Abstract
The protein kinases IRAK [IL-1 (interleukin 1) receptor-associated kinase] 1 and 4 play key roles in a signalling pathway by which bacterial infection or IL-1 trigger the production of inflammatory mediators. In the present study, we demonstrate that IRAK1 and IRAK4 phosphorylate Pellino isoforms in vitro and that phosphorylation greatly enhances Pellino’s E3 ubiquitin ligase activity. We show that, in vitro, Pellino 1 can combine with the E2 conjugating complex Ubc13 (ubiquitin-conjugating enzyme 13)–Uev1a (ubiquitin E2 variant 1a) to catalyse the formation of K63-pUb (Lys63-linked polyubiquitin) chains, with UbcH3 to catalyse the formation of K48-pUb chains and with UbcH4, UbcH5a or UbcH5b to catalyse the formation of pUb-chains linked mainly via Lys11 and Lys48 of ubiquitin. In IRAK1−/− cells, the co-transfection of DNA encoding wild-type IRAK1 and Pellino 2, but not inactive mutants of these proteins, induces the formation of K63-pUb–IRAK1 and its interaction with the NEMO [NF-κB (nuclear factor κB) essential modifier] regulatory subunit of the IKK (inhibitor of NF-κB kinase) complex, a K63-pUb-binding protein. These studies suggest that Pellino isoforms may be the E3 ubiquitin ligases that mediate the IL-1-stimulated formation of K63-pUb–IRAK1 in cells, which may contribute to the activation of IKKβ and the transcription factor NF-κB, as well as other signalling pathways dependent on IRAK1/4.
Keywords: E3 ligase, inflammation, interleukin-1, interleukin 1 receptor-associated kinase (IRAK), Pellino, ubiquitin, ubiquitin-conjugating enzyme 13 (Ubc13)
Introduction
During infection by bacteria, bacterial products engage TLRs (Toll-like receptors) in immune cells, triggering the activation of signalling pathways that lead to the production of pro-inflammatory cytokines, interferons and chemokines. These inflammatory mediators are released into the circulation, where they mount responses to combat the invading pathogen. An initial event in signalling via the IL-1R [IL-1 (interleukin-1) receptor] and all TLRs, except for TLR3, is the recruitment to the receptors of a complex that includes the adaptor protein MyD88 (myeloid differentiation factor 88) [1,2] and two protein kinases, termed IRAK (IL-1R-associated kinase) 1 and 4 [3,4]. IRAK4 then activates IRAK1, allowing the latter to autophosphorylate at multiple sites [5]. This induces the dissociation of IRAK1/IRAK4 from MyD88 [6] and their interaction with an E3 ubiquitin ligase, termed TRAF6 [TNF (tumour necrosis factor)-associated factor 6] [7]. The next step in signalling is reported to involve the covalent attachment of K63-pUb (Lys63-linked polyubiquitin) chains to TRAF6, which is thought to be mediated by TRAF6 itself and E2 conjugating complexes, such as Ubc13 (ubiquitin-conjugating enzyme 13)–Uev1a (ubiquitin E2 variant 1a) [8,9]. The K63-pUb–TRAF6 can then recruit the TAK1 [TGFβ (transforming growth factor β)-activated kinase 1] complexes because the nuclear zinc-finger motifs present at the C-termini of the TAB (TAK1-binding protein) 2 and TAB3 regulatory subunits interact with K63-pUb chains [10]. This is believed to induce the dimerization, autophosphorylation and activation of TAK1, which then activates IKKβ {IκB [inhibitor of NF-κB (nuclear factor κB)] kinase β} and MKKs (mitogen-activated protein kinase kinases), switching on downstream signalling pathways that stimulate the production of inflammatory mediators [9,11,12].
We recently found that IL-1 also induces a rapid and striking formation of K63-pUb–IRAK1 and an interaction with both NEMO (NF-κB essential modifier), a regulatory subunit of the IKK complex, and ABIN2 (A20-binding inhibitor of NF-κB 2), a regulatory subunit of the Tpl2 (tumour progression locus 2) kinase (also called Cot) (M. Windheim and P. Cohen, unpublished work). The co-localization of these protein kinases to K63-pUb scaffolds may facilitate the IKKβ-catalysed activation of Tpl2, enabling Tpl2 to switch on MKK1/MKK2 and hence ERK (extracellular-signal-regulated kinase) 1 and 2, which is critical for the production of pro-inflammatory cytokines, such as TNF. These findings suggested that the formation of K63-pUb–IRAK1, as well as the formation of K63-pUb–TRAF6, may be important for downstream signalling and raised the question of the identity of the E3 ubiquitin ligase and E2 conjugating complexes that catalyse the formation of K63-pUb–IRAK1.
IRAK1 and IRAK4 are known to interact with three closely related mammalian proteins, termed Pellino 1, Pellino 2 and Pellino 3 [13–17]. These proteins appear to be important for signalling by IL-1, because, for example, siRNA (small interfering RNA) knockdown of Pellino 1 has been reported to impair the activation of NF-κB [18]. Similarly the TGFβ-induced protein SMAD6 binds to Pellino 1, disrupts its interaction with IRAK1/IRAK4 and prevents signalling by IL-1 [19]. Recently, a RING (really interesting new gene)-like domain was identified near the C-terminus of Pellino 1, 2 and 3, and these Pellino isoforms were shown to induce the polyubiquitination of IRAK1 when co-transfected with this protein kinase in cells. In contrast, Pellino mutants in which a cysteine and a histidine residue in the RING-like domain had been mutated did not induce the polyubiquitination of IRAK1 [20,21]. Taken together, these experiments suggested that the Pellino isoforms may be E3 ubiquitin ligases. The co-transfection of wild-type IRAK1, but not a catalytically inactive mutant of IRAK1, decreased the electrophoretic mobility of Pellino isoforms, which could be reversed by treatment with a protein serine/threonine phosphatase, suggesting that Pellino isoforms are phosphorylated by IRAK1, or by a kinase activated by IRAK1, under these conditions. Since wild-type IRAK1 interacted more strongly with Pellino isoforms than catalytically inactive IRAK1 in these experiments, it was suggested that the role of Pellino phosphorylation might be to enhance its interaction with IRAK1 [20,21].
In the present study, we have analysed the phosphorylation of Pellino isoforms by IRAK1 in vitro. We show that not only IRAK1, but also IRAK4, phosphorylates Pellino isoforms and that phosphorylation strikingly enhances their E3 ubiquitin ligase activity. We also show that Pellino 1 can act in vitro with a number of E2 conjugating complexes to form K63-pUb chains (Ubc13–Uev1a), K48-pUb chains (UbcH3) or polyubiquitin chains linked via more than one lysine residue of ubiquitin (UbcH4, UbcH5a or UbcH5b). We show further that the co-transfection of IRAK1 with Pellino 2 in cells induces the formation of K63-pUb–IRAK1 almost exclusively, and that co-transfection with Pellino 2 alone causes the endogenous IRAK1 to undergo Lys63-linked polyubiquitination. These experiments suggest that Pellinos may be the E3 ubiquitin ligases that mediates the formation of K63-pUb–IRAK1 in vivo.
