status was first determined by ELISA (fig. S8A) and confirmed by immunoprecipitation of hRo60–Y RNA immune complexes (fig. S8B). Twenty anti-Ro60–positive patient sera selected based on Ro60 ortholog reactivity by ELISA were subjected to immunoprecipitation of P. prop Ro60–YrlA RNA, which again demonstrated cross-reactivity in most patients (Fig. 6D).
Fig. 6. Human lupus sera immunoprecipitate YrlA RNA–containing RNPs from P. prop lysates.
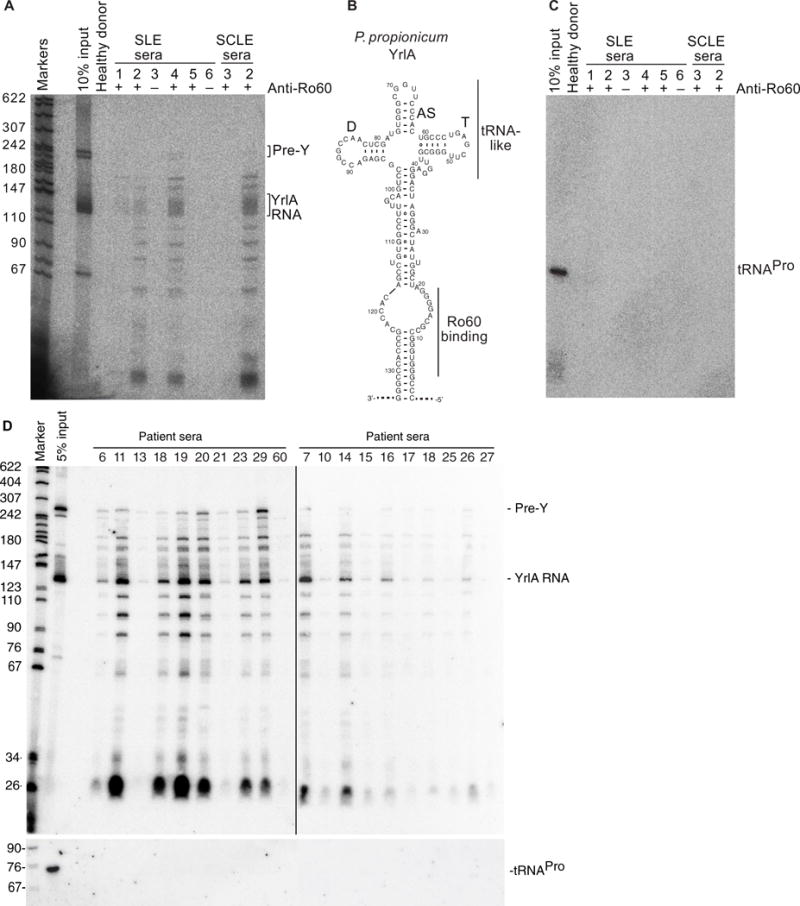
(A) After incubating P. prop lysates with human sera, RNAs in immunoprecipitates were extracted and subjected to Northern blotting to detect YrlA. First lane, molecular size markers (nucleotides). Total P. prop, total RNA extracted from the input lysate. Serum from a healthy donor is shown in the next lane. Six SLE sera and two SCLE sera are labeled with + or − representing the anti-Ro60 IgG antibody status by ELISA. (B) Predicted structure of P. prop YrlA. (C) As a negative control, the blot was reprobed for P. prop tRNA- proline-GGG. (D) Lupus sera from independent Harvard patient cohort (listed by identification numbers).
In contrast, none of the patient sera from the Yale cohort coimmuno-precipitated the B. theta YrlA RNA (fig. S9A), although YrlA RNA was detected in total RNA extracted from B. theta at the late log phase of bacterial growth (fig. S9B). To explore whether human antibodies could bind recombinant B. theta Ro60 protein (BtRo60) in the absence of YrlA RNA, Western blots were performed. hRo60, BtRo60, and B. theta lysates were run on a protein gel and blotted with serum from human anti-Ro60–positive lupus patients (Fig. 7, A and B). Lysates from two other commensal bacteria, Propionibacterium acnes and Roseburia intestinalis, which do not contain Ro60 orthologs, were used as negative controls. Anti-Ro60–positive patients had serum antibody binding to both hRo60 (60 kDa) and recombinant BtRo60 (~56.5 kDa), as well as a positive ~60-kDa band in the lane containing the total B. theta bacterial lysate. Translation of BtRo60 can initiate at one of three methionines, giving rise to potential proteins of 57.7, 57.6, and 56.5 kDa. The recombinant protein, which migrates faster than the bands detected in extracts, corresponds to the shortest version, suggesting that in vivo, the larger species may predominate. Protein bands of varying sizes were seen in the P. acnes lysate as might be expected, given the role of P. acnes in acne pathogenesis. None of the sera bound to proteins of the gut symbiont R. intestinalis. These results indicate that despite the lack of antibodies to B. theta RNPs, anti-Ro60–positive lupus patients from two independent patient cohorts had serum antibodies that bound the BtRo60 ortholog protein.
Fig. 7. Anti-Ro60–positive SLE sera bind the recombinantly expressed BtRo60 ortholog by Western blot.
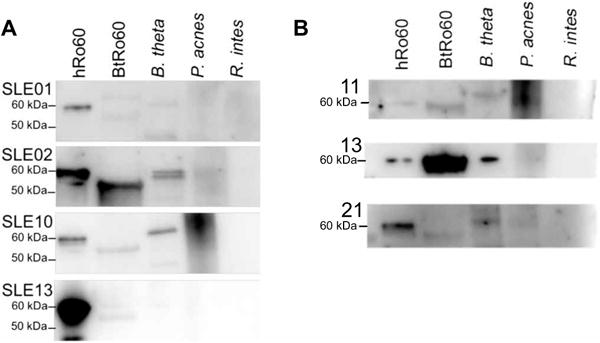
(A) Sera from four Yale SLE patients with positive serol-ogies for anti-hRo60 IgG were subjected to Western blotting with recombinant hRo60 (60kDa), recombinant BtRo60 (~56.5 kDa) and bacterial lysates from B. theta, as well as two skin/gut commensal strains that do not carry Ro60 orthologs, P. acnes and R. intestinalis (R. intes). (B) Sera from three Harvard SLE patients with positive serologies for anti-hRo60 IgG were subjected to Western blotting as in (A).
