Abstract
Adult mammalian cardiomyocyte regeneration after injury is thought to be minimal. Mononuclear diploid cardiomyocytes (MNDCMs), a relatively small subpopulation in the adult heart, may account for the observed degree of regeneration, but this has not been tested. We surveyed 120 inbred mouse strains and found that the frequency of adult mononuclear cardiomyocytes was surprisingly variable (>7-fold). Cardiomyocyte proliferation and heart functional recovery after coronary artery ligation both correlated with pre-injury MNDCM content. Using genome-wide association, we identified Tnni3k as one gene that influences variation in this composition and demonstrated that Tnni3k knockout resulted in elevated MNDCM content and increased cardiomyocyte proliferation after injury. Reciprocally, overexpression of Tnni3k in zebrafish promoted cardiomyocyte polyploidization and compromised heart regeneration. Our results corroborate the relevance of MNDCMs in heart regeneration. Moreover, they imply that intrinsic heart regeneration is not limited nor uniform in all individuals, but rather is a variable trait influenced by multiple genes.
The adult mammalian heart is not generally thought to have significant capacity to regenerate. Instead, heart injury is thought to result in permanent loss of myocardium combined with fibrotic scarring and declining function, along with progression to heart failure if damage is extensive. However, in several other contexts, the myocardium is proliferative and can sustain growth and repair. For example, car-diomyocytes are proliferative during embryonic development in all species. In mice, proliferative capacity gradually ceases over the first postnatal week1. Accordingly, neonatal (postnatal day (P) 1) mouse hearts fully regenerate after resection, but this regenerative capacity is lost by P7 (ref. 2). In contrast to mammals, zebrafish3 and newts4 maintain efficient cardiac regenerative capacity throughout their life span. Neonatal mice and adult zebrafish both regenerate myocardium through proliferation of preexisting cardiomyocytes5. Although normal turnover and postinjury regeneration are both highly limited in the adult mammalian heart, it is clear that new cardiomyocytes do arise, albeit at a very low frequency6,7. Most studies concur that most of these new cardiomyocytes come from preexisting cardio-myocytes7,8, although it has not yet been possible to identify the sub-population of adult cardiomyocytes that retains proliferative ability and that accounts for this activity.
A notable difference between the mostly nonproliferative car-diomyocytes of the adult mammalian heart and the proliferation-competent cardiomyocytes described in the examples above is their total DNA content. During embryonic heart development across all species, including in rodents1,9 and human10, all cardiomyocytes are mononuclear and diploid. In zebrafish11 and newt4, almost all car-diomyocytes remain mononuclear and diploid throughout life. In contrast, mammalian cardiomyocytes mostly become binucleated and/or polyploid after birth. In mice, this occurs during the first post-natal week1, coinciding with their loss of proliferative and regenerative capacity. A similar process occurs in the human heart, although with uncertain timing after birth12,13. Notably, a small subpopula-tion of MNDCMs persists in the adult mammalian heart. The limited number of these cells and the limited degree of heart regeneration that has been observed in most studies6,7,14,15 have led to speculation that MNDCMs may be a population of proliferative cardiomyocytes in the adult heart7,14,16–19, but this has yet to be tested.
To address these questions, we performed an extensive comparison across inbred mouse strains for the frequency of adult MNDCMs in the uninjured state. Contrary to current thought, we show that the frequency of MNDCMs is not uniformly small across all individuals, but rather is highly variable. We demonstrate that the frequency of this subpopulation predicts regenerative capacity and, through genome-wide association, identify Tnni3k as a gene that controls the size of the MNDCM population and the ability of cardiomyocytes to proliferate after injury. Our observations challenge the notion that heart regeneration in mammals is minimal and instead suggest that the capacity to regenerate after injury is a complex and variable trait dependent on an individual’s genetic background.
RESULTS
Frequency of MNDCMs in the adult mammalian heart is a variable phenotype
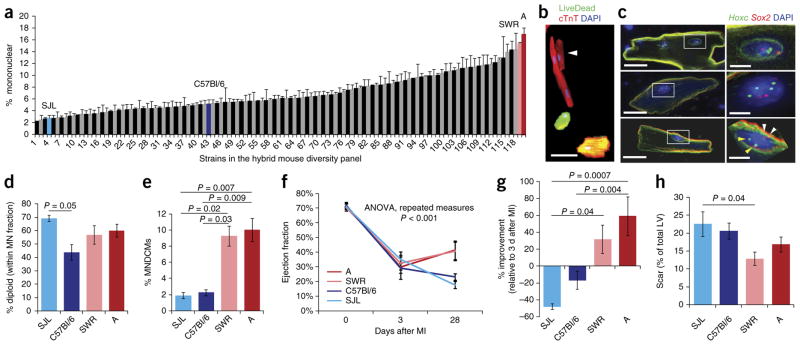
In mice, the transition from the mononuclear diploid condition that typifies embryonic cardiomyocytes to the binucleated condition of most adult cardiomyocytes occurs during the first postnatal week1. We considered that this process, and therefore the percentage of MNDCMs in the adult mouse heart, might be a variable trait subject to the combined influence of a number of polymorphic genes. To address this model, we surveyed 120 inbred mouse strains drawn from the Hybrid Mouse Diversity Panel (HMDP)20. Hearts from female mice 8–10 weeks old were collagenase digested, and ventricular cells were isolated and stained for evaluation (Fig. 1). Mononuclear cardiomyocytes tended to be smaller than their binu-cleated counterparts (Fig. 1b). Across mouse strains, we observed a >7-fold range (2.3–17.0%) in the percentage of mononuclear car-diomyocytes (Fig. 1a and Supplementary Table 1), which is a surprisingly large degree of variation. The median frequency for the measured strains was 6.1%, consistent with previous studies1. Of note, C57Bl/6, the most commonly used inbred strain in studies of heart biology, has a below-average frequency (5.2%). Since other variables (sex, age, diet, environment) were controlled, the most plausible explanation for variability between strains is genetic background. Indeed, the broad-sense heritability of the trait (a measure of intra- versus inter-strain variation21) was 71.1%, which indicates a very high genetic component. The observation that the distribution is roughly continuous implies the involvement of multiple genes, each with polymorphic alleles that contribute varying quantitative effect. The narrow-sense heritability of the trait (an indication of nonaddi-tive effects21) was 59.2%, which suggests a moderate degree of epi-static interaction among these genes.
Figure 1.
Variation in mononuclear diploid cardiomyocyte content predicts outcome after injury. (a) Distribution of 120 inbred mouse strains of the HMDP for frequency of mononuclear cardiomyocytes in the naive adult heart. See supplementary table 1 for full list of strains, including n-values. (b) Single-cell ventricular suspension stained with the vital dye LiveDead (green), for cardiac troponin T (cTnT) (red) and with DAPI (blue), identifying a small mononuclear cardiomyocyte (white arrowhead) next to a larger binucleated cardiomyocyte, with two dead cardiomyocytes also evident. Scale bar represents 50 μm. (c) DNA FISH using probes from the Hoxc (chr15, green; yellow arrowheads) and Sox2 (chr3, red; white arrowheads) loci; nuclei are stained with DAPI. Top, a binucleated cardiomyocyte with two diploid nuclei. Middle, a mononuclear cardiomyocyte with a tetraploid nucleus. Bottom, a mononuclear cardiomyocyte with a diploid nucleus. Scale bars in the cell view (left) represent 20 μm; scale bars in the nuclear view (right) represent 5 μm. (d) Quantification for four strains of the percentage of mononuclear (MN) cardiomyocytes that had diploid nuclei as assessed by FISH. n = 3 mice for all strains. One-way ANOVA P = 0.048; F = 4.14; P value on graph is Bonferroni correction. (e) Calculation of percentage of MNDCMs across four strains. One-way ANOVA P = 0.0027; P values on graph are Tukey HSD post hoc. (f) Ejection fraction measured by echocardiography on four strains at baseline, 3 d after myocardial infarction (MI) caused by coronary artery ligation and 28 d after ligation. See supplementary tables 2 and 3 for complete statistics. Strain A (n = 5 mice), SWR (n = 8 mice), C57Bl/6 (n = 4 mice) and SJL (n = 7 mice). (g) Percentage improvement 1 month after injury, calculated by (Ejection fraction at 28 d – Ejection fraction at 3 d)/(Ejection fraction at 3 d) for the four strains. One-way ANOVA P = 0.0004; F = 9.20; P values on graph are Bonferroni correction. (h) Quantification of the scar area 1 month after injury, represented as a percentage of the total left ventricle (LV) for the four strains. Strain A (n = 6), SWR (n = 8), C57Bl/6 (n = 4) and SJL (n = 7). One-way ANOVA P = 0.04; F = 3.29; P value on graph is Bonferroni corrected. All error bars are s.e.m.
