ABSTRACT
The family of polyomaviruses, which cause severe disease in immunocompromised hosts, has expanded substantially in recent years. To accommodate measurement of IgG seroresponses against all currently known human polyomaviruses (HPyVs), including the Lyon IARC polyomavirus (LIPyV), we extended our custom multiplex bead-based HPyV immunoassay and evaluated the performance of this pan-HPyV immunoassay. The VP1 proteins of 15 HPyVs belonging to 13 Polyomavirus species were expressed as recombinant glutathione S-transferase (GST) fusion proteins and coupled to fluorescent Luminex beads. Sera from healthy blood donors and immunocompromised kidney transplant recipients were used to analyze seroreactivity against the different HPyVs. For BK polyomavirus (BKPyV), the GST-VP1 fusion protein-directed seroresponses were compared to those obtained against BKPyV VP1 virus-like particles (VLP). Seroreactivity against most HPyVs was common and generally high in both test populations. Low seroreactivity against HPyV9, HPyV12, New Jersey PyV, and LIPyV was observed. The assay was reproducible (Pearson's r2 > 0.84, P < 0.001) and specific. Weak but consistent cross-reactivity between the related viruses HPyV6 and HPyV7 was observed. The seroresponses measured by the GST-VP1-based immunoassay and a VP1 VLP-based enzyme-linked immunosorbent assay were highly correlated (Spearman's ρ = 0.823, P < 0.001). The bead-based pan-HPyV multiplex immunoassay is a reliable tool to determine HPyV-specific seroresponses with high reproducibility and specificity and is suitable for use in seroepidemiological studies.
KEYWORDS: immunoassay, immunology, polyomavirus, seroepidemiology
INTRODUCTION
The Polyomaviridae family is a group of double-stranded DNA viruses that infect a broad spectrum of hosts, including humans. The number of identified human polyomaviruses (HPyVs) has substantially increased over recent years and currently includes 13 Polyomavirus species, which are listed in Table 1, including full virus names and abbreviations (1). A novel polyomavirus recently identified in human skin samples, named Lyon IARC polyomavirus (LIPyV), has not yet been assigned to a Polyomavirus species (2).
TABLE 1.
Nomenclature, origins, and GenBank accession numbers of the HPyVs used in the multiplex immunoassay
| Species | Virus (abbreviation) | Original tissue (disease) | GenBank accession no. | Reference |
|---|---|---|---|---|
| HPyV1 | BK polyomavirus (BKPyV) | Urine | JF894228 | 37 |
| HPyV2 | JC polyomavirus (JCPyV) | Brain (PML) | NC_001699 | 38 |
| HPyV3 | Karolinska Institutet polyomavirus (KIPyV) | Nasopharynx | NC_009238 | 39 |
| HPyV4 | Washington University polyomavirus (WUPyV) | Nasopharynx | NC_009539 | 40 |
| HPyV5 | Merkel cell polyomavirus (MCPyV) | Skin (Merkel cell carcinoma) | JF812999 | 5 |
| HPyV6 | Human polyomavirus 6 (HPyV6) | Skin | NC_014406 | 28 |
| HPyV7 | Human polyomavirus 7 (HPyV7) | Skin | NC_014407 | 28 |
| HPyV8 | Trichodysplasia spinulosa polyomavirus (TSPyV) | Skin (TS spicule) | NC_014361 | 6 |
| HPyV9 | Human polyomavirus 9 (HPyV9) | Serum | NC_015150 | 41 |
| HPyV10 | Malawi polyomavirus (MWPyV) | Stool | NC_018102 | 42 |
| HPyV10 | Human polyomavirus 10 (HPyV10) | Skin (anal condyloma) | JX262162 | 43 |
| HPyV11 | Saint Louis polyomavirus (STLPyV) | Stool | NC_020106 | 44 |
| HPyV12 | Human polyomavirus 12 (HPyV12) | Liver | NC_020890 | 34 |
| HPyV13 | New Jersey polyomavirus (NJPyV) | Muscle | NC_024118 | 10 |
| Unassigned | Lyon IARC polyomavirus (LIPyV) | Skin | NC_034253 | 2 |
Several HPyVs are associated with severe disease, such as BK polyomavirus (BKPyV), which is associated with nephropathy and hemorrhagic cystitis; JC polyomavirus (JCPyV), which is associated with progressive multifocal leukoencephalopathy (PML); TS polyomavirus (TSPyV), which is associated with a dysplastic hair follicle disorder called trichodysplasia spinulosa; and MC polyomavirus (MCPyV), which is associated with Merkel cell carcinoma (3–6). An association between HPyV6 and HPyV7 and pruritic and dyskeratotic dermatosis has recently been proposed (7). In addition, HPyV7 might be involved in thymomagenesis (8, 9). New Jersey polyomavirus (NJPyV) likely can cause vasculitis, myositis, and retinal blindness (10).
The seroprevalence of well-studied polyomaviruses, for instance, BKPyV and JCPyV, is generally high and comparable among geographically different populations (11–16). Primary HPyV infections usually occur in childhood and are followed by asymptomatic persistent infection throughout life, sometimes accompanied by little virus shedding (12). Though HPyV infection is widespread and its pathology is diverse, symptomatic or manifest HPyV infections are rare and usually limited to the immunocompromised and the elderly (17). For most HPyVs, symptomatic infection occurs when the persistent virus is no longer controlled by the immune system, a phenomenon often referred to as virus reactivation. However, for some HPyVs primary infection coincident with severe immunosuppression has been proposed to be the driver of symptomatic disease (18).
Although knowledge of the prevalence of HPyV infections is increasing, little is known about the incidence and transmission of infection, in particular, of the recently identified HPyVs, such as Saint Louis polyomavirus (STLPyV), HPyV12, NJPyV, and LIPyV. One way of filling this knowledge gap is to develop HPyV species-specific serology.
