Abstract
Heat shock protein 70 (Hsp70) and Hsp90 are molecular chaperones that play essential roles in tumor growth by stabilizing pro-survival client proteins. However, although the development of Hsp90 inhibitors has benefited from the identification of clients, such as Raf-1 proto-oncogene, Ser/Thr kinase (RAF1), that are particularly dependent on this chaperone, no equivalent clients for Hsp70 have been reported. Using chemical probes and MDA-MB-231 breast cancer cells, we found here that the inhibitors of apoptosis proteins, including c-IAP1 and X-linked inhibitor of apoptosis protein (XIAP), are obligate Hsp70 clients that are rapidly (within ∼3–12 h) lost after inhibition of Hsp70 but not of Hsp90. Mutagenesis and pulldown experiments revealed multiple Hsp70-binding sites on XIAP, suggesting that it is a direct, physical Hsp70 client. Interestingly, this interaction was unusually tight (∼260 nm) for an Hsp70–client interaction and involved non-canonical regions of the chaperone. Finally, we also found that Hsp70 inhibitor treatments caused loss of c-IAP1 and XIAP in multiple cancer cell lines and in tumor xenografts, but not in healthy cells. These results are expected to significantly accelerate Hsp70 drug discovery by providing XIAP as a pharmacodynamic biomarker. More broadly, our findings further suggest that Hsp70 and Hsp90 have partially non-overlapping sets of obligate protein clients in cancer cells.
Keywords: chemical biology, chaperone, cancer biology, protein–protein interaction, heat shock protein 90 (Hsp90)
Introduction
In cancer, high levels of the chaperones Hsp70 and Hsp90 are associated with poor prognosis and resistance to chemotherapeutics (1–4). These chaperones are not oncogenes themselves; rather, they are thought to create a permissive environment by binding to multiple proteins and stabilizing them (5), resulting in “non-oncogene addiction” (6, 7). Indeed, a hallmark of Hsp90 inhibition is that a subset of these proteins (sometimes termed “clients”) becomes unstable and is degraded after treatment (8, 9). This response is so robust that loss of specific clients, such as Akt1 or Raf-1, is routinely used as a surrogate biomarker of Hsp90 target engagement (10). Indeed, medicinal chemistry campaigns often take advantage of this property to help guide the optimization of Hsp90 inhibitors (11, 12). In other words, the relative ability of a molecule to reduce the levels of Hsp90 clients is used to guide structure–activity relationships. However, no equivalent client for Hsp70 has yet been reported. Rather, Hsp90 clients are often used to estimate activity on Hsp70, making it unclear whether the compound acts strictly through that chaperone.
Hsp70 is composed of an N-terminal nucleotide-binding domain (NBD),3 a substrate-binding domain (SBD), and C-terminal disordered region (13). In its ATP-bound state, Hsp70 has a poor affinity for clients, but upon ATP hydrolysis, the SBD adopts a tight-binding conformation (14, 15). In mammals, there are two major Hsp70 family members in the cytosol: Hsc70 (HSPA8) and Hsp70 (HSPA1A). For both proteins, cycling between the tight- and weak-binding states is further regulated by co-chaperones, such as J proteins (also called Hsp40s), that increase the rate of nucleotide hydrolysis and nucleotide exchange factors that promote release of ADP (16, 17). Clients of Hsp70s contain short stretches of non-polar amino acids within extended polypeptides (18–20) and hydrophobic “patches” within partially folded conformations (21–23). These client interactions often involve a hydrophobic cleft in the β-basket subdomain of the SBD (24). The physical features of Hsp70 clients are likely to be found in many unfolded and metastable proteins (25–27), such that the theoretical client pool is large.
Although Hsp70 and Hsp90 often work together to stabilize shared clients (28), such as steroid hormone receptors (29), the two chaperones have no sequence or structural similarity. Hsp90 is an obligate dimer that uses its N-terminal and middle domains to interact with partially folded and near-native states (30). Hsp90 has been reported to bind a more restricted set of ∼200 kinases and transcription factors (5), but it also binds to some intrinsically disordered proteins, including Tau (31, 32). Much of what we know about how Hsp70 and Hsp90 bind to clients comes from pioneering work on glucocorticoid receptor (29). In that system, Hsp70 uses its SBD to bind to newly synthesized glucocorticoid receptor during primary folding, whereas Hsp90 is recruited to bind the later, partially folded states. Because of these differences in chaperone structure and binding preference, it seems possible that some clients might be more dependent on one chaperone than the other for stability.
Recent work has yielded first-generation Hsp70 inhibitors, which have proven to be useful chemical probes (16). For example, analogs of MKT-077 (including JG-98) stabilize the ADP-bound state of Hsc70 and Hsp70 (33) and have anti-proliferative activity in cell-based and animal models of breast cancer (34, 35). Likewise, pifithrin-μ (PES) and its analogs bind to the SBD of Hsp70 and have anti-proliferative activity in multiple cancer cells (36–38). In both cases, the compounds have been found to be relatively selective for Hsp70 based on pulldown studies and genetics (39, 40). Moreover, both JG-98 and PES are less toxic to normal, non-transformed cells, consistent with the “addiction” of cancer cells to Hsp70. Together, these experiments have helped solidify the proposed role of Hsp70 in tumorigenesis (4). However, further development of Hsp70 inhibitors has been limited by a lack of pharmacodynamic biomarkers. In particular, the reported Hsp70 clients, such as Raf-1 (34), are also Hsp90 clients, so it is not clear whether their degradation is due to inhibition of just Hsp70 or a combination of the chaperones.
The availability of Hsp90 inhibitors, such as 17-DMAG, was essential in discovering the clients of Hsp90 (5, 41). In that work, the inhibitors were identified first and then used to reveal that many kinases and transcription factors rely on that chaperone. Inspired by this history, we wanted to take advantage of the new Hsp70 inhibitors to learn whether any reported chaperone clients might be relatively more sensitive to Hsp70 versus Hsp90. Based on recent reports (35), we selected a handful of candidate clients and found that the inhibitor of apoptosis proteins (IAPs), such as c-IAP1 and XIAP, appear to be obligate clients of Hsp70 that are less reliant on Hsp90. This was a particularly interesting result because IAPs are important mediators of cell survival signaling and are overexpressed in many cancers (42). The IAP family includes c-IAP1, c-IAP2, and XIAP, and it is defined by the inclusion of one or more baculoviral IAP repeat (BIR) domain (43, 44). The BIR domains bind to caspases and prevent apoptotic signaling (45) and are bona fide drug targets in their own right. Here, we show that treatment with Hsp70 inhibitors leads to rapid and dramatic loss of the IAPs in MDA-MB-231 breast cancer cells. To understand this relationship in more detail, we explored the interaction between XIAP and Hsp70 in vitro and found that the chaperone binds to multiple sites within the BIR2 and BIR3 domains. Mutagenesis and NMR studies suggested that the interaction is tight and not canonical; it seemed to involve regions outside of Hsp70's expected binding cleft. Together, these results suggest that IAPs are direct clients of Hsp70, and they might be candidate biomarkers of Hsp70. Moreover, these results illuminate surprising differences between how cancer cells rely on Hsp70 and Hsp90.
