Abstract
Interactions between the gut microbiota and the host are important for health, where dysbiosis has emerged as a likely component of mucosal disease. The specific constituents of the microbiota that contribute to mucosal disease are not well defined. The authors sought to define microbial components that regulate homeostasis within the intestinal mucosa. Using an unbiased, metabolomic profiling approach, a selective depletion of indole and indole-derived metabolites was identified in murine and human colitis. Indole-3-propionic acid (IPA) was selectively diminished in circulating serum from human subjects with active colitis, and IPA served as a biomarker of disease remission. Administration of indole metabolites showed prominent induction of IL-10R1 on cultured intestinal epithelia that was explained by activation of the aryl hydrocarbon receptor. Colonization of germ-free mice with wild-type Escherichia coli, but not E. coli mutants unable to generate indole, induced colonic epithelial IL-10R1. Moreover, oral administration of IPA significantly ameliorated disease in a chemically induced murine colitis model. This work defines a novel role of indole metabolites in anti-inflammatory pathways mediated by epithelial IL-10 signaling and identifies possible avenues for utilizing indoles as novel therapeutics in mucosal disease.
The mammalian gastrointestinal tract plays host to trillions of microbes, collectively termed the microbiota, where a critical mutualism exists within the intestinal mucosa. The microbiota contributes significantly to gut homeostasis but can also contribute to establishing and maintaining mucosal disease.1 Intestinal mucosal surfaces act as primary barriers to microbial invasion, where commensal bacteria work in a dynamic and intimate interaction with the gut epithelium and influence host cellular and immune responses.2 Inflammatory bowel disease (IBD) is a chronic inflammatory disease of the gastrointestinal tract that comprises Crohn disease and ulcerative colitis (UC). It is known that IBD is caused by interactions between genetic and environmental factors, and results in perturbations of the microbiota, though precisely how microbial factors affect gut homeostasis and immune response have not been extensively explored.3, 4, 5
The significance of IL-10 signaling is well established in IBD. This anti-inflammatory cytokine, which signals through the IL-10 receptor ligand-binding subunit (IL-10R1), is induced during inflammation and attenuates excessive production of proinflammatory mediators in various cell types, including intestinal epithelial cells (IEC).6, 7 Functional IL-10 signaling is associated with enhanced mucosal barrier function and results in maintenance and homeostasis of the epithelia.7 Previous work, including our own, has shown that the epithelial IL-10R1 contributes fundamentally to resistance to intestinal inflammation and represents a component of epithelial innate immunity originally described to be induced by cytokines such as interferon-γ (IFN-γ).8 For instance, mice deficient in IL-10 or IL-10 receptor develop spontaneous severe colitis, and mice conditionally lacking intestinal epithelial IL-10R1 show increased susceptibility to colitis.7, 9, 10
Metabolomic analysis has revealed that gut bacteria impact host immunity through a variety of metabolites, including indole metabolites,11 which originate from the microbial metabolism of tryptophan. Indole-3-propionic acid (IPA) and indole-3-aldehyde (IAld), which are tryptophan metabolites produced by intestinal bacteria, are known for their intercellular signaling activity. Further, IAld has recently been identified as an aryl hydrocarbon receptor (AHR) ligand.12 AHR is a ligand-dependent transcription factor activated by a variety of synthetic and biological molecules that plays an important role in immunological response and inhibition of inflammation.13 AHR contributes to immune homeostasis through various methods, including T-cell differentiation and Th17 development,14, 15 as well as the up-regulation of IL-22 production.16 Recently, several studies have shown that the host microbiota provides a consistent source of endogenous AHR ligands with distinct effects on immune homeostasis.16, 17, 18 However, the precise role of indole metabolites in the gastrointestinal tract remains elusive. Together, these data lead us to hypothesize that microbial-derived indole metabolites promote intestinal homeostasis through AHR-mediated regulation of the IEC IL-10R1.
Materials and Methods
Animal Studies
For metabolomics analysis, C57BL/6-129 mice were administered 3% (wt/vol) dextran sodium sulfate (DSS) (molecular weight 36,000 to 50,000; MP Biochemicals, Burlingame, CA) in drinking water for 5 days, followed by a 2-day recovery period. Normal tap water was returned during these 2 days before tissue collection. Control mice were maintained on tap water for 7 days. In subsequent animal experiments, 8– to 12-week–old C57BL/6 mice were administered water or 2.5% DSS ad libitum for 9 days. This administration approach varied slightly to account for a change in death susceptibility to DSS. DSS was then removed and mice were allowed to recover for 2 days before euthanasia. For IPA treatment experiments, the addition of IPA at 0.1 mg/mL was administered to water- and DSS-treated animals. Mice were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research Protocols, and all animal work was approved by the Institutional Animal Care and Use Committee of the University of Colorado.
Cell Lines and shRNA Knockdown
Human T84 IEC were cultured in 1:1 Dulbecco's modified Eagle's medium–Ham's F12 with 2.5 mmol/L l-glutamine and 10% fetal bovine serum. Cells were maintained at 37°C with 5% CO2. Transepithelial electrical resistances were monitored using an EVOM2 Voltohmmeter (World Precision Instruments, Sarasota, FL). Cytokines were purchased from R&D Systems (Minneapolis, MN). IAld, IPA, and AHR inhibitor, CH-223191, were obtained from Sigma-Aldrich (St. Louis, MO). 6-Formylindole[3,2-b] carbazole was obtained from Tocris Bioscience (Bristol, UK). All compounds were used at indicated concentrations. Human intestinal organoids (HIOs) were derived following previously described methods19 and kindly provided by Dr. Jason Spence (University of Michigan, Ann Arbor, MI). HIOs were maintained by embedding in Matrigel (BD Biosciences, San Jose, CA) and applying Advanced DMEM-F12 medium (Invitrogen, Carlsbad, CA) containing 1X B27 supplement (Invitrogen), 1X GlutaMAX (Life Technologies, Carlsbad, CA), 10 μmol/L HEPES, 10% penicillin/streptomycin, 100 ng/mL rhNoggin (R&D Systems), 100 ng/mL epidermal growth factor (R&D Systems), and approximately 500 ng/mL R-Spondin1 (RSPO1). RSPO1 was obtained from conditioned media collected from a HEK293 cell line that was stably transfected and zeocin-selected for the RSPO1 expression vector. The medium was changed every 2 to 4 days, and HIOs were transferred to fresh Matrigel once a week until they reached approximately 0.5 mm to 1 mm in size for experiments and RNA isolation. Lentiviral particles encoding shRNA directed against aryl hydrocarbon receptor nuclear translocator (ARNT; TRC MISSION shRNA, University of Colorado Functional Genomics Facility) were used to transduce T84 cells using standard protocols. Stable integration was achieved by puromycin selection at 6 μg/mL. Knockdown was confirmed by real-time quantitative PCR analysis, indicating 80% to 85% depletion of ARNT levels.
RNA Isolation and Real-Time PCR
Total RNA was extracted from cells using TRIzol (Invitrogen) and from tissue using RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was prepared using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time PCR to measure transcripts was performed in 1 × Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) using an ABI 7300 thermocycler (Applied Biosystems). Fold-change in expression of target mRNA relative to β-actin mRNA was calculated as previously described.20 Total RNA from HIOs was isolated using Direct-zol RNA Miniprep Kit (Zymo Research Corp, Irvine, CA). Fold-change in expression of mRNA transcript from HIO experiments was calculated relative to HSPCB (heat shock protein 90 α family class B member 1). Human and mouse primer sequences are listed in Table 1.