In general, two viral protein 1 (VP1) antigen expression and presentation methods are used to measure HPyV seroreactivity. One is based on insect cell-expressed VP1 assembled into VP1 virus-like particles (VLP). The other, used in this study, is based on bacterially expressed glutathione S-transferase (GST)-VP1 fusion proteins. Here we aimed to extend our present HPyV bead-based immunoassay measuring IgG seroresponses against the VP1 major capsid protein of HPyVs belonging to the species Human polyomavirus 1 (BKPyV), 5 (MCPyV), 6 (HPyV6), 7 (HPyV7), 8 (TSPyV), and 9 (HPyV9) (12) to HPyVs belonging to the species Human polyomavirus 2 (JCPyV), 3 (Karolinska Institutet polyomavirus [KIPyV]), 4 (Washington University polyomavirus [WUPyV]), 10 (Malawi polyomavirus [MWPyV] and HPyV10), 11 (STLPyV), 12 (HPyV12), and 13 (NJPyV) and LIPyV. MWPyV and HPyV10 belong to the same species and were both included because they differ at eight amino acid positions in VP1, of which three might be located in immunogenic loops important for antigen recognition (15, 19).
The performance of this new pan-HPyV multiplex immunoassay was evaluated in this study by measuring seroreactivity in two pilot populations and by determining the reproducibility and specificity of the assay. The GST-VP1 fusion protein bead-based assay was also compared with a VP1 VLP-based serological assay for BKPyV.
MATERIALS AND METHODS
Human polyomavirus serology assays.
IgG seroreactivities against VP1 were measured using a customized Luminex xMAP assay, as previously described, albeit expanded to include all currently known HPyVs (12, 16, 20). In short, synthetic DNA sequences of VP1 (Table 1) (gBlocks; IDT, San Jose, CA, USA), either wild type (JCPyV, KIPyV, WUPyV, HPyV12, NJPyV, LIPyV) or codon optimized (MWPyV, HPyV10, STLPyV), were cloned into pGEX-5x-3 vectors (GE Healthcare Life Sciences, Chicago, IL, USA) and expressed as GST-VP1.tag fusion proteins in Escherichia coli BL21 Rosetta bacteria. Expression of each newly expressed GST-VP1 fusion protein was analyzed by glutathione-Sepharose 4B purification and SDS-PAGE (10%) separation, followed by Coomassie staining.
The GST-VP1.tag fusion protein was subsequently coupled to glutathione-casein-linked polystyrene beads (Bio-Rad Laboratories, Hercules, CA, USA.) Each bead is color coded by fluorescent dyes, which allows distinction between the different analytes in a single well. The coupling of the complete GST-VP1.tag fusion protein to the bead was verified on a Bio-Plex apparatus using mouse anti-tag antibodies (1:100; a kind gift from M. Pawlita), followed by anti-mouse immunoglobulin-phycoerythrin for detection (1:250; Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) (which were incubated for 30 min each in the dark at room temperature).
In the HPyV multiplex immunoassay, serum samples (1:100) were incubated for 1 h in blocking buffer (1 mg/ml casein, 0.5% polyvinyl alcohol, 0.8% polyvinylpyrrolidone, 2.5% Super ChemiBlock [Chemicon International, Billerica, MA, USA], 2 mg/ml GST bacterial lysate in phosphate-buffered saline) to suppress potential nonspecific binding to the beads or to GST (20, 21). In the meantime, the GST-VP1 fusion proteins were coupled to glutathione-casein-linked polystyrene beads and the serum samples were subsequently incubated with the mixture of GST-VP1 beads (for 1 h in the dark at room temperature). For detection of a VP1-directed human IgG response, biotinylated goat anti-human IgG (H+L) was used (1:1,000; Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA), followed by streptavidin–R-phycoerythrin (SAPE; 1:1,000; Invitrogen, Waltham, MA, USA) (which were incubated for 30 min each in the dark at room temperature). As a positive control, a serially diluted mixture of four serum samples with known seroreactivity against various polyomaviruses was included in each test run (12). The seroreactivity was measured in a Bio-Plex 100 analyzer (Bio-Rad Laboratories, Hercules, CA, USA). Specific seroreactivity was defined by subtracting the median fluorescence intensity (MFI) values for both a blank sample and beads coupled to an irrelevant GST fusion protein (simian virus 40 small T antigen).
For a comparison between the in-house GST-VP1-based immunoassay and the VP1-VLP enzyme-linked immunosorbent assay (ELISA), 396 serum samples were analyzed in both assays for BKPyV IgG detection, as described previously (22). Our assay uses the VP1 protein from BKPyV genotype Ib1, while the VLP ELISA uses the VP1 protein from BKPyV genotype Ib2 (with 98.6% VP1 amino acid similarity existing between genotypes Ib2 and Ib1) (22).
HPyV12 and NJPyV VP1 seroreactivity confirmation.
To demonstrate the antigenicity of the HPyV12 and NJPyV GST-VP1 antigens used, two synthetic peptides (HPyV12 VP1 [VPKSVTDVTAKIQC] and NJPyV VP1 [SIHPNDIAKLPEED]) were generated (GenScript, Nanjing, China) and used to immunize rabbits. These peptides were chosen on the basis of their expected antigenicity in VP1 (15, 19) and the low amino acid similarity with other HPyV VP1 proteins. The polyclonal rabbit antisera raised against these peptides were used in a 1:100 dilution for the recognition of GST-HPyV12- and NJPyV VP1-coupled beads (which were incubated for 30 min in the dark at room temperature). Detection was performed with anti-rabbit immunoglobulin-biotin (1:1,000; Dako, Santa Clara, CA, USA) and SAPE (which were incubated for 30 min each in the dark at room temperature).
Competition analysis.
To gain further insight into cross-reactivity, VP1 antigen competition experiments were performed, as described previously (12). Serum samples with known seroreactivity were serially diluted from 1:100 to 1:409,600 and incubated with regular blocking buffer containing either GST or the GST-VP1 fusion proteins (∼2 mg/ml). For this purpose, only serum samples with measured seroreactivity above 5,000 MFI at a 1:100 serum dilution were selected.
Study population.
For evaluation of the HPyV multiplex serology assay, anonymized serum samples from a cohort of 87 healthy blood donors (HBD) (23) and a cohort of 65 immunocompromised kidney transplant recipients (KTR) (24) were tested. The participants gave written informed consent, and the study adhered to the Declaration of Helsinki principles.
Statistical analysis.
Squared Pearson correlation coefficients (r2) were calculated to determine intertest reliability. The correlation between the assessed HPyVs was further examined by calculating Spearman rank correlation coefficients (ρ). Statistical analysis was performed in IBM SPSS Statistics (version 23) software. When necessary, the significance level (α = 0.05) was adjusted according to the Bonferroni method for multiple comparisons.