Tri- and tetra-nucleated adult cardiomyocytes also exist. In our analysis, we also scored the frequency of cardiomyocytes with three or more nuclei (Supplementary Table 1) and observed that this too was a variable trait, and one that did correlate, though not strongly, with the frequency of mononuclear cardiomyocytes (Supplementary Fig. 1a).
Strains A and SWR had a high mononuclear content, while strains C57Bl/6 and SJL had a moderately low to very low mononuclear content (Fig. 1a). We selected these for further studies. Mononuclear tetraploid cardiomyocytes are present in mice and are more common in human than binuclear cardiomyocytes22. To assess ploidy within the mononuclear cardiomyocyte fraction of strains A, C57Bl/6, SJL and SWR, we conducted DNA fluorescence in situ hybridization using two independent autosomal probes (Fig. 1c). Across these four strains, 40–70% of the mononuclear cardiomyocyte nuclei were diploid (Fig. 1d), which suggests a modest degree of variation in this specific feature. These data allowed us to calculate the MNDCM frequency of the four strains and confirmed that strains SJL (1.9%) and C57Bl/6 (2.3%) have a low content and strains SWR (9.3%) and A (10.0%) have a high content of this population in the normal adult heart (Fig. 1e).
MNDCM frequency predicts functional recovery after injury
We used echocardiography to evaluate heart function in mice of the four inbred strains before and after permanent coronary artery ligation (Fig. 1f). We did not observe any difference in pre-injury functional parameters despite the variation between these strains in MNDCM composition. All strains exhibited a similar degree of functional impairment 3 d after injury, indicating a similar degree of infarction. At 1 month, ejection fraction had significantly worsened in SJL and stabilized in C57Bl/6, as in many past observations14,23. In contrast, the two high-MNDCM strains (A and SWR) showed improved function at day 28 compared to immediately after injury (Fig. 1f,g). The differences between the high strains and low strains combined, both at day 28 in absolute terms and when compared to day 3, were even more striking (Supplementary Table 2). Analysis of the individual echocardiographic components revealed less severe thinning of the anterior wall, less severe ventricular dilation and proportionally improved systolic contraction in A and SWR relative to C57Bl/6 and SJL (Supplementary Fig. 1b–e and Supplementary Table 3). Measurement of the scar in histological sections revealed a trend toward less scar after injury in strains with a higher initial level of MNDCMs (Fig. 1h and Supplementary Fig. 1f). To our knowledge, this degree of natural functional recovery and morphological normalization in mice is unprecedented. Collectively, these observations show that declining heart function is not the inevitable outcome of mammalian adult heart injury. Furthermore, these results support the premise that MNDCM frequency is a good surrogate to predict improved postinjury outcome.
MNDCM content corresponds to cardiomyocyte regeneration after injury
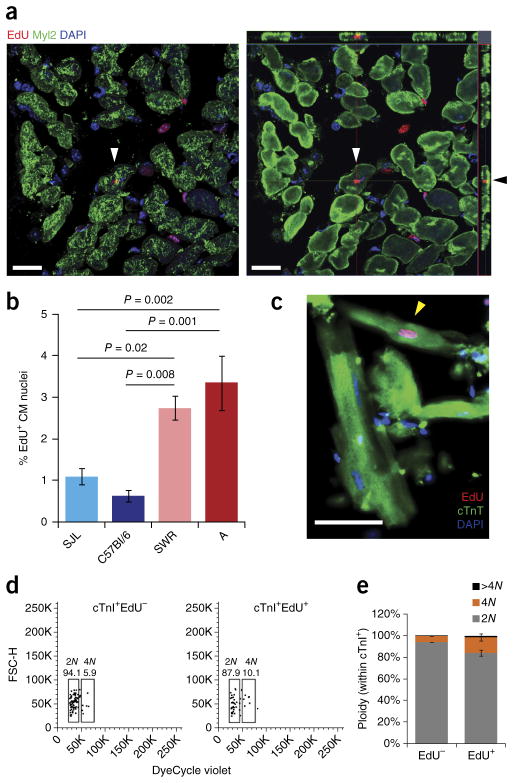
It is likely that several processes influence long-term functional and morphological outcome after heart injury, including cardiomyocyte proliferation, cardiomyocyte hypertrophy, collagen deposition by fibroblasts (scarring), regrowth of coronary vasculature and perhaps many others. To more explicitly consider the contribution of cardio-myocyte regeneration, 8-week-old mice were subjected to permanent coronary ligation as above. We injected them with 5-ethynyl-2′-deoxyuridine (EdU) once per day on days 4–10 after injury, isolated hearts at 1 month, and sectioned and stained the hearts for EdU and the ventricular cardiomyocyte marker Myl2 (Fig. 2a). We observed a striking correlation between the preinjury MNDCM percentage and the postinjury percentage of EdU+ border zone cardiomyocytes (Fig. 2b). To confirm this result, we used an independent mitotic marker, phospho-histone H3, quantified 11 d after injury in strains A and C57Bl/6, and again observed a >2-fold difference in putative mitotic cardiomyocytes proportional to the pre-injury MNDCM frequency (Supplementary Fig. 1g,h).
Figure 2.
Mononuclear diploid cardiomyocytes proliferate after injury. (a) Confocal image of an EdU+ (red) and Myl2+ (green) cardiomyocyte (white arrowheads). Left panel is the flattened z-stack. Right panel is the orthogonal view. Black arrowhead points to the same cell, indicating Myl2+ cytoplasm above and below the EdU+ nucleus. Scale bars represent 20 μm. (b) Quantification of percentage of EdU+ cardiomyocyte (CM) nuclei in the border zone of the left ventricle for the four strains. Strain A (n = 6 mice), SWR (n = 8 mice), C57Bl/6 (n = 4 mice) and SJL (n = 7 mice). One-way ANOVA P = 0.0002; F = 10.51; P values on graph are Bonferroni corrected. (c) A Myl2+EdU+ mononuclear cardiomyocyte (yellow arrowhead) identified in a ventricular cell preparation from a mouse exposed to EdU after coronary artery ligation. Scale bar represents 50 μm. (d) Flow cytometry plots (forward scatter, FSC-H, vs. DNA dye, DyeCycle Violet) of ventricular nuclei isolated from 1 strain A mouse exposed to EdU after coronary artery ligation. The different nuclear populations (cTnI+, cardiomyocyte; EdU− or EdU+) and their ploidy (2N, 4N) and quantifications are indicated. Plots and values shown are one example of three independent experiments. (e) Average quantification of ploidy across nuclear populations from 3 independent strain A mice (as in d). All error bars represent s.e.m.
Cardiomyocyte labeling with EdU can result from DNA replication followed by cytokinesis, resulting in proliferation, or followed by cell-cycle interruption before cytokinesis, resulting in polyploidy or multinucleation. Increased nucleation and ploidy are associated with hypertrophy, a nonproliferative volumetric increase24,25. It is difficult in histological sections to reliably determine whether EdU+ car-diomyocytes are mononuclear or multinuclear, let alone the nuclear ploidy. Thus, to distinguish a proliferative vs. hypertrophic response, we performed coronary artery ligation in strain A mice and exposed them to continuous EdU in the postinfarction recovery period. Ventricular single-cell suspensions were then stained for EdU and Myl2 (Fig. 2c). We observed 81.0% (±2.3%; n = 3 mice, 224 cells counted) of EdU-positive cardiomyocytes to be mononuclear. These mononuclear cardiomyocytes could be diploid or tetraploid. To address this point, we isolated ventricular nuclei from additional animals, stained cardiomyo-cyte nuclei using cardiac troponin I (cTnI) as described previously15,26, and evaluated the ploidy of cardiomyocyte nuclei by flow cytometry. There was a modest increase in the percentage of tetraploid nuclei within the cardiomyocyte EdU+ subpopulation compared to the EdU− population, presumably arising by endoreplication without karyokinesis, but 83.9% (±2.8%) of the cTnI+EdU+ cardiomyocyte nuclei were diploid (Fig. 2d,e). These observations suggest that approximately two-thirds of strain A cardiomyocytes undergoing DNA synthesis after injury complete both karyokinesis and cytokinesis, evidence of a true regenerative event. Previous lineage studies concluded that most proliferation-labeled cardiomyocytes arise from preexisting cardiomyocytes7,8, which discounts the possibility that our observations can be explained by proliferation of a progenitor followed by cardiomyocyte differentiation. Fusion of cardiomyocytes with hematopoietic or other cell types can occur27–29, but this results in binuclear rather than mononuclear cells. Although we cannot formally exclude the possibility of DNA replication by preexisting binuclear or tetraploid cardiomyocytes with two subsequent rounds of division into MNDCMs, our observations are consistent with a proliferative response by MNDCMs that divide to generate more MNDCMs. A related analysis described below also supports this interpretation.