Table 1.
Primer Sequences for Real-Time Quantitative PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Human | ||
| ACTB | 5′-CCTGGCACCCAGCACAAT-3′ | 5′-GCCGATCCACACGGAGTACT-3′ |
| HSPCB | 5′-TCTGGGTATCGGAAAGCAAGCC-3′ | 5′-GTGCACTTCCTCAGGCATCTTG-3′ |
| IL10R1 | 5′-CACATCCTCCACTGGACACC-3′ | 5′-CAAGGTCACTGCGGTAAGGT-3′ |
| SOCS3 | 5′-GGCCACTCTTCAGCATCTC-3′ | 5′-ATCGTACTGGTCCAGGAACTC-3′ |
| CYP1A1 | 5′-ACCCGCCACCCTTCGACAGTTC-3′ | 5′-TGCCCAGGCGTTGCGTGAGAAG-3′ |
| CYP1B1 | 5′-CTGGCACTGACGACGCCAAGA-3′ | 5′-TGGTCTGCTGGATGGACAGC-3′ |
| MDR1 | 5′-AACGGAAGCCAGAACATTCC-3′ | 5′-AGGCTTCCTGTGGCAAAGAG-3′ |
| Mouse | ||
| Actb | 5′-TACGGATGTCAACGTCACAC-3′ | 5′-AAGAGCTATGAGCTGCCTGA-3′ |
| Il10r1 | 5′-CCCATTCCTCGTCACGATCTC-3′ | 5′-TCAGACTGGTTTGGGATAGGTTT-3′ |
Western Blot Analysis
Whole-cell and tissue lysates were extracted in Tris-Lysis buffer on ice and disrupted by sonication. Protein content was quantified using BCA protein assay reagent (Thermo Fisher Scientific, Waltham, MA) and 30 μg of whole-cell or tissue extract was boiled in Laemmli buffer in reducing conditions subjected to SDS-PAGE. The SDS-PAGE was transferred onto a polyvinylidene difluoride membrane and probed for IL-10R1 using a rabbit polyclonal IL-10RA antibody (dilution 1:1000; Thermo Fisher Scientific) and β-actin (dilution 1:10,000; Abcam, Cambridge, UK).
Metabolomic Analysis
Distal colon tissue (1 cm) was collected from control and DSS-treated mice. Tissues were flash-frozen and placed at −80°C. Global metabolomics was performed by Metabolon, Inc. (Durham, NC). Briefly, tissue samples were thawed and weighed in tared cryovials, and 80% ice-cold methanol was added at a ratio of 75 μL of solvent per mg of sample, then incubated overnight at 4°C to extract biochemicals. Internal standards were included to control for extraction efficiency. Following methanol extraction, colon samples were processed and analyzed as previously described.21
HPLC Metabolite Analysis
Indole derivatives were quantified in mouse and human samples using reversed-phase high-performance liquid chromatography with electrochemical coulometric array detection (EC-HPLC) (CoulArray; Thermo Fisher Scientific). Archived human serum samples from patients diagnosed with IBD or from healthy individuals were obtained under research protocols approved by the Colorado Multi-Institutional Review Board. Study participant demographics are listed in Table 2. Tissue and serum samples were extracted in 80% methanol, and protein precipitate was removed by centrifugation at 15,000 × g. Separation was achieved using an Acclaim Polar Advantage II C18 column (Thermo Fisher Scientific) at a flow rate of 1 mL/minute on a gradient of 10% to 55% acetonitrile in 50 mmol/L sodium phosphate buffer (pH 3), containing 0.42 mmol/L octanesulphonic acid as an ion-pairing agent. Calibration curves were composed by performing linear regression analysis of the peak area versus the analyte concentration. The data were quantified using the peak area in comparison to standards.
Table 2.
Study Participant Demographics
| Characteristic | Control | UC remission | Ulcerative colitis |
|---|---|---|---|
| Number | 20 | 20 | 15 |
| Sex | |||
| Male | 11 | 12 | 13 |
| Female | 9 | 8 | 2 |
| Age, years | 58.6 ± 10.9 | 46.5 ± 18.6 | 40.4 ± 13.4 |
| Disease location | |||
| Left-sided | NA | 6 | 1 |
| Extensive | NA | 14 | 12 |
| Proctitis | NA | 0 | 2 |
NA, not applicable.
Bacterial Strains and Gnotobiotic Colonization Experiments
Escherichia coli (E. coli) strains were obtained from GE Dharmacon (Lafayette, CO). An E. coli K12 parent strain [E. coli BW25113; wild type (WT)], along with strains containing a deletion of tnaA (E. coli JW3686; ΔtnaA) and tnaB (E. coli JW5619; ΔtnaB) were used for experiments. All strains were grown in Lysogeny broth (10 g/L tryptone, 10 g/L NaCl, 5 g/L yeast extract) or on Lysogeny broth–agar plates supplemented with appropriate antibiotics (kanamycin, 50 μg/mL), if needed. Cultures were grown overnight at 37°C to stationary phase and tested for the presence of indole by the addition of 200 μL of Kovac's reagent (Sigma-Aldrich) to 2 mL bacterial culture. For T84 cell experiments, cultures were grown overnight at 37°C to stationary phase. Briefly, cultures were spun, supernatants collected, and afterward serially diluted and placed onto cells for 24 hours. Bacterial supernatants used for EC-HPLC analysis were placed through a 0.22-μm filter before the run. For gnotobiotic colonization experiments, 5- to 8-week–old germ-free C57BL/6 mice were gavaged with a 100-μL bacterial suspension for mono-association (109 colony forming units of each bacterial strain collected from a stationary-phase culture and re-suspended in phosphate-buffered saline). Mice were colonized for 2 weeks before euthanasia. Fresh fecal pellets were collected periodically, weighed, homogenized, and then serially diluted in phosphate-buffered saline phosphate-buffered saline for plating to determine bacterial colony forming units per g of feces.
Histology
Colon samples were fixed in 10% neutral buffered formalin and paraffin embedded before staining with hematoxylin and eosin. All histologic quantitation was performed blinded by the same individual (S.P.C.) using a scoring system previously described.22 Briefly, the three independent parameters measured were severity of inflammation (0 to 3: none, slight, moderate, severe), extent of injury (0 to 3: none, mucosal, mucosal and submucosal, transmural), and crypt damage (0 to 4: none, basal 1/3 damaged, basal 2/3 damaged, only surface epithelium intact, entire crypt and epithelium lost). The score of each parameter was multiplied by a factor reflecting the percentage of tissue involvement (×1: 0% to 25%, ×2: 26% to 50%, ×3: 51% to 75%, ×4: 76% to 100%), and all numbers were summed. Maximum possible score was 40.
Quantification of Cytokines in Colon Tissue
For cytokine analysis, colon tissue was extracted in Tris-Lysis buffer by sonication and protein homogenates were quantified using BCA protein assay reagent (Thermo Fisher Scientific). Tissue concentrations of cytokines were measured using a proinflammatory cytokine screen (Meso Scale Discovery, Rockville, MD). Assays were performed according to the manufacturer's instructions. Cytokine concentrations were normalized to total protein concentration.