Results
Hsp70 inhibition results in rapid degradation of XIAP
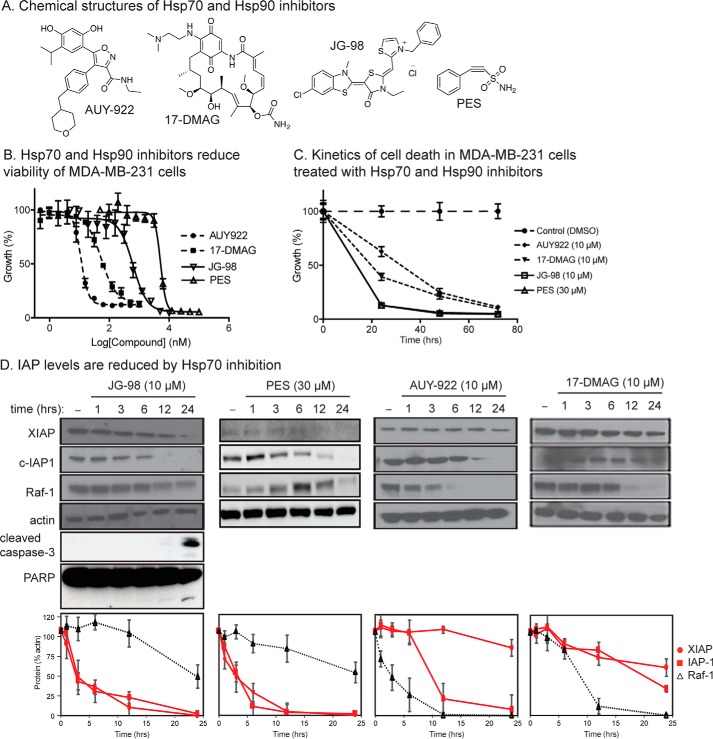
Chemical inhibitors of Hsp70 have been reported to enhance turnover of a number of proteins, including IAP-1, XIAP, Raf-1, tau, androgen receptor and others (35, 40, 46). However, it is not clear whether any of them might be relatively selective for Hsp70 compared with Hsp90. To address this question, we first examined the levels of XIAP, c-IAP1, and Raf-1 after treatment with either Hsp70 or Hsp90 inhibitors. These studies employed MDA-MB-231 breast cancer cells because growth of these cells had previously been shown to be sensitive to both Hsp70 and Hsp90 inhibitors (34). Moreover, we initially focused on XIAP and c-IAP1, rather than other putative clients, based on serendipitous observations made during recent studies of necroptosis (59). Finally, to provide greater confidence in the results, we used two structurally distinct inhibitors of each chaperone. For Hsp70, we used PES and JG-98, and we used AUY-922 and 17-DMAG as Hsp90 inhibitors (see Fig. 1A). In the first experiments, MTT assays were used to confirm that all of the inhibitors have anti-proliferative activity at the expected EC50 values (Fig. 1B). These experiments allowed us to use each of the compounds at a concentration that ensured maximal activity (10 μm for JG-98, AUY-922, and 17-DMAG and 30 μm for PES). Next, we characterized the kinetics of cell death by performing MTT assays at 24, 48, and 72 h after treatment. We found that both JG-98 and PES caused relatively rapid responses, with 50% cell proliferation lost by ∼10 h and more than 80% by 24 h (Fig. 1C). In contrast, the response to AUY-922 and 17-DMAG took a longer period of time, with 72+ h required to reduce growth by 80%. The results of these kinetic experiments were important in our search for Hsp70 clients because we were most interested in those clients that are degraded prior to extensive loss of cell viability. More explicitly, we considered it likely that non-specific client degradation, triggered by downstream caspase activation and/or other proteolytic events, might be confounding at later times; whereas the bona fide clients should be direct physical interaction partners.
Figure 1.
IAPs are selectively destabilized by Hsp70 inhibition. A, chemical structures of inhibitors. PES and JG-98 inhibit Hsp70, whereas AUY-922 and 17-DMAG inhibit Hsp90. B, Hsp70 and Hsp90 inhibitors reduce the growth of MDA-MB-231 cells, as measured by MTT assays. C, kinetics of Hsp70 and Hsp90 inhibitor-mediated anti-proliferative activity. Results are the average of experiments performed in triplicate. Error bars represent S.E. D, destabilization of IAPs occurs after treatment with Hsp70 inhibitors. MDA-MB-231 cells were treated for the indicated times. The blots shown are representative of at least two independent experiments. The error bars represent S.E.
With these criteria in mind, we performed Western blottings of the candidate proteins at 0, 1, 3, 6, 12, and 24 h after treatment. From these experiments, we confirmed (47) that Raf-1 is a selective client of Hsp90 (Fig. 1D). Specifically, the levels of Raf-1 were significantly (>95%) decreased by 12 h after treatment with either 17-DMAG or AUY-922. In contrast, treatment with these inhibitors tended to have a less pronounced effect on cIAP-1 and XIAP levels. In contrast, treatment with either of the Hsp70 inhibitors JG-98 or PES caused a dramatic loss of cIAP-1 and XIAP at relatively early time points, although Raf-1 was largely spared (Fig. 1D). This effect was particularly strong in the first 6 h after treatment, when c-IAP1 and XIAP levels were reduced ∼75% and Raf-1 levels were only reduced ∼20%. Thus, XIAP and c-IAP1 seemed to be relatively selective clients of Hsp70 but not Hsp90.
To better understand the mechanism of IAP loss in response to Hsp70 inhibitors, we first tested whether activation of caspase activity might be involved by blotting the JG-98-treated MDA-MB-231 cell lysates for cleaved caspase-3 and poly(ADP-ribose) polymerase (PARP). Interestingly, cleaved caspase-3 and PARP only emerged 12–24 h after treatment (Fig. 1D), which is substantially after the levels of XIAP and c-IAP1 were reduced. This finding is consistent with a recent result, in which removal of the RING domain of XIAP was found to block turnover by JG-98 (59). Thus, inhibition of Hsp70 seems to initiate the normal turnover of IAPs rather than triggering loss through caspase activation.
Hsp70 and XIAP form a complex in vitro and in cells
We next wanted to determine whether Hsp70 physically interacts with the IAPs. In these studies, we focused on XIAP (Fig. 2A) as a representative member of the IAP family because of a wealth of available structural data (44). Using co-immunoprecipitations, we found that endogenous XIAP was bound to Hsp70 in MDA-MB-231 cell lysate (Fig. 2B). To understand whether this interaction might be direct, we first used a computational algorithm to predict possible Hsp70-binding sites in full-length human XIAP (18). This approach suggested that XIAP contains nine putative binding sites, all but two of which are located within the BIR2 and BIR3 domains (see Fig. 2A). To test this prediction, the region of XIAP(120–356) corresponding to the BIR2 and BIR3 domains was purified, and its binding to recombinant human Hsp70 was measured by ELISA. We found that Hsp70 bound XIAP(120–356) with a KD = 260 ± 20 nm (Fig. 2C). Consistent with binding in the BIR2 and BIR3 domains, addition of a Smac mimetic (48) blocked interactions with Hsp70 (Fig. 2D). Also, addition of JG-98 (10 μm) weakened the apparent affinity of Hsp70 for XIAP(120–356) (see Fig. 2D). Thus, XIAP appears to be a direct client of Hsp70 in vitro and in cells, and its interaction with Hsp70 is mediated by the BIR2/3 domains.