RESULTS
Expression and coupling of HPyV VP1 to polystyrene beads.
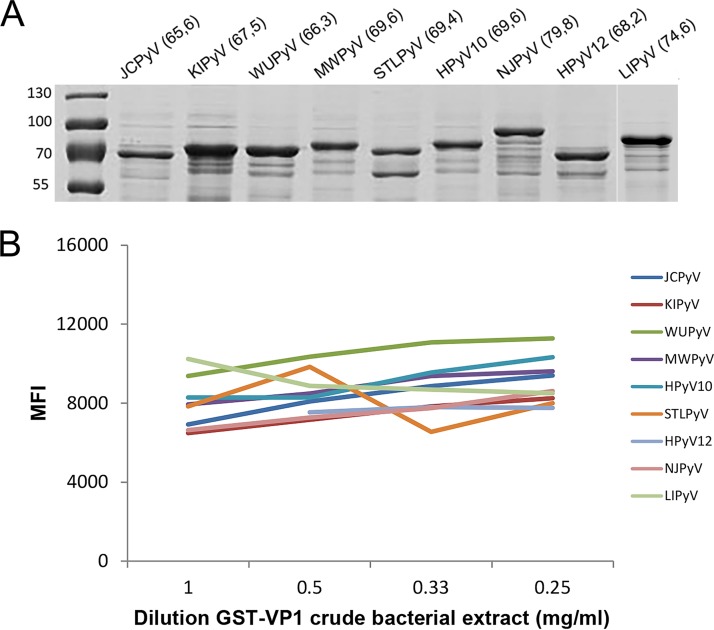
To extend the in-house multiplex immunoassay to all currently known HPyVs, the VP1 genes of JCPyV, KIPyV, WUPyV, MWPyV, HPyV10, STLPyV, HPyV12, NJPyV, and LIPyV were individually cloned and expressed as GST-VP1 fusion proteins. Expression of glutathione-purified GST-VP1 fusion proteins was checked by Coomassie-stained SDS-PAGE and found to be comparable for all HPyVs (Fig. 1A). GST-VP1-containing crude bacterial extracts were purified and coupled to the glutathione-casein cross-linked beads. A tag sequence was included at the C terminus of each GST-VP1 fusion protein to check for efficient antigen binding and saturation of the beads. This was shown in a dilution series of GST-VP1-containing crude bacterial extracts (Fig. 1B). For convenience, it was decided to use a dilution of ∼1 mg/ml of each GST-VP1 crude extract in the HPyV VP1 multiplex immunoassay.
FIG 1.
Expression and coupling of HPyV VP1 to polystyrene beads. (A) Coomassie-stained SDS-PAGE gel showing glutathione-purified GST-VP1 bacterial lysates of JCPyV, KIPyV, WUPyV, MWPyV, STLPyV, HPyV10, NJPyV, HPyV12, and LIPyV. Numbers in parentheses display the molecular masses (in kilodaltons) of the GST-VP1 fusion proteins. The molecular masses (in kilodaltons) of the PageRuler prestained protein ladder (Thermo Fisher Scientific, Waltham, MA, USA) are indicated on the left. The lane for LIPyV was added at a later date. (B) Purification and coupling of GST-VP1.tag fusion proteins of JCPyV, KIPyV, WUPyV, MWPyV, HPyV10, STLPyV, HPyV12, NJPyV, and LIPyV to glutathione-casein cross-linked beads. GST-VP1-containing crude bacterial extracts were serially diluted (1 to 0.25 mg/ml). GST-VP1.tag coupling, detected by using anti-tag antibodies followed by anti-mouse immunoglobulin-phycoerythrin antibodies, is depicted as the median fluorescence intensity (MFI), measured in a Bio-Plex 100 analyzer.
Antigenicity of GST-VP1 in the HPyV multiplex immunoassay.
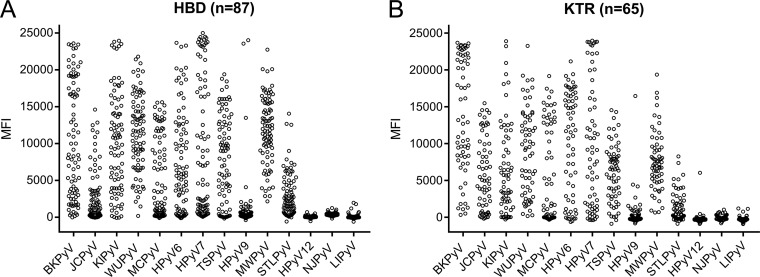
Serum samples from HBD and immunocompromised KTR were tested to analyze the performance of the HPyV multiplex immunoassay. A broad range of seroreactivities that spanned the entire dynamic range of the assay (0 to 25,000 MFI units) was observed. Overall, comparable results were obtained for both sample sets (Fig. 2A and B). The measured seroreactivities against HPyV9, HPyV12, NJPyV, and LIPyV were generally lower than those against most other HPyVs, with the exception of some outliers.
FIG 2.
Seroresponses against each GST-HPyV VP1 antigen measured in the multiplex immunoassay. Seroreactivity was measured in a cohort of healthy blood donors (HBD; n = 87) (A) and a cohort of kidney transplant recipients (KTR; n = 65) (B). The results are depicted as the median fluorescence intensity (MFI), measured in a Bio-Plex 100 analyzer. Each circle represents one serum sample.
To ensure the antigenicity of the HPyV12 and NJPyV VP1 preparations, polyclonal rabbit antisera were raised against specific HPyV12- and NJPyV-derived immunogenic peptides. These rabbit antisera recognized the relevant HPyV VP1 antigen (see Fig. S1 in the supplemental material), demonstrating the ability of our assay to detect HPyV12 and NJPyV antibody reactivity.
Reproducibility of the HPyV multiplex immunoassay.