Gnotobiotic mouse monocolonization with Ro60 commensals
Although the Yale lupus sera did not immunoprecipitate BtRo60, the Western blots showing patient sera binding to BtRo60 may be biologically relevant, because binding of hRo60 antibodies to the human protein can occur in the absence of Y RNAs (35, 36). Thus, Ro60 antibodies may exist that cross-react with only the YrlA RNA–free form of the BtRo60. Because autoantibodies, including against Ro60, are found in a low frequency and titer in the general population (37, 38), cross-reactivity with some Ro60 orthologs may contribute to benign autoreactivity that may only spread to pathogenic epitopes depending on the HLA haplotype of the individual and the Ro60-directed T helper cell responses that we also demonstrated to be cross-reactive with B. theta. To study the general concept of commensal ortholog cross-reactivity in vivo, we next monocolonized germ-free (GF) wild- type C57Bl/6 mice with the common gut commensal, B. theta (n = 15). After 3 or 5 months of monocolonization, sera from all mice were tested and found to be positive for anti-hRo60 IgG by ELISA as compared to age- and sex-matched GF C57Bl/6 mice (Fig. 8A). Subsets of these mice were also treated orally with 0.1% imiquimod (IMQ) or 1 to 2% dextran sulfate sodium salt or the combination of both to promote inflammation and barrier irritation, but the anti-hRo60 IgG titers of these animals were not different from untreated animals (table S5), which is why they were treated as one cohort. Although the mouse anti-Ro60 antibodies did not immunoprecipitate hRo60 from cell extracts, none of the sera were positive for anti-insulin antibodies by ELISA, which were used as a negative control to test for nonspecific polyreactivity (table S5). C57Bl/6 mice monocolonized with a different gut commensal, Enterococcus gallinarum, were also negative for anti-Ro60 antibodies by ELISA (table S5). These data support selective B cell cross-reactivity between a commensal Ro60 ortholog and hRo60 in vivo.
Fig. 8. Monocolonization of the GF mouse gut with B. theta leads to hRo60 T and B cell responses.
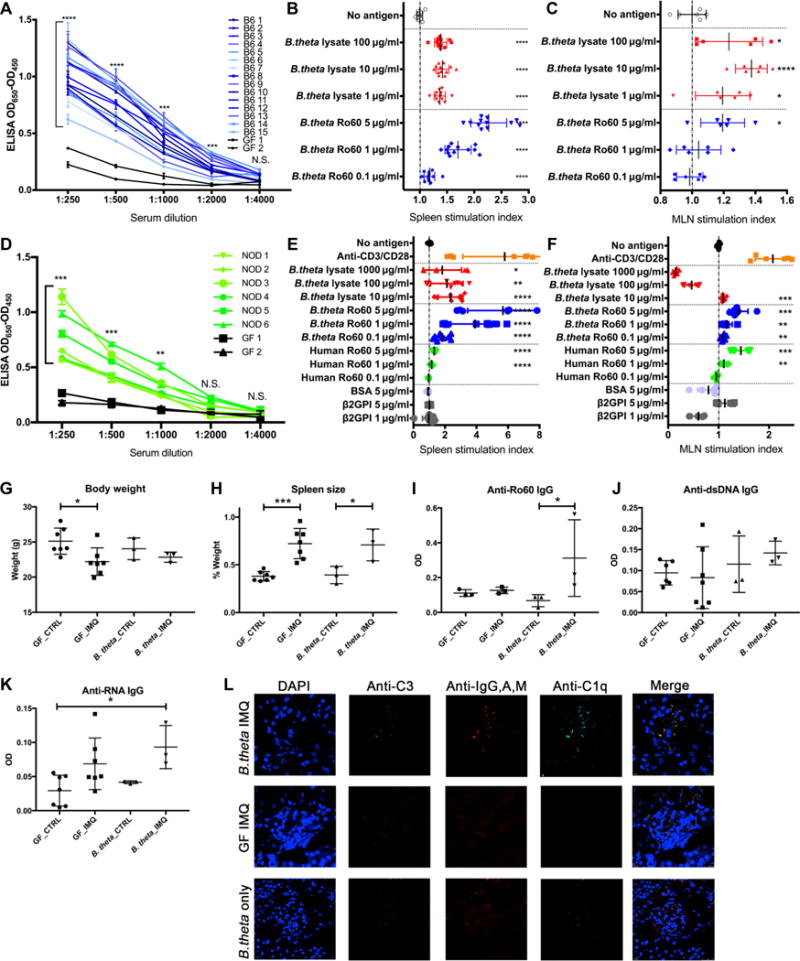
(A) hRo60 ELISA of B. theta monocolo-nized C57Bl/6 mice (n = 15) after 3 to 5 months of coloni-zation compared to GF age-and sex-matched controls of the C57Bl/6 strain (n = 2). X axis shows twofold serial dilutions of sera from 1:250 to 1:4000, with the mean and SD of duplicate measurements plotted for each mouse. (B) Proliferation, measured by an ATP release assay, of cells from MLNs and (C) spleens from B. theta–monocolonized C57Bl/6 mice, stimulated with B. theta lysate or recombinant BtRo60 protein for 72 hours in triplicate. Each point represents the SI (proliferation divided by the mean background proliferation from cells with no antigen) of MLNs and spleens pooled by cage. P values were calculated using two-sample t tests. (D) hRo60 ELISA of B. theta– monocolonized, autoimmune- prone NOD mice (n = 6) after 2 weeks of colonization, compared to two age-matched GF mice (6-week-old C57Bl/6 females). X axis shows twofold serial dilutions of sera from 1:250 to 1:4000, with the mean and SD of duplicate measurements plotted for each mouse. (E) Proliferation assay of cells from MLNs and spleens (F) from monocolonized NOD mice stimulated with B. theta lysate, BtRo60, hRo60, and control proteins (BSA and β2GP1, an unrelated autoantigen) for 72 hours in triplicate as above. (G to L) Eight-week-old GF C57Bl/6 mice were monocolonized with B. theta followed by topical treatment with or without a TLR7 agonist (IMQ) for 8 weeks. Monocolonized mice were compared to age- and sex-matched GF C57Bl/6 mice with and without IMQ (n = 3 to 7 in each of the four groups). (G and H) Comparison of body weight and relative spleen weight in B. theta–monocolonized mice with and without IMQ versus GF controls. (I to K) Anti-hRo60, anti-dsDNA, and anti- RNA IgG ELISAs of B. theta–monocolonized and GF mice with or without IMQ after 8 weeks of treatment. B. theta monocolonization together with IMQ. (L) Renal immunofluorescence of B. theta–monocolonized and GF mice treated with or without IMQ. Immunofluorescence of kidneys was performed on kidney tissue from B. theta–monocolonized mice with or without IMQ and GF with IMQ. Anti-C3 (green), anti-C1q (teal), and anti-IgG, anti-IgA, and anti-IgM (red). OD, optical density; N.S., not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-sample t test.