Figure 2.
Hsp70 binds XIAP. A, domains of human XIAP. B, XIAP is co-immunoprecipitated with Hsp70 in MCF7 lysates. Results are representative of experiments performed in triplicate. C, XIAP(120–356) binds Hsp70 in vitro, as measured by ELISA. Addition of JG-98 (10 μm) modestly weakens the interaction. Results are the average of three independent experiments performed in triplicate. Error bars are S.E. D, Smac mimetic SM164 blocks binding of Hsp70 to XIAP(120–356) by ELISA. Results are the average of three independent experiments performed in triplicate. Error bars represent S.E. E, purified Hsc70 and Hsp90a compete with Hsp72 for binding to XIAP in the ELISA format. Results are the average of triplicates, and error bars represent S.E. IP, immunoprecipitation; IB, immunoblot.
We next wondered why Hsp90 inhibitors might have a less pronounced effect on XIAP stability. One possibility is that Hsp90 might not bind to this client. Hsp90 and Hsp70 are known to bind similar or even overlapping sites in other clients. To understand whether this might be the case for XIAP, we tested the ability of recombinant human Hsp90α to compete with Hsp70 for binding to XIAP(120–356) in the ELISA. Strikingly, Hsp90α was a relatively strong competitor, with an IC50 of 227 ± 138 nm (Fig. 2E). Similarly, the highly conserved Hsp70 family member, Hsc70 (HSPA8), also competed with an IC50 of 302 ± 196 nm. Thus, Hsp90 binds XIAP, although inhibitors do not seem to trigger degradation.
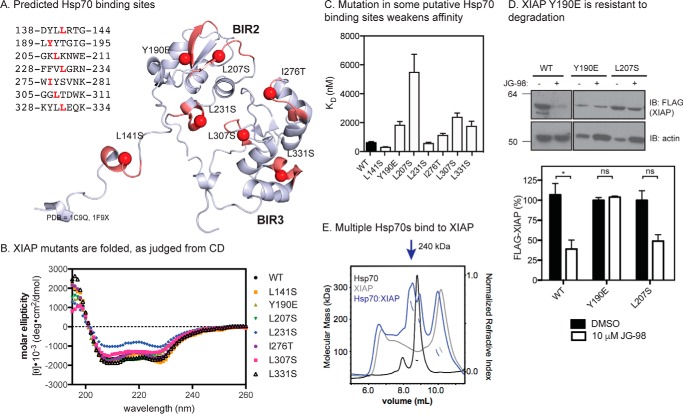
To further explore the interaction of XIAP with Hsp70, we created point mutations to disrupt each of the seven putative binding sites in XIAP(120–356) (Fig. 3A). Other than the possible exception of L231S, each of the mutants was folded to the same extent as wildtype, as judged by circular dichroism (CD) (Fig. 3B). In the ELISA platform, we found that Y190E, L207S, L307S, and L331S all had weaker affinity for Hsp70, with dissociation constants ranging from 2-fold to greater than 10-fold higher than the wildtype protein (Fig. 3C). To test whether this difference in affinity would translate into resistance to Hsp70 inhibitors, we expressed full-length XIAP (XIAPWT, XIAPY190E, and XIAPL207S) in HeLa cells and measured protein stability after treatment with JG-98. Consistent with the model, we found that XIAPY190E was difficult to overexpress, likely because of reduced Hsp70 binding, and that its levels were not sensitive to JG-98 (Fig. 3D). The other mutant, XIAPL207S was not as dramatically affected, suggesting that some sites might be more functionally important for stability than others.
Figure 3.
Hsp70 binds multiple sites in the BIR2 and BIR3 domains. A, location of predicted Hsp70-binding sites on BIR2 and BIR3 (Protein Data Bank codes 1C9Q and 1F9X) are shown in red. B, CD spectra for XIAP(120–356) wildtype and mutant proteins. Each reported spectrum is the average of six scans, subtracting the signal acquired for buffer alone. C, binding of XIAP(120–356) point mutants to Hsp70, as measured by ELISA. Results are the average of three independent experiments performed in triplicate. Error bars are S.E. D, XIAPY190E is partially resistant to degradation in response to JG-98. HeLa cells over-expressing the indicated XIAP point mutations were treated for 24 h. Blots are representative of two independent experiments, and quantification is from the average of biological replicates. Error bars are S.E. *, p < 0.06; ns, not significant. E, Hsp70 binds to XIAP and forms a stable, multimeric complex, as analyzed by SEC-MALS. Molecular weight standards were BSA and β-amylase. Results are representative of duplicates.
The ELISA results suggested that Hsp70 may bind to multiple sites on XIAP(120–356) because none of the individual mutations were sufficient to completely block the interaction. To examine this possibility, we incubated Hsp70 with XIAP(120–356) and analyzed the complex by size-exclusion chromatography and multiangle light scattering (SEC-MALS). Consistent with the other binding studies, we found that Hsp70 and XIAP(120–356) formed a stable, multimeric complex (Fig. 3E). Moreover, based on the apparent molecular mass of ∼240 kDa, we estimate that approximately three Hsp70s may be bound.
Interaction with XIAP involves non-canonical interactions with Hsp70
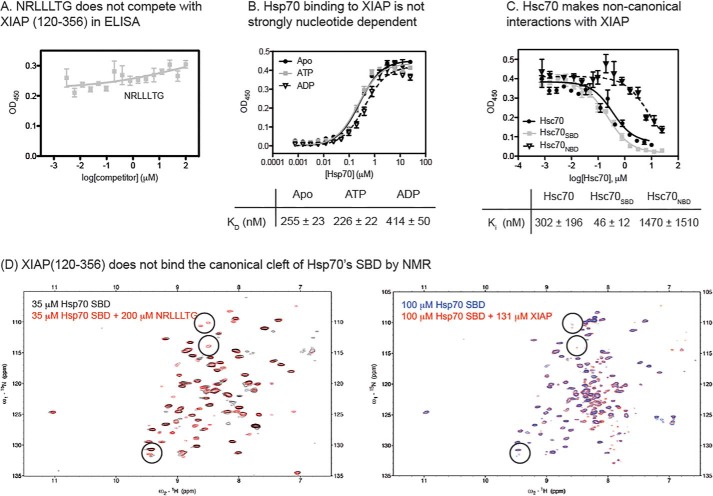
Next, we asked whether XIAP(120–356) binds to Hsp70 in the chaperone's canonical SBD. As a first test, we titrated a known SBD ligand, the NRLLLTG peptide, into the ELISA experiment. Surprisingly, we found that NRLLLTG was not able to compete with XIAP(120–356) for binding to Hsp70 (Fig. 4A), suggesting that the XIAP(120–356) was not binding the canonical cleft. A growing number of non-canonical interactions with Hsp70 have been discovered (49, 50), and these interactions often do not fit the normal profile for nucleotide dependence. In other words, the interaction is not always tighter in the ADP-bound state (20). Similarly, we found that the affinity of XIAP(120–356) for Hsp70 did not follow the canonical behavior; its affinity for ADP-bound or apo-Hsp70 was 1.5-fold worse than in the ATP-bound state Hsp70 (Fig. 4B). This result was consistent with our observation that the apparent affinity of Hsp70 for XIAP(120–356) was slightly weakened, not strengthened, by addition of JG-98 (see Fig. 2D), as this compound is known to stabilize the ADP-bound state (51). Some non-canonical interactions with Hsp70 are reported to involve contacts outside the SBD (49). To probe whether XIAP(120–356) might share this feature, we purified truncated mutants of Hsc70 that are composed of either the NBD (Hsc70NBD) or SBD (Hsc70SBD) alone and tested them for the ability to compete with full-length Hsp70 for binding to XIAP(120–356). Unlike what would be expected for a strictly canonical interaction, we found that both Hsc70NBD and Hsc70SBD competed with full-length Hsp70 for binding to XIAP(120–356) (Fig. 4C), although the Hsc70NBD interaction seemed to be significantly weaker. Finally, we titrated NRLLLTG or XIAP(120–356) into samples of 15N-labeled Hsc70SBD (residues 395-508, lacking the C-terminal subdomain). This NMR-based assay has previously (50) been used to measure binding of canonical clients, such as NRLLLTG, to the hydrophobic cleft, and it is based on the appearance of characteristic cross-peaks upon “rigidification” of the β-subdomain. Consistent with previous observations, we found that adding NRLLLTG to Hsc70SBD caused the appearance of ∼ 15 new cross-peaks (Fig. 4D). Because the peptide is not labeled, these cross-peaks are likely due to “rigidification” of the SBD after binding to the canonical hydrophobic cleft. Consistent with the ELISA studies, XIAP(120–356) failed to produce these characteristic peaks, again suggesting that it is a non-canonical client. Together, these results all suggest that XIAP makes non-canonical contacts with Hsp70 and that the NBD is involved.