The reproducibility of the HPyV multiplex assay was determined by calculating the squared Pearson's correlation coefficients between repeated measurements while using beads independently coupled to VP1 fusion proteins. These analyses were highly reproducible, with r2 values ranging from 0.84 to 0.98 (Fig. S2A to J). Furthermore, we compared the use of different fluorescent beads for the same GST-VP1 fusion protein, which was tested for three HPyVs (BKPyV, KIPyV, and HPyV10) and revealed reproducible results, with r2 values ranging from 0.77 to 0.95 (Fig. S3A to C). A historical comparison between seroresponses obtained in 2013 for six of the current HPyV targets with those for the HBD population revealed highly reproducible results (r2 range, 0.71 to 0.97) (Fig. S4A to F) (12).
Specificity of the HPyV multiplex immunoassay.
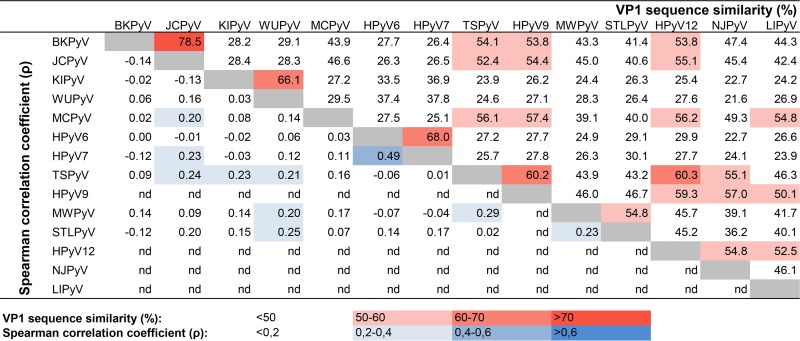
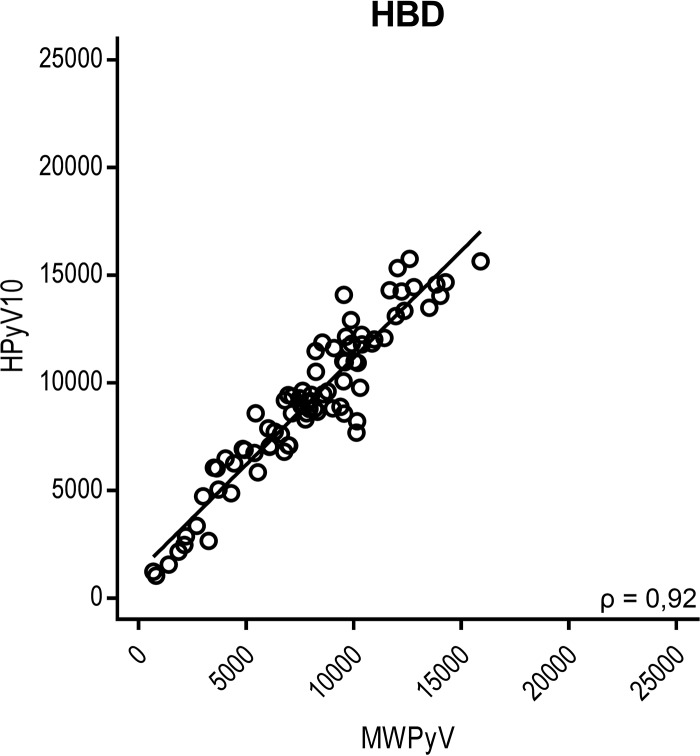
Due to the VP1 amino acid sequence similarity between different HPyV species, varying between 21.6% and 78.5% (Fig. 3, red values at top right), one might expect epitope sharing and therefore a certain degree of cross-reactivity among (related) HPyVs. To evaluate this, a correlation matrix of the HPyV seroresponses was generated (Fig. S5), and Spearman rank correlation coefficients were calculated for each HPyV combination for the HBD population (Fig. 3, blue values at bottom left). The KTR population showed comparable data (not shown). The lack of measured seroreactivity against HPyV9, HPyV12, NJPyV, and LIPyV did not allow a meaningful correlation analysis, and these viruses were therefore excluded from this analysis. Overall, we observed little correlation between the seroreactivities determined against the individual HPyVs. A moderate correlation between HPyV6 and HPyV7 was observed in both the HBD population (Spearman's ρ = 0.49, P < 0.001) and the KTR population (Spearman's ρ = 0.44, P < 0.001). Despite 78.5% VP1 amino acid sequence similarity between BKPyV and JCPyV, no correlation between these types was measured (Spearman's ρ = −0.14, P = 0.19). Between species (HPyV10) members MWPyV and HPyV10, a high correlation was observed (Spearman's ρ = 0.92, P < 0.001), which can be explained by their high VP1 amino acid sequence similarity (98%) (Fig. 4).
FIG 3.
Summary of observed cross-reactivity between individual HPyV VP1 antigens. The data in the upper right show the percent VP1 sequence similarity based on a pairwise alignment obtained using Geneious software (version 10.0.9) with default ClustalW settings. The data in the lower left show Spearman correlation coefficients (ρ) calculated on the basis of the seroresponses measured against VP1 of the HPyV types tested in the HBD cohort. nd, Spearman correlation coefficients were not determined for these HPyVs.
FIG 4.
Comparison of seroreactivity between MWPyV and HPyV10, both of which belong to Polyomavirus species 10. MWPyV and HPyV10 seroreactivities were measured in a cohort of healthy blood donors (HBD). Results are depicted as the median fluorescence intensity (MFI), measured in a Bio-Plex 100 analyzer, for MWPyV on the x axis and for HPyV10 on the y axis. The Spearman correlation coefficient is depicted. Each circle represents one serum sample, and the line represents the results of linear regression analyses.