Quantification and Statistical Analysis
Data are expressed as means ± SEM. Statistical analyses were performed with GraphPad Prism software version 7.0 (GraphPad Software, La Jolla, CA) using a two-tailed unpaired t-test for direct comparisons, and one-way or two-way analysis of variance with Tukey's test for multiple comparisons. Statistical differences are reported as significant when P < 0.05.
Results
Tryptophan-Indole Metabolism Is Altered in Murine and Human Colitis
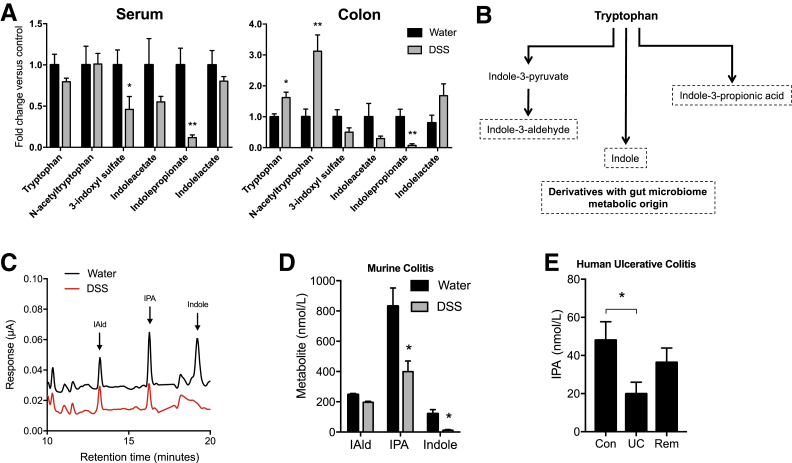
To better understand microbe-derived factors that contribute to intestinal homeostasis, a comprehensive, unbiased screen of serum and colonic tissue metabolites was performed from healthy and DSS colitic mice. Mice were administered DSS via drinking water, and serum and colonic tissue were collected during peak disease (day 7). Of the microbe-derived metabolites, most notable were decreases in tryptophan-indole metabolites in both serum and colons of colitic mice compared with mice administered water alone (Figure 1A). Tryptophan is metabolized through various pathways, including the indole pathway, resulting in derivatives that are produced from metabolism by gut microbiota (Figure 1B). Colitis profoundly altered tryptophan metabolism, specifically revealing a selective decrease in indole metabolites. To validate this mass spectrometry–based metabolite screen, an HPLC-based protocol was developed using electrochemical detection methods (Figure 1C). Eight- to 10-week–old C57BL/6 mice were administered water or 2.5% DSS ad libitum for 9 days. DSS was then removed and mice were allowed to recover for 2 days before euthanasia. Serum indole metabolites were profiled by EC-HPLC (Figure 1D). Serum indole and IPA levels were significantly decreased in actively colitic animals (P < 0.05). A marked decrease in serum IAld was also observed (P = 0.09). It is notable that overall, tryptophan levels in DSS colitis actually increase (Figure 1A), suggesting that the findings of indole depletion are not a result of diminished tryptophan absorption.
Figure 1.
Tryptophan metabolism is altered in murine and human colitis. A: Relative levels of indole metabolites in serum and whole colon tissue of mice receiving either water or 3% dextran sodium sulfate (DSS) for 7 days. Metabolites were measured by liquid chromatography–mass spectrometry and gas chromatography–mass spectrometry analysis. B: Condensed pathway of tryptophan metabolism to indole and indole derivatives. C: EC-HPLC analysis of indole metabolites in serum of mice receiving either water or 2.5% DSS for 9 days. D: Concentrations of indole metabolites in serum of mice receiving either water or 2.5% DSS for 9 days. E: Indole-3-propionic acid (IPA) levels in serum samples from healthy controls (Con), subjects with active ulcerative colitis (UC), and subjects with UC in remission (Rem) profiled by EC-HPLC. Data are expressed as means ± SEM (A, D, and E). n = 5 (A, mice per treatment group, and D, water treatment); n = 10 (D, DSS treatment); n = 20 (E, control and remission); n = 15 (E, UC). ∗P < 0.05, ∗∗P < 0.01, t-test and one-way analysis of variance. IAld, indole-3-aldehyde.
Guided by results of indole depletion in active murine colitis, we attempted to translate our results to human patients. Here, the EC-HPLC method was used to quantify various indole metabolites in serum samples from patients with UC. For these purposes, serum samples from healthy controls (n = 20), subjects with active UC (n = 15), and subjects with UC in remission (n = 20) were profiled (Table 2). This analysis revealed that serum IPA was deceased by nearly 60% in subjects with active UC compared with healthy controls (P < 0.05) (Figure 1E). Notably, this IPA deficiency normalized in UC patients in remission, implicating IPA as both a biomarker for active UC as well as an indicator of disease remission in human UC. Based on these findings, the role of indole and metabolites IPA and IAld on intestinal epithelial function was further studied.
Indole Metabolites Induce IL-10R1 Expression on Intestinal Epithelia and Improve Barrier Formation
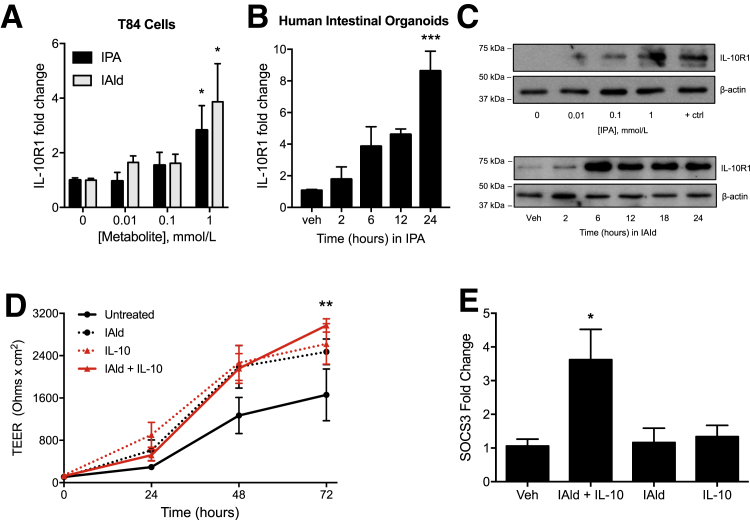
We have previously shown that tryptophan metabolites are important regulators of IL-10R1 in IEC.23 Guided by our unbiased metabolomic profile of serum and colon tissue from healthy and DSS-colitic mice, and our recent work identifying the epithelial interleukin-10 receptor as the dominant signature for resolution of inflammation in DSS colitis,6, 7 it was hypothesized that indole-containing tryptophan metabolites regulate intestinal homeostasis via regulation of epithelial IL-10R1 expression. To examine the induction of IL-10R1 in response to indole metabolites, T84 IEC were exposed to IPA or IAld for 6 hours; real-time quantitative PCR of IL-10R1 transcript levels showed a concentration-dependent induction of IL-10R1 (Figure 2A). These findings were not limited to colonic cancer cell lines. HIOs are complex, three-dimensional spheroid tissues derived from human pluripotent stems cells19 and contain the majority of functional epithelial cell types (ie, enterocytes, goblet, Paneth, enteroendocrine, intestinal stem cells) and structures (ie, brush borders of microvilli, crypt-like structures, and a mesoderm layer) comprising the human small intestine.24 HIOs treated with IPA exhibited increasing levels of IL-10R1 transcript, with significant induction at 24 hours (P < 0.001) (Figure 2B). Further analysis revealed a prominent induction of IL-10R1 protein expression in T84 IEC exposed to IPA and IAld (Figure 2C).