Figure 4.
XIAP(120–356) binds Hsp70 through partially non-canonical interactions. A, canonical peptide, NRLLLTG, does not compete with XIAP(120–356) for binding to Hsc70, as measured by ELISA. B, Hsp70 binding to XIAP(120–356) is not classically nucleotide-dependent. The affinity is slightly tighter in the presence of ATP, rather than ADP. C, full-length Hsc70, Hsc70NBD, and Hsc70SBD compete with Hsp70 for binding to XIAP(120–356), as measured by ELISA. All results shown are averages of three independent experiments performed in triplicate, and error bars represent the S.E. Binding data were fit to the Langmuir binding isotherm, and competition data were fit to the Hill equation. D, 15N-1H TROSY-HSQC spectra of 35 μm Hsp70 SBD(395–507) without (black) or with (red) 200 μm NRLLLTG and 15N-1H TROSY-HSQC spectra of 100 μm Hsp70 SBD(395–507) without (blue) or with (red) 131 μm XIAP(120–356).
Stability of IAPs is responsive to inhibitors of Hsp70 but not Hsp90
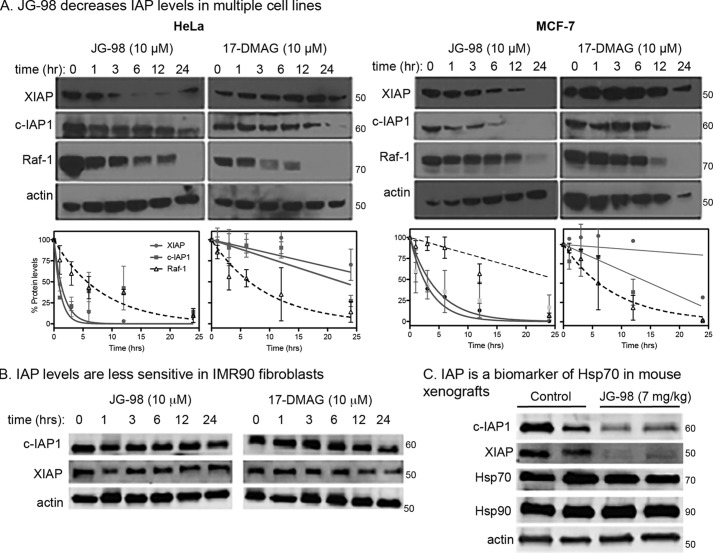
Our experiments in MDA-MB-231 cells showed that Hsp70 inhibitors induce degradation of XIAP and c-IAP1. Thus, we envisioned that IAPs might be useful surrogates for Hsp70 target engagement, similar to how Raf-1 is used to monitor pharmacological inhibition of Hsp90 (11). However, we wanted to test this relationship in additional cell lines. Similar to what we found in MDA-MB-231 cells, we found that the IAPs in HeLa and MCF7 cells were selectively degraded in response to JG-98, but not 17-DMAG, at early time points (∼6 h) (Fig. 5A). Next, we wanted to test whether IAP stability was sensitive to Hsp70 inhibition in normal healthy cells. This experiment was important because clients often depend on chaperone only in a cancer cell-specific context (4). Consistent with this idea, JG-98 did not reduce levels of XIAP or c-IAP1 in IMR90 normal human lung fibroblasts (Fig. 5B). Finally, we wanted to understand whether IAPs might be surrogate biomarkers of Hsp70 engagement in vivo. Accordingly, we established MCF7 tumor xenografts in mice and treated with JG-98, using a previously reported dosing scheme (35). In this model, daily i.p. injections of JG-98 (7 mg/kg/day) begin to show a therapeutic effect on tumor volume 4–6 days after the initiation of treatment. When we isolated tumors from these treated animals, we found that the lysates had significantly lower levels of XIAP and c-IAP1 (Fig. 5C) when compared with those from mock-treated animals. Together, these results suggest that XIAP and c-IAP1 are candidate biomarkers of Hsp70 in cells and animals.
Figure 5.
IAPs are potential biomarkers of Hsp70 inhibition in cancer cells. A and B, IAPs are de-stabilized by Hsp70 inhibition in HeLa (left) and MCF7 (right) cells (A), but not in IMR90 cells (B). Representative blots of at least two independent biological replicates are shown. Protein levels are quantified, and results shown are averages of two independent experiments. Error bars represent S.E. C, treatment with JG-98 results in loss of IAP protein levels in MCF-7 mouse xenografts.
Discussion
There is growing interest in Hsp70 inhibitors as potential anti-cancer therapeutics (3, 52, 53), and the development of such molecules would benefit from the identification and characterization of selective biomarkers. Particularly sensitive Hsp70 clients could be used as surrogates for target engagement in cancer cells, facilitating hit-to-lead medicinal chemistry campaigns. In addition, such clients could potentially be used in animal models or in the clinic to develop relationships between pharmacokinetics and pharmacodynamics. Importantly, the ideal client should be more sensitive to inhibition of Hsp70 than Hsp90 to allow for the rapid discrimination of their relative contributions. Here, we found that degradation of c-IAP1 and XIAP occurs rapidly (∼6 h) and dramatically (∼50–80%) after treatment with Hsp70 inhibitors in MDA-MB-231 cells. Furthermore, the levels of these proteins did not seem to be responsive to Hsp90 inhibitors; for example, treatment with AUY-922 only reduced XIAP levels by ∼25% in 24 h. Despite the fact that Hsp90 inhibitors had little effect on IAP levels, this chaperone did seem to bind relatively tightly to XIAP in vitro and to share Hsp70-binding sites. These chaperones are known to work together in the folding and stability of many clients (29), but to our knowledge, no clients have been experimentally shown to be clearly reliant on Hsp70 but not Hsp90. Future work will be directed at better understanding the structural differences between how Hsp90 and Hsp70 interact with XIAP to reveal the molecular mechanisms that make this client different from previously studied ones. This question is also motivated by the fact that IAPs themselves are validated targets for cancer therapy (54), so ways of reducing their levels might be directly beneficial in cancer.
