Abstract
Tripartite motif (TRIM) proteins comprise a large family of RING-type ubiquitin E3 ligases that regulate important biological processes. An emerging general model is that TRIMs form elongated antiparallel coiled-coil dimers that prevent interaction of the two attendant RING domains. The RING domains themselves bind E2 conjugating enzymes as dimers, implying that an active TRIM ligase requires higher-order oligomerization of the basal coiled-coil dimers. Here, we report crystal structures of the TRIM23 RING domain in isolation and in complex with an E2-ubiquitin conjugate. Our results indicate that TRIM23 enzymatic activity requires RING dimerization, consistent with the general model of TRIM activation.
INTRODUCTION
Tripartite motif (TRIM) proteins comprise a large family of E3 ligases, with about 100 different human proteins identified1. TRIM proteins regulate many different cellular processes and pathways, and in particular have important roles in antiviral defense, activation of the innate immune response, inflammation, and development of cancer. TRIMs share a common domain organization – called the tripartite or RBCC motif – which consists of an N-terminal RING domain, followed by one or two B-box domains, a coiled-coil domain, and a C-terminal domain, which is variable across the protein family2 (Fig. 1A). The RING domain functions as a ubiquitin (Ub) E3 ligase and cooperates with E2 enzymes to generate polyUb chains. Recent studies have shown that despite considerable sequence divergence, the coiled-coil domains of different TRIM proteins share a common quaternary fold3–6. Two subunits pack in an anti-parallel manner, forming an elongated rod-shaped scaffold with a RING domain on each end; the two RING domains are separated by 17–20 nm and so cannot directly interact with each other. On the other hand, other studies have shown that the isolated RING domains of TRIM5α, TRIM25, and TRIM32 engage E2-Ub conjugates and catalyze Ub conjugation as close-packed dimers7–9. Taken together, these observations imply that the catalytically active form of TRIMs comprises at least 2 coiled-coil mediated dimers (4 protein molecules). Thus, the polyubiquitination activity of a TRIM protein requires higher-order assembly or oligomerization beyond its basal, coiled-coil mediated dimeric quaternary fold.
Figure 1.
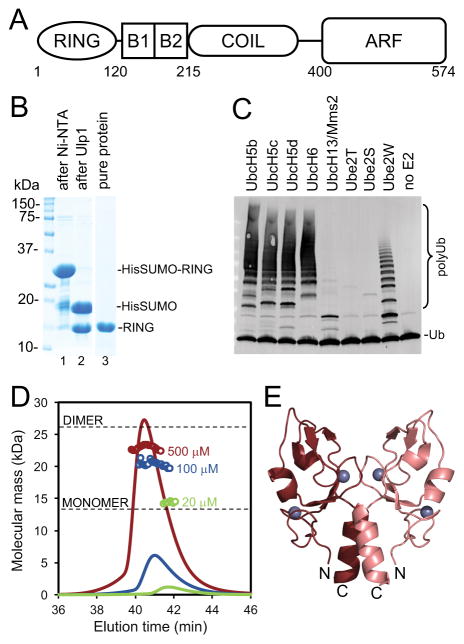
(A) Schematic diagram of the domain organization of TRIM23 with approximate residue boundaries. (B) Coommassie-stained SDS-PAGE gels showing purification of the TRIM23 RING domain, after Ni-NTA step (lane 1), cleavage with Ulp1 to remove the tag (lane 2), and the final sample (lane 3). (C) Immunoblot with anti-Ub antibody showing reaction products after 20 min incubation at 37 °C. Reactions contained E1 (100 nM), indicated E2s (1 μM, except UbcH13/Mms2 which was at 0.25 μM), TRIM23 RING (5 μM), Ub (40 μM), and Mg-ATP (5 mM). (D) SEC-MALS analysis of the purified TRIM23 RING domain. Curves show the refractive index signal, and open circles show the calculated masses of the eluting components. Black horizontal lines indicate the monomer and dimer masses calculated from the protein sequence. (E) Crystal structure of the TRIM23 RING domain dimer. Zinc atoms are shown as gray spheres, and the N and C-termini of each subunit are indicated.
The above requirement for RING activation is exploited by the cell to regulate when, where, and how TRIM proteins synthesize polyUb chains. One mechanism is exemplified by TRIM5α, which inhibits replication of HIV-1 and other retroviruses in a ubiquitin-dependent manner10. TRIM5α assembles into a hexagonal network that surrounds the HIV-1 capsid, and this network promotes dimerization of the associated RING domains7,11,12. The activated RING domains synthesize both self-attached and free polyUb chains to inhibit viral replication13–15. Another example is TRIM25, whose catalytic activity is induced by multivalent binding to its ubiquitination substrate, RIG-I9. These examples illustrate how a general biochemical property can be translated into different pathway-specific mechanisms to control a common enzymatic activity.