Figure 2.
Indole metabolites improve barrier formation and induce IL-10R1 on intestinal epithelia. A: Real-time quantitative PCR (qPCR) of IL-10R1 transcript levels in T84 cells treated with indole-3-propionic acid (IPA) or indole-3-aldehyde (IAld) at varying concentrations for 6 hours. B: Human intestinal organoids treated with 1 mmol/L IPA over 24 hours. C: Western blot analysis of IL-10R1 levels in T84 cells treated with IPA at varying concentrations for 6 hours or in response to 1 mmol/L IAld over 24 hours. D: Transepithelial electrical resistance of T84 cells treated with IAld (1 μmol/L), IL-10 (10 ng/mL), or a combination of both was measured over 72 hours. E: qPCR of SOCS3 transcript in T84 cells treated with IAld for 12 hours, followed by treatment with IL-10 for 6 hours. Data are expressed as the average TEER ± SEM (D) and as means ± SEM (A, B, and E). n = 3 (D, samples, and E, experiments). ∗P < 0.05, t-test, ∗∗P < 0.01 versus untreated cells, two-way analysis of variance, and ∗∗∗P < 0.001, t-test. +ctrl, 6-formylindole[3,2-b] carbazole at 1 μmol/L; TEER, transepithelial electrical resistance; Veh, vehicle.
The influence of the metabolite IAld on barrier formation in T84 cell monolayers was further examined, as our previous work has demonstrated that IL-10 signaling is important in IEC barrier development and maintenance.7, 23 Cells exposed to both IAld and IL-10 exhibited significantly increased barrier formation at 72 hours compared with untreated cells as measured by transepithelial electrical resistances (P < 0.01) (Figure 2D). Further, IAld in combination with IL-10 (10 ng/mL) significantly induced suppressor of cytokine signaling 3 (SOCS3), an IL-10–responsive gene that has been found to be protective in intestinal inflammation25 (P < 0.05) (Figure 2E). From this perspective, it is notable that SOCS3 was not induced by IL-10 alone as a result of nearly undetectable levels of IL-10R1 at baseline.6, 7 Overall, these results identify indole metabolite–dependent induction of the epithelial IL-10R1 as a target pathway for mucosal homeostasis.
Loss of Indole-Dependent IL-10R1 Induction in Cells Lacking ARNT
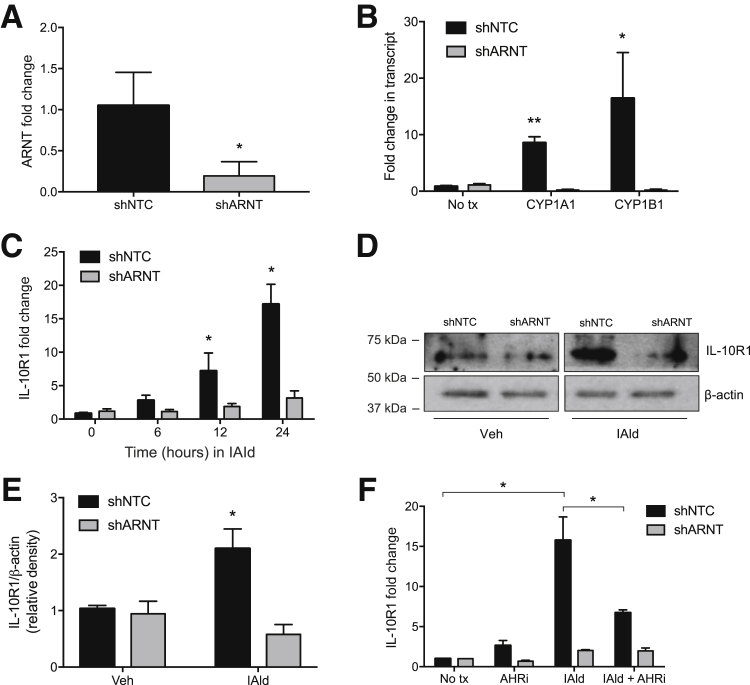
The mechanism of indole signaling in intestinal epithelia was studied next. Others have shown that tryptophan metabolites function as endogenous ligands for AHR.26 To determine the relative contribution of AHR to indole signaling, shRNA knockdown was used to deplete the AHR dimeric partner ARNT, and IL-10R1 induction was examined as an endpoint. Lentiviral shRNA-mediated knockdown of ARNT (shARNT) in T84 IEC resulted in significantly reduced ARNT mRNA expression (81% ± 5% decrease; P < 0.01) relative to cells containing a nontemplate control (shNTC) (Figure 3A). A broader examination of classical AHR-ARNT target genes26, 27, 28 revealed that transcript levels of cytochrome P450 family 1 subfamily A member 1 (CYP1A1) and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) were significantly increased in shNTC cells treated with IAld compared with untreated shNTC cells. Both targets were also significantly increased in the IAld-treated shNTC compared with shARNT T84 IEC following IAld treatment (P < 0.01, P < 0.05, respectively) (Figure 3B). Analysis of these ARNT-deficient cells revealed a prominent decrease in IAld-dependent induction of IL-10R1 transcript and protein (P < 0.05) (Figure 3, C–E). The AHR small-molecule inhibitor, CH-223191, serves as a specific AHR antagonist. shNTC and shARNT T84 IEC were exposed to IAld (1 mmol/L), CH-223191 (10 μmol/L), or in combination for 24 hours. IAld treatment alone significantly increased IL-10R1 expression at 24 hours. CH-223191 significantly reduced IAld-mediated IL-10R1 induction, supporting the hypothesis that IAld modulates IL-10R1 expression via an AHR-mediated mechanism (P < 0.05) (Figure 3F). These results suggest that the AHR pathway is involved in the indole metabolite–dependent expression of IL-10R1 in IEC.
Figure 3.
IL-10R1 expression is not induced by cells lacking aryl hydrocarbon receptor (AHR) binding partner aryl hydrocarbon receptor nuclear translocator (ARNT). A: Lentiviral shRNA-mediated knockdown of ARNT (shARNT) in T84 intestinal epithelial cells relative to T84 cells containing a nontemplate control (shNTC). B: Transcript levels of cytochrome P450 family 1 subfamily A member 1 (CYP1A1) and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) by real-time quantitative PCR (qPCR) in shNTC and shARNT T84 cells following indole-3-aldehyde (IAld) treatment for 12 hours. C: qPCR analysis of IL-10R1 expression in shNTC and shARNT T84 cells treated with 1 mmol/L IAld for 24 hours. D: Western blot analysis of IL-10R1 levels in ARNT knockdown T84 cells. Confluent monolayers of T84 cells were treated with IAld for 24 hours. E and F: Protein expression was quantified by densitometry (E) and normalized to β-actin. (F) qPCR of IL-10R1 transcript levels in shNTC and shARNT T84 cells treated with AHR inhibitor (AHRi, 10 μmol/L), IAld (1 mmol/L), or both for 24 hours. Data are expressed as means ± SEM (A–C, E, and F). n = 3. ∗P < 0.05, ∗∗P < 0.01 versus untreated shNTC, t-test. tx, treatment; Veh, vehicle.