Why does treatment with an Hsp70 inhibitor lead to a reduction in the levels of XIAP? There are still many important mechanistic details to uncover, but it seems that treatment with JG-98 weakens the affinity of Hsp70 for XIAP by more than 5-fold in vitro. Furthermore, it is known that the RING domains are important for normal XIAP turnover and that the RING domain is critical for turnover in response to JG-98. Thus, we speculate that Hsp70 might contribute to keeping XIAP in an inactive state, such that release would favor auto-ubiquitination and degradation, possibly correlating with changes in oligomerization. Because Hsp70 inhibitors of different chemical classes bind different locations and “trap” different nucleotide states, it will be interesting to use a broader panel of probes to understand the relationships between client affinity and turnover in the XIAP system.
A major goal of this work was to characterize the protein–protein interaction (PPI) between putative clients and Hsp70. We considered it important to understand the physical contacts Hsp70 makes with possible biomarkers, similar to the detailed information available for binding of other obligate clients to Hsp90 (55). This is important because many putative clients are expected to be indirectly degraded during cell death, and such clients might therefore not be the best choice as biomarkers. Accordingly, we focused on the potential interaction between XIAP and Hsp70 and confirmed that these two proteins bind to each other in vitro and in cells. We were initially surprised to find that the measured affinity of Hsp70 for XIAP was relatively tight (∼230 nm), compared with the affinity for many other clients (typically low to mid-micromolar). This tight affinity may be due, in part, to avidity effects from binding to multiple sites. Mutagenesis and SEC-MALS studies suggested that as many as three Hsp70s may bind to XIAP(120–356). Multiple Hsp70 molecules have also been shown to bind other clients, including denatured rhodanese (21, 56), so multivalency seems to underlie the interaction with a subset of clients. Second, the relatively tight affinity of the XIAP–Hsp70 complex might be a product of the non-canonical nature of the PPI. We found that binding to XIAP does not follow the normal nucleotide dependence and that it involves at least one additional contact within the Hsp70's NBD. Furthermore, the NMR titrations clearly showed that the NRLLLTG peptide, but not XIAP(120–356), binds the canonical hydrophobic cleft of the SBD. Currently, the Hsp70 NBD is not fully assigned, so the specific binding site of XIAP(120–356) is not clear. However, there are intriguing chemical shift perturbations in the XIAP(120–356) treated samples (see Fig. 4D) that might lead to additional insights. We speculate that XIAP could be a good model for understanding the location of non-canonical contacts. In addition, although the exact site(s) of the non-canonical interaction(s) are not known, recent work has shown that phosphoserines bind to the α-helical lid of Hsp70 (49), so we posit that secondary contacts here may contribute to XIAP binding. More generally, our results suggest that IAPs might serve as good models for probing this category of poorly understood Hsp70 interactions. We speculate that non-canonical interactions may make a major, unresolved contribution to Hsp70-mediated quality control and that new model systems will have a substantial impact on our knowledge of the system.
Experimental procedures
Reagents and general methods
Antibodies used are as follows: XIAP (Enzo Life Sciences ADI-AAM-050-E); c-IAP1 (Enzo Life Sciences ALX-803-335-C100); β-actin (AnaSpec AS-54591); FLAG (Sigma F1804); Hsp70 (Santa Cruz Biotechnology sc-137239 and sc-33575); Raf-1 (Santa Cruz Biotechnology sc-133); goat anti-mouse HRP (AnaSpec 28173); goat anti-rabbit HRP (AnaSpec 28177); and goat anti-rat HRP (Santa Cruz Biotechnology sc-2006). JG-98 was synthesized according to previously described methods (34), and PES, AUY-922, and 17-DMAG were purchased from Millipore, Selleckchem, and LC Laboratories, respectively. All other biological reagents were purchased from Sigma unless otherwise noted. All spectroscopic measurements were obtained with a SpectraMax M5 microplate reader (Molecular Devices).
Plasmids and site-directed mutagenesis
XIAP mutants were prepared using the QuikChange site-directed mutagenesis kit (Stratagene). The following mutants were engineered into the human XIAP(120–356) gene in the pet28a vector: L141S, Y190E, L207S, L231S, I276T, L307S, and L331S. Wildtype and mutant XIAP(120–356) constructs all contained additional C202A/C213G mutations for stability. N-terminally FLAG-tagged, full-length XIAP in pCMV6 was obtained from GeneArt (Invitrogen).
Protein expression and purification
All His-tagged Hsp70 proteins (HSPA1A, HSPA8, Hsc70NBD(1–383), and Hsc70SBD(395–509)) were purified as described previously (57) using batch purification with Ni-NTA resin (Novagen) and subsequent cleavage of the His tag with tobacco etch virus protease. Hsp70, Hsc70, and Hsc70NBD were further purified using an ATP column, whereas Hsc70SBD underwent gel-filtration chromatography on a Superdex 75 16/60 column (GE Healthcare). Hsc70 (HSPA8) domain truncations were used in Fig. 4, A and C, because we have found that they are more stable than the corresponding Hsp70 (HSPA1A) domains. WT His-tagged XIAP(120–356) and its mutants were batch-purified with Ni-NTA resin and eluted with 400 mm imidazole. DTT was added to 10 mm, and XIAP(120–356) was further purified by gel-filtration chromatography on a Superdex 75 16/60 column (GE Healthcare) in 20 mm Tris buffer containing 200 mm NaCl, 50 μm zinc acetate, and 1 mm DTT, pH 7.5. Fractions containing XIAP(120–356), as assessed by SDS-PAGE, were pooled and concentrated, and DTT was added to 10 mm before storing at −80 °C. The bicinchoninic acid (BCA) assay kit (Thermo Fisher Scientific, Inc.) was used to determined protein concentration, and protein purities were estimated at >90% by SDS-PAGE and Q-TOF LC-MS (Agilent).
Tissue culture, viability assays, and transfections
MCF-7 and HeLa cells (ATCC) were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. MDA-MB-231 cells (ATCC) were maintained in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and non-essential amino acids. Cells were used at low passage, typically less than 20. No further validation of the cell lines was performed. If indicated, cell viability was determined using the MTT assay as described previously (34). XIAP pCMV6 plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Western blotting
Cell extracts were prepared in chilled RIPA buffer (50 mm Tris, pH 8, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) unless otherwise indicated. Protein concentration was determined by the BCA assay, and 20 μg of total protein was separated by SDS-PAGE on 10% Mini-PROTEAN TGX gel (Bio-Rad) and transferred to PVDF membrane (Thermo Fisher Scientific Inc). Membranes were blocked with 5% milk in TBS, 0.05% Tween for 1 h at room temperature, incubated with primary antibodies overnight at 4 °C, washed with TBS, 0.05% Tween, and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Membranes were developed using chemiluminescence (ECL Prime, GE Healthcare). Xenograft experiments were carried out as described (35, 58).