Experimental
DNA constructs
Pellino 1 (GenBank® accession number AJ278859) was amplified from human liver total RNA by RT (reverse transcription)–PCR using Superscript III (Invitrogen). The resulting product was cloned into pSC-b (Stratagene) and sequenced to completion. The insert was then digested with BamHI and NotI and cloned into the same sites in pGEX6P-1 to produce pGEX6P-1-Pellino 1 for expression in Escherichia coli. Pellino 2 (GenBank® accession number BC009476) was cloned in a similar fashion and was ligated into the same sites in pCMV-HA-1 to form pCMV-HA-Pellino 2 for transfection into mammalian cells. Pellino 3b (GenBank® accession number AF487457) was amplified from IMAGE EST (expressed sequence tag) 5198999 using KOD Hot Start Polymerase (Novagen), subcloned into pSC-b, sequenced and ligated into the same sites in pGEX6P-1 to produce pGEX6P-1-Pellino 3b. IRAK1 (GenBank® accession number AAC41949.1) was amplified by RT–PCR, and the fragment was subcloned into the NotI sites of pFBGST, for expression in insect Sf21 cells, and into pCMV-HA-2 for expression in mammalian cells. IRAK4 (GenBank® accession number AAH13316.1) was amplified from IMAGE EST 4287014 and ligated into the BamHI/NotI sites of pFBHTb for expression in insect Sf21 cells. UbcH3 (GenBank® accession number NM_004359) was amplified from IMAGE EST 3942735, UbcH5a (GenBank® accession number NM_003338) from IMAGE EST 4305900 and UbcH5b (GenBank® accession number NM_003339) from IMAGE genomic library 5533590, digested with BamHI and NotI and ligated into the same sites in pET28a for expression in E. coli. UbcH4 (GenBank® accession number BAA91697) was amplified from IMAGE EST 3835609 and ligated into the NotI site of pET28a.
NEMO (GenBank® accession number BC050612) was amplified from IMAGE EST 6062527 and subcloned into the BamHI and NotI sites of pCMVtag3b for expression in mammalian cells. The mutant NEMO[D311N], was made by following the QuikChange™ site-directed mutagenesis method (Stratagene), but using KOD Hot Start DNA Polymerase. All DNA constructs were sequenced by the Sequencing Service (MRC Protein Phosphorylation Unit).
Protein expression and purification
GST (glutathione transferase)–IRAK1 and His6-tagged IRAK4 were expressed as active enzymes in insect Sf21 cells. GST–IRAK1 was purified from the cell lysates by affinity-purification on glutathione–Sepharose (GE Healthcare) and IRAK4 on Ni-NTA (Ni2+-nitrilotriacetate)–agarose. GST–Pellino 1 and GST–Pellino 3b were expressed in E. coli and purified on glutathione–Sepharose, and the GST was cleaved with PreScission protease to release the untagged Pellino 1 and 3b. The purified proteins were dialysed against 50 mM Tris/HCl (pH 7.5), 270 mM sucrose, 150 mM NaCl, 0.1 mM EGTA, 0.1% 2-mercaptoethanol, 1 mM benzamidine and 0.2 mM PMSF, and stored in aliquots at − 80°C. UbcH3, UbcH4, UbcH5a and UbcH5b were expressed in E. coli as His6-tagged proteins and were purified on Ni-NTA–agarose. Ubc13–Uev1a, E1 and ubiquitin mutants were expressed and purified as described in [22]. The protein phosphatase encoded by bacteriophage λgt10 was obtained from New England Biolabs, and wild-type ubiquitin was from Sigma.
Antibodies
Anti-FLAG and anti-HA (haemagglutinin) were purchased from Sigma, anti-IRAK1 and anti-NEMO were from Santa Cruz, anti-myc from Roche, anti-ubiquitin was from DakoCytomation, rabbit, mouse and sheep-specific secondary antibodies conjugated to HRP (horseradish peroxidase) were from Pierce and Protein G (HRP-conjugated) was from Bio-Rad.
Cell culture and transfection
HEK-293 (human embryonic kidney-293) cells stably expressing the IL-1R, but deficient in IRAK1 (termed here IRAK1−/− cells) [23] (a gift from Dr Xiaoxia Li, Department of Immunology, Lerner Research Institute, Cleveland, OH, U.S.A.), were cultured in DMEM (Dulbecco’s modified Eagle’s medium) with 10% (v/v) foetal calf serum. The cells were transfected at 60–70% confluence using polyethyleneimine and a total of 7.5 μg of DNA for each transfection [24]. After 24 h, the medium was replaced with DMEM without serum and then left for a further 16 h to avoid stimulation by any agonists that might have been present in the serum. The cells were extracted in ice-cold lysis buffer [50 mM Tris/HCl (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% (v/v) Triton X-100, 1 mM Na3VO4, 50 mM NaF, 5 mM sodium pyrophosphate, 0.27 M sucrose, 50 mM iodoacetamide, 10 mM sodium 2-glycerophosphate, 0.2 mM PMSF and 1 mM benzamidine). Cell lysates were clarified by centrifugation at 20000 g for 15 min at 4°C, and aliquots of the supernatants were quick-frozen in liquid nitrogen and stored at − 80°C. Protein concentrations were determined by the Bradford method.
Immunoprecipitation
Cell lysate (0.5 mg of protein) was incubated for 45 min at 4°C with 1–2 μg of antibody. Washed Protein G-Dynabeads (10 μl of Dynabeads per 1 μg of antibody) were added and incubated for a further 2–3 h with continuous stirring. The Dynabeads were collected by magnetic separation, the supernatant was discarded, and the beads were washed twice with 1 ml of lysis buffer and once with 1 ml of 10 mM Tris/HCl (pH 8.0).
Immunoblotting
A total of 20–30 μg of cell lysate protein or immunoprecipitates was denatured by incubation for 10 min at 75°C in 1% SDS, subjected to SDS/PAGE on 3–12% (w/v) gradient gels and transferred on to nitrocellulose membranes. The membranes were incubated for 30 min with Tris/HCl (pH 7.5), 0.15 M NaCl and 0.5% Tween (buffer A) containing 5% (w/v) BSA, immunoblotted for 2 h at 20°C in the same buffer and then overnight with the primary antibodies indicated. Antibodies were diluted to 0.2 μg · ml−1 for anti-NEMO and anti-IRAK-1, 1 μg · ml−1 for anti-FLAG and 2 μg · ml−1 for anti-HA and anti-myc. The blots were washed three times with buffer A and incubated with secondary HRP-conjugated antibodies in 5% (w/v) BSA. After three further washes in buffer A, the signal was detected with the ECL® (enhanced chemiluminescence) reagent (GE Healthcare).
Analysis of ubiquitination by MS
Ubiquitinated IRAK1 produced by co-transfection with Pellino 2 in IRAK1-deficient IL-1R cells was separated by SDS/PAGE, stained with colloidal Coomassie Blue and digested with trypsin [25]. The polybiquitination chain architecture of tryptic digests was analysed by LC (liquid chromatography)–MS on an Applied Biosystems 4000 Q-Trap mass spectrometer with MRM (multiple reaction monitoring) for the seven possible tryptic peptides derived from polyubiquitin chains. The Q1 and Q3 masses for the seven linkages chosen to trigger an MRM event above 200 counts were Lys6 (698.4/276.1), Lys11 (801.4/1002.5), Lys27 (701.0/215.1), Lys29 (408.7/503.3), Lys33 (546.6/740.4), Lys48 (487.6/232.1) and Lys63 (748.7/288.2). The intensity of each ubiquitin-linkage peptide was determined from the extracted ion chromatogram of each MRM using Analyst 1.4.1 software. In the experiments where IRAK1 was co-expressed with Pellino 2 and isolated by immunoprecipitation, the tryptic digests were analysed by both MRM and LC–MS on a ThermoElectron LTQ-orbitrap system coupled to a Dionex 3000 HPLC system. The extracted ion chromatograms for the ubiquitin-linkage peptides were performed using Qual Browser in Xcalibur software.
MS/MS spectra were searched using Mascot (Matrix Science) run on a local server allowing for Gly-Gly or Leu-Arg-Gly-Gly attached to a ubiquitinated lysine residue. The mass tolerances used were ± 1 Da for 4000 Q-Trap experiments and ± 20 p.p.m. for orbitrap experiments.
