Summary
Human Cytomegalovirus (HCMV), a leading cause of congenital birth defects, forms an unusual cytoplasmic virion maturation site termed the “Assembly Compartment” (AC). Here, we show that the AC also acts as a microtubule-organizing center (MTOC) wherein centrosome activity is suppressed and Golgi-based MT nucleation is enhanced. This involved viral manipulation of discrete functions of MT plus-end binding (EB) proteins. In particular, EB3, but not EB1 or EB2, was recruited to the AC and was required to nucleate MTs that were rapidly acetylated. EB3-regulated acetylated MTs were necessary for nuclear rotation prior to cell migration, maintenance of AC structure and for optimal virus replication. Independently, a myristoylated peptide that blocked EB3-mediated enrichment of MT regulatory proteins at Golgi regions of the AC also suppressed acetylated MT formation, nuclear rotation and infection. Thus, HCMV offers new insights into the regulation and functions of Golgi-derived MTs, and the therapeutic potential of targeting EB3.
Keywords: microtubule organizing center, end-binding protein, Golgi, nuclear rotation, virus infection
eTOC Blurb
Live-cell imaging reveals the dynamic nature of the Human Cytomegalovirus (HCMV) assembly compartment (AC), which acts as a Golgi-derived microtubule-organizing center. Specific recruitment of microtubule plus-end binding EB3 facilitates acetylated MT formation at the AC and nuclear rotation prior to cell migration. EB3 targeting with myristoylated peptides suppresses infection.
Introduction
Human Cytomegalovirus (HCMV) infects 60–100% of adults depending on geographical location (Mocarski et al., 2007). Currently there is no vaccine or cure and once infected, individuals harbor lifelong latent reservoirs of HCMV that periodically reactivate. Although innocuous to most healthy individuals, HCMV can cause life-threatening complications in immunosuppressed individuals. In pregnant mothers, HCMV can cross the placenta with devastating effects. Every year in the US alone, HCMV induces hundreds of cases of perinatal mortality and leaves thousands with serious physical and developmental defects, yet awareness of this is alarmingly low (Manicklal et al., 2013).
HCMV is a β-herpesvirus with an unusually protracted replication cycle spanning several days (Mocarski et al., 2007). During this time, HCMV forms a unique yet poorly understood cytoplasmic Assembly Compartment (AC) (Alwine, 2012). Only recently has fixed-cell imaging revealed that the AC is a restructured Golgi surrounded by several other host organelles (Das and Pellett, 2011; Das et al., 2007; Rebmann et al., 2016; Sanchez et al., 2000a). The AC forms intimate connections with the nucleus resulting in a pinched “kidney bean” shape characteristic of infection. This connectivity allows virus particles to bud from the nucleus into the AC to mature. Notably, microtubules (MTs) emanate from the AC and maintain its structure (Sanchez et al., 2000a), while the MT motor dynein recruits specific host and viral components to the AC (Clippinger and Alwine, 2012; Indran et al., 2010). However, we have no knowledge of how the AC behaves in living cells, or its potential functions beyond serving as a site for virus maturation.
MTs form through the assembly of α/β-tubulin subunits into polarized filaments through minus-end seeding at MTOCs such as the centrosome (Petry and Vale, 2015). Central to MT nucleation are γ-tubulin proteins, which assemble into higher-order γ-Tubulin Ring Complexes (γ-TuRCs) that bind α-tubulin. While the centrosome is the primary MTOC in many cell types, several non-centrosomal nucleation sites exist. These include the Golgi, discussed in more detail below, that contribute to the broader complexity of MT networks in different cell types (Sanders and Kaverina, 2015). After nucleating, MT plus-ends undergo rapid phases of growth, pause and de-polymerization, generating dynamic arrays that radiate from MTOCs. The dynamic behavior and function of MTs is regulated by specialized end-binding proteins (EBs), comprising three family members (Akhmanova and Steinmetz, 2015). EB1 and EB3 are structurally similar and can form hetero- or homo-dimers, while the more divergent EB2 forms only homodimers (Komarova et al., 2009). EB dimers specifically recognize GTPtubulin that is transiently present at growing MT plus-ends, and thereby track growing MT tips (Maurer et al., 2011; Morrison et al., 1998; Rickard and Kreis, 1990). EBs influence MT behavior both through direct effects on polymerization and by recruiting other plus-end tracking proteins (+TIPs) (Dixit et al., 2009; Komarova et al., 2009; Komarova et al., 2005; Tirnauer and Bierer, 2000; Vitre et al., 2008; Zhang et al., 2015). Although many +TIPs can bind MTs, they utilize CAP-Gly or SxIP motifs to bind EB proteins in order to accumulate at MT plusends where they influence MT dynamics or engagement with cellular structures (Akhmanova and Steinmetz, 2015; Honnappa et al., 2009). In response to signaling cues often at specific subcellular sites, +TIPs can mediate the capture and stabilization of MTs. While dynamic MTs typically have half-lives lasting minutes, stable MTs persist for several hours allowing them to accumulate distinguishing post-translation modifications (PTMs) (Janke and Bulinski, 2011). This includes tubulin detyrosination on the outer filament surface, which does not impart stability but allows stable MTs to be recognized by specific motors. In contrast, tubulin acetylation in the inner lumen confers mechanical strength (Portran et al., 2017; Xu et al., 2017).
Here, live-cell imaging reveals the dynamic behavior of the HCMV AC and its ability to control host cell remodeling by acting as an MTOC whose nucleating activity is predominantly Golgi-based. Through transcriptional and posttranscriptional control of different EBs, HCMV exploits different MT subsets for different purposes. In particular, EB3 is recruited to the AC to form acetylated MTs that control both nuclear rotation and AC structure. siRNAs or myristoylated peptides targeting EB3 highlight its specific role in these events, revealing novel functions and regulatory mechanisms for the AC and Golgi-derived MTs, and identifying new targets to suppress infection.
Results
The AC is a dynamic structure that controls host cell behavior
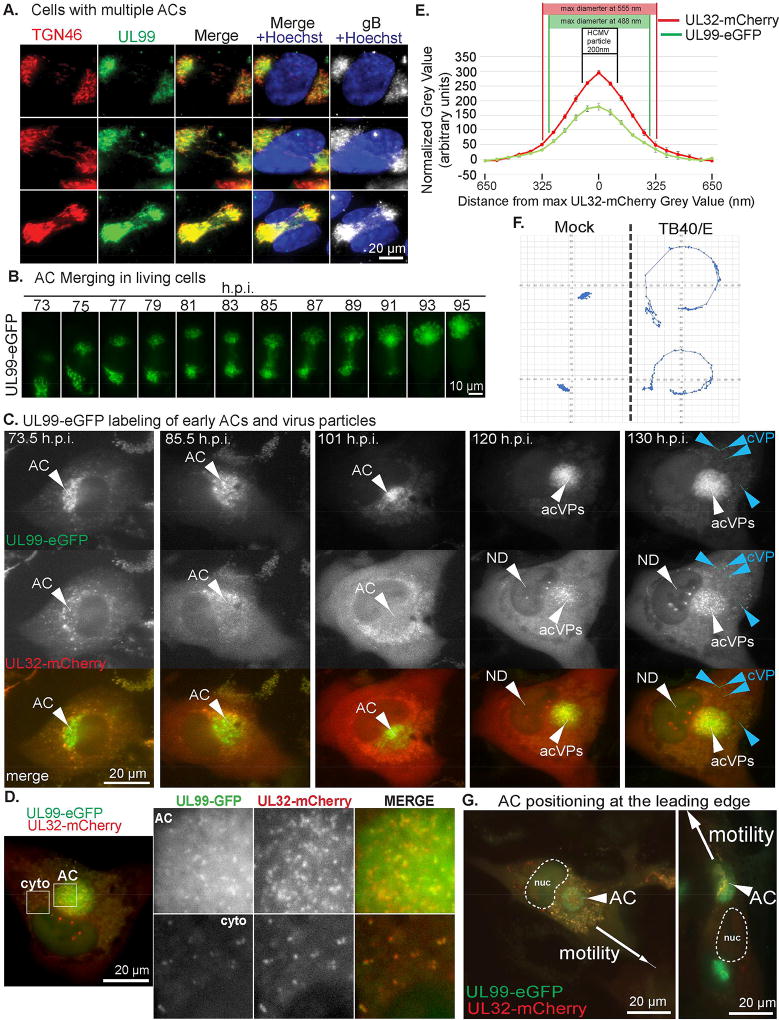
To image the AC in living cells we fluorescently tagged two HCMV proteins; UL99 and UL32. UL99 localizes to trans-Golgi vesicles and labels the AC, and also binds virus particles (Moorman et al., 2010; Sanchez et al., 2000a; Sanchez et al., 2000b). UL32 localizes to the nucleus very early (1–2d.p.i.) but becomes predominantly cytoplasmic thereafter, associating with clathrin-containing compartments recruited to the AC. At later stages (~5–6d.p.i.) UL32 is detected in the nucleus again, incorporating into new virus particles that then bud into the AC (Moorman et al., 2010; Sampaio et al., 2005; Sanchez et al., 2000a). We engineered the HCMV clinical strain, TB40/E to express either UL99-eGFP or UL32-mCherry. To validate these viruses, we first infected primary normal human dermal fibroblasts (NHDFs) with TB40/E-UL99-eGFP. Cultures were fixed at different days post-infection (d.p.i.) and stained for the trans-Golgi marker, TGN46. In line with fixed imaging of laboratory adapted strains (Moorman et al., 2010), UL99-eGFP labeled the Golgi that reorganized into an AC between 2–5 d.p.i. (Figure S1A). Notably, 15% of infected cells imaged (65/431) contained two distinct or weakly inter-connected UL99-eGFP and TGN46-positive structures (Figure 1A). Staining for the viral glycoprotein gB confirmed that both structures contained other viral proteins and were likely nascent ACs. However, live cell imaging of UL99-eGFP-labeled structures revealed that in 100% of cases when two nascent ACs were present in a cell, they merged into one (Figure 1B; Movie S1A). As such, multiple ACs observed in a single cell in fixed images, here and evident in other studies (Buchkovich et al., 2010; Das et al., 2007), represent intermediate stages of merging. This seemed to ensure that only one AC resided in cells as infection progressed but also bore a striking resemblance to Golgi reorientation in migrating fibroblasts (Wu et al., 2016), discussed later.
Figure 1. AC dynamics and nuclear rotation during HCMV infection.
(A) NHDFs were infected with TB40/E-UL99-eGFP at MOI 0.5 for 4d. Fixed cells were stained for GFP, TGN46 and gB. Nuclei were stained with Hoechst. Note, both Golgi structures in each cell stain for UL99-eGFP and gB, and appear to have weak connections. (B) Time lapse images (Movie S1A) of NHDFs infected with TB40E-UL99-eGFP at MOI 0.5 imaged 3–5d.p.i. illustrating AC merging. (C) Time lapse images (Movie S1B) of NHDFs co-infected with TB40/E-UL99-eGFP and TB40/E-UL32-mCherry at MOI 0.5 imaged 3–5d.p.i. UL99 labels the AC. UL32 domains form in the nucleus (ND) prior to the appearance of virus particles in the AC (acVPs) and then in the cytoplasm (cVP). (D) Still (Movie S1B) showing virus particles in the AC and cytoplasm (cyto) co-labeled with UL99-eGFP and UL32-mCherry. (E) Distribution of fluorescent intensity for UL32-mCherry and UL99-eGFP particles in D. measured using line-scan analysis. n = 5 (64 particles). Both peaks are in the size range of HCMV particles (indicated), and drop rapidly outside this size-range. (F) NHDFs expressing NLS-mCherry mock infected or infected with TB40E-UL99-eGFP at MOI 0.5 and imaged 3–5d.p.i. Displacement of NLS-mCherry-labeled nuclei (Movie S2) was measured. 2 representative examples per group are shown. (G) Time lapse images (Movie S3) of NHDFs co-infected with TB40/E-UL99-eGFP and TB40/E-UL32-mCherry at MOI 0.5. Arrows indicate the direction of cell migration. nuc=nucleus.
We next co-infected NHDFs with TB40/E-UL99-eGFP and TB40/E-UL32-mCherry followed by two-color live cell imaging. In line with fixed images (Sanchez et al., 2000a), between 3–5 d.p.i. UL32-mCherry labeled cytoplasmic structures that gradually accumulated around the UL99-eGFP-labeled Golgi (Figure 1C; Movie S1B). As infection progressed, UL32-mCherry-positive domains also formed in the nucleus, followed by the appearance of small uniform structures in the AC and cytoplasm. These co-labeled with UL99-eGFP and their size was consistent with HCMV virions (Figure 1D–E; Movie S1B). This order series of events occurred in all cells imaged and is in line with predictions from prior studies using fixed samples. This established UL99-eGFP as an appropriate marker of the AC from its earliest stages of formation.
While imaging the AC we noted that infected cell nuclei rotated (Movie S1B). To visualize nuclei, we generated NHDFs stably expressing mCardinal fused to a Nuclear Localization Signal (mCardinal-NLS) and either mock-infected or infected them with TB40/E-UL99-eGFP. Live cell imaging over 3–5 d.p.i. confirmed nuclear rotation in infected cells (Movie S2). Notably, mCherry-NLS signal in infected cells became progressively weaker over time. This did not occur in uninfected cells, suggesting it was not due to photobleaching but instead reflected increased nuclear porosity in HCMV-infected cells (Buchkovich et al., 2010). To measure the extent of nuclear rotation we used nucleoli and the “nuclear pinch” as reference points. Results showed that ~80% (53/66) of infected cell nuclei exhibited persistent rotation >180° (Figure 1F). This net range underestimates the extent of nuclear rotation, as often nuclei reversed directionality or rotated ~720° (see below). By contrast, uninfected-cell nuclei exhibited no discernible rotation and translocated in the direction of cell motility.
Nuclear rotation in infected cells ceased prior to cell migration (Figure 1G; Movie S3A–B). Before migration, in 95% (57/60) cells imaged the AC was either already positioned at or moved toward the leading edge (between the nucleus and direction of migration) of cells. This demonstrated that the AC does not always remain at the nuclear “pinch”, and suggested it might also regulate cell migration later in infection. In these later stages the AC continued to exhibit merging behaviors. If the AC lost integrity it rapidly “regrouped” while if two ACs encountered one another, presumably upon cell fusion, they merged (Movie S3C–D). Although these later events arose from diverse, uncontrollable circumstances making them difficult to quantify, they illustrated the continued dynamic behavior of the AC throughout infection. Overall, AC merging, nuclear rotation and AC positioning at the leading edge all suggested the AC might function as an MTOC.