To determine activation of T cells by Ro60, proliferation of cells collected from mesenteric lymph nodes (MLNs) and spleens was measured 72 hours after stimulation with bacterial lysate or recombinant protein. BtRo60 induced cellular proliferation from splenocytes in a dose-dependent manner (Fig. 8B), strongly supporting that mono-colonization led to a systemic T cell–dependent anti-Ro60 immune response. Whole bacterial lysates from cultured B. theta also led to highly significant (P < 0.0001) proliferation, albeit at lower levels than to recombinant protein. MLN cells also proliferated in response to higher concentrations of BtRo60 as well as whole bacterial lysate (Fig. 8C).
GF autoimmune-prone nonobese diabetic (NOD) mice were then monocolonized with B. theta to determine whether the Ro60 ortholog responses were different and possibly pathogenic with a genetic background that can lead not only to type 1 diabetes but also to lupus and Sjögren’s syndrome under certain circumstances (39, 40). After only 2 weeks of monocolonization and before the onset of diabetes, sera were positive for anti-hRo60 IgG antibodies by ELISA and spleen and MLN cells from NOD mice proliferated vigorously in response to BtRo60 in a dose-dependent manner (Fig. 8, D to F). The purified cells also proliferated in response to recombinant hRo60 in a dose- dependent manner, suggesting ortholog cross-reactivity because the mice had no previous exposure to the human antigen. Cells from MLN were approximately equally responsive to hRo60 and BtRo60, whereas splenic cells had a similar response to hRo60 with a more robust proliferation response to BtRo60, possibly because of the proximity of the MLN to the source of the antigen. Intriguingly, MLN incubation with high-dose B. theta lysates led to inhibition, reminiscent of the tolerogenic response to B. theta in the human gut (41). This suggests that an inflammatory response to BtRo60 may be dampened by the natural high-density colonization with this common human gut commensal and may explain the heterogeneity seen in responses to different Ro60 orthologous commensals as above. Notably, MLN and spleen cells did not proliferate in response to the control protein bovine serum albumin (BSA) or the unrelated human autoantigen β2-glycoprotein-I (B2GP1) (42). Together, these data support com-mensal Ro60 ortholog cross-reactivity as an instigator of hRo60 auto-reactivity in vivo. During the study period, however, none of the mice developed overt autoimmunity.
To this end, we decided to use an inducible model for lupus in the GF setting and then test whether B. theta monocolonization leads to signs of autoimmunity. We therefore repeated the monocolonization in GF C57Bl/6 mice followed by 8 weeks of topical treatment with IMQ, a TLR7 agonist that has been shown to induce a lupus-like syndrome in mice under specific pathogen–free (SPF) conditions (43, 44). After 8 weeks of monocolonization with or without IMQ treatment (until 16 weeks of age), sera and organs were collected from all mice for autoimmune studies (Fig. 8, G to L). The sera were tested for anti- hRo60, anti–double-stranded DNA (dsDNA), and anti-RNA IgG levels by ELISA and compared to IMQ-treated age- and sex-matched GF C57Bl/6 mice. B. theta monocolonization plus IMQ led to increased levels of anti- hRo60 compared to all other conditions but did not enhance other lupus autoantibody titers, likely due to the short study period of 8 weeks that does not yet allow for epitope spreading, which takes years in humans (1, 44). Despite this, lupus nephritis–like immune complex deposition composed of C1q, C3, IgG, IgA, and IgM (45–47) was detectable in 88% of all glomeruli of the IMQ-treated B. theta–monocolonized mice (37 of 42 glomeruli) compared to only 4 of 49 glomeruli involved in IMQ-treated GF mice (Fig. 8L shows representative images of immunofluorescence), indicating a systemic autoimmune effect. Last, we performed flow cytometric analysis of immune cell subsets from gut and peripheral tissues to determine the effects of B. theta monocolonization on these tissues. A significant increase of CD11b+MHCII+ monocytes (P < 0.05) and CD11b+CD11c+MHCII+ dendritic cells (DCs; P < 0.01) was noted that was enhanced further by IMQ (fig. S10, A to C), suggesting that antigen-presenting cells are recruited to the gut and may interact with B. theta in vivo to present commensal Ro60 antigens. Small intestinal plasmacytoid DCs (pDCs)—a major source of type I IFN in SLE (48)—were increased as well by B. theta monocolonization alone (fig. S10, D and E). B. theta can induce both anti-inflammatory regulatory T cells (Tregs) in the colon and proin-flammatory TH17 in the ileum (41, 49), but its effects on Tregs in secondary lymphoid organs are not clear. We unexpectedly found that B. theta reduced Treg numbers in the spleen independently of IMQ treatment (fig. S10, F and G), creating an immunostimulatory environment also in the periphery that may allow for antigen-specific immune responses to Ro60, as shown in Fig. 8 (A to F). Together, these in vivo data in wild-type and autoimmune-prone mice demonstrate that B and T cell responses to Ro60 occur after monocolonization with a human gut commensal that produces Ro60 orthologs and that monocoloni-zation with a bacterial Ro60 ortholog enhances systemic lupus–like disease in a lupus mouse model.
DISCUSSION
In vivo studies have shown that T and B cell–directed anti-Ro60 responses drive epitope spreading (11) and might thus initiate the chronic autoimmune response in SLE patients independently from flares. In addition, anti-Ro60 antibodies are considered pathogenic in cutaneous and neonatal lupus (5–7). Among extrinsic factors, infectious agents such as viruses have been implicated in the pathogenesis of lupus either as innate stimuli, cross-reactive agents, or sources for exogenous nucleic acids that promote flares (50). The constant triggers of this lifelong disease remain elusive, particularly in those individuals without any evidence of pathogenic infection. Notably, in several murine models of autoimmunity, commensal bacteria have been shown to represent potential chronic triggers or perpetuators (51). Because anti-Ro60 autoantibodies are present in asymptomatic individuals years before onset of disease (3), a chronic trigger such as permanent colonization with autoreactivity-inducing commensals is plausible. In support of a chronic stimulus, long-term Ro60 humoral autoimmunity is maintained by rapid turnover of continuously activated B cell clones (18). Our data suggest that this response may be driven by Ro60 orthologs encoded in human commensal bacteria. The fact that Ro60 orthologs bind bacterial Y RNA and thus could trigger TLR7 if recognized by cross- reactive autoantibodies may add fuel to the “autoimmunity fire.”