In this study, we analyzed human TRIM23 in order to determine if this protein follows the model of catalytic activation described above. In contrast to the TRIM5α and TRIM25 RING domains that were observed to dimerize only in complex with E2 or E2-Ub7–9, the isolated TRIM23 RING domain had a marked, although weak propensity to dimerize independently. We report crystal structures of the TRIM23 RING domain, both alone and in complex with a stable E2-Ub conjugate. In both structures, the RING domain adopts the same dimeric quaternary fold, indicating that TRIM23 also requires RING dimerization to catalyze polyUb synthesis. Our studies therefore support a general model wherein higher-order oligomerization or assembly regulates catalytic activation of TRIM proteins.
METHODS
Recombinant protein production
The TRIM23 RING domain was sub-cloned from a full-length construct (OpenBio 4811707) into a pET vector with a His6-SUMO leader sequence. The fusion protein was expressed in E. coli BL21(DE3) cells through IPTG induction, and initially purified on Ni-NTA resin (Qiagen). The His6-SUMO tag was removed by overnight cleavage with Ulp1 protease, and the RING was purified to homogeneity by ion exchange and size exclusion. Pure protein was exchanged into buffer (20 mM Tris, pH 7.5, 0.1 M NaCl, 1 μM ZnCl2, 1 mM Tris(2-carboxyethyl)phosphine (TCEP)) during the sizing step. UbcH5b C85K/S22R conjugated to Ub through an isopeptide bond was prepared as described for UbcH139. Ubiquitination reagents (E1, E2, Ub) were either prepared in-house or purchased from UBPBio.
Biochemical characterization
Ubiquitination assays were performed as described for the TRIM25 RING9, with 5 μM E3 and 1 μM E2. SEC-MALS analysis was carried out by injecting 100 μL of purified protein and then developing on a Superdex 200 column (GE Healthcare) equilibrated with running buffer (20 mM Tris, pH 7.5, 0.1 M NaCl, 1 mM TCEP). Particle size and refractive index data were collected with a Dionex UltiMate3000 system (ThermoFisher) equipped with an in-line miniDAWN TREOS static light scattering detector (Wyatt Technology) and Optilab T-rEX differential refractometer (Wyatt Technology).
Structure determination
Crystals were grown at 17 °C in hanging drops by mixing equal volumes of protein and precipitant. The RING domain alone (stock concentration = 8.8 mg/mL) crystallized in 0.1 M Tris, pH 8.5, 25% (w/v) PEG 3,350, and the RING/E2-Ub complex (7.5 mg/mL) crystallized in 0.1 M Bis-Tris, pH 5.5, 0.2 M CaCl2, 17% (w/v) PEG 3,350. Crystals were cryoprotected with 25% (v/v) ethylene glycol and flash-frozen in liquid nitrogen. Diffraction data were collected at SER-CAT beamline 22ID at the Advanced Photon Source and processed using HKL200016. The RING structure was solved by molecular replacement with a polyalanine homology model built with SWISS-MODEL17. The RING/E2-Ub structure was solved by molecular replacement with a polyalanine version of TRIM25 RING/UbcH13-Ub structure9. Structure refinement and model building were performed with PHENIX18 and Coot19. Figures were prepared with PyMol (Delano Scientific). Structure statistics are provided in Table 1. Coordinates and structure factors were deposited at the RCSB PDB database with ID codes 5VZV (RING) and 5VZW (RING/E2-Ub).
Table 1.
Statistics for the TRIM23 RING domain crystal structures.