The HCMV AC acts as a Golgi-derived MTOC
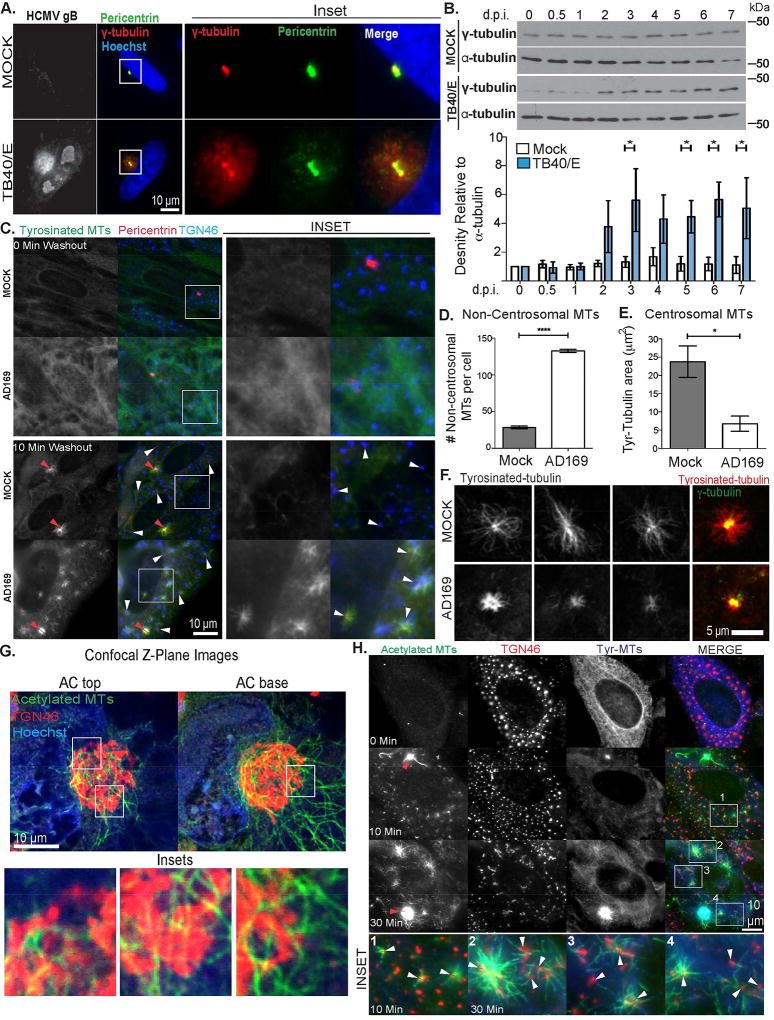
Although MTs emanate from the AC (Sanchez et al., 2000a), the true nature of their nucleation and function remains poorly understood. We therefore stained fixed samples of mock or HCMV-infected NHDFs for two key MT nucleation factors. Imaging detected bright γ-tubulin and pericentrin puncta at centrosomes in uninfected cells and within ACs (Figure 2A). However, centrosomes in infected cells appeared “split” as shown in prior reports (Hertel and Mocarski, 2004). Moreover, γ-tubulin and pericentrin were broadly distributed throughout the AC in regions positive for TGN46 (Figure 2A and S1B). This was not observed in uninfected cells since the Golgi does not normally contain high levels of nucleating material (Sanders and Kaverina, 2015). Western Blot (WB) analysis of mock or infected cell lysates revealed that HCMV also increased γ-tubulin abundance around the time of AC formation (Figure 2B). This suggested HCMV increased the availability of nucleating material to potentially form new MTOCs within the AC.
Figure 2. The HCMV AC is a Golgi-derived MTOC.
(A–B) NHDFs were mock infected or infected with TB40/E at MOI 3. (A) Cells were fixed 5.d.p.i. and stained for γ-tubulin, pericentrin and gB. Nuclei were stained with Hoechst. (B) Cell lysates were prepared at the indicated times and analyzed by WB. Lower: Densitometry analysis of γ-tubulin relative to α-tubulin. n = 3; unpaired two-tailed t-test, *p<0.05. (C–F) NHDFs were mock-infected or infected with AD169 at MOI 3 for 3d. Cells were treated with 10µM nocodazole for 8h before washout for the indicated time. (C) Samples were stained for tyrosinated tubulin, TGN46 and pericentrin. Red arrows indicate centrosomes, white arrows indicate Golgi fragments. Insets show non-centrosomal nucleation sites. (D) The number of new MTs at non-centrosomal sites 10 min post-washout was quantified. n = 3 (>65 cells), bars = s.e.m., unpaired two-tailed t-test, ****p = 0.0001. (E–F) Tyrosinated MTs at centrosomes 10 min post-nocodazole washout imaged using confocal microscopy. (E) Area occupied by tyrosinated MTs nucleated at centrosomes was measured. n = 3 (>45 cells), bars = s.e.m., unpaired two-tailed t-test, *p = 0.0240. (F) Representative examples of MTs at centrosomes, including a bright γ-tubulin merge used to identify centrosomes. (G) NHDFs were infected with TB40/E at MOI 1 for 5d. Fixed cells were stained for acetylated tubulin and TGN46, along with hoechst. Samples were imaged using confocal microscopy. Top and bottom Z-plane images of the AC are shown. Insets show acetylated MTs localized at TGN sites. (H) NHDFs were infected and treated with nocodazole as in C. Samples were stained for acetylated and tyrosinated (tyr) tubulin, along with TGN46. Red arrows indicate centrosomes, white arrows in insets indicate Golgi fragments.
To test this directly, we performed nocodazole washout assays to identify sites of MT nucleation. Nocodazole disrupts the Golgi in uninfected cells and the AC in infected cells (Chabin-Brion et al., 2001; Sanchez et al., 2000a). While this prevented us from imaging MT growth from an intact AC, this effect was advantageous as it spatially separated the centrosome from surrounding Golgi sites and avoided uncertainty over the origin of new MTs. Staining for TGN46, pericentrin and tyrosinated tubulin, which detects dynamic MTs, confirmed that nocodazole depolymerized MTs and dispersed Golgi and AC structures (Figure 2C). Quantitation revealed that new MTs almost exclusively originated from the centrosome in uninfected cells (Figure 2C–D). In infected cells, new MTs originated from Golgi-based sites distributed throughout the cell due to AC disruption. Notably, uninfected or infected NHDFs contained no detectable detyrosinated MTs beyond centrosome asters (Figure S1C). As such, detyrosination does not affect the use of tyrosinated tubulin antibody to image MTs in NHDFs. Confocal imaging and quantification of new MTs further demonstrated that centrosomes in uninfected cells nucleated MTs more efficiently than in infected cells (Figure 2E–F). This suggested that HCMV overturned the normal dominance of the centrosome to favor Golgi-based nucleation.
A characteristic of Golgi-derived MTs is their rapid acetylation (Chabin-Brion et al., 2001; Efimov et al., 2007; Rios et al., 2004; Sanders and Kaverina, 2015). We therefore stained uninfected or infected samples for acetylated tubulin and TGN46. Imaging and quantification revealed that beyond singular asters, uninfected NHDFs contained very few acetylated MTs (Figure S1C–D). However, HCMV induced the formation of acetylated MTs that co-localized with TGN46-positive regions of the AC (Figure 2G and Movie S4), suggesting Golgi domains nucleated these MT subsets. To test this, we again performed nocodazole washout assays staining for acetylated tubulin and TGN46. Although centrosomes stained brightly for acetylated tubulin they were associated with relatively few acetylated filaments, while abundant arrays of acetylated MTs emanated from TGN46-positive structures dispersed throughout the cell (Figure 2H). Cumulatively, these findings demonstrated that Golgi regions of the AC nucleated new MTs that were quickly acetylated.
Differential regulation and localization of EB proteins during HCMV infection
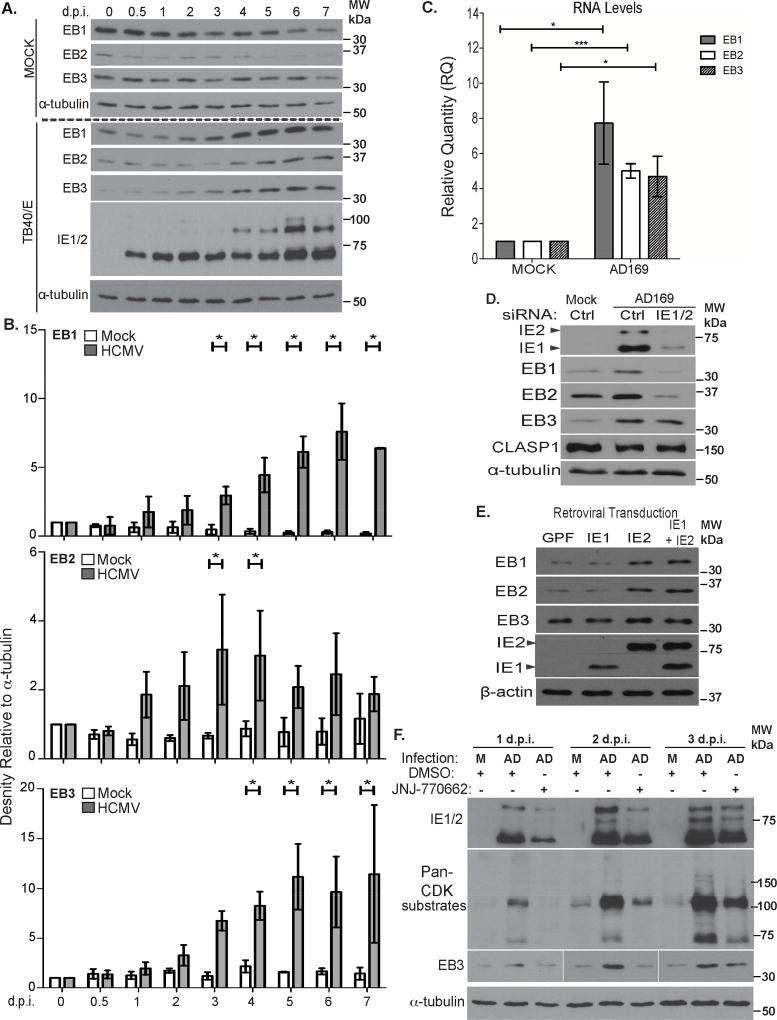
Formation of MTs at Golgi sites was recently shown to require EB proteins (Yang et al., 2017). Therefore, we tested whether HCMV influenced their abundance or localization. WB analysis over a timecourse of infection (Figure 3A–B) revealed that the abundance of EB1 and EB2 gradually declined in mock-infected NHDFs, likely as cells became quiescent, while levels of other proteins remained unchanged. However, in infected NHDFs all three EB proteins increased in abundance by 2–3 d.p.i. Similar increases in EB proteins were observed with independent HCMV strains, the timing of which correlated with differences in each strain’s replication kinetics (Figure S2A). Alongside protein increases, qRT-PCR revealed that HCMV increased the transcript abundance of each EB relative to mock-infected cells (Figure 3C). The relative increases in transcript and protein levels for EB1 and EB2 largely corresponded but increases in EB3 protein were greater than those of its transcript. This suggested that EB3 might be additionally regulated at post-transcriptional levels.
Figure 3. HCMV increases EB protein levels.
(A) Growth-arrested NHDFs were mock-infected or infected with TB40/E at MOI 3 for the indicated times. Cell lysates were analyzed by WB. (B) Densitometry analysis of EB1, EB2 and EB3 relative to α-tubulin. n = 3; bars = s.e.m.;, *p<0.05, unpaired two-tailed t-test (C) Growth-arrested NHDFs were mock-infected or infected at MOI 3 for 4d. EB transcript levels were measured using qRT-PCR. EB levels were normalized to uninfected controls (arbitrarily set to 1). n = 3; bars = s.e.m; *p<0.05, ***p<0.001, unpaired two-tailed t-test. (D) NHDFs were treated with control or IE1/2 siRNA prior to infection with AD169 at MOI 3 for 3d. Cell lysates were analyzed by WB for the indicated proteins. (E) NHDFs were transduced with retroviruses encoding GFP or IE proteins. Samples were analyzed by WB. (F) NHDFs were infected with AD169 at MOI 3 in the presence of DMSO or CDK1 inhibitor (JNJ-770662), re-dosing daily, for the indicated times. Samples were analyzed by WB.
See also Figure S2.
Increases in EB levels coincided with expression of the viral transcriptional activator, Immediate Early protein 2 (IE2) in all three HCMV strains tested (Figure 3A and S2A). To test if IE2 was involved in regulating EBs we suppressed IE2 expression in infected cells using siRNAs. HCMV IE1 and IE2 proteins are encoded by alternatively spliced transcripts of the major immediate early (MIE) gene (Mocarski et al., 2007). NHDFs were treated with control non-targeting or MIE (IE1/2) siRNAs prior to infection with HCMV. WB analysis showed that IE1/2 depletion prevented increases in EB1 and EB2 by HCMV (Figure 3D). IE1/2 siRNAs did not reduce EB abundance in uninfected cells (Figure S2B), demonstrating that these changes were not off-target effects. Moreover, IE1/2 depletion did not block EB3 increases (Figure 3D), supporting the idea that EB3 was regulated at additional levels. To test if IE2 was sufficient to increase EB levels, we transduced NHDFs with lentiviruses encoding GFP control or IE proteins and found that IE2, but not IE1, was sufficient to increase EB1 and EB2 abundance (Figure 3E). However, EB3 expression was not detectably affected. As such, independent siRNA and expression approaches supported the notion IE2 enhanced EB1 and EB2 accumulation, likely at the transcriptional level although changes in mRNA stability cannot be ruled out. Both approaches also suggested additional modes of regulation for EB3 were involved, beyond simply increasing EB3 transcript levels.
EB3 stability is controlled by Cyclin Dependent Kinase 1 (CDK1)-mediated phosphorylation (Ban et al., 2009), and HCMV activates CDK1 (Hertel et al., 2007; Sanchez et al., 2003). To test the contribution of CDKs to EB3 accumulation, NHDFs were mock infected or infected in the presence of CDK1 inhibitor, JNJ-7706621. WB analysis of time-courses showed that in cells infected with AD169, where EB3 increases begin 1–2d.p.i. (Figure S2A), JNJ-7706621 suppressed HCMV-induced CDK activation as determined by pan-CDK1 substrate antibody, and prevented increases in EB3 at 1 and 2 d.p.i. (Figure 3F). Although JNJ-7706621 modestly affected IE2 expression, siRNA approaches above showed that reduced IE2 was not sufficient to affect EB3 increases. As such, JNJ-7706621 demonstrated that CDK, or the combined action of IE2 and CDK were required to increase EB3. However, by 3d.p.i. both CDK substrate phosphorylation and increases in EB3 abundance became insensitive to JNJ-7706621, despite daily inhibitor replenishment. Notably, HCMV encodes a CDK homolog, pUL97 that is resistant to host CDK inhibitors (Hertel et al., 2007). As such, our findings establish that CDK activity regulates EB3 abundance during infection, but that the viral CDK homolog likely takes over from host kinases once infection is established. Overall, these findings established that HCMV uses multiple strategies to increase the abundance of EB proteins.