Tnni3k contributes to MNDCM frequency variation
The genetic basis of natural variation is inherited polymorphisms that influence the expression or function of genes relevant to the trait of interest. Any single inbred mouse strain is likely to contain both positively and negatively acting polymorphisms that influence that phenotype. With adequate strain phenotype information, genes that are relevant to variation in complex traits can be identified through genome-wide association. Indeed, the inbred strains of the HMDP were chosen because the high density SNP genotypes for each that are needed for genome-wide association are already available20. This approach represents an unbiased forward genetic strategy, and the genes identified by association analysis need not be part of any preconceived mechanism.
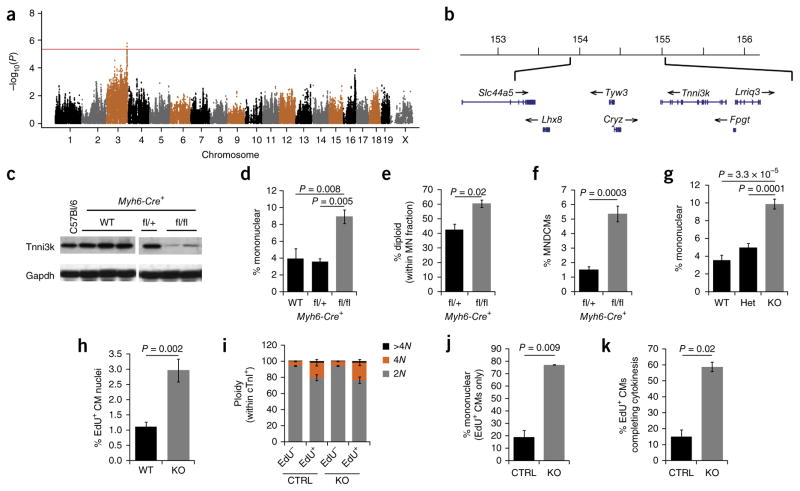
Using an efficient mixed model association30 to correct for relatedness between the 120 strains, we conducted a genome-wide association analysis for the trait of mononuclear cardiomyocyte frequency. The association algorithm calculates a probability value for each SNP based on its quantitative association with the measured phenotype, which results in a Manhattan plot that implicates regions of the genome associated with variation in the trait. Several peaks surpassed the suggestive threshold of P < 10−4, and a prominent peak on chromosome 3 surpassed the statistically significant threshold of P < 4 × 10−6 (Fig. 3a). An expanded view of this region of chromosome 3 (Fig. 3b) implicated a 1.2-Mb domain containing seven protein-coding genes, two uncharacterized ESTs, two pseudogenes and no known micro-RNAs. Of these, Tnni3k, which encodes an understudied kinase that, to the extent known, is cardiomyocyte-specific in expression31, became our lead candidate for analysis. A known T/C polymorphism (rs49812611) in a splice junction of the Tnni3k gene has previously been described32; the T allele is present in C57Bl/6 mice and supports normal gene expression, whereas the C allele causes improper splicing and near elimination of protein expression. A third hap-lotype reduces Tnni3k expression because of a linked but different polymorphism33. The distribution of strains across these haplotypes (Supplementary Fig. 2a) yields several findings. First, the difference in mononuclear cardiomyocyte frequency (mean 8.1% vs. 5.4%), the large number of strains polymorphic at rs49812611, and the total number of tested strains account for the high statistical significance of this locus in the Manhattan plot. At the same time, it is also clear that all subgroups include a substantial range of values. This is expected, and it implies additional loci that influence the overall phenotype. This analysis also showed that the hypomorphic C allele of rs49812611 is associated with a higher level of mononuclear cardiomyocytes (Supplementary Fig. 2a). Accordingly, Tnni3k expression across the strains by microarray analysis was negatively correlated with mono-nuclear cardiomyocyte frequency (bicor −0.30; P = 0.006). As the C allele results in greatly reduced protein expression (for example, as in strain A; Supplementary Fig. 2b), it is predicted to be recessive. Indeed, the mononuclear cardiomyocyte frequency in F1 mice from a cross of strains C57Bl/6 and A was 4.7 ± 1.0% (n = 4), which is equivalent to the (dominant) C57Bl/6 phenotype. Neither strains homozygous for the natural C allele nor engineered Tnni3k knockout mice, described below, had any gross perturbation of heart anatomy or function (see baseline echocardiography measurements in Fig. 1f and Supplementary Figs. 1b–e and 2c–f); thus, the Tnni3k gene is not required for normal viability.
Figure 3.
Tnni3k influences mononuclear diploid cardiomyocyte population size and cellular regenerative capacity after injury. (a) Manhattan plot of the genome-wide association analysis performed on the data for 120 HMDP strains shown in Figure 1a and supplementary table 1. Red line indicates the boundary for statistical significance (P = 4 × 10−6). (b) Chromosomal map of the 1.2-Mb locus on chromosome 3 corresponding to the most significant peak identified by genome-wide association analysis. Coordinates are for mouse genome build 37. (c) Immunoblot for Tnni3k and Gapdh on adult atrial tissue isolated from C57Bl/6 control and Myh6-Cre+ mice with varying copy numbers of the Tnni3k fl allele; WT, wild type. Both panels are from the same blot and exposed for the same length of time. (d) Frequency of mononuclear cardiomyocytes in Myh6-Cre+ mice with varying copy numbers of the Tnni3k fl allele. n = 4 mice for all genotypes. One-way ANOVA P = 0.003; F = 12.01; P values on graph are Bonferroni corrected. (e) Quantification of the number of diploid nuclei found within the mononuclear cardiomyocyte population, assessed by DNA FISH. n = 3 mice for both genotypes. Two-tailed Student t-test. (f) Calculation of MNDCM percent in the Myh6-Cre+, Tnni3k fl/+ genotype vs. the Tnni3k fl/fl genotype. Two-tailed Student t-test; t = 11.6. (g) Frequency of mononuclear cardiomyocytes in mice with varying copy numbers of Tnni3k null allele (global knockout). WT (n = 4 mice), heterozygote (het; n = 7 mice), knockout (KO; n = 4 mice). One-way ANOVA P = 2.43 × 10−5; F = 29.25; P values on graph are Bonferroni correction. (h) Quantification of EdU+ cardiomyocyte (CM) nuclei in the border zone of the left ventricle 1 month after coronary artery ligation in Tnni3k knockout (KO, n = 7) and WT (n = 6) littermates. Cardiomyocytes were identified as Myl2+ as in Figure 2a. Two-tailed Student t-test. (i) Distribution of cTnI+ nuclear ploidy as assessed by flow cytometry. KO (n = 4), control (CTRL; n = 2 heterozygotes and 1 WT). (j) Quantification of EdU+ cardiomyocytes that were mononuclear after infarction in both CTRL (n = 1 heterozygote and 1 WT) and KO (n = 2). Two-tailed Student t-test. (k) Calculation of the number of EdU+ cardiomyocytes completing cell division based on the analyses in i and j. Two-tailed Student t-test; t = 7.08. All error bars represent s.e.m.
Tnni3k knockout mice have increased MNDCM content
We crossed a conditional loss-of-function (floxed) Tnni3k allele (Tnni3kfl) that was generated and thereafter maintained on an inbred C57Bl/6 background34 with two different Cre recombinase lines, both of which were also on an inbred C57Bl/6 background. First, we crossed with cardiomyocyte-specific Myh6-Cre; conditional Myh6-Cre; Tnni3kfl/fl mice had a near complete elimination of Tnni3k protein (Fig. 3c; the residual expression likely reflects lack of complete recombination driven by Myh6-Cre). These conditional knockout mice were viable and seemingly normal. Second, we crossed Tnni3kfl with Sox2-Cre to generate a germline-deleted allele and then crossed the deleted Tnni3k allele to homozygosity while eliminating Sox2-Cre. Mice globally lacking Tnni3k were viable and seemingly normal; most notably, there were no measurable differences in echocardio-graphic parameters between uninjured knockout and wild-type littermates (Supplementary Fig. 2c–f; baseline measurements). In both mutants (conditional and global), we measured a 2.5-fold increase in the mononuclear cardiomyocyte frequency and a threefold elevation in the MNDCM population (Fig. 3d–g). Because these comparisons were conducted on a C57Bl/6 inbred strain background, the change in cardiomyocyte composition can be unambiguously assigned to the manipulation of this single gene. This outcome confirms that variation in adult MNDCM frequency, which clearly is a polygenic phenotype, can be deconstructed into individual polymorphic genes, one of which is Tnni3k.