| RING | RING/E2-Ub | |
|---|---|---|
| PDB ID | 5VZV | 5VZW |
| Diffraction Data | ||
| Beamline | APS 22ID | APS 22ID |
| Wavelength, Å | 1.000 | 1.000 |
| Space group | C2221 | P212121 |
| Unit cell | ||
| dimensions, Å | a = 91.7, b = 142.5, c = 45.4 | a = 52.8, b = 109.3, c = 123.0 |
| angles, ° | α = β = γ = 90 | α = β = γ = 90 |
| Resolution range, Å | 50-1.80 (1.86-1.80) | 50-2.28 (2.38-2.28) |
| Rmerge/Rpim | 0.10 (0.96) / 0.03 (0.43) | 0.12 (1.2) / 0.03 (0.44) |
| Mean I/σ<I> | 17.8 (1.2) | 20.1 (1.3) |
| Completeness, % | 90.5 (45.7) | 99.9 (98.6) |
| Average redundancy | 12.5 (4.6) | 13.7 (7.8) |
| Wilson B-factor, Å2 | 17.1 | 58.8 |
| Refinement | ||
| Resolution range | 38.57-1.81 (1.86-1.81) | 47.58-2.28 (2.34-2.28) |
| No. of unique reflections | 20,670 (100) | 33,020 (1,872) |
| Reflections in free set | 1,998 (11) | 1,855 (120) |
| Rwork/Rfree, % | 18.6 (27.9) / 22.1 (28.4) | 20.7 (27.5) / 24.1 (39.0) |
| No. of nonhydrogen atoms | ||
| protein | 3,660 | 4,685 |
| zinc | 6 | 4 |
| water | 163 | 51 |
| Average B-factor, Å2 | ||
| protein | 26.6 | 66.9 |
| zinc | 16.4 | 52.5 |
| water | 32.8 | 53.5 |
| Coordinate deviations | ||
| bond lengths, Å | 0.004 | 0.003 |
| bond angles, ° | 0.697 | 0.569 |
| Ramachandran favored/outliers, % | 100 / 0 | 98.5 / 0 |
| MolProbity clashscore | 1.10 | 1.40 |
Values in parenthesis are for the highest resolution shell.
RESULTS AND DISCUSSION
The human TRIM23 RING domain (residues 1-123) was expressed as a polyhistidine-tagged SUMO-fusion protein and purified to homogeneity after cleavage of the leader sequence (Fig. 1B). The purified RING domain was functional, and upon screening a panel of E2 conjugating enzymes was found to catalyze polyubiquitination reactions with the following E2s: UbcH5b, UbcH5c, UbcH5d, UbcH6, UbcH13/Mms2, and Ube2W (Fig. 1C). Previous studies have shown that the equivalent RING domains of TRIM5α and TRIM25 were monomeric in isolation7–9, whereas the TRIM32 RING was dimeric8. We therefore analyzed the TRIM23 RING domain by using SEC-MALS (size exclusion chromatography coupled with multi-angle light scattering) to determine its oligomeric state (Fig. 1D). When injected at 20 μM concentration, the protein eluted as a single peak (green curve), with a calculated molecular weight (MW) close to that of a monomer (14.3 kDa, expected MW from sequence = 13.1 kDa). When injected at higher concentrations (100 μM, blue and 500 μM, maroon), the peak eluted at progressively earlier times with correspondingly higher apparent MW closer to that of a dimer. These results indicated that the purified TRIM23 RING domain was under dynamic monomer-dimer equilibrium in solution. On the basis of the above data, we estimate that the dimer dissociation constant is around 100–200 μM. Thus, like TRIM32 but unlike TRIM5α and TRIM25, the TRIM23 RING domain can independently dimerize in solution.
We obtained well-diffracting crystals of the isolated RING domain from primary crystallization screens. The refined structure consisted of 3 molecules in the asymmetric unit, with each monomer having well-defined electron density spanning residues 28–103. Of the three chains in the asymmetric unit, two formed a non-crystallographic dimer, whereas the third dimerized with its symmetry-related mate. The crystallographically distinct dimers were very similar with a root mean square deviation (RMSD) of 0.39 Å over equivalent Cα atoms and 0.43 Å over backbone (N, Cα, C, and O) atoms.
The TRIM23 RING dimer is organized around a 2-helix bundle made up of residues that are C-terminal to the canonical zinc-coordinating folds in primary sequence (Fig. 1E). Additional contacts are contributed by N-terminal residues, which insert hydrophobic sidechains into the grooves between the two α-helices. Thus, the N-terminal and C-terminal residues that flank the zinc lobes make up a “pedestal” upon which the zinc lobes sit. This arrangement is very similar to the structures of E2-bound RING dimers from other TRIM proteins, except that in those cases the “pedestal” is a 4-helix bundle because the N-terminal residues also fold into an α-helix7–9. More importantly, these results suggested that the dimer made by the TRIM23 RING domain in solution is already in the same configuration as the E2-bound dimer.