We also tested HCMV’s effects on EB protein localization. While EB1 and EB3 track MT plus-ends, the more divergent EB2 exhibits weaker plus-end-specific localization (Komarova et al., 2009). In line with this, EB1 and EB3 formed “comets” indicative of plus-end tracking on MTs in both uninfected and infected NHDFs, while EB2 was more broadly distributed along the MT lattice (Figure S2C–E). Notably, while EB1 comets localized throughout infected cells and near the AC, EB3 comets intensely concentrated around γ-tubulin-positive regions of the AC (Figure S2C–F). As such, EB1 and EB3 exhibited differences in both regulation and localization in infected cells, suggesting they might perform discrete functions.
EB1 and EB3 play distinct roles in HCMV infection
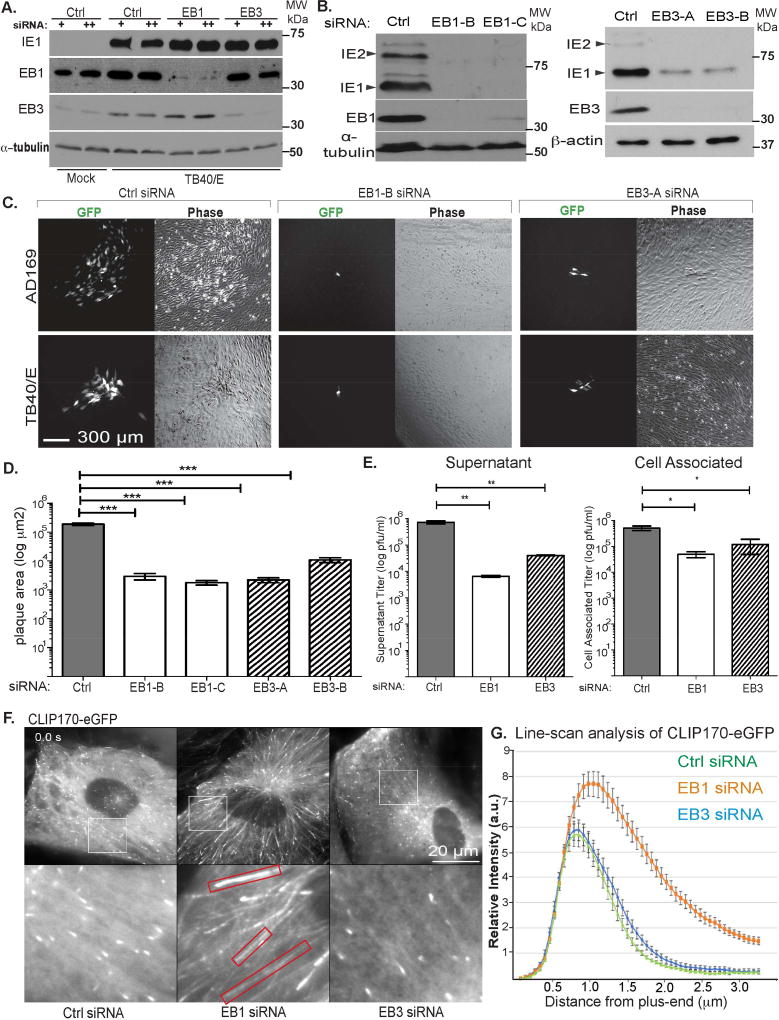
We next examined the role of EB proteins in HCMV infection. EB1 is required for some viruses to reach the nucleus and establish infection (Jovasevic et al., 2015; Sabo et al., 2013). Similar effects on early HCMV infection would confound our understanding of their roles in later processes. To avoid this, we exploited HCMV’s protracted lifecycle by reducing siRNA pre-treatment time such that the establishment of infection was unaffected. Using this approach, viral IE protein production was not affected despite efficient depletion of EBs, or by secondary dosing of siRNAs at 3d.p.i. (Figure 4A). The fact this approach did not affect the establishment of infection was further evidenced by the formation of ACs and stable MTs, shown later. Using this approach, we assessed effects of EB depletion on HCMV replication using independent siRNAs targeting EB1 or EB3. In cultures infected at low multiplicity of infection (MOI) for 12d, WB analysis of viral protein accumulation (Figure 4B) or imaging and measurement of plaque sizes (Figure 4C–D) revealed that both EB1 and EB3 were required for efficient HCMV spread. We also assayed effects of EB1 or EB3 depletion on the production of virus in single round high MOI infections. Titration of infectious virus produced at 7d.p.i. revealed a 100-fold and 14-fold reduction in supernatant virus, and 10-fold and 4-fold reduction in cell-associated virus in EB1- or EB3-depleted cells, respectively (Figure 4E). These findings suggested EB1 and EB3 might regulate HCMV replication by distinct mechanisms.
Figure 4. EB1 and EB3 play distinct roles in HCMV replication.
(A) NHDFs were treated with siRNA 30h prior to infection (+) or with a second siRNA treatment at 3d.p.i. (++). Cultures were infected at MOI 3 for 5d. Lysates were analyzed by WB for the indicated proteins. Note, IE1/2 are expressed because early infection is not affected using this siRNA treatment strategy. (B–D) NHDFs were treated with siRNAs as in A. and infected with TB40/E-eGFP at MOI 0.001 for 14d. (B) Cells were lysed and analyzed by WB for the indicated proteins. Note, IE1/2 are reduced because of reduced HCMV spread in EB1 or EB3 depleted cultures. (C) Phase and fluorescent images of plaques for TB40/E or AD169. (D) Plaque sizes in C for TB40/E were measured. n = 3 (>22 plaques); bars = s.e.m. ***p=0.0005, unpaired two-tailed t-test. (E) NHDFs were infected with TB40/E at MOI 3. Cell supernatants and cell-associated virus were harvested at 7d.p.i. and titrated on NHDFs. n = 3; bars = s.e.m; *p<0.05, **p<0.01, unpaired two-tailed t-test. (F) Still images (Movie S5A) of NHDFs expressing eGFP-CLIP170, treated with siRNAs as in A and infected with TB40/E-UL32-mCherry at MOI 1 for 5d. Red boxes highlight elongated eGFPCLIP170 tracks in EB1-depleted cells. (G) Line-scan analysis of eGFP-CLIP170 intensity distribution from the microtubule plus-end in F. n = 10 (>150 MT linescans); bars = s.e.m.
See also Movie S5A.
To determine whether EBs had different roles in infection, we tested how EB1 or EB3 depletion affected MT behavior. NHDFs expressing CLIP170-eGFP, a +TIP widely used to visualize MT growth, were treated with control or EB-targeting siRNAs followed by infection with HCMV. Imaging at 4.d.p.i. revealed that while EB3 depletion had no detectable effect on the number or behavior of CLIP170-eGFP comets, EB1 depletion caused a re-distribution of CLIP170 along the MT lattice (Figure 4F; Movies S5A). Line-scan analysis of CLIP170-eGFP signal intensity and distribution further confirmed its broader distribution along MTs in EB1-depleted cells (Figure 4G), suggesting the accumulation of CLIP170 at growing MTs was specifically regulated by EB1.
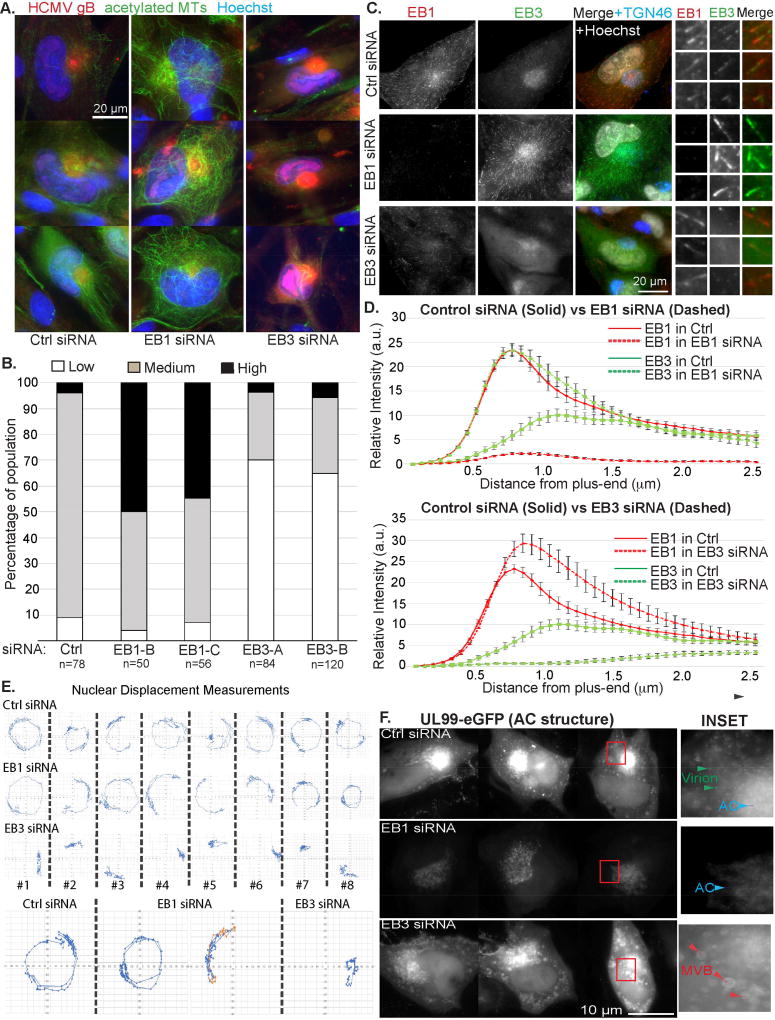
We next tested whether EBs were required for the formation of acetylated MTs by HCMV. Staining showed that EB1 depletion increased acetylated MTs, while EB3 depletion suppressed their formation compared to controls (Figure 5A). These effects were quantified scoring cells as 1) lacking acetylated MTs (none), 2) having acetylated MTs extending from the AC (medium), 3) having extensive acetylated arrays filling the cytoplasm (high). This quantification approach confirmed that EB1 depletion increased, while EB3 depletion decreased the extent of acetylated MT formation by HCMV (Figure 5B). EB1 and EB3 compete for binding to MT plus-ends (Komarova et al., 2009), which potentially explained their differential effects on the formation of acetylated MTs. In line with this, the staining intensity of EB3 comets at the AC increased in EB1-depleted cells compared with controls, while EB1 did not localize to the AC in EB3-depleted cells (Figure 5C). This suggested EB1 could not functionally substitute for EB3 in infected cells. Moreover, line-scan analysis showed that in controls, EB1 formed bright tracks ahead of EB3 (Figure 5D). EB3 formed longer and brighter comets upon EB1 depletion, while EB3 depletion resulted in brighter and longer EB1 tracks. This demonstrated that EB1 and EB3 compete for binding to MT plus-ends in HCMV-infected cells, and that EB1 depletion enhanced EB3 activity and localization the AC. The levels of acetylated MTs in each condition correlated with the extent of EB3 localization to the AC and MT plus-ends. Overall, this suggested that both EB1 and EB3 contributed to efficient infection, but did so through effects on distinct MT subpopulations.
Figure 5. EB3 regulates acetylated MTs, nuclear rotation and AC structure.
(A–D) NHDFs were treated with siRNAs and infected with TB40/E at MOI 1 for 5d. (A) Fixed samples were stained for acetylated tubulin and TGN46. Nuclei were stained with Hoechst. (B) Percentage of cells containing low (as in EB3 siRNA panels in A.), medium (as in Ctrl siRNA panels) or high (as in EB1 siRNA panels) levels of acetylated MTs. n = as indicated. (C) Fixed samples were stained for EB1, EB3 and TGN46. Enlarged insets show EB1 and EB3 comets. (D) Line-scan analysis of EB1 or EB3 comet intensity and distribution in samples in C. Note, loss of one EB increases MT tip binding by the other. Top: Distributions of EB1 (red) or EB3 (green) in control siRNA (solid) or EB1 siRNA (dashed) samples. Bottom: Distributions of EB1 (red) or EB3 (green) in control siRNA (solid) or EB3 siRNA (dashed) samples. n = 5 (>125 MT linescans); bars = s.e.m. (E–F) NHDFs were treated with siRNAs and infected with TB40/E-UL99-eGFP at MOI 0.5. (E) Effects on nuclear rotation: Cells were imaged at 2 frames per hour between 3–5d.p.i. Time lapse images (Movie S5B) were used to measure nuclear rotation. Bottom: Examples of rotations including changes in direction (red and orange). (F) Effects on AC structure: Still images (Movie S6) from faster frame rate analysis of UL99-eGFP showing different AC architectures in control versus EB1 or EB3-depleted cells. Insets use non-linear scaling to show details within the bright AC region, and highlight cytoplasmic virions and MVBs.
See also Movies S5B–S6.
Acetylated MTs control nuclear rotation and AC structure
We examined whether EB1 or EB3 influenced nuclear rotation or AC formation. NHDFs were treated with control or EB-targeting siRNAs followed by infection with TB40/E-UL99-eGFP. Live cell imaging over 3–5d.p.i together with nuclear displacement measurements revealed that in control or EB1 siRNA-treated samples ~86% (37/43) nuclei rotated >180° (Figure 5E; Movie S5B). These involved cases of >360° unidirectional rotations or 180° rotations with reversed directionality. By contrast, other than limited general displacement similar to mock-infected cells (Figure 1G), nuclear rotation could not be detected in EB3-depleted cells (Figure 5E). Using faster frame rate imaging, in control siRNA-treated cells UL99-eGFP-labeled ACs consisted of a dense core while smaller structures and uniform-sized particles, likely virions (Figure 1E), moved bi-directionally in the cytoplasm (Figure 5F; Movie S6). By contrast, in EB1-depleted cells the AC was diffuse and fewer motile organelles and particles could be detected in the cytoplasm. In EB3-depleted cells the AC also appeared disorganized but the cytoplasm contained large structures that resembled multi-vesicular bodies (MVBs). These results demonstrated that depletion of EB1 or EB3 resulted in distinct alterations to AC structure, while EB3 was also required for HCMV-induced nuclear rotation.