Bacterial Indole Production Induces IL-10R1
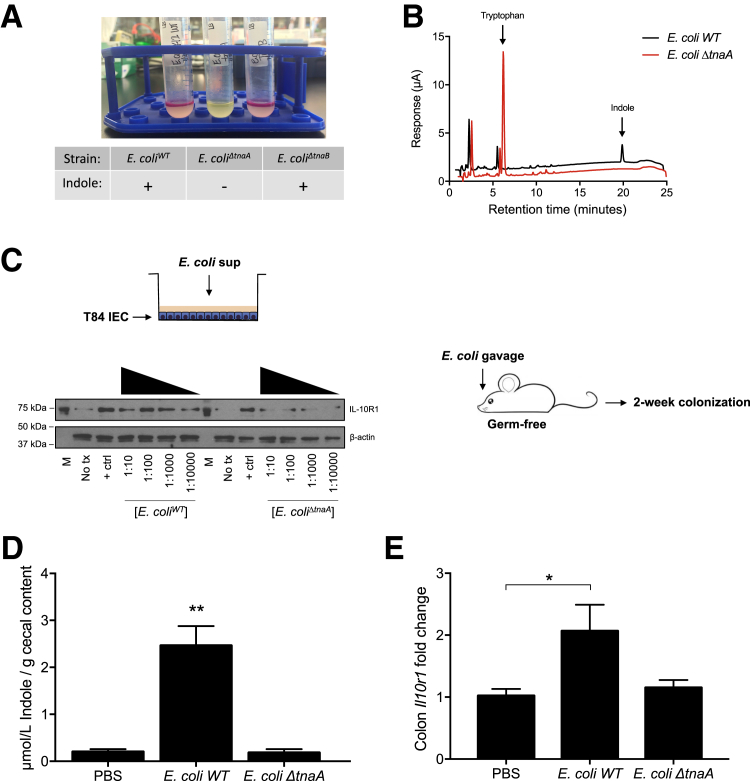
Whether microbe-derived sources of indole could similarly regulate epithelial target genes was examined next. Indole production was targeted in E. coli. Indole is synthesized from tryptophan by microbial tryptophanase.29 Using Kovac's reagent to test for the presence of indole, an E. coli K12 strain (E. coli BW25113; WT) clearly produced indole (Figure 4A), whereas deletion of the tryptophanase gene (ΔtnaA) revealed a lack of detectable indole. Conversely, deletion of one of the tryptophan transporters (ΔtnaB) did not compromise indole production (Figure 4A). To validate these observations, the presence of indole in cell-free supernatants of bacteria was examined by EC-HPLC. Although E. coli WT bacteria exhibited readily detectable levels of indole, metabolism of tryptophan to indole was completely abolished in E. coli ΔtnaA (Figure 4B). To test the activity of this microbial-derived indole, cell-free supernatants from E. coli WT and ΔtnaA mutant were serially diluted and exposed to T84 IEC for 24 hours. Supernatants from E. coli WT readily induced IL-10R1 protein expression, whereas minimal IL-10R1 induction was observed in cells exposed to E. coli ΔtnaA supernatant (Figure 4C).
Figure 4.
Bacterial indole production induces IL-10R1. A: Indole production test of cultures from E. coli K12 WT, tnaA, and tnaB mutants; plus sign indicates presence of indole. B: EC-HPLC chromatogram of supernatants collected from E. coli K12 WT and E. coli ΔtnaA. C: T84 cells were treated for 24 hours with serially diluted supernatants from E. coli WT or E. coli ΔtnaA cultures grown to stationary phase, followed by media replenishment. Germ-free mice were colonized with E. coli WT or E. coli ΔtnaA for 2 weeks followed by euthanasia. D and E: Indole concentrations in cecal contents were validated by HPLC in vehicle and colonized mice (D), and RNA was extracted from colons for real-time quantitative PCR analysis of Il10r1 expression (E). Data are expressed as means ± SEM (D and E). n = 5 (D and E, PBS and E. coli ΔtnaA); n = 4 (D and E, E. coli WT). ∗P < 0.05, ∗∗P < 0.01, t-test. + ctrl, interferon-γ at 10 ng/mL; M, molecular marker; PBS, phosphate-buffered saline; sup, supernatant; tx, treatment.
These findings were extended to an in vivo model. The function of indole-producing bacteria was examined on epithelial IL-10R1 in germ-free mice. Germ-free mice were colonized with E. coli WT or E. coli ΔtnaA for 2 weeks followed by euthanasia. Fresh fecal pellets were collected periodically for plating to ensure equal colonization, quantified as bacterial colony forming units per gram of feces (data not shown). Indole concentrations in cecal contents were validated by HPLC in vehicle and colonized animals. Mice monocolonized with E. coli WT displayed significantly increased cecal indole, whereas negligible levels were measured in phosphate-buffered saline– and E. coli ΔtnaA–gavaged animals (P < 0.01) (Figure 4D). Further, colons were harvested and RNA extracted for real-time quantitative PCR analysis. This strategy revealed that mice monocolonized with E. coli WT displayed significantly increased colonic il10r1 expression, whereas no significant il10r1 induction was observed in colon tissue of mice colonized with E. coli ΔtnaA mutant (P < 0.05) (Figure 4E). Taken together, these in vitro and in vivo results strongly support the hypothesis that microbe-derived indoles promote epithelial homeostasis.
IPA Improves DSS Colitis Outcomes
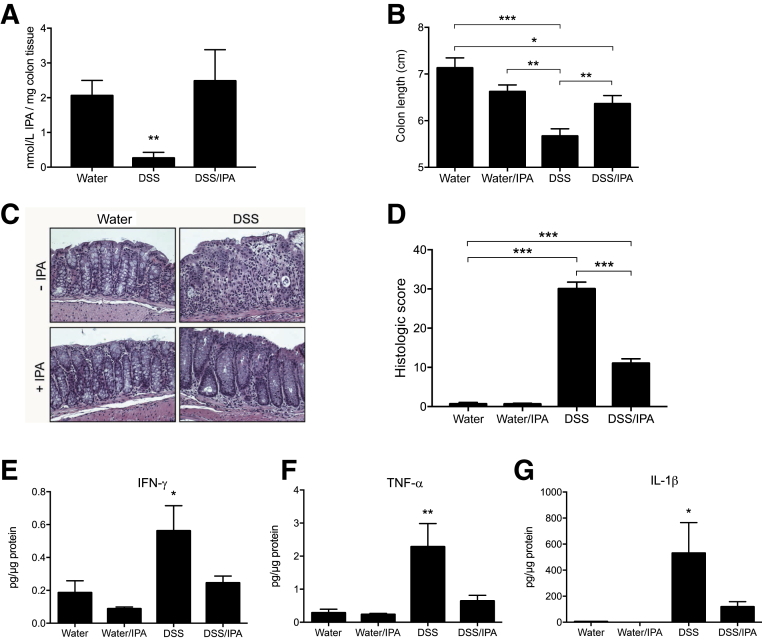
Given the observation that indole metabolites are significantly decreased in active colitis, the therapeutic potential of IPA was tested in murine colitis outcomes. For these purposes, mice were administered either normal drinking water, DSS (2.5% wt/vol), or a combination of DSS and IPA (0.1 mg/mL) in drinking water for 9 days, followed by DSS removal and a 2-day recovery. To verify that oral administration normalized IPA levels, colonic IPA was quantified by EC-HPLC. This analysis revealed that although DSS colitic mice displayed significantly lower levels of IPA, oral administration of IPA normalized colonic levels of this metabolite (P < 0.01) (Figure 5A). Under these treatment conditions, DSS/IPA-treated animals displayed significantly less reduction in colon length, a marker of intestinal inflammation, compared with DSS-treated animals (P < 0.05) (Figure 5B). Histologic analysis revealed that DSS/IPA-treated mice displayed attenuated inflammatory infiltration and decreased loss of architecture in comparison with mice treated with DSS alone, which displayed pronounced loss of epithelium and tissue architecture (Figure 5C). These differences resulted in greater histopathologic severity scores (P < 0.001) (Figure 5D). Tissue cytokine levels were also examined in these mice. Proinflammatory cytokine levels were dramatically increased in DSS-treated mice, specifically IFN-γ (P < 0.05) (Figure 5E), tumor necrosis factor-α (TNF-α) (P < 0.01) (Figure 5F), and IL-1β (P < 0.05) (Figure 5G), whereas cytokine levels in DSS/IPA-treated mice were similar to mice administered water alone. Similarly, IL-10 levels were significantly increased in DSS-treated mice, though IPA administration did not alter levels of this cytokine (Supplemental Figure S1). Further, IPA administration increased transcript and protein levels of AHR target genes (Supplemental Figure S2). Taken together, these results indicate that therapeutic normalization of IPA during active colitis attenuates disease and promotes intestinal homeostasis.