Results
Pellino 1 is an E3 ubiquitin ligase that is activated by IRAK1
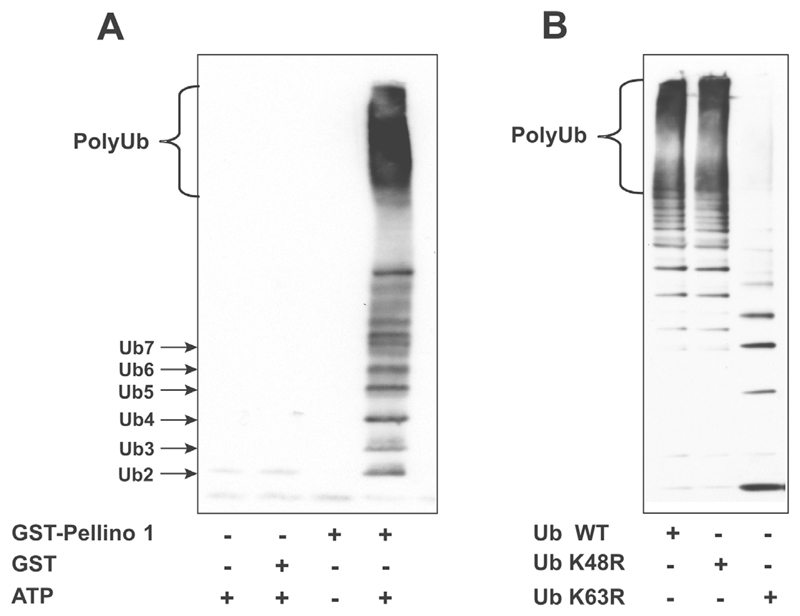
Previous studies had demonstrated that IRAK1 became polyubiquitinated when it was co-transfected with DNA encoding Pellinos 1, 2 and 3, suggesting that the Pellino isoforms are E3 ligases [20,21]. To check whether Pellino 1 was really an E3 ubiquitin ligase and that in co-transfection experiments it did not induce the polyubiquitination of IRAK1 indirectly by interacting with and activating a different E3 ligase, we carried out ubiquitination assays in vitro in the presence of Ubc13–Uev1a, an E2 conjugating complex known to specify the formation of K63-pUb chains. These experiments established that Pellino 1 was indeed an E3 ligase that could mediate the formation of polyubiquitin chains in the presence of Ubc13–Uev1a, E1, ubiquitin and MgATP (Figure 1A). Moreover, the polyubiquitin chains formed in the presence of Pellino 1 and Ubc13–Uev1a were predominantly linked via Lys63, because they were not formed when wild-type ubiquitin was replaced by ubiquitin[K63R] (Figure 1B). While the present paper was in preparation, Butler et al. [26] also reported that all three Pellino isoforms can act with Ubc13–Uev1a to form K63-pUb chains in vitro.
Figure 1. Pellino 1 is an E3 ubiquitin ligase which induces the formation of K63-pUb chains in the presence of Ubc13–Uev1a.
(A) A mixture of 0.1 μM E1, 1 μM Ubc13–Uev1a, 1 μM GST–Pellino 1 or 1 μM GST, 0.1 mM ubiquitin (Ub), 5 mM MgCl2 and 2 mM ATP in 50 mM Tris/HCl (pH 7.5) was incubated for 60 min at 30°C in a total volume of 0.02 ml. The reaction was stopped by the addition of SDS, and the products were resolved by SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted with an anti-ubiquitin antibody. Ub2, Ub3, Ub4, etc. denote ubiquitin dimers, trimers, tetramers, etc. (B) The reaction was carried out as in (A), but in the presence of wild-type (WT) ubiquitin, ubiquitin[K48R] or ubiquitin[K63R]. Ubiquitination reactions were carried out for 60 min and then analysed as in (A).
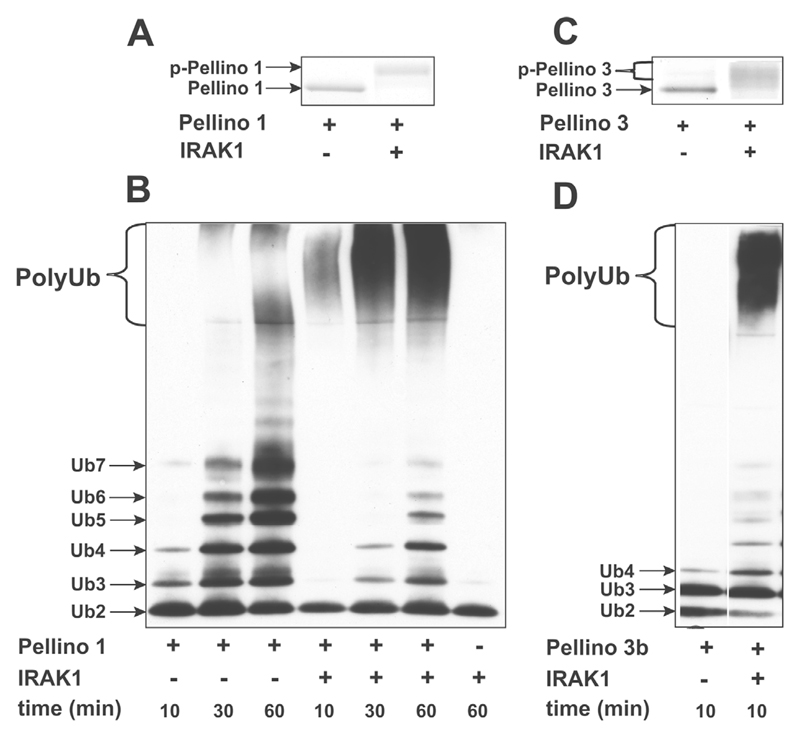
It has also been reported that Pellino isoforms become phosphorylated when DNA encoding these proteins was cot-ransfected with DNA encoding IRAK1 [20,21]. To investigate whether Pellino 1 was phosphorylated directly by IRAK1, or whether the phosphorylation of Pellino isoforms observed in co-transfection experiments was catalysed by another protein kinase that was activated by IRAK1, we investigated whether IRAK1 was capable of phosphorylating Pellino 1 in vitro. These experiments demonstrated that, in the presence of MgATP, IRAK1 induced a decrease in the electrophoretic mobility of Pellino 1, indicative of phosphorylation (Figure 2A). Moreover, incubation of Pellino 1 with MgATP and IRAK1 caused a dramatic enhancement of its E3 ligase activity. For example, after incubation for 10 or 30 min with E1, Ubc13–Uev1a, ubiquitin and MgATP, the unphosphorylated form of Pellino 1 was able to induce the formation of small ubiquitin oligomers only, whereas, following phosphorylation by IRAK1, far larger polyubiquitin chains migrating near the top of the SDS/polyacrylamide gels were formed over the same time period (Figure 2B).
Figure 2. IRAK1 phosphorylates Pellino 1 and enhances its ubiquitin ligase activity.
(A, C) A sample of 1 μM Pellino 1 (A) or1 μM Pellino 3b (C) was incubated for 60 min at 30°C with MgATP in the absence (−) or presence (+) of 1 μM GST–IRAK1 in 50 mM Tris/HCl (pH 7.5). The reactions were denatured in SDS and subjected to SDS/PAGE, and the gels were stained with Coomassie Blue. p-, phosphorylated protein. (B, D) A mixture of 1 μM Pellino 1 (B) or 1 μM Pellino 3b (D), 1 μM Ubc13–Uev1a, 0.1 mM ubiquitin, 5 mM MgCl2 and 2 mM ATP, with (+) or without (−)1 μM GST–IRAK1, was incubated for 30 min at 30°C, and the ubiquitination reactions initiated by the addition of 0.1 μM E1. The reactions were stopped after a further 10, 30 or 60 min by the addition of SDS, subjected to SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted with anti-ubiquitin antibody. Ub, ubiquitin.