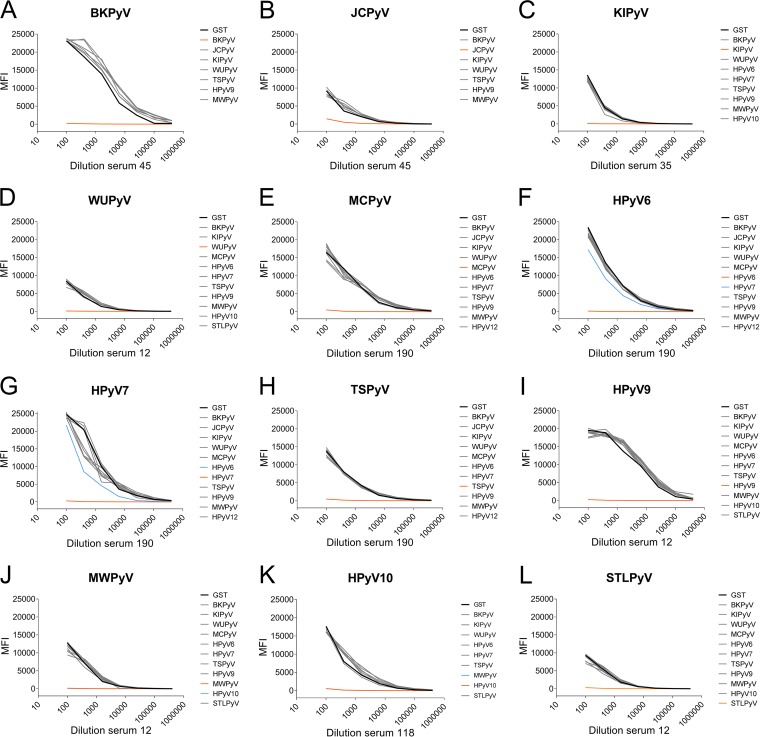
To gain more insight into cross-reactivity, antigen competition experiments were performed. In these experiments reactive serum samples were titrated and preincubated with soluble GST-VP1 of various HPyVs before being exposed to antigenic beads coated with the relevant HPyV VP1. Figure 5 shows some examples of the results of these analyses. A complete overview of the selected serum samples tested in this way can be found in Fig. S6. Overall, little competition between VP1 antigens from HPyVs belonging to different species was observed. Preincubation with JCPyV VP1 did not show a reduction in BKPyV seroreactivity in three out of four experiments (Fig. 5A and S6A4 and A5); however, in one competition experiment, a substantial reduction was seen (Fig. S6A3). Vice versa, preincubation with BKPyV VP1 reduced JCPyV seroreactivity in two out of four competition experiments (Fig. S6B1 and B3). Between closely related species HPyV6 and HPyV7, partial antigen competition indicative of limited cross-reactivity was observed (Fig. 5F and G and S6F1, F2, and G2). As expected, HPyV10 species members MWPyV and HPyV10 showed high levels of cross-reactivity in this analysis (Fig. 5J and K). Interestingly, in three out of six HPyV10 competition experiments, preincubation with MWPyV VP1 did not block HPyV10 seroreactivity (Fig. S6K1, K4, and K5). A summary of the results of the competition experiments is shown in Table 2.
FIG 5.
Analysis of cross-reactivity of polyomavirus seroresponses by VP1-specific competition. Titrated serum samples were preincubated with crude bacterial extract containing GST alone (black), with GST-VP1 of the autologous HPyV (orange), or with the nontarget heterologous HPyVs (gray). Blue lines indicate competition by VP1 other than the target analyte. Results are depicted as median fluorescence intensity (MFI), measured in a Bio-Plex 100 analyzer and shown for the seroresponses measured for BKPyV (A), JCPyV (B), KIPyV (C), WUPyV (D), MCPyV (E), HPyV6 (F), HPyV7 (G), TSPyV (H), HPyV9 (I); MWPyV (J), HPyV10 (K), and STLPyV (L).
TABLE 2.
Summary of cross-reactivity observed among HPyV-VP1 antigens in individual serum samples
| Source of VP1 antigen | Cross-reactivitya |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BKPyV | JCPyV | KIPyV | WUPyV | MCPyV | HPyV6 | HPyV7 | TSPyV | HPyV9 | MWPyV | HPyV10 | STLPyV | |
| BKPyV | ++++ | ++ | − | − | − | − | − | − | − | − | − | − |
| JCPyV | + | ++++ | − | − | − | − | − | − | − | − | − | − |
| KIPyV | − | − | ++++ | − | − | − | − | − | − | − | − | − |
| WUPyV | − | − | − | ++++ | − | − | − | − | − | − | − | − |
| MCPyV | − | − | − | − | ++++ | − | − | − | − | − | − | − |
| HPyV6 | − | − | − | − | − | ++++ | ++ | − | − | − | − | − |
| HPyV7 | − | − | − | − | − | ++ | ++++ | − | − | − | − | − |
| TSPyV | − | − | − | − | − | − | − | ++++ | − | − | − | − |
| HPyV9 | − | − | − | − | − | − | − | − | ++++ | − | − | − |
| MWPyV | − | − | − | − | − | − | − | − | − | ++++ | +++ | − |
| HPyV10 | − | − | − | − | − | − | − | − | − | ++++ | ++++ | − |
| STLPyV | − | − | − | − | − | − | − | − | − | − | − | ++++ |
Arbitrary interpretation of the VP1 competition observed in the experiments whose results are shown in Fig. 5 and Fig. S6 in the supplemental material. −, no reduction; +, slight reduction; ++, moderate reduction; +++, high reduction; ++++, complete reduction.
Comparison between the GST-VP1-based HPyV multiplex immunoassay and a VP1 VLP-based ELISA.
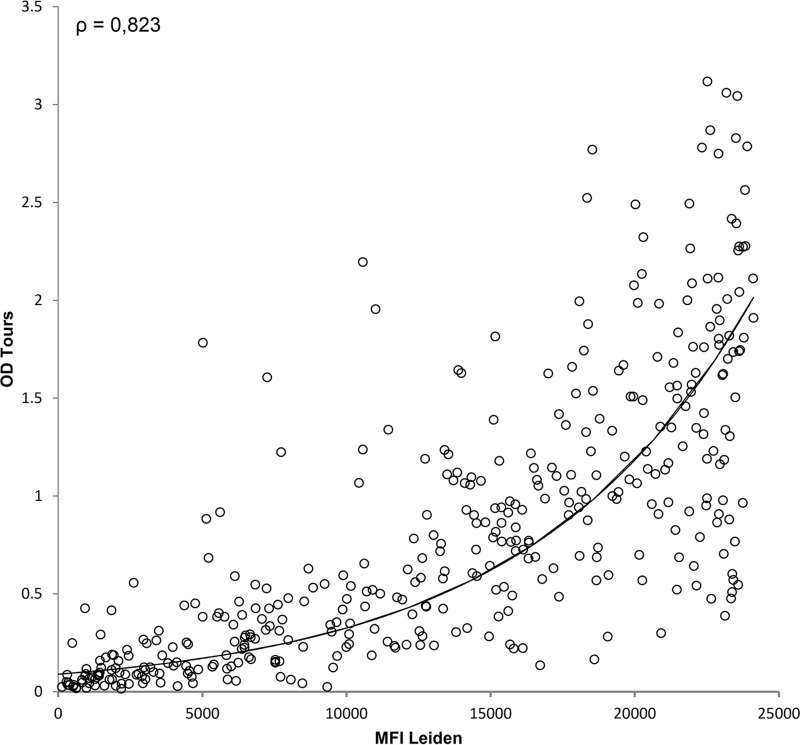
To learn more about the antigenicity of the GST-VP1 fusion proteins that we used, we compared the seroresponses measured for BKPyV in our method to those obtained with a VLP-based ELISA. Although differences in especially the presentation of conformational epitopes were anticipated, the BKPyV seroreactivities measured by both methods were quite similar (Fig. 6) (22). A high Spearman correlation coefficient (ρ = 0.823, P < 0.001) was observed between the optical density (OD) values obtained with the VP1 VLP ELISA and the MFI values obtained with the GST-VP1 immunoassay.