Figure 5.
Indole-3-propionic acid (IPA) improves dextran sodium sulfate (DSS) colitis outcomes. Eight- to 10-week–old C57BL/6 mice were administered water or 2.5% DSS ± IPA ad libitum for 9 days. DSS was then removed and mice were allowed to recover for 2 days as predetermined before euthanasia. A: Concentration of IPA in colons of mice receiving DSS ± IPA for up to 9 days. B: Colon length measured at time of euthanasia. C: Representative hematoxylin and eosin–stained colonic sections isolated from water-treated, water/IPA-treated, DSS-treated, and DSS/IPA-treated mice. D: Histologic score of hematoxylin and eosin–stained colonic sections. E–G: Colon tissue was homogenized and cytokines were measured in protein lysates by Meso Scale Discovery analysis [interferon-γ (IFN-γ; E); tumor necrosis factor–α (TNF-α; F); and IL-1β (G)]. Data are expressed as means ± SEM (A, B, D, and E–G). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, two-way analysis of variance and t-test. Original magnification, ×20.
Discussion
Recent efforts to define the complex nature of IBD have focused on the contribution of the host microbiota. It is now established that altered microbial composition, termed dysbiosis, is strongly associated with IBD30 and other intestinal diseases.31 Such dysbiosis is thought to alter the immune response and influence epithelial function, resulting in increased intestinal permeability.32 The particular components of the microbiome that promote disease and distinguish phenotypes are not well understood. In the current work, we sought to identify and characterize microbial-derived metabolites that contribute to colonic disease and homeostasis.
Immunometabolism is an area of significant interest in mucosal inflammation.33 Guided by an unbiased metabolomic profile of serum and colon tissue from healthy and DSS-colitic mice, a significant shift in tissue-associated, microbiota-derived indole derivatives was identified. This study shows that tryptophan does not decrease in murine colitis. In fact, tryptophan levels significantly increased in colons of colitic mice. On the basis of these findings, we believe the observed decrease of indole metabolites to be a result of a selective depletion of tryptophan-indole metabolites due to dysbiosis, rather than poor absorption of tryptophan.
Indole is produced from the action of microbial tryptophanase on tryptophan to produce indole, pyruvate, and ammonia via a β-elimination reaction.34 Microbes that express tryptophanase have the capacity to utilize tryptophan as a source of nitrogen, carbon, and energy.35 The tnaA gene, encoding tryptophanase, is highly conserved among Gram-negative indole-producing species, including Citrobacter, Morganella, Klebsiella, Providencia, and Haemophilus influenzae type b.36 The presence of tnaA is strongly associated with virulence of H. influenzae.37 At present, we do not know the nature of indole deficiencies associated with active inflammation, though our work is consistent with a study demonstrating that bacterial fermentation metabolites are less abundant in patients with IBD compared with healthy controls.38 Additionally, Wlodarska et al39 have recently shown that microbes of IBD patients have a reduced ability to cleave intestinal mucins and metabolize tryptophan. Such a precedent exists for other microbiota-derived metabolites. For example, studies investigating dysbiosis in IBD have identified lower concentrations of luminal short chain fatty acids and significant depletion of butyrate-producing organisms (eg, specific Faecalibacterium and Roseburia genera) with active colonic disease.40, 41, 42 It is therefore likely that the observed decreases in indole metabolism reflect inflammation-associated dysbiosis.
Some evidence exists that microbial-derived indole and indole metabolites could be anti-inflammatory. For instance, Shimada et al43 demonstrated that indole administration to germ-free mice increased the expression of some epithelial tight junction proteins and attenuated weight loss in a DSS colitis model. No mechanisms were evaluated to describe these changes elicited by indole. Moreover, Venkatesh et al44 have shown that IPA is a ligand for the pregnane X receptor (PXR) and promotes intestinal barrier integrity through down-regulation of epithelial TNFα, induction of MDR1, and regulation of epithelial junctional complexes. It is interesting to note that PXR, a member of the superfamily of nuclear receptors, is under consideration as a novel drug target in IBD.45 Further, Wlodarska et al39 have shown that the metabolite indoleacrylic acid promotes intestinal barrier function and mitigates inflammatory responses through down-regulation of genes involved in inflammation and oxidative stress. Conversely, it is also notable that some indole derivatives have also been shown to be nephrotoxic. For example, Devlin et al46 recently identified a widely distributed family of tryptophanases produced by commensal Bacteroides within the gut microbiota. They demonstrate that microbe-derived indoles are modified by the host to generate toxic renal levels of indoxyl sulfate and that colonization with tryptophanase-deficient Bacteroides decreases systemic levels of indoxyl sulfate that may be renal protective.
We translated these findings of shifts in indole metabolism from murine colitis to human subjects. This analysis revealed, for the first time, that serum indole metabolites in human subjects with active UC revealed a remarkable similarity to mice with active colitis, with the exception that human subjects were selectively deficient in IPA and not indole or IAld. The reason for this selective loss of IPA in humans is not known. It should be noted that indole and indole metabolites are produced by different pathways and different bacteria. Because IPA is an indole derivative conjugated to propionate, and propionate is among the short chain fatty acids that become depleted in active human IBD,40 it is possible that this selective depletion of IPA is explained by an IBD-associated dysbiosis that results from the combined depletion of both short chain fatty acid– and indole-producing microbiota. Further studies will be necessary to define the nature of this observation.