Co-immunoprecipitation
Cell extracts were prepared in chilled lysis buffer (50 mm Tris, pH 8, 150 mm NaCl, 1 mm ATP, 10 mm KCl, 5 mm Mg(OAc)2, 1% Nonidet P-40) supplemented with protease inhibitor mixture (Roche Applied Science). The total protein concentration was adjusted to 5 mg of protein in 1 ml of cell extract. PureProteome protein G magnetic beads (Millipore) were incubated with 6 μg of the appropriate antibody or non-specific mouse IgG (Santa Cruz Biotechnology) for 30 min at room temperature with mixing, followed by antibody cross-linking with bis(sulfosuccinimidyl) suberate (Thermo Fisher Scientific, Inc.) for 1 h at room temperature with mixing. The cross-linking reaction was quenched with 1 m Tris, pH 7.5, for 1 h at room temperature with mixing. Meanwhile, equal 100-μl samples of cell lysate were pre-cleared by incubation with 50 μl of protein G beads for 1 h at room temperature with mixing. Protein complexes were immunoprecipitated by incubation of the pre-cleared lysate (1 mg of total protein per immunoprecipitation) with 50 μl of antibody–cross-linked protein G beads for 1 h at room temperature with mixing. The immunocomplexes were washed three times with 500 μl of wash buffer (PBS, pH 7.4, 0.1% Tween 20) and eluted with 0.1 m glycine, pH 2.6. Proteins were visualized by Western blotting.
ELISA
XIAP(120–356) was non-covalently immobilized in the wells of a clear, flat-bottom 96-well plate (Thermo Fisher Scientific, Inc.) by incubating 100 μl of 100 nm XIAP(120–356) in immobilization buffer (20 mm MES, pH 5.2) overnight at 37 °C. XIAP(120–356) was removed from the wells, and the wells were washed with 3× 150 μl of TBS supplemented with 0.05% Tween (TBS-T). Each wash was incubated, with gentle rocking, for 3 min at room temperature. Following washing, 30 μl of Hsp70 was added at the indicated concentrations in binding buffer (25 mm HEPES, pH 7.4, 40 mm KCl, 8 mm MgCl2, 100 mm NaCl, 0.01% Tween), supplemented with 1 mm nucleotide and 1 mm DTT. The plates were incubated at room temperature for 24 h with gentle rocking. Solutions of Hsp70 were removed, and each well was washed as before and blocked with 100 μl of 5% milk in TBS-T for 5 min at room temperature. The plates were developed using 50 μl each of Hsp70 primary antibody (1:5000 in TBS-T) and an HRP-conjugated secondary antibody (1:5000 in TBS-T) and washed with TBS-T between each 1-h incubation at room temperature. Binding was detected using the TMB substrate kit (Cell Signaling Technology), and the absorbance was read at 450 nm. Data were analyzed using GraphPad Prism software and fit to the Langmuir binding isotherm (Y = BmaxX/[KD + X]).
Size-exclusion chromatography and multiangle light scattering
XIAP(120–356) and Hsp70 were subjected to size-exclusion chromatography using a KW-803 silica resin column (Shodex Group) with an AKTA micro-HPLC (GE Healthcare) in 20 mm Tris buffer, pH 7.5, containing 200 mm NaCl, 5 mm MgCl2, 10 mm KCl, 50 μm zinc acetate, 1 mm DTT. Separated samples were then analyzed for light scattering using a DAWN HELEOS II MALS detector and for protein concentration using the change in refractive index measured by a Optilab rEX. Molecular weights of species contained in the SEC peaks were calculated using ASTRA VI software (Wyatt Technology).
Circular dichroism
CD spectra of XIAP(120–356) and mutants were acquired on a J-715 spectropolarimeter (Jasco Inc.) using a 1 mm pathlength quartz cuvette, subtracting the CD signal acquired for buffer alone (10 mm sodium phosphate, pH 7.6, 100 mm NaF, 50 μm zinc acetate, 0.5 mm DTT). Data were converted to mean residue ellipticity (degrees cm−1 dmol−1) according to the equation Θ = Ψ/(1000 nlc), where Ψ is the CD signal in degrees; n is the number of amides; l is the path length in centimeters; and c is the concentration in decimoles/ml. Each spectrum reported is the average of six scans.
NMR spectroscopy
NMR-based binding studies were performed as described previously (50). Briefly, 13C-15N Hsc70 SBD (residues 395–508) was expressed in BL21 cells in M9 minimal media supplemented with 15NH4Cl, purified, and refolded from 4 m GdHCl on a Ni-NTA column. This protein was dialyzed into 25 mm Tris, 50 mm NaCl, 1 mm EDTA, 2 mm DTT, 0.02% sodium azide, 5% D2O, protease inhibitors, pH 7.2. Then, 15N−1H TROSY-HSQC spectra with either NRLLLTG or XIAP(120–356) were recorded at 30 °C for 1 h, using a Bruker 600 MHz NMR spectrometer with cryoprobe.
Author contributions
L. C. C., S. R. S., A. K. M., and J. E. G. conceptualization; L. C. C. and H. S. data curation; L. C. C., H. S., S. R. S., and J. E. G. formal analysis; L. C. C., H. S., S. R. S., E. T., C. J., and E. R. Z. investigation; L. C. C., H. S., E. T., C. J., and E. R. Z. methodology; L. C. C., H. S., E. R. Z., D. R. S., A. K. M., and J. E. G. writing-original draft; L. C. C., H. S., E. T., C. J., E. R. Z., D. R. S., A. K. M., and J. E. G. writing-review and editing; E. R. Z., D. R. S., A. K. M., and J. E. G. supervision; J. E. G. funding acquisition.
Acknowledgments
The XIAP(120–356) pet28a vector was a kind gift from Dr. Jeanne Stuckey. SM164 was provided by Dr. Shaomeng Wang. We thank Michael Y. Sherman for the xenograft tumor samples and members of the Gestwicki and Mapp laboratories for helpful discussions.
This work was supported in part by National Institutes of Health Grant NS059690 (to J. E. G. and E. R. P. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- NBD
- nucleotide-binding domain
- IAP
- inhibitor of apoptosis protein
- XIAP
- X-linked inhibitor of apoptosis protein
- SBD
- substrate-binding domain
- PES
- pifithrin-μ
- PARP
- poly(ADP-ribose) polymerase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- SEC-MALS
- size-exclusion chromatography-multiangle light scattering
- BIR
- baculoviral IAP repeat
- PPI
- protein–protein interaction
- Ni-NTA
- nickel-nitrilotriacetic acid
- DMEM
- Dulbecco's modified Eagle's medium
- 17-DMAG
- 17-dimethylaminoethylamino-17-demethoxygeldanamycin.