FIG 6.
Comparison between the GST-VP1 bead-based assay and the VLP-based ELISA for BKPyV. The seroreactivities of kidney transplantation donors (n = 396) were measured by both the bead-based GST-VP1 immunoassay and the VP1 VLP ELISA for BKPyV. Each circle represents one serum sample, and the black line indicates the correlation between the bead-based measurement (MFI) and the ELISA (OD). (Adapted with permission from reference 22.)
DISCUSSION
Based on the performed evaluation, the broad HPyV multiplex immunoassay described in this report provides highly reproducible and species-specific serological data. Cross-recognizing antibody detection is sometimes seen between related HPyV species, especially between HPyV6 and HPyV7, which was observed in other studies as well (25). The mean correlation calculated between JCPyV and BKPyV seroreactivity was very low. Nevertheless, some serum samples clearly demonstrated cross-reactivity between these two clinically relevant HPyVs. This observation deserves further study, since individual seroresponses against JCPyV and perhaps against BKPyV as well (26) are used for patient risk assessment regarding serious complications of HPyV-induced infection, for example, PML (27). However, a limited role for cross-reactivity between HPyV6 and HPyV7 serology (28) and between JCPyV and BKPyV serology (15, 19, 29, 30) has also been described.
Apart from the cross-reactivity between two related HPyV species pairs that has been described before for other serological platforms, the GST-VP1 bead-based pan-HPyV assay seems to be a reliable tool for seroepidemiological HPyV studies. To what extent this assay measures neutralizing antibodies was not investigated in this study, but the high correlation between the BKPyV serological data obtained with this assay and those obtained with the VP1 VLP-based ELISA suggests that the GST-VP1 fusion proteins presented on glutathione-casein-coupled beads express conformational epitopes. This was also previously suggested by highly comparable HPyV seroprevalence data obtained worldwide and independently with VP1 VLP and GST-VP1 fusion proteins, for example, for TSPyV (11, 12, 24, 31, 32).
The high intraspecies cross-reactivity observed between MWPyV and HPyV10 did not come as a surprise and probably resulted from their high VP1 amino acid sequence similarity. Nevertheless, seroreactivity toward HPyV10 was not always abolished after preincubation with the MWPyV VP1, indicating a subtle difference between some epitopes of MWPyV and HPyV10, which could be explained by the fact that three of the eight amino acid differences between MWPyV and HPyV10 might be located within the antigenic loops (15, 19). The overall high degree of similarity between the seroreactivity profiles of MWPyV and HPyV10, however, suggests no need for separate measurements for both viruses when testing larger cohorts. The lack of samples seroreactive against HPyV9, HPyV12, NJPyV, and LIPyV did not allow a thorough analysis of potential cross-reactivity for these HPyVs. As a general remark, the possibility of cross-reactivity by antibodies against yet unknown HPyVs cannot be excluded.
The aim of this study was to evaluate the abilities of the assay and not to determine seroprevalence. As such, no seronegative cutoff determination was performed. Seroreactivity against most HPyVs was high in both the immunosuppressed KTR and the HBD cohorts. The observed seroreactivity profile of HPyV9 was lower than that of other polyomaviruses, in line with the findings presented in previous publications, including ours (11, 12, 14, 33).
We observed limited seroreactivity against HPyV12, NJPyV, and LIPyV. For comparison, to date no other serological data are available for NJPyV and LIPyV. For HPyV12, one study reported a seroprevalence of 15 to 33% in healthy adults (34). Based on our observed seroreactivity against HPyV12, presented in Fig. 2, we assume that in Dutch populations the seroprevalence of HPyV12 is low. On the basis of the VP1 amino acid sequence alignment, it was recently suggested that the translation initiation site of HPyV12 VP1 is located 48 nucleotides (16 amino acids) downstream of the 5′ end of the VP1 open reading frame (35). We also analyzed the antigenicity of this shorter GST-HPyV12 VP1 fusion protein and noticed no difference in HPyV12 seroreactivity (not shown). (After submission of the manuscript, the discoverers of HPyV12 published data that convincingly show that HPyV12 is, in fact, a shrew-derived virus [36], suggesting that HPyV12 does not circulate among humans and explaining the lack of HPyV12 seroreactivity found in our cohorts.)
To our knowledge, infection with NJPyV has been described only once, in an immunocompromised kidney-pancreas transplant patient fleeing through sewage water following Superstorm Sandy (10). Supported by the prompt recognition of NJPyV VP1 by the rabbit polyclonal serum raised against NJPyV VP1 peptides, we are confident that our assay is capable of measuring seroresponses against NJPyV. Therefore, we interpret the lack of detectable seroresponses to be an indication that this polyomavirus does not represent a human polyomavirus but, rather, represents a zoonotic polyomavirus that was introduced into humans under exceptional conditions. Alternatively, the lack of NJPyV seroresponses could suggest a difference in geographical spread for NJPyV between North America and Europe, which is rather unusual for (human) polyomaviruses. LIPyV also showed a low seroreactivity profile, suggesting the possibility of environmental contamination of LIPyV in the original skin sample. A larger seroprevalence study could help to elucidate this issue.