A small peptide targeting EB3 suppresses HCMV replication
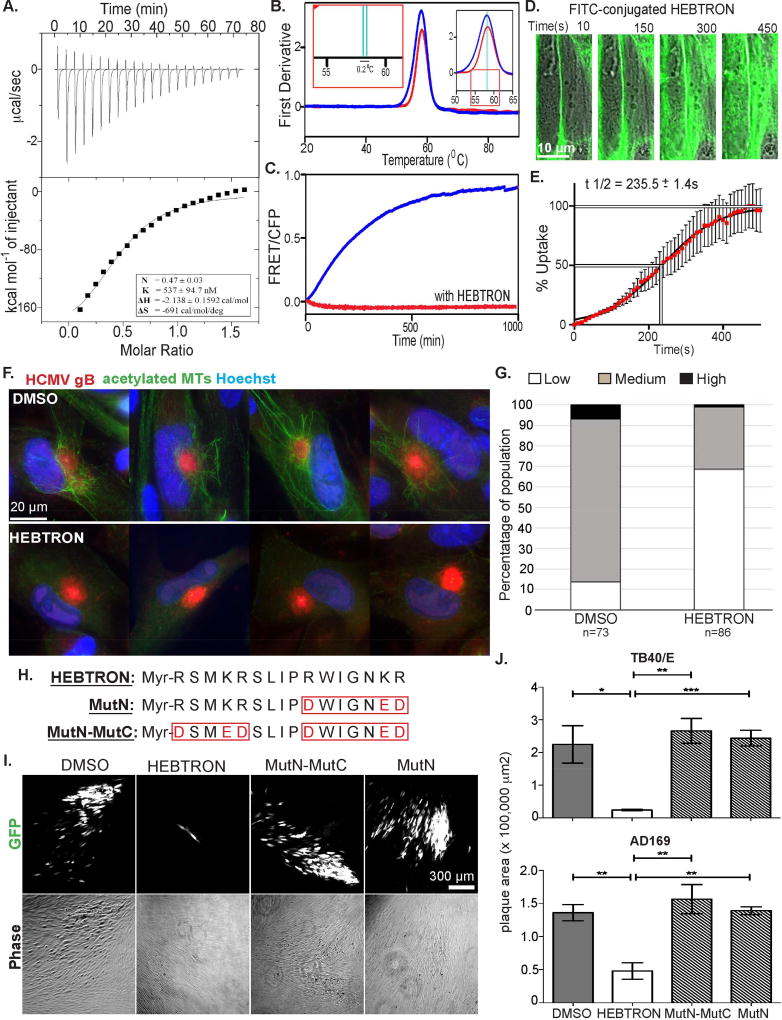
Our data suggested EB3 might be a “druggable” target to suppress HCMV replication. Peptide aptamers containing SxIP motifs specifically bind EB1 or EB3 due to variations in their flanking amino acid sequence, and displace their interacting +TIPs (Lesniewska et al., 2014). We designed a myristoylated peptide (Myr-RSMKRSLIPRWIGNKR, dubbed HEBTRON) to target EB3 and first characterized it in vitro. Using Drug Affinity Responsive Target Stability (DARTS) assays wherein drug binding protects target proteins from proteolytic degradation by thermolysin, we found that HEBTRON protected purified recombinant EB3 (Figure S3A). This was specific to HEBTRON as an EB1-specific peptide did not prevent EB3 proteolysis (Figure S3A). Using isothermal titration calorimetry (ITC) to measure binding affinity further revealed an exothermic binding with the dissociation constant (KD) of 537 ± 94.7 nM and enthalpy (ΔH) of −2.138 ± 0.1592 cal/mol (Figure 6A and S3B–C), indicating a strong and stable interaction of HEBTRON with EB3. The measured binding stoichiometry (N) of 0.47 ± 0.03 suggested formation of a 1:2 complex, in line with HEBTRON binding both EB3 subunits of dimers.
Figure 6. Characterization of HEBTRON.
(A) ITC of HEBTRON binding to purified EB3 C-terminus (200–281aa). KD, binding enthalpy, and stoichiometry were calculated from changes in heat upon binding of HEBTRON to the protein using the "one set of sites" binding model. (B) Thermal unfolding of full-length EB3 alone (blue) or with HEBTRON (red) using NanoDSF. The first derivative of the 350/330 nm (peak) defines transitions between folded (left) to unfolded (right) states. Magnified insets show 0.2°C temperature shift (green lines) for the complex. This suggests HEBTRON stabilizes dimers. (C) FRET signal after mixing full-length EB3-CFP and EB3-YFP without (blue) or with HEBTRON (red). FRET indicates formation of YFP/CFP-EB3 dimers, blocked by HEBRTON at 1:1 molar ratio. (D) NHDFs were treated with 10µM Myr-FITC-conjugated HEBTRON. Time lapse images were taken and overlaid with DIC images at the indicated times. (E) % uptake of Myr-FITC-conjugated HEBTRON in NHDFs over time was determined by the normalized fluorescence intensity of the cell. n = 3; assaying 32 cells. Bars = s.e.m. (F) NHDFs treated with DMSO or 25µM HEBTRON were infected with TB40/E at MOI 1 for 5d. Fixed cells were stained for acetylated tubulin and gB. (G) % DMSO or HEBTRON-treated cells in F containing acetylated MTs. n = as indicated. (H) Sequence of HEBTRON, MutN or MutN-MutC. (I) NHDFs treated with DMSO or 25µM HEBTRON, Mut-N or MutN-MutC were infected with TB40/E-eGFP at MOI 0.001 for 12d. Representative phase and fluorescent images of plaques. (J) Cells were treated and infected with TB40/E or AD169 as in I. Plaque areas are shown. n = 3 (>40 and >50 plaques. respectively); bars = s.e.m.; p*<0.05, **p<0.01, ***p<0.001, unpaired two-tailed t-test.
See also Figure S3.
We next tested the effects of HEBTRON on EB3 dimer stability using nano Differential Scanning Fluorimetry (nanoDSF). In this assay, thermal unfolding of proteins is measured to identify melting temperature (Tm). Compared with EB3 alone, a 0.2°C peak shift was detected with HEBTRON present (Figure 6B). This suggested HEBTRON increased EB3 dimer stability. To independently test this, we used a FRET-based assay in which preassembled YFP-EB3 dimers were mixed with CFP-EB3 dimers. In solution, input homodimers dissociate and form mixed species of dimers over time, resulting in FRET signal when YFP/CFP tags are proximal within heterodimers (De Groot et al., 2010). In the absence of HEBTRON increasing FRET was observed over time with YFP/CFP-EB3 dimer formation (Figure 6C). However, HEBTRON blocked FRET signal indicating that it stabilized input EB3 dimers.
We next determined uptake of myristoylated HEBTRON by cultured NHDFs. Confocal time lapse imaging showed that 5’6-fluorescein(FITC)-conjugated HEBTRON initially labeled the plasma membrane but was observed uniformly distributed throughout the cytosol within 7–8 minutes, with t1/2 uptake of 235.5 ± 1.4s (Figure 6D–E). We next tested whether HEBTRON affected MT behavior during infection. NHDFs expressing CLIP170-eGFP were treated with vehicle (DMSO) or 25µM HEBTRON followed by infection with HCMV. Time lapse imaging combined with line-scan analysis of CLIP170-eGFP intensity and distribution showed that HEBTRON had no detectable effect on CLIP170-eGFP behavior (Figure S3D). However, when fixed samples were stained for acetylated tubulin, imaging and quantification using approaches described above demonstrated that HEBTRON suppressed acetylated MT formation by HCMV (Figure 6F–G).
To understand how it functioned, we examined the effects of HEBTRON and two HEBTRON mutants containing amino acid substitutions in either one (MutN) or both (MutN-MutC) SLIP-flanking regions (Figure 6H). We first assessed their effects on plaque formation by TB40/E-GFP or AD169-GFP in low MOI spreading assays. Phase and fluorescence imaging at 12d.p.i. along with plaque size measurements showed that HEBTRON suppressed HCMV spread compared with DMSO control, while both HEBTRON mutants had no effect (Figure 6I–J). To further test its antiviral effects, NHDFs were treated with DMSO or 25µM HEBTRON before low MOI infection with an unrelated poxvirus, Vaccinia Virus (VacV). Imaging and measurements of plaque sizes formed by VacV expressing B5-GFP (Figure S3E–F) or WB analysis of VacV protein accumulation (Figure S4G) revealed HEBTRON had no significant effect on VacV spread. As such, HEBTRON exhibited specific antiviral effects toward HCMV that were dependent on guiding amino acids.
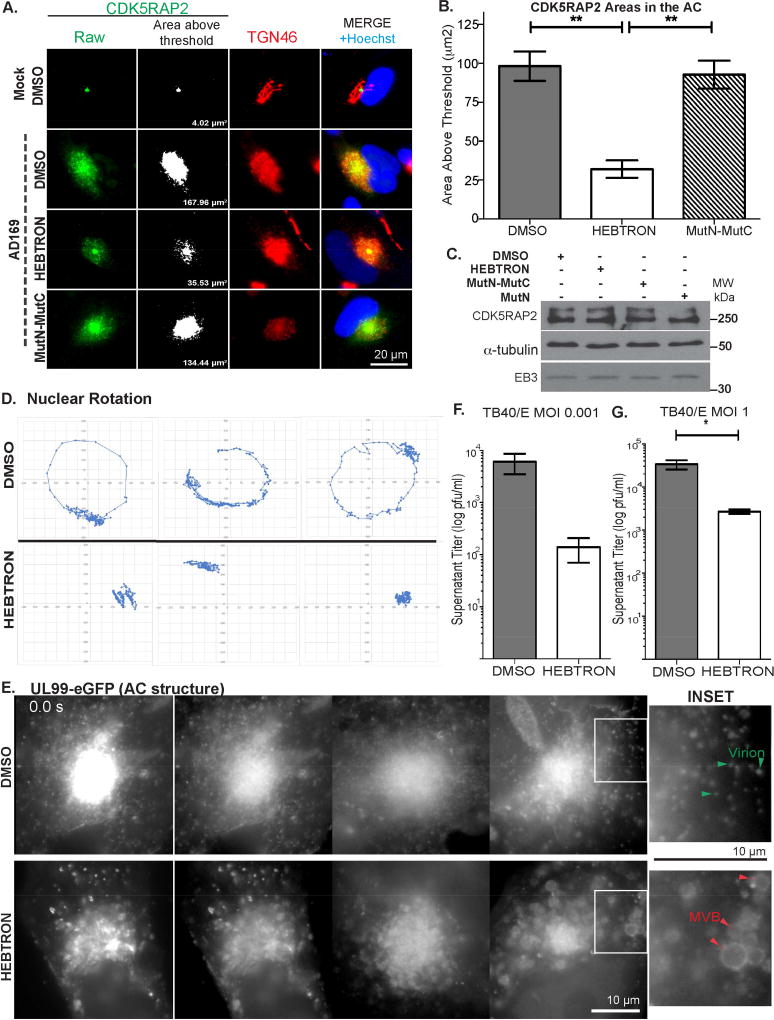
As published aptamers displace +TIPs from EB proteins (Lesniewska et al., 2014), we tested whether HEBTRON affected the localization of representative proteins known to utilize an SxIP motif to bind EB3; Tastin and CDK5RAP2 (Jiang et al., 2012). IF imaging of infected cells showed that HEBTRON did not affect EB3 localization to the AC compared with DMSO controls (Figure S3H). However, staining DMSO-treated cells revealed that Tastin localized in the same area as EB3, behind EB1 on MT plus-ends, and that HEBTRON displaced Tastin from MT plus-ends (Figure S4A–B). Notably, the second SxIP-interacting protein, CDK5RAP2 not only binds EB proteins but also stimulates the nucleating activity of γ-TuRC (Choi et al., 2010; Fong et al., 2009; Wang et al., 2010; Wu et al., 2016). CDK5RAP2 was present at centrosomes in uninfected NHDFs, but upon infection it localized throughout Golgi regions of the AC (Figure 7A). Imaging and measurements threshold brightness revealed that HEBTRON, but not the MutN-MutC form of HEBTRON, significantly reduced CDK5RAP2 recruitment to the AC (Figure 7B). HEBTRON did not decrease CDK5RAP2 or EB3 abundance (Figure 7C), demonstrating that HEBTRON specifically affected CDK5RAP2 enrichment in Golgi regions. Cumulatively, these data supported the notion that HEBTRON suppressed infection by interfering with EB3-mediated enrichment of factors such as CDK5RAP2 at non-centrosomal sites within the AC. This, combined with recent reports that EB proteins function in MT minus-end organization and nucleation at the Golgi (Yang et al., 2017), suggested that EB3 likely regulated the nucleation of MTs at the AC. To test this, we performed nocodazole washout assays on EB3-depleted or HEBTRON-treated cells infected with HCMV. Staining samples for α-tubulin, acetylated tubulin and TGN46 revealed that in control siRNA or DMSO-treated cells new MTs formed at Golgi regions dispersed throughout the cytoplasm, and these MTs were acetylated (Figure S4C–D). By contrast, formation of new MTs at Golgi fragments was suppressed in either EB3 depleted or HEBTRON-treated cells.
Figure 7. HEBTRON blocks recruitment of CDK5RAP2, nuclear rotation, and HCMV replication.
(A–B) NHDFs treated with DMSO or 25µM HEBTRON or MutN-MutC were infected at MOI 1 for 5d. (A) Fixed cells were stained for TGN46 and CDK5RAP2. Example of measurements of CDK5RAP2-positive area (based on fluorescence above intensity threshold). (B) Area measurements of CDK5RAP2 at the AC above threshold intensity. n = 3 (>90 cells), bars = s.e.m; **p<0.01, unpaired two-tailed t-test. (C) HEBTRON does not affect CDK5RAP2 or EB3 abundance. NHDFs treated with DMSO or 25µM HEBTRON, MutN or MutN-MutC were infected with AD169 at MOI 3 for 5d. Lysates were analyzed by WB. (D–E) NHDFs treated with DMSO or 25µM HEBTRON were infected with TB40/E-UL99-eGFP at MOI 0.5. (D) Time lapse imaging was performed 3–5d.p.i (Movie S7A) and nuclear displacement was measured. Representative traces are shown. (E) Time lapse imaging of the AC at 5d.p.i. (Movie S7B). Still images show AC structure in DMSO or HEBTRON-treated cells. Insets and non-linear scaling show details within the bright AC. Arrows indicate virus particles and MVBs. (F–G) NHDFs treated with DMSO or 25µM HEBTRON were infected with TB40/E at the indicated MOI. Infectious virus in culture supernatants was titrated. (F) Spreading assay infections at MOI 0.001 for 12d. n = 2; bars = s.e.m. (G) Single cycle infections at MOI 1 for 7d. n = 3; bars = s.e.m. *p<0.05, unpaired two-tailed t-test.
See also Figure S3 and S4 and Movie S7.
To further determine how HEBTRON suppressed HCMV replication we tested whether it affected nuclear rotation or AC structure. NHDFs were treated with DMSO or 25µM HEBTRON followed by infection with TB40/E-UL99-eGFP. While time lapse imaging over 3–5d.p.i. detected nuclear rotation >180° in ~71% (36/51) DMSO-treated cells, rotations could not be detected in cultures treated with HEBTRON (Figure 7D; Movie S7A). Furthermore, consistent with our results in siRNA-treated cells (Figure 5F), imaging of UL99-labeled AC structures at 5d.p.i. revealed that DMSO-treated cells contained a dense AC core while the cytoplasm was filled with UL99-positive structures, many of which were uniform in size and exhibited bi-directional motility suggestive of HCMV particles (Figure 7E; Movie S7B). By contrast, HEBTRON treatment resulted in aberrant ACs and large structures resembling MVBs in the cytoplasm, similar to effects of EB3 depletion.