Tnni3k knockout mice have improved cardiomyocyte proliferation after injury
We evaluated Tnni3k control vs. mutant mice, all on an inbred C57Bl/6 background, for their responses to adult heart injury. Following the same EdU injection regimen used above, we observed a near threefold greater percentage of EdU-labeled cardiomyocytes in the injury border zone in mutants compared to controls (Fig. 3h). It is noteworthy that the fold difference in EdU-labeled cells after injury was equivalent to the fold difference in MNDCMs before injury. This demonstration provides further support for the model that MNDCMs are activated by, and initiate the proliferative response to, adult heart injury.
To estimate how many of the EdU+ cells had completed cell division, we next assessed cardiomyocyte nucleation and ploidy after injury, just as performed on strain A mice in Figure 2c–e. In bulk ventricular nuclear preparations, the cTnI+EdU+ double-positive population was twice as large in knockout animals (0.75 ± 0.04%, n = 4) compared to control littermates (0.36% ± 0.02%, n = 3; P < 0.001), consistent with our observations quantified in sections. As in strain A (Fig. 2e), there was a modest increase in tetraploid nuclei within the cardiomyocyte EdU+ subpopulation, suggesting that approximately 20% of adult cardiomyocytes activated by injury to initiate DNA synthesis become tetraploid; however, most cardiomyocyte nuclei in both control and Tnni3k knockout mice were diploid (Fig. 3i). In dissociated single-cell preparations, EdU-labeled cardiomyocytes from Tnni3k knockout mice were primarily mononuclear, whereas those of control mice were predominantly binucleated (Fig. 3j). As discussed above, we interpret EdU+ diploid nuclei in mononuclear cardiomyocytes to indicate a proliferative event. Taking these results together, we estimate that 58.4 ± 3.0% of EdU-labeled cardiomyocytes in Tnni3k knockouts completed cyto-kinesis, compared to only 14.8 ± 4.4% in control mice (Fig. 3k). This analysis confirms that Tnni3k influences cardiomyocyte proliferation after adult heart injury.
Comparing control versus Tnni3k-null mice by echocardiography, we did not measure a significant improvement in heart function after infarction (Supplementary Fig. 2c–f; P = 0.79). Similarly, histological analysis of scar area 1 month after injury did not show a significant difference between genotypes (Supplementary Fig. 2i; P = 0.18). In both cases, the magnitude of effect for this single-gene manipulation (based on a 5% MNDCM content (Fig. 3f), which is half the MNDCM content of strains A and SWR (Fig. 1e) was expected to be modest. The noise and shallow dynamic range inherent in both analyses would make detection of a modest effect difficult to detect. Evaluation of cardiomyocyte size by WGA staining 1 month after injury showed no difference between knockout and wild-type littermates (Supplementary Fig. 2g,h), though, notably, this analysis was performed as an average of all cardio-myocytes of the left ventricle, was therefore disproportionately influenced by the much more abundant binucleated cardiomyocyte population, and was not specific to those cardiomyocytes that had incorporated EdU.
Overexpression of Tnni3k in zebrafish increases cardiomyocyte ploidy and impairs regeneration
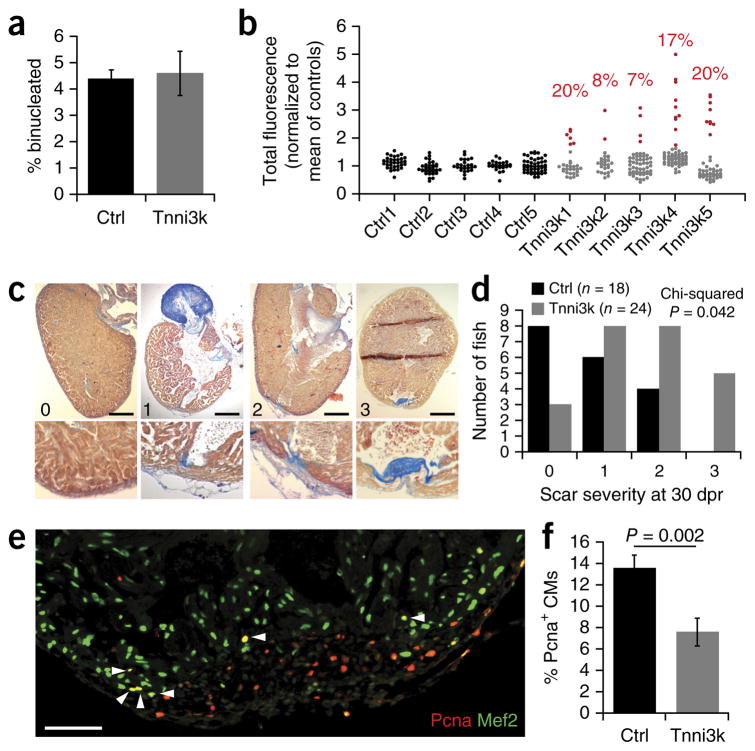
In contrast to mammals, zebrafish are able to fully regenerate after adult heart injury3, and almost all zebrafish cardiomyocytes are mononuclear and diploid35. Manipulations that impair zebrafish heart regeneration result in a persistent scar in the ventricle wall. Zebrafish have a tnni3k gene, although its expression and function have not been studied. We transgenically overexpressed mouse Tnni3k (mTnni3k) in zebrafish cardiomyocytes using a cmlc2 promoter (Supplementary Fig. 2j). Transgenic mTnni3k expression did not result in an increase in binucleated cardiomyocytes compared to cmlc2:GFP controls (Fig. 4a). To address cardiomyocyte nuclear ploidy, we approximated nuclear DNA content in single-cell preparations by DAPI fluorescence signal. Cardiomyocyte nuclei from control fish clustered tightly around a modal value of fluorescence intensity, whereas there was a prominent expansion to higher fluorescence in the corresponding population from cmlc2:mTnni3k fish (Fig. 4b). Using a strict cutoff threshold (3 s.d. from the mean of controls), which defined all control cardiomyocyte nuclei as diploid, we estimate that transgene expression resulted in 14.4 ± 2.8% of cardiomyocytes becoming polyploid (P < 0.001). This demonstrates that expression of mouse Tnni3k increases polyploidy in zebrafish cardiomyocytes just as it does in mouse cardiomyocytes.
Figure 4.
Overexpression of Tnni3k induces polyploidization and impairs regeneration in zebrafish hearts. (a) Quantification of the percent of binucleated cardiomyocytes in single-cell suspensions isolated from adult control (Ctrl; n = 5) and mouse Tnni3k transgenic (n = 5) zebrafish hearts. (b) Total fluorescence calculated from DAPI-stained cardiomyocyte nuclei across single-cell suspensions isolated from control and mTnni3k transgenic adult zebrafish. Red points represent nuclei that had a total fluorescence intensity greater than 3 s.d. above the mean of control preparations and are therefore presumed to be polyploid. Values indicated in red above each sample are the percentage of nuclei presumed to be polyploid. Mean of all Tnni3k samples = 14.4 ± 2.8% compared to 0% from control samples (P < 0.001; two-tailed Student t-test). (c) Examples of zebrafish hearts sectioned and stained for acid fuchsin orange G (AFOG) 30 d post-ventricular resection (dpr) with scores “0” (no scar, fully regenerated), “1” (small residual scar on either the endocardial or epicardial surface), “2” (small transmural scar) or “3” (large transmural scar) indicated. Scale bars represent 200 μm. (d) Quantification of scar severity at 30 dpr across control (Ctrl, n = 18) and Tnni3k transgenic (n = 24) fish. Chi-squared test P = 0.042. (e) An example image of the injury border zone stained with Pcna (red) and Mef2 (green) at 7 dpr. White arrowheads indicate double-positive cardiomyocyte nuclei. Scale bar represents 100 μm. (f) Quantification of Pcna+ cardiomyocyte (CM) nuclei in the border zone of 7-dpr heart sections represented as a percent of the total number of Mef2+ cardiomyocyte nuclei. Ctrl (n = 11 fish) and Tnni3k transgenic (n = 10 fish). Two-tailed Student t-test. Error bars represent s.e.m.
Adult fish carrying the cmlc2:mTnni3k transgene were seemingly normal. We used a scale of scar severity to evaluate the extent of heart regeneration 30 d after apex resection (Fig. 4c,d). Control fish mostly regenerated efficiently, with few fish showing a prominent scar. In contrast, transgenic fish showed a statistically significant elevation in the degree of scarring, which is indicative of impaired regeneration. This failure to regenerate was at least partially due to impairment in cardiomyocyte proliferation as determined by quantification of proliferating cell nuclear antigen (Pcna)-positive cardiomyocytes 7 d after resection (Fig. 4e,f). These results confirm that MNDCM content is associated with cardiomyocyte proliferation and heart regeneration in both mouse and zebrafish, even though the two start at opposite ends of the spectrum of regenerative competence.