We first showed that high homology exists between the major T and B cell epitopes within hRo60 and commensal Ro60 orthologs, and that human subjects including SLE/SCLE patients are colonized with Ro60 orthologous commensals in different niches without overt dysbiosis of the overall composition of the microbiota. Sera from anti- Ro60– positive lupus patients, but not anti-Ro60–negative controls, preferentially coimmunoprecipitated Ro60 RNPs from a skin/oral Ro60 ortholog–containing commensal P. prop. Given that we identified commensal bacterial reactivity also with autoreactive human memory T cell clones generated with a T cell library assay (32), T cell cross-reactivity is functionally demonstrated. The cytokine secretion patterns of the T cell clones kept in culture suggest a heterogeneous functional profile as predicted based on pathogen-specific memory T cell clones (34). Our animal studies using gnotobiotic models showed that Ro60 ortholog cross-reactivity could also occur in vivo. Particularly, systemic T cell proliferation to recombinantly expressed commensal and hRo60 proteins supports this concept.
On the basis of the current data, we hypothesize that patients with genetic autoimmune susceptibility who are colonized with Ro60- positive commensals mount an aberrant cross-reactive immune response. For example, HLA class II–related polymorphisms could lead to presentation of bacterial Ro60 antigens with high linear or conformational homology to hRo60. Supportive of such a combinatorial effect was that T cell clones from one SLE patient responded to P. prop but not B. theta. Besides gradual differences in homology with hRo60, the immunoregulatory versus stimulatory potential of different commensals may also account for these findings. We therefore propose that P. prop could be a prime candidate not only as a trigger for systemic serum autoantibodies but also as a local driver of anti-Ro60–induced pathology in cutaneous lupus. It is notable that the anatomical niche of the genus (Propionibacteria) within sebaceous regions of the chest and back is highly similar to the common areas of UV-exposed, lesional skin in cutaneous lupus, and that we detected P. prop within SCLE lesional skin. Although photosensitive areas are generally affected by SCLE, photoprotected areas can also be involved, suggesting that commensal colonization patterns could play a role in the absence of UV exposure. UV light is known to not only trigger keratinocyte apoptosis and exposure of endogenous Ro60 on the cell surface in apoptotic blebs (52) but also induce antimicrobial peptides (53). These peptides are well known to cause dysbiosis via alterations of the community structure of microbiota (54). It is thus plausible that a vicious cycle of microbial Ro60 ortholog production, UV exposure and anti-Ro60 antibody binding to first microbial, and then hRo60 promotes cutaneous manifestations in lupus patients and Ro60-positive commensals sustain systemic auto-antibodies (fig. S11).
Although our data support the importance of commensal orthologs in lupus pathogenesis, there are several limitations to our study. First, Ro60 ortholog–targeted microbial community profiling of a larger population of lupus patients from different geographic regions was not performed to demonstrate relevance for patients outside the New England area. Second, we have not extensively quantified the Ro60 ortholog levels in lesional and nonlesional skin from a cohort of SCLE patients, which might allow for development of novel bio-markers related to cutaneous or systemic Ro60 autoimmunity. It would be particularly insightful to survey also predisease cohorts before onset of SLE (1) for the presence of serum reactivity to Ro60 commensals identified in this study. Third, we have not defined and mutated the anchor residues of the recognized peptides of the cross-reactive T cell clones, which would allow for additional cross-reactivity studies. Fourth, we have not cloned B cells and tested monoclonal anti- hRo60 antibodies from SLE patients for Ro60 ortholog cross-reactivity. Last, we have not rederived HLA-DR3–transgenic animals GF to test whether gut or skin monocolonization with the Ro60 bacteria defined in this study leads to autoimmune manifestations in vivo in humanized mice.
Sjögren’s syndrome is also closely linked with anti-Ro60 antibodies and could potentially be triggered by other oral Ro60 commensal species cross-reacting with different hRo60 epitopes important in this syndrome. C. amyc was described to colonize the lacrimal duct and could therefore represent a potential candidate driving sicca symptoms in Sjögren’s syndrome (55, 56). Our human studies in lupus, together with more mechanistic experiments and formal demonstration of cross-reactivity on a molecular level (57), might eventually lead to development of Ro60 microbiota-targeted therapies that would represent an alternative approach to treating or preventing lupus and possibly also manifestations of Sjögren’s syndrome.
In summary, our current studies uncovered commensal Ro60 or-tholog reactivity as a mechanism to induce autoreactivity to the Ro60 autoantigen in lupus. We established a functional link between common members of the human microbiota and a primordial autoimmune response in humans. Persistent, commensal-induced autoreactivity mediated by microbial orthologs could represent a novel paradigm to explain parts of the pathogenesis of not only lupus but also other chronic immune-mediated diseases. Given the enormous richness of the com-mensal proteome (58), we expect the existence of other commensal orthologs of autoantigens. This paradigm could have implications for early prevention and novel microbiota-targeted therapies for autoimmunity in the future.
MATERIALS AND METHODS
Study design
This was a pilot proof-of-concept study using observational human subject data, ex vivo human sample experiments, and in vivo mouse experiments to demonstrate the ability of Ro60 ortholog bacteria to induce autoimmunity. Primary data are located in table S5, and details are described in Supplementary Materials and Methods. All human subject protocols were approved by the Yale Human Investigations Committee and in accordance with the Declaration of Helsinki. A signed document of informed consent was obtained from all study subjects. Exclusion criteria were as follows: ongoing chronic infection, antibiotic or probiotic use in the last 90 days, topical antibiotic or antimicrobial use in the last 7 days, bathing or tooth brushing in the last 8 hours, major gastrointestinal surgery in the last 5 years, gastrointestinal bleeding history, inflammatory bowel disease, bulimia or anorexia nervosa, morbid obesity, uncontrolled diabetes mellitus, malignancy in the past year, and known excessive alcohol use. Power calculations were performed using a known microbiome reference set. Assuming an SD of 8% for a commensal genus in each group, the power to detect a difference between groups with a 0.05 significance level would be 80% with n = 10 in each group. Lupus patients (n = 18; 16 with SLE and 2 with SCLE) and age-matched (±5 years) and sex-matched healthy controls (n = 11) were enrolled over a 2-year period. All study subjects completed up to three study visits for the collection of detailed health and diet histories, whole blood, and oral, skin, and fecal microbi-ota sampling following a larger microbiome study protocol at Yale (ClinicalTrials.gov identifier: NCT02394964).