Finally, we measured HEBTRON’s effects on infectious virus production. In low MOI spreading assays, 25µM HEBTRON suppressed the release of infectious HCMV approximately ~100-fold compared with DMSO controls (Figure 7F). In single-round high MOI infections virus release was suppressed ~13-fold (Figure 7G), similar to effects of EB3 depletion above. Overall, HEBTRON not only validated siRNA-based observations but provided additional insights into the underlying mechanism by which EB3 regulated MT formation at the AC to facilitate HCMV replication.
Discussion
Here, we reveal the dynamic behavior of the HCMV AC and its ability to act as a Golgi-based MTOC by impairing centrosome activity and exploiting functions specific to EB3. Moreover, how HCMV remodels host MT organization at the AC adds to our broader understanding of non-centrosomal MTOCs.
There are several notable features to how MTs are generated at the Golgi in uninfected cells, and how these are coopted at the AC. Nucleation material such as γ-tubulin is normally less concentrated at the Golgi than at the more dominant centrosome (Rios et al., 2004; Sanders and Kaverina, 2015; Wu et al., 2016; Yang et al., 2017). However, experimental depletion of centrosomes greatly enhances the nucleation activity of the Golgi (Yang et al., 2017), suggesting that centrosomes sequester much of the cells valuable nucleating material from other sites. In this regard, it is notable that HCMV impairs centrosome function and as others report (Hertel and Mocarski, 2004), appears to cause centrosome “splitting”. HCMV blocks cell division but pushes the cell into a “pseudo-mitotic” state that promotes viral DNA replication (Hertel et al., 2007; Hertel and Mocarski, 2004). Deregulation of the cell cycle and centrosome organization may release nucleating factors such as γ-tubulin, pericentrin and CDK5RAP2 that we find become enriched at Golgi regions during infection. Our data also suggests that other processes beyond centrosome deregulation are involved. These include increased abundance and availability of proteins such as γ-tubulin. We also observe that the enrichment of CDK5RAP2 at Golgi sites is dependent on EB3 and is therefore also an actively controlled process of redistribution of MT nucleating factors. CDK5RAP2 is known to bind and translocate with EB1 (Fong et al., 2009), but has >4-fold higher affinity for EB3 (Jiang et al., 2012). CDK5RAP2 is particularly noteworthy as it enhances the nucleating activity of γ-TuRCs and may be particularly important at weaker, non-centrosomal sites (Choi et al., 2010; Wang et al., 2010; Wu et al., 2016; Yang et al., 2017). Whether both EB1 and EB3 can control CDK5RAP2 distribution in uninfected cells remains to be tested, but our data show that HCMV specifically exploits EB3 to enrich factors at Golgi regions.
It was notable that the enrichment of CDK5RAP2 and formation of acetylated MTs was blocked by HEBTRON, demonstrating this process involved SxIP-interacting proteins. Recent studies have shown that EB1 and EB3 are not only important for the polymerization of MT plus-ends, but also play a particularly important role in the attachment and organization of MT minus-ends at Golgi sites (Yang et al., 2017). This too involves SxIP-interacting proteins such as Myomegalin, a core component of the GM130-AKAP450 complex that regulates Golgi-based MT nucleation (Efimov et al., 2007; Rios et al., 2004; Rivero et al., 2009; Roubin et al., 2013; Wang et al., 2014; Wu et al., 2016; Yang et al., 2017). Moreover, in these studies it was found that both EB proteins and CDK5RAP2 were required for MT nucleation at the Golgi (Yang et al., 2017). Our data suggests that at least in the context of the remodeled Golgi that becomes the AC, EB3 also uses SxIP-based interactions to mediate the redistribution of factors such as CDK5RAP2 to Golgi sites. Hence, our findings are in line with the emerging model whereby EB proteins play a key role in MT minus-end nucleation and growth at non-centrosomal sites.
It is also notable that HCMV specifically exploited EB3, but not EB1 to generate acetylated MTs. This diverges from Golgi-based MT formation that utilizes both EB1 and EB3 (Yang et al., 2017), and potentially reflects differences in the behavior of the remodeled Golgi or CDK-dependent phosphorylation of EB3 during infection. Alternatively, the hyper-activate nature of Golgi-based MT nucleation during HCMV infection may make differences between the functions of EB1 and EB3 more readily detectable. Indeed, although structurally similar and often functionally interchangeable, examples of diversification amongst EB proteins have emerged. This involves differences in N-terminal domains and a limited number of specific binding partners for each family member (Akhmanova and Steinmetz, 2015; Geraldo et al., 2008; Hsieh et al., 2007). For example, EB1 regulates spindle orientation while EB3 is required for daughter cell reattachment during mitosis, when EB3 stability is specifically increased (Ban et al., 2009; Ferreira et al., 2013). Here again, HCMV’s induction of a pseudo-mitotic state may underlie how it specifically exploits EB3. EB3 also plays specific roles in Focal Adhesion and Adherens Junction formation, ciliogenesis, and myoblast and epithelial apico-basal elongation (Bazellieres et al., 2012; Geyer et al., 2015; Komarova et al., 2012; Schroder et al., 2011; Straube and Merdes, 2007). While in many systems these functional differences can be subtle, in HCMV-infected cells robust divergence is evident; EB1 and EB3 specifically regulate CLIP170 behavior and acetylated MTs, respectively, with each contributing differently to AC structure and the extent of HCMV replication.
The dynamic behavior of the AC, critical to efficient virus maturation, likely maintains its integrity during cellular reorganization and subsequent cell migration. However, there are also striking similarities to the dynamics of Golgi reorientation in migrating cells, which like the AC can traverse the nucleus as smaller fragments that reassemble on the side of the leading edge (Wu et al., 2016). This plays a key role in polarizing the cell for migration through relocalization of vesicles and organelles, as well as control of cell adhesion (Maninova et al., 2013; Vinogradova et al., 2009; Wu et al., 2016). Our data demonstrate that MTs formed at the AC also control nuclear rotation prior to cell migration. Nuclear movement, which involves elements of rotation and the distinct process of front-rear repositioning, helps generate cell polarity in migrating cells (Maninova et al., 2013; Zhu et al., 2017). MTs contribute to this by directly pushing the nucleus or by enabling force exertion by motors associated with the nuclear envelope (Daga et al., 2006; Hui et al., 2016; Levy and Holzbaur, 2008; Maninova et al., 2013; Szikora et al., 2013; Wu et al., 2011; Zhao et al., 2012). While much attention has focused on centrosomes, growing MTs push oocyte nuclei independently of centrosomes during establishment of the Drosophila dorsal-ventral axis (Zhao et al., 2012), hinting at potential roles for non-centrosomal MTs in at least some contexts. Indeed, nuclear rotation requires considerable force and recent work revealed that α-tubulin acetylation confers mechanical strength to MTs (Portran et al., 2017; Xu et al., 2017). Given that Golgi-derived MTs are rapidly acetylated and may be the primary source of acetylated MT subsets in many cell types (Chabin-Brion et al., 2001; Rivero et al., 2009; Sanders and Kaverina, 2015), our data suggest Golgi-derived MTs may be underappreciated regulators of nuclear rotation.
Although nuclear rotation has been observed in several systems, precisely why it happens remains unclear and challenging to address. Rotation of nuclei is a fundamental part of intracellular reorganization during polarization and migration (Gundersen and Worman, 2013; Maninova et al., 2013). Nuclear rotation may be central to how HCMV remodels its host cell to replicate and spread, in part through polarizing the cell for migration. ACs also exhibited structural abnormalities in EB3-depleted or HEBTRON-treated cells, suggesting that in order to maintain their structure ACs may need acetylated MTs to tether to the nucleus. This tight coupling to a dynamic AC may in turn cause nuclei to rotate. Rotation has also been suggested to contribute to chromosome organization during meiosis (Christophorou et al., 2015). Given the pseudomitotic state of infected cells it is also possible that the role of Golgi-derived MTs in nuclear rotation is specific to mitotic processes coopted by HCMV, rather than migration-related cell polarization discussed above. Notably, HCMV forms discrete nuclear replication compartments that may be organized, similar to chromosomes, by rotating the nucleus, or be positioned relative to sites of AC tethering to the nucleus for efficient virus budding into the AC. Future studies will hopefully reveal precisely why nuclei rotate in different biological contexts, but our findings suggest this is a fundamental aspect of HCMV infection driven by EB3-regulated MTs.
While much remains to be learned from this and other systems, findings here reveal the dynamic behavior the HCMV AC and its potential to instruct our broader understanding of the regulation and function of non-centrosomal MTs, all in a biological context of infection by a clinically important pathogen. Our data also show that these virus-controlled processes can be exploited therapeutically. Targeting host proteins like EB3 with highly specialized functions is an attractive approach to avoid the emergence of drug-resistance commonly associated with therapeutics targeting evolutionarily adaptable viral proteins.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Derek Walsh (derek.walsh@northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cells
Validated and certified primary Normal Human Dermal Fibroblasts (NHDFs) isolated from human male neonatal foreskin were purchased from Lonza (CC-2509). HEK-293-T, HEK-293-A, VERO and BSC-40 cells were from Dr. Ian Mohr, NYU. Phoenix-Ampho cells were purchased from ATCC. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Fisher Scientific) supplemented with 5% Fetal Bovine Serum (FBS), 2 mM L-Glutamine and penicillin-streptomycin and maintained at 37°C, 5% CO2. Where indicated, confluent cultures of NHDFs were growth-arrested by washing three times in PBS before being maintained in DMEM supplemented with 0.2% Fetal Bovine Serum (FBS), 2 mM L-Glutamine and penicillin-streptomycin for 72 h.
Viruses
HCMV strain AD169 and bacterial artificial chromosome (BAC)-derived HCMV strains AD169, TB40/E and FIX expressing GFP reporters were grown on NHDFs until >90 cytopathic effect was observed. Cells and medium were collected and freeze-thawed to release virus. Cell debris was removed by centrifugation and virus was titrated by serial dilution on NHDFs and plaque counting. Generation of HCMV TB40/E expressing UL99-eGFP or UL32-mCherry, as well as viral expression vectors is described in Method Details. VacV-B5-GFP was grown on BSC-40 cells until >90% cypopathic effect was observed. Cells and medium were collected and freeze-thawed to release virus. Cell debris was removed by centrifugation and virus titrated on BSC-40 cells.
METHOD DETAILS
Viruses and cell transduction
HCMV BAC-derived strain TB40/E (clone 4) was used to generate either TB40/E expressing UL99-eGFP or UL32-mCherry using galK BAC recombineering protocols and transformation in SW105 E. coli. The galK ORF was inserted into UL99 or UL32 locus in BAC-TB40/E (clone 4) using the following primers: UL99-GalK-5': 5’-CAA CGT CCA CCC ACC CCC GGG ACA AAA AAG CCC GCC GCC CCC TTG TCC TTT CCT GTT GAC AAT TAA TCA TCG GCA-3’ UL99-GalK-3': 5’-GTG TCC CAT TCC CGA CTC GCG AAT CGT ACG CGA GAC CTG AAA GTT TAT GAG TCA GCA CTG TCC TGC TCC TT-3’ UL32-Galk-5': 5’-CCG TGC AGA ACA TCC TCC AAA AGA TCG AGA AGA TTA AGA AAA CGG AGG AAC CTG TTG ACA ATT AAT CAT CGG CA-3’ UL32-Galk-3': 5’-CGT CAC TAT CCG ATG ATT TCA TTA AAA AGT ACG TCT GCG TGT GTG TTT CTT CAG CAC TGT CCT GCT CCT T-3’. Positive clones were selected on GalK plates and further validated on McConkey plates. Insertion of the cassette was confirmed by PCR. Positive clones were then selected and grown in LB-chloramphenicol at 32°C overnight. Cultures were then diluted and grown to an OD600 0.5–0.7. Competent bacteria were electroporated with PCR products generated as follows: UL99-eGFP-5': 5’-CAACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCCCCTTGTCCTTT GTGAGCAAGGGCGAGGAGCTGTTCACCG-3’ coupled with UL99-eGFP-3':5’-GTGTCCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAG TTACTTGTACAGCTCGTCCATGCCGAGAGT-3’ using eGFP as a template, or UL32-Rev-5': 5’-CCG TGC AGA ACA TCC TCC AAA AGA TCG AGA AGA TTA AGA AAA CGG AGG AAA TGG TGA GCA AGG GCG AGG AG-3’ coupled with UL32 Rev-3': 5’-CGT CAC TAT CCG ATG ATT TCA TTA AAA AGT ACG TCT GCG TGT GTG TTT CTT TAC TTG TAC AGC TCG TCC ATG CCG-3’ using mCherry as a template.
Recombinant clones were selected for loss of GalK expression by screening on 2-Deoxy galactose/Glycerol containing plates. Clones were further validated by PCR analysis and then analyzed by sequencing. Virus stocks were grown and titrated on NHDFs as described above.
GFP-CLIP170, GFP, HCMV IE1 and HCMV IE2 retroviral expression vectors were produced by transfecting Phoenix-Ampho cells with pBABE-puro vectors described below. Cell culture medium was changed 24 h after transfection. Supernatant containing virus was then collected at 48 h and 72 h post-transfection and filtered through a 0.45µM filter.
For stable expression of eGFP-CLIP170 or NLS-mCardinal sub-confluent NHDFs were transduced with a pBABE-puro-AA-GFP-CLIP170-derived or pBABE-puro-AA-NLS-mCardinal-derived retroviral vector in the presence of polybrene. Four hours after transduction, NHDFs were washed with PBS and fresh cell culture media was added. 24 h after transduction, pools of stably expressing cells were generated by selecting with either 0.8 µg/ml puromycin (for eGFP-CLIP170 and NLS-mCardinal). Following selection, cells were maintained in growth medium containing either 160 ng/ml puromycin. For transient expression of GFP or IE1/2 NHDFs were transduced with retroviral vectors described above and processed at the indicated times.
RNA interference (RNAi) and inhibitors
siRNAs were obtained from Life Technologies (Thermo Fisher Scientific); See Key Resources Table for details. siRNAs were transfected as described previously (Jovasevic et al., 2015). To avoid effects on early HCMV infection, cells were transfected with 150 pmol/ml siRNA using RNAiMax (Invitrogen) and 30 h later infected with HCMV at the indicated MOI. When examining late stages of infection, 3 d.p.i. cells were transfected with siRNA for a second time to counter HCMV-induced increases in EB expression. In the case of spreading assays, this was done without changing cell culture medium. Infected cultures were then processed for imaging, western blot analysis or titration of virus production as described below. To suppress viral IE1/2 expression, confluent NHDFs were treated with 300 pmol/ml siRNA. 1 day later, cells were infected with HCMV AD169 at MOI 3. Lysates were collected at 3 d.p.i. and analyzed by Western blotting as described below. Aurora Kinase/CDK inhibitor JNJ-7706621 (Calbiochem) was dissolved in DMSO and used at a final concentration of 3 µM. Medium was replaced with fresh DMSO-or inhibitor-containing medium every 24h. HEBTRON and mutN-mutC, mutN controls were dissolved in DMSO and is described in detail below. To test effects of HEBTRON on infection, confluent NHDFs were pretreated for 1 h with DMSO control or 25 µM HEBTRON in culture medium without penicillin/streptomycin, followed by infection with HCMV TB40/E (GFP) at the indicated MOI. Cell culture media with DMSO or 25 µM HEBTRON was replaced every 48h. To avoid removing newly produced virus at later stages, at 6 d.p.i. a 10× stock of DMSO or HEBTRON was added directly to the cell culture medium such that the final concentration added was 25µM. At the indicated time-points infected cultures were then processed for imaging as described below.