DISCUSSION
A major conclusion from our analysis is the new perspective that heart injury does not inexorably lead to a permanent loss of myocardium and diminished function. Although this is generally thought to be the only outcome in mice and in humans, only a very small fraction of the natural genetic diversity of the mouse population has previously been subjected to direct experimental evaluation. Our results show that the percentage of MNDCMs before injury, as well as the extent of endogenous cellular regeneration and functional recovery after injury, can be redefined as variable traits subject to the influence of multiple genes. If 120 inbred mouse strains have a >7-fold range of variation in mononuclear cardiomyocyte content (Fig. 1a; >5-fold variation in MNDCM content (Fig. 1e)), it is expected that the much more genetically diverse human population would have an even larger degree of variation in this parameter. Indeed, natural or engineered loss of Tnni3k promotes higher MNDCM content in mice (as shown here), and the ExAC36 and DiscovEHR37 collections indicate that many heterozygous TNNI3K loss-of-function alleles exist in the human population, with some individuals identified to carry homozygous loss-of-function alleles. Additionally, while regeneration cannot be studied directly in humans, patients with initially similar degrees of infarction are known to have substantially different extents of ventricular remodeling and long-term outcomes38,39, and some of this divergence might be explained by genetic variation. These observations imply that natural regeneration of heart tissue occurs in mammals, albeit to a substantially variable degree based on genetic background.
Tnni3k had particularly high statistical prominence in the association analysis for frequency of mononuclear cardiomyocytes, but it is obvious that this trait is influenced by many genes. Indeed, although strain SJL has the same low level of Tnni3k expression as A and SWR (Supplementary Fig. 2b), it is among the lowest in mono-nuclear content whereas the others are the two highest (Fig. 1a), clearly demonstrating that Tnni3k is not the sole determinant of this feature. The Manhattan plot analysis (Fig. 3a) implicates several suggestive loci other than Tnni3k, and by refining the analysis other regions of the genome become more significant (data not shown). Notably, none of the significant or suggestive peaks we have observed so far overlap with genes that have been identified in previous studies to influence heart regeneration. This may be because the latter group were studied as candidate genes with a preconceived role in mind, whereas our forward genetic approach is unbiased. In addition, our primary phenotype was the percentage of mononuclear cardiomyocytes, whereas past studies have not evaluated this feature except perhaps as a correlate of their gene-specific manipulations. Ultimately, of course, we expect that genes evaluated through both approaches will converge into a unified genetic scheme of heart regeneration.
Our results support the premise that MNDCMs are one of the populations, if not the primary population, that accounts for proliferation in the adult heart after injury. A more definitive demonstration would require a cell lineage marker specific for MNDCMs, which is not yet available. In principle, two alternative populations may also or alternatively contribute to heart regeneration: non-cardiomyocyte progenitors that might differentiate into cardiomyocytes (discussed above) and mature polyploid cardiomyocytes that might dedifferen-tiate and become mitotically active40. It should be emphasized that models of regeneration invoking progenitor differentiation, dediffer-entiation of polyploid cardiomyocytes or proliferation of MNDCMs are not mutually exclusive.
Cardiomyocyte polyploidization is thought to occur through a modification in the cell cycle machinery that controls karyoki-nesis and/or cytokinesis25, although further insight is lacking. As the transition to the binucleated and/or polyploid state primarily occurs during the first postnatal week in mouse, we predict that the effect of Tnni3k manifests during this time. Indeed, Tnni3k protein abundance increases during this period (Supplementary Fig. 2k), suggesting that Tnni3k’s genetic function may be regulated in part by expression. In adult mice, injury activates cardiomyo-cytes to undergo DNA replication. In Tnni3k-expressing control C57Bl/6 mice, most such events led to either binucleation or tetra-ploidy, with relatively little proliferation (Fig. 3h–k). In contrast, in C57Bl/6-background Tnni3k knockout mice or in strain A mice that naturally lack Tnni3k, most EdU incorporation culminated in proliferation (Figs. 2 and 3). These divergent outcomes might simply reflect the relative levels of MNDCMs before injury. A related explanation might be that Tnni3k influences the propensity of injury-activated adult cardiomyocytes to interrupt cell cycle progression before cytokinesis. To the extent that the latter explanation may be relevant in the injured adult heart, we suggest that this is simply the recapitulation of the same process that occurs naturally during the early postnatal period. In either case, in mice, where most cardio-myocytes are binucleated, Tnni3k seems to preferentially influence cytokinesis, whereas Tnni3k overexpression in zebrafish promoted polyploidization rather than binucleation (Fig. 4a,b). Human car-diomyocytes, which are primarily mononuclear tetraploid, would also appear more prone to stalling before karyokinesis. A more general interpretation might therefore be that Tnni3k limits car-diomyocyte progression through the cell cycle, in the neonate or after adult injury, with arrest preferentially occurring before either karyokinesis or cytokinesis depending on species, and likely also depending on the involvement of many other polymorphic genes that converge with Tnni3k.
At present, we do not have direct insight to explain how Tnni3k participates in the process that sets the MNDCM percentage. Strong evidence indicates that oxidative stress is a major driver of the post-natal transition of cardiomyocytes the postmitotic state41. Previous studies implicated Tnni3k in responding to adult oxidative stress34, perhaps suggesting that Tnni3k similarly mediates cardiomyocyte response to oxidative stress experienced at birth. Some Tnni3k-interacting proteins have been identified31,42, but their in vivo relevance to Tnni3k function or to polyploidization has not yet been established. Furthermore, upstream processes that control Tnni3k kinase activity are unknown, and no direct kinase substrates have yet been confirmed in vivo. Tnni3k protein localizes to the sar-comere and may modulate the contractile apparatus43, although how this might relate to the persistence of MNDCMs after birth is likewise unknown.
A question that we cannot yet address is whether an increase in MNDCMs has any functional consequence in the uninjured heart. Adult mononuclear cardiomyocytes are generally smaller than binucleated cardiomyocytes (Fig. 1b) and may, if they resemble fetal cardiomyocytes, have a different metabolic profile or a less mature contractile apparatus, all of which might confer different mechanical or physiological properties. Echocardiographic comparison of mice before injury did not reveal differences (Fig. 1f and Supplementary Fig. 2c), although this is not the only means of addressing the question. Similarly, we do not yet know the upper limit of MNDCMs that the mammalian heart can accommodate. Two human families with conduction system disease (tachyarrhythmia) have been identified to carry nonsynonymous coding region point mutations in the TNNI3K gene44,45. If TNNI3K is associated with cardiomyocyte cell cycle progression, one possibility is that interference with the process of becoming polyploid, such as perhaps occurs in these two families, might lead to functional defects such as tachyarrhythmia. More broadly, these observations raise the fundamental question why such a high proportion of mammalian cardiomyocytes become binucleated or polyploid in the postnatal period, even if this comes with an apparent loss of regenerative ability after injury.
ONLINE METHODS
Mice
All animal experiments were performed following procedures reviewed by the Institutional Animal Care and Use Committee of the University of Southern California. All mouse strains of the Hybrid Mouse Diversity Panel (HMDP) were originally purchased from Jackson Laboratory, Bar Harbor, Maine; the “/J” designation for these mice has been omitted throughout the text. Tnni3k floxed mice were maintained on a C57Bl/6 background and only crossed to lines (Sox2-Cre or Myh6-Cre) that had been similarly maintained on a C57Bl/6 background. All experiments related to mononuclear cardiomyocyte content in HMDP strains used virgin females 8–10 weeks of age. Experiments related to Tnni3k knockouts used both males and females 8–10 weeks of age. There were no statistical differences between genders. Numbers of animals used for each experiment are compiled in Supplementary Table 1, the figure legends and the source data.
Single-cell ventricular suspensions (mouse)
Hearts were digested with 1 mg/mL collagenase type II via Langendorff retroaortic perfusion15. After digestion, atria and valves were removed and ventricular tissue alone was triturated in Kruftbrühe (KB) solution15, filtered through a 250-μm mesh, stained with LiveDead Fixable (ThermoFisher, L10120) for 20 min at room temperature and then fixed in 2% PFA at room temperature for 15 min. Fixed ventricular cell suspensions were stained for cTnT (1:1,000, Abcam ab8295) overnight at 4 °C followed by goat anti-mouse secondary (1:500, ThermoFisher A11001) and DAPI. Cell suspensions were then pipetted across a slide and coverslipped. Cardiomyocyte nucleation was quantified on an Olympus BX41 fluorescence microscope with a 20× objective. Only live car-diomyocytes were counted for mono-, bi-, tri- and tetranucleation; at least 200 cells were counted per heart. An unpaired, two-tailed Student t-test was used to assess statistical significance when only two groups were being compared. One-way ANOVA with Bonferroni correction was used to assess statistical significance across >2 groups.