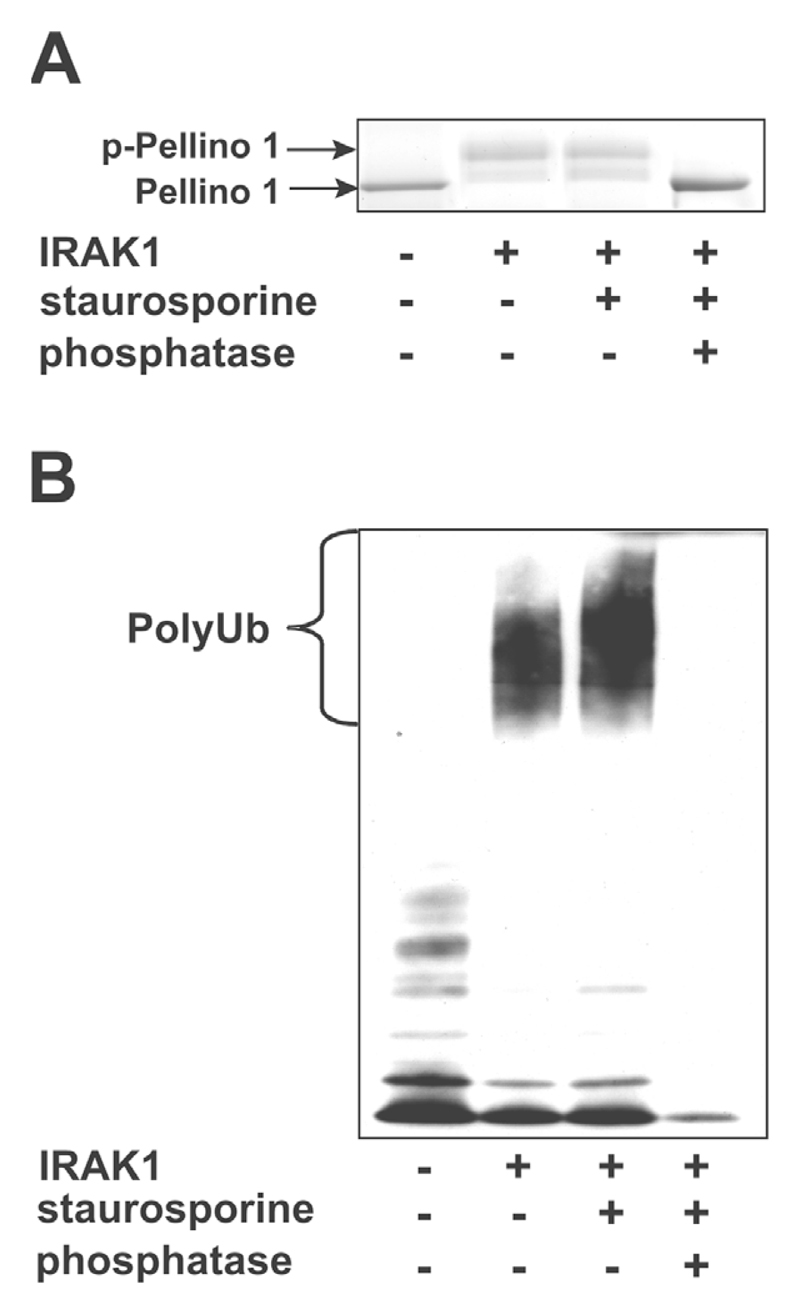
To establish that the activation of Pellino had resulted from phosphorylation, we stopped the IRAK1-catalysed phosphorylation with the non-specific protein kinase inhibitor staurosporine and then added the protein serine/threonine phosphatase produced by bacteriophage λgt10. Incubation with the phosphatase increased the electrophoretic mobility of Pellino 1 to that of the unphosphorylated bacterially expressed protein (Figure 3A) and abolished its E3 ligase activity (Figure 3B). The finding that λ-phosphatase treatment reduced the E3 ligase activity of Pellino 1 to below the level of the bacterially expressed protein suggested that the slight activity of Pellino 1 purified from E. coli was due to trace phosphorylation, presumably catalysed by a bacterial kinase.
Figure 3. The E3 ligase activity of Pellino 1 is regulated by reversible phosphorylation.
(A) A1 μM Pellino 1 sample was incubated with MgATP in 50 mM Tris/HCl (pH 7.5) without (−) or with (+) 0.5 μM GST–IRAK1 as in Figure 2. After 30 min, the reactions were supplemented with (+) or without (−) staurosporine (Calbiochem), which was added to a final concentration of 75 μM to inhibit IRAK1. The reactions were then incubated for 30 min without (−) or with (+) bacteriophage λ-phosphatase (100 units). p-, phosphorylated. (B) The reaction was carried out as in (A), except that the E3 ligase activity of each sample was assessed by incubation for 5 min at 30°C with Ubc13–Uev1a, E1 and ubiquitin as in Figure 2(B).
Pellino 3b is also phosphorylated and activated by IRAK1 in vitro
We carried out further studies in which Pellino 1 was replaced by Pellino 3b. These experiments showed that, similarly to Pellino 1, the electrophoretic mobility of Pellino 3b was decreased upon incubation with MgATP and IRAK1 (Figure 2C) and this was accompanied by a great increase in its associated E3 ligase activity (Figure 2D). We did not carry out these experiments with Pellino 2 because we were unable to express it in E. coli.
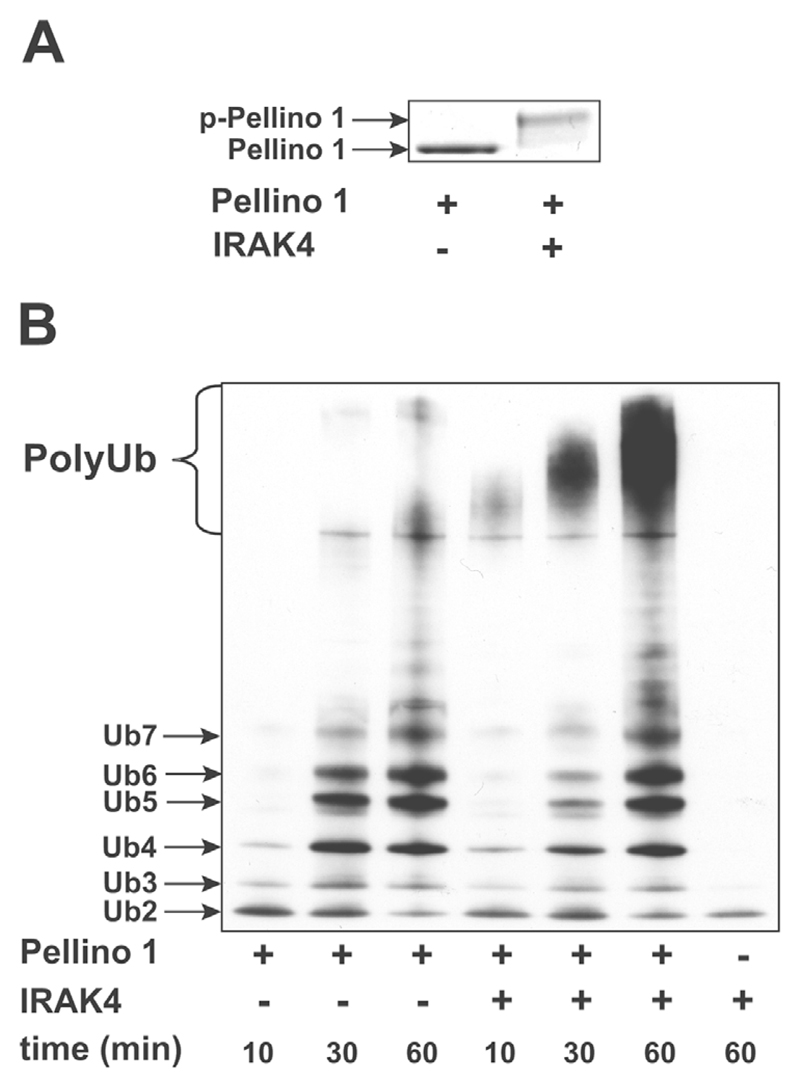
Pellino 1 is also phosphorylated and activated by IRAK4 in vitro
Since Pellino isoforms have been shown to interact with IRAK4, as well as IRAK1 [18], we investigated whether Pellino 1 was also a substrate for IRAK4 in vitro. These experiments showed that IRAK4 phosphorylated (Figure 4A) and enhanced the E3 ligase activity (Figure 4B) of Pellino 1 in vitro.