Generation and Characterization of HEBTRON
Expression and purification of EB3
Preparation of (His6)-YFP-EB3, (His6)-CFP-EB3, (His6)-EB3 and EB3-C terminus (200–281aa) was described previously (Geyer et al., 2015; Komarova et al., 2012). (His6)-tagged recombinant proteins were expressed in Escherichia coli strain BL21 (DE3) (Stratagene). The expression constructs for (His6)-EB3 and EB3-C terminus contained a tobacco etch virus (TEV) protease cleavage site immediately following the His6 tag for efficient removal. Bacteria were grown at 37°C in LB medium containing 50 µg/ml kanamycin. Wh en the OD600 reached 0.6–0.7, protein synthesis was induced by addition of isopropyl 1-thio-β-D-galactopyranoside (IPTG) to a final concentration of 250 µM. After 4 h at 30°C, bacterial pellets were isolated and sonicated (4 × 1 min) in medium comprising 150 mM NaCl, 5 mM 2-mercaptoethanol, 2 mM CaCl2, 10 mM imidazole, 2 mM PMSF, 25 mM Tris, pH 7.4.
For affinity purification, (His6)-EB proteins were purified using Ni-NTA beads (Thermo Scientific) as previously described (Geyer et al., 2015). Ni-NTA beads (1 ml) in a 20 ml column (Bio-Rad) were equilibrated with 50 bed-volumes of binding buffer (25 mM Tris, pH 7.4, 300 mM NaCl, 5 mM 2-mercaptoethanol, 2 mM PMSF). Bacterial lysate (50 ml) was then added to the column, followed by washing (150 bed-volumes of wash buffer, ~75 ml). The protein-bound beads were washed with phosphate-buffered saline (PBS) supplemented with 2 mM CaCl2 and protease inhibitor cocktail (Sigma) and stored in the same buffer.
For gel-filtration, after washing the Ni-NTA beads, recombinant His6-EB3 was eluted by addition of wash buffer containing 150 mM imidazole. Peak elution fractions were pooled, exchanged to imidazole-free buffer using PD-10 desalting columns (GE Life Sciences), and concentrated with an Amicon Ultra-15 with 10 kDa cut-off concentrator unit (Millipore, Inc.). The His6 tag was removed by addition of 1.5% (w/w) recombinant TEV protease and incubation at °C for 16 hr. For further polishing, EB3 proteins were then subjected to gel filtration chromatography over tandem Superdex 200 HR 10/30 columns connected in series and controlled by an AKTA FPLC (GE Life Sciences). Peak fractions containing EB3 proteins were then pooled and concentrated as described above.
Peptide synthesis and purification
Peptides were synthesized using the stepwise solid-phase method by 9-fluorenylmethoxycarbonyl (Fmoc) chemistry on Wang resin (AnaSpec, Fremont, CA, USA) with a 12-channel multiplex peptide synthesizer (Protein Technologies, Tucson, AZ, USA) according to the manufacturer’s procedures. Detachment of peptide from the resin and removal of the side chain protection groups were done by incubating the resin with a mixture of trifloroacetic acid (TFA):Thioanisole:Water:Phenol:1,2-ethanedithio (82.5:5:5:5:2.5 v/v) for 2 hours.
The crude peptide was purified on a preparative Kinetex reversed-phase C18 column, 150 × 21.1 mm (Phenomenex, Torrance, CA, USA) using a BioCad Sprint (Applied Biosystems, Foster City, CA, USA). A flow rate of 30 mL/min with solvent A (0.1% TFA in deionized water) and solvent B (0.1% TFA in acetonitrile) was used. The column is equilibrated with 5% solvent B before sample injection. Elution is performed with a linear gradient from 5% solvent B to 100% solvent B in 60 min. The absorbance of the column effluent is monitored at 214 nm, and peak fractions are pooled and lyophilized.
The pure peptide fraction is identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) or electrospray ionization mass spectrometry (ESI-MS) and lyophilized.
Synthesis of N-myristoylated peptides
Chloroform (2.5 ml) was added to ~250 mg of Myr-anhydride and mixed until Myr-anhydride was dissolved. 2.5 ml of Dimethylformamide (DMF) is added following 50 µmole peptide on the resin. The mixture was kept at 60°C for 1 h and then filtered and washed with 50 ml hot chloroform:DMF (1:1).
Isothermal titration calorimetry
HEBTRON and EB3(200–281) were dialyzed into PBS. Titration experiments were performed using a VP-ITC Microcalorimeter (MicroCal LLC, Northampton, MA) at 25°C. For titration, 25–27 aliquots (5 µL each) of 60 µM EB3 (200–281) were injected into the ITC cell containing 1.4 mL of 4 µM HEBTRON. Each titration was preceded by a single 2µL injection to address diffusion artifacts. Two reference titrations were run. One of these titrations controlled for protein dilution effects and the other controlled for the peptide dilution effects. The reference data were subtracted from the protein titration data points. The integrated heat values were analyzed using the ‘Origin 7.0’-software as well as the Scientist 3.0 software (Micromath Scientific Software). Data were fit using the ‘One set of sites’ model, to yield the dissociation constant (Kd), the stoichiometry, the enthalpic and entropic contributions to the Gibbs free energy of complex formation.
Analysis of EB3 Dimerization
FRET measurements were carried out using a PHERAstar FS (BMG LABTECH Inc., Cary, NC) microplate reader equipped with a FRET module for CFP and YFP pair (Komarova et al., 2012). (His6)-YFP-EB3 and (His6)-CFP-EB3 were mixed in equimolar concentration. In the experiments with HEBTRON, the protein and the peptide were mixed in 1:1 molar ratio. CFP and FRET fluorescence were recorded using simultaneous dual emission at λ= 480 nm and 530 nm, respectively, and excitation at λ= 420 nm.
DARTS using purified full length (His6)EB3
100 nM of recombinant (His6)-EB3 was incubated with 100 nM HEBTRON or the EB1-specific peptide for 2h at 4°C, followed by proteolysis with 1µg of thermolysin at 37°C for 1 hr. To stop proteolysis, 0.5 M EDTA (pH 8.0) was added to each sample at a 1:10 ratio. The protein was detected using anti-EB3 antibody by Western blotting.
NanoDSF
Unfolding of EB3 alone or in presence of HEBTRON was measured by detecting the temperature-dependent change in tryptophan fluorescence at λ=330nm and 350nm using the Prometheus NT.Plex (NanoTemper). The temperature gradient was set in a range from 20°C to 90°C. Melting temperatures were calculated from the maximum of the first derivative of the fluorescence ratios (F350/F330). For this, an eighth order polynomial fit was calculated for the transition region and then the first derivative of the fit was formed. The peak position at melting temperature was determined from the first derivative.
HEBTRON Uptake Assays
NHDFs were grown on glass bottom dishes for 2 days at 37 °C and 5% CO2. Cells were kept at 37 °C for the experiment. The 5’ 6-FAM (Fluorescein)-conjugated Myr-HEBTRON was added at a concentration of 10 µM directly to the media during imaging. Images were acquired every 10 seconds for 10 minutes in both the FITC and DIC channels (to mark cell boundaries) using a Zeiss LSM 880 confocal microscope equipped with a 63× 1.4 NA oil objective. The fluorescent intensity inside each cell for each time point was measured using ImageJ and normalized to the initial fluorescent background intensity inside the cell. The normalized fluorescent intensity was plotted over time. The half time to maximum uptake as well as the maximum uptake in the cells was found by fitting a Sigmoidal dose response curve to the data using GraphPad Prism 7.
Virus production and spreading assays
To determine effects on virus production, cells were treated with siRNAs or inhibitors and infected as indicated. Culture medium was then removed, centrifuged at 4°C for 3 min at 3,000 rpm to remove cell debris and supernatants collected. To determine viral titers, a 1:10 dilution series was generated for each virus supernatant and used to infect NHDFs seeded on 12-well plates using four technical replicates for each dilution. At 14 d.p.i. GFP-positive plaques were counted at the appropriate dilution using a Leica DMI6000B-AFC microscope with a 37°C InVivo environmental chamber. Phase imaging was also used to examine Cytopathic Effect (CPE) to ensure no infectious virus was present that did not retain GFP expression.
To determine effects of siRNAs on plaque sizes, NHDFs were treated and infected as outlined above before phase and fluorescent (GFP channel) images of plaques were acquired using a Leica DMI6000B-AFC microscope with a 37°C InVivo environmental chamber, 10× objective, X-Cite XLED1 illumination, ORCA FLASH 4.0 cMOS camera and Metamorph software (Molecular devices) using the multi-dimensional acquisition function. Plaque areas were determined by using the threshold area function of the FIJI. To determine the effect of HEBTRON treatment on virus plaque sizes, NHDFs seeded in 12-well plates were pretreated with DMSO, 25 µM HEBTRON or 25 µM HEBTRON controls for 1 h. Cultures were then infected with either HCMV AD169-GFP or TB40/E-GFP, VACV-B5-GFP at MOI 0.001. For slower-replicating HCMV, cell culture media containing DMSO or 25 µM HEBTRON or controls was replaced every 48h. Images were acquired at the indicated times and analyzed as described above. 50 pixel rolling ball background subtraction was used to reduce autofluorescence from the cell culture vessel evident in VACV-B5-GFP plaque images using FIJI.
Western blotting (WB)
For Western blot analysis, cells were lysed in laemmli buffer (62.5 mM Tris-HCl at pH 6.8, 2% SDS, 10% glycerol, 0.7 M β-mercaptoethanol), followed by boiling for 3 min. Samples were resolved using reducing 10% polyacrylamide Tris-glycine SDS PAGE. Resolved proteins were transferred to a nitrocellulose membrane (GE Healthcare Life Sciences) at 57 V for 60 min (Mini Trans-Blot system, Bio-Rad), washed in Tris-Buffered Saline (TBS) containing 0.1% Tween (TBS-T) and blocked (5% non-fat milk TBS-T) before incubating with primary antibodies diluted in 3% BSA TBS-T overnight at 4°C. For antibodies used see Key Resources Table 1. Membranes were washed with TBS-T and incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (GE Healthcare Life Sciences) diluted 1:3,000 in TBS-T containing 5% non-fat milk for 1 h at room temperature. Membranes were then washed in TBS-T and incubated with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) before exposure to x-ray film. Densitometry was performed on scanned films using Gel Analyzer in FIJI.
Immunofluorescence and Live Cell Imaging
For immunofluorescence microscopy, confluent monolayers of NHDFs were grown on glass coverslips in a 12-well plate. Cells were treated with siRNAs or inhibitors and infected as indicated. Cells were then rinsed in PBS and fixed in ice-cold methanol for 7 min on ice. Samples were then rinsed in PBS and blocked in PBS containing 10% human serum and 0.25% (w/v) saponin for 1 h at 37°C. Samples were incubated with primary antibody diluted in PBS containing 10% human serum and 0.025% (w/v) saponin overnight at 4°C. The next day, samples were washed in PBS containing 0.025% (w/v) saponin, then incubated with appropriate Alexa Fluor-conjugated secondary antibodies diluted in PBS containing 10% human serum and 0.025% (w/v) saponin for 1 h at room temperature. Samples were stained with Hoechst 33342 where indicated, washed in PBS containing 0.025% (w/v) saponin, then washed with water, dried and mounted on a glass slide using FluroSave (Calbiochem). For antibody details see Key Resources Table 1. Wide-field images were acquired using a Leica DMI6000B-AFC microscope using a 100× objective (HC PL APO 100×/1.44NA OIL), X-Cite XLED1 illumination, ORCA FLASH 4.0 cMOS camera and Metamorph software using the multi-dimensional acquisition function to ensure use of the same acquisition settings for all collected images. Confocal images were acquired using a motorized spinning-disc confocal microscope (Leica DMI 6000B), Yokogawa CSU-X1 A1 confocal head and MetaMorph software using the multi-dimensional acquisition function. Confocal z-stacks were acquired at 0.2 µm intervals. All images were analyzed using Metamorph and compiled using the FIJI. All images within a given dataset were processed equivalently.
For nocodazole wash-out assays, NHDFs were seeded onto glass coverslips and grown to confluence. Cultures were then mock infected or infected with HCMV strain AD169 at MOI 3. At 3 d.p.i. culture medium was changed to DMEM supplemented with 10 µM nocodazole for 8 h to completely depolymerize MTs. Cells were then washed with PBS and incubated in normal culture medium for 20 min. Cells were then fixed in ice cold methanol, stained with the indicated antibodies for immunofluorescence and imaged as described above. Areas of centrosomal tyrosinated tubulin staining after nocodazole washout were determined using the threshold area function on single confocal slices obtained at the centrosomal focal plain, indicated by γ-tubulin staining. Identical thresholds were used for all conditions. Non-centrosomal microtubules were quantified using the cell counter plugin for FIJI, centrosomal microtubules were excluded from analysis using pericentrin staining. CDK5RAP2 staining area was determined for infected cells with consistent gB staining using the threshold area function of the FIJI.
For live-cell imaging, immediately prior to imaging culture medium was changed Leibovitz's L-15 Medium without phenol red (Thermo Fisher Scientific) supplemented with 2 mM L-Glutamine and 5% Fetal Bovine Serum (FBS). Cultures then transferred to a Leica DMI6000B-AFC microscope with a 37°C InVivo environmental chamber and allowed to equilibrate for 30 min. For single condition experiments, cells were seeded on 35 mm glass-bottom dishes (MatTek). Where multiple conditions were compared, cells were seeded on cell view four compartment 35 mm glass-bottom dish (Greiner Bio-One) to ensure the same experimental conditions between comparisons. For analysis of microtubule dynamics, NHDFs stably expressing eGFP-CLIP170 were imaged at 500 ms intervals for 1 min using a 100× objective (HC PL APO 100×/1.44NA OIL), X-Cite XLED1 illumination and an ORCA FLASH 4.0 cMOS camera with Metamorph software.