Therapeutic administration of oral IPA was protective in a murine model of colitis. Animals that received IPA not only exhibited fewer physical signs of disease, but also had significantly less damage to crypt structure and restricted inflammatory infiltration. Indoles and other tryptophan metabolites have been demonstrated to function as AHR ligands.26, 47, 48 It is likely that as AHR ligands, these molecules directly influence the epithelial/immune cell axis. For example, Zelante et al12 showed that IAld drives AHR-dependent IL-22 production and mucosal protection. Others have shown innate lymphoid cell AHR responses to be protective in inflammation through the production of IL-22.16 It is also noteworthy that intraepithelial lymphocytes localize in response to AHR stimulation by dietary ligands,49 including specific CD4+CD8αα+ subsets of intraepithelial T cells.50 The activity of AHR ligands on intestinal epithelia is less well understood. We recently demonstrated that similar to indole metabolites, other tryptophan-derived metabolites, including kynurenine, regulate epithelial IL-10R1 activity in an AHR-dependent manner.23 Such activity was associated with protection in colitis models and promoted epithelial wound healing. These varied ligands suggest a promiscuous ligand-binding pocket of AHR that is associated with agonistic activity.51 Moreover, these results suggest redundant mechanisms to maintain expression of functional epithelial IL-10 receptors, which have been shown to be essential for the development and maintenance of barrier function.7
Finally, it is known that colitis occurs as the result of immune cell responses to microbiota in susceptible hosts.52 These interactions are complex, involving multiple cell types and a spectrum of metabolites. In this regard, the contribution of cell types beyond the epithelium (eg, immune cells) in regulation by bacterial-derived metabolites in DSS cannot be ruled out. Rather, it is likely that these metabolites have a plethora of actions on various cell types in the mucosa. In our studies, we have limited this analysis to the epithelium to allow for a deeper and more mechanistic understanding of these signaling responses. These studies provide strong evidence for the role of microbiota-derived indole metabolites in anti-inflammatory pathways mediated by IL-10 signaling in the intestinal epithelium. These findings present new insight to our understanding of host-microbial communication within the mucosa and identify possible avenues for utilizing indoles as novel therapeutics in mucosal disease.
Acknowledgments
We thank Dr. Jason Spence and Sha Huang (University of Michigan, Ann Arbor, MI) for developing the human intestinal organoids from human pluripotent stem cells.
E.E.A., D.J.Ko., and S.P.C. designed the study; E.E.A., J.M.L., D.J.Ka., E.L.C., C.J.K., and K.D.B. performed experiments; M.E.G. provided human serum samples; B.R.J., S.T.W., and D.J.Ko. provided germ-free mice, human intestinal organoids, and corresponding in vitro experiments; E.E.A. and S.P.C. wrote the manuscript; and all authors read and edited the manuscript.
Footnotes
Supported by NIH grants DK1047893, DK50189, DK095491, DK103639, and DK103712 (S.P.C.) and VA Merit grant BX002182 (S.P.C.). Funding for generating and maintaining the human intestinal organoids was provided by the Bill and Melinda Gates Foundation (D.J. Ko.).
D.J.Ko. and S.P.C. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.01.011.
Contributor Information
Douglas J. Kominsky, Email: douglas.kominsky@montana.edu.
Sean P. Colgan, Email: sean.colgan@ucdenver.edu.
Supplemental Data
Indole-3-propionic acid (IPA) up-regulates IL-10R1 in dextran sodium sulfate (DSS) colitis. Eight- to 10-week–old C57BL/6 mice were administered water or 2.5% DSS ± IPA ad libitum for 9 days. DSS was then removed and mice were allowed to recover for 2 days as predetermined prior to euthanasia. A: Real-time quantitative PCR of Il10r1 transcript levels in colon tissue. B: IL-10 levels in colon tissue protein lysates were measured by Meso Scale Discovery analysis. Data are expressed as means ± SEM. n = 5 to 10 mice/group. ∗P < 0.05.
Indole-3-propionic acid (IPA) induces aryl hydrocarbon receptor (AHR) target genes in vivo. A and B: Transcript levels of cytochrome P450 family 1 subfamily A member 1 (CYP1A1) (A) and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) (B) were measured in colon tissue of 8- to 10-week–old C57BL/6 mice administered water or 2.5% dextran sodium sulfate (DSS) ± IPA ad libitum for 9 days. C: Further, colon tissue levels of IL-6 were measured in protein lysates by Meso Scale Discovery analysis. Data are expressed as means ± SEM. n = 5 to 10 mice/group. ∗P < 0.05.
References
- 1.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muniz L.R., Knosp C., Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol. 2012;3:310. doi: 10.3389/fimmu.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Huttenhower C., Kostic A.D., Xavier R.J. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelhardt K.R., Grimbacher B. IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr Top Microbiol Immunol. 2014;380:1–18. doi: 10.1007/978-3-662-43492-5_1. [DOI] [PubMed] [Google Scholar]
- 7.Kominsky D.J., Campbell E.L., Ehrentraut S.F., Wilson K.E., Kelly C.J., Glover L.E., Collins C.B., Bayless A.J., Saeedi B., Dobrinskikh E., Bowers B.E., MacManus C.F., Muller W., Colgan S.P., Bruder D. IFN-gamma-mediated induction of an apical IL-10 receptor on polarized intestinal epithelia. J Immunol. 2014;192:1267–1276. doi: 10.4049/jimmunol.1301757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colgan S.P., Parkos C.A., Matthews J.B., D'Andrea L., Awtrey C.S., Lichtman A., Delp C., Madara J.L. Interferon-γ induces a surface phenotype switch in intestinal epithelia: downregulation of ion transport and upregulation of immune accessory ligands. Am J Physiol. 1994;267(Pt 1):C402–C410. doi: 10.1152/ajpcell.1994.267.2.C402. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn R., Lohler J., Rennick D., Rajewsky K., Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhry A., Samstein R.M., Treuting P., Liang Y., Pils M.C., Heinrich J.M., Jack R.S., Wunderlich F.T., Bruning J.C., Muller W., Rudensky A.Y. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., Carvalho A., Puccetti P., Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Hubbard T.D., Murray I.A., Bisson W.H., Lahoti T.S., Gowda K., Amin S.G., Patterson A.D., Perdew G.H. Adaptation of the human aryl hydrocarbon receptor to sense microbiota-derived indoles. Sci Rep. 2015;5:12689. doi: 10.1038/srep12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascanfroni I.D., Takenaka M.C., Yeste A., Patel B., Wu Y., Kenison J.E., Siddiqui S., Basso A.S., Otterbein L.E., Pardoll D.M., Pan F., Priel A., Clish C.B., Robson S.C., Quintana F.J. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-alpha. Nat Med. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura A., Naka T., Nohara K., Fujii-Kuriyama Y., Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteleone I., Rizzo A., Sarra M., Sica G., Sileri P., Biancone L., MacDonald T.T., Pallone F., Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141:237–248. doi: 10.1053/j.gastro.2011.04.007. 248.e1. [DOI] [PubMed] [Google Scholar]
- 17.Gaitanis G., Magiatis P., Stathopoulou K., Bassukas I.D., Alexopoulos E.C., Velegraki A., Skaltsounis A.L. AhR ligands, malassezin, and indolo[3,2-b]carbazole are selectively produced by Malassezia furfur strains isolated from seborrheic dermatitis. J Invest Dermatol. 2008;128:1620–1625. doi: 10.1038/sj.jid.5701252. [DOI] [PubMed] [Google Scholar]
- 18.Vlachos C., Schulte B.M., Magiatis P., Adema G.J., Gaitanis G. Malassezia-derived indoles activate the aryl hydrocarbon receptor and inhibit Toll-like receptor-induced maturation in monocyte-derived dendritic cells. Br J Dermatol. 2012;167:496–505. doi: 10.1111/j.1365-2133.2012.11014.x. [DOI] [PubMed] [Google Scholar]