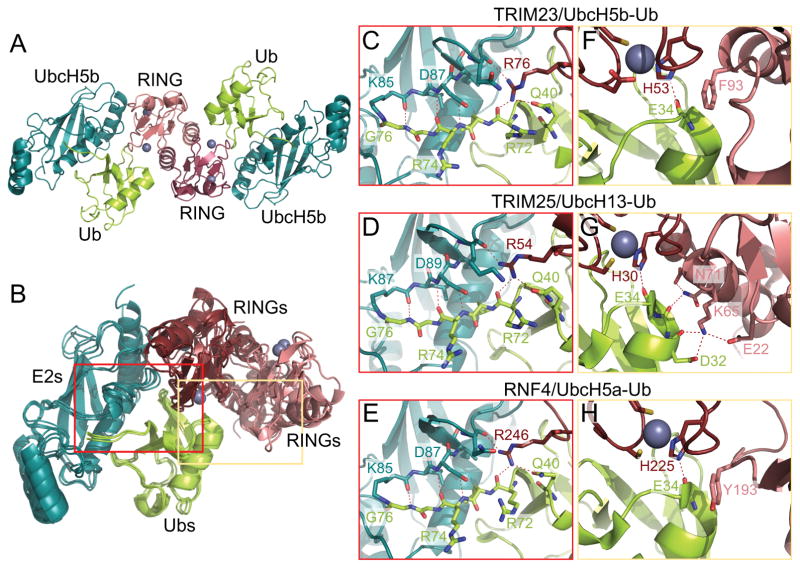
To confirm that the E2-bound dimer is similar to the uncomplexed dimer, we also crystallized the TRIM23 RING domain in complex with UbcH5b conjugated to Ub. To prevent loss of the Ub moiety, we used an established strategy of stably conjugating Ub to the E2 enzyme through an isopeptide linkage, instead of the labile, native thioester20. Despite local structural differences, isopeptide-linked E2-Ub conjugates have been shown to recapitulate their catalytically primed structures when crystallized with RING E3 ligases8,9,20. Our final refined structure contained one RING dimer flanked by two UbcH5b-Ub conjugates in the asymmetric unit (Fig. 2A). This quaternary arrangement is very similar to equivalent RING/E2 and RING/E2-Ub structures of TRIM5α7, TRIM258,9, and the more distantly related RNF420 and BIRC721 (Fig. 2B shows a superposition of the TRIM23 RING/E2-Ub structure with the TRIM25 and RNF4 complexes).
Figure 2.
(A) Crystal structure of the TRIM23 RING domain dimer in complex with UbcH5b-Ub. (B) Superposition of the TRIM23, TRIM259, and RNF420 RING/E2-Ub structures. (C–E) Conserved interactions between the first RING domain, E2, and Ub, shown for TRIM23 (C), TRIM25 (D), and RNF4 (E). (F–H) Second set of RING/Ub interactions, shown for TRIM23 (F), TRIM25 (G), and RNF4 (H). These panels also show the conserved hydrogen bond between a zinc-coordinating histidine (His53 in TRIM23) in the first RING and the Ub backbone. Landmark residues are labeled and hydrogen bonds are indicated by dashed lines.
The unbound and E2-bound TRIM23 RINGs were highly similar, with average RMSD of 0.36 Å over equivalent Cα atoms and 0.37 Å over backbone atoms. Indeed, there were remarkably few differences between the bound and unbound states, apart from side chain rotamers buried within the contact sites. These results imply that the TRIM23 RING dimer has a relatively rigid quaternary fold, and that the E2 and Ub-binding surfaces are “pre-made” prior to E2-Ub binding.
The two RING domains have distinct interactions with the E2-Ub conjugate and cooperate in facilitating the ubiquitination reaction. One TRIM23 RING binds both the E2 and Ub moieties and provides an “arginine finger” (Arg76) to coordinate a hydrogen bond network that packs the Ub C-terminus against a shallow groove leading to the UbcH5b active site (Fig. 2C). This set of interactions is highly conserved, as seen in crystal structures of divergent dimeric RINGs in complex with various E2-Ub conjugates8,9,20,21 (e.g., TRIM25 in Fig. 2D and RNF4 in Fig. 2E). On the other hand, the second RING domain interacts only with the Ub moiety, and this set of interactions is less conserved. In TRIM25, the second interface consists of a hydrogen bond network mediated by hydrophilic residues8,9 (Fig. 2G). In contrast, the TRIM23 interface consists of a pi-stacking interaction between the sidechain of Phe93 and the Ub peptide bond between Glu34 and Gly35 (Fig. 2F). Interestingly, similar pi-stacking interactions involving a RING hydrophobic sidechain and the Ub backbone are found in structures of more distantly related RING domains, BIRC7 (phenylalanine)21 and RNF4 (tyrosine)20 (Fig. 2H). Mutation of these residues abrogate ubiquitination by these RING domains20,21, and we expect the same effect for TRIM23.