CLIP-170-eGPF plus-end line-scan analysis was performed using the line-scan function of Metamorph on single frames from time lapse images of polymerizing microtubules only in TB40/E UL32-mCherry infected cells. EB1 and EB3 line-scan were obtained from fixed cells as above with intensities for EB1 and EB3 acquired simultaneously from individual microtubules in 2-channel stacks. All line-scans were first normalized and aligned before analysis using Microsoft Excel. For live-cell imaging to determine AC maturation dynamics, a multi-position stage (was used to acquire images once every 30 mins from multiple infected cells and across multiple sections of compartmentalized dishes over the indicated imaging periods. For faster frame-rate imaging of virion trafficking and AC behavior, images were acquired at 500 ms intervals for 1 min. All live-cell image stacks were analyzed using Metamorph and compiled using the FIJI. UL32-mCherry/UL99-eGFP labelling of virus particles was analyzed simultaneously using the line-scan function of Metamorph drawn across individual virus particles in 2-channel stacks generated from single frames of time lapse images of AC maturation of cells infected with both viruses. The maximum point of UL32-mCherry fluorescence were aligned for all virus particles before analysis using Microsoft Excel.
In all figures and movies, representative examples are shown. Cartoons of live cell images were generated using Adobe Illustrator.
Tracking nuclear rotation
Two approaches to tracking nuclear rotation were used. In the first, nuclear reorientation was manually tracked using two fixed points on the nuclear membrane of infected cells using the Manual Tracking plugin of Fiji. The first point was the center of the unique nuclear “pinch” generated by HCMV (or the point most proximal to the AC if the pinch was yet to form). The second point was on the nuclear membrane directly opposite the first point. These coordinates were center corrected by deducting the coordinates of the first point from the second point to visualize rotation around a single, fixed point. Degrees of rotation were derived from the inverse tan trigonometric function using center corrected coordinates. The second approach to tracking nuclear rotation involved labeling nuclei in NHDFs stably expressing mCardinal fused to a nuclear localization signal (NLS), which enabled the tracking of nuclear movement in both infected and uninfected cells. Nuclear movement was measured as described above, this time using far-red fluorescent nuclei. For uninfected cells, the two points used to track rotation were directly opposite each other at the center of the elongated side of the nuclear membrane.
RNA Isolation and RT-qPCR
To determine the relative expression of EB transcripts between mock and HCMV infected cells, growth-arrested NHDFs on 60mm dishes were either mock infected or infected with HCMV AD169 at MOI 3. Cells were collected in Trizol (Invitrogen) at 3 d.p.i. or 4 d.p.i. and RNA was isolated using RNeasy kit (Qiagen). cDNA was generated from 0.1 µg of RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche). Real-Time quantitative PCR (RT-qPCR) was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems) and FastStart Universal SYBR Green Master (Rox). For details of primers used to amplify POLR2L, EB1, EB2 and EB3 see Key Resources Table 1. Quantification of RNA was performed using the comparative Ct method using POLR2L as a stable endogenous control in HCMV infected cells. Fold changes were determined by average Ct comparison to mock infected cells. Data was generated from 3 independent biological replicates for each point.
Retroviral expression vectors used a modified version of pBABE-puro in which unique AgeI, NdeI and ApaI restriction sites were added to the major cloning site by ligating the annealed oligos 5’-ATAGCCGGCACCGGTACATATGGGCCCGGATCCATA-3’ 5’-TATGGATCCGGGCCCATATGTACCGGTGCCGGCTAT-3’ into NgoMIV and BamHI sites to produce pBABE-puro-AA.
To generate pBABE-puro-AA-eGFP-CLIP170, pC1-eGFP-CLIP170 was digested with AgeI and ApaI restriction enzymes. The eGFP-CLIP170 fragment was gel purified and ligated into pBABE-puro-AA. To generate the HCMV IE1 expression construct, IE1 was amplified from cDNA isolated from NHDFs infected with HCMV AD169 for 2 d using a forward primer with an AgeI site 5’-ATACATATGACCGGTACCATGGAGTCCTCTGCCAAGAGAAAG-3’ and reverse primer with an EcoRI site 5’-ATACATATGGAATTCTTACTGGTCAGCCTTGCTTCTAG -3’ The amplicon was digested with AgeI and EcoRI and ligated into pBABE-puro-AA to produce pBABE-puro-AA-IE1. To generate the HCMV IE2 expression construct, IE2 was amplified from pcDNA3.1(+)-IE2-HA (gift of Mirko Trilling) using a forward primer with an AgeI site 5’-ATACATATGACCGGTACCATGGAGTCCTCTGCCAAGAGAAAG-3’ and reverse primer with an EcoRI site 5’-ATACATATGGAATTCTTACTGAGACTTGTTCCTCAGG-3’ The amplicon was digested with AgeI and EcoRI and ligated into pBABE-puro-AA to produce pBABE-puro-AA-IE2. To generate the GFP expression construct, GFP was amplified from pC1-GFP-CLIP-170 using a forward primer with an NgoMIV site 5’-ATAGCCGGCCTCGAGATGGTGAGCAAGGGCGA-3’ and reverse primer with an EcoRI site 5’-ATAGAATTCTTACTTGTACAGCTCGTCCATGCCG-3’ The amplicon was digested with NgoMIV and EcoRI then ligated into pBABEpuro-AA to produce pBABE-puro-AA-GFP.
To generate pBABE-PURO-AA-NLS-mCardinal, mCardinal was amplified from pmCardinal-Rab4a-7 using a forward primer with an AgeI site and the SV40 NLS (indicated in lower case) 5'ATACATATGACCGGTACCATGACTGCTccaaagaagaagcgtaaggtaATGGTGAG CAAGGGCGAGGAGCTGATC-3' and reverse primer with a BamHI site 5'-TATCATATGGGATCCTTACTTGTACAGCTCGTCCATGCC-3' The amplicon was digested with AgeI and BamHI then ligated into pBABE-puro-AA to produce pBABE-puro-AA-NLS-mCardinal.
All cloning followed standard cloning procedures and all construct inserts and flanking regions were verified by sequencing at the Northwestern University Sequencing Core Facility (NUSeq). Restriction enzyme sites are shown in underlined bold.
Quantification and Statistical Analyses
GraphPad Prism 7 was used to graph and analyze data. One-way ANOVA were used to compare multiple conditions to a control, whereas individual comparisons used a unpaired student’s two-tailed t test where applicable. See figure legends for details of each analysis and definitions of statistical significance.
KEY RESOURCES TABLE
Supplementary Material
Movie S1: The dynamics of the AC. Related to Figure 1C–E
Part A: Early ACs merge into a singular structure. NHDFs were infected with TB40/E-UL99-eGFP at MOI 0.5. Images were acquired at 2 frames per hour over the indicated times post-infection using a Leica DMi6000B widefield microscope. In all cases where two UL99-positive ACs were observed they eventually merged.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
PartB: The behavior of UL32-mCherry and UL99-eGFP labeled compartments in HCMV-infected cells. NHDFs were co-infected with TB40/E-UL32-mCherry and TB40/E-UL99-eGFP at MOI 0.5. Images were acquired at 2 frames per hour over the indicated times post-infection using a Leica DMi6000B widefield microscope. Lower panels show individual channels in grey for increased contrast (Left: UL99-eGFP; Right: UL32-mCherry). UL99-eGFP labels early ACs. UL32-mCherry labels cytoplasmic structures that accumulate around the UL99-eGFP-labeled AC. Note also, nuclear rotation is observed in infected cells. As infection progresses, UL32-mCherry-positive domains appear in the nucleus followed by the appearance of UL32-mCherry and UL99-eGFP labeled particles in the AC and cytoplasm.
Time is in hours post infection (h.p.i.).
Scale Bar = 10µm.
NHDFs expressing mCardinal-NLS were either mock infection or infected with TB40/E-UL99-eGFP at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times post-infection. Two examples of each event are provided in the top and bottom panels for mock and infected cells. Note that NLS signal declines in infected cells, in line with reports that infected cell nuclei become more permeable.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
Left Panels: The AC positions itself at the leading edge prior to cell migration. NHDFs were co-infected with TB40/E-UL99-eGFP and TB40/E-UL32-mCherry at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times in hours post infection (h.p.i.). Panel 1 shows two cells in which, after nuclear rotation, the AC positions itself at the leading edge and appears to drag the nucleus in the direction of cell motility. Panel 2 shows an example of how, after nuclear rotation, the AC leaves the nuclear “pinch” to position at the leading edge and again appears to drag the nucleus in the direction of migration.
Right Panels: ACs continue to merge under different circumstances during later stages of infection. NHDFs were infected with TB40/E-UL99-eGFP at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times in hours post infection (h.p.i.). Panel 3 shows an AC that splits during cell migration but regroups. Panel 4 illustrates the merging of two ACs, presumably as a result of cell fusion. To aid in viewing the merge event, UL99-eGFP signal was converted to a color-intensity heat map. At the imaging mid-point each infected cell contains an AC of equivalent intensity. However, the two ACs merge into a singular more intensely colored single AC.
Scale Bar = 20µm.
NHDFs were infected with TB40/E at MOI 1 for 5d. Fixed cells were stained for acetylated tubulin and TGN46. Nuclei were stained with Hoechst. Confocal Z-stacks were generated at 0.2 µm intervals.
Part A: Effects of EB1 or EB3 depletion on eGFP-CLIP170 tracking in HCMV-infected cells. Depletion of EB1 results in CLIP170-eGFP comets that pause or elongate down the length of growing MTs. NHDFs expressing eGFP-CLIP170 were treated with control, EB1 or EB3 siRNAs and infected with TB40/E (MOI 1). Images were acquired at 4 d.p.i. at 2 frames per second using a Leica DMi6000B widefield microscope. Insets and event mark arrows highlight the mix of elongated (green arrow) and pausing (red arrows) eGFP-CLIP170 populations in EB1 depleted cells.
Time is in seconds (s).
Scale Bar = 20µm.
PartB: Effects of EB1 or EB3 depletion on nuclear rotation in HCMV-infected cells. EB3 depletion blocks nuclear rotation in infected cells. NHDFs were treated with control, EB1 or EB3 siRNAs and infected with TB40/E-UL99-eGFP (MOI 0.5). Images were acquired at 2 frames per hour using a Leica DMi6000B widefield microscope.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
NHDFs were treated with control, EB1 or EB3 siRNAs and infected with TB40/E-UL99-eGFP (MOI 0.5). At 7.d.p.i. cultures were imaged at 2 frames per second using a Leica DMi6000B widefield microscope. Three examples are provided for each condition. Left panels show the overall structure of the AC. Right panels show zoomed images of the corresponding left panel showing the behavior of UL99-eGFP-postive structures and particles in the cytoplasm.
Time is in seconds (s).
Scale Bar = 30µm.
Part A: Effects of HEBTRON on nuclear rotation in HCMV-infected cells. Live-cell images of NHDFs treated with vehicle (DMSO) (top three panels) or HEBTRON (bottom three panels) and infected with TB40/E-UL99-eGFP (MOI 0.5). Cultures were imaged at 2 frames per hour using a Leica DMi6000B widefield microscope.
The central DMSO panel provides an example of an initial nuclear rotation through 180°, followed by rotations in alternating directions rather than progression through a full 360°.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
Part B: Effects of HEBTRON on AC structure and virus particle trafficking. Live-cell images of NHDFs treated with vehicle (DMSO) or HEBTRON and infected with TB40/E-UL99-eGFP (MOI 0.5). At 7.d.p.i. cultures were imaged at 2 frames per second using a Leica DMi6000B widefield microscope. Vertical white bar separates a single linearly scaled movie from four examples using non-linear scaling.
Left Panel Set: Linearly scaled movies show a dense AC core in DMSO-treated cells.
Right Panel Sets: Non-linear scaling was used to remove saturation in the center of the dense AC of vehicle-treated samples in order to reveal details of trafficking within the AC itself. To illustrate this, the first panel set corresponds to the Linearly Scaled Panel Set on the far left.
Time is in seconds (s).
Scale Bar = 10µm.
Highlights.
HCMV assembly compartments (ACs) are dynamic Golgi-derived MTOCs
Viral IE2 protein and CDK regulation increase host EB protein levels for MT control
AC structure and nuclear rotation are controlled by EB3-regulated acetylated MTs
A cell-permeable peptide targeting EB3 suppresses HCMV replication
Acknowledgments
We thank Daniel Streblow, Mirko Trilling and Bernard Moss for reagents, and Wyatt Strutz and NanoTemper Technologies, Inc, for help with NanoDSF. This work was funded by grants from the National Institutes of Health to E.M. (R01AI101080), V.G. (R01CA188427), Y.K. (R01HL103922) and D.W. (P01GM105536), and through the NIH-funded Third Coast Center For AIDS Research (CFAR) (P30AI117943).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
D.J.P designed, performed and analyzed experiments. A.B., K.K., V.G. and Y.K. designed and characterized HEBTRON. M.N. and E.A.M. constructed and characterized recombinant viruses. D.W. and Y.K. conceived the project and interpreted data. D.W., Y.K., D.J.P. and E.A.M. wrote the manuscript.
Declaration of Interests
Y.K. and D.W. filed an invention disclosure on the use of HEBTRON as an antiviral. The authors declare no competing financial interests.
References
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Alwine JC. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 2012;8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban R, Matsuzaki H, Akashi T, Sakashita G, Taniguchi H, Park SY, Tanaka H, Furukawa K, Urano T. Mitotic regulation of the stability of microtubule plus-end tracking protein EB3 by ubiquitin ligase SIAH-1 and Aurora mitotic kinases. J Biol Chem. 2009;284:28367–28381. doi: 10.1074/jbc.M109.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellieres E, Massey-Harroche D, Barthelemy-Requin M, Richard F, Arsanto JP, Le Bivic A. Apico-basal elongation requires a drebrin-E-EB3 complex in columnar human epithelial cells. J Cell Sci. 2012;125:919–931. doi: 10.1242/jcs.092676. [DOI] [PubMed] [Google Scholar]
- Buchkovich NJ, Maguire TG, Alwine JC. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol. 2010;84:7005–7017. doi: 10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Liu P, Sze SK, Dai C, Qi RZ. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191:1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou N, Rubin T, Bonnet I, Piolot T, Arnaud M, Huynh JR. Microtubule-driven nuclear rotations promote meiotic chromosome dynamics. Nat Cell Biol. 2015;17:1388–1400. doi: 10.1038/ncb3249. [DOI] [PubMed] [Google Scholar]
- Clippinger AJ, Alwine JC. Dynein mediates the localization and activation of mTOR in normal and human cytomegalovirus-infected cells. Genes Dev. 2012;26:2015–2026. doi: 10.1101/gad.196147.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daga RR, Yonetani A, Chang F. Asymmetric microtubule pushing forces in nuclear centering. Curr Biol. 2006;16:1544–1550. doi: 10.1016/j.cub.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Das S, Pellett PE. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol. 2011;85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Vasanji A, Pellett PE. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol. 2007;81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot CO, Jelesarov I, Damberger FF, Bjelic S, Scharer MA, Bhavesh NS, Grigoriev I, Buey RM, Wuthrich K, Capitani G, et al. Molecular insights into mammalian end-binding protein heterodimerization. J Biol Chem. 2010;285:5802–5814. doi: 10.1074/jbc.M109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, Holzbaur EL. Microtubule plus-end tracking by CLIP-170 requires EB1. Proc Natl Acad Sci U S A. 2009;106:492–497. doi: 10.1073/pnas.0807614106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JG, Pereira AJ, Akhmanova A, Maiato H. Aurora B spatially regulates EB3 phosphorylation to coordinate daughter cell adhesion with cytokinesis. J Cell Biol. 2013;201:709–724. doi: 10.1083/jcb.201301131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Hau SY, Kho YS, Jia Y, He L, Qi RZ. Interaction of CDK5RAP2 with EB1 to track growing microtubule tips and to regulate microtubule dynamics. Mol Biol Cell. 2009;20:3660–3670. doi: 10.1091/mbc.E09-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo S, Khanzada UK, Parsons M, Chilton JK, Gordon-Weeks PR. Targeting of the F-actin-binding protein drebrin by the microtubule plus-tip protein EB3 is required for neuritogenesis. Nat Cell Biol. 2008;10:1181–1189. doi: 10.1038/ncb1778. [DOI] [PubMed] [Google Scholar]
- Geyer M, Huang F, Sun Y, Vogel SM, Malik AB, Taylor CW, Komarova YA. Microtubule-Associated Protein EB3 Regulates IP3 Receptor Clustering and Ca(2+) Signaling in Endothelial Cells. Cell Rep. 2015;12:79–89. doi: 10.1016/j.celrep.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel L, Chou S, Mocarski ES. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 2007;3:e6. doi: 10.1371/journal.ppat.0030006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel L, Mocarski ES. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. J Virol. 2004;78:11988–12011. doi: 10.1128/JVI.78.21.11988-12011.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Hsieh PC, Chang JC, Sun WT, Hsieh SC, Wang MC, Wang FF. p53 downstream target DDA3 is a novel microtubule-associated protein that interacts with end-binding protein EB3 and activates beta-catenin pathway. Oncogene. 2007;26:4928–4940. doi: 10.1038/sj.onc.1210304. [DOI] [PubMed] [Google Scholar]
- Hui TH, Zheng F, Lin Y, Fu C. The linear and rotational motions of the fission yeast nucleus are governed by the stochastic dynamics of spatially distributed microtubules. J Biomech. 2016;49:1034–1041. doi: 10.1016/j.jbiomech.2016.02.017. [DOI] [PubMed] [Google Scholar]
- Indran SV, Ballestas ME, Britt WJ. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J Virol. 2010;84:3162–3177. doi: 10.1128/JVI.01776-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- Jiang K, Toedt G, Montenegro Gouveia S, Davey NE, Hua S, van der Vaart B, Grigoriev I, Larsen J, Pedersen LB, Bezstarosti K, et al. A Proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr Biol. 2012;22:1800–1807. doi: 10.1016/j.cub.2012.07.047. [DOI] [PubMed] [Google Scholar]
- Jovasevic V, Naghavi MH, Walsh D. Microtubule plus end-associated CLIP-170 initiates HSV-1 retrograde transport in primary human cells. J Cell Biol. 2015;211:323–337. doi: 10.1083/jcb.201505123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, Lansbergen G, Galjart N, Grosveld F, Borisy GG, Akhmanova A. EB1 and EB3 control CLIP dissociation from the ends of growing microtubules. Mol Biol Cell. 2005;16:5334–5345. doi: 10.1091/mbc.E05-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova YA, Huang F, Geyer M, Daneshjou N, Garcia A, Idalino L, Kreutz B, Mehta D, Malik AB. VE-cadherin signaling induces EB3 phosphorylation to suppress microtubule growth and assemble adherens junctions. Mol Cell. 2012;48:914–925. doi: 10.1016/j.molcel.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniewska K, Warbrick E, Ohkura H. Peptide aptamers define distinct EB1- and EB3-binding motifs and interfere with microtubule dynamics. Mol Biol Cell. 2014;25:1025–1036. doi: 10.1091/mbc.E13-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicklal S, Emery VC, Lazzarotto T, Boppana SB, Gupta RK. The "silent" global burden of congenital cytomegalovirus. Clin Microbiol Rev. 2013;26:86–102. doi: 10.1128/CMR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maninova M, Klimova Z, Parsons JT, Weber MJ, Iwanicki MP, Vomastek T. The reorientation of cell nucleus promotes the establishment of front-rear polarity in migrating fibroblasts. J Mol Biol. 2013;425:2039–2055. doi: 10.1016/j.jmb.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Maurer SP, Bieling P, Cope J, Hoenger A, Surrey T. GTPgammaS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs) Proc Natl Acad Sci U S A. 2011;108:3988–3993. doi: 10.1073/pnas.1014758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski ES, Shenk T, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams & Wilkins; 2007. pp. 2701–2772. [Google Scholar]
- Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics. 2010;9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EE, Wardleworth BN, Askham JM, Markham AF, Meredith DM. EB1, a protein which interacts with the APC tumour suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 1998;17:3471–3477. doi: 10.1038/sj.onc.1202247. [DOI] [PubMed] [Google Scholar]
- Petry S, Vale RD. Microtubule nucleation at the centrosome and beyond. Nat Cell Biol. 2015;17:1089–1093. doi: 10.1038/ncb3220. [DOI] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Thery M, Nachury MV. Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol. 2017;19:391–398. doi: 10.1038/ncb3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebmann GM, Grabski R, Sanchez V, Britt WJ. Phosphorylation of Golgi Peripheral Membrane Protein Grasp65 Is an Integral Step in the Formation of the Human Cytomegalovirus Cytoplasmic Assembly Compartment. MBio. 2016;7 doi: 10.1128/mBio.01554-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. Identification of a novel nucleotide-sensitive microtubule-binding protein in HeLa cells. J Cell Biol. 1990;110:1623–1633. doi: 10.1083/jcb.110.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 2004;118:323–335. doi: 10.1016/j.cell.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubin R, Acquaviva C, Chevrier V, Sedjai F, Zyss D, Birnbaum D, Rosnet O. Myomegalin is necessary for the formation of centrosomal and Golgi-derived microtubules. Biol Open. 2013;2:238–250. doi: 10.1242/bio.20123392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe. 2013;14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio KL, Cavignac Y, Stierhof YD, Sinzger C. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol. 2005;79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol. 2000a;74:975–986. doi: 10.1128/jvi.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, McElroy AK, Spector DH. Mechanisms governing maintenance of Cdk1/cyclin B1 kinase activity in cells infected with human cytomegalovirus. J Virol. 2003;77:13214–13224. doi: 10.1128/JVI.77.24.13214-13224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez V, Sztul E, Britt WJ. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J Virol. 2000b;74:3842–3851. doi: 10.1128/jvi.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AA, Kaverina I. Nucleation and Dynamics of Golgi-derived Microtubules. Front Neurosci. 2015;9:431. doi: 10.3389/fnins.2015.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder JM, Larsen J, Komarova Y, Akhmanova A, Thorsteinsson RI, Grigoriev I, Manguso R, Christensen ST, Pedersen SF, Geimer S, et al. EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J Cell Sci. 2011;124:2539–2551. doi: 10.1242/jcs.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Merdes A. EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Curr Biol. 2007;17:1318–1325. doi: 10.1016/j.cub.2007.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szikora S, Gaspar I, Szabad J. 'Poking' microtubules bring about nuclear wriggling to position nuclei. J Cell Sci. 2013;126:254–262. doi: 10.1242/jcs.114355. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol. 2000;149:761–766. doi: 10.1083/jcb.149.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T, Miller PM, Kaverina I. Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle. 2009;8:2168–2174. doi: 10.4161/cc.8.14.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Coquelle FM, Heichette C, Garnier C, Chretien D, Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wu T, Shi L, Zhang L, Zheng W, Qu JY, Niu R, Qi RZ. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J Biol Chem. 2010;285:22658–22665. doi: 10.1074/jbc.M110.105965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang C, Qi RZ. A newly identified myomegalin isoform functions in Golgi microtubule organization and ER-Golgi transport. Journal of Cell Science. 2014;127:4904–4917. doi: 10.1242/jcs.155408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, de Heus C, Liu Q, Bouchet BP, Noordstra I, Jiang K, Hua S, Martin M, Yang C, Grigoriev I, et al. Molecular Pathway of Microtubule Organization at the Golgi Apparatus. Dev Cell. 2016;39:44–60. doi: 10.1016/j.devcel.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Wu J, Lee KC, Dickinson RB, Lele TP. How dynein and microtubules rotate the nucleus. J Cell Physiol. 2011;226:2666–2674. doi: 10.1002/jcp.22616. [DOI] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Thery M, Nachury MV. Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science. 2017;356:328–332. doi: 10.1126/science.aai8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wu J, de Heus C, Grigoriev I, Liv N, Yao Y, Smal I, Meijering E, Klumperman J, Qi RZ, et al. EB1 and EB3 regulate microtubule minus end organization and Golgi morphology. J Cell Biol. 2017 doi: 10.1083/jcb.201701024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, Nogales E. Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell. 2015;162:849–859. doi: 10.1016/j.cell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Graham OS, Raposo A, St Johnston D. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 2012;336:999–1003. doi: 10.1126/science.1219147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu R, Antoku S, Gundersen GG. Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr Biol. 2017;27:3097–3110. e3095. doi: 10.1016/j.cub.2017.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: The dynamics of the AC. Related to Figure 1C–E
Part A: Early ACs merge into a singular structure. NHDFs were infected with TB40/E-UL99-eGFP at MOI 0.5. Images were acquired at 2 frames per hour over the indicated times post-infection using a Leica DMi6000B widefield microscope. In all cases where two UL99-positive ACs were observed they eventually merged.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
PartB: The behavior of UL32-mCherry and UL99-eGFP labeled compartments in HCMV-infected cells. NHDFs were co-infected with TB40/E-UL32-mCherry and TB40/E-UL99-eGFP at MOI 0.5. Images were acquired at 2 frames per hour over the indicated times post-infection using a Leica DMi6000B widefield microscope. Lower panels show individual channels in grey for increased contrast (Left: UL99-eGFP; Right: UL32-mCherry). UL99-eGFP labels early ACs. UL32-mCherry labels cytoplasmic structures that accumulate around the UL99-eGFP-labeled AC. Note also, nuclear rotation is observed in infected cells. As infection progresses, UL32-mCherry-positive domains appear in the nucleus followed by the appearance of UL32-mCherry and UL99-eGFP labeled particles in the AC and cytoplasm.
Time is in hours post infection (h.p.i.).
Scale Bar = 10µm.
NHDFs expressing mCardinal-NLS were either mock infection or infected with TB40/E-UL99-eGFP at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times post-infection. Two examples of each event are provided in the top and bottom panels for mock and infected cells. Note that NLS signal declines in infected cells, in line with reports that infected cell nuclei become more permeable.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
Left Panels: The AC positions itself at the leading edge prior to cell migration. NHDFs were co-infected with TB40/E-UL99-eGFP and TB40/E-UL32-mCherry at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times in hours post infection (h.p.i.). Panel 1 shows two cells in which, after nuclear rotation, the AC positions itself at the leading edge and appears to drag the nucleus in the direction of cell motility. Panel 2 shows an example of how, after nuclear rotation, the AC leaves the nuclear “pinch” to position at the leading edge and again appears to drag the nucleus in the direction of migration.
Right Panels: ACs continue to merge under different circumstances during later stages of infection. NHDFs were infected with TB40/E-UL99-eGFP at MOI 1. Images were acquired using a Leica DMi6000B widefield microscope at 2 frames per hour over the indicated times in hours post infection (h.p.i.). Panel 3 shows an AC that splits during cell migration but regroups. Panel 4 illustrates the merging of two ACs, presumably as a result of cell fusion. To aid in viewing the merge event, UL99-eGFP signal was converted to a color-intensity heat map. At the imaging mid-point each infected cell contains an AC of equivalent intensity. However, the two ACs merge into a singular more intensely colored single AC.
Scale Bar = 20µm.
NHDFs were infected with TB40/E at MOI 1 for 5d. Fixed cells were stained for acetylated tubulin and TGN46. Nuclei were stained with Hoechst. Confocal Z-stacks were generated at 0.2 µm intervals.
Part A: Effects of EB1 or EB3 depletion on eGFP-CLIP170 tracking in HCMV-infected cells. Depletion of EB1 results in CLIP170-eGFP comets that pause or elongate down the length of growing MTs. NHDFs expressing eGFP-CLIP170 were treated with control, EB1 or EB3 siRNAs and infected with TB40/E (MOI 1). Images were acquired at 4 d.p.i. at 2 frames per second using a Leica DMi6000B widefield microscope. Insets and event mark arrows highlight the mix of elongated (green arrow) and pausing (red arrows) eGFP-CLIP170 populations in EB1 depleted cells.
Time is in seconds (s).
Scale Bar = 20µm.
PartB: Effects of EB1 or EB3 depletion on nuclear rotation in HCMV-infected cells. EB3 depletion blocks nuclear rotation in infected cells. NHDFs were treated with control, EB1 or EB3 siRNAs and infected with TB40/E-UL99-eGFP (MOI 0.5). Images were acquired at 2 frames per hour using a Leica DMi6000B widefield microscope.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
NHDFs were treated with control, EB1 or EB3 siRNAs and infected with TB40/E-UL99-eGFP (MOI 0.5). At 7.d.p.i. cultures were imaged at 2 frames per second using a Leica DMi6000B widefield microscope. Three examples are provided for each condition. Left panels show the overall structure of the AC. Right panels show zoomed images of the corresponding left panel showing the behavior of UL99-eGFP-postive structures and particles in the cytoplasm.
Time is in seconds (s).
Scale Bar = 30µm.
Part A: Effects of HEBTRON on nuclear rotation in HCMV-infected cells. Live-cell images of NHDFs treated with vehicle (DMSO) (top three panels) or HEBTRON (bottom three panels) and infected with TB40/E-UL99-eGFP (MOI 0.5). Cultures were imaged at 2 frames per hour using a Leica DMi6000B widefield microscope.
The central DMSO panel provides an example of an initial nuclear rotation through 180°, followed by rotations in alternating directions rather than progression through a full 360°.
Time is in hours post infection (h.p.i.).
Scale Bar = 20µm.
Part B: Effects of HEBTRON on AC structure and virus particle trafficking. Live-cell images of NHDFs treated with vehicle (DMSO) or HEBTRON and infected with TB40/E-UL99-eGFP (MOI 0.5). At 7.d.p.i. cultures were imaged at 2 frames per second using a Leica DMi6000B widefield microscope. Vertical white bar separates a single linearly scaled movie from four examples using non-linear scaling.
Left Panel Set: Linearly scaled movies show a dense AC core in DMSO-treated cells.
Right Panel Sets: Non-linear scaling was used to remove saturation in the center of the dense AC of vehicle-treated samples in order to reveal details of trafficking within the AC itself. To illustrate this, the first panel set corresponds to the Linearly Scaled Panel Set on the far left.
Time is in seconds (s).
Scale Bar = 10µm.