Genome-wide association analysis (mouse)
120 strains of the HMDP were assessed for their percentage mononuclear cardiomyocytes (n = 2–8 animals per strain) as described above. Strain averages were arcsin transformed to normalize the distribution of the data. Association testing of each single nucleotide polymorphism was performed in the R software package as described46. P ≤ 4 × 10−6 is significant. Heritability was calculated as described21.
Bicor analysis of Tnni3k
A microarray analysis was performed on ventricular RNA (both chambers) collected from many strains in the HMDP46. These data were used to determine the correlation (bicor) between Tnni3k expression and frequency of mononuclear cardiomyocytes across strains. Bicor calculation and statistics were performed as described47.
DNA FISH (mouse)
Single-cell suspensions were plated in cardiomyocyte plating medium containing M199 plus 1 mg/ml 2,3-butanedione monoxime, 1% BSA and Pen-Strep onto glass coverslips coated with 0.1% polylysine and 10 μg/mL laminin (Sigma, L2020). Within 2 h of plating, cells were fixed with 4% PFA for 10 min at room temperature, washed in 0.075 M KCl for 10 min at 37 °C and treated with 3:1 methanol:acetic acid for 10 min at room temperature. After fixation, cells were treated with 0.5% Triton-X (in 2× SSC) for 10 min at room temperature, 50 μg/ml proteinase K (in 20 μM Tris-HCl pH7.5) for 8 min at room temperature and 100 μg/mL DNase-free RNase A (in 2× SSC) for 30 min at 37 °C. Washes in 2× SSC were performed between each treatment. Coverslips were then dehydrated in ice-cold 70%, 80%, 95%, 100% ethanol for 2 min each. Coverslips were air dried briefly then denatured together with probes at 76 °C in 50% formamide, 50% 2× SSC for 4 min before 48 h incubation at 37 °C in a humid chamber. After hybridization, coverslips were washed three times in 2× SSC and once each in 1× SSC, 1× SSC plus DAPI (5 min) and 1× SSC all prewarmed to 42 °C. DNA FISH probes were generated from BAC clones using fluorescently labeled nucleotides as described48. The BAC clones were Hoxc-488 (RP23-473J19; chr15), Sox2-555 (RP23-2B8; chr3), Hoxb-488 (RP23-290I2; chr11) and Hoxd-555 (RP24-398B4; chr2), and two were used at a time. Ploidy of mononuclear cardiomyocytes alone was assessed using a 63× oil objective on a Zeiss Axioimager fluorescence microscope. To be counted, ploidy had to be confirmed by two probes labeling distinct autosomes. Males 8 weeks old were used to assess ploidy of Myh6-Cre+;Tnni3kfl/fl compared to Myh6-Cre+;Tnni3kfl/+. Females 8 weeks old were used to assess ploidy of strains SJL, C57Bl/6, SWR and A. An unpaired, two-tailed Student t-test was used to assess statistical significance when only two groups were compared. One-way ANOVA with Bonferroni correction was used to assess statistical significance across >2 groups.
Surgeries (mouse) and EdU administration
Permanent ligation of the left artery descending (LAD) was performed under 2–4% isoflurane anesthesia as described49. Mice were administered oral meloxicam for pain immediately upon waking and for 2 d following surgery. For EdU+ cardiomyocyte quantifications in tissue sections, animals were injected with 10 mg/kg EdU (in sterile saline) intraperitoneally once daily for 7 d (days 4–10 after infarction). Animals were euthanized 30 d after infarction. For EdU+ cardiomyocyte quantifications on single-cell preparations or nuclear isolation, Alzet micro-osmotic pumps (model 1002) carrying 5 mg of EdU (50% DMSO, 50% saline) were implanted subcutaneously in the interscapular region on day 5 after infarction under isoflurane sedation and were removed on day 18 also under isoflurane sedation. A 48-h chase period was allowed after removal of the pump before euthanasia to ensure that any cell divisions initiated on day 18 had time to complete. For phospho-histone H3 cardiomyocyte quantifications in tissue sections, a separate cohort of mice underwent surgery, were not exposed to EdU during recovery, and were euthanized 11 d after infarction.
Echocardiography
Parameters of heart morphology and function were evaluated by echocardiography at the USC Molecular Imaging Core under 2% isoflurane anesthesia on uninjured animals at 7 weeks of age and then on animals at 3 d and 28 d after myocardial infarction. Images were captured with a VisualSonics Vevo2100 Ultrasound and data analyzed with Vevo2100 software package. Left ventricular diastolic and systolic dimensions, stroke volume and ejection fraction (EF) were measured from M-mode conversions of long- and short-axis views by a reader blinded to animal genotype. Animals that had an ejection fraction ≥60% at day 3 or died before the collection of the day 28 time point were excluded from the study. Percent improvement of function was calculated as follows: (day 28 EF – day 3 EF)/(day 3 EF). Multiple statistical tests were used, including one-way ANOVA with repeated measures, one-way ANOVA with Bonferroni correction to assess statistical significance across groups at individual time points, paired t-tests to assess statistical significance across time points within a single group, and Student t-tests to assess significance when combining high strains and low strains each into one group.
Histology (mouse)
Hearts were hung on a Langendorff apparatus, perfused retroaortically with 4% PFA for 15 min and further fixed overnight at 4 °C in 4% PFA. Fixed tissue was embedded in OCT after sucrose treatment and sectioned on a Leica CM1900 cryostat at 8 μm. Twelve series containing sections ~200 μm apart were collected for every heart.
Trichrome stain and analysis (mouse)
Cryosections of heart tissue were postfixed in Bouin’s solution at room temperature overnight. Masson’s tri-chrome stain was otherwise performed according to manufacturer′s protocol (Sigma). Five sections in the mid-papillary region of the left ventricle (~200 μm apart from each other) were selected for imaging and analysis. Pictures of each section were taken on a Nikon SMZ-U dissection scope with a Spot camera, holding all exposure and colorimetric variables constant. Left ventricles were digitally defined out and pasted on a black background (Supplementary Fig. 1f). Area of scar represented as a percentage of the total area of the left ventricle was calculated using FIJI imaging software. Briefly, the “Analyze → Measure” tool used to calculate area of a selected region was calibrated using an image of a metric ruler taken at the same magnification (1.0×) as the tissue sections. The “Image → Adjust → Color threshold” tool was used to select regions of interest. All colors were selected when measuring the total area of the left ventricle and only hues between 114 and 206 were used to calculate the area of the scar (blue). All other hue, saturation and brightness adjustments were held constant across samples. Analysis was performed by a blinded investigator. An unpaired Student t-test was used to assess statistical significance when only two groups were being compared. One-way ANOVA with Bonferroni correction was used to assess statistical significance across >2 groups.
Immunofluorescence and analysis (mouse)
Cryosections of heart tissue (30 d after infarction) were stained with the Click-it EdU kit (ThermoFisher; C10339) according to manufacturer’s protocol, followed by rabbit anti-Myl2 (1:200, Abcam ab79935), Alexa Fluor secondary antibody (1:500, ThermoFisher A11008) and DAPI using standard procedures. Separately, tissue sections from 11 d after infarction were treated for 30 min at room temperature with 0.03% Sudan black B (SBB) dissolved in 70% ethanol. After SBB incubation, sections were stained for rabbit anti-phospho-histone H3 (1:500, EMD Biosciences 06-570) and goat anti-cardiac troponin C (1:500, Abcam ab30807) using otherwise standard procedures (Alexa Fluor donkey anti-rabbit 546, ThermoFisher A10040 and Alexa Fluor donkey anti-goat 488, ThermoFisher A11055, respectively). Pictures were taken with a 20× objective using an Olympus BX41 fluorescence microscope and Spot Pursuit camera. Ten total pictures were taken for each heart. Regions were randomly selected based only on the inclusion of scar (Myl2 negative) being on the border of the image, therefore limiting the quantification to cardiomyocytes of the border zone. Quantifications of EdU+ (or pHH3+) cardiomyocyte nuclei were performed using FIJI imaging software by first tagging every DAPI+ nucleus that fell within a Myl2+ cell and then assessing whether any of those were EdU+; ~500 total cardiomyocyte nuclei were counted per heart. Pictures were taken and analysis was performed by a blinded investigator. While quantifications were performed on pictures taken from a standard fluorescence scope, EdU+ nuclei inclusion within a Myl2+ cell was later confirmed by confocal microscopy (Fig. 2a; Zeiss LSM800). An unpaired Student t-test was used to assess statistical significance when only two groups were being compared. One-way ANOVA with Bonferroni correction was used to assess statistical significance across >2 groups.