In summary, our structural analyses indicate that the TRIM23 RING domain is catalytically active as a dimer. Since full-length TRIM23 is expected to also form an elongated coiled-coil dimer with two RINGs on opposite ends, enzymatic activation will require self-association of at least two dimers in a higher-order oligomer or assembly state. In other TRIM proteins, RING activation is triggered by multivalent interactions with ubiquitination substrates or higher-order assembly platforms7,9,12,15. TRIM23 itself is not yet well-characterized, but has been reported to function in multiple signaling pathways and to ubiquitinate different substrates to regulate antiviral responses and cell differentiation22–24. It is likely that the mode of activation described here has an important role in defining the pathway specificity and timely occurrence of TRIM23-mediated polyubiquitination.
Acknowledgments
We thank Yueping Wan for assistance with cloning and bacterial growth. This study was funded by NIH grant R01-GM112508 (OP). JMW was supported by a postdoctoral NIH fellowship (F32-GM115007). DMD performed this study while on study leave from Wrocław University of Science and Technology, Poland.
References
- 1.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez JG, Okreglicka K, Chandrasekaran V, Welker JM, Sundquist WI, Pornillos O. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proc Natl Acad Sci U S A. 2014;111:2494–2499. doi: 10.1073/pnas.1318962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y, Cheng M, Chen YG, Gao N, Yu H, Wang L, Li W, Yang M. Structural insights into the TRIM family of ubiquitin E3 ligases. Cell Res. 2014;24:762–765. doi: 10.1038/cr.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstone DC, Walker PA, Calder LJ, Coombs PJ, Kirkpatrick J, Ball NJ, Hilditch L, Yap MW, Rosenthal PB, Stoye JP, Taylor IA. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci U S A. 2014;111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinert C, Morger D, Djekic A, Grutter MG, Mittl PR. Crystal structure of TRIM20 C-terminal coiled-coil/B30. 2 fragment: implications for the recognition of higher order oligomers. Sci Rep. 2015;5:10819. doi: 10.1038/srep10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yudina Z, Roa A, Johnson R, Biris N, de Souza Aranha Vieira DA, Tsiperson V, Reszka N, Taylor AB, Hart PJ, Demeler B, Diaz-Griffero F, Ivanov DN. RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 2015;12:788–797. doi: 10.1016/j.celrep.2015.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35:1204–1218. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez JG, Chiang JJ, Sparrer KM, Alam SL, Chi M, Roganowicz MD, Sankaran B, Gack MU, Pornillos O. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 2016;16:1315–1325. doi: 10.1016/j.celrep.2016.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 11.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5α protein. Proc Natl Acad Sci U S A. 2011;108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner JM, Roganowicz MD, Skorupka K, Alam SL, Christensen D, Doss G, Wan Y, Frank GA, Ganser-Pornillos BK, Sundquist WI, Pornillos O. Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5α. Elife. 2016;5:e16309. doi: 10.7554/eLife.16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher AJ, Christensen DE, Nelson C, Tan CP, Schaller T, Lehner PJ, Sundquist WI, Towers GJ. TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 2015;34:2078–2095. doi: 10.15252/embj.201490361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell EM, Weingart J, Sette P, Opp S, Sastri J, O’Connor SK, Talley S, Diaz-Griffero F, Hirsch V, Bouamr F. TRIM5α-mediated ubiquitin chain conjugation is required for inhibition of HIV-1 reverse transcription and capsid destabilization. J Virol. 2015;90:1849–1857. doi: 10.1128/JVI.01948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grütter MG, Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 17.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole E, Groves I, MacDonald A, Pang Y, Alcami A, Sinclair J. Identification of TRIM23 as a cofactor involved in the regulation of NF-κB by human cytomegalovirus. J Virol. 2009;83:3581–3590. doi: 10.1128/JVI.02072-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe M, Takahashi H, Saeki Y, Ozaki T, Itoh S, Suzuki M, Mizushima W, Tanaka K, Hatakeyama S. The E3 ubiquitin ligase TRIM23 regulates adipocyte differentiation via stabilization of the adipogenic activator PPARγ. Elife. 2015;4:e05615. doi: 10.7554/eLife.05615. [DOI] [PMC free article] [PubMed] [Google Scholar]