The V4 regions of rDNA extracted from human skin, oral, and fecal swabs were sequenced to determine the composition of the micro-biota and subjected to qPCR to validate the presence of target species. Human sera were used to immunoprecipitate bacterial Ro60. T cell libraries were generated using one out of three visits from each human subject studied for each library. Researchers were not blinded.
Three cohorts of GF mice were monocolonized with B. theta. Serum, lymph node, spleen, gut, and kidney samples were collected after termination of the experiment. Serum was used for Ro60 ELISA, lymph nodes and spleens were used for proliferation assays in response to bacteria and proteins, spleens and gut tissues were used to isolate cells for flow cytometry, and kidneys were used for immunofluores-cence. One monocolonized C57Bl/6 animal was found dead before the end of the study and excluded from analysis. No samples were otherwise excluded.
Bacteria and culture conditions
C. amyc strain SK46 was obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI), and single colonies were grown in Brain Heart Infusion Medium broth (BD Difco) aerobically at 37°C. P. prop strain F0230 was obtained from BEI and grown in Modified Reinforced Clostridial Medium (BD Difco) anaerobically at 37°C. A primary human isolate of B. theta (59) was grown in Gut Micro-biota Medium anaerobically at 37°C to an OD600 (optical density at 600 nm) value of 1.6 to 1.8 for all experiments (late log phase, see fig. S5). A. mass strain F0489 was obtained from BEI and grown anaerobically in Actinomyces Broth (BD Difco) at 37°C. Additional details are available in Supplementary Materials and Methods.
T cell cloning, tetramers, and proliferation assay
Human PBMCs were isolated from whole blood and immunomagnetically separated by CD19, CD14, and CD4 (STEMCELL Technologies). Viable CD4+ T cell were sorted into CCR6− memory (CD45RA−CD45RO+ CD25−CCR6−) and CCR6+ memory (CD45RA−CD45RO+CD25−CCR6+) populations of at least 97% purity for use in T cell library assays as previously described (32). Library screening was carried out by stimulation of 250,000 T cells per well with irradiated (45 Gy) autologous monocytes (~25,000 per well). The monocytes were pulsed for 3 hours with recombinant mouse Ro60 (100 μg/ml) or hRo60 protein or TT (1 μg/ml). Positive control wells were expanded and then restimulated at a ratio of 1:10 with irradiated monocytes that were pulsed with either whole, heat-killed bacteria or commensal mimic peptides. Negative control wells contained monocytes only to assess any background signal. After 64 hours, culture supernatants were removed for cytokine measurement using a bead-based immunoassay (LEGENDplex, BioLegend). Cell proliferation was measured either by measuring [3H]-thymidine incorporation on a scintillation β-counter (PerkinElmer) or alternatively by nonradioactive ATP measurement using the ATPlite Kit (PerkinElmer), as indicated in the figure legends. Additional details are available in Supplementary Materials and Methods.
Immunoprecipitation and Northern blotting
After lysing P. prop bacteria by cryogenic grinding, the lysates were incubated with human sera, followed by incubation with Protein A Sepharose CL-4B (Pharmacia). Beads were washed and RNA was extracted using phenol/chloroform/isoamyl alcohol (50:50:1), fractionated in 6% polyacrylamide/8 M urea gels, transferred to Hybond (Amersham), and RNA cross-linked to the membrane as described (60). Hybridization with [γ-32P]ATP-labeled oligonucleotides was performed as described (61). Oligonucleotides were as follows: P. prop YrlA RNA, 5′-ATCCCTGATAACCGATCCCCTGCGG-3′ and 5′-CAACCTCCT-GATCCCTGATAAC-3′; P. prop tRNAPro, 5′-TTGTCGGGCTGACAG-GATTTG-3′. Additional details are available in Supplementary Materials and Methods.
Mice
Animal care and handling was approved by the Yale Institutional Animal Care and Use Facility and in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. GF mice were housed in gnotobiotic isolators, and monocolonization was achieved by oral gavage with 0.2 ml of thawed B. theta at ~1.3 × 109 cells/ml in 20% glycerol, followed by confirmation after 2 weeks by Sanger sequencing of the 16S region of DNA extracted from the fecal pellet. Four male and 12 female 6-week-old C57Bl/6 mice were monocolonized for 3 months (n = 7) or 5 months (n = 8) followed by terminal blood collection and organ collection. A second group of C57Bl/6 GF mice was treated with topical IMQ three times weekly from age 8 to 16 weeks to mirror an inducible lupus model, as previously described under SPF conditions (44). Fourteen C57Bl/6 mice remained GF, and 6 mice were monocolonized with B. theta. Half of each group was treated with topical IMQ and housed in gnotobiotic isolators. Three 11-week-old male and three 6-week-old female GF NOD mice were monocolonized and sacrificed after 2 weeks. Serum anti-Ro60 IgG was measured by ELISA. Cells from MLNs and spleen were plated at 100,000 cells per well and stimulated with B. theta, Ro60, or control proteins. Proliferation was measured after 72 hours using an ATP assay. For flow cytometric analysis, cells from spleen and small intestinal lamina propria were stained on the surface with fluorescently conjugated antibodies specific to CD3ε, CD4, CD8, CD45, CD45R (B220), PDCA-1, CD11c, CD11b, and MHCII and intracellularly stained with fluorescently conjugated antibody specific for FoxP3 (BD Biosciences). Kidneys were immunofluorescently stained with anti-IgG/IgA/IgM, anti-C1q, and anti-C3 (Abcam). Additional details are available in Supplementary Materials and Methods.
Statistical analysis
Plotting of data and statistical analysis were performed using GraphPad Prism software. Unless otherwise stated, statistical significance was determined by the unpaired two-tailed Student’s t test, and differences were considered statistically significant if P < 0.05. P values are represented using * for P < 0.05, ** for P < 0.01, *** for P < 0.001, and **** for P < 0.0001. Shannon-Weiner diversity index and the false discovery rate were calculated using R software (62).
Supplementary Material
Fig. S1. Sequence alignment of full-length hRo60 and selected commensal orthologs.
Fig. S2. Coimmunoprecipitation of lupus study subject sera confirmed anti-Ro60 antibody status.
Fig. S3. α-Diversity represented by box plots of the Shannon-Weiner diversity index.
Fig. S4. SCLE skin eruption.
Fig. S5. TT-reactive CD4+ T cell clone from a healthy donor generated by a CD4 T cell library assay.
Fig. S6. Ro60-negative SLE patient CD4+ T cells lack reactivity to hRo60 protein.
Fig. S7. Cytokine concentrations (pg/ml) of supernatants from the cross-reactive T cell clone from Fig. 5 measured using a bead-based immunoassay.