WGA stain and analysis of cell size
Cryosections of heart tissue (30 d after infarction) were stained with Alexa Fluor 488–conjugated wheat germ agglutinin (WGA; ThermoFisher W11261) according to the manufacturer’s protocol with a single exception: sections were incubated in WGA-488 for 1 h at 37 °C rather than 10 min. 20× pictures were taken on a Zeiss LSM800 confocal microscope specifically targeting regions of the left ventricle where cross-sectioned cardiomyocytes were found, avoiding longitudinally sectioned cells. FIJI software was used to trace the outline of the cardiomyocyte based on the WGA stain and area was calculated in pixels. At least 200 cells were analyzed per animal. Analysis was performed by a blinded investigator. An unpaired, two-tailed Student t-test was used to assess statistical significance.
Nuclear isolation and flow cytometry (mouse)
Nuclei were isolated using an adaptation of the Nuclear Isolation Kit (Sigma catalog number NUC201) protocol. Briefly, hearts isolated from animals administered EdU via an osmotic pump (described above) were digested via Langendorff perfusion with 1 mg/ml collagenase type II. After digestion, ventricles were removed, diced and snap frozen in liquid nitrogen. Ten milliliters of lysis buffer plus 1 mM DTT (with Triton omitted) were added to frozen tissue and tissue homogenized first with an Omni TH115 homogenizer for 10 s on low speed followed by 10 strokes with a loose-fitting Dounce homogenizer and 10 strokes with a tight-fitting Dounce homogenizer. Homogenized tissue was strained through a 70-μm mesh, followed by two 40-μm meshes. Twenty milliliters of 2 M sucrose buffer (plus 1 mM DTT) was added to each sample and mixed by inversion. In an ultracentrifuge tube, 10 ml of 2 M sucrose buffer was added to the bottom of the tube and the sample (now 30 ml) was poured gently on top. Samples were spun in a Beckman Coulter ultracentrifuge, rotor SW-28, at 28,000 RPM for 1 h at 4 °C. Nuclei pelleted to the bottom of the tube. Sucrose was removed and pellets were resuspended in 5% BSA in PBS. Nuclei were fixed with 2% PFA for 10 min at room temperature. Fixed nuclei were stained for preconjugated cTnI–Alexa Fluor 647 (BD Biosciences 564409) or IgG2a–Alexa Fluor 647 (BD Biosciences, 558713) as described26. Nuclei were then permeabilized with 0.1% Triton and stained for EdU according to the supplier’s procedure and with DyeCycle Violet (ThermoFisher V35003). All washes were performed with 5% BSA in PBS and all spins were performed at 600–800g on a standard benchtop centrifuge.
FACS was performed on a BD Aria II flow cytometer equipped with 488, 561 and 640 nm lasers and configured with a 70-μm nozzle. Nuclei were identified based on forward and side scatter profile and positivity for DyeCycle Violet. Gates were set based on fluorescence minus one (FMO) controls, isotype staining (cTnI) and/or uninjected control hearts (EdU). For sorting of 2N and 4N nuclei, gates were set on contour plots of cTnI−EdU− cells. Plots were prepared with FlowJo software (Version X, FlowJo, LLC). Nuclei from the cTnI+EdU+ 4N and >4N gates were sorted into 5% BSA/PBS, pipetted onto slides and visualized under a microscope. For the flow analysis performed on strain A, it was determined that 35% of 4N nuclei were in fact doublets (one EdU+ nucleus and one EdU− nucleus; n = 65 nuclei observed); and 85% of >4N nuclei were in fact clusters (only one EdU+ nucleus with ≥1 negative nucleus; n = 20). In the case of Tnni3k knockout and control preparations, 23% of 4N nuclei were doublets (n = 60 nuclei observed) and 42% of >4N nuclei were clusters (n = 12). These numbers were used to correct the percentages on the FACS plots for their respective experiments.
Immunoblots
Atrial or ventricular tissue was snap frozen in liquid nitrogen then homogenized with an OMNI TH homogenizer in RIPA buffer. Immunoblotting was performed by standard protocols using anti-Tnni3k (1:1,000, Abcam ab111140), anti-Gapdh (1:5,000, Cell Signaling Technology 2118s) and HRP-coupled secondary (1:10,000, Jackson ImmunoResearch sc2040) antibodies. The immunoblot across Tnni3k genotypes (Fig. 2c) was performed twice with multiple animals for each genotype included for each blot. The immunoblot across strains (Supplementary Fig. 2b) was performed three times. The immunoblot across developmental ages (Supplementary Fig. 2k) was performed three times. Uncropped image files can be found in Supplementary Data.
Zebrafish
All animal experiments were performed following procedures reviewed by the Institutional Animal Care and Use Committee of the University of Southern California. Transgenic zebrafish (Danio rerio) lines were generated by standard protocols50. Briefly, the full-length mouse Tnni3k cDNA sequence was PCR-amplified, confirmed by sequencing and inserted into pDONR221 via Gateway cloning (ThermoFisher) to form pME-Tnni3k, which was combined with p5E-cmlc2, p3E-2a-nlsGFP-polyA and pDestTol2pA2 to generate cmlc2:Tnni3k-2a-nlsGFP-polyA. p5E-cmlc2, pME-nlsGFP, p3E-polyA and pDestTol2pA2 were assembled to make cmlc2:nlsGFP:pA. Transgenic zebrafish lines carrying these constructs were produced via Tol2-mediated transgenesis, and stable transgenic F1 and F2 progeny were used for heart regeneration and nuclear analyses. Fish used for experimental analysis were at least 3 months of age and at least 2.5 cm long from mouth to tail base.
Single-cell ventricular suspensions and nucleation and ploidy analyses (zebrafish)
Pools of three ventricles from naive control fish (cmlc2:nlsGFP) and Tnni3k transgenics (cmlc2:Tnni3k-2a-nlsGFP, abbreviated as cmlc2:mTn-ni3k in the text) were digested in 5 mg/ml collagenase type II and 5 mg/ml collagenase type IV as described51. Following stopping buffer washes, cells were filtered through a 250-μm mesh and fixed with 2% PFA for 15 min at room temperature. Cell suspensions were stained for mouse anti-Myh1e (1:50, DSHB MF-20) with an Alexa Fluor secondary (1:500, ThermoFisher A11001) and DAPI using standard procedures. Cells were pipetted across a slide and coverslipped. Cardiomyocyte nucleation was quantified on an Olympus BX41 fluorescence microscope with a 20× objective. ~180 cells were counted per pool. To estimate ploidy, pictures of single-cell suspensions were captured using a Spot camera holding all exposure settings constant. Fluorescence intensity of DAPI nuclear stain was measured using FIJI software. Briefly, nuclei were identified and outlined with a standard threshold requirement for all samples. A histogram of each nucleus was generated and total fluorescence was calculated as area under the curve, thereby accounting for both size of the nucleus and fluorescence intensity. Total fluorescence was normalized to the mean value of the control samples. Quantifications were performed by an investigator blinded to the genotypes of the fish samples. An unpaired, two-tailed Student t-test was used to assess statistical significance.
Resection surgery (zebrafish)
Control and Tnni3k transgenic fish were subjected to ventricle resection under 4.2% tricaine anesthesia as previously performed3,52. Fish were allowed to recover for 30 d before euthanasia and heart dissection.
Histology and AFOG stain (zebrafish)
Hearts were fixed overnight in 4% PFA, dehydrated and paraffin embedded, and then serial sectioned on a Leica RM2135 microtome at 7 μm and stained with AFOG as described3,52. Severity of scar was assessed by a scoring system: 0 = no visible scar; 1 = mild blue stain on either the epicardial or endocardial surface (but not transmural); 2 = small blue collagen stain extending from endocardial to epicardial surface (transmural); 3 = larger or thick transmural blue collagen stain. Two independent blinded investigators assessed control and mTnni3k fish for scar severity. Statistical significance was assessed by a chi-squared test.