Figure 4. IRAK4 phosphorylates Pellino 1 and enhances its E3 ligase activity.
The experiment was carried out as described in Figure 2, except that GST–IRAK1 was replaced by His6 –IRAK4. (A) SDS/PAGE of Pellino 1 before and after phosphorylation by IRAK4. p-, phosphorylated. (B) The formation of polyubiquitin chains before and after phosphorylation of Pellino 1 by IRAK4. Ub, ubiquitin.
Pellino 1 operates as an E3 ligase with several E2 conjugating complexes
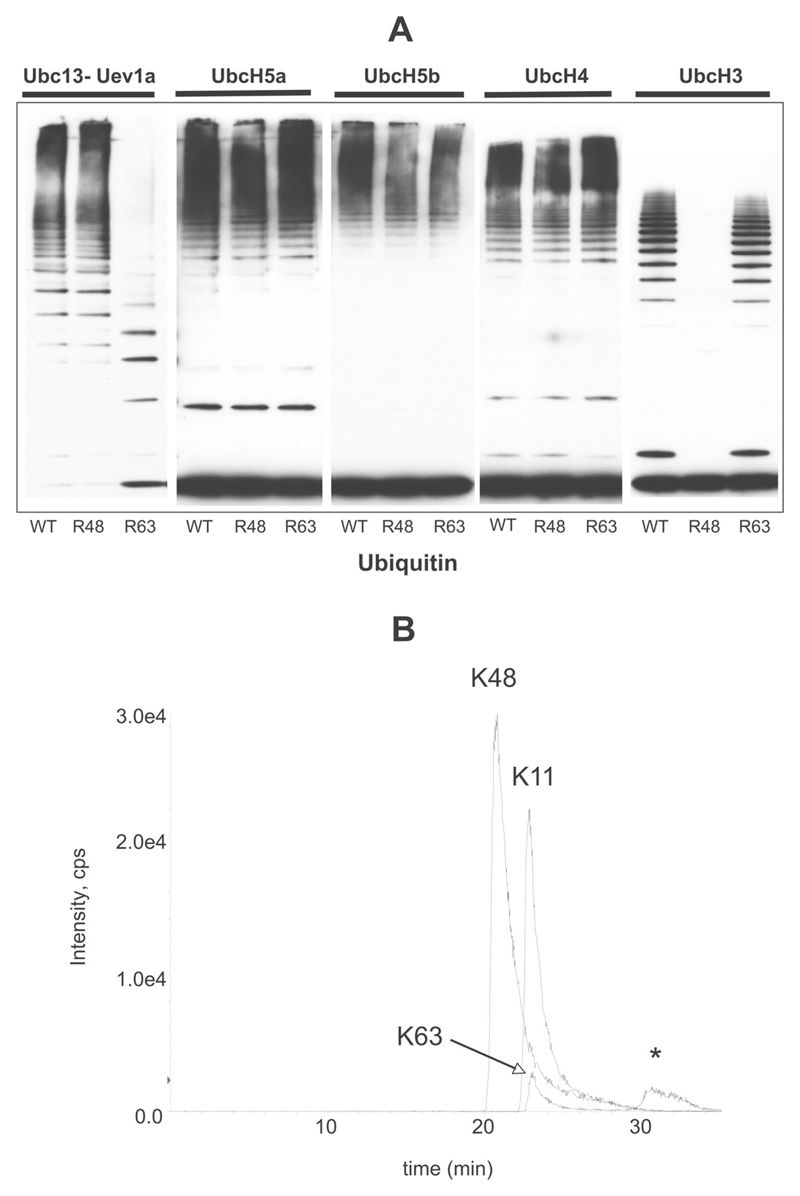
We next examined whether Pellino 1 was capable of inducing the formation of polyubiquitin chains in the presence of E2 conjugating complexes other than Ubc13–Uev1a. These experiments showed that Pellino 1 could also combine with UbcH3, UbcH4, UbcH5a and UbcH5b to produce polyubiquitin chains (Figure 5A). Further experiments using ubiquitin mutants in which either Lys48 or Lys63 were changed to arginine showed that, in the presence of UbcH3, Pellino 1 induced the formation of K48-pUb chains specifically, in contrast with the K63-pUb chains produced in the presence of Ubc13–Uev1a. In the presence of UbcH4, UbcH5a and UbcH5b, Pellino 1 induced the formation of polyubiquitin chains that were not linked exclusively via either Lys48 or Lys63 (Figure 5A). Direct identification of ubiquitination sites by MS showed further that, in the presence of UbcH5a (Figure 5B), UbcH5b or UbcH4 (results not shown) and wild-type ubiquitin, Pellino 1 mainly induced the formation of polyubiquitin chains linked via Lys48 and Lys11 of ubiquitin, with a small number of chains linked via Lys63.
Figure 5. Pellino 1 can operate as an E3 ubiquitin ligase with several different E2 conjugating complexes.
(A) GST–Pellino 1 was incubated with the E2 conjugating complexes indicated, in the presence of wild-type ubiquitin (WT), ubiquitin[K48R] (R48) or ubiquitin[K63R] (R63). The ubiquitination reactions were carried out as described in the legend to Figure 1. The reaction products were resolved by SDS/PAGE and immunoblotted with an anti-ubiquitin antibody. (B) IRAK1 was ubiquitinated in the presence of Pellino 1, wild-type ubiquitin and the E2 ligase UbcH5a. The area of the stained gel from 90 to 250 kDa in (A) (lane 4) was digested with trypsin, and ubiquitin-linkage analysis was performed by LC–MS on a 4000 Q-Trap mass spectrometer with MRM of the seven possible tryptic peptides from ubiquitin containing the characteristic Gly-Gly (gg) linkage to lysine residues 6, 11, 27, 29, 33, 48 or 63. The extracted ion chromatograms for the Lys11 (TLTGKggTITLEVEPSTIENVK), Lys48 (LIFAGKggQLEDGR) and Lys63 (TLSDYNIQKggESTLHLVLR) tryptic peptides are shown. The peak annotated with an asterisk was determined to be a false positive for Lys63. e4 = × 104.
The co-expression of IRAK1 and Pellino 2 in HEK-293 cells induces the formation of K63-pUb–IRAK1
Using HEK-293 cells deficient in IRAK1, but stably expressing IL-1R (termed IRAK1−/− cells), we co-expressed DNA encoding either wild-type IRAK1 or a catalytically inactive mutant (IRAK1[K239A]) together with DNA encoding either wild-type Pellino 2 or Pellino 2[H371S/C373S] in which two residues in the RING-like domain had been mutated. The equivalent mutations in Pellino 1 abolished its E3 ligase activity in vitro ([20], and results not shown). Pellino 2 was used in these experiments because it was expressed much more efficiently than Pellino 1 using the transfection protocol employed in the present study.