Fig. S8. Anti-Ro60 antibody status of Harvard cohort lupus subjects.
Fig. S9. YrlA RNA is not detected in immunoprecipitates from B. theta using human lupus sera.
Fig. S10. B. theta monocolonization of GF mice induces gut and systemic immune changes.
Fig. S11. Schematic of proposed mechanism of how Ro60 bacteria trigger and sustain autoimmunity.
Table S1. Commensal bacterial Ro60 orthologs identified by in silico methods.
Table S2. Lupus study subject clinical data.
Table S3. Healthy control study subject clinical data.
Table S4. Efficiency and specificity of bacterial Ro60 qPCR primers.
Table S5. Primary data.
Acknowledgments
We thank all patients who have participated in this study as well as K. DeFrancesco and I. Matos for patient recruitment at the Yale Center for Clinical Investigation. We also thank J. Sterpka for technical assistance, L. Wen for providing GF NOD mice, N. Palm for use of an anaerobic chamber, M. Bosenberg for the dermatopathology samples, and S. Lewis for efforts to produce recombinant Ro60 protein. We thank W. Kwok (Benaroya Research Institute) for tetramer synthesis.
Funding: This work was supported in part by grants from the NIH (K08AI095318, R01AI118855, R01 GM073863; T32AI07019), the Yale Rheumatic Diseases Research Core (NIH P30 AR053495), the Women’s Health Research at Yale, the O’Brien Center at Yale (NIH P30DK079310), the Arthritis National Research Foundation, the Arthritis Foundation, and the Lupus Research Institute. T.M.G. was supported by NIH training grant 5T32AR007016-41. K.H. was supported by NIH training grant T32GM007223 and by an NSF Predoctoral Fellowship. M.B. was supported by a Ruth L. Kirschstein National Research Service Award (F32 ES026227). The Yale Center for Clinical Investigation is supported by the Clinical and Translational Science Awards grant number UL1 RR024139 from the National Center for Research Resources, the National Center for Advancing Translational Science, and the NIH Roadmap for Medical Research. This work was also partly supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (Xinguo Chen, K.H., M.B., S.S., and S.L.W.) and by the NIH PREP program R25GM104553 (A.J.I.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/10/434/eaan2306/DC1
Materials and Methods
References (63–77)
Author contributions: S.S., P.D., and S.L.W. created the list of Ro60 orthologs; T.M.G. cross-referenced them with the commensal databases and created the phylogenetic tree; and S.S. identified YrlA RNAs. T.M.G., C.D., and J.G. led human subjects protocols and recruitment, and C.D., S.C.R., and C.K. assisted in human sample processing. K.H.C. provided sera and clinical information of Harvard cohort. T.M.G. and W.E.R. performed 16S sequencing and analysis. Anaerobic bacterial cultures were done by T.M.G. and C.D. T.M.G., S.C.R., and C.K. performed qPCR. Xinguo Chen, K.H., and M.B. expressed and purified recombinant bacterial Ro60 proteins. Xinguo Chen expressed and purified the human protein from baculovirus-infected cells with assistance from T.M.G. C.D. generated the hRo60 protein produced in mammalian cells. ELISA was designed by T.M.G. and C.D. and also performed by C.K. and Xindi Chen. T cell cloning, single-cell sorting, tetramer studies, T cell proliferation, and TCR sequencing were done by C.D. Human immunoprecipitation was done by M.B. Bacterial immunoprecipitation and Northern blots were done by Xinguo Chen with assistance from T.M.G. Western blotting was performed by C.D. 16S rRNA FISH experiments were performed by C.D. Immunofluorescence was performed by C.D. and A.J.I. Mouse experiments were performed by T.M.G. with help from S.M.V., W.E.R., and C.K. IMQ mouse experiments were performed by A.J.I. and D.Z.R. M.G. provided human subjects recruitment and experimental design. A.L.G. assisted in experiment design and critical review of the manuscript. M.A.K., T.M.G., C.D., and S.L.W. wrote the manuscript. M.A.K. and S.L.W. conceived the study and provided experiment design and guidance.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Human sera of lupus patients from Massachusetts are available from Yale University under a material transfer agreement with Brigham and Women’s Hospital. The 16S sequencing data have been deposited in the European Nucleotide Archive, accession PRJEB24742.
REFERENCES AND NOTES
- 1.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, Harley JB. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 2.Sontheimer RD, McCauliffe DP, Zappi E, Targoff I. Antinuclear antibodies: Clinical correlations and biologic significance. Adv Dermatol. 1992;7:3–52. discussion 53. [PubMed] [Google Scholar]
- 3.Heinlen LD, McClain MT, Ritterhouse LL, Bruner BF, Edgerton CC, Keith MP, James JA, Harley JB. 60 kD Ro and nRNP a frequently initiate human lupus autoimmunity. PLOS ONE. 2010;5:e9599. doi: 10.1371/journal.pone.0009599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izmirly PM, Llanos C, Lee LA, Askanase A, Kim MY, Buyon JP. Cutaneous manifestations of neonatal lupus and risk of subsequent congenital heart block. Arthritis Rheum. 2010;62:1153–1157. doi: 10.1002/art.27333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sim S, Wolin SL. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip Rev RNA. 2011;2:686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolin SL, Reinisch KM. The Ro 60 kDa autoantigen comes into focus: Interpreting epitope mapping experiments on the basis of structure. Autoimmun Rev. 2006;5:367–372. doi: 10.1016/j.autrev.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Clancy RM, Alvarez D, Komissarova E, Barrat FJ, Swartz J, Buyon JP. Ro60-associated single-stranded RNA links inflammation with fetal cardiac fibrosis via ligation of TLRs: A novel pathway to autoimmune-associated heart block. J Immunol. 2010;184:2148–2155. doi: 10.4049/jimmunol.0902248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung T, Pratt GA, Sundararaman B, Townsend MJ, Chaivorapol C, Bhangale T, Graham RR, Ortmann W, Criswell LA, Yeo GW, Behrens TW. The Ro60 autoantigen binds endogenous retroelements and regulates inflammatory gene expression. Science. 2015;350:455–459. doi: 10.1126/science.aac7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Smith JD, Shi H, Yang DD, Flavell RA, Wolin SL. The Ro autoantigen binds misfolded U2 small nuclear RNAs and assists mammalian cell survival after UV irradiation. Curr Biol. 2003;13:2206–2211. doi: 10.1016/j.cub.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Quinn AM, Wolin SL. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–782. [PMC free article] [PubMed] [Google Scholar]