References
- 1. Ciocca D. R., and Calderwood S. K. (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10, 86–103 10.1379/CSC-99r.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Workman P., Burrows F., Neckers L., and Rosen N. (2007) Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann. N.Y. Acad. Sci. 1113, 202–216 10.1196/annals.1391.012 [DOI] [PubMed] [Google Scholar]
- 3. Brodsky J. L., and Chiosis G. (2006) Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem. 6, 1215–1225 10.2174/156802606777811997 [DOI] [PubMed] [Google Scholar]
- 4. Powers M. V., Clarke P. A., and Workman P. (2009) Death by chaperone: HSP90, HSP70 or both? Cell Cycle 8, 518–526 10.4161/cc.8.4.7583 [DOI] [PubMed] [Google Scholar]
- 5. Taipale M., Krykbaeva I., Koeva M., Kayatekin C., Westover K. D., Karras G. I., and Lindquist S. (2012) Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 10.1016/j.cell.2012.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagel R., Semenova E. A., and Berns A. (2016) Drugging the addict: non-oncogene addiction as a target for cancer therapy. EMBO Rep. 17, 1516–1531 10.15252/embr.201643030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weinstein I. B. (2002) Cancer. Addiction to oncogenes–the Achilles heal of cancer. Science 297, 63–64 10.1126/science.1073096 [DOI] [PubMed] [Google Scholar]
- 8. Trepel J., Mollapour M., Giaccone G., and Neckers L. (2010) Targeting the dynamic HSP90 complex in cancer. Nat. Rev. 10, 537–549 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sherman M. Y., and Gabai V. L. (2015) Hsp70 in cancer: back to the future. Oncogene 34, 4153–4161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blagg B. S., and Kerr T. D. (2006) Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med. Res. Rev. 26, 310–338 10.1002/med.20052 [DOI] [PubMed] [Google Scholar]
- 11. Matts R. L., Brandt G. E., Lu Y., Dixit A., Mollapour M., Wang S., Donnelly A. C., Neckers L., Verkhivker G., and Blagg B. S. (2011) A systematic protocol for the characterization of Hsp90 modulators. Bioorg. Med. Chem. 19, 684–692 10.1016/j.bmc.2010.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vasko R. C., Rodriguez R. A., Cunningham C. N., Ardi V. C., Agard D. A., and McAlpine S. R. (2010) Mechanistic studies of Sansalvamide A-amide: an allosteric modulator of Hsp90. ACS Med. Chem. Lett. 1, 4–8 10.1021/ml900003t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mayer M. P. (2013) Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38, 507–514 10.1016/j.tibs.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 14. Swain J. F., Dinler G., Sivendran R., Montgomery D. L., Stotz M., and Gierasch L. M. (2007) Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol. Cell 26, 27–39 10.1016/j.molcel.2007.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zuiderweg E. R., Bertelsen E. B., Rousaki A., Mayer M. P., Gestwicki J. E., and Ahmad A. (2013) Allostery in the Hsp70 chaperone proteins. Top. Curr. Chem. 328, 99–153 10.1007/128_2012_323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Assimon V. A., Gillies A. T., Rauch J. N., and Gestwicki J. E. (2013) Hsp70 protein complexes as drug targets. Curr. Pharm. Des. 19, 404–417 10.2174/1381612811306030404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rauch J. N., and Gestwicki J. E. (2014) Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J. Biol. Chem. 289, 1402–1414 10.1074/jbc.M113.521997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rüdiger S., Germeroth L., Schneider-Mergener J., and Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 10.1093/emboj/16.7.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mayer M. P., Schröder H., Rüdiger S., Paal K., Laufen T., and Bukau B. (2000) Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7, 586–593 10.1038/76819 [DOI] [PubMed] [Google Scholar]
- 20. Rüdiger S., Buchberger A., and Bukau B. (1997) Interaction of Hsp70 chaperones with substrates. Nat. Struct. Biol. 4, 342–349 10.1038/nsb0597-342 [DOI] [PubMed] [Google Scholar]
- 21. Sekhar A., Rosenzweig R., Bouvignies G., and Kay L. E. (2015) Mapping the conformation of a client protein through the Hsp70 functional cycle. Proc. Natl. Acad. Sci. U.S.A. 112, 10395–10400 10.1073/pnas.1508504112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mashaghi A., Bezrukavnikov S., Minde D. P., Wentink A. S., Kityk R., Zachmann-Brand B., Mayer M. P., Kramer G., Bukau B., and Tans S. J. (2016) Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539, 448–451 10.1038/nature20137 [DOI] [PubMed] [Google Scholar]
- 23. Marcinowski M., Höller M., Feige M. J., Baerend D., Lamb D. C., and Buchner J. (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 10.1038/nsmb.1970 [DOI] [PubMed] [Google Scholar]
- 24. Pellecchia M., Montgomery D. L., Stevens S. Y., Vander Kooi C. W., Feng H. P., Gierasch L. M., and Zuiderweg E. R. (2000) Structural insights into substrate binding by the molecular chaperone DnaK. Nat. Struct. Biol. 7, 298–303 10.1038/74062 [DOI] [PubMed] [Google Scholar]
- 25. Srinivasan S. R., Gillies A. T., Chang L., Thompson A. D., and Gestwicki J. E. (2012) Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome. Mol. Biosyst. 8, 2323–2333 10.1039/c2mb25145k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calloni G., Chen T., Schermann S. M., Chang H. C., Genevaux P., Agostini F., Tartaglia G. G., Hayer-Hartl M., and Hartl F. U. (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 1, 251–264 10.1016/j.celrep.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 27. Clerico E. M., Tilitsky J. M., Meng W., and Gierasch L. M. (2015) How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427, 1575–1588 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wegele H., Müller L., and Buchner J. (2004) Hsp70 and Hsp90–a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44 10.1007/s10254-003-0021-1 [DOI] [PubMed] [Google Scholar]
- 29. Pratt W. B., and Toft D. O. (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133 10.1177/153537020322800201 [DOI] [PubMed] [Google Scholar]
- 30. Li J., Soroka J., and Buchner J. (2012) The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 1823, 624–635 10.1016/j.bbamcr.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 31. Thompson A. D., Scaglione K. M., Prensner J., Gillies A. T., Chinnaiyan A., Paulson H. L., Jinwal U. K., Dickey C. A., and Gestwicki J. E. (2012) Analysis of the τ-associated proteome reveals that exchange of hsp70 for hsp90 is involved in τ degradation. ACS Chem. Biol. 7, 1677–1686 10.1021/cb3002599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karagöz G. E., Duarte A. M., Akoury E., Ippel H., Biernat J., Morán Luengo T., Radli M., Didenko T., Nordhues B. A., Veprintsev D. B., Dickey C. A., Mandelkow E., Zweckstetter M., Boelens R., Madl T., and Rüdiger S. G. (2014) Hsp90-τ complex reveals molecular basis for specificity in chaperone action. Cell 156, 963–974 10.1016/j.cell.2014.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rousaki A., Miyata Y., Jinwal U. K., Dickey C. A., Gestwicki J. E., and Zuiderweg E. R. (2011) Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 411, 614–632 10.1016/j.jmb.