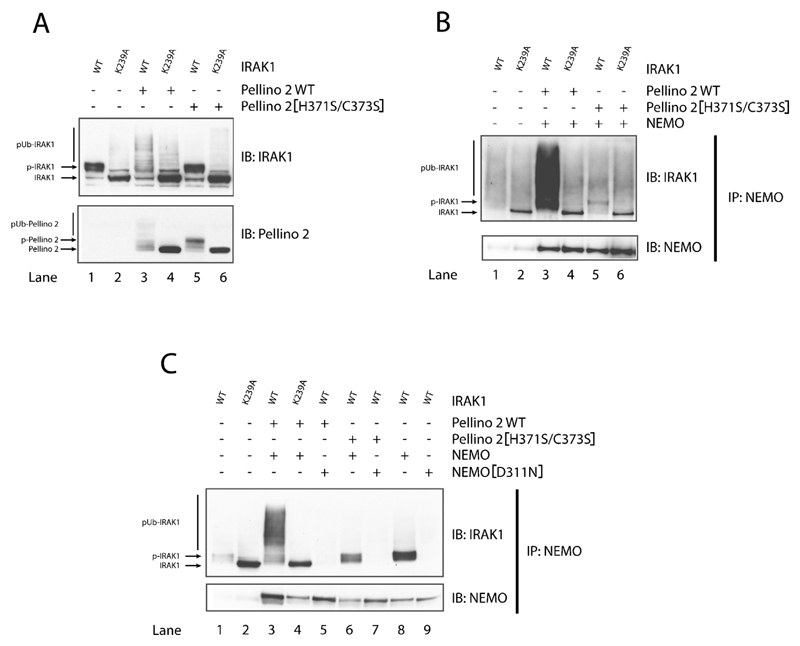
The transfected wild-type IRAK1 migrated more slowly than IRAK1[K239A], presumably as a consequence of autophosphorylation (Figure 6A, upper panel, lanes 1 and 2). When co-transfected with wild-type Pellino 2, wild-type IRAK1 was converted into (ubiquitinated) species of even lower electrophoretic mobility (Figure 6A, upper panel, lane 3), but this did not occur if wild-type Pellino 2 was replaced by Pellino 2[H371S/C373S] (Figure 6A, upper panel, lanes 5). The mobility of IRAK1[K239A] was not altered by co-transfection with either wild-type Pellino 2 (Figure 6A, upper panel, lanes 4) or Pellino 2[H371S/C373S] (Figure 6A, upper panel, lanes 6). The electrophoretic mobility of Pellino 2[H371S/C373S] decreased when it was co-transfected with wild-type IRAK1 (Figure 6A, lower panel, lane 5), but not IRAK1[K239A] (Figure 6A, lower panel, lane 6), indicative of phosphorylation. However, conversion of Pellino 2 into (ubiquitinated) species of even lower mobility was only observed when wild-type Pellino 2 was co-transfected with wild-type IRAK1 (Figure 6A, lower panel, lane 3), and not with IRAK1[K239A] (Figure 6A, lower panel, lane 4). Taken together, these results are consistent with the experiments performed with Pellino 1 and Pellino 3b in vitro and indicate that, when co-transfected into cells, wild-type IRAK1 phosphorylates wild-type Pellino 2, activating its E3 ligase function which can then induces the polyubiquitination of IRAK1 and Pellino 2 itself.
Figure 6. Phosphorylation and polyubiquitination of IRAK1 and Pellino 2 induced by co-transfection of both proteins in IRAK1−/− cells.
Cells in 10-cm-diameter dishes were transfected with (+) or without (−) vectors expressing DNA encoding FLAG-tagged wild-type IRAK1 (WT) or IRAK1[K239A] with (+) or without (−) vectors expressing HA-tagged wild-type Pellino 2 or Pellino 2[H371S/C373S] and (in B, C) with myc-tagged wild-type NEMO or myc-tagged NEMO[D311N]. After 24 h, the cells were deprived of serum for 16 h and proteins were extracted with 0.5 ml of lysis buffer. (A) Cell lysates (20 μg of protein) were denatured in 1% SDS, subjected to SDS/PAGE, transferred on to nitrocellulose membranes and immunoblotted (IB) with anti-IRAK1 and anti-HA (to detect Pellino 2). (B, C) As for (A), except that NEMO was immunoprecipitated (IP) from 0.5 mg of cell lysate protein with anti-NEMO and the immunoprecipitates immunoblotted with anti-IRAK1 and anti-Myc (to detect NEMO). p-, phosphorylated.
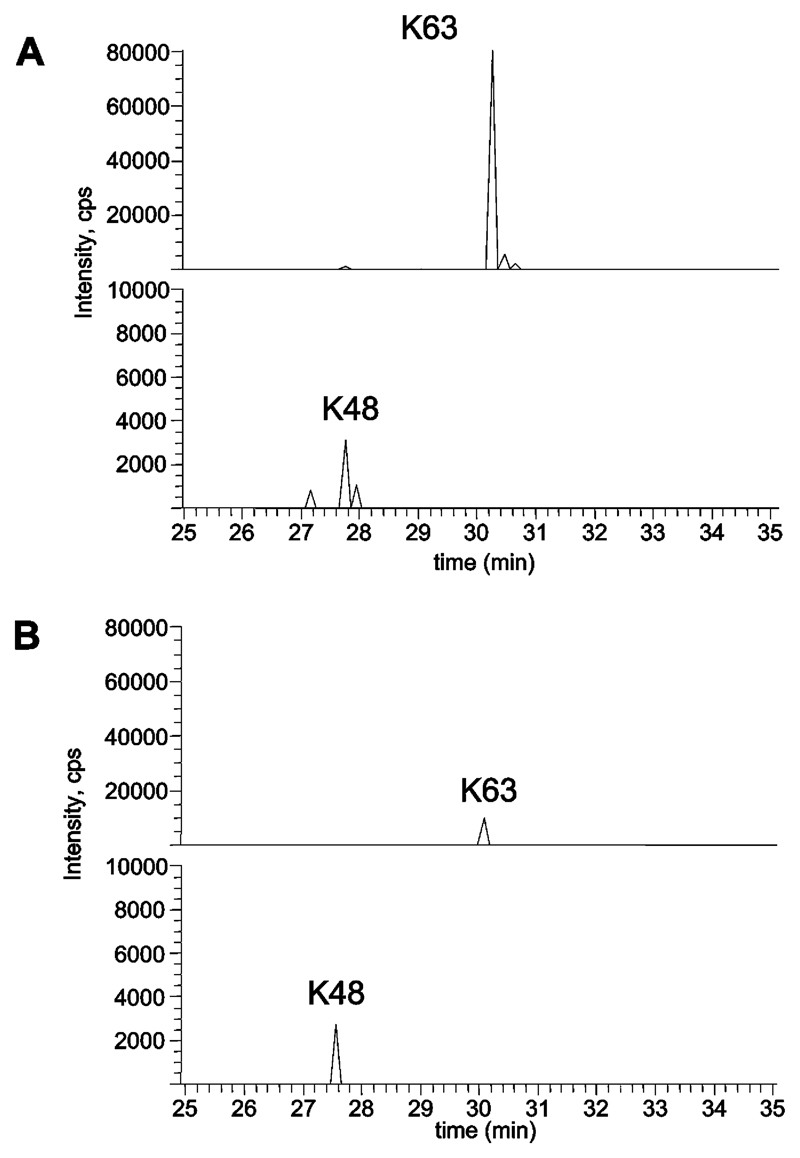
To identify the type of polyubiquitin chain produced when DNA encoding wild-type IRAK1 and wild-type Pellino 2 were co-transfected, IRAK1 was immunoprecipitated from the cell extracts, and, after SDS/PAGE, the region of the polyacrylamide gel corresponding to the polyubiquitinated IRAK1 species (Figure 6A, upper panel, lane 3) was excised. Following incubation with trypsin, the digest was subjected to MS. These experiments showed that the polyubiquitin chains attached to IRAK1 were linked via Lys63 almost exclusively (Figure 7A, upper panel), with only traces of IRAK1 linked via Lys48 (Figure 7A, lower panel; note the different scale on the y-axis). When DNA encoding wild-type IRAK1 was transfected into IRAK1–/– cells with DNA encoding Pellino 2[H371S/C373S], the IRAK1 was also polyubiquitinated, but to a much lesser extent. This was presumably catalysed by an endogenous E3 ligase present in the IRAK1–/– cells, perhaps a Pellino isoform(s). Again, the IRAK1 was mainly ubiquitinated via Lys63 of ubiquitin with only minor ubiquitination via Lys48 (Figure 7B).
Figure 7. Identification of ubiquitin linkages in polyubiquitinated IRAK1/Pellino 2.