- 11.Deshmukh US, Lewis JE, Gaskin F, Kannapell CC, Waters ST, Lou Y-h, Tung KSK, Fu SM. Immune responses to Ro60 and its peptides in mice. I. The nature of the immunogen and endogenous autoantigen determine the specificities of the induced autoantibodies. J Exp Med. 1999;189:531–540. doi: 10.1084/jem.189.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xue D, Shi H, Smith JD, Chen X, Noe DA, Cedervall T, Yang DD, Eynon E, Brash DE, Kashgarian M, Flavell RA, Wolin SL. A lupus-like syndrome develops in mice lacking the Ro 60-kDa protein, a major lupus autoantigen. Proc Natl Acad Sci USA. 2003;100:7503–7508. doi: 10.1073/pnas.0832411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago-Raber ML, Baudino L, Izui S. Emerging roles of TLR7 and TLR9 in murine SLE. J Autoimmun. 2009;33:231–238. doi: 10.1016/j.jaut.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Sim S, Wurtmann EJ, Feke A, Wolin SL. Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA. 2014;20:1715–1724. doi: 10.1261/rna.047241.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR, Jr, Hendrix RW, Hatfull GF. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/s0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 16.Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, Wardemann H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4+ memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindop R, Arentz G, Bastian I, Whyte AF, Thurgood LA, Chataway TK, Jackson MW, Gordon TP. Long-term Ro60 humoral autoimmunity in primary Sjögren’s syndrome is maintained by rapid clonal turnover. Clin Immunol. 2013;148:27–34. doi: 10.1016/j.clim.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 19.McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med. 2005;11:85–89. doi: 10.1038/nm1167. [DOI] [PubMed] [Google Scholar]
- 20.Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T cell epitope mimicry between Sjögren’s syndrome Antigen A (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin Immunol. 2014;152:1–9. doi: 10.1016/j.clim.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42:D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Routsias JG, Tzioufas AG, Sakarellos-Daitsiotis M, Sakarellos C, Moutsopoulos HM. Epitope mapping of the Ro/SSA60KD autoantigen reveals disease-specific antibody-binding profiles. Eur J Clin Invest. 1996;26:514–521. doi: 10.1046/j.1365-2362.1996.186316.x. [DOI] [PubMed] [Google Scholar]
- 24.Rioux JD, Goyette P, Vyse TJ, Hammarström L, Fernando MMA, Green T, De Jager PL, Foisy S, Wang J, de Bakker PIW, Leslie S, McVean G, Padyukov L, Alfredsson L, Annese V, Hafler DA, Pan-Hammarström Q, Matell R, Sawcer SJ, Compston AD, Cree BA, Mirel DB, Daly MJ, Behrens TW, Klareskog L, Gregersen PK, Oksenberg JR, Hauser SL. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci USA. 2009;106:18680–18685. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hevia A, Milani C, López P, Cuervo A, Arboleya S, Duranti S, Turroni F, González S, Suárez A, Gueimonde M, Ventura M, Sánchez B, Margolles A. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5:e01548–14. doi: 10.1128/mBio.01548-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puhvel SM, Reisner RM, Amirian DA. Quantification of bacteria in isolated pilosebaceous follicles in normal skin. J Invest Dermatol. 1975;65:525–531. doi: 10.1111/1523-1747.ep12610239. [DOI] [PubMed] [Google Scholar]
- 28.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell A, Patrick S, Dongyou L. Propionibacterium. In: Liu D, editor. Molecular Detection of Human Bacterial Pathogens. CRC Press; 2011. pp. 137–154. [Google Scholar]
- 30.Alekseyenko AV, Perez-Perez GI, De Souza A, Strober B, Gao Z, Bihan M, Li K, Methe BA, Blaser MJ. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiger R, Duhen T, Lanzavecchia A, Sallusto F. Human naive and memory CD4+ T cell repertoires specific for naturally processed antigens analyzed using libraries of amplified T cells. J Exp Med. 2009;206:1525–1534. doi: 10.1084/jem.20090504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Y, Goods BA, Raddassi K, Nepom GT, Kwok WW, Love JC, Hafler DA. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Transl Med. 2015;7:287ra274. doi: 10.1126/scitranslmed.aaa8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, Lanzavecchia A, Sallusto F. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347:400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 35.Wolin SL, Steitz JA. The Ro small cytoplasmic ribonucleoproteins: Identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci USA. 1984;81:1996–2000. doi: 10.1073/pnas.81.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venables PJ, Smith PR, Maini RN. Purification and characterization of the Sjögren’s syndrome A and B antigens. Clin Exp Immunol. 1983;54:731–738. [PMC free article] [PubMed] [Google Scholar]
- 37.Gaither KK, Fox OF, Yamagata H, Mamula MJ, Reichlin M, Harley JB. Implications of anti-Ro/Sjögren’s syndrome A antigen autoantibody in normal sera for autoimmunity. J Clin Invest. 1987;79:841–846. doi: 10.1172/JCI112892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth AJ, Ooi JD, Hess JJ, van Timmeren MM, Berg EA, Poulton CE, McGregor J, Burkart M, Hogan SL, Hu Y, Winnik W, Nachman PH, Stegeman CA, Niles J, Heeringa P, Kitching AR, Holdsworth S, Jennette JC, Preston GA, Falk RJ. Epitope specificity determines pathogenicity and detectability in ANCA-associated vasculitis. J Clin Invest. 2013;123:1773–1783. doi: 10.1172/JCI65292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silveira PA, Baxter AG. The NOD mouse as a model of SLE. Autoimmunity. 2001;34:53–64. doi: 10.3109/08916930108994126. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Nakagawa Y, Purushotham KR, Humphreys-Beher MG. Functional changes in salivary glands of autoimmune disease-prone NOD mice. Am J Physiol. 1992;263:E607–E614. doi: 10.1152/ajpendo.1992.263.4.E607. [DOI] [PubMed] [Google Scholar]
- 41.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe–host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuwana M. Beta2-glycoprotein I: Antiphospholipid syndrome and T-cell reactivity. Thromb Res. 2004;114:347–355. doi: 10.1016/j.thromres.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 43.Gong L, Wang Y, Zhou L, Bai X, Wu S, Zhu F, Zhu YF. Activation of toll-like receptor-7 exacerbates lupus nephritis by modulating regulatory T cells. Am J Nephrol. 2014;40:325–344. doi: 10.1159/000368204. [DOI] [PubMed] [Google Scholar]