The comparison between a VLP-based ELISA and the bead-based assay showed a clear monotonic relationship, despite the different methods in which conformational epitopes are presented by both assays (Fig. 6). A close look at the kinetics of each assay reveals a large dynamic range of the bead-based assay, which has a seemingly increased sensitivity compared to that of the ELISA for the detection of seroresponses in the lower reactivity range. For the purpose of seroepidemiology, we believe that serological testing using HPyV VP1 expressed as a GST fusion protein or as a VP1 VLP yields equally useful results. For individual use, for instance, to predict the risk of developing polyomavirus-related disease such as PML, additional analyses and assay validation are necessary.
In conclusion, the custom-made pan-HPyV multiplex immunoassay is a reliable tool for determination of HPyV-specific seroprevalences. It measures HPyV-specific IgG seroreactivities with high reproducibility and specificity, can easily be extended in case of new HPyV discoveries, and can potentially be combined with other (viral) antigens of interest.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported and funded by Sanquin Blood Supply Foundation, a not-for-profit organization.
We declare no conflict of interest.
S.K., E.V.D.M., A.T., and M.C.W.F. conceived and designed the experiments. S.K., E.V.D.M., and A.T. performed the experiments. S.K. and E.V.D.M. analyzed the data. S.K., E.V.D.M., H.F.W., H.L.Z., and M.C.W.F. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01566-17.
REFERENCES
- 1.Calvignac-Spencer S, Feltkamp MCW, Daugherty MD, Moens U, Ramqvist T, Johne R, Ehlers B. 2016. A taxonomy update for the family Polyomaviridae. Arch Virol 161:1739–1750. doi: 10.1007/s00705-016-2794-y. [DOI] [PubMed] [Google Scholar]
- 2.Gheit T, Dutta S, Oliver J, Robitaille A, Hampras S, Combes J-D, McKay-Chopin S, Le Calvez-Kelm F, Fenske N, Cherpelis B, Giuliano AR, Franceschi S, McKay J, Rollison DE, Tommasino M. 2017. Isolation and characterization of a novel putative human polyomavirus. Virology 506:45–54. doi: 10.1016/j.virol.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purighalla R, Shapiro R, McCauley J, Randhawa P. 1995. BK virus infection in a kidney allograft diagnosed by needle biopsy. Am J Kidney Dis 26:671–673. doi: 10.1016/0272-6386(95)90608-8. [DOI] [PubMed] [Google Scholar]
- 4.Astrom KE, Mancall EL, Richardson EP. 1958. Progressive multifocal leuko-encephalopathy; a hitherto unrecognized complication of chronic lymphatic leukaemia and Hodgkin's disease. Brain J Neurol 81:93–111. doi: 10.1093/brain/81.1.93. [DOI] [PubMed] [Google Scholar]
- 5.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Meijden E, Janssens RWA, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MCW. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 6:e1001024. doi: 10.1371/journal.ppat.1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen KD, Lee EE, Yue Y, Stork J, Pock L, North JP, Vandergriff T, Cockerell C, Hosler GA, Pastrana DV, Buck CB, Wang RC. 2017. Human polyomavirus 6 and 7 are associated with pruritic and dyskeratotic dermatoses. J Am Acad Dermatol 76:932–940.e3. doi: 10.1016/j.jaad.2016.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rennspiess D, Pujari S, Keijzers M, Abdul-Hamid MA, Hochstenbag M, Dingemans A-M, Kurz AK, Speel E-J, Haugg A, Pastrana DV, Buck CB, De Baets MH, zur Hausen A. 2015. Detection of human polyomavirus 7 in human thymic epithelial tumors. J Thorac Oncol 10:360–366. doi: 10.1097/JTO.0000000000000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keijzers M, Rensspiess D, Pujari S, Abdul-Hamid MA, Hochstenbag M, Dingemans A-M, Kurz AK, Haugg A, Maessen JG, De Baets MH, zur Hausen A. 2015. Expression of pRb and p16INK4 in human thymic epithelial tumors in relation to the presence of human polyomavirus 7. Diagn Pathol 10:201. doi: 10.1186/s13000-015-0418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, Kapoor A, Briese T, Faust PL, Lipkin WI. 2014. Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. J Infect Dis 210:1595–1599. doi: 10.1093/infdis/jiu250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossai A, Waterboer T, Nelson HH, Michel A, Willhauck-Fleckenstein M, Farzan SF, Hoen AG, Christensen BC, Kelsey KT, Marsit CJ, Pawlita M, Karagas MR. 2016. Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol 183:61–69. doi: 10.1093/aje/kwv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Meijden E, Bialasiewicz S, Rockett RJ, Tozer SJ, Sloots TP, Feltkamp MCW. 2013. Different serologic behavior of MCPyV, TSPyV, HPyV6, HPyV7 and HPyV9 polyomaviruses found on the skin. PLoS One 8:e81078. doi: 10.1371/journal.pone.0081078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Šroller V, Hamšíková E, Ludvíková V, Vochozková P, Kojzarová M, Fraiberk M, Saláková M, Morávková A, Forstová J, Němečková Š. 2014. Seroprevalence rates of BKV, JCV, and MCPyV polyomaviruses in the general Czech Republic population. J Med Virol 86:1560–1568. doi: 10.1002/jmv.23841. [DOI] [PubMed] [Google Scholar]
- 14.Šroller V, Hamšíková E, Ludvíková V, Musil J, Němečková Š, Saláková M. 2016. Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors. J Med Virol 88:1254–1261. doi: 10.1002/jmv.24440. [DOI] [PubMed] [Google Scholar]
- 15.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. 2009. Association of Merkel cell polyomavirus-specific antibodies with Merkel cell carcinoma. J Natl Cancer Inst 101:1510–1522. doi: 10.1093/jnci/djp332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feltkamp MCW, Kazem S, van der Meijden E, Lauber C, Gorbalenya AE. 2013. From Stockholm to Malawi: recent developments in studying human polyomaviruses. J Gen Virol 94:482–496. doi: 10.1099/vir.0.048462-0. [DOI] [PubMed] [Google Scholar]