IRAK1 was co-expressed with either wild-type Pellino 2 (A) or Pellino 2[H371S/C373S] (B) and immunoprecipitated with an anti-IRAK1 antibody. The immunoprecipitates were separated by SDS/PAGE and the region of the gel from 250 to 90 kDa was digested with trypsin and analysed by LC–MS on an LTQ-orbitrap system. The extracted ion chromatograms for the Lys63 (K63, upper panel) or Lys48 (K48, lower panel) ubiquitin peptides (containing the Gly-Gly signature) from the tryptic digest of either IRAK1 co-expressed with wild-type Pellino 2 (A) or from IRAK1 co-expressed with Pellino 2[H371S/C373S] (B) are shown.
If the transfected IRAK1 undergoes Lys63-linked polyubiquitination, then it should be capable of binding to proteins, such as NEMO, that interact with K63-pUb chains specifically [27,28]. We therefore repeated the experiment presented in Figure 6(A), except that the DNA encoding IRAK1 and Pellino 2 was also co-transfected with DNA encoding myc–NEMO. After pulling down myc–NEMO from the cell extracts with an anti-NEMO antibody, we examined whether IRAK1 was bound to NEMO. These experiments demonstrated that the polyubiquitinated IRAK1 formed by co-transfection with Pellino 2 did indeed associate with NEMO (Figure 6B, upper panel, lane 3), but not if wild-type IRAK1 was replaced by IRAK1[K239A] (Figure 6B, lane 4) or if wild-type Pellino 2 was replaced by Pellino 2[H371S/C373S] (Figure 6B, lane 5). In the experiments where NEMO was co-transfected with catalytically inactive IRAK1[K239A], the inactive non-ubiquitinated IRAK1 was found in the NEMO immunoprecipitates (Figures 6B, lanes 4 and 6, and 6C, lanes 2 and 4), but this interaction was non-specific, since it was also observed in cells where NEMO was not transfected (Figure 6B, lane 2)
To establish further that IRAK1 had bound to NEMO by virtue of the interaction of NEMO with the K63-pUb chains linked covalently to IRAK1, we repeated the experiment shown in Figure 6(B), but also using NEMO[D311N], a mutant which cannot bind K63-pUb chains. This experiment demonstrated that wild-type NEMO (Figure 6C, lane 3), but not NEMO[D311N] (Figure 6C, lane 5) was able to bind to the polyubiquitinated wild-type IRAK1 generated by co-transfection with Pellino 2. When wild-type IRAK1 was co-transfected with E3 ligase-inactive Pellino, some wild-type IRAK1 was found in the NEMO immunoprecipitates, presumably as a result of some IRAK1 ubiquitination mediated by endogenous Pellino.
Discussion
In the present study, we have established not only that Pellino isoforms are E3 ubiquitin ligases, but also that Pellino E3 ligase activities are greatly enhanced after phosphorylation by either IRAK1 or IRAK4 in vitro. Co-transfection experiments in which IRAK1 and Pellino 2 were expressed in IRAK1–/– cells showed that wild-type IRAK1 became ubiquitinated when co-transfected with wild-type Pellino 2, but not if a catalytically inactive mutant of IRAK1 was co-transfected with wild-type Pellino 2 or if wild-type IRAK1 was co-transfected with an E3 ligase-deficient mutant of Pellino 2. These results were consistent with the observations made in vitro, namely that Pellino isoforms are activated by IRAK1-catalysed phosphorylation and that, once activated, can ubiquitinate IRAK1 in cells.
The human genome is thought to encode over 500 ubiquitin E3 ligases and approx. 50 E2 conjugating complexes, indicating that, on average, each E2 conjugating complex must operate in conjunction with 10–20 E3 ligases. Conversely, E3 ligases are able to interact with many E2s. In the present study, we found that Pellino 1 was able to induce the formation of polyubiquitination chains with the five E2 conjugating complexes that we tested in vitro. In the presence of Ubc13–Uev1a, Pellino induced the formation of K63-pUb chains, while, in the presence of UbcH3, K48-pUb chains were formed specifically. In the presence of UbcH4, UbcH5a and UbcH5b, the polyubiquitin chains formed were linked via Lys48 and Lys11 with minor ubiquitination via Lys63. These observations were consistent with the notion that E2 conjugating complexes direct the specificity of polyubiquitin chain formation and raised the question of which type of polyubiquitin chain became linked covalently to IRAK1 when it was co-transfected with Pellino 2 in cells.
The dominant form of ubiquitinated IRAK1 produced by co-transfection with Pellino 2 contained polyubiquitin chains linked via Lys63 of ubiquitin, as determined by MS or the ability to interact with NEMO, a protein that binds K63-pUb chains preferentially over K48-pUb chains [27,28]. Moreover the NEMO[D311N] mutant, which is unable to bind K63-pUb chains, was unable to interact with the polyubiquitinated IRAK1 produced by co-transfection with Pellino 2. These findings are consistent with other recent studies from our laboratory, which have shown that the endogenous IRAK1 undergoes Lys63-linked polyubiquitination within 5–10 min of stimulating HEK-293 cells that stably express the IL-1R with IL-1 (M. Windheim and P. Cohen, unpublished work). This suggests that one or more Pellino isoforms may be the E3 ligases that mediate the IL-1-induced formation of K63-pUb–IRAK1 in cells. However, further studies are needed to establish whether this is the case and whether Ubc13–Uev1a is the relevant E2 conjugating enzyme.
A catalytically inactive mutant of IRAK1 has been reported to interact with Pellino isoforms less well than wild-type IRAK1 [20], suggesting that the phosphorylation of Pellino may enhance its interaction with IRAK1. However, we did not observe a major difference between the amount of Pellino immunoprecipitated with wild-type and inactive IRAK1 in our co-transfection experiments (A. Ordureau, unpublished work).
IRAK1 and IRAK4 have both been reported to form a complex in cells with Pellino isoforms, and we have shown that both protein kinases are capable of phosphorylating and activating Pellino 1 and 3b in vitro. Very recently, Butler et al. [26] reported that IRAK4 could stimulate a weak polyubiquitination of Pellino 1 and Pellino 2 and an extensive polyubiquitination of Pellino 3 in co-transfection experiments, but whether IRAK4 exerted these effects directly, or indirectly by activating the endogenous IRAK1, was unclear. Our findings raise the question of whether it is IRAK1 or IRAK4 or both protein kinases that catalyse the phosphorylation of different Pellino isoforms in vivo. Mice have been generated that do not express either IRAK1 [29,30] or IRAK4 [31] or express a catalytically inactive mutant of IRAK4 instead of the wild-type protein [32,33], and potent small-molecule inhibitors of IRAK4 have been developed [34] that may help to address this problem. However, as IRAK4 is the kinase that activates IRAK1 (reviewed in [35]), and IRAK1 may be needed to form the IRAK4–IRAK1–Pellino complex, such studies may not be definitive. The generation of mice that express a catalytically inactive mutant of IRAK1 instead of the wild-type kinase will clearly be necessary to solve this problem.
The IRAK1 protein was reported to disappear within 10 min of stimulating MRC-5 breast cancer cells with IL-1 [36], and we have made similar observations in the HEK-293 cells used in the present study (M. Windheim and P. Cohen, unpublished work). The original observations led to the suggestion that the role of IRAK1 polyubiquitination was to prime the protein for destruction by the proteasome and it was reported that three proteasome inhibitors, LLL, IEAL and LLNV, slightly delayed the IL-1-induced disappearance of IRAK1. However, we have found that MG-132, a more potent and specific proteasome inhibitor, has no effect on the IL-1-induced disappearance of IRAK1 in HEK-293 cells, although it blocks the IL-1-stimulated degradation of IκBα mediated by the proteasome in the same cells (M. Windheim and P. Cohen, unpublished work). These results are consistent with our finding that IL-1 induces the Lys63-linked polyubiquitination of IRAK1 and not Lys48-linked polyubiquitination, and indicate that IL-1 triggers the disappearance of IRAK1 by another mechanism that has still to be identified.