- 44.Yokogawa M, Takaishi M, Nakajima K, Kamijima R, Fujimoto C, Kataoka S, Terada Y, Sano S. Epicutaneous application of toll-like receptor 7 agonists leads to systemic autoimmunity in wild-type mice: A new model of systemic lupus erythematosus. Arthritis Rheumatol. 2014;66:694–706. doi: 10.1002/art.38298. [DOI] [PubMed] [Google Scholar]
- 45.Jost SA, Tseng LC, Matthews LA, Vasquez R, Zhang S, Yancey KB, Chong BF. IgG, IgM, and IgA antinuclear antibodies in discoid and systemic lupus erythematosus patients. Scientific World Journal. 2014;2014:171028. doi: 10.1155/2014/171028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricker DM, Hebert LA, Rohde R, Sedmak DD, Lewis EJ, Clough JD. Serum C3 levels are diagnostically more sensitive and specific for systemic lupus erythematosus activity than are serum C4 levels. The Lupus Nephritis Collaborative Study Group. Am J Kidney Dis. 1991;18:678–685. doi: 10.1016/s0272-6386(12)80609-3. [DOI] [PubMed] [Google Scholar]
- 47.Bock M, Heijnen I, Trendelenburg M. Anti-C1q antibodies as a follow-up marker in SLE patients. PLOS ONE. 2015;10:e0123572. doi: 10.1371/journal.pone.0123572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganguly D, Haak S, Sisirak V, Reizis B. The role of dendritic cells in autoimmunity. Nat Rev Immunol. 2013;13:566–577. doi: 10.1038/nri3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, Kasper DL, Benoist C, Mathis D. Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci USA. 2016;113:E8141–E8150. doi: 10.1073/pnas.1617460113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper GS, Gilbert KM, Greidinger EL, James JA, Pfau JC, Reinlib L, Richardson BC, Rose NR. Recent advances and opportunities in research on lupus: Environmental influences and mechanisms of disease. Environ Health Perspect. 2008;116:695–702. doi: 10.1289/ehp.11092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruff WE, Kriegel MA. Autoimmune host-microbiota interactions at barrier sites and beyond. Trends Mol Med. 2015;21:233–244. doi: 10.1016/j.molmed.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Felton S, Navid F, Schwarz A, Schwarz T, Gläser R, Rhodes LE. Ultraviolet radiation-induced upregulation of antimicrobial proteins in health and disease. Photochem Photobiol Sci. 2013;12:29–36. doi: 10.1039/c2pp25158b. [DOI] [PubMed] [Google Scholar]
- 54.Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M. Temporal stability and composition of the ocular surface microbiome. Sci Rep. 2017;7:9880. doi: 10.1038/s41598-017-10494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SH, Oh DH, Jung JY, Kim JC, Jeon CO. Comparative ocular microbial communities in humans with and without blepharitis. Invest Ophthalmol Vis Sci. 2012;53:5585–5593. doi: 10.1167/iovs.12-9922. [DOI] [PubMed] [Google Scholar]
- 57.Birnbaum ME, Dong S, Garcia KC. Diversity-oriented approaches for interrogating T-cell receptor repertoire, ligand recognition, and function. Immunol Rev. 2012;250:82–101. doi: 10.1111/imr.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Doré J, Ehrlich SD, MetaHIT Consortium. Bork P, Wang J. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 59.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- 61.Tarn WY, Yario TA, Steitz JA. U12 snRNA in vertebrates: Evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- 62.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2011. http://www.R-project.org/ [Google Scholar]
- 63.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bateman A, Kickhoefer V. The TROVE module: A common element in Telomerase, Ro and Vault ribonucleoproteins. BMC Bioinformatics. 2003;4:49. doi: 10.1186/1471-2105-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I, Bork P. Interactive tree of life iTOL v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.The PyMOL Molecular Graphics System, Version 1.8 Schrodinger, LLC.
- 67.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers PCR-SSP in 2 hours: An alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39:225–235. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 69.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58:572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 73.Patel P, Garson JA, Tettmar KI, Ancliff S, McDonald C, Pitt T, Coelho J, Tedder RS. Development of an ethidium monoazide–enhanced internally controlled universal 16S rDNA real-time polymerase chain reaction assay for detection of bacterial contamination in platelet concentrates. Transfusion. 2012;52:1423–1432. doi: 10.1111/j.1537-2995.2011.03484.x. [DOI] [PubMed] [Google Scholar]
- 74.Deutscher SL, Harley JB, Keene JD. Molecular analysis of the 60-kDa human Ro ribonucleoprotein. Proc Natl Acad Sci USA. 1988;85:9479–9483. doi: 10.1073/pnas.85.24.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM. Structural insights into RNA quality control: The Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raddassi K, Kent SC, Yang J, Bourcier K, Bradshaw EM, Seyfert-Margolis V, Nepom GT, Kwok WW, Hafler DA. Increased frequencies of myelin oligodendrocyte glycoprotein/MHC class II-binding CD4 cells in patients with multiple sclerosis. J Immunol. 2011;187:1039–1046. doi: 10.4049/jimmunol.1001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Millican DS, Bird IM. Preparation of single-stranded antisense cDNA probes by asymmetric PCR. Methods Mol Biol. 1998;105:337–350. doi: 10.1385/0-89603-491-7:337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequence alignment of full-length hRo60 and selected commensal orthologs.
Fig. S2. Coimmunoprecipitation of lupus study subject sera confirmed anti-Ro60 antibody status.
Fig. S3. α-Diversity represented by box plots of the Shannon-Weiner diversity index.
Fig. S4. SCLE skin eruption.
Fig. S5. TT-reactive CD4+ T cell clone from a healthy donor generated by a CD4 T cell library assay.
Fig. S6. Ro60-negative SLE patient CD4+ T cells lack reactivity to hRo60 protein.
Fig. S7. Cytokine concentrations (pg/ml) of supernatants from the cross-reactive T cell clone from Fig. 5 measured using a bead-based immunoassay.
Fig. S8. Anti-Ro60 antibody status of Harvard cohort lupus subjects.
Fig. S9. YrlA RNA is not detected in immunoprecipitates from B. theta using human lupus sera.
Fig. S10. B. theta monocolonization of GF mice induces gut and systemic immune changes.
Fig. S11. Schematic of proposed mechanism of how Ro60 bacteria trigger and sustain autoimmunity.
Table S1. Commensal bacterial Ro60 orthologs identified by in silico methods.
Table S2. Lupus study subject clinical data.
Table S3. Healthy control study subject clinical data.
Table S4. Efficiency and specificity of bacterial Ro60 qPCR primers.
Table S5. Primary data.