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X., Srinivasan S. R., Connarn J., Ahmad A., Young Z. T., Kabza A. M., Zuiderweg E. R., Sun D., and Gestwicki J. E. (2013) Analogs of the allosteric heat shock protein 70 (Hsp70) inhibitor, MKT-077, as anti-cancer agents. ACS Med. Chem. Lett. 4, 10.1021/ml400204n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colvin T. A., Gabai V. L., Gong J., Calderwood S. K., Li H., Gummuluru S., Matchuk O. N., Smirnova S. G., Orlova N. V., Zamulaeva I. A., Garcia-Marcos M., Li X., Young Z. T., Rauch J. N., Gestwicki J. E., et al. (2014) Hsp70-Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 74, 4731–4740 10.1158/0008-5472.CAN-14-0747,10.1158/1538-7445.AM2014-4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaiser M., Kühnl A., Reins J., Fischer S., Ortiz-Tanchez J., Schlee C., Mochmann L. H., Heesch S., Benlasfer O., Hofmann W. K., Thiel E., and Baldus C. D. (2011) Antileukemic activity of the HSP70 inhibitor pifithrin-μ in acute leukemia. Blood Cancer J. 1, e28 10.1038/bcj.2011.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leu J. I., Pimkina J., Frank A., Murphy M. E., and George D. L. (2009) A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 36, 15–27 10.1016/j.molcel.2009.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leu J. I., Zhang P., Murphy M. E., Marmorstein R., and George D. L. (2014) Structural basis for the inhibition of HSP70 and DnaK chaperones by small-molecule targeting of a C-terminal allosteric pocket. ACS Chem. Biol. 9, 2508–2516 10.1021/cb500236y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taguwa S., Maringer K., Li X., Bernal-Rubio D., Rauch J. N., Gestwicki J. E., Andino R., Fernandez-Sesma A., and Frydman J. (2015) Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell 163, 1108–1123 10.1016/j.cell.2015.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang A. M., Miyata Y., Klinedinst S., Peng H. M., Chua J. P., Komiyama T., Li X., Morishima Y., Merry D. E., Pratt W. B., Osawa Y., Collins C. A., Gestwicki J. E., and Lieberman A. P. (2013) Activation of Hsp70 reduces neurotoxicity by promoting polyglutamine protein degradation. Nat. Chem. Biol. 9, 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mimnaugh E. G., Chavany C., and Neckers L. (1996) Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J. Biol. Chem. 271, 22796–22801 10.1074/jbc.271.37.22796 [DOI] [PubMed] [Google Scholar]
- 42. Arnaud L. T., Myeku N., and Figueiredo-Pereira M. E. (2009) Proteasome-caspase-cathepsin sequence leading to tau pathology induced by prostaglandin J2 in neuronal cells. J. Neurochem. 110, 328–342 10.1111/j.1471-4159.2009.06142.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Birnbaum M. J., Clem R. J., and Miller L. K. (1994) An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J. Virol. 68, 2521–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hinds M. G., Norton R. S., Vaux D. L., and Day C. L. (1999) Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat. Nat. Struct. Biol. 6, 648–651 10.1038/10701 [DOI] [PubMed] [Google Scholar]
- 45. Eckelman B. P., Salvesen G. S., and Scott F. L. (2006) Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 7, 988–994 10.1038/sj.embor.7400795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abisambra J., Jinwal U. K., Miyata Y., Rogers J., Blair L., Li X., Seguin S. P., Wang L., Jin Y., Bacon J., Brady S., Cockman M., Guidi C., Zhang J., Koren J., et al. (2013) Allosteric heat shock protein 70 inhibitors rapidly rescue synaptic plasticity deficits by reducing aberrant τ. Biol. Psychiatry 74, 367–374 10.1016/j.biopsych.2013.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tillotson B., Slocum K., Coco J., Whitebread N., Thomas B., West K. A., MacDougall J., Ge J., Ali J. A., Palombella V. J., Normant E., Adams J., and Fritz C. C. (2010) Hsp90 (heat shock protein 90) inhibitor occupancy is a direct determinant of client protein degradation and tumor growth arrest in vivo. J. Biol. Chem. 285, 39835–39843 10.1074/jbc.M110.141580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sun H., Stuckey J. A., Nikolovska-Coleska Z., Qin D., Meagher J. L., Qiu S., Lu J., Yang C. Y., Saito N. G., and Wang S. (2008) Structure-based design, synthesis, evaluation, and crystallographic studies of conformationally constrained Smac mimetics as inhibitors of the X-linked inhibitor of apoptosis protein (XIAP). J. Med. Chem. 51, 7169–7180 10.1021/jm8006849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morozova K., Clement C. C., Kaushik S., Stiller B., Arias E., Ahmad A., Rauch J. N., Chatterjee V., Melis C., Scharf B., Gestwicki J. E., Cuervo A. M., Zuiderweg E. R., and Santambrogio L. (2016) Structural and biological interaction of hsc-70 protein with phosphatidylserine in endosomal microautophagy. J. Biol. Chem. 291, 18096–18106 10.1074/jbc.M116.736744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rauch J. N., Zuiderweg E. R., and Gestwicki J. E. (2016) Non-canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) co-chaperones are important for client release. J. Biol. Chem. 291, 19848–19857 10.1074/jbc.M116.742502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young Z. T., Rauch J. N., Assimon V. A., Jinwal U. K., Ahn M., Li X., Dunyak B. M., Ahmad A., Carlson G. A., Srinivasan S. R., Zuiderweg E. R., Dickey C. A., and Gestwicki J. E. (2016) Stabilizing the Hsp70-τ complex promotes turnover in models of tauopathy. Cell Chem. Biol. 23, 992–1001 10.1016/j.chembiol.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Garrido C., Schmitt E., Candé C., Vahsen N., Parcellier A., and Kroemer G. (2003) HSP27 and HSP70: potentially oncogenic apoptosis inhibitors. Cell Cycle 2, 579–584 [PubMed] [Google Scholar]
- 53. Evans C. G., Chang L., and Gestwicki J. E. (2010) Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 53, 4585–4602 10.1021/jm100054f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hunter A. M., LaCasse E. C., and Korneluk R. G. (2007) The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis 12, 1543–1568 10.1007/s10495-007-0087-3 [DOI] [PubMed] [Google Scholar]
- 55. Kirschke E., Goswami D., Southworth D., Griffin P. R., and Agard D. A. (2014) Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157, 1685–1697 10.1016/j.cell.2014.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kellner R., Hofmann H., Barducci A., Wunderlich B., Nettels D., and Schuler B. (2014) Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc. Natl. Acad. Sci. U.S.A. 111, 13355–13360 10.1073/pnas.1407086111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang L., Thompson A. D., Ung P., Carlson H. A., and Gestwicki J. E. (2010) Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK). J. Biol. Chem. 285, 21282–21291 10.1074/jbc.M110.124149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li X., Colvin T., Rauch J. N., Acosta-Alvear D., Kampmann M., Dunyak B., Hann B., Aftab B. T., Murnane M., Cho M., Walter P., Weissman J. S., Sherman M. Y., and Gestwicki J. E. (2015) Validation of the Hsp70-Bag3 protein–protein interaction as a potential therapeutic target in cancer. Mol. Cancer Ther. 14, 642–648 10.1158/1535-7163.MCT-14-0650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Srinivasan S. R., Cesa L. C., Li X., Julien O., Zhuang M., Shao H. Chung J., Maillard I., Wells J. A., Duckett C. S., and Gestwicki J. E. (2017) Heat shock protein 70 (Hsp70) suppresses RIP1-dependent apoptotic and necroptotic cascades. Mol. Cancer Res. 16, 58–68 10.1186/s12943-017-0630-y [DOI] [PMC free article] [PubMed] [Google Scholar]