Pcna stain and quantification (zebrafish)
Sections were stained for mouse anti-Pcna (1:200, ThermoFisher 13-3900) paired with goat anti-mouse Alexa Fluor 546 (ThermoFisher A11030), rabbit anti-Mef2 (1:50, Santa Cruz Biotech sc313) paired with goat anti-rabbit Alexa Fluor 488 (ThermoFisher A11008), and DAPI following standard procedures. 20× images were captured on an Olympus BX41 fluorescence microscope and Spot Pursuit camera. Three total pictures were taken for each heart, each being ~80–100 μm apart. Quantifications of EdU+ cardiomyocyte nuclei were performed using FIJI imaging software by first tagging every DAPI+Mef2+ nucleus and then assessing whether any of those were Pcna+; ~500 total cardiomyocyte nuclei were counted per heart. Pictures were taken and analysis was performed by a blinded investigator. Statistical significance was assessed by an unpaired, two-tailed, Student t-test.
Data availability
SNP data relevant to the HMDP are publically available from The Center for Genome Dynamics at The Jackson Laboratory and can be found at http://cgd.jax.org/datasets/diversityarray.shtml. The microarray data used to perform the bicor analysis are accessible from Gene Expression Omnibus with accession code GSE48760. Source data are provided online for Figures 1d,f–h, 2b,e, 3d,e,g–j and 4a,f and Supplementary Figures 1b–e,h and 2c–f,h,i.
Supplementary Material
Acknowledgments
M.P. was funded by CIRM Training Grant TG2-01161, an award from the University of Southern California’s Provost Office and NHLBI NRSA 1F32HL124932. Zebrafish work was supported in part by a Doerr Family Foundation award provided to M.P. and L.B. S.R.K. was supported by K08HL121191. Purchase and husbandry of HMDP strains was supported by grants HL123295 and HL114437 to A.J.L. and grant NS083265 to T.M.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.P., L.B., B.V.H., P.G. and A.S. conducted laboratory analyses. C.D.R. contributed bioinformatics insight and analysis, and M.D. provided FISH probes. S.P. and T.I.F. provided Tnni3k conditional mice. M.D., Y.H., Y.Y. and H.S. gave technical guidance. H.A. and A.J.L. provided insight into the use of the HMDP, and A.J.L. provided most of the HMDP mice. J.G.C., C.-L.L., T.M. and A.J.L. provided conceptual advice. S.R.K. read and interpreted echocardiographic data and provided conceptual advice. The overall project was conceived by, data were interpreted by, and the manuscript was written by M.P. and H.M.S.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 2.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 5.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali SR, et al. Existing cardiomyocytes generate cardiomyocytes at a low rate after birth in mice. Proc Natl Acad Sci USA. 2014;111:8850–8855. doi: 10.1073/pnas.1408233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 10.Xavier-Vidal R, Mandarim-de-Lacerda CA. Cardiomyocyte proliferation and hypertrophy in the human fetus: quantitative study of the myocyte nuclei. Bull Assoc Anat (Nancy) 1995;79:27–31. [PubMed] [Google Scholar]
- 11.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mollova M, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci USA. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye L, et al. Cardiomyocytes in young infants with congenital heart disease: a three-month window of proliferation. Sci Rep. 2016;6:23188. doi: 10.1038/srep23188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 15.Malliaras K, et al. Cardiomyocyte proliferation and progenitor cell recruitment underlie therapeutic regeneration after myocardial infarction in the adult mouse heart. EMBO Mol Med. 2013;5:191–209. doi: 10.1002/emmm.201201737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, et al. Adolescent feline heart contains a population of small, proliferative ventricular myocytes with immature physiological properties. Circ Res. 2007;100:536–544. doi: 10.1161/01.RES.0000259560.39234.99. [DOI] [PubMed] [Google Scholar]
- 17.Kühn B, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 18.Liao HS, et al. Cardiac-specific overexpression of cyclin-dependent kinase 2 increases smaller mononuclear cardiomyocytes. Circ Res. 2001;88:443–450. doi: 10.1161/01.res.88.4.443. [DOI] [PubMed] [Google Scholar]
- 19.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett BJ, et al. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20:281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett BJ, et al. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 2015;11:e1005711. doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brainard RE, et al. High fat feeding in mice is insufficient to induce cardiac dysfunction and does not exacerbate heart failure. PLoS One. 2013;8:e83174. doi: 10.1371/journal.pone.0083174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orr-Weaver TL. When bigger is better: the role of polyploidy in organogenesis. Trends Genet. 2015;31:307–315. doi: 10.1016/j.tig.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zebrowski DC, Engel FB. The cardiomyocyte cell cycle in hypertrophy, tissue homeostasis, and regeneration. Rev Physiol Biochem Pharmacol. 2013;165:67–96. doi: 10.1007/112_2013_12. [DOI] [PubMed] [Google Scholar]
- 26.Bergmann O, et al. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res. 2011;317:188–194. doi: 10.1016/j.yexcr.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Fukata M, et al. Contribution of bone marrow-derived hematopoietic stem/progenitor cells to the generation of donor-marker+ cardiomyocytes in vivo. PLoS One. 2013;8:e62506. doi: 10.1371/journal.pone.0062506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JMF, et al. Circulating cells contribute to cardiomyocyte regeneration after injury. Circ Res. 2015;116:633–641. doi: 10.1161/CIRCRESAHA.116.304564. [DOI] [PubMed] [Google Scholar]
- 30.Ghazalpour A, et al. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, et al. Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin I. J Mol Med (Berl ) 2003;81:297–304. doi: 10.1007/s00109-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler FC, et al. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009;5:e1000647. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lodder EM, et al. Dissection of a quantitative trait locus for PR interval duration identifies Tnni3k as a novel modulator of cardiac conduction. PLoS Genet. 2012;8:e1003113. doi: 10.1371/journal.pgen.1003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vagnozzi RJ, et al. Inhibition of the cardiomyocyte-specific kinase TNNI3K limits oxidative stress, injury, and adverse remodeling in the ischemic heart. Sci Transl Med. 2013;5:207ra141. doi: 10.1126/scitranslmed.3006479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi K, Poss KD. Cardiac regenerative capacity and mechanisms. Annu Rev Cell Dev Biol. 2012;28:719–741. doi: 10.1146/annurev-cellbio-101011-155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dewey FE, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:1549. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 38.Taylor GJ, et al. Predictors of clinical course, coronary anatomy and left ventricular function after recovery from acute myocardial infarction. Circulation. 1980;62:960–970. doi: 10.1161/01.cir.62.5.960. [DOI] [PubMed] [Google Scholar]
- 39.White HD, et al. Left ventricular end-systolic volume as the major determinant of survival after recovery from myocardial infarction. Circulation. 1987;76:44–51. doi: 10.1161/01.cir.76.1.44. [DOI] [PubMed] [Google Scholar]
- 40.D’Uva G, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 41.Puente BN, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, et al. AOP-1 interacts with cardiac-specific protein kinase TNNI3K and down-regulates its kinase activity. Biochemistry (Mosc ) 2007;72:1199–1204. doi: 10.1134/s0006297907110053. [DOI] [PubMed] [Google Scholar]
- 43.Tang H, Xiao K, Mao L, Rockman HA, Marchuk DA. Overexpression of TNNI3K, a cardiac-specific MAPKKK, promotes cardiac dysfunction. J Mol Cell Cardiol. 2013;54:101–111. doi: 10.1016/j.yjmcc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theis JL, et al. TNNI3K mutation in familial syndrome of conduction system disease, atrial tachyarrhythmia and dilated cardiomyopathy. Hum Mol Genet. 2014;23:5793–5804. doi: 10.1093/hmg/ddu297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xi Y, et al. Whole exome sequencing identifies the TNNI3K gene as a cause of familial conduction system disease and congenital junctional ectopic tachycardia. Int J Cardiol. 2015;185:114–116. doi: 10.1016/j.ijcard.2015.03.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rau CD, et al. High-density genotypes of inbred mouse strains: improved power and precision of association mapping. G3 (Bethesda) 2015;5:2021–2026. doi: 10.1534/g3.115.020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao E, et al. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ Res. 2010;107:1445–1453. doi: 10.1161/CIRCRESAHA.110.223925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 51.Sander V, Suñe G, Jopling C, Morera C, Izpisua Belmonte JC. Isolation and in vitro culture of primary cardiomyocytes from adult zebrafish hearts. Nat Protoc. 2013;8:800–809. doi: 10.1038/nprot.2013.041. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y, et al. Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One. 2013;8:e67266. doi: 10.1371/journal.pone.0067266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SNP data relevant to the HMDP are publically available from The Center for Genome Dynamics at The Jackson Laboratory and can be found at http://cgd.jax.org/datasets/diversityarray.shtml. The microarray data used to perform the bicor analysis are accessible from Gene Expression Omnibus with accession code GSE48760. Source data are provided online for Figures 1d,f–h, 2b,e, 3d,e,g–j and 4a,f and Supplementary Figures 1b–e,h and 2c–f,h,i.