The observation that IL-1 stimulates the formation of a complex between IRAK1 and TRAF6 and the formation of K63-pUb–IRAK1 (M. Windheim and P. Cohen, unpublished work) as well as K63-pUb–TRAF6 [8,9] raises the question of the relative roles of these two Lys63-polyubiquitinated proteins in triggering downstream signalling events. A working hypothesis is that K63-pUb–IRAK1/TRAF6 may act as a scaffold for the assembly of the TAK1–IKKβ–Tpl2 kinase cascade. Thus K63-pUb–TRAF6, which may be triggered by IRAK2 [37], is reported to interact with and induce the activation of the TAK1 complex [9], while we have shown that K63-pUb–IRAK1 interacts with NEMO and ABIN2 (M. Windheim and P. Cohen, unpublished work), which are the regulatory subunits of the IKK and Tpl2 complexes respectively. These observations suggest that TAK1, IKKβ and Tpl2 may colocalize in IL-1-stimulated cells by virtue of their interaction with the K63-pUb–IRAK1·K63-pUb–TRAF6 complex, which may facilitate the activation of IKKβ by TAK1 and the activation of Tpl2 by IKKβ.
Acknowledgments
This study was supported by the UK Medical Research Council (MRC), AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Merck and Co., Merck-Serono and Pfizer. P. C. is a Royal Society Research Professor and H. S. and A. O. are the recipients of Wellcome Trust and MRC Prize Studentships respectively. We thank Xiaolia Li and George Stark (Department of Molecular Genetics, Lerner Research Institute, Cleveland, OH, U.S.A.) for generously providing the IRAK1–/– cells, Gursant Kular (MRC Protein Phosphorylation Unit) for purification of IRAK4, the Protein Production team of the Division of Signal Transduction Therapy at Dundee for expression and purification of other proteins used, and the DNA Sequencing Service (http://www.dnaseq.co.uk) for sequencing clones.
Abbreviations used
- ABIN2
A20-binding inhibitor of nuclear factor κB 2
- DMEM
Dulbecco’s modified Eagle’s medium
- EST
expressed sequence tag
- GST
glutathione transferase
- HA
haemagglutinin
- HEK-293
human embryonic kidney-293
- HRP
horseradish peroxidase
- IκB
inhibitor of nuclear factor κB
- IKK
IκB kinase
- IL-1
interleukin 1
- IL-1R
IL-1 receptor
- IRAK
IL-1R-associated kinase
- K63-pUb
Lys63-linked polyubiquitin
- LC
liquid chromatography
- MKK
mitogen-activated protein kinase kinase
- MRM
multiple reaction monitoring
- MyD88
myeloid differentiation factor 88
- NEMO
nuclear factor κB essential modifier
- NF-κB
nuclear factor κB
- Ni-NTA
Ni2+-nitrilotriacetate
- RING
really interesting new gene
- RT
reverse transcription
- TAK1
transforming growth factor β-activated kinase 1
- TAB
TAK1-binding protein
- TGFβ
transforming growth factor β
- TLR
Toll-like receptor
- TNF
tumour necrosis factor
- Tpl2
tumour progression locus 2
- TRAF6
TNF-receptor associated factor 6
- Ubc
ubiquitin-conjugating enzyme
- Uev1a
ubiquitin E2 variant 1a
References
- 1.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 2.Wesche H, Henzel WJ, Shillinglaw W, Li S, Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7:837–847. doi: 10.1016/s1074-7613(00)80402-1. [DOI] [PubMed] [Google Scholar]
- 3.Martin M, Bol GF, Eriksson A, Resch K, Brigelius-Flohe R. Interleukin-1-induced activation of a protein kinase co-precipitating with the type I interleukin-1 receptor in T cells. Eur J Immunol. 1994;24:1566–1571. doi: 10.1002/eji.1830240717. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kollewe C, Mackensen A, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin M. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem. 2003;279:5227–5236. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol. 2002;22:7158–7167. doi: 10.1128/MCB.22.20.7158-7167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 8.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen Z. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Deng L, Hong M, Akkaraju G, Inoue J, Chen Z. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 10.Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–548. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O, Akira S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Mira-Arbibe L, Ulevitch RJ. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopolysaccharide. J Leukocyte Biol. 2000;68:909–915. [PubMed] [Google Scholar]
- 13.Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–161. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- 14.Rich T, Allen RL, Lucas AM, Stewart A, Trowsdale J. Pellino-related sequences from Caenorhabditis elegans and Homo sapiens. Immunogenetics. 2000;52:145–149. doi: 10.1007/s002510000249. [DOI] [PubMed] [Google Scholar]
- 15.Resch K, Jockusch H, Schmitt-John T. Assignment of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet Cell Genet. 2001;92:172–174. doi: 10.1159/000056895. [DOI] [PubMed] [Google Scholar]
- 16.Jensen LE, Whitehead AS. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J Immunol. 2003;171:1500–1506. doi: 10.4049/jimmunol.171.3.1500. [DOI] [PubMed] [Google Scholar]
- 17.Grosshans J, Schnorrer F, Nüsslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)–IRAK–tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278:10952–10956. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 19.Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, Hong S, Kim IH, Kim SJ, Park SH. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- 20.Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 21.Schauvliege R, Janssens S, Beyaert R. Pellino proteins: novel players in TLR and IL-1R signalling. J Cell Mol Med. 2007;11:453–461. doi: 10.1111/j.1582-4934.2007.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation: crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Commane M, Jiang Z, Stark GR. IL-1-induced NFκB and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK) Proc Natl Acad Sci USA. 2001;98:4461–4465. doi: 10.1073/pnas.071054198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, Mackintosh C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler MP, Hanly JA, Moynagh PN. Kinase active IRAKs promote polyubiquitination and degradation of the Pellino family: direct evidence for Pellino proteins being E3 ubiquitin ligases. J Biol Chem. 2007;282:29729–29737. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- 27.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 28.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Kanakaraj P, Schafer PH, Cavender DE, Wu Y, Ngo K, Grealish PF, Wadsworth SA, Peterson PA, Siekierka JJ, Harris CA, Fung-Leung WP. Interleukin (IL)-1 receptor-associated kinase (IRAK) requirement for optimal induction of multiple IL-1 signaling pathways and IL-6 production. J Exp Med. 1998;187:2073–2079. doi: 10.1084/jem.187.12.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas JA, Allen JL, Tsen M, Dubnicoff T, Danao J, Liao XC, Cao Z, Wasserman SA. Impaired cytokine signaling in mice lacking the IL-1 receptor-associated kinase. J Immunol. 1999;163:978–984. [PubMed] [Google Scholar]
- 31.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, Mirtsos C, Takada H, Wakeham A, Itie A, Li S, et al. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–756. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 32.Kawagoe T, Sato S, Jung A, Yamamoto M, Matsui K, Kato H, Uematsu S, Takeuchi O, Akira S. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J Exp Med. 2007;204:1013–1024. doi: 10.1084/jem.20061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koziczak-Holbro M, Joyce C, Gluck A, Kinzel B, Muller M, Tschopp C, Mathison JC, Davis CN, Gram H. IRAK-4 kinase activity is required for interleukin-1 (IL-1) receptor- and toll-like receptor 7-mediated signaling and gene expression. J Biol Chem. 2007;282:13552–13560. doi: 10.1074/jbc.M700548200. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Liu J, Sudom A, Ayres M, Li S, Wesche H, Powers JP, Walker NP. Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper. Structure. 2006;14:1835–1844. doi: 10.1016/j.str.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell Signalling. 2007 doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Yamin TT, Miller DK. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- 37.Keating SE, Maloney GM, Moran EM, Bowie AG. IRAK-2 participates in multiple Toll-like receptor signaling pathways to NFκB via activation of TRAF6 ubiquitination. J Biol Chem. 2007;282:33435–33443. doi: 10.1074/jbc.M705266200. [DOI] [PubMed] [Google Scholar]