- 19.McCracken K.W., Howell J.C., Wells J.M., Spence J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albrecht E., Waldenberger M., Krumsiek J., Evans A.M., Jeratsch U., Breier M., Adamski J., Koenig W., Zeilinger S., Fuchs C., Klopp N., Theis F.J., Wichmann H.E., Suhre K., Illig T., Strauch K., Peters A., Gieger C., Kastenmuller G., Doering A., Meisinger C. Metabolite profiling reveals new insights into the regulation of serum urate in humans. Metabolomics. 2014;10:141–151. doi: 10.1007/s11306-013-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieleman L.A., Palmen M.J., Akol H., Bloemena E., Pena A.S., Meuwissen S.G., Van Rees E.P. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin Exp Immunol. 1998;114:385–391. doi: 10.1046/j.1365-2249.1998.00728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanis J.M., Alexeev E.E., Curtis V.F., Kitzenberg D.A., Kao D.J., Battista K.D., Gerich M.E., Glover L.E., Kominsky D.J., Colgan S.P. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017;10:1133–1144. doi: 10.1038/mi.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M., Shroyer N.F., Wells J.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki A., Hanada T., Mitsuyama K., Yoshida T., Kamizono S., Hoshino T., Kubo M., Yamashita A., Okabe M., Takeda K., Akira S., Matsumoto S., Toyonaga A., Sata M., Yoshimura A. CIS3/SOCS3/SSI3 plays a negative regulatory role in STAT3 activation and intestinal inflammation. J Exp Med. 2001;193:471–481. doi: 10.1084/jem.193.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard T.D., Murray I.A., Perdew G.H. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab Dispos. 2015;43:1522–1535. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo R., Matthews J. High-resolution genome-wide mapping of AHR and ARNT binding sites by ChIP-Seq. Toxicol Sci. 2012;130:349–361. doi: 10.1093/toxsci/kfs253. [DOI] [PubMed] [Google Scholar]
- 28.Mathieu M.C., Lapierre I., Brault K., Raymond M. Aromatic hydrocarbon receptor (AhR).AhR nuclear translocator- and p53-mediated induction of the murine multidrug resistance mdr1 gene by 3-methylcholanthrene and benzo(a)pyrene in hepatoma cells. J Biol Chem. 2001;276:4819–4827. doi: 10.1074/jbc.M008495200. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.H., Wood T.K., Lee J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015;23:707–718. doi: 10.1016/j.tim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Manichanh C., Borruel N., Casellas F., Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 31.Butto L.F., Haller D. Dysbiosis in intestinal inflammation: cause or consequence. Int J Med Microbiol. 2016;306:302–309. doi: 10.1016/j.ijmm.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Sartor R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 33.Kominsky D.J., Keely S., MacManus C.F., Glover L.E., Scully M., Collins C.B., Bowers B.E., Campbell E.L., Colgan S.P. An endogenously anti-inflammatory role for methylation in mucosal inflammation identified through metabolite profiling. J Immunol. 2011;186:6505–6514. doi: 10.4049/jimmunol.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newton W.A., Snell E.E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci U S A. 1964;51:382–389. doi: 10.1073/pnas.51.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hopkins F.G., Cole S.W. A contribution to the chemistry of proteids: part II. The constitution of tryptophane, and the action of bacteria upon it. J Physiol. 1903;29:451–466. doi: 10.1113/jphysiol.1903.sp000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.H., Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 37.Martin K., Morlin G., Smith A., Nordyke A., Eisenstark A., Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Preter V., Machiels K., Joossens M., Arijs I., Matthys C., Vermeire S., Rutgeerts P., Verbeke K. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015;64:447–458. doi: 10.1136/gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 39.Wlodarska M., Luo C., Kolde R., d'Hennezel E., Annand J.W., Heim C.E., Krastel P., Schmitt E.K., Omar A.S., Creasey E.A., Garner A.L., Mohammadi S., O'Connell D.J., Abubucker S., Arthur T.D., Franzosa E.A., Huttenhower C., Murphy L.O., Haiser H.J., Vlamakis H., Porter J.A., Xavier R.J. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37.e6. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., Ballet V., Claes K., Van Immerseel F., Verbeke K., Ferrante M., Verhaegen J., Rutgeerts P., Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 41.Eeckhaut V., Machiels K., Perrier C., Romero C., Maes S., Flahou B., Steppe M., Haesebrouck F., Sas B., Ducatelle R., Vermeire S., Van Immerseel F. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- 42.Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Dore J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 43.Shimada Y., Kinoshita M., Harada K., Mizutani M., Masahata K., Kayama H., Takeda K. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One. 2013;8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S., Fleet J.C., Kortagere S., Mukherjee P., Fasano A., Le Ven J., Nicholson J.K., Dumas M.E., Khanna K.M., Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng J., Shah Y.M., Gonzalez F.J. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devlin A.S., Marcobal A., Dodd D., Nayfach S., Plummer N., Meyer T., Pollard K.S., Sonnenburg J.L., Fischbach M.A. Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe. 2016;20:709–715. doi: 10.1016/j.chom.2016.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heath-Pagliuso S., Rogers W.J., Tullis K., Seidel S.D., Cenijn P.H., Brouwer A., Denison M.S. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry. 1998;37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 48.Jin U.H., Lee S.O., Sridharan G., Lee K., Davidson L.A., Jayaraman A., Chapkin R.S., Alaniz R., Safe S. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol. 2014;85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y., Innocentin S., Withers D.R., Roberts N.A., Gallagher A.R., Grigorieva E.F., Wilhelm C., Veldhoen M. Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation. Cell. 2011;147:629–640. doi: 10.1016/j.cell.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 50.Cervantes-Barragan L., Chai J.N., Tianero M.D., Di Luccia B., Ahern P.P., Merriman J., Cortez V.S., Caparon M.G., Donia M.S., Gilfillan S., Cella M., Gordon J.I., Hsieh C.S., Colonna M. Lactobacillus reuteri induces gut intraepithelial CD4+CD8alphaalpha+ T cells. Science. 2017;357:806–810. doi: 10.1126/science.aah5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bessede A., Gargaro M., Pallotta M.T., Matino D., Servillo G., Brunacci C. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belkaid Y., Hand T.W. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Indole-3-propionic acid (IPA) up-regulates IL-10R1 in dextran sodium sulfate (DSS) colitis. Eight- to 10-week–old C57BL/6 mice were administered water or 2.5% DSS ± IPA ad libitum for 9 days. DSS was then removed and mice were allowed to recover for 2 days as predetermined prior to euthanasia. A: Real-time quantitative PCR of Il10r1 transcript levels in colon tissue. B: IL-10 levels in colon tissue protein lysates were measured by Meso Scale Discovery analysis. Data are expressed as means ± SEM. n = 5 to 10 mice/group. ∗P < 0.05.
Indole-3-propionic acid (IPA) induces aryl hydrocarbon receptor (AHR) target genes in vivo. A and B: Transcript levels of cytochrome P450 family 1 subfamily A member 1 (CYP1A1) (A) and cytochrome P450 family 1 subfamily B member 1 (CYP1B1) (B) were measured in colon tissue of 8- to 10-week–old C57BL/6 mice administered water or 2.5% dextran sodium sulfate (DSS) ± IPA ad libitum for 9 days. C: Further, colon tissue levels of IL-6 were measured in protein lysates by Meso Scale Discovery analysis. Data are expressed as means ± SEM. n = 5 to 10 mice/group. ∗P < 0.05.