- 18.van der Meijden E, Horváth B, Nijland M, de Vries K, Rácz EK, Diercks GF, de Weerd AE, Groningen MCC, van der Blij-de Brouwer CS, van der Zon AJ, Kroes ACM, Hedman K, van Kampen JJA, Riezebos-Brilman A, Feltkamp MCW. 2016. Primary polyomavirus infection, not reactivation, as the cause of trichodysplasia spinulosa in immunocompromised patients. J Infect Dis 215:1080–1084. doi: 10.1093/infdis/jiw403. [DOI] [PubMed] [Google Scholar]
- 19.Moens U, Van Ghelue M, Song X, Ehlers B. 2013. Serological cross-reactivity between human polyomaviruses. Rev Med Virol 23:250–264. doi: 10.1002/rmv.1747. [DOI] [PubMed] [Google Scholar]
- 20.Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M. 2005. Multiplex human papillomavirus serology based on in situ-purified glutathione S-transferase fusion proteins. Clin Chem 51:1845–1853. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]
- 21.Waterboer T, Sehr P, Pawlita M. 2006. Suppression of non-specific binding in serological Luminex assays. J Immunol Methods 309:200–204. doi: 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Wunderink HF, van der Meijden E, van der Blij-de Brouwer CS, Mallat MJK, Haasnoot GW, van Zwet EW, Claas ECJ, de Fijter JW, Kroes ACM, Arnold F, Touzé A, Claas FHJ, Rotmans JI, Feltkamp MCW. 2016. Pretransplantation donor-recipient pair seroreactivity against BK polyomavirus predicts viremia and nephropathy after kidney transplantation. Am J Transplant 17:161–172. doi: 10.1111/ajt.13880. [DOI] [PubMed] [Google Scholar]
- 23.van der Meijden E, Wunderink HF, van der Blij-de Brouwer CS, Zaaijer HL, Rotmans JI, Bavinck JNB, Feltkamp MCW. 2014. Human polyomavirus 9 infection in kidney transplant patients. Emerg Infect Dis 20:991–999. doi: 10.3201/eid2006.140055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Meijden E, Kazem S, Burgers MM, Janssens R, Bavinck JNB, de Melker H, Feltkamp MCW. 2011. Seroprevalence of trichodysplasia spinulosa-associated polyomavirus. Emerg Infect Dis 17:1355–1363. doi: 10.3201/eid1708.110114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicol JTJ, Robinot R, Carpentier A, Carandina G, Mazzoni E, Tognon M, Touzé A, Coursaget P. 2013. Age-specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and trichodysplasia spinulosa-associated polyomavirus. Clin Vaccine Immunol 20:363–368. doi: 10.1128/CVI.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wunderink HF, van der Meijden E, van der Blij-de Brouwer CS, Zaaijer HL, Kroes ACM, van Zwet EW, Rotmans JI, Feltkamp MCW. 2017. Stability of BK polyomavirus IgG seroreactivity and its correlation with preceding viremia. J Clin Virol 90:46–51. doi: 10.1016/j.jcv.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Bloomgren G, Richman S, Hotermans C, Subramanyam M, Goelz S, Natarajan A, Lee S, Plavina T, Scanlon JV, Sandrock A, Bozic C. 2012. Risk of natalizumab-associated progressive multifocal leukoencephalopathy. N Engl J Med 366:1870–1880. doi: 10.1056/NEJMoa1107829. [DOI] [PubMed] [Google Scholar]
- 28.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viscidi RP, Clayman B. 2006. In Serological cross reactivity between polyomavirus capsids, p 73–84. In Ashan N. (ed), Polyomaviruses and human diseases. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 30.Egli A, Infanti L, Dumoulin A, Buser A, Samaridis J, Stebler C, Gosert R, Hirsch HH. 2009. Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 199:837–846. doi: 10.1086/597126. [DOI] [PubMed] [Google Scholar]
- 31.Fukumoto H, Li T-C, Kataoka M, Hasegawa H, Wakita T, Saeki H, Suzuki T, Katano H. 2015. Seroprevalence of trichodysplasia spinulosa-associated polyomavirus in Japan. J Clin Virol 65:76–82. doi: 10.1016/j.jcv.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 32.Chen T, Mattila PS, Jartti T, Ruuskanen O, Söderlund-Venermo M, Hedman K. 2011. Seroepidemiology of the newly found trichodysplasia spinulosa-associated polyomavirus. J Infect Dis 204:1523–1526. doi: 10.1093/infdis/jir614. [DOI] [PubMed] [Google Scholar]
- 33.Antonsson A, Pawlita M, Feltkamp MCW, Bouwes Bavinck JN, Euvrard S, Harwood CA, Naldi L, Nindl I, Proby CM, Neale RE, Waterboer T. 2013. Longitudinal study of seroprevalence and serostability of the human polyomaviruses JCV and BKV in organ transplant recipients. J Med Virol 85:327–335. doi: 10.1002/jmv.23472. [DOI] [PubMed] [Google Scholar]
- 34.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. doi: 10.1371/journal.pone.0058021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norkiene M, Stonyte J, Ziogiene D, Mazeike E, Sasnauskas K, Gedvilaite A. 2015. Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast. BMC Biotechnol 15:68. doi: 10.1186/s12896-015-0187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gedvilaite A, Tryland M, Ulrich RG, Schneider J, Kurmauskaite V, Moens U, Preugschas H, Calvignac-Spencer S, Ehlers B. 2 November 2017. Novel polyomaviruses in shrews (Soricidae) with close similarity to human polyomavirus 12. J Gen Virol. doi: 10.1099/jgv.0.000948. [DOI] [PubMed] [Google Scholar]
- 37.Gardner S, Field A, Coleman D, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 297:1253–1257. doi: 10.1016/S0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 38.Padgett B, Zurhein G, Walker D, Eckroade R, Dessel B. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 297:1257–1260. doi: 10.1016/S0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 39.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MAA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J Virol 81:4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 3:e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kühn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J Virol 85:4586–4590. doi: 10.1128/JVI.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Identification of MW polyomavirus, a novel polyomavirus in human stool. J Virol 86:10321–10326. doi: 10.1128/JVI.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. 2012. Complete genome sequence of a tenth human polyomavirus. J Virol 86:10887. doi: 10.1128/JVI.01690-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. doi: 10.1016/